Abstract
DNA-based gene therapy has considerable therapeutic potential, but the challenges associated with delivery continue to limit progress. Messenger RNA (mRNA) has the potential to provide for transient production of therapeutic proteins, without the need for nuclear delivery and without the risk of insertional mutagenesis. Here we describe the sustained delivery of therapeutic proteins in vivo in both rodents and non-human primates via nanoparticle-formulated mRNA. Nanoparticles formulated with lipids and lipid-like materials were developed for delivery of two separate mRNA transcripts encoding either human erythropoietin (hEPO) or factor IX (hFIX) protein. Dose-dependent protein production was observed for each mRNA construct. Upon delivery of hEPO mRNA in mice, serum EPO protein levels reached several orders of magnitude (>125 000-fold) over normal physiological values. Further, an increase in hematocrit (Hct) was established, demonstrating that the exogenous mRNA-derived protein maintained normal activity. The capacity of producing EPO in non-human primates via delivery of formulated mRNA was also demonstrated as elevated EPO protein levels were observed over a 72-h time course. Exemplifying the possible broad utility of mRNA drugs, therapeutically relevant amounts of human FIX (hFIX) protein were achieved upon a single intravenous dose of hFIX mRNA-loaded lipid nanoparticles in mice. In addition, therapeutic value was established within a hemophilia B (FIX knockout (KO)) mouse model by demonstrating a marked reduction in Hct loss following injury (incision) to FIX KO mice.
Introduction
The application of nucleic acids for therapeutic use has been of interest to the scientific community for decades.1 While viral systems have shown great promise and recent success, challenges associated with safety, manufacturing and potency continue to persist.2 Non-viral systems have many potential advantages but have proven challenging to translate.3 In general, this work has predominantly focused on the use of plasmid DNA (gene therapy),4, 5, 6 small interfering RNA (RNA interference),7, 8, 9, 10 antisense oligonucleotides, microRNA (translation repression)11, 12, 13, 14, 15 and aptamers.16, 17, 18 Messenger RNA (mRNA) has garnered much less attention during this time, perhaps due to the widespread consideration of its transient nature, and only recently has been publicized to have therapeutic value in the treatment of various diseases.19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30
Systemic mRNA therapy (MRT) is a new approach to the delivery of therapeutic proteins in vivo, using biosynthetic mRNA transcripts as the source for therapeutic protein.20, 21, 22, 23 Protein production derived from such exogenous mRNA takes advantage of the endogenous protein translational machinery within the body along with all of the endogenous post-translational modifications and processing machinery that are present in human cells and tissues. Theoretically, this approach could enable the treatment of many diseases, including those in which protein-based therapeutics are unattainable, such as large, transmembrane proteins; heavily processed, post-translationally modified proteins; or proteins requiring cytoplasmic delivery. In addition to this, MRT could provide flexibility with respect to delivery within the body. The specific site of delivery and subsequent translation need not be within the target organ for therapeutic treatment. Protein production derived from the delivered mRNA can remain intracellular or can be secreted to treat the respective disease systemically. Finally, MRT has potential advantages over other nucleic acid protein replacement platforms such as gene therapy, including the lack of requirement for nuclear localization and elimination of insertional mutagenesis risks.
As with almost all nucleic acid-based therapeutics, intracellular delivery is required to achieve function. Over the years, many different modalities have been examined, including viral vectors,31, 32 polymers,33, 34, 35, 36, 37, 38, 39, 40 lipid nanoparticles (LNPs)41, 42, 43 and covalent conjugation.44, 45, 46, 47 Here we develop therapeutic LNPs, using lipid and lipid-like materials for the delivery of mRNA to hepatocytes. mRNA is notoriously labile, especially under biological environments such as exposure to endo- and exonucleases present within plasma.48 Encapsulation of RNA within LNPs can provide for sustained RNA stability.49 In general, LNPs are formulated with either a cationic or ionizable lipid or lipidoid, several helper lipids and a nucleic acid payload, which can facilitate accumulation in specific sites within the body by manipulation of size and/or inclusion of targeting agents.49, 50, 51, 52, 53, 54, 55 While considerable progress has been made in the clinical development of small interfering RNA delivery systems, the intravenous delivery of LNP-encapsulated mRNA in primates has not been demonstrated.56, 57, 58, 59, 60, 61, 62, 63, 64
Here we describe the first example of efficacy in a human disease mouse model using a lipidoid-based LNP formulation for systemic delivery of mRNA, in vivo. A formulation containing the cationic lipidoid C12-200 was developed for the delivery of in vitro-synthesized mRNA. LNPs containing mRNA constructs for either human erythropoietin (hEPO) or human factor IX (hFIX) were developed. Delivery of the mRNA to hepatocytes was confirmed using in situ hybridization (ISH) methods. Robust protein production from both exogenously synthesized mRNA transcripts was observed and quantified in multiple species, including non-human primates. Further, pharmacodynamic effects were assessed and proof of efficacy was established within a hemophilia B (FIX knockout (KO)) disease model.65
Results
We examined the potential of LNP-formulated mRNA for two therapeutic proteins, hEPO and hFIX, as well as the utility of FIX in a relevant disease model. Quantitative measurement demonstrated that the desired proteins derived from exogenous human mRNAs were delivered at supraphysiological levels via LNP formulations. Protein secretion resulted in potent pharmacodynamic effects as well as therapeutic efficacy in a KO mouse model.
In vivo hEPO protein production
To evaluate the ability of mRNA-encapsulated lipidoid nanoparticles to facilitate the delivery of mRNA, we monitored both hEPO mRNA as well as hEPO protein levels in the serum over a 1-week time period. This was performed as a single-dose administration (1.0 mg kg−1 based on encapsulated mRNA) given intravenously. All formulations were well tolerated in the mice at the given dose with no observable adverse events.
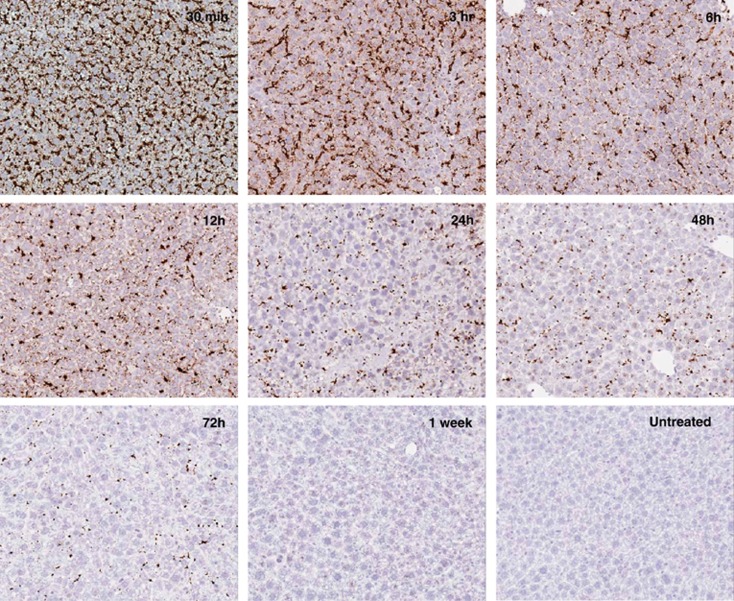
ISH methods were used to evaluate the delivery of hEPO mRNA to the liver. As represented in Figure 1, a strong positive signal for hEPO mRNA was detected from the earliest time point within the experiment (30 min post administration). hEPO mRNA was detected in both the sinusoidal cells as well as hepatocytes within treated mouse livers with no cross-reactivity observed for endogenous mouse EPO transcripts (Figure 1). mRNA was still present at detectable levels within the hepatocytes up to 72 h after a single dose.
