Abstract
The misincorporation of 2′-deoxyribonucleotides (dNs) into RNA has important implications for the function of non-coding RNAs, the translational fidelity of coding RNAs and the mutagenic evolution of viral RNA genomes. However, quantitative appreciation for the degree to which dN misincorporation occurs is limited by the lack of analytical tools. Here, we report a method to hydrolyze RNA to release 2′-deoxyribonucleotide-ribonucleotide pairs (dNrN) that are then quantified by chromatography-coupled mass spectrometry (LC-MS). Using this platform, we found misincorporated dNs occurring at 1 per 103 to 105 ribonucleotide (nt) in mRNA, rRNAs and tRNA in human cells, Escherichia coli, Saccharomyces cerevisiae and, most abundantly, in the RNA genome of dengue virus. The frequency of dNs varied widely among organisms and sequence contexts, and partly reflected the in vitro discrimination efficiencies of different RNA polymerases against 2′-deoxyribonucleoside 5′-triphosphates (dNTPs). Further, we demonstrate a strong link between dN frequencies in RNA and the balance of dNTPs and ribonucleoside 5′-triphosphates (rNTPs) in the cellular pool, with significant stress-induced variation of dN incorporation. Potential implications of dNs in RNA are discussed, including the possibilities of dN incorporation in RNA as a contributing factor in viral evolution and human disease, and as a host immune defense mechanism against viral infections.
INTRODUCTION
The potential for misincorporation of ribonucleotides (rN) into DNA and 2′-deoxyribonucleotides (dN) into RNA has long been recognized as a possible consequence of the ability of polymerases to discriminate among components of the highly dynamic nucleotide pool in prokaryotic and eukaryotic cells in different environments (1–3). A key discriminatory feature here is the 2′-OH in RNA that plays critical roles in protein–RNA recognition (4), stabilization of RNA structures (5–7), RNA packing (8) and RNA-mediated catalysis (9), among other processes. A single dN substitution at random or specific sites in RNA can significantly affect these processes (5,9–11) by perturbing both single- and double-stranded RNA structures (5). The incorporation of dN into RNA can have a variety of deleterious consequences for cells and RNA viruses. For example, it has been shown that mutation of residues involved in discrimination against dNTPs in RNA polymerases can cause growth defects and lethality in cells and viruses (12,13). Furthermore, dNs inserted by cellular and viral RNA polymerases impede further extension of a transcript (14–16). Beyond the obvious miscoding effects of dN in mRNA, incorporation of dN at specific sites in rRNAs and tRNA has been shown to significantly alter critical steps in translation (10,17–22), including tRNA binding to the ribosome, translocation and elongation. Such alterations may trigger ribosomal frameshifting (23) or result in the production of prematurely terminated peptides, which has been implicated in neurodegenerative and other diseases (24). dN in RNA also has implications for replication of viral RNA genomes and reverse-transcription of retroviruses, with in vitro studies revealing problems with initiation and completion of these two processes when RNA-dependent RNA polymerases (RdRPs) and HIV-1 reverse transcriptase were presented with dN-containing RNA templates (15,25–29). The presence of dN in viral RNA genomes thus has the potential to both alter the rate of viral evolution by mutation and cause genome truncation that is associated with persistent viral infections (30).
In spite of the many potentially deleterious consequences of the presence of dN in coding and non-coding RNAs, this phenomenon is largely unexplored in vivo due to a lack of appropriate tools for quantitative analysis of dN in RNA. Our understanding of the mechanisms contributing to dN incorporation into RNA is based on numerous in vitro studies of RNA polymerase incorporation of dNs during RNA synthesis and the potential for dN incorporation in cells with defined nucleotide pool imbalances. In general, the concentrations of ribonucleotide triphosphates (NTP) range from ∼1000 to ∼3000 μM in cells while 2′-deoxynucleotide triphosphates (dNTP) are ∼10-fold lower in concentration, ranging from ∼100 to ∼300 μM (examples in Table 1) (31–34). Large shifts in these concentrations can occur under a variety of genetic and environmental conditions, such as loss of nucleoside diphosphate kinase (Ndk) E. coli (Table 1), an enzyme that converts (d)NDPs to their corresponding triphosphates (35). For reasons that are not entirely clear, loss of ndk disrupts the (d)NTP pool balance to cause large increases in dCTP concentrations (33). In the context of RNA polymerase activity, the active sites of most, if not all, RNA polymerases have evolved to efficiently exclude dNTPs during RNA synthesis (1). However, dNTP exclusion is not absolute (1). Kinetic studies (14,16,36–38) have revealed that the degree of selectivity for insertion of rNTPs over dNTPs into RNA varies from ∼30- to ∼5000-fold, depending upon the RNA polymerase and the rNTP/dNTP pair examined. The probability of dN incorporation during RNA synthesis is reduced by the fact that the concentrations of dNTPs in vivo are lower than the concentrations of rNTPs by orders-of-magnitude (3) (Table 1). There is also evidence that DNase I can cleave dN-containing RNA in vitro (39) that raises the possibility of a surveillance mechanism in vivo.
Table 1. Concentrations of ribonucleotides and 2′-deoxyribonucleotides in E. coli.
| (d)NNP | Wild-type1 (μM) | Wild-type2 (μM) | Δndk3 (μM) |
|---|---|---|---|
| ATP | 3560 | ||
| ADP | 116 | ||
| dATP | 181 | 205 | 110 |
| CTP | 325 | ||
| dCTP | 184 | 475 | 7455 |
| GTP | 1660 | ||
| GDP | 203 | ||
| dGTP | 92 | 95 | 440 |
| UTP | 667 | ||
| UDP | 54 | ||
| dTTP | 256 | 420 | 580 |
Here, we present the development and application of a method to quantify dN incorporation into any form of RNA from any organism. The platform combines alkaline hydrolysis of RNA to yield dNrN dinucleotides that are then quantified by chromatography-coupled tandem mass spectrometry (LC-MS/MS). Analysis of coding and non-coding RNAs from human, yeast and bacterial cells, and an RNA virus, reveals highly abundant dN incorporation that varies as a function of the identity of the polymerase, environmental stresses, and nucleotide pool balance. The results have implications for viral evolution and human disease.
MATERIALS AND METHODS
Cell and virus culture
CCRF-SB human B lymphoblasts (a gift from Dr Jianzhu Chen, Singapore-MIT Alliance for Research and Technology) were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin and 50 μg/ml streptomycin at 37°C and 5% CO2. The cells were collected by centrifugation at 350 xg for 10 min at 4°C. Saccharomyces cerevisiae strain W1588-4C (a gift from Dr Graham C. Walker, Massachusetts Institute of Technology) was grown exponentially in YPDA medium (1% yeast extract, 2% peptone, 2% dextrose, with 0.004% adenine hemisulfate) at 30°C with shaking at 200 rpm. The Δndk mutant strain of Escherichia coli W3110 was constructed by transduction using bacteriophage P1 vir. The mutant gene was obtained from the Keio Collection (40) in the form of a gene knockout carrying a kanamycin gene cartridge. The mutant gene was transduced into E. coli W3110 using a selection for kanamycin resistance. The resistance cartridge was then flipped out using flp sequences surrounding the kanamycin cartridge and flp recombinase supplied from the plasmid pCP20 (41). All E. coli strains were grown exponentially in LB broth at 37°C with shaking (220 rpm) to stationary phase. The cells were collected by centrifugation (4000 xg at 4°C) and washed once with ice-cold PBS. All cells were stored at −80°C until total RNA extraction. The DENV-2 strain TSV01 was prepared as described previously (42). Briefly, genome-length viral RNA (vRNA) was in vitro transcribed from full-length cDNA plasmid linearized by ClaI. The transcribed RNA is then electroporated into BHK-21 cells, the transfected cells were resuspended in 20 ml of DMEM medium and cultured at 30°C with 5% CO2 for 5 days to produce virus. The virus titer was measured by plaque assay. To purify virus particles, TSVOI was propagated in mosquito C6/36 cells. Briefly, C6/36 cells were infected with TSVOI at a multiplicity of infection (MOI) of 0.1 and incubated at 28°C for 4–5 days. Culture fluids were then harvested, virus particles were precipitated by 8% PEG8000 and purified through 10–30% potassium tartrate gradient. Purified viral particles were kept frozen at −80°C until use.
Methyl methanesulfonate treatment
Treatment of S. cerevisiae W1588-4C cells with 0.01% methyl methanesulfonate (MMS) was started when the O.D. of the cultures reached ∼0.17. After 3 h treatment, the cells were collected by centrifugation (4500 xg at 4°C) and washed once with ice-cold deionized water.
