In Arabidopsis homografts, proteins carrying organelle-targeting signals move from companion cells to sieve elements and traffic across the graft union to target the correct organelle in the root.
Abstract
In addition to moving sugars and nutrients, the phloem transports many macromolecules. While grafting and aphid stylectomy experiments have identified many macromolecules that move in the phloem, the functional significance of phloem transport of these remains unclear. To gain insight into protein trafficking, we micrografted Arabidopsis thaliana scions expressing GFP-tagged chloroplast transit peptides under the 35S promoter onto nontransgenic rootstocks. We found that plastids in the root tip became fluorescent 10 d after grafting. We obtained identical results with the companion cell-specific promoter SUC2 and with signals that target proteins to peroxisomes, actin, and the nucleus. We were unable to detect the respective mRNAs in the rootstock, indicating extensive movement of proteins in the phloem. Outward movement from the root protophloem was restricted to the pericycle-endodermis boundary, identifying plasmodesmata at this interface as control points in the exchange of macromolecules between stele and cortex. Intriguingly, signals directing proteins to the endoplasmic reticulum and Golgi apparatus from membrane-bound ribosomes were not translocated to the root. It appears that many organelle-targeting sequences are insufficient to prevent the loss of their proteins into the translocation stream. Thus, nonspecific loss of proteins from companion cells to sieve elements may explain the plethora of macromolecules identified in phloem sap.
INTRODUCTION
The phloem is a remarkable conduit that connects distant organs of a plant (Turgeon and Wolf, 2009; Ham and Lucas, 2014). In addition to having a major role in solute transport, the phloem functions in the movement of several macromolecules, including RNAs and proteins (Molnar et al., 2010; Turgeon and Wolf, 2009; Haroldsen et al., 2012; Turnbull and Lopez-Cobollo, 2013). Recently, the true extent of macromolecular trafficking in the phloem has begun to emerge. For example, in Arabidopsis thaliana plants parasitized by Cuscuta, over 9000 mRNA species were identified to move from host to pathogen, representing approximately half the Arabidopsis transcriptome (Kim et al., 2014). In a recent grafting study between different Arabidopsis ecotypes, over 2000 genes were found to produce mobile RNA transcripts, while proteomic data for the grafted plants suggested that some of these had been translated at their destination (Thieme et al., 2015). In addition to mRNAs, phloem sap is replete with diverse array of proteins, many of which appear to play no obvious role in long-distance signaling (Kehr, 2006; Batailler et al., 2012; Turnbull and Lopez-Cobollo, 2013). Collectively, these data reveal a prolific movement of macromolecules in the phloem. A central question concerning the appearance of macromolecules in phloem sap is: How many enter the translocation stream by default rather than design? In a previous study, we showed that a range of soluble protein-GFP fusions were able to enter the translocation stream and move to the root tip after translation in companion cells (Stadler et al., 2005). All fusion proteins examined were partially unloaded from the root protophloem. However, only free GFP (27 kD) was able to move basipetally toward the root tip following unloading. These data suggested that many soluble proteins synthesized in companion cells (CCs) enter the sieve element (SE) nonspecifically and that protein movement into the translocation stream may be a default pathway unless proteins are strongly anchored within either the CC or SE following their translation (Stadler et al., 2005). Although not tested directly, it is also possible that such a default pathway operates for the numerous mRNAs present in CCs. In a recent study, Calderwood et al. (2016) suggested that mRNA movement in the phloem may be directly related to mRNA abundance and half-life within CCs. Against this background, it is clear that many macromolecular signals generated in CCs play important roles in long-distance signaling (Kim et al., 2001; Haywood et al., 2005; Kragler, 2010; reviewed in Ham and Lucas, 2014). A much studied example is the movement of FLOWERING LOCUS T (FT), which is translated in CCs and moves to the shoot apex to induce flowering (Mathieu et al., 2007; reviewed in Ham and Lucas, 2014; Turnbull and Lopez-Cobollo, 2013).
Here, we show that numerous GFP-tagged proteins, destined for intracellular organelles in the shoot, enter the translocation stream and move across a graft union. This phenomenon was observed routinely for proteins translated on cytoplasmic ribosomes but not for those translated on endoplasmic reticulum (ER)-bound ribosomes. Those proteins that crossed the graft union were unloaded laterally from the root protophloem where they were targeted to the correct subcellular address. None of the proteins crossed the boundary between the pericycle and endodermis, suggesting that the size exclusion limit (SEL) of plasmodesmata at this interface is an important regulator of macromolecular exchange between the stele and cortex.
Our results show that organelle-targeted proteins are lost routinely to the translocation stream following their translation in the cytoplasm of source CCs. Significantly, these proteins do not remain confined to the phloem but are unloaded laterally into cells of the stele. We suggest that cells around the root protophloem poles ensure that the terminal SEs of the phloem do not become occluded by extensive protein trafficking. Our data reveal that both soluble and targeted proteins are lost constitutively to the translocation stream, making the challenge of identifying unique systemic phloem signals a difficult challenge for the future.
