Abstract
Spinal and bulbar muscular atrophy (SBMA, also known as Kennedy's disease) is one of nine neurodegenerative disorders that are caused by expansion of polyglutamine-encoding CAG repeats. Intracellular accumulation of abnormal proteins in these diseases, a pathological hallmark, is associated with defects in protein homeostasis. Enhancement of the cellular proteostasis capacity with small molecules has therefore emerged as a promising approach to treatment. Here, we characterize a novel curcumin analog, ASC-JM17, as an activator of central pathways controlling protein folding, degradation and oxidative stress resistance. ASC-JM17 acts on Nrf1, Nrf2 and Hsf1 to increase the expression of proteasome subunits, antioxidant enzymes and molecular chaperones. We show that ASC-JM17 ameliorates toxicity of the mutant androgen receptor (AR) responsible for SBMA in cell, fly and mouse models. Knockdown of the Drosophila Nrf1 and Nrf2 ortholog cap ‘n’ collar isoform-C, but not Hsf1, blocks the protective effect of ASC-JM17 on mutant AR-induced eye degeneration in flies. Our observations indicate that activation of the Nrf1/Nrf2 pathway is a viable option for pharmacological intervention in SBMA and potentially other polyglutamine diseases.
Introduction
The expansion of CAG trinucleotide repeats encoding polyglutamine tracts in unrelated proteins causes nine neurodegenerative disorders, including spinal and bulbar muscular atrophy (SBMA, also known as Kennedy's disease), Huntington's disease and six spinocerebellar ataxias (1). At present, effective disease-modifying treatment is not available for this family of diseases. In SBMA, a polyglutamine expansion in the androgen receptor (AR) leads to lower motor neuron degeneration and muscle weakness (2). The mutation in AR interferes with a variety of cellular processes, including transcription (3), axonal transport (4), mitochondrial function (5) and splicing (6).
Misfolding and abnormal accumulation of the mutant protein, a hallmark of polyglutamine diseases, reflects imbalances in the cellular network that governs protein synthesis, folding, transport and degradation. This network is controlled by multiple signaling pathways to minimize damage to macromolecules and organelles in response to extrinsic and intrinsic stressors (7). The heat shock and antioxidant responses are critical components of the cellular stress mechanism and influence protein homeostasis (proteostasis) primarily through the regulation of gene expression. The heat shock response is mediated by heat shock factor 1 (Hsf1), which increases the expression of molecular chaperones that assist protein folding (8). Several Hsf1 targets, such as Hsp70, Hsp40 and Hsp27, are modifiers of polyglutamine toxicity in disease models (9–12). The antioxidant response is a cellular defense mechanism that protects against the damaging consequences of oxidative stress through transcriptional control of antioxidant enzymes by Nrf2, also referred to as nuclear factor (erythroid-derived 2)-like 2 (NFE2L2), which belongs to the cap ‘n’ collar family of basic leucine zipper transcription factors (13). Recently, the antioxidant pathway has been shown to play a role in the regulation of components of the ubiquitin–proteasome system (UPS) through a related factor, Nrf1/NFE2L1 (14,15). The UPS is a major system for the clearance of short-lived and abnormal proteins in both the nucleus and the cytoplasm. Protein degradation by the UPS involves the covalent attachment of ubiquitin to specific targets by a series of enzymatic reactions, followed by proteolysis of the ubiquitylated substrates by the proteasome, a multi-subunit complex consisting of a 20S proteolytic core and 19S regulatory particles (16). The proteasome is the final destination of UPS substrates and can be rate-limiting under proteotoxic stress. It is not settled whether proteasomes are able to efficiently degrade proteins containing long polyglutamine tracts (17,18). However, substantial evidence in animal models suggests that increasing UPS-mediated clearance of the mutant AR has a protective outcome (19–21).
Modulation of the proteostasis network with small molecules is a desirable goal for treating polyglutamine diseases, because it offers the opportunity both to decrease levels of the mutant proteins and to alleviate their pathogenic effects. One candidate for proteostasis regulation is curcumin, a naturally occurring polyphenol with pleiotropic biological properties, including antioxidant, anti-inflammatory and neuroprotective activities (22,23). It is currently under investigation for a variety of disorders, including Alzheimer's disease (24); however, low potency and bioavailability limit its efficacy in clinical applications. A structural analog, dimethylcurcumin (ASC-J9), was evaluated in SBMA based on its ability to inhibit ligand-dependent activity of the AR in cultured cells (25), which is required for full disease manifestation (26,27). ASC-J9 was shown to reduce accumulation of polyglutamine-expanded AR and attenuate the SBMA phenotype in mice (28). However, a replication study of ASC-J9 with randomization and administration after disease onset did not show a robust effect (unpublished data).
To improve the delivery and optimize the efficacy of this class of compounds, we tested a new, orally bioavailable curcumin analog, ASC-JM17 [(1E,4Z,6E)-4-(cyclobutylmethyl)-1,7-bis(3,4-dimethoxyphenyl)-5-hydroxyhepta-1,4,6-trien-3-one], in cell and animal models of SBMA and investigated its mechanism of action. We found that ASC-JM17 activates Nrf1, Nrf2 and Hsf1, thereby enhancing the capacity of cellular proteostasis. The protective effect of ASC-JM17 in a fly model of SBMA is dependent on the Drosophila ortholog of Nrf1 and Nrf2, cap ‘n’ collar isoform-C (CncC). Our findings highlight a key role for this pathway in counteracting mutant AR toxicity, and establish ASC-JM17 as a potential treatment for SBMA and other polyglutamine diseases.
Results
ASC-JM17 promotes UPS-mediated degradation of the mutant AR
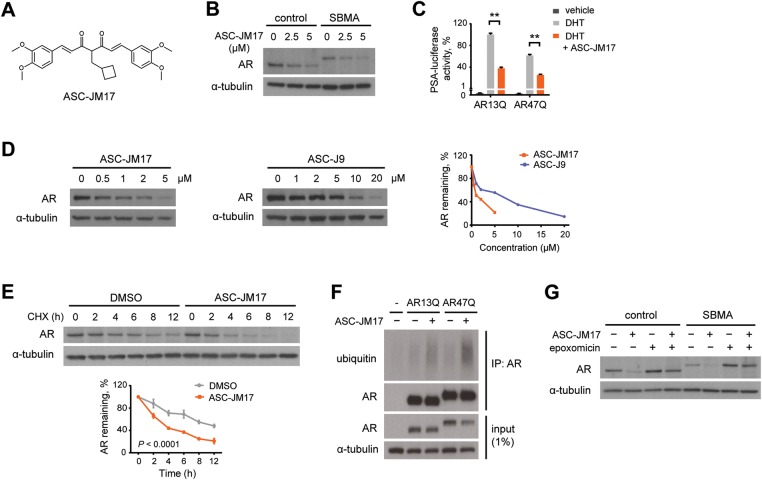
The synthetic curcumin analog ASC-JM17 (Fig. 1A) is structurally related to ASC-J9 (28). We found that ASC-JM17 has similar physicochemical properties and toxicity, but greater metabolic stability and exposure with oral administration in mice compared with ASC-J9.
Figure 1.
