Abstract
The ST-segment and adjacent T-wave (ST-T wave) amplitudes of the electrocardiogram are quantitative characteristics of cardiac repolarization. Repolarization abnormalities have been linked to ventricular arrhythmias and sudden cardiac death. We performed the first genome-wide association meta-analysis of ST-T-wave amplitudes in up to 37 977 individuals identifying 71 robust genotype–phenotype associations clustered within 28 independent loci. Fifty-four genes were prioritized as candidates underlying the phenotypes, including genes with established roles in the cardiac repolarization phase (SCN5A/SCN10A, KCND3, KCNB1, NOS1AP and HEY2) and others with as yet undefined cardiac function. These associations may provide insights in the spatiotemporal contribution of genetic variation influencing cardiac repolarization and provide novel leads for future functional follow-up.
Introduction
Duration of cardiac repolarization has been previously studied by genome-wide association studies (GWAS) and led to the discovery of 35 associated loci (1). However, abnormalities of repolarization are not limited to changes in duration, but are also captured by changes in amplitudes (2). Ventricular repolarization occurs in a geometric pattern across the heart. This process entails differences in timings of recovery across the ventricular wall and between regions of the left ventricle after ventricular depolarization. The cellular basis for the origins of the ST-segment and adjacent T wave (ST-T wave) amplitudes is still under debate, but is generally thought to arise as a consequence of electrical heterogeneities that exist within the ventricular myocardium (3). It is thought that the ST-T wave begins when the epicardial cells start to recover ahead of the midwall and endocardial cells. Electrical currents flow from the midmyocardial and endocardial regions toward the epicardium, resulting in a gradual rise in current, until the epicardium is fully repolarized and the currents have reached maximal intensity (T-top) (4). Continuous indices of T-wave amplitudes are not only relevant in patients with myocardial infarction (5), ischemic cardiomyopathy (6) and a malignant form of ER (7), but also in non-diseased population cohorts. In the general or healthy population, inversions of the ST-T wave are strongly associated with adverse outcome (8–10), but also minor depressions or elevations in ST-T-wave amplitudes are predictors of cardiovascular mortality (11–13) and disease (10,14).
Deviations of ST-T-wave amplitudes can be indicative of a variety of cardiac pathologies, including myocardial ischemia, ventricular hypertrophy, long-QT syndrome, ER and Brugada syndrome (2,8,12,15–17). For clinical use, phenotypes are dichotomized based on optimal sensitivity and specificity to predict worse outcome. However, there is no evidence that the underlying biology of ST-T wave is truly binary. Therefore, we hypothesized that common genetic variation involved in the biology of quantitative ST-T waves traits might provide additional biological insights into the (patho)physiological mechanisms of repolarization. GWAS have proven to be a powerful and unbiased tool to identify novel mechanisms and pathways.
Here, we aim to identify key genetic loci associated with the heart's repolarization phase during the ST-T wave of the electrocardiogram (ECG) to further advance our knowledge on biological factors regulating cardiac repolarization and thereby provide new gene targets for future study.
Results
Genome-wide discovery analysis and replication
We performed genome-wide meta-analyses in 15 943 subjects of European descent on the ST-T wave (Supplementary Material, Table S1), with up to 2 316 136 directly genotyped and imputed autosomal single nucleotide polymorphisms (SNPs). Phenotypes for the ST-segment amplitudes and T-wave amplitudes were derived from leads used in the clinic for diagnosing Brugada syndrome (17) (Septal, V1 and V2) and early repolarization (ER) (16–18) (lateral: I, aVL, V5, V6 and inferior: II, III, aVF). Aiming to capture additional information available on the ECG during cardiac repolarization, we also investigated the other lead recordings (Anterior: V3, V4 and lead aVR) that are presumed to have anatomical meaning (Supplementary Material, Table S2). This resulted in 10 phenotypes representing the ST-T-wave amplitude: 5 ST-segment amplitudes and 5 T-wave amplitudes (Fig. 1A). We estimated that studying 10 correlated ST-T wave phenotypes was equal to studying eight independent phenotypes (see the ‘Materials and Methods’ section); therefore, the threshold of genome-wide significance was set to P< 5 × 10−8/8 (P < 6.25 × 10−9). We considered a threshold of P < 1 × 10−6 to be suggestive of association and used this threshold to prioritize SNPs for replication. The genome-wide meta-analysis across the 10 ST-T-wave phenotypes identified a total of 36 loci to pass the suggestive threshold for association with at least one of the ST-T wave phenotypes. There was no evidence for inflation of test statistics in the final meta-analysis (Supplementary Material, Table S3) or significant heterogeneity. Of the 36 loci, there were multiple loci were associated with one or more 10 ST-T-wave phenotypes: totaling 95 genotype–phenotype associations (P < 1 × 10−6, Supplementary Material, Table S4).
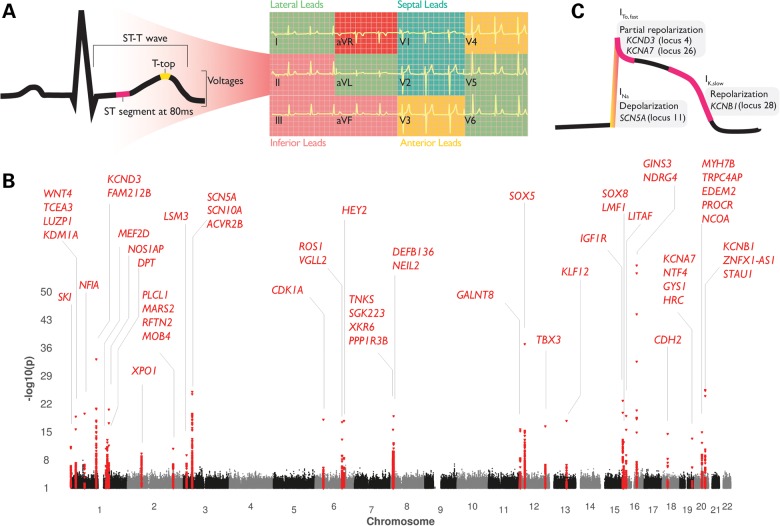
Figure 1.
(A) We conducted genome-wide analyses of ST-segment amplitudes at 80 ms after J-point and T-wave (top) amplitudes reflecting temporal patterns in the cardiac cycle during the repolarization phase. In total, 12 phenotypes were defined by taking the sum of the ST-T-wave amplitudes in the lateral (I, aVL, V5 and V6), inferior (II, III and aVF), septal (V1 and V2), anterior (V3 and V4) leads and lead aVR. These lead groups cover the combination of leads with presumed anatomical meaning of the heart, and those that are used in the clinic for diagnosing Brugada syndrome (17) and ER (16–18). (B) Genome-wide association analyses of all ST-T wave traits identified 71 significant genotype–phenotype associations in 28 genetic loci (2 MB). The x-axis represents the chromosomal position for each SNP, which was assigned the lowest P-value across the 12 traits; the y-axis represents the −log10 of the P-value for association. Twenty-seven loci were significant for ST-T-wave amplitudes. (C) Four genes overlapping four loci are directly involved in the cardiac action potential; SNPs near SCN5A were associated with ST-segment and T-top amplitudes whereas the loci containing potassium channel-coding genes KCND3, KCNA7 and KCNB1 were primarily associated with ST-segment amplitudes.
Next, we performed replication testing of the 95 genotype–phenotype associations (P < 1 × 10−6) in 22 034 independent individuals derived from 7 independent cohorts (Supplementary Material, Table S5), identifying 71 genotype–phenotype associations in 28 independent loci to be replicated (P < 0.01) and become genome-wide significantly associated (Fig. 1B, Table 1 and Supplementary Material, Table S4 and Fig. S1).
