Abstract
The Justinianic Plague, which started in the sixth century and lasted to the mid eighth century, is thought to be the first of three historically documented plague pandemics causing massive casualties. Historical accounts and molecular data suggest the bacterium Yersinia pestis as its etiological agent. Here we present a new high-coverage (17.9-fold) Y. pestis genome obtained from a sixth-century skeleton recovered from a southern German burial site close to Munich. The reconstructed genome enabled the detection of 30 unique substitutions as well as structural differences that have not been previously described. We report indels affecting a lacl family transcription regulator gene as well as nonsynonymous substitutions in the nrdE, fadJ, and pcp genes, that have been suggested as plague virulence determinants or have been shown to be upregulated in different models of plague infection. In addition, we identify 19 false positive substitutions in a previously published lower-coverage Y. pestis genome from another archaeological site of the same time period and geographical region that is otherwise genetically identical to the high-coverage genome sequence reported here, suggesting low-genetic diversity of the plague during the sixth century in rural southern Germany.
Keywords: Yersinia pestis, ancient DNA, justinianic plague, whole genome, reconstruction
Introduction
The bacterium Yersinia pestis has been infecting humans for over 5,000 years (Rasmussen et al. 2015) and is thought responsible for at least three known historic plague pandemics. The first was the sixth- to AD eighth-century Justinianic pandemic, the second started with the infamous Black Death, claiming the lives of up to 50% of the European population during the 14th century (Benedictow 2004), and the last plague pandemic began in late 19th century China, seeding many of the plague foci that exist globally today (Pollitzer 1954; World Health Organization 2004). At present, plague is classified as a reemerging infectious disease in certain endemic regions and remains a public health problem with reservoirs on nearly every major continent (World Health Organization 2004).
Historical records suggest that the first known outbreak of the Justinianic Plague occurred between 541 and AD 543 in Egypt and spread throughout the eastern Roman Empire and its neighbors (Little 2007; Stathakopoulos 2004). Contemporary accounts indicate massive mortality caused by the disease that might have contributed to the weakening and the eventual decline of the eastern Roman Empire (Little 2007; Mitchell 2014). The epidemic itself returned in about 18 waves over a period of 200 years until it disappeared in Europe and the near East in the middle of the 8th century for yet unknown reasons (Stathakopoulos 2004).
Apparent discrepancies in epidemiological patterns between the modern and the historical pandemics have led scholars to suggest that etiological agents other than Y. pestis may have been responsible for the early and later medieval pandemics (Cohn 2008; Duncan and Scott 2005; Scott and Duncan 2001; Twigg 1984). Molecular evidence obtained from ancient plague victims, however, has established Y. pestis as at least one of the causative agents for both historical pandemics (Bos et al. 2011; Haensch et al. 2010; Harbeck et al. 2013; Schuenemann et al. 2011; Wagner et al. 2014; Wiechmann and Grupe 2005). Nevertheless, the differences in epidemiology such as the apparently much faster geographical spread of the historical pandemics compared with the modern third pandemic (Christakos, et al. 2007; Cohn 2008; Kanaroglou and Delmelle 2015; Maddicott 1997) still need to be addressed. The geographic reach and mortality impact of individual waves as well as of the Justinianic pandemic as a whole remain unknown. Environmental and behavioral factors as well as genetic factors in the host, vector or pathogen have been known to alter the disease dynamics in modern plague outbreaks (Duplantier et al. 2005; Enscore et al. 2002; Guiyoule et al. 1997; Keim and Wagner 2009; Parmenter et al. 1999; Schmid et al. 2015; Xu et al. 2014). The characterization of historical Y. pestis strains and a comparison to extant strains may well shed light on the role of the evolving bacterial genetic structure in forming these notable epidemiological differences. Moreover, a robust genomic description from the early stages of the Justinianic pandemic affords opportunities to trace and understand the evolution of one of humanity’s most devastating pathogens over a period of deep time, with insights that may illuminate the evolutionary trajectories of other like organisms.
Despite their genetic resemblance Y. pestis and its closest relative the enteric soil- and water-borne Yersinia pseudotuberculosis differ greatly in pathogenicity and transmission (Achtman et al. 1999). Furthermore, the Y. pestis genome is characterized by structural variation caused by frequent intragenomic rearrangements due to abundant insertion sequences, high expansion through horizontal gene transfer from other bacteria and bacteriophages and considerable gene decay that is evident from the large number of pseudogenes in the genome (Guiyoule et al. 1994; Parkhill et al. 2001; Zinser et al. 2003). Comparing the genome structure of historical Y. pestis strains to those of extant strains and to that of Y. pseudotuberculosis could offer insights into key evolutionary changes, such as those in virulence associated genes, through time.
A genome reconstructed from two sixth-century Justinianic Plague victims unearthed in Aschheim, southern Germany, estimated at 7.6-fold coverage, has recently been reported and used to characterize the Y. pestis strain responsible for the Justinianic Plague (Wagner et al. 2014). Phylogenetic analysis of this genome placed the Justinianic branch between two extant basal Y. pestis strains isolated from rodent populations in China, thus suggesting a Chinese origin for the Justinianic lineage. The Justinianic strain was found to have no extant descendants. In addition, nonsynonymous substitutions and one deletion were identified and were suggested to play a role in virulence changes (Wagner et al. 2014).
Here we present a high-coverage Y. pestis genome (17.9-fold) from the time of the Justinianic Plague, reconstructed from a victim buried during the sixth century in another early medieval settlement, Altenerding, southern Germany, approximately 20 km from Aschheim. The coverage and quality of the data enabled a robust phylogenetic and structural analysis of the Justinianic Y. pestis strain. Comparison of the high-coverage genome to the previously published lower-coverage genome suggests that both represent a single strain. Our analysis confirms the phylogenetic placement previously suggested; however, we identify 19 out of 176 chromosomal substitutions previously reported in the lower-coverage genome as false positives. Fourteen of these false positives were purported as unique substitutions defining the Justinianic branch. In addition, our high-quality data permitted the identification of novel substitutions and structural polymorphisms that are unique to the Justinianic Y. pestis strain.
Results and Discussion
To identify Justinianic Plague victims that could potentially be used for Y. pestis genome reconstructions, teeth extracted from 20 skeletons unearthed from double burials in a sixth-century Bavarian burial ground in Altenerding, southern Germany were screened with a conventional polymerase chain reaction (PCR) assay (Seifert et al. 2013). Two adults (AE1175 female/AE1176 male) in the same double burial (Fig. 1, supplementary table S1, Supplementary Material online) generated amplification products of the expected size using primers specific for Y. pestis. An additional qPCR assay amplifying the Y. pestis specific plasminogen activator (pla) gene revealed that only individual AE1175, with 90 copies per µl could be a candidate for subsequent Y. pestis genome capture (supplementary fig. S1, Supplementary Material online).
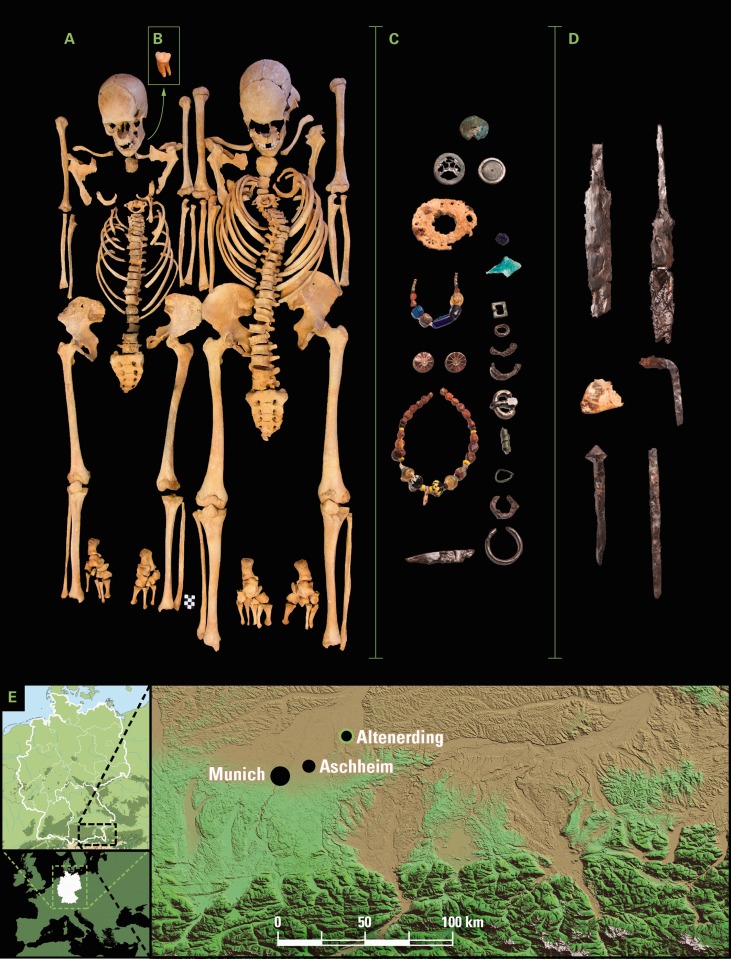
Fig. 1.
