Abstract
DNA methylation, a well-studied epigenetic mark, is important for gene regulation in adulthood and for development. Using genetic and epigenetic approaches, the present study aimed at evaluating the effects of heat stress and copper exposure during zebrafish early embryogenesis when patterns of DNA methylation are being established, a process called reprogramming. Embryos were exposed to 325 μg Cu/L from fertilization (<1 h post fertilization - hpf) to 4 hpf at either 26.5 °C or 34 °C, followed by incubation in clean water at 26.5 °C till 96 hpf. Significant increased mortality rates and delayed hatching were observed following exposure to combined high temperature and Cu. Secondly, both stressors, alone or in combination, significantly upregulated the expression of de novo DNA methyltransferase genes (dnmt3) along with no differences in global cytosine methylation level. Finally, Cu exposure significantly increased the expression of metallothionein (mt2) and heat shock protein (hsp70), the latter being also increased following exposure to high temperature. These results highlighted the sensitivity of early embryogenesis and more precisely of the reprogramming period to environmental challenges, in a realistic situation of combined stressors.
In eukaryotes, DNA methylation is an important epigenetic mechanism involved in the regulation of gene expression and appears to be crucial for proper development of organisms1. DNA methylation occurs primarily through the transfer of a methyl group to the 5-position of cytosine residues in a CpG dinucleotide context and is catalyzed by a group of enzymes, the DNA methyltransferases (DNMTs). In mammals, there are three enzymatically active DNMTs, namely DNMT1, DNMT3a and DNMT3b. DNMT1 is maintaining methylation after each cell division, whereas DNMT3a and DNMT3b are two functionally related proteins that are responsible for de novo methylation during development and differentiation2. Due to genome duplication, zebrafish (Danio rerio), a well-established model in toxicology and developmental biology, has 8 different dnmt genes, of which 6 are most likely de novo DNMTs as they are most similar to the mammalian DNMT3 members3.
Recent studies in mammals, fish, and some invertebrate species have demonstrated that a variety of environmental stimuli, including exposure to contaminants, physiological stress and nutritional deficits, alters DNA methylation at either a global level or on selected genes4,5,6. In zebrafish, previous studies have primarily focused on DNA methylation changes triggered by environmental pollutants, such as arsenic, benzo[a]pyrene, 17α ethinylestradiol and methylmercury, in embryos, larvae or adults7,8. It has been further suggested that the DNA methylome is most susceptible to environmental challenges during early embryogenesis and particularly when patterns of DNA methylation are being established, a process called reprogramming6. Zebrafish is one of the few non-mammalian vertebrates in which DNA methylation reprogramming has been investigated. Significant differences in genome DNA methylation status have been found in zebrafish early embryos and germ cells. The oocyte DNA is globally hypomethylated compared to the one of sperm. After fertilization, zebrafish exhibit moderate bulk DNA demethylation, with de novo methylation starting at the 32-cell stage at 13/4 h post-fertilization (hpf). The methylation level is then restored by sphere stage at 4 hpf to levels observed in sperm9,10,11,12. It is also noteworthy that in zebrafish the paternal DNA methylation pattern is maintained throughout early embryogenesis while the relatively hypomethylated maternal DNA is reprogrammed to a pattern similar to that of the sperm (i.e., zebrafish inherit the DNA methylome from sperm)11,12. Currently, alterations in DNA methylation due to environmental exposures during reprogramming have received little attention. Martin et al. reported severe phenotypical changes and lower global DNA methylation levels in zebrafish exposed to 5-azacytidine, a demethylating agent, from 1 to 24 hpf13. Shorter windows of exposure from 1 to 4 hpf appeared to be critical and resulted in similar results, whereas exposure from 6 to 24 hpf showed almost no effects on morphology. Interestingly, this sensitive early stage window corresponds with the dynamic demethylation and remethylation events that occur during development and indicates that this time window must be included in toxicity testing7. More recently, McGee et al. exposed zebrafish embryos to different flame retardant chemicals during early development. They reported a critical window of exposure from 0.75 hpf to 2 hpf, which induced mortality and malformations at 96 hpf and delayed remethylation of the zygotic genome14.
Accordingly, the present study aimed at evaluating the effects of two widespread environmental stress factors that frequently co-occur in aquatic systems, i.e., temperature and copper exposure, during the DNA methylation reprogramming, a presumed sensitive period of zebrafish development. Copper (Cu) is an essential micronutrient for all organisms, but, in excess, it can cause detrimental effects on fish from the cellular to population level15. In zebrafish, Johnson et al. reported that exposure of embryos to a range of copper concentrations from 50 to 1000 μg/L resulted in mortality, hatching inhibition, impairment of larval development and lateral line dysfunction16. It is also admitted that metal toxicity generally increases with rising temperature in aquatic species17,18. For instance, rising temperature was found to increase nickel toxicity in zebrafish embryos19, copper susceptibility in fathead minnow Pimephales promelas20 and cadmium toxicity in the European bullhead Cottus gobio21. Besides, although epigenetic changes following exposure to multiple stressors remain to date unknown, several studies suggested that thermal stress22,23,24 and metals exposure4,25,26 may result in an altered DNA methylation pattern. For instance, Zhou et al. demonstrated that exposure to copper, zinc, lead, cadmium or a metal mixture for 48 h increased global DNA methylation in the liver of the goldfish Carassius auratus27.
In order to test whether the reprogramming event is sensitive to the effects of a combination of environmental stressors, zebrafish embryos were exposed to Cu from fertilization (before complete hardening of the protective chorion) to 4 hpf (sphere stage) at either 26.5 °C or 34 °C, followed by incubation in clean water at 26.5 °C to 96 hpf. We then quantified global DNA methylation and the expression of the 6 dnmt3 genes in 96 hpf larvae. The analysis was completed by measuring the expression of genes encoding for well-known stress-induced proteins, namely metallothionein (mt2) and heat shock proteins (hsp70 and hsp90). Using genetic and epigenetic approaches, this research aimed to refine the current knowledge on the sensitivity of early embryogenesis and more precisely of the reprogramming period to environmental challenges.
