Abstract
The genetic basis of phenotypic changes in extreme environments is a key but rather unexplored topic in animal evolution. Here we provide an exemplar case of evolution by relaxed selection in the Somalian cavefish Phreatichthys andruzzii that has evolved in the complete absence of light for at least 2.8 million years. This has resulted in extreme degenerative phenotypes, including complete eye loss and partial degeneration of the circadian clock. We have investigated the molecular evolution of the nonvisual photoreceptor melanopsin opn4m2, whose mutation contributes to the inability of peripheral clocks to respond to light. Our intra- and inter-species analyses suggest that the ‘blind' clock in P. andruzzii evolved because of the loss of selective constraints on a trait that was no longer adaptive. Based on this change in selective regime, we estimate that the functional constraint on cavefish opn4m2 was relaxed at ∼5.3 Myr. This implies a long subterranean history, about half in complete isolation from the surface. The visual photoreceptor rhodopsin, expressed in the brain and implicated in photophobic behavior, shows similar evolutionary patterns, suggesting that extreme isolation in darkness led to a general weakening of evolutionary constraints on light-responsive mechanisms. Conversely, the same genes are still conserved in Garra barreimiae, a cavefish from Oman, that independently and more recently colonized subterranean waters and evolved troglomorphic traits. Our results contribute substantially to the open debate on the genetic bases of regressive evolution.
Introduction
Phreatichthys andruzzii is a cavefish that inhabits the subterranean waters of central Somalia and shows an extreme level of adaptation to the dark environment. The adult fishes completely lack eyes, pigments and scales and their metabolic rate is extremely reduced. In particular, the eye is totally regressed 36 h post fertilization and 1 month later there remains only a rudimentary cyst, with loss of the optic nerves and strongly reduced optic lobes (Berti et al., 2001; Stemmer et al., 2015). This degeneration is rather premature compared, for example, with what is known for the blind Mexican cavefish Astyanax mexicanus. In this Mexican species, small eye primordia develop during embryogenesis, grow for a while, and only later arrest in development, degenerate and sink into the orbits (Wilkens and Strecker, 2003; Jeffery, 2009). Other important biological peculiarities of P. andruzzii connected to the absence of light involve the circadian clock, a physiological timing mechanism that allows organisms to anticipate and adapt to the day–night cycle. Our previous studies showed significant regression of this mechanism in terms of light sensitivity. When exposed to cycles of light and dark, the cavefish lacks behavioral rhythmicity, and the expression of a set of clock-regulated and light-regulated genes is also arrhythmic. Interestingly, the clock of P. andruzzii does not respond to light, but is still sensitive to other timing signals, such as food availability that are possibly more important in the dark environment (Cavallari et al., 2011). The regression of the light-related pathway is extended to the mechanism of light perception, in particular to some nonvisual photoreceptors, melanopsin (opn4m2) and teleost multiple tissue opsin. In P. andruzzii, loss-of-function (LOF) mutations in the genes coding for these peripheral photoreceptors abolish their ability to respond to light. Consequently, the downstream cascade in the light-responsive pathway is not induced, and the peripheral circadian clocks are light insensitive (Cavallari et al., 2011).
The extreme troglomorphism developed by P. andruzzii in the absence of light is probably the result of both complete and long-term isolation. Although the water table where these fish live is very superficial (from some meters to tens of centimeters), for the sake of simplicity we will refer to it as ‘the cave'. P. andruzzii cave populations probably originated from normal-eyed ancestors that lived in surface water when Somalia had a wetter climate. As a consequence of increases in North East African aridity after 2.8 Myr that led to the desertification at the surface (deMenocal, 1995), P. andruzzii ancestors that colonized the cave environment remained isolated in the dark of the phreatic waters. This complete isolation is because of the disappearance of surface populations, unlike some other species that maintain some level of gene flow with surface-dwelling populations (Li and He, 2009; Kruckenhauser et al., 2011). The extent to which gene flow occurs between surface- and cave-dwelling forms is still debated for A. mexicanus, the model species in cavefish evolution (Bradic et al., 2012; Gross, 2012; Strecker et al., 2012). Another key factor that determines the extent of regression is time. Mutations that render the gene nonfunctional may require a certain time to enter the population (1/(2Nμ) generations on the average if Nμ is small), and, if they do get fixed, the time will be 4N generations. Thus, the integrity of a gene, and its ability to code for a functional protein, may persist for a long period purely because of chance alone, unless selection favors degeneration. In this case the fixation time depends inversely on the selective coefficient and directly on the logarithm of the population size (Kimura and Ohta, 1969). The period of time that the Somalian caves have been inhabited by populations of P. andruzzii is unknown, although the desertification of the surface environment, which occurred at ∼2.8 Myr, gives us an approximate lower limit for the colonization event. Adaptation to constant darkness probably started before the complete isolation of the Somalian cavefish, although the time required to reach this extreme level of troglomorphism is still unknown. Inferences on the pattern of molecular evolution of proteins involved in the development of troglomorphic traits can be exploited to reconstruct the evolutionary history of P. andruzzii. For example, environmental change such as the colonization of a new environment often eliminates or weakens a source of selection that was formerly important for the maintenance of a particular trait but then subsequently lost its adaptive value (Lahti et al., 2009). Proteins contributing to determining the trait start to evolve neutrally, accumulating amino acid substitutions and LOF mutations that will eventually impair their function (Motoo, 1983). An alternative hypothesis is that losing the trait may directly or indirectly increase the fitness of an organism in the new environment (Espinasa and Espinasa, 2008). In this case, after a LOF mutation gets fixed, the gene loses its ability to code for a functional protein and its neutral evolution begins. Opposing hypotheses invoking neutral mutation or natural selection have been advanced to explain regressive evolution in subterranean animals, and Astyanax cavefish, in particular (Poulson and White, 1969; Wilkens, 1988; Culver and Wilkens, 2000; Jeffery, 2005, 2009; Wilkens, 2010).
If new environmental conditions mean relaxation of natural selection on a specific trait, the time when the trait was freed of constraint can be put in relation with the time since colonization of the new habitat (Wang et al., 2004; Zhao et al., 2010). In the case of P. andruzzii, characterizing the molecular evolution of a mutated photoreceptor gene and estimating when selection became relaxed would provide insight into the time of cave colonization. This will in turn shed light on the mechanisms underlying regressive evolution in this species.