ASC-JM17 reduces AR protein through enhanced degradation by the UPS. (A) Chemical structure of ASC-JM17. (B) Effect of ASC-JM17 on AR protein levels. Fibroblasts derived from a healthy control and an SBMA patient with 68 CAG repeats were treated with 10 nm DHT and indicated concentrations of ASC-JM17, and analyzed by western blotting. (C) Effect of ASC-JM17 on AR transcriptional activity. PC12 cells expressing AR with wild-type (AR13Q) or expanded (AR47Q) polyglutamine tracts were incubated with ethanol vehicle or DHT (10 nm), with and without 5 µm ASC-JM17. PSA-luciferase activity was measured using the dual luciferase assay (n = 3). Data are expressed as mean ± SEM. **P < 0.01 (two-way ANOVA). (D) Patient fibroblasts were treated with indicated concentrations of ASC-JM17 or ASC-J9 and analyzed by western blotting. AR band intensities were quantified and normalized to α-tubulin. (E) Patient fibroblasts were treated with 10 µm cycloheximide (CHX), with or without 5 µm ASC-JM17 and harvested at indicated time points. The graph shows quantification of AR band intensities normalized to α-tubulin (n = 3). Data are expressed as mean ± SEM (two-way ANOVA). (F) PC12 cells transfected with AR13Q, AR47Q or empty vector were treated with 5 µm ASC-JM17 or DMSO vehicle. AR was immunoprecipitated from whole-cell extracts and its ubiquitylation status was assessed using the ubiquitin antibody FK2. (G) Fibroblasts were incubated with 5 µm ASC-JM17 in the presence or absence of 100 nm epoxomicin.
We investigated the effect of ASC-JM17 on endogenous AR protein levels in fibroblasts obtained from a healthy control and from an SBMA patient with a 68 CAG repeat (29). Treatment with the compound reduced steady-state levels of both normal and polyglutamine-expanded AR variants in the presence of dihydrotestosterone (DHT; Fig. 1B). This decrease in protein level was accompanied by reduced transcriptional activity of the AR in a luciferase-based transactivation assay (Fig. 1C). ASC-JM17 also reduced AR levels in the absence of ligand (Supplementary Material, Fig. S1). We found ASC-JM17 to be effective in reducing mutant AR protein at lower concentrations than ASC-J9 (Fig. 1D). To address whether the ASC-JM17-induced decrease in AR is due to increased degradation, we performed turnover experiments in the patient fibroblasts using cycloheximide, which blocks new protein synthesis. We found that ASC-JM17 significantly reduces the half-life of polyglutamine-expanded AR compared with dimethyl sulfoxide (DMSO) vehicle (Fig. 1E). ASC-JM17 induced ubiquitylation (Fig. 1F) and promoted proteasome-mediated clearance of the wild-type and mutant AR, which could be blocked by the specific proteasome inhibitor epoxomicin (Fig. 1G). Our observations show that ASC-JM17 accelerates the clearance of the AR through the UPS.
ASC-JM17 activates the antioxidant response and increases proteasome activity through Nrf1
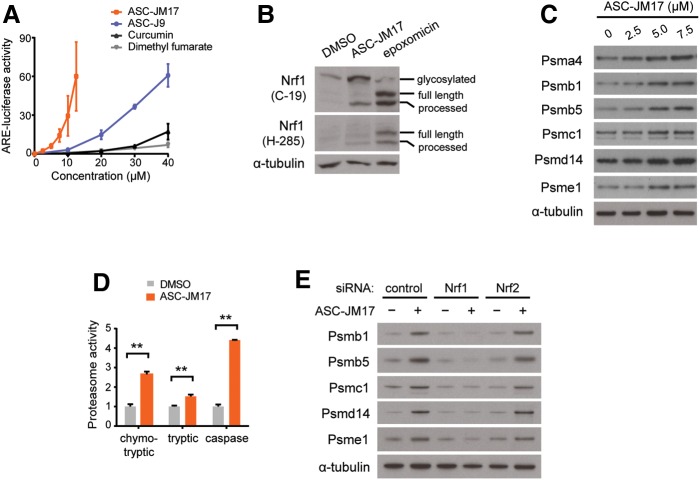
We examined whether ASC-JM17 elicits the antioxidant response in cells. For this, we used a reporter assay with a construct that contains four copies of an antioxidant response element (ARE) upstream of the firefly luciferase open reading frame. We found that ASC-JM17 increases the activity of this reporter, indicating that the compound indeed induces the antioxidant pathway (Fig. 2A). ASC-JM17 increased luciferase activity at considerably lower concentrations than ASC-J9 and other known activators of the antioxidant pathway, curcumin and dimethyl fumarate, indicating that ASC-JM17 is more potent than the other agents tested in activating the antioxidant response.
Figure 2.
ASC-JM17 increases levels of active proteasomes through Nrf1. (A) Effect of ASC-JM17 on the antioxidant response. PC12 cells transfected with ARE-luciferase were treated with increasing concentrations of ASC-JM17, ASC-J9, curcumin or dimethyl fumarate, and analyzed using the dual luciferase assay. Data are expressed as mean ± SEM (n = 3). (B) PC12 cells were treated with ASC-JM17 or epoxomicin and analyzed by western blotting using Nrf1 antibody clones C-19 and H-285. (C) Expression of proteasome subunits in PC12 cells following treatment with indicated concentrations of ASC-JM17. (D) Proteasome activity was measured in PC12 cells after incubation with ASC-JM17. Data are expressed as mean ± SEM (n = 3). **P < 0.01 (Student's t-test). (E) Expression of proteasome subunits in MCF7 cells following transfection with indicated siRNAs and incubation with ASC-JM17.
The ARE-luciferase reporter can be activated by both Nrf1 and Nrf2 (30). We first examined Nrf1, which mediates a feedback mechanism to increase the synthesis of new proteasome complexes in response to proteasome inhibition (14). Nrf1 is synthesized as an endoplasmic reticulum-associated form and rapidly degraded under basal conditions. Upon activation, full-length Nrf1 is cleaved and releases a smaller fragment that translocates into the nucleus to regulate gene expression (31). To test whether ASC-JM17 activates Nrf1, we used two well-characterized antibodies that recognize different epitopes in Nrf1. C-19 recognizes an epitope in the carboxy-terminus and detects the full-length protein as well as a smaller, 75-kDa fragment corresponding to the cleaved form, whereas H-285 recognizes an epitope located in the central part of the protein and preferentially detects cleaved Nrf1 in proteasome inhibitor-treated cells (15). Using these antibodies, we detected cleaved, active Nrf1 in extracts from cells following proteasome inhibition with epoxomicin or ASC-JM17 treatment (Fig. 2B). In addition, we found that ASC-JM17 increases the expression of proteasome components corresponding to the 20S core (Psma4, Psmb1 and Psmb5) as well as the 19S regulatory particle (Psmc1 and Psmd14; Fig. 2C and Supplementary Material, Fig. S2). ASC-JM17 also increased the expression of Psme1, a constituent of the 11S regulatory cap which forms part of the immunoproteasome, suggesting that the compound may also induce structural rearrangements of proteasome complexes (Fig. 2C and Supplementary Material, Fig. S2). To assess whether the increase in proteasome subunits reflects an increase in proteolytic function, we measured the catalytic activity of the proteasome in extracts from cells treated with ASC-JM17. The chymotrypsin-like, trypsin-like and caspase-like activities were significantly increased following incubation with ASC-JM17 (Fig. 2D). We found that the treatment of cells with ASC-J9 or curcumin also leads to an increase in the chymotrypsin-like proteasome activity, but at much higher concentrations and to a lesser extent (Supplementary Material, Fig. S3). ASC-JM17-dependent induction of proteasome subunits was abolished by RNAi-mediated suppression of Nrf1, but not of Nrf2 (Fig. 2E and Supplementary Material, Fig. S4). Our findings show that ASC-JM17 increases proteasome levels and activity through Nrf1.