The Altenerding plague burials. (A) Adult woman AE1175 (left) and adult man AE1176 (right) excavated at Altenerding and found positive for presence of Y. pestis. (B) The third molar sampled from individual AE1175 from which the Altenerding high-coverage genome was obtained. (C) Grave goods of individual AE1175 typical of the middle of the sixth century (© Archaeological State Collection Munich), Grave goods not shown true to scale (Detailed description in the historical and archaeological supplementary section). (D) Grave goods of individual AE1176 (© Archaeological State Collection Munich), Grave goods not shown true to scale (detailed description in the historical and archaeological supplementary section). (E) Geographical map specifying the location of the archaeological site “Altenerding”. The other site, where Y. pestis has been identified and its genome has been previously reconstructed (Wagner et al. 2014), is the cemetery of Aschheim, which is located approximately 20 km southwest of Altenerding.
Radiocarbon dating suggests the two individuals died between 426 and AD 571 cal (95.4% probability at 2 sigma); grave goods point to a burial in or after ∼530–570 (supplementary table S2, Supplementary Material online and Supplementary archaeological and historical information). According to historical sources, the Justinianic Plague struck Europe at least three times during this time period. The first known wave raged across the Mediterranean world from 541 to AD 543; further big waves occurred from 558 to ca. 590 (Stathakopoulos 2004). The incomplete written records do not mention plague in this region of Germany (see Supplementary archaeological and historical information). Since it is unclear whether the second big wave reached western Europe, these victims most likely died in the first or third wave of plague. In any case, the Altenerding genome dates to the first decades of the 200-year long pandemic. How the pathogen reached southern Germany is at present unknown: further evidence will be needed to clarify whether it traveled across the Alps from the Mediterranean as suggested for the Black Death (Carmichael 2014) or from France and western Germany. Alternatively, it could have traveled up the Danube from the east.
For genomic analysis, the DNA extract of individual AE1175 was converted into Illumina DNA double indexed libraries following established protocols (Kircher,et al. 2012; Meyer and Kircher 2010). Shotgun sequencing of the nonenriched library produced 5,024,997 reads of which only 0.91% mapped to the human genome (hg19) and 0.11% mapped to the Y. pestis reference chromosome (CO92), indicating that targeted DNA enrichment would be required for genome-level analysis. Target enrichment was performed using DNA array hybridization. The DNA array capture probes were designed using the genome of Y. pseudotuberculosis as a template in order to avoid ascertainment bias for a specific Y. pestis strain. In addition, the Y. pestis (CO92) pCD1 and pMT1 plasmids were included. The array hybridization was performed following an established protocol (Hodges et al. 2009).
Sequencing of the enriched library produced in 62,126,715 total reads of which 11.3% mapped to the Y. pestis genome (9.69% to the chromosome), with an average length of 67 bp. The average genome coverage was 17.9-fold and 91.5% of the chromosome was covered at least 5-fold. Mapping to the pCD1 and to the pMT1 plasmid references resulted in an average 52-fold coverage with 94% of the positions covered at least 5-fold and an average of 77-fold coverage with 94.19% covered at least 5-fold, respectively (supplementary fig. S12, Supplementary Material online). The copy numbers of the plasmids can be roughly estimated by dividing the average coverage of plasmids by that of the chromosome. The estimated copy numbers were 3 for the pCD1 plasmid and 4.3 for the pMT1 plasmid. The following differences between the three loci may bias this estimate by affecting the average coverage of each locus: 1—mappability of the reference sequences, 2—base composition may alter the representation of sequenced reads (Benjamini and Speed 2012), 3—DNA degradation processes (Briggs et al. 2007) 4—average read length, and 5—DNA enrichment bias. The average read lengths are similar between the three loci and range between 61 to 68.7 bp (supplementary table S3, Supplementary Material online). To address the mappability and the base composition bias, artificial reads were generated from the CO92 genome reference sequence (with read length of 100 bp and tiling density of 1 bp). The artificial reads were mapped to the CO92 genome reference with the same mapping parameters used for the enriched library. Regions of 5,000 bp with 100% mappability (covered 100-fold) and a “G + C” content of around 48% were randomly chosen for each locus and an average coverage of the Y. pestis enriched reads in each of these three regions was calculated (supplementary table S3, Supplementary Material online). Dividing the average coverage of the corresponding regions in the plasmids by that of the chromosome resulted in a similar copy number estimate of 2.87 for the pCD1 plasmid and 2.55 for the pMT1 plasmid. Since DNA enrichment bias could affect the calculated copy number, it should be considered as a rough estimate testing whether the plasmid copy numbers in the sixth century strain are in general agreement with the previous estimates from modern Y. pestis strains (Parkhill et al. 2001).
Prior to array capture, the library was treated to prevent erroneous substitution assignments originating in DNA damage (Uracil DNA glycolsylase [UDG] treatment) (Briggs et al. 2010). The DNA substitution plot of the library not treated with UDG mapped to the Y. pestis reference, displayed a pattern expected for ancient molecules (Briggs et al. 2007) with C to T substitution frequency of 21.3% at the first 5’ position and G to A substitution of 20.7% at the first 3’ position (supplementary fig. S2, Supplementary Material online). Similar patterns were observed for endogenous human DNA thus supporting the authenticity of the ancient bacterium (supplementary fig. S3, Supplementary Material online). In addition, in-solution enrichment of human mitochondrial DNA was performed following established protocols (Maricic, et al. 2010), resulting in 5,292-fold coverage. The mtDNA haplotype inferred from the obtained consensus sequence match the H1f1a haplogroup, which is common in European populations.