RNA extraction
The total RNA from CCRF-SB pellets was directly extracted with Trizol reagent (Life Technologies), according to the manufacturer's protocol. For yeast and E. coli, lysis was performed with lyticase (Sigma) and lysozyme (Fluka), respectively, before total RNA was extracted with Trizol reagent as described earlier. Dengue virus genomic RNA was extracted from purified viral particles using Trizol reagent (42). The poly(A)-tailed RNA in human and yeast cells was isolated from the total RNA using a Fasttrack MAG Maxi mRNA isolation kit (Life Technologies) following the manufacturer's protocol. All RNA samples were used immediately or stored at −80°C.
Size-exclusion chromatography
The size-exclusion chromatography (SEC) of cellular total RNA and viral RNA was performed with a Bio SEC-5 1000 Å (i.d. 7.8 mm; length 300 mm), a Bio SEC3 300 Å (i.d. 7.8 mm, length 300 mm) or a Bio SEC-5 2000 Å (i.d. 7.8 mm; length 300 mm) column (Agilent Technologies), with 100 mM ammonium acetate as the mobile phase (43) (Supplementary Figure S1A). Isocratic separations were performed at 0.5 ml/min under partially denaturing conditions at 60°C on an Agilent 1200 HPLC system. The preliminary separations of cellular total RNA were performed on a Bio SEC-5 1000 Å column. Further separations (up to 3 rounds) of large rRNA: 28S and 18S rRNA for CCRF-SB, 25S and 18S rRNA for yeast, and 23S and 16S rRNA for E. coli; and small rRNA: 5.8S and/or 5S rRNA, and tRNA were performed on a Bio SEC-5 1000 Å and Bio SEC3 300 Å column, respectively. Separation of DENV-2 vRNA genome was performed with a Bio SEC-5 2000 Å column.
Analysis of purified RNA
The SEC-purified cellular and viral RNAs were concentrated and desalted with Amicon Ultracel 10k MWCO spin columns (Millipore). The quality of each RNA species was subsequently assessed using an Agilent Bioanalyzer (Agilent Technologies). RNA 6000 Nano or Pico chips were used to evaluate large cellular rRNA, poly(A) RNA and vRNA (Supplementary Figures S2A, S3A), whereas small rRNA and tRNA were evaluated on Small RNA chips (Supplementary Figure S4A).
RNA hydrolysis
Isolated RNA (typically 5 μg) was incubated in 50 mM potassium hydroxide at 100°C for 40 min following a published protocol for quantitatively hydrolyzing RNA (44). The digestion mixture was then neutralized by addition of acetic acid. Subsequent dephosphorylation was carried out by addition of 20 μl of 100 mM Tris-HCl (pH 8.0) and 20 U of bacterial alkaline phosphatase (BAP; Invitrogen) at 37°C for 2 h. The enzymes were then removed by extraction with chloroform:isoamyl alcohol 24:1 (Fluka). The matrix in the aqueous layer was removed by solid-phase extraction with Strata-X polymeric reversed phase cartridges (Phenomenex), washing with 1 ml of deinoized water and subsequent elution with 3 ml of methanol. The eluate was concentrated and subjected to LC-MS/MS analysis. Using CCRF-SB 28S rRNA as substrate, the alkaline hydrolysis efficiency was assessed and BAP concentration optimized by quantifying canonical ribonucleosides with reversed-phase HPLC (Supplementary Figure S5A). Technical duplicates were performed.
LC-MS/MS analysis of dNrN
Using synthetic standards (Midland Certified Reagent Company, Texas, USA), we defined the retention times for the set of all 16 possible dNrN dinucleotides on a Thermo Hypersil GOLD aQ column (100 × 1 mm, 3 μm) mounted in an Agilent 1200 capillary HPLC system that was coupled to an Agilent 6460 triple quadrupole mass spectrometer. The elution was conducted at 25°C and a flow rate of 8 μl/min, with a gradient of 100% buffer A (0.1% formic acid in water) and 0% buffer B (0.1% formic acid in methanol) for 5 min, followed by 0% to 40% buffer B over a period of 40 min. The mass spectrometer was operated with an electrospray ionization source in positive mode with the following parameters: gas temperature, 300°C; gas flow, 10 l/min; nebulizer, 40 psi; sheath gas temperature, 200°C; sheath gas flow, 7 l/min; and capillary voltage, 4000 V. Dynamic multiple reaction monitoring (MRM) mode was used for detection of product ions derived from the precursor ions for all 16 dNrN dinucleotides (see Supplementary Figure S6A for the MS/MS spectra) and 4 canonical nucleosides, with all instrument parameters optimized for maximal sensitivity for dNrN dinucleotides only (retention time in min, precursor ion m/z, product ion m/z, fragmentor voltage, collision energy): dCrC, 29.3, 533, 112, 110 V, 29 V; dCrU, 30.6, 534, 112, 110 V, 9 V; dCrA, 31.2, 557, 136, 110 V, 26 V; dCrG, 30.1, 573, 152, 120 V, 35 V; dTrC, 35.8, 548, 112, 120 V, 13 V; dTrU, 35.4, 549, 97, 90 V, 33 V; dTrA, 35.6, 572, 136, 140 V, 26 V; dTrG, 34.3, 588, 152, 140 V, 35 V; dArC, 32.6, 557, 112, 110 V, 21 V; dArU, 33.1, 558, 136, 110 V, 13 V; dArA, 33.5, 581, 136, 110 V, 40 V; dArG, 31.7, 597, 152, 110 V, 35 V; dGrC, 32.6, 573, 112, 100 V, 29 V; dGrU, 33.8, 574, 152, 80 V, 5 V; dGrA, 33.0, 597, 136, 90 V, 33 V; dGrG, 32.2, 613, 152, 110 V, 29 V; rC, 9.9, 244, 112, 85 V, 10 V; rU, 15.2, 245, 113, 85 V, 10 V; rA, 23.2, 268, 136, 85 V, 10 V; rG, 24.9, 284, 152, 85 V, 10 V. Analysis of dNrN dinucleotides in human and E. coli RNA was performed on this instrument with the above settings, while an Agilent 6490 Triple Quad was used to analyze dNrN dinucleotides in yeast and DENV-2 RNA in MRM mode with the following parameters: gas temperature, 50°C; gas flow, 11 l/min; nebulizer, 20 psi; sheath gas temperature, 300°C; sheath gas flow, 12 l/min; fragmentor voltage, 380 V; and capillary voltage, 1800 V. A Thermo Hypersil GOLD aQ column (100 × 1 mm, 3 μm) coupled to an Agilent 1290 HPLC system and the mass spectrometer was used with the elution conducted at 25°C at a flow rate of 20 μl/min, with a gradient of 0% buffer B to 50% buffer B over a period of 25 min. The same molecular transitions were used for detection of all 16 possible dNrN dinucleotides and 4 canonical rNs, with all instrument parameters optimized for maximal sensitivity for the dNrN dinucleotides (retention time in min, collision energy): dCrC, 15.1, 32 V; dCrU, 15.3, 10 V; dCrA, 17.2, 30 V; dCrG, 16.6, 26 V; dTrC, 18.1, 24 V; dTrU, 17.8, 24 V; dTrA, 18.1, 32 V; dTrG, 17.4, 26 V; dArC, 16.7, 32 V; dArU, 16.7, 14 V; dArA, 17.2, 32 V; dArG, 16.6, 32 V; dGrC, 16.5, 32 V; dGrU, 17.0, 12 V; dGrA, 16.6, 32 V; dGrG, 16.3, 32 V; rC, 4.5, 10 V; rU, 5.3, 10 V; rA, 11.1, 10 V; rG, 11.8, 10 V.