RESULTS
Chloroplast Fusion Proteins Are Translocated across a Graft Union
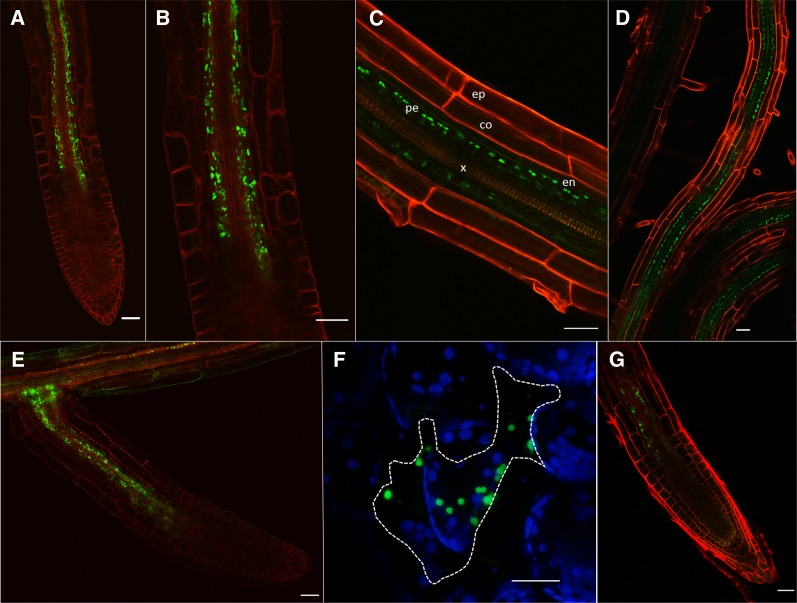
In our initial experiments, we examined whether chloroplast-targeted proteins could cross a graft union and enter the root from the scion. We grafted scions expressing the transit peptide for the chloroplast protein ferredoxin-NADP+ oxidoreductase (FNR; Mulo, 2011) fused to GFP (tpFNR-GFP; Mr 35 kD), driven by the 35S promoter, onto nontransgenic rootstocks (Figure 1B). At 10 d after grafting (dag), a fluorescent signal was present close to the root meristem. This pattern was observed in 100% of homografts (n = 50). Confocal examination revealed that plastids in cells surrounding the protophloem expressed GFP (Figure 2A). The fluorescent signal was present in files of cells parallel to the protophloem but did not extend apically toward the root meristem (Figure 2B). Optical sections of the root revealed that labeled plastids were restricted to cells of the stele, including the pericycle, but not in the endodermis or cortex (Figure 2C). As the roots continued to elongate, an increasing number of cells within the stele showed GFP expression, a reflection of the continued unloading of the protein near the root tip (Figure 2D). When lateral roots formed (8 to 10 dag) the fluorescent plastid signal was also associated with the terminal protophloem elements of the emerging root (Figure 2E). To examine whether tpFNR-GFP could gate plasmodesmata and move between epidermal cells, we bombarded a transient expression vector containing this sequence onto leaves of Nicotiana benthamiana. All bombardments showed cell-autonomous expression of the fusion protein (Figure 2F; n = 100 cells), indicating that this protein does not increase the SEL of plasmodesmata. When scions expressing tpFNR-GFP from the SUC2 promoter (Stadler et al., 2005) were grafted onto wild-type rootstock, we found unloading of the fusion protein around the terminal root protophloem in an identical pattern to that observed with the 35S promoter (c.f. Figures 2A and 2G). As the 35S promoter is expressed in CCs (Juchaux-Cachau et al., 2007; Corbesier et al., 2007; Mathieu et al., 2007), the most likely origin of the mobile fusion protein observed in roots was from CCs adjacent to the mature SEs in the scion.
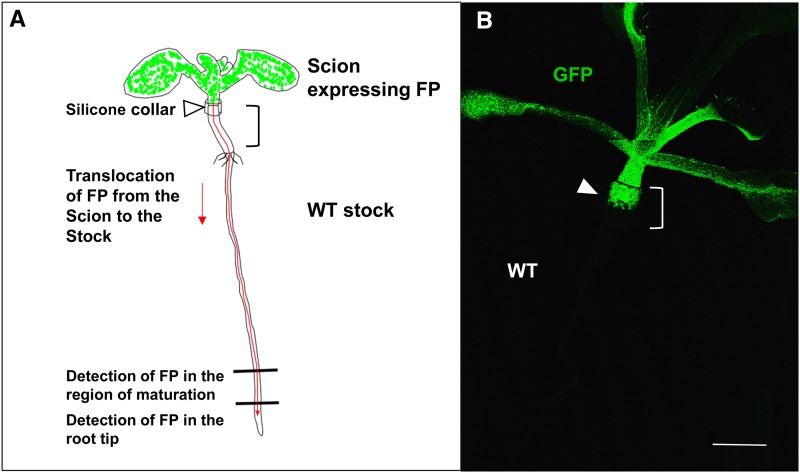
Figure 1.
Experimental Grafting System.
(A) Transgenic scions expressing fluorescent protein (FP) fusions were grafted onto nontransgenic rootstocks using a plastic collar. Ten days after grafting the roots were examined for the FP.
(B) Fluorescence of the scion at the graft interface (the position of the collar is bracketed; the arrowhead indicates the graft junction). Bar = 1 mm.
Figure 2.
Translocation of tpFNR-GFP from Scion to Rootstock.
(A) At 10 dag a strong fluorescent signal was observed around the terminal protophloem sieve elements.
(B) Enlargement of (A) showing fluorescent plastids around the phloem poles.
(C) In the unloading zone of the root, fluorescent plastids are restricted to the stele. ep, epidermis; co, cortex; en, endodermis; pe, pericycle; x, xylem.
(D) As roots continued to elongate, the fluorescent signal remained confined to the stele.
(E) Emerging lateral root showing fluorescence around the phloem poles.
(F) Bombardment of tpFNR into single leaf epidermal cells (dotted lines) failed to show movement into surrounding cells.
(G) Expression of tpFNR-GFP from the SUC2 promoter showed an identical pattern of fluorescence expression observed with the 35S promoter.
Bars = 30 µm.
