Abstract
Many viruses bypass canonical cap-dependent translation in host cells by using internal ribosomal entry sites (IRESs) on their transcripts that hijack initiation factors to assemble initiation complexes. However, it is currently unknown how IRES RNAs recognize initiation factors that have no endogenous RNA binding partners; as a prominent example, the IRES of Encephalomyocarditis virus (EMCV) interacts with the HEAT-1 domain of the initiation factor 4G (eIF4G). Here we report the solution structure of the J-K region of this IRES and show that its stems are precisely organized to position protein recognition bulges. This multi-site interaction mechanism works on an all-or-nothing principle, where all domains are required. This pre-organization is accomplished by an ‘adjuster module’ — a pentaloop motif that acts as a dual-sided docking station for basepair receptors. Since subtle changes to the orientation abrogate protein capture, our study highlights how a viral RNA acquires affinity for a target protein.
RNA viruses use varied mechanisms to manipulate the host cell translational machinery in order to efficiently produce viral proteins for replication. One such mechanism utilizes IRES in the untranslated region of the transcript to subvert ribosomes from canonical eukaryotic cap-dependent translation1. In fact, many RNA viruses rely on IRES-dependent translation as the sole mechanism for protein synthesis, which allows for protein production even when the host translation is down regulated. Increasing evidence suggests that cellular mRNAs may also capitalize on this alternate translation mechanism; and some estimates suggest that up to 10% of cellular mRNAs may use IRESs to regulate processes such as development and responses to cellular stress2–4.
In general, the IRESs are either configured to interact directly with the 40S ribosomal subunit5–7, or first engage initiation factors to begin ribosomal initiation complex assembly. The IRESs of EMCV and Foot-and-mouth disease virus (FMDV) in the Picornaviridae family are prototypes for understanding the latter mechanism both because these IRESs are highly efficient and are thus used widely in biotechnological applications8, and because the protein factors they use are similar to those used by cellular IRESs9,10. Moreover, FMDV is responsible for major economic losses in the livestock industry11. IRESs are also attractive targets for chemotherapeutic inhibition12 and the detailed characterization of conserved functional elements in these picornavirus IRESs would facilitate the development of such inhibitors.
As a first step towards assembling ribosomes, the EMCV IRES element is designed to directly interact with the first of the three HEAT domains – HEAT-1 – of eukaryotic initiation factor 4G (eIF4G)13,14. This interaction is enhanced by the presence of eIF4A, which directly interacts with eIF4G HEAT-115. The J-K region of EMCV IRES (G680-C787), which is composed of two stem loops (J and K domains) bifurcating from a base stem (St domain) and a highly conserved stretch of a six adenosine (A-rich region) linker between the K and St domains16, is responsible for specifically mediating the interaction with the HEAT-1 domain of eIF4G (Figs. 1a,b)17. The A-rich region and internal loops in the J, K, and St domains are conserved features in both EMCV and FMDV, although they belong to different genera in the Picornaviridae family18. Mutations to these regions have been shown to be highly detrimental to the translational efficiency19–23, demonstrating a critical role of this region in recruiting ribosomes. However, very little structural and mechanistic information exists for how the J-K region engages the HEAT-1 domain of eIF4G. Here we set out to solve the solution structure of the J-K region and characterize its interaction with the HEAT-1 domain by using solution nuclear magnetic resonance (NMR), small angle X-ray scattering (SAXS), and biochemical studies, to understand how IRESs are configured to capture host initiation factors.
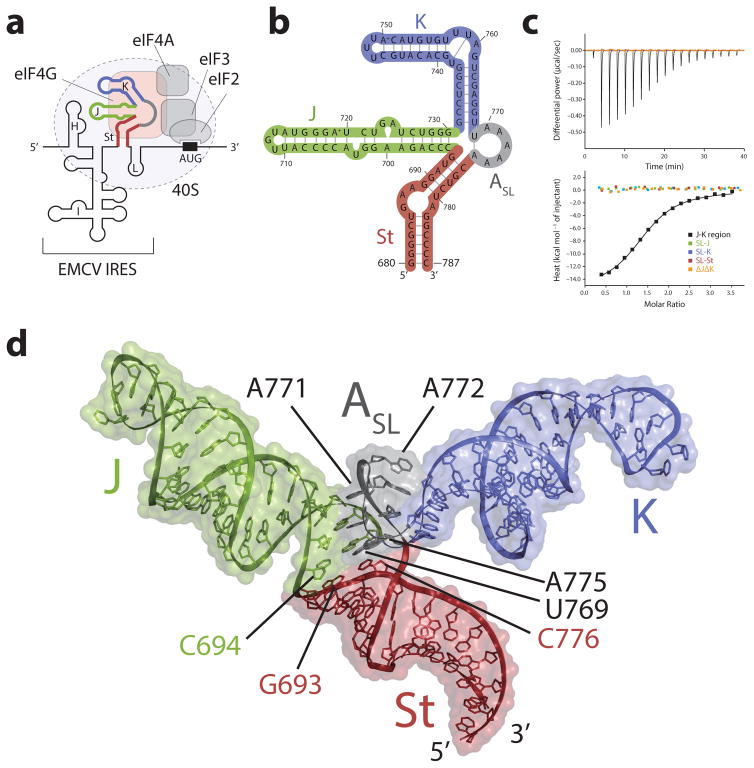
Figure 1. Structure and function of the J-K region of the EMCV IRES RNA.