Results
Mortality rate and hatching efficiency
Figure 1 shows the proportion of eggs and embryos that died during the experiment (black bars), the proportion of embryos that stayed alive (dark grey bars), that hatched (i.e., larvae, light grey bars) and finally proportion of larvae that died (spotted dark grey bars). Mortality typically occurred within the first 24 hpf. After that period, the survival rate was stable until 96 hpf. Temperature, Cu exposure and their interaction significantly (P < 0.001) influenced mortality rate at 4 hpf and 24 hpf (Table 1). At 4 hpf, no mortality was recorded in groups held at control temperature, whereas we observed an increased mortality rate in groups held at 34 °C (4.4 ± 2.3% in unexposed group and 24.2 ± 7.0% in Cu-exposed group). At 24 hpf, mortality rate was increased following exposure to combined stressors with values reaching 48.0 ± 10.4%.
Figure 1. Proportion of eggs and embryos that died during the 96 hours experiment (black bars), proportion of embryos that stayed alive (dark grey bars), that hatched (i.e., larvae, light grey bars) and proportion of larvae that died (spotted dark grey bars) following exposure of zebrafish embryo from <1 to 4 hpf to 325 μg Cu/L at either 26.5 °C or 34 °C.
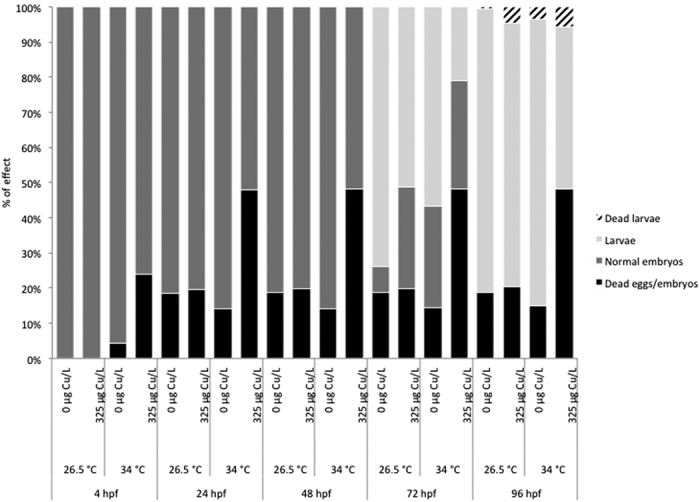
Table 1. Cumulative percent mortality and hatching of zebrafish embryos/larvae following exposure to 325 μg Cu/L at either 26.5 °C or 34 °C from <1 to 4 hpf.
| 26.5 °C |
34 °C |
||||
|---|---|---|---|---|---|
| 0 μg/L | 325 μg/L | 0 μg/L | 325 μg/L | ||
| Cumulative percent mortality | 4 hpf | 0.0 ± 0.0c | 0.0 ± 0.0c | 4.4 ± 2.3b | 24.2 ± 7.0a |
| 24 hpf | 19.0 ± 3.1b | 19.8 ± 2.7b | 14.0 ± ± 3.7b | 48.0 ± 10.4a | |
| 48 hpf | 19.1 ± 3.0 | 20.0 ± 2.5 | 14.1 ± 3.7 | 48.4 ± 10.4 | |
| 72 hpf | 19.1 ± 3.0 | 20.1 ± 2.5 | 14.5 ± 3.9 | 48.4 ± 10.4 | |
| 96 hpf | 19.2 ± 2.9 | 20.6 ± 2.7 | 14.8 ± 4.3 | 48.4 ± 10.4 | |
| Cumulative percent hatching | 48 hpf | 0.7 ± 0.5 | 0.0 ± 0.0 | 0.1 ± 0.2 | 0.1 ± 0.2 |
| 72 hpf | 91.1 ± 3.0 | 65.2 ± 10.0 | 65.6 ± 14.1 | 41.3 ± 11.2 | |
| 96 hpf | 99.1 ± 1.2 | 93.8 ± 2.4 | 95.9 ± 2.2 | 89.5 ± 4.6 | |
Data are represented as mean ± S.E.M. (n = 6).
Different letters (a–c) mean significant (P < 0.001) interaction effect between treatments within a given time period.
A peak of hatching was observed at 72 hpf in the control group (average hatching rate of 91.1 ± 3.0%), although some embryos started to hatch at 48 hpf. No interaction effect on the hatching success was observed. However, hatching rate was significantly affected at 72 hpf and 96 hpf by temperature (P < 0.001 at 72 hpf and P < 0.01 at 96 hpf) and Cu exposure (P < 0.001) (Table 1). Both factors significantly reduced hatching rate in an independent way. Overall, delayed embryo hatching was observed following exposure to high temperature and Cu from <1 to 4 hpf. In fact, at 72 hpf, average hatching rates of 91.1 ± 3.0% were observed in control group compared to 41.3 ± 11.2% in Cu-exposed group at 34 °C; at 96 hpf, average hatching rates of 99.1 ± 1.2% were observed in control group compared to 89.5 ± 4.6% in Cu-exposed group at 34 °C.
DNA methylation
The global DNA methylation levels in 96 hpf larvae following exposure to 325 μg Cu/L at either 26.5 °C or 34 °C from <1 to 4 hpf is shown in Fig. 2. The average percentage of methylated cytosines ranged between 1.12 ± 0.16% and 1.35 ± 0.21% over all treatments. There was no significant difference in global cytosine methylation levels between the four groups.
Figure 2. Level of methylated cytosines in 96 hpf zebrafish larvae following exposure to 325 μg Cu/L at either 26.5 °C or 34 °C from <1 to 4 hpf.
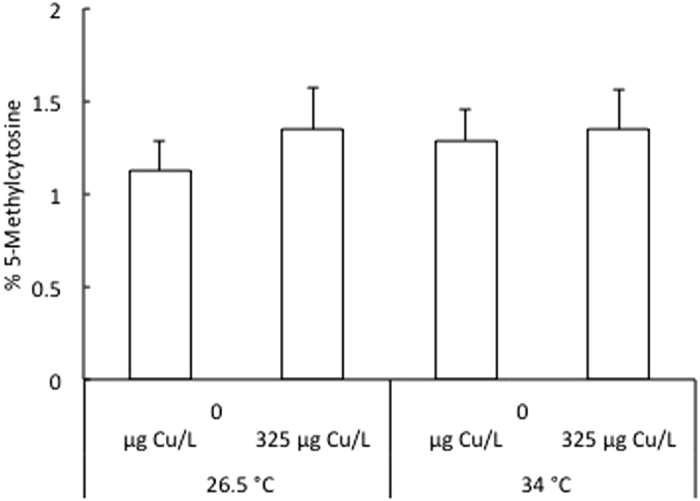
Data are represented as mean ± S.E.M. (n = 6 pools, 20 embryos/pool).
