Abstract
Despite many methodological advances that have facilitated investigation of Mycobacterium tuberculosis pathogenesis, analysis of essential gene function in this slow-growing pathogen remains difficult. Here, we describe an optimized CRISPR-based method to inhibit expression of essential genes based on the inducible expression of an enzymatically inactive Cas9 protein together with gene-specific guide RNAs (CRISPR interference). Using this system to target several essential genes of M. tuberculosis, we achieved marked inhibition of gene expression resulting in growth inhibition, changes in susceptibility to small molecule inhibitors and disruption of normal cell morphology. Analysis of expression of genes containing sequences similar to those targeted by individual guide RNAs did not reveal significant off-target effects. Advantages of this approach include the ability to compare inhibited gene expression to native levels of expression, lack of the need to alter the M. tuberculosis chromosome, the potential to titrate the extent of transcription inhibition, and the ability to avoid off-target effects. Based on the consistent inhibition of transcription and the simple cloning strategy described in this work, CRISPR interference provides an efficient approach to investigate essential gene function that may be particularly useful in characterizing genes of unknown function and potential targets for novel small molecule inhibitors.
INTRODUCTION
CRISPR-Cas9 based methods for gene editing in eukaryotic cells have advanced rapidly since the first functional and mechanistic insights into this programmable endonuclease-mediated bacterial immunity system (1–3). Though CRISPR biotechnology derives from bacterial systems for degrading foreign DNA, the development of CRISPR-based systems for genome editing and control of gene expression in bacteria has been relatively limited (1,4,5).
In mycobacteria, several well-developed methods allow for deletion of non-essential genes (6–9). The study of essential genes, which comprise approximately 20% of theMycobacterium tuberculosis genome and which encode the proteins targeted by all current anti-tuberculars, has been more challenging. Protein depletion systems have been used, but these require the addition of tag-encoding sequences to the 3′ end of coding sequences in the M. tuberculosis chromosome (10,11). In addition to the time required to add the sequences encoding these tags to genes of interest, these insertions may alter the expression of downstream genes in the same operon as the targeted gene and the tag may interfere with the function of the protein, even in the absence of degradation of the targeted protein. Further, substantial variation in the extent of depletion has been observed with degradation-tag protein depletion systems (10).
An alternative approach, genetic depletion has also been used with some success, with regulated expression of the gene of interest from an inducible promoter in a strain in which native expression has been abrogated, typically by deletion. This approach, however, requires construction of a merodiploid strain. A major limitation of this approach is that achieving physiologic levels of the desired mRNA can be challenging with the inducible promoter systems available for use in M. tuberculosis (12–14).
CRISPR interference (CRISPRi) (15) has the potential to overcome some of these limitations to allow more facile analysis of essential genes in M. tuberculosis. This approach incorporates expression of a small guide RNA (sgRNA) to target the gene of interest in a bacterial cell in which a catalytically inactive Cas9 protein (dCas9) is expressed. The sgRNA binds specifically to the target gene via a short (12–20 nucleotide (nt)) sequence that is homologous to a sequence in the gene, and recruits dCas9 to the site via the ‘handle’ sequence. The result is interference with either transcription initiation or elongation, depending on the target site chosen, leading to lower levels of RNA of the gene targeted (15). An initial report of CRISPRi in mycobacteria was recently published, demonstrating that this approach can work (16). Although this report provided some data demonstrating CRISPRi in M. tuberculosis, most of the data characterized CRISPRi activity in the non-pathogenic rapid grower Mycobacterium smegmatis.
In this work, we describe an optimized system for CRISPR-dCas9-mediated RNA interference in M. tuberculosis, including (i) a vector for expression of dCas9 at levels that do not interfere with bacterial growth but are sufficient to achieve consistent, physiologically relevant inhibition of gene expression, and (ii) a vector that allows simple directional cloning of annealed primers that contain the appropriate sequence for targeting the gene of interest, and (iii) expression of dcas9 and sgRNAs from inducible Tet repressor (TetR)-regulated promoters that can be titrated to achieve a range of inhibition of expression of target gene. We demonstrate potent and sustained knockdown of expression of several essential M. tuberculosis genes, and the ability to modulate the extent of transcription inhibition. We illustrate the value of this approach in several types of phenotypic analysis to gain insight into essential gene function in M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains, culture conditions and growth curves
Bacterial strains used in this study were Escherichia coli DH5α, M. smegmatis mc2-155 and M. tuberculosis H37Rv. Escherichia coli DH5α was used for cloning and vector construction and was cultured in Luria–Bertani medium supplemented with antibiotics when appropriate (50 μg/ml kanamycin or 200 μg/ml hygromycin). Mycobacterium smegmatis and M. tuberculosis were cultured in Middlebrook 7H9 broth supplemented with 0.5% albumin, 0.2% dextrose, 0.085% NaCl, 0.2% glycerol and 0.05% Tween 80 (M-ADN-Tw), and with antibiotics when appropriate (25 μg/ml kanamycin or 50 μg/ml hygromycin). Mycobacteria were grown at 37°C with shaking at 120 rpm. For induction of sgRNA and dcas9 expression, M. smegmatis and M. tuberculosis cultures were supplemented with anhydrotetracycline (aTc) to achieve a final concentration of 200 ng/ml. Addition of aTc was repeated every 48 h to maintain induction of dcas9 and sgRNAs for experiments that extended beyond 48 h.
To monitor growth with and without induction of CRISPRi, M. tuberculosis cells containing the integrated dcas9-expressing vector plus the replicating vector expressing a gene-specific sgRNA or a control sequence that is not homologous to any M. tuberculosis sequence, were inoculated from log phase cultures into M-ADN-Tw medium to a theoretical OD600 of 0.001. Cultures were incubated at 37°C with shaking at 120 rpm. When the OD600 reached 0.1–0.2 the cultures were split into equal volumes and further incubated with or without the addition of aTc to induce expression of dcas9 and the sgRNA. aTc was added to a final concentration of 200 ng/ml every 48 h and cultures were grown for 4–6 days.
Construction of vectors for regulated expression of dcas9 and sgRNAs, and selection of sgRNA protospacer sequences
pRH2502, a vector expressing an inactive version of Streptococcus pyogenes cas9, was constructed by assembling two DNA blocks in which the cas9 DNA sequence had been optimized for expression in M. tuberculosis (GenScript) (Supplementary Figure S1). Mutations to eliminate Cas9 enzymatic activity (dcas9) were incorporated into this sequence at codon 10 (Asp to Ala) and codon 820 (His to Ala). The assembled dcas9 DNA was cloned into a mycobacterial integrating vector to obtain pRH2502, in which dcas9 is expressed from a TetR-regulated uvtetO promoter (Supplementary Figure S2) (17,18).