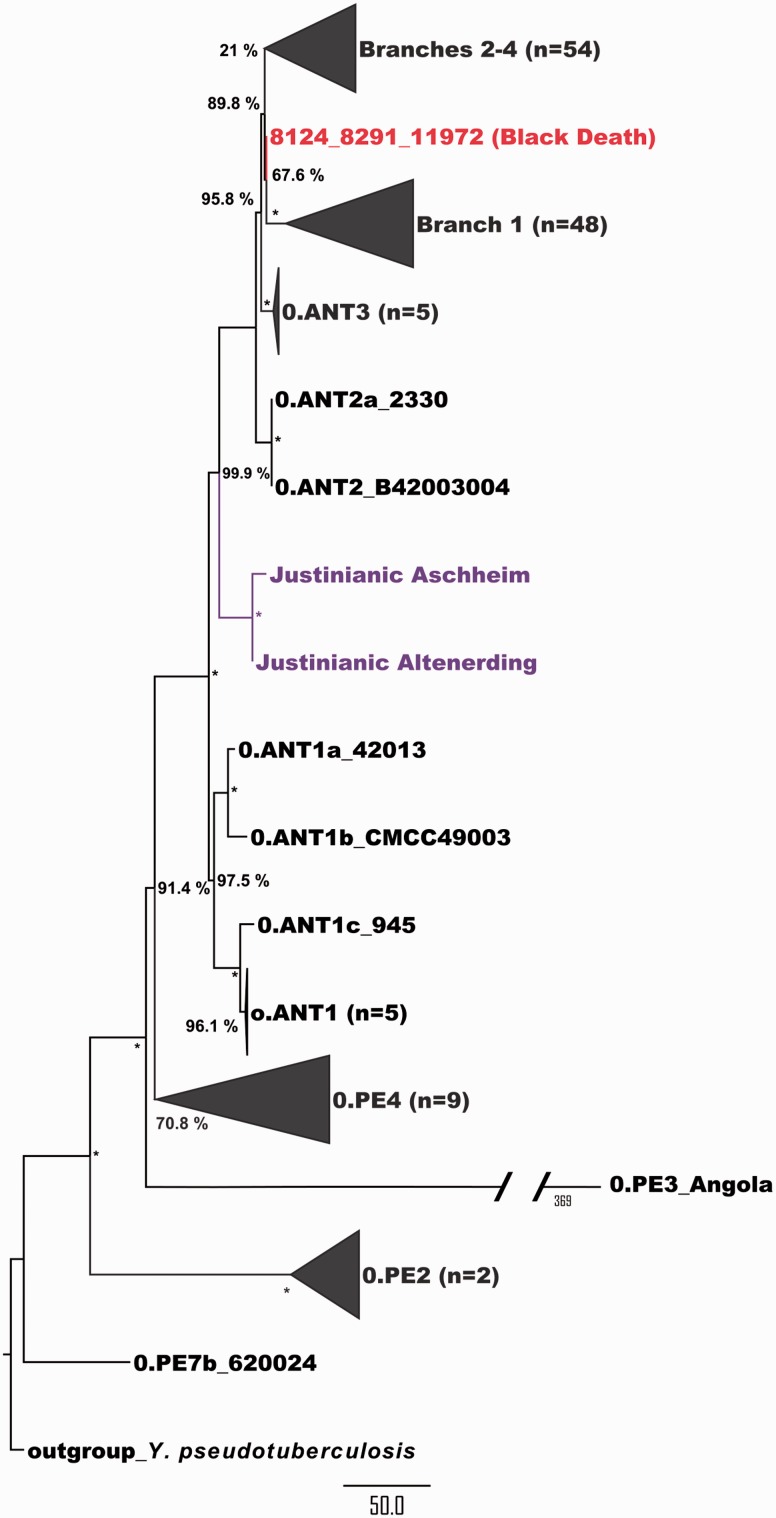
A mapping of 133 Y. pestis genomes to the CO92 reference genome resulted in 3,086 single nucleotide polymorphisms (SNPs) identified for the entire data set excluding the Y. pseudotuberculosis out-group. The number of SNPs ranged between 26 and 889 per genome (supplementary table S4, Supplementary Material online). For the ancient Y. pestis genome obtained from individual AE1175 (from now on referred to as the Altenerding genome) a total of 157 chromosomal SNPs were identified compared to the reference (supplementary table S5, Supplementary Material online) and 11 SNPs for the plasmids (supplementary table S6, Supplementary Material online). As input for phylogenetic analysis we used an alignment of 2,603 variable positions after removal of all positions in the alignment with missing data. The phylogenetic reconstructions (Fig.2, supplementary figs. S4–S6, Supplementary Material online) resemble previously obtained phylogenies (Cui et al. 2013) and confirm the placement of the branch leading to the Justinianic strain on Branch 0 between two modern strains isolated from Chinese rodents (0.ANT1 and 0.ANT2) as reported previously (Wagner et al. 2014). However, our phylogeny indicates that the Justinianic strain was more genetically divergent than previously suggested. The genetic distance of the 14th-century Black Death strain and some of the extant strains isolated in China (e.g., 0.ANT1 and 0.ANT2) from the root of the tree is shorter than the much older Justinianic strain. This observation is counterintuitive, as ancient lineages should have fewer substitutions than their modern counterparts since they have had less time to accumulate genetic changes. Thus, the previously observed variation in mutation rates on different branches of the Y. pestis phylogeny (Cui et al. 2013; Wagner et al. 2014) is even more pronounced for the Justinianic strain than suggested before. The rate variation in Y. pestis has been attributed to an increased speed of SNP fixation as a result of higher replication rates of the bacteria per unit of time during epidemics and outbreaks compared with periods of enzootic disease (Cui et al. 2013).
Fig. 2.
Maximum parsimony tree: maximum parsimony analysis of 1418 nucleotide positions from genomes of 133 Y. pestis strains. All positions containing missing data were eliminated. Bootstrap values are indicated next to the nodes and bootstrap values of 100% are indicated by an asterisk. The tree is rooted using the genome of Yersinia pseudotuberculosis (strain IP32953). Branches leading to isolates from the historical pandemics are colored red and purple representing the second and first pandemics, respectively. Monophyletic nodes have been collapsed and are represented by a triangle. The number of isolates in a collapsed node is indicated in brackets. Our analysis suggests the specifically derived SNPs represented by the Aschheim branch to be false positives, supporting that the Aschheim and the Altenerding genomes likely represent the same bacterial strain.
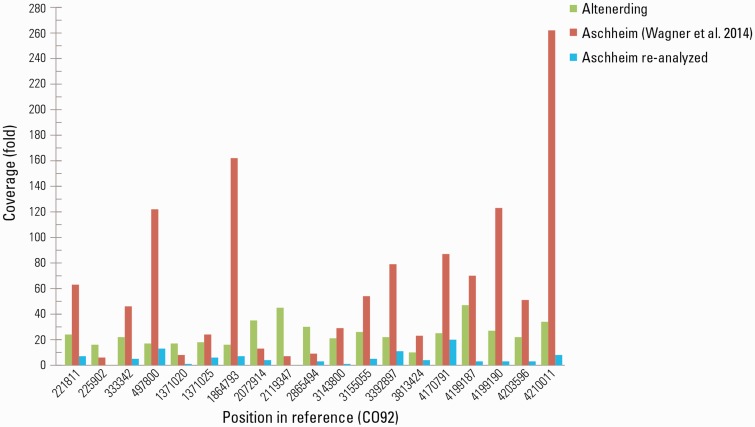
The Aschheim cemetery and the Altenerding cemetery are located about 20 km apart and the two burials are dated to a similar time period (McCormick 2015; McCormick Forthcoming; Wagner et al. 2014). For ancient genome comparison, we reanalyzed the raw data from the Aschheim individual A120 genome (Wagner et al. 2014). When mapped to the Y. pestis reference chromosome, the reads from this reanalysis produced 3.9-fold average coverage with 31.08% of the CO92 reference chromosome covered at least 5-fold and 86% of the reference chromosome covered at least once; this is in contrast to an average of 7.6-fold coverage for 91.5% of the chromosome reported by Wagner et al. 2014. Only 55.51% of the reanalyzed Aschheim genome was covered at least three times when mapped to the CO92 reference, compared with 92.32% covered at least three times in the Altenerding genome when mapped against the same reference with the same parameters. Of the 176 chromosomal SNPs reported by Wagner et al. 2014, 66 were not called in the Altenerding genome (supplementary table S7, Supplementary Material online). Of these, 47 were excluded from our analysis since they fell in noncore regions. Noncore regions, which have been defined elsewhere, contain deletions or duplications and are therefore more prone to erroneous mapping (Cui et al. 2013; Morelli et al. 2010). We inspected the remaining 19 SNPs to determine whether they indeed represent variation between the two genome sequences. Reads spanning a 150 bp flanking region on either side of each SNP from both the Altenerding genome and the reanalyzed Aschheim genome were considered (supplementary fig. S7, Supplementary Material online). Visual inspection revealed that these SNPs did not fulfill our criteria for SNP calling (coverage of more than five times and presence in at least 90% of the sequences obtained). The regions surrounding most of these positions show a pattern of abnormal coverage peaks (Fig.3, supplementary fig. S8, Supplementary Material online) in which the SNP position is often heterozygous and located at the end of reads. A significant drop in coverage is often observed next to the SNP position, resulting in a lack of overlapping reads to support the SNP. These regions are therefore prone to erroneous SNP calls and the visual inspection suggests the reads containing the SNPs are not authentic to the Y. pestis ancient strain. The abnormal coverage peaks seem not to be artifacts from the SNP capture, since similar coverage peaks are also apparent in our reanalysis of the non-SNP captured data for these libraries (supplementary figs. S8 and S9 Supplementary Material online). In addition, our reanalysis revealed some of these 19 SNP positions to be identical to the reference sequence, whereas others had a coverage that is below the threshold set for SNP calling (supplementary fig. S7, Supplementary Material online). This analysis suggests that these 19 SNPs reported previously for the Aschheim genome are false positives and do not represent a true variation between the two genomes. Of these 19 SNPs, 14 were reported as derived SNPs, 2 of which were reported as nonsynonymous nucleotide changes (supplementary table S8, Supplementary Material online). In an additional analysis, we performed SNP calling for the reanalyzed Aschheim genome and identified 8 derived SNPs not shared with other Y. pestis genomes, including the Altenerding genome reported here (supplementary fig. S10, Supplementary Material online). Visualization of these SNPs revealed a similar pattern of abnormal coverage peaks and heterozygosity, suggesting that they too are likely false positives. Two of these positions (position 1,371,025 and 3,392,897) are among the 19 false positives described above, three positions (positions 362,357, 3,956,001 and 4,575,345) were manually removed from the final analysis in Wagner et al. 2014 following a visual inspection by the authors, and the remaining three were not identified in the published analysis of this genome (Wagner et al. 2014). This analysis reveals a repeated and problematic pattern in the Aschheim data set, which could in some cases, cause false SNP calls even under stringent SNP calling parameters. This pattern is exceedingly problematic for low-coverage genomes where nonendogenous reads could dominate positions where few or no endogenous reads mapped.