Figure 1.
Detection of exogenous hEPO mRNA via ISH. Positive staining was observed in hepatocytes as well as sinusoidal cells. Strong detection was observed out to 72 h with remnant staining 1 week post administration.
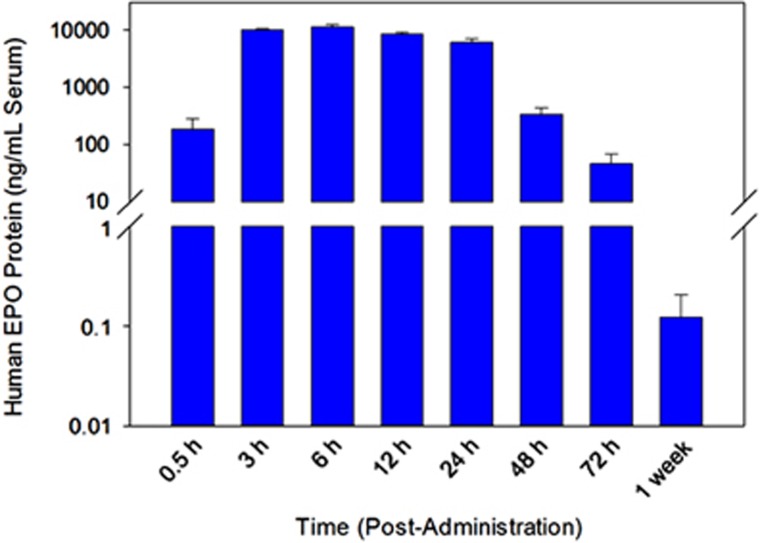
As expected for a secreted protein, hEPO was detected in the serum at various time points after administration of hEPO mRNA-loaded C12-200 lipidoid nanoparticles (Figure 2). hEPO protein was present at markedly higher quantities than physiological levels in a normal, healthy mouse at almost every time point examined, yielding a maximum serum concentration at ~6 h post administration. As shown in Figure 2 and listed in Table 1, upon treatment with a 1.0-mg kg−1 dose of EPO mRNA-encapsulated LNPs, ~11 μg of hEPO protein per ml of serum can be produced. This is several orders of magnitude (>125 000-fold) over the normal human physiological level of EPO (average normal levels reported to be 31.5–150 pg ml−1, average ~90 pg ml−1).66 Measurable levels of protein (~47 ng ml−1, Table 1) were still observed at 72 h with an appreciable increase over normal physiological levels. This indicates that after 3 days, levels ~520-fold over baseline physiological quantities were maintained. Further, 1 week after a single dose of exogenous hEPO mRNA, hEPO protein was present at slightly above normal physiological levels (~120 pg ml−1 serum). These data demonstrate the ability of organs, which internalize LNP-delivered mRNA to act as a ‘depot' for the production (and secretion) of hEPO protein.
Figure 2.
Quantification of secreted hEPO protein levels as measured via enzyme-linked immunosorbent assay (N=4 mice per group, error bars represent s.d.). The protein detected is a result of its production from EPO mRNA delivered intravenously via a single dose of LNPs (1.0 mg kg−1 encapsulated hEPO mRNA) over time (1 week). Aliquots of mouse serum were collected at the designated time points.
Table 1. Concentrations of secreted human EPO protein over time as measured via ELISA analysis (as depicted in Figure 1).
| Time post administration (h) | Secreted human EPO protein (ng ml−1) |
|---|---|
| 0.50 | 188 |
| 3 | 10 125 |
| 6 | 11 296 |
| 12 | 8521 |
| 24 | 6114 |
| 48 | 341 |
| 72 | 47.0 |
| 1 week | 0.12 |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; EPO, erythropoietin.
EPO mRNA (1.0 mg kg−1) was delivered via C12-200-based lipidoid nanoparticles. Values are depicted as ng of human EPO protein per ml of serum.
This experiment also revealed the rapid onset of protein production derived from such delivery. Approximately 190 ng ml−1 of hEPO protein can be observed within 30 min of dosing C12-200-based lipidoid nanoparticles delivering hEPO mRNA. Therefore, an ~2000-fold increase over normal physiological levels could be achieved in less than 1 h. Further, by 3 h, levels of hEPO protein reached >112 000-fold over normal.
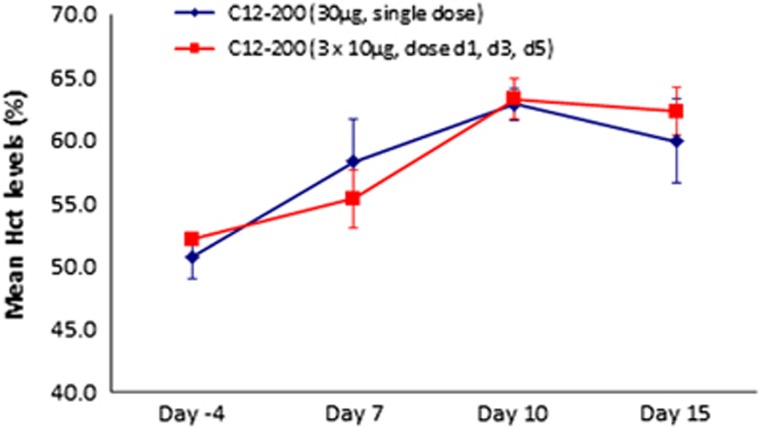
To verify that the protein produced via mRNA-loaded lipidoid nanoparticles was active, we measured hematocrit (Hct) levels in mice following delivery of EPO mRNA. Hct changes were monitored over a 15-day period as described above (Figure 3, Table 2). Two separate dosing regimens were given to wild-type mice: a single, bolus injection of 30 μg encapsulated hEPO mRNA (~1.0 mg kg−1) and three separate 10-μg injections on days 1, 3 and 5. As depicted in Figure 3, over the course of the study, both dosing regimens produced a significant increase in Hct (~20% change), further demonstrating that hEPO protein produced from a tissue depot is physiologically active using EPO mRNA nanoparticles.
Figure 3.
Hct levels in mice (N=4, error bars represent s.e.m.) treated with either single intravenous dose of hEPO mRNA-loaded LNPs or three injections (d1, d3 and d5). Whole-blood samples were taken before injection (day −4), day 7, day 10 and day 15.
Table 2. Hct levels of each group over a 15-day observation period.
| Formulation | Dose (μg per animal) |
Hct levels, mean (%)±s.e.m. |
|||
|---|---|---|---|---|---|
| Day −4 | Day 7 | Day 10 | Day 15 | ||
| C12-200 | 30 | 50.8±1.8 | 58.3±3.3 | 62.8±1.3 | 59.9±3.3 |
| C12-200 | 10 (× 3) | 52.2±0.5 | 55.3±2.3 | 63.3±1.6 | 62.3±1.9 |
Abbreviation: Hct, hematocrit.
