Abstract
Similar to the inherent clinical heterogeneity of most, if not all, lymphoma entities, the genetic landscape of these tumors is markedly complex in the majority of cases, with a rapidly growing list of recurrently mutated genes discovered in recent years by next-generation sequencing technology. Whilst a few genes have been implied to have diagnostic, prognostic and even predictive impact, most gene mutations still require rigorous validation in larger, preferably prospective patient series, to scrutinize their potential role in lymphoma diagnostics and patient management. In selected entities, a predominantly mutated gene is identified in almost all cases (e.g. Waldenström’s macroglobulinemia/lymphoplasmacytic lymphoma and hairy-cell leukemia), while for the vast majority of lymphomas a quite diverse mutation pattern is observed, with a limited number of frequently mutated genes followed by a seemingly endless tail of genes with mutations at a low frequency. Herein, the European Expert Group on NGS-based Diagnostics in Lymphomas (EGNL) summarizes the current status of this ever-evolving field, and, based on the present evidence level, segregates mutations into the following categories: i) immediate impact on treatment decisions, ii) diagnostic impact, iii) prognostic impact, iv) potential clinical impact in the near future, or v) should only be considered for research purposes. In the coming years, coordinated efforts aiming to apply targeted next-generation sequencing in large patient series will be needed in order to elucidate if a particular gene mutation will have an immediate impact on the lymphoma classification, and ultimately aid clinical decision making.
Introduction
Most, if not all, lymphoma subtypes display considerable heterogeneity in their clinical and pathological characteristics, intricately linked to the underlying biological heterogeneity.1 Indeed, the genomic landscape of lymphomas is remarkably diverse, although an increasing number of ‘shared’ genetic lesions have recently emerged, affecting similar mechanisms and processes that in certain instances are important for ‘generic’ cell homeostasis (e.g. DNA repair) while in others they are ‘lymphocyte-specific’ (e.g. antigen receptor signaling) (Figure 1).
Figure 1.
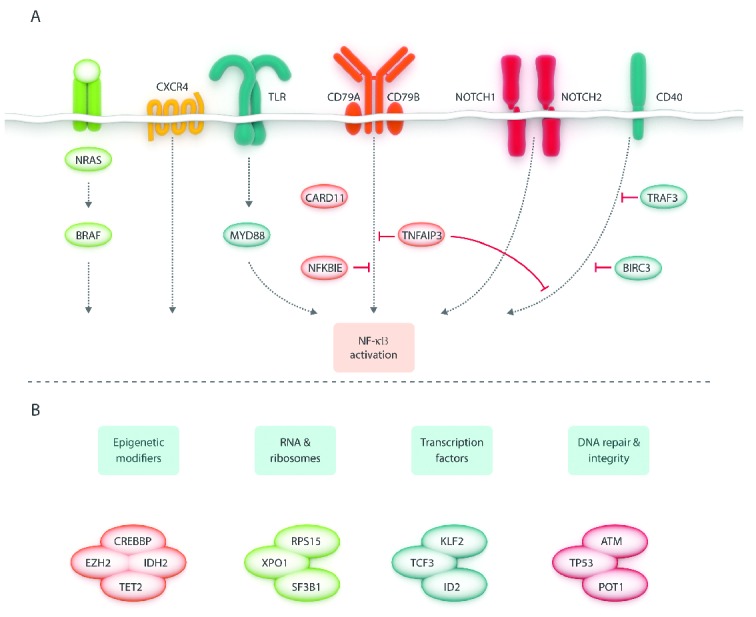
Schematic overview of recurrently affected pathways (A) and processes (B) in B-cell lymphomas. Based on references listed in Table 1.
Thanks to next-generation sequencing (NGS), it has become possible to appreciate the panorama of recurrently affected genes that contribute to disease pathogenesis and/or evolution, at least in major lymphoma subtypes. Mounting evidence suggests that certain gene mutations have diagnostic, prognostic and/or predictive impact. However, for most mutations, functional in vitro validation and confirmation in larger patient series are warranted in order to fully elucidate their role in the pathobiology of a particular lymphoma as well as their relevance for routine diagnostics.