With this aim, we first tested the hypothesis that the lack of response to light of the circadian clock of P. andruzzii observed by Cavallari et al. (2011) results from a change in selective regime on the light-responsive mechanism, as expected when the phenotypic trait is no longer adaptive. In particular, we investigated the molecular evolution of the nonvisual photoreceptor melanopsin opn4m2, whose premature stop codon accounts at least in part for the inability of peripheral clocks to respond to light. To address the question of whether other light-related mechanisms are also undergoing degeneration, we also tested for a change in the rate of evolution on the visual receptor rhodopsin (rho) that is expressed in the brain of P. andruzzii, is still functional and is probably involved in photophobic behavior (Tarttelin et al., 2012). These gene sequences were also analyzed in another blind cavefish, Garra barremiae, from Oman that much more recently colonized subterranean waters and evolved troglomorphic traits (Kruckenhauser et al., 2011). The two cavefish show an extremely similar osteology (Banister, 1984), and a recent phylogenetic study proposed that Phreatichthys should be treated as a junior synonym of Garra (Yang et al., 2012). Similar to P. andruzzii, the eyes of G. barreimiae are submitted to ontogenetic regression. Although the juveniles develop eyes, they are completely reduced during growth to the adult stage, and hence the adults have no externally visible eyes, and the optic lobes are not fully developed (Banister, 1984). The time of divergence between the two cavefish species was estimated using a fossil-calibrated phylogeny of mitochondrial cytochrome b (cytb) sequences of 148 teleost species. This divergence time and the results of our selection analyses were used to date when selection became relaxed on opn4m2. Based on our results we conclude that the change in selective regime that followed the colonization of the new environment predates the desertification of Somalia by ∼2.5 million years. This indicates that the complete isolation of P. andruzzii may have occurred several million years after cave colonization.
Materials and methods
Cavefish samples
The P. andruzzii and G. barreimiae individuals typed in this study are described in Table 1 (Berti and Messana, 2010; Kruckenhauser et al., 2011).
Table 1. Description of Phreatichthys andruzzii and Garra barreimiae individuals analyzed in this study with sampling localities.
| Species | Individual IDs | Locality | Latitude/longitude | Phenotypes |
|---|---|---|---|---|
| P. andruzziia | PA07 Cavefish | Bud Bud (Somalia) | 04°11'19''N/46°28'27''E | Troglomorphic |
| PA11 Cavefish | Gheriale (Somalia) | 04°08'22''N/46°29'05''E | Troglomorphic | |
| G. barreimiaeb | Gbar215 | Wadi Al Falahi (Oman) | 23°04'50''N/57°21'14''E | Surface |
| Gbar216 | Wadi Al Falahi (Oman) | 23°04'50''N/57°21'14''E | Surface | |
| Gbar223 Cavefish | Hoti Pit (Oman) | 23°06'10''N/57°22'12''E | Troglomorphic | |
| Gbar224 Cavefish | Hoti Pit (Oman) | 23°06'10''N/57°22'12''E | Troglomorphic | |
| Gbar284 | Wadi Misfat (Oman) | 23°08'52''N/57°18'57''E | Surface |
Hoti Pit is a cave adjacent to the Al Hoota cave described in Kruckenhauser et al. (2011).
Molecular techniques
The genomic DNA of two specimens from two different sampling sites of P. andruzzii (Bud Bud and Gheriale) and of five specimens from three sampling locations of G. barreimiae was extracted using DNAeasy blood and tissue kit (Qiagen, Venlo, The Netherlands) following the manufacturer's protocol. Part of the opn4m2 gene (1187 bp spanning nucleotides from 40 to 1226, premature stop codon position: nucleotide 886, CDS length: 1503 in zebrafish ENSDARG00000007553) and the entire rhodopsin (rho) gene were amplified and Sanger sequenced in each of the 7 individuals (Supplementary Methods).
To estimate the divergence time between P. andruzzii and G. barreimiae, the mitochondrial cytb gene was amplified and sequenced in both species with primers Glu32-fwd 5′-GACTTGAAGAACCACCGTTGT-3′ and Thr1-rev 5′-GATCTTCGGATTACAAGACCG-3′ (Supplementary Methods). A fragment of 903 bp of P. andruzzii from the ATG start codon was aligned with the entire coding region of G. barreimiae cytb and with previously published cytb sequences available in GenBank (see the section ‘Analysis of divergence time between P. andruzzii and G. barreimiae') and used for analysis.
For the three genes, the coding sequences were translated into their respective protein sequences and aligned using ClustalW available in MEGA version 6 (Tamura et al., 2013) with default parameters. Once aligned, the protein sequences were translated back to their respective nucleotide sequences that were used for the following analyses.
Estimation of the functional impact of nonsynonymous substitutions
For both the opn4m2 and rho genes, we identified amino acids at a specific position in only one of the five cyprinid lineages of the tree shown in Figures 1a and b (Phreatichthys andruzzii, Garra barreimiae, Danio rerio, Cyprinus carpio and Rutilus rutilus or Carassius auratus, accession numbers are listed in Supplementary Table 1). We considered those amino acids to be the result of a nonsynonymous substitution that, at present, characterizes only that specific branch (hereafter ‘species-specific'). As the aim of the analysis was to compare the effect of fixed nonsynonymous substitutions that differ among species, in the case of P. andruzzii and G. barreimiae polymorphic nonsynonymous substitutions were not considered. We predicted the functional impact of each species-specific amino acid change using two different approaches, both based on the degree of conservation of specific residues in evolutionary related proteins.
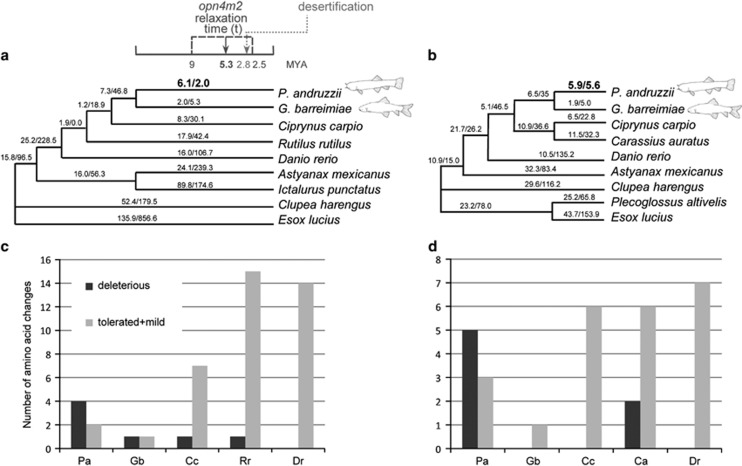
Figure 1.
The two species trees used for CODEML analyses of (a) melanopsin (opn4m2) and (b) rhodopsin (rho) genes. Branch lengths are not drawn to scale. The two numbers shown along each branch are the maximum likelihood estimates of the number of nonsynonymous and synonymous substitutions (N*dN and S*dS) along that branch (CODEML, free ratio model). Dates indicated with the dark gray arrow and dashed lines are inferred from the molecular genetic evidence of this study (opn4m2 relaxation time), whereas for the shift in climatic condition (desertification) we referred to deMenocal (1995). Melanopsin (c) and rhodopsin (d) species-specific amino acid changes are more likely to be deleterious in P. andruzzii (Pa) compared with changes in other cyprinid species (Ca, Carassius auratus; Cc, Ciprynus carpio; Dr, Danio rerio; Gb, Garra barreimiae; Pa, Phreatichthys andruzzii; Rr, Rutilus rutilus).
First, we used the Coding SNP Analysis tool (Thomas et al., 2006) based on the PANTHER database (http://pantherdb.org/) that calculates a score called ‘substitution position-specific evolutionary conservation' (subPSEC). This score corresponds to the negative logarithm of the probability ratio of the wild-type and mutant amino acids at a particular position. PANTHER subPSEC scores are converted to a probability of deleterious effect on protein function; a subPSEC of −3 is the cutoff point for functional significance and corresponds to a Pdeleterious of 0.5 (50% probability that the substitution is deleterious).