RESULTS
Development of the alkaline hydrolysis LC-MS/MS method for dNrN analysis
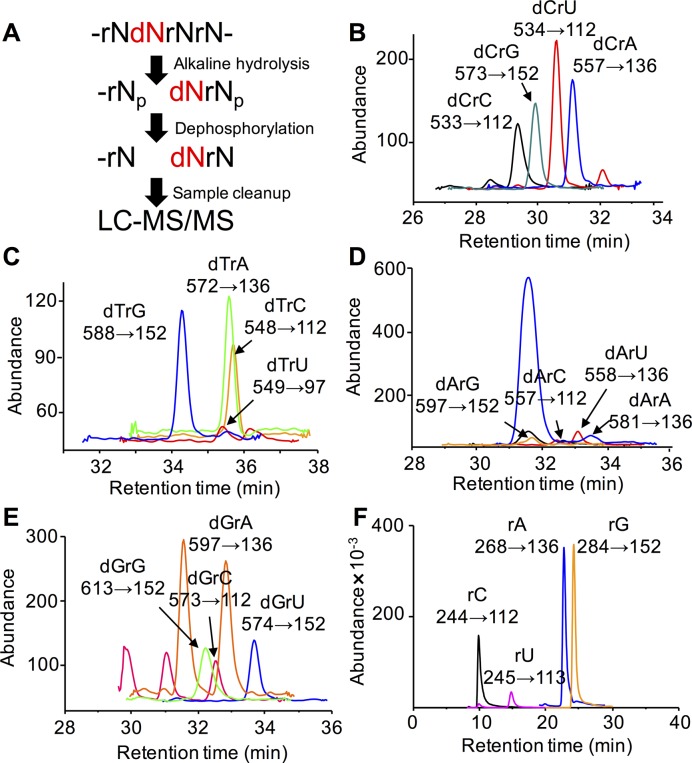
To quantitatively define the extent of dN misincorporation into RNA, we developed a highly specific, sensitive and quantitative technique that combines alkaline hydrolysis to release dNrN dinucleotides for LC-MS/MS quantification. The method relies on the well established role of the 2′-OH in the alkaline hydrolysis of RNA, with the lack of a 2′-OH in DNA rendering it resistant to alkaline hydrolysis (45,46). Alkaline hydrolysis of dN-containing RNA will thus yield a limit digest of dNrN dinucleotides with a 3′-terminal phosphate, along with 2′-O-methyl-rN-containing dinucleotides that are also alkali-resistant (47), and an abundance of canonical ribonucleoside 3′-monophosphates (Supplementary Figure S5A). To facilitate separation and detection in subsequent LC-MS/MS analysis, the 3′-phosphates were removed with BAP to afford a mixture of ribonucleosides and dNrN dinucleotides. The alkaline hydrolysis conditions used here have been shown to afford quantitative hydrolysis of RNA (44), which was borne out in analyses of human CCRF-SB 28S rRNA based on the full recovery of rNs after dephosphorylation (Supplementary Figure S5B and C). We also optimized BAP concentration for dephosphorylation of the nucleotides resulting from alkaline hydrolysis of RNA (Supplementary Figure S5C). Using synthetic dNrNs, we optimized the HPLC retention times (Figure 1) and collision-induced dissociation (CID) molecular transitions (Figure 1, Supplementary Figure S6) to achieve limits of detection for the 16 dNrNs ranging from 6 to 720 amol using an Agilent 6490 QQQ instrument and 0.2 to 11 fmol using an Agilent 6460 QQQ instrument (Supplementary Table S1). An analysis of E. coli W3110 23S rRNA shown in Supplementary Figure S7 demonstrates the identification of all dNrNs and rNs based on their specific CID molecular transitions and retention times relative to synthetic standards. Using external calibration curves to quantify individual dNrNs (Supplementary Figure S8), the LC-MS/MS method provided a highly precise approach to quantify misincorporation of dN into any RNA species and to assess stress-induced changes in the misincorporation.
Figure 1.
Analysis of dNrN dinucleotides by alkaline hydrolysis-LC-MS/MS. (A) Workflow for the method. (B-F) Illustration of the analytical method with E. coli W3110 23S rRNA, showing HPLC elution profiles and MS/MS transitions (X→Y) for (B) dCrN dinucleotides, (C) dTrN dinucleotides, (D) dArN dinucleotides, (E) dGrN dinucleotides and (F) canonical rNs.
The frequency of dNs in eukaryotic, prokaryotic and viral RNA
The first application of the dNrN analytical method involved quantification of dN in coding and non-coding RNAs from a range of organisms: human cells, S. cerevisiae, two E. coli strains and dengue virus. As context for interpreting the results of these analyses, we note in Table 2 that mRNA in eukaryotes is synthesized by RNA Pol II, with 3′ poly(A) tailing by poly(A) polymerase (PAP) and 3′ poly(U) tailing by poly(U) polymerase (PUP) or 3′ terminal uridylyl transferase (TUTase), while 28S (human), 25S rRNA (yeast), 18S and 5.8S rRNAs are synthesized by RNA Pol I, and 5S rRNA and tRNA are synthesized by RNA Pol III, with 5′ CCA tailing by tRNA nucleotidyltransferase (NTR). All RNAs in E. coli are synthesized by a single RNA polymerase (RNAP), while the dengue NS5 protein is the source of RNA polymerase activity in this virus (Table 2).
Table 2. Levels of dNs in cellular and viral RNA species1.
| Organism | RNA species | dC per 106 nt | dT per 106 nt | dA per 106 nt | dG per 106 nt | Total dN/106 nt | Polymerase |
|---|---|---|---|---|---|---|---|
| Human CCRF-SB | poly(A) RNA | 2.1 ± 0.6 | 0.7 ± 0.5 | 5.3 ± 1.0 | 8.9 ± 1.4 | 17 ± 3.5 | Pol II + PAP + TUTase |
| 28S rRNA | 0.9 ± 0.2 | 0.2 ± 0.06 | 0.3 ± 0.06 | 1.1 ± 0.3 | 2.6 ± 0.6 | Pol I | |
| 18S rRNA | 1.6 ± 0.6 | 0.2 ± 0.08 | 0.6 ± 0.1 | 1.0 ± 0.4 | 3.3 ± 1.3 | Pol I | |
| 5.8S rRNA | 2.8 ± 0.8 | 0.5 ± 0.3 | 0.8 ± 0.3 | 1.2 ± 0.5 | 5.3 ± 1.8 | Pol I | |
| 5S rRNA | 4.3 ± 0.6 | 0.6 ± 0.09 | 0.9 ± 0.3 | 5.3 ± 0.6 | 11 ± 2 | Pol III | |
| tRNA | 6.3 ± 1.4 | 1.0 ± 0.1 | 0.8 ± 0.3 | 2.4 ± 0.4 | 11 ± 2 | Pol III + NTR | |
| S. cerevisiae W1588-4C | poly(A) RNA | 12 ± 3 | 8.9 ± 2.1 | 2000 ± 950 | 1100 ± 440 | 3200 ± 1400 | Pol II + PAP + TUTase |
| 25S rRNA | 6.1 ± 2.8 | 16 ± 4 | 96 ± 18 | 132 ± 58 | 250 ± 83 | Pol I + PAP | |
| 18S rRNA | 8.9 ± 1.9 | 12 ± 0.8 | 85 ± 5 | 55 ± 12 | 161 ± 20 | Pol I + PAP | |
| 5.8S rRNA | 10 ± 3 | 4.6 ± 0.6 | 64 ± 5 | 87 ± 13 | 165 ± 21 | Pol I + PAP | |
| 5S rRNA | 19 ± 4 | 6.7 ± 1.4 | 55 ± 8 | 96 ± 23 | 176 ± 36 | Pol III + PAP | |
| tRNA | 43 ± 11 | 17 ± 1 | 90 ± 13 | 128 ± 38 | 278 ± 63 | Pol III + NTR + PAP | |
| S. cerevisiae W1588-4C + MMS | poly(A) RNA | 23 ± 5 | 15 ± 2 | 1,200 ± 260 | 697 ± 169 | 2000 ± 440 | Pol II + PAP + TUTase |
| 25S rRNA | 32 ± 6 | 21 ± 4 | 90 ± 6 | 132 ± 16 | 276 ± 33 | Pol I + PAP | |
| 18S rRNA | 26 ± 7 | 14 ± 2 | 100 ± 22 | 136 ± 16 | 276 ± 47 | Pol I + PAP | |
| 5.8S rRNA | 26 ± 6 | 7.2 ± 1.4 | 64 ± 7 | 71 ± 13 | 168 ± 28 | Pol I + PAP | |
| 5S rRNA | 47 ± 18 | 10 ± 2 | 50 ± 8 | 182 ± 79 | 290 ± 108 | Pol III + PAP | |
| tRNA | 157 ± 44 | 34 ± 5 | 116 ± 9 | 357 ± 57 | 663 ± 115 | Pol III + NTR + PAP | |
| E. coli DH5α | 23S rRNA | 31 ± 8 | 12 ± 2 | 9.9 ± 3.8 | 54 ± 13 | 108 ± 27 | RNAP |
| 16S rRNA | 19 ± 8 | 13 ± 5 | 11 ± 2 | 50 ± 23 | 93 ± 38 | RNAP | |
| 5S rRNA | 24 ± 17 | 6.8 ± 5.2 | 10 ± 7 | 36 ± 18 | 76 ± 47 | RNAP | |
| tRNA | 32 ± 12 | 22 ± 4 | 12 ± 6 | 36 ± 10 | 101 ± 32 | RNAP | |
| E. coli W3110 | 23S rRNA | 54 ± 5 | 6.3 ± 0.6 | 7.5 ± 0.7 | 44 ± 3 | 111 ± 9 | RNAP |
| 16S rRNA | 45 ± 8 | 5.0 ± 0.8 | 5.4 ± 1.3 | 37 ± 11 | 92 ± 21 | RNAP | |
| 5S rRNA | 65 ± 14 | 5.3 ± 1.4 | 6.4 ± 1.0 | 41 ± 7 | 118 ± 23 | RNAP | |
| tRNA | 65 ± 7 | 7.2 ± 1.2 | 4.1 ± 1.2 | 31 ± 4 | 107 ± 14 | RNAP | |
| E. coli W3110 Δndk | 23S rRNA | 203 ± 32 | 9.4 ± 0.7 | 5.1 ± 1.0 | 70 ± 19 | 288 ± 53 | RNAP |
| 16S rRNA | 167 ± 37 | 8.5 ± 0.9 | 5.4 ± 1.4 | 32 ± 4 | 213 ± 43 | RNAP | |
| 5S rRNA | 345 ± 51 | 8.5 ± 2.0 | 8.0 ± 1.2 | 55 ± 7 | 416 ± 61 | RNAP | |
| tRNA | 157 ± 46 | 9.9 ± 2.6 | 3.5 ± 0.4 | 26 ± 5 | 196 ± 54 | RNAP | |
| DENV-2 virus | RNA genome | 171 ± 36 | 39 ± 21 | 103 ± 31 | 455 ± 95 | 768 ± 183 | RdRP |
1Values (as dN per 106 nt) represent mean ± SD for three independent cultures for all cell lines, and for three technical replicates of a single culture for DENV-2 virus. PAP, poly(A) polymerase; TUTase, 3′ terminal uridylyl transferase; NTR, tRNA nucleotidyltransferase.
