Abstract
Bacterial‐derived compounds from the intestinal microbiome modulate host mucosal immunity. Identification and mechanistic studies of these compounds provide insights into host–microbial mutualism. Specific Lactobacillus reuteri strains suppress production of the proinflammatory cytokine, tumor necrosis factor (TNF), and are protective in a mouse model of colitis. Human‐derived L. reuteri strain ATCC PTA 6475 suppresses intestinal inflammation and produces 5,10‐methenyltetrahydrofolic acid polyglutamates. Insertional mutagenesis identified the bifunctional dihydrofolate synthase/folylpolyglutamate synthase type 2 (folC2) gene as essential for 5,10‐methenyltetrahydrofolic acid polyglutamate biosynthesis, as well as for suppression of TNF production by activated human monocytes, and for the anti‐inflammatory effect of L. reuteri 6475 in a trinitrobenzene sulfonic acid‐induced mouse model of acute colitis. In contrast, folC encodes the enzyme responsible for folate polyglutamylation but does not impact TNF suppression by L. reuteri. Comparative transcriptomics between wild‐type and mutant L. reuteri strains revealed additional genes involved in immunomodulation, including previously identified hdc genes involved in histidine to histamine conversion. The folC2 mutant yielded diminished hdc gene cluster expression and diminished histamine production, suggesting a link between folate and histadine/histamine metabolism. The identification of genes and gene networks regulating production of bacterial‐derived immunoregulatory molecules may lead to improved anti‐inflammatory strategies for digestive diseases.
Keywords: Colitis, folate, folC2, histamine, immunomodulation, Lactobacillus reuteri.
Introduction
Lactobacillus reuteri is a vertebrate symbiont found in the gastrointestinal (GI) tract of a variety of mammalian species and considered indigenous to the human gut (Reuter 2001; Walter et al. 2011). Several L. reuteri strains are probiotics, “viable microorganisms that confer a health benefit to the host when administered in adequate amounts” (FAO/WHO, 2006). Selective deficiencies of intestinal lactobacilli have been described in patients with inflammatory bowel disease (IBD) (Giaffer et al. 1991; Vigsnaes et al. 2012; Zella et al. 2011), and oral or intrarectal supplementation with various probiotic Lactobacillus species effectively ameliorates intestinal inflammation in patients with pouchitis (Bibiloni et al. 2005; Gupta et al. 2000) and in rodent colitis models (Foligne et al. 2006; Holma et al. 2001; Moller et al. 2005; Pena et al. 2005; Schreiber et al. 2009; Satish Kumar et al. 2015; Liu et al. 2011; Peran et al. 2007). In vitro studies have demonstrated that lactobacilli possess species‐specific, potent immunosuppressive activities, such as modulation of murine dendritic cell‐induced differentiation of Th1 and Th2 cells (Christensen et al. 2002), increasing production of IL‐10 from dendritic cells and macrophages (Livingston et al. 2010; Bleau et al. 2010), inhibiting production of TNF from lipopolysaccharide (LPS)‐stimulated monocytes (Kim et al. 2008), and driving development of IL‐10‐producing regulatory T cells (Smits et al. 2005; Zhao et al. 2013). The immunosuppressive functions of probiotics, like L. reuteri, could be harnessed to make new therapeutics for chronic autoimmune or inflammatory disorders.
The reduction in biologically active, circulating TNF by neutralizing antibodies has been an effective treatment strategy for patients with IBD (Peyrin‐Biroulet 2010) and trinitrobenzene sulfonic acid (TNBS)‐challenged rats (Triantafillidis et al. 2005). Anti‐TNF strategies, however, are complicated by secondary deficiencies in antimycobacterial immunity and possible sensitization or development of antibodies to these therapies (Hoentjen and van Bodegraven 2009; Jauregui‐Amezaga et al. 2013; Ungar et al. 2014). Additionally, anti‐TNF therapies have been associated with reactivation of hepatitis B virus and a slightly increased risk of melanoma (Chebli et al. 2014). These side effects make anti‐TNF strategies less desirable as long‐term therapeutics. In contrast to systemic antibody‐based strategies, luminal bacteria in the intestine may be able to suppress inflammation and proinflammatory cytokine activities in a gut‐specific manner. Bacterial‐derived, cell‐free culture supernatants of human‐derived L. reuteri ATCC PTA 6475 (6475) and L. reuteri CRL1098 suppressed TNF production by primary monocyte‐derived macrophages from patients with Crohn's disease and activated myeloid cell lines (Lin et al. 2008; Pena et al. 2005) and activated peripheral blood mononuclear cells (Mechoud et al. 2012). L. reuteri 6475 biofilms were capable of suppressing TNF production by LPS‐activated monocytoid cells (Jones and Versalovic 2009). A combination of L. reuteri 6475 and L. paracasei reduced colonic TNF as well as intestinal inflammation in an IL‐10‐deficient, Helicobacter hepaticus‐induced IBD mouse model (Pena et al. 2005), and a mixture of four L. reuteri strains was protective in a dextran sodium sulfate (DSS)‐induced colitis rat model (Schreiber et al. 2009). These previously published results suggest that L. reuteri strains may be effective immunoregulatory probiotics, and autochthonous components of the gut microbiome may affect the biology of the mucosal immune system.
Probiotic supernatants and cell‐derived factors inhibit cytokine production and suppress inflammatory signaling in macrophages and other immune cells (Grangette et al. 2005; Thomas and Versalovic 2010), but a paucity of bacterial genes and products required for immunomodulation have been identified (Grangette et al. 2005; Yasuda et al. 2008). Recently the biogenic amine, histamine, was identified as a TNF‐inhibitory factor produced by L. reuteri 6475. Histamine is produced by the decarboxylation of l‐histidine, and L. reuteri‐mediated histamine production can be increased by histidine supplementation in the growth medium (Thomas et al. 2012). L. reuteri also synthesizes the essential B‐complex vitamin, folate, when a precursor para‐aminobenzoic acid (pABA), is provided in the medium (Spinler et al. 2014; Rossi et al. 2011; Santos et al. 2008). In selected microorganisms, folate may catalyze one‐carbon units into histidine, suggesting that folate may be involved in histidine biosynthesis (Broquist 1957). Crosstalk between these microbial metabolic pathways may result in the coregulation of histamine and folate biosynthesis. Additionally, in eukaryotic cells, it is known that oxidation of amino acids, including histidine, is linked to folate metabolism. Folate plays a key role in the reduction in NAD+ to NADH and NADP+ to NADPH in the oxidation–reduction reactions necessary for one‐carbon metabolism (Brosnan et al. 2015). Similar reactions may occur in prokaryotes, reinforcing the possible coregulation of histamine and folate biosynthesis in L. reuteri.
Greater understanding of bacterial immunomodulatory gene networks and mechanistic studies of immunomodulatory compounds should improve selection of effective probiotics for specific therapeutic applications. In this era of microbiome science, functional linkages between different microbial metabolic pathways may elucidate mechanisms of probiosis and immunoregulation by gut microbes. The goal of this study was to identify immunomodulatory genes and regulatory networks present in TNF‐inhibitory L. reuteri 6475. These studies demonstrated a novel role for the dihydrofolate synthase/folylpolyglutamate synthase gene type 2 (folC2) in TNF suppression and colitis attenuation, and demonstrate a potentially important link between folate metabolism and histamine production.
Experimental Procedures
Bacterial strains and culture conditions
All bacterial strains and plasmids used in this study are described in Table S2. L. reuteri strains ATCC PTA 6475, ATCC 6475 folC2::pORI28, ATCC 6475 folC::pORI28, and ATCC 55730, are referred to as strains 6475, 6475::folC2, 6475::folC, and 55730, respectively. L. reuteri strains were cultured for 24 h at 37°C in an anaerobic workstation (MACS MG‐500, Microbiology International, Frederick, MD) supplied with a mixture of 10% CO2, 10% H2, and 80% N2 for 16–18 h in de Man–Rogosa–Sharpe (MRS) medium (Difco, Franklin Lakes, NJ), and then inoculated into a defined medium, LDMIII (OD600 adjusted to 0.1), which has been described previously (Jones and Versalovic 2009). At stationary phase (24 h), the cells were pelleted (4000g, 10 min). Insertion mutants were cultured in the presence of 10 μg/mL erythromycin.
Construction folC2 and folC insertion mutants (L. reuteri 6475::folC2 and 6475::folC)
Bifunctional dihydrofolate synthase/folylpolyglutamyl synthase type 2 (folC2) and bifunctional dihydrofolate synthase/folylpolyglutamyl synthase (folC) genes were identified in the whole draft genome sequence of L. reuteri 6475 (GenBank NZ_ACGX02000001‐007; HMPREF0536_11260 and HMPREF0536_10555, respectively). Inactivation of these genes was achieved by site‐specific integration of plasmid pORI28 into the L. reuteri 6475 chromosome as described previously (Walter et al. 2005). Briefly, internal gene fragments were amplified by PCR (outlined in Table S2) and directionally cloned into pORI28. Site‐specific homologous recombination of target‐specific pORI28 vectors was performed as detailed by Walter et al. (2005). Site‐specific insertional mutagenesis was confirmed by dideoxy DNA sequencing.
Tetrahydrofolic acid compound analysis by MALDI mass spectrometry
Cell pellets normalized by weight from wild‐type and mutant L. reuteri strains were washed with ice‐cold PBS and water. Folates were extracted by washing with 50% acetonitrile/0.1% v/v TFA. The cell suspension was centrifuged for 10 min, 4000g at 4°C. Samples were prepared with α‐cyano‐4‐hydroxy‐cinnamic acid as MALDI matrix, spotted onto a sample plate, dried, and analyzed on a prOTOF2000 MALDI mass spectrometer (PerkinElmer/Sciex, Boston, MA) or on a SimulTOF Combo 200 MALDI mass spectrometer (Virgin Instruments Marlborough, MA). For MS/MS fragmentation analysis, the same MALDI sample plate was subsequently transferred into a self‐built MALDI quadrupole ion trap that was based on a modified LCQ DECA iontrap (Thermo Waltham, MA) (Krutchinsky et al. 2001). Tetrahydrofolate ions were fragmented with a 4 Da selection window, 30% collision energy, a fixed ion trap injection time of 200 msec, and a 2–10‐hz laser repetition rate.