Fig. 3.
Coverage of the false positive substitutions called for the Aschheim genome by Wagner et al., 2014 compared with the reanalyzed Aschheim genome and the Altenerding genome. Fold coverage was determined for each position where substitutions were erroneously reported in the Aschheim genome (Wagner et al. 2014). Values for the Altenerding genome, values published in Wagner et al. 2014 for the Aschheim genome and the values for the reanalyzed Aschheim genome are colored green, red, and blue, respectively. Most of the false positive positions in the Aschheim genome contain abnormal coverage peaks, significantly higher than the average coverage of the corresponding genome.
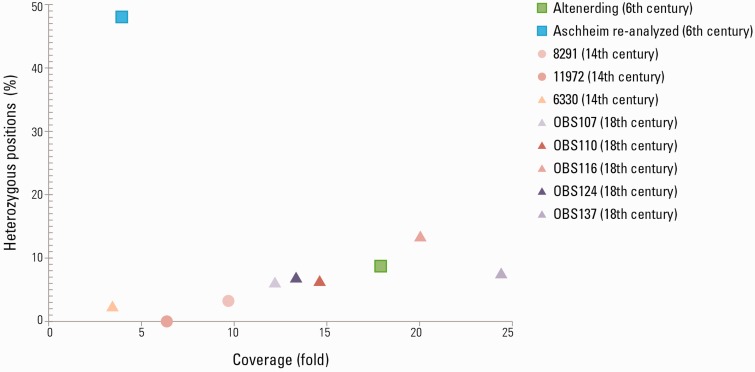
Our reanalysis of the Aschheim genome resulted furthermore in a high percentage of heterozygous positions. A position was considered heterozygous when reads mapping to the position showed more than one allele, which is unexpected in a haploid organism. We identified a total of 48.05% heterozygous positions where SNPs were identified (111 heterozygous positions out of 231 SNPs) in the reanalyzed Aschheim genome compared with 8.72% (15 heterozygous positions out of 172 SNPs) in the Altenerding genome (Fig.4). The Aschheim enriched libraries were reported by Wagner et al., 2014 to be UDG treated, thus, damage that is typical of ancient DNA should be absent from the raw data and should not affect heterozygosity levels. The possibility that the A120 individual was infected with multiple strains of Y. pestis that exhibit a genetic distance as high as 48.05% from each other at polymorphic positions is unlikely. Furthermore, the genome-wide SNP allele frequency plot of the reanalyzed Aschheim genome does not show a bimodal pattern or any other pattern consistent with an infection with multiple strains as observed in other pathogens (Bos et al. 2014)( supplementary fig. S11, Supplementary Material online). It is, therefore more likely that the data contain a high percentage of reads that are either nontarget or are damaged in a nonstandard way for ancient molecules. There are several possible causes for a high percentage of heterozygous positions in a reconstructed ancient genome. One possibility is contamination of the sample by a phylogenetically close organism. Sequenced reads originating from the genome of such an organism could erroneously map to regions of the reference sequence that are conserved among bacterial species. It should be noted, however, that Y. pestis is the best match for these reads in the current Genbank database. Other explanations could be related to sample processing, including the accumulation of PCR errors due to extensive amplification of low-template libraries or artifacts created as a result of genome-wide capture processes. Identification of substitutions in low-coverage genomes necessitates a low-minimal coverage thus, making these analyses prone to false positive SNP calls. Awareness of the limitations arising from low-coverage data and attention to parameters such as heterozygosity are important for reliable future analyses.
Fig. 4.
Comparison of coverage and heterozygous positions between ancient Y. pestis genomes. The average coverage calculations and the calling of heterozygous positions were done against the CO92 chromosome (NC_003143.1) as a reference. The percentage of heterozygous positions was calculated out of the total number of SNPs called for each genome, using a mapping quality threshold of 30 and a coverage threshold of 5 reads. The raw data published in Wagner et al. 2014, Bos et al. 2011, and Bos et al. 2016 were reanalyzed and used for calculating the coverage and the percentage of heterozygous positions for the Aschheim Justinianic genome and the genomes from the second pandemic, respectively. The Black Death genome 8124 was excluded from this analysis since the coverage was not sufficient for calling of heterozygous positions. The Aschheim Justinianic genome contains a high percentage of heterozygous positions compared with the other ancient Y. pestis genomes. Heterozygous positions called for a genome of the haploid Y. pestis indicate nonendogenous reads.
Excluding the above putative false positive SNPs, no difference was found between the Aschheim genome and the Altenerding genome reported here, supporting that both genomes represent the same bacterial strain and that the infected individuals were victims of the same outbreak. Previous studies have reported the presence of multiple Y. pestis strains in modern plague outbreaks (Guiyoule et al. 1994; Shivaji, et al. 2000), for example, in the case of the 1980s and 1990s plague outbreak in the Ambositra region of Madagascar (Guiyoule et al. 1997). However, our findings point to a low-genetic diversity of the bacterium in this rural region of southern Germany during the sixth century. Future investigations of other Justinianic material may reveal the range of the outbreak, and allow us to comment on the paths traveled by the disease and its rate of transmission.
SNP calling for the Altenerding genome resulted in the detection of 63 substitutions unique to the Justinianic strain (supplementary table S9, Supplementary Material online), 30 of which were not previously reported. Of these novel unique SNPs, 14 are nonsynonymous (supplementary table S10, Supplementary Material online), 3 of which are located in genes that are thought to be associated with plague virulence: the nrdE, fadJ, and pcp genes (table 1). The nrdE gene codes for the subunit alpha of a protein catalyzing Deoxynucleotide Triphosphate (dNTP) synthesis. This gene is strongly transcriptionally induced in human plasma infected by Y. pestis (Chauvaux et al. 2007) and is upregulated in the bubo during infection by Y. pestis in rats (Sebbane et al. 2006). The fadJ gene codes for the FadJ protein that is involved in fatty acid oxidation and is transcriptionally upregulated in Y. pestis infected cells at 37 °C (Chauvaux et al. 2011). The pcp gene (also referred to as pcpY and slyB) codes for an outer membrane lipoprotein. A study of the Y. pestis transcriptional profile has shown that a homolog of the pcp was transcribed 5.5-fold higher in a lcrG deleted strain of Y. pestis compared with the wild type strain. lcrG is a negative regulator for secretion of Yersinia outer membrane proteins (Yops), suggesting a role for pcp in the type 3 secretion system (Du et al. 2009) which is a known virulence determinant (Perry and Fetherston 1997).
Table 1.