Table 1.
All sentinel SNP associations per ST-T wave phenotype
| No. | Region | SNP | Trait | LD (r2) with Sentinel SNP(†) in Locus | EA (FRQ)/NEA | β (SE) | Pmeta | N | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1p36.33 | rs260505 | T.ant | † | A (0.49)/C | 0.05 (0.01) | 1.30E-11 | 37775 | SKIn |
| 2 | 1p36.12 | rs2072944 | T.sep | 0.37 | T (0.55)/C | 0.05 (0.01) | 1.70E-13 | 37821 | LUZP1n, KDM1An, WNT4e |
| 2 | 1p36.12 | rs2298632 | ST.sep | † | T (0.47)/C | −0.07 (0.01) | 4.40E-18 | 37753 | TCEA3ne |
| 3 | 1p31.3 | rs2207792 | T.aVR | – | A (0.66)/G | −0.06 (0.01) | 3.30E-13 | 37799 | NFIAnd |
| 3 | 1p31.3 | rs2207792 | T.inf | † | A (0.66)/G | 0.07 (0.01) | 3.00E-20 | 37810 | NFIAnd |
| 4 | 1p13.2 | rs12145374 | ST.ant | – | A (0.76)/C | 0.11 (0.01) | 5.90E-33 | 37753 | KCND3ng, FAM212Bd |
| 4 | 1p13.2 | rs12145374 | ST.sep | – | A (0.76)/C | 0.07 (0.01) | 1.20E-14 | 37753 | KCND3ng, FAM212Bd |
| 4 | 1p13.2 | rs12145374 | ST.aVR | – | A (0.76)/C | −0.12 (0.01) | 1.40E-31 | 37784 | KCND3ng, FAM212Bd |
| 4 | 1p13.2 | rs12145374 | ST.lat | † | A (0.76)/C | 0.12 (0.01) | 8.20E-34 | 37771 | KCND3ng, FAM212Bd |
| 4 | 1p13.2 | rs12145374 | ST.inf | – | A (0.76)/C | 0.09 (0.01) | 6.00E-18 | 37770 | KCND3ng, FAM212Bd |
| 5 | 1q22 | rs10908505 | ST.aVR | † | A (0.66)/T | −0.05 (0.01) | 1.70E-11 | 37784 | MEF2Dngd |
| 6 | 1q23.3 | rs12567315 | T.ant | 0.42 | A (0.19)/G | −0.06 (0.01) | 4.60E-11 | 37775 | NOS1APn |
| 6 | 1q23.3 | rs4657178 | T.inf | † | T (0.24)/C | −0.06 (0.01) | 1.90E-11 | 37810 | NOS1APn |
| 7 | 1q24.2 | rs545833 | ST.ant | † | T (0.26)/C | −0.06 (0.01) | 7.00E-18 | 37753 | DPTn |
| 7 | 1q24.2 | rs511187 | ST.inf | 0.79 | A (0.72)/G | 0.06 (0.01) | 2.60E-12 | 37770 | DPTn |
| 7 | 1q24.2 | rs511187 | ST.aVR | 0.79 | A (0.72)/G | −0.07 (0.01) | 3.10E-17 | 37784 | DPTn |
| 7 | 1q24.2 | rs511187 | ST.lat | 0.79 | A (0.72)/G | 0.07 (0.01) | 7.20E-18 | 37771 | DPTn |
| 8 | 2p15 | rs7576036 | ST.sep | † | T (0.54)/C | 0.05 (0.01) | 2.80E-10 | 37753 | XPO1n |
| 9 | 2q33.1 | rs1866666 | T.aVR | † | T (0.49)/C | −0.05 (0.01) | 1.80E-11 | 37799 | PLCL1nced, MARS2e, RFTN2e, MOB4d |
| 10 | 3p25.1 | rs4684185 | T.aVR | † | T (0.35)/C | −0.05 (0.01) | 7.10E-10 | 37799 | LSM3n, TMEM43d |
| 11 | 3p22.2 | rs7638909 | ST.sep | 0.02 | T (0.76)/G | −0.06 (0.01) | 2.00E-11 | 37753 | SCN5And, ACVR2Bd |
| 11 | 3p22.2 | rs9851710* | T.inf | 0.03 | A (0.66)/C | 0.08 (0.01) | 9.30E-19 | 37810 | SCN10An, SCN5Ad, ACVR2Bd |
| 11 | 3p22.2 | rs7633988 | T.ant | 0.02 | A (0.71)/T | 0.06 (0.01) | 3.80E-11 | 37775 | SCN10An, SCN5Ad, ACVR2Bd |
| 11 | 3p22.2 | rs6783110 | T.lat | 0.94 | A (0.42)/G | 0.06 (0.01) | 3.70E-12 | 30699 | SCN10Anc, SCN5Ad, ACVR2Bd |
| 11 | 3p22.2 | rs4076737 | T.aVR | 0.93 | T (0.57)/G | 0.07 (0.01) | 1.20E-20 | 37799 | SCN10Anc, SCN5Ad, ACVR2Bd |
| 11 | 3p22.2 | rs4076737 | ST.inf | 0.93 | T(0.57)/G | −0.07 (0.01) | 8.30E-20 | 37770 | SCN10Anc, SCN5Ad, ACVR2Bd |
| 11 | 3p22.2 | rs6801957 | ST.aVR | † | T (0.43)/C | −0.08 (0.01) | 1.10E-25 | 37784 | SCN10Anc, SCN5Ad, ACVR2Bd |
| 11 | 3p22.2 | rs6801957 | ST.lat | – | T (0.43)/C | 0.06 (0.01) | 8.50E-16 | 37771 | SCN10Anc, SCN5Ad, ACVR2Bd |
| 12 | 6p21.31 | rs7756236 | ST.ant | † | A (0.76)/G | −0.06 (0.01) | 1.30E-14 | 37753 | CDKN1Aned |
| 13 | 6q22.2 | rs210966 | ST.lat | – | C (0.28)/G | 0.07 (0.01) | 5.10E-16 | 37771 | ROS1n, VGLL2d |
| 13 | 6q22.2 | rs210966 | ST.aVR | † | C (0.28)/G | −0.07 (0.01) | 4.00E-18 | 37784 | ROS1n, VGLL2d |
| 13 | 6q22.2 | rs172409 | ST.inf | 0.41 | A (0.54)/C | −0.05 (0.01) | 6.40E-11 | 30708 | ROS1n, VGLL2d |
| 14 | 6q22.31 | rs9388451 | ST.ant | † | T (0.52)/C | −0.06 (0.01) | 2.00E-18 | 37753 | HEY2ned |
| 14 | 6q22.31 | rs9388451 | T.ant | – | T (0.52)/C | −0.05 (0.01) | 3.60E-13 | 37775 | HEY2ned |
| 15 | 8p23.1 | rs1458942 | ST.aVR | † | A (0.60)/G | 0.06 (0.01) | 1.10E-13 | 37784 | TNKSnd, SGK223d, XKR6d, PPP1R3Bd |
| 16 | 8p23.1 | rs7011924 | ST.aVR | † | A (0.40)/G | 0.07 (0.01) | 1.30E-19 | 37784 | DEFB136n, NEIL2e |
| 16 | 8p23.1 | rs7011924 | ST.inf | – | A (0.40)/G | −0.07 (0.01) | 3.90E-19 | 37770 | DEFB136n, NEIL2e |
| 17 | 12p13.32 | rs7953399 | ST.sep | 0.95 | A (0.61)/G | −0.04 (0.01) | 2.10E-09 | 37753 | GALNT8n |
| 17 | 12p13.32 | rs2286582 | ST.ant | † | T (0.40)/C | 0.06 (0.01) | 2.40E-16 | 37753 | GALNT8n |
| 18 | 12p12.1 | rs10842350 | T.lat | – | A (0.54)/G | −0.09 (0.01) | 4.60E-37 | 37759 | SOX5ng |
| 18 | 12p12.1 | rs10842350 | T.aVR | † | A (0.54)/G | 0.09 (0.01) | 1.30E-37 | 37799 | SOX5ng |