Assessment of TNF inhibition by ELISA
Bacterial supernatants from a 24 h LDMIII culture were filter‐sterilized using polyvinylidene fluoride membrane filters (0.22 μm pore size, Millipore, Bedford, MA) and size‐fractionated with Amicon Ultra‐15 centrifugal filter units using ultracel‐3 membrane (Millipore). The filtrate was speed vacuum‐dried and resuspended in RPMI medium. All supernatants were normalized by volume to OD600 = 1.5. Supernatants and cell pellet washes were tested for their ability to modulate TNF production. In vitro experiments were performed with THP‐1 cells (human monocytoid cell line, ATCC number TIB‐202, ATCC, Manassas, VA) maintained in RPMI (ATCC) and heat‐inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C, with 5% CO2. THP‐1 cells (5 × 104 cells) were stimulated to produce TNF by the addition of 100 ng/mL Pam3Cys‐SKKKK x 3 HCl (EMC Microcollections, Tüebingen, Germany) as previously described (Pena et al. 2004). L. reuteri supernatant or cell pellet wash was added to the activated THP‐1 cells (5% v/v). Plates were incubated at 37°C and 5% CO2 for 3.5 h. THP‐1 cells were pelleted (3000g, 5 min, 4°C), and quantitative ELISAs were used to determine TNF quantities in THP‐1 cell supernatants according to the manufacturer's instructions (R&D Systems, Minneapolis, MN).
TNF gene expression studies by qPCR
THP‐1 cells were treated with L. reuteri cell‐free supernatant and PCK as described above. RNA isolation was performed with the AllPrep DNA/RNA mini kit from Qiagen (Valencia, CA) according to manufacturer's instructions. RNA quantity and quality was assessed, and only RNA with RIN greater than or equal to 9 was used in the subsequent assays. Gene expression was analyzed using the RT2 Profiler PCR Array (innate and adaptive immune response) from Qiagen according to manufacturer's instructions. In brief, cDNA was prepared from purified RNA with the RT2 first strand kit. The cDNA was added to RT2 SYBR green mastermix, and aliquoted into the 96‐well RT2 Profiler PCR Array of interest. All PCR reactions were performed using the Stratagene Mx3005P PCR System. Cycling parameters were as follows: program 1; One cycle of 95°C for 10 min, program 2; 40 two‐step cycles of 95°C for 15 sec, 60°C for 1 min, program 3; hold at 4°C. Fluorescence was detected after the extension step in each cycle. The 2−ΔΔCT method was used to calculate relative changes in gene expression.
Transcriptomics comparisons of L. reuteri mutants
L. reuteri 6475, 6475::folC2, and 6475::folC were cultured in LDMIII to stationary phase (24 h). For expression analyses, three biological replicates were performed with dye‐swap experiments for each strain/mutant. Following mRNA isolation, cDNA synthesis, labeling, and hybridization were performed as previously described (Wall et al. 2007; Yang et al. 2005). Information regarding the microarray platforms is at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under GEO platform GPL754. The complete set of microarray data for 6475::folC2 (formerly known as 6475::thfs) and 6475::folC (formerly known as 6475::thfs2) can be found under the GEO series accession GSE32971 and GSE32972, respectively. Microarray data analysis was performed as previously described (Maguin et al. 1992) utilizing the array package in R2.12.1. The gene set of interest (GSI) was the list of genes potentially contributing to the immunomodulatory phenotype of strain 6475. The GSI was refined by analyzing subsequent layers of gene expression data. The GSEA v2.07 analysis was performed using the online analysis tool (http://www.broadinstitute.org/gsea/index.jsp). The 402 genes significantly up‐regulated in stationary phase of 6475 that were missing a homolog or not significantly up‐regulated in stationary phase of 55,730 were used as the input Gene Set. The ranked list of 6475::folC2 expression pattern via fold change was used as the preranked input. A weighted enrichment statistic was calculated using 1000 permutations. The core enriched and down‐regulated 6475::folC2 genes were further refined by removing genes significantly down‐regulated in 6475::folC. These potential immunoregulatory genes were sorted by a combined DEDS statistic which placed equal weight on 6475::folC2 and 6475 stationary phase linear fold changes (Yang et al. 2005). DEDS represents the selected differential expression measures from the previous analyses as a multivariate point cloud with multiple dimensions associated with different input statistics. The scalar distance to the most extreme combination of these input statistics, following permutation, is the final metric by which genes are ranked and displayed here.
Gene expression studies of the L. reuteri hdc cluster
L. reuteri 6475 and 6475::folC2 were grown as described above in LDMIII or LDMIII + 4 mg/mL l‐histidine. At 16 h post‐inoculation, the cultures were harvested. RNA isolation and cDNA synthesis from total RNA were performed as previously described (Thomas et al. 2012). Expression of the hdcA, hdcB, hdcP, and narI genes was analyzed using quantitative real‐time PCR. All primers were designed using the Universal Probe Library Assay Design Center (Roche Applied Science, Indianapolis, IN) and are described in Table S2. The RNA polymerase β‐subunit (rpoB) gene was used as a reference gene. PCR reactions were set up using 2× FastStart Universal Probe Master (Rox) (Roche Applied Science) and the cDNA described above, with final concentrations of 200 nmol/L for each primer and 100 nmol/L for each probe. All PCR reactions were performed using the ViiA 7 Real‐Time PCR System (Life Technologies, Carlsbad, CA). Cycling parameters were as follows: program 1; One cycle of 25°C for 2 min, one cycle of 95°C for 10 min, program 2; 50 three‐step cycles of 95°C for 15 sec, 60°C for 1 min and 72°C for 1 min, program 3; hold at 4°C. Fluorescence was detected after the extension step in each cycle. The 2−ΔΔCT method was used to calculate relative changes in gene expression.
Quantification of histamine by ELISA
Wild‐type L. reuteri 6475 and 6475::folC2 were grown as described above in LDMIII or LDMIII + 4 mg/mL l‐histidine. Cultures were harvested at 24 h, centrifuged (1500g), and filter‐sterilized with 0.22‐μm PVDF filters. Histamine concentrations were determined as previously described (Thomas et al. 2012) using the Histamine ELISA kit (Neogen, Lexington, KY). Absorbance was measured with a Spectramax 340PC (Molecular Devices, Sunnyvale, CA), and data were analyzed using GraphPad Prism 5 software. Data were corrected with values obtained from the background control.
Preparation of bacterial supernatants and administration to mice
Bacterial supernatants were prepared as described for the TNF inhibition bioassay above, filter‐sterilized, and concentrated 20× with speed vacuum drying. Administration to mice was as described previously (Hemarajata et al. 2013). In brief, each mouse received two intraperitoneal injections of bacterial supernatant or medium control, with the first dose at 18 h before TNBS rectal enema (described below) and the second dose at 2 min before TNBS enema. All mouse experiments were performed in a Specific Pathogen‐Free (SPF) animal facility, according to an Institutional Animal Care and Use Committee (IACUC)‐approved mouse protocol at Baylor College of Medicine, Houston, TX.
Induction of acute colitis using TNBS rectal enema
Female Balb/c mice (45‐day old) were received from Harlan Laboratories (Houston, TX) and maintained under specific pathogen‐free conditions. Animals were provided standard chow and water and allowed to feed ad libitum under a 12 h daylight cycle. Mice acclimated post‐shipment for 10 d. Induction of TNBS colitis, determination of colitis severity, and protection conferred by probiotic compounds was performed according to established protocols with minor modifications as described previously (Foligne et al. 2006; Hemarajata et al. 2013). In brief, mice were anesthetized by constant isoflurane inhalation. A 5% v/v TNBS (Sigma‐Aldrich, St. Louis, MO) solution in water was diluted with equal volume of absolute ethanol and administered intrarectally via catheter at a dose of 100 mg/kg body weight, 4 cm distal to the anus. Mice were kept head down in a vertical position for 2 min after enema to ensure complete retention of enema in the colon. Procedure control mice received 50% ethanol in PBS as an enema and two IP injections of the medium control. Colitis‐positive mice received a TNBS enema and two IP injections of the medium control, while treated mice received a TNBS enema and two IP injections of the prepared bacterial supernatant. Mice were weighed immediately prior to TNBS enema and again 48 h after TNBS enema. Percent weight loss was calculated based on differences between these measurements.
Macroscopic assessment of TNBS‐induced colitis
Colons were collected 48 h after induction of TNBS colitis and opened longitudinally. Colonic inflammation and damage were determined according to the Wallace criteria (Morris et al. 1989). In brief, the grading scale was: Score 0: normal/healthy appearance; Score 1: focal hyperemia, slight thickening, and no ulcers; Score 2: hyperemia, prominent thickening, and no ulcers; Score 3: ulceration with inflammation at one site; Score 4: ulceration with inflammation at two or more sites; Score 5: major sites of damage extending >1 cm; Score 6–10: when area of damage extends >2 cm, the score is increased by each additional cm of tissue involvement. Each colon was scored blindly by one individual.
Plasma measurements of mouse serum amyloid protein A (SAA)
Blood samples were collected from mice via cardiac puncture, stored with anticoagulant, and centrifuged (10 min, 17000g) to isolate plasma. SAA levels in plasma were measured using ELISA kits from ALPCO (Salem, NH) according to the manufacturer's instructions.
Results
Identification of 5,10‐CH = THF polyglutamate compounds produced by TNF‐inhibitory L. reuteri 6475
L. reuteri cell pellets from stationary phase cultures were treated with 0.1% trifluoroacetic acid (TFA)‐acidified water to collect a concentrated solution of extracellular compounds loosely associated with the bacterial cell surface. TFA‐treated cell pellets from TNF‐inhibitory L. reuteri 6475 and the non‐TNF‐inhibitory strain 55730 were analyzed by matrix‐assisted laser desorption/ionization time‐of‐flight (MALDI‐TOF) mass spectrometry. Differences in the composition of TFA‐treated cell pellets were observed. Multiple peaks (labeled MGlun) differing by m/z 129, the expected mass of glutamate (Glu), were identified in strain 6475 (Fig. 1A). Two of these peaks were also identified in strain 55730, but the peak intensity was much less compared to strain 6475 (Fig. 1B). These peaks were also identified in bacterial cell‐free culture supernatants from strains 6475 and 55730 (data not shown). The observed masses did not match the predicted masses for simple polyglutamate homopolymers, indicating the presence of a covalently linked compound (M). MS/MS fragmentation analysis in both L. reuteri strains 6475 and 55730 at m/z 1101.4 (MGlu5) (Fig. 1C) indicated that the core compound M was 5,10‐methenyltetrahydrofolic acid (5,10‐CH = THF) with a covalently linked, polyglutamate homopolymer tail (5,10‐CH = THF polyglutamate). Neither 5,10‐CH = THF nor 5,10‐CH = THF polyglutamate were produced by the folC2 mutant (Fig. 1D), and only 5,10‐CH = THF is produced by the folC mutant (Fig. 1E). The structure of 5,10‐CH = THF is depicted in Fig. 1E.