Second, we applied the SIFT sequence prediction tool, interrogating three different databases: UniRef90, UniProt-SwissProt and UniProt-TrEMBL (http://sift.jcvi.org/) (Kumar et al., 2009). Probability values of ⩽0.05 indicate that the substitution would not be phenotypically tolerated (deleterious).
Overall, the substitutions were considered ‘deleterious' if three tests out of four predicted a deleterious effect, ‘mild' if two out of four predicted a deleterious effect and, otherwise ‘tolerated'. We tested whether the P. andruzzii or G. barreimiae observed proportion of deleterious and tolerated/mild substitutions differ significantly from those of the other species by means of a χ2 test of independence with Yates' correction.
Detecting selection (and selection relaxation) acting on particular lineages
To analyze the selective constraints on opn4m2 and rho, we estimated the nonsynonymous/synonymous rate ratio (ω=d(N)/d(S)), under different evolutionary models in a likelihood framework. This ratio is <1 under purifying selection, approaches 1 under neutral rates of evolution and is >1 under positive selection. For both genes, we analyzed the coding sequence of nine teleost fishes (Figures 1a and b), including those of P. andruzzii (showing a premature stop codon at nucleotide 886 of the opn4m2 sequence) and G. barreimiae (uninterrupted open reading frame) from this study. In particular, we used the sequence of P. andruzzii from Bud Bud (PA07), and of G. barreimiae from the Hoti Pit cave (Gbar223 and Gbar224 are identical and homozygotes at both genes, see Supplementary Table 2). Orthologous sequences were retrieved using BLAST protein: we considered melanopsin protein sequences with query cover >90% and excluded isoforms and low-quality proteins (default algorithm parameters, database: non-redundant protein sequences, organism: Teleostei). As the melanopsin of P. andruzzii is truncated, the known orthologous sequence of zebrafish was used as a query. The respective coding sequences were obtained from GenBank and aligned using the MAFFT online version with default settings (Katoh and Standley, 2013). An alignment of rhodopsin coding sequences was obtained with the same procedure, including, when available, the same species of the melanopsin alignment (see Supplementary Table 1 for Ensembl or GenBank IDs). Codons showing indels were not considered in the analyses.
Maximum likelihood trees for the coding sequences of melanopsin and rhodopsin were estimated using the program RAxML 8.1.2 (Stamatakis, 2014) and the GTR+gamma substitution model. The inferred topologies are shown in Figures 1a and b and were used as input (unrooted tree) for the following tests for selection. Three different approaches were used: Branch models (Yang, 1998), Clade models (Bielawski and Yang, 2004; Weadick and Chang, 2012) and Branch-site models (Zhang et al., 2005). Codon frequencies were approximated using the F3x4 calculation. Each model was fit to the data multiple times from different starting ω or k (the transition/transversion rate ratio) values to avoid local optima.
In particular, we considered the next Branch models: the simplest model assumes the same ratio for all branches in the phylogeny (‘one-ratio' model); a second and nested model assumes that the branches of interest (in our case P. andruzzii or G. barreimiae) have a dN/dS ratio (ωPa or ωGb) that is different from the background ratio ω0 (‘two-ratio model', both ω estimated). We also considered a two-ratio model where ω was fixed at 1 in the lineage of P. andruzzii or G. barreimiae, as we are specifically interested in testing for relaxed molecular evolution in dark conditions. Finally, the ‘free-ratio' model that assumes an independent dN/dS ratio for each branch in the phylogeny was used to calculate branch-specific rate of variation. The analyses were performed using the CODEML module of PAML package (Yang, 2007).
Although Branch models assume that ω varies across branches of the phylogeny, but that it is invariant across sites, Clade models and Branch-site models incorporate among-site variation in selective constraint. We tested for the signature of divergent selective constraint at specific sites between the foreground branch (either P. andruzzii or G. barreimiae) and the background branches (the rest of the phylogeny) using the Clade model C (CmC). This model assumes three site classes, Class 0 and Class 1 corresponding to codons that evolve under purifying selection (0<ω0<1) and neutral evolution (ω1=1), respectively. These two site classes are assumed to be shared between foreground and background branches, whereas the selection pressure at the third class can differ between the background and the foreground (ω2≠ω3). M2a_rel (Weadick and Chang, 2012) assumes three classes of sites, 0<ω0<1, ω1= 1, and ω2>0, shared by background and foreground branches, and was used as a null model to test for the presence of divergently selected sites. A Bayes empirical Bayes approach was used to identify specific codons with high posterior probability of being in the divergent class (Yang et al., 2005).
Clade models and Branch-site models similarly assume different classes of sites (three and four, respectively), the first two (0<ω0<1 and ω1= 1) shared between the background and the foreground. However, in Branch-site models the final two classes correspond to sites that are under purifying selection or neutral evolution in the background branches, but positive selection (ω2>1) in the foreground branches. The null model to test for the presence of positively selected sites assumes ω2=1.
The likelihoods of different models were then compared by means of likelihood ratio tests (LRTs) of hypotheses through comparison of nested statistical models. The LRT was calculated as twice the difference in the log-likelihoods values between models. The significance of the LRT statistic was determined by using the χ2 distribution where the degrees of freedom equal the difference between the number of estimated parameters in the two nested models (Tables 2 and 3).
Table 2. Log-likelihood values (lnL), parameter estimates and likelihood ratio tests for various Branch-based models testing for heterogeneous selection pressures for melanopsin and rhodopsin.
| Branch model | ω0 | ωPa | ωGb | lnLa | npb | H0 | H1 | 2ΔlnLc | d.f. | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Melanopsin (opn4m2) | ||||||||||
| A. One ratio: all branches have the same ω | 0.08 | ω0 | ω0 | −5800.498 | 17 | |||||
| B. One ratio: all branches have the same ω=1 | 1 | ω0 | ω0 | −6407.852 | 16 | B | A | 1214.708 | 1 | 3.88E−266 |
| C. Two ratio: (ω0), ωPa | 0.077 | 1.189 | ω0 | −5794.580 | 18 | A | C | 11.836 | 1 | 0.0006 |
| D. Two ratio: (ω0), ωPa =1 | 0.077 | 1 | ω0 | −5794.597 | 17 | D | C | 0.033 | 1 | 0.856 |
| E. Two ratio: (ω0), ωGb | 0.079 | ω0 | 0.167 | −5800.181 | 18 | A | E | 0.634 | 1 | 0.426 |
| F. Free ratio (each branch has one ω) | Variable by branchd | 1.127 | 0.137 | −5777.164 | 31 | C | F | 34.832 | 13 | 0.001 |
| Rhodopsin (rho) | ||||||||||
| A. One ratio: all branches have the same ω | 0.055 | ω0 | ω0 | −4379.978 | 17 | |||||
| B. One ratio: all branches have the same ω=1 | 1 | ω0 | ω0 | −4971.150 | 16 | B | A | 1182.344 | 1 | 4.19E−259 |
| C. Two ratio (ω0), ωPa | 0.053 | 0.209 | ω0 | −4378.015 | 18 | A | C | 3.926 | 1 | 0.048 |
| D. Two ratio: (ω0), ωPa =1 | 0.053 | 1 | ω0 | −4379.183 | 17 | D | C | 2.337 | 1 | 0.126 |
| E. Two ratio: (ω0), ωGb | 0.054 | ω0 | 0.110 | −4379.770 | 18 | A | E | 0.414 | 1 | 0.520 |
| F. Free ratio (each branch has one ω) | Variable by branchd | 0.199 | 0.072 | −4365.531 | 31 | C | F | 24.968 | 13 | 0.023 |
Abbreviations: Gb, Garra barreimiae; H0 and H1, null and alternative hypothesis; Pa, Phreatichthys andruzzii.