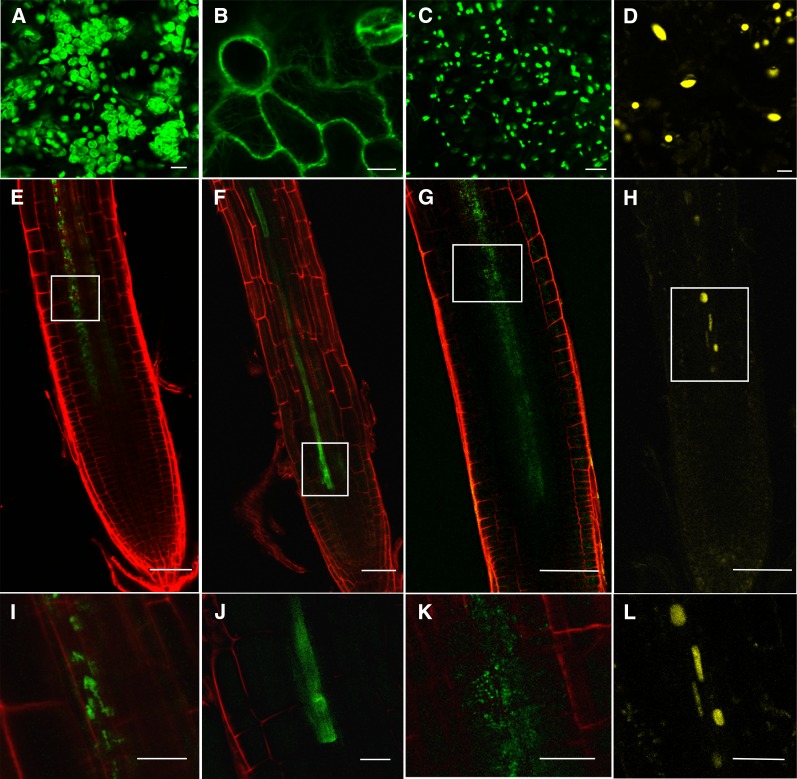
We also examined whether additional chloroplast signals fused to GFP could move across a graft union when expressed from the 35S promoter. We grafted scions expressing the reporter gene fused to transit peptides for RecA homolog1 (CT-GFP; Mr 33 kD), Rubisco subunit 1a (RBCS1a; CP-eGFP, Mr 37 kD), and plastocyanin (tpPC-eGFP, Mr 36 kD) onto wild-type rootstocks. At 10 dag, fluorescent plastids were observed adjacent to the terminal protophloem sieve elements in the root (movement of CP-eGFP shown in Figure 3A; Table 1). The exception was the transit peptide for CT-GFP that, despite being smaller (Mr 33 kD) than some of the other transit peptides, was not detected in any of the roots following grafting (Table 1).
Figure 3.
Translocation of Organelle-Targeted Fusion Proteins from Scion to Rootstock.
Comparison of the fluorescent signals from the transgenic scions ([A] to [D]; bars = 10 µm) with the nontransgenic rootstocks ([I] to [L]; bars = 30 µm). Phloem translocation across a graft union of CP-GFP (chloroplast) (E), FABD-GFP (actin) (F), A5-GFP (peroxisome) (G), and H2B-YFP (nucleus) markers (H) (bars = 50 µm). The boxed regions of the root are shown at higher magnification in the lowest panels.
Table 1. Properties of Protein Fusions Used in This Study.
| Fusion Protein | FP | Size (kD) | Promoter | Targeted Organelle(s) | Present in Grafted Root Tip of Col | Frequency | Reference |
|---|---|---|---|---|---|---|---|
| TP of RecA homolog1: CT-GFP | S65T-mGFP4 | ∼33 | 35S | Chloroplast | No | 100%; n = 20 | Köhler et al. (1997) |
| TP of RBCS1a: CP-eGFP | eGFP | ∼37 | 35S | Chloroplast | Yes | 100%; n = 27 | Unpublished |
| TP of FNR: tpFNR-eGFP | eGFP | ∼35 | 35S | Chloroplast | Yes | 100%; n = 50 | Marques et al. (2003) |
| TP of plastocyanin: tpPC-eGFP | eGFP | ∼36 | 35S | Chloroplast | Yes | 100%; n = 7 | Marques et al. (2003) |
| A5-eGFP | eGFP | – | 35S | Peroxisome | Yes | 100%; n = 32 | Cutler et al. (2000) |
| FABD2-GFP | S65T-GFP | ∼67 | 35S | Actin | Yes | 67%; n = 29 | Ketelaar et al. (2004) |
| H2B-YFP | mYFP | ∼42 | 35S | Nucleus | Yes | 57%; n = 42 | Federici et al. (2012) |
| RTNLB6-GFP | sGFP | ∼57 | 35S | ER | No | 100%; n = 5 | Knox et al. (2015) |
| HDEL-GFP | mGFP4 | ∼28 | 35S | ER lumen | No | 100%; n = 15 | Haseloff et al. (1997) |
| STtmd-GFP | GFP | ∼33 | 35S | Golgi apparatus | No | 100%; n = 14 | Boevink et al. (1998) |
TP, transit peptide; FP, fluorescent protein.
Additional Organelle Signals
Chloroplast proteins are translated on cytoplasmic ribosomes before delivery to the outer chloroplast envelope by HSP70 and associated chaperones (Lee at al., 2013). We tested whether additional proteins, destined for other organelles, might behave in the same way as the chloroplast transit peptides. We grafted scions expressing fluorescent reporters with targeting signals for peroxisomes (A5-eGFP), nucleus (H2B-YFP), and F-actin binding domain (FABD2-GFP) onto nontransgenic rootstocks. We compared the fluorescence pattern observed in the transgenic scions with the nontransgenic rootstocks (Figure 3). In all of these cases, we detected the equivalent labeled substructures in stelar cells adjacent to the protophloem (Figures 3A to 3D, Table 1). Some of the fusion proteins we employed also encoded a significant region of the targeted protein (Table 1). The largest of these was FABD2-GFP (67 kD) that, in addition to crossing the graft union, was also unloaded from the protophloem (Figure 3B).
We next examined whether proteins translated on ER-bound ribosomes, destined for the endomembrane system, could cross the graft union and be unloaded. We grafted lines expressing HDEL-GFP (ER lumen), reticulon 6 (RTNLB6)-GFP (ER membrane) and sialyl transferase (ST) transmembrane domain-GFP (Golgi apparatus) onto nontransgenic rootstocks. However, we were unable to detect a fluorescent signal in the root for any of these fusion proteins at 10 dag (Table 1).