Figure 1.
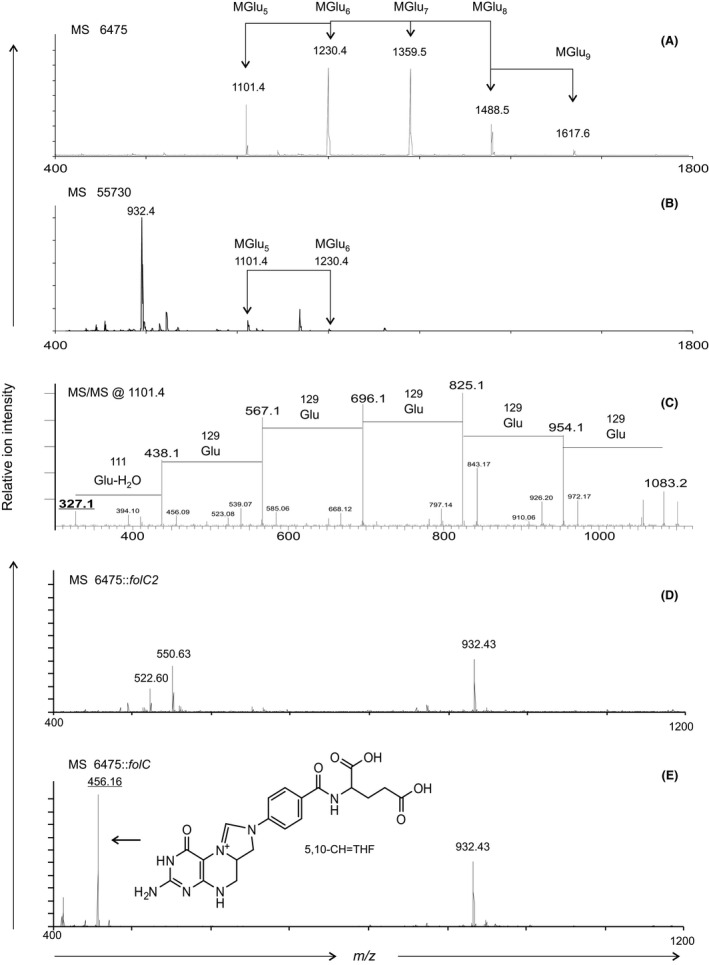
MALDI‐MS detection of 5,10‐CH = THF polyglutamates in TFA‐treated L. reuteri cell pellets. Peaks indicating the presence of 5,10‐CH = THF polyglutamate (arrows, MGlun) are marked for strain (A) 6475 and (B) 55730. The subscripted number (n) after MGlu indicates the number of glutamate residues present in the polyglutamate tail, not counting the intrinsic glutamate of folic acid. (C) MALDI ion trap MS/MS fragmentation analysis of MGlu5 at m/z 1101.4 confirms the presence of a folate compound, consistent with the lightest MS/MS fragment ion at m/z values 327.1. (D) Inactivation of folC2 in 6475 inhibited the production of 5,10‐CH = THF compounds as monitored by MALDI MS. (E) The 6475::folC mutant produced 5,10‐CH = THF (structure depicted), but no 5,10‐CH = THF polyglutamates of any chain length. The unidentified ion at m/z 932.4 was consistently observed after TFA treatment of all L. reuteri cell pellets. All spectra were scaled to comparable maximum intensity levels. TFA, trifluoroacetic acid; 5,10‐CH = THF, 5,10‐methenyltetrahydrofolic acid.
The folC2 gene in L. reuteri is required for human TNF suppression
Targeted mutagenesis was used to construct mutants defective in bifunctional dihydrofolate synthase/folylpolyglutamate synthase type 2 (folC2) and bifunctional dihydrofolate synthase/folylpolyglutamate synthase (folC), 6475::folC2 and 6475::folC, respectively. The ability of the mutants to modulate TNF levels was compared to wild‐type strain 6475. Wild‐type L. reuteri 6475 TFA‐treated cell pellet washes containing 5,10‐CH = THF and 5,10‐CH = THF polyglutamate significantly inhibited TNF production by activated human monocytoid cells stimulated with a Toll‐like receptor 2 (TLR2) agonist (P < 0.05) (Fig. 2). Inactivation of folC had no significant effect on TNF inhibition by strain 6475 (Fig. 2); however, inactivation of folC2 in strain 6475 resulted in abrogation of the TNF‐inhibitory phenotype (Fig. 2), suggesting that folC2 is a part of the immunomodulatory gene network. Similar results were obtained with the bacterial cell‐free culture supernatants of the same L. reuteri strains (Fig. S1A), indicating that the immunomodulins were actively secreted and associated with the bacterial cell surface. In TLR2‐activated human monocytoid cells, TNF gene expression was decreased by L. reuteri 6475 compared to the medium control (Fig. S1B), confirming prior results that L. reuteri suppressed human TNF at the transcriptional level (Lin et al. 2008). The 6475::folC2 mutant did not suppress TNF gene expression compared to the medium control (Fig. S1B), indicating that the folC2 gene was important for human TNF suppression by L. reuteri 6475 in vitro.
Figure 2.
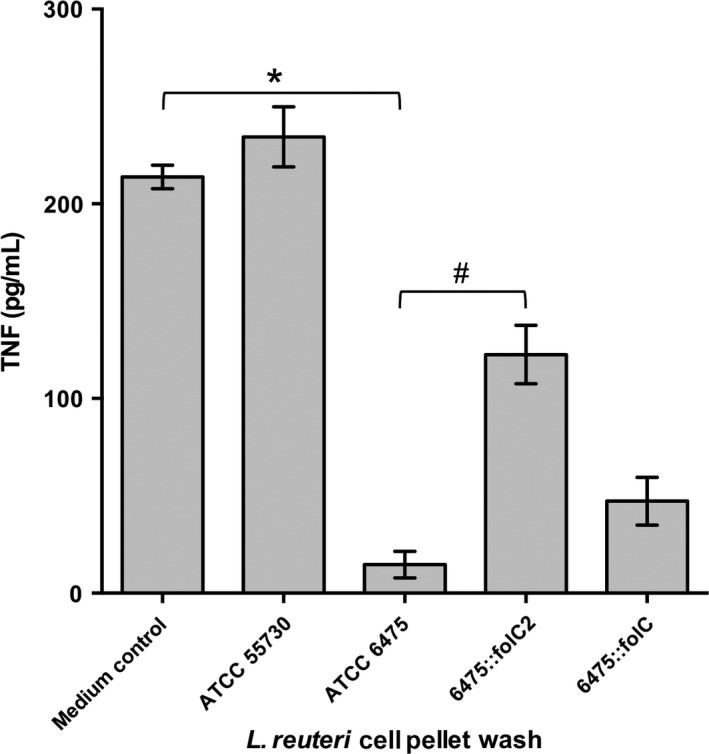
The folC2 gene contributed to the TNF‐suppressive ability of L. reuteri 6475. TFA‐treated cell pellets (normalized to cell pellet weight of 1 g) from various L. reuteri strains were tested for the ability to inhibit TNF by TLR2‐activated THP‐1 cells. THP‐1 cells were treated with 100 ng/mL PCK (TLR2 agonist) in the presence of L. reuteri for 3.5 h, and TNF production was monitored by ELISA. Wild‐type 6475 significantly inhibited TNF compared to medium control. The 6475::folC2 mutant yielded reduced capability to inhibit TNF production compared to wild‐type 6475. There was no significant difference between 6475 and 6475::folC strains. Data were analyzed with one‐way analysis of variance with Bonferroni's multiple comparison test correction, mean ± SD, n = 3, *P < 0.05 compared to medium control # P < 0.05 compared to 6475.TLR2, Toll‐like receptor 2; TNF, tumor necrosis factor.
FolC2 is necessary for 5,10‐CH = THF and 5,10‐CH = THF polyglutamate production and immunomodulation
Production of 5,10‐CH = THF and 5,10‐CH = THF polyglutamate in L. reuteri 6475::folC2 and 6475::folC was determined by MALDI‐TOF mass spectrometry. Analysis revealed the absence of both 5,10‐CH = THF and 5,10‐CH = THF polyglutamate in the 6475::folC2 mutant, which lacks the TNF‐inhibitory phenotype (Fig. 1D). Analysis of the 6475::folC mutant, which retains the TNF‐inhibitory phenotype demonstrated the presence of 5,10‐CH = THF (m/z 456.2), but not 5,10‐CH = THF polyglutamate (Fig. 1E). Strain 6475::folC inhibited TNF production (Fig. 2 and S1A), indicating that polyglutamylation of 5,10‐CH = THF was not necessary for TNF inhibition by L. reuteri.
Additional L. reuteri genes potentially involved in immunomodulation were identified by comparative transcriptomics between wild‐type and mutant L. reuteri strains 6475 and 55730, representing the two known human clades II and VI, respectively (Spinler et al. 2014) (Fig 3A). L. reuteri immunomodulins (TNF‐inhibitory factors) were only detected in strain 6475 cultures grown to stationary phase (Lin et al. 2008). The gene expression profile of strain 6475 in stationary phase (24 h) was compared to the same strain in early log phase (8 h), and 461 significantly up‐regulated genes (P < 0.05) were identified as genes potentially important for immunomodulin production (Saulnier et al. 2011). These 461 up‐regulated genes comprised the initial gene set of interest. L. reuteri 55730 does not inhibit TNF, allowing the gene set of interest to be restricted to up‐regulated genes that were unique to strain 6475 or not significantly up‐regulated in strain 55730 in stationary phase (402 total genes). A Gene Set Enrichment Analysis (GSEA) demonstrated that these 402 genes were significantly enriched in 6475::folC2 down‐regulated genes (P < 0.001, Fig. 3B) (Subramanian et al. 2005). Analysis stringency was increased by including only “core enrichment” genes, genes that contributed most to the GSEA enrichment score (168 genes). The list of immunoregulatory genes was further restricted by selecting only those genes not significantly down‐regulated in 6475::folC (24 h) versus wild‐type 6475 (24 h), since the folC mutant retained the ability to inhibit TNF production. The comparative transcriptomics analysis revealed a total of 125 potential immunoregulatory genes out of 1974 protein‐coding genes (Fig. 3A and 3C). These 125 genes (approximately 6% of the genome) were (1) significantly up‐regulated in strain 6475 grown to stationary phase, (2) not present or not significantly up‐regulated in strain 55730 grown to stationary phase, (3) significantly down‐regulated in 6475::folC2, (4) part of the GSEA “core enrichment,” and (5) not significantly down‐regulated in 6475::folC (Table S1). The 10 genes with the greatest fold change in 6475 stationary phase and 6475::folC2 versus wild‐type 6475 (stringent selection) are listed in Table 1. The gene encoding the histidine/histamine antiporter (hdcP) was included in this exclusive list of top 10 genes and the gene encoding histidine decarboxylase pyruvoyl type A (hdcA) was included in the final list of 125 genes following GSEA. The hdc gene cluster was identified by global gene expression studies including folate pathway mutants, and mechanistic studies probing the ability of Lactobacillus‐generated histamine to suppress the proinflammatory human cytokine TNF (Thomas et al. 2012).