| 19 | 12q24.21 | rs10850409 | T.sep | † | A (0.27)/G | 0.07 (0.01) | 4.30E-17 | 34881 | TBX3n |
| 20 | 13q22.1 | rs728926 | T.lat | † | T (0.38)/C | −0.07 (0.01) | 2.00E-18 | 37759 | KLF12n |
| 20 | 13q22.1 | rs728926 | T.aVR | – | T (0.38)/C | 0.06 (0.01) | 1.50E-16 | 37799 | KLF12n |
| 21 | 15q26.3 | rs7174918 | ST.ant | 0.74 | T (0.29)/C | −0.06 (0.01) | 6.90E-16 | 37753 | IGF1Rnd |
| 21 | 15q26.3 | rs3803476 | T.sep | – | A (0.37)/G | −0.05 (0.01) | 2.00E-14 | 37821 | IGF1Rnd |
| 21 | 15q26.3 | rs3803476 | ST.sep | † | A (0.37)/G | −0.07 (0.01) | 1.80E-23 | 37753 | IGF1Rnd |
| 22 | 16p13.3 | rs7192150 | T.aVR | † | A (0.40)/G | 0.05 (0.01) | 4.30E-13 | 37799 | LMF1ne, SOX8gd |
| 23 | 16p13.13 | rs7191330 | T.aVR | 0.95 | T (0.46)/C | −0.08 (0.01) | 3.10E-16 | 30731 | LITAFn |
| 23 | 16p13.13 | rs735951 | ST.ant | – | A (0.47)/G | 0.06 (0.01) | 9.50E-17 | 37753 | LITAFn |
| 23 | 16p13.13 | rs735951 | ST.aVR | – | A (0.47)/G | −0.07 (0.01) | 1.70E-18 | 37784 | LITAFn |
| 23 | 16p13.13 | rs735951 | ST.lat | † | A (0.47)/G | 0.07 (0.01) | 1.20E-18 | 37771 | LITAFn |
| 24 | 16q21 | rs4784938 | ST.lat | 0.7 | A (0.74)/G | 0.08 (0.01) | 4.80E-21 | 37771 | GINS3n |
| 24 | 16q21 | rs4784939 | ST.ant | – | T (0.36)/C | −0.09 (0.01) | 4.80E-41 | 37753 | GINS3n |
| 24 | 16q21 | rs4784939 | T.ant | – | T (0.36)/C | −0.06 (0.01) | 1.00E-17 | 37775 | GINS3n |
| 24 | 16q21 | rs4784939 | T.sep | – | T (0.36)/C | −0.08 (0.01) | 4.10E-29 | 37821 | GINS3n |
| 24 | 16q21 | rs4784939 | ST.sep | † | T (0.36)/C | −0.11 (0.01) | 3.00E-57 | 37753 | GINS3n |
| 24 | 16q21 | rs8057901 | T.aVR | 0.93 | A (0.63)/G | −0.06 (0.01) | 4.90E-11 | 30731 | NDRG4n |
| 24 | 16q21 | rs8057901 | ST.aVR | 0.93 | A (0.63)/G | −0.08 (0.01) | 1.60E-21 | 30713 | NDRG4n |
| 24 | 16q21 | rs8057901 | ST.inf | 0.93 | A (0.63)/G | 0.07 (0.01) | 1.30E-16 | 30708 | NDRG4n |
| 24 | 16q21 | rs9940062 | T.inf | 0.94 | T (0.68)/G | 0.07 (0.01) | 1.20E-10 | 30738 | NDRG4n |
| 25 | 18q12.1 | rs8083566 | ST.lat | – | A (0.07)/C | −0.09 (0.01) | 4.30E-10 | 37771 | CDH2nd |
| 25 | 18q12.1 | rs8083566 | ST.inf | – | A (0.07)/C | −0.11 (0.02) | 1.70E-12 | 37770 | CDH2nd |
| 25 | 18q12.1 | rs8083566 | ST.aVR | † | A (0.07)/C | 0.12 (0.01) | 3.60E-15 | 37784 | CDH2nd |
| 26 | 19q13.33 | rs11673003 | ST.lat | † | A (0.90)/G | −0.09 (0.01) | 5.10E-14 | 37771 | KCNA7nc, NTF4n, GYS1d, HRCd |
| 27 | 20q11.22 | rs6087666 | ST.inf | † | A (0.20)/G | −0.07 (0.01) | 3.10E-15 | 37770 | TRPC4APne, EDEM2ce, MYH7Bcd, NCOAd |
| 27 | 20q11.22 | rs6088738 | ST.aVR | 0.55 | A (0.21)/G | 0.06 (0.01) | 5.90E-13 | 37784 | EDEM2ne, PROCRe, MYH7Bcd, NCOAd |
| 28 | 20q13.13 | rs11907908 | ST.ant | 0.83 | T (0.07)/C | −0.12 (0.01) | 8.00E-18 | 37753 | ZNFX1 (−AS1)n, STAU1d |
| 28 | 20q13.13 | rs6019750 | ST.lat | 1 | C (0.93)/G | 0.13 (0.01) | 4.30E-23 | 37771 | KCNB1n, STAU1d |
| 28 | 20q13.13 | rs2202261 | ST.inf | – | A (0.93)/G | 0.12 (0.01) | 4.40E-18 | 37770 | KCNB1n, STAU1d |
| 28 | 20q13.13 | rs2202261 | ST.aVR | † | A (0.93)/G | −0.14 (0.01) | 5.60E-26 | 37784 | KCNB1n, STAU1d |
There are 71 genome-wide significant genotype–phenotype associations clustered in 28 genetic loci (based on a 2 MB locus definition, see the ‘Materials and Methods’ section). Gene superscripts indicate the method of identification: n, nearest gene or nearby gene (within 10 kb of the SNP); g, Grail’ d, Depict; e, eQTL; c, coding SNP (non-synonymous). rs9851710 denoted by an asterisk (*) indicates that this SNP is an independent, secondary, association for ST.aVR (rs9851710 conditioned on rs6801957: β (SE) per minor allele = 0.13 (0.0204), P = 2.38 × 10−10).
The majority of loci that show multiple genotype–phenotype associations in the same locus consist of sentinel SNPs that are either the same SNP or in LD with each other, as indicated by the LD (r2) column in Table 1. Conditional analysis within the discovery set revealed a secondary signal [rs9851710 conditioned on rs6801957: β (SE) per minor allele = 0.13 (0.0204), P = 2.38 × 10−10] in the SCN5A/SCN10A locus to be associated with ST aVR amplitudes. This SNP has also been identified to be associated with other ST-T phenotypes (see Table 1), suggesting that there are 29 independent signals within the 28 loci.
Some loci were predominantly associated with ST-segment amplitudes (locus 4, 5, 16, 26 and 28) while others were predominantly associated with T-wave amplitudes (locus 6, 18 and 29). The test statistics for every combination of genotype–phenotype association are summarized in Supplementary Material, Table S6.