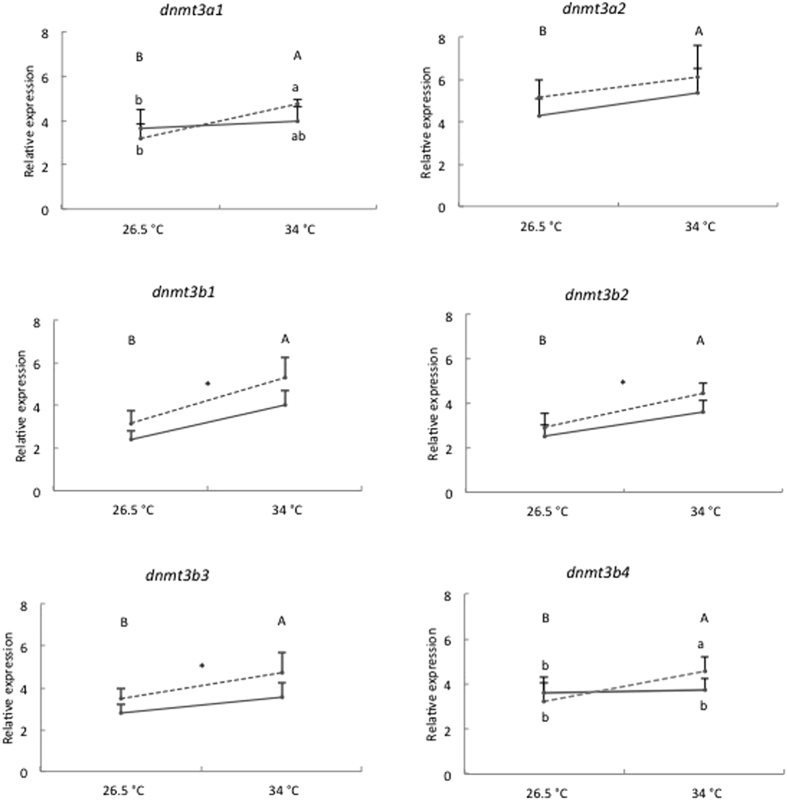
The expression profiles of the 6 dnmt3 genes in 96 hpf larvae following exposure to 325 μg Cu/L at either 26.5 °C or 34 °C from <1 to 4 hpf are depicted in Fig. 3. In our study, we followed the new nomenclature based on molecular evolution and phylogenetic analysis of dnmt3 paralogues proposed by Campos et al.28. Temperature and Cu exposure, alone or in interaction, significantly affected the mRNA expression of dnmt3 genes (Table 2). First, the mRNA expression of all dnmt3 was significantly increased by exposure to high temperature, regardless of Cu exposure. The increase ranged from 22% for dnmt3a2 and dnmt3b4 to 68% for dnmt3b1. Second, the mRNA expression of dnmt3b1, dnmt3b2 and dnmt3b3 was significantly influenced by exposure to Cu, independently of heat stress. Exposure to 325 μg Cu/L significantly increased the mRNA expression of dnmt3b1, dnmt3b2 and dnmt3b3 by 33%, 20% and 29%, respectively. Finally, we observed an interaction effect of temperature and Cu exposure for the expression of dnmt3a1 and dnmt3b4. Overall, their expressions were increased following exposure to both stressors. For instance, we observed an increased dnmt3b4 expression in groups held at 34 °C (3.71 ± 0.40 in unexposed group compared to 4.56 ± 0.41 in Cu-exposed group), whereas no significant changes were recorded in groups held at 26.5 °C.
Figure 3. Relative expression of dnmt3a1, dnmt3a2, dnmt3b1, dnmt3b2, dnmt3b3 and dnmt3b4 in 96 hpf zebrafish larvae following exposure to 0 μg Cu/L (solid line) or 325 μg Cu/L (dashed line) at either 26.5 °C or 34 °C from <1 to 4 hpf.
Data are represented as mean ± S.E.M (n = 6 pools, 30 embryos/pool). The expression is normalized to geometric mean of ef1a and actb. Different capital letters (A,B) indicate significant differences between temperature groups independent of copper exposure. Asterisk (*) along a line indicates a significant effect of copper exposure independent of temperature. In case of a significant interaction (dnmt3a1 and dnmt3b4) between temperature and copper exposure, different small letters (a,b) indicate significant differences between treatments.
Table 2. Two-way ANOVA performed for relative expression of dnmt3 genes in 96 hpf zebrafish larvae following exposure to 325 μg Cu/L at either 26.5 °C or 34 °C from <1 to 4 hpf, testing the effect of temperature treatment and copper exposure.
| Temperature | Copper | Temperature × Copper | |
|---|---|---|---|
| dnmt3a1 | P < 0.01 | P > 0.05 | P < 0.05 |
| dnmt3a2 | P < 0.05 | P > 0.05 | P > 0.05 |
| dnmt3b1 | P < 0.001 | P < 0.001 | P > 0.05 |
| dnmt3b2 | P < 0.001 | P < 0.01 | P > 0.05 |
| dnmt3b3 | P < 0.01 | P < 0.01 | P > 0.05 |
| dnmt3b4 | P < 0.05 | P > 0.05 | P < 0.05 |
Given are P values of each gene.
Transcriptional expression of stress-related genes
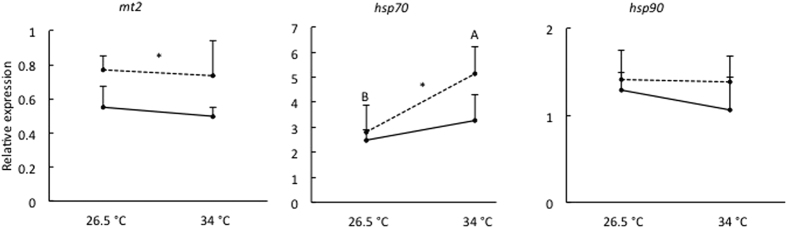
The expression profiles of mt2, hsp70 and hsp90 genes in 96 hpf larvae following exposure to 325 μg Cu/L at either 26.5 °C or 34 °C from <1 to 4 hpf are depicted in Fig. 4. We did not observe any interaction between temperature and Cu exposure for the expression of studied stress genes. However, the mRNA expression of mt2 and hsp70 was significantly affected by Cu exposure (P < 0.01 and P < 0.05, respectively), regardless of heat stress. Exposure to 325 μg Cu/L significantly increased the mRNA expression of mt2 and hsp70 by 44% and 37%, respectively. Further, hsp70 mRNA expression was significantly influenced by temperature (P < 0.01), independently of Cu exposure. Exposure to 34 °C significantly increased hsp70 mRNA expression by 59%. The mRNA expression of hsp90 was not significantly affected by temperature or Cu exposure and ranged between 1.06 ± 0.38 and 1.42 ± 0.33 over all treatments.
