Abstract
Ten Eleven Translocation (TET) protein-catalyzed 5mC oxidation not only creates novel DNA modifications, such as 5hmC, but also initiates active or passive DNA demethylation. TETs’ role in the crosstalk with specific histone modifications, however, is largely elusive. Here, we show that TET2-mediated DNA demethylation plays a primary role in the de novo establishment and maintenance of H3K4me3/H3K27me3 bivalent domains underlying methylated DNA CpG islands (CGIs). Overexpression of wild type (WT), but not catalytic inactive mutant (Mut), TET2 in low-TET-expressing cells results in an increase in the level of 5hmC with accompanying DNA demethylation at a subset of CGIs. Most importantly, this alteration is sufficient in making de novo bivalent domains at these loci. Genome-wide analysis reveals that these de novo synthesized bivalent domains are largely associated with a subset of essential developmental gene promoters, which are located within CGIs and are previously silenced due to DNA methylation. On the other hand, deletion of Tet1 and Tet2 in mouse embryonic stem (ES) cells results in an apparent loss of H3K27me3 at bivalent domains, which are associated with a particular set of key developmental gene promoters. Collectively, this study demonstrates the critical role of TET proteins in regulating the crosstalk between two key epigenetic mechanisms, DNA methylation and histone methylation (H3K4me3 and H3K27me3), particularly at CGIs associated with developmental genes.
INTRODUCTION
Covalent modifications of genomic DNA and histones constitute the biochemical foundation of epigenetic regulation (1). Methylation at the 5-position of cytosine (5mC) is the primary covalent modification found on genomic DNA. It is known to influence genomic imprinting, X-chromosome inactivation, gene expression, genome stabilization, cell differentiation and embryonic development (2,3). Similarly, differential histone modifications within the nucleosome have instrumental effects on the remodeling of chromatin structure as well as the aforementioned cellular and developmental processes (4,5).
It is believed that DNA methylation and the coordinated modification of histones function both independently and in conjunction to regulate cellular processes and to determine the final outcome of biological events (6). This is seen with the ability of 5mC to recruit ‘5mC readers’ such as methylated CpG binding protein (MeCP2) and its associated histone modifying and remodeling complexes. These act to reconfigure the underlying chromatin structure and establish a repressive chromatin state suitable for stable gene silencing (7). On the other hand, histone modifications have also been shown to regulate DNA methylation. For example, an unmethylated K4 residue on Histone H3 can be recognized by DNMT3L, which is a core component of enzymatic complex that recruits DNA de novo methyltransferases, DNMT3A and DNMT3B (8). In contrast, histone H3K4 trimethylation (H3K4me3) prevents the DNA methyltransferase complex from accessing CGIs by blocking the binding of DNMT3L. This ensures that CGIs remain free of 5mC, leading to the activation of gene transcription (9).
While H3K4me3 is generally associated with active transcription, H3K27me3 most often accompanies transcriptional repression (10,11). Interestingly, many developmental genes in pluripotent embryonic stem (ES) cells possess what are called ‘bivalent domains,’ which are characterized by the co-existence of H3K4me3 and H3K27me3 (12,13). Bivalent domains are believed to poise genes for future activation or repression. In response to differentiation cues, they eventually resolve into either H3K4me3 or H3K27me3 monovalent chromatin structures (12).
Recent studies have suggested that DNA methylation plays a critical role in the regulation of histone methylation and establishment of bivalent domains (14,15). H3K27me3 has been found to be widely distributed throughout the whole genome (16–18). However, its methyltransferase, the PRC2 complex, is primarily localized to unmethylated CGIs (11,19). Furthermore, almost all of the genomic H3K4me3 is localized to unmethylated CGIs (20). Therefore, it is no surprise that bivalent domains are predominately confined to unmethylated CGIs (21,22). Recent studies have demonstrated that introduction of unmethylated exogenous CGIs is sufficient to establish bivalent domains (23–25). Collectively, these findings suggest that an intricate relationship exists between the methylation status of CGIs, the state of H3K4me3 and H3K27me3 and the establishment and regulation of bivalent domains. Still, there are large gaps in our knowledge pertaining to the following fundamental questions: (i) Is there an epistatic order between DNA methylation and histone modification—who is the ‘chicken’ and who is the ‘egg’; and (ii) Is there a cellular factor(s), which acts as a modulator in commissioning the crosstalk between the status of DNA methylation and the establishment of bivalent domains at CGIs?
An important protein family involved in the modulation of DNA methylation is the Ten Eleven Translocation (TET) proteins. They are responsible for the oxidation of 5mC into 5-hydroxymethylcytosine (5hmC) as well as 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (26–28). 5caC can undergo excision by thymine-DNA glycosylase (TDG) and is then replaced by an unmethylated cytosine via the base excision repair (BER) pathway (27). These findings suggest that TET protein-catalyzed 5mC oxidation not only generates new epigenetic marks (5hmC, 5fC and 5caC), but also initiates active or passive DNA demethylation (29). Despite efforts, little is known about the dynamic regulation of Tet-mediated DNA demethylation, and how the potential consequences of dynamic changes in the state of 5mC may impact the reconfiguration of histone modification patterns at the demethylated CGIs.
In this study, we employed both ‘gain-of-function’ and ‘loss-of-function’ approaches to investigate the possible functions of TET proteins in regulating the dynamics of histone H3K4 and K27 methylation status and the establishment of bivalent domains underlying the associated CGIs. Our data uncovered that TET2 has a direct role in regulating the crosstalk between DNA methylation and histone methylation (H3K4me3 and H3K27me3) at CGIs. This finding provides evidence to support a model in which TET2-catalyzed 5mC oxidation is a prerequisite for the demethylation of methylated CGIs. This commissions the transition of a 5mC-repressive chromatin state to an H3K4me3/H3K27me3-bivalent domain, thereby poising for the reactivation of genes whose promoters were previously methylated at CGIs.