Mice were either dosed as a single, 30-μg intravenous injection, or three 10-μg intravenous injections, every other day.
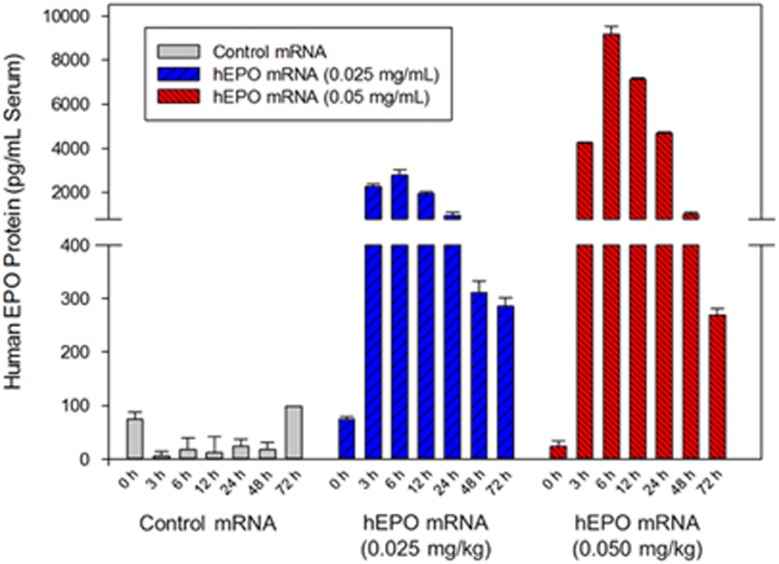
To further evaluate the potential of this formulation, we dosed three male cynomolgus monkeys (~4 kg in body weight) and monitored for hEPO protein production via serum analysis over time (Figure 4). Each monkey received a single bolus, intravenous injection via the ear vein of either hEPO mRNA LNPs (two monkeys) or control LNP encapsulating nonsense mutated, untranslatable mRNA (one monkey). The control formulation was dosed at 0.025 mg kg−1 while the two treated monkeys were dosed at 0.025 and 0.050 mg kg−1. All formulations were well tolerated by the monkeys with no clinical signs and normal physiological liver enzyme levels (alanine transaminase/aspartate transaminase) observed (Supplementary Table 1).
Figure 4.
hEPO protein levels observed over 72 h time course in cynomolgus monkeys. Monkeys were either dosed at 0.025 or 0.050 mg kg−1. Control formulation (untranslatable) was dosed at 0.025 mg kg−1. Error bars represent replicates of analysis per sample.
While accounting for dose differences, the pharmacokinetic profile of EPO production in the cynomolgus monkeys shows striking similarity to what was observed in mice (Figure 2; Table 1). Serum EPO levels reach their maximum 6 h post administration, while still detectable after 72 h at as low a dose of 0.025 mg kg−1. Monkeys treated with a dose of 0.050 mg kg−1 demonstrated hEPO protein levels of ~9000 pg ml−1 (6 h time point), which results in an increase over physiological levels by over 100-fold.
In sum, these data clearly demonstrate the ability to effectively deliver mRNA to the liver when administering mRNA-loaded LNPs. The exogenous mRNA was efficiently processed by the host translational machinery to produce a fully formed, functional protein. Further, this depot effect provides large quantities of desired protein secreted systemically resulting in extended pharmacokinetic plasma exposure profiles and substantial pharmacodynamic effects within these mice.
In vivo hFIX protein production
To examine the potential of mRNA nanoparticles for the treatment of hemophilia B, we formulated hFIX mRNA-loaded LNPs and evaluated their activity in vivo. Successful delivery of hFIX mRNA upon intravenous injection of a single 1.0 mg kg−1 dose was determined by ISH (Supplementary Figure 1). Staining patterns of hFIX mRNA within both sinusoidal cells and hepatocytes were equivalent to what was observed with EPO formulations (Figure 1). Again, no cross-reactivity was observed for endogenous mouse FIX transcripts within the liver. The strongest signal occurs at the earliest time point analyzed (30 min) and diminishes over the course of 3 days, with detectable mRNA still present at 72 h post administration.
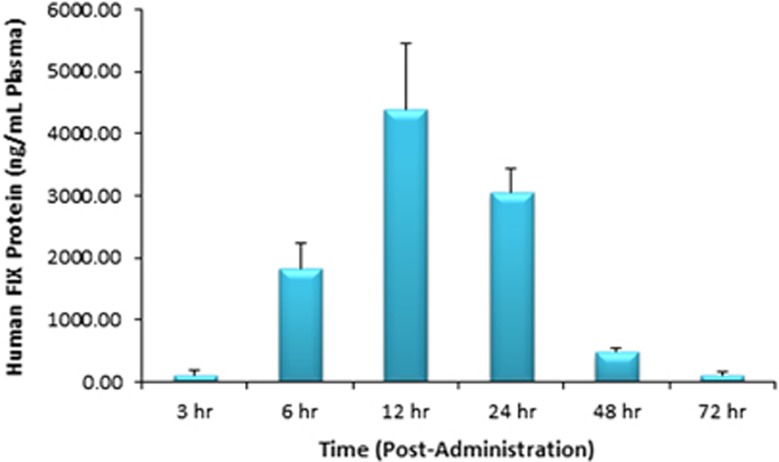
A pharmacokinetic analysis over 72 h showed that MRT-derived FIX protein could be detected at all time points tested (Figure 5). The peak plasma concentration was observed at 12 h post injection with a value of ~4.4 μg of FIX protein per ml of plasma. This robust protein production followed by secretion into the blood stream for a sustained blood residence time represents another successful example of the depot effect upon application of LNP-mediated MRT.
Figure 5.
Quantification of secreted hFIX protein levels measured using enzyme-linked immunosorbent assay (N=4 mice per group, error bars represent s.d.). FIX protein is produced from FIX mRNA delivered via nanoparticles (1.0 mg kg−1 hFIX mRNA per single intravenous dose, based on encapsulated mRNA). FIX protein is monitored in plasma through 72 h.
To further establish the potential for MRT as a viable therapeutic modality, we applied our hFIX mRNA-loaded LNPs in a hemophilia B mouse model.65 FIX KO mice were treated with either saline or C12-200-based hFIX mRNA-loaded LNPs (0.25 and 0.50 mg kg−1). FIX serum levels, activity and subsequent efficacy were assessed in this mouse model.