ASC-JM17 increases the expression of antioxidant enzymes through Nrf2
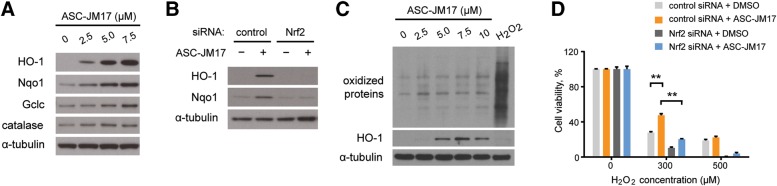
Having established that ASC-JM17 activates Nrf1, we asked whether it also activates Nrf2, which governs the expression of antioxidant enzymes controlling cellular redox potential and stress resistance. Under basal conditions, Nrf2 activity is antagonized by Keap1, which mediates its ubiquitin-dependent degradation in the cytoplasm. Oxidative or electrophilic stress leads to Keap1 inactivation, and stabilization and nuclear translocation of Nrf2 (13). We found that ASC-JM17 treatment increases expression of the antioxidant enzymes HO-1, Nqo1, Gclc and catalase in cultured cells (Fig. 3A and Supplementary Material, Fig. S5). Knockdown of Nrf2 prevented the induction of HO-1 and Nqo1 in response to ASC-JM17 (Fig. 3B). To test whether Nrf2 activation is a consequence of oxidative stress induction by ASC-JM17, we analyzed the oxidation status of cellular proteins after treatment with this compound or hydrogen peroxide. While hydrogen peroxide caused global oxidative damage but failed to induce HO-1 expression, ASC-JM17 did not affect the oxidation status of cellular proteins (Fig. 3C), suggesting that Nrf2 activation is not a downstream consequence of oxidative stress.
Figure 3.
ASC-JM17 increases the expression of antioxidant enzymes via Nrf2. (A) Expression of antioxidant enzymes in PC12 cells following treatment with indicated concentrations of ASC-JM17. (B) PC12 cells were transfected with indicated siRNAs, treated with ASC-JM17 and analyzed by western blotting. (C) PC12 cells were treated with indicated concentrations of ASC-JM17 for 16 h, or with 1 mm H2O2 for 30 min, which induces oxidative stress in cells. The oxidation status of total cellular proteins was visualized by western blotting after derivatization with 2,4-dinitrophenyl hydrazine. HO-1 protein levels were also analyzed. (D) MCF7 cells transfected with indicated siRNAs were pretreated with 2.5 µm ASC-JM17 for 16 h, followed by indicated concentrations of H2O2 for 4 h. Cell viability was assessed using the XTT assay. Data are expressed as mean ± SEM (n = 3). **P < 0.01 (one-way ANOVA).
To investigate the significance of Nrf2 activation by ASC-JM17, we exposed cells to 300 or 500 µm hydrogen peroxide following pretreatment with ASC-JM17 or DMSO vehicle. We found that the protective effect of ASC-JM17 against the moderate oxidative stress obtained with 300 µm hydrogen peroxide was significantly reduced after siRNA-mediated knockdown of Nrf2 (Fig. 3D). Therefore, Nrf2 activation by ASC-JM17 increases cellular defense mechanisms to counteract oxidative damage.
ASC-JM17 induces the heat shock response through Hsf1
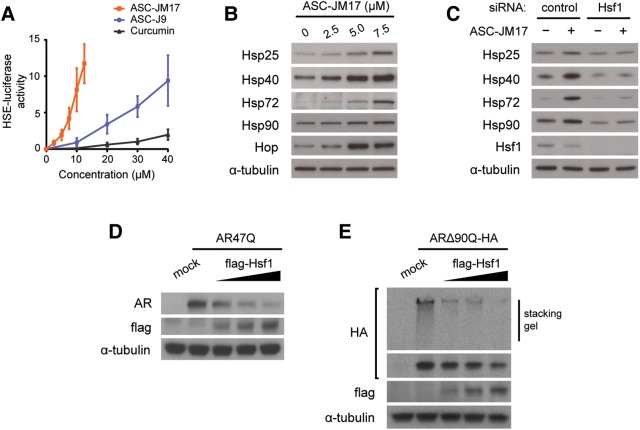
We investigated whether ASC-JM17 activates the heat shock response, which facilitates proteasomal targeting of the mutant AR (20,21). We found that ASC-JM17 induces the activity of a luciferase reporter driven by heat shock element (HSE; Fig. 4A), and augments heat shock protein and co-chaperone expression in cell culture (Fig. 4B). RNAi-mediated silencing of Hsf1 abolished the induction of heat shock proteins by ASC-JM17 (Fig. 4C).
Figure 4.
ASC-JM17 activates the heat shock response via Hsf1, which influences mutant AR protein levels and aggregation. (A) Effect of ASC-JM17 on the heat shock response. PC12 cells transfected with HSE-luciferase were treated with increasing concentrations of ASC-JM17, ASC-J9 or curcumin, and analyzed using the dual luciferase assay. Data are expressed as mean ± SEM (n = 3). (B) Expression of heat shock proteins in PC12 cells following treatment with indicated concentrations of ASC-JM17. (C) MCF7 cells were transfected with indicated siRNAs, treated with ASC-JM17 and analyzed by western blotting. (D and E) Effect of Hsf1 overexpression on protein levels (D) and aggregation (E) of polyglutamine-expanded AR. PC12 cells were transfected with AR47Q or an HA-tagged amino-terminal fragment of the mutant AR (ARΔ90Q-HA) and increasing concentrations of flag-Hsf1.
This led us to investigate whether Hsf1 influences levels of the polyglutamine-expanded AR in cultured cells. We found that Hsf1 overexpression decreases steady-state levels of full-length mutant AR in the presence of DHT (Fig. 4D), and reduces aggregation of an amino-terminal AR fragment with a 90-residue polyglutamine tract (Fig. 4E). Our results support a model, in which ASC-JM17-dependent activation of Hsf1 promotes AR degradation through transcriptional induction of heat shock proteins.