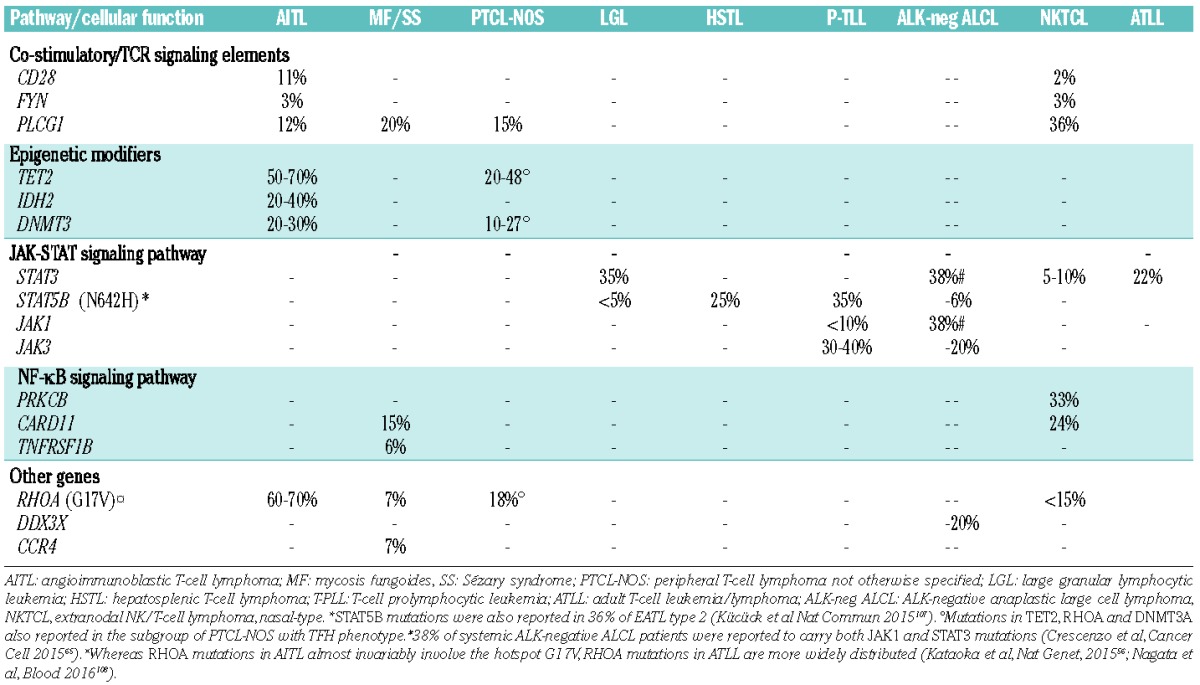
In few circumstances, a single recurrent mutation is identified in almost all cases of a given lymphoma and predominates by far in the genomic landscape of that particular tumor, e.g. the MYD88L265P mutation in Waldenström’s Macroglobulinemia (WM)/lymphoplasmacytic lymphoma (LPL)2 and the BRAFV600E mutation in hairy-cell leukemia (HCL).3 However, for the great majority of lymphomas, including chronic lymphocytic leukemia (CLL),4 diffuse large B-cell lymphoma (DLBCL),5 follicular lymphoma (FL),6 mantle cell lymphoma (MCL),7 Burkitt lymphoma (BL),8 splenic marginal zone lymphoma (SMZL)9 and most peripheral T-cell lymphoma (PTCL) subtypes,10–13 NGS studies have revealed a quite diverse and complex mutation pattern, with a limited number of frequently mutated genes accompanied by a long tail of genes with low-frequency mutations. In addition, while some genes are biased to certain lymphoma entities, e.g. SF3B1 mutations in CLL,14,15 KLF2 mutations in SMZL,16–18 ID3 and TCF3 mutations in BL,19 STAT3 mutations in large granular lymphocyte (LGL) leukemia,11 and RHOA mutations in angioimmunoblastic T-cell lymphoma (AITL),20,21 other genes found recurrently mutated, such as genes involved in DNA repair, epigenetic modification and regulation of transcription (Figure 1), can be detected in multiple subtypes (Table 1) and even in other cancer types.
Table 1A.
Mutation frequencies in different B-cell lymphoma entities.
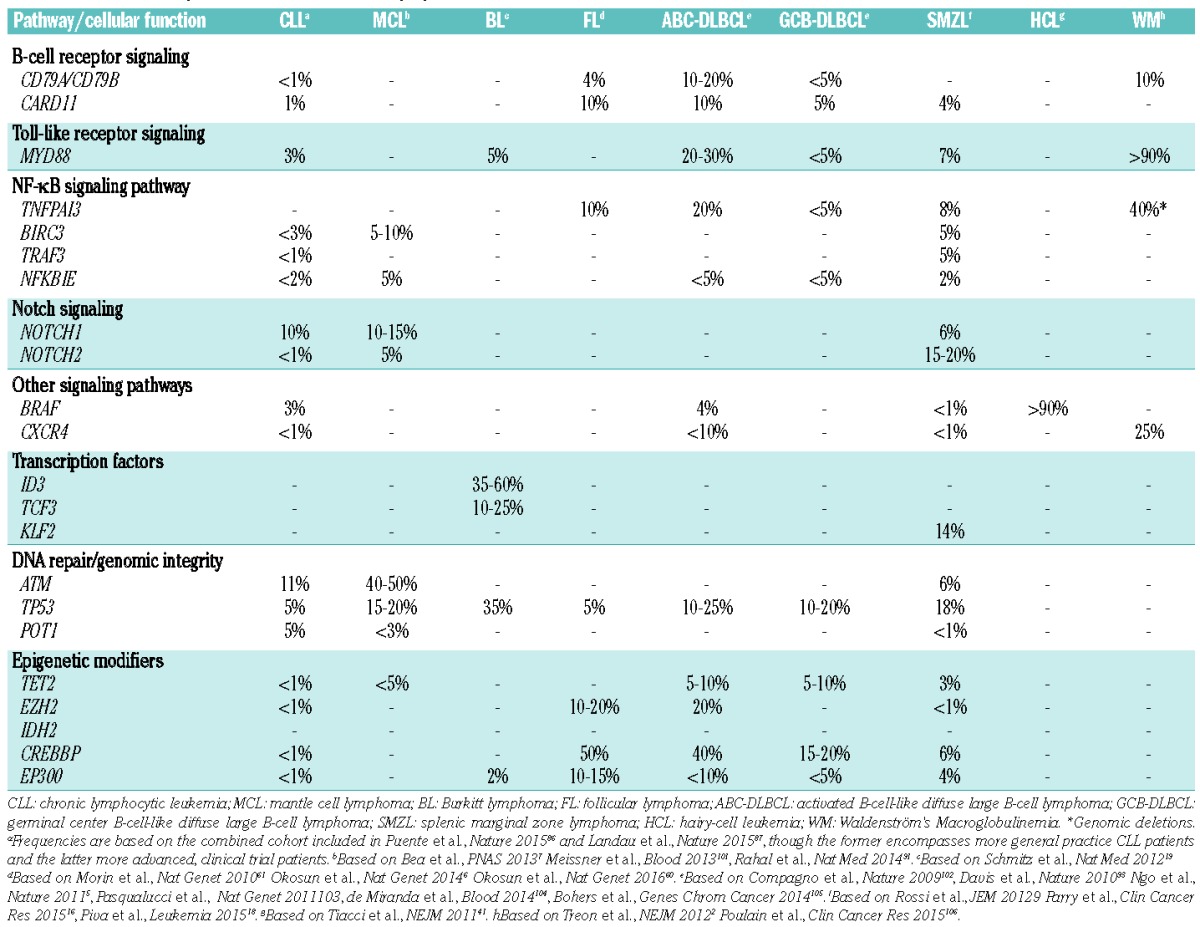
Table 1B.