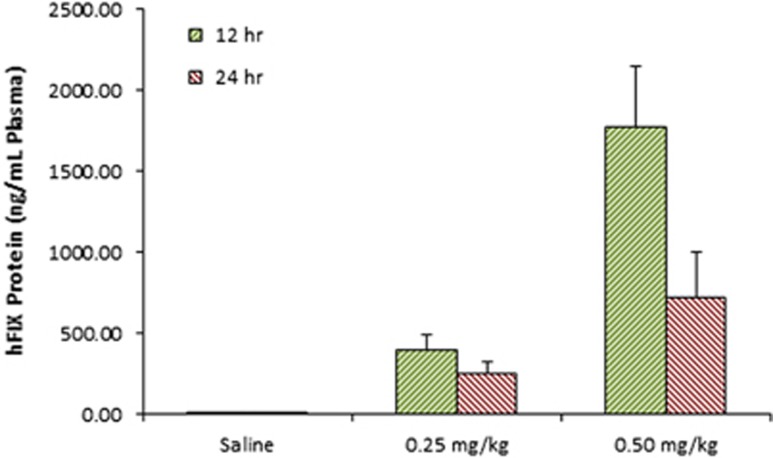
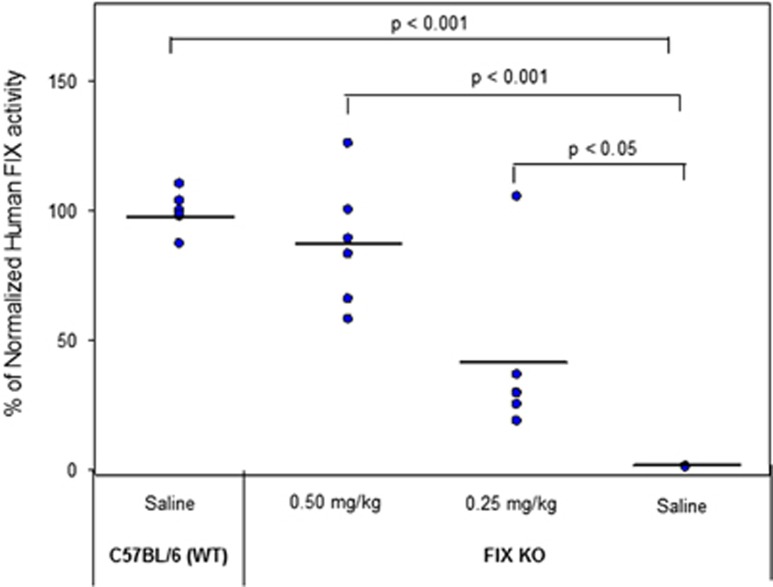
Plasma levels of hFIX protein were measured 12 and 24 h post administration (Figure 6). Plasma FIX levels for a dose of 0.25 and 0.50 mg kg−1 in the KO mice reached 400 ng ml−1 and 1.77 μg ml−1, respectively, 12 h post administration. All groups were tested for FIX activity using a commercially available chromogenic assay and are represented in Figure 7. The wild-type (C57Bl/6) saline-treated group was normalized to represent 100% activity of FIX protein. The FIX KO saline-treated group showed trace amounts of FIX activity (~1.15%) of wild-type levels. This is consistent with, although slightly lower than, the 8% residual activity reported previously as measured via an alternate method.65 Upon treatment of the mice with hFIX mRNA, we observed a marked increase in activity resulting in 40.9% and 87% of wild-type levels for the 0.25 and 0.50 mg kg−1 groups, respectively.
Figure 6.
Quantification of secreted hFIX protein levels in FIX KO mice (N=5, error bars represent s.d.) after single, intravenous dose (0.25 and 0.50 mg kg−1, based on encapsulated mRNA). FIX protein is measured in plasma 12 and 24 h post administration. Saline represents control group.
Figure 7.
FIX protein activity as measured by a chromogenic assay. Controls include saline-treated wild-type (C57BL/6) and FIX KO cohorts. All values are normalized to wild-type FIX activity values.
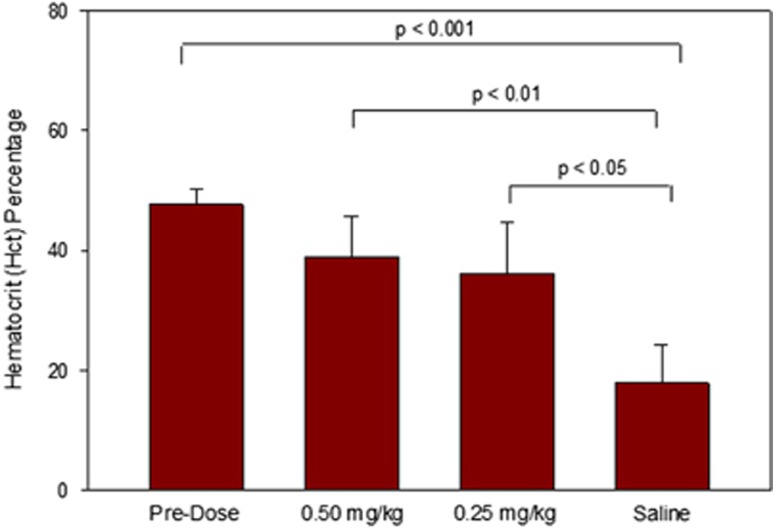
Furthermore, given the high FIX levels observed, we expected such doses to be therapeutic. We therefore hypothesized that upon application of an incision wound, prolonged bleeding would occur in the untreated KO mice as opposed to hFIX mRNA-treated cohorts. These effects could be measured via changes in Hct levels (that is, lower levels were a result of extended blood loss). To achieve this, at ~12 h post administration, the mice were subjected to a small surgical incision on the back of the animal. This time point was selected to represent the maximum observed hFIX protein plasma levels from the pharmacokinetic data previously obtained in wild-type mice. As represented in Figure 8, pre-treatment Hct levels averaged at ~47.7%. Final Hct readings were taken from blood samples obtained 12 h post incision (24 h post administration). Saline-treated FIX KO mice had a final Hct level of 17.9% representing a 62.5% overall decrease from pre-treatment levels. Mice that were treated with hFIX mRNA had much more sustained Hct levels measured at 36.1% and 39.0% for 0.25 and 0.50 mg kg−1 dosing, respectively. The overall drop in Hct for the treated groups was significantly inhibited (24.3% and 18.2% overall decrease for 0.25 and 0.50 mg kg−1, respectively) as compared with the saline treatment, which can be attributed to the presence of active FIX protein to restore clotting and prevent further bleeding.
Figure 8.
Hct levels of blood from FIX KO mice (N=5, error bars represent s.d.) 12 h post incision (24 h post injection). Mice were either dosed with 0.25 or 0.50 mg kg−1 of C12-200-based hFIX mRNA-loaded LNPs. Control FIX KO mice were treated with saline.
Discussion
Here we describe the successful application of two separate mRNA-loaded lipidoid nanoparticle systems toward hepatocyte deposition of exogenous mRNA and subsequent high-level protein production sustained over several days, creating a depot system. Such a system is presumed to be the result of uptake and clearance mechanisms with independent kinetic constants. These include the kinetics of cellular uptake of the LNPs, the kinetics of intracellular compartment escape (endosomal/lysosomal), the kinetics of release of the exogenous mRNA, the half-life of the mRNA, as well as the rate of translation of the mRNA into protein. Further, with respect to secreted proteins, such processes may occur in multiple organs to which the LNP was distributed, resulting in contribution toward the total circulating pool of protein produced from the exogenous mRNA. Previous studies using this system (not reported) have demonstrated that >80% of the packaged mRNA is deposited in the liver. Moreover, an additional complicating factor concerns possible differences in the accessibility and function of the endogenous translational machinery that could vary with different organs and is beyond the scope of this work.