mRNA Analysis
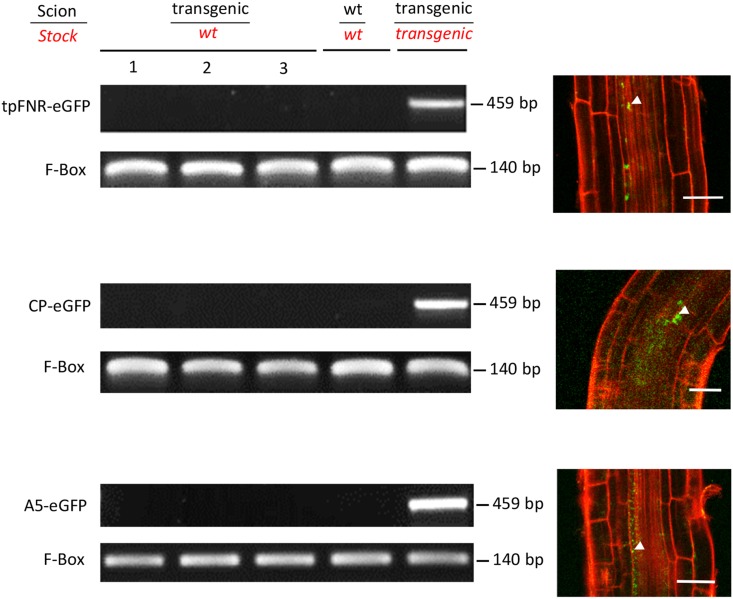
Using RT-PCR, we examined the nontransgenic rootstocks of 18 to 24 graft partners for evidence of mRNA trafficking. In this experiment, we used two chloroplast signal peptides (tpFNR-eGFP and CP-eGFP) and a peroxisomal signal sequence-fused GFP (A5-eGFP), all of which showed consistent movement across the graft union (Table 1). However, we were not able to detect the mRNA of any of these fusion proteins in roots at 5 weeks after grafting (Figure 4), suggesting that mobile proteins are the likely source of fluorescent signals in the developing root tissues. To confirm that protein expression was visible at this time point, we examined the root tips under the confocal microscope. For all three graft partners, we observed a clear fluorescent signal adjacent to the protophloem, although the signal was weaker than at 10 dag (c.f. Figures 2 and 4).
Figure 4.
Mobility of Transgene Transcripts in Grafted Arabidopsis.
RT-PCR of different graft combinations at 5 weeks after grafting to detect the respective mRNAs present in the rootstocks when scions expressed tpFNR-eGFP, CP-eGFP (chloroplast), or A5-eGFP (peroxisome) protein signals (sampled tissue highlighted in red italics). Corresponding images of protein localization in the root at 5 weeks after grafting are shown to the right. Bars = 50 µm.
Bioinformatic and Statistical Analysis
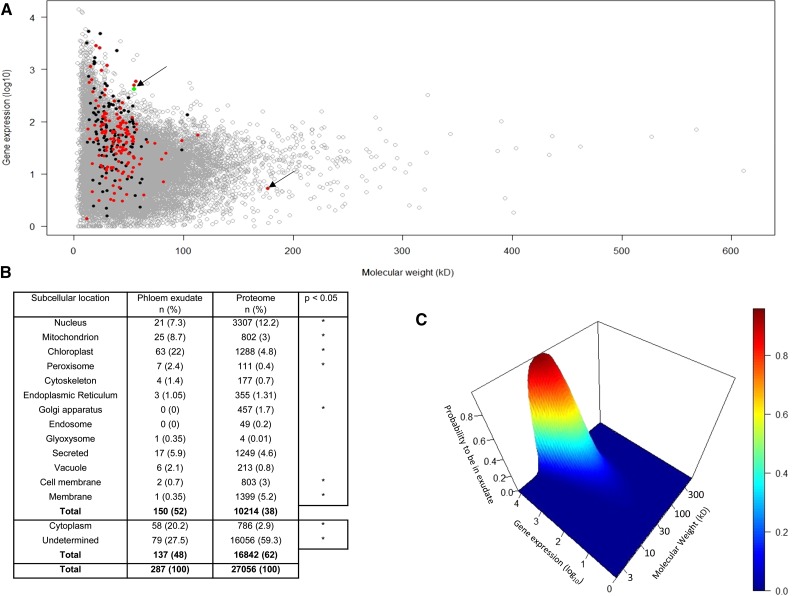
In our grafting experiments, GFP fusions were expressed under the strong promoters 35S and SUC2, raising the possibility that protein overexpression in CCs may have contributed to loss of fusion proteins to the SE. To address this issue, we examined published data relating to the profile of proteins found in the translocation stream, an approach independent of expression of GFP fusions. We conducted a bioinformatic analysis of data on the occurrence of mRNAs in phloem tissue and proteins in the phloem exudate in relation to their corresponding molecular mass (data derived from Deeken et al., 2008; Batailler et al., 2012). We separated phloem-mobile proteins with known organelle-targeting sequences from those without such sequences (Figure 5A). In total, 150 proteins (52%) detected in phloem exudate were shown to have organelle-targeting sequences. The relative distribution of these proteins among different subcellular organelles, compared with the Arabidopsis proteome, is shown in Figure 5B. The main difference lies in the proportion of proteins allocated to organelles, in particular chloroplasts, mitochondria, and peroxisomes. This probably reflects the unique protein composition of the CC.
Figure 5.
Bioinformatic and Statistical Analysis of Proteins Found in Phloem Exudate, Taking into Account Their Molecular Weight, Gene Expression, and Subcellular Location.
(A) Bioinformatic analysis showing relationship between proteins expressed in the phloem and those detected specifically in phloem exudate. The majority of phloem-mobile proteins cluster in the size range 20 to 70 kD. The outlying arrows indicate a 179-kD chloroplast-targeted protein and the green dot corresponding to SUC2 expression. Data were derived from Deeken et al. (2008) and Batailler et al. (2012). Proteins with organelle-targeting sequences (red) are discriminated from those without such signals (black).
(B) Relative allocation of proteins from phloem exudate and the Arabidopsis proteome to different subcellular organelles and structures (*P < 0.05).
(C) Probability of proteins to be found in exudate according to gene expression and molecular mass. Gene expression and molecular mass are shown in base-10 logarithm.
The gene expression levels in the phloem were not significantly different from other phloem-mobile proteins that lacked targeting sequences (P = 0.07; nonparametrical statistical test), which rules out the possibility that mobile proteins with an organelle-targeting sequence are found in the phloem exudate only at high levels of gene expression. The data also reveal that the majority of proteins entering the translocation stream cluster in the size range 20 to 70 kD, suggesting that molecular mass, or more specifically Stokes radius (Dashevskaya et al., 2008), may govern the passage between CC and SE. This was confirmed using a logistic regression model that examined the impact of both protein size (kD) and transcript abundance on the likelihood of a given protein to be found in phloem exudate (Figure 5C). The model shows that for proteins below 70 kD there is an exponential-like relationship between gene expression level and protein size, i.e., the more abundantly a protein is expressed, the more likely it is to enter the translocation stream. Above 70 kD, the probability of a protein entering SEs declines dramatically, consistent with a simple diffusive model based on the SEL of the pore-plasmodesmata that connect SEs and CCs (Stadler et al., 2005). A small number of proteins detected in phloem exudate exceeded 70 kD, one example being a chloroplast-targeted protein (AT5G04140; 179 kD; Figure 5A, arrow).