MATERIALS AND METHODS
Constructs
pPB-CAG-EBNXN and pCMV-PBase vectors were kind gifts from Sanger Institute (30,31). pPB-CAG-ires-Pac was generated by ligating ires-Pac into the multiple-cloning sites (MCS) of pPB-CAG-EBNXN. Wild type TET2 (TET2-WT) and its catalytically inactive mutant (TET2-Mut) cDNA with Flag-HA tag (in POZ vector) were subcloned into pPB-CAG-ires-Pac.
Cell culture
The 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin (Invitrogen). Mock, TET2-WT and TET2-Mut stable 293T cells were obtained by co-transfection of 293T cells with pPB-CAG-ires-Pac, pPB-CAG-TET2-WT or pPB-CAG-TET2-Mut and pCMV-PBase. After 2 μg/ml puromycin (Amresco) screening for 1 month, stable cell lines were selected and identified by Western blotting, immunofluorescence and dot blotting.
For DNMT inhibitor treatment, 293T cells were seeded at a density of 4 × 104 cells/cm2 and treated with 5-aza-2′-deoxycytidine (5-Aza-CdR, Sigma A3656) at a final concentration of 10 μM for 72 h.
Wild type and Tet1/2 DKO mouse ES cells, kindly provided by Dr Guoliang Xu (Shanghai Institutes of Biochemistry and Cell Biology), were cultured in gelatin-coated dishes with mouse ES medium (DMEM, 15% FBS, 1% NEAA, 2 mM Glutamine, 0.1 mM beta-Mercaptoethanol, 100 U/ml penicillin/streptomycin, 1000 U/ml LIF and 3 μM chir99021).
Western blotting
Cells were lysed using 1x SDS loading buffer (50 mM Tris-HCl pH6.8, 10% glycerol, 2% SDS, 0.05% bromophenol blue and 1% 2-mercaptoethanol). Western blotting was performed as described in Molecular Cloning II.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from 5 × 106 freshly harvested cells using TRIzol (Invitrogen) according to the manufacturer's protocol. Extracted RNA was quantified by Nanodrop 2000 based on the value of A260, and the ratio of A260/A280 was 1.85–1.95. PrimeScript™ RT reagent Kit with gDNA Eraser (TAKARA) was used for reverse transcription. One microgram of RNA was dealt with gDNA Eraser in 10 μl reaction system for 5 min at room temperature. 10 μl of RNA mixture was added to 10 μl of reverse-transcription reaction solution and reacted for 15 min at 37°C. Reverse-transcription reaction was terminated by heating for 5 s at 85°C. Transcript levels of candidate genes were measured using quantitative real-time PCR, which was carried out in an iQ5 real-time PCR cycler (Bio-Rad) using the SYBR Green reagent (TAKARA). A total of 20 μl cDNA was diluted to 200 μl by TE buffer (10 mM Tris pH 8.0, 1 mM EDTA). One microliter of diluted cDNA was used for one qPCR reaction. One microgram of RNA was diluted to 200 μl by TE buffer. One microliter of diluted RNA was used as negative control template for qPCR reaction. Primer sequences are described in Supplementary Table S1.
DNA Dot blotting
Genomic DNA (gDNA) was extracted from cells using the DNeasy Blood and Tissue Kit (Qiagen). Denatured gDNA was spotted on a nitrocellulose membrane (Whatman) and cross-linked by UVC irradiation (Hoefer). The membrane was then blocked with 5% milk in TBS-Tween 20 and incubated with anti-5hmC antiserum (1:10 000) (Active motif). After incubation with a horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Beyond time), the membrane was washed with TBS-Tween 20. DNA signal was detected using a Western blotting analysis system (Kodak), by means of enhanced chemilluminescence (GE-Healthcare).
HPLC-MS/MS
The relative abundance of 5hmC and 5mC in gDNA was analyzed by HPLC-MS/MS as previously described (27,32).
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and permeabilized using 0.1% Triton X-100 diluted in phosphate buffered saline (PBS). DNA was denatured using 2N HCl for 30 min, and neutralized for 10 min using 100 mM Tris-HCl pH 8.0. After blocking in 5% serum for 1 h at room temperature, cells were incubated with anti-5hmC antibody (1:5000) (Active Motif) or anti-Flag antibody (1:1000) (Sigma) overnight at 4°C. After washing, cells were incubated with Cy3-labeled anti-rabbit IgG (Beyond time), and the nuclei were labeled using DAPI (Sigma). Fluorescence was observed and recorded using Leica microscopy system.
Sequenom MassARRAY
Genomic DNA was treated using EZ DNA Methylation Kit™ (Zymo Research). Target genes were amplified for 45 cycles using the Hotstart PCR enzyme (TAKARA). The PCR products were treated with shrimp alkaline phosphatase (SAP) (Sequenom), processed using Mass CLEAVE Reaction (Sequenom) and mapped for the MALDI-TOF mass. Primer sequences are described in Supplementary Table S2.
Chromatin immunoprecipitation (ChIP)
Adherent cells were crosslinked using 1% formaldehyde at room temperature for 10 min, and the reaction was stopped using 0.125 M glycine. Crosslinked cells were suspended with lysis buffer (50 mM Hepes, pH 7.4, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycholate, 0.5 mM PMSF) and protease inhibitor cocktail (Roche). Chromatin was sheared using sonication (Bioruptor) to 200–500 bp, and the fragments were incubated with 2–5 μg of the respective antibody (H3K4me3 antibody, CST #9751; H3K27me3 antibody, CST #9733; H3K4me1 antibody, ActiveMotif #39297; H3K4me2 antibody, Millipore #07-030; H3K9me2 antibody, Abcam #1220; H3K9me3, ActiveMotif #39161) at 4°C for 4 h. Protein A/G beads were added to the samples and incubated at 4°C for 2 h. The beads were washed with lysis buffer, high salt buffer (50 mM Tris, pH 7.4, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate), LiCl buffer (20 mM Tris, pH 7.4, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate) and TE buffer (10 mM Tris PH8.0, 1 mM EDTA). The beads were resuspended with elution buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS). The eluate was supplemented with NaCl (at a final concentration of 200 mM) and incubated at 65°C for 5 h. The crosslink-reversed ChIP product was then treated using RNase A and Proteinase K. The final sample was extracted using QIAquick PCR Purification Kit (Qiagen). Enrichment of H3K4me3 or H3K27me3 at certain gene loci were validated using quantitative PCR analysis of the ChIP products. Primer sequences are described in Supplementary Table S3.