Observations of hepatocellular mRNA delivery and protein production were consistent between the two mRNA constructs. The first mRNA transcript delivered using this approach was designed to produce hEPO protein. EPO is a hormone, which has long been used as a therapeutic with applications in anemia and oncology.67 The majority of these drugs have been in the protein form itself, serving as a hormone replacement/supplement therapy. Recently, it has been demonstrated that naked, modified EPO mRNA can be administered subdermally to mice with favorable pharmacodynamic effects.20, 21 We applied a synthetic hEPO mRNA transcript toward LNP-based delivery for enrichment in the liver. As demonstrated by ISH techniques, rapid deposition in the liver was achieved, which was shown by intense staining in both sinusoidal cells and hepatocytes within 30 min in the livers of treated mice. Furthermore, detectable hEPO mRNA was observed for up to 72 h after a single, intravenous administration.
High levels of protein production were observed following a single treatment of hEPO MRT in wild-type mice as quantified and depicted in Figure 2. Mouse serum levels reached >125 000-fold normal physiological levels of EPO protein (>11.2 μg EPO per ml of serum) within the first 6 h of administration. As noted above, rapid onset of protein production can be observed within 30 min of administration while levels reached several orders of magnitude above normal levels by 3 h post dose. Such observations indicate the potential use of MRT as a protein production ‘depot' therapy. In the example presented here, a single 1.0 mg kg−1 produces a sustained exposure of hEPO protein that is detectable for 1 week post administration.
Hct levels were monitored before and post treatment with EPO MRT. Two dosing regimens were applied to wild-type mice consisting of either a single, 30 μg bolus intravenous injection or three 10 μg hEPO mRNA-loaded C12-200-based LNPs dosed every other day (days 1, 3 and 5). Both dosing regimens resulted in an ~20% change in Hct as represented in Figure 3. These findings confirm that hormonal activity is maintained within the secreted, MRT-derived hEPO protein upon encapsulation and delivery of the respective mRNA via LNPs.
The potential of this formulation was further demonstrated by the application of LNP-encapsulated hEPO mRNA in non-human primates. A pharmacokinetic profile of serum hEPO protein levels was obtained in male cynomolgus monkeys when administered at two separate doses (0.025 and 0.050 mg kg−1, Figure 4) that was remarkably similar to the results seen in mice. Serum levels of EPO within both dose regimens could be detected up to 72 h post administration within maximum levels reached by 6 h (matching trends observed in mice vide supra). Control formulations consisting of an untranslatable (nonsense mutated, non-protein producing) mRNA construct resulted in no EPO protein detection as anticipated. All formulations were well tolerated in all species studied.
We used a second mRNA transcript (hFIX) using this LNP delivery formulation to demonstrate the broad potential applicability of the technology. The application of FIX MRT can be used for the treatment of hemophilia B, a recessive, X-linked rare genetic blood coagulation disorder predominantly caused by mutations in the FIX gene.68, 69, 70 Patients suffering from this disease have an inability to form proper blood clotting, resulting in varying degrees of bleeding episodes as well as other complications. The only currently approved treatment modality for this disease is the direct infusion of FIX enzyme, either from recombinant or plasma-derived sources.68, 69 Patients suffering from hemophilia B have been classified based on the percentage of functional FIX protein within the body: >5–40% of normal physiological levels is defined as mild; 1–5% of normal physiological levels is defined as moderate; <1% of normal physiological levels is defined as severe.71 The relative ratio of each classification level within the patient population has not been fully determined, however it has been reported that up to ~70% of patients suffer from moderate to severe forms of the disease.72, 73
hFIX mRNA was encapsulated in an identical fashion to that described above using hEPO mRNA C12-200-based LNPs. Encapsulation efficiency as well as particle size was consistent with the hEPO mRNA formulations. A pharmacokinetic study to assess FIX protein production (and subsequent secretion) was performed similarly to the EPO MRT study described above. Wild-type mice were treated with a 1.0-mg kg−1 single, intravenous dose of C12-200-based hFIX mRNA-loaded LNPs, and were analyzed at selected time points. To confirm specific delivery and enrichment of hepatic cells, ISH methods were used. Similar staining patterns and amounts were observed when delivering hFIX mRNA via LNPs as the previous EPO study (vide supra, Supplementary Figure 1). Upon quantitative assessment of plasma hFIX protein levels within the treated mice, we observed sustained protein production, with subsequent secretion, for 72 h after a single, intravenous administration. Again, high-level protein production resulted in maximum plasma levels reaching ~4.40 μg of hFIX protein per ml within 12 h of dosing (Figure 5). While normal physiological levels of FIX protein are relatively high (2–3 μg ml−1 plasma), the observed FIX protein production in wild-type mice via FIX MRT can be considered to be well within therapeutic levels.74 More specifically, moderate to severe patients represent only up to 5% active protein available yet constitute up to ~70% of the patient population.72, 73 This equates to ~100 ng FIX protein per ml plasma maximum. On the basis of a single 1.0-mg kg−1 dose of hFIX mRNA-loaded LNPs, we achieve ~40-fold higher than this level. Therefore, a significant portion of the patient population may be candidates for a possible future treatment with this modality.
To establish the possible therapeutic value of FIX MRT, we applied this LNP technology toward a hemophilia B mouse model.65 This model entails a FIX transgenic KO, which has been reported to have minimal residual FIX activity. Traditional measures of efficacy in hemophilia models involve comparisons of bleeding times or volumes upon tail snip.75, 76 However, such methods are sensitive to many variables, and protocols vary considerably within the literature.77, 78 We sought to establish an alternative measure for efficacy that would provide a surrogate for FIX protein function. To this end, we measured Hct levels pre and post incision to serve as a reflection of the degree of bleeding, which occurred within treated and untreated animals.79 Mice deficient in FIX protein would demonstrate prolonged bleeding episodes upon injury, which could be directly compared with those mice, which have been pre-treated with FIX MRT.
FIX KO mice were treated with a single dose of FIX mRNA-loaded C12-200 nanoparticles at either 0.25 or 0.50 mg kg−1. Plasma levels of hFIX protein were measured at 12 and 24 h post administration (Figure 6). Plasma levels of FIX protein at 12 h reached ~1.8 μg ml−1 plasma and 400 ng ml−1 plasma for 0.50 and 0.25 mg kg−1 doses, respectively. These FIX concentrations correspond to 90% (0.50 mg kg−1) and 20% (0.25 mg kg−1) of normal physiological levels and are well within therapeutic range. This is reflected both in the activity data and the Hct observations. Detectable levels were still observed 24 h post administration at 724 and ~250 ng ml−1 for the respective doses. Protein levels observed in treated wild-type mice were equivalent for identical doses (Supplementary Figure 2).
Further, FIX activity measurements were conducted on all treated and untreated mice. Figure 7 represents a comparison of activity of treated mice as compared with normal FIX activity levels in untreated, wild-type mice (normalized to 100%). A single 0.50-mg kg−1 dose of FIX MRT resulted in a restoration of activity to 87% of wild-type levels. This level far surpasses the range of ‘mild' defined as 5–40% activity.71 Moreover, a single dose of only 0.25 mg kg−1 provided activity of ~40% of wild-type levels. These increases in activity levels are statistically significant when compared with the saline group, although one mouse demonstrated abnormally high activity (105% of wild-type levels) in the lower-dose cohort (0.25 mg kg−1).