Suppression of polyglutamine toxicity in Drosophila by ASC-JM17 requires CncC
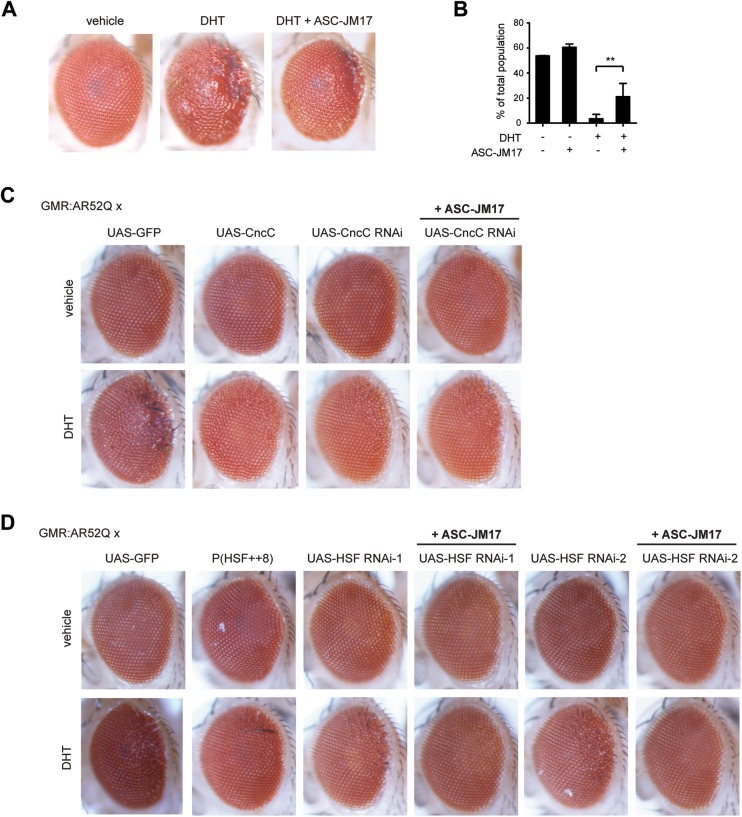
We explored the effect of ASC-JM17 in a Drosophila melanogaster model of SBMA, which recapitulates the DHT-dependent toxicity of the disease (27). The flies express full-length human AR with an expanded polyglutamine tract (AR52Q) under the control of a UAS element and a GMR-GAL4 driver, which targets transgene expression to the eye. Flies reared in the presence of DHT develop a degenerative eye phenotype, which is rescued by ASC-JM17 (Fig. 5A).
Figure 5.
ASC-JM17 rescues mutant AR toxicity in Drosophila. (A) Eye phenotypes of adult GMR:UAS-AR52II flies following treatment with vehicle, DHT or DHT and ASC-JM17. (B) Viability of Elav > AR52Q flies reared in the presence or absence of DHT and ASC-JM17 was scored over the course of 10 days. **P < 0.01 (χ2 test). (C) Effect of CncC overexpression or knockdown on the DHT-dependent eye phenotype in GMR:UAS-AR52II flies. (D) Effect of HSF overexpression or knockdown in GMR:UAS-AR52II flies. Two independent HSF RNAi lines were evaluated.
We also investigated the effect of ASC-JM17 on DHT-induced lethality in adult flies with pan-neuronal expression of AR52Q driven by Elav-GAL4. To exclude the effect of impaired fertility, we used an internal balancer control of the second chromosome (Cyo-GFP) as a phenotypic marker. The addition of DHT reduced the number of AR52Q flies from an expected frequency of ∼50 to ∼4% of the total population, demonstrating the ligand-dependent toxicity of AR52Q expression in neuronal tissues. The population frequency of AR52Q flies was significantly augmented by ASC-JM17 treatment (Fig. 5B).
Next, we exploited the Drosophila model to decipher the individual contributions of the antioxidant pathway and the heat shock response to the protective effect of ASC-JM17. To this end, we investigated whether a genetic interaction exists between CncC, the Drosophila ortholog of Nrf1 and Nrf2 (32) and mutant AR toxicity in the fly model. For this, we crossed GMR:UAS-AR52Q flies with CncC overexpression or RNAi lines. CncC overexpression considerably reduced DHT-induced eye degeneration, whereas the RNAi lines retained degeneration, indicating that elevated levels of CncC can impose a protective effect against mutant AR toxicity. ASC-JM17 treatment did not rescue the eye phenotype in the CncC RNAi line (Fig. 5C), suggesting that the antioxidant pathway is required for the protective effect of the compound. We also tested for a genetic interaction between the Drosophila ortholog of Hsf1, HSF and AR-induced eye degeneration. Similar to CncC, HSF overexpression reduced the DHT-dependent eye phenotype. Interestingly, ASC-JM17 treatment completely restored the eye phenotype to that of vehicle-treated control flies in the two independent HSF RNAi lines tested (Fig. 5D). While our findings emphasize the potential of both CncC and HSF as modifiers of polyglutamine toxicity in the fly model, they identify the Nrf1- and Nrf2-dependent antioxidant pathways, but not Hsf1 as a critical mediator of ASC-JM17 effects.
ASC-JM17 ameliorates the disease phenotype in AR97Q mice
We tested the efficacy of ASC-JM17 in a mouse model of SBMA that carries a transgene consisting of the polyglutamine-expanded, full-length human AR (AR97Q). Male AR97Q mice typically develop disease manifestations such as weight loss, muscle atrophy and weakness, at ∼8 weeks of age (26). Similar to other SBMA mouse models, these transgenic animals have neuronal and muscle pathology (19,26).
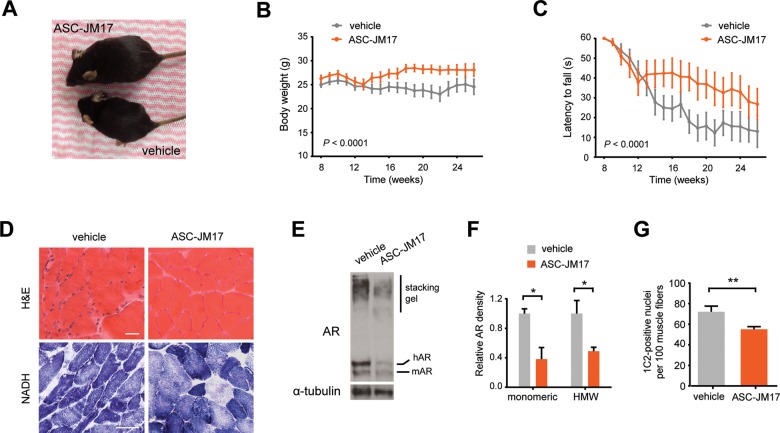
A randomized cohort of male AR97Q mice was treated with either vehicle or ASC-JM17 by gavage daily for 16 weeks starting at 10 weeks of age, after the disease onset. In contrast to previous preclinical interventional studies of SBMA, where treatment was started well before the onset of weakness, we opted to start after the onset of disease manifestations, so that the findings would be more relevant to the treatment of symptomatic SBMA patients. With this experimental design, we did not detect an overall effect of the compound on survival (Supplementary Material, Fig. S6A), but we found that ASC-JM17 administration significantly rescued weight loss (Fig. 6A and B) and improved motor function (Fig. 6C) in symptomatic animals compared with vehicle control. Post hoc analysis excluding mice that were most severely affected at treatment onset (>5% weight loss) showed that ASC-JM17 increased the median survival by ∼10 weeks (Supplementary Material, Fig. S6B). Histopathological examination of skeletal muscle cross-sections following hematoxylin and eosin (H&E) or nicotinamide adenine dinucleotide (NADH) staining revealed that ASC-JM17 markedly ameliorates the neurogenic and myogenic features of muscle atrophy in AR97Q mice (Fig. 6D). We also found that the compound reduces levels and accumulation of the mutant AR protein in mouse tissues (Fig. 6E–G).