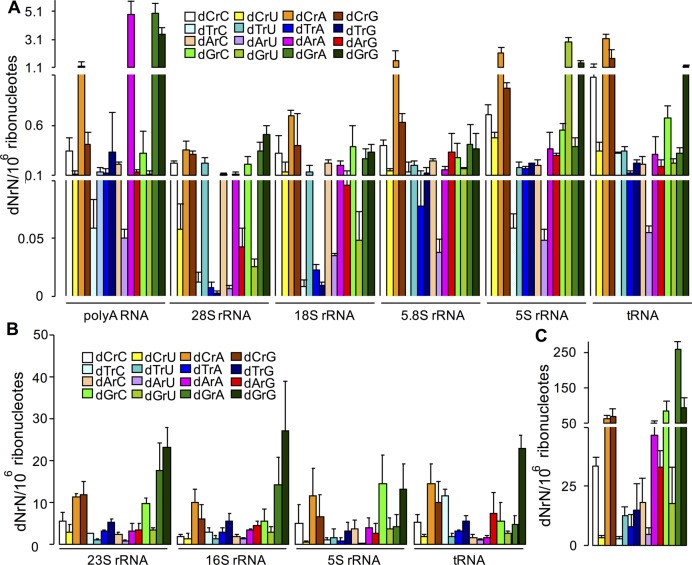
The first analysis involved poly(A)-tailed RNA (mainly mRNA), rRNA and tRNA from cultured human lymphoblasts CCRF-SB (Figure 2A). The levels of dNrNs were found to range from ∼0.01−1.6 per 106 rNs in the three rRNA species made by Pol I, with a similar patterns of dNrN frequency (Table 2). In contrast, the Pol III-mediated 5S rRNA and tRNAs contain misincorporated dNs at levels ∼2- to 4-fold higher than the Pol I rRNAs. Interestingly, the patterns of dNrN frequency are similar for Pol I and Pol II rRNAs. On the other hand, while poly(A) RNA, comprised mainly of mRNA, has a slightly higher dN misincorporation than 5S rRNA and tRNA, it contains substantially more dArA and dGrA than other RNAs. This suggests that the high levels of these two dinucleotides derive from the poly(A) tails in mRNA, which is consistent with the relatively low discrimination of dATP and ATP by human PAPγ (hPAPγ) (48). Moreover, poly(A) tails in some human transcripts contain a high percentage of guanine ribonucleotides, such as β-actin mRNA and truncated and polyadenylated 28S rRNA (49). However, we are not certain about the contribution of more recently discovered PUP and TUTase to the burden of misincorparated dN into pol(A) tails (50–54). Several interesting trends are apparent when comparing all of the human RNA species, including the consistently high levels of dCrA, the highly variable levels of dGrU (e.g. 5S rRNA versus 28S rRNA) and the consistently low levels of dArU (Figure 2).
Figure 2.
Quantitation of dN misincorporation in coding and non-coding RNAs. dNrN dinucleotides were quantified by alkaline hydrolysis LC-MS/MS in different RNA species. (A) Human CCRF-SB cells: poly(A) RNA, rRNA and tRNA. (B) E. coli DH5α rRNA and tRNA. (C) DENV-2 virus RNA genome. Values represent mean ± SD for three independent cultures for CCRF-SB and DH5α, and for three technical replicates of a single culture for DENV-2.
We next analyzed dN misincorporation into RNAs from S. cerevisiae and E. coli as well as the RNA genome of dengue virus type 2 (DENV-2), a member of the Flavivirus family. In S. cerevisiae, all rRNA and tRNA species contain similar levels of dNs (∼160−280 dN per 106 rNs), which is an order-of-magnitude lower than the dN levels in poly(A) RNA (Figure 3A; Table 2). These dN frequencies are generally one to two orders-of-magnitude higher than those in human RNA. Interestingly, all RNA species in S. cerevisiae share similar dNrN distribution patterns. For example, dArA and dGrA are enriched in all RNA species, especially in the poly(A) RNA (Figure 3A). This can be explained by several factors. First, S. cerevisiae PAP, like human PAP, can use 2′-dATP as a substrate, but more efficiently than human PAP (55,56). Second, a small percentage of some rRNAs in S. cerevisiae, including 25S rRNA, contain poly(A) tails (57) and both guanine and uracil have been detected in the poly(A) tails of S. cerevisiae mRNA and 25S rRNA (49).
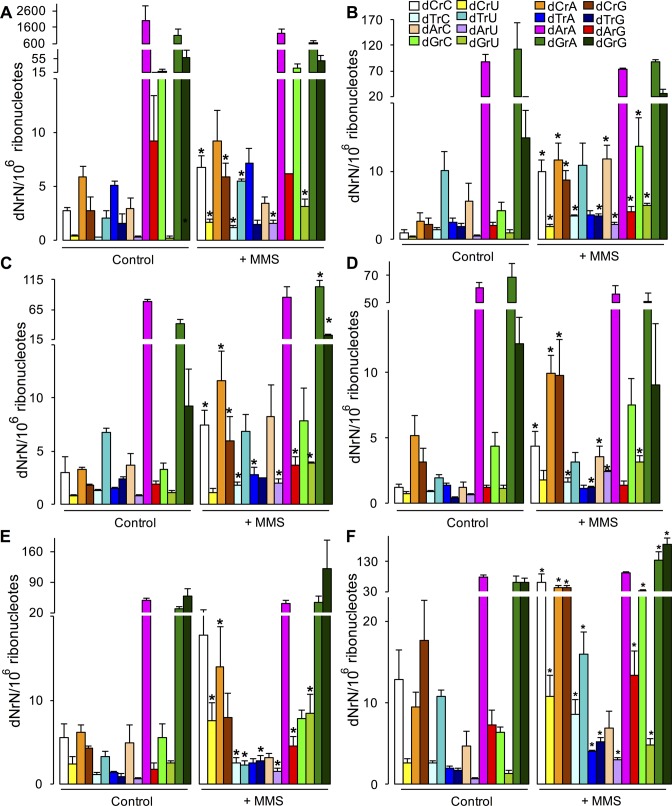
Figure 3.
Exposure to methyl methanesulfonate (MMS) increases dN misincorporation in RNA in Saccharomyces cerevisiae. dNrN dinucleotides were quantified by alkaline hydrolysis LC-MS/MS in different RNA species from yeast exposed to MMS. (A) poly(A) RNA. (B) 25S rRNA. (C) 18S rRNA. (D) 5.8S rRNA. (E) 5S rRNA. (G) tRNA. Values represent mean ± SD for three biological replicates. * P < 0.05, two-tailed unpaired Student's t test.
The dN frequency and patterns of dNrNs are similar in all rRNA and tRNA species in E. coli DH5α W3110 (Figures 2B and 4; Table 2). This is not surprising in light of the fact that all rRNAs and tRNAs are synthesized by a single RNAP. Further, the dN frequency in both E. coli strains is an order-of-magnitude higher than that in human CCRF-SB cells and is comparable to that in S. cerevisiae. The dN frequency in the RNA genome of DENV-2 virus is remarkably high at ∼800 per 106 rNs, with dG as the most abundant dN (Figure 2C; Table 2).