The best-fit model is underlined.
ln(likelihood) value.
Number of parameters.
Twice the difference of ln(likelihood) between the two models compared.
(N*dN) and (S*dS) values are shown in Figures 1a and b.
Analysis of divergence time between P. andruzzii and G. barreimiae
Divergence time between P. andruzzii and G. barreimiae was estimated using the Bayesian implementation Phylobayes 3.3f (Lartillot et al., 2009). We used a dating strategy involving multiple calibration points based on 20 fossil constraints obtained from the literature (Near et al., 2012; Wang et al., 2012; Supplementary Table 3) coupled with a root prior for the origin of Neopterygii of 360 Myr and a permissive s.d. of 20 Myr (Near et al., 2012). The data set included the first 903 nucleotide positions of the cytb gene for 148 Teleost species (26 of which were cyprinids, including the specimens of P. andruzzii and G. barreimiae that were sequenced in this work) and an Amiid (Amia calva) as outgroup (Supplementary Tables 3 and 4). Details of the analysis are described in the Supplementary Methods.
Dating the relaxation of selection on the opn4m2 gene in P. andruzzii
To estimate when the functional constraint on the P. andruzzii opn4m2 gene became relaxed, we followed a method described in Zhao et al. (2010), based on the rate of nucleotide substitution across the tree of Figure 1a. According to this method, we assumed that the level of selective constraint observed in the other branches of the phylogeny became suddenly and completely relaxed in the P. andruzzii branch t Myr. Time of relaxation of selection t was estimated as:
where dPa is the total number of nonsynonymous substitutions per site for the P. andruzzii branch, d0 is the total number of nonsynonymous substitutions per site for all the branches of the phylogeny except the P. andruzzii branch, D0 is the total number of synonymous and nonsynonymous substitutions for the same branches and DGb is the total number of synonymous and nonsynonymous substitutions for the G. barreimiae branch. dPa, d0, D0 and DGb were obtained running a free-ratio model in PAML that assumes a different dN/dS ratio for each branch in the tree. The ω is the dN/dS ratio before the functional relaxation, that is, the background ratio in a two-ratio Branch model of PAML where P. andruzzii is set as foreground (Branch model C, Table 2). T is the estimated divergence time between P. andruzzii and G. barreimiae. Specifically, we estimated the divergence time between the two cavefish at 15.1 Myr, with a 95% support interval between 7.4 and 26.7 Myr. This age is the average of the point estimates obtained in the three Phylobayes analyses, and the support interval is conservatively computed using the lowest and the highest limits of the three 95% highest posterior density intervals, jointly considered (Supplementary Table 5; Rota-Stabelli et al., 2013). Random sampling from a normal approximation of the posterior distribution of T produced by Phylobayes was used to estimate the uncertainty of t.
Results
Genetic variation of opn4m2 and rho within and between P. andruzzii and G. barreimiae
In P. andruzzii the deletion at position 815 (codon 272) of opn4m2 that causes a frameshift leading to a premature stop codon 24 amino acids downstream (Cavallari et al., 2011) was observed in each of the four chromosomes typed in this study. Within P. andruzzii we observed three polymorphic sites, all of them synonymous (Supplementary Table 2). The opn4m2 gene showed four polymorphic positions in surface and cave individuals of G. barreimiae (six and four chromosomes, respectively), two of which are nonsynonymous and carried by surface individuals (Supplementary Table 2). Conversely, the two individuals from the cave are identical and homozygous at this locus. Comparing the two species, we observed seven synonymous and eight nonsynonymous fixed differences.
Within P. andruzzii the rho gene shows two synonymous and four nonsynonymous polymorphic positions, whereas no variation was identified within G. barreimiae. Comparing the two species, we observed eight synonymous and eight nonsynonymous fixed differences. When the rho gene was first characterized in P. andruzzii, an in-frame insertion of 3 bp was identified in comparison with zebrafish, with the cavefish showing an additional valine at amino acid position 349 (Tarttelin et al., 2012). As the total length of the protein is 355 amino acids, the valine insertion might not have major phenotypic consequences. In addition, this protein region is rather repetitive, and this may have caused recurrent insertion and deletion, as also supported by the lack of three amino acids in G. barreimiae compared with P. andruzzii (or two in comparison with zebrafish) (Supplementary Table 2).
Predictions of the functional impact of nonsynonymous mutations
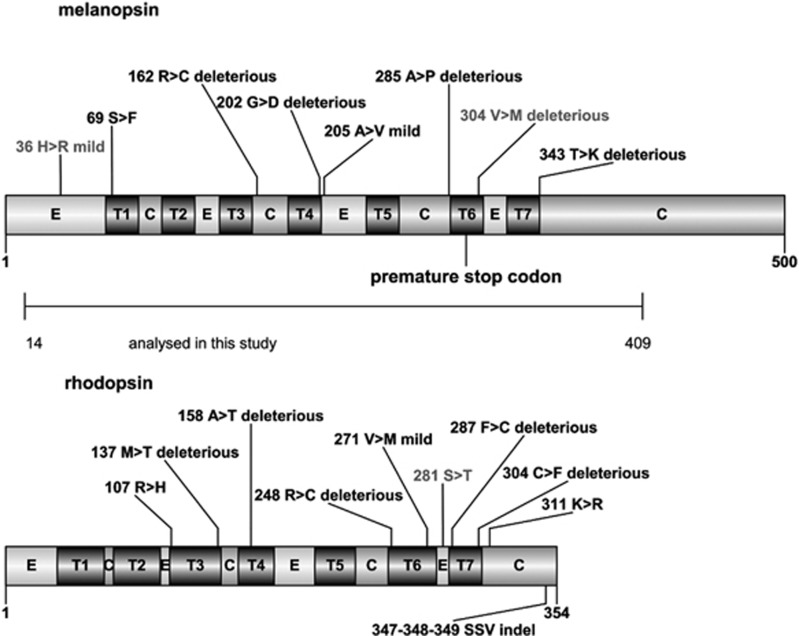
We predicted the effects of the species-specific amino acid changes for the five species of cyprinids shown in Figures 1a and b for both the opn4m2 and rho genes. We observed a significant excess of nonsynonymous substitutions predicted to be deleterious in the opn4m2 gene of P. andruzzii compared with the other cyprinid species (χ2 test with Yates' correction P<0.005) (Figures 1c, d and 2 and Supplementary Table 6). One of the six amino acid changes was located downstream of the premature stop codon (K343T, deleterious). Conversely, the melanopsin of G. barreimiae showed only two species-specific amino acid changes, one mild and one deleterious, and the six amino acid changes shared by the two cavefish are almost completely tolerated. These results are expected given that P. andruzzii melanopsin is truncated and its photoreceptor activity is abolished (Cavallari et al., 2011), thus purifying selection should no longer preserve the protein structure. However, the significant excess of nonsynonymous substitutions predicted to be deleterious in P. andruzzii compared with the other cyprinid species is less expected for the functional rhodopsin (χ2 test with Yates's correction P<0.005) (Tarttelin et al., 2012). Conversely, G. barreimiae showed only one species-specific amino acid substitution, and this change showed a predicted tolerated effect (Figures 1c, d and 2 and Supplementary Table 6).