Figure 6.
ASC-JM17 ameliorates the disease phenotype in AR97Q mice and reduces mutant AR accumulation in vivo. (A) Picture of two representative AR97Q mice from the same litter treated with 120 mg/kg of ASC-JM17 or vehicle for 6 weeks. (B and C) Body weight (B) and hang time (C) of AR97Q mice treated with ASC-JM17 or vehicle daily from 10 to 26 weeks of age (n = 15 per group). Data are expressed as mean ± SEM (two-way ANOVA). (D) Representative cross-sections of quadriceps muscle from AR97Q mice following treatment with vehicle or ASC-JM17 for 6 weeks. Sections were processed with H&E or NADH staining. Scale bar: 50 μm. (E) AR protein levels in quadriceps muscle of AR97Q mice. (F) Quantification of the monomeric and high molecular weight (HMW) AR signal shown in (E) (n = 3). Data are expressed as mean ± SEM. *P < 0.05 (Student's t-test). (G) Quantification of 1C2-positive nuclei in quadriceps muscle of AR97Q mice (n = 3 per group). Data are expressed as mean ± SEM. **P < 0.01 (Student's t-test).
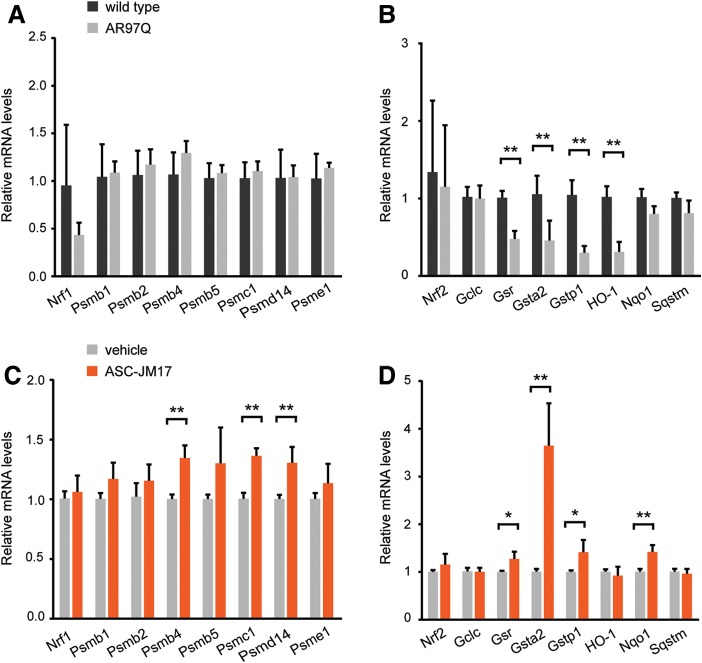
Next, we explored whether the presence of the mutant AR affects expression profiles of Nrf1 and Nrf2 target genes in mouse tissues. While Nrf1 targets were unchanged in quadriceps muscle at 16 weeks of age, several Nrf2 target genes (Gsr, Gsta2, Gstp1 and HO-1) tested were significantly reduced in AR97Q mice compared with non-transgenic littermates (Fig. 7A and B), suggesting that the cellular redox balance may be compromised in the context of mutant AR expression. The treatment of AR97Q mice with ASC-JM17 for 6 weeks significantly elevated the expression of a number of genes downstream of Nrf1 (Psmb4, Psmc1 and Psmd14) and Nrf2 (Gsr, Gsta2 and Nqo1) in AR97Q mice compared with vehicle-treated animals at the 16-week time point (Fig. 7C and D). This indicates that oral administration of ASC-JM17 leads to activation of the antioxidant response in vivo.
Figure 7.
ASC-JM17 increases the expression of Nrf1 and Nrf2 target genes in AR97Q mice. (A and B) Gene expression analysis of Nrf1 (A) and Nrf2 (B) targets in AR97Q mice and non-transgenic littermates (n = 4 per group). Total RNA was extracted from quadriceps muscle of 16-week-old mice. All transcript levels were normalized to Gusb. Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01 (Student's t-test). (C and D) Gene expression analysis of Nrf1 (C) and Nrf2 (D) and downstream targets in AR97Q mice treated with ASC-JM17 or vehicle for 6 weeks.
Discussion
In this study, we demonstrate that the small molecule ASC-JM17 counteracts polyglutamine toxicity in cell and animal models of SBMA by inducing the antioxidant and heat shock responses, and thus targets multiple pathogenic processes associated with the polyglutamine expansion in cells, including aggregation, protein accumulation and oxidative damage. We propose that enhancement of mutant AR degradation by curcumin analogs, previously attributed to selective effects on AR co-factor interaction (28), is linked to modulation of proteostasis.
The function and stability of the AR, a heat shock protein client, are under the control of the Hsp90/Hsp70-based chaperone machinery (33). Activation of Hsp70 chaperone function, or inhibition of Hsp90, targets the mutant AR for proteasome-mediated degradation and reduces its toxicity in cell and animal models (10,21,34). The beneficial effects of Hsp90 inhibitors in polyglutamine disease models are at least in part mediated through induction of the heat shock response, which increases the expression of molecular chaperones (21,35). Pharmacological induction of chaperones alleviates the phenotype in SBMA mouse models (36,37). In line with this, it has been shown that Hsf1 levels influence the extent and distribution of mutant AR accumulation in SBMA mice (38). We found that overexpression of Hsf1 reduces aggregation of the mutant AR and suppresses its toxicity in Drosophila. Not all studies are in agreement with a protective role for Hsf1 activation in counteracting polyglutamine accumulation (39). However, our findings confirm previous reports, which identified Hsf1 as a modifier of polyglutamine toxicity. Nevertheless, it appears that induction of the heat shock response is not necessary for ASC-JM17-mediated mitigation of the SBMA phenotype; instead, we found that activation of the antioxidant response is required for the protective effect of ASC-JM17 in the fly model. Constitutive activation of this pathway, achieved through overexpression of CncC, prevented AR-induced eye degeneration, whereas knockdown of this gene, but not Hsf1, blocked the beneficial effect of ASC-JM17. Upregulation of the CncC-mediated antioxidant response has also been shown to ameliorate α-synuclein toxicity in a fly model of Parkinson's disease (40), indicating that chemical activators of this pathway may be relevant for the treatment of not only SBMA but also other neurodegenerative disorders.
In mammalian cells, the antioxidant response is mediated mainly by Nrf1 and Nrf2, which are both activated by ASC-JM17 treatment. Despite overlapping specificities in binding AREs, the functions of these transcription factors appear to be non-redundant, as mice with the corresponding genetic deletions show distinct phenotypes. While Nrf1 knockout results in embryonic lethality at mid-gestation (41,42), Nrf2-deficient mice are viable but develop neurodegeneration (43,44).
Nrf1 is a promising target for therapeutic intervention in neurodegenerative diseases due to its ability to enhance proteasome activity (14). It has been shown that polyglutamine aggregation and toxicity are reduced in Caenorhabditis elegans that overexpress the proteasome subunit Rpn6/Psmd11 (45). Moreover, a small-molecule inhibitor of Usp14, a proteasome-associated deubiquitylating enzyme, leads to enhancement of proteasome activity and accelerates the clearance of polyglutamine-expanded ataxin-3 as well as several other disease-linked proteins (46). Nrf1 is activated by proteasome inhibitors and stimulates the expression of UPS components as a result of feedback regulation (14). Although the mechanism underlying activation and proteolytic cleavage of Nrf1 is not fully understood, our findings demonstrate that this process can be triggered in the absence of proteasome inhibition.