(a) Schematic diagram of the translation initiation complex formed by the EMCV IRES. The various structural domains in the EMCV IRES, H-L, are labeled. The J-K region (depicted with thick line colored red, green, blue and gray) directly interacts with eIF4G (orange) of the host cell, recruiting the ribosomal 40S subunit to the AUG start codon via multiple protein-protein interactions that involve the other translation factors eIFs 4A, 3, and 2. (b) NMR derived secondary structure of the J-K region. The four domains of the J-K region, J, K, St, and ASL are highlighted in green, blue, red, and gray, respectively. (c) Isothermal titration calorimetry (ITC) results where the reconstituted eIF4G HEAT-1 domain was titrated into the full-length J-K region (black) and its isolated segments SL-J (green), SL-K (blue), SL-St (red), and ΔJΔK (orange). (d) Solution structure of the J-K region with surface representation. The J, K, St, and ASL domains are colored green, blue, red, and gray respectively.
Results
Concerted J-K domain action needed to capture HEAT-1
As a first step towards identifying the atomic determinants of the eIF4G recognition by the J-K region of EMCV IRES, we prepared a series of truncated constructs corresponding to the three stems of the J-K region (SL-J, SL-K, SL-St) and the A-rich region (ΔJΔK) (Fig. 1b and Supplementary Figs. 1a,2a,3,4a), to be used for solution state biophysical studies. NMR assignments showed that all of the constructs correctly reflected the structures present in the full-length J-K region (Supplementary Figs 1–4 and Supplementary Note). However, none of these isolated fragments measurably interacted with the HEAT-1 domain as demonstrated by the isothermal titration calorimetry (ITC) (Fig. 1c, see also Supplementary Figs. 5 and Supplementary Note). In fact, tight binding (Kd = 1.3 ± 0.1 μM) was observed only for the full-length J-K region, implicating an unusual protein-RNA recognition mechanism, wherein the interaction involves the stoichiometric binding of the HEAT-1 domain to an epitope defined by the collective contributions of two or more domains of the J-K region.
The overall solution structure of the J-K region
We then solved the solution structure of the full-length J-K region by NMR combined with SAXS (Figs. 1d,2,3, and Table 1. See Supplementary Figs 1–4,6a and Supplementary Note for detailed description). The J-K region of the EMCV IRES adopts a Y-shape structure where the St domain is the base of the Y, and the J and K domains form the two prongs. Unexpectedly, we found that the intervening A-rich region folds into a very short stem loop (referred here as ASL domain) where residues A770-A774 adopt a pentaloop structure, and U769-A775 close the loop as a single stem basepair, as evidenced by the typical NOE signals from A775 H2 to U769 H3, C776 H1′, and A770 H1′ (Fig. 1d,4a, and Supplementary Fig. 4e). Both the J and the ASL domains are supported by the terminal basepair (G693-C776) of the St domain by partial coaxial stacking interactions as evidenced by NOEs between G693 and C694 and C776 and A775, respectively (Fig. 4a and Supplementary Fig. 4f). The K domain, on the other hand, does not interact with either J or the St domain but is held in place by interactions with the ASL domain (see below). Thus overall, while the ASL domain has a perpendicular orientation with respect to the St domain, it is tightly sandwiched between the minor grooves of the J and K domains (Fig. 4c).
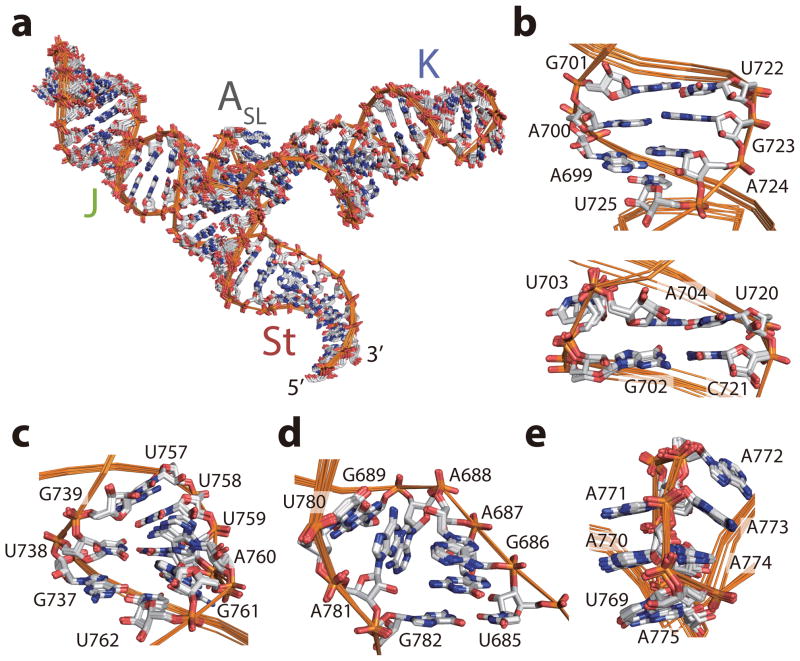
Figure 2. Ensembles of the low-energy structures.
(a–e) Ensembles of the ten low-energy conformers of the J-K region (a), and the bulge regions of the J domain (b), the K domain (c), the St domain (d), and the ASL domain (e), respectively.
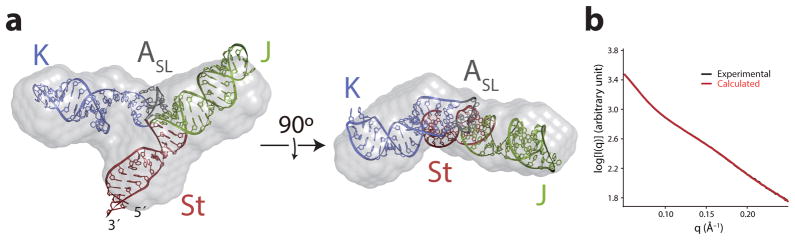
Figure 3. Small angle X-ray scattering of the J-K region.