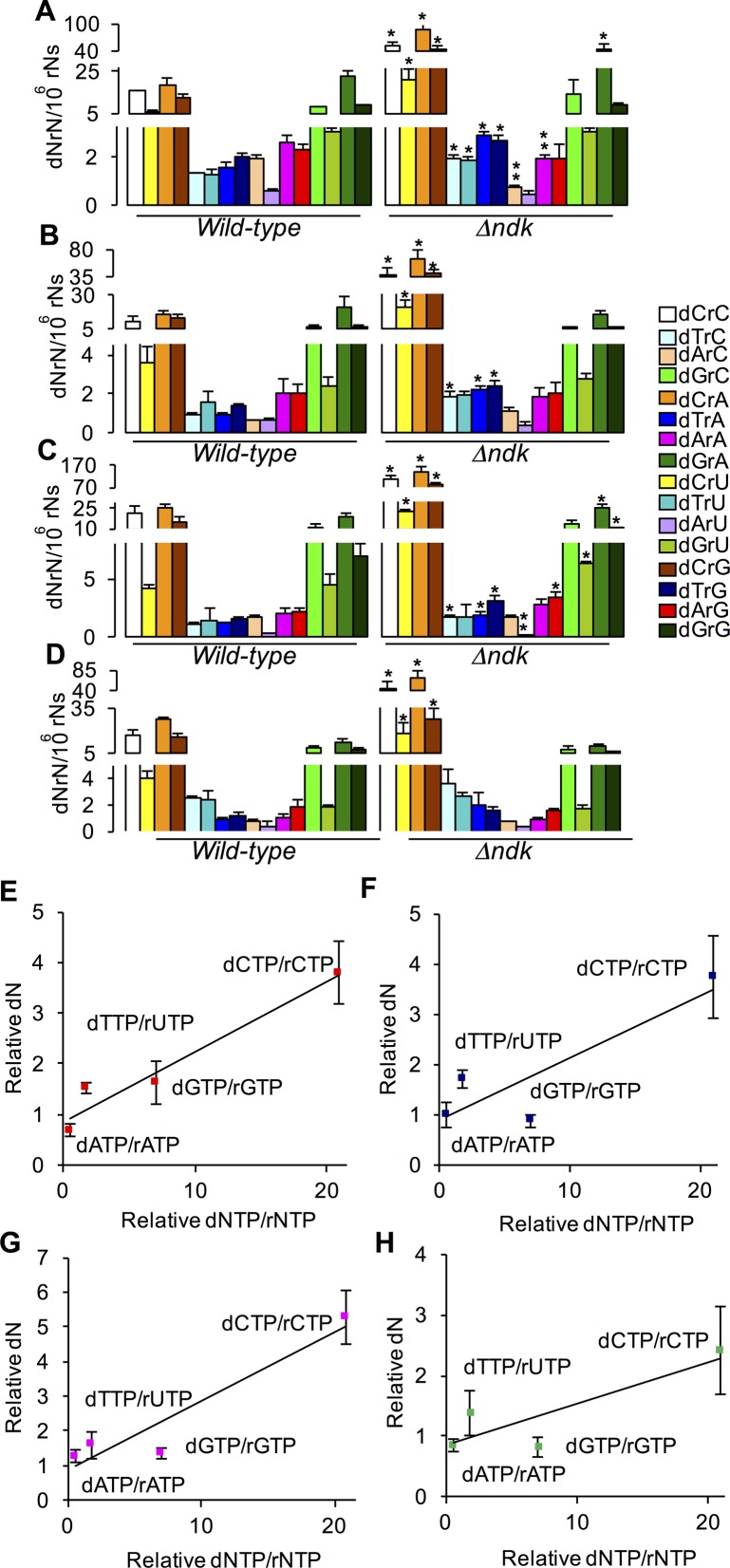
Figure 4.
dNrN levels and dNTP/rNTP ratios in wild-type and Δndk E. coli W3110. dNTP/rNTP ratios in WT and Δndk E. coli W3110 strains were taken from Lu et al. (33). (A−D) dNrN levels in (A) 23S rRNA, (B) 16S rRNA, (C) 5S rRNA and (D) tRNA. Values represent mean ± SD for three independent cultures. * and ** represent statistically significant increases and decreases, respectively; P < 0.05, two-tailed unpaired Student's t-test. (E−H) Correlation between misincorporated dN and nucleotide pool concentrations. ‘Relative dN’: ratio of dNrN in RNA from E. coli Δndk to that in WT E. coli; ‘Relative dNTP/rNTP’: ratio of dNTP/rNTP in E. coli Δndk to that in WT E. coli: (E) 23S rRNA, (F) 16S rRNA, (G) 5S rRNA and (H) tRNA.
Determinants of dN levels in RNA
Three major determinants of dN misincorporation into RNA are the type of RNA polymerase, the sequence context and the composition of the nucleotide pool. Regarding the role of the RNA polymerase, the data in Table 2 for variation in dN frequency in the different types of RNA and in different organisms correlates well with the in vitro discrimination efficiencies of bacterial, yeast and viral RNA polymerases (14,16,36,38). To investigate the role of nucleotide pool composition in dN misincorporation frequency, we quantified dNrN levels in an E. coli strain lacking NDK, a key enzyme in the control of cellular concentrations of (d)NTPs. The (d)NTP pool in ndk-deficient E. coli strains has a significant increase in the dCTP concentration and a smaller decrease in dATP concentration (32,33). As shown in Figure 4, while there were significant increases up to 1.7-fold in the levels of several dN misincorporated into the various RNA species, dC misincorporation increased by 2.4- to 5.3-fold in the bacteria lacking ndk (Figure 4; Supplementary Table S2). This is consistent with the greatly elevated level of dCTP in Δndk E. coli (33). There are parallel, but less consistent, decreases in dA content in the ndk mutant compared to the WT cells, possibly due to the decreases in dATP concentrations caused by loss of Ndk activity (32,33). To further assess the role of nucleotide pool composition in dN misincorporation, we calculated the ratio of dNTPs to rNTPs for each dNTP–rNTP pair from the various nucleoside triphosphate pool concentrations determined by Liu et al. in wild-type and Δndk E. coli strains (33) (Supplementary Table S2) and plotted these ratios against the ratio of misincorporated dN in the Δndk cells relative to wild-type cells. As shown in Figure 4E–H, there is a generally linear relationship between the level of dNTPs and the dN misincorporation frequency, which illustrates the importance of the nucleotide pool composition in misincorporation of dN into RNA.
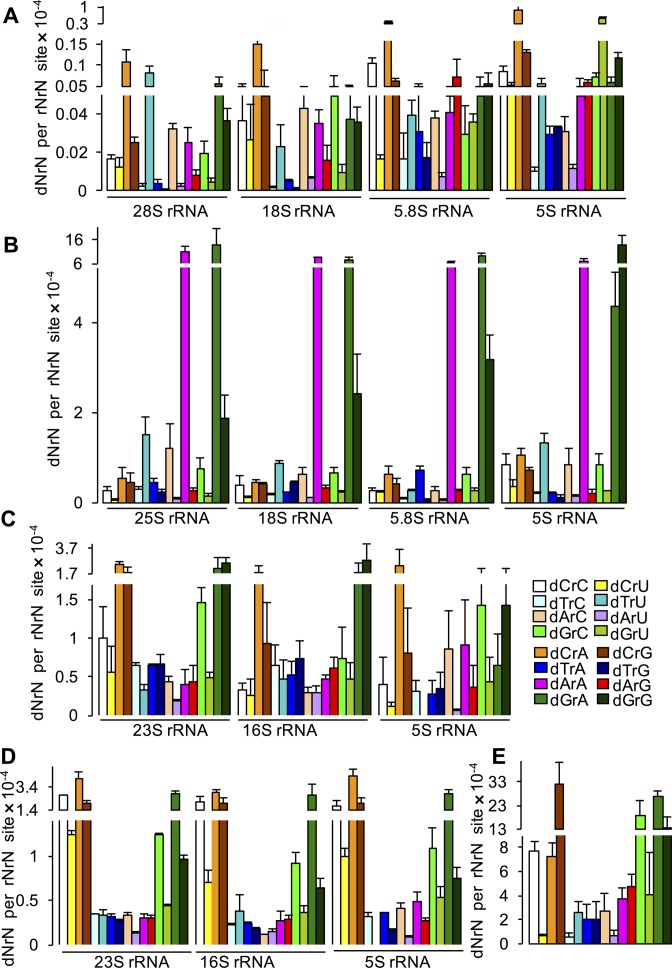
To explore the effects of sequence context on dN misincorporation, we quantified the effect of the identity of the rN positioned 3′ to the misinserted dN on the level of the dN. Here, we calculated the relative abundance of each rNrN sequence in rRNAs in human, S. cerevisiae, and E. coli K-12 cells and in the RNA genome of DENV-2 virus (Supplementary Figure S9) and used these values to normalize the level of the corresponding dNrN (Figure 5). The normalized dNrN values (dNrN/rNrN) correct the absolute abundance of the misincorporated dN for the frequency of the corresponding rNrN motif in each type of RNA. Supplementary Table S3B shows Z-scores (i.e. the degree of deviation from the mean) that quantify biases the frequencies of the normalized dNrN values in each RNA species. A survey of the data reveals that dC is among the most frequently misincorporated dNs when the 3′ nucleotide is rA in all types of rRNA in human cells and E. coli (Figure 5A, C and D; Supplementary Table S3B), while dCrG is the most abundant context for dC misincorporation in DENV-2 (Figure 5E; Supplementary Table S3B). Similarly, dG is misincorporated frequently next to rA in rRNAs in yeast and E. coli. Interestingly, the two E. coli strains show significant differences in nearest neighbor effects on the misincorporation, as illustrated by the differences in dGrA and dGrG (Figure 5C and D; Supplementary Table S3). In contrast, dArA, dGrA and dGrG dominate the purine misincorporation risk in yeast and dGrA in DENV-2 genome.