Nrf2 controls the expression of antioxidant and detoxification enzymes and thereby protects cells from the damaging consequences of oxidative stress. The potential of this pathway in counteracting neurodegeneration is supported by the finding that overexpression of Nrf2 decreases neuronal loss in a mouse model of amyotrophic lateral sclerosis (47). Several reports have indicated impairment of the cellular response to oxidative stress in polyglutamine diseases (5,48), and hence Nrf2 activators, which belong to diverse classes of chemical compounds, are promising agents for treating this family of disorders. For example, sulforaphane reduces polyglutamine toxicity in cultured cells (49), and dimethyl fumarate, used clinically in the treatment of multiple sclerosis, ameliorates disease manifestations in mouse models of Huntington's disease (50). While our study does not yet allow dissection of the individual contributions of Nrf1 and Nrf2, it warrants further investigation into the antioxidant pathway in SBMA and other polyglutamine diseases, and points to a new candidate for therapeutic intervention.
Materials and Methods
Chemicals
ASC-JM17, ASC-J9 and curcumin were provided by AndroScience Corporation. For cell culture and Drosophila experiments, the compounds were dissolved in DMSO and kept at −20°C for long-term storage. For the mouse study, ASC-JM17 was prepared as a liquid formulation at 20 mg/g in corn oil. Briefly, N,N-dimethylacetamide (Sigma) was added to ASC-JM17 and heated to 55°C until dissolved, then d-α-tocopherol polyethylene glycol 1000 succinate (Isochem), Cremophor RH 40 (BASF) and corn oil (Biomedicals) were added sequentially. The preparation was heated to 42°C for 30 min, vortexed and stored in glass vials in the dark for up to 1 week. DHT, cycloheximide and dimethyl fumarate were from Sigma-Aldrich, epoxomicin from Enzo Life Sciences and hydrogen peroxide from EMD Millipore.
Cell culture
Human fibroblasts derived from an SBMA patient and unaffected control have been described (29); other cell lines were obtained from ATCC. Cell culture reagents were obtained from Life Technologies unless indicated otherwise. Fibroblasts and MCF7 cells were maintained in DMEM containing 10% fetal bovine serum in a humidified chamber at 37°C and 5% CO2. PC12 cells were cultured in DMEM supplemented with 10% horse serum and 5% fetal bovine serum. Transfection of plasmid DNA or siRNAs was performed using Lipofectamine 2000 according to the manufacturer's instructions. Pools of siRNAs targeting human Hsf1 (sc-35611), Nrf1 (sc-43575) and Nrf2 (sc-37030), as well as negative control siRNA (sc-37007), were obtained from Santa Cruz Biotechnology. For viability measurements, cells were analyzed with the Cell Proliferation Kit II (Roche Diagnostics) according to the manufacturer's instructions.
Protein analysis
For western analysis of whole-cell extracts, cultured cells were lysed directly in Laemmli sample buffer and processed for polyacrylamide gel electrophoresis. For detection of ubiquitylated AR, PC12 cells were transfected with pEF-AR13Q or -AR47Q and incubated with and without ASC-JM17 in the presence of 10 nm DHT for 16 h. Following treatment with 100 nm epoxomicin, the cells were lysed in RIPA buffer (150 mm NaCl, 50 mm Tris, 2 mM EDTA, 1% sodium deoxycholate, 0.5% Triton X-100, 0.1% sodium dodecyl sulfate and protease inhibitors) and AR was immunoprecipitated using a polyclonal rabbit antibody (sc-816, Santa Cruz Biotechnology) and protein G-coupled dynabeads (Life Technologies). For assessment of protein oxidation, cells were pelleted in phosphate-buffered saline and resuspended in 300 mm NaCl, 50 mm Tris–HCl pH 7.5, 0.5% Triton X-100, 50 mm DTT and protease inhibitor cocktail (Roche). Derivatization of carbonyl groups with 2,4-dinitrophenyl hydrazine was performed using the OxyBlot kit (EMD Millipore). For detection of AR in mouse tissues, quadriceps muscle was dissected, snap-frozen in liquid nitrogen and processed in RIPA buffer on ice using a polytron homogenizer. Tissue homogenates were sonicated, pre-cleared (4000 g for 10 min at 4°C) and total protein concentration was determined using the Bradford reagent (Biorad). Samples were separated on 8% or 4–12% polyacrylamide gels, transferred to PVDF membranes (all Life Technologies) and probed with relevant primary antibodies. Western blots were visualized with peroxidase-linked secondary antibodies (R&D Systems) and chemiluminescence reagent (Perkin Elmer). For quantification of western blot data, band densities were measured using the ImageJ software (NIH). The following antibodies were used: AR clones N-20 (sc-816) and H-280 (sc-13062), Hsf1 (sc-13516), Nrf1 clones C-19 (sc-721) and H-285 (sc-13031), and Nrf2 (sc-722) were from Santa Cruz Biotechnology; flag (2368), Psmb5 (11903) and Psmd14 (4197) from Cell Signaling Technology; Nqo1 (ab34173), Gclc (ab41463), catalase (ab16731), Psma4 (ab119419) and Psmc1 (ab140450) from Abcam; Psmb1 (A303-873A) from Bethyl Laboratories; ubiquitin FK2 (BML-PW8810), Hsp25 (ADI-SPA-801), Hsp72 (ADI-SPA-810), Hsp90 (ADI-SPA-830), Hop (ADI-SRA-1500), HO-1 (ADI-OSA-110) and Psme1 (BML-PW8185) from Enzo Life Sciences; α-tubulin (T6199) from Sigma-Aldrich.
Luciferase assays
A construct consisting of firefly luciferase under the control of a minimal promoter with androgen-response elements from prostate-specific antigen, PSA-luc (from Amilcar Flores-Morales), was used for AR transactivation experiments and transfected along with expression plasmids encoding AR13Q or AR47Q, or empty vector. Activation of the heat shock and antioxidant responses was quantified by means of the reporter constructs pGL4.37 and pGL4.41 (Promega), respectively. For all reporter assays, firefly luciferase constructs were co-transfected with a plasmid encoding constitutively expressed renilla luciferase (pGL4.74) at a ratio of 10 : 1, and luciferase measurements were performed using the Dual luciferase assay kit (both Promega). Chymotryptic, tryptic and caspase-like activities of the proteasome were quantified using the cell-based Proteasome-Glo assay (Promega) according to the manufacturer's instructions and a Victor3 1420-050 plate reader (Perkin Elmer).