(a) Overlay of the structure of the J-K region with the SAXS ab initio envelope structure (gray). J, K, St, and ASL domains are colored as in Fig. 1. (b) Overlay of the experimental SAXS profile (black) and that back-calculated from the structure (red). I is scattering intensity, q is proportional to the scattering angle (q = 4sinθ/λ, where 2θ is the angle between the incident x-ray beam and the detector, and λ is x-ray wavelength in Å).
Table 1.
NMR and refinement statistics for the J-K region
| J-K | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance restraints | |
| Total NOE | 830 |
| Intraresidue | 471 |
| Inter-residue | 359 |
| Sequential (|i – j| = 1) | 330 |
| Nonsequential (|i – j| > 1) | 29 |
| Hydrogen bonds | 300 |
| Total dihedral-angle restraints | 773 |
| Base pair | 0 |
| Sugar pucker | 194 |
| Backbone | 579 |
| Based on A-form geometry | 773 |
| Structure statistics | |
| Violations (mean ± s.d.) | |
| Distance constraints (Å) | 0.19 ± 0.01 |
| Dihedral-angle constraints (°) | 7.04 ± 2.05 |
| Max. dihedral-angle violation (°) | 12.77 ± 1.47 |
| Max. distance-constraint violation (Å) | 0.64 ± 0.03 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.010 ± 0.001 |
| Bond angles (°) | 0.69 ± 0.07 |
| Impropers (°) | 0.48 ± 0.01 |
| Average pairwise r.m.s. deviation (Å)a | |
| All RNA heavy | 0.73 |
| All nucleotides | 0.76 |
Pairwise r.m.s. deviation was calculated among ten refined structures.
Figure 4. Structure and long-range interactions of the ASL domain.
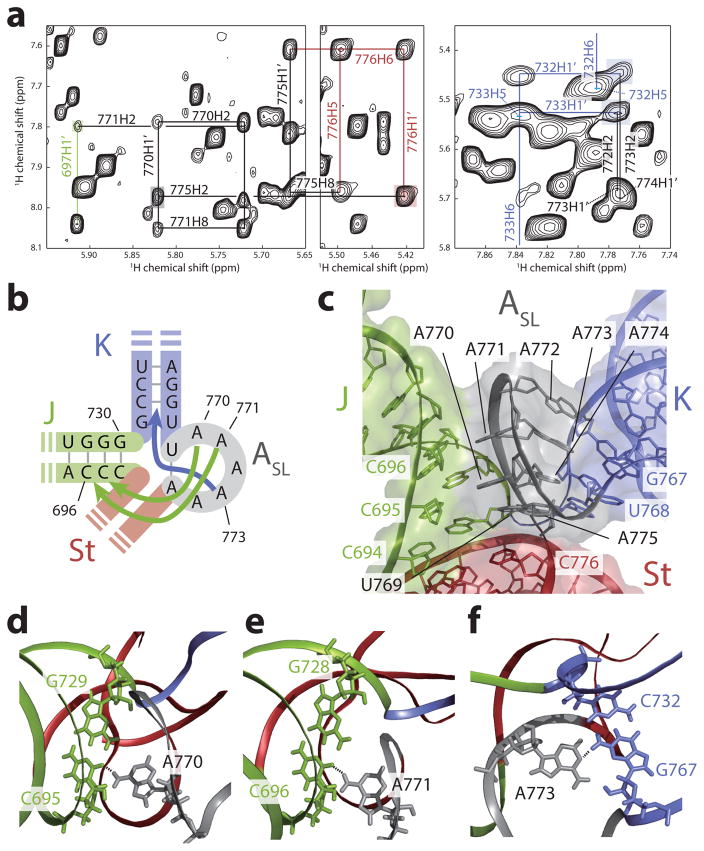
(a) Regions of 1H-1H NOESY spectra of [u-2H, 1H-Ade/Cyt] ΔJΔK construct. Long range NOE signals indicating a basepair between U769 and A775 are highlighted in gray and red. NOE signals showing long range interactions of the J and the K domain with the ASL domain are highlighted in green and cyan, respectively. (c) Schematic representation of base triples. A770:C695-G729 and A771:C696-G728 are formed between the ASL and J domains, whereas A773:C732-G767 is formed between the ASL and K domains. (d) Structure of the ASL domain. A770 and A771 are stacked together pointing toward the J domain (green) whereas the stretch A772-A773-A774 are stacked together pointing toward the K domain (blue). A775, A770, and A771 are in the same register with C694:G730, C695:G729, and C696:G728 basepairs in the J domain, whereas A772, A773, and A774 are in the same register with C733:G766, C732:G767, and G731:U768 in the K domain, respectively. (e–g) Structures of the base triples. Base triples A770:C695-G729 (e), A771:C696-G728 (f) and A773:C732-G767 (g) are shown. Hydrogen bonds between base triples are depicted with dashed lines.