Mutation frequencies in different T-cell lymphoma entities.
Considering the genetic heterogeneity of lymphomas, also highlighted by diverse results reported in hitherto published NGS studies, it will require large-scale initiatives encompassing thousands of patients to clarify if a specific gene mutation has an impact on the current lymphoma classification/diagnostics and aids clinical decision-making, including therapy selection and response prediction.
In an effort to summarize the current status of this ever-evolving field and provide guidelines regarding the clinical relevance of recent genetic findings, we have established the European Expert Group on NGS-based Diagnostics in Lymphomas (EGNL), with expertise in hematopathology, molecular pathology, clinical genetics, hematology, and oncology. The EGNL Group is supported by the European Research Initiative on CLL (ERIC; Scientific Working Group within the European Hematology Association, EHA) and the European Association for Haematopathology (EAHP).
As our first task, we took advantage of the published literature on lymphomas to identify recurrently mutated genes with a potential clinical relevance that were reported in at least two independent studies; other genomic aberrations, e.g. translocations and copy number aberrations, may also be clinically relevant, though they are not discussed in this review. As the next step, we grouped the identified genes into the following categories based on: i) immediate impact on treatment decisions, ii) diagnostic impact, ii) prognostic impact, iv) potential clinical impact in the near future, or v) interest for research purposes only. In the end, only few genes were deemed to have direct therapeutic or diagnostic implications, while a sizeable proportion of genes were judged as prognostic and/or with a potential role for patient management within the next few years (Table 2). Though this latter category is evidently difficult to define, we decided to include genes for which recent evidence strongly suggests a diagnostic and/or predictive role, sometimes limited to retrospective studies, or for which targeted therapy is under development. In the following sections, we outline our arguments for including a particular gene in one of these categories; highlighted genes in each category are summarized in Table 2.
Table 2.
Categorization of gene mutations based on current evidence levels.

I. Genes with immediate impact on treatment decisions
Currently, there are few genetic lesions with documented impact on therapy selection and patient management in patients with lymphoma. This category is best exemplified by TP53 aberrations in CLL.22 Such aberrations are due to (i) deletions of chromosome 17p (covering the TP53 gene) seen in 5–10% of patients at diagnosis, which are often associated with TP53 mutations on the remaining allele; and, (ii) in a small fraction of CLL patients (3–6%), mutations within the TP53 gene only.23–25 Both types of aberrations (i.e. 17p-deletions and TP53 mutations) are equally adverse in CLL, portending for refractoriness to standard chemoimmunotherapy and poor overall survival.26–29 Notably, such patients experience major clinical benefit by the recently approved novel agents targeting B-cell receptor signaling, namely, the Bruton tyrosine kinase inhibitor ibrutinib and the phosphatidylinositol 3-kinase delta inhibitor idelalisib that are approved for the treatment of patients carrying either of these lesions;30 however, these patients still constitute a high-risk group with an increased risk of disease recurrence with time.
On these grounds, the assessment of TP53 status is essential for clinical decision-making. Hence, sequencing of the TP53 gene is now recommended for all CLL patients, in addition to FISH analysis, before the start of any line of therapy (except in the palliative situation).31,32 Indeed, testing for TP53 aberrations should be considered companion diagnostics for signaling inhibitor treatment in CLL, in order to identify patients with aberrant TP53 for whom the new targeted therapies, described above, represent the current treatment of choice. Furthermore, the fact that the frequency of TP53 mutations gradually increases as the disease becomes more aggressive and chemorefractory supports the need to repeat TP53 mutation analysis prior to any subsequent line of chemotherapy.31,32 Of note, it has also been demonstrated that TP53 microclones at diagnosis, i.e. subclones with a low-allelic burden detected only by NGS but not by Sanger sequencing, were selected by repeated rounds of chemoimmunotherapy and conferred a poor outcome similar to clonal TP53 mutations.33,34 If this finding is confirmed within the context of prospective clinical trials including signaling inhibitors, it is very likely that NGS-based protocols will become the recommended method for TP53 mutation screening in routine clinical practice.
