Abstract
Background:
Histone deacetylase inhibitors (HDACis) like vorinostat are promising radiosensitisers in prostate cancer, but their effect under hypoxia is not known. We investigated gene expression associated with radiosensitisation of normoxic and hypoxic prostate cancer cells by vorinostat.
Methods:
Cells were exposed to vorinostat under normoxia or hypoxia and subjected to gene expression profiling before irradiation and clonogenic survival analysis.
Results:
Pretreatment with vorinostat led to radiosensitisation of the intrinsically radioresistant DU 145 cells, but not the radiosensitive PC-3 and 22Rv1 cells, and was independent of hypoxia status. Knockdown experiments showed that the sensitisation was not caused by repression of hypoxia-inducible factor HIF1 or tumour protein TP53. Global deregulation of DNA repair and chromatin organisation genes was associated with radiosensitisation under both normoxia and hypoxia. A radiosensitisation signature with expression changes of 56 genes was generated and valid for both conditions. For eight signature genes, baseline expression also correlated with sensitisation, showing potential as pretreatment biomarker. The hypoxia independence of the signature was confirmed in a clinical data set.
Conclusions:
Pretreatment with HDACi may overcome radioresistance of hypoxic prostate tumours by similar mechanisms as under normoxia. We propose a gene signature to predict radiosensitising effects independent of hypoxia status.
Keywords: hypoxia, radiosensitisation, HDACi, gene signature, DNA repair, chromatin organisation, HIF1, TP53
Relapse is a major clinical problem in the radiotherapy of localised, high-risk prostate cancer, where more than half of the patients will have a biochemical relapse within 5 years (Cooper and Sanfilippo, 2015). Strategies for radiosensitisation are therefore highly warranted, especially in patients with hypoxic tumours, as hypoxia is both frequent (Parker et al, 2004; Carnell et al, 2006; Ragnum et al, 2015) and an important cause of treatment failure (Milosevic et al, 2012; Turaka et al, 2012) in this disease. Histone deacetylease inhibitors (HDACis) can block subsets of isoenzymes belonging to the HDAC superfamily, and inhibitors targeting class I and II HDACs have emerged as promising epigenetic drugs for radiosensitisation (Ree et al, 2010; Smits et al, 2014). Several of them, including vorinostat and valproic acid (VPA), are currently evaluated in combination trials with radiotherapy (Groselj et al, 2013), and a phase 1 trial has been completed on prostate cancer (NCT00670553). Discovery of biomarkers for selection of patients to the combination trials is crucial, but no biomarkers exist or are even ready for clinical testing (Smits et al, 2014). A better understanding of the mechanisms underlying the radiosensitising effect is needed (Groselj et al, 2013). In particular, only few studies report on a possible sensitisation of hypoxic cells (Saelen et al, 2012), and current knowledge of the molecular changes by HDACi under hypoxia is scarce and based on studies of individual candidate proteins (Kong et al, 2006; Yu et al, 2012).
Class I/II HDACis act by increasing acetylation of histones and other proteins, leading to alterations in the cellular gene expression program, impaired cell proliferation and induction of apoptosis (Bose et al, 2014). Downregulation of DNA repair enzymes, alterations in chromatin structure and cell cycle perturbations may affect the cellular radiosensitivity and have been proposed to be involved in the radiosensitising effect (Groselj et al, 2013) that also appears to be dependent on the mutation status of the tumour suppressor protein TP53 (Chen et al, 2009, 2011). The HDACis may further repress signalling through the hypoxia-inducible factor 1 (HIF1) (Chen and Sang, 2011), an important transcription factor in the pathogenesis of prostate cancer that may play a role in the radiation response (Moeller and Dewhirst, 2006). Under hypoxia, downregulation of the HIF1 α-subunit (HIF1A) and selected HIF1 target genes by HDACis has been demonstrated (Kong et al, 2006; Yu et al, 2012). However, a clear and comprehensive picture of the molecular effect of the inhibitors under hypoxia and its relationship to radiosensitivity is still lacking.
The present work was conducted to better understand the potential of HDACis as radiosensitiser in hypoxic prostate cancer. We aimed to compare the radiosensitising properties of vorinostat in prostate cancer cell lines grown under normoxic and hypoxic conditions and identify a gene expression signature associated with a possible sensitising effect. Three prostate cancer cell lines with different intrinsic radiosensitivity were treated with vorinostat before radiation. To identify genes with a role in vorinostat-mediated radiosensitisation, a global gene expression analysis was performed where we addressed the importance of HIF1 and TP53 repression and searched for differentially affected DNA repair and chromatin organisation genes across the cell lines.
Materials and methods
Cell cultures
The prostate cancer cell lines DU 145, PC-3 and 22Rv1 were included. Cell line identity was confirmed by STR/DNA profiling using PowerPlex 21 (Promega, Madison, WI, USA) by Eurofins Genomics (Ebersberg, Germany). All cell lines are classified as androgen independent (van Bokhoven et al, 2003). The DU 145 has mutation at both alleles of TP53 and shows strong protein expression, PC-3 expresses no TP53 because of a truncated translation product, whereas 22Rv1 has heterozygote mutation and shows weak expression of wild-type TP53 (van Bokhoven et al, 2003). The DU 145 has been shown to be more radioresistant than PC-3 and 22Rv1 (Leith et al, 1993; Hennessey et al, 2013). The cells were kept in RPMI-1640 medium (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin and 1% L-glutamine, incubated at 37 °C in humidified room air with 5% CO2 and tested for mycoplasma on a routine basis. Automatic cell counting was performed using the Coulter Counter Z2 (Beckman Coulter, Brea, CA, USA).
Lentiviral production and knockdown of HIF1A and TP53
Lentiviral-based vectors for RNA interference-mediated gene silencing (FSVsi) consisted of an U6 promoter for expression of short-hairpin RNAs (shRNAs) against HIF1A (5′-CCAGTTATGATTGTGAAGTTA-3′) or TP53 (5′-GTCCAGATGAAGCTCCCAGAA-3′) and the Venus variant of yellow fluorescent protein (YFP) under the control of an SV40 promoter for monitoring transduction efficiency. Oligonucleotides to produce plasmid-based shRNA were cloned into the FSVsi vector using AgeI–BamHI restriction sites. Lentiviral particles were produced in 293T human embryonic kidney cells co-transfected by the calcium phosphate method as previously described (Li et al, 2012) with the above plasmid plus plasmids coding for the envelope and the packaging systems, VSV-G and D8.9, respectively.
The 293T cells were allowed to produce lentiviral particles for 3–4 days. Culture medium was then collected, centrifuged for 10 min at 2500 r.p.m., filtered through a 0.45 mm filter (Millipore, Bedford, MA, USA) and concentrated by centrifugation through a filter column of 100 kDa (VWR International LLC, West Chester, PE, USA) for 1 h at 4000 r.p.m. The DU 145 cells were incubated overnight in the presence of medium containing lentiviral particles. Then, medium was replaced with fresh medium and cells were incubated for two additional days to allow endogenous protein knockdown.
Treatments
Cells were seeded in triplicates 12 h before treatment using 6-well plates and T25 cell flasks with sealable caps (both from Nunc, Penfield, NY, USA) for the normoxia and hypoxia treatment arm, respectively. Vorinostat (Cayman Chemical Company, Ann Arbor, MI, USA) was diluted in DMSO and added to the medium at a dose of 1 μM for 24 h before irradiation. This treatment condition has been reported to induce histone hyperacetylation in prostate cancer cell lines (Kortenhorst et al, 2008), and corresponding drug concentration has been found achievable in patients (Ramalingam et al, 2007).