Estimation of synonymous and nonsynonymous substitution rates and detection of natural selection on cavefish lineages
We used maximum likelihood estimates of parameters for each branch of the trees shown in Figures 1a and b under the CODEML free-ratio model to calculate the number of nonsynonymous and synonymous substitutions per lineage. The P. andruzzii branch shows a higher number of estimated nonsynonymous substitutions compared with synonymous ones. This unique pattern may be the result of a different relative rate of evolution (neutral or adaptive) of this cavefish lineage relative to the rest of the tree. To formally test this hypothesis, we compared a series of evolutionary models in the likelihood framework. The results obtained from Branch model analyses are summarized in Table 2. We found that the average ω across the opn4m2 tree is 0.08 and differs significantly from 1 (model A vs B, LRT, P<10−60), as expected if purifying selection is in general acting on the protein. A two-ratio model with P. andruzzii as the foreground branch (estimated ωPa=1.189) compared with the rest of the tree (estimated ω0=0.077) fits the data better than the one-ratio (model C vs A, LRT, P<10−3). This indicates neutral evolution of the CDS, as the neutral model with ωPa=1 fitted the data better than the previous (model C vs D, LRT, P=0.856), suggesting an evident relaxation of natural selection on this protein. In the other blind cavefish, G. barreimiae, opn4m2 did not show a significantly different rate of evolution compared with the rest of the tree (ωGb: 0.167; ω0: 0.079; model E vs A, LRT, P=0.426), indicating that the protein is conserved in this lineage. The free-ratio model (F) fits the data significantly better than the two-ratio model (C), where P. andruzzii is the foreground and the rest of the species are the background (model F vs C, LRT, P=0.001). This indicates that the dN/dS ratios are indeed different among background lineages (Figure 1a).
Exactly the same pattern was observed for rhodopsin. In P. andruzzii the protein seems to have lost the signature for selective constraint, whereas in G. barreimiae it is under purifying selection. In this case, however, the level of constraint across the tree (ω=0.055) seems more pronounced than for melanopsin (ω=0.08), and the estimated ωPa is lower (0.209), although the model with ωPa=1 is still preferred (model C vs D, LRT, P=0.126).
To test for divergent selection across the phylogeny incorporating among-site variation in selective constraint, we applied Clade model analyses on both the opn4m2 and rho data sets (Table 3). P. andruzzii branch showed a signature of significant divergent selection at ∼60% of the melanopsin sites, with ω2=0.014 on the background lineages and ω3(Pa)=1.37 on the cavefish lineage (CmC_Pa vs M2a_rel, LRT, P<10−3). Of the 392 analyzed codons, 22 had a posterior probability of >0.70 to come from the class of divergent sites (Bayes empirical Bayes analysis under the CmC_Pa model). The 22 sites were rather homogeneously distributed across the protein sequence, and included all the six amino acid changes specific to P. andruzzii (Table 4 and Supplementary Table 6). Conversely, G. barreimiae opn4m2 did not show signature of site-specific divergence from the background (CmC_Gb vs M2a_rel, LRT, P=0.343). The results obtained from Clade model analyses of rho failed to identify a class of divergent sites, although the P. andruzzii trend was close to significance (ω2=0.001 and ω3Pa=0.059; CmC_Pa vs M2a_rel, LRT, P=0.06).
Table 3. Log-likelihood values (lnL), parameter estimates and likelihood ratio tests for Clade model analyses of the melanopsin and rhodopsin data set.
| Clade model |
Site class 0 |
Site class 1 |
Site class 2 |
lnLa | npb | 2ΔlnLc | d.f. | P-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ω0 | p0 | ω1 | p1 | ω2, ω3 | p2 | ||||||
| Melanopsin (opn4m2) | |||||||||||
| CmC_Pa | 0.180 | 0.375 | 1 | 0.019 | ω2=0.014 ω3(Pa)= 1.370 | 0.606 | −5706.616 | 21 | 21.721 | 1 | 3.154E−06 |
| CmC_Gb | 0.017 | 0.614 | 1 | 0.019 | ω2=0.184 ω3(Gb)= 0.484 | 0.367 | −5717.026 | 21 | 0.8999 | 1 | 0.343 |
| M2a_rel | 0.017 | 0.617 | 1 | 0.019 | ω2= ω3=0.187 | 0.365 | −5717.476 | 20 | |||
| Rhodopsin (rho) | |||||||||||
| CmC Pa | 0.147 | 0.329 | 1 | 0.028 | ω2= 0.0013 ω3(Pa)= 0.059 | 0.643 | −4279.794 | 21 | 3.420 | 1 | 0.064 |
| CmC Gb | 0.154 | 0.314 | 1 | 0.027 | ω2=0.0031 ω3(Gb)= 0.0 | 0.659 | −4281.461 | 21 | 0.086 | 1 | 0.770 |
| M2a_rel | 0.152 | 0.317 | 1 | 0.028 | ω2= ω3=0.0028 | 0.655 | −4281.504 | 20 | |||
ln(likelihood) value.
Number of parameters.
Twice the difference of ln(likelihood) between the two models compared.
Table 4. Melanopsin sites identified by Bayes empirical Bayes analysis as coming from the class of divergent sites under the CmC_Pa model (PP cutoff=70%).
| Amino acid site | PP | Protein domain |
|---|---|---|
| 34 | 0.73301 | Extracellular |
| 37 | 0.83092 | Extracellular |
| 45 | 0.81544 | Extracellular |
| 50 | 0.712 | Extracellular |
| 69a | 0.9634 | Transmembrane 1 |
| 160 | 0.77478 | Transmembrane 3 |
| 162a | 0.78929 | Cytoplasmic |
| 188 | 0.70835 | Transmembrane 4 |
| 192 | 0.70139 | Transmembrane 4 |
| 202a | 0.73608 | Transmembrane 4 |
| 205a | 0.76531 | Extracellular |
| 235 | 0.70811 | Transmembrane 5 |
| 261 | 0.72722 | Cytoplasmic |
| 265 | 0.73131 | Cytoplasmic |
| 271 | 0.7328 | Cytoplasmic |
| 274 | 0.79893 | Cytoplasmic |
| 285a | 0.76222 | Cytoplasmic |
| 343a | 0.81869 | Transmembrane 7 |
| 366 | 0.70826 | Cytoplasmic |
| 370 | 0.71896 | Cytoplasmic |
| 376 | 0.8297 | Cytoplasmic |
| 397 | 0.78226 | Cytoplasmic |
Abbreviation: PP, posterior probability.