Derived nonsynonymous SNPs compared with CO92.
| Position (CO92 chromosome) | CO92 | Altenerding | Codon Change | A.A. Change | Gene ID | Gene Name | Gene Function |
|---|---|---|---|---|---|---|---|
| 260148a | C | T | CCG to TCG | P to S | YPO0257 | Type III secretion protein | |
| 271114a | C | A | CTT to ATT | L to I | YPO0270 | Type III secretion system protein (iron–sulfur binding protein) | |
| 557841a | C | T | CGT to CAT | R to H | YPO0517 | hepA (RapA) | ATP-dependent helicase (transcription regulator) |
| 727741 | G | A | GAA to AAA | E to K | YPO0668 | parE | DNA to poisomerase IV subunit B |
| 1067966 | C | A | GGC to TGC | G to C | YPO0966 | Kinase | |
| 1296743a | C | T | GTG to ATG | V to M | YPO1150 | bioA | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase (part of the Biotin operon) |
| 1434752a | C | A | GAT to TAT | D to Y | YPO1275 | spr | Outer membrane lipoprotein (Murein hydrolase) |
| 1530658 | C | A | AGA to ATA | R to I | YPO1363 | Virulence factor | |
| 1609461a | T | C | ACA to GCA | T to A | YPO1417 | Iron–sulfur binding protein | |
| 1754708 | C | T | CCA to CTA | P to L | YPO1539 | galU | UTP-glucose-1-phosphate uridylyltransferase |
| 2352174a | T | G | GTG to GGG | V to G | YPO2071 | DEAD box family helicase | |
| 2977542a | C | A | AGC to ATC | S to I | YPO2649 | nrdE | Ribonucleotide-diphosphate reductase subunit alpha |
| 3078807a | C | A | CGT to AGT | R to S | YPO2747 | fadJ (faoA) | Multifunctional fatty acid oxidation complex subunit alpha |
| 3360963a | A | C | ACC to CCC | T to P | YPO3008 | Two-component sensor histidine kinase (TCSs) | |
| 3360984a | C | T | CAT to TAT | H to Y | YPO3008 | TCSs | |
| 3500922 | T | G | GTG to GGG | V to G | YPO3141 | tesB | Acyl-CoA thioesterase |
| 3535148a | G | T | GCT to TCT | A to S | YPO3171 | apbA (panE) | 2-dehydropantoate 2-reductase (vitamin B5 biosynthesis) |
| 3560088 | G | A | CCA to TCA | P to S | YPO3199 | Short chain dehydrogenase | |
| 3568597 | C | T | GGA to GAA | G to E | YPO3205 | phoB | Phosphate regulon transcriptional regulator |
| 4066494a | C | T | GTT to ATT | V to I | YPO3646 | pcp (pcpY, slyB) | Outer membrane lipoprotein |
| 4307755 | G | A | GCG to GTG | A to V | YPO3839 | ||
| 4460688 | C | T | CGG to CAG | R to Q | YPO3963 | Sugar transport system permease | |
| pCD1 | |||||||
| 29959 | A | G | AAC to AGC | N to S | YPCD1.41 | yscO | Type III secretion apparatus component |
| 50462 | T | C | AAA to GAA | K to E | YPCD1.71c | yopJ | Targeted effector protein |
| 66608 | C | T | CTT to TTT | L to F | YPCD1.92 |
aDerived nonsynonymous SNPs detected in the Altenerding genome that were not called in Wagner et al. 2014 for the Aschheim genome.
Insertion and deletion analysis revealed 16 regions present in the Justinianic strain that were absent in the CO92 reference (supplementary table S11, Supplementary Material online). One such region is the 15.6 kb DFR4 locus (Radnedge et al. 2002) that was also present in other historic epidemic strains from the Black Death and post-Black Death periods including the previously published Justinianic genome (Bos et al. 2016; Wagner et al. 2014). Another such region is a 1122bp long locus that includes parts of the glpFKX operon that enables aerobic glycerol fermentation. Loss of capacity to ferment glycerol is shared by several Y. pestis “branch 1” strains, including CO92, due to a deletion in the glpD gene. Furthermore, additional deletions in the glpFKX operon are often present (Achtman et al. 1999; Motin et al. 2002). The aerobic glycerol fermentation pathway has been suggested to upregulate Y. pestis biofilm formation in the flea gut. In addition, a negative selection against glpK activity in the presence of dysfunctional glpD has been suggested (Willias, et al. 2014; Zinser et al. 2003). The fact that the glpFKX operon is pseudogenized in some Y. pestis strains but not all (Tong et al. 2005) indicates that its loss is not adaptive to the shift from an environmental and enteric life-style to one of specialized rodent-flea transmission, but is rather due to either genetic drift or a selective advantage specific to the glpFKX pseoudogenized strains. The aerobic glycerol fermentation pathway is induced during different phases of the Y. pestis infectious cycle (Motin et al. 2004; Zhou et al. 2006). Thus, its loss in CO92 should be further explored to evaluate its potential role in regulating biofilm formation and its subsequent effect on the bacterial fitness and disease dynamics.
In an attempt to identify unique structural differences we compared the Altenerding genome to the phylogenetically close strains 0.ANT1b_CMCC49003 and 0.ANT2a_2330. Three deletions unique to the Justinianic strain were observed. A 1334 bp long unique deletion (positions 1010683-1012016 in the Y. pseudotuberculosis reference) in the metabolic pheA gene, which functions in L-phenylalanine biosynthesis, was detected. This deletion may account for an obligatory nutritional requirement for L-phenylalanine in the Justinianic strain similar to those exhibited in other Y. pestis strains (Brubaker 1991; Parkhill et al. 2001). Further we detected a 1074 bp unique deletion (positions 2563546-2564619 in the CO92 reference) interrupting the CO92 annotated gene YPO2283. YPO2283 is homologous to the Escherichia coli galR gene and has the characteristics of a lacl family transcription regulator. However, the gene’s function in Y. pestis is unknown (Carnoy et al. 2002; Marceau 2005). Finally, a unique deletion of 195 bp (positions 3462627-2462821 in the Y. pseudotuberculosis reference) was located in the range of the metabolic celB gene (also called chbC) that codes for the Phosphotransferase (PTS) system cellobiose-specific IIC component. This gene has been shown to be upregulated in the flea compared with in vitro conditions (Vadyvaloo et al. 2010). The presence of these unique deletions in the Justinianic strain call for further functional studies on the proteins coded by the above genes to examine whether and in what way the deletions affect bacterial physiology and disease infectious cycle.
In summary, we have demonstrated the presence of Y. pestis in a second early medieval rural site in southern Germany where no historical source records it, expanding the number of sites known to have been afflicted by plague and reinforcing the time placement of this strain in the early waves of the 200-year-long pandemic. Our reconstructed high-coverage genome permitted a more thorough and reliable analysis than was previously possible. Through this we identified novel substitutions and structural polymorphisms in this early Y. pestis strain, located in genomic regions such as the glpFKX operon (Motin et al. 2004; Zhou et al. 2006) as well as in genes such as nrdE, fadJ, and pcp that have been suggested as plague virulence factors (Chauvaux et al. 2007; Sebbane et al. 2006). Substitutions reported in the previously published Justinianic lower-coverage genome that were here found to be erroneous highlight the importance of using high-quality ancient pathogen genomes for future analysis when they are available, as well as following strict criteria of authenticity and genome quality. Such criteria include average coverage, evenness of coverage, and heterozygosity estimates in addition to the standard criteria such as DNA damage and fragmentation patterns. Exhaustive sampling of ancient material from suspected plague foci of various periods and geographical locations is needed to increase availability of high-quality data sets and to deepen our understanding of the disease, its evolution and its human impact. In addition, the unique genetic features identified here call for functional studies that would explore their role in terms of Y. pestis physiology and adaptation.