Other electrocardiographic traits
Previous genome-wide associations have studied other ECG indices (1,19–23) such as QRS duration, PR interval, heart rate and QT interval. We intersected these previously identified loci with our ST-T-wave loci and found overlap (within 2 MB) in 13 loci. Lead SNPs from previous and current findings in 4 loci were in low LD (r2 < 0.02), suggesting that 19 of the current loci are novel associations with the ECG (Supplementary Material, Table S7).
Heritability estimates
The 28 identified sentinel SNPs collectively explained between 1.6% (T-wave septal) and 5.1% (ST-segment aVR) of the observed phenotypic variance (Supplementary Material, Table S8). Familial heritability estimates in the Erasmus Rucphen Family (ERF) study varied between h2 = 30% (T-wave inferior) and h2 = 42% (ST septal) suggesting additional genetic variants and other mechanisms that remain to be discovered (Supplementary Material, Table S9). The variation in proportion of variance associated with covariates varied more widely; between r2 = 0.09 (ST-segment inferior) and r2 = 0.36 (ST-segment septal).
Identification of candidate genes and pathway analyses
In total, we prioritized 54 candidate genes in the 28 genetic loci (Table 1) that could play a causal role in ST-T-wave amplitudes based on several criteria, as described in the ‘Materials and Methods’ section. Thirty-four genes were prioritized based on the proximity criteria, 5 genes contained one or more non-synonymous SNPs in high LD with a lead SNP (Supplementary Material, Table S10), 12 genes had a top-eQTL SNP in high LD with a lead SNP (Supplementary Material, Table S11), 65 genes were selected by the data-driven expression prioritized integration for complex trait (DEPICT) analysis of which 25 were within genome-wide significant loci (at false discovery rate ≤ 5%; Supplementary Material, Table S12) and 3 genes are based on literature mining using GRAIL (Supplementary Material, Table S13). The top five keywords retrieved from GRAIL were ‘muscle’, ‘channel’, ‘transcription’, ‘heart’ and ‘channels’.
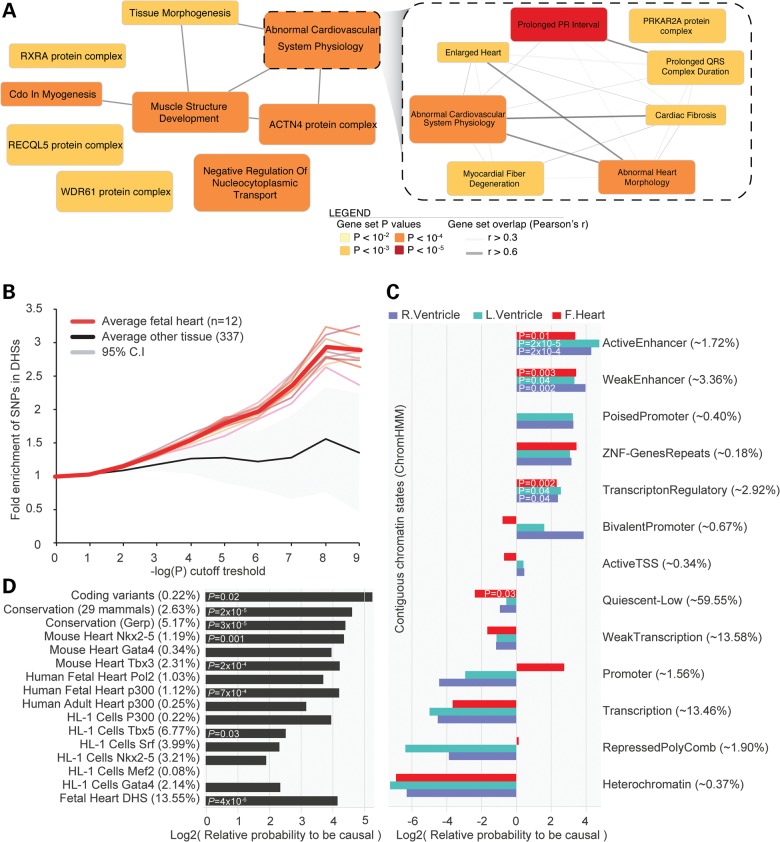
The DEPICT method (24) was applied to subsequently offer insight into the biological pathways underlying changes in ST-T-wave amplitudes of the ECG. DEPICT identified 56 reconstituted gene sets, which grouped into 9 distinct clusters. The cluster containing the most significantly enriched reconstituted gene set (‘VCL protein complex’, a protein–protein interaction network centered on the Vinculin gene product, P = 6.00 × 10−6) represented various protein complexes likely related to Fascia Adherens, while the second most significant gene cluster ‘Abnormal Cardiovascular System Physiology’ contains reconstituted gene set known to be important for cardiac repolarization (Fig. 2 and Supplementary Material, Table S14).
Figure 2.
(A) DEPICT identified 56 significantly enriched gene-sets relevant for ST-T-wave amplitudes. (B) SNPs were significantly more enriched in DHSs of fetal heart tissue (n = 12) compared with other tissue and cells (n = 337), across the full range of P-values of the discovery meta-analyses (genome wide), suggesting that functionality of regulatory DNA elements may underlie some of the associations. (C and D) Next, we performed a meta-analysis of the 28 identified ST-T wave loci using 1000 Genomes imputed data, for this 1000G variants needed to be in LD r2 > 0.1 with the HapMap sentinel SNP. Subsequent prioritization of potential causal annotations in these loci also suggested that regions of DHS in fetal heart are possibly underlying the associations as well as cardiac transcription factors, conserved regions (exonic), active and weak enhancers. While regions that are transcribed, tightly packed (heterochromatin) or function as promoters in the ventricles may be less important for the biological mechanisms of genetic variants that are associated with ST-T-wave amplitudes. Subtle difference are present between the ventricles and fetal heart which could suggest that promoters that overlap potential causal ST-T wave SNPs may be active in the fetal heart but repressed in the ventricles. Percentages between parentheses indicate the amount of SNPs in the 28 loci overlapping with the annotation. Conservation (GERP and 29 mammals), DHS of fetal heart, enhancers of the left ventricle and Tbx3 bound regions were used to prioritize potential causal SNPs in the 28 loci.
Role of regulatory DNA and 1000 genomes imputation
ST-T-wave associated SNPs were 3-fold enriched in Dnase1 Hypersensitivity sites (DHSs) from human fetal heart (Fig. 2), which was significantly higher compared with 337 other cell types and tissues (P = 0.0004, Z-score statistics) in line with earlier observations (25,26). We then performed fine mapping of all 71 genome-wide significant genotype–phenotype associations (Table 1) in prevention of renal and vascular end-stage disease (Prevend) and Lifelines imputed with 1000 genomes (1000G), covering more of the known common variants in humans. We observed that 64 (of 71) associations were assigned to a different lead SNP, that 63 of these associations were more significant in the 1000G imputed data and effect estimates were higher for 58 of these associations (Supplementary Material, Table S16). To facilitate future functional experiments toward the identification of causal variants and their underlying biological mechanisms, we prioritized potential causal SNPs using the probabilistic framework of Probabilistic Annotation INTegratOR (PAINTOR) (27). PAINTOR determined the significance of each annotation (Fig. 2) and used the five most significant annotations (conservation scores, DHS of fetal heart, enhancers of the left ventricle and Tbx3 bound regions) to prioritize potential causal SNPs in the 28 loci (27). This yielded 315 SNPs in the 99% confidence set and 96 SNPs in the 95% confidence set (Supplementary Material, Table S17).