Amino acid changes specific of Phreatichthys andruzzii branch. The amino acid changes predicted to be deleterious by Panther and SIFT analyses are underlined (Supplementary Table 6).
The possibility that the divergence shown by P. andruzzii melanopsin is due to positive selection was tested using Branch-site models. Branch-site model A estimated that all the sites come from the third and fourth classes (that is, purifying selection or neutral evolution on the background and positive selection on the foreground), whereas no sites are consistently under purifying selection or neutral evolution on background and foreground branches. Under this model, 7% of sites evolve neutrally (ω1=1) and 93% are under purifying selection on the background lineages (estimated ω0=0.061), and the estimated ω for these sites in P. andruzzii is ωPa=1.135. The constrained null model with ωPa fixed at 1 fitted the data better than Branch-site model A (LRT, P=0.45).
In summary, the results of Branch models and Clade models consistently suggest that selective constraint was relaxed on melanopsin in P. andruzzii. The ω3(Pa)=1.37 estimated by CmC may indicate the action of weak positive selection as well, a scenario that was not supported by Branch-site models. Rhodopsin showed a signature of relaxation in P. andruzzii under different Branch models, whereas CmC showed a similar trend, but the divergence between the cavefish lineage and the background was not significant. Both proteins were under selective constraint in G. barreimiae.
Dating the relaxation of selection on the melanopsin photoreceptor
To estimate when the selective pressure on P. andruzzii melanopsin became relaxed, we used two sources of information: the estimated rates of synonymous and nonsynonymous substitutions across the lineages of the tree shown in Figure 1a and the estimated divergence time between P. andruzzii and G. barreimiae (T). Posterior estimates using Phylobayes indicate a mean divergence time between the two cavefish species of 15.1 Myr, although with a large range of uncertainty when considering 95% posterior densities (7.4–26.7 Myr) (Supplementary Table 5 and Supplementary Figures 1 and 2).
The divergence time between the two cavefish, T, and its 95% interval were used to calculate when the functional constraint on the P. andruzzii opn4m2 gene became relaxed. The frequency distribution of relaxation time ranges from 2.5 to 9.0 Myr, with mean and median of 5.3 Myr (Figure 1a).
Discussion
Statistically supported evidence for relaxed selection on melanopsin and rhodopsin photoreceptors
Studies on the mechanism of regressive evolution often start from the identification of a regressed phenotypic trait, connect the trait to a specific environmental condition and then test for relaxed selection on candidate genes possibly involved in the phenotype. If relaxation of selection is the best model to explain the observed data, then the existence of a causal relationship between the new environment, the genotype and the regressive phenotype is inferred. For example, Niemiller et al. (2013) studied regressive evolution in the amblyopsid blind cavefish from North America investigating whether the candidate gene rhodopsin showed evidence of relaxed selection. The authors concluded that the loss of functional constraints caused parallel inactivation of the protein in different cave lineages, suggesting multiple independent cave colonization events. Similarly, Veilleux et al. (2013) studied the evolution of the opn1sw gene, coding for visual receptors sensitive to diurnal short wavelengths, in lemurs living in a nocturnal environment. The ‘regressive' hypothesis was that monochromatic primates may have lost the dichromatic color vision as a result of the relaxation of selective constraints on genes such as opn1sw, as the ability to perceive short wavelengths would be useless in nocturnal animals. The study concluded that selection on opn1sw was weakened or relaxed in animals living in closed canopy rainforests, although the gene, and thus dichromacy, was still under strong purifying selection in similar species living in a brighter environment.
In our study, we did not have to hypothesize that gene may have lost the selective constraints that, in turn, may have led to the regression of the phenotype. Instead, we started from the results of comparative functional analysis of the circadian clock that proved how specific LOF mutations on photoreceptors caused the impairment of an important physiological and adaptive mechanism (Cavallari et al., 2011). Specifically, the LOF mutation, the gene, the phenotype and the environmental condition that led to the regression were already well defined. Having this information available, we could test whether the experimentally proven relationship between an event of pseudogenization and a regressive phenotype arose by relaxation of natural selection in a new environment. Our results suggest that the melanopsin protein is under neutral evolution in P. andruzzii, most probably as a consequence of the loss of selective constraint on this photoreceptor in the dark environment. Although this conclusion may sound obvious given that the opn4m2 gene is actually a pseudogene, statistically supported evidence of relaxed selection have rarely been found in similar studies (Leys et al., 2005; Li and He, 2009; Zhao et al., 2010), possibly because statistical tests have little power if the change in selective regime is relatively recent. To generalize our conclusions, we tested for the presence of relaxed selection in a second protein, the widely studied photoreceptor rhodopsin. We expected this protein to be under purifying selection in P. andruzzii for the following reasons: (1) in this cavefish the rho gene shows an intact open reading frame that encodes a full-length opsin photoreceptor, (2) functional experiments have proven its ability to respond to light and (3) behavioral experiments suggest that it may be involved in negative phototaxis (Tarttelin et al., 2012). Indeed, in analyzing this gene in both cave and surface G. barreimiae individuals (10 chromosomes) we did not observe a single nucleotide change within the species (Supplementary Table 2), and comparison with other cyprinid species showed only one species-specific amino acid change that should not affect the function of the protein (Figure 2). Conversely, P. andruzzii showed both intra- and inter-specific variation in amino acids, most of which with a predicted disrupting effect (Figure 2), although crucial amino acid residues for the photoreception are conserved, such as a lysine (K296) in the seventh transmembrane domain acting as a Schiff's base linkage to the chromophore and the ERY[W] motif (134–136), necessary to bind the α-subunit of transducine (Tarttelin et al., 2012). We believe that this observation, together with the statistical support obtained by a model for neutral evolution of the rho gene on P. andruzzii branch (Table 2), reinforce our conclusion that relaxed selection operates on light-responsive mechanisms in this species.
Figure 2.
Graphic representation of the two proteins analyzed in this study. Species-specific nonsynonymous fixed substitutions of P. andruzzii (black) and G. barreimiae (gray) are indicated together with their predicted effect. C, cytoplasmic domain; E, extracellular domain; T, transmembrane domain.
