Abstract
microRNAs (miRNAs) play critical roles in various biological processes including cell proliferation, development, and host defense. However, the molecular mechanism for miRNAs in regulating bacterial-induced inflammation remains largely unclear. Here we report that miR-301b augments pro-inflammatory response during pulmonary infection and caffeine (CAF) suppresses miR-301b’s effect and thereby augmenting respiratory immunity. LPS treatment or Pseudomonas aeruginosa infection induces miR-301b expression via a TLR4/MyD88/NF-κB pathway. Importantly, CAF decreases miR-301b expression through negative regulation of the cAMP/PKA/NF-κB axis. Further, c-Myb is identified as a target of miR-301b, which positively modulates anti-inflammatory cytokines IL-4 and TGF-β1, but negatively regulates pro-inflammatory cytokines MIP-1α and IL-17A. Moreover, repression of miR-301b results in increased transcription of c-Myb and elevated levels of neutrophil infiltration, thereby alleviating infectiou symptoms in mice. These findings reveal miR-301b as a new controller of inflammatory response by repressing c-Myb function to inhibit anti-inflammatory response to bacterial infection, representing a novel mechanism for balancing inflammation.
Keywords: caffeine, inflammation, microRNA, c-Myb, bacterial infection
Introduction
microRNAs (miRNAs) are a class of small non-coding RNAs and regulate gene expression at the post-transcriptional levels. Literature suggests that a single miRNA can play multiple roles in influencing divergent pathways by binding to different target mRNAs at 3′-untranslated regions (UTR)1. miRNAs have been implicated earlier in many cellular processes, including but not limited to development, cellular proliferation, apoptosis, and necrosis 2,3. More recent, findings indicate that miRNAs may modulate immune responses at different stages, including production of cytokines/chemokines, expression of adhesion and costimulatory molecules, and release of exosomes 4–7. Several TLRs/NF-κB-responsive miRNAs are shown to be abundantly expressed, which may be tailored to harness the TLRs/NF-κB pathway by interfering with their target gene expression 5,6,8. miRNAs may also provide feedback regulation of NF-κB signaling to maintain homeostasis 8. Importantly, a number of miRNAs have been implicated in downregulating inflammatory responses, which help control overzealous inflammation during host defense against pathogens and other processes both in vitro and in vivo 9,10.
As a metabolic stimulant, caffeine (CAF) enhances performance by reducing physical fatigue and drowsiness. CAF exerts either positive or negative effects on neurons or other cells beneficial for patients with Parkinson’s disease and cancer 11,12. CAF has also been tested for treatment of bronchopulmonary dysplasia in premature infants 13. However, excessive ingestion of CAF may lead to psychiatric diseases including mild anxiety, jitteriness, and insomnia 14. Previous studies revealed that CAF activates innate immune response in tumor models 15 or suppresses TNF-α via cAMP/PKA pathway in vitro 16. CAF could also negatively affect carcinogenesis through regulation of DNA damage and repair 17. More recently, CAF has been linked to regulation of miRNAs in cardiovascular diseases 18,19, but has not been linked to lung diseases. Particularly, CAF has not been demonstrated to participate in respiratory inflammatory response against microorganisms.
We aimed to explore roles of CAF in host defense and in particular defined a regulatory mechanism exerted by a novel miRNA, namely miR-301b in pulmonary infection. Using miScript PCR System Screening, we analyzed the expression of a set of miRNAs in alveolar macrophages (AMs) upon CAF treatment 20. miR-301b, miR-301a, and miR-15a are significantly suppressed, while only miR-301b is determined to be responsive and play roles in regulating inflammation during bacterial infection. We observed less neutrophil infiltration upon Pa infection at the early time in lungs of miR-301b mimics transfected mice. Neutrophils are normally associated with heightened inflammatory response and exacerbated lung injury in acute infection models 21–23. We then dissected the molecular mechanism by which miR-301b impacts neutrophil infiltration and inflammatory response. Our results indicate that CAF negatively regulated inflammatory response in bacterial infection by inhibiting miR-301b and augmenting expression of the miR-301b target c-Myb.
Results
miR-301b expression is altered by CAF both in vitro and in vivo
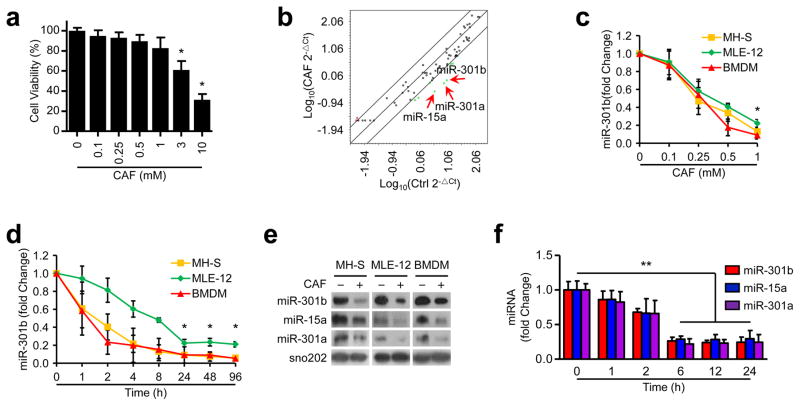
CAF is implicated in infection and inflammation. To search for miRNAs that may be regulated by CAF, we used MH-S cells to perform array based miRNA profiling. Since CAF treatment at higher concentrations (>3 mM) for 24 h affected cell viability (Fig. 1a), a safer concentration (1 mM) was used in the following experiments. CAF suppresses the expression of several miRNAs, among which miR-301b, 301a, and 15a are the most significantly inhibited (>10 fold change) (Fig. 1b, Supplementary Table 1). The expression of miR-301b, 301a, and 15a in MH-S, MLE-12, primary murine bone marrow-derived macrophages (BMDM) was similarly suppressed by CAF in a dose- and time-dependent manner as determined by qRT-PCR (Fig. 1c, d, Supplementary Fig. 1a, b, and Table 2). The levels of miRNA expression upon CAF treatment were then vigorously validated by Northern blotting analysis (Fig. 1e). To elucidate the physiological relevance of underlying miRNA alterations, we used a mouse model to evaluate the expression of miR-301b, 301a, and 15a in the lung from mice treated with CAF by qRT-PCR, which was also decreased after CAF treatment (Fig. 1f). These findings indicate the regulatory role of CAF in miR-301b, 301a, and 15a expression in respiratory systems.
Figure 1. CAF decreases miR-301b, 301a, and 15a expression.
(a) MH-S cell viability was examined by MTT assay after treatment with CAF at different concentrations for 24 h. (b) Expression of miRNAs was measured using miScript PCR assay in MH-S cells with or without CAF treatment (1 mM, 24 h). The lines indicated genes with >4 fold change among subsets. (c) miR-301b expression in MH-S, MLE-12, and BMDM cells was determined by qRT-PCR 24 h post CAF treatment. (d) qRT-PCR validation of miR-301b expression in cells treated with CAF (1 mM). (e) Northern blot showing the expression of miR-301b, 15a, and 301a in cells treated with LPS (100 ng/ml, 6 h). The above data are representative of 3 independent experiments. Panel a, c, d, means±SD from triplicates. (f) miRNA expression in mouse lungs after CAF treatment (50 mg/kg, i.v., n=3, means±SD). *, p<0.05, **, p<0.01. One-way ANOVA with Tukey’s post hoc.