Discussion
Amplitudes of the ST segment and T wave are important traits that are associated with abnormal heart rhythm, conduction disturbances and ventricular arrhythmias. In this study, we performed the first GWAS of ST-T-wave amplitudes of the ECG in up to 37 977 individuals. Traits were defined according to the combination of ECG leads presumed to have anatomical meaning or used in the clinic for diagnosing Brugada syndrome and early repolarization (16–18). ST-T-wave traits were moderately heritable (h2 = 30–42%) while the proportion of variance associated with covariates (gender, body mass index and age) varied considerably (9–36%) strongly supporting the genetic background of these traits. We identified 28 genome-wide significant loci for ST-T-wave amplitudes and a set of 54 candidate genes.
A recent GWAS on Brugada syndrome revealed the association of two loci that are shared by our ST-T-wave loci (28). One of these signals in the SCN5A/SCN10A locus is in complete LD with our sentinel SNP (rs10428132, r2 = 0.96 with rs6801957) and rs9388451 near HEY2, is a ST-T-wave sentinel SNP; suggesting that Brugada syndrome susceptibility loci share a common genetic background with ST-T-wave traits. One of the strongest associated GWAS signals for all amplitudes of the ST segment was in the KCND3 gene (locus 4). KCND3 encodes the Kv4.3 α-subunit that conducts the cardiac fast transient outward K+ current (ITo,f). This current is prominent in Phase 1 of the action potential and contributes to the ‘notch’ of the cardiomyocyte's action potential (2). Mutations in KCND3 have been implicated with increased risk of sudden cardiac death (29,30), as well as mutations in SCN5A (2) and SCN10A (31). We specifically identified loci containing potassium channels KCND3 (locus 4), KCNB1 (locus 28) and KCNA7 (locus 26) to be predominantly associated with amplitudes of the ST segment and not with the T wave, suggesting that these potassium channels are activated during the ST segment of the repolarization phase and may have clinical relevance (16,32,33). Future studies are required to examine the potential role of the other ST-T-wave SNPs for increased arrhythmogenic risk and sudden death through an altered cardiac repolarization.
In addition to HEY2, a transcriptional regulator of cardiac electrical function in the right ventricular outflow tract (28), we identified six loci containing genes with strong evidence of being directly involved in cardiac tissue development via transcriptional regulatory pathways. Locus 5 contains a myocyte enhancer factor-2 (MEF2D) important for cardiac muscle morphogenesis and heart looping (34,35). Loci 18 and 22 contain two transcription factors from the SOX family, SOX5 and SOX8. SOX5 is important for a functioning heart and involved in correct Wnt signaling (36). This locus has previously been associated with the PR interval and resting heart rate, but the identified variant in the current study is in low LD (r2 = 0.01), suggesting multiple molecular-genetic mechanisms at this locus to influence heart function. Transcripts of SOX8 are concentrated in the subendothelial mesenchym of the whole outflow tract (37) in which N-Cadherin (38) (encoded by CDH2, locus 25), the transcription factor Tbx3 (39) (TBX3, locus 19) and WNT4 (40) (specific for the endocardial endothelial cushion development, part of the Wnt pathway) in locus 2 also play essential developmental roles, among other cardiac cell lineages. Other compelling cardiac genes are: SKI, for which mutations (1p36 deletion- and Sprintzen–Goldberg syndrome) are characterized by heart defects, brain abnormalities and muscle tone (hypotonia) in infancy (41); TMEM43 (locus 10) in which mutations cause arrhythmogenic right ventricular cardiomyopathy and Emery–Dreifuss muscular dystrophy; the sarcomeric gene MYH7B and miR499 (locus 27), which regulates a multitude of cardiac mRNA and microRNAs and promotes ventricular specification; and GALNT1, a glycosyltransferases that is required for normal heart valve development and cardiac function by regulating the extracellular matrix and altering conserved signaling pathways that regulate cell proliferation during heart development (42). We also identified a number of less well-characterized genes, which should be explored in further research.
Five of the 28 ST-T-wave sentinel SNPs were in LD with SNPs associated with QT duration (1), suggesting that ST-T-wave amplitudes provides additional information on cardiac repolarization. The known functions of the candidate genes and our pathway analyses suggest that the ST-T-wave amplitudes are influenced and regulated by a wide variety of molecular mechanisms, including ion-channels, structural proteins and cardiac transcription factors. However, the finding that protein complexes related to Fascia Adherens are most enriched in our pathway analyses hints that some of the biology underlying ST-T waves is less well captured in well-established pathways and better represented by data-driven (and not manually curated) pathways.
Ventricular repolarization is a complex process. To capture this process, we chose to apply a selection of composite ECG parameters with the aim of gaining more insight into the biological processes underlying the ST-T wave. These surface ECG parameters are not specific enough to identify the exact anatomical region of the heart; e.g. the septal leads (V1 and V2) may also include aspects of the right ventricular wall activity. However, there are no data available of more precise measurements such as could be derived from more sophisticated surface ECG equipment or intra-cardiac ECG measurements.
In summary, we present a large number of genome-wide significant loci robustly associated with cardiac repolarization parameters, some with compelling biological basis for their association and a number of loci not previously implicated in cardiac function. The identified loci and selected genes have the potential to aid future studies that are focused on risk stratification or on molecular mechanisms underlying cardiac repolarization and diseases of cardiac repolarization; to facilitate these studies, we have made our results (including the genome-wide association) publicly available.
Materials and Methods
Study populations
The discovery phase included participants of the Prevend study and the Lifelines study cohort. Both are community-based cohort studies from the northern part of the Netherlands. The replication phase included participants of the Atherosclerosis Risk in Communities study (ARIC), Cardiovascular Health Study (CHS), Young Finns Study (YFS), ERF study, Rotterdam study I, II and III (RS I, II and III). Detailed descriptions of each cohort are given in the supplementary material. Characteristics of participants are summarized in Supplementary Material, Tables S1 and S5. Detailed Information on genotyping methods, quality control of SNPs, imputation and statistical analysis for each cohort is summarized in Supplementary Material, Table S18.
Phenotype modeling
Summing correlated variables when expecting that genetic variants are associated to multiple of these variables will increase the power for detection; with genetic variant (Snp), and traits (Ampitudei), consider the correlation: Corr(Snp, Ampitude1 + Ampitude2) = Corr(Snp, Ampitude1) + Corr(Snp, Ampitude2) + 2 × Corr(Ampitude1, Ampitude2) > Corr(Snp, Ampitude1) + Corr(Snp, Ampitude2), hence if Corr(Ampitude1, Ampitude2) > 0, the traits Ampitudei measure the same latent variable (L). Therefore, phenotypes for the ST-segment amplitudes 80 ms after J-point and T-wave amplitudes were defined by taking the sum of the lateral (I, aVL, V5 and V6), inferior (II, III and aVF), septal (V1 and V2), anterior (V3 and V4) leads and lead aVR. Individuals were excluded for bundle branch block or QRS duration > 120 ms, atrial fibrillation, flutter, history of myocardial infarction or electronic pacemaker rhythm and when available, heart failure and ECG altering medication. Also participants with extreme measurements (more than ±4SD from mean) were excluded on a per phenotype basis.
Statistical analyses
To control for multiple testing of the 10 phenotypes while accounting for the correlation between them, we performed an eigenvalue decomposition of the correlation matrix (Supplementary Material, Table S2) of the phenotypes. The variance of the eigenvalues [Var (λobs) = 2.36] was used to estimate the effective number of independent phenotypes tested (43). Our findings indicate that studying 10-related ST-T-wave traits is equivalent to analysis of 8 independent phenotypes. We, therefore, adopt αa = 5 × 10−8/8 = 6.25 × 10−9 as threshold for declaring genome-wide significance in order to correct for the effective number of independent phenotypes studied.