Figure 3.
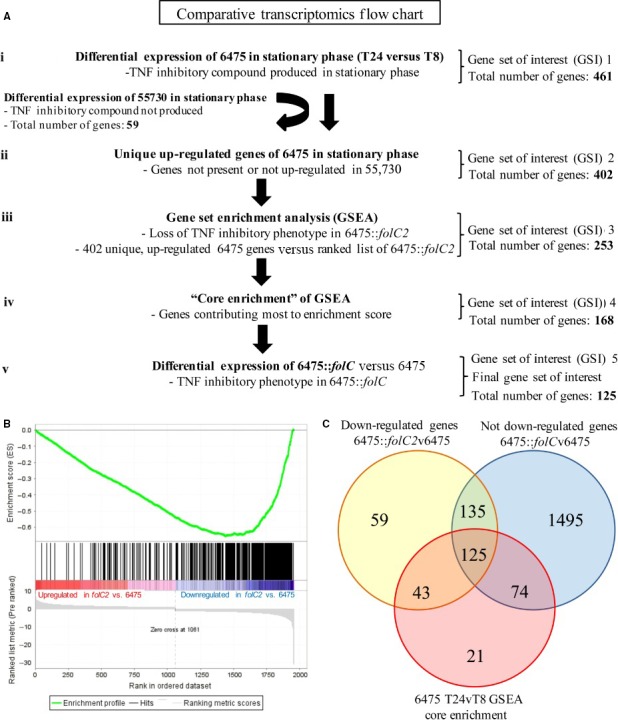
Comparative transcriptomics analysis revealed gene set encoding potential immunomodulins. (A) Flowchart of the comparative transcriptomic analysis employed in this study. N = 3 for each wild‐type and mutant L. reuteri strain that was included in the comparative analysis. (B) GSEA showed significant enrichment of strain 6475 genes up‐regulated in stationary phase in the down‐regulated genes of 6475::folC2. (C) The yellow circle indicates genes significantly down‐regulated in 6475::folC2 that were up‐regulated in wild‐type 6475 (stationary phase). The blue circle indicates genes not down‐regulated in 6475::folC that were up‐regulated in wild‐type 6475 (stationary phase). The red circle indicates genes that were included in the GSEA “core enrichment.” The overlap between these three gene sets revealed a final gene set of interest including 125 genes potentially involved in immunomodulin production by L. reuteri 6475. GSEA, A Gene Set Enrichment Analysis
Table 1.
Bacterial genes of interest
| Gene ID | Description | Functional Group |
|---|---|---|
| NT01LR1336 | Esterase | Central intermediary metabolism |
| NT01LR1981 | Lr1016 | Unclassified |
| NT01LR0282 | Conserved hypothetical protein | Hypothetical protein |
| NT01LR1905 | Conserved membrane protein | Cell envelope |
| NT01LR0849 | Hypothetical protein | Hypothetical protein |
| NT01LR0279 | Conserved hypothetical protein | Hypothetical protein |
| NT01LR1786 | Respiratory nitrate reductase, γ subunit | Energy metabolism |
| NT01LR0128 | Amidohydrolase family, putative | Central intermediary metabolism |
| NT01LR1242 | Histidine/histamine antiporter | Transport and binding proteins |
| NT01LR1034 | Hypothetical protein | Hypothetical protein |
1The differential expression via distance synthesis (DEDS) statistic represents the selected differential expression measures from the previous analyses as a multivariate point cloud on as many dimensions as there are input statistics. The scalar distance to the most extreme combination of these input statistics, following permutation, was the final metric by which genes are ranked and displayed here.
Expression of the hdc gene cluster and histamine production were diminished in L. reuteri 6475::folC2
Comparative transcriptomics analysis identified 125 genes that may be responsible for immunomodulation by L. reuteri 6475 (Table 1 and Table S1). Two identified genes, hdcA and hdcP, are L. reuteri genes known to be involved in conversion of histidine to histamine, a compound that suppresses TNF production by myeloid cells (Thomas et al. 2012). Quantitative RT‐PCR validated the comparative transcriptomics studies by confirming changes in gene expression in 6475::folC2 for three of the 10 genes listed in Table 1: hdcA, hdcP, and respiratory nitrate reductase gamma subunit (narI). All three genes had diminished gene expression in the 6475::folC2 mutant compared to wild‐type 6475 (Fig. 4A). Expression of the complete hdc gene cluster (hdcA, hdcB, and hdcP) was examined in wild‐type L. reuteri and 6475::folC2. Expression of the entire hdc gene cluster was increased in wild‐type L. reuteri 6475 grown in l‐histidine‐supplemented medium compared to unsupplemented medium (Fig. 4B), while expression was not significantly changed in 6475::folC2 in the presence of additional histidine (Fig. 4C). Histamine production by L. reuteri 6475 and 6475::folC2 strains was measured with a histamine‐specific ELISA. Production of histamine was significantly increased in L. reuteri 6475 when grown in medium supplemented with l‐histidine (Fig. 4D). The folC2 mutant produced significantly less histamine compared to wild‐type L. reuteri even in the presence of media supplementation with l‐histidine (Fig. 4D).
Figure 4.
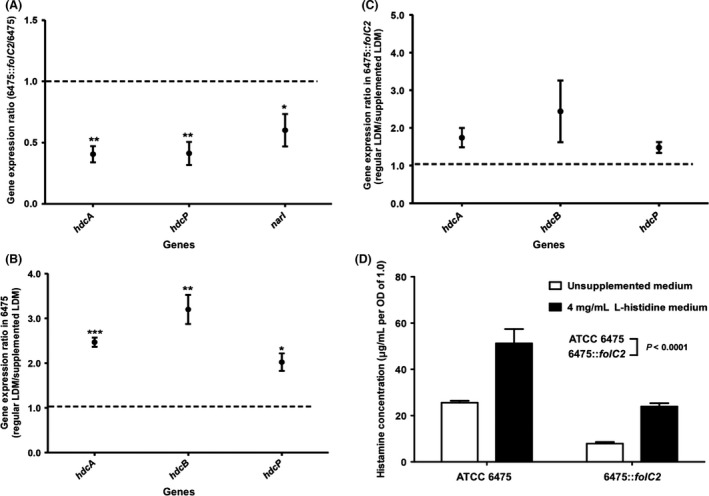
Inactivation of the folC2 gene resulted in repression of the hdc gene cluster. (A) Quantitative real‐time PCR yielded evidence of decreased expression of hdcA and hdcP genes in the L. reuteri 6475::folC2 mutant compared to wild‐type L. reuteri 6475. The gene narI, which was suggested by the comparative transcriptomics analysis to be down‐regulated in the folC2 mutant, was also repressed in 6475::folC2. Expression ratios of each gene (folC2 mutant vs. wild‐type) were calculated, and results represent the mean ± SD, n = 3, **P < 0.01, *P < 0.05 compared to the theoretical mean of 1.0. (B) Quantitative real‐time PCR demonstrated increased expression of all three hdc genes, hdcA, hdcB, and hdcP, when L. reuteri 6475 was grown in medium supplemented with l‐histidine compared to unsupplemented medium. Expression ratios of each gene (l‐histidine‐supplemented vs. unsupplemented) were calculated. Results represent the mean ± SD, n = 3, ***P < 0.005, **P < 0.01, *P < 0.05 compared to the theoretical mean of 1.0. (C) Quantitative real‐time PCR demonstrated no significant changes in expression of all three hdc genes when L. reuteri 6475::folC2 was grown in medium supplemented with l‐histidine compared to unsupplemented medium. (D). Inactivation of the folC2 gene resulted in decreased histamine production. Quantification of secreted L. reuteri‐derived histamine by a histamine‐specific ELISA demonstrated decreased histamine production in the folC2 mutant compared to wild‐type L. reuteri 6475 even when grown in l‐histidine‐supplemented medium. Data were analyzed by two‐way ANOVA. Results represent the mean ± SD, n = 3, P < 0.0001 compared to wild‐type 6475.
The folC2 gene contributes to suppression of intestinal inflammation by L. reuteri 6475 in vivo
To investigate whether the folC2 gene contributes to anti‐inflammatory effects in vivo, L. reuteri strain 6475 and 6475::folC2 bacterial cell‐free supernatants were tested in a TNBS‐induced mouse model of acute colitis. An 8‐week‐old female inbred Balb/c mice received two intraperitoneal (IP) injections of concentrated bacterial supernatant (18 h apart) followed by induction of colitis by TNBS instillation. Mice that received IP injections of the medium control and were challenged with TNBS (colitis‐positive mice) or phosphate‐buffered saline (PBS) (colitis‐negative mice) were studied as controls. Weight loss, which reflects the overall health status of mice, and a Wallace scoring system, which assesses the relative extent of macroscopic colon injury and inflammation, were measured to evaluate colitis severity. Serum amyloid A (SAA), an acute phase protein that serves as a plasma biomarker of intestinal mucosal inflammation in mice and correlates with severity of colitis, (de Villiers et al. 2000; Uhlar and Whitehead 1999) was quantified in plasma. Colitis‐negative mice showed no evidence of spontaneous disease (Fig. 5A–B). Colitis‐positive mice developed moderate colitis characterized by weight loss and macroscopic intestinal inflammation (Fig. 5A–B). As expected, these mice had significantly elevated concentrations of SAA (P < 0.001) compared to colitis‐negative mice (Fig. 5C). IP injection with L. reuteri 6475 supernatant reduced macroscopic inflammation (Fig. 5B). Treatment with L. reuteri 6475 supernatant also resulted in diminished weight loss 48 h post‐treatment compared to colitis‐positive mice (Fig. 5A), and reduced quantities of SAA in peripheral blood serum (Fig. 5C). Treatment with bacterial supernatants from the L. reuteri 6475::folC2 strain did not attenuate colitis as indicated by no significant change in body weight or macroscopic colonic inflammation (Fig. 5A–B). Additionally, the 6475::folC2 supernatant did not reduce SAA compared to colitis‐positive mice (Fig. 5C).