CAF inhibits miR-301b through cAMP/PKA/NF-κB signaling
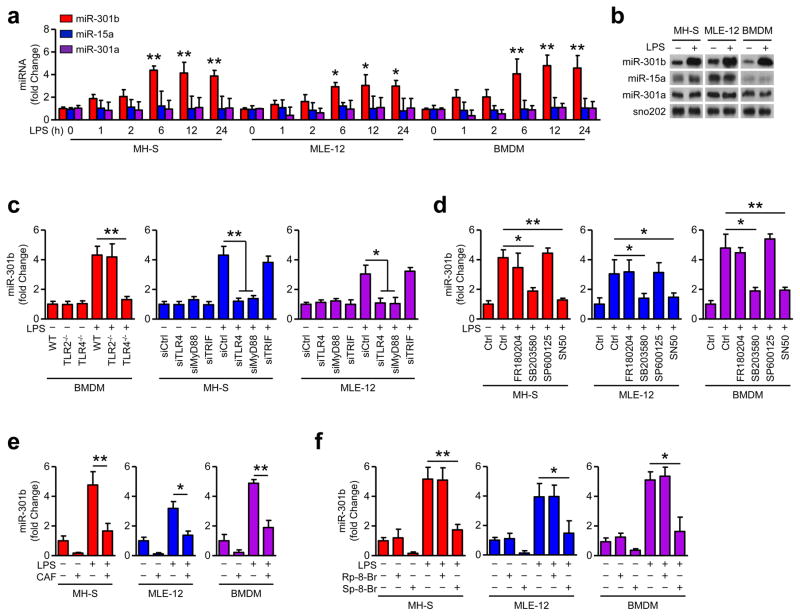
miR-301b was increased during LPS treatment in a time-dependent manner, while miR-301a and miR-15a were not (Fig. 2a). Northern blotting was performed and validated the induction of miR-301b by LPS priming (Fig. 2b). We determined whether toll-like receptors (TLRs) play a role in miR-301b expression. LPS-induced miR-301b was markedly inhibited by depleting TLR4 in BMDM, but was not affected by TLR2 depletion (Fig. 2c, Supplementary Fig. 2a). After MyD88 knock-down, LPS-induced miR-301b expression was reduced but was not affected by TRIF knock-down, indicating that the induction of miR-301b is likely MyD88-dependent (Fig 2c, Supplementary Fig. 2a). To identify the relevant pathway that regulates miR-301b expression, we pretreated MH-S, MLE-12, and BMDM cells with a set of inflammatory regulatory inhibitors: FR180204 (ERK inhibitor), SB203580 (p38 inhibitor), SP600125 (JNK inhibitor) and SN50 (NF-κB inhibitor), respectively 20. We found that inhibition of p38 and NF-κB suppressed LPS-induced miR-301b expression (Fig. 2d). These findings indicate that miR-301b expression may be regulated by a TLR4/p38/NF-κB axis.
Figure 2. CAF inhibits LPS-induced miR-301b expression by regulating cAMP/PKA/CREM/NF-κB axis.
(a) LPS (100 ng/ml) induces miR-301b expression in MH-S, MLE-12, and BMDM cells in a time dependent manner as determined by qRT-PCR. (b) Northern blot showing the expression of miR-301b, 15a, and 301a in cells treated with LPS (100 ng/ml, 6 h). (c) qRT-PCR of LPS-induced miR-301b expression in BMDM from TLR4−/− and TLR-2−/− mice, or in MH-S and MLE-12 cells with TLR4, MyD88 or TRIF knockdown. (d) Cells were cultured for 6 h with LPS in combination with FR180204 (0.3 μM), SB203580 (0.5 μM), SP600125 (90 nM) and SN50 (10 μM). miR-301b was analyzed by qRT-PCR. (e) After CAF pretreatment for 24 h, LPS (100 ng/ml) was added for another 6 h incubation and miR-301b was detected by qRT-PCR. (f) Cells were treated with LPS in combination with Sp-8-Br (2.5 μM) and Rp-8-Br (4 μM) for 6 h and miR-301b was analyzed by qRT-PCR. Panel a, c–f, means+SD from triplicate; b, representative of 3 independent experiments. *, p<0.05, **, p<0.01. One-way ANOVA with Tukey’s post hoc.
To pinpoint the mechanistic feature of this signaling axis, we used cell culture models and surprisingly observed that CAF inhibited LPS-induced miR-301b expression in vitro (Fig. 2e). Pre-incubation with CAF for 24 h led to the strongest inhibition in miR-301b in MLE-12, MH-S, and BMDM cells (Supplementary Fig. 2b). CAF was previously reported to indirectly preserve cAMP by inhibiting phosphodiesterase, an enzyme that degrades cAMP and thus activating downstream PKA 24. Next, cells were pretreated with Sp-8-Br (activator of cAMP-dependent PKA) and Rp-8-Br (PKA inhibitor), respectively 25. Sp-8-Br was found to inhibit LPS-induced miR-301b expression (Fig. 2f). Phosphorylation of PKA and CREB-1 was increased upon CAF treatment, while no further increase occurred upon LPS treatment (Supplementary Fig. 2c). A previous report revealed that PKA activating agents inhibited the NF-κB-dependent reporter gene expression 26. While LPS increased phosphorylation of p65 (p-p65), CAF decreased p-p65 as assessed by immunoblotting (Supplementary Fig. 2c). We found that CREB-1 was translocated into the nucleus upon CAF treatment, suggesting distinct roles of CREB and NF-κB as downstream transcription factors of PKA (Supplementary Fig. 2d). Supplementary Fig. 2e, f shows that p65 knockdown inhibited miR-301b expression, while knockdown of PKA or CREB-1 had no effects on miR-301b expression after LPS challenge. Additionally, PKA knockdown was found to restore the suppressed miR-301b expression after CAF treatment (Supplementary Fig. 2g). Together, our data suggest that CAF regulates miR-301b expression through a cAMP signaling axis.
miR-301b modulates bacterium-induced inflammatory responses in vivo
Both mRNA and protein levels of IL-4, IL-6, TNF-α, and TGF-β1 were found to be increased upon LPS treatment determined by qRT-PCR and ELISA (Supplementary Fig. 3a, b). To assess the regulatory mechanism in inflammation regulation, chemically synthesized mimics (301b-m) or inhibitors (301b-i) were transfected into cells. Having verified transfection efficiencies, we studied the effects of these mimics or inhibitors on inflammatory cytokines compared to negative controls (NS-m or NS-i) (Supplementary Fig. 4a). Anti-inflammatory cytokines (IL-4 and TGF-β1) induced by LPS treatment were inhibited by 301b-m but increased by 301b-i as compared to negative controls as determined by qRT-PCR or ELISA (Supplementary Fig. 3a, b). However, the pro-inflammatory cytokines (IL-6 and TNF-α) were not changed significantly by manipulating miR-301b (Supplementary Fig. 3a, b). Additionally, the protein levels of IL-4 and TGF-β1 in MLE-12 cells were also lower in cells overexpressing 301b-m than those in control cells, whereas 301b-i increased these anti-inflammatory cytokines as assessed by western blotting (Supplementary Fig. 4b). We next found that the conditioned medium of MLE-12 cells (treated with miR-301-m) decreased the migration capabilities of MH-S cells by approximately 30% using Boyden chamber assay (Supplementary Fig. 4c). 301b-m transfection did not affect viability of MH-S cells (Supplementary Fig. 4d), but decreased their phagocytosis ability (Supplementary Fig. 4e, f). Collectively, these findings suggest that miR-301b may selectively down-regulate the expression of anti-inflammatory genes as well as phagocytic activities.