Mutations within TP53 have been described in most other lymphoid malignancies besides CLL, albeit with varying frequency. Although they have been linked to poor clinical outcome in DLBCL,35 SMZL16 and MCL,36,37 this information does not currently have any impact on treatment decisions or follow-up strategies for the individual lymphoma patient.
II. Genes of diagnostic potential
Few lymphoma entities show a predominating, recurrent mutation, such as: the hotspot MYD88L265P mutation in more than 90% of WM/LPL,2,38–40 the hotspot BRAFV600E mutation in ~90% of HCL,41–43 and the STAT3 mutations detected in up to 40% of LGL leukemia.11,44 None of these mutations are pathognomonic of (i.e. exclusive to) a particular entity and can also be found in other lymphomas, though generally at lower frequencies (Table 1). For instance, the MYD88L265P mutation is detected in a significant fraction of DLBCL of the activated B-cell-like subtype (ABC DLBCL),5 as well as in primary cutaneous,45 the central nervous system46 and testicular large B-cell lymphomas,47 but also in a minority of patients with CLL4,48–50 and SMZL.16,51 As another example, STAT3 mutations have been reported, albeit rarely, in immune, mainly hypoplastic, bone marrow failure characterizing a subset of patients with severe aplastic anemia or myelodysplastic syndrome.52 Moreover, some gene mutations are mainly found in a specific lymphoma entity at a relatively high frequency, whereas they are rare in other subtypes, e.g. ID3 and TCF3 mutations in BL,19 KLF2 in SMZL,16–18 SF3B1 mutations in CLL,14,15 RHOA mutations in AITL and other PTCL with a follicular helper T cell (TFH) phenotype,13,20,21,53–59 and, very recently, the novel somatic mutations in RRAGC encoding a Rag GTPase protein (RagC) that were enriched in FL (16%) but were absent in other mature B-cell lymphomas.60
That said, a pattern has started to emerge in recent years where certain lymphoma entities ‘share’ common types of genetic events affecting selected pathways or biological mechanisms (Figure 1). For instance, germinal center derived B-cell malignancies, such as DLBCL of the germinal center B-cell-like type (GCB DLBCL) and FL, appear to have higher frequencies of aberrations in epigenetic-related genes (e.g. EZH2, CREPPB, TET2, IDH2),61,62 while other B-cell lymphomas demonstrate common mutations of members in the NOTCH, B-cell receptor (BcR), and NF-κB signaling pathways (Table 1).63,64 Similarly, different PTCL subtypes, such as mycosis fungoides and Sézary syndrome, AITL and other TFH-derived PTCL, and adult T-cell leukemia/lymphoma (ATLL) share frequent alterations affecting both epigenetic regulation and T-cell receptor (TCR) signaling, whereas mutations of members of the JAK-STAT signaling pathway (STAT3, STAT5B) are shared by several cytotoxic T-cell or NK-cell lymphomas, such as T-LGL, nasal NK/T-cell lymphomas, hepatosplenic T-cell lymphoma, enteropathy-associated T-cell lymphoma (type 2) and ALK-negative anaplastic large cell lymphomas11,56,65,66 (Table 1).
Subgroups of patients within a lymphoma entity may also display differential mutation profiles, initially shown for the GCB vs. ABC subtypes of DLBCL (Table 1) and, more recently, also demonstrated in other lymphomas e.g. HCL, where cases expressing the IGHV4-34 gene in the clonotypic B-cell receptor lack the canonical BRAFV600E, and instead display enrichment for MAP2K1 mutations with an overall similar profile to the HCL-variant.67,68 Similarly, a subgroup of SMZLs with IGHV1-2 expressing B-cell receptor commonly harbors inactivating KLF2 mutations and 7q deletions.16,17 Potentially, this might aid future diagnostics aiming to distinguish these subtypes, however the utility and applicability of this approach have to be studied further.
In summary, presently, MYD88, BRAF, ID3, TCF3, STAT3, STAT5B, RHOA, TET2, and IDH2 mutations are the only genes that can be considered as a complement to the current set-up for lymphoma diagnostics. However, the list is rapidly expanding as evidenced by the case of RRAGC mutations, for example.
III. Genes with prognostic potential
Several gene mutations have been associated with clinical outcome in various lymphoma subtypes. In CLL, besides TP53 and ATM mutations, which are both known to confer poor prognosis, recent high-throughput NGS studies have revealed recurrent mutations within NOTCH1, SF3B1, and BIRC3 for example, that were reported to be associated with poor clinical outcome with higher frequencies in relapsing/treatment-refractory CLL and in Richter’s syndrome.48,69–79 More recent studies have also identified additional gene mutations that may confer a worse outcome in CLL, e.g. NKFBIE, EGR2, and RPS15, although they have been studied less.78–79 In a recent multi-center study conducted within ERIC, sequencing of TP53, NOTCH1, SF3B1, BIRC3 and MYD88 was performed in a large patient series (totaling 3,490 patients), revealing that TP53 and SF3B1 mutations, but not NOTCH1 mutations, remained as independent prognostic markers of shorter time to first treatment in multivariate analysis, even amongst patients expressing unmutated IGHV genes.50 A few published clinical trials have also pointed to a prognostic and even predictive role of SF3B1 and NOTCH1 mutations in CLL,28 where, in particular, the latter confers resistance to the anti-CD20 monoclonal antibodies rituximab29 and ofatumumab,80 however, this needs further exploration and validation. Currently, as a new ERIC project, a dedicated NGS-based gene panel (including 11 genes) has been designed with the purpose to test different targeted enrichment techniques and evaluate inter-center variability and reproducibility.