Structural features of the ASL domain
The configuration of the pentaloop in ASL domain is partly similar to a reported GAAA tetraloop structure24 (a GNRA type tetraloop, see reference 25 for a review), with an important exception in that the G is replaced by a A-A dinucleotide stack. Correspondingly, in the pentaloop, the chain turns between residues A771 and A772, and the two base stacks, A770-A771 and A772-A774, take on opposing orientations within the loop, with the former facing the minor groove of J domain and the later facing the minor groove of the K domain (Figs. 4b,c). The observed NOE signals between H2 protons of A774, A773 and A772 and H1′ protons of G731, C732, and C733, respectively, placed the A772-A774 stack in the same register as the first three basepairs of the K domain (Fig. 4a, and Supplementary Fig. 4g). Similarly, long range NOE signals placed the A770-A771 stack, along with the closing U769-A775 basepair, in register with the first three basepairs of the J domain (see Supplementary Figs. 4h–j). Most importantly, we observed evidence for ‘loop:receptor’ like interactions25: namely, NOE signals between the H2 protons of A770, A771 and A773 and imino protons G729, G728, and G767 indicated formation of A770:C695-G729, A771:C696-G728 and A773:C732-G767 base triples between ASL and the J and K domains, respectively (Figs 4d–f, and Supplementary Fig. 4j).
This ‘dual-sided’ loop:receptor interaction leads to the tight wedging of the ASL domain deep in between the minor grooves of the J and K domains, which requires a 90° turn in the backbone chain between U769 of ASL domain and U768 of the K domain. To allow for this structure, a number of accommodations are made: residue U769 adopts an intermediate syn conformation and the base of the K domain is widened by the presence of a highly conserved terminal G-U basepair. The latter is an especially intriguing feature: in a canonical GAAA-receptor interaction, a C-G basepair would be required at the terminal position in the K domain to allow for triple base formation with the last adenosine25. Thus, while the triple base with the penultimate adenosine takes place, the interaction with the last adenosine is forgone in the ASL-K domain interaction to accommodate the tight turn, and the required hydrogen bonding contributions are likely compensated for by triple base interactions with the J domain.
Interaction of the J-K region with the HEAT-1 domain
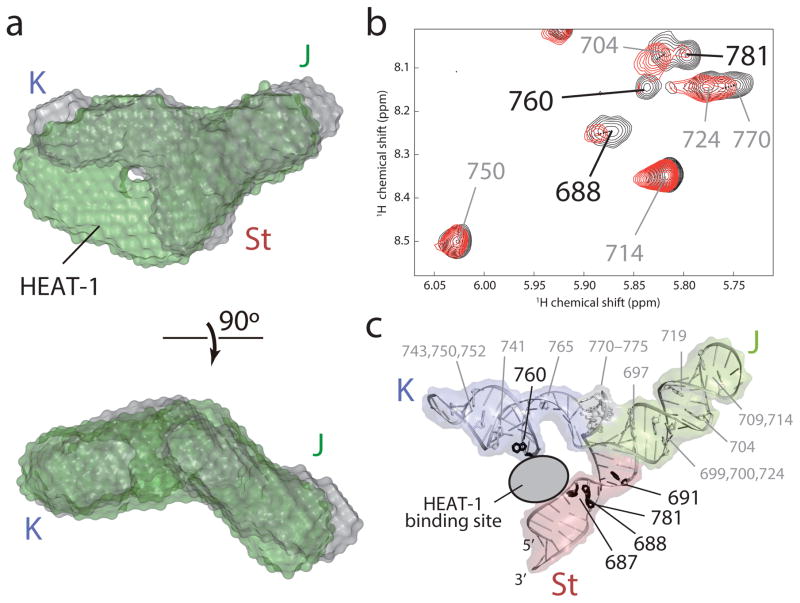
To understand how this unique structure engages the HEAT-1 domain of eIF4G, we first performed SAXS studies of the J-K region both in the absence and presence of the HEAT-1 domain26,27. Interestingly, the reconstructed ab initio SAXS envelope demonstrated that there are no major structural rearrangements in the overall architecture of the IRES upon protein binding, and extra density corresponding to the molecular size of the HEAT-1 domain was observed only between the K and St domains, indicating that binding occurs in a cleft between these domains (Fig. 5a and Supplementary Fig. 6a). Since isolated K and St domains cannot engage the protein (Fig. 1c), this result suggests that the capture of the HEAT-1 domain occurs through the concerted docking of these motifs onto the HEAT-1 domain. Previous biochemical analyses had indeed implicated the bulge regions of the K and St domains in protein binding; however, the dominant interaction was believed to occur via ASL domain13,21–23,28. Surprisingly, the SAXS density assigned to the ASL domain remained unperturbed, suggesting that it is not directly involved in protein contacts. To confirm our findings, we performed NMR titration of substoichiometric amounts of the HEAT-1 domain into an A-protonated J-K region sample because the ASL loop, and the J, K and St domain bulges are all adenosine-rich (Fig. 1b). While we observed substantial intensity reductions of specific adenosine residues in the bulges of the K and the St domain, the intensities of the adenosine residues in the J and ASL domain remained unaffected, thus confirming the interaction surfaces indicated by SAXS (Figs. 5b,c and Supplementary Fig. 6b).
Figure 5. Interaction of the J-K region with the HEAT-1 domain of eIF4G.
(a) Overlay of the SAXS ab initio envelope structure of the J-K region (gray) with that in complex with the HEAT-1 domain (green). The top panel is shown from the same perspective as in Fig. 3a, left panel. (b) 1H-1H NOESY spectra of [u-2H, {H1′,H2′,H2,H8}-Ade] J-K region in the absence (black) and substoichiometric presence (red) of the HEAT-1 domain. Assignments of the intraresidual H1′–H8 NOE signals are labeled. Residues whose NOE signal intensities were substantially reduced by the addition of the HEAT-1 domain are labeled in black. (c) Mapping of the adenosine residues in (b) onto the structure of the J-K region. Adenosine residues whose signal intensities are substantially reduced upon addition of the HEAT-1 domain are colored black whereas the others are white.