The humidified hypoxia chamber In Vivo2 200 (Ruskinn Technology, Brigend, UK) with integrated gas concentration measurements was used to create a model system for tumour hypoxia. Cells were exposed to 0.2% O2 and 5% CO2 for 24 h before irradiation. This oxygen level has been shown to cause radioresistance and alter DNA damage signalling of prostate cancer cell lines (Chan et al, 2007; Stewart et al, 2011). To avoid reoxygenation of hypoxia-treated cells before irradiation, the sealable caps on cell flasks were closed inside the hypoxia chamber before transport to the irradiation machine. Single-fraction irradiation of 2 or 5 Gy was delivered by a 160 kV X-ray generator (Faxitron, Tucson, AZ, USA) with a dose rate of 1 Gy min−1.
Parallel normoxic and hypoxic cell cultures without exposure to irradiation were harvested at 70–80% confluence for analysis of cell cycle distribution, gene expression and protein expression. To avoid reoxygenation, samples from the hypoxia treatment arm were harvested inside the hypoxia chamber using reagents that had been exposed to 0.2% O2 for 1–2 h, except for the ice-cold methanol used for cell fixation.
Clonogenic assay
Clonogenic assay was performed to measure surviving fraction of the cells after vorinostat treatment, knockdown of HIF1A or TP53 and/or irradiation as described previously (Saelen et al, 2012). Fresh medium without vorinostat or solvent was added to the cells after treatment. The cells were incubated under normoxia in 6-well plates (normoxic treatment arm) or unsealed T25 cell flasks (hypoxia treatment arm) for 10–14 days before aspiration of medium, fixation in ice-cold methanol and staining with 0.05% crystal violet. Colonies of >50 cells were considered to stem from clonogenic cells and counted. Plating efficiencies (mean±s.e.m.) were 67±3% (DU 145), 70±7% (PC-3) and 32±2% (22Rv1).
Cell cycle distribution
Cell cycle distributions following combinations of vorinostat and hypoxia treatments were analysed in an LSRII flow cytometer (BD Biosciences, Mountain View, CA, USA) using the detergent-trypsin method developed by Vindelov et al (1983). Approximately 2 × 106 cells were washed with PBS, and nuclei were prepared and stained with propidium iodide as previously described (Vindelov et al, 1983) using 900 μl of solution A and 750 μl of solutions B and C. Cell doublets were excluded from analysis in the BD FACSDiva software (BD Biosciences, Franklin Lakes, NJ, USA) by using area and width of the DNA signal. Fraction of cells in the different cell cycle phases was assessed using ModFit version 5.11 (Verity Software House, Topsham, ME, USA) and FlowJo (Tree Star Inc., Ashland, OR, USA).
Illumina gene expression analysis
Gene expression analysis of three biological replicates of each cell line was performed using HumanHT-12 v4 bead arrays from Illumina (San Diego, CA, USA) with ∼48 000 probes. Total RNA was extracted with the QiaShredder and RNeasy Mini Kits (Qiagen, Oslo, Norway) following the manufacturer's instructions. Quality control of RNA was performed with Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA amplification, cRNA synthesis, hybridisation, scanning and quantile normalisation were performed as previously described (Halle et al, 2011). Log2-transformed data were used in further analysis. The data have been deposited to the Gene Expression Omnibus (GEO) repository (GSE80657). Direct HIF1 and TP53 target genes were identified in the data sets by using a list of 276 HIF1 and 129 TP53 targets (Riley et al, 2008; Mole et al, 2009; Xia et al, 2009). Target genes of mutant TP53 may differ from those depicted in the latter list, but no alternative list was available.
Protein expression analysis
Whole-cell protein lysates were prepared using Mammalian Protein Extraction Reagent (Thermo Scientific, Oslo, Norway) supplemented with phosphatase and protease inhibitors. To ensure complete disruption of cells, sonication was performed. After 2–3 h of incubation at 4 °C, the lysates were centrifuged at 15 000 r.p.m. for 15 min. The supernatant was aspirated and protein concentration was measured using a bicinchoninic acid assay (Thermo Scientific). Protein expression levels were evaluated by western blots, applying 20 μg protein onto 4–20% SDS gels and overnight incubation at 4 °C with the antibodies anti-acetylated histone H3 (polyclonal; Merck Millipore, Billerica, MA, USA), anti-total histone H3 (clone A3S; Merck Millipore), anti-HIF1 A (clone 54; BD), anti-EPAS1 (HIF2α) (polyclonal; Novus Biologicals, Littleton, CO, USA) and anti-TP53 (polyclonal, sc-6243; Santa Cruz, Dallas, TX, USA). The mouse monoclonal anti-γ-tubulin (anti-TUBG1) (clone GTU-88; Sigma-Aldrich) or anti-MCM7 (clone DCS-141, Santa Cruz) was used as loading control. Secondary antibodies were peroxidase-conjugated goat anti-rabbit and donkey anti-mouse (both from Jackson ImmunoResearch Laboratories, West Grove, MA, USA). For visualisation, chemiluminescence with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) or LumiGLO Chemiluminescent Substrate (KPL, Gaithersburg, MD, USA) was used.
Clinical data
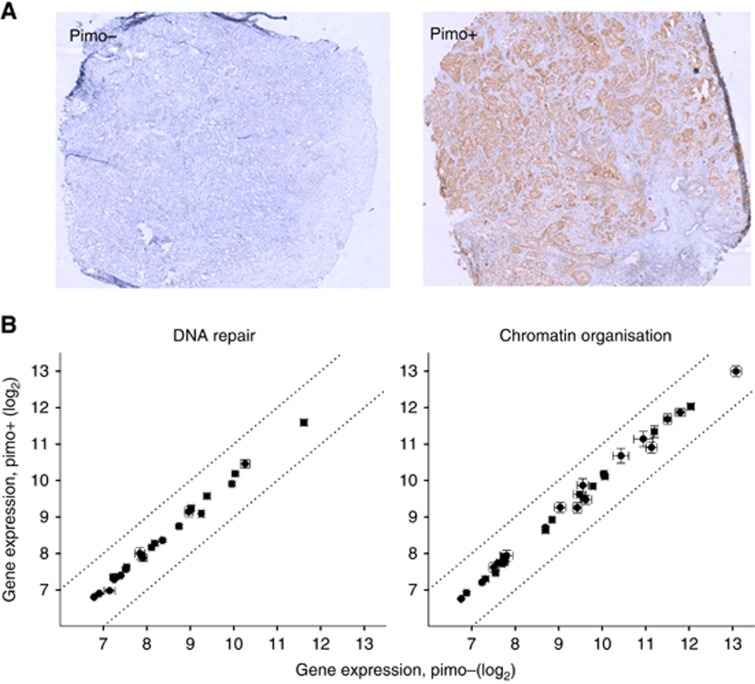
Associations between gene expression and hypoxia in prostate tumours were investigated in a cohort of 39 patients recruited to our ongoing FuncProst-study (NCT01464216). Clinical information, analysis methods, gene expression and hypoxia data have been presented in previous work (Ragnum et al, 2015). All patients had localised, intermediate or high-risk disease and were subjected to prostatectomy the day after i.v. administration of the hypoxia marker pimonidazole hydrochloride (Hydroxyprobe Inc., Burlington, MA, USA). Gene expression profiles were assessed with HumanHT-12 v4 bead arrays (Illumina) and hypoxia was measured by immunohistochemistry of pimonidazole (1 : 50, Hydroxyprobe Inc.) in fresh frozen punch biopsies taken from the surgical specimen. For the immunohistochemistry, antigen retrieval was performed with citrate buffer (pH 6.0). Envision System (Dako, Glostrup, Denmark) followed by counterstaining with haematoxylin, was used for visualisation. Percentage of pimonidazole-positive malignant gland staining in the histological sections was scored from 0 to 5 (0: 0% 1: 1–10% 2: 11–50% 3: 51–90% 4: 91–100% and 5: 100%). The tumours were divided into a more and less hypoxic group based on a median score of 2, implying a pimonidazole-positive percentage above 10% for the more hypoxic tumours. The gene expression profiles are available in the GEO repository (GSE55935).