For expression of sgRNAs, we constructed a vector in which the sgRNA is expressed from a TetR-regulated smyc promoter (Pmyc1tetO) (pRH2521) (18) (Supplementary Figure S2). To allow simple, directional cloning of any sequence into this vector we used a strategy similar to that of Hwang et al (19), by incorporating two Type IIS restriction enzyme (BbsI) sites into these vectors. When cut by BbsI, two different 4 nt overhangs are generated in the vector and the BbsI sites are eliminated (Supplementary Figure S3). To express sgRNAs that target specific genes in M. tuberculosis, two complementary oligonucleotides containing 20 bases homologous to the target sequence plus 4 bases at the 5′ end of each oligonucleotide that are complementary to the overhangs that result from BbsI digestion of the vectors, were annealed, phosphorylated and cloned into the BbsI-digested vector (Supplementary Figure S3). All oligonucleotides were designed so that the expressed sgRNA contains a 20 bp sequence that is complementary to the non-template strand of the target gene. These vectors were designed so that the transcription start site is the first base of the complementary sequence. In each case the chromosomal sequence that is targeted directly follows the sequence CCX on the non-template strand, i.e. the reverse complement of the NGG protospacer adjacent motif (PAM) sequence. This vector also expresses a codon-optimized tetR expressed from the mycobacterial imyc promoter (13,18). These vectors were transformed into M. tuberculosis cells into which pRH2502 (for dcas9 expression) had been previously introduced.
To select 20 nt sequences in genes of interest as protospacers to incorporate into sgRNAs, we first identified PAMs (NGG) in the promoter region, 5′ untranslated region (UTR) and coding sequence of these genes. We then searched the M. tuberculosis genome sequence to determine whether the identified sequences were unique. The oligonucleotides used to generate sgRNA constructs to target-specific genes are shown in Supplementary Table S2.
RNA Isolation, 5′-RACE and RT-qPCR
To identify transcription start sites (TSS) of M. tuberculosis genes for which the TSS was not known, we performed 5′-rapid amplification of cDNA ends (5′-RACE) as previously described (20). For analysis of expression of dcas9 and repression of genes of interest targeted by sgRNAs, strains were grown with or without aTc as described above. Samples from M. tuberculosis cultures were taken every 24 h for isolation of RNA, cells were harvested by centrifugation, re-suspended in TRI Reagent (Molecular Resource Center) and mechanically disrupted with 0.1 mm Zirconia beads in a MagNA Lyser instrument (Roche) by agitating the samples 4–6 times at 7000 rpm for 45 s. RNA was extracted, purified and treated with Turbo DNAse (Thermo Fisher) twice (20). Quantity and quality of RNA was determined by measuring absorbance at 260 and 280 nm using a Nanodrop instrument (Thermo Fisher) and the absence of significant contaminating genomic DNA was confirmed by qPCR of the RNA without reverse transcription.
Reverse transcription (RT) and quantitative PCR of cDNA (RT-qPCR) were performed using the Quanta Biosciences qScript cDNA synthesis kit and the PerfeCTaSYBR green Supermix, respectively. qPCR was performed on an Applied Biosystems 7300 real time PCR system with Seqeunce Detection Software version v1.4.0, as previously described (20). The RT reaction was performed with 100 ng RNA isolated and purified as described above in a final reaction volume of 20 μl. Integrity of each RNA sample was determined using the 3′:5′ method in which qPCR of each cDNA was performed using two primer pairs, one of which amplifies a sequence from the 5′ end of the reference gene sigA and the other from the 3′ end of the gene (21). All 3′:5′ ratios exceeded 0.2, consistent with intact non-degraded RNA. Primers for RT-qPCR were designed using NCBI primer blast online tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast) to generate amplicons of 130–150 nt (Supplementary Table S2). Amplification efficiency of all M. tuberculosis primer pairs was determined to be >95% based on the slope of a standard curve of serial dilutions of cDNA. Data were analyzed using the ΔΔCT method with the M. tuberculosis reference gene sigA as the control (22). Biological duplicate samples were analyzed for each target sequence. Significance of the differences between the expression of uninduced versus aTc-induced cultures were determined at each time point using an unpaired t-test (Prism 5.0, GraphPad Software).
Western blotting
Mycobacterium tuberculosis broth cultures were harvested, washed with PBS plus 0.05% Tween 80, resuspended in TRI Reagent (Molecular Resource Center) and mechanically disrupted as described above. Proteins were extracted following the manufacturer's protocol and solubilized in 9.5 M urea/2% CHAPS, pH 9.1 (23,24). For western blotting, 10 μg of protein was resolved by SDS-PAGE followed by transfer to a PVDF membrane, which was probed with primary rabbit polyclonal sera against PknB (1:3000) followed by anti-rabbit secondary antibody (1:10 000) (Cell Signaling Technology). The blot was incubated with LumiGLO chemiluminescent substrate (Cell Signaling Technology) and membrane-bound antibody was detected using a Kodak Image Station (Carestream Health).
Antibiotic susceptibility determination
Mycobacterium tuberculosis H37Rv expressing dcas9 and either the control sgRNA or sgRNAs targeting inhA or dfrA were tested for drug susceptibility using the Microplate Alamar Blue Assay (MABA) (25). 100 μl of 7H9 medium containing 2X the desired concentration of the specific drug (isoniazid or methotrexate) was added to wells of a 96-well plate. M. tuberculosis strains were grown at 37°C with shaking (120 rpm) to an OD600 between 0.6 and 0.8 and then diluted to a theoretical OD600 of 0.001 in M-ADN-Tw culture medium. 100 μl of this diluted M. tuberculosis was added to each well. To obtain CRISPRi-mediated repression of gene expression aTc was added to a final concentration of 200 ng/ml every 48 h. Plates were incubated at 37°C for 6 days, at which point 20 μl of Alamar Blue (Invitrogen) + 12.5 μl of 20% Tween 80 were added to each well. Plates were incubated at 37°C for an additional 18–24 h and then read in a fluorescent plate reader (Ex 540 nm/Em 595 nm) (Tecan). The IC50 was defined as the lowest drug concentration that reduced fluorescence by 50% compared to control wells with no inhibitor.
Microscopy
To examine effects on cell morphology resulting from CRISPRi of select genes, cells were grown in M-ADN-Tw medium, harvested, washed in PBS with 0.05% Tween 80, and fixed in 4% paraformaldehyde in PBS for 2 h. Fixed cells were pelleted, washed and resuspended in PBS plus 0.05% Tween 80. Since M. tuberculosis cells tend to clump together, the cell suspension was briefly sonicated in a Bioruptor UCD-200 sonication water bath (Diagenode, Belgium) for 10 s at medium setting. 5–10 μl of the cell suspension was spotted on a 1% agarose coated glass slide to immobilize the cells. A coverslip was placed on the slide, and the cells were imaged immediately. The cell morphology was visualized using a Nikon Eclipse TE2000-E inverted microscope fitted with a 100x Plan Achromatic phase contrast oil-immersion objective with a 1.25 numerical aperture. Photographs were collected with a Hamamatsu ORCA-AG CCD camera, using IPLab imaging software. Cell length and width were measured by using ObjectJ, an ImageJ plugin, according to instructions provided on the ObjectJ website (https://sils.fnwi.uva.nl/bcb/objectj/). The length of the FtsZ-inhibited branched bacteria was determined by measuring the distance between the tips of the two longest branches. The length of the filamentous or rod-shaped cells was determined by measuring the distance between the cell poles. Cell length and width in ftsZ+25 sgRNA containing cells with and without aTc induction were compared using an unpaired t-test (Prism 5.0, GraphPad Software).