At 12 h post administration, a group of mice were subjected to a small incision (~1 cm) in the thoracic region of each mouse followed by suturing while a separate group was killed. After 12 more hours (24 h total post administration), the injured mice were killed and Hct levels were measured and compared with pre-treatment baseline values. As depicted in Figure 8, a substantial decrease in Hct was observed in saline-treated KO mice as compared with pre-treatment levels (17.9% (saline) vs 47.7% (pre-treatment)). This decrease in Hct was inhibited upon treatment with FIX MRT as a function of dose (36.1% and 39.0% for 0.25 and 0.50 mg kg−1, respectively). Wild-type mice were treated in an identical fashion with respect to both dose and incision with no change in Hct levels observed. Such production of active FIX protein after a single dose of FIX MRT demonstrates therapeutic potential relevant for enabling treatment of severe through mild classifications of hemophilia B patient population.
Summary
In summary, we have established successful encapsulation and delivery of mRNA constructs to the liver using C12-200-based LNP technology. Hepatocyte deposition and detection of active mRNA was observed for two separate transcripts, hEPO and hFIX mRNA. Furthermore, high level protein production (μg ml−1) and secretion sustained for up to 1 week post injection was achieved. Active MRT-derived protein was confirmed through pharmacodynamic measures (increase in Hct (EPO)) and therapeutic means (prevention of blood loss) in a hemophilia B (FIX KO) disease model. These examples demonstrate the applicability of formulated mRNA to treat not only organs of direct LNP uptake but also sites within the body, which benefit from the prolonged exposure of MRT-derived protein secreted into the bloodstream.
Materials and methods
Lipid materials
The formulations described herein consisted of a multi-component lipid mixture of specific ratios using a combination of lipidoid, helper lipids and PEGylated lipids designed to encapsulate mRNA therapeutic molecules. C12-200 was synthesized as previously described.61 DOPE (1,2-dioleyl-sn-glycero-3-phosphoethanolamine, Avanti Polar Lipids, Alabaster, AL, USA) and cholesterol (Sigma, St Louis, MO, USA) served as helper lipids within the nanoparticle. The PEGylated lipid selected was a dimyristoyl glycerol–polyethylene glycol (PEG) analog with a PEG molecular weight of ~2000 Da (DMG-PEG-2K) purchased from NOF (White Plains, NY, USA).
mRNA material
hEPO and hFIX were synthesized by in vitro transcription via T7 RNA polymerase from a plasmid DNA template encoding the gene using unmodified nucleotides, which was followed by the addition of a 5′-cap structure (Cap 1) and a 3′-poly(A) tail of ~250 nucleotides in length as determined by gel electrophoresis.80 Fixed 5′- and 3′-untranslated regions were constructed to flank the coding sequences of each mRNA. The control mRNA consisted of a nonsense mutated coding sequence designed to prevent protein production.
Formulation protocol
Example 1
Aliquots of 50 mg ml−1 ethanolic solutions of C12-200, DOPE, cholesterol and DMG-PEG2K were mixed and diluted with ethanol to 3 ml final volume. Separately, an aqueous buffered solution (10 mM citrate/150 mM NaCl, pH 4.5) of hEPO mRNA was prepared from a 1-mg ml−1 stock. The lipid solution was injected rapidly into the aqueous mRNA solution and shaken to yield a final suspension in 20% ethanol. The resulting nanoparticle suspension was filtered, diafiltrated with 1 × phosphate-buffered saline (pH 7.4), concentrated and stored at 2–8 °C. Final concentration=0.20 mg ml−1 EPO mRNA (encapsulated). % Encapsulation=75% % recovery=81% Zave=91 nm (Dv(50)=75 nm); polydispersity=0.14.
Example 2
Aliquots of 50 mg mL−1 ethanolic solutions of C12-200, DOPE, cholesterol and DMG-PEG2K were mixed and diluted with ethanol to 3 ml final volume. Separately, an aqueous buffered solution (10 mM citrate/150 mM NaCl, pH 4.5) of hFIX mRNA was prepared from a 1-mg ml−1 stock. The lipid solution was injected rapidly into the aqueous mRNA solution and shaken to yield a final suspension in 20% ethanol. The resulting nanoparticle suspension was filtered, diafiltrated with 1 × phosphate-buffered saline (pH 7.4), concentrated and stored at 2–8 °C. Final concentration=0.20 mg ml−1 FIX mRNA (encapsulated). % Encapsulation=78% % recovery=79% Zave=86 nm (Dv(50)=69 nm); polydispersity=0.16.
In vivo animal models
EPO and FIX pharmacokinetic studies used 6- to 8-week-old male mice (CD-1). For FIX efficacy experiments, 10- to 12-week-old male mice (C57BL/6J) were used as wild-type reference controls. Male FIX KO mice (8–18 weeks old) were used for efficacy studies.65 All mice were treated via tail vein injection. Male cynomolgus monkeys (~4 kg) were treated with a single intravenous injection as a total dose volume of 1 ml via ear vein injection.
Select FIX KO mice were subjected to incision to determine efficacious treatment. At 12 h post dose administration, animals were anesthetized with isoflurane before surgery. With the animals in a prone position, the incision site (previously shaved) was aseptically prepared using betadine followed by alcohol. The animals were subjected to a small (~1.0 cm) surgical skin incision at the dorsal thoracic region and the incision was closed with surgical sutures.
Analysis of protein produced via intravenously delivered mRNA-loaded nanoparticles
Injection protocol
All pharmacokinetic studies were performed using male CD-1 mice of ~6–8 weeks of age at the beginning of each experiment. Samples were introduced by a single bolus tail vein injection of the specified dose of encapsulated EPO or FIX mRNA-loaded LNPs. Mice were killed and perfused with saline at the designated time points.
Isolation of organ tissues for analysis
The liver of each mouse was collected and stored in either 10% neutral buffered formalin or snap-frozen and stored at −80 °C for analysis.
Isolation of plasma/serum for analysis
All animals were killed by CO2 asphyxiation at respective time points post administration (±5%) followed by thoracotomy and terminal cardiac blood collection. Whole blood (maximal obtainable volume) was collected via cardiac puncture on killed animals. For serum collection, whole blood was placed into serum separator tubes, allowed to clot at room temperature for at least 30 min, centrifuged at 22±5 °C at 9300 g for 10 min and extracted. For plasma collection, whole blood was placed into either lithium heparin tubes or citrate-coated tubes and processed to plasma. For interim blood collections, ~40–50 μl of whole blood were collected via facial vein puncture or tail snip. Samples collected from non-treatment animals were used as a baseline level for comparison to study animals.