The study was reviewed and approved by The Regional Committee for Medical and Health Research Ethics in Southeast Norway. All patients provided informed consent for the use of their data in the analysis.
Statistics
Statistical analyses were performed with the SigmaStat software version 3.5 (Systat software, Erkrath, Germany) or with R, version 3.1.1 (www.r-project.org). Differences between treatment groups were evaluated with two-sided Student's t-test under conditions of normal distribution or with Mann–Whitney test when this criterion was not met. Pearson correlation analyses were performed to search for correlations in gene expression data. A significance level of 0.05 was used unless otherwise indicated.
The Linear Models for Microarray Data (LIMMA) software (Smyth, 2004) was applied to find differentially expressed genes between groups using Benjamini–Hochberg correction for multiple testing (Benjamini and Hochberg, 1995) with an adjusted (adj) P-value (false discovery rate) of <0.01 considered to be significant. This strict adj P-value cutoff provided an appropriate number of genes for the analysis. Gene ontology (GO) analysis was performed with the DAVID version 6.7 software (Huang da et al, 2009), where the same multiple testing correction was included and with adj P-value of 0.1 as significance level. Pathway analyses of HIF1 and TP53 target genes were carried out by allowing proteins encoded by the target genes with adj P<0.01 from the LIMMA analysis and a >1.5-fold up- or downregulation to generate a network of their known protein interactions from an integrated set of 10 interaction databases (Razick et al, 2008). In the networks, each interaction had at least one Medline citation, was experimentally validated and was a physical binding protein interaction. All interaction partners were proteins encoded by the differentially expressed genes with adj P<0.01 from the LIMMA analysis. The Cytoscape software (Shannon et al, 2003) was used to visualise the networks.
Results
Alterations in surviving fractions and cell cycle distributions by vorinostat
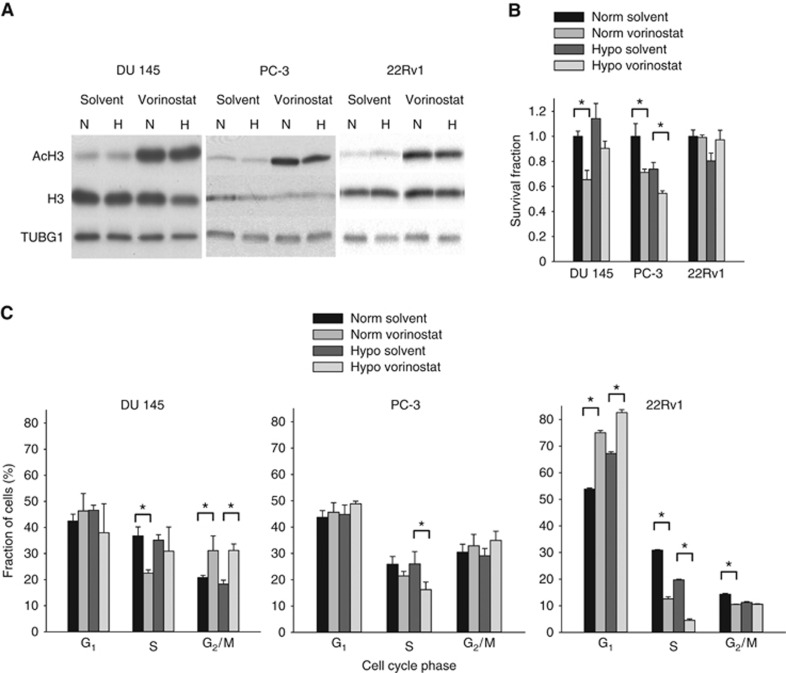
Vorinostat treatment caused hyperacetylation of histone H3 in all cell lines under both normoxia and hypoxia (Figure 1A), showing that the treatment had the expected effect on histone H3. A significant decrease in surviving fraction by vorinostat was observed in normoxic DU 145 and PC-3 cells (P=0.002 and P=0.025, respectively; Figure 1B), but not in 22Rv1. Under hypoxia, a minor decrease in survival fraction was found only in PC-3 (P=0.027; Figure 1B). Thus, at a dose of 1 μM, vorinostat appeared to have similar cytotoxic effect in DU 145 and PC-3 under both normoxia and hypoxia, whereas 22Rv1 was more resistant.
Figure 1.
Vorinostat-mediated changes in histone acetylation, survival fraction and cell cycle distribution. (A) Western blots of acetylated histone H3 (AcH3) and total histone H3 under normoxia and following combinations of vorinostat (1 μM, 24 h) and hypoxia exposure (0.2% O2, 24 h) in DU 145, PC-3 and 22Rv1 prostate cancer cell lines. TUBG1 was included as loading control. H=hypoxia; N=normoxia. (B) Survival fraction under normoxia and following combinations of vorinostat (1 μM, 24 h) and hypoxia exposure (0.2% O2, 24 h) in DU 145, PC-3 and 22Rv1 prostate cancer cell lines. The data for combinations of vorinostat and hypoxia exposure were normalised to the survival fraction under normoxia. (C) Fraction of cells in different cell cycle phases under normoxia and following combinations of vorinostat (1 μM, 24 h) and hypoxia exposure (0.2% O2, 24 h) in DU 145, PC-3 and 22Rv1 prostate cancer cell lines. (B and C) Columns represent mean values from three independent experiments, and bars depict s.e.m. *Statistically significant difference.
The effect of vorinostat on cell cycle distribution also differed across the cell lines and the same tendency was observed in the normoxic and hypoxic treatment arms (Figure 1C; Supplementary Figure S1). In DU 145, fraction of cells in G2- and M-phase increased by ∼10% (normoxia, P=0.034; hypoxia, P=0.002), whereas no significant change in G2/M was observed in PC-3. In 22Rv1, vorinostat primarily increased the G1 fraction (normoxia, P<0.001; hypoxia, P=0.005). Hypoxia treatment alone had no significant effects on the cell cycle distribution in DU-145 and PC-3, but led to an increase in G1 fraction in 22Rv1 (P<0.001).