DISCUSSION
Numerous studies over the last decade have shown that the phloem translocation stream is replete with mRNAs and proteins (Turnbull and Lopez-Cobollo, 2013). The appearance of a diverse array of macromolecules in the phloem is intriguing, giving rise to the suggestion that the phloem functions as an “information superhighway” (Jorgensen et al., 1998). It is clear that many systemic macromolecular signals are involved in development and defense (reviewed in Ham and Lucas, 2014). A much studied example is the flowering signal, FT, a protein translated in CCs and transported to the shoot meristem where it activates the flowering response (Wigge, 2011; Turnbull and Lopez-Cobollo, 2013). To date, the pathway taken by FT from the terminal protophloem to the shoot meristem is not clear (Corbesier et al., 2007), and FT may initiate a downstream signal cascade that leads to flowering (Wigge, 2011). Other developmental long-distance macromolecules are thought to be mRNAs. For example, BEL1-type homeodomain proteins are thought to function as long-distance signals involved in tuberization (Banerjee et al., 2006), while the Mouse ears (me) mRNA affects leaf development in tomato (Solanum lycopersicum; Kim et al., 2001). The extent to which these mRNAs are translated in sink tissues remains unknown (Spiegelman et al., 2013).
During pathogen attack, protein signals enter the translocation stream and subsequently prime distant tissues against invading pathogens, inducing systemic-acquired resistance (Fu and Dong, 2013). In most of these instances, the signals are produced in the CC before they enter the SE. A common feature of both developmental and pathogen-induced signals is that they are produced within a discrete time window, in response to either environmental change (e.g., photoperiod; Turnbull and Lopez-Cobollo, 2013) or sudden pathogen attack (Fu and Dong, 2013). Thus, one could envisage a scenario in which the movement of protein signals in the phloem is regulated by the timing of their translation in CCs. However, not all proteins and mRNAs detected in phloem sap have obvious signaling functions and many soluble proteins may enter the SE constitutively. In the study of Stadler et al. (2005), a range of soluble proteins entered the SE from the CC when expressed from the SUC2 promoter and were translocated to the root. Only free GFP (27 kD) was unloaded into all root tissues, but larger fusion proteins were also able to leave the protophloem and enter a distinct post-phloem domain (Stadler et al., 2005). Recently, Calderwood et al. (2016) proposed that a default pathway, based on transcript abundance and decay within CCs, might operate for several phloem-mobile mRNA species. However, Calderwood et al. (2016) also identified a subset of transcripts that were mobile but whose movement could not be explained by abundance alone. More recently, Zhang et al. (2016) showed that tRNA-related sequences may trigger mRNA movement into the translocation stream, providing a potential explanation for the large number of endogenous transcripts reported to move across graft unions. Our present data suggest that molecular mass, in addition to transcript abundance, is a major determinant for the entry of proteins into the SE. Interestingly, a small number of proteins detected in phloem exudate were significantly larger than the 70 kD cutoff we observed here. Such large proteins merit further study as their molecular mass would predict that they are too large to pass from CC to SE by simple diffusion. Thus, specific subsets of proteins and mRNAs may enter the phloem by a specific, unidentified, route. The final entry of macromolecules into the phloem will depend on passage through the specialized pore-plasmodesmata that connect the SEs and CCs (Oparka and Turgeon, 1999) where the SEL of these pores is the ultimate determinant for nonspecific passage into the SE.
The CC contains a full complement or organelles, including plastids (Lalonde et al., 2001). Our study shows that the transit/signal sequences responsible for directing proteins to organelles in CCs are insufficiently strong to prevent protein loss to the translocation stream. The 35S promoter is expressed strongly in CCs (Juchaux-Cachau et al., 2007; Corbesier et al., 2007; Mathieu et al., 2007), as is the SUC2 promoter. For example, FT driven from the 35S promoter induces flowering in an identical fashion to that seen with the CC promoter, SUC2 (Mathieu et al., 2007), suggesting that the level of 35S expression in CCs is sufficiently high to promote FT movement into SEs. In epidermal cells, it appears that the subcellular targeting of a protein expressed from the 35S promoter may prevent its movement through plasmodesmata to adjacent cells (Crawford and Zambryski, 2000), suggesting that protein targeting signals in these cells are sufficiently strong to prevent diffusion to adjacent cells. However, this observation would not appear to hold true for proteins translated in CCs. It could be argued that the strong promoters we used here (e.g., 35S) enhanced phloem entry by virtue of increasing protein expression levels in CCs. We do not have data relating to proteins expressed under native CC promoters, other than SUC2. However, our bioinformatics analysis of published data showed clearly that for proteins up to 70 kD, there is an exponential relationship between transcript abundance and appearance of the respective protein in exudate.
Significantly, we found that fusion proteins translated on cytoplasmic ribosomes were able to enter the SE while those translated on ER-bound ribosomes were not. Of the chloroplast fusions we tested, all but CT-GFP moved across the graft union. The reason for non-movement of this protein is unclear. Its targeting sequence may be sufficiently strong to retain it within the CC or, like some other proteins described recently, it may use the secretory pathway for targeting to the chloroplast (Villarejo et al., 2005). In our experimental study, GFP fusions up to 67 kD (FABD2-GFP) entered the translocation stream following translation within CCs, close to the predicted molecular cutoff of 70 kD we observed in our bioinformatics study. In our grafting studies, we used GFP-fusion proteins where the addition of the GFP moiety would have added significantly to the molecular mass. Also, the GFP fluorophore potentially may have masked internal protein signals that interact with plasmodesmata. We think it is very unlikely that the large number of diverse proteins present in phloem exudate each contain a signal that interacts with the pore-plasmodesmata between CC and SE, but rather that the organelle-targeting sequences for proteins expressed in CCs are insufficiently strong to prevent their entry into the translocation stream.