Residuals of ST-T-wave amplitudes were calculated using general linear regression models to adjust for age, gender and body mass index and standardized to a mean of zero and a standard deviation of one. GWAS analyses in Prevend and Lifelines of 2 316 136 genotyped or imputed SNPs (Supplementary Material, Table S18) were performed on the standardized residuals using an additive genetic model in PLINK (v.1.07). Phenotypes were normally distributed (Supplementary Material, Fig. S2). Test statistics from each cohort were then corrected for their respective genomic control inflation factor to adjust for residual population sub-structure and meta-analyzed using the inverse-variance method. SNPs with minor allele frequency < 1% (weighted average across cohorts) were removed. Variants were considered to be independent if the pair-wise LD (r2) was <0.1 and if they were separated by at least 1 MB; this was defined a ‘locus’. We selected one sentinel SNP (the most-significantly associated SNP in a locus) for each genotype–phenotype combination.
For conditional analyses, we repeated the primary association analysis for each trait while conditioning on the trait-specific genome-wide significant sentinel SNPs by adding the SNP genotypes as covariates. Association results for each study were again combined by inverse variance weighting.
Replication of the significant genotype–phenotype associations (P < 6.25 × 10−9) and associations that did not exceed this threshold, but were suggestive (6.25 × 10−9 < P <1 × 10−6), was performed in the RS I, II and III, ERF, ARIC, CHS and YFS and combined using fixed-effects meta-analysis by inverse variance weighting (Supplementary Material, Table S18). The pre-specified statistical significance threshold for heterogeneity by Cochran's Q (Phet) was Phet < 0.0007 to account for multiple testing. Inverse variance weighting was used to determine meta-P-values for combined discovery and replication data. An association was considered replicated if the direction of effect was concordant with discovery, replication P < 0.01 and meta-P < 6.25 × 10−9.
Heritability estimates were calculated in the ERF study using the ‘tdist’ function in the SOLAR software, including gender, age and body mass index as covariates.
Data-driven expression prioritized integration for complex traits
DEPICT systematically identifies the most likely causal gene at a given associated locus, tests gene sets for enrichment in associated SNPs, and identifies tissues and cell types in which genes from associated loci are highly expressed [see Pers et al. (24) for a detailed description]. For this work, we ran DEPICT on 140 independently associated loci (association P < 10−5; PLINK parameters, ‘--clump-p1 1e-5 --clump-kb 500 --clump-r2 0.05’) resulting in 103 independent, autosomal DEPICT loci containing 363 genes (loci overlapping with the major histocompatibility complex region are by default excluded in DEPICT). We have extended the locus definition used in DEPICT (LD r2 > 0.5) with 100 kb at either side of the loci, because several genes that may be important for cardiac repolarization were outside the default DEPICT locus boundaries (e.g. SCN5A, TMEM43 and VGLL2). The gene set enrichment results for this slightly extended locus definition were similar to the results based on the default locus definition used in DEPICT (Supplementary Material, Table S19).
Identification of candidate genes
We prioritized candidate genes based on nearby genes: we considered the nearest gene and any other gene located within 10 kb of the sentinel SNP. Coding variants: for identification of coding variants and LD calculations, we used the 1000G Project data set (March 2012 release) in the European populations. We considered genes that harbor non-synonymous SNPs in LD with the ST-T-wave SNPs at r2 > 0.8. GRAIL analyses: we carried out a literature analysis by employing the GRAIL text-mining algorithm, a statistical tool that identifies subsets of genes with known functional interrelationships based on PubMed abstracts. We carried out the analysis using the 2006 data set to avoid confounding by subsequent GWAS discovery. The DEPICT method (see above), and expression QTL (eQTL) analyses in cis, we search for eQTLs (sentinel SNPs or SNPs in LD, r2 > 0.8, HapMap r27) in an eQTL data set that was compiled from the summary statistics of various studies and tissues (see Supplementary Material, Table S11). We only considered eQTLs that were in LD (r2 > 0.8, HapMap r27) with the sentinel SNP and reached a P-value cut-off of at least P < 0.05/(28 loci × 17 eQTL studies).
Imputation using 1000G
Genome positions from the Prevend and Lifelines genotypes were converted from hg18 to hg19 using the UCSC LiftOver tool. Genome-wide genotype imputation was performed with SHAPEIT (v2) and IMPUTE2 (v2.3.0) using the complete 1000 Genomes v3, March 2012 haplotypes Phase I integrated variant set release as reference panel.
Functional information
We overlapped SNPs with data from the ENCODE project (44) and Roadmap Epigenomics Program (45), conservation across mammals, various cardiac transcription factor measured by ChIP-Seq and contiguous annotations of the human fetal heart, left ventricle and right ventricle as determined by ChromHMM (46) (Supplementary Material).
Prioritization of potentially causal variants and enrichment of DNA elements
For the prioritization of genetic variants for the future functional follow-up and insight of the underlying DNA elements that might be relevant for causal ST-T-wave amplitude variants, we employed the PAINTOR framework (27). In short, this method allows us to prioritize genetic variants in each of the 28 significant associated loci by an integrating strength of association and annotations of functional DNA elements to estimate the probability for each variant to be causal, but also investigate which annotations are potentially causal.