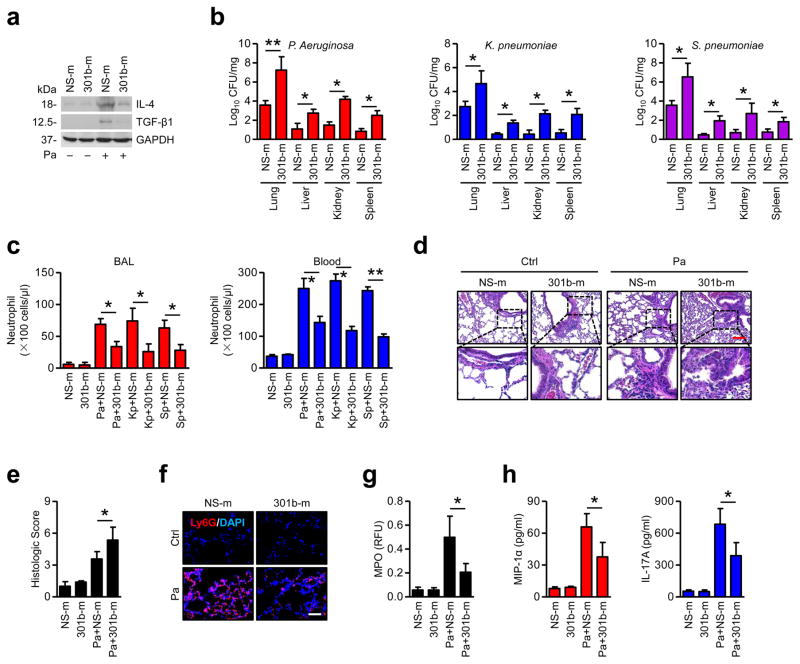
To determine whether the in vitro phenomenon similarly occurs in vivo and can be expanded to other bacteria, we established acute respiratory infections in cells and mice with different bacteria, P. aeruginosa (Pa), K. pneumoniae (Kp) and S. pneumoniae (Sp). These bacterial infections all induced miR-301b expression both in vitro and in vivo, but did not induce miR-15a and miR-301a (Supplementary Fig. 5a). To further study the physiological impact of miR-301b, systemic administration of miR-301b was performed to determine bacterium-induced gene expression in vivo. Likewise, expression of miR-301b in the lungs was consistent with in vitro data (Supplementary Fig. 5b). 24 h after bacterial infection, IL-4 and TGF-β1 in BAL were decreased by 301b-m, but IL-6 and TNF-α were not affected (Supplementary Fig. 5c). The lungs of mice infected with Pa were immunoblotted to detect IL-4 and TGF-β1, which showed similar results to ELISA data (Fig. 3a). Interestingly, 301b-m injection resulted in higher bacterial burdens than in the NS-m group, while effects of 301b-i were opposite to those of the mimics, which confirmed the physiological relevance in pulmonary infection (Fig. 3b, Supplementary Fig. 5d). To dissect whether miR-301b contributes to inflammatory cell infiltration, we performed hematoxylin and eosin (H&E) staining and found that 301b-m overexpression markedly inhibited neutrophil recruitment upon bacterial infection (Fig. 3c). Further, more severe lung injury was observed in 301b-m-treated mice, whereas less lung injury was observed in 301b-i-treated mice after Pa infection (Fig. 3d, e and Supplementary Fig. 5e). 301b-m was found to inhibit neutrophil recruitment upon Pa infection (Fig. 3e). In addition, myeloperoxidase (MPO) assay demonstrated that neutrophil infiltration was decreased in 301b-m transfected mice (Fig. 3g). Neutrophil mobilization and recruitment to the lung is usually associated with release of chemokines/cytokines to BAL. We next performed ELISA to detect MIP-1α and IL-17A, which showed similar reduction (Fig. 3h). These findings are consistent with in vitro data about phagocyte migration, suggesting that phagocyte recruitment and phagocytosis may be impacted by enhanced miR-301b levels. Thus the inhibitory effects of miR-301b on host defense may be due to effects on neutrophil recruitment to the lung.
Figure 3. Enforced expression of miR-301b suppresses bacterium-induced anti-inflammatory responses in vivo.
(a) Mice (n=3) were i.v. injected with NS-m or 301b-m (50 μg/mouse, 24 h), then infected with Pa (1×107 CFU), Kp (1×105 CFU) or Sp (5×106 CFU) for 24 h. Lungs from mice infected with Pa as above were collected and homogenized for immunoblotting for IL-4 and TGF-β1. (b) Organs from mice were collected and homogenized for CFU assay. (c) Neutrophils in BAL and blood were counted by H&E staining. (d, e) Lungs were embedded in formalin for H&E staining and lung injuries were double-blind scored. Scale bar=100 μm. Insets showing tissue injury and inflammatory influx. (f) Lungs were embedded in formalin for immunostaining with a Ly6G antibody to show neutrophil infiltration. Scale bar=50 μm. (g, h) After treatment as above, MPO and ELISA assays were performed to test myeloperoxidase activities in lung homogenates or MIP-1α and IL-17A levels in BAL, respectively. Panel a, d, e, f, representative from triplicate samples; b, c, g, h, data are from 3 mice/group, means+SD. *, p<0.05, **, p<0.01. One-way ANOVA with Tukey’s post hoc.
c-Myb is a target of miR-301b
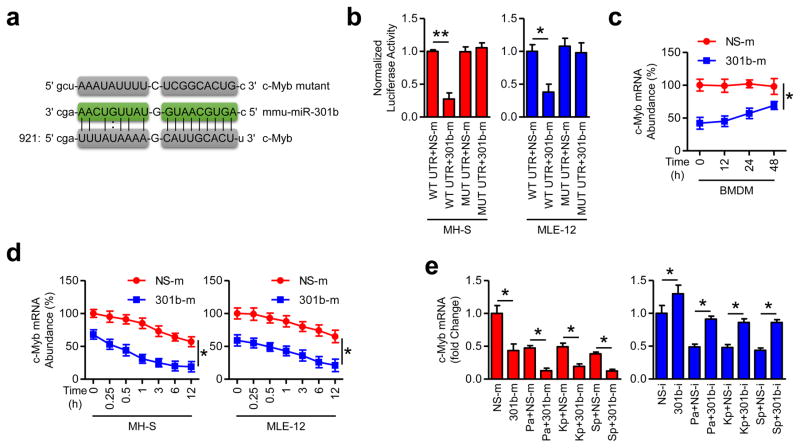
c-Myb possesses a conserved miR-301b seed sequence in its 3′UTR with a high mirSVR score (Fig. 4a), and was demonstrated to be a critical player in immune responses 27. We experimentally confirmed the prediction and showed that 301b-m inhibited the activity of a luciferase reporter containing c-Myb 3′UTR, while the activity of luciferase reporters containing the mutant 3′UTR of c-Myb was not significantly altered (Fig. 4b, Supplementary Table 3). Further, c-Myb mRNA abundancy in BMDM from mice transfected with 301b-m was lower than that in NS-m group (Fig. 4c). Similarly, 301b-m decreased c-Myb mRNA levels (Fig. 4d). Moreover, in vivo data showed that bacterial infection resulted in decreased c-Myb abundance, which was further decreased by 301b-m, but this decrease could be reversed by 301b-i (Fig. 4e). These data collectively identify c-Myb as a bona fide target of miR-301b.
Figure 4. c-Myb is a functional target of miR-301b.