Figure 5.
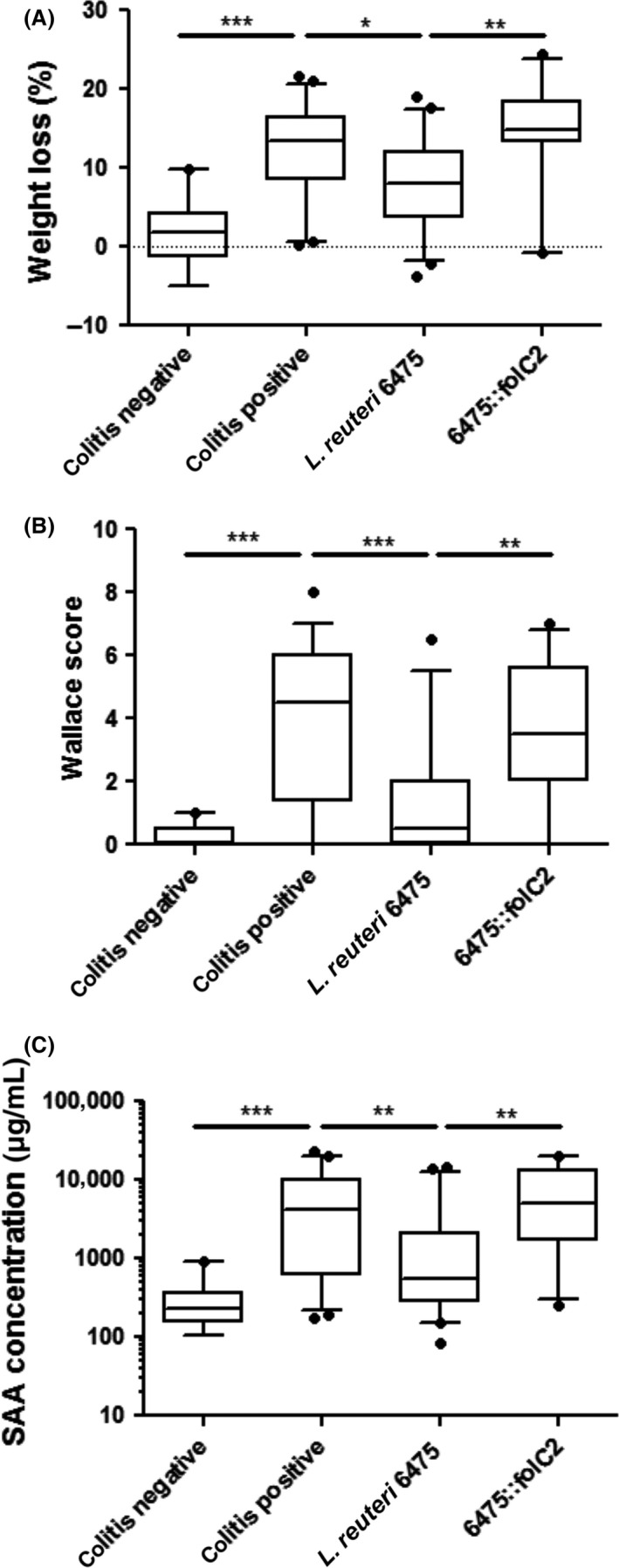
The folC2 gene was essential for colitis attenuation and anti‐inflammatory activity of L. reuteri 6475 in vivo. (A) Supernatant from L. reuteri 6475 significantly decreased weight loss in mice challenged with intrarectal TNBS, whereas 6475::folC2 did not have such effects. Differences in weight loss are shown as percent weight loss 48 h after induction of TNBS colitis. (B) Supernatant from L. reuteri 6475 significantly decreased colonic macroscopic injury in mice challenged with TNBS, whereas 6475::folC2 did not have such effects. Differences in colonic macroscopic injury are shown as Wallace score. Data presented as mean ± SEM. (C) Supernatant from L. reuteri 6475 significantly decreased plasma SAA concentrations in mice challenged with TNBS, whereas 6475::folC2 did not have such effects. Plasma levels of SAA, an indicator of inflammation, were measured by ELISA. N = 19, 50, 47, and 26 for colitis‐negative, colitis‐positive, ATCC 6475, and 6475::folC2, respectively. Statistical analyses were performed using GraphPad Prism (GraphPad Inc., La Jolla,CA). Data were presented using box and whisker plots showing the median and 5th and 95th percentiles. Statistical significance was assessed by nonparametric Kruskal–Wallis test. Differences between experimental groups are reported as mean fold difference ± SEM, ***P < 0.001, **P < 0.01, *P < 0.05.SAA, Serum amyloid A; TNBS, trinitrobenzene sulfonic acid.
Discussion
Probiotic Lactobacillus species secrete a variety of organic compounds that may regulate host immune responses (Lin et al. 2008). Our results show that specific strains of L. reuteri are capable of producing abundant 5,10‐CH = THF and 5,10‐CH = THF polyglutamate compounds. This report clarifies the contributions of the folC and folC2 genes to folate metabolism in Firmicutes. While detection of 5,10‐CH = THF and polyglutamylated 5,10‐CH = THF have been reported in other lactic acid bacteria cultures (Sybesma et al. 2003), this study demonstrates that production of these compounds in human‐derived L. reuteri strains correlates with the ability to inhibit TNF production in vitro. Mutagenesis of the L. reuteri folC2 gene and subsequent lack of 5,10‐CH = THF is associated with loss of TNF‐inhibitory activity in vitro. L. reuteri 6475 attenuates murine TNBS‐induced colitis and the folC2 gene is necessary for L. reuteri's protective and anti‐inflammatory activities in vivo. Additional studies demonstrate decreased L. reuteri hdc gene expression and histamine production in 6475::folC2, linking the pathways responsible for 5,10‐CH = THF production to histamine production and immunomodulation (Fig. 6A–B).
Figure 6.
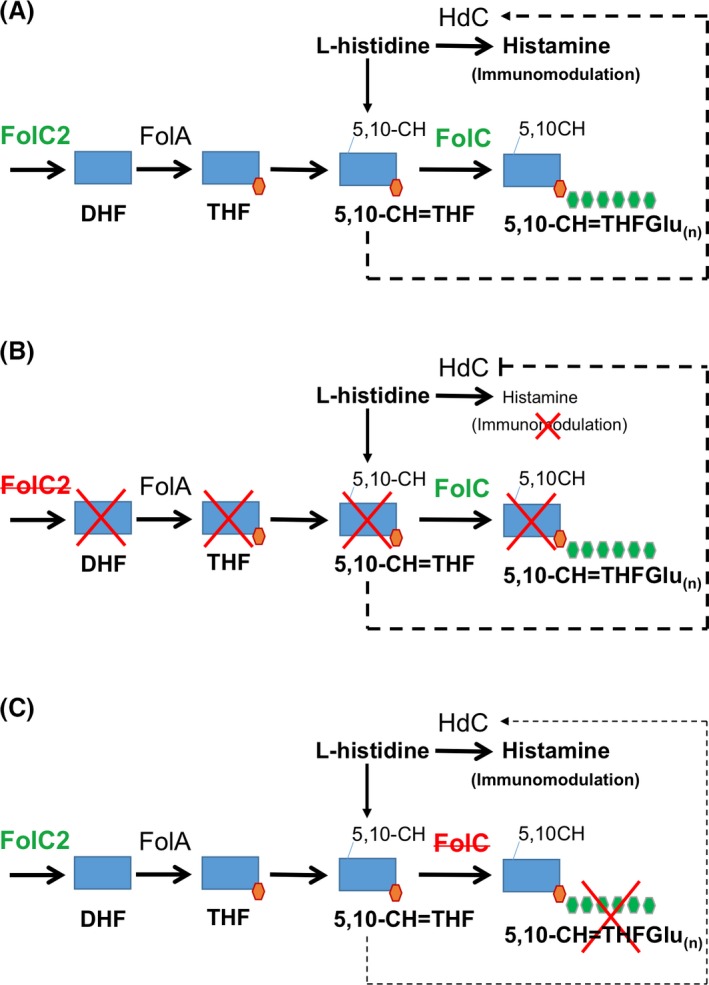
Folate synthesis appears to be linked to histamine production in L. reuteri 6475. (A) Folate synthesis in wild‐type L. reuteri 6475 with FolC2 necessary for production of dihydrofolate and FolC responsible for addition of a polyglutamate tail to tetrahydrofolate. Folate synthesis contributes to histamine production and the anti‐inflammatory effect of L. reuteri. (B) Inhibition of folate synthesis in L. reuteri 6475::folC2 leads to reduced histamine production and loss of anti‐inflammatory activity. (C) Inhibition of folate polyglutamylation in L. reuteri 6475::folC does not impact histamine production and L. reuteri anti‐inflammatory activity is preserved. Hdc, histidine decarboxylase.