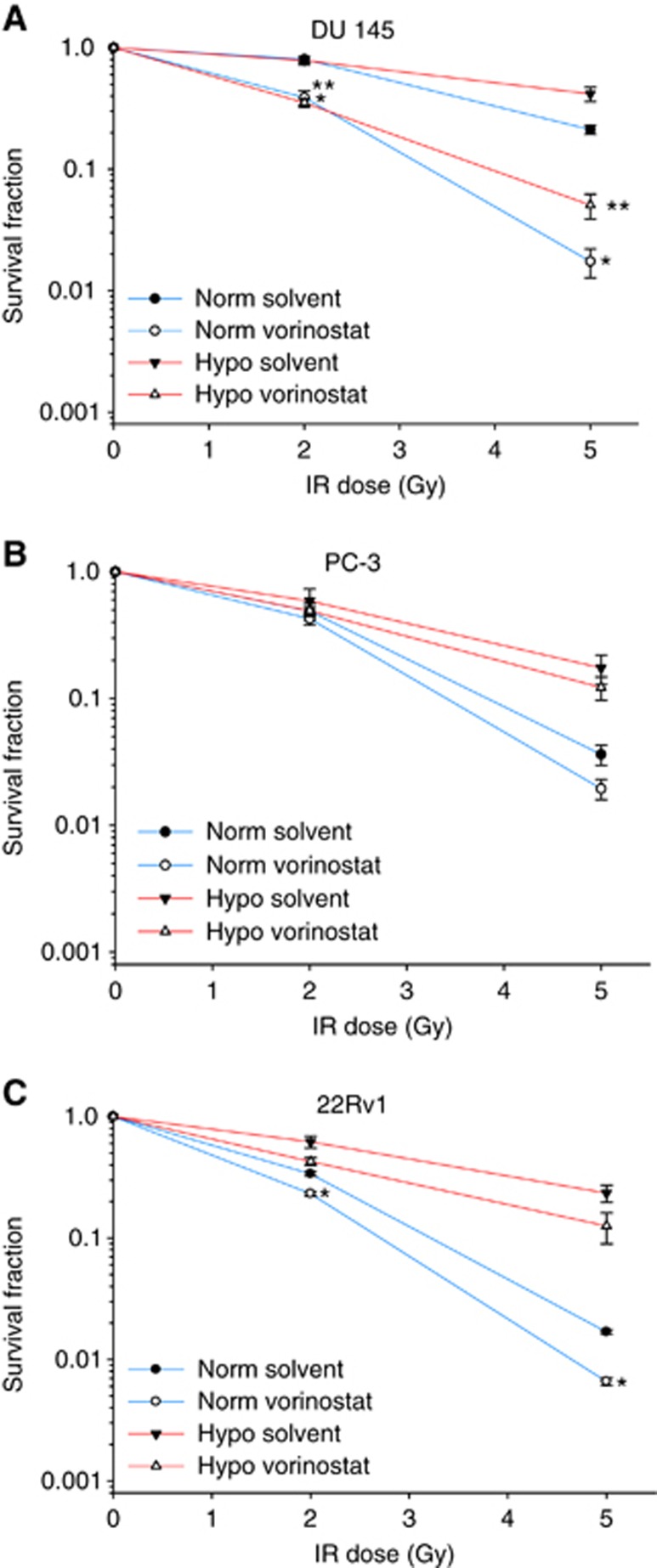
Vorinostat-mediated radiosensitisation
At single doses of 2 and 5 Gy, DU 145 was the most radioresistant cell line under normoxia followed by PC-3 and 22Rv1, in accordance with previous work (Leith et al, 1993; Hennessey et al, 2013), and similar results were found for the hypoxic condition (Figure 2). Pretreatment with vorinostat under normoxia decreased the surviving fraction of DU 145 cells at both 2 and 5 Gy (P<0.001; Figure 2A), whereas a much smaller, although significant, effect was seen in 22Rv1 (P<0.001; Figure 2C). No significant radiosensitisation was observed in PC-3 (Figure 2B). Thus, combined treatment with vorinostat made DU 145 cells equal or more radiosensitive than the intrinsically radiosensitive PC-3 and 22Rv1 treated with radiation alone at both radiation doses.
Figure 2.
Radiosensitising effect of vorinostat. Survival fraction following irradiation (IR) under normoxia and combinations of vorinostat (1 μM, 24 h) and hypoxia exposure (0.2% O2, 24 h) in DU 145 (A), PC-3 (B) and 22Rv1 (C) prostate cancer cell lines. Points represent mean value from at least three independent experiments, and bars depict s.e.m. *Statistically significant difference compared with normoxia solvent control; **statistically significant differences compared with hypoxia solvent control.
A pronounced radiosensitisation of DU 145 by vorinostat was also seen under hypoxia (P<0.001; Figure 2A), whereas no significant effects were found in PC-3 or 22Rv1 (Figure 2B and C). In particular, pretreatment with vorinostat rendered hypoxic DU 145 cells more radiosensitive than normoxic DU 145 cells without pretreatment and hypoxic PC-3 or 22Rv1 cells with or without pretreatment (Figure 2). The radiosensitising effect of vorinostat therefore appeared to differ considerably across the cell lines, and was significant under both normoxia and hypoxia only in the most radioresistant DU 145.
HIF1 and TP53 repression in relation to radiosensitisation
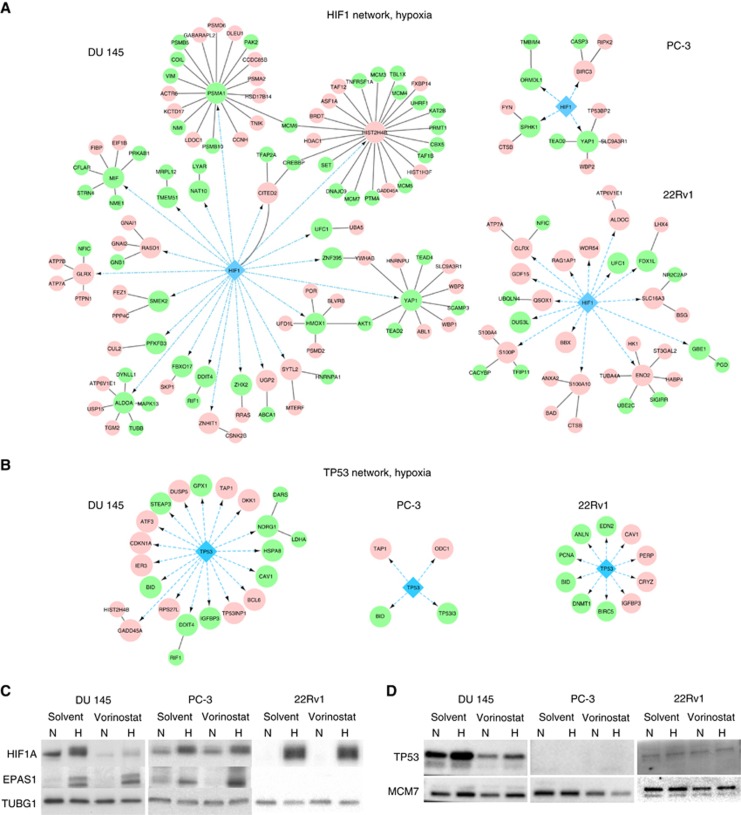
To investigate mechanisms underlying the radiosensitising effect of vorinostat in DU 145, we first explored whether repression of the HIF1 and TP53 transcription factors could be involved for one or both of the normoxic and hypoxic conditions. Global gene expression analysis revealed a significant change (adj P<0.01) in the expression of numerous genes by vorinostat, including several HIF1 and TP53 targets, in normoxic and hypoxic DU 145 and 22Rv1, whereas fewer genes were affected in PC-3 (Supplementary Figure S2). In general, the HIF1 and TP53 targets showed a greater change in DU 145 than in the other cell lines, as demonstrated by network analysis of the targets altered by >1.5-fold (Figure 3A and B and Supplementary Figures S3 and S4). In addition, HIF1A and TP53 protein were downregulated by vorinostat under normoxia and hypoxia in DU 145, but not in the others (Figure 3C and D). Vorinostat induced no major changes in EPAS1 (HIF2α) protein expression in any cell line. Altogether, it seemed that vorinostat repressed HIF1 and TP53 in DU 145, but had less effect in PC-3 and 22Rv1.
Figure 3.