(a) c-Myb 3′UTR contains one predicted miR-301b binding domain with predicted duplex formations between c-Myb 3′UTR (bottom) and miR-301b (middle). The site of target mutagenesis (top) is also indicated. (b) Luciferase reporter assay evaluating c-Mab targeting with the WT or mutated 3′UTR reporter constructs (WT UTR or MUT UTR) of c-Myb in MH-S and MLE-12 cells that were then transfected with NS-m or 301b-m, respectively. (c) Mice (n=3) were i.v. injected with NS-m or 301b-m (50 μg/mouse, 24 h). BMDM were collected for c-Myb mRNA abundance determination at indicated times by qRT-PCR. (d) MH-S and MLE-12 cells were transfected with NS-m and 301-m for 24 h, and c-Myb mRNA was detected at different time points by qRT-PCR. (e) Mice (n=3) were i.v. injected with NS-m, 301b-m, NS-i or 301b-i (50 μg/mouse, 24 h), infected with Pa (1×107 CFU), Kp (1×105 CFU) or Sp (5×106 CFU) for 24 h, and c-Myb mRNA abundance determined by qRT-PCR. Panel b, d, means±SD from triplicate; c, e, Means±SD from 3 mice. *, p<0.05, **, p<0.01. One-way ANOVA with Tukey’s post hoc.
c-Myb negatively modulates infection-induced anti-inflammatory responses
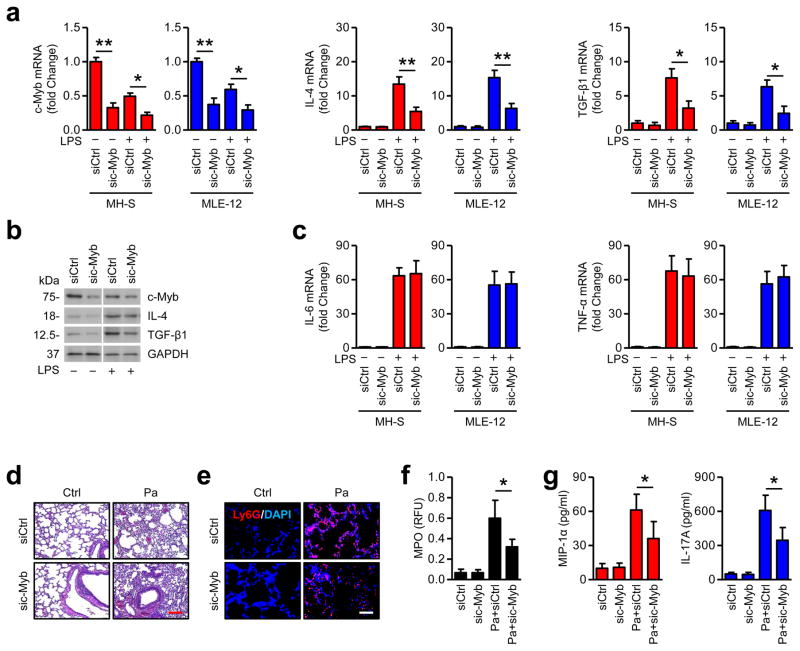
To determine whether c-Myb plays a role in infection-induced inflammatory responses, we knocked down c-Myb in MH-S and MLE-12 cells (Fig. 5a). Interestingly, c-Myb mRNA was decreased upon LPS stimulation, which is negatively correlated with miR-301b (Fig. 5a). Anti-inflammatory cytokines (IL-4 and TGF-β1) were partially reduced (50%~60%) by c-Myb knockdown (Fig. 5a, b). However, pro-inflammatory cytokines (IL-6 and TNF-α) did not change after c-Myb knocked down (Fig. 5c). Likewise, anti-inflammatory cytokines were inhibited in BAL fluids of c-Myb knockdown mice after bacterial (Pa, Kp or Sp) infection (Supplementary Fig. 6a). However, IL-6 or TNF-α release was not altered by c-Myb knockdown (Supplementary Fig. 6a). Further, bacterial burdens were higher in c-Myb knockdown mice locally and systemically than in control mice (Supplementary Fig. 6b). Moreover, histological analysis showed that Pa infection caused more severe tissue injury and less neutrophil infiltration in c-Myb knockdown mice (Fig. 5d, e). MPO assays and ELISA also showed decreased myeloperoxidase activity and anti-inflammatory response in c-Myb knockdown mice, which confirmed the above observations (Fig. 5f, g). Overall results imply that c-Myb is a functional target of miR-301b in bacterial-induced inflammatory responses.
Figure 5. c-Myb plays a role in mediating infection-induced inflammatory responses.
(a) MH-S and MLE-12 cells were transfected with Ctrl siRNA or c-Myb siRNA for 24 h and were treated with LPS (100 ng/ml) for 6 h. mRNA of c-Myb, IL-4, and TGF-β1 was measured by qRT-PCR. (b) c-Myb, IL-4, and TGF-β1 protein was assessed in MLE-12 cells from above by immunoblotting. (c) mRNA levels of IL-6 and TNF-α were determined in the MH-S and MLE-12 cells by qRT-PCR. (d) Mice (n=3) were i.v. injected with Ctrl siRNA or c-Myb siRNA (50 μg/mouse, 24 h) and were infected with Pa (1×107 CFU) for 24 h. Lungs were evaluated by H&E staining. Scale bar=100 μm. (e) Lungs were evaluated by immunostaining with a Ly6G antibody to show neutrophil infiltration. Scale bar=50 μm. (f, g) After treatment as above, MPO assay and ELISA were performed to determine myeloperoxidase activities in lung homogenates or MIP-1α and IL-17A levels in BAL. Panel a, c, means+SD from triplicates; b, representative from triplicate samples; d, e, representative of the 3 mice; f, g, means+SD from 3 mice. *, p<0.05, **, p<0.01. One-way ANOVA with Tukey’s post hoc.
CAF decreases the susceptibility to bacterial infection in vivo
To define the direct role of CAF in bacterial invasion, we set out to elucidate how CAF administration improves cytokine production. We observed that CAF significantly decreased miR-301b levels and partially increased c-Myb mRNA levels within several hours either before or after infection (Supplementary Fig. 7a). Interestingly, 6 h CAF pretreatment followed by bacterial infection for 24 h led to the strongest miR-301b inhibition and highest c-Myb mRNA level in the tested times (Supplementary Fig. 7a). To verify whether CAF has a broad role in inflammatory responses, we used primary human alveolar macrophages to recapitulate the above results. CAF was found to prevent LPS-induced p65 nuclear translocation, while the CREB-1 nuclear translocation was only changed following CAF treatment (Supplementary Fig. 8a). miRNA database shows that hsa-miR-301b has two high scored binding sites with human MYB mRNA (Supplementary Fig. 8b). We found that both hsa-miR-301b and hsa-miR-301a were decreased by CAF, while only hsa-miR-301b was increased by LPS stimulation (Supplementary Fig. 8c). Similar results were obtained in primary human AMs as seen in murine cells, showing that CAF increased MYB expression and enhanced LPS-induced IL-4 production (Supplementary Fig. 8d).