RESULTS
Vectors for CRISPRi in mycobacteria
We obtained custom-synthesized DNA (GenScript) encoding cas9 of Streptococcus pyogenes (39.1% GC) that was codon-optimized for expression in M. tuberculosis (65.6% GC) (26) (Supplementary Figure S1). Two mutations, Asp10Ala and His820Ala, were incorporated into this sequence to express a catalytically inactive dCas9 protein (15). This codon-optimized dcas9 was cloned under control of the aTc-inducible uv15tetO promoter in an integrating mycobacterial vector (17,18) (Supplementary Figure S2).
For expression of small guide RNAs (sgRNAs), sequences containing all but the gene-specific targeting sequences of the sgRNA were cloned into a mycobacterial replicating vector that expresses a TetR protein optimized for mycobacteria (13) in which expression of the sgRNA was driven from the TetR-regulated Pmyc1tetO promoter (Supplementary Figure S2) (18). To allow simple directional cloning of any gene-targeting sequence into this vector we used a strategy similar to that of Hwang et al. (19), incorporating two Type IIS restriction enzyme (BbsI) sites into the vectors. To obtain sgRNAs that target specific sequences in the mycobacterial chromosome, two complementary oligonucleotides corresponding to the targeted sequence plus sequences complementary to the overhangs that result from BbsI digestion of the vectors, were annealed, phosphorylated and cloned into the BbsI-digested vector. The vector is designed so that transcription of the sgRNA starts at the first nt of the sequence homologous to the sequence in the target gene (Supplementary Figure S3).
Testing CRISPRi in Mycobacterium smegmatis
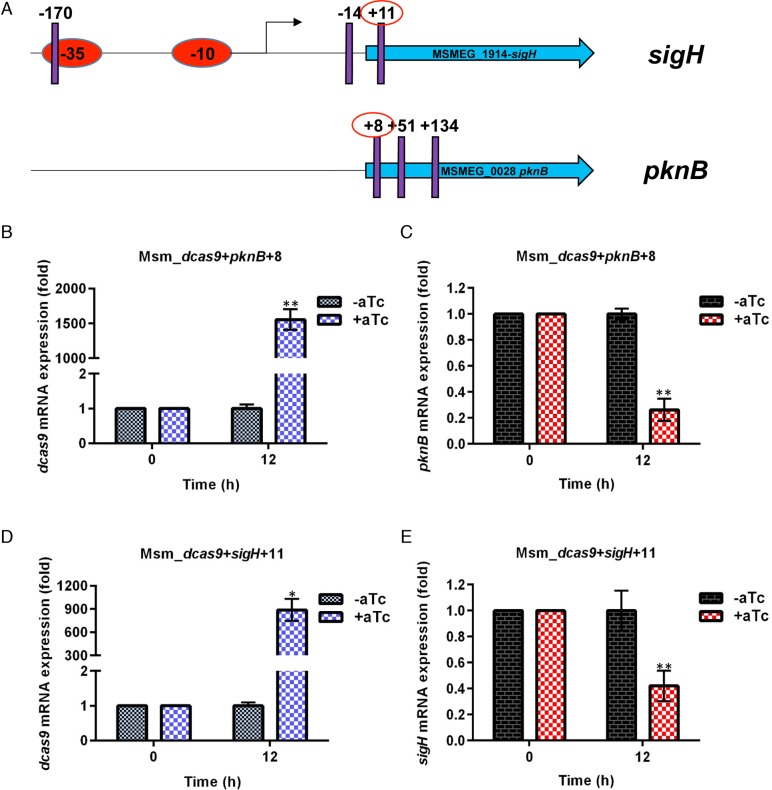
We first tested the CRISPRi vectors for their ability to inhibit gene expression in the rapid-growing non-pathogenic species M. smegmatis. These initial experiments targeted genes encoding the alternative sigma factor sigH, which regulates the expression of a large number of genes in response to oxidative and other stresses (20,27,28), and the essential Ser/Thr protein kinase pknB, which regulates cell shape and cell wall synthesis (29–31). As shown in Figure 1, for both genes we chose 3 target sequences. For sigH, which is the first gene in its operon, these included a site in the -35 element of the promoter, a site in the 5′-UTR between the transcription start site and the initiation codon and a sequence in the 5′ end of the coding sequence. For pknB, which is the last gene in a 5-gene operon, we targeted three sites in the coding sequence. We obtained extremely high levels of dcas9 in the presence of the inducer aTc in each case. Using aTc-induced expression of these sgRNAs to target these two genes we observed decreased expression, achieving up to 60–80% knockdown. Figure 1 shows the results for the construct that achieved the greatest depletion for each gene.
Figure 1.
CRISPRi of sigH and pknB in M. smegmatis. (A) Position of target sequences, shown by purple vertical lines. Numbering refers to the first nucleotide in the target sequence relative to annotated start of coding sequence. Circled sgRNAs are those for which expression data are shown. The arrow shows the position of the transcription start site. (B and D) Expression of dcas9 determined by RT-qPCR with and without induction of dcas9 and the sgRNA indicated. (C and E) Expression of pknB and sigH, respectively, with and without induction. The significance of the difference in gene-specific RNA between samples from induced and uninduced cultures is indicated by asterisks above the bar for the induced sample (*P < 0.01; **P < 0.001; ***P < 0.0001).