Similar gene panel-based efforts have also been performed for SMZL, revealing a high frequency of TP53, KLF2, NOTCH2, TNFAIP3, and MYD88 mutations,17,18 and demonstrating NOTCH2 and TP53 mutations as independent markers of short treatment-free and overall survival, respectively.16 Nonetheless, caution is required given the retrospective nature of the published studies and the overall rarity of SMZL raising concerns about potential selection biases.
MCL is characterized by a relatively high number of recurrent secondary genomic aberrations, but few of them have demonstrated additional prognostic value. From recent NGS studies, the prognostic value of NOTCH1/NOTCH2 mutations was recently highlighted7,81 in addition to TP53 defects.
In DLBCL, the prognostic impact of MYC translocations critically depends on the second hit, with cases harboring TP53 mutation and MYC translocation showing the worst overall survival, followed by those cases carrying MYC and BCL2 translocation.82,83 Indeed, DLBCL with TP53 mutation and MYC translocation is a newly recognized subset of ‘double-hit’ lymphoma, accounting for one-third of MYC translocation positive DLBCL. Hence, it is pivotal to perform TP53 mutation screening, in addition to the detection of BCL2 translocation in MYC translocation positive DLBCL, in order to distinguish the double-hit DLBCL from those with an isolated MYC translocation.84
Among PTCL, TET2 mutations in AITL patients were reported to associate with a more aggressive clinical presentation and a shorter progression-free survival,53 whereas in a recent study of NK-TCL DDX3X mutations conferred a particularly poor prognosis.85
Considering the significant differences in mutation frequencies observed between different studies, best exemplified by DLBCL, large-scale efforts are now needed, in particular for rarer entities, to fully understand which genes are the most relevant and will retain independent prognostic impact in relation to other known clinical/molecular markers. This is particularly relevant in light of recent studies86–88 pointing to a prognostic relevance of particular combinations of mutations and/or an increasing number of ‘driver mutations’. In addition, the time point of mutation screening may also differ depending on the disease entity and response to therapy.
IV. Genes with potential clinical impact in the near future
Recent studies in different lymphoma subtypes have highlighted relevant associations between certain recurrent gene mutations and response to treatment. The most striking example is perhaps offered by MYD88 and CXCR4 mutations, which were shown to affect responses to ibrutinib in WM. Indeed, patients with MYD88L265PCXCR4WT (with WT indicating wild-type) status had 100% overall response rate (91% major response rate) as opposed to 86% (62%) and 71% (29%) for MYD88L265PCXCR4WHIM and MYD88WTCXCR4WT patients, respectively.89 Although these results were obtained from a small cohort of patients with WM, they offer a tantalizing glimpse into the future of lymphoma treatment with novel companion diagnostics tailored to novel therapeutic agents.
From recent studies, we have also learnt that mutations within the BTK (C481S) and PLCG2 genes may emerge in CLL patients relapsing after and/or refractory to ibrutinib treatment, and we foresee that the assessment of these genes may soon be incorporated in the diagnostic set-up.90 Preliminary results also indicate that the divergent responses of patients with MCL or DLBCL to ibrutinib may be linked to distinct profiles of genomic aberrations, e.g. BIRC3 mutations in MCL91 and isolated MYD88 mutation in DLBCL92 as well as mutations downstream of BTK, such as activating mutations of CARD11 in DLBCL,93,94 may make these tumors resistant to BTK inhibition.
Recent reports of genes linked to particular physiological processes have revealed new types of mechanisms that may be suitable for targeted therapy. For mutant BRAF, the inhibitor vemurafenib is already in clinical trials in relapsed/refractory HCL, demonstrating high activity even among heavily pre-treated patients.3,95 New types of promising inhibitors have also been developed for EZH2 (a histone methyl transferase),96 SF3B1 (a splicing factor),97 NOTCH198 and IDH2,99 and in some cases have already entered early phase clinical trials. The high frequency of TET2 and/or DNMT3A mutations in AITL and other TFH−derived PTCL may also support the rationale to use demethylating agents as an alternative way to treat patients, supported by the results of a recent single report.100 More generally, targeting certain epigenetic abnormalities or alterations in pathways frequently involved in patients with PTCL (TCR, JAK-STAT, NF-κB; Table 2) represents an attractive approach, also taking into consideration the overall poor outcome of the majority of such patients with conventional chemotherapy-based approaches.
In conclusion, we expect that the list of potential therapeutic targets will expand quickly in coming years, once markers have been functionally validated and new compounds have been discovered.
V. Genes for research purposes only
Finally, for most recurrently mutated genes identified thus far, we do not yet understand their functional role and/or their clinical association, either alone or in the presence of cooperating events. These genes might still be of interest, however, before large-scale studies are performed, it will not be possible to discern their potential contribution to disease pathobiology. Having said that, and similar to category IV above, this subgroup of genes will be a ‘moving target’ depending on the evidence level ascertained for a particular mutation in the coming years, and genes may be taken out if a certain type of mutation is judged to have minor impact for a specific lymphoma entity.