Fly stocks and phenotypic characterization
Drosophila were reared on food containing 1 mm DHT or 1% ethanol vehicle, with or without 50 nm ASC-JM17. For the viability assay, virgin female homozygous Elav > Gal4X flies were mated to male UAS-hAR52QII/Cyo-GFPII flies (27) to produce either Elav > AR52Q or Elav;Cyo-GFP progeny. Upon the presence of pupae within the mating vial, parental flies were removed and emerging adult progeny were scored for the presence or absence of the Cyo-GFP phenotypic marker over the course of 10 days. Approximately 100 Elav > AR52Q flies were scored for each treatment group. For the evaluation of eye phenotypes, homozygous virgin female GMR:UAS-hAR52QII flies were crossed to males from the following lines: UAS-GFP, UAS-CncCIII, UAS-CncCRNAi (kindly provided by Dirk Bohmann), P(HSF++8) or UAS-HSFRNAi (Bloomington Drosophila Stock Center, #5490, 27070, 41583). Eye phenotypes of anesthetized flies were scored using an Leica MZ APO or M205C stereomicroscope and photographed with a Leica DFC320 digital camera. At least three crosses were performed for each genotype and the effects of drug treatment evaluated.
AR97Q mouse line and drug treatment
All procedures were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (8th ed., National Academies Press, Revised 2011), and have been approved by the NINDS Animal Care Committee. The transgenic AR97Q mouse model has been described (26). Experiments were performed in male F1 animals derived from crossing the AR97Q line in a C57Bl6 background to BDF1. The mice were randomized to receive either ASC-JM17 (120 mg/kg) or the formulation without added drug daily via the oral route by gavage starting at 10 weeks of age and were treated for 16 weeks for behavioral studies, or for 6 weeks for biochemical and histological analyses. The dose and treatment regimen were selected based on pilot studies in rodents to maximize systemic exposure in the absence of toxicity with daily administration of the compound. Body weight and grip strength using the hang wire grip test were recorded weekly by investigators blinded to the identity of the treatment (51). For muscle histology, quadriceps muscles were snap-frozen in isopentane and cut into 8-μm thick sections, which were either processed with H&E or NADH staining, or fixed in 4% formaldehyde and probed with polyglutamine-specific 1C2 antibody (EMD Millipore). 1C2 signal was visualized using biotinylated mouse IgG (R&D Systems), streptavidin-HRP and 3,3′-diaminobenzidine substrate (Life Technologies), and nuclei were counterstained using hematoxylin. For each mouse, the number of 1C2-positive nuclei was determined in at least 500 muscle fibers in randomly selected views.
Gene expression
Total RNA was extracted from frozen quadriceps muscle using Trizol and reverse-transcribed using the High Capacity cDNA Reverse Transcription kit according to the manufacturer's instructions. Transcript levels were determined by real-time PCR using Taqman reagents with an ABI9900 Sequence Detector System using the threshold cycle method and β-glucuronidase as a reference gene. Taqman assays were performed with the following probes (all from Life Technologies): Gclc (Mm00802655_m1), Gsr (Mm00439154_m1), Gsta2 (Mm03019257_g1), Gstp1 (Mm04213618_gH), Gusb (Mm01197698_m1), HO-1 (Mm00516005_m1), Nqo1 (Mm01253561_m1), Nrf1 (Mm00599712_m1), Nrf2 (Mm00477784_m1), Psmb1 (Mm00650840_m1), Psmb2 (Mm00449477_m1), Psmb4 (Mm01263563_m1), Psmb5 (Mm01615821_g1), Psmc1 (Mm04213282_g1), Psmd14 (Mm00451955_m1), Psme1 (Mm00650858_g1) and Sqstm (Mm00448091_m1).
Statistical analysis
For statistical comparisons, data sets were analyzed using two-tailed Student's t-test or two-way ANOVA with the Graphpad Prism software (version 6), except where indicated otherwise. A value of P≤ 0.05 was considered significant.
Supplementary Material
Funding
This work was supported by the Intramural Research Program of the NINDS, NIH (1ZIA NS003038); NIH grants U01NS069515 (C.C.-Y.S.) and NS053825 (J.P.T.) and the Swedish Research Council (N.P.D.). Further support comes from the KI-NIH Graduate Partnership Program in Neuroscience (L.C.B.) and the Association Française contre les Myopathies (AFM) Telethon (C.R.).
Supplementary Material
Acknowledgements
We thank Amilcar Flores-Morales and Dirk Bohmann for reagents and Drosophila stocks, respectively. We also thank Hiroaki Adachi for assistance with the AR97Q mouse line.
Conflict of Interest statement. C.C.-Y.S. was an officer and shareholder at AndroScience Corp. when this collaborative study was conducted.
References
- 1.Orr H.T., Zoghbi H.Y. (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci., 30, 575–621. [DOI] [PubMed] [Google Scholar]
- 2.La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. (1991) Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature, 352, 77–79. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman A.P., Harmison G., Strand A.D., Olson J.M., Fischbeck K.H. (2002) Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet., 11, 1967–1976. [DOI] [PubMed] [Google Scholar]
- 4.Katsuno M., Adachi H., Minamiyama M., Waza M., Tokui K., Banno H., Suzuki K., Onoda Y., Tanaka F., Doyu M. et al. (2006) Reversible disruption of dynactin 1-mediated retrograde axonal transport in polyglutamine-induced motor neuron degeneration. J. Neurosci., 26, 12106–12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranganathan S., Harmison G.G., Meyertholen K., Pennuto M., Burnett B.G., Fischbeck K.H. (2009) Mitochondrial abnormalities in spinal and bulbar muscular atrophy. Hum. Mol. Genet., 18, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z., Wang A.M., Robins D.M., Lieberman A.P. (2009) Altered RNA splicing contributes to skeletal muscle pathology in Kennedy disease knock-in mice. Dis. Mod. Mech., 2, 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. (2008) Adapting proteostasis for disease intervention. Science, 319, 916–919. [DOI] [PubMed] [Google Scholar]
- 8.Pirkkala L., Nykanen P., Sistonen L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J., 15, 1118–1131. [DOI] [PubMed] [Google Scholar]
- 9.Adachi H., Katsuno M., Minamiyama M., Sang C., Pagoulatos G., Angelidis C., Kusakabe M., Yoshiki A., Kobayashi Y., Doyu M. et al. (2003) Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J. Neurosci., 23, 2203–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey C.K., Andriola I.F., Kampinga H.H., Merry D.E. (2002) Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum. Mol. Genet., 11, 515–523. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi Y., Kume A., Li M., Doyu M., Hata M., Ohtsuka K., Sobue G. (2000) Chaperones Hsp70 and Hsp40 suppress aggregate formation and apoptosis in cultured neuronal cells expressing truncated androgen receptor protein with expanded polyglutamine tract. J. Biol. Chem., 275, 8772–8778. [DOI] [PubMed] [Google Scholar]
- 12.Wyttenbach A., Sauvageot O., Carmichael J., Diaz-Latoud C., Arrigo A.P., Rubinsztein D.C. (2002) Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum. Mol. Genet., 11, 1137–1151. [DOI] [PubMed] [Google Scholar]
- 13.Kensler T.W., Wakabayashi N., Biswal S. (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol., 47, 89–116. [DOI] [PubMed] [Google Scholar]
- 14.Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., Deshaies R.J. (2010) Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell, 38, 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sha Z., Goldberg A.L. (2014) Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol., 24, 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 17.Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A.L. (2004) Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell, 14, 95–104. [DOI] [PubMed] [Google Scholar]