Orientations of the domains and relation to function
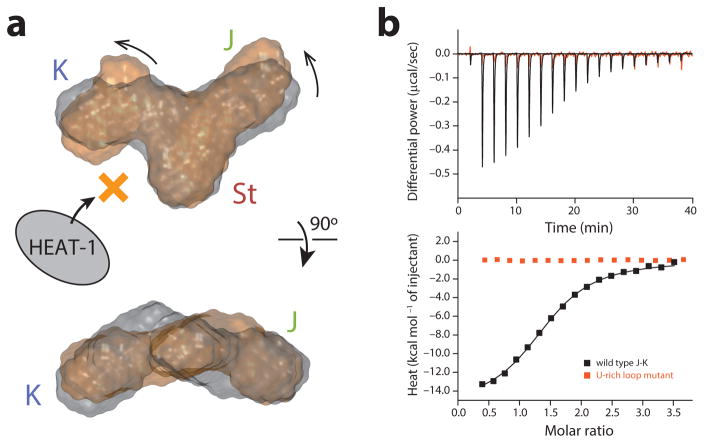
Next, we wanted to test the role of the ASL and J domain that do not appear to contact the HEAT-1 domain. Data at minimally elevated temperature (45 °C), which maintains secondary structure but causes partial loss of long-range tertiary interactions29, showed the presence of multiple conformations for residues at the junction (for example, U768 and U769) (Supplementary Fig. 6c), despite being involved in canonical base-pairing interactions. This suggests that the role of the ASL motif is to restrain the inherent conformational heterogeneity at the junction of the J-K region. In order to investigate this hypothesis, we mutated the A-rich loop to a U-rich loop. NMR and SAXS analyses of this construct showed the structures of the individual J, K, and St domains to remain unaffected, but the overall topology of the molecule was slightly perturbed by the U-rich loop linker (Fig. 6a and Supplementary Fig. 6d). Briefly, while the coaxial stacking of the J domain over the St domain was preserved, the orientation of the K domain bulge changed with respect to the St domain bulge. This subtle reorientation, however, completely abrogated HEAT-1 domain binding, demonstrating the critical role of the ASL–K inter-domain interaction for accurate placement of the K domain-binding pocket relative to the pocket in the St domain (Fig. 6b). This structural role played by the ASL domain explains all of the previous analyses, and shows that there is a limited degree of plasticity present in the relative organization of the protein recognition interface in the J-K region.
Figure 6. Structural basis for the ASL domain as an adjuster module.
(a) SAXS ab initio structures of the wild-type J-K region (gray) and the U-rich loop mutant (orange). (b) ITC results of the interaction of the wild type J-K region and the U-rich loop mutant with the HEAT-1 domain.
Since the J domain also does not contact the HEAT-1 domain directly, we next wanted to test the functional significance of this domain. We thus made both an A771U mutant and a corresponding C696A-G729U mutant, designed to abrogate the A771:C696-G728 base triple formation between the ASL and the J domain (Fig. 4e). Intriguingly, both of these constructs also lost the ability to engage the HEAT-1 domain (Supplementary Fig. 6e). NMR analyses of the C696A-G729U mutant showed that the ASL domain interacts with the K domain but loses the ability to engage the J domain in this mutant (Supplementary Fig. 6f). These studies indicate that the ASL domain acts as an ‘adjuster module’, and uses the J domain interaction as an anchoring point for the correct placement of the K domain relative to the St domain.
Identifying interaction surfaces on the HEAT-1 domain
Lastly, we set out to understand why the EMCV IRES requires multiple domains that are appropriately positioned for capturing a single molecule of the HEAT-1 domain. Both the K and St domain bulges contain adenosine residues poised to interact with the HEAT-1 domain residues (Fig. 2c,d). In general, exposed adenosine residues are known to interact with aromatic residues in proteins, and in fact, statistical studies show that such stacking interactions contribute to more than 90% of surface contacts in protein-RNA interactions30. However, although the importance of basic residues of HEAT-1 domain has been previously demonstrated31, the contributions of the few aromatic residues (F777, F812, F949, and F978), which are potentially available for interaction, have not yet been tested (Supplementary Fig. 7a). Thus, we prepared four mutants of the HEAT-1 domain, F777A, F812A, F949A, and F978A, and investigated their ability to interact with the J-K region by using ITC (Supplementary Fig. 7b). While F777A and F949A did not affect the RNA binding, F812A and F978A exhibited a complete loss of the hallmark 1:1 binding in the ITC experiment. These data, together with the previous chemical footprinting and the mutagenesis data22,31,32, allowed us to dock the HEAT-1 domain crystal structure into the SAXS envelope, wherein F777 and F949 are positioned away from the IRES RNA, while F812 and F978 are appropriately positioned near the St and the K domain, respectively (Supplementary Fig. 7c), illustrating that the distally orientated bulges in the J-K stem contact limited and scattered aromatic residues on the HEAT-1 domain.