Figure 4. Relative expression of mt2, hsp70 and hsp90 in 96 hpf zebrafish larvae following exposure to 0 μg Cu/L (solid line) or 325 μg Cu/L (dashed line) at either 26.5 °C or 34 °C from <1 to 4 hpf.
Data are represented as mean ± S.E.M. (n = 6 pools, 30 embryos/pool). The expression is normalized to geometric mean of ef1a and actb. Different capital letters (A,B) indicate significant differences between temperature groups independent of Cu exposure. Asterisk (*) along a line indicates a significant effect of Cu exposure independent of temperature.
Discussion
Our study was designed to investigate whether the DNA methylation reprogramming that occurs during zebrafish early development is sensitive to environmental stress factors. According to Kamstra et al.7, this is an extremely important time window to include in studies dealing with the effects of environmental challenges. Epigenetic modifications early in life are proposed to underlie the development of an adverse adult phenotype. Overall, both heat stress and copper exposure applied during reprogramming were particularly stressful and resulted in adverse developmental consequences on zebrafish, as indicated by high mortality, delayed hatching and increased expression of several stress-related genes. Further, we observed changes in the expression of de novo DNA methyltransferases (dnmt3a and 3b) but no differences in global methylation level.
Aquatic organisms such as fish are, in most cases, exposed to multiple stressors that are either natural or anthropogenically introduced into their environment. Temperature, a potent physical stressor, and copper, a toxic chemical stressor when in excess, were addressed in the present study. Several studies have investigated the possible interaction between temperature and copper, but these were mainly conducted with adult fish. For instance, Perschbacher found an inverse relationship between temperature and copper toxicity in channel catfish, Ictalurus punctatus29. In his experiments, copper was less toxic to catfish if applied at increasing temperatures. In contrast, Furuta et al. observed no significant effect of temperature on copper toxicity in Japanese flounder, Paralichthys olivaceus, and red sea bream, Pagrus major30. More recently, Lapointe et al. reported synergistic effects between heat stress and copper exposure in fathead minnows Pimephales promelas20. In our study, both heat stress and copper exposure resulted in increased mortality rates and delayed embryo hatching, suggesting adverse developmental consequences on zebrafish. We highlighted increased mortality of Cu-exposed embryos with increasing temperature (significant interaction), while both stressors independently influenced the hatching success. To date, there is no study considering the interaction effects of these two stressors on zebrafish embryogenesis, although a few studies have investigated temperature effects on the toxicity of other metals, i.e., cadmium31 and nickel19. Looking first at the effects of copper, a previous study reported that exposure of zebrafish embryos to copper from fertilization to 72 hpf resulted in increased mortality between 5 and 24 hpf as well as decreased hatching success16. These observations are quite consistent with our results. During embryonic development, a strong influence of metals is often observed immediately after fertilization, particularly before hardening of the protective chorion. This structure is highly permeable before hardening and does not fully protect the embryo against metal penetration; thus, metals may exert direct toxic influence upon developing embryos32,33. Further, several mechanisms have been proposed to explain how copper delays the hatching process, e.g., inhibition of the hatching enzyme activity (chorionase), interference with the energy-requiring muscular movements necessary for the embryo to rupture the chorion, and alteration of oxygen uptake by the embryo33,34. However, these mechanisms are still unclear, and more than one or all of these processes may contribute to the observed delay in hatching. Secondly, zebrafish embryos normally develop within a temperature range of approximately 25–33 °C, and it was suggested that exposure to temperatures above or below these limits may produce abnormalities35,36. For instance, an increase in the developmental rates of zebrafish with increasing temperatures has been reported in several papers37. Schnurr et al. have recently assessed the thermal breadth of zebrafish embryonic development38. The authors observed that embryos developed successfully to hatching from 22 to 32 °C, but suffered sharp increase in mortality outside of this range. Further, the rate of embryonic development increased gradually from 22 to 34 °C, beyond which the rate dropped. Another study reported increased mortality and a higher number of developmental anomalies in embryos exposed for 2–3 h to 34–35 °C during early cleavage stages, which is similar to our results39.
Increasing evidence shows that environmental challenges can affect normal development of organisms by altering DNA methylation patterns. Additionally, the DNA methylation reprogramming after fertilization is a dynamic mechanism that is essential for early development and which is considered to be sensitive to environmental stressors6,7,40. Dnmt3a and dnmt3b are required for this process, and the inactivation of both genes causes a complete failure in the genome-wide methylation41. In this context, the present study investigated whether heat stress and copper exposure applied during reprogramming affect DNA methylation patterns by altering the expression of dnmt3 genes in 96 hpf larvae. Altogether, we demonstrated that both stressors, alone or in combination, alter dnmt3 gene expression without changes in global cytosine methylation. The absence of effects on the global scale does not mean that DNA methylation modifications do not occur. The DNA methylation assay used in this study only tested for a global shift in methylated cytosines content and does not detect changes at specific loci. This is in line with several recent studies that have reported locus-specific methylation modifications and no changes in global methylation level in zebrafish embryos42,43,44. For instance, Bouwmeester et al. observed changes in methylation levels in the promoter regions of three selected genes vasa, vtg1 and cyp19a2 but no differences in global methylation level after exposure of embryos to several environmental contaminants from 0 to 72 hpf43.