Figure 5.
dNrN frequencies normalized to rNrN occurrences in rRNAs and the DENV-2 RNA genome. The absolute abundance of dNrN was normalized by dividing by the frequency of the corresponding rNrN dinucleotide sequence in (A) human rRNAs (B) yeast rRNAs, (C) E. coli DH5α rRNAs, (D) E. coli W3110 rRNAs and (E) DENV-2 RNA genome. Values represent mean ± SD for either three biological or technical replicates. The values for dTrU in 5S rRNA are not shown in (C) and (D) because of the absence of rUrU in the reported sequence of E. coli 5S rRNA.
The next question concerned the relative roles of nucleotide pool concentration and local sequence context in determining polymerase-induced dN misincorporation. If dN misincorporation is driven only by nucleotide pool concentration, then the normalized data in Figure 5 should be identically proportional to the absolute dNrN levels in Figure 2. However, if some sequence contexts affect the polymerase misincorporation more than others, then there should be a discrepancy between the absolute dNrN value and the rNrN-normalized data in relation to other sequence contexts. This is illustrated by visual comparisons of absolute dNrN values in Figures 2 and 3 to the sequence-normalized values in Figure 5, which reveals sequence context effects that vary according to the organism and the RNA polymerase. For example, dCrG and dCrA have similar absolute levels in DENV-2 RNA (Figure 2C), while the normalized value for dCrG is 4- to 5-fold higher than that for dCrA (Figure 5D). This shows that DENV-2 NS5 polymerase is sensitive to sequence context in selectively incorporating dC next to rG. To address this problem quantitatively across all sequence contexts, we calculated Z-scores for the various dNrN values in each RNA species in Figures 2 and 3 and plotted these scores against the Z-scores for the associated dNrN/rNrN values. The resulting plots shown in Supplementary Figure S10 reveal that, by and large, the magnitude of the normalized misincorporation frequency correlates directly with the absolute level for a dNrN, which is consistent with a stronger influence of polymerase than sequence context on dN misincorporation. However, Supplementary Figure S10 also shows that there are several clear deviations from this relationship, such as the dCrG and dCrA context effects in DENV-2 and dGrG in human 28S rRNA, which point to a strong influence of sequence context on dN insertion. Overall, the results point to varying and significant influences by the RNA polymerase, the nucleotide pool composition and the sequence context as determinants of dN frequency in vivo.
Stress-induced misincorporation of dNs into RNA
The studies with Δndk E. coli revealed an important role for nucleotide pool imbalances in the frequency of dN misincorporation into RNA. We next sought to assess dN misincorporation in a eukaryotic system based on stress-induced changes in the nucleotide pool. Ribonucleotide reductases (RNR) play a central role in dNTP synthesis and altered RNR activity has been shown to cause large, mutagenic changes in dNTP levels in the nucleotide pool (31,42,58,59). Since Begley and coworkers have demonstrated significant up-regulation of RNR activity in yeast exposed to MMS (32,35), we sought to quantify the effects of MMS exposure on dN levels in S. cerevisiae RNA. As shown in Figure 3, MMS treatment caused a significant ∼2- to ∼3-fold increase in dN misincorporation in all RNA species examined, including poly(A) RNA, rRNAs and tRNA. dC and dT were the most consistently affected across all RNA species with increases also noted for some dA and dG misincorporations.
DISCUSSION
By combining alkaline hydrolysis with LC-MS/MS analysis, we were able to develop a highly specific and sensitive method to identify and quantify dN misincorporation in any type of RNA. Application of this method to human cells, yeast, bacteria and an RNA virus has provided the first quantitative insights into the in vivo contributions of nucleotide pool balance, RNA polymerase and local sequence context as determinants of dN misincorporation in RNA, and then how stress disrupts these factors to alter misincorporation. The frequency of dN misincorporation into RNA at ∼1 per 105 nucleotides in mammalian cells parallels other forms of RNA damage, such as 8-oxoguanosine (10−5/nt), 1,N6-ethenoadenosine (10−8/nt) and xanthosine (10−7/nt) (60). The results have implications for eukaryotic and prokaryotic pathophysiology and viral evolution.
This highly specific method utilizes the selective hydrolysis of rNs in RNA under alkaline conditions, which allows the dNs in RNA to be liberated as dNrN dinucleotides. Thus, the presence of contaminating DNA in the purified RNA species, which is inevitable and would obviously lead to artifacts in quantifying dNs in RNA in nuclease-based approaches, would not affect dN quantification using our approach. In the analysis of the released dNrNs, it is important to keep in mind that alkaline hydrolysis of RNA containing 2′-O-methylated ribonucleotides also produces dinucleotides containing the methylated ribonucleotide at the 5′-end, which protects the phosphodiester bond from cleavage the same way the lack of a 2′-OH does in dNrN. Previous attempts to quantify dN misincorporation in RNA have involved in vitro analyses using gel shift, in vitro transcription, or radiolabel incorporation assays that provide information on the rNTP/dNTP discrimination efficiencies for RNA polymerases (14,16,36–38). The major limitation of the alkaline hydrolysis/LC-MS/MS assay is the loss of other sequence context information for the misincorporated dN. One possible approach to mapping these dN in RNA is to use limited hydrolysis at alkaline pH to nick RNA at each ribonucleotide, with the misincorporated dN and 2′-O-methylated ribonucleotides protected from hydrolysis. Reverse transcription using a hybridized primer or a primer to a ligated linker, with subsequent sequencing analysis, would produce a one-nucleotide resolution ladder of RNA fragments lacking the protected site (47).
While in vitro studies predicted that the identity of the RNA polymerase and the relative concentrations of nucleotide pool components would affect the in vivo frequency and identity of misincorporated dNs in RNA (14,16,36–38), the role of RNA sequence context in dN insertion emerges here as a significant complicating factor in understanding the dN misincorporation phenomenon. The results of the in vivo analyses presented here establish that both polymerase and pool factors contribute to the dN misincorporation, with a surprisingly large range of misincorporation frequencies in different organisms (Table 2) and very significant biases in both the identity of the misincorporated dN and its location in terms of nearest neighbor. Indeed, the relative differences in dN frequencies among different RNA species in the human, yeast and bacterial cells are consistent with the discrimination efficiencies established in vitro for the different types of RNA polymerases in each organism (14,16,36–38,61). Similarly, our observation of a general increase in dN misincorporation frequency in moving from eukaryotic cells to bacteria to viruses is also consistent with species-specific RNA polymerase behaviors determined in vitro (14,16,36–38,61). As a matter awaiting further study, the molecular basis for biased dN incorporation due to sequence context may arise from both local primary or secondary RNA structural influences.