CRISPRi in M. tuberculosis
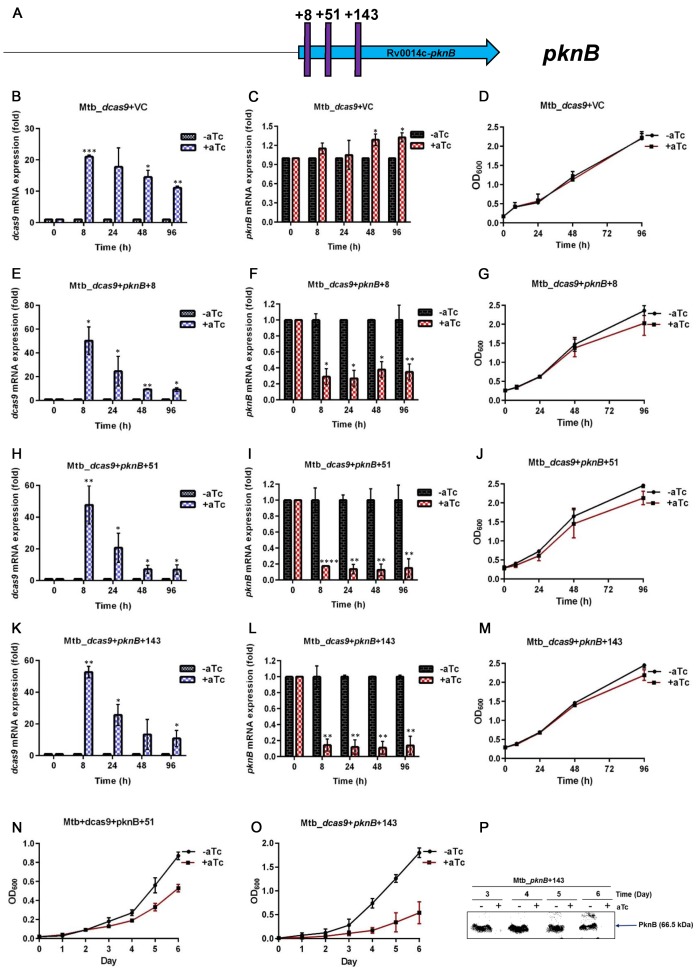
Based on these successful pilot experiments in M. smegmatis, we investigated this gene depletion system in depth in the important global pathogen M. tuberculosis. We first assessed the effects of expressing a sgRNA vector that did not include a gene-specific targeting sequence. Induced and un-induced cultures were grown and samples were taken for measurement of growth (OD600) and for RNA isolation. Using this construct we observed ∼20-fold induced expression of dcas9 at 8 h after addition of aTc to a final concentration of 200 ng/ml, followed by a steady decrease to ∼11-fold induced expression at 96 h, despite addition of a second aliquot of aTc at 48 h (Figure 2). Importantly, there was no difference in growth between the induced and un-induced strains and, as expected, no decrease in pknB expression was observed with induction of the strain containing this sgRNA control vector (Figure 2).
Figure 2.
CRISPRi of pknB in M. tuberculosis at serial time points. (A) Position of target sequences, shown by purple vertical lines. Numbering refers to the first nucleotide in the target sequence relative to the annotated start of the pknB coding sequence. (B, E, H, K) Expression of dcas9 with and without induction of dcas9 and the sgRNA indicated. (C, F, I, L) Expression of pknB with and without induction of dcas9 and the sgRNA indicated. (D, G, J, M) Growth curves of culture with and without induction of dcas9 and the sgRNA indicated. (N, O) Extended growth curves. (P) Immunoblot of total protein from the last two time points shown in panel (O) using rabbit polyclonal anti-PknB antibody. VC, vector control. The significance of the difference in gene-specific RNA between samples from induced and uninduced cultures is indicated by asterisks above the bar for the induced sample (*P < 0.01; **P < 0.001; ***P < 0.0001).
We then examined the effect of expressing sgRNAs with gene-specific sequences, starting with pknB, an essential Ser/Thr protein kinase of M. tuberculosis. Because M. tuberculosis pknB is at the 3′ end of an operon, we targeted 3 sites in the coding sequence (Figure 2). Each sgRNA construct was transformed into M. tuberculosis into which the dcas9-expressing integrating vector had been introduced. Expression of dcas9 and pknB were determined by RT-qPCR at serial time points in cultures induced with 200 ng/ml aTc and in control cultures to which no inducer was added. We observed strongly induced expression of dcas9 in these experiments, though less than we observed with the same construct in M. smegmatis. For each sgRNA we observed dcas9 induction of 50–60-fold relative to the un-induced control at 8 h, decreasing to ∼10–20-fold at 96 h, despite addition of a second aliquot of aTc at 48 h to achieve a concentration of 200 ng/ml. For the induced strains expressing sgRNAs targeting the +51 and +143 sequences we obtained 80–90% reduction of pknB RNA abundance compared to the un-induced strains; the +8 sequence-targeting sgRNA was slightly less effective, achieving 60–70% reduction in pknB mRNA (Figure 2). For each strain, we observed slower growth of the induced CRISPRi strain starting at 48–96 h. This result is similar to genetic experiments that show decreased growth compared to wild type starting at 3–4 days after onset of pknB depletion (31). To determine whether this growth defect was more evident with longer incubation, we performed growth curves starting at OD600 0.1 and compared growth from induced and un-induced cultures expressing the +51 and +143 sgRNAs over 6 days. We observed decreased growth in the induced strains, starting between days 2 and 3, with increasing separation of the growth curves through day 6 (Figure 2).These results demonstrate the ability of CRISPRi to achieve substantial inhibition of gene expression in M. tuberculosis sufficient to cause a clear growth phenotype.
The experiments described above focused on measurement of gene-specific mRNA based on the mechanism of CRISPRi, i.e. inhibition of transcription. To verify that decreased mRNA resulted in decreased protein corresponding to the gene being targeted, we performed western blotting using an anti-PknB antibody. As expected and consistent with the growth defect we observed, we found that depletion of pknB mRNA by CRIPSRi resulted in marked depletion of PknB protein (Figure 2).
Using CRISPRi to investigate phenotypes of essential M. tuberculosis genes
We expect that major uses of a CRISPRi system in M. tuberculosis will be to investigate essential genes of unknown function, and to identify or verify targets of small molecule inhibitors of growth identified in whole cell screens. To determine the utility of this system for these purposes, we selected four additional essential genes to test. Two (inhA (Rv1484) and dfrA (Rv2763c)) are the targets of the small molecule drugs isoniazid and methotrexate, respectively, and two others (wag31 (Rv2145c) and ftsZ (Rv2150c)) have both growth and morphologic phenotypes when they are genetically depleted in M. smegmatis (32–35). Only for ftsZ had the transcriptional start sites (TSSs) been previously determined experimentally (36). We therefore determined the TSS for inhA, dfrA and wag31 using 5′-RACE as previously described (Supplementary Figure S4) (20). We then designed 3 sgRNA-expressing constructs for each gene, based on the location of PAM sequences in the promoter, 5′-UTR and coding sequences (Supplementary Figure S4). As described above, these vectors were transformed into M. tuberculosis containing the integrating vector expressing dcas9 and effects on gene expression and growth, with and without induction were determined. Samples were obtained at 24 and 48 h to measure gene-specific mRNA and OD600 was measured every 24 h from 0 to 96 h. In each case, we observed induction of dcas9 at 24 and 48 h, ranging from approximately 20-fold up to 65-fold relative to the uninduced level (Figures 3 and 4). For each construct, fold induction of dcas9 was stable between 24 and 48 h.
Figure 3.