Enzyme-linked immunosorbent assay analysis
Quantification of EPO protein was performed following procedures reported for hEPO enzyme-linked immunosorbent assay analysis kit (Quantikine IVD, R&D Systems, Catalog # Dep-00; Minneapolis, MN, USA). Positive controls used consisted of ultrapure and tissue culture grade recombinant hEPO protein (R&D Systems, Catalog # 286-EP and 287-TC, respectively). Blood samples were taken at designated time points and processed as described above. Detection was monitored via absorption (450 nm) on a Molecular Devices Flex Station instrument (Sunnyvale, CA, USA). Quantification of FIX protein was performed following procedures reported for hFIX ELISA kit (AssayMax, Assay Pro, Catalog # EF1009-1; St Charles, MO, USA). High specificity for the human forms of each respective protein was obtained with no cross-reactivity observed in mouse samples (EPO/FIX) and minimal cross-reactivity observed in monkey serum samples (EPO).
Hct analysis
Whole-blood samples were collected in heparinized micro-capillary blood collection tubes and centrifuged at 10 000–15 000 g for 5 min at ambient temperature. Packed cell volume was calculated vs total volume to obtain Hct values.
FIX activity assay
FIX activity measurements in plasma samples were performed using the Rossix Factor IX Activity Chromogenic Assay (ROX Factor IX 900020, Mölndal, Sweden) according to the manufacturer's instructions. Samples were collected 12 h post dose administration.
hEPO and hFIX ISH assay
The detection of exogenous hEPO mRNA and hFIX mRNA was performed by Advanced Cell Diagnostics (ACD, Hayward, CA, USA) using proprietary technology involving a probe design strategy that allows simultaneous signal amplification and background suppression to achieve single-molecule visualization while preserving tissue morphology. Tissues and cells mounted on slides are first pre-treated (antigen retrieval) to prepare for hybridization. Oligonucleotide target probes (up to 20 probe pairs) are hybridized to the RNA in the sample, followed by a series of steps and washes designed to amplify the signal. The probe sets were designed to not cross-react with mouse, rat, pig and rhesus monkey.
Acknowledgments
We thank Kari Harbert for her work on breeding the factor IX knockout mouse colony, and Kelly Daly for dose administration on all mouse studies. We also acknowledge Lisa Louth and Cindy Bell for activity assay experiments. FIX KO mouse colony was obtained at Jackson Laboratories via licensing through the University of North Carolina, Chapel Hill.
Footnotes
Supplementary Information accompanies this paper on Gene Therapy website (http://www.nature.com/gt)
Dedicated to the memory of Brad Guild.
FD, BG and MWH are inventors on patent applications filed in 2009 and 2011 covering delivery of mRNA and those such as EPO and Factor IX (Shire Pharmaceuticals). The remaining authors declare no conflict of interest.
Supplementary Material
References
- Alvarez-Salas LM. Nucleic acids as therapeutic agents. Curr Top Med Chem 2008; 8: 1379–1404. [DOI] [PubMed] [Google Scholar]
- Vannucci L, Lai M, Chiupessi F, Ceccherrini-Nerri L, Pistello M. Viral vectors: a look back and ahead on gene transfer technology. New Microbiol 2013; 36: 1–22. [PubMed] [Google Scholar]
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev 2014; 15: 541–555. [DOI] [PubMed] [Google Scholar]
- O'Reilly M, Kohn DB, Bartlett J, Benson J, Brooks PJ, Byrne BJ et al. Gene therapy for rare diseases: summary of National Institutes of Health Workshop. Hum Gene Ther 2013; 24: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotrim AP, Baum BJ. Gene therapy: some history, applications, problems and prospects. Toxicol Pathol 2008; 36: 97–103. [DOI] [PubMed] [Google Scholar]
- Wirth T, Parker N, Ylä-Hertualla S. History of gene therapy. Gene 2013; 525: 162–169. [DOI] [PubMed] [Google Scholar]
- Davidson BL, McCray PB Jr. Current prospects for rna interference-based therapies. Nat Rev 2011; 12: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev 2007; 59: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 2004; 3: 318–329. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Dasaradhi PVN, Mohmmed A, Malhotra P, Bhatnagar RJ, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev 2005; 67: 657–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboul-Fadl T. Antisense oligonucleotides: the state of the art. Curr Med Chem 2005; 12: 2193–2214. [DOI] [PubMed] [Google Scholar]
- Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther 2002; 1: 347–355. [PubMed] [Google Scholar]
- Chan JHP, Lim S, Wong WSF. Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol 2006; 33: 533–540. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. MicroRNA therapeutics: a new niche for antisense therapeutics. Trends Mol Med 2006; 12: 99–101. [DOI] [PubMed] [Google Scholar]
- Kaur G, Roy I. Therapeutic applications of aptamers. Expert Opin Investig Drugs 2008; 17: 43–60. [DOI] [PubMed] [Google Scholar]
- Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol 2006; 10: 282–289. [DOI] [PubMed] [Google Scholar]
- Germer K, Leonard M, Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol Biol 2013; 4: 27–40. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Current prospects for mRNA gene delivery. Eur J Pharm Biopharm 2009; 71: 484–489. [DOI] [PubMed] [Google Scholar]
- Kormann MSD, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 2011; 29: 154–159. [DOI] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 2012; 20: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays LE, Ammon-Treiber S, Mothes B, Alkhaled M, Rottenberger J, Muller-Hermelink ES et al. Modified Foxp3 mRNA protects against asthma through an IL-10-dependent mechanism. J Clin Invest 2013; 123: 1216–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L, Lui KO, Von Gise A, Ma Q, Ebina W, Ptasek LM et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 2013; 31: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Itaka K, Kondo K, Yamasoba T, Kataoka K. Treatment of neurological disorders by introducing mRNA in vivo using polyplex nanomicelles. J Control Release 2015; 201: 41–48. [DOI] [PubMed] [Google Scholar]
- Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett 2015; 15: 7300–7306. [DOI] [PubMed] [Google Scholar]
- Dong Y, Dorkin JR, Wang W, Chang PH, Webber MJ, Tang BC et al. Poly(glycoamidoamine) brushes formulated nanomaterials for systemic siRNA and mRNA delivery in vivo. Nano Lett 2016; 16: 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su H-H, Yang Y, Hu Y, Zhang L, Blancafort P et al. Systemic delivery of modified mrna encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol Ther 2013; 21: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide B, Carralot JP, Reese A, Scheel B, Eigentler TK, Hoerr I et al. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 2008; 31: 180–188. [DOI] [PubMed] [Google Scholar]
- Fotin-Mleczek M, Zanzinger K, Heidenreich R, Lorenz C, Thess A, Duchardt KM et al. Highly potent mRNA based cancer vaccines represent an attractive platform for combination therapies supporting an improved therapeutic effect. J Gene Med 2012; 14: 428–439. [DOI] [PubMed] [Google Scholar]
- Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA 2012; 109: 14604–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 2008; 21: 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel MA, Weinstein JR, Schaffer DV. Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Ther 2012; 19: 694–700. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethyleneimine (PEI)-complexed siRNA in vivo. Gene Ther 2005; 12: 461–466. [DOI] [PubMed] [Google Scholar]