MeDIP and hMeDIP
DNA was sonicated to 200–500 bp fragments, denatured in boiling water and immediately cooled on ice. Fragmented DNA was added to IP buffer (10 mM Na-Phosphate pH 7.0, 140 mM NaCl, 0.05% Triton X-100). 5hmC/ 5mC antibody (5hmC antibody, Active Motif #39769; 5mC antibody, Active Motif #39649) was added to the sample and incubated at 4°C for 4 h. Protein A/G beads were added to each reaction and incubated at 4°C for 2 h. The beads were washed using IP buffer for three times. The sample was resuspended in Proteinase K digestion buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS, included 2 μl protease K) and incubated at 50°C for 2 h. The final sample was extracted using QIAquick PCR Purification Kit. Enrichment of 5mC or 5hmC at certain gene loci were validated by quantitative PCR analysis of hMeDIP or MeDIP products. The primers are same as those used in ChIP-qPCR (see Supplementary Table S3).
Next-generation sequencing and data processing
HMeDIP, MeDIP and ChIP-seq libraries were submitted for sequencing on the Illumina Genome Analyzer IIx (GAIIx) platform. Illumina SCS v2.5 software was used for image analysis and base-call steps. Demultiplex of sequencing data was done by configureBclToFastq.pl in the CASAVA (Illumina, v1.6) package according to the specific barcode sequences (6-bp) at 5′-end of each sequence read. Sequencing reads were mapped onto the reference genome (NCBI Build UCSC hg19 and mm9) using the Bowtie (v0.12.7) algorithm. Unique and monoclonal reads were used for further analysis. Duplicate reads were removed with the SAM tools. Since the average DNA fragment length used in hMeDIP-seq, MeDIP-seq and ChIP-seq data sets was 300 bp, each sequence was extended to 300 bp in the mapping process. The sequencing data have been deposited in the Gene Expression Omnibus database under accession number GSE72481.
Identification of DHMRs and DMRs
The approach used for the identification of DHMRs and DMRs has been described in our recent paper (33). Briefly, 5hmC peaks were identified from the pooled hMeDIP-seq data by MACS (v1.4) at P < 1e-5 and FDR < 0.05. For the quantitative analysis of 5hmC, the densities of 5hmC peaks in Mock, TET2-WT and TET2-Mut 293T cells were normalized using their respective input. The threshold of DHMRs between TET2-WT and Mock was set at 4-fold (log2density ratio ≥ 2 or ≤ −2) for the identification of ‘DNA de-hydroxymethylation (loss of 5hmC)’ regions and ‘de novo DNA hydroxymethylation (gain of 5hmC)’ regions. Similar approach was used for 5mC quantitative analysis.
Identification of H3K4me3, H3K27me3 and bivalent peaks
Significant histone mark enrichments were identified using MACS software from the aligned results. H3K4me3 peaks were identified by MACS (v1.4) at P < 1e-5 and FDR < 0.05. H3K27me3 peaks were identified by MACS2.0 using Broad Peak mode at q-value = 0.05. Regions with both H3K4me and H3K27me3 histone mark enrichment were defined as bivalent domains.
Density distribution analysis
The distributions of 5mC, 5hmC, H3K4me3 and H3K27me3 densities at certain regions or genomic features (CGIs or bivalent domains) were analyzed using our previously developed algorithm (34).
Heatmap analysis
The density matrixes of 5mC, 5hmC, H3K4me3 and H3K27me3 in 293T cells and mouse ES cells were generated for all CGIs (CGI center ± 10 kb) based on a 50 bp length bin as previously described (34). The orders of CGIs were sorted by their 5mC densities on CGI center ± 2 kb region of Mock 293T cells or wild type mouse ES cells and then clustered using Cluster 3.0 and Java Treeview program.
Gene ontology (GO) analysis
GO analysis for genes with ‘gain of 5hmC’ DHMRs, ‘loss of 5mC’ DMRs, or ‘gain of bivalency’ regions in TET2-WT 293T cells were performed by the database for annotation, visualization and integrated discovery (DAVID) website (35).