The loss of specificity by the mutations at F812 and F978 as measured by ITC provided strong evidence for the location of the binding interface between the HEAT-1 domain and the J-K region, which is consistent with the SAXS data. In vivo, however, the interaction with the initiation factor eIF4A is known to enhance binding of eIF4G for the EMCV IRES, probably by adding additional contacts to the IRES and/or by modulating the J-K–HEAT-1 complex15. We thus evaluated the significance of the J-K–HEAT-1 interface in a functional context by using toeprinting assay, which is used to show the enhancement of the affinity by eIF4A. Here we used eIF4G1 (a.a. 736-1115), which contains the HEAT-1 and a section of the linker between the first two HEAT domains, and eIF4G1 (a.a. 653-1599), which contains all three HEAT domains through the C terminus, and investigated how their complex formation is affected by the F812A, F978A, and F812A/F978A mutations (Supplementary Fig. 7d–f). Although noticeable differences were not observed between the wild type and the mutants when only the eIF4G variants were added to the EMCV IRES in the condition of the toeprinting assays, differences were observed when eIF4A was added with eIF4G. The enhancement of the affinity by eIF4A, evidenced by the decrease of the toeprint for the full-length cDNA for the wild types, was weakened by the two mutants, and their effects were additive (Supplementary Fig. 7d,e). This pattern of altered activity was also observed in the 48S formation assay on the EMCV IRES, where both eIF4G binding to the J-K region and the 48S formation were decreased by the mutations, suggesting the synergy of eIF4G and eIF4A for IRES binding and the importance of the two phenylalanine residues in the functional contexts (Supplementary Fig. 7f).
Discussion
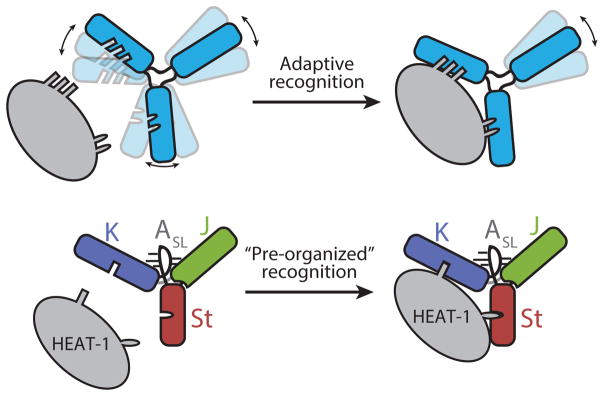
Overall, our study highlights how viruses adapt RNA structures to capture host proteins — even ones that are not conventional RNA binders (Fig. 7). In general, biomolecular complex formations occur through adaptive recognition of the binding partners33,34. Although there is an energetic cost associated with losing conformational entropy upon complex formation, conformational variations in molecules nevertheless exist to provide opportunities for optimizing intermolecular interactions. In stark contrast, our study shows that loss of tertiary structure in the J-K region, which increases the degree of conformational freedom, prohibits HEAT-1 domain recognition, despite all of the binding interfaces still being intact. Consistent with this idea, the HEAT-1 domain interaction with the J-K region is predominantly enthalpically driven (ΔH = −15.6 kcal/mol and ΔS = −25.4 cal/mol/K). The inability of the A771U and C696A-G729U mutant IRESs to overcome the entropic barrier is likely due to the limited number of intermolecular contacts possible with the HEAT-1 domain. Indeed, the HEAT-1 domain of eIF4G, which canonically interacts with other protein initiation factors, has very few exposed aromatic residues available for interaction (Supplementary Fig. 7a). The viral IRES is elegantly designed to overcome this limitation: one, it contains interacting bulges in distal domains for capturing remote binding surfaces, and two, these domains are pre-organized in a fixed configuration by the adjuster module to allow for precise docking onto the sparsely distributed binding patches without the loss of entropy.
Figure 7. Two modes of protein recognition by RNA.
Top: adaptive recognition, bottom: ‘pre-organized’ recognition that is employed by the J-K region of EMCV IRES to capture the HEAT-1 domain. See text for details.
Furthermore, our initial studies on establishing the interaction sites on the HEAT-1 domain have identified two aromatic residues (F812 and F978) that may play a role in interacting with the distal bulges on the K and St domains, respectively. While F812 is available for the IRES interaction upon eIF4A interaction, the importance of F978 is intriguing because this site is also known to interact with the N-terminal domain of eIF4A. However, our previous study has shown that it is the C-terminal domain of eIF4A that predominantly interacts with the HEAT-1 domain, while the N-terminal domain has minor contribution to the HEAT-1 binding35. In fact, the crystal structure of the yeast eIF4A complexed with the middle domain of eIF4G (homologous to the HEAT-1 domain in human eIF4G) shows very small buried surface area at the N-terminal domain interaction site and is consistent with the minor importance of this interaction36. Thus our current study suggests a potential change in the interaction between eIF4A and eIF4G, where the N-terminal domain interaction may be forgone upon J-K region encounter.
Online methods
Purification of RNA samples
Plasmids for RNA transcription of the J-K region, SL-J, SL-K, and SL-St were constructed by inserting PCR-amplified DNA sequences containing the T7 promoter, an insert sequence, and a SmaI or DraI linearization site, into pUC57 or pUC19 plasmid, by using BamHI and EcoRI restriction sites. The RNA sequences are: J-K: 5′-GGGGCUGAAGGAUGCCCAGAAGGUACCCCAUUGUAUGGGAUCUGAUCU GGGGCCUCGGUGCACAUGCUUUACAUGUGUUUAGUCGAGGUUAAAAAAC GUCUAGGCCCC-3′, SL-J: 5′-gggCAGAAGGUACCCCAUUGUAUGGGAUCUGAUCUGccc-3′, SL-K: 5′-gggCUCGGUGCACAUGCUUUACAUGUGUUUAGUCGAGccc-3′, SL-St: 5′-GGGCUGAAGGAUGGAGACGUCUAGGCCC-3′, where the non-native nucleotides to enhance the transcription by T7 RNA polymerase are shown in the lower case. The plasmids for RNA transcription of ΔJΔK and U-rich loop mutant were generated by PCR based mutagenesis, using the J-K plasmid as a template. The sequences are: ΔJΔK: 5′-GGGGCUGAAGGAUGCCCAGAGAGAUCUGGGGCCUCGGGAGAUCGAGGU UAAAAAACGUCUAGGCCCC-3′, U-rich loop mutant: 5′-GGGGCUGAAGGAUGCCCAGAAGGUACCCCAUUGUAUGGGAUCUGAUCU GGGGCCUCGGUGCACAUGCUUUACAUGUGUUUAGUCGAGGUUUUUUGUC GUCUAGGCCCC-3′. These plasmids were used as templates for PCR to amplify the regions containing the T7 promoter and sequence of interest. For J-K, ΔJΔK, and their mutants, the 5′-terminal two nucleotides of the reverse primer were 2′-O-methylated in order to suppress the heterogeneity at the 3′ end of the transcripts. The PCR products were extracted by phenol/chloroform/isoamylalcohol and precipitated by isopropanol. RNA samples were obtained by in vitro transcription using T7 RNA polymerase and the template DNA amplified by PCR. After transcription, RNA samples were heat denatured and purified by using urea-denaturing polyacrylamide gels. For isotopically labeled samples, nucleotides with appropriate isotopic labeling (Cambridge isotope laboratory) were used in in vitro transcription.