Supplementary Material
Funding
PREVEND: PREVEND genetics is supported by the Dutch Kidney Foundation (grant E033), the EU Project grant GENECURE (FP-6 LSHM CT 2006 037697), the National Institutes of Health (grant 2R01LM010098), The Netherlands Organization for Health Research and Development (NWO-Groot grant 175.010.2007.006, NWO VENI grant 916.761.70, ZonMw grant 90.700.441) and the Dutch Inter University Cardiology Institute Netherlands (ICIN). N.V. is supported by the Netherlands Heart Foundation (grant NHS2010B280). LifeLines: The LifeLines Cohort Study, and generation and management of GWAS genotype data for the LifeLines Cohort Study is supported by the Netherlands Organization of Scientific Research NWO (grant 175.010.2007.006), the Economic Structure Enhancing Fund (FES) of the Dutch government, the Ministry of Economic Affairs, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the Northern Netherlands Collaboration of Provinces (SNN), the Province of Groningen, University Medical Center Groningen, the University of Groningen, Dutch Kidney Foundation and Dutch Diabetes Research Foundation. L.F. is supported by the Netherlands Organization for Scientific Research (NWO VENI grant 916.10.135) and a Horizon Breakthrough grant from the Netherlands Genomics Initiative (grant 92519031. The research leading to these results has received funding from the European Community's Health Seventh Framework Programme (FP7/2007-2013) under grant agreement 259867. RS I, II and III: Akzo Nobel, Alzheimer's Association, Astra Pharmaceutial N.V., AstraZeneca, Bayer AG, Blinden-penning Foundation, Amsterdam, Brain Foundation of the Netherlands, Bristol-Myers Squibb, Center of Medical Systems Biology (CMSB), Dutch Diabetes Research Foundation, Dutch Kidney Foundation, Dutch Arthritis Association, Elise Mathilde Foundation, Maarn, Erasmus Medical Center, Erasmus University Rotterdam, European Commission, Foundation for Helping the Blind, The Hague, Foundation for the Ophthalmic Diseased, Rotterdam, Foundation G. Ph. Verhagen, General Electric Healthcare, Glaxo Smith Kline, International Foundation Alzheimer's Research, Inspectorate for Healthcare, Janivo Foundation, K.F. Hein Foundation, Merck Sharp and Dohme, Haarlem, Municipality of City of Rotterdam, National Epilepsy Fund, National Health Fundraising Foundation, National Institute on Aging, NIH, Bethesda, MD, USA, National Society for the Blind and Visually Impaired (LSBS), Netherlands Foundation for the Blind and Visually Handicapped, Netherlands Heart Foundation, Netherlands Institute for Health Sciences (Nihes), Netherlands Ophthalmic Research Institute, Netherlands Organisation for Health Research and Development (ZonMw), Netherlands Organisation for Scientific Research (NWO), Netherlands Society for the Prevention of Blindness, Netherlands Thrombosis Foundation, Novo Nordisk, Numico Research B.V., OOG Foundation, The Hague, N.V. Organon, Oxagen, Optimix Foundation, Amsterdam, Physicotherapeutic Institute, Prinses Beatrix Foundation, Procter and Gamble, Research Institute for Diseases in the Elderly (RIDE), Rotterdam Foundation for Ophthalmic Research, Rotterdam Foundation for the Interests of the Blind, St Laurens Institute, Rotterdam, Topcon Europe B.V., Trust Fund Erasmus University Rotterdam, Unilever, Van Leeuwen Van Lignac Foundation, Rotterdam. ERF: The ERF study was supported by grants from the Netherlands Organization for Scientific Research (NWO; Pioneer grant), Erasmus Medical Center, the Centre for Medical Systems Biology (CMSB) and the Netherlands Kidney Foundation. We are grateful to all patients and their relatives, general practitioners and neurologists for their contributions and to P. Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work and P. Snijders for his help in data collection. ARIC: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402 and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by grant number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. CHS: Cardiovascular Health Study: this CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086 and NHLBI grants HL080295, HL087652, HL105756, HL103612 and HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org/. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124 and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The Young Finns Study has been financially supported by the Academy of Finland: grants 286284 (T.L.), 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi) and 41071 (Skidi); the Social Insurance Institution of Finland; Kuopio, Tampere and Turku University Hospital Medical Funds (grant X51001 for T.L.); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research (T.L.); Finnish Cultural Foundation; Tampere Tuberculosis Foundation (T.L.); Emil Aaltonen Foundation (T.L.) and Yrjö Jahnsson Foundation (T.L.). Funding to pay the Open Access publication charges for this article was provided by the Marie Sklodowska-Curie GF grant (661395) from N. Verweij.
Supplementary Material
Acknowledgements
We thank Behrooz Alizadeh, Annemieke Boesjes, Marcel Bruinenberg, Noortje Festen, Ilja Nolte, Lude Franke, Mitra Valimohammadi for their help in creating the GWAS database, and Rob Bieringa, Joost Keers, René Oostergo, Rosalie Visser, Judith Vonk for their work related to data-collection and validation. The authors are grateful to the study participants, the staff from the LifeLines Cohort Study and Medical Biobank Northern Netherlands, and the participating general practitioners and pharmacists. LifeLines Scientific Protocol Preparation: Rudolf de Boer, Hans Hillege, Melanie van der Klauw, Gerjan Navis, Hans Ormel, Dirkje Postma, Judith Rosmalen, Joris Slaets, Ronald Stolk, Bruce Wolffenbuttel; LifeLines GWAS Working Group: Behrooz Alizadeh, Marike Boezen, Marcel Bruinenberg, Noortje Festen, Lude Franke, Pim van der Harst, Gerjan Navis, Dirkje Postma, Harold Snieder, Cisca Wijmenga and Bruce Wolffenbuttel. The authors acknowledge the services of the LifeLines Cohort Study, the contributing research centres delivering data to LifeLines and all the study participants.
Conflict of Interest statement. None declared.
References
- 1.Arking D.E., Pulit S.L., Crotti L., van der Harst P., Munroe P.B., Koopmann T.T., Sotoodehnia N., Rossin E.J., Morley M., Wang X. et al. (2014) Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet., 46, 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nerbonne J.M., Kass R.S. (2005) Molecular physiology of cardiac repolarization. Physiol. Rev., 85, 1205–1253. [DOI] [PubMed] [Google Scholar]
- 3.Antzelevitch C. (2006) Cellular basis for the repolarization waves of the ECG. Ann. N. Y. Acad. Sci., 1080, 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberger A.L., Mirvis D.M. (2011) In Bonow R.O., Mann D.L., Zipes D.P., Libby P. (eds), Braunwald's Heart Disease: A Textbook of Cardiovascular Medicine. Elsevier Health Sciences, in press, US, Philadelphia, pp. 126–167. [Google Scholar]
- 5.Hasan M.A., Abbott D., Baumert M. (2013) Beat-to-beat QT interval variability and T-wave amplitude in patients with myocardial infarction. Physiol. Meas., 34, 1075–1083. [DOI] [PubMed] [Google Scholar]
- 6.Al-Zaiti S.S., Fallavollita J.A., Canty J.M., Carey M.G. (2015) The prognostic value of discordant T waves in lead aVR: a simple risk marker of sudden cardiac arrest in ischemic cardiomyopathy. J. Electrocardiol., 48, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roten L., Derval N., Maury P., Mahida S., Pascale P., Leenhardt A., Jesel L., Deisenhofer I., Kautzner J., Probst V. et al. (2015) Benign vs malignant inferolateral early repolarization: focus on the T wave. Heart Rhythm, in press, doi:10.1016/j.hrthm.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Anttila I., Nikus K., Nieminen T., Jula A., Salomaa V., Reunanen A., Nieminen M.S., Lehtimaki T., Virtanen V., Kahonen M. (2011) Relation of positive T wave in lead aVR to risk of cardiovascular mortality. Am. J. Cardiol., 108, 1735–1740. [DOI] [PubMed] [Google Scholar]
- 9.Aro A.L., Anttonen O., Tikkanen J.T., Junttila M.J., Kerola T., Rissanen H.A., Reunanen A., Huikuri H.V. (2012) Prevalence and prognostic significance of T-wave inversions in right precordial leads of a 12-lead electrocardiogram in the middle-aged subjects. Circulation, 125, 2572–2577. [DOI] [PubMed] [Google Scholar]
- 10.Rautaharju P.M., Kooperberg C., Larson J.C., LaCroix A. (2006) Electrocardiographic predictors of incident congestive heart failure and all-cause mortality in postmenopausal women: the Women's Health Initiative. Circulation, 113, 481–489. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki T., Myers J., Froelicher V.F. (2005) Prognostic importance of isolated T-wave abnormalities. Am. J. Cardiol., 95, 300–304. [DOI] [PubMed] [Google Scholar]