Vorinostat-mediated changes in HIF1 and TP53 signalling. (A and B) First-degree protein interaction networks of the direct HIF1 (A) and TP53 (B) target genes with >1.5-fold change following vorinostat exposure (1 μM, 24 h) under hypoxic conditions (0.2% O2, 24 h) in DU 145, PC-3 and 22Rv1 prostate cancer cell lines. Large circles indicate direct interaction partners of HIF1 and TP53. The first-degree interaction partners (smaller circles) were selected among the genes significantly changed by vorinostat (adj P<0.01, LIMMA). Green and red colour denotes downregulation and upregulation by vorinostat, respectively. Blue arrows indicate regulatory DNA–protein interactions and black lines indicate protein–protein interactions. (C and D) Western blots of HIF1A, EPAS1 (HIF2α) and TP53 under normoxia and following combinations of vorinostat (1 μM, 24 h) and hypoxia (0.2% O2, 24 h) exposure in DU 145, PC-3 and 22Rv1 prostate cancer cell lines. TUBG1 (C) and MCM7 (D) were included as loading controls. The experiment was repeated three times with similar results (data not shown). H=hypoxia; N=normoxia.
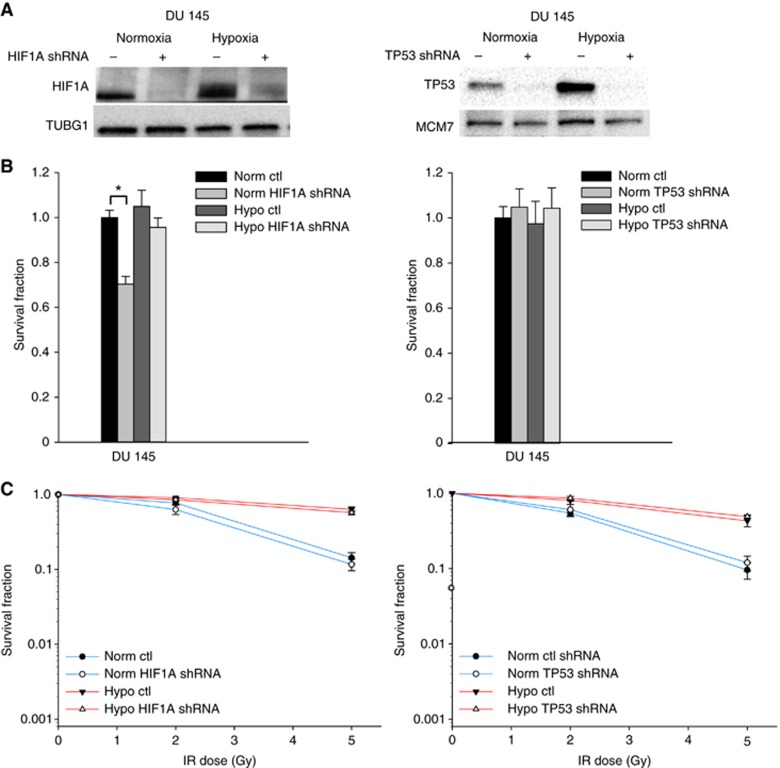
Mutant TP53 has been shown to have oncogenic function in many cancer cell lines, including DU 145 (Zhu et al, 2011). Both HIF1 and mutant TP53 repression could therefore be a possible cause of the vorinostat-mediated radiosensitisation of DU 145. To test this hypothesis, the radiosensitising effect of vorinostat under normoxia and hypoxia was studied in HIF1A and mutant TP53 knockdown DU 145 cells, for which downregulation of the proteins was confirmed (Figure 4A). Knockdown of HIF1A decreased the surviving fraction of normoxic, unirradiated cells by 30% (P=0.001), whereas this knockdown under hypoxia or mutant TP53 knockdown had no significant effect on survival (Figure 4B). Neither HIF1A nor mutant TP53 knockdown led to any significant change in the survival of DU 145 cells upon 2 or 5 Gy irradiation (Figure 4C). Repression of HIF1A or mutant TP53 could therefore not explain the vorinostat-mediated radiosensitisation of DU 145.
Figure 4.
Radiosensitising effect of HIF1A and mutant TP53 knockdown. (A) Western blots of HIF1A and TP53 under normoxia and following hypoxia exposure (0.2% O2, 24 h) in DU 145 prostate cancer cell line without (−) and with (+) lentiviral-mediated knockdown of HIF1A and mutant TP53. TUBG1 and MCM7 were included as loading controls. The experiment was repeated three times with similar results (data not shown). (B) Survival fraction under normoxia and hypoxia (0.2% O2, 24 h) in DU 145 prostate cancer cell line without (ctl) and with lentiviral mediated knockdown of HIF1A (HIF1A shRNA) and mutant TP53 (TP53 shRNA). The data for hypoxia and knockdown were normalised to the survival fraction under normoxia. Columns represent mean values from three independent experiments, and bars depict s.e.m. *Statistically significant difference. (C) Survival fraction following irradiation (IR) under normoxia and hypoxia exposure (0.2% O2, 24 h) in DU 145 prostate cancer cell line without (ctl) and with lentiviral-mediated knockdown of HIF1A (HIF1A shRNA) and mutant TP53 (TP53 shRNA). Points represent mean value from at least three independent experiments, and bars depict s.e.m.
Expression of DNA repair and chromatin organisation genes in relation to radiosensitisation
Unsupervised GO analysis of all genes differentially expressed by vorinostat was performed to identify DNA repair and chromatin organisation genes that could play a role in radiosensitisation. The analysis revealed six main biological processes enriched in DU 145 and 22Rv1, including cellular stress response with its subgroup response to DNA damage stimulus, DNA repair and chromatin organisation (Supplementary Table S1). Approximately the same processes were identified for the normoxic and hypoxic condition. In PC-3, only RNA processing and cell cycle were significantly enriched.
The DNA repair and chromatin organisation genes from the GO analysis of DU 145 were potential mediators of the radiosensitising effect, and their expression changes by vorinostat were compared with those in PC-3 and 22Rv1. Many genes were altered similarly in all three cell lines and are therefore probably not key mediators of the effect. These included the double-strand break repair gene CHEK1 that was significantly downregulated under both normoxia and hypoxia. Totally, 51 DNA repair and 77 chromatin organisation genes were significantly more up- or downregulated in DU 145 than in either PC-3 or 22Rv1 in at least one of the normoxic or hypoxic conditions (Supplementary Tables S2 and S3). Thus, they were candidate genes in radiosensitisation of DU 145.
Further support for a role of the candidate genes in radiosensitisation was provided by comparing the expression changes in DU 145 with the baseline expression levels (without vorinostat treatment) in the intrinsically radiosensitive PC-3 and 22Rv1. A significant inverse correlation was found between the expression changes in DU 145 under normoxia and hypoxia and the baseline difference between this cell line and PC-3 or 22Rv1, both for the DNA repair genes (normoxia, r=−0.40, P<0.001; hypoxia, r=−0.26, P=0.03) and chromatin organisation genes (normoxia, r=−0.19, P=0.04; hypoxia, r=−0.26, P=0.03). Hence, vorinostat seemed to change the expression of the candidate DNA repair and chromatin organisation genes in DU 145 towards the baseline level of PC-3 and 22Rv1 cells and therefore towards a more radiosensitive phenotype.