In previous studies, it has been suggested that exudate proteins with strong organelle-targeting sequences may be artifacts of sample preparation, emanating from nonphloem tissues near the cut ends of stems or petioles (Schobert et al., 1998; Lin et al., 2009) or resulting from sudden pressure release of the phloem during wounding (Oparka and Turgeon, 1999). Similarly, it has been argued that some of the proteins detected by aphid stylectomy might be artifactual as they have no obvious signaling or protein turnover functions within SEs (Atkins et al., 2011). Our current data suggest that such mobile proteins may not be anomalies but rather represent the routine transfer of small proteins (<70 kD) from CC to SE.
In our GFP-fusion studies, all of the proteins that entered the translocation stream were able to leave the root protophloem and target the appropriate organelle in stelar cells. Our data suggest that postphloem macromolecular trafficking is restricted to the pericycle-endodermis boundary. We do not have data relating to the numerous proteins detected in phloem exudate, but it seems likely that many of these might also be restricted to the stele. However, plant viruses are able to cross this boundary (Valentine et al., 2004), as are endogenous transcription factors, such as SHORT ROOT (Gallagher et al., 2004), that are translated in the stele. Therefore, it appears that plasmodesmata at this interface must be regulated to allow macromolecular exchange between stele and cortex.
What is the significance of constitutive protein and mRNA trafficking in the phloem? Loss of macromolecules to the translocation stream may be an inevitability of the design of the SE-CC complex, where the pore-plasmodesmata connecting these cells have a high SEL (Stadler et al., 2005). One of the principal functions of the phloem is to move solutes from source to sink regions of the plants. For pressure flow to operate, a turgor gradient is required along the axial transport pathway (Froelich et al., 2011; De Schepper et al., 2013) with the removal of solutes in sink tissues. The continuous loss of macromolecules to the translocation stream might cause a potential hindrance to flow in SEs if proteins and mRNAs were not removed from the translocation stream. Our data indicate that proteins entering the SE constitutively from the CC are removed from the protophloem at its terminus, ensuring that mass flow and unloading of solutes is unimpeded. Thus, a protein destined for a plastid in a leaf CC may eventually end up in a root pericycle cell. The fate of soluble proteins that leave the protophloem is currently unknown and represents a challenge for future research on this topic. Similarly, it remains to be shown if all the mobile mRNA species detected in phloem exudate (Calderwood et al., 2016; Zhang et al., 2016) enter this postphloem domain.
Our data suggest that many macromolecules below 70 kD enter the phloem by default (Stadler et al., 2005), a view supported by our bioinformatics survey (Figure 5). When Arabidopsis plants are parasitized by Cuscuta, about half the transcriptome of the host enters the parasite via the phloem (Kim et al., 2014). From a signaling point of view, it seems unlikely that this level of trafficking is significant, but rather represents a large-scale exchange of macromolecules between the two species, similar to the movement of proteins across a graft union reported here. When a plant is parasitized by Cuscuta, and in situations where phloem sap is collected, macromolecules are intercepted in transit and will be unable to reach the postphloem domain associated with the terminal phloem elements. Thus, their presence in the translocation stream does not necessarily imply a function in sink tissues. Among the vast number of proteins and mRNA species found in phloem sap, it is very likely that some are generated by design rather than by default (Calderwood et al., 2016; Zhang et al., 2016). Identifying such systemic signals against the background of “flotsam” generated by CCs may prove a difficult task for the future.
METHODS
Plant Material, Growth Conditions, and Grafting
Seeds of Arabidopsis thaliana, ecotype Columbia, and all the transgenic lines listed in Table 1 were surface sterilized in an 8% bleach and 1% Tween 20 solution. After five washes in distilled water, these were either sown on soil or plated on Petri dishes containing Murashige and Skoog (MS) basal salts, 1.2% agar, and 0.2% sucrose, pH 5.7, and stratified in darkness for 2 to 3 d at 4°C. Seedlings were then grown with plates oriented vertically at 23°C under long days (18 h light/6 h dark; intensity, 100 µmol m−2 s−1).
After 5 to 7 d, seedlings were grafted following the hypocotyl-grafting procedure of Turnbull et al. (2002) consisting of a transverse cut and butt alignment with silicon collars. The seedlings were cut transversely in the upper region of the hypocotyl with ultrafine microknives (Interfocus; no. 10315-12). Scions were grafted onto wild-type stocks using a short silicon collar for support on MS agar plates. The grafts were left to grow under long days with the plates still oriented vertically until new lateral roots of the stocks were fully established (∼10 d). The grafts were imaged between 5 dag and 5 weeks after grafting, at which point the tissue was collected for total nucleic acid extraction (TNA).
Plasmid Construct and Plant Transformation
For the construction of the AtSUC2 promoter, tpFNR-eGFP, 938 bp of AtSUC2 promoter was PCR amplified from the pES1 cloning vector (Stadler et al., 2005) using the primers 5′-AACAGCTATGACCATGATTACGC-3′ and 5′-ATATCTCGAGTTGACAAACCAAGAAAGTAAG-3′, while the tp-FNR-eGFP insert was PCR amplified from the pGreenII109 plasmid (courtesy of Martin Schattat) with the primers 5′-ATATCTCGAGATTCTTCCAATCATCGTACTC-3′ and 5′-ATATGAGCTCGCCGCTTTACTTGTACAG-3′. The resulting HindIII-XhoI and XhoI-SacI fragments, respectively, were then ligated into pES1 pretreated with HindIII and SacI to remove the AtSUC2/GFP construct. Successful clones were selected on kanamycin LB plates, yielding pES1-tpFNR. The insert was sequenced using the primers 5′-AGCTATGACCATGATTACGC-3′, 5′-ACCCTACGCTATAGACACAGC-3′, and 5′-AAGCTCCTCCGTCATTTC-3′. The plasmid was then used to transform electro-competent Agrobacterium tumefaciens (strain Agl1). Arabidopsis plants, ecotype Col-0, were floral dipped as described by Clough and Bent (1998). Seedlings were selected on MS media with 50 µg/mL kanamycin.
Biolistic Bombardment
Up to 5 µg of the pGreenII109 plasmid containing the tp-FNR-eGFP insert was CaCl2 precipitated onto 1.25 mg of 1-μm gold particles (Bio-Rad Laboratories) and resuspended in 100 μL ethanol. Five-microliter aliquots were bombarded onto leaves of 2-week-old Arabidopsis plantlets using a biolistic particle delivery system (PDS-1000/He; Bio-Rad Laboratories) at 1100 p.s.i. The plants were returned to their growth conditions and monitored at 5 and 10 d postbombardment by confocal microscopy (see below).