The high rate of misincorporation of dNs into the RNA genome of DENV-2 (Table 2) suggests a potential mechanism that contributes to the high frequency of mutations in RNA-based viruses. Mutation serves as an adaptive mechanism for RNA viruses during infections and can increase viral pathogenicity, with the magnitude of detrimental or lethal effects of the mutations mitigated by mechanisms that confer mutational robustness (i.e. the ability to buffer mutational effects on phenotype) (62). In general, RNA viruses have high mutation rates, with rates of substitution mutations ranging from 10−6 to 10−4 per nucleotide per cell infection (63) and ∼2% sequence variation among viruses in the same host during an infection(64). The main driver of this high mutation rate is likely the error-prone nature of the NS5 RNA-dependent RNA polymerase (65). While it is unclear to what extent the RNA-dependent RNA polymerase selects the wrong RTP (direct mutation) or misinserts a dNTP (indirect mutation), the exceptionally high dN frequency in the DENV-2 RNA genome—∼7 dN in ∼11 000 nt (∼10−3/nt)—likely contributes to polymerase infidelity, which is supported by recent structure-function studies (66). RNA-dependent RNA polymerases have been shown to copy DNA templates albeit with significantly lower efficiency and with evidence for a requirement of 2′-OH groups at positions near the transcription start site (67). While further study is needed to define the relative contributions of the sugar and base mispairing in viral mutagenesis, dNs in RNA may also contribute to viral evolution by mechanisms other than single-nucleotide substitution. It is well established that premature termination and pausing of the RNA polymerase or reverse transcriptase can trigger RNA recombination (68). Given that dN in an RNA template can cause pausing/premature termination during viral transcription, viral RNA genome replication and reverse transcription (15,25–29) and dN misincorporated into nascent RNA during viral RNA synthesis impedes further extension (15), it is possible that dNs in RNA can contribute to RNA recombination. Additionally, dN misincorporation in RNA may contribute to low genome GC content in RNA-based viruses (69). This is supported by the large bias (by 5.8-fold) in dC+dG incorporation over dT+dA incorporation in DENV-2 genome (Figure 5E). dN misincorporation in RNA may also contribute to the repression of CpG usage in RNA-based viruses (69), with dCrG as the most abundant context for dNrN errors and rCrG as the most under-represented dinucleotide in the DENV-2 genome (Figure 5E, Supplementary Figure S9D). dNs in RNA may thus play a role in the evolution of genome compression and genome novelty of RNA-based viruses (70).
As illustrated here with the E. coli Δndk mutant, the composition of the nucleotide pool plays a significant role in the risk of inserting dN into RNA, with effects likely to be felt in all organisms, including viruses. The stress imposed by viral infection of a human cell may be associated with shifts in nucleotide pool balance that favor dN incorporation into replicating viral RNA genomes. The nucleotide pool imbalance could compound the high error rates of viral RNA-dependent RNA polymerases to further increase the rate of viral mutagenesis. The 10-fold higher concentrations of rNTPs than dNTPs in the intracellular nucleotide pool (3) also likely account for the fact that the frequency of dN in RNA in yeast (this study) is lower than the misincorporation of rN into DNA observed by Kunkel et al. (2). Again, cell stress can alter the nucleotide pool balance and thus alter dN and rN misincorporations. The genes encoding RNR, a protein that is key to dNTP synthesis in all organisms and tightly regulates pool dNTP concentrations, are inducible at the level of transcription in all organisms, including E. coli, S. cerevisiae and humans, by agents that damage DNA, such as UV light and MMS, or by agents that block DNA synthesis (58). This is consistent with our observation that MMS treatment results in increased levels of dNs in coding and non-coding RNA species in S. cerevisiae (Figure 3).
Viral RNA-dependent RNA polymerases are far more error-prone than eukaryotic DNA-dependent RNA polymerases, likely due to the absence of several polymerase-specific mechanisms to control fidelity. For example, RNA PolII in yeast has a ‘trigger loop’ that contributes significantly to the discrimination between NTPs and dNTPs (13). Once incorporated into the nascent RNA, dNs often inhibit further extension of RNA transcripts in vitro for both cellular and viral RNA polymerases (14–16). However, successful elongation past dN leads to a relatively stable transcript. The nucleolytic proofreading activity of eukaryotic RNA polymerases (71) appears to be more sensitive to base pairing incompatibility rather than sugar identity, with RNA polymerases removing incorporated dNs with an incorrect base in vitro as efficiently as incorporated rNs with the same incorrect base (72). This suggests that incorporated dNs with the correct base may not be subject to removal by RNA polymerases and further that most, if not all, dNs present in cellular RNA have the correct base. Indeed, the level of base misincorporations in bacterial and eukaryotic RNA species is estimated to be around 10−5 per nt or lower (71), which is approximately one order-of-magnitude lower than the dN frequencies in various coding and noncoding RNA species in bacteria and yeast (Table 2).
The consequences of dN misincorporation into RNA involve translational errors in all organisms and mutations in viral RNA genomes. The presence of a single dN in mRNA can cause mis-translation in the form of truncation of the peptide at a frequency of 30% to 50% (10,64,66). The dN likely also causes miscoding and insertion of the incorrect amino acid, as do many other RNA modifications such as m5C (64), though the effect of dNs on translational recoding has not received significant attention. The potential for a link between dN misincorporation into RNA and human diseases is apparent in studies demonstrating markedly elevated (up to ∼2000-fold) dNTP concentrations in human immunodevelopment diseases caused by inherited deficiencies in enzymes responsible for the salvage or degradation of dNTPs (73,74). It has also been established that poly(A) tail length contributes to translational regulation and is implicated in a number of physiological and pathological processes (75). Given that incorporation of dNs into the poly(A) tails of RNA by PAP can inhibit further tail extension (55), it is possible that variation in poly(A) tail lengths may be driven by dN incorporation. For example, we observed higher dN levels in poly(A) RNA in yeast compared to humans, which correlated with the shorter mean poly(A) tail length in yeast than in human cells (76). Interestingly, recent studies showed that uridylation occurs on oligoadenylated mRNAs in yeast, Arabidopsis and human cells (50–52). TUTases selectively insert UTP over the other NTPs (53), however it remains unknown if 2′-dNTPs are substrates of TUTases although crystal structures suggest that they can discriminate ribose sugar against 2′-deoxyribose sugar (54).
Our results suggest that dNs in RNA are present at appreciable levels in all organisms, with strong dependence on the RNA polymerase, nucleotide pool balance and environmental stresses. The analytical method developed here will find utility in defining the relationship between misincorporation of dN in RNA and biological endpoints such as disease pathophysiology and viral evolution.
Supplementary Material
Acknowledgments
The authors thank Prof. Graham Walker and Prof. Jianzhu Chen from the Massachusetts Institute of Technology for generously sharing S. cerevisiae W1588-4C and human CCRF-SB cells, respectively.
Footnotes
Present addresses:
Yok Hian Chionh, TyChan, Pte. Ltd., Singapore 068898.