In addition to measuring the global DNA methylation levels, we measured the expression of all dnmt3 genes aiming to associate their expression profiles with DNA methylation changes. Previous studies have indicated that zebrafish dnmt3 genes exhibit distinct spatio-temporal expression patterns during embryonic development28,42,45,46,47. For instance, Aluru et al. have recently reported that dnmt3b genes are highly expressed in early stages of development and dnmt3a genes are more abundant in later stages, suggesting distinct roles for dnmt3a and 3b genes during development42. Takayama et al. observed tissue-specific expression of dnmt3a genes during later stages of development in zebrafish, suggesting a role for these genes in tissue differentiation47. Other studies reported localization of dnmt3b genes to specific regions of the developing zebrafish, as for instance dnmt3b1 in the aorta-gonad-mesonephros and caudal hematopoietic tissue regions47 and dnmt3b2 in the brain, particularly in the tectum3, suggesting important roles in hematopoietic stem cell differentiation and neurogenesis, respectively. However, the specific roles of these genes in establishing de novo methylation patterns remain to date unknown. In the present study, we observed heat-induced upregulation of all dnmt3 genes and copper-induced upregulation of dnmt3b1, dnmt3b2 and dnmt3b3. Further, transcription levels of dnmt3a1 and dnmt3b4 were affected by copper exposure in a temperature-dependent way (significant interaction). The effects of heat stress and copper exposure on dnmt3 gene expression levels observed in this study suggest that the establishment of DNA methylation patterns could potentially be impacted. A few recent studies have shown differences in the expression patterns of dnmt3 genes following heat stress or pollutant exposure during zebrafish embryogenesis. Campos et al. observed that embryonic temperature influenced dnmt3a and dnmt3b transcript levels in a developmental stage-specific manner28. The authors have further reported differential responses of dnmt3 paralogues to embryonic temperature, suggesting that they play different roles in thermal epigenetic regulation of gene expression during early development. A direct comparison with our data is tricky since the experimental conditions were different and the dnmt3 expression during early development was reported to be very dynamic. Moreover, the oldest embryos were sampled at 80.8 hpf (protruding mouth stage) in Campos’ experiment, while we sampled zebrafish after 96 hpf. At 80.8 hpf, they didn’t observe significant differences between embryos exposed to 27 °C and 31 °C for any dnmt3 genes. More recently, Aluru et al. reported that exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) during zebrafish early development selectively alters the expression of dnmt genes, and these effects were developmental stage-specific42. The authors also reported changes in methylation levels in promoters of aryl hydrocarbon receptor (AHR) target genes but no differences in global methylation or hydroxymethylation levels.
Metallothioneins (MTs) are low molecular weight, cysteine-rich metal-binding proteins that play essential biological roles in metal homeostasis and detoxification48. Heat shock proteins (HSPs) are a set of evolutionary conserved proteins that are constitutively expressed and function as molecular chaperones in protein biogenesis and homeostasis49. As stress response proteins, MTs and HSPs confer cellular protection against a wide variety of stress factors and can be induced at the transcription level by diverse environmental stressors, including toxic metals and temperature50,51. Moreover, previous studies suggested that MTs52 and HSPs36 play significant roles during normal embryo development in zebrafish. In our study, we observed that exposure to copper increased the transcription level of mt2 and hsp70 in zebrafish larvae. Concomitantly, the hsp70 transcript level was also increased in response to thermal stress. Further, none of the applied stresses was able to affect the transcription level of hsp90. Similar results were previously observed in various fish species exposed either to heat or copper20,53,54. In zebrafish, several studies also reported that heat shock and/or copper exposure resulted in increased expression of MTs and/or HSPs in different adult tissues55,56 or in embryos36,57. Overall, our gene expression results indicate that applied conditions were stressful to zebrafish larvae, thereby suggesting the susceptibility of reprogramming period to environmental challenges.
To conclude, our study is the first to report that the DNA methylation reprogramming is sensitive to the effects of environmental stressors in zebrafish embryos. As a matter of fact, we have shown that exposure to elevated temperature and/or copper during this critical developmental period resulted in high mortality, delayed hatching and increased expression of several stress genes, i.e., mt2 and hsp70. Furthermore, the expression of dnmt3 genes suggests possible impact on the establishment of DNA methylation patterns. In the future it would be interesting to focus on DNA methylation levels of selected genes (e.g., mt2 and hsp70) as well as on genome-wide methylation pattern and the relationships with gene expression. Further exploration is also needed to examine the adult phenotype of exposed embryos and to analyze molecular mechanisms that potentially link epigenetic effects and altered phenotypes.
Materials and Methods
Ethics statement
All zebrafish husbandry and experimental procedures were performed in accordance with the Belgian animal protection standards and were approved by the University of Namur Local research Ethics Committee (14 224 KE).
Zebrafish embryo collection and exposure
AB line wild-type zebrafish (Danio rerio) were maintained in a recirculating ZebTec housing system (Techniplast) at 28 °C with a 12:12 h (light/dark) photoperiod. Conductivity was maintained at approximately 500 μS/cm, pH at 7.2 and dissolved oxygen at 95% saturation. Fish were fed ad libitum three times daily (twice on flake and once on Artemia nauplii). Males and females were placed together in breeding aquaria the night before spawning in a ratio of 3:2. The next morning, eggs were collected within 30 min of spawning, pooled from several breeding tanks, washed with ZebTec system water to remove debris and then randomly transferred into polypropylene beakers.
Zebrafish embryos were exposed to CuCl2 (Sigma-Aldrich 751944) at nominal concentrations of 0 and 325 μg/L from <1 hpf (before complete hardening of the protective chorion) to 4 hpf (sphere stage) at either 26.5 °C or 34 °C. The tested treatments were chosen based on previous studies16,37. Six replicates for each treatment were used and each replicate consisted of a polypropylene beaker containing 150 ml of the respective treatment solutions and 166 ± 21 embryos. After the exposure, the embryos were rinsed thoroughly with fresh ZebTec system water and incubated at 26.5 °C until 96 hpf. Half of the water in each beaker was replaced every day to remove the wastes and dead embryos. Mortality rate and hatching efficiency were recorded daily until 96 hpf. At 96 hpf, larvae were washed, collected and stored at −80 °C until DNA and RNA were isolated.
Total Cu concentration in the exposure water was measured using an Inductively Coupled Plasma-Mass Spectrometer (ICPMS, Elan DRC II). The accuracy of analytical methods was checked using certified reference standards (ICP/MS Multi-Element Standards ICP-MS-QC2-1, ACCUStandard). The mean concentration and standard deviation in the 325 μg/L treatment were 280.4 ± 13.6 μg/L.