CRISPRi of inhA and dfrA in M. tuberculosis. (A, D, G, J) Expression of dcas9 with and without induction of dcas9 and the sgRNA indicated. (B, E, H, K) Expression of inhA or dfrA with and without induction of dcas9 and the sgRNA indicated. (C, F, I, L) Growth curves of culture with and without induction of dcas9 and the sgRNA indicated. (M, N) Inhibition of fluorescence in the MABA at different concentrations of methotrexate (M) or isoniazid (N) with induction of the sgRNAs indicated, compared to vector control. The significance of the difference in gene-specific RNA between samples from induced and uninduced cultures is indicated by asterisks above the bar for the induced sample (*P < 0.01; **P < 0.001; ***P < 0.0001).
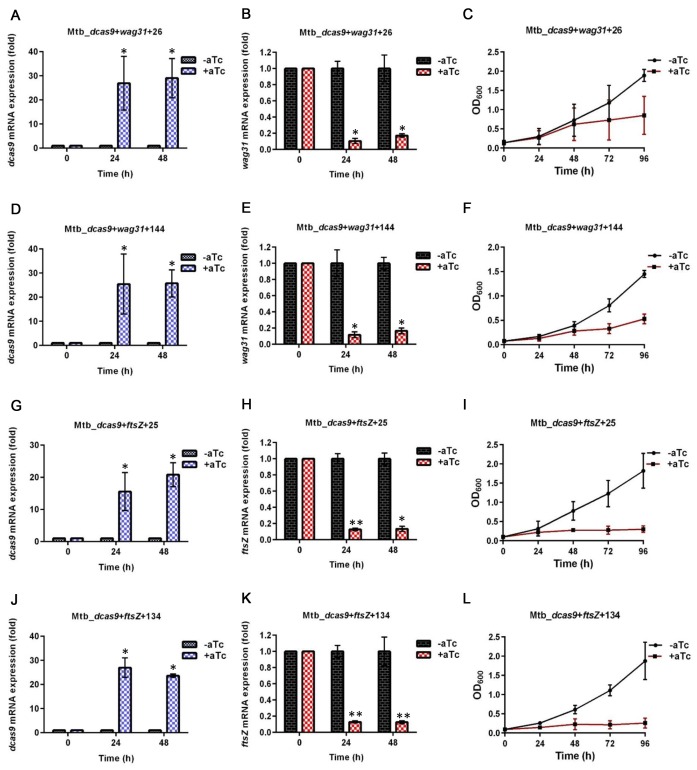
Figure 4.
CRISPRi of wag31 and ftsZ in M. tuberculosis. (A, D, G, J) Expression of dcas9 with and without induction of dcas9 and the sgRNA indicated. (B, E, H, K) Expression of wag31 or ftsZ with and without induction of dcas9 and the sgRNA indicated. (C, F, I, L) Growth curves of culture with and without induction of dcas9 and the sgRNA indicated. The significance of the difference in gene-specific RNA between samples from induced and uninduced cultures is indicated by asterisks above the bar for the induced sample (*P < 0.01; **P < 0.001; ***P < 0.0001).
For both inhA and dfrA, two of three sgRNAs achieved 80–90% knockdown of specific mRNA at 24 and 48h following induction (Figure 3) while the third sgRNA achieved 10–50% inhibition (data not shown). For both genes, the two most effective RNA depletion constructs resulted in decreased growth starting at 48–72 h following induction. We also determined whether the depletion of these RNAs resulted in increased susceptibility to chemical inhibitors, as would be expected from depletion of InhA or DhfR proteins. Using the MABA (25), we determined that the IC50 of isoniazid for the strains expressing both sgRNAs that decreased inhA transcript abundance was 4-fold lower (0.01 μg/ml) than the IC50 of isoniazid for wild type (0.04 μg/ml) (Figure 3 and Supplementary Figure S5).
Similar results were obtained with two sgRNAs that optimally inhibited transcription of dfrA. Specific transcript abundance was reduced by 80–90% in both strains when the sgRNA was induced and a growth defect was observed (Figure 3). In the MABA assay, induction of both sgRNAs targeting dfrA led to increased susceptibility to the dihydrofolate reductase inhibitor methotrexate. The methotrexate IC50 decreased from 150 μg/mL in wild type and the un-induced samples to 19.8 μg/ml in the strain expressing the sgRNA that targets the 5′ UTR, and to 75 μg/ml in the strain in which the sgRNA targets the dfrA promoter (Figure 3 and Supplementary Figure S5).
We then used CRISPRi to target the essential genes wag31 (DivIVA), which is required for peptidoglycan synthesis at the growing cell pole in mycobacteria (34,37), and ftsZ, which is required for cell division (35). Using two different sgRNA constructs targeting the 5′ end of the wag31 coding sequence, we again obtained 80–90% reduction in gene-specific RNA in strains in which either construct was induced (Figure 4). Consistent with the severe growth defect seen with genetic depletion of wag31 in M. smegmatis (34), we observed nearly complete inhibition of growth in both M. tuberculosis strains expressing sgRNAs targeting wag31, starting at 24–48h following induction of these sgRNAs. For ftsZ, we obtained approximately 90% depletion with both sgRNAs in the induced strains and saw a severe growth phenotype, with essentially no growth following induction of the sgRNA in either strain (Figure 4).
Depletion of ftsZ or wag31 in M. smegmatis has been shown to cause changes in cell morphology (34,37,38). These include elongation of ftsZ-depleted cells and asymmetric polar bulging, followed by rounding and eventual lysis, of wag31-depleted cells. We therefore examined the morphology of M. tuberculosis cells in which these mRNAs were depleted by CRISPRi. Depletion of ftsZ resulted in marked elongation of cells at 24–48 h of sgRNA induction, consistent with the phenotype observed with genetic depletion of ftsZ in M. smegmatis (35). We also observed very large, multiply branched cells (Figure 5), indicating both failure of cell division and aberrant initiation of peptidoglycan synthesis. This novel phenotype suggests a critical role for ftsZ both in the localized peptidoglycan synthesis required to form the septum, and for localization of cell wall synthesis at the new cell pole following cytokinesis. Depletion of wag31 also showed clear morphologic phenotypes. We observed initial shortening of cells starting at 48h, followed by rounding and asymmetric bulging of cells after 72–96 h of depletion (Figure 5). This phenotype indicates that in M. tuberculosis, as in M. smegmatis, Wag31 is essential for polar, or more precisely sub-polar, cell wall synthesis (34,37).
Figure 5.
Morphology of cells in which ftsZ or wag31 have been depleted. (A–C) Phase contrast microscopy M. tuberculosis cells containing vectors for expression of dcas9 and the ftsZ+25 sgRNA, without aTc (A) or with addition of aTc to 200 ng/ml (B and C). In each case photographs were taken at 48 h. Panel (C) shows selected images of large, multiply branched cells. (D) Measurement of cell size at 48h of CRISPRi with ftsZ+25 sgRNA. (E and F) Phase contrast microscopy M. tuberculosis cells containing vectors for expression of dcas9 and the wag31+26 sgRNA, without aTc addition (E) or with aTc addition to 200 ng/ml (F). Pictures in both panels were taken after 96h incubation. The significance of the difference in cell length and width between samples from induced and uninduced cultures is indicated by asterisks above the bar for the induced sample (*P < 0.01; **P < 0.001; ***P < 0.0001; ****P < 0.00001).