This manuscript provides insights into genes and pathways involved in folate metabolism by human‐associated microbes. Varied chain length 5,10‐CH = THF polyglutamates (up to nine glutamate residues) were identified in cell wall‐associated compounds and secreted factors of TNF‐inhibitory L. reuteri strain 6475. Production of 5,10‐CH = THF and 5,10‐CH = THF polyglutamate of any chain length by strain 6475 required folC2. In contrast, 6475::folC maintained production of 5,10‐CH = THF but without the polyglutamate tail. Based on these results, folC2 appears to be necessary for production of 5,10‐CH = THF, and folC appears to encode the enzyme involved in polyglutamylation of 5,10‐CH = THF in L. reuteri. To date, activities of FolC2 have not been well characterized in lactobacilli. FolC2 and FolC are not orthologs and are included in different Clusters of Orthologous Groups (COGs), COG1478 and COG0285, respectively (de Crecy‐Lagard 2014). However, both FolC and FolC2 are considered to be bifunctional enzymes with dihydrofolate synthase and folylpolyglutamate synthase activity (Bognar and Shane 1986; Toy and Bognar 1990; Kimlova et al. 1991; Murata et al. 2000). Dihydrofolate synthase is needed for the synthesis of dihydrofolate by coupling glutamate with dihydropteroate. Conversion of dihydrofolate to tetrahydrofolate (THF) is performed as an intermediate reaction by dihydrofolate reductase, which is encoded by the folA gene (Fig. 6A). Presence of folA is ubiquitous in Lactobacillus strains (Magnusdottir et al. 2015), and consequently should not play a major role in differential folate production in lactobacilli. Folylpolyglutamate synthase adds a glutamyl tail to THF (Fig. 6A). Many lactobacilli possess folC in their genome, however, prevalence of folC2 is restricted to fewer species (PATRIC database, https://www.patricbrc.org/). Whether or not these genes are fully functional in all species and strains is currently unknown. FolC2 is found in human‐derived Lactobacillus strains (L. reuteri, L. iners), as well as in lactobacilli commonly found in fermented foods (L. plantarum, L. delbrueckii, L. sakei, L. pentosus, L. buchneri, and L. hilgardii). These genes have been poorly characterized at the functional level and more research is needed in this area.
The 5,10‐CH = THF compound that we detected in L. reuteri 6475 can be synthesized in two successive reactions from THF (Fig. 6A). THF is converted to 10‐formyl tetrahydrofolate (10‐CHO = THF) via formate tetrahydroligase. 10‐CHO = THF is then transformed into 5,10‐CH = THF by the bifunctional enzyme methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase. The genes encoding these two enzymes are present in the L. reuteri strains included in this report. The conversion of 10‐CHO = THF into 5,10‐CH = THF also occurs spontaneously at a very low pH (Eto and Krumdieck 1980; Arnold and Reilly 2000), however, our data suggest that 5,10‐CH = THF was synthesized by L. reuteri.
The generation of tetrahydrofolate compounds by L. reuteri strains contributes to immunoregulation (TNF suppression) through interconnected metabolic pathways involved in folate and histidine metabolism. Even if lactobacilli can produce abundant, long‐chain 5,10‐CH = THF, the TNF‐inhibitory phenotype relies primarily on the ability of specific strains to convert l‐histidine to histamine and several histamine metabolites (Spinler et al. 2014). Folate compounds mediate the interconversion of serine and glycine, and play a role in histidine biosynthesis and catabolism (Broquist 1957). The folC2 gene may exert its immunomodulatory effects by regulating the production of histamine, a known L. reuteri immunomodulin (Figs 6A and B). The folC gene plays a role in adding the polyglutamate tail to 5,10‐CH = THF, but unlike folC2, this gene does not affect the production of histamine nor the ability of L. reuteri to suppress production of proinflammatory cytokines (Fig. 6C). Histamine production as well as the expression of genes involved in histamine production were decreased in the 6475::folC2 mutant, and parallel changes in 5,10‐CH = THF and histamine production suggest that folate metabolism and specifically the folC2 gene may be important for the conversion of l‐histidine to histamine. The synchronous changes in gene expression affecting histamine and folate metabolism link these two pathways, contributing to the immunomodulatory phenotype of L. reuteri. Further studies will continue to elucidate biologic connections between histamine, a known TNF‐inhibitory factor, l‐histidine metabolism, and other compounds produced by L. reuteri 6475 including 5,10‐CH = THF.
Tetrahydrofolic acid and its derivatives (5,10‐CH = THF, 10‐CHO = THF) are essential cofactors that facilitate the transfer of single‐carbon units from donor molecules into important biosynthetic pathways leading to methionine, purine, and pyrimidine biosynthesis (Ragsdale 2008; Fowler 2001). Histidine is a known end product of purine biosynthesis (Allen et al. 2002). Additionally, the oxidation of amino acids, including histidine, has been linked to the role of folate in generating reducing agents, NADH and NADPH, in eukaryotes (Brosnan et al. 2015). Our gene expression data suggest that this function of folate reviewed by Brosnan et al. may also occur in prokaryotes. In the folC2 mutant, there is down‐regulation of several genes encoding enzymes that require NADPH as a substrate (i.e., thioredoxin reductase, nitrate reductase, methionine‐S‐reductase). These enzymes are involved in oxidative stress response, making it possible that the redox state of L. reuteri plays a role in its immunosuppressive function. Preliminary studies demonstrate that growth of L. reuteri in anaerobic versus aerobic conditions (different redox states) affects folate production and immunomodulation. The studies presented here were performed under standard anaerobic conditions. When L. reuteri is grown under aerobic conditions, the composition of folate compounds is different and the ability of L. reuteri to inhibit TNF production is reduced (data not shown). Further studies are needed to understand the metabolic pathways of L. reuteri and how they are regulated to produce histamine and 5,10‐CH = THF, resulting in the immunosuppression phenotype of L. reuteri.
Secreted factors produced by wild‐type L. reuteri 6475 can significantly ameliorate the intestinal pathology and inflammation in a TNBS‐induced mouse model of colitis. Previous studies demonstrated that delivery of pharmacological agents via the IP route is an efficient and successful method, especially when the target is within the peritoneal cavity (Chaudhary et al. 2010). We administered L. reuteri‐derived secreted factors in a concentrated form inside the peritoneal cavity and observed significantly diminished ulceration, local inflammation, and mucosal biomarkers of inflammation. Our study demonstrates that secreted factors produced by L. reuteri are important for preventing colitis, enabling alternative strategies that could supplant or supplement delivery of intact viable bacteria or bacterial colonization in the gut. These findings have important implications for future human clinical trials. Studies of various L. reuteri mutants in this model have provided insights into candidate genes that may be essential for suppression of intestinal inflammation by the model commensal L. reuteri strain 6475. In this study, we demonstrated the role of the Lactobacillus gene, folC2, as a potential key regulatory gene in the microbiome involved in protection from or amelioration of colitis. Our data support the conclusion that L. reuteri strain 6475 protects mammals from severe intestinal inflammation by production and secretion of potent immunosuppressive compounds locally in the gut lumen.
Deficiencies in bacterial‐derived micronutrients such as folate can lead to immune dysregulation highlighting the interdependence between diet, commensal bacteria, and the host mucosal immune system (Spencer and Belkaid 2012). Diminished TNF production by M1 macrophages has been observed when these cells were grown in the presence of folic acid (Samaniego, 2014). Immune cells of the GI tract such as regulatory T cells express high levels of the folate receptor 4 (FR4) (Yamaguchi et al. 2007). Interestingly, it has been demonstrated that folic acid can prevent TNBS‐induced colitis by maintaining regulatory T cells via suppression of apoptosis and subsequent prevention of colonic inflammation. Mice fed a folic acid‐free diet had significantly increased colonic inflammation, weight loss, and mortality rate after exposure to TNBS compared to mice fed a normal chow diet (Kinoshita et al. 2012). Protection against colitis could be restored if FR4‐expressing regulatory T cells were transferred into folic acid‐free diet mice prior to induction of colitis (Kinoshita et al. 2012). Folate is also able to activate mucosal‐associated invariant T cells (MAIT) in the GI tract by acting as antigens on infected cells (Chua and Hansen 2012; Kjer‐Nielsen et al. 2012). In our TNBS‐induced colitis studies, decreased production of 5,10‐CH = THF by L. reuteri 6475::folC2 may contribute to the diminished protective effect by reduced TNF suppression as well as loss or dysfunction of regulatory T cells in the colon.
A comprehensive understanding of gene networks and gene regulation in beneficial gut microbes is critical to understanding the interaction of bacterial metabolites with the host. This knowledge may enhance the ability of the scientific community to select or engineer commensal gut bacterial strains that can suppress mucosal inflammation. Combining a genome‐scale metabolic model of Bacteroides thetaiotaomicron (iAH991) with a mouse metabolic model demonstrated the essential host–microbe symbiosis that occurs in the GI tract (Heinken et al. 2013). Modeling metabolic interactions between a gut microbe and its host has enabled identification of metabolites that are exchanged between the two organisms, influences on growth fitness for both organisms, and the ability of commensal bacteria to rescue lethal enzyme deficiencies in the host (Heinken et al. 2013). The recent identification of multiple biological pathways and genes involved in suppression of proinflammatory cytokine production in L. reuteri provides opportunities for combining such discoveries in future therapies and disease prevention strategies. For example, the potential dietary contribution of the amino acid l‐histidine to bacterial histamine generation in combination with methods to enhance production of tetrahydrofolic acid compounds may result in nutritional and immunomodulatory benefits for the mammalian host. As metabolic pathways and modules become linked together in human microbiome research (Hmp 2012), nutritional and medical interventions may promote healthy whole body metabolism and immune function in partnership with the gut microbiome. Future probiotic strategies will benefit from the identification of biochemical compounds and genes required for healthy intestinal physiology and probiotic‐mediated immunomodulation.
Conflict of Interest
JV receives unrestricted research support from BioGaia AB.
Supporting information
Figure S1. FolC2 was necessary for suppression of TNF production at protein and mRNA levels. (A) L. reuteri cell‐free supernatants (normalized to an OD600 of 1.5) were tested for the ability to inhibit TNF production by TLR2‐activated THP‐1 cells. THP‐1 cells were treated with 100 ng/mL PCK (TLR2 agonist) in the presence of L. reuteri for 3.5 h and TNF production was monitored by ELISA. As seen with the cell pellets, wild‐type 6475 significantly inhibited TNF compared to medium control. The 6475::folC2 mutant yielded significantly reduced ability to inhibit TNF production compared to wild‐type 6475. There was no significant difference between 6475 and 6475::folC in terms of effects on human TNF production. Data were analyzed with one‐way analysis of variance with Bonferroni's multiple comparison test correction, mean ± SD, n = 3, *P < 0.05 compared to medium control # P < 0.05 compared to 6475. (B) TNF gene expression was determined in THP‐1 cells treated with a TLR2 agonist plus medium control, 6475, or 6475::folC2 cell‐free supernatants. Quantitative real‐time PCR demonstrated down‐regulation of human TNF gene expression by L. reuteri strain 6475. No significant effects on human TNF gene expression were seen when THP‐1 cells were treated with 6475::folC2. Gene expression data were normalized using five housekeeping genes, b2 m, hprt1, rpl13A, gapdh, and actb. Expression ratios of tnf (L. reuteri strain/medium control) were calculated, and results represent the mean ± SD, n = 3, *P < 0.05 compared to the theoretical mean of 1.0.