Materials and Methods
Prescreening of Samples
A total of 20 individuals from the Altenerding site, located in Bavaria, Germany, were included in the Y. pestis prescreening. The cemetery was used continuously between the first half of the fifth century and until the first half of the seventh century, with around 1,450 graves (Hakenbeck 2011; Losert and Pleterski 2003). The 20 individuals were buried in 10 double burials. Two teeth were sampled per individual and tested twice in independent PCRs for replication. The samples were prepared as described in Sofeso et al. 2012 (Sofeso et al. 2012) and DNA extraction was performed following the protocol C of Yang et al. 1998, modified by Wiechmann and Grupe 2005, using up to 0.40 g of tooth powder. All pre-PCR steps were performed in the aDNA facility of the ArchaeoBioCenter, University of Munich (Seifert et al. 2013). Conventional PCR targeting 133 bp of the Y. pestis specific plasminogen activator (pla) gene on the pPCP1 plasmid was performed on both samples of each individual, respectively, as described in Seifert et al. 2013, using the primers Y.-pest_F and Y.-pest_R2 (Tomaso et al. 2003). All PCRs were performed with extraction blanks and PCR blanks. Individuals AE1175 and AE1176 buried in a shared grave were found positive for Y. pestis DNA in the prescreening. Two new teeth were removed from each individual: AE1175 (lower- and upper- M3 teeth) and AE1176 (two molars from the mandible) for further screening.
Radiocarbon and Archaeological Dating
Metacarpal bones from individuals AE1175 and AE1176 were radiocarbon dated at the CEZ Archaeometry gGmbH, Mannheim, Germany. In addition, an archaeological date was assessed based on the grave furnishing (Supplementary archaeological and historical information).
DNA Extraction
DNA was sampled from each tooth by drilling the dental pulp. Approximately 50 mg of drilled powder was used for DNA extraction. DNA was extracted following established protocols for archaeological skeletal tissue (Rohland and Hofreiter 2007). Extraction was performed in a dedicated aDNA laboratory located at the University of Tuebingen. A negative control was included.
PLA Assay
Additional screening of all extracts was performed to assess preservation of Y. pestis DNA, using a previously described qPCR assay (pla assay) (Schuenemann et al. 2011) specific to the plasminogen activator (pla) gene that is located on the high copy pPCP1 plasmid of Y. pestis (Parkhill et al. 2001). The pla assay was performed using a Roche Lightcycler 480 machine. Standards were diluted in 10-fold serial dilutions to 2.23 copies/µl in TET buffer (10 mM Tris, 1 mM Ethylenediaminetetraacetic acid (EDTA), and 0.05% Tween). PCRs were performed in 20 µl reactions consisting of 1 unit of 10X PCR buffer II, 2.5 mM MgCl2, 250 µM each dNTP, 5% Dimethyl sulfoxide (DMSO), 0.75 mg/ml Bovine serum Albumin (BSA), 300 nM each primer (Schuenemann et al. 2011), 1 unit of 20X EvaGreen dye (Biotium), 0.05 U/µl Amplitaq gold DNA polymerase (Applied Biosystems) and 2 µl of DNA extract.
Library Preparation for Sequencing
A 10 µl aliquot of each extract was converted into an Illumina double stranded (ds) DNA library. In addition, a 60 µl aliquot of each extract was converted into a concentrated ds Illumina DNA library, following established protocols (Meyer and Kircher 2010), with replacement of the final MinElute (Qiagen) purification by an 80 °C incubation for 20 minutes to denature the Bst polymerase. The concentrated libraries were pretreated with a UDG enzyme followed by endonuclease VIII to remove deaminated cytosines in this way preventing erroneous substitution assignments originating in DNA damage (Briggs et al. 2010). A negative control for library preparation was included for each library type. Libraries were quantified using an IS7 and IS8 primer quantification assay (Lightcycler 480 Roche) (Meyer and Kircher 2010), using the DyNAmo SYBR Green qPCR kit (Thermo Scientific). Each library was double indexed following established protocols in 1 to 12 parallel 100 µl reactions (Kircher, et al. 2012). Indexed products for each library were then pooled and purified over MinElute columns (Quiagen), eluted in 50 µl TET buffer (10 mM Tris, 1 mM EDTA, and 0.05% Tween), and quantified using the above quantification assay with IS5 and IS6 primers (Meyer and Kircher 2010). Five microliters of MinElute purified product was subsequently amplified in 100µl reactions using AccuPrime Taq DNA polymerase (Life technologies) following the manufacturer’s protocol with 0.3 µM each primer. Products were again MinElute purified and quantified using the Agilent 2100 Bioanalyzer DNA 1000 chip. A 10 nM pool of all the libraries not treated with UDG was prepared for shotgun sequencing. The UDG treated concentrated libraries were used for downstream Y. pestis array capture enrichment. In addition, both UDG treated and non-UDG treated libraries were used for downstream mitochondrial in-solution target enrichment. The sequenced data generated from the human mitochondrial DNA enriched libraries was later used to estimate the preservation of human DNA in the sample and to assess the level of ancient human DNA damage as another means of ancient DNA authentication.
Mitochondrial Target Enrichment
Target enrichment of human mitochondrial DNA was performed by in-solution bead capture of the pooled libraries, following established protocols (Maricic, et al. 2010) and using baits generated from modern human mitochondrial DNA.
Y. Pestis Array Enrichment
Probes for the Y. pestis array capture were designed using an in-house probe design software. In order to avoid ascertainment bias for a specific Y. pestis strain, the genome of the Y. pseudotuberculosis outgroup (GenBank accession NC_006155) was used as a template for probe design. In addition, the Y. pestis (CO92) pCD1 (70 kb) and pMT1 (100 kb) plasmids were included (GenBank accession NC_003131 and NC_003134, respectively). Probes were designed with 5 bp tiling density across the Y. pseudotuberculosis template and 6 bp tiling density across the CO92 pMT1 and pCD1 plasmids template. A total of 976,658 probes were used on an Agilent 1-million feature array.
Array hybridization was performed following established protocols (Hodges et al. 2009), with two nights of incubation at 65 °C. Postharvest the 490 µl eluate was reamplified to generate a 20 µg pool of purified product for serial capture on an array identical to the one used for the first round of capture. Products were reamplified and purified as described above and made into a 10 nM pool for high-throughput sequencing. In addition, the purified product from the first round of capture was also made into a 10 nM pool and sequenced on in the same way.
Sequencing
Library pools were sequenced on the Illumina Hiseq 2500 platform using two index reads (2*100 + 7 + 7 cycles) following the manufacturer’s protocol. Deindexing was performed by sorting all sequences by their P7 and P5 combinations, using the CASAVA software version 1.8. Adapters were clipped from all reads and forward and reverse reads were merged into single sequences if they overlapped by at least 11 bp (Kircher, et al. 2011). Reads that could not be merged were discarded. All reads were filtered for a length of at least 30 bp.
Analysis of Sequenced Reads
Merged and filtered reads from the two rounds of serial Y. pestis capture were pooled and mapped via BWA version 0.7.12 (Li and Durbin 2009) to the CO92 Y. pestis reference chromosome (GenBank accession NC_003143) and to the pCD1 plasmid (GenBank accession NC_003131) and the pMT1 plasmid (GenBank accession NC_003134). To avoid cross-mapping of reads from other organisms, the mapping stringency of BWA was increased to 0.1 (−n parameter). Duplicate reads were removed and remaining reads were filtered for a minimal mapping quality of 37 using Samtools version 0.1.19. Mean coverage and coverage across the reference were calculated and plotted using QualiMap version 2.1. The same mapping tools and parameters were used for all data sets in the study. A set of published Y. pestis genomes that have been described elsewhere (Cui et al. 2013) was used, together with the published ancient Black Death Y. pestis genome (Bos et al. 2011), a reanalyzed A120 genome (Wagner et al. 2014), the new Altenerding genome, and the genome of strain IP32953 of Y. pseudotuberculosis as an out-group (GenBank accession NC_006155). For complete genomes, artificial reads were generated by a tiling approach with read length of 100 bp and tiling density of 1 bp. Genomes used for analyses are listed in supplementary table S4, Supplementary Material online.
Ancient DNA Damage Assessment
The level of DNA damage was determined for the reads mapping to the human mitochondrial reference and for the reads mapping to the Y. pestis reference, using mapDamage version 2.0 (Jonsson et al. 2013) with standard parameters.