Titration of CRISPRi
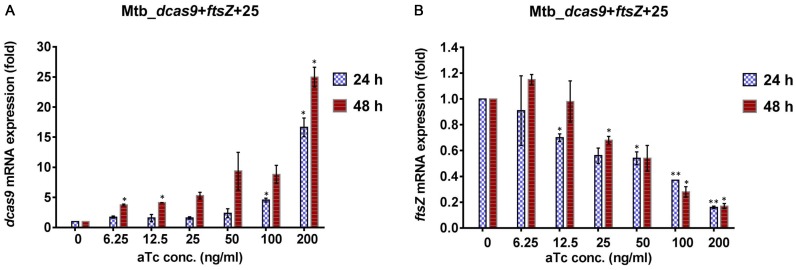
In testing the ability of CRISPRi to block expression of essential genes, we used 200 ng/ml of aTc to achieve and maintain maximal induction of dcas9 and the sgRNA. In some situations, however, it may be desirable to achieve more limited inhibition of gene expression. We therefore examined expression of ftsZ in M. tuberculosis cells expressing dcas9 and an ftsZ-specific sgRNA that were treated with a range of aTc concentrations. We observed a clear dose-response between the aTc concentration in the culture and both the extent of dcas9 expression, and the extent to which ftsZ mRNA was depleted (Figure 6). No effect was observed at the lowest concentration (6.25 ng/ml) but at higher aTc concentrations we saw a progressive increase in dcas9 expression at both 24 and 48 h, which correlated with greater ftsZ mRNA depletion. This result indicates that it is possible to titrate the extent of inhibition of gene expression to achieve a level that is optimal for experiments that may require moderate versus more potent knockdown of specific gene expression.
Figure 6.
Titration of inducer for CRISPRi of ftsZ in M. tuberculosis. (A) Expression of dcas9 at 24 and 48 h of induction with different concentrations of aTc. (B) Expression of ftsZ at 24 and 48 h of induction with different concentrations of aTc. The significance of the difference in gene-specific RNA in samples from cultures treated with different concentrations of aTc compared to untreated samples at the same time point is indicated by asterisks above the bar for the induced samples (*P < 0.01; **P < 0.001; ***P < 0.0001).
Off-target effects of CRISPRi in M. tuberculosis
A major challenge in the use of CRISPR nuclease technology in eukaryotes has been the potential for off-target effects, i.e. cleavage at sites other than the one for which the sgRNA was designed. Several approaches to identify and minimize these effects have been investigated including targeted approaches to detect predicted off-target sites and unbiased genome-wide methods (Reviewed in (39)). Though particularly problematic for gene deletion and gene editing, off-target effects on gene expression could also affect interpretation of target gene function in CRISPRi experiments.
We therefore attempted to investigate whether significant off-target effects were observed in strains where we had achieved effective inhibition of expression of the target gene. Current unbiased genome-wide methods to identify off-target effects of CRISPR gene editing require cleavage of genomic DNA by enzymatically active Cas9, and tagging or marking of cleaved sites to allow enrichment and sequencing of these sites (reviewed in (39)). There is no established method for analysis of off-target effects of CRISPRi of essential genes, because inhibition of expression of an essential gene will affect expression of many other genes. We therefore undertook a targeted approach based on the sequence-dependence of sgRNA gene targeting and observations that off-target cleavage by Cas9 can occur at sites with as many as five to six mismatches compared to the protospacer sequence in the target gene (39). To do this, we searched for sequences throughout the M. tuberculosis genome that were similar to the 20-base sequence in the target gene that is targeted by each sgRNA. Using the search pattern function of the Tuberculist website (http://genolist.pasteur.fr/Tuberculist), we searched the M. tuberculosis H37Rv genome sequence for exact matches and for sequences that differed by up to five bases from the 20 bases of the protospacer sequence the plus the adjacent 3-base PAM sequence in the target gene (Supplementary Table S1).
For the 16 sgRNAs that showed strong gene-specific inhibition in M. tuberculosis, four target sequences had no matches in the M. tuberculosis genome that differed by five or fewer bases from the PAM plus 20 nucleotide target gene sequence. For five sgRNAs we found sequences that differed by four bases, two of which had an adjacent PAM, and for one sgRNA we found a sequence that differed by two bases, but which lacked a PAM. For nine sgRNAs, we identified sites that differed by 5 nts from the target sequence, of which three had an adjacent PAM sequence. We measured expression of all off-target genes containing a sequence that differed by 2 or 4 nts from the target sequence regardless of whether there was an adjacent PAM. We also measured expression of the three genes that contained a sequence that differed by 5 nt and that had an adjacent PAM. No significant RNA depletion was observed for any of these off-target genes (Supplementary Figure S6).
Based on its mechanism of action we anticipated that CRISPRi targeting a gene in a multicistronic operon would have polar effects on genes that were 3′ of the target gene in the same operon. To address this experimentally, we designed four sgRNAs to target pknA, a gene in the same operon as, and immediately 5′ of pknB. Induction of two of these constructs resulted in decreased pknA expression, comparable to the decrease observed with other genes shown above (Supplementary Figure S7). As expected, similar decreases in pknB expression were seen in these strains, indicating that targeting dCas9 to pknA had a polar effect on the downstream gene pknB. Growth defects similar to those seen with CRISPRi targeting of pknB were also seen.
DISCUSSION
In this work, we describe an efficient CRISPR-based system for specific inhibition of gene expression in M. tuberculosis. Using this approach we consistently achieved >80% decreased expression of several essential genes, sufficient to demonstrate growth phenotypes, altered drug susceptibility and morphologic phenotypes. Our ability to rapidly analyze gene depletion phenotypes of each of the six essential M. tuberculosis genes that we targeted demonstrates the power of this approach. Though many of these genes have been partially characterized, much of the prior work has been done in the rapid-growing non-pathogen M. smegmatis (18,34,35) because of the difficulties of achieving mRNA or protein depletion in M. tuberculosis using previously available tools.
In each experiment, we obtained induction of dcas9 in M. tuberculosis at 24 and 48h, ranging from approximately 20-fold to 65-fold above the uninduced level. In the pknB experiment where expression was monitored from 8–96h we saw a progressive decline in dcas9 expression, despite a second addition of aTc at 48 h. The reason for the decline in dcas9 expression is not known, but may reflect generalized decreased gene expression at later growth stages resulting from growth inhibition. Despite this, we saw persistent inhibition of expression of pknB RNA and PknB protein (Figure 2). We also saw effects on growth and morphology at 4–6 days following induction of dcas9 and sgRNAs targeting several other essential genes (Figures 3–5), in each case with addition of aTc every 48 h.