Besides neutrality (Wilkens, 1988, 2010), other theories have been proposed to explain how regressive evolution may happen (Espinasa and Espinasa, 2008). In particular, two main hypotheses have been explored: first, positive selection directly eliminates traits, either because the reduction in function is itself beneficial or because it allows the preservation of energy (Protas et al., 2007; Lang et al., 2012); second, positive selection indirectly causes the loss of a trait as a by-product of the gain of a new adaptive function, through mechanisms such as genetic hitchhiking or pleiotropy (Jeffery, 2009). It may be argued that the neutral scenario supported by our results as the most plausible explanation for the evolution of a ‘blind' circadian clock does not definitively rule out alternative possibilities. In particular, the Branch models implemented in CODEML were originally designed to detect adaptive evolution by positive selection on a specific lineage (Yang, 1998), although it is very commonly used in the literature for cases of relaxed selection (Li and He, 2009; Niemiller et al., 2013; Zhao et al., 2010; Feng et al., 2014; Marková et al., 2014). As only a few sites may be affected by positive selection in an otherwise conserved background, this approach of averaging rates over sites and over time may have little power (Yang and Nielsen, 2002). If that was also the case for the opn4m2 gene of P. andruzzii, a lack of power of the test may be the reason why ωPa=1.189 resulted not significantly higher than 1 (Table 2). To overcome this limitation, we analyzed our data sets with Clade models that account for heterogeneity across sites. The results show that ∼60% of the analyzed melanopsin sites are evolving at a different rate in P. andruzzii compared with the rest of the tree (Tables 3 and 4). The estimated ωPa=1.37 would indicate that selective constraint was relaxed on this lineage, although the action of weak positive selection cannot be ruled out. However, the hypothesis that a class of sites is evolving under positive selection in P. andruzzii was not supported by a third approach based on Branch-site models. In addition, it is difficult to imagine a direct or indirect adaptive explanation for the inactivation of this melanopsin photoreceptor. In the case of eye regression, the energy-saving hypothesis was not completely dismissed, but still lacks experimental support. In particular, the burden posed by the development and the maintenance of the eye on the organism's survival in the cave is still unclear (Sadoglu, 1967; Protas et al. 2007; Espinasa and Espinasa, 2008). Such a direct benefit seems even less convincing in the case of a single nonvisual photoreceptor that is unlikely to contribute to eye maintenance. Furthermore, its downstream signaling pathway is still sensitive to other external signals and, therefore, still partially functional (Cavallari et al., 2011). As for the indirect outcome of positive selection, at present, improvements in other physiological mechanisms due to the loss of melanopsin function are hard to envisage. In addition, the selection coefficient should be high enough to counteract the effect of genetic drift in populations that presumably have rather limited effective population sizes (Bradic et al., 2012). As at this point no credible selective hypothesis can be formulated or tested, thus we are confident to conclude that neutrality explains the regressive evolution of melanopsin in P. andruzzii.
Divergence time between the two cavefish P. andruzzii and G. barreimiae
The divergence time between the two cavefish P. andruzzii and G. barreimiae was required to infer when the selective pressure on the opn4m2 gene became relaxed in P. andruzzii. Using a Bayesian approach and the cytb as a marker, Colli et al. (2008) found that divergence times at 95% highest posterior density interval were 8.3–11.5 Myr for the subclade including P. andruzzii and G. barreimiae and 0.8–2.0 Myr for the epigean and hypogean G. barreimiae. However, Kruckenhauser et al. (2011) estimated a split below 1 Myr between the two forms of G. barreimiae, and their experiments indicate that the sequences of the cytb gene used by Colli et al. (2008) may be derived partly from the functional cytb gene and partly from nuclear pseudogenes (numts (copies of mitochondrial genes that have been translocated into the nuclear genome)). For this reason, in the present study we resequenced the cytb gene in both G. barreimiae and P. andruzzii cavefish and analyzed this together with cytb sequences from an additional 146 species (Supplementary Figures 1 and 2). We provide a posterior mean estimate of 15.1 Myr and 95% highest posterior density interval of 7.4–26.7 Myr for the divergence between the two species, an estimate that turned out to be robust to changes in tree prior (Birth-Death or Uniform), and replacement model (GTR or CAT) (Supplementary Table 5).
Estimating the time of the Somalian cave colonization
If the absence of light typical of the cave environment caused the relaxation of selective pressure on the melanopsin protein, estimating when the molecule was freed from constraint would allow us to infer when the cave was colonized. After concluding that the melanopsin in P. andruzzii shows a signature of relaxation of natural selection, we could assume a neutral rate of evolution for this genomic sequence, and we used this information to estimate for how long the cavefish has been in the dark environment. We can confidently assume that P. andruzzii populations have been isolated under the Somalian desert at least since the desertification of the surface, but what happened before the colonization and the isolation in the cave is still unknown. Our results based on the rate of evolution of the opn4m2 gene across a phylogeny of teleost species suggest that the colonization may have happened at ∼5.3 Myr, with an interval that ranges from 2.5 to 9.0 Myr (Figure 1a). If, on the other hand, positive selection drove the LOF to fixation, the neutral evolution would have started only after the pseudogenization event, and would not coincide with the colonization of the cave. However, at present this possibility is not supported by any evidence. In addition, the fact that the LOF was found on both chromosomes of each of the two individuals from different sampling localities suggests that the pseudogenization happened only once. In this case, the inactivation of the gene would have been already present in the ancestral populations that then colonized different parts of the hydrographic system. This is supported by the analysis of the genetic variation of 30 allozyme loci indicating the presence of a genetic structure between sampling localities and a respective low or absent level of gene flow between them (Sbordoni et al., 1996). However, the possibility that the historic locality of certain cave populations may have been much closer together than they are presently should be considered. We cannot exclude that the fixation of the LOF predates the colonization of the subterranean environment, but this alternative scenario seems quite unlikely. Conversely, no line of evidence from this and previous studies on this species support multiple independent colonization events of the dark environment, as observed in other species (Strecker et al., 2012; Bradic et al., 2013).
Conclusions
In summary, our results suggest that P. andruzzii represents an excellent model to study regressive evolution in constant darkness, as the long time spent in the absence of light allowed an extreme level of genetic and phenotypic changes and adaptations. Our results suggest that in the case of this species, the evolutionary framework in which the circadian clock lost its ability to be synchronized by light is very likely neutral regressive evolution due to loss of selective constraint. The evolution of remarkable troglomorphic traits can be due to a long history of divergence of P. andruzzii from similar cyprinid species living in internal water of the same continent, a rather old colonization of the phreatic system under the Somalian desert, and the total absence of gene flow with surface populations given the disappearance of the epigean forms. Although clearly further genetic or genomic analysis will be required to obtain a higher resolution and improve the confidence of our time estimation of this process, this study represents the first key step to reconstructing the evolutionary history of this fascinating species using genetic information.
Data archiving
Sequence data have been submitted to GenBank: KU926693, KU738784, KT953343–KT953348, KU302709–KU302711.