SNP Calling
Comparative SNP typing was performed on mapped data using the UnifiedGenotyper of the Genome Analysis Toolkit (GATK) (DePristo et al. 2011). vcf files (variant call format) were produced for the set of 133 genomes. The “EMIT_ALL_SITES” option was set to generate calls for both variant and nonvariant sites. A custom Java program (MultiVCFanalyzer) was applied to the generated vcf files to analyze and filter the detected SNPs and to produce a multiple sequence alignment of all positions for which a SNP was called in at least one of the strains in the complete data set. A SNP was called if GATK called a homozygous SNP, if the position was covered by at least 5 reads and the quality of the GATK call was at least 30. If GATK called a heterozygous SNP, we called the SNP allele if it was supported by at least 5 reads with a quality of at least 30 and if the fraction of reads containing the SNP was at least 90%. If a SNP call was not possible, the reference base was called instead if the respective thresholds were reached. If neither a reference call nor a SNP call was possible based on these thresholds, the “N” character was inserted in the SNP alignment. We omitted genomic regions that were defined in previous studies as noncore regions (Cui et al. 2013; Morelli et al. 2010). In addition, we excluded tRNA regions, rRNAs, and annotated repetitive elements.
Comparison of SNPs called for the Aschheim genome by Wagner et al. 2014 to SNP Calls in the Altenerding Genome
SNPs called in Wagner et al. 2014 for the A120 Aschheim genome were compared with the SNPs called for the Altenerding genome and to SNPs called in the reanalyzed A120 Aschheim genome. In order to inspect discrepancies in SNP calling between the compared genomes, relevant positions were visualized using the IGV gene browser (Robinson et al. 2011).
Phylogenetic Analysis
For phylogenetic analysis, the SNP alignment generated from the vcf files was used as input for phylogenetic reconstruction software MEGA6 (Tamura et al. 2013). All alignment columns that contained missing values (i.e., “N”) were excluded from the analysis (complete deletion) in order to avoid any bias resulting from different genomic coverage between samples. Phylogenies were constructed from a total of either 2603 nucleotide positions (phylogenies excluding A120) or 1418 nucleotide positions (phylogenies including A120). Both maximum-likelihood and maximum-parsimony topologies were generated. Bootstrap support values were obtained over 1,000 replicates. The phylogenetic trees were edited in FigTree version 1.4.2.
SNP Effect Analysis
SNPs that were detected in the Altenerding Justinianic strain were investigated for their effect on protein-coding genes using SnpEff (Cingolani et al. 2012). Genes were annotated according to the annotation in the CO92 Y. pestis reference genome. SnpEff was applied to the complete set of SNPs using standard parameters, except that the size of the upstream and downstream regions of genes in which SNPs are classified was set to 100 nt.
Indel Analysis
In order to identify regions in the Altenerding genome that are missing in CO92, we performed a second mapping of the captured Altenerding reads to the CO92 reference as described above but with the mapping quality filtering parameter set to 0 (−q) in order to retain also unspecifically mapping reads. We extracted all reads that did not map to CO92 and mapped them to the Y. pseudotuberculosis reference as described above. All covered regions longer than 250 bp were visually examined using the IGV gene browser.
In order to investigate regions that were uniquely deleted in the Altenerding Justinianic strain, noncovered regions in the mapping to Y. pseudotuberculosis reference were listed and cross-referenced with a list of noncovered regions in the mapping of two phylogenetically close strains to the Y. pseudotuberculosis reference: 0.ANT1b_CMCC49003 which is basal to the Justinianic strains and 0.ANT2a_2330 (Cui et al. 2013) which is more derived than the Justinianic strains. Regions that were covered in the two 0.ANT strains were filtered for length higher than 100 bp and visually inspected using the IGV gene browser. In addition, these regions were cross-referenced with the probes used for enrichment to make sure they are not missing due to capture bias.
Supplementary Material
Supplementary figures S1–S12 and tables S1–S11 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Julian Parkhill and Sophia David from the Sanger Institute, Cambridge, UK for their useful comments on data analyses and on early drafts. We thank Dr Verena Schünemann and the other members of the Institute for Archaeological Sciences, Tuebingen University as well as the members of the Department of Archaeogenetics of the Max Planck Institute for the Science of Human History for their comments and advice. We thank Stephen Clayton, Alexander Immel, and Ofer Habushi for their help and support with data analysis. We thank Annette Günzel for graphical support. We thank the city of Erding for funding the prescreening of the samples from Altenerding. This work was supported by the European Research Council starting grant APGREID (to J.K.).
References
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 96(24):14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedictow OJ. 2004. The Black Death, 1346-1353: the complete history. England: Boydell & Brewer. [Google Scholar]
- Benjamini Y, Speed TP. 2012. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 40(10):e72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos KI, Herbig A, Sahl J, Waglechner N, Fourment M, Forrest SA, Klunk J, Schuenemann VJ, Poinar D, Kuch M. 2016. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. eLife 5:e12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, Comas I, Forrest SA, Bryant JM, Harris SR, Schuenemann VJ. 2014. Pre-Columbian mycobacterial genomes reveal seals as a source of new world human tuberculosis. Nature 514(7523):494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, et al. 2011. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478(7370):506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prufer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci. 104(37):14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Paabo S. 2010. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38(6):e87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker RR. 1991. Factors promoting acute and chronic diseases caused by Yersiniae. Clin Microbiol Rev. 4(3):309–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael AG. 2014. Plague persistence in western Europe: A hypothesis. The Medieval Globe 1(1):8. [Google Scholar]
- Carnoy C, Floquet S, Marceau M, Sebbane F, Haentjens-Herwegh S, Devalckenaere A, Simonet M. 2002. The superantigen gene ypm is located in an unstable chromosomal locus of Yersinia pseudotuberculosis. J Bacteriol. 184(16):4489–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvaux S, Dillies M, Marceau M, Rosso M, Rousseau S, Moszer I, Simonet M, Carniel E. 2011. In silico comparison of Yersinia pestis and Yersinia pseudotuberculosis transcriptomes reveals a higher expression level of crucial virulence determinants in the plague bacillus. Int J Med Microbiol. 301(2):105–116. [DOI] [PubMed] [Google Scholar]
- Chauvaux S, Rosso ML, Frangeul L, Lacroix C, Labarre L, Schiavo A, Marceau M, Dillies MA, Foulon J, Coppee JY, et al. 2007. Transcriptome analysis of Yersinia pestis in human plasma: An approach for discovering bacterial genes involved in septicaemic plague. Microbiology 153(Pt 9):3112–3124. [DOI] [PubMed] [Google Scholar]
- Christakos G, Olea RA, Yu H. 2007. Recent results on the spatiotemporal modeling and comparative analysis of Black Death and Bubonic Plague epidemics. Public Health 121(9):700–720. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn S,K. 2008. Epidemiology of the Black Death and successive waves of plague. Med Hist. 52:74–100. [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, et al. 2013. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci. 110(2): 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Tan Y, Yang H, Qiu J, Qin L, Wang T, Liu H, Bi Y, Song Y, Guo Z. 2009. Gene expression profiling of Yersinia pestis with deletion of lcrG, a known negative regulator for Yop secretion of type III secretion system. Inter J Med Microb 299(5):355–366. [DOI] [PubMed] [Google Scholar]
- Duncan CJ, Scott S. 2005. What caused the black death? Postgrad Med J. 81(955):315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplantier J, Duchemin J, Chanteau S, Carniel E. 2005. From the recent lessons of the Malagasy foci towards a global understanding of the factors involved in plague reemergence. Vet Res. 36(3):437–453. [DOI] [PubMed] [Google Scholar]
- Enscore RE, Biggerstaff BJ, Brown TL, Fulgham RE, Reynolds PJ, Engelthaler DM, Levy CE, Parmenter RR, Montenieri JA, Cheek JE. 2002. Modeling relationships between climate and the frequency of human plague cases in the southwestern United States, 1960-1997. Am J Trop Med Hyg. 66(2):186–196. [DOI] [PubMed] [Google Scholar]
- Guiyoule A, Grimont F, Iteman I, Grimont PA, Lefevre M, Carniel E. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J Clin Microbiol. 32(3):634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiyoule A, Rasoamanana B, Buchrieser C, Michel P, Chanteau S, Carniel E. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J Clin Microbiol. 35(11):2826–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haensch S, Bianucci R, Signoli M, Rajerison M, Schultz M, Kacki S, Vermunt M, Weston DA, Hurst D, Achtman M. 2010. Distinct clones of Yersinia pestis caused the Black Death. PLoS Pathogens 6(10):e1001134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck S. 2011. Local, regional and ethnic identities in early Medieval cemeteries in Bavaria. Firenze: All'Insegna del Giglio.