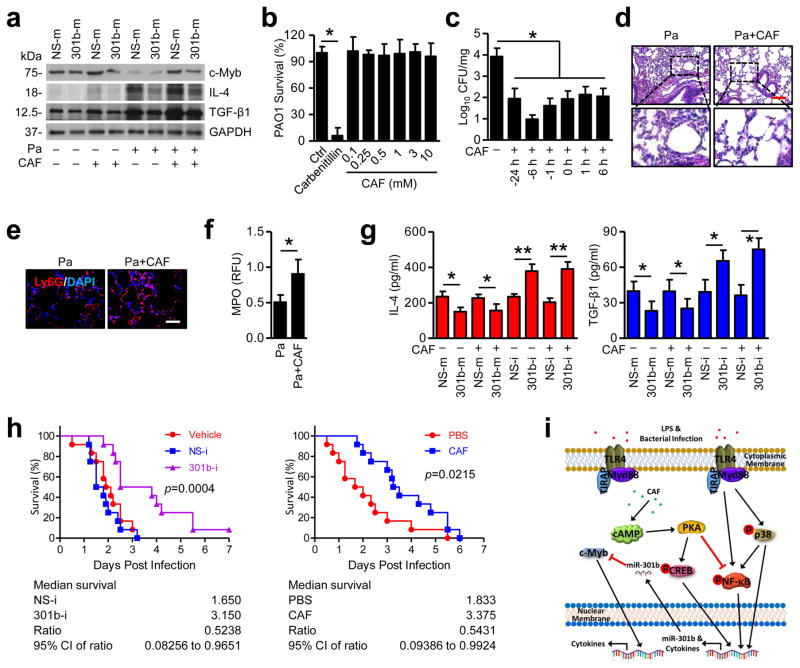
Next, we focused on Pa infection to determine whether CAF has effects on inflammatory responses in physiological context. IL-4 and TGF-β1 secretion in BAL fluids was found to be negatively correlated with miR-301b, while positively correlated with c-Myb when mice were administrated with CAF at different times; and IL-10 and TNF-α were not altered (Supplementary Fig. 9a). In addition, 24 h post Pa infection, the lungs (6 h pretreated with CAF group) showed similar results to ELISA data (Fig. 6a). We next performed in vitro bacterial growth assays, demonstrating that CAF did not have obvious anti-bacterial effects (Fig. 6b). However, CAF was able to inhibit bacterial growth in vivo, particularly with 6 h CAF pretreatment (Fig. 6c). Furthermore, CAF significantly alleviated lung injury and increased neutrophil infiltration (Fig. 6d, e, and Supplementary Fig. 10a, b). MPO assay showed similar increase in neutrophil MPO activity (Fig. 6f). CAF was also found to increase MIP-1α and IL-17A levels in mouse BAL, while CAF alone has no function in inflammatory response without infection (Supplementary Fig. 10c, d). We also noted no significant tissue injury in the liver and kidney (Supplementary Fig. 10e). However, infection-induced lower c-Myb expression in all these tissues, whereas CAF counteracted this damaging effect by increasing c-Myb expression (Supplementary Fig. 10f). Lower CAF treatment did not induce significant neutrophil infiltration in BAL (Supplementary Fig. 10g) and had limited effects on infection mitigation reflected by bacterial burdens (Supplementary Fig. 10h). To determine a potential causal relationship, we established a gain-of-function setting using miR-301b mimics. After CAF treatment and subsequent infection, we found that CAF has no impact on levels of IL-4 and TGF-β1 in BAL (Fig. 6g). These data suggest that CAF exerts its inflammatory regulation by impacting levels of miR-301b. 301b-i significantly attenuated mouse mortality after Pa infection (Fig. 6h). Systemic delivery of CAF decreased mortality following bacterial infection (Fig. 6h). To determine whether manipulation of CAF and miR-301b affects the adaptive immunity, we stained an adaptive immunity marker (CD4+ T cell) in lung sections. CAF or miR-301b itself could not induce CD4+ T cell recruitment in lungs without infection (Supplementary Fig. 11a). However, Pa infection in both early and late phases increased CD4+ T cell recruitment to the lungs, which was further increased by CAF but decreased by 301b-m (Supplementary Fig. 11b). Further, IL-2 in BAL showed similar patterns of CD4+ T cell recruitment (Supplementary Fig. 11c). Further, we evaluated neutrophils in BAL and found that CAF played a role in the recruitment of neutrophils upon bacterial infection, which may result in stronger bacterial clearance at early times. However, miR-301b plays an opposing role in this infection process (Supplementary Fig. 11d, e), suggesting that neutrophils at early phases may benefit host defense while late phases may be detrimental. Nevertheless, our data indicate that the adaptive immunity particularly the Th2 lineage may affect later processes of infection.
Figure 6. CAF and 301b-i decrease susceptibility and mortality after pulmonary bacterial infection.
(a) Mice (n=3) were infected with Pa (1×107 CFU) for 24 h. CAF (50 mg/kg, i.v.) was injected at indicated time (pre-treatment strategy or post-infection treatment). Lungs were immunoblotted to detect c-Myb, IL-4, and TGF-β1 protein. (b) Same amounts of PAO1 were cultured in LB liquid medium with Carbenicillin (50 μg/ml) or different concentrations of CAF overnight. Same amounts of culture mediums were plated for CFU counting. (c) CFU was also performed using lung homogenates from mice infected with PAO1 as above. (d) Lungs from mice infected with Pa (24 h) and CAF (6 h pretreatment) were evaluated by H&E staining. Scale bar=100 μm. (e) Lungs were evaluated by immunostaining with a Ly6G antibody to show neutrophil infiltration. Scale bar=50 μm. (f) After treatment as above, MPO assays were performed to test myeloperoxidase activities in lung homogenates. (g) Mice were pretreated with miR-301b mimics or inhibitors. 24 h later, mice were infected with Pa combined with CAF treatment as above. Inflammatory cytokines in BAL were assayed using ELISA. (h) Kaplan-Meier survival curves (n=12) of Pa-infected mice. Mice were i.v. injected with vehicle, NS-i or 301b-i (50 μg/mouse, 24 h), PBS or CAF (6 h pretreatment), the mice were challenged with Pa (1×107 CFU). Survival was determined up to 7 days. Log-rank test. (i) The schematic diagram showing how miR-301b is regulated and how it affects immune responses against bacterial infection. Panel a, d, e, representative of 3 mice; b, means+SD from triplicate; c, f, g, means+SD from 3 mice. *, p<0.05. One-way ANOVA with Tukey’s post hoc.
The overall findings reveal a central role of CAF and miR-301b in bacterial-induced immune response. Modulation of miR-301b may be a novel strategy to regulate the c-Myb pathway by altering inflammation to reduce lung and other organ damage, and thus may ultimately be beneficial for patients with acute infection (Fig. 6i). We found 10 other miRNAs (e.g., miR-301a) with the potential to bind to the same or surrounding sites in c-Myb 3′UTR, which are very similar to miR-301b (Supplementary Fig. 12a). We used miR-301a, miR-302b 20 and miR-15a as controls to establish the specific role of miR-301b. Although both 301a-m and 302b-m showed the potential to target c-Myb 3′UTR, only 301a-m played a similar role in LPS-induced IL-4 release in MLE-12 cells (Supplementary Fig. 12b, c). Overall results confirmed that miR-301b serves as a novel regulator of inflammatory response upon bacterial infection.
Discussion
We reveal a critical role of miR-301b in affecting inflammatory response against infection, which is negatively regulated by CAF. Utilizing both in vitro and in vivo infection models, we show that CAF down-regulates miR-301b and thereby increasing neutrophil infiltration and anti-inflammatory cytokines (IL-4 and TGF-β1). Neutrophils are normally increased along with increased lung injury 22; however, early PMN may be beneficial for bacterial clearance but late phases may not as excessive PMN accumulation can result in life-threatening tissue injury 21,23. miR-301b may inhibit the initial recruitment of neutrophils for immune response against bacterial infection. We also found that miR-301b, by targeting c-Myb, inhibits anti-inflammatory cytokines, leading to excessive inflammation and impaired host defense. In contrast, CAF counteracts miR-301b’s effects on inflammatory response, repressing overzealous pro-inflammatory response in order to maintain homeostasis and prevent potential tissue damage and septic shock.