Imaging
Grafts were imaged using a Leica SP2 confocal laser scanning microscope (Leica Microsystems) with either a ×10 (HCXPL FLUOTAR; Leica Microsystems) or a ×20 water-immersion lens (HCX PLAPO CS; Leica Microsystems).
Total Nucleic Acid Extraction and cDNA Synthesis
The rootstocks of 18 to 24 grafts were pooled into three biological replicates for each transgenic line. The roots were harvested immediately below the root collar of 5-week-old grafts. This was performed under a stereomicroscope (Leica; Wild M3C) to prevent tissue contamination from the scion. Grafts showing the formation of adventitious roots above the graft junction were disregarded. TNA (DNA and RNA) was extracted using the modified protocol of White and Kaper (1989).
TNA samples were used for cDNA synthesis. Three micrograms of TNA was treated using a TURBO DNA-free kit (Ambion). One microgram of DNA-free TNA was then reverse transcribed using a RevertAid first-strand cDNA synthesis kit (Thermo Scientific). The presence of the eGFP coding sequence was analyzed by PCR using primers 5′-ACGGCGTGCAGTGCTTC-3′ and 5′-CCATGTGATCGCGCTTC-3′. F-box gene (At5g15710) specific primers were used as cDNA quality and loading controls from Lilly et al. (2011).
Bioinformatic and Statistical Analysis
Gene expression data for Arabidopsis phloem tissue were found in the GSE10247 Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) data set for 22,746 proteins (Deeken et al., 2008). Their corresponding molecular masses and subcellular locations were obtained from Uniprot (http://www.uniprot.org) using the retrieve/ID mapping tool. This led to identification of 21,072 proteins with a unique transcript ID. Mean expression level was computed for each protein. The two files were merged to obtain both mean expression and molecular weight for 21,072 phloem proteins. In this set of proteins, we retrieved 264 phloem exudate proteins from among the 287 identified by Batailler et al. (2012).
A logistic regression in a Bayesian framework with a noninformative prior distribution was used to determine whether the probability of a protein to be found in a given location was significantly different between exudate proteins (n = 287) and other proteins of the Arabidopsis proteome (n = 27,056). The Arabidopsis proteome was downloaded from Uniprot (http://www.uniprot.org). The analysis was performed in turn for each of the 15 possible subcellular locations. For each model, the response binary variable was the location (yes/no) and the explicative variable, the group (proteins from the phloem exudate/proteome without proteins from the phloem exudate). If 1 belonged to the 95% credible interval of the odds ratio, the probabilities to be in a subcellular location for proteins in the phloem exudate and for the other proteins were not significantly different. In the inverse case, the difference was significant at the 5% level. The Bayesian analysis was performed using the rjags R package available at https://sourceforge.net/projects/mcmc-jags/.
Gene expression distributions of proteins with and without targeting sequences were compared using Wilcoxon test, also known as Mann-Whitney test (R command wilcox.test). The impact of molecular mass and gene expression on the probability of a protein to be found in the phloem was studied using a logistic second degree polynomial regression model taking into account the interaction between both variables, after base-10 logarithm transformation, to obtain normal distributions. Statistical analysis was performed using R language accessible at https://cran.r-project.org (version 3.2.2).
Accession Numbers
All proteins used in the bioinformatics study are listed in the supplemental information of Batailler et al. (2012).
Acknowledgments
Work in K.J.O.’s laboratory is supported by the BBSRC. D.S.G.P. acknowledges receipt of a Principal’s Career Development/BBSRC DTG studentship (BB/F017073/1). A.M. is a Chancellor’s Fellow at the University of Edinburgh. We thank the following colleagues who generously supplied transgenic lines expressing fluorescent proteins: Martin Schattat (tpFNR-GFP and tpPC-eGFP), Guo-Zhang Wu (CP-GFP), Maureen Hanson (CT-GFP), Chris Hawes (st-GFP, HDEL-GFP), Kirsten Knox (RTLNB6-GFP), Patrick Hussey (FABD2-GFP), Imogen Sparkes (A5-GFP), and Jim Haseloff (H2B-YFP).
AUTHOR CONTRIBUTIONS
D.S.G.P. designed and conducted experiments and analyzed data. M.-P.G. conducted bioinformatics and statistical analysis. A.M. designed experiments and analyzed results. K.J.O. designed experiments and wrote the manuscript.
Glossary
- CC
companion cell
- SE
sieve element
- SEL
size exclusion limit
- ER
endoplasmic reticulum
- dag
days after grafting
- MS
Murashige and Skoog
- TNA
total nucleic acid extraction
Footnotes
Articles can be viewed without a subscription.