- 18.Pratt G., Rechsteiner M. (2008) Proteasomes cleave at multiple sites within polyglutamine tracts: activation by PA28gamma(K188E). J. Biol. Chem., 283, 12919–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palazzolo I., Stack C., Kong L., Musaro A., Adachi H., Katsuno M., Sobue G., Taylor J.P., Sumner C.J., Fischbeck K.H. et al. (2009) Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron, 63, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokui K., Adachi H., Waza M., Katsuno M., Minamiyama M., Doi H., Tanaka K., Hamazaki J., Murata S., Tanaka F. et al. (2009) 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum. Mol. Genet., 18, 898–910. [DOI] [PubMed] [Google Scholar]
- 21.Waza M., Adachi H., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G. (2005) 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat. Med., 11, 1088–1095. [DOI] [PubMed] [Google Scholar]
- 22.Alavez S., Vantipalli M.C., Zucker D.J., Klang I.M., Lithgow G.J. (2011) Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature, 472, 226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calamini B., Silva M.C., Madoux F., Hutt D.M., Khanna S., Chalfant M.A., Saldanha S.A., Hodder P., Tait B.D., Garza D. et al. (2012) Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol., 8, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brondino N., Re S., Boldrini A., Cuccomarino A., Lanati N., Barale F., Politi P. (2014) Curcumin as a therapeutic agent in dementia: a mini systematic review of human studies. Sci. World J., 2014, 174282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsu H., Xiao Z., Ishida J., Nagai M., Wang H.K., Itokawa H., Su C.Y., Shih C., Chiang T., Lee Y. et al. (2002) Antitumor agents. 217. Curcumin analogues as novel androgen receptor antagonists with potential as anti-prostate cancer agents. J. Med. Chem., 45, 5037–5042. [DOI] [PubMed] [Google Scholar]
- 26.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. (2002) Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron, 35, 843–854. [DOI] [PubMed] [Google Scholar]
- 27.Nedelsky N.B., Pennuto M., Smith R.B., Palazzolo I., Moore J., Nie Z., Neale G., Taylor J.P. (2010) Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron, 67, 936–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z., Chang Y.J., Yu I.C., Yeh S., Wu C.C., Miyamoto H., Merry D.E., Sobue G., Chen L.M., Chang S.S. et al. (2007) ASC-J9 ameliorates spinal and bulbar muscular atrophy phenotype via degradation of androgen receptor. Nat. Med., 13, 348–353. [DOI] [PubMed] [Google Scholar]
- 29.Grunseich C., Kats I.R., Bott L.C., Rinaldi C., Kokkinis A., Fox D., Chen K.L., Schindler A.B., Mankodi A.K., Shrader J.A. et al. (2014) Early onset and novel features in a spinal and bulbar muscular atrophy patient with a 68 CAG repeat. Neuromuscul. Disord., 24, 978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Crouch D.H., Yamamoto M., Hayes J.D. (2006) Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J., 399, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radhakrishnan S.K., den Besten W., Deshaies R.J. (2014) p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife, 3, e01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimberg K.B., Beskow A., Lundin D., Davis M.M., Young P. (2011) Basic leucine zipper protein Cnc-C is a substrate and transcriptional regulator of the Drosophila 26S proteasome. Mol. Cell. Biol., 31, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratt W.B., Gestwicki J.E., Osawa Y., Lieberman A.P. (2015) Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol., 55, 353–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A.M., Miyata Y., Klinedinst S., Peng H.M., Chua J.P., Komiyama T., Li X., Morishima Y., Merry D.E., Pratt W.B. et al. (2013) Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol., 9, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sittler A., Lurz R., Lueder G., Priller J., Lehrach H., Hayer-Hartl M.K., Hartl F.U., Wanker E.E. (2001) Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum. Mol. Genet., 10, 1307–1315. [DOI] [PubMed] [Google Scholar]
- 36.Katsuno M., Sang C., Adachi H., Minamiyama M., Waza M., Tanaka F., Doyu M., Sobue G. (2005) Pharmacological induction of heat-shock proteins alleviates polyglutamine-mediated motor neuron disease. Proc. Natl. Acad. Sci. USA, 102, 16801–16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik B., Nirmalananthan N., Gray A.L., La Spada A.R., Hanna M.G., Greensmith L. (2013) Co-induction of the heat shock response ameliorates disease progression in a mouse model of human spinal and bulbar muscular atrophy: implications for therapy. Brain, 136, 926–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo N., Katsuno M., Adachi H., Minamiyama M., Doi H., Matsumoto S., Miyazaki Y., Iida M., Tohnai G., Nakatsuji H. et al. (2013) Heat shock factor-1 influences pathological lesion distribution of polyglutamine-induced neurodegeneration. Nat. Comm., 4, 1405. [DOI] [PubMed] [Google Scholar]
- 39.Bersuker K., Hipp M.S., Calamini B., Morimoto R.I., Kopito R.R. (2013) Heat shock response activation exacerbates inclusion body formation in a cellular model of Huntington disease. J. Biol. Chem., 288, 23633–23638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barone M.C., Sykiotis G.P., Bohmann D. (2011) Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson's disease. Dis. Mod. Mech., 4, 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan J.Y., Kwong M., Lu R., Chang J., Wang B., Yen T.S., Kan Y.M. (1998) Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J., 17, 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farmer S.C., Sun C.W., Winnier G.E., Hogan B.L., Townes T.M. (1997) The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev., 11, 786–798. [DOI] [PubMed] [Google Scholar]
- 43.Chan K., Lu R., Chang J.C., Kan Y.W. (1996) NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA, 93, 13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubbs A.F., Benkovic S.A., Miller D.B., O'Callaghan J.P., Battelli L., Schwegler-Berry D., Ma Q. (2007) Vacuolar leukoencephalopathy with widespread astrogliosis in mice lacking transcription factor Nrf2. Am. J. Pathol., 170, 2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilchez D., Morantte I., Liu Z., Douglas P.M., Merkwirth C., Rodrigues A.P., Manning G., Dillin A. (2012) RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature, 489, 263–268. [DOI] [PubMed] [Google Scholar]
- 46.Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P. et al. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature, 467, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vargas M.R., Johnson D.A., Sirkis D.W., Messing A., Johnson J.A. (2008) Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J. Neurosci., 28, 13574–13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin Y.N., Yu Y.V., Gundemir S., Jo C., Cui M., Tieu K., Johnson G.V. (2013) Impaired mitochondrial dynamics and Nrf2 signaling contribute to compromised responses to oxidative stress in striatal cells expressing full-length mutant huntingtin. PLoS ONE, 8, e57932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Hettinger C.L., Zhang D., Rezvani K., Wang X., Wang H. (2014) Sulforaphane enhances proteasomal and autophagic activities in mice and is a potential therapeutic reagent for Huntington's disease. J. Neurochem., 129, 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellrichmann G., Petrasch-Parwez E., Lee D.H., Reick C., Arning L., Saft C., Gold R., Linker R.A. (2011) Efficacy of fumaric acid esters in the R6/2 and YAC128 models of Huntington's disease. PLoS ONE, 6, e16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinaldi C., Bott L.C., Chen K.L., Harmison G.G., Katsuno M., Sobue G., Pennuto M., Fischbeck K.H. (2012) Insulin like growth factor (IGF)-1 administration ameliorates disease manifestations in a mouse model of spinal and bulbar muscular atrophy. Mol. Med., 18, 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.