How to move forward in the diagnostic field using NGS?
As mentioned, targeted NGS allows us to select and test many genes and samples simultaneously, and this approach has now been validated for CLL, SMZL and DLBCL, amongst others. In addition, targeted NGS permits a high sequence read depth, an important factor for studying minor subclones and thus clonal heterogeneity, and can also be adapted to formalin-fixed, paraffin-embedded (FFPE) tumor material, which in reality is the source of material for most diagnostic lymphoma specimens.
To gain further insight into NGS-based diagnostics in lymphoma, the EGNL Group has participated in the design of a lymphoma gene panel, compatible with FFPE material, that includes 30 recurrently mutated genes. Our ambition now is to perform a multi-center validation study using a defined number of matched FFPE/fresh-frozen lymphoma specimens to set the technical requirement for NGS-based diagnostics (including metrics such as sequence coverage, specificity, sensitivity and reproducibility of the technique). Once validated, large-scale international collaborative efforts can be conducted for each lymphoma entity, in particular within ongoing or planned treatment studies, to fully understand how to adapt and implement NGS-based diagnostics in day-to-day routine diagnostics.
Acknowledgments
This work was supported by the Swedish Cancer Society, the Swedish Research Council, and The Lion’s Cancer Research Foundation, Uppsala, Sweden; the Special Program in Molecular Clinical Oncology, 5 × 1000 No. #9965 and 10007, AIRC, Milan, Italy, and Ricerca Finalizzata RF-2010-2318823 and RF-2011-02349712, Ministero della Salute, Rome, Italy; the Institut National du Cancer (INCa) and Association pour la Recherche contre le Cancer (ARC), France; the Spanish Ministry of Economy SAF2015-64885-R and Generalitat de Catalunya 2009SGR992, Spain; H2020 “AEGLE, An analytics framework for integrated and personalized healthcare services in Europe”, and “MEDGENET” (No. 692298) by the EU. EC is an ICREA-Academia researcher of the Generalitat de Catalunya, Spain.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/101/9/1002
References
- 1.Swerdlow S, Campo E, Harris N, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues: Lyon, France: IARC Press, 2008. [Google Scholar]
- 2.Treon SP, Xu L, Yang G, et al. MYD88 L265P somatic mutation in Waldenstrom’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. [DOI] [PubMed] [Google Scholar]
- 3.Tiacci E, Park JH, De Carolis L, et al. Targeting Mutant BRAF in Relapsed or Refractory Hairy-Cell Leukemia. N Engl J Med. 2015;373(18):1733–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okosun J, Bodor C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bea S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110(45):18250–18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter J, Schlesner M, Hoffmann S, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. [DOI] [PubMed] [Google Scholar]
- 9.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366(1):95–96. [DOI] [PubMed] [Google Scholar]
- 11.Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odejide O, Weigert O, Lane AA, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171–175. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. [DOI] [PubMed] [Google Scholar]
- 16.Parry M, Rose-Zerilli MJ, Ljungstrom V, et al. Genetics and Prognostication in Splenic Marginal Zone Lymphoma: Revelations from Deep Sequencing. Clin Cancer Res. 2015;21(18):4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clipson A, Wang M, de Leval L, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177–1185. [DOI] [PubMed] [Google Scholar]
- 18.Piva R, Deaglio S, Fama R, et al. The Kruppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia. 2015;29(2):503–507. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo HY, Sung MK, Lee SH, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(4):371–375. [DOI] [PubMed] [Google Scholar]
- 21.Palomero T, Couronne L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stilgenbauer S. Prognostic markers and standard management of chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2015;2015(1):368–377. [DOI] [PubMed] [Google Scholar]
- 23.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 24.Malcikova J, Smardova J, Rocnova L, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114(26):5307–5314. [DOI] [PubMed] [Google Scholar]
- 25.Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28(29):4473–4479. [DOI] [PubMed] [Google Scholar]
- 26.Trbusek M, Smardova J, Malcikova J, et al. Missense mutations located in structural p53 DNA-binding motifs are associated with extremely poor survival in chronic lymphocytic leukemia. J Clin Oncol. 2011;29(19):2703–2708. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez D, Martinez P, Wade R, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29(16):2223–2229. [DOI] [PubMed] [Google Scholar]
- 28.Oscier DG, Rose-Zerilli MJ, Winkelmann N, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121(3):468–475. [DOI] [PubMed] [Google Scholar]
- 29.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247–3254. [DOI] [PubMed] [Google Scholar]
- 30.Stilgenbauer S, Furman RR, Zent CS. Management of chronic lymphocytic leukemia. Am Soc Clin Oncol Educ Book. 2015;164–175. [DOI] [PubMed] [Google Scholar]
- 31.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pospisilova S, Gonzalez D, Malcikova J, et al. ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26(7):1458–1461. [DOI] [PubMed] [Google Scholar]