RESULTS AND DISCUSSION
TET2 overexpression induces DNA demethylation of hypermethylated CGIs
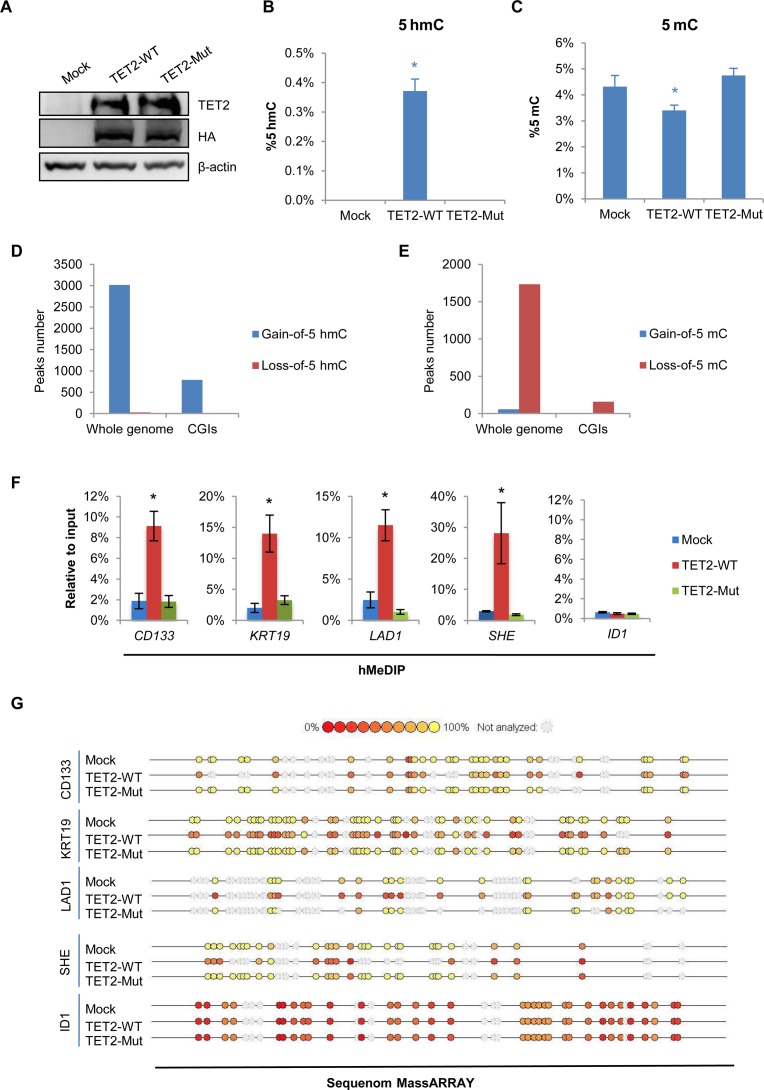
To determine the functional role of TET2-catalyzed 5mC oxidation in DNA demethylation and histone bivalent domain patterning, we established 293T cells, which stably overexpress wild-type TET2 (TET2-WT) or catalytically inactive mutant TET2 (TET2-Mut). This was accomplished using the PiggyBac (PB) transposon system (Figure 1A and Supplementary Figure S1A) (30,31). As previously reported (32), low expression of endogenous TET2 in 293T cells makes it the ideal model to investigate the resultant ‘gain of function’ effect. Overexpression of TET2-WT but not TET2-Mut resulted in a dramatic elevation of the global 5hmC level in 293T cells (Figure 1B and Supplementary Figure S1B and S1C). Concurrently, a significant decrease in 5mC in TET2-WT 293T cells was detected using a quantitative HPLC-MS/MS approach (Figure 1C).
Figure 1.
TET2 overexpression induces DNA demethylation of hypermethylated CGIs. (A) Western blot analysis of TET2 expression in Mock, TET2-WT and TET2-Mut 293T cells. β-Actin was used as loading control. (B–C) Total 5hmC (B) and 5mC (C) in Mock, TET2-WT and TET2-Mut 293T cells were detected by mass spectrometry. Data are presented as mean ± SD (n = 3). *P < 0.05. (D–E) Number of peaks in whole genome and CGIs gaining 5hmC (left) or losing 5mC (right) in 293T cells overexpressing TET2-WT. (F) hMeDIP-qPCR analysis for 5hmC at four representative hypermethylated CGI promoters. The ID1 CGI promoter was used as a negative control. Data are presented as mean ± SD (n = 3). *P < 0.05. (G) Representative data of Sequenom MassARRAY analysis of ‘5mC + 5hmC’ at CpG sites within four representative hypermethylated CGI promoters. The ID1 CGI promoter was used as a negative control.
Next, we profiled the DNA hydroxymethylomes (5hmC) and methylomes (5mC) of Mock, TET2-WT and TET2-Mut 293T cells and identified the differential hydroxymethylated regions (DHMRs) and differential methylated regions (DMRs). This was carried out using the comparative (h)MeDIP-seq method we recently developed (33). A total of 3018 ‘gain-of-5hmC’ DHMRs (de novo DNA hydroxymethylation) and 1732 ‘loss-of-5mC’ DMRs (DNA demethylation) were identified in 293T cells upon TET2-WT overexpression (Figure 1D and E). Strikingly, 793 ‘gain-of-5hmC’ DHMRs and 159 ‘loss-of-5mC’ DMRs were located at CGIs (Figure 1D and E). In contrast, the TET2-Mut overexpressing cells exhibit little, if any, change in 5hmC or 5mC status on within these corresponding CGIs (Supplementary Figure S2A and B).
To validate these findings, we examined the changes in 5hmC and 5mC levels at selected hypermethylated CGI promoters (CD133, KRT19, LAD1 and SHE) in response to TET2 overexpression. Analysis by hMeDIP-qPCR revealed that overexpression of TET2-WT, but not TET2-Mut, resulted in a drastic increase in 5hmC at these promoters (Figure 1F). However, assessment of the same gene promoters using MeDIP-qPCR revealed only a slight change in the levels of 5mC (Supplementary Figure S3). This may be attributed to highly methylated CGIs exceeding the linear detection range of MeDIP-qPCR. Such limitations have been previously reported for MeDIP (36). We, therefore, employed the Sequenom MassARRAY approach in order to more closely and precisely profile the change in methylation status at these CGIs. This technique is a bisulfite-based method, which allows for the examination of ‘5mC + 5hmC’ percentage at base pair resolution (37). Sequenom MassARRAY data showed a significant decrease in the percentage of ‘5mC + 5hmC’ within these CGIs in TET2-WT but not TET2-Mut 293T cells (Figure 1G). Given the increase of 5hmC on these CGIs, we conclude that the decrease of ‘5mC + 5hmC’ is due to the site-specific DNA demethylation mediated by exogenously introduced TET2 enzyme. Taken together, our data suggest that TET2 overexpression induces not only DNA hydroxymethylation (gain-of-5hmC) but also DNA demethylation (loss-of-5mC) at these specific methylated CGIs.