Table S1. Final Gene Set of Interest – 125 genes potentially involved in immunomodulation by wild‐type 6475.
Table S2. Bacterial strains, vectors, and primers used in this study.
Acknowledgments
This work was supported by the National Institutes of Health to JV, including the National Institute of Diabetes, Digestive and Kidney Diseases (R01 DK065075), Texas Medical Center Digestive Disease Center (P30 DK56338), National Cancer Institute (U01 CA170930), and National Center for Complementary and Alternative Medicine (R01 AT004326). Additionally, the use of facilities at City of Hope was partially supported by the National Institutes of Health, National Cancer Institute (P30 CA33572).We thank Eamonn Connolly (BioGaia AB, Stockholm) for providing the L. reuteri strains, Oscar Ayala and Sujata Gosh for culturing the strains, Fan Zhang for his efforts in designing the TNBS mouse model, and Toni‐Ann Mistretta and Ruth Ann Luna for assistance with the statistical analysis of the data.
MicrobiologyOpen 2016; 5(5): 802–818
References
- Allen, S. , Zilles J. L., and Downs D. M.. 2002. Metabolic flux in both the purine mononucleotide and histidine biosynthetic pathways can influence synthesis of the hydroxymethyl pyrimidine moiety of thiamine in Salmonella enterica . J. Bacteriol. 184:6130–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, R. J. , and Reilly J. P.. 2000. Observation of tetrahydrofolylpolyglutamic acid in bacteria cells by matrix‐assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 281:45–54. [DOI] [PubMed] [Google Scholar]
- Bibiloni, R. , Fedorak R. N., Tannock G. W., Madsen K. L., Gionchetti P., Campieri M., et al., 2005. VSL#3 probiotic‐mixture induces remission in patients with active ulcerative colitis. Am. J. Gastroenterol. 100:1539–1546. [DOI] [PubMed] [Google Scholar]
- Bleau, C. , Monges A., Rashidan K., Laverdure J. P., M. Lacroix , Van Calsteren M. R., et al., 2010. Intermediate chains of exopolysaccharides from Lactobacillus rhamnosus RW‐9595M increase IL‐10 production by macrophages. J. Appl. Microbiol. 108:666–675. [DOI] [PubMed] [Google Scholar]
- Bognar, A. L. , and Shane B.. 1986. Bacterial folylpoly(gamma‐glutamate) synthase‐dihydrofolate synthase. Methods Enzymol. 122:349–359. [DOI] [PubMed] [Google Scholar]
- Broquist, H. P. 1957. Evidence for the involvement of folic acid in histidine synthesis in microorganisms. Arch. Biochem. Biophys. 70:210–216. [DOI] [PubMed] [Google Scholar]
- Brosnan, M. E. , MacMillan L., Stevens J. R., and Brosnan J. T.. 2015. Division of labour: how does folate metabolism partition between one‐carbon metabolism and amino acid oxidation? Biochem. J. 472:135–146. [DOI] [PubMed] [Google Scholar]
- Chaudhary, K. , Haddadin S., Nistala R., and Papageorgio C.. 2010. Intraperitoneal drug therapy: an advantage. Curr. Clin. Pharmacol. 5:82–88. [DOI] [PubMed] [Google Scholar]
- Chebli, J. M. , Gaburri P. D., Chebli L. A., da Rocha Ribeiro T. C., Pinto A. L., Ambrogini Junior O., et al. 2014. A guide to prepare patients with inflammatory bowel diseases for anti‐TNF‐alpha therapy. Med. Sci. Monit. 20:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, H. R. , Frokiaer H., and Pestka J. J.. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 168:171–178. [DOI] [PubMed] [Google Scholar]
- Chua, W. J. , and Hansen T. H.. 2012. Immunology: vitamins prime immunity. Nature 491:680–681. [DOI] [PubMed] [Google Scholar]
- de Crecy‐Lagard, V. 2014. Variations in metabolic pathways create challenges for automated metabolic reconstructions: examples from the tetrahydrofolate synthesis pathway. Comput. Struct. Biotechnol. J. 10:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto, I. , and Krumdieck C. L.. 1980. Determination of three different pools of reduced one‐carbon‐substituted folates. 1. A study of the fundamental chemical reactions. Anal. Biochem. 109:167–184. [DOI] [PubMed] [Google Scholar]
- FAO/WHO . 2006. Probiotics in Food: health and nutritional properties and guidelines for evaluation. Report of the Joint Food and Agriculture Organization (FAO) of the United Nations / World Health Organization (WHO) Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Report of the Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food.
- Foligne, B. , Nutten S., Steidler L., Dennin V., Goudercourt D., Mercenier A., et al. 2006. Recommendations for improved use of the murine TNBS‐induced colitis model in evaluating anti‐inflammatory properties of lactic acid bacteria: technical and microbiological aspects. Dig. Dis. Sci. 51:390–400. [DOI] [PubMed] [Google Scholar]
- Fowler, B. 2001. The folate cycle and disease in humans. Kidney Int. Suppl. 78:S221–229. [DOI] [PubMed] [Google Scholar]
- Giaffer, M. H. , Holdsworth C. D., and Duerden B. I.. 1991. The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J. Med. Microbiol. 35:238–243. [DOI] [PubMed] [Google Scholar]
- Grangette, C. , Nutten S., Palumbo E., Morath S., C. Hermann , Dewulf J., et al., 2005. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl Acad. Sci. USA 102:10321–10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P. , Andrew H., Kirschner B. S., and Guandalini S.. 2000. Is Lactobacillus GG helpful in children with Crohn's disease? Results of a preliminary, open‐label study. J. Pediatr. Gastroenterol. Nutr. 31:453–457. [DOI] [PubMed] [Google Scholar]
- Heinken, A. , Sahoo S., Fleming R. M., and Thiele I.. 2013. Systems‐level characterization of a host‐microbe metabolic symbiosis in the mammalian gut. Gut. Microbes 4:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata, P. , Gao C., Pflughoeft K. J., Thomas C. M., Saulnier D. M., Spinler J. K., et al. 2013. Lactobacillus reuteri‐specific immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri . J. Bacteriol. 195:5567–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmp, H. M. P. C. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoentjen, F. , and van Bodegraven A. A.. 2009. Safety of anti‐tumor necrosis factor therapy in inflammatory bowel disease. World J. Gastroenterol. 15:2067–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holma, R. , Salmenpera P., Lohi J., Vapaatalo H., and R. Korpela . 2001. Effects of Lactobacillus rhamnosus GG and Lactobacillus reuteri R2LC on acetic acid‐induced colitis in rats. Scand. J. Gastroenterol. 36:630–635. [DOI] [PubMed] [Google Scholar]
- Jauregui‐Amezaga, A. , Turon F., Ordas I., Gallego M., F. Feu , Ricart E., et al. 2013. Risk of developing tuberculosis under anti‐TNF treatment despite latent infection screening. J. Crohns. Colitis 7:208–212. [DOI] [PubMed] [Google Scholar]
- Jones, S. E. , and Versalovic J.. 2009. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti‐inflammatory factors. BMC Microbiol. 9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. G. , Kim N. R., Gim M. G., Lee J. M., Lee S. Y., Ko M. Y., et al., 2008. Lipoteichoic acid isolated from Lactobacillus plantarum inhibits lipopolysaccharide‐induced TNF‐alpha production in THP‐1 cells and endotoxin shock in mice. J. Immunol. 180:2553–2561. [DOI] [PubMed] [Google Scholar]
- Kimlova, L. J. , Pyne C., Keshavjee K., Huy J., G. Beebakhee , and Bognar A. L.. 1991. Mutagenesis of the folC gene encoding folylpolyglutamate synthetase‐dihydrofolate synthetase in Escherichia coli . Arch. Biochem. Biophys. 284:9–16. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M. , Kayama H., Kusu T., Yamaguchi T., J. Kunisawa , Kiyono H., et al., 2012. Dietary folic acid promotes survival of Foxp3 + regulatory T cells in the colon. J. Immunol. 189:2869–2878. [DOI] [PubMed] [Google Scholar]
- Kjer‐Nielsen, L. , Patel O., Corbett A. J., Le Nours J., B. Meehan , Liu L., et al., 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491:717–723. [DOI] [PubMed] [Google Scholar]
- Krutchinsky, A. N. , Kalkum M., and Chait B. T.. 2001. Automatic identification of proteins with a MALDI‐quadrupole ion trap mass spectrometer. Anal. Chem. 73:5066–5077. [DOI] [PubMed] [Google Scholar]
- Lin, Y. P. , Thibodeaux C. H., Pena J. A., Ferry G. D., and Versalovic J.. 2008. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c‐Jun. Inflamm. Bowel Dis. 14:1068–1083. [DOI] [PubMed] [Google Scholar]
- Liu, Y. W. , Su Y. W., Ong W. K., Cheng T. H., and Tsai Y. C.. 2011. Oral administration of Lactobacillus plantarum K68 ameliorates DSS‐induced ulcerative colitis in BALB/c mice via the anti‐inflammatory and immunomodulatory activities. Int. Immunopharmacol. 11:2159–2166. [DOI] [PubMed] [Google Scholar]
- Livingston, M. , Loach D., Wilson M., Tannock G. W., and Baird M.. 2010. Gut commensal Lactobacillus reuteri 100‐23 stimulates an immunoregulatory response. Immunol. Cell Biol. 88:99–102. [DOI] [PubMed] [Google Scholar]
- Magnusdottir, S. , Ravcheev D., de Crecy‐Lagard V., and I. Thiele . 2015. Systematic genome assessment of B‐vitamin biosynthesis suggests co‐operation among gut microbes. Front. Genet. 6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin, E. , Duwat P., Hege T., Ehrlich D., and Gruss A.. 1992. New thermosensitive plasmid for gram‐positive bacteria. J. Bacteriol. 174:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechoud, M. A. , Mateos M. V., de Valdez G. F., Villena J., Salvador G. A., and Rodriguez A. V.. 2012. Lactobacillus reuteri CRL1098 soluble factors modulate tumor necrosis factor alpha production in peripheral blood mononuclear cells: involvement of lipid rafts. Int. Immunopharmacol. 14:446–453. [DOI] [PubMed] [Google Scholar]
- Moller, P. L. , Paerregaard A., Gad M., Kristensen N. N., and Claesson M. H.. 2005. Colitic scid mice fed Lactobacillus spp. show an ameliorated gut histopathology and an altered cytokine profile by local T cells. Inflamm. Bowel Dis. 11:814–819. [DOI] [PubMed] [Google Scholar]