Though we focused on essential genes of at least partially known function, our ability to achieve sustained inhibition of gene expression in M. tuberculosis allowed us to identify a novel phenotype in ftsZ-depleted cells. Whereas a genetic depletion strategy in M. smegmatis previously demonstrated elongated filamentous cells (18), we observed not only elongated cells but also very large cells with multiple branches as early as 48 h of CRISPRi-mediated inhibition of ftsZ in M. tuberculosis. A previous study demonstrated that a small proportion of M. tuberculosis cells grown in activated macrophages, in which ftsZ was found not to be localized to the mid-cell, showed lateral budding that would likely lead to branching over time (40). Our findings together with this published result suggest that ftsZ plays a critical role not only in formation of the divisome, but also in localizing new cell wall synthesis to the septum and to the new cell pole after cell division. This interpretation is consistent with several reports that have shown physical or functional interactions of mycobacterial FtsZ with proteins involved in cell wall synthesis (41–44).
The CRISPRi system described here has a number of features that will make it extremely useful for the analysis of essential gene function in M. tuberculosis. These include (i) simple, unidirectional cloning of sequences homologous to the target gene into an E. coli-mycobacterial shuttle vector, (ii) aTc-induced expression of codon-optimized dcas9 at levels that do not inhibit growth but that result in sustained, physiologically relevant inhibition of target gene expression when expressed with a gene-specific sgRNA, (iii) the ability to design sgRNAs containing target gene sequences that differ sufficiently from sequences elsewhere in the M. tuberculosis genome to minimize the likelihood of off-target effects, (iv) the ability to compare CRISPRi-inhibited expression to native levels of expression from the chromosomal gene, in contrast to genetic depletion strains where expression from an inducible promoter often differs markedly from native levels of expression and (v) no disruption of the chromosomal locus or the native gene, as is required for protein degradation tag strategies. These advantages should make CRISPRi a preferred approach for investigating non-essential genes as well essential genes.
Despite these advantages, there is at least one important limitation to this approach, the polar effect on expression of genes that are 3′ of, and in the same operon as, the target gene. Though in some cases, e.g. where the function of an operon is being investigated, this may be acceptable, in many situations it is desirable to be able to investigate the function of a single gene. In many cases, polar effects from CRISPRi of genes 3′ of the target gene in an operon could be addressed by heterologous expression of the downstream genes, however achieving appropriate levels of expression can be difficult, as noted above.
Another potential limitation of CRISPRi is off-target effects. Though our testing of potential off-target sites for several sgRNAs did not reveal gene-specific RNA depletion at these sites, this result does not indicate that off-target effects may not occur in some instances. Our approach of scanning the genome for sequences similar to the candidate target sequence, however, should allow investigators to design sgRNAs to avoid off-target effects in most cases by choosing target sequences that are not similar (>5 mismatches) to other sequences in the M. tuberculosis genome. If a target sequence in a gene of interest that is dissimilar to all other M. tuberculosis sequences cannot be identified, verifying that the off-target sequences lack an adjacent PAM should preclude dCas9 binding and inhibition of gene expression at that site. In cases where off-target sequences that have an intact PAM adjacent to a sequence with five or fewer mismatches to the protospacer being targeted cannot be avoided, these can be considered potential sites for off-target inhibition of gene expression, which can be tested experimentally.
In every case where we designed and tested two to four sgRNAs, most constructs achieved highly effective (80–90%) inhibition of gene expression. The reason for the lower activity of the minority of constructs is not apparent. Each sgRNA construct incorporated 20 bases of sequence homologous to a sequence in the target gene, and met known criteria for effective sequence targeting, i.e. each sgRNA was designed to bind to the non-template DNA strand, and each selected target was immediately adjacent to a PAM sequence. It is possible that specific sequences are more efficient at displacing the template strand to allow binding of the sgRNA to the non-template strand. A recent report identified more effective gene targeting in eukaryotes when the last 4 nts of the protospacer sequence were purines (45). In our limited number of sgRNAs, this preference was not apparent, e.g. the one protospacer sequence in which all four 3′ nts were purines (pknB+8) achieved slightly less inhibition than two others (pknB+51 and pknB+143) in which 3 of 4 nts were pyrimidines. We observed effective repression of gene expression with constructs that targeted the promoter region, the 5′-UTR and the coding sequence, suggesting that interference with either transcription initiation or elongation can be effective, as described for E. coli (15).
Tools for the study of M. tuberculosis, including more efficient gene deletion techniques, better inducible promoters and new protein depletion systems have markedly improved our ability to investigate this important human pathogen. CRISPRi adds a powerful approach to this complement of methods to investigate M. tuberculosis. In particular, this simple and efficient method to inhibit the expression of essential genes of unknown function has the potential both to accelerate the investigation of M. tuberculosis pathogenesis and to facilitate the development of novel drugs for M. tuberculosis, e.g. by allowing the characterization of novel drug targets and by identifying or verifying targets of novel small molecule inhibitors identified in whole cell screens.
Supplementary Material
Acknowledgments
The authors thank Rebekah DePew for initial work on dcas9 synthesis and vector construction, Paula Watnick for use of her microscope, and members of the Husson laboratory for helpful discussions.
Footnotes
Present addresses:
Jared D. Sharp, Patheon, Inc., Princeton, NJ 08540, USA.
Ranfei Xu, Harvard College, Cambridge, MA 02138, USA.