TET2 overexpression promotes the establishment of bivalent domains at hypermethylated CGIs and activates gene transcription
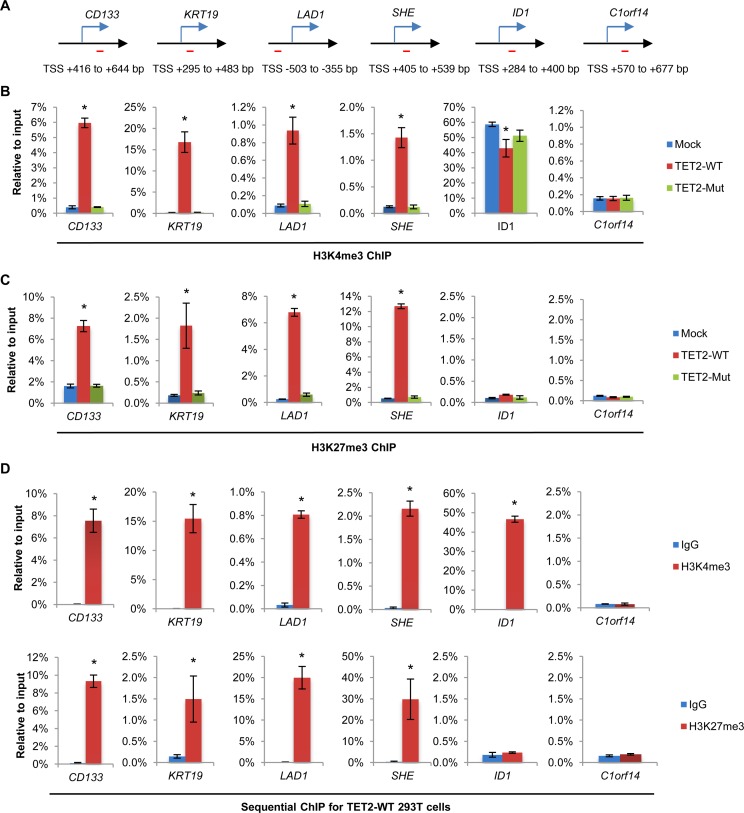
Previous studies have suggested a crosstalk between DNA methylation and histone modification (6). Seeing as the overexpression of TET2 in 293T cells dramatically shapes DNA methylation, we set out to examine whether histone methylation within these regions were also altered. ChIP-qPCR was carried out on the CD133, KRT19, LAD1 and SHE promoters within CGIs. Under normal conditions, these promoters are hypermethylated and lack H3K4me3 and H3K27me3. ChIP-qPCR data revealed a significant gain of both de novo H3K4me3 and H3K27me3 marks at the examined CGIs in TET2-WT but not TET2-Mut 293T cells (Figure 2A–C). H3K4me2 also increased dramatically at these CGIs, whereas other histone modifications such as H3K4me1, H3K9me2 and H3K9me3 showed only a slight increase (Supplementary Figure S4). It should be noted that these ChIP amplicons are all located in regions away from the TSS. Indeed, ChIP-qPCR analysis of histone H3 excluded the possibility that the observed changes in histone methylation on H3 are due to changes in nucleosome occupancy at these CGI loci (Supplementary Figure S5). Sequential-ChIP analysis further confirmed that the H3K4me3 and H3K27me3 at these gene promoters were bona fide bivalent domains (Figure 2D). Furthermore, the de novo generation of the bivalent domains is loci specific; Western Blot analysis showed no significant changes in the global levels of H3K4me3 and H3K27me3 in response to TET2 overexpression (Supplementary Figure S6). Given that DNA methylation within promoter regions is associated with transcriptional silencing (38), we asked whether the overexpression of TET2 would, consequently, affect the expression of these genes. RT-qPCR revealed a marked elevation in the levels of mRNA of these genes in TET2-WT 293T cells but not in TET2-Mut 293T cells in comparison with Mock control cells (Supplementary Figure S7). This indicates that TET2 overexpression is able to de-repress these genes in a manner that is dependent on the catalytic activity of the enzyme. It should be noted that although we observed apparent derepression of these hypermethylated genes upon TET2 overexpression, their expression levels are still at the lower end. Therefore, it is very likely that the transcription activation of these genes occurs in a small fraction of the TET2 overexpressing cells.
Figure 2.
TET2 overexpression promotes the establishment of bivalent domains at hypermethylated CGIs and activates gene transcription. (A) Schematic diagram of the amplified region of ChIP primers in the representative genes. (B) H3K4me3 ChIP-qPCR analysis of the hypermethylated CGI promoters in Mock, TET2-WT and TET2-Mut 293T cells. Data are presented as mean ± SD (n = 3). *P < 0.05. (C) H3K27me3 ChIP-qPCR analysis of the hypermethylated CGI promoters in mock, TET2-WT and TET2-Mut 293T cells. Data are presented as mean ± SD (n = 3). *P < 0.05. (D) Sequential ChIP-qPCR analysis of the hypermethylated CGI promoters in TET2-WT 293T cells using anti-H3K4me3 (upper panel) as the 1st round ChIP and anti-H3K27me3 (bottom panel) antibodies in the 2nd round ChIP. The % IP in the 2nd ChIP was plotted relative to input in the eluate from the first round ChIP. Data are presented as mean ± SD (n = 3). *P < 0.05.
It has been reported that unmethylated CGIs facilitate the establishment of H3K4me3. This occurs through the recruitment of the SET1A/B complex, which is mediated by the CGI binding protein, CFP1 (39). A recent report from Reddington and colleagues also showed that DNA hypomethylation could induce redistribution of H3K27me3 and PRC2 (40). In addition, unmethylated CpG dinucleotides have been identified as a key modulator for PRC2 recruitment and activation (25,41). Our observations suggest that local DNA demethylation —prompted by active TET2 at methylated CGIs—is the likely mechanism leading to the de novo formation of a bivalent state of histone modification. A previous report suggested that DNMT inhibitor (5-Aza-CdR), which induces DNA demethylation, influences gene expression and chromatin structures in various cancer cell lines (42). To ascertain the possibility that DNA demethylation is sufficient in establishing bivalent domains underlying methylated CGIs, we chemically induced DNA demethylation by treating 293T cells with 5-Aza-CdR. Treatment with 5-Aza-CdR further confirmed that DNA demethylation is sufficient in inducing the formation of bivalent domains, which may proceed further to result in transcriptional activation of the target genes in a fraction of the cells (Supplementary Figure S8A–C). Importantly, sequential-ChIP analysis confirmed that 5-Aza-CdR treatment induced bona fide bivalent domains (Supplementary Figure S8D).
Collectively, we conclude that the de novo formation of bivalent domains in TET2-WT 293T cells is likely due to DNA demethylation at CGIs targeted by ectopically expressed TET2 protein. Our data support a model in which the overexpression of TET2 prompts the demethylation of methylated CGI promoters, triggering the regeneration of a bivalent chromatin organization.