- Harbeck M, Seifert L, Hänsch S, Wagner DM, Birdsell D, Parise KL, Wiechmann I, Grupe G, Thomas A, Keim P. 2013. Yersinia pestis DNA from skeletal remains from the 6th century AD reveals insights into Justinianic Plague. PLoS Pathogens 9(5):e1003349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Gordon DB, Brizuela L, McCombie WR, Hannon GJ. 2009. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 4(6):960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. 2013. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29(13):1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaroglou P, Delmelle E. 2015. Spatial analysis in health geography. United Kingdom: Ashgate Publishing. [Google Scholar]
- Keim PS, Wagner DM. 2009. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nature Rev. Microbiol. 7(11):813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Heyn P, Kelso J. 2011. Addressing challenges in the production and analysis of Illumina sequencing data. BMC Genomics 12:382. 2164-12-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40(1):e3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little LK. 2007. Plague and the end of antiquity: The pandemic of 541–750. UK: Cambridge University Press in association with the American Academy in Rome. [Google Scholar]
- Losert H, Pleterski A. 2003. Altenerding in Oberbayern. Struktur Des frühmittelalterlichen gräberfeldes und “Ethnogenese” der Bajuwaren. Berlin, Bamberg, Ljubljana. Berlin: Scrîpvaz/Založba ZRC.
- Maddicott J. 1997. Plague in seventh-century England. Past and Present 156(1):7–54. [Google Scholar]
- Marceau M. 2005. Transcriptional regulation in Yersinia: An update. Curr Issues Mol Biol. 7(2):151–178. [PubMed] [Google Scholar]
- Maricic T, Whitten M, Pääbo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PloS One 5(11):e14004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick M. 2015. Tracking mass death during the fall of rome’s empire (I). J Roman Archaeol. 28:325–357. [Google Scholar]
- McCormick M. (forthcoming). Tracking mass death during the fall of rome's empire (II). J Roman Archaeol. 29. [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010(6): pdb. prot5448. [DOI] [PubMed] [Google Scholar]
- Mitchell S. 2014. A history of the later Roman Empire, AD 284-641. United Kingdom: John Wiley & Sons. [Google Scholar]
- Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, et al. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 42(12):1140–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motin VL, Georgescu AM, Elliott JM, Hu P, Worsham PL, Ott LL, Slezak TR, Sokhansanj BA, Regala WM, Brubaker RR, et al. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J Bacteriol. 184(4):1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motin VL, Georgescu AM, Fitch JP, Gu PP, Nelson DO, Mabery SL, Garnham JB, Sokhansanj BA, Ott LL, Coleman MA, et al. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J Bacteriol. 186(18):6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MTG, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413(6855):523–527. [DOI] [PubMed] [Google Scholar]
- Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, Gage KL. 1999. Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am J Trop Med Hyg. 61(5):814–821. [DOI] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. 1997. Yersinia pestis-etiologic agent of plague. Clin Microbiol Rev. 10(1):35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitzer R. 1954. Plague. Geneva: WHO; 409–482. [Google Scholar]
- Radnedge L, Agron PG, Worsham PL, Andersen GL. 2002. Genome plasticity in Yersinia pestis. Microbiology 148(Pt 6):1687–1698. [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Allentoft ME, Nielsen K, Orlando L, Sikora M, Sjögren K, Pedersen AG, Schubert M, Van Dam A, Kapel CMO. 2015. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163(3):571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Hofreiter M. 2007. Ancient DNA extraction from bones and teeth. Nat Protoc. 2(7):1756–1762. [DOI] [PubMed] [Google Scholar]
- Schmid BV, Buntgen U, Easterday WR, Ginzler C, Walloe L, Bramanti B, Stenseth NC. 2015. Climate-driven introduction of the Black Death and successive plague reintroductions into Europe. Proc Natl Acad Sci USA. 112(10):3020–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann VJ, Bos K, DeWitte S, Schmedes S, Jamieson J, Mittnik A, Forrest S, Coombes BK, Wood JW, Earn DJ, et al. 2011. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc Natl Acad Sci USA. 108(38):E746–E752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S, Duncan CJ. 2001. Biology of plagues: evidence from historical populations. United Kingdom: Cambridge University Press. [Google Scholar]
- Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci USA. 103(14):5526–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert L, Harbeck M, Thomas A, Hoke N, Zöller L, Wiechmann I, Grupe G, Scholz HC, Riehm JM. 2013. Strategy for sensitive and specific detection of Yersinia pestis in skeletons of the Black Death pandemic. PloS One 8(9):e75742.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaji S, Bhanu NV, Aggarwal R. 2000. Identification of Yersinia pestis as the causative organism of plague in India as determined by 16S rDNA sequencing and RAPD-based genomic fingerprinting. FEMS Microbiol Lett. 189(2):247–252. [DOI] [PubMed] [Google Scholar]
- Sofeso C, Vohberger M, Wisnosky A, Päffgen B, Harbeck M. 2012. Verifying archaeological hypotheses: Investigantions on origin and genealogical lineages of a privileged society in Upper Bavaria from Imperial Roman times (Erding, Kletthamer Feld) In: Burger J, Kaiser E, Schier W, editors. Population dynamics in pre- and Early History. New approaches by using stable isotopes and genetics. proceedings of the conference in Berlin, March 24-26, 2010. Berlin/Boston: De Gruyter; p. 113–130. [Google Scholar]
- Stathakopoulos DC. 2004. Famine and pestilence in the late Roman and early Byzantine Empire: A systematic survey of subsistence crises and epidemics. United Kingdom: Ashgate. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaso H, Reisinger EC, Dahouk S, Frangoulidis D, Rakin A, Landt O, Neubauer H. 2003. Rapid detection of Yersinia pestis with multiplex real‐time PCR assays using fluorescent hybridisation probes. FEMS Immunol Med Microbiol. 38(2):117–126. [DOI] [PubMed] [Google Scholar]
- Tong Z, Zhou D, Song Y, Zhang L, Pei D, Han Y, Pang X, Li M, Cui B, Wang J. 2005. Pseudogene accumulation might promote the adaptive microevolution of Yersinia pestis. J Med Microbiol. 54(3):259–268. [DOI] [PubMed] [Google Scholar]
- Twigg G. 1984. The Black Death: a biological reappraisal. London: Batsford Academic and Educational. [Google Scholar]
- Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathogens 6(2):e1000783.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DM, Klunk J, Harbeck M, Devault A, Waglechner N, Sahl JW, Enk J, Birdsell DN, Kuch M, Lumibao C. 2014. Yersinia pestis and the Plague of Justinian 541–543 AD: a genomic analysis. Lancet Infect Dis. 14(4):319–326. [DOI] [PubMed] [Google Scholar]
- Wiechmann I, Grupe G. 2005. Detection of Yersinia pestis DNA in two early Medieval skeletal finds from Aschheim (upper Bavaria, 6th century AD). Am J Phys Anthropol. 126(1):48–55. [DOI] [PubMed] [Google Scholar]
- Willias SP, Chauhan S, Motin VL. 2014. Functional characterization of Yersinia pestis aerobic glycerol metabolism. Microb Pathog. 76:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2004. Human plague in 2002 and 2003. Wkly Epidemiol Rec. 79(301):6. [PubMed] [Google Scholar]
- Xu L, Stige LC, Kausrud KL, Ben Ari T, Wang S, Fang X, Schmid BV, Liu Q, Stenseth NC, Zhang Z. 2014. Wet climate and transportation routes accelerate spread of human plague. Proc Biol Sci. 281(1780):20133159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. 1998. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 105(4):539–543. [DOI] [PubMed] [Google Scholar]
- Zhou D, Han Y, Qiu J, Qin L, Guo Z, Wang X, Song Y, Tan Y, Du Z, Yang R. 2006. Genome-wide transcriptional response of Yersinia pestis to stressful conditions simulating phagolysosomal environments. Microb Infect. 8(12):2669–2678. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Schneider D, Blot M, Kolter R. 2003. Bacterial evolution through the selective loss of beneficial genes. trade-offs in expression involving two loci. Genetics 164(4):1271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.