- 33.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malcikova J, Stano-Kozubik K, Tichy B, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29(4):877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu-Monette ZY, Wu L, Visco C, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120(19):3986–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordstrom L, Sernbo S, Eden P, et al. SOX11 and TP53 add prognostic information to MIPI in a homogenously treated cohort of mantle cell lymphoma–a Nordic Lymphoma Group study. Br J Haematol. 2014;166(1):98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delfau-Larue MH, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood. 2015;126(5):604–611. [DOI] [PubMed] [Google Scholar]
- 38.Poulain S, Roumier C, Decambron A, et al. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood. 2013;121(22):4504–4511. [DOI] [PubMed] [Google Scholar]
- 39.Varettoni M, Arcaini L, Zibellini S, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom’s macroglobulinemia and related lymphoid neoplasms. Blood. 2013;121(13):2522–2528. [DOI] [PubMed] [Google Scholar]
- 40.Rossi D. Role of MYD88 in lymphoplasmacytic lymphoma diagnosis and pathogenesis. Hematology Am Soc remato Educ Program. 2014;2014(1):113–118. [DOI] [PubMed] [Google Scholar]
- 41.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiacci E, Schiavoni G, Forconi F, et al. Simple genetic diagnosis of hairy cell leukemia by sensitive detection of the BRAF-V600E mutation. Blood. 2012;119(1):192–195. [DOI] [PubMed] [Google Scholar]
- 43.Arcaini L, Zibellini S, Boveri E, et al. The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood. 2012;119(1):188–191. [DOI] [PubMed] [Google Scholar]
- 44.Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham-Ledard A, Cappellen D, Martinez F, Vergier B, Beylot-Barry M, Merlio JP. MYD88 somatic mutation is a genetic feature of primary cutaneous diffuse large B-cell lymphoma, leg type. J Invest Dermatol. 2012;132(8):2118–2120. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Aguilar A, Idbaih A, Boisselier B, et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res. 2012;18(19):5203–5211. [DOI] [PubMed] [Google Scholar]
- 47.Kraan W, van Keimpema M, Horlings HM, et al. High prevalence of oncogenic MYD88 and CD79B mutations in primary testicular diffuse large B-cell lymphoma. Leukemia. 2014;28(3):719–720. [DOI] [PubMed] [Google Scholar]
- 48.Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28(1):108–117. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Trillos A, Pinyol M, Navarro A, et al. Mutations in TLR/MYD88 pathway identify a subset of young chronic lymphocytic leukemia patients with favorable outcome. Blood. 2014;123(24):3790–3796. [DOI] [PubMed] [Google Scholar]
- 50.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329–336. [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Lopez A, Curiel-Olmo S, Mollejo M, et al. MYD88 (L265P) somatic mutation in marginal zone B-cell lymphoma. Am J Surg Pathol. 2015;39(5):644–651. [DOI] [PubMed] [Google Scholar]
- 52.Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013;122(14):2453–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemonnier F, Couronne L, Parrens M, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–1469. [DOI] [PubMed] [Google Scholar]
- 54.Cairns RA, Iqbal J, Lemonnier F, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SH, Kim JS, Kim J, et al. A highly recurrent novel missense mutation in CD28 among angioimmunoblastic T-cell lymphoma patients. Haematologica. 2015;100(12):e505–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nature genetics. 2015;47(11):1304–1315. [DOI] [PubMed] [Google Scholar]
- 57.Vaque JP, Gomez-Lopez G, Monsalvez V, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014;123(13):2034–2043. [DOI] [PubMed] [Google Scholar]
- 58.Ungewickell A, Bhaduri A, Rios E, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47(9):1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Ni X, Covington KR, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47(12):1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okosun J, Wolfson RL, Wang J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nature Genet. 2016;48(2):183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98(12):1912–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossi D, Ciardullo C, Gaidano G. Genetic aberrations of signaling pathways in lymphomagenesis: revelations from next generation sequencing studies. Semin Cancer Biol. 2013;23(6):422–430. [DOI] [PubMed] [Google Scholar]
- 64.Sutton LA, Rosenquist R. The complex interplay between cell-intrinsic and cell-extrinsic factors driving the evolution of chronic lymphocytic leukemia. Semin Cancer Biol. 2015;34(22–35. [DOI] [PubMed] [Google Scholar]
- 65.Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicolae A, Xi L, Pittaluga S, et al. Frequent STAT5B mutations in gammadelta hepatosplenic T-cell lymphomas. Leukemia. 2014;28(11):2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xi L, Arons E, Navarro W, et al. Both variant and IGHV4-34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood. 2012;119(14):3330–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waterfall JJ, Arons E, Walker RL, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46(1):8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schnaiter A, Paschka P, Rossi M, et al. NOTCH1, SF3B1, and TP53 mutations in fludarabine-refractory CLL patients treated with alemtuzumab: results from the CLL2H trial of the GCLLSG. Blood. 2013;122(7):1266–1270. [DOI] [PubMed] [Google Scholar]
- 70.Rossi D, Bruscaggin A, Spina V, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118(26):6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cortese D, Sutton LA, Cahill N, et al. On the way towards a ‘CLL prognostic index’: focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia. 2014;28(3):710–713. [DOI] [PubMed] [Google Scholar]