Genome-wide analysis of the de novo formation of bivalent domains underlying CGIs in response to TET2 action
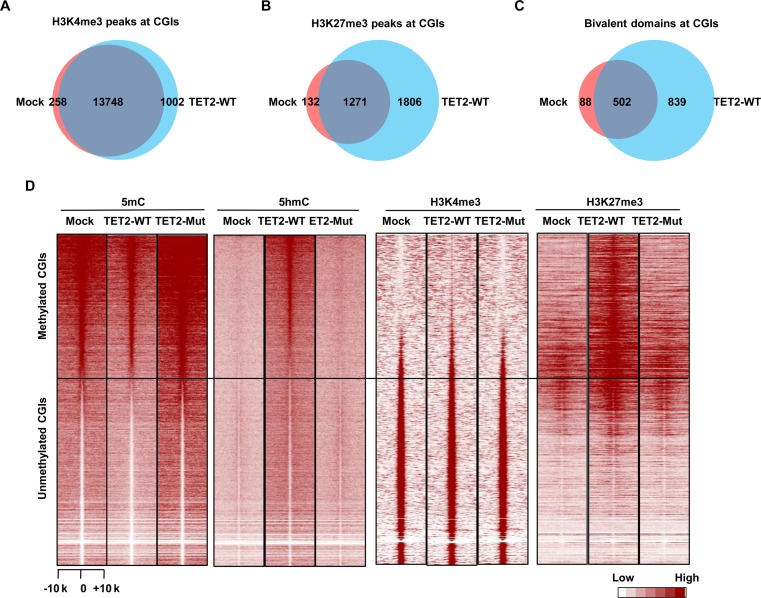
To comprehensively grasp the functional role of TET2 in directing the de novo formation of bivalent domains, we analyzed the genome-wide patterns of H3K4me3 and H3K27me3 in Mock, TET2-WT and TET2-Mut 293T cells. Combined analysis of these ChIP-seq data sets identified 987 new bivalent domains in TET2-WT over-expressed cells (Supplementary Figures S9 and S10). Strikingly, the majority of the new bivalent domains (839/987) were located at CGIs (Figure 3A–C). Furthermore, the ‘gain of bivalency’ occurred at methylated CGIs that had undergone DNA demethylation (Figure 3D). The overexpression of TET2-Mut had no significant effect on the patterns of H3K4me3 and H3K27me3 (Figure 3D and Supplementary Figure S11). Gene ontology (GO) functional enrichment analysis showed that genes, which experienced this ‘gain of bivalency," were most significantly enriched for developmental function, especially those concerning the nervous system and cancer development (Supplementary Figure S12A). Intriguingly, most ‘gain-of-bivalency’ genes in TET2-WT 293T cells are also bivalent genes in human ES cells (Supplementary Figure S12B).
Figure 3.
Genome-wide analysis of the de novo formation of bivalent domains underlying CGIs in response to TET2 action. (A) Venn diagram showing the overlapping of H3K4me3 peaks at CGIs between Mock and TET2-WT 293T cells. (B) Venn diagram showing the overlapping of H3K27me3 peaks at CGIs between Mock and TET2-WT 293T cells. (C) Venn diagram showing the overlapping of bivalent domains at CGIs between Mock and TET2-WT 293T cells. (D) Heatmap of 5mC, 5hmC, H3K4me3 and H3K27me3 density at CGIs in Mock, TET2-WT and TET2-Mut 293T cells. CGIs were ranked according to the 5mC density in Mock 293T cells.
The data gathered from our genome-wide analyses provide additional support to the model we have proposed. By introducing exogenous TET2-WT into TET2-low-expressing cells, we have observed both DNA hydroxymethylation and demethylation at methylated CGIs. The alteration of these DNA epigenomic marks leads to significant gain of H3K4me3/H3K27me3 bivalency.
Depletion of TET1/2 impairs bivalent domains of mouse ES cells
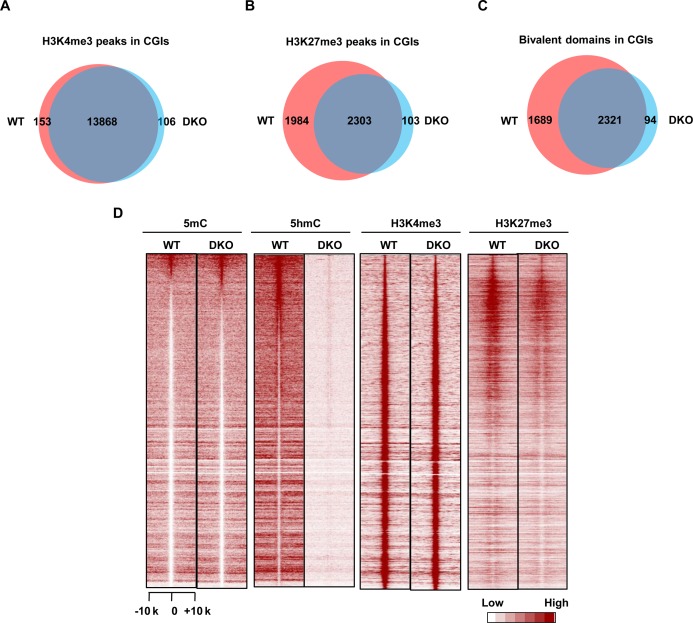
Mouse ES cells are characterized by abundant bivalent domains and high-level expression of Tet1 and Tet2 proteins, which are associated with and regulate genes affiliated with differentiation and development (12,34,43). We hypothesized that the depletion of endogenous Tet1/2 in mouse ES cells would impair the maintenance of proper chromatin bivalency. To investigate this, we compared genome-wide H3K4me3 and H3K27me3 patterns within CGIs in wild type and Tet1/2-double knockout (DKO) mouse ES cells. ChIP-seq analysis showed no significant H3K4me3 pattern changes in the Tet1/2-DKO ES cells (Figure 4A). This finding is appropriate, as the majority of CGIs are free of DNA methylation and are actively transcribed (20,44). We did, however, observe impairment in the level of H3K27me3 at CGIs associated with poised developmental genes (Figure 4B), leading to the loss of a substantial portion (42.1%, 1689 out of 4010) of bivalent domains (Figure 4C).