Global DNA methylation
Genomic DNA was isolated from zebrafish larvae (96 hpf; n = 6 pools, 20 larvae/pool) using DNeasy Blood & Tissue Kit (Qiagen) according to manufacturer’s protocol. DNA samples were treated with RNase A (Qiagen) to remove RNA contaminant. The DNA concentration and the quality of samples were assessed by spectrophotometry (NanoDrop 2000c Spectrophotometer, Thermo Scientific) and agarose gel electrophoresis. Quantification of global DNA methylation was examined by the colorimetric MethylFlashTM Methylated DNA Quantification Kit (Epigentek Group Inc.) following the manufacturer’s instructions. The analysis was performed in duplicates with 100 ng of genomic DNA per sample. Absorption at 450 nm was determined. The percentage of methylated cytosines was calculated using the formula described by the manufacturer.
Quantitative real-time PCR (qPCR)
The expression of 9 target genes was assessed by qPCR: 6 are involved in de novo DNA methylation (dnmt3a1, dnmt3a2, dnmt3b1, dnmt3b2, dnmt3b3 and dnmt3b4) and three play important roles in cell protection against environmental stressors (mt2, hsp70 and hsp90). Total RNA was isolated from zebrafish larvae (96 hpf; n = 6 pools, 30 larvae/pool) with Extract-All Reagent (Eurobio) and DNase treated (DNA-free™ Kit, Ambion) by following the manufacturers’ protocol. The RNA concentration and the quality of samples were assessed by spectrophotometry (NanoDrop 2000c Spectrophotometer, Thermo Scientific) and agarose gel electrophoresis. Complementary DNA was synthesized from 1 μg total RNA using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific). Specific primers were selected from the literature or newly designed using Primer3Plus software and are listed in Table 3. Whenever possible, primers were designed to span at least one intron/exon border to avoid amplification of potential contaminating genomic DNA. Quantitative PCR was performed in a total volume of 20 μl (5 μl of cDNA, 2.5 μl of each primer at 5 μM, 10 μl Master Mix 2x) with SYBR Green chemistry (Applied Biosystems) on a StepOnePlus Real-Time PCR System (Applied Biosystems). The PCR conditions used were 1 cycle at 95 °C for 10 min and 40 cycles at 95 °C for 15 s and 58–61.8 °C depending on gene for 1 min (Table 3). This was followed by a melting curve analysis to verify the specificity of the PCR products. Two technical replicates were used for each sample. No reverse transcriptase and no template controls were included on each plate to ensure the absence of background contamination. Gene expression data were calculated using the relative standard curve method58 and normalized by geometric averaging of beta-actin (actb) and elongation factor alpha (ef1a). actb and ef1a were selected among several candidate housekeeping genes that remain stable over zebrafish development mentioned in the literature59,60, and after validation of their expression in our samples.
Table 3. List of specific primers used for quantitative real-time PCR.
| Gene | Accession | Primer (5′–3′) | Size (bp) | E (%) | Ta | Reference | |
|---|---|---|---|---|---|---|---|
| dnmt3a1 | AB196917 | F | GCTAAGTTTGGTAAAGTGCGG | 103 | 96 | 58 °C | 28 |
| R | GGATGTCCTCCTTATCATTCA | ||||||
| dnmt3a2 | AB196919 | F | TAGGAAAGGCTTGTTTGAGGG | 157 | 105 | 59 °C | 28 |
| R | GCGTGAGATGTCTTTCTTGTC | ||||||
| dnmt3b1 | AB196915 | F | GTGCCTCTGGGATGGATAAA | 174 | 98 | 60 °C | Newly designed |
| R | GCTCTGTGCACCACAGGATA | ||||||
| dnmt3b2 | AB196918 | F | CGCTACATTGCCTCTGAGA | 113 | 95 | 58 °C | 28 |
| R | GCCAGATGTTTCCTAGTGATG | ||||||
| dnmt3b3 | AB196914 | F | GCTCAGGTGCTGCTTTTTGTC | 152 | 95 | 61 °C | 28 |
| R | TTTTTGAATCTGTGCTTTGCTG | ||||||
| dnmt3b4 | AB196916 | F | TCATTGACTTGGGGGAAGAG | 172 | 97 | 60 °C | Newly designed |
| R | TAATCATCATGGCTGCTGCT | ||||||
| mt2 | BC152694 | F | AAGCTCTTTGTGGATACTCTCTGG | 112 | 94 | 60.8 °C | Newly designed |
| R | GCAGGTAGTACACTGGCAATTAGTG | ||||||
| hsp70 | AB062116 | F | CATCGACGCCAACGGG | 191 | 95 | 60 °C | 61 |
| R | CCAGGGAGTTTTTAGCAGAAATCTT | ||||||
| hsp90 | BC075757 | F | GGTACCAAAGTCATTCTCCACCTT | 154 | 97 | 61.8 °C | Newly designed |
| R | CCTCAAGATCCACCTCTTTTTCTC | ||||||
| actb | BC165823 | F | CCAACACTGTATTGTCTGGTGGTA | 203 | 101 | 61 °C | Newly designed |
| R | GTACTCCTGCTTGCTAATCCACAT | ||||||
| ef1a | AY422992 | F | GGTACTACTCTTCTTGATGCCCTTG | 200 | 98 | 60.8 °C | Newly designed |
| R | GACTTGACCTCAGTGGTTACATTG |
For each gene, its GenBank accession number, amplicon size (bp), amplification efficiency (E) and annealing temperature (Ta) are indicated.
Statistical analysis
Data were expressed as mean ± S.E.M. Data were log-transformed when the conditions of normality and/or homogeneity of variance were not fully filled, or arcsine-square-root-transformed if expressed in percents. Differences between groups were analyzed using two-way analysis of variance (ANOVA 2) followed by a multiple comparison Tukey’s HSD test at a 5% significant level. All tests were performed using the Statistica 5.5 software (StatSoft, Tulsa, OK, USA).
Additional Information
How to cite this article: Dorts, J. et al. DNA methyltransferases and stress-related genes expression in zebrafish larvae after exposure to heat and copper during reprogramming of DNA methylation. Sci. Rep. 6, 34254; doi: 10.1038/srep34254 (2016).