- 72.Del Giudice I, Rossi D, Chiaretti S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97(3):437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rossi D, Fangazio M, Rasi S, et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood. 2012;119(12):2854–2862. [DOI] [PubMed] [Google Scholar]
- 74.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8):1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weissmann S, Roller A, Jeromin S, et al. Prognostic impact and landscape of NOTCH1 mutations in chronic lymphocytic leukemia (CLL): a study on 852 patients. Leukemia. 2013;27(12):2393–2396. [DOI] [PubMed] [Google Scholar]
- 76.Villamor N, Conde L, Martinez-Trillos A, et al. NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia. 2013;27(5):1100–1106. [DOI] [PubMed] [Google Scholar]
- 77.Mansouri L, Cahill N, Gunnarsson R, et al. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia. Leukemia. 2013;27(2):512–514. [DOI] [PubMed] [Google Scholar]
- 78.Ljungstrom V, Cortese D, Young E, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mansouri L, Sutton LA, Ljungstrom V, et al. Functional loss of IkappaBepsilon leads to NF-kappaB deregulation in aggressive chronic lymphocytic leukemia. J Exp Med. 2015;212(6):833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tausch E, Beck P, Schlenk R, et al. NOTCH1 mutation and treatment outcome in CLL patients treated with Chlorambucil (Chl) or Ofatumumab-Chl (O-Chl): Results from the Phase III Study COMPLEMENT 1 (OMB110911). Blood. 2013;122(21):527 abstr. [Google Scholar]
- 81.Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. [DOI] [PubMed] [Google Scholar]
- 82.Schiefer AI, Kornauth C, Simonitsch-Klupp I, et al. Impact of Single or Combined Genomic Alterations of TP53, MYC, and BCL2 on Survival of Patients With Diffuse Large B-Cell Lymphomas: A Retrospective Cohort Study. Medicine. 2015;94(52):e2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gebauer N, Bernard V, Gebauer W, Thorns C, Feller AC, Merz H. TP53 mutations are frequent events in double-hit B-cell lymphomas with MYC and BCL2 but not MYC and BCL6 translocations. Leuk Lymphoma. 2015;56(1):179–185. [DOI] [PubMed] [Google Scholar]
- 84.Clipson A, Barrans S, Zeng N, et al. The prognosis of MYC translocation positive diffuse large B-cell lymphoma depends on the second hit. J Pathol. 2015;1(3):125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang L, Gu ZH, Yan ZX, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47(9):1061–1066. [DOI] [PubMed] [Google Scholar]
- 86.Puente XS, Bea S, Valdes-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. [DOI] [PubMed] [Google Scholar]
- 87.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guieze R, Robbe P, Clifford R, et al. Presence of multiple recurrent mutations confers poor trial outcome of relapsed/refractory CLL. Blood. 2015;126(18):2110–2117. [DOI] [PubMed] [Google Scholar]
- 89.Treon SP, Tripsas CK, Meid K, et al. Ibrutinib in previously treated Waldenstrom’s macroglobulinemia. N Engl J Med. 2015;372(15):1430–1440. [DOI] [PubMed] [Google Scholar]
- 90.Woyach JA, Furman RR, Liu TM, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370(24):2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rahal R, Frick M, Romero R, et al. Pharmacological and genomic profiling identifies NF-kappaB-targeted treatment strategies for mantle cell lymphoma. Nature Med. 2014;20(1):87–92. [DOI] [PubMed] [Google Scholar]
- 92.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nature Med. 2015;21(8):922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pettirossi V, Santi A, Imperi E, et al. BRAF inhibitors reverse the unique molecular signature and phenotype of hairy cell leukemia and exert potent antileukemic activity. Blood. 2015;125(8):1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beguelin W, Popovic R, Teater M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Larrayoz M, Blakemore SJ, Dobson RC, et al. The SF3B1 inhibitor spliceostatin A (SSA) elicits apoptosis in chronic lymphocytic leukaemia cells through downregulation of Mcl-1. Leukemia. 2016;30(2):351–360. [DOI] [PubMed] [Google Scholar]
- 98.Lopez-Guerra M, Xargay-Torrent S, Rosich L, et al. The gamma-secretase inhibitor PF-03084014 combined with fludarabine antagonizes migration, invasion and angiogenesis in NOTCH1-mutated CLL cells. Leukemia. 2015;29(1):96–106. [DOI] [PubMed] [Google Scholar]
- 99.Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. [DOI] [PubMed] [Google Scholar]
- 100.Cheminant M, Bruneau J, Kosmider O, et al. Efficacy of 5-azacytidine in a TET2 mutated angioimmunoblastic T cell lymphoma. Br J Haematol. 2015;168(6):913–916. [DOI] [PubMed] [Google Scholar]
- 101.Meissner B, Kridel R, Lim RS, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121(16):3161–3164. [DOI] [PubMed] [Google Scholar]
- 102.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Miranda NF, Georgiou K, Chen L, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124(16):2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bohers E, Mareschal S, Bouzelfen A, et al. Targetable activating mutations are very frequent in GCB and ABC diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2014;53(2):144–153. [DOI] [PubMed] [Google Scholar]
- 106.Poulain S, Roumier C, Venet-Caillault A, et al. Genomic Landscape of CXCR4 Mutations in Waldenström Macroglobulinemia. Clin Cancer Res. 2016;22(6):1480–1488. [DOI] [PubMed] [Google Scholar]
- 107.Küçük C, Jiang B, Hu X, Zhang W, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from -T or NK cells. Nat Commun. 2015;6:6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nagata Y, Kontani K, Enami T, et al. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood. 2016;127(5):596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]