Mechanically, CAF inhibits 3, 5-cyclic nucleotide phosphodiesterase (cAMP-PDE), an enzyme that converts cAMP into non-cyclic AMP. Thus cAMP diffuses through the cell and acts as a “secondary messenger” to activate PKA, which in turn suppresses the activity of NF-κB. NF-κB has been reported to play fundamental roles in initiating expression of many miRNAs, including miR-146, let-7, miR-21, miR-302b, etc. 5–7,20. We found that increased cAMP down-regulates NF-κB but does not affect TNF-α upon infection. This might be because TNF-α release is predominantly related to MAPK signaling upon bacterial infection 28,29. This pro-inflammatory response is so strong that it is difficult to differentiate CAF effects from the drastic response to bacterial infection. We demonstrated that the expression of miR-301b after bacterial infection is dependent on the transcriptional activity of NF-κB. Chemical inhibitors of NF-κB or siRNA interference of NF-κB component (p65) markedly decreased infection-induced miR-301b. Previous reports demonstrated that miR-301b promotes cell invasiveness by targeting TP63 in pancreatic carcinoma cells 30. However, no study has linked miR-301b to regulation of inflammatory responses against infection. Therefore our study has extended the biological significance and function of miR-301b in control of inflammation.
Prior work has characterized miR-155, miR-146a, and 132 as critical regulators in innate immunity and autoimmune inflammation 6,8,31–34. We have recently identified miR-302b as a regulator for anti-Pseudomonal inflammatory response by targeting IRAK4 20. Thus far, miR-301b’s biological function is largely unknown, with only a reported role in colon cancer 35,36. Thus, our study has dramatically extended the function of miR-301b by adding it to the growing list of miRNAs for regulating inflammation.
The transcription factor c-Myb has been recognized as a proto-oncogene during cellular proliferation 37. It has also been shown to control multiple steps of lymphocyte development and fetal hepatic hematopoiesis 4,38. Recently, c-Myb was reported to modulate miR-143 and play central and yet complex roles in DNA damage, DNA repair, cancer cell migration and invasion 39. However, no study has linked c-Myb to modulation of anti-inflammatory cytokines. Anti-inflammatory cytokines are a series of immune regulatory molecules, which work to balance inflammation during severe bacterial infection, preventing excessive pro-inflammatory responses that can cause severe SIRS and multiple organ damage syndrome (MODS) 40.
Our study demonstrated that miR-301b binds the 3′UTR of c-Myb, and that c-Myb in turn regulates down-stream inflammatory responses. Several anti-inflammatory cytokines, such as IL-4 and TGF-β1, were negatively regulated by miR-301b mimics upon LPS or bacterial infection both in vitro and in vivo, which requires the molecular interaction of miR-301b with c-Myb mRNA. Furthermore, CAF prevented mice from suffering severe organ injury by repressing miR-301b and thereby increasing levels of anti-inflammatory cytokines in both LPS and infection models.
The exact mechanism of action of CAF on immune cells has been debated for some time 17. Some studies suggest that CAF inhibits cyclic AMP (cAMP)-phosphodiesterase, resulting in increased cAMP levels and activation of protein kinase A (PKA) 16. The cAMP-PKA signaling axis plays multiple roles in regulating complex cellular networks, including blood development, Th2 immunity, and inflammation 24,41. Since it seems that CAF affects Th2 lineage, CAF and miR-301b manipulation may influence the adaptive immunity. The adaptive immunity needs longer time to develop and is orchestrated by different mechanisms from the innate immunity. Nevertheless, our data suggest that Th2 lineage may affect later processes, when the adaptive immunity is activated. Actually, CAF and miR-301b pre-treatment could be seen as an earlier process of the adaptive immunity, which then affects inflammatory response. CAF may be an important regulator for both miRNAs and signaling proteins, such as c-Myb, but the underlying mechanisms are complex. In addition, CAF may possess multiple other unknown pharmacologic effects and require further investigation.
This report reveals a novel inflammatory regulator, miR-301b, which directly interacts with its target c-Myb to finely maintain a balance between two types of inflammatory factors. As mentioned above, down-regulation of hyper-inflammation may be particularly beneficial for patients with severe infection and sepsis shock. In summary, we have identified a novel function of CAF in preferentially regulating anti-inflammatory response in bacterial infection through repression of miR-301b expression. This pathway takes effect through regulation of c-Myb, which is targeted by miR-301b and influences NF-κB-mediated cytokine production. Down-regulation of miR-301b by CAF is negatively regulated via cAMP/PKA/NF-κB signaling. Finally, augmenting c-Myb up-regulates anti-inflammatory cytokine release, leading to a subdued inflammatory response. Our findings reveal miR-301b and CAF as new regulators in inflammatory response, which may be tailored to alleviate sepsis-mediated tissue injury.
Material and methods
Ethics statement
Animal experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of North Dakota, School of Medicine (Assurance Number: A3917-01). Dissections and injections were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Mouse
TLR4−/− and TLR2−/− mice were provided by Dr. Jyotika Sharma at University of North Dakota, Grand Forks, ND. These mice were based on C57BL/6J genetic background, and normal age and sex matched C57BL/6J mice were used as wild-type (WT) controls. Mice were kept and bred in the animal facility at the University of North Dakota 42. Animal experiments were performed with randomization. We used 12 mice for survival assays, it will give an effective power of better than 0.9 (when alpha is 0.1). Other experiments used at least 3 mice for getting better statistic results.
Cells
MLE-12 and MH-S cells were obtained from ATCC and cultured in HITES medium (MLE-12) and RPMI 1640 medium (MH-S) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT) and 100 U/ml of penicillin/streptomycin (Life Technologies, Rockville, MD) antibiotics in a 37°C incubator with 5% CO2. The cell lines were authenticated by phenotypic analysis of critical treats of the cells and they were tested for mycoplasma. Mouse alveolar macrophage (AM) cells were isolated from the lung by bronchoalveolar lavage (BAL). In brief, the trachea was cannulated with a 20-gauge catheter; 0.9 ml BAL buffer was instilled, flushed four times, and retrieved. A total of 3.0 ml BALF was retrieved from each mouse and cytospin slides prepared with 0.5 ml BALF were analyzed by H&E staining (Fisher, Rockford, IL) to enumerate leukocyte subtypes based on their cellular and nuclear morphological properties. After centrifugation at 1500 rpm, AM cells were resuspended and cultured in RPMI 1640 medium supplemented with 10% fetal bovine.
Infection Experiments
P. aeruginosa strain PAO1 WT was provided by Dr. S. Lory (Harvard Medical School, Boston, MA). PAO1-GFP strain was obtained from Dr. G. Pier (Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) 43. K. pneumoniae strain Kp WT (ATCC 43816 serotype II) was provided by Dr. V. Miller (University of North Carolina, Chapel Hill, NC). S. pneumoniae strain was bought from ATCC (49619, Serotype 19F). LPS purified by phenol extraction from E. coli serotype 0111:B4 was bought from Sigma (St. Louis, MO). Caffeine was also bought from Sigma (Powder, ReagentPlus, #C0750).