- Morris, G. P. , Beck P. L., Herridge M. S., Depew W. T., Szewczuk M. R., and Wallace J. L.. 1989. Hapten‐induced model of chronic inflammation and ulceration in the rat colon. Gastroenterology 96:795–803. [PubMed] [Google Scholar]
- Murata, T. , Bognar A. L., Hayashi T., Ohnishi M., K. Nakayama , and Terawaki Y.. 2000. Molecular analysis of the folC gene of Pseudomonas aeruginosa . Microbiol. Immunol. 44:879–886. [DOI] [PubMed] [Google Scholar]
- Pena, J. A. , Li S. Y., Wilson P. H., Thibodeau S. A., A. J. Szary , and Versalovic J.. 2004. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl. Environ. Microbiol. 70:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena, J. A. , Rogers A. B., Ge Z., Ng V., Li S. Y., Fox J. G., et al. 2005. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus‐induced inflammatory bowel disease in interleukin‐10‐deficient mice. Infect. Immun. 73:912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peran, L. , Sierra S., Comalada M., Lara‐Villoslada F., E. Bailon , Nieto A., et al., 2007. A comparative study of the preventative effects exerted by two probiotics, Lactobacillus reuteri and Lactobacillus fermentum, in the trinitrobenzenesulfonic acid model of rat colitis. Br. J. Nutr. 97:96–103. [DOI] [PubMed] [Google Scholar]
- Peyrin‐Biroulet, L. 2010. Anti‐TNF therapy in inflammatory bowel diseases: a huge review. Minerva Gastroenterol. Dietol. 56:233–243. [PubMed] [Google Scholar]
- Ragsdale, S. W. 2008. Catalysis of methyl group transfers involving tetrahydrofolate and B(12). Vitam. Horm. 79:293–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43–53. [PubMed] [Google Scholar]
- Rossi, M. , Amaretti A., and Raimondi S.. 2011. Folate production by probiotic bacteria. Nutrients 3:118–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaniego, R. , Palacios B. S., Domiguez‐Soto A., Vidal C., A. Salas , T. Matsuyama , et al. 2014. Macrophage uptake and accumulation of folates are polarization‐dependent in vitro and in vivo and are regulated by activin A. J. Leukoc. Biol. 95:797–808. [DOI] [PubMed] [Google Scholar]
- Santos, F. , Wegkamp A., de Vos W. M., Smid E. J., and J. Hugenholtz . 2008. High‐Level folate production in fermented foods by the B12 producer Lactobacillus reuteri JCM1112. Appl. Environ. Microbiol. 74:3291–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satish Kumar, C. S. , Kondal Reddy K., Reddy A. G., A. Vinoth , Ch S. R., Boobalan G., et al. 2015. Protective effect of Lactobacillus plantarum 21, a probiotic on trinitrobenzenesulfonic acid‐induced ulcerative colitis in rats. Int. Immunopharmacol. 25:504–510. [DOI] [PubMed] [Google Scholar]
- Saulnier, D. M. , Santos F., Roos S., Mistretta T. A., J. K. Spinler , Molenaar D., et al., 2011. Exploring metabolic pathway reconstruction and genome‐wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS ONE 6:e18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, O. , Petersson J., Phillipson M., Perry M., Roos S., and Holm L.. 2009. Lactobacillus reuteri prevents colitis by reducing P‐selectin‐associated leukocyte‐ and platelet‐endothelial cell interactions. Am. J. Physiol. Gastrointest. Liver Physiol. 296:G534–542. [DOI] [PubMed] [Google Scholar]
- Smits, H. H. , Engering A., van der Kleij D., de Jong E. C., Schipper K., van Capel T. M., et al., 2005. Selective probiotic bacteria induce IL‐10‐producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell‐specific intercellular adhesion molecule 3‐grabbing nonintegrin. J. Allergy Clin. Immunol. 115:1260–1267. [DOI] [PubMed] [Google Scholar]
- Spencer, S. P. , and Belkaid Y.. 2012. Dietary and commensal derived nutrients: shaping mucosal and systemic immunity. Curr. Opin. Immunol. 24:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinler, J. K. , Sontakke A., Hollister E. B., Venable S. F., Oh P. L., Balderas M. A., et al., 2014. From prediction to function using evolutionary genomics: human‐specific ecotypes of Lactobacillus reuteri have diverse probiotic functions. Genome Biol. Evol. 6:1772–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., et al., 2005. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc. Natl Acad. Sci. USA 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sybesma, W. , Starrenburg M., Tijsseling L., Hoefnagel M. H., and Hugenholtz J.. 2003. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl. Environ. Microbiol. 69:4542–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C. M. , and Versalovic J.. 2010. Probiotics‐host communication: modulation of signaling pathways in the intestine. Gut. Microbes 1:148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C. M. , Hong T., van Pijkeren J. P., Hemarajata P., Trinh D. V., Hu W., et al., 2012. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 7:e31951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toy, J. , and Bognar A. L.. 1990. Cloning and expression of the gene encoding Lactobacillus casei folylpoly‐gamma‐glutamate synthetase in Escherichia coli and determination of its primary structure. J. Biol. Chem. 265:2492–2499. [PubMed] [Google Scholar]
- Triantafillidis, J. K. , Papalois A. E., Parasi A., Anagnostakis E., Burnazos S., Gikas A., et al., 2005. Favorable response to subcutaneous administration of infliximab in rats with experimental colitis. World J. Gastroenterol. 11:6843–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlar, C. M. , and Whitehead A. S.. 1999. Serum amyloid A, the major vertebrate acute‐phase reactant. Eur. J. Biochem. 265:501–523. [DOI] [PubMed] [Google Scholar]
- Ungar, B. , Chowers Y., Yavzori M., Picard O., Fudim E., Har‐Noy O., et al., 2014. The temporal evolution of antidrug antibodies in patients with inflammatory bowel disease treated with infliximab. Gut 63:1258–1264. [DOI] [PubMed] [Google Scholar]
- Vigsnaes, L. K. , Brynskov J., Steenholdt C., Wilcks A., and Licht T. R.. 2012. Gram‐negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef. Microbes 3:287–297. [DOI] [PubMed] [Google Scholar]
- de Villiers, W. J. , Varilek G. W., de Beer F. C., Guo J. T., and Kindy M. S.. 2000. Increased serum amyloid a levels reflect colitis severity and precede amyloid formation in IL‐2 knockout mice. Cytokine 12:1337–1347. [DOI] [PubMed] [Google Scholar]
- Wall, T. , Bath K., Britton R. A., Jonsson H., Versalovic J., and Roos S.. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall‐altering esterase. Appl. Environ. Microbiol. 73:3924–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J. , Chagnaud P., Tannock G. W., Loach D. M., F. Dal Bello , Jenkinson H. F., et al., 2005. A high‐molecular‐mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J. , Britton R. A., and Roos S.. 2011. Host‐microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl Acad. Sci. USA 108(Suppl 1):4645–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T. , Hirota K., Nagahama K., Ohkawa K., T. Takahashi , Nomura T., et al. 2007. Control of immune responses by antigen‐specific regulatory T cells expressing the folate receptor. Immunity 27:145–159. [DOI] [PubMed] [Google Scholar]
- Yang, Y. H. , Xiao Y., and Segal M. R.. 2005. Identifying differentially expressed genes from microarray experiments via statistic synthesis. Bioinformatics 21:1084–1093. [DOI] [PubMed] [Google Scholar]
- Yasuda, E. , Serata M., and Sako T.. 2008. Suppressive effect on activation of macrophages by Lactobacillus casei strain Shirota genes determining the synthesis of cell wall‐associated polysaccharides. Appl. Environ. Microbiol. 74:4746–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zella, G. C. , Hait E. J., Glavan T., Gevers D., Ward D. V., Kitts C. L., et al. 2011. Distinct microbiome in pouchitis compared to healthy pouches in ulcerative colitis and familial adenomatous polyposis. Inflamm. Bowel Dis. 17:1092–1100. [DOI] [PubMed] [Google Scholar]
- Zhao, H. M. , Huang X. Y., Zuo Z. Q., Pan Q. H., Ao M. Y., Zhou F., et al., 2013. Probiotics increase T regulatory cells and reduce severity of experimental colitis in mice. World J. Gastroenterol. 19:742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. FolC2 was necessary for suppression of TNF production at protein and mRNA levels. (A) L. reuteri cell‐free supernatants (normalized to an OD600 of 1.5) were tested for the ability to inhibit TNF production by TLR2‐activated THP‐1 cells. THP‐1 cells were treated with 100 ng/mL PCK (TLR2 agonist) in the presence of L. reuteri for 3.5 h and TNF production was monitored by ELISA. As seen with the cell pellets, wild‐type 6475 significantly inhibited TNF compared to medium control. The 6475::folC2 mutant yielded significantly reduced ability to inhibit TNF production compared to wild‐type 6475. There was no significant difference between 6475 and 6475::folC in terms of effects on human TNF production. Data were analyzed with one‐way analysis of variance with Bonferroni's multiple comparison test correction, mean ± SD, n = 3, *P < 0.05 compared to medium control # P < 0.05 compared to 6475. (B) TNF gene expression was determined in THP‐1 cells treated with a TLR2 agonist plus medium control, 6475, or 6475::folC2 cell‐free supernatants. Quantitative real‐time PCR demonstrated down‐regulation of human TNF gene expression by L. reuteri strain 6475. No significant effects on human TNF gene expression were seen when THP‐1 cells were treated with 6475::folC2. Gene expression data were normalized using five housekeeping genes, b2 m, hprt1, rpl13A, gapdh, and actb. Expression ratios of tnf (L. reuteri strain/medium control) were calculated, and results represent the mean ± SD, n = 3, *P < 0.05 compared to the theoretical mean of 1.0.
Table S1. Final Gene Set of Interest – 125 genes potentially involved in immunomodulation by wild‐type 6475.
Table S2. Bacterial strains, vectors, and primers used in this study.