Acknowledgments
We acknowledge Giorgio Bertorelle and Lino Ometto for important scientific input and discussion, Helmut Sattmann and Elisabeth Haring for help with sample collection and critical discussions, Barbara Däubl for technical assistance, and Francesca Tassi and Silvia Guglielmi for helping with figures. This project was partly funded within the SYNTHESYS framework (project no. AT-TAF-3369).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Banister KE. (1984). A subterranean population of Garra barreimiae (Teleostei: Cyprinidae) from Oman, with comments on the concept of regressive evolution. J Nat Hist 18: 927–938. [Google Scholar]
- Berti R, Durand JP, Becchi S, Brizzi R, Keller N, Ruffat G. (2001). Eye degeneration in the blind cave-dwelling fish Phreatichthys andruzzii. Can J Zool 79: 1278–1285. [Google Scholar]
- Berti R, Messana G. (2010) Subterranean fishes of Africa. In: Trajano E, Bichuette ME, Kapoor BG (eds), Biology of Subterranean Fishes. Science Publishers: Enfield, NH, pp 357–395. [Google Scholar]
- Bielawski JP, Yang Z. (2004). A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol 59: 121–132. [DOI] [PubMed] [Google Scholar]
- Bradic M, Beerli P, García-de León FJ, Esquivel-Bobadilla S, Borowsky RL. (2012). Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus). BMC Evol Biol 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradic M, Teotónio H, Borowsky RL. (2013). The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol Biol Evol 30: 2383–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallari N, Frigato E, Vallone D, Fröhlich N, Lopez-Olmeda JF, Foà A et al. (2011). A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol 9: e1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colli L, Paglianti A, Berti R, Gandolfi G, Tagliavini J. (2008). Molecular phylogeny of the blind cavefish Phreatichthys andruzzii and Garra barreimiae within the family Cyprinidae. Environ Biol Fishes 84: 95–107. [Google Scholar]
- Culver DC, Wilkens H. (2000) Critical review of the relevant theories of the evolution of subterranean animals. In: Wilkens H, Culver DC, Humphreys WF (eds). Ecosystems of the World. Subterranean Ecosystems Vol. 30, Elsevier: Amsterdam, pp 381–398. [Google Scholar]
- deMenocal PB. (1995). Plio-Pleistocene African climate. Science 270: 53–59. [DOI] [PubMed] [Google Scholar]
- Espinasa M, Espinasa L. (2008). Losing sight of regressive evolution. Evol Educ Outreach 1: 509–516. [Google Scholar]
- Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. (2014). Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol 6: 1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB. (2012). The complex origin of Astyanax cavefish. BMC Evol Biol 12: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR. (2005). Adaptive evolution of eye degeneration in the Mexican blind cavefish. J Hered 96: 185–196. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. (2009). Regressive evolution in Astyanax cavefish. Annu Rev Genet 43: 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. (1969). The average number of generations until fixation of a mutant gene in a finite population. Genetics 61: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoo K. (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press: Cambridge. [Google Scholar]
- Kruckenhauser L, Haring E, Seemann R, Sattmann H. (2011). Genetic differentiation between cave and surface-dwelling populations of Garra barreimiae (Cyprinidae) in Oman. BMC Evol Biol 11: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Lahti DC, Johnson N a, Ajie BC, Otto SP, Hendry AP, Blumstein DT et al. (2009). Relaxed selection in the wild. Trends Ecol Evol 24: 487–496. [DOI] [PubMed] [Google Scholar]
- Lang M, Murat S, Clark AG, Gouppil G, Blais C, Matzkin LM et al. (2012). Mutations in the neverland gene turned Drosophila pachea into an obligate specialist species. Science 337: 1658–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009). PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288. [DOI] [PubMed] [Google Scholar]
- Leys R, Cooper SJB, Strecker U, Wilkens H. (2005). Regressive evolution of an eye pigment gene in independently evolved eyeless subterranean diving beetles. Biol Lett 1: 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He S. (2009). Relaxed purifying selection of rhodopsin gene within a Chinese endemic cavefish genus Sinocyclocheilus (Pisces: Cypriniformes). Hydrobiologia 624: 139–149. [Google Scholar]
- Marková S, Searle JB, Kotlík P. (2014). Relaxed functional constraints on triplicate α-globin gene in the bank vole suggest a different evolutionary history from other rodents. Heredity (Edinb) 113: 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP et al. (2012). Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA 109: 13698–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemiller ML, Fitzpatrick BM, Shah P, Schmitz L, Near TJ. (2013). Evidence for repeated loss of selective constraint in rhodopsin of amblyopsid cavefishes (teleostei: Amblyopsidae). Evolution (NY) 67: 732–748. [DOI] [PubMed] [Google Scholar]
- Poulson TL, White WB. (1969). The cave environment. Science 165: 971–981. [DOI] [PubMed] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. (2007). Regressive evolution in the Mexican Cave Tetra, Astyanax mexicanus. Curr Biol 17: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O, Daley AC, Pisani D. (2013). Molecular timetrees reveal a cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol 23: 392–398. [DOI] [PubMed] [Google Scholar]
- Sadoglu P. (1967). The selective value of eye and pigment loss in mexican cave fish. Evolution (NY) 21: 541–549. [DOI] [PubMed] [Google Scholar]
- Sbordoni MC, Matthaeis E, Mattoccia M, Berti R, Sbordoni V. (1996). Genetic variability and differentiation of hypogean Cyprinid fishes from Somalia. J Zool Syst Evol Res 34: 75–84. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Schuhmacher L-N, Foulkes NS, Bertolucci C, Wittbrodt J. (2015). Cavefish eye loss in response to an early block in retinal differentiation progression. Development 142: 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker U, Hausdorf B, Wilkens H. (2012). Parallel speciation in Astyanax cave fish (Teleostei) in Northern Mexico. Mol Phylogenet Evol 62: 62–70. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarttelin EE, Frigato E, Bellingham J, Di Rosa V, Berti R, Foulkes NS et al. (2012). Encephalic photoreception and phototactic response in the troglobiont Somalian blind cavefish Phreatichthys andruzzii. J Exp Biol 215: 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A et al. (2006). Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res 34: W645–W650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veilleux CC, Louis EE, Bolnick DA. (2013). Nocturnal light environments influence color vision and signatures of selection on the OPN1SW opsin gene in nocturnal lemurs. Mol Biol Evol 30: 1420–1437. [DOI] [PubMed] [Google Scholar]
- Wang X, Thomas SD, Zhang J. (2004). Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet 13: 2671–2678. [DOI] [PubMed] [Google Scholar]
- Wang X, Gan X, Li J, Mayden RL, He S. (2012). Cyprinid phylogeny based on Bayesian and maximum likelihood analyses of partitioned data: implications for Cyprinidae systematics. Sci China Life Sci 55: 761–773. [DOI] [PubMed] [Google Scholar]
- Weadick CJ, Chang BSW. (2012). An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol 29: 1297–1300. [DOI] [PubMed] [Google Scholar]
- Wilkens H. (1988) Evolution and genetics of Epigean and Cave Astyanax fasciatus (Characidae, Pisces). In: Hecht MK, Wallace B (eds), Evolutionary Biology. Springer US: Boston, MA, pp 271–367. [Google Scholar]
- Wilkens H. (2010). Genes, modules and the evolution of cave fish. Heredity (Edinb) 105: 413–422. [DOI] [PubMed] [Google Scholar]
- Wilkens H, Strecker U. (2003). Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): genetic evidence from reduced eye-size and pigmentation. Biol J Linn Soc 80: 545–554. [Google Scholar]
- Yang Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang L, Arunachalam M, Sado T, Levin BA, Golubtsov AS, Freyhof J et al. (2012). Molecular phylogeny of the cyprinid tribe Labeonini (Teleostei: Cypriniformes). Mol Phylogenet Evol. 65: 362–379. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. (2002). Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yang J-R, Xu H, Zhang J. (2010). Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol Biol Evol 27: 2669–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.