- 12.Tan S.Y., Engel G., Myers J., Sandri M., Froelicher V.F. (2008) The prognostic value of T wave amplitude in lead aVR in males. Ann. Noninvasive Electrocardiol., 13, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarafshar S., Wong M., Singh N., Aggarwal S., Adhikarla C., Froelicher V.F. (2013) Resting ST amplitude: prognosis and normal values in an ambulatory clinical population. Ann. Noninvasive Electrocardiol., 18, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rautaharju P.M., Prineas R.J., Wood J., Zhang Z.M., Crow R., Heiss G. (2007) Electrocardiographic predictors of new-onset heart failure in men and in women free of coronary heart disease (from the Atherosclerosis in Communities [ARIC] Study). Am. J. Cardiol., 100, 1437–1441. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Timothy K.W., Vincent G.M., Lehmann M.H., Fox J., Giuli L.C., Shen J., Splawski I., Priori S.G., Compton S.J. et al. (2000) Spectrum of ST-T-wave patterns and repolarization parameters in congenital long-QT syndrome: ECG findings identify genotypes. Circulation, 102, 2849–2855. [DOI] [PubMed] [Google Scholar]
- 16.Tikkanen J.T., Anttonen O., Junttila M.J., Aro A.L., Kerola T., Rissanen H.A., Reunanen A., Huikuri H.V. (2009) Long-term outcome associated with early repolarization on electrocardiography. New Engl. J. Med., 361, 2529–2537. [DOI] [PubMed] [Google Scholar]
- 17.Priori S.G., Wilde A.A., Horie M., Cho Y., Behr E.R., Berul C., Blom N., Brugada J., Chiang C.E., Huikuri H. et al. (2013) HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm, 10, 1932–1963. [DOI] [PubMed] [Google Scholar]
- 18.Haissaguerre M., Derval N., Sacher F., Jesel L., Deisenhofer I., de Roy L., Pasquie J.L., Nogami A., Babuty D., Yli-Mayry S. et al. (2008) Sudden cardiac arrest associated with early repolarization. N. Engl. J. Med., 358, 2016–2023. [DOI] [PubMed] [Google Scholar]
- 19.Kolder I.C., Tanck M.W., Bezzina C.R. (2012) Common genetic variation modulating cardiac ECG parameters and susceptibility to sudden cardiac death. J. Mol. Cell. Cardiol., 52, 620–629. [DOI] [PubMed] [Google Scholar]
- 20.Marjamaa A., Oikarinen L., Porthan K., Ripatti S., Peloso G., Noseworthy P.A., Viitasalo M., Nieminen M.S., Toivonen L., Kontula K. et al. (2012) A common variant near the KCNJ2 gene is associated with T-peak to T-end interval. Heart Rhythm, 9, 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Hoed M., Eijgelsheim M., Esko T., Brundel B.J., Peal D.S., Evans D.M., Nolte I.M., Segre A.V., Holm H., Handsaker R.E. et al. (2013) Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet., 45, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong K.W., Shin D.J., Lee S.H., Son N.H., Go M.J., Lim J.E., Shin C., Jang Y., Oh B. (2012) Common variants in RYR1 are associated with left ventricular hypertrophy assessed by electrocardiogram. Eur. Heart J., 33, 1250–1256. [DOI] [PubMed] [Google Scholar]
- 23.Verweij N., Mateo Leach I., van den Boogaard M., van Veldhuisen D.J., Christoffels V.M., Hillege H.L., van Gilst W.H., Barnett P., de Boer R.A., van der Harst P. (2014) Genetic Determinants of P Wave Duration and PR Segment. Circ. Cardiovasc. Genet., 7, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pers T.H., Karjalainen J.M., Chan Y., Westra H.J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T. et al. (2015) Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun., 6, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verweij N., Mateo Leach I., van den Boogaard M., van Veldhuisen D.J., Christoffels V.M., Hillege H.L., van Gilst W.H., Barnett P., de Boer R.A., van der Harst P. (2014) Genetic determinants of P wave duration and PR segment. Circ. Cardiovasc. Genet., 7, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kichaev G., Yang W.Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. (2014) Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet., 10, e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bezzina C.R., Barc J., Mizusawa Y., Remme C.A., Gourraud J.B., Simonet F., Verkerk A.O., Schwartz P.J., Crotti L., Dagradi F. et al. (2013) Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet., 45, 1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giudicessi J.R., Ye D., Kritzberger C.J., Nesterenko V.V., Tester D.J., Antzelevitch C., Ackerman M.J. (2012) Novel mutations in the KCND3-encoded Kv4.3 K+ channel associated with autopsy-negative sudden unexplained death. Hum. Mutat., 33, 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giudicessi J.R., Ye D., Tester D.J., Crotti L., Mugione A., Nesterenko V.V., Albertson R.M., Antzelevitch C., Schwartz P.J., Ackerman M.J. (2011) Transient outward current (I(to)) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Heart Rhythm, 8, 1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu D., Barajas-Martinez H., Pfeiffer R., Dezi F., Pfeiffer J., Buch T., Betzenhauser M.J., Belardinelli L., Kahlig K.M., Rajamani S. et al. (2014) Mutations in SCN10A are responsible for a large fraction of cases of Brugada syndrome. J. Am. Coll. Cardiol., 64, 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinner M.F., Porthan K., Noseworthy P.A., Havulinna A.S., Tikkanen J.T., Muller-Nurasyid M., Peloso G., Ulivi S., Beckmann B.M., Brockhaus A.C. et al. (2012) A meta-analysis of genome-wide association studies of the electrocardiographic early repolarization pattern. Heart Rhythm, 9, 1627–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benito B., Guasch E., Rivard L., Nattel S. (2010) Clinical and mechanistic issues in early repolarization of normal variants and lethal arrhythmia syndromes. J. Am. Coll. Cardiol., 56, 1177–1186. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y., Kuhl S.J., Pfister A.S., Cizelsky W., Denk S., Beer-Molz L., Kuhl M. (2014) Comparative analysis reveals distinct and overlapping functions of Mef2c and Mef2d during cardiogenesis in Xenopus laevis. PLoS One, 9, e87294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebastian S., Faralli H., Yao Z., Rakopoulos P., Palii C., Cao Y., Singh K., Liu Q.C., Chu A., Aziz A. et al. (2013) Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev., 27, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li A., Ahsen O.O., Liu J.J., Du C., McKee M.L., Yang Y., Wasco W., Newton-Cheh C.H., O'Donnell C.J., Fujimoto J.G. et al. (2013) Silencing of the Drosophila ortholog of SOX5 in heart leads to cardiac dysfunction as detected by optical coherence tomography. Hum. Mol. Genet., 22, 3798–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montero J.A., Giron B., Arrechedera H., Cheng Y.C., Scotting P., Chimal-Monroy J., Garcia-Porrero J.A., Hurle J.M. (2002) Expression of Sox8, Sox9 and Sox10 in the developing valves and autonomic nerves of the embryonic heart. Mech. Dev., 118, 199–202. [DOI] [PubMed] [Google Scholar]
- 38.Luo Y., High F.A., Epstein J.A., Radice G.L. (2006) N-cadherin is required for neural crest remodeling of the cardiac outflow tract. Dev. Biol., 299, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sizarov A., Devalla H.D., Anderson R.H., Passier R., Christoffels V.M., Moorman A.F. (2011) Molecular analysis of patterning of conduction tissues in the developing human heart. Circ.. Arrhythmia Electrophysiol., 4, 532–542. [DOI] [PubMed] [Google Scholar]
- 40.Alfieri C.M., Cheek J., Chakraborty S., Yutzey K.E. (2010) Wnt signaling in heart valve development and osteogenic gene induction. Dev. Biol., 338, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu X., Zhang Y., Wang J., Yang J.F., Yang Y.F., Tan Z.P. (2013) 576 kb deletion in 1p36.33-p36.32 containing SKI is associated with limb malformation, congenital heart disease and epilepsy. Gene, 528, 352–355. [DOI] [PubMed] [Google Scholar]
- 42.Tian E., Stevens S.R., Guan Y., Springer D.A., Anderson S.A., Starost M.F., Patel V., Ten Hagen K.G., Tabak L.A. (2015) Galnt1 is required for normal heart valve development and cardiac function. PLoS One, 10, e0115861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nyholt D.R. (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet., 74, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. (2012) The accessible chromatin landscape of the human genome. Nature, 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. et al. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol., 28, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ernst J., Kellis M. (2015) Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol., 33, 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.