Generation of a hypoxia-independent gene expression signature of radiosensitisation
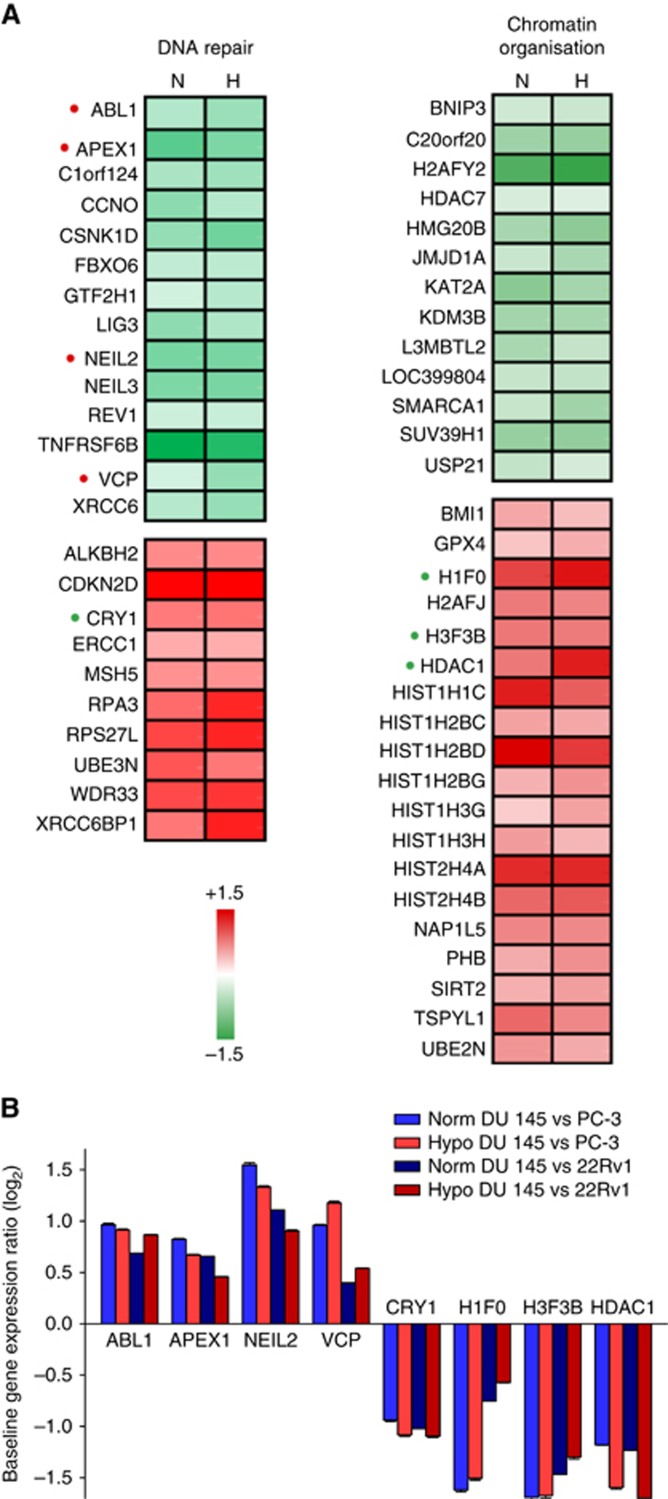
All candidate genes were regulated in the same direction by vorinostat under normoxia and hypoxia (Supplementary Tables S2 and S3). Moreover, the expression level differed little between the normoxic and hypoxic conditions regardless of cell line, except for BNIP3 in chromatin organisation (Supplementary Figure S5). In general, the candidate genes were therefore apparently not regulated by hypoxia. Based on this observation, we could define a radiosensitisation signature with the expression changes of 56 of the most altered candidate genes in DU 145 compared with PC-3 and 22Rv1; 24 genes in DNA repair and 32 in chromatin organisation. These genes were significantly altered by vorinostat (adj P<0.01, LIMMA) in DU 145 and showed a larger change in this cell line than in the others (P<0.05, t-test) under both normoxia and hypoxia (Figure 5A and Supplementary Tables S2 and S3). The signature included both up- and downregulated genes, reflecting a pronounced deregulation of the DNA repair and chromatin organisation processes in association with the vorinostat-mediated radiosensitisation.
Figure 5.
Gene expression signature of radiosensitisation. (A) Change in expression by vorinostat in DU 145 prostate cancer cell line under normoxia (N) and hypoxia (H) for 24 DNA repair genes (left) and 32 chromatin organisation genes (right). Genes that were significantly altered by vorinostat in DU 145 under either normoxia or hypoxia (adj P<0.01, LIMMA, three experiments) and significantly more altered in DU 145 than in PC-3 or 22Rv1 (P<0.05, t-test) under both normoxia and hypoxia were selected. Genes with a significantly higher or lower expression (adj P<0.01, LIMMA, three experiments) in DU 145 than in both PC-3 and 22Rv1 at baseline (i.e., before vorinostat treatment) under normoxic and hypoxic conditions are marked with a red or green dot, respectively. (B) Baseline expression ratio between DU 145 and PC-3 and 22Rv1 for the eight genes marked with a red or green dot in (A). Columns represent mean value of three independent experiments, and bars depict s.e.m.
To investigate whether any of the signature genes could serve as pretreatment biomarker for prediction of a radiosensitising effect already before the onset of vorinostat treatment, we search for baseline gene expression characteristics of the radioresistant DU 145 phenotype, for which a radiosensitising effect was observed, as compared with the radiosensitive PC-3 and 22Rv1 phenotype with no radiosensitisation. This comparison identified a subset of eight signature genes; ABL1, APEX1, CRY1, NEIL2 and VCP in DNA repair and H1F0, H3F3B and HDAC1 in chromatin organisation, for which a significant difference (P<0.01, LIMMA) in baseline expression was seen between DU 145 and both PC-3 and 22Rv1 (Figure 5B). Hence, a high expression of ABL1, APEX1, NEIL2 and VCP and low expression of CRY1, H1F0, H3F3B and HDAC1 represented a pretreatment predictor of a radioresistant phenotype for sensitisation with vorinostat regardless of hypoxia status.
To confirm that the radiosensitisation signature, including the eight biomarker genes, was also independent of hypoxia in a clinical setting, we compared the expression level of the 56 genes between pimonidazole-positive and -negative prostate tumours in patients (Figure 6). No difference between the two groups was seen for any of the genes, consistent with our cell line results.
Figure 6.
Expression of signature genes in relation to hypoxia in prostate tumours in patients. (A) Histological section of tumour biopsy from two different prostate cancer patients, showing negative (pimo−) and positive (pimo+) pimonidazole staining. (B) Expression of signature genes in pimo+ vs pimo− tumours. Data of 24 DNA repair genes and 32 chromatin organisation genes are shown in the left and right panel, respectively. Points represent mean value of 21 pimo+ and 18 pimo− tumours, bars depict s.e.m. and stippled lines indicate limits for two-fold difference between the pimo+ and pimo− groups.
Discussion
Pretreatment of prostate cancer cells with clinically relevant doses of the class I/II HDACi vorinostat led to significant radiosensitisation under both normoxic and hypoxic conditions. This effect was observed in the most radioresistant DU 145 cell line that became as sensitive as the radiosensitive PC-3 and 22Rv1 lines. Our results encourage further development of drugs inhibiting class I/ II HDACs for use in combination with radiation to counteract the poor response of the most radioresistant prostate tumours and, in particular, to overcome a major problem in radiotherapy caused by hypoxia. Approval by the Food and Drug Administration (FDA) and ongoing testing of vorinostat in clinical studies (Groselj et al, 2013) make this drug especially interesting for the exploration of radiosensitising properties. Moreover, new drugs are in pipeline, although none of those tested in patients have so far demonstrated superior clinical activity or more favourable toxicity profiles (West and Johnstone, 2014). Regardless of cell line and radiosensitisation, HDACi treatment led to deregulation of HIF1, TP53, DNA repair and chromatin organisation genes. However, to the best of our knowledge, our work is the first to identify a gene expression signature associated with a radiosensitising effect of the HDACi treatment and demonstrate its independence of hypoxia status.