Purification of the HEAT-1 domain
Human eIF4G1 HEAT-1 domain (residue number 746-992) was expressed and purified as described previously37, with some modifications. HEAT-1 domain was expressed in E.coli BL21(DE3) strain with N-terminal GB1 and Hisx6 tags. After disrupting the cells by sonication, polyethyleneimine (Sigma Aldrich) were added to the final concentration of 0.1% and the lysate was incubated at 4°C for 30 min with mild agitation. After centrifugation at 14,000xg for 30 min, the supernatant was applied to the TALON cellthru resion (Cloneteq), and the resin was washed extensively with a buffer containing 5 mM imidazole. The protein was eluted with a buffer containing 200 mM imidazole. After cleaving the tag with TEV protease, the tag and protease were removed by passing through the TALON resin.
NMR
For NMR experiments, the RNA samples were dissolved in a buffer containing 10 mM potassium phosphate (pH 6.4) and 10 mM NaCl. All NMR experiments were acquired by using Bruker 600, 700, 750, 800, or 900 MHz instruments equipped with cryogenic probes. Spectra for observing non-exchangeable protons were collected at 298, 308, and 318K in 99.96% D2O, whereas those for exchangeable protons were at 278 and 283K in 10% D2O. For NOESY experiments, mixing times were set to 200 ms except the ones of [u-2H, u-1H-Ade/Cyt] ΔJΔK and [u-2H, u-1H-Ade/Gua] ΔJΔK, where two spectra were acquired at the mixing times of 200 and 500 ms for each sample. Assignments for non-exchangeable 1H and 13C signals of SL-J, SL-K, and SL-St were obtained by analyzing two-dimensional 1H-1H NOESY recorded with non-labeled samples, two-dimensional 13C-HMQC, and three-dimensional 13C-edited HMQC-NOESY spectra recorded with non-uniform sampling38. Assignments of ΔJΔK were obtained first by transferring assignments of the SL-J, SL-K, and SL-St to the regions that are shared with them, and then by analyzing 1H-1H 2D NOESY spectra of fully protonated, [u-2H, {H1′,H2′,H2,H8}-Ade]-labeled, [u-2H, u-1H-Ade/Cyt]-labeled, and [u-2H, u-1H-Ade/Gua]-labeled samples. Assignments of the adenosine 1H signals of the J-K regions were obtained by comparing the 1H-1H 2D NOESY spectra of the isolated segments with that of [u-2H, {H1′,H2′,H2,H8}-Ade]-labeled J-K.
NMR titration experiments were conducted in buffer containing 20 mM Hepes-NaOH (pH 6.4), 150 mM NaCl, 1 mM MgCl2, and 2 mM DTT. Two 1H-1H 2D NOESY spectra of [u-2H, {H1′,H2′,H2,H8}-Ade]-labeled J-K were acquired at the mixing time of 200 ms and at 35°C, in the absence and presence of the natural abundance HEAT-1 domain at the molar ratio of 0.2 against the J-K region.
Isothermal titration calorimetry
Prior to all isothermal titration calorimetry (ITC) experiments, the HEAT-1 domain and RNAs were dissolved into ITC buffer (20 mM Hepes-NaOH (pH 6.5), 150 mM NaCl, 2 mM MgCl2, 1 mM 2-mercaptoethanol), unless otherwise indicated. For each ITC experiment, reaction heats (μcal s−1) were measured for 2 μl titrations of 150–250 μM of HEAT-1 domain into 10–20 μM of RNA at 25°C using an iTC-200 machine (MicroCal). Titration curves were analyzed by using ORIGIN (OriginLab).
Small angle X-ray scattering
SAXS data were obtained at SIBYLS beamline of Advanced Light Source at Lawrence Berkeley National Laboratory. Measurements were performed in buffer containing 50 mM Hepes-NaOH (pH 6.5) and 150 mM NaCl, and 2% glycerol for RNA samples, and in buffer containing 50 mM Hepes-NaOH (pH 6.5) and 150 mM NaCl, 2% glycerol, 2 mM MgCl2, and 1 mM DTT for RNA-protein complex samples, unless otherwise indicated. Data sets that were collected at the exposure time of 3 s were used for further analyses after confirming that no saturation was observed. The scattering intensity from the solute was obtained by subtracting the background scattering from the sample scattering. The scattering intensities at q = 0 Å−1 [I(0)], as determined by Guinier analysis, were compared between four different concentrations (0.5, 1.0, 2.0, and 4.0 mg/mL) to detect possible inter-particle interactions. Data were analyzed by using ScÅtter software, and the ab-initio envelope structures were reconstructed by using DAMMIF/DAMMIN software.