Acknowledgments
The authors thank R. Biondo from Laboratory of Oceanology – MARE Center, University of Liège (Liège, Belgium) for valuable help during chemical analysis. This study was supported by the Belgian Science Policy Office (Belspo) AquaStress project (IUAPVII/64/Sorgeloos), including a postdoctoral fellowship to J. Dorts. Supports were also provided by the FNRS-FRS (Fonds de la Recherche Scientifique) grant No. 2.4635.11 (Transgenerational Impacts of Pollutants).
Footnotes
Author Contributions Study conception and design: D.J., K.P. and S.F. Acquisition of data: D.J., F.E., S.E. and F.E. Analysis and interpretation of data: D.J. Drafting of manuscript: D.J. Critical revision: K.P. and S.F. Each author read and approved the final manuscript.
References
- Goll M. G. & Halpern M. E. DNA methylation in zebrafish. Prog. Mol. Biol. Transl. Sci. 101, 193–218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002). [DOI] [PubMed] [Google Scholar]
- Shimoda N., Yamakoshi K., Miyake A. & Takeda H. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev. Dyn. 233, 1509–1516 (2005). [DOI] [PubMed] [Google Scholar]
- Vandegehuchte M. B. & Janssen C. R. Epigenetics and its implications for ecotoxicology. Ecotoxicology 20, 607–624 (2011). [DOI] [PubMed] [Google Scholar]
- Vandegehuchte M. B. & Janssen C. R. Epigenetics in an ecotoxicological context. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 764–765, 36–45 (2014). [DOI] [PubMed] [Google Scholar]
- Head J. A. Patterns of DNA methylation in animals: an ecotoxicological perspective. Integr. Comp. Biol. 54, 77–86 (2014). [DOI] [PubMed] [Google Scholar]
- Kamstra J. H., Aleström P., Kooter J. M. & Legler J. Zebrafish as a model to study the role of DNA methylation in environmental toxicology. Environ. Sci. Pollut. Res. Int. 22, 16262–16276 (2015). [DOI] [PubMed] [Google Scholar]
- Williams T. D., Mirbahai L. & Chipman J. K. The toxicological application of transcriptomics and epigenomics in zebrafish and other teleosts. Brief. Funct. Genomics 13, 157–171 (2014). [DOI] [PubMed] [Google Scholar]
- Mhanni A. A. & McGowan R. A. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev. Genes Evol. 214, 412–417 (2004). [DOI] [PubMed] [Google Scholar]
- Fang X., Corrales J., Thornton C., Scheffler B. E. & Willett K. L. Global and gene specific DNA methylation changes during zebrafish development. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 166, 99–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok M. E., Nix D. A., Parnell T. J. & Cairns B. R. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. C., Laforest L., Akimenko M. A. & Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev. Biol. 206, 189–205 (1999). [DOI] [PubMed] [Google Scholar]
- McGee S. P. & Cooper E. M. Early Zebrafish Embryogenesis Is Susceptible to Developmental TDCPP Exposure. Environmental health perspectives 120, 1585–1591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okocha R. & Adedeji O. Overview of copper toxicity to aquatic life. Report and Opinion 4, 57–68 (2012). [Google Scholar]
- Johnson A., Carew E. & Sloman K. A. The effects of copper on the morphological and functional development of zebrafish embryos. Aquat. Toxicol. 84, 431–438 (2007). [DOI] [PubMed] [Google Scholar]
- Sokolova I. & Lannig G. Interactive effects of metal pollution and temperature on metabolism in aquatic ectotherms: implications of global climate change. Climate Res. 37, 181–201 (2008). [Google Scholar]
- Holmstrup M. et al. Interactions between effects of environmental chemicals and natural stressors: a review. Sci. Total Environ. 408, 3746–3762 (2010). [DOI] [PubMed] [Google Scholar]
- Scheil V. & Köhler H.-R. Influence of nickel chloride, chlorpyrifos, and imidacloprid in combination with different temperatures on the embryogenesis of the zebrafish Danio rerio. Arch. Environ. Contam. Toxicol. 56, 238–243 (2009). [DOI] [PubMed] [Google Scholar]
- Lapointe D., Pierron F. & Couture P. Individual and combined effects of heat stress and aqueous or dietary copper exposure in fathead minnows (Pimephales promelas). Aquat. Toxicol. 104, 80–85 (2011). [DOI] [PubMed] [Google Scholar]
- Dorts J. et al. Proteasome and antioxidant responses in Cottus gobio during a combined exposure to heat stress and cadmium. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 155, 318–324 (2012). [DOI] [PubMed] [Google Scholar]
- Campos C., Valente L. M. P., Conceição L. E. C., Engrola S. & Fernandes J. M. O. Temperature affects methylation of the myogenin putative promoter, its expression and muscle cellularity in Senegalese sole larvae. Epigenetics 8, 389–397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzitallab P. et al. Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogenetic Artemia model. FASEB Journal 28, 3552–3563 (2014). [DOI] [PubMed] [Google Scholar]
- Skjærven K. H., Hamre K., Penglase S., Finn R. N. & Olsvik P. A. Thermal stress alters expression of genes involved in one carbon and DNA methylation pathways in Atlantic cod embryos. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 173C, 17–27 (2014). [DOI] [PubMed] [Google Scholar]
- Baccarelli A. & Bollati V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 21, 243–251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.-F., Choudhuri S. & Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: a review of the literature. J. Appl. Toxicol. 32, 643–653 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou X., Zhu G., Jilisa M. & Sun J. Influence of Cu, Zn, Pb, Cd and their heavy metal ion mixture on the DNA methylation level of the fish (Carassius auratus). China Environ. Sci. 21, 4–7 (2001). [Google Scholar]
- Campos C., Valente L. M. P. & Fernandes J. M. O. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene 500, 93–100 (2012). [DOI] [PubMed] [Google Scholar]
- Perschbacher P. W. Temperature effects on acute copper toxicity to juvenile channel catfish Ictalurus punctatus. Aquaculture 243, 225–228 (2005). [Google Scholar]