The HDACi treatment had a large effect on HIF1 and TP53 in DU 145. Previous work has shown that HIF1 may play a role in the cellular radiation response, although both radioprotective and radiosensitising effects have been demonstrated, and the underlying mechanisms are not clarified (Moeller et al, 2007). Moreover, various lines of evidence indicate that mutations in TP53 may lead to gain of function and resistance to anticancer treatment, including radiation (Oren and Rotter, 2010). However, it appeared from our knockdown experiments that neither HIF1 nor TP53 repression alone could explain the radiosensitising effects of HDAC inhibition. It is possible that a HDACi-mediated cell cycle redistribution could have played a role in the radiosensitisation of DU 145. The HDACi treatment caused accumulation of DU 145 cells in G2 and M phases, which are the most radiosensitive phases (Joiner and van der Kogel, 2009), whereas no such accumulation was observed in PC-3 and 22Rv1. However, this accumulation led to an increase in the G2/M fraction of only 10%, and this is not enough to explain the entire difference of approximately one order of magnitude in radiosensitisation between DU 145 and the other cell lines.
By comparing HDACi-mediated gene expression changes on a global scale, significant differences in the expression of numerous DNA repair and chromatin organisation genes across the cell lines were identified, most likely contributing to radiosensitisation in a collective manner. This hypothesis was supported by the finding that HDACi treatment changed the expression of these genes in DU 145 towards the level in the more radiosensitive PC-3 and 22Rv1. Many of the same genes were affected by vorinostat under both normoxia and hypoxia, and only few of them were responsive to hypoxia exposure alone as confirmed in a clinical data set. It therefore seems that the mechanisms underlying the radiosensitising effect are independent on the hypoxia status of the tumour cells. This observation is important for the use of HDACi in the clinic, as pretreatment identification of the hypoxic tumours would not be necessary.
The 56-gene signature may provide information about the mechanisms underlying the sensitising effect. In particular, the signature contained downregulated genes with a regulatory role in DNA repair pathways, such as genes encoding the ubiquitin-selective segregase VCP and the ABL tyrosine kinase. These proteins orchestrate double-strand break repair by regulating proteins like TP53BP1, BRCA1, RAD51 and RAD52 (Colicelli, 2010; Meerang et al, 2011). Notably, genes encoding these downstream proteins have been found to be downregulated by class I/II HDACis in previous work (Lee et al, 2010; Kachhap et al, 2010), but their regulators, VCP and ABL1, have not been mentioned. In addition, the signature contained downregulated genes involved in the first steps of base excision repair (Curtin, 2012), including genes encoding the multifunctional DNA repair enzyme APEX1 and the members of the Nei-like protein family, NEIL2 and NEIL3, suggesting that deregulation of this pathway also plays a role. In accordance with this, the importance of APEX1 for the radiosensitivity of DU 145 has been demonstrated by knockdown experiments (Skvortsova et al, 2008), although the radiosensitising effect was not as large as observed in our study. The genes EEPD1 and CRY1 were among the upregulated DNA repair genes and appear to be involved in DNA damage sensing and stabilisation of genotoxic stress (Papp et al, 2015; Wu et al, 2015).
The chromatin organisation genes included the downregulated H2AFY2 and upregulated H3F3B genes, encoding the variant histone H2A (macroH2A) and H3 (H3.3), respectively, suggesting reorganisation of the chromatin in DU 145 by HDACi towards a more open and transcriptionally active structure (Melters et al, 2015). Hence, increased acetylation of histones by HDACis results in loosening of chromatin and better accessibility for replication and transcription. It has been shown that the number of double-strand breaks after irradiation is enhanced by chromatin relaxation possibly because of more accessible DNA attack sites for the radicals (Lavelle and Foray, 2014). Chromatin reorganisation by HDACi treatment may therefore have increased the radiation-induced DNA damage in DU 145 cells. Interpretation of the chromatin organisation data is, however, difficult, as many genes, including HDAC1, may act as both tumour suppressor gene and oncogene, depending on cell type and state (West and Johnstone, 2014), and also affect the cellular repair capacity (Robert et al, 2011).
Monitoring expression changes of the signature genes during HDACi treatment may indicate a possible radiosensitisation before the onset of radiotherapy, and might be of clinical value. In addition, a subset of eight genes showed differences in baseline expression across the cell lines and might be useful in the initial treatment planning to decide upon a possible pretreatment with HDACi. Hence, high baseline expression of DNA repair genes (ABL1, VCP, APEX1, NEIL2) in DU 145 as compared with PC-3 and 22Rv1 suggests higher repair capacity of the former cell line, whereas low H3F3B expression may indicate a close chromatin structure. It could be speculated that these baseline characteristics play a role in the intrinsic radioresistance of DU 145 and that targeting of these characteristics by HDACi is an efficient way to radiosensitise the cells. The association between baseline expression and radioresistance could be tested retrospectively in stored tumour specimens from prostate cancer patients treated with radiotherapy and with known outcome data. Moreover, for the HDACi-mediated radiosensitisation, it would be of interest to evaluate pretreatment expression level of the 8 genes as well as the expression changes during HDACi pretreatment of all 56 genes in specimens collected in combination trials with radiotherapy.
Acknowledgments
Financial support was provided by the European Union 7th Framework Program (Grant No. 222741-METOXIA), the Norwegian Cancer Society (Grant No. 107438 - PR-2007-0179) and The South-Eastern Norway Regional Health Authority (Grant No. 2015020).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- Bose P, Dai Y, Grant S (2014) Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol Ther 143(3): 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell DM, Smith RE, Daley FM, Saunders MI, Bentzen SM, Hoskin PJ (2006) An immunohistochemical assessment of hypoxia in prostate carcinoma using pimonidazole: implications for radioresistance. Int J Radiat Oncol Biol Phys 65: 91–99. [DOI] [PubMed] [Google Scholar]
- Chan N, Milosevic M, Bristow RG (2007) Tumor hypoxia, DNA repair and prostate cancer progression: new targets and new therapies. Future Oncol 3: 329–341. [DOI] [PubMed] [Google Scholar]
- Chen S, Sang N (2011) Histone deacetylase inhibitors: the epigenetic therapeutics that repress hypoxia-inducible factors. J Biomed Biotechnol 2011: 197946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wong JY, Wong P, Radany EH (2011) Low-dose valproic acid enhances radiosensitivity of prostate cancer through acetylated p53-dependent modulation of mitochondrial membrane potential and apoptosis. Mol Cancer Res 9(4): 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wong P, Radany E, Wong JY (2009) HDAC inhibitor, valproic acid, induces p53-dependent radiosensitization of colon cancer cells. Cancer Biother Radiopharm 24(6): 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J (2010) ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal 3(139): re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper BT, Sanfilippo NJ (2015) Concurrent chemoradiation for high-risk prostate cancer. World J Clin Oncol 6(4): 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin NJ (2012) DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 12(12): 801–817. [DOI] [PubMed] [Google Scholar]
- Groselj B, Sharma NL, Hamdy FC, Kerr M, Kiltie AE (2013) Histone deacetylase inhibitors as radiosensitisers: effects on DNA damage signalling and repair. Br J Cancer 108(4): 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle C, Lando M, Svendsrud DH, Clancy T, Holden M, Sundfør K, Kristensen GB, Holm R, Lyng H (2011) Membranous expression of ectodomain isoforms of the epidermal growth factor receptor predicts outcome after chemoradiotherapy of lymph node-negative cervical cancer. Clin Cancer Res 17: 5501–5512. [DOI] [PubMed] [Google Scholar]
- Hennessey D, Martin LM, Atzberger A, Lynch TH, Hollywood D, Marignol L (2013) Exposure to hypoxia following irradiation increases radioresistance in prostate cancer cells. Urol Oncol 31: 1106–1116. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Joiner M, van der Kogel AJ (2009) Basic Clinical Radiobiology Taylor & Francis Group: Boca Raton, FL, USA.