References
- Atkins C.A., Smith P.M., Rodriguez-Medina C. (2011). Macromolecules in phloem exudates--a review. Protoplasma 248: 165–172. [DOI] [PubMed] [Google Scholar]
- Banerjee A.K., Chatterjee M., Yu Y., Suh S.G., Miller W.A., Hannapel D.J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18: 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batailler B., Lemaître T., Vilaine F., Sanchez C., Renard D., Cayla T., Beneteau J., Dinant S. (2012). Soluble and filamentous proteins in Arabidopsis sieve elements. Plant Cell Environ. 35: 1258–1273. [DOI] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447. [DOI] [PubMed] [Google Scholar]
- Calderwood A., Stanislav Kopriva S., Morris R.J. (2016). Transcript abundance explains mRNA mobility data in Arabidopsis thaliana Plant Cell 28: 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Crawford K.M., Zambryski P.C. (2000). Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10: 1032–1040. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashevskaya S., Kopito R.B., Friedman R., Elbaum M., Epel B.L. (2008). Diffusion of anionic and neutral GFP derivatives through plasmodesmata in epidermal cells of Nicotiana benthamiana. Protoplasma 234: 13–23. [DOI] [PubMed] [Google Scholar]
- Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G., Hedrich R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55: 746–759. [DOI] [PubMed] [Google Scholar]
- De Schepper V., De Swaef T., Bauweraerts I., Steppe K. (2013). Phloem transport: a review of mechanisms and controls. J. Exp. Bot. 64: 4839–4850. [DOI] [PubMed] [Google Scholar]
- Federici F., Dupuy L., Laplaze L., Heisler M., Haseloff J. (2012). Integrated genetic and computation methods for in planta cytometry. Nat. Methods 9: 483–485. [DOI] [PubMed] [Google Scholar]
- Froelich D.R., Mullendore D.L., Jensen K.H., Ross-Elliott T.J., Anstead J.A., Thompson G.A., Pélissier H.C., Knoblauch M. (2011). Phloem ultrastructure and pressure flow: Sieve-Element-Occlusion-Related agglomerations do not affect translocation. Plant Cell 23: 4428–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Gallagher K.L., Paquette A.J., Nakajima K., Benfey P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14: 1847–1851. [DOI] [PubMed] [Google Scholar]
- Ham B.-K., Lucas W.J. (2014). The angiosperm phloem sieve tube system: a role in mediating traits important to modern agriculture. J. Exp. Bot. 65: 1799–1816. [DOI] [PubMed] [Google Scholar]
- Haroldsen V.M., Szczerba M.W., Aktas H., Lopez-Baltazar J., Odias M.J., Chi-Ham C.L., Labavitch J.M., Bennett A.B., Powell A.L.T. (2012). Mobility of transgenic nucleic acids and proteins within grafted rootstocks for agricultural improvement. Front. Plant Sci. 3: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V., Yu T.-S., Huang N.-C., Lucas W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42: 49–68. [DOI] [PubMed] [Google Scholar]
- Jorgensen R.A., Atkinson R.G., Forster R.L.S., Lucas W.J. (1998). An RNA-based information superhighway in plants. Science 279: 1486–1487. [DOI] [PubMed] [Google Scholar]
- Juchaux-Cachau M., Landouar-Arsivaud L., Pichaut J.-P., Campion C., Porcheron B., Jeauffre J., Noiraud-Romy N., Simoneau P., Maurousset L., Lemoine R. (2007). Characterization of AgMaT2, a plasma membrane mannitol transporter from celery, expressed in phloem cells, including phloem parenchyma cells. Plant Physiol. 145: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J. (2006). Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 57: 767–774. [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149. [DOI] [PubMed] [Google Scholar]
- Kim G., LeBlanc M.L., Wafula E.K., dePamphilis C.W., Westwood J.H. (2014). Plant science. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 345: 808–811. [DOI] [PubMed] [Google Scholar]
- Kim M., Canio W., Kessler S., Sinha N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289. [DOI] [PubMed] [Google Scholar]
- Knox K., Wang P., Kriechbaumer V., Tilsner J., Frigerio L., Sparkes I., Hawes C., Oparka K. (2015). Putting the squeeze on plasmodesmata: a role for reticulons in primary plasmodesmata formation. Plant Physiol. 168: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler R.H., Cao J., Zipfel W.R., Webb W.W., Hanson M.R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042. [DOI] [PubMed] [Google Scholar]
- Kragler F. (2010). RNA in the phloem: A crisis or a return on investment? Plant Sci. 178: 99–104. [Google Scholar]
- Lalonde S., Franceschi V.R., Frommer W.B. (2001). Companion cells. eLS, 10.1038/npg.els.0002087.
- Lee D.W., Jung C., Hwang I. (2013). Cytosolic events involved in chloroplast protein targeting. Biochim. Biophys. Acta 1833: 245–252 (BBA). [DOI] [PubMed] [Google Scholar]
- Lilly S.T., Drummond R.S., Pearson M.N., MacDiarmid R.M. (2011). Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol. Plant Microbe Interact. 24: 294–304. [DOI] [PubMed] [Google Scholar]
- Lin M.K., Lee Y.J., Lough T.J., Phinney B.S., Lucas W.J. (2009). Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol. Cell. Proteomics 8: 343–356. [DOI] [PubMed] [Google Scholar]
- Marques J.P., Dudeck I., Klösgen R.B. (2003). Targeting of EGFP chimeras within chloroplasts. Mol. Genet. Genomics 269: 381–387. [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875. [DOI] [PubMed] [Google Scholar]
- Mulo P. (2011). Chloroplast-targeted ferredoxin-NADP+ oxidoreductase (FNR): Structure, function and location. Biochim. Biophys. Acta 1807: 927–934. [DOI] [PubMed] [Google Scholar]
- Oparka K.J., Turgeon R. (1999). Sieve elements and companion cells-traffic control centers of the phloem. Plant Cell 11: 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert C., Baker L., Szederkenyi J., Großmann P., Komor E., Hayashi H. (1998). Identification of immunologically related proteins in sieve-tube exudate collected from monocotyledonous and dicotyledonous plants. Planta 206: 245–252. [Google Scholar]
- Spiegelman Z., Golan G., Wolf S. (2013). Don’t kill the messenger: Long-distance trafficking of mRNA molecules. Plant Sci. 213: 1–8. [DOI] [PubMed] [Google Scholar]
- Stadler R., Lauterbach C., Sauer N. (2005). Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 139: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme C.J., Rojas-Triana M., Stecyk E., Schudoma C., Zhang W., Yang L., Miñambres M., Walther D., Schulze W.X., Paz-Ares J., Scheible W.-R., Kragler F. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Turnbull C.G.N., Booker J.P., Leyser H.M.O. (2002). Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32: 255–262. [DOI] [PubMed] [Google Scholar]
- Turnbull C.G.N., Lopez-Cobollo R.M. (2013). Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytol. 198: 33–51. [DOI] [PubMed] [Google Scholar]
- Turgeon R., Wolf S. (2009). Phloem transport: cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 60: 207–221. [DOI] [PubMed] [Google Scholar]
- Valentine T., Shaw J., Blok V.C., Phillips M.S., Oparka K.J., Lacomme C. (2004). Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol. 136: 3999–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7: 1224–1231. [DOI] [PubMed] [Google Scholar]
- White J.L., Kaper J.M. (1989). A simple method for detection of viral satellite RNAs in small plant tissue samples. J. Virol. Methods 23: 83–93. [DOI] [PubMed] [Google Scholar]
- Wigge P.A. (2011). FT, a mobile developmental signal in plants. Curr. Biol. 21: R374–R378. [DOI] [PubMed] [Google Scholar]
- Zhang W., Thieme C.J., Kollwig G., Apelt F., Yang L., Winter N., Andresen N., Walther D., Kragler F. (2016). tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]