Sladjana Prisic, University of Hawaii at Manoa, Honolulu, HI 96822, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R03 AI113281 and R01 AI099204 to R.N.H.]. Funding for open access charge: National Institutes of Health [R03 AI113281 and R01 AI099204 to R.N.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright A.V., Nuñez J.K., Doudna J.A. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Peters J.M., Silvis M.R., Zhao D., Hawkins J.S., Gross C.A., Qi L.S. Bacterial CRISPR: accomplishments and prospects. Curr. Opin. Microbiol. 2015;27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelicic V., Jackson M., Reyrat J.M., Jacobs W.R., Gicquel B., Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tufariello J., Bardarov S., Hatfull G., Larsen M., Bardarov S., Chan J., Pavelka M.S., Sambandamurthy V., Jacobs W.R. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 8.Tufariello J.M., Malek A.A., Vilchèze C., Cole L.E., Ratner H.K., González P.A., Jain P., Hatfull G.F., Larsen M.H., Jacobs W.R. Enhanced specialized transduction using recombineering in Mycobacterium tuberculosis. mBio. 2014;5 doi: 10.1128/mBio.01179-14. doi:10.1128/mBio.01179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy K.C., Papavinasasundaram K., Sassetti C.M. Mycobacterial recombineering. Methods Mol. Biol. 2015;1285:177–199. doi: 10.1007/978-1-4939-2450-9_10. [DOI] [PubMed] [Google Scholar]
- 10.Wei J.-R., Krishnamoorthy V., Murphy K., Kim J.-H., Schnappinger D., Alber T., Sassetti C.M., Rhee K.Y., Rubin E.J. Depletion of antibiotic targets has widely varying effects on growth. Proc. Natl. Acad .Sci. U.S.A. 2011;108:4176–4181. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.-H., O'Brien K.M., Sharma R., Boshoff H.I.M., Rehren G., Chakraborty S., Wallach J.B., Monteleone M., Wilson D.J., Aldrich C.C., et al. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19095–19100. doi: 10.1073/pnas.1315860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forti F., Crosta A., Ghisotti D. Pristinamycin-inducible gene regulation in mycobacteria. J. Biotechnol. 2009;140:270–277. doi: 10.1016/j.jbiotec.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Klotzsche M., Ehrt S., Schnappinger D. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 2009;37:1778–1788. doi: 10.1093/nar/gkp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolly G.S., Boldrin F., Sala C., Dhar N., Hartkoorn R.C., Ventura M., Serafini A., Mckinney J.D., Manganelli R., Cole S.T. Assessing the essentiality of the decaprenyl-phospho-d-arabinofuranose pathway in Mycobacterium tuberculosis using conditional mutants. Mol. Microbiol. 2014;92:194–211. doi: 10.1111/mmi.12546. [DOI] [PubMed] [Google Scholar]
- 15.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhary E., Thakur P., Pareek M., Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 17.Lee M.H., Hatfull G.F. Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J. Bacteriol. 1993;175:6836–6841. doi: 10.1128/jb.175.21.6836-6841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrt S., Guo X.V., Hickey C.M., Ryou M., Monteleone M., Riley L.W., Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.-R.J., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp J.D., Singh A.K., Park S.T., Lyubetskaya A., Peterson M.W., Gomes A.L.C., Potluri L.-P., Raman S., Galagan J.E., Husson R.N. Comprehensive definition of the SigH regulon of Mycobacterium tuberculosis reveals transcriptional control of diverse stress responses. PLoS ONE. 2016;11:e0152145. doi: 10.1371/journal.pone.0152145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 22.Arany Z.P. In: Current Protocols in Human Genetics. Haines JL, Korf BR, Morton CC, Seidman CE, Seidman JG, Smith DR, editors. Hoboken, NJ: John Wiley & Sons, Inc; 2001. [Google Scholar]
- 23.Man T.-K., Li Y., Dang T.A., Shen J., Perlaky L., Lau C.C. Optimising the use of TRIzol-extracted proteins in surface enhanced laser desorption/ ionization (SELDI) analysis. Proteome Sci. 2006;4:3. doi: 10.1186/1477-5956-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prisic S., Hwang H., Dow A., Barnaby O., Pan T.S., Lonzanida J.A., Chazin W.J., Steen H., Husson R.N. Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol. Microbiol. 2015;97:263–280. doi: 10.1111/mmi.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins L., Franzblau S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 27.Raman S., Song T., Puyang X., Bardarov S., Jacobs W.R., Husson R.N. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 2001;183:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manganelli R., Voskuil M.I., Schoolnik G.K., Dubnau E., Gomez M., Smith I. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang C.-M., Abbott D.W., Park S.T., Dascher C.C., Cantley L.C., Husson R.N. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gee C.L., Papavinasasundaram K.G., Blair S.R., Baer C.E., Falick A.M., King D.S., Griffin J.E., Venghatakrishnan H., Zukauskas A., Wei J.-R., et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci. Signal. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chawla Y., Upadhyay S.K., Khan S., Nagarajan S.N., Forti F., Nandicoori V.K. Protein Kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 2014;289:13858–13875. doi: 10.1074/jbc.M114.563536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen M.H., Vilchèze C., Kremer L., Besra G.S., Parsons L., Salfinger M., Heifets L., Hazbon M.H., Alland D., Sacchettini J.C., et al. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol. Microbiol. 2002;46:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 33.White E.L., Ross L.J., Cunningham A., Escuyer V. Cloning, expression, and characterization of Mycobacterium tuberculosis dihydrofolate reductase. FEMS Microbiol. Lett. 2004;232:101–105. doi: 10.1016/S0378-1097(04)00038-2. [DOI] [PubMed] [Google Scholar]
- 34.Kang C.-M., Nyayapathy S., Lee J.-Y., Suh J.-W., Husson R.N. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology (Reading, Engl.) 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 35.Dziadek J., Rutherford S.A., Madiraju M.V., Atkinson M.A.L., Rajagopalan M. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology (Reading, Engl) 2003;149:1593–1603. doi: 10.1099/mic.0.26023-0. [DOI] [PubMed] [Google Scholar]
- 36.Kiran M., Maloney E., Lofton H., Chauhan A., Jensen R., Dziedzic R., Madiraju M., Rajagopalan M. Mycobacterium tuberculosis ftsZ expression and minimal promoter activity. Tuberculosis (Edinb.) 2009;89(Suppl. 1):S60–S64. doi: 10.1016/S1472-9792(09)70014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Méniche X., Otten R., Siegrist M.S., Baer C.E., Murphy K.C., Bertozzi C.R., Sassetti C.M. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc. Natl .Acad. Sci. U.S.A. 2014;111:E3243–E3251. doi: 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plocinska R., Martinez L., Gorla P., Pandeeti E., Sarva K., Blaszczyk E., Dziadek J., Madiraju M.V., Rajagopalan M. Mycobacterium tuberculosis MtrB sensor kinase interactions with FtsI and Wag31 proteins reveal a role for MtrB distinct from that regulating MtrA activities. J. Bacteriol. 2014;196:4120–4129. doi: 10.1128/JB.01795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai S.Q., Joung J.K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauhan A., Madiraju M.V.V.S., Fol M., Lofton H., Maloney E., Reynolds R., Rajagopalan M. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 2006;188:1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta P., Dasgupta A., Singh A.K., Mukherjee P., Kundu M., Basu J. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 2006;62:1655–1673. doi: 10.1111/j.1365-2958.2006.05491.x. [DOI] [PubMed] [Google Scholar]
- 42.Sureka K., Hossain T., Mukherjee P., Chatterjee P., Datta P., Kundu M., Basu J. PLOS ONE: Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS ONE. 2010;5:e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Plocinski P., Ziolkiewicz M., Kiran M., Vadrevu S.I., Nguyen H.B., Hugonnet J., Veckerle C., Arthur M., Dziadek J., Cross T.A., et al. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J. Bacteriol. 2011;193:3246–3256. doi: 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plocinski P., Arora N., Sarva K., Blaszczyk E., Qin H., Das N., Plocinska R., Ziolkiewicz M., Dziadek J., Kiran M., et al. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J. Bacteriol. 2012;194:6398–6409. doi: 10.1128/JB.01005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.