- Lynn DM, Langer R. Degradable poly(beta-amino esters): synthesis, characterization and self-assembly with plasmid DNA. J Am Chem Soc 2000; 122: 10761–10768. [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL et al. Dynamic polyconujugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA 2007; 104: 12982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc 2002; 124: 8–9. [DOI] [PubMed] [Google Scholar]
- Murthy N, Campbell J, Fausto N, Hoffman AS, Stayton PS. Design and synthesis of pH-responsive polymeric carriers that target uptake and enhance the intracellular delivery of oligonucleotides. J Control Release 2003; 89: 365–374. [DOI] [PubMed] [Google Scholar]
- Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev 2006; 58: 467–486. [DOI] [PubMed] [Google Scholar]
- Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm 2009; 6: 659–668. [DOI] [PubMed] [Google Scholar]
- Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev 2009; 61: 710–720. [DOI] [PubMed] [Google Scholar]
- Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc Natl Acad Sci USA 2011; 108: 12996–13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P, Monck M, Lee D, Ludkovski O, Leng EC, Clow K et al. Stabilized plasmid-lipid particles for systemic gene therapy. Gene Ther 2000; 7: 1867–1874. [DOI] [PubMed] [Google Scholar]
- Heyes J, Palmer L, Chan K, Giesbrecht C, Jeffs L, MacLachlan I. Lipid encapsulation enables the effective systemic delivery of polyplex plasmid DNA. Mol Ther 2007; 15: 713–720. [DOI] [PubMed] [Google Scholar]
- Oishi M, Sasaki S, Nagasaki Y, Kataoka K. pH-responsive oligodeoxynucleotide (ODN)-poly(ethylene glycol) conjugate through acid-labile β-thiopropionate linkage: preparation and polyion complex micelle formation. Biomacromolecules 2003; 4: 1426–1432. [DOI] [PubMed] [Google Scholar]
- Burcovich B, Veronese FM, Zarylova V, Bonora GM. Branched polyethyleneglycol (bPEG) conjugated antisense oligonucleotides. Nucleosides Nucleotides 1998; 17: 1567–1570. [Google Scholar]
- Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bionconjug Chem 2009; 20: 5–14. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol 2007; 25: 1149–1157. [DOI] [PubMed] [Google Scholar]
- Tsui NBY, Ng EKO, Lo YMD. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem 2002; 48: 1647–1653. [PubMed] [Google Scholar]
- Lin Q, Chen J, Zhang Z, Zheng G. Lipid-based nanoparticles in the systemic delivery of siRNA. Nanomedicine 2014; 9: 105–120. [DOI] [PubMed] [Google Scholar]
- Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther 2010; 18: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XB, Lee RJ. Tumor-selective targeted delivery of genes and antisense oligodeoxyribonucleotides via the folate receptor. Adv Drug Deliv Rev 2004; 56: 1193–1204. [DOI] [PubMed] [Google Scholar]
- Li SD, Chono S, Huang L. Efficient gene siliencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release 2008; 126: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CW, Lu DW, Yeh MK, Shiau CY, Chiang CH. Novel RGD-lipid conjugate-modified liposomes for enhancing siRNA delivery in human retinal pigment epithelial cells. Int J Nanomed 2011; 6: 2567–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill J, Singhania R, Burgess M, Allavena R, Wu S, Blumenthal A et al. Efficient biodistribution and gene silencing in the lung epithelium via intravenous liposomal delivery of siRNA. Mol Ther Nucleic Acids 2013; 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K. Intracellular targeting delivery of liposomal drugs to solid tumors based on EPR effects. Adv Drug Deliv Rev 2011; 63: 161–169. [DOI] [PubMed] [Google Scholar]
- Tam YYC, Chen S, Cullis PR. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics 2013; 5: 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol 2010; 28: 172–178. [DOI] [PubMed] [Google Scholar]
- Koltover I, Salditt T, Radler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 1998; 281: 78–81. [DOI] [PubMed] [Google Scholar]
- Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed 2012; 51: 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release 2005; 107: 276–287. [DOI] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol 2008; 26: 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR et al. Lipid-like materials for low-dose in vivo gene silencing. Proc Natl Acad Sci USA 2010; 107: 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup CJ, Sahay G et al. Rapid discovery of potent siRNA-lipid-nanoparticles. J Am Chem Soc 2012; 134: 6948–6951. [DOI] [PubMed] [Google Scholar]
- Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol 2013; 31: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HF, Maeda N, Smithies O, Straight DL, Stafford DW. A coagulation factor-IX deficient mouse model for human hemophilia B. Blood 1997; 90: 3962–3966. [PubMed] [Google Scholar]
- Gardner DG, Shoback D. Normal hormone reference ranges. In Shanahan JF, Boyle PJ (eds) Greenspan's Basic and Clinical Endocrinology, 9th edn The McGraw-Hill Companies: China, 2011, Appendix 825. [Google Scholar]
- Bunn HF. Erythropoietin. Cold Spring Harb Perspect Med 2013; 3: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande P. Haemophilia B: Christmas disease. Expert Opin Pharmacother 2005; 6: 1517–1524. [DOI] [PubMed] [Google Scholar]
- Coppola A, Di Capua M, Di Minno MND, Di Palo M, Marrone E, Lerano P et al. Treatment of hemophilia: a review of current advances and ongoing issues. J Blood Med 2010; 1: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini M, Frattini F, Crestani S, Sissa C, Bonfanti C. Treatment of hemophilia B: focus on recombinant factor IX. Biologics 2013; 7: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White GC, Rosendaal F, Aledort LM, Lusher JM, Rothschild C, Ingerslev J. Definitions in Hemophilia. Thromb Haemost 2001; 85: 560. [PubMed] [Google Scholar]
- Franchini M, Frattini F, Crestani S, Bonfanti C. Hemophilia B: current pharmacotherapy and future directions. Expert Opin Pharmacother 2012; 13: 2053–2063. [DOI] [PubMed] [Google Scholar]
- Soucie JM, Miller CH, Kelly FM, Payne AB, Creary M, Bockenstedt PL et al. A study of prospective surveillance for inhibitors among persons with haemophilia in the United States. Haemophilia 2014; 20: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler EJ, Davie EW. The Molecular Basis of Blood Diseases 3rd edn 2001, p 691.
- Broze Jr GJ, Zheng-Feng Y, Lasky N. A tail vein bleeding time model and delayed bleeding in hemophiliac mice. Thromb Haemost 2001; 85: 747–748. [PubMed] [Google Scholar]
- Sambrano GR, Weiss EJ, Zheng Y-W, Huang W, Coughlin SR. Role of thrombin signalling in platelets in haemostasis and thrombosis. Nature 2001; 413: 74–78. [DOI] [PubMed] [Google Scholar]
- Greene TK, Schiviz A, Hoellriegl W, Poncz M, Muchitsch EM. Towards a standardization of the murine tail bleeding model. J Thromb Haemost 2010; 8: 2820–2822. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jennings NL, Dart AM, Du XJ. Standardizing a simpler, more sensitive and accurate tail bleeding assay in mice. World J Exp Med 2012; 2: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui T, Reheman A, Ni H, Gross PL, Yin F, Monroe D et al. Abnormal hemostasis in a knock-in mouse carrying a variant of factor IX with impaired binding to collagen type IV. J Thromb Haemost 2009; 7: 1843–1851. [DOI] [PubMed] [Google Scholar]
- Fechter P, Brownlee GG. Recognition of mRNA cap structures by viral and cellular proteins. J Gen Virol 2005; 86: 1239–1249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.