Megan E. McBee, TyChan, Pte. Ltd., Singapore 068898.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [ES022858]; Singapore-MIT Alliance for Research and Technology with a grant from the National Research Foundation of Singapore. Funding for open access charge: National Institutes of Health (NIH) [ES022858]; Singapore-MIT Alliance for Research and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Joyce C.M. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundstrom E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 4.Bahadur R.P., Zacharias M., Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindqvist M., Sarkar M., Winqvist A., Rozners E., Stromberg R., Graslund A. Optical spectroscopic study of the effects of a single deoxyribose substitution in a ribose backbone: implications in RNA-RNA interaction. Biochemistry. 2000;39:1693–1701. doi: 10.1021/bi992055n. [DOI] [PubMed] [Google Scholar]
- 6.Plavec J., Thibaudeau C., Chattopadhyaya J. How does the 2′-hydroxy group drive the pseudorotational equilibrium in nucleoside and nucleotide by the tuning of the 3′-gauche effect. J. Am. Chem. Soc. 1994;116:6558–6560. [Google Scholar]
- 7.Thibaudeau C., Plavec J., Garg N., Papchikhin A., Chattopadhyaya J. How does the electronegativity of the substituent dictate the strength of the gauche effect. J. Am. Chem. Soc. 1994;116:4038–4043. [Google Scholar]
- 8.Doudna J.A., Cate J.H. RNA structure: Crystal clear. Curr. Opin. Struct. Biol. 1997;7:310–316. doi: 10.1016/s0959-440x(97)80045-0. [DOI] [PubMed] [Google Scholar]
- 9.Fedor M.J., Williamson J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 10.Fahlman R.P., Olejniczak M., Uhlenbeck O.C. Quantitative analysis of deoxynucleotide substitutions in the codon-anticodon helix. J. Mol. Biol. 2006;355:887–892. doi: 10.1016/j.jmb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Pan T., Loria A., Zhong K. Probing of tertiary interactions in RNA: 2′-Hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gohara D.W., Crotty S., Arnold J.J., Yoder J.D., Andino R., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 2000;275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan C.D., Jin H., Zhang I.L., Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin B., Whelan S.P. Sensitivity of the polymerase of vesicular stomatitis virus to 2′ substitutions in the template and nucleotide triphosphate during initiation and elongation. J. Biol. Chem. 2014;289:9961–9969. doi: 10.1074/jbc.M113.542761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunelle J.L., Shaw J.J., Youngman E.M., Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl-tRNA substrate. RNA. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erlacher M.D., Lang K., Wotzel B., Rieder R., Micura R., Polacek N. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J. Am. Chem. Soc. 2006;128:4453–4459. doi: 10.1021/ja0588454. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg J.S., Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phelps S.S., Jerinic O., Joseph S. Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol. Cell. 2002;10:799–807. doi: 10.1016/s1097-2765(02)00686-x. [DOI] [PubMed] [Google Scholar]
- 21.Schmeing T.M., Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 22.Youngman E.M., He S.L., Nikstad L.J., Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol. Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Harger J.W., Meskauskas A., Dinman J.D. An 'integrated model' of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 24.Murphy R.M. Peptide aggregation in neurodegenerative disease. Annu. Rev. Biomed. Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- 25.Kim M.J., Zhong W., Hong Z., Kao C.C. Template nucleotide moieties required for de novo initiation of RNA synthesis by a recombinant viral RNA-dependent RNA polymerase. J. Virol. 2000;74:10312–10322. doi: 10.1128/jvi.74.22.10312-10322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjith-Kumar C.T., Gutshall L., Kim M.J., Sarisky R.T., Kao C.C. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 2002;76:12526–12536. doi: 10.1128/JVI.76.24.12526-12536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel R.W., Bellon L., Beigelman L., Kao C.C. Use of DNA, RNA and chimeric templates by a viral RNA-dependent RNA polymerase: Evolutionary implications for the transition from the RNA to the DNA world. J. Virol. 1999;73:6424–6429. doi: 10.1128/jvi.73.8.6424-6429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudima S.O., Kazantseva E.G., Kostyuk D.A., Shchaveleva I.L., Grishchenko O.I., Memelova L.V., Kochetkov S.N. Deoxyribonucleotide-containing RNAs: a novel class of templates for HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:4614–4618. doi: 10.1093/nar/25.22.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tayon R., Jr, Kim M.J., Kao C.C. Completion of RNA synthesis by viral RNA replicases. Nucleic Acids Res. 2001;29:3576–3582. doi: 10.1093/nar/29.17.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barr J.N., Fearns R. How RNA viruses maintain their genome integrity. J. Gen. Virol. 2010;91:1373–1387. doi: 10.1099/vir.0.020818-0. [DOI] [PubMed] [Google Scholar]
- 31.Buckstein M.H., He J., Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaaper R.M., Mathews C.K. Mutational consequences of dNTP pool imbalances in E. coli. DNA Repair (Amst) 2013;12:73–79. doi: 10.1016/j.dnarep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Q., Zhang X., Almaula N., Mathews C.K., Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 34.Milo R., Jorgensen P., Moran U., Weber G., Springer M. BioNumbers–the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacombe M.L., Milon L., Munier A., Mehus J.G., Lambeth D.O. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000;32:247–258. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y., Eckstein F., Padilla R., Sousa R. Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry. 1997;36:8231–8242. doi: 10.1021/bi962674l. [DOI] [PubMed] [Google Scholar]
- 37.Smidansky E.D., Arnold J.J., Reynolds S.L., Cameron C.E. Human mitochondrial RNA polymerase: evaluation of the single-nucleotide-addition cycle on synthetic RNA/DNA scaffolds. Biochemistry. 2011;50:5016–5032. doi: 10.1021/bi200350d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svetlov V., Vassylyev D.G., Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paddock G.V., Heindell H.C., Salser W. Deoxysubstitution in RNA by RNA polymerase in vitro: A new approach to nucleotide sequence determinations. Proc. Natl. Acad. Sci. U.S.A. 1974;71:5017–5021. doi: 10.1073/pnas.71.12.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong H., Chang D.C., Hua M.H., Lim S.P., Chionh Y.H., Hia F., Lee Y.H., Kukkaro P., Lok S.M., Dedon P.C., et al. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012;8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chionh Y.H., Ho C.H., Pruksakorn D., Ramesh Babu I., Ng C.S., Hia F., McBee M.E., Su D., Pang Y.L., Gu C., et al. A multidimensional platform for the purification of non-coding RNA species. Nucleic Acids Res. 2013;41:e168. doi: 10.1093/nar/gkt668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bock R.M. Alkaline hydrolysis of RNA. Methods Enzymol. 1967;12:224–228. [Google Scholar]
- 45.Berns K.I., Thomas C.A. A study of single polynucleotide chains derived from T2 and T4 bacteriophage. J. Mol. Biol. 1961;3:289–300. [Google Scholar]
- 46.Vinograd J., Morris J., Davidson N., Dove W.F. The buoyant behavior of viral and bacterial DNA in alkaline CsCl. Proc. Natl. Acad. Sci. U.S.A. 1963;49:12–17. doi: 10.1073/pnas.49.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maden B.E. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 48.Kyriakopoulou C.B., Nordvarg H., Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J. Biol. Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- 49.Slomovic S., Fremder E., Staals R.H., Pruijn G.J., Schuster G. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc. Natl. Acad. Sci. U.S.A. 2009;107:7407–7412. doi: 10.1073/pnas.0910621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159:1365–1376. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sement F.M., Ferrier E., Zuber H., Merret R., Alioua M., Deragon J.M., Bousquet-Antonelli C., Lange H., Gagliardi D. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013;41:7115–7127. doi: 10.1093/nar/gkt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rissland O.S., Mikulasova A., Norbury C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stagno J., Aphasizheva I., Aphasizhev R., Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin G., Doublie S., Keller W. Determinants of substrate specificity in RNA-dependent nucleotidyl transferases. Biochim. Biophys. Acta. 2008;1779:206–216. doi: 10.1016/j.bbagrm.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhelkovsky A., Helmling S., Bohm A., Moore C. Mutations in the middle domain of yeast poly(A) polymerase affect interactions with RNA but not ATP. RNA. 2004;10:558–564. doi: 10.1261/rna.5238704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhelkovsky A., Helmling S., Moore C. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuai L., Fang F., Butler J.S., Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elledge S.J., Zhou Z., Allen J.B., Navas T.A. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 59.Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 60.Prestwich E.G., Mangerich A., Pang B., McFaline J.L., Lonkar P., Sullivan M.R., Trudel L.J., Taghizedeh K., Dedon P.C. Increased levels of inosine in a mouse model of inflammation. Chem. Res. Toxicol. 2013;26:538–546. doi: 10.1021/tx300473n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert M., Lazzarini R.A. In vitro transcription of vesicular stomatitis virus. Incorporation of deoxyguanosine and deoxycytidine, and formation of deoxyguanosine caps. J. Biol. Chem. 1982;257:2968–2973. [PubMed] [Google Scholar]
- 62.Lauring A.S., Frydman J., Andino R. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 2013;11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanjuan R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parameswaran P., Charlebois P., Tellez Y., Nunez A., Ryan E.M., Malboeuf C.M., Levin J.Z., Lennon N.J., Balmaseda A., Harris E., et al. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J. Virol. 2012;86:8546–8558. doi: 10.1128/JVI.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Ann. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 66.Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., Peersen O.B. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015;89:275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang C., Mammen M.P., Jr, Chinnawirotpisan P., Klungthong C., Rodpradit P., Nisalak A., Vaughn D.W., Nimmannitya S., Kalayanarooj S., Holmes E.C. Structure and age of genetic diversity of dengue virus type 2 in Thailand. J. Gen. Virol. 2006;87:873–883. doi: 10.1099/vir.0.81486-0. [DOI] [PubMed] [Google Scholar]
- 68.Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng X., Virk N., Chen W., Ji S., Sun Y., Wu X. CpG usage in RNA viruses: data and hypotheses. PLoS One. 2013;8:e74109. doi: 10.1371/journal.pone.0074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belshaw R., Pybus O.G., Rambaut A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007;17:1496–1504. doi: 10.1101/gr.6305707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sydow J.F., Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 72.Zenkin N., Yuzenkova Y., Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 73.Kretschmer S., Wolf C., Konig N., Staroske W., Guck J., Hausler M., Luksch H., Nguyen L.A., Kim B., Alexopoulou D., et al. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann. Rheum. Dis. 2014;74:e17. doi: 10.1136/annrheumdis-2013-204845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin D.W., Jr, Gelfand E.W. Biochemistry of diseases of immunodevelopment. Annu. Rev. Biochem. 1981;50:845–877. doi: 10.1146/annurev.bi.50.070181.004213. [DOI] [PubMed] [Google Scholar]
- 75.Weill L., Belloc E., Bava F.A., Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 76.Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.