Figure 4.
Depletion of Tet1/2 reduces H3K27me3 enrichment on bivalent domains in mouse ES cells. (A) Venn diagram showing the overlapping of H3K4me3 peaks at CGIs between WT and Tet1/2 DKO mouse ES cells. (B) Venn diagram showing the overlapping of H3K27me3 peaks at CGIs between WT and Tet1/2 DKO mouse ES cells. (C) Venn diagram showing the overlapping of bivalent domains at CGIs between WT and Tet1/2 DKO mouse ES cells. (D) Heatmap of 5mC, 5hmC, H3K4me3 and H3K27me3 density at CGIs in WT and Tet1/2 DKO mouse ES cells.
We next profiled the DNA methylomes and hydroxymethylomes of wild type and Tet1/2-DKO ES cells and examined the genome-wide correlation between 5hmC/5mC change and the alteration of bivalency. Deletion of Tet1/2 led to a significant decrease in the levels of H3K27me3 at bivalent domain promoters (Figure 4D and Supplementary Figure S13). Reciprocally, a significant increase in 5mC was also observed (Supplementary Figure S13). Importantly, the inverse alteration of H3K27me3 and 5mC can be further verified by ChIP- and MeDIP-qPCR (Supplementary Figure S14). These findings suggest that endogenous Tet1/2 proteins contribute to the maintenance of bivalency and DNA hypomethylation at a specific subset of CGIs in pluripotent ES cells. We should note that the remaining 57.9% (2321/4010) of bivalent domains, which have little to no 5hmC/5mC change, still remained intact in the Tet1/2-DKO ES cells. This possibly indicates that ES cells contain multiple mechanisms to maintain both bivalency and CGI hypomethylation (45). Nevertheless, the correlation between gain of DNA methylation and loss of bivalency in mES cells is in agreement with our observations in TET2-overexpressing 293T cells. Taken together, these findings suggest that TET-mediated DNA demethylation is a prerequisite for the de novo establishment and maintenance of bivalent domains at a specific subset of methylated CGIs.
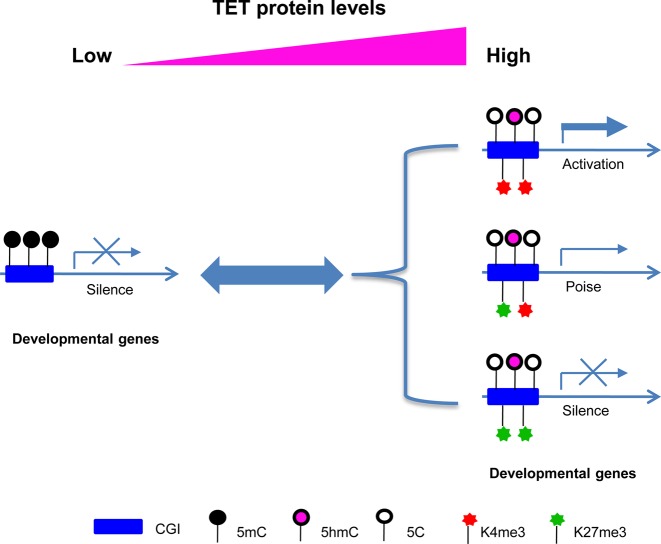
In sum, the present study provides compelling evidence that TET enzymes are one of the primary factors involved in the reprogramming of de novo ‘bivalent histone code’ at hypermethylated CGIs. Our data support a new model in which TET2 functions as a critical regulator of chromatin structure and gene transcription through its role in DNA demethylation and, subsequent, establishment of bivalency at CGIs (Figure 5). This regulation not only reprograms the histone code (at least H3K4me3 and H3K27me3) but also favors the loading of RNA polymerase, reactivating the transcription of genes associated with methylated CGI promoters. Consistent with our findings, several groups have also reported that TET1 overexpression could alter the genome-wide distribution of 5hmC and 5mC as well as the gene expression in 293T cells (46,47). Moreover, Reddington et al. reported that, in the absence of DNA methylation, Polycomb spreads all over the genome, leading to a genome-wide redistribution of H3K27me3 (40). Finally, Deplus et al. reported that TET2/TET3 might influence H3K4me3 through interactions with OGT and the direct recruitment of SET1/COMPASS H3K4 methyltransferase (48). Since TET2-Mut overexpression has no significant effect on H3K4me3 establishment at the representative hypermethylated CGIs, our study provides further mechanistic explanations related to these early findings. We propose that TET2-induced H3K4me3/H3K27me3 establishment at these CGIs not only requires a direct recruiting (for H3K4) or a default (for H3K27) mechanism for histone methyltransferase complexes, but also requires active TET-mediated CGI demethylation prior to the loading of the methyltransferases. Nonetheless, these studies as well as our present study support the notion that TET enzymes function as important physiological players in the fine-tuning of DNA methylation and histone modification. Perturbation of the interplay between these two key epigenetic pathways is expected to deregulate cell processes including proper development and cell differentiation, lending to the occurrence of detrimental diseases, such as cancer (49,50).
Figure 5.
A hypothetic model that illustrates the functional role of TET proteins in regulating the crosstalk between DNA methylation and histone H3K4 and H3K27 methylation.