Toeprinting assay
Substitutions were introduced into a vector for expression of eIF4G1(653-1599)14 by NorClone Biotech Laboratories (London, Ontario, Canada). Recombinant wild type and mutant forms of eIF4G1(653-1599) were purified after expression in Escherichia coli BL21(DE3), and toeprinting of EMCV IRES–eIF4G complexes was done as described15.
Structure calculation
Initial structures of the four isolated segments (St-J, St-K, St-St, and ΔJΔK) were calculated as described previously by using manually assigned restraints in CYANA. Standard torsion angle restraints were used for regions of A-helical geometry, allowing for ± 50° deviations from ideality. Standard hydrogen bonding restraints and cross-helix phosphate-phosphate restraints were employed for A-helical regions.
The CYANA-minimized structures of each isolated domain with lowest target function were created as angular files and merged to create a structure of the full-length J-K region. The structure was first minimized by using CYANA to create a coordinate file, and then subjected to a refinement in xplor-nih with SAXS restraint and NMR restraints used in CYANA except for the phosphate-phosphate distances39. Molecular images were generated with PyMOL (http://www.pymol.org).
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM047467 (to G.W.) and R01 AI51340 (to C.H.). This work was also supported by Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowships for Research Abroad, Daiichi-Sankyo Foundation of Life Science, and the Naito Foundation (to S.I.). We would also like to thank the Department of Energy (DOE) Integrated Diffraction Analysis (IDAT) grant contract number DE-AC02-05CH11231 for the SAXS data acquisition. The authors wish to thank Dr. M. Durney for many helpful discussions.
Footnotes
Accession codes
Atomic coordinates of the J-K region, SL-J, SL-K, SL-St, and ΔJΔK have been deposited in the Protein Data Bank under accession codes 2NBX, 2NBY, 2NBZ, 2NC0, and 2NC1, respectively. Chemical shifts of the J-K region, SL-J, SL-K, SL-St, and ΔJΔK have been deposited in the Biological Magnetic Resonance Data Bank under accession codes 25996, 25997, 25998, 25999, and 26000, respectively.
Author contributions
S.I., C.U.T.H., V.M.D’S, and G.W. conceived and designed the experiments. S.I. purified the samples and performed the NMR, ITC, and SAXS experiments. S.I. and V.M.D’S did the structural analyses. P.K. conducted the toeprinting assay. S.I., C.U.T.H., V.M.D’S and G.W. interpreted the data and wrote the manuscript.
References
- 1.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gebauer F, Hentze MW. Genetics. IRES unplugged. Science. 2016;351:228. doi: 10.1126/science.aad8540. [DOI] [PubMed] [Google Scholar]
- 3.Weingarten-Gabbay S, et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351 doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 4.Xue S, et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashem Y, et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkov YA, Palmenberg AC. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques. 2006;41:283–284. 286. doi: 10.2144/000112243. 288 passim. [DOI] [PubMed] [Google Scholar]
- 9.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci U S A. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spriggs KA, et al. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol Cell Biol. 2009;29:1565–1574. doi: 10.1128/MCB.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight-Jones TJ, Rushton J. The economic impacts of foot and mouth disease - what are they, how big are they and where do they occur? Prev Vet Med. 2013;112:161–173. doi: 10.1016/j.prevetmed.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dibrov SM, et al. Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J Med Chem. 2014;57:1694–1707. doi: 10.1021/jm401312n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duke GM, Hoffman MA, Palmenberg AC. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 18.Lozano G, Martinez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol. 2015;12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Bassili G, et al. Sequence and secondary structure requirements in a highly conserved element for foot-and-mouth disease virus internal ribosome entry site activity and eIF4G binding. J Gen Virol. 2004;85:2555–2565. doi: 10.1099/vir.0.80106-0. [DOI] [PubMed] [Google Scholar]
- 20.Clark AT, Robertson ME, Conn GL, Belsham GJ. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J Virol. 2003;77:12441–12449. doi: 10.1128/JVI.77.23.12441-12449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman MA, Palmenberg AC. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Correll CC, Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiore JL, Nesbitt DJ. An RNA folding motif: GNRA tetraloop-receptor interactions. Q Rev Biophys. 2013;46:223–264. doi: 10.1017/S0033583513000048. [DOI] [PubMed] [Google Scholar]
- 26.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes FE, Schwartz CR, Tainer JA, Rambo RP. Methods for using new conceptual tools and parameters to assess RNA structure by small-angle X-ray scattering. Methods Enzymol. 2014;549:235–263. doi: 10.1016/B978-0-12-801122-5.00011-8. [DOI] [PubMed] [Google Scholar]
- 28.Kaminski A, Jackson RJ. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theimer CA, Wang Y, Hoffman DW, Krisch HM, Giedroc DP. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J Mol Biol. 1998;279:545–564. doi: 10.1006/jmbi.1998.1812. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcotrigiano J, et al. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schutz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akabayov SR, Wagner G. Backbone resonance assignment of the HEAT1-domain of the human eukaryotic translation initiation factor 4GI. Biomol NMR Assign. 2014;8:89–91. doi: 10.1007/s12104-013-9459-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J Biomol NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwieters CD, Clore GM. Using small angle solution scattering data in Xplor-NIH structure calculations. Prog Nucl Magn Reson Spectrosc. 2014;80:1–11. doi: 10.1016/j.pnmrs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.