After culturing in Luria-Bertani (LB) broth at 37°C with vigorous shaking overnight, bacteria were centrifuged at 6000 rpm for 5 min, and then resuspended in 5 ml fresh LB broth to allow growing till mid-logarithmic phase. The concentration of the bacteria was counted by reading at OD600 (1 OD=1×109 cells/ml) 44. After anesthesia (40 mg/kg ketamine), mice were infected with 1×107 colony-forming units (CFU, suspended in 50 μl PBS) of Pa, or 1×105 CFU of Kp by intranasal instillation. The mice were sacrificed when they were moribund. Survival was determined using Kaplan Meier curve. After BAL, the lung, liver, kidney and spleen were excised for homogenization for RNA extraction or western blotting, or fixed in 10% formalin for histological or immunohistochemical staining. For cells, before infection, they were washed once with PBS, and replaced with antibiotic-free medium immediately. Cells were infected by bacteria at multiplicity of infection (MOI) of 10:1 (bacteria: cells) and then washed 3 times with PBS to remove the floating bacteria at indicated time.
Transfection
TLR4, MyD88, TRIF, PKA, p65, CREB-1 and c-Myb siRNA were bought from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). miRNA negative control (NS-m, CN-001000-01-05), miR-301a mimics (301a-m, C-310482-05), miR-301b mimics (301b-m, C-310775-03), miR-302b mimics (302b-m, C-310669-05) miRNA inhibitor negative control (NS-i, IN-001005-01), or miR-301b inhibitor (301b-i, IH-310775-04) were bought from Dharmacon Inc. (Chicago, IL). MH-S or MLE-12 cells were respectively transfected with miRNA or siRNA in serum-free medium using LipofectAmine 2000 reagent (Invitrogen, Grand Island, NY) following the manufacturer’s instruction for 48 hours. Mice were i.v. administered with vehicle (in vivo-jetPEI, Polyplus-transfection Inc., New York, NY), NS-m, 301b-m, NS-i, or 301b-i (50 μg/mouse) 24 h before bacteria challenge following the manufacturer’s instruction 20.
Measurement of miRNA and mRNA expression
Total RNAs were isolated using TRIzol reagent (Invitrogen, Grand Island, NY) following the manufacturer’s instructions. RNAs were eluted in RNase-free water and stored at −80°C. For non-radioactive northern blot, DIG-modified miR-301b, miR-301a, miR-15a and sno202 probes were synthesized by Integrated DNA Technologies (Coralville, IA) 20. Northern blot was performed following a published protocol described previously 45. Real-time PCR profiling of miRNA were performed using a SYBR Green-based, miScript PCR System (Qiagen, Valencia, CA). The expression of other mRNAs was detected using QuantiTect SYBR Green RT-PCR Kit (Qiagen, Valencia, CA). The separate well 2-ΔΔCt cycle threshold method was used to determine relative quantities of individual miRNAs or mRNA, and these were expressed as fold changes respectively. GAPDH was used as control throughout the manuscript.
Histological Analysis
Organs were fixed in 10% formalin by a routine histologic procedure 46. 5 μl of BAL and blood were applied evenly on microscope slides. After H&E staining (Thermo fisher), the numbers of neutrophils were counted using a light microscope. The formalin-fixed tissues were used for H&E staining or immunohistochemical staining to examine tissue damage or Ly6G expression post treatment.
Immunoblotting
Samples taken from either cells or tissues of mice after experimental treatment were lysed with RIPA buffer (30 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol, and complete cocktail (Life technologies, Grand Island, NY) and phosphatases (Sigma, St. Louis, MO). Lysates were centrifuged at 14000×g for 15 min, the supernatants were collected and the concentration was quantitated. The samples were boiled for 10 min, and equal amount was applied to 12% SDS-polyacrylamide minigels and electrophoresed. The proteins in the gel were then transferred to nitrocellulose filter membranes (Thermofisher, Rockford, IL). Horseradish peroxidase (HRP)-linked secondary antibody (Rockland, Gilbertsville, PA) and X-ray film (Kodak) were used for exposure 47,48. Rabbit polyclonal IgG antibody anti-GAPDH, c-Myb, MyD88, TLR4, PKA, pPKA (Thr 198), CREB-1, pCREB-1 (Ser 133), pp65 (Ser 536), TGF-β1, goat polyclonal IgG antibody anti-IL-4, and mouse monoclonal IgG antibody anti-p65 were bought from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal IgG antibody anti-TRIF were bought form Abcam (Cambridge, MA).
Cytokine Profiling and Assays
Cytokine concentrations in the first 0.6 ml BAL fluid collected at the indicated times after infection were measured by standard ELISA kits following the manufacturer’s instructions (eBioscience Inc., San Diego, CA). 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyltetrazolium bromide (MTT) assay, were carried out following the manufacturer’s instructions. Lung tissues were homogenized and equal amount of proteins was used for myeloperoxidase (MPO) assays.
Confocal Laser Scanning Microscopy
AM, MH-S and MLE-12 cells were cultured in glass bottom dishes (MatTek, Ashland, MA). The fluorescence images were obtained by LSM 510 Meta confocal microscope (Carl Zeiss Micro Imaging, Thornwood, NY) 46. For immunostaining, the cells were fixed in 3.7% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS and incubated with blocking buffer containing 2% BSA for 30 min. Cells were incubated with primary antibodies at 1/500 dilution in blocking buffer for 1 h and washed three times. After incubation with appropriate fluorophore-conjugated secondary antibodies, the cover slips were mounted on slides with Vectashield mounting medium. The images were captured and processed using the software provided by the manufacturer 20,49.
Migration Assay
MLE-12 cells were transfected with NS-m or 301b-m and treated with LPS. The culture medium was collected and placed in the lower chamber. MH-S cells were loaded into the Boyden chamber with an 8 μm porous membrane (Corning) in 100 μl RPMI 1640 medium supplied with 1% serum and incubated at 37°C for 4 h. Cells were removed from the upper side of membranes, and nuclei of migratory cells on the lower side of the membrane were stained with crystal violet. MH-S cells that attached to the filter were counted by light microscopy.
Luciferase Reporter Assay
The sequences of the miRNAs and c-Myb used in this work can be found from the public database microRNA.org (miRanda) that revealed c-Myb as a representative target of miR-301b (higher scores indicating stronger binding). The luciferase assay report plasmid was designed with adding a sequence of c-Myb 3‘UTR to Luciferase Reporter Assay System (Promega, Madison, WI). The designed plasmids were transfected to MLE-12 in 6 well plate for 24 h. Then treated with miR-301b mimics or control mimics separately. Cell lysates were performed for luciferase activity analysis following the manufacturer’s instructions 20.
Statistical Analysis
Each experiment was conducted in triplicate or repeated at least 3 times. The differences in outcomes of mice are presented as percent or amount changes compared with the control after treatment. Data were analyzed by one-way ANOVA (Tukey’s post hoc) using Graphpad software with means ± SD. The survival rate was calculated using Kaplan-Meier curve from a log-rank test 50. Differences were accepted as significant at p<0.05.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health (AI109317-01A1 and AI101973-01) to Dr. Min Wu; This work was also supported by grants from the National 973 Basic Research Program of China (2013CB911300), the Chinese NSFC (81225015 and 81430071), and Sichuan Science-Technology Innovative Research Team for Young Scientist (2013TD0001) to Dr. Canhua Huang. We thank S. Abrahamson of the UND imaging core for help with confocal imaging.
Footnotes
Author Contributions
Conceived and designed the experiments: XL SH CH MW. Performed the experiments: XL SH RL XZ. Analyzed the data: XL SH RL XZ SZ MY YY YW. Contributed reagents/materials/analysis tools: CH MW. Wrote the paper: XL SH CH MW.