Global DNA hypomethylation and CGI-specific hypermethylation are well-reported epigenetic landscapes in cancer cells (51). Recent studies have revealed that in cancer, ∼75% of abnormally hypermethylated CGI genes were marked by CGI-specific bivalent chromatin in either ES or adult stem/progenitor cells (52). More interestingly, the expression and/or activity of TET family genes are down-regulated in almost all cancer types (53,54). Our previous work, as well as that of others, has identified the ‘loss of 5hmC’ as a hallmark of many cancers (53–55). Here, we have shown that the overexpression of TET2 induces both DNA hydroxymethylation and DNA demethylation at a subset of hypermethylated CGIs. Moreover, this study shows that the overexpression of TET2 can reprogram a specific subset of hypermethylated CGI promoters into a bivalent chromatin structure, amenable to transcriptional de-repression of these hypermethylated genes. Treatment with 5-Aza-CdR further confirmed that DNA demethylation is sufficient in inducing the formation of bivalent domains and switching on a set of genes silenced by DNA methylation. Collectively, these findings provide novel insight into the regulatory function of TET proteins in the reconfiguration of the chromatin structure of genes silenced by DNA hypermethylation and shed light on how the ‘loss-of-function’ of TET family genes contributes to characteristic abnormalities observed in human diseases, such as cancers. In light of this, future investigation will be of clinical and therapeutic importance to address the role of TET2-mediated DNA demethylation in the reconfiguration of the chromatin structure of tumor suppressor genes silenced by DNA hypermethylation. Furthermore, future studies should also explore whether this reconfiguration may act to reactivate tumor suppressor genes, thereby reverting cancerous phenotypes.
Supplementary Material
Acknowledgments
The authors thank Clark Yin for reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31200966 and 81272295]; NIH [R01GM112062 and R01CA194302]. Funding for open access charge: National Natural Science Foundation of China [31200966].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bernstein B.E., Meissner A., Lander E.S. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Meissner A. Guiding DNA methylation. Cell Stem Cell. 2011;9:388–390. doi: 10.1016/j.stem.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 5.Lennartsson A., Ekwall K. Histone modification patterns and epigenetic codes. Biochim et Biophys. Acta. 2009;1790:863–868. doi: 10.1016/j.bbagen.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 7.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 8.Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 11.Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez J., Munoz M., Vives L., Frangou C.G., Groudine M., Peinado M.A. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19809–19814. doi: 10.1073/pnas.0810133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmer F., Brinkman A.B., Assenov Y., Matarese F., Kaan A., Sabatino L., Villanueva A., Huertas D., Esteller M., Lengauer T., et al. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics. 2012;7:1355–1367. doi: 10.4161/epi.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young M.D., Willson T.A., Wakefield M.J., Trounson E., Hilton D.J., Blewitt M.E., Oshlack A., Majewski I.J. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39:7415–7427. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauler F.M., Sloane M.A., Huang R., Regha K., Koerner M.V., Tamir I., Sommer A., Aszodi A., Jenuwein T., Barlow D.P. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 20.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendenhall E.M., Koche R.P., Truong T., Zhou V.W., Issac B., Chi A.S., Ku M., Bernstein B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch M.D., Smith A.J., De Gobbi M., Flenley M., Hughes J.R., Vernimmen D., Ayyub H., Sharpe J.A., Sloane-Stanley J.A., Sutherland L., et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer L.M., Tahiliani M., Rao A., Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan L., Shi Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusa K., Rad R., Takeda J., Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hamalainen R., Cowling R., Wang W., Liu P., Gertsenstein M., et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H., Zhang X., Clark E., Mulcahey M., Huang S., Shi Y.G. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 33.Tan L., Xiong L., Xu W., Wu F., Huang N., Xu Y., Kong L., Zheng L., Schwartz L., Shi Y., et al. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res. 2013;41:e84. doi: 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A.J., Zheng L., Zhang H., Huang S., et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson M.D., Stirzaker C., Statham A.L., Coolen M.W., Song J.Z., Nair S.S., Strbenac D., Speed T.P., Clark S.J. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res. 2010;20:1719–1729. doi: 10.1101/gr.110601.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrich M., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubeler D., Lorincz M.C., Cimbora D.M., Telling A., Feng Y.Q., Bouhassira E.E., Groudine M. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 2000;20:9103–9112. doi: 10.1128/mcb.20.24.9103-9112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clouaire T., Webb S., Skene P., Illingworth R., Kerr A., Andrews R., Lee J.H., Skalnik D., Bird A. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddington J.P., Perricone S.M., Nestor C.E., Reichmann J., Youngson N.A., Suzuki M., Reinhardt D., Dunican D.S., Prendergast J.G., Mjoseng H., et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14:R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan W., Wu T., Fu H., Dai C., Wu H., Liu N., Li X., Xu M., Zhang Z., Niu T., et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 42.Mayor R., Munoz M., Coolen M.W., Custodio J., Esteller M., Clark S.J., Peinado M.A. Dynamics of bivalent chromatin domains upon drug induced reactivation and resilencing in cancer cells. Epigenetics. 2011;6:1138–1148. doi: 10.4161/epi.6.9.16066. [DOI] [PubMed] [Google Scholar]
- 43.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fouse S.D., Shen Y., Pellegrini M., Cole S., Meissner A., Van Neste L., Jaenisch R., Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams K., Christensen J., Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grosser C., Wagner N., Grothaus K., Horsthemke B. Altering TET dioxygenase levels within physiological range affects DNA methylation dynamics of HEK293 cells. Epigenetics. 2015;10:819–833. doi: 10.1080/15592294.2015.1073879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin C., Lu Y., Jelinek J., Liang S., Estecio M.R., Barton M.C., Issa J.P. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014;42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deplus R., Delatte B., Schwinn M.K., Defrance M., Mendez J., Murphy N., Dawson M.A., Volkmar M., Putmans P., Calonne E., et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., Zhang L., Mao S.Q., Li Z., Chen J., Zhang R.R., Wu H.P., Gao J., Guo F., Liu W., et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Dawlaty M.M., Breiling A., Le T., Barrasa M.I., Raddatz G., Gao Q., Powell B.E., Cheng A.W., Faull K.F., Lyko F., et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 52.Easwaran H., Johnstone S.E., Van Neste L., Ohm J., Mosbruger T., Wang Q., Aryee M.J., Joyce P., Ahuja N., Weisenberger D., et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H., Liu Y., Bai F., Zhang J.Y., Ma S.H., Liu J., Xu Z.D., Zhu H.G., Ling Z.Q., Ye D., et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W., Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids. 2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lian C.G., Xu Y., Ceol C., Wu F., Larson A., Dresser K., Xu W., Tan L., Hu Y., Zhan Q., et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.