- Furuta T., Iwata N. & Kikuchi K. Effects of fish size and water temperature on the acute toxicity of copper for Japanese flounder, Paralichthys olivaceus, and red sea bream, Pagrus major. J. World Aquacult. Soc. 39, 766–773 (2008). [Google Scholar]
- Hallare A. V., Schirling M., Luckenbach T. & Ko H. & riebskorn, R. Combined effects of temperature and cadmium on developmental parameters and biomarker responses in zebrafish (Danio rerio) embryos. J. Therm. Biol. 30, 7–17 (2005). [Google Scholar]
- Gellert G. & Heinrichsdorff J. A. N. Effect of age on the susceptibility of zebrafish eggs to industrial wastewater. Water Res. 35, 3754–3757 (2001). [DOI] [PubMed] [Google Scholar]
- Jezierska B., Lugowska K. & Witeska M. The effects of heavy metals on embryonic development of fish (a review). Fish Physiol. Biochem. 35, 625–640 (2009). [DOI] [PubMed] [Google Scholar]
- Palmer F. B., Butler C. A., Timperley M. H. & Evans C. W. Toxicity to embryo and adult zebrafish of copper complexes with two malonic acids as models for dissolved organic matter. Environ. Toxicol. Chem. 17, 1538–1545 (1998). [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- Krone P. H., Evans T. G. & Blechinger S. R. Heat shock gene expression and function during zebrafish embryogenesis. Semin. Cell Dev. Biol. 14, 267–274 (2003). [DOI] [PubMed] [Google Scholar]
- López-Olmeda J. F. & Sánchez-Vázquez F. J. Thermal biology of zebrafish (Danio rerio). J. Therm. Biol. 36, 91–104 (2011). [Google Scholar]
- Schnurr M. E., Yin Y. & Scott G. R. Temperature during embryonic development has persistent effects on metabolic enzymes in the muscle of zebrafish. J. Exp. Biol. 217, 1370–1380 (2014). [DOI] [PubMed] [Google Scholar]
- Schirone R. C. & Gross L. Effect of temperature on early embryological development of the zebrafish, Brachydanio rerio. J. Exp. Zool. 169, 43–52 (1968). [Google Scholar]
- Faulk C. & Dolinoy D. C. In Environmental Epigenomics in Health and Disease: Epigenetics and Human Health (eds Jirtle R. L. & Tyson F. L.) Ch. 4, 77–97 (Springer, 2013). [Google Scholar]
- Chen T., Ueda Y., Dodge J. E., Wang Z. & Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23, 5594–5605 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru N. et al. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 284, 142–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester M. C. et al. Zebrafish embryos as a screen for DNA methylation modifications after compound exposure. Toxicol. Appl. Pharmacol. 291, 84–96 (2016). [DOI] [PubMed] [Google Scholar]
- Olsvik P. A. et al. Impacts of TCDD and MeHg on DNA methylation in zebrafish (Danio rerio) across two generations. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 165, 17–27 (2014). [DOI] [PubMed] [Google Scholar]
- Rai K. et al. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J. Biol. Chem. 285, 4110–4121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. H., Collins T. M. & McGowan R. A. Expression of the dnmt3 genes in zebrafish development: similarity to Dnmt3a and Dnmt3b. Dev. Genes Evol. 220, 347–353 (2011). [DOI] [PubMed] [Google Scholar]
- Takayama K., Shimoda N., Takanaga S., Hozumi S. & Kikuchi Y. Expression patterns of dnmt3aa, dnmt3ab, and dnmt4 during development and fin regeneration in zebrafish. Gene Expr. Patterns 14, 105–110 (2014). [DOI] [PubMed] [Google Scholar]
- Kagi J. H. & Schaffer A. Biochemistry of metallothionein. Biochemistry 27, 8509–8515 (1988). [DOI] [PubMed] [Google Scholar]
- Fink A. L. Chaperone-mediated protein folding. Physiol. Rev. 79, 425–449 (1999). [DOI] [PubMed] [Google Scholar]
- Feder M. E. & Hofmann G. E. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282 (1999). [DOI] [PubMed] [Google Scholar]
- Bourdineaud J. P., Baudrimont M., Gonzalez P. & Moreau J. L. Challenging the model for induction of metallothionein gene expression. Biochimie 88, 1787–1792 (2006). [DOI] [PubMed] [Google Scholar]
- Chen W.-Y. et al. Expression of metallothionein gene during embryonic and early larval development in zebrafish. Aquat. Toxicol. 69, 215–227 (2004). [DOI] [PubMed] [Google Scholar]
- Said Ali K., Ferencz A., Nemcsók J. & Hermesz E. Expressions of heat shock and metallothionein genes in the heart of common carp (Cyprinus carpio): effects of temperature shock and heavy metal exposure. Acta Biol. Hung. 61, 10–23 (2010). [DOI] [PubMed] [Google Scholar]
- Zhu B., Liu L., Li D.-L., Ling F. & Wang G.-X. Developmental toxicity in rare minnow (Gobiocypris rarus) embryos exposed to Cu, Zn and Cd. Ecotoxicol. Environ. Saf. 104C, 269–277 (2014). [DOI] [PubMed] [Google Scholar]
- Airaksinen S. et al. Stressor-dependent regulation of the heat shock response in zebrafish, Danio rerio. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 134, 839–846 (2003). [DOI] [PubMed] [Google Scholar]
- Craig P. M., Hogstrand C., Wood C. M. & McClelland G. B. Gene expression endpoints following chronic waterborne copper exposure in a genomic model organism, the zebrafish, Danio rerio. Physiol. Genomics 40, 23–33 (2009). [DOI] [PubMed] [Google Scholar]
- Chan K. M., Ku L. L., Chan P. C.-Y. & Cheuk W. K. Metallothionein gene expression in zebrafish embryo-larvae and ZFL cell-line exposed to heavy metal ions. Mar. Environ. Res. 62, S83–S87 (2006). [DOI] [PubMed] [Google Scholar]
- Larionov A., Krause A. & Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6, 62 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Shuying X. & Jihua Y. Identification of Novel Reference Genes Suitable for qRT-PCR Normalization with Respect to the Zebrafish Developmental. Plos one 11, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Li C., Zeng Q., Agrawal I., Zhu X. & Gong Z. Genome-Wide Identification of Suitable Zebrafish Danio Rerio Reference Genes for Normalization of Gene Expression Data by RT-qPCR. J. Fish Biol. 88, 2095–2110 (2016). [DOI] [PubMed] [Google Scholar]
- Gonzalez P., Baudrimont M., Boudou A. & Bourdineaud J.-P. Comparative effects of direct cadmium contamination on gene expression in gills, liver, skeletal muscles and brain of the zebrafish (Danio rerio). Biometals 19, 225–235 (2006). [DOI] [PubMed] [Google Scholar]