References
- 1.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer research. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- 3.Marques-Rocha JL, et al. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2015;29:3595–3611. doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- 4.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Zhao JL, et al. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. Journal of immunology. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 9.Siddle KJ, et al. bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS genetics. 2015;11:e1005064. doi: 10.1371/journal.pgen.1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwerk J, Savan R. Translating the Untranslated Region. Journal of immunology. 2015;195:2963–2971. doi: 10.4049/jimmunol.1500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamza TH, et al. Glutamate receptor gene GRIN2A, coffee, and Parkinson disease. PLoS genetics. 2014;10:e1004774. doi: 10.1371/journal.pgen.1004774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosendahl AH, et al. Caffeine and Caffeic Acid Inhibit Growth and Modify Estrogen Receptor and Insulin-like Growth Factor I Receptor Levels in Human Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1877–1887. doi: 10.1158/1078-0432.CCR-14-1748. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt B, et al. Caffeine therapy for apnea of prematurity. The New England journal of medicine. 2006;354:2112–2121. doi: 10.1056/NEJMoa054065. [DOI] [PubMed] [Google Scholar]
- 14.Spaeth AM, Goel N, Dinges DF. Cumulative neurobehavioral and physiological effects of chronic caffeine intake: individual differences and implications for the use of caffeinated energy products. Nutrition reviews. 2014;72(Suppl 1):34–47. doi: 10.1111/nure.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu G, et al. Inhibition of adenoma progression to adenocarcinoma in a 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis model in A/J mice by tea polyphenols and caffeine. Cancer research. 2006;66:11494–11501. doi: 10.1158/0008-5472.CAN-06-1497. [DOI] [PubMed] [Google Scholar]
- 16.Horrigan LA, Kelly JP, Connor TJ. Caffeine suppresses TNF-alpha production via activation of the cyclic AMP/protein kinase A pathway. International immunopharmacology. 2004;4:1409–1417. doi: 10.1016/j.intimp.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Lou Y, et al. Oral caffeine during voluntary exercise markedly inhibits skin carcinogenesis and decreases inflammatory cytokines in UVB-treated mice. Nutrition and cancer. 2013;65:1002–1013. doi: 10.1080/01635581.2013.812224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pisano F, et al. Combination of miRNA499 and miRNA133 exerts a synergic effect on cardiac differentiation. Stem cells. 2015;33:1187–1199. doi: 10.1002/stem.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tritsch E, et al. An SRF/miR-1 axis regulates NCX1 and annexin A5 protein levels in the normal and failing heart. Cardiovascular research. 2013;98:372–380. doi: 10.1093/cvr/cvt042. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, et al. MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nature communications. 2014;5:3619. doi: 10.1038/ncomms4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grailer JJ, Kalbitz M, Zetoune FS, Ward PA. Persistent neutrophil dysfunction and suppression of acute lung injury in mice following cecal ligation and puncture sepsis. Journal of innate immunity. 2014;6:695–705. doi: 10.1159/000362554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nouailles G, et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. The Journal of clinical investigation. 2014;124:1268–1282. doi: 10.1172/JCI72030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perl M, et al. Role of activated neutrophils in chest trauma-induced septic acute lung injury. Shock. 2012;38:98–106. doi: 10.1097/SHK.0b013e318254be6a. [DOI] [PubMed] [Google Scholar]
- 24.Lefkimmiatis K, Leronni D, Hofer AM. The inner and outer compartments of mitochondria are sites of distinct cAMP/PKA signaling dynamics. The Journal of cell biology. 2013;202:453–462. doi: 10.1083/jcb.201303159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aandahl EM, et al. Inhibition of antigen-specific T cell proliferation and cytokine production by protein kinase A type I. Journal of immunology. 2002;169:802–808. doi: 10.4049/jimmunol.169.2.802. [DOI] [PubMed] [Google Scholar]
- 26.Ollivier V, Parry GC, Cobb RR, de Prost D, Mackman N. Elevated cyclic AMP inhibits NF-kappaB-mediated transcription in human monocytic cells and endothelial cells. The Journal of biological chemistry. 1996;271:20828–20835. doi: 10.1074/jbc.271.34.20828. [DOI] [PubMed] [Google Scholar]
- 27.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim MX, et al. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infection and immunity. 2014;82:4789–4801. doi: 10.1128/IAI.02279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoong P, Pier GB. Immune-activating properties of Panton-Valentine leukocidin improve the outcome in a model of methicillin-resistant Staphylococcus aureus pneumonia. Infection and immunity. 2012;80:2894–2904. doi: 10.1128/IAI.06360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funamizu N, et al. MicroRNA-301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells. International journal of oncology. 2014;44:725–734. doi: 10.3892/ijo.2014.2243. [DOI] [PubMed] [Google Scholar]
- 31.Shaked I, et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu LF, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagos D, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nature cell biology. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 35.Wang YX, et al. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. Journal of digestive diseases. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 36.Reid JF, et al. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Molecular cancer research : MCR. 2012;10:504–515. doi: 10.1158/1541-7786.MCR-11-0342. [DOI] [PubMed] [Google Scholar]
- 37.Thompson CB, Challoner PB, Neiman PE, Groudine M. Expression of the c-myb proto-oncogene during cellular proliferation. Nature. 1986;319:374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- 38.Mucenski ML, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, et al. c-MYB regulates cell growth and DNA damage repair through modulating MiR-143. FEBS letters. 2015;589:555–564. doi: 10.1016/j.febslet.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Banchereau J, Pascual V, O’Garra A. From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nature immunology. 2012;13:925–931. doi: 10.1038/ni.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz MF, et al. Biomechanical forces promote blood development through prostaglandin E2 and the cAMP-PKA signaling axis. The Journal of experimental medicine. 2015 doi: 10.1084/jem.20142235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Priebe GP, et al. Construction and characterization of a live, attenuated aroA deletion mutant of Pseudomonas aeruginosa as a candidate intranasal vaccine. Infection and immunity. 2002;70:1507–1517. doi: 10.1128/IAI.70.3.1507-1517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, et al. Lyn Delivers Bacteria to Lysosomes for Eradication through TLR2-Initiated Autophagy Related Phagocytosis. PLoS pathogens. 2016;12:e1005363. doi: 10.1371/journal.ppat.1005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SW, et al. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic acids research. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, Pasula R, Smith PA, Martin WJ., 2nd Mapping alveolar binding sites in vivo using phage peptide libraries. Gene therapy. 2003;10:1429–1436. doi: 10.1038/sj.gt.3302009. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Wu M, Nelson DE, Pasula R, Martin WJ., 2nd Alpha-1-antitrypsin expression in the lung is increased by airway delivery of gene-transfected macrophages. Gene therapy. 2003;10:2148–2152. doi: 10.1038/sj.gt.3302121. [DOI] [PubMed] [Google Scholar]
- 48.Wisniowski PE, et al. Vitronectin protects alveolar macrophages from silica toxicity. American journal of respiratory and critical care medicine. 2000;162:733–739. doi: 10.1164/ajrccm.162.2.9808015. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Ye Y, Zhou X, Huang C, Wu M. Atg7 Enhances Host Defense against Infection via Downregulation of Superoxide but Upregulation of Nitric Oxide. Journal of immunology. 2015;194:1112–1121. doi: 10.4049/jimmunol.1401958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu M, et al. Host DNA repair proteins in response to P. aeruginosa in lung epithelial cells and in mice. Infection and immunity. 2011;79:75–87. doi: 10.1128/IAI.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.