- Kachhap SK, Rosmus N, Collis SJ, Kortenhorst MS, Wissing MD, Hedayati M, Shabbeer S, Mendonca J, Deangelis J, Marchionni L, Lin J, Hoti N, Nortier JW, DeWeese TL, Hammers H, Carducci MA (2010) Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS One 5(6): e11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J (2006) Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Mol Cell Biol 26(6): 2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortenhorst MS, Zahurak M, Shabbeer S, Kachhap S, Galloway N, Parmigiani G, Verheul HM, Carducci MA (2008) A multiple-loop, double-cube microarray design applied to prostate cancer cell lines with variable sensitivity to histone deacetylase inhibitors. Clin Cancer Res 14: 6886–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle C, Foray N (2014) Chromatin structure and radiation-induced DNA damage: from structural biology to radiobiology. Int J Biochem Cell Biol 49: 84–97. [DOI] [PubMed] [Google Scholar]
- Lee JH, Choy ML, Ngo L, Foster SS, Marks PA (2010) Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci USA 107(33): 14639–14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leith JT, Quaranto L, Padfield G, Michelson S, Hercbergs A (1993) Radiobiological studies of PC-3 and DU-145 human prostate cancer cells: X-ray sensitivity in vitro and hypoxic fractions of xenografted tumors in vivo. Int J Radiat Oncol Biol Phys 25: 283–287. [DOI] [PubMed] [Google Scholar]
- Li M, Husic N, Lin Y, Snider BJ (2012) Production of lentiviral vectors for transducing cells from the central nervous system. J Vis Exp, (63)e4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hubscher U, Meyer H, Ramadan K (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 13(11): 1376–1382. [DOI] [PubMed] [Google Scholar]
- Melters DP, Nye J, Zhao H, Dalal Y (2015) Chromatin dynamics in vivo: a game of musical chairs. Genes (Basel) 6(3): 751–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic M, Warde P, Ménard C, Chung P, Toi A, Ishkanian A, McLean M, Pintilie M, Sykes J, Gospodarowicz M, Catton C, Hill RP, Bristow R (2012) Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin Cancer Res 18: 2108–2114. [DOI] [PubMed] [Google Scholar]
- Moeller BJ, Dewhirst MW (2006) HIF-1 and tumour radiosensitivity. Br J Cancer 95(1): 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller BJ, Richardson RA, Dewhirst MW (2007) Hypoxia and radiotherapy: opportunities for improved outcomes in cancer treatment. Cancer Metastasis Rev 26: 241–248. [DOI] [PubMed] [Google Scholar]
- Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ (2009) Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem 284(25): 16767–16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Rotter V (2010) Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp SJ, Huber AL, Jordan SD, Kriebs A, Nguyen M, Moresco JJ, Yates JR, Lamia KA (2015) DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. Elife 4: e04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C, Milosevic M, Toi A, Sweet J, Panzarella T, Bristow R, Catton C, Catton P, Crook J, Gospodarowicz M, McLean M, Warde P, Hill RP (2004) Polarographic electrode study of tumor oxygenation in clinically localized prostate cancer. Int J Radiat Oncol Biol Phys 58: 750–757. [DOI] [PubMed] [Google Scholar]
- Ragnum HB, Vlatkovic L, Lie AK, Axcrona K, Julin CH, Frikstad KM, Hole KH, Seierstad T, Lyng H (2015) The tumour hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br J Cancer 112(2): 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, Potter DM, Argiris AE, Zwiebel JA, Egorin MJ, Belani CP (2007) Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res 13: 3605–3610. [DOI] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM (2008) iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree AH, Dueland S, Folkvord S, Hole KH, Seierstad T, Johansen M, Abrahamsen TW, Flatmark K (2010) Vorinostat, a histone deacetylase inhibitor, combined with pelvic palliative radiotherapy for gastrointestinal carcinoma: the Pelvic Radiation and Vorinostat (PRAVO) phase 1 study. Lancet Oncol 11: 459–464. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9(5): 402–412. [DOI] [PubMed] [Google Scholar]
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, Foiani M (2011) HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 471(7336): 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelen MG, Ree AH, Kristian A, Fleten KG, Furre T, Hektoen HH, Flatmark K (2012) Radiosensitization by the histone deacetylase inhibitor vorinostat under hypoxia and with capecitabine in experimental colorectal carcinoma. Radiat Oncol 7: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsova I, Skvortsov S, Stasyk T, Raju U, Popper BA, Schiestl B, von Guggenberg E, Neher A, Bonn GK, Huber LA, Lukas P (2008) Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics 8(21): 4521–4533. [DOI] [PubMed] [Google Scholar]
- Smits KM, Melotte V, Niessen HE, Dubois L, Oberije C, Troost EG, Starmans MH, Boutros PC, Vooijs M, van Engeland M, Lambin P (2014) Epigenetics in radiotherapy: where are we heading? Radiother Oncol 111(2): 168–177. [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, e-pub ahead of print 12 February 2004. [DOI] [PubMed] [Google Scholar]
- Stewart GD, Nanda J, Katz E, Bowman KJ, Christie JG, Brown DJ, McLaren DB, Riddick AC, Ross JA, Jones GD, Habib FK (2011) DNA strand breaks and hypoxia response inhibition mediate the radiosensitisation effect of nitric oxide donors on prostate cancer under varying oxygen conditions. Biochem Pharmacol 81: 203–210. [DOI] [PubMed] [Google Scholar]
- Turaka A, Buyyounouski MK, Hanlon AL, Horwitz EM, Greenberg RE, Movsas B (2012) Hypoxic prostate/muscle PO2 ratio predicts for outcome in patients with localized prostate cancer: long-term results. Int J Radiat Oncol Biol Phys 82: e433–e439. [DOI] [PubMed] [Google Scholar]
- van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS (2003) Molecular characterization of human prostate carcinoma cell lines. Prostate 57(3): 205–225. [DOI] [PubMed] [Google Scholar]
- Vindelov LL, Christensen IJ, Nissen NI (1983) A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 3(5): 323–327. [DOI] [PubMed] [Google Scholar]
- West AC, Johnstone RW (2014) New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 124(1): 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Lee SH, Williamson EA, Reinert BL, Cho JH, Xia F, Jaiswal AS, Srinivasan G, Patel B, Brantley A, Zhou D, Shao L, Pathak R, Hauer-Jensen M, Singh S, Kong K, Wu X, Kim HS, Beissbarth T, Gaedcke J, Burma S, Nickoloff JA, Hromas RA (2015) EEPD1 rescues stressed replication forks and maintains genome stability by promoting end resection and homologous recombination repair. PLoS Genet 11(12): e1005675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL (2009) Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA 106: 4260–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Mi J, Wang Y, Wang A, Tian X (2012) Regulation of radiosensitivity by HDAC inhibitor trichostatin A in the human cervical carcinoma cell line Hela. Eur J Gynaecol Oncol 33(3): 285–290. [PubMed] [Google Scholar]
- Zhu H, Mao Q, Lin Y, Yang K, Xie L (2011) RNA interference targeting mutant p53 inhibits growth and induces apoptosis in DU145 human prostate cancer cells. Med Oncol 28(Suppl 1): S381–S387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.