Abstract
The discovery of novel specific ribosome-associated factors challenges the assumption that translation relies on standardized molecular machinery. In this work, we demonstrate that Tma108, an uncharacterized translation machinery-associated factor in yeast, defines a subpopulation of cellular ribosomes specifically involved in the translation of less than 200 mRNAs encoding proteins with ATP or Zinc binding domains. Using ribonucleoparticle dissociation experiments we established that Tma108 directly interacts with the nascent protein chain. Additionally, we have shown that translation of the first 35 amino acids of Asn1, one of the Tma108 targets, is necessary and sufficient to recruit Tma108, suggesting that it is loaded early during translation. Comparative genomic analyses, molecular modeling and directed mutagenesis point to Tma108 as an original M1 metallopeptidase, which uses its putative catalytic peptide-binding pocket to bind the N-terminus of its targets. The involvement of Tma108 in co-translational regulation is attested by a drastic change in the subcellular localization of ATP2 mRNA upon Tma108 inactivation. Tma108 is a unique example of a nascent chain-associated factor with high selectivity and its study illustrates the existence of other specific translation-associated factors besides RNA binding proteins.
INTRODUCTION
As intermediates between DNA and proteins, mRNAs are the prime targets for the specific gene expression controls. Decades of research have demonstrated that multiple layers of fine tuning regulation mechanisms determine unique fates for mRNA molecules. In addition to the transcription and chromatin-remodeling factors required for the production of specific sets of mRNAs, post-transcriptional events allow for precise spatio-temporal controls. Indeed, a new picture of gene regulation has emerged in which RNA-binding proteins and small RNAs bind to mRNA molecules to define RNA regulons with distinct subcellular localizations, stabilities and/or translation efficiencies (1,2). These specific post-transcriptional controls are generally believed to result in the loading of the ribosome, which is assumed to be a standardized molecular machine acting as an automaton in translating the genetic code.
However, recent studies have revealed some heterogeneity in translation machinery-associated proteins (3–5) which supports the hypothesis of specialized control of translation, mediated by ribosomal complexes with unique compositions and specific activities. Ribosomal particles combined to various additional factors could associate with defined subsets of the cellular mRNAs and have specific translational properties (translation efficiency, sub-cellular localization, nascent chain quality control). This is supported by the observation that accessory proteins, like RNA-binding proteins or nascent chain-associated factors are recruited by the translation machinery and participate in the specialized co-translational control required for the synthesis of a specific subset of proteins. For example, Rack1, a core component of the ribosome, has been shown to bind various partners such as transmembrane receptors, the miRNA-induced silencing complex (miRISC) and Scp160, an RNA-binding protein (6). Such interactions may contribute to the targeting of the ribosome to the cell membrane, miRNA translation inhibition and selective translation of specific mRNAs associated with RNA-binding proteins. Similarly, proteins involved in protein quality control can be recruited to the ribosomal complex during translation to eliminate aberrant transcripts and nascent chains (7,8) or to facilitate the proper folding of specific nascent proteins and prevent their aggregation (9).
Localized mRNAs are good models to study specific translational controls since site-specific translation requires the coordination of translation and mRNA targeting (10). ATP2 mRNA codes for the β-subunit of ATP synthase and is a long-standing model used to investigate the localization to the mitochondria of diverse nuclear-encoded mRNAs translated in close proximity to this organelle (11–15).In the present work, we hypothesize that a particular ribosomal complex composition might regulate ATP2 mRNA localization to its translation site. To address this question, we took advantage of the recent proteomic identification of several uncharacterized Translation Machinery-Associated factors (TMAs) in yeast that could constitute good candidates for specific ribosomal partners (16). We observed that the inactivation of Tma108 significantly impacted the sub-cellular localization of ATP2 mRNA. Immunoprecipitation experiments further demonstrated that Tma108 defined a sub-population of ribosomes with high selectivity for a set of 174 mRNAs, including that of ATP2. However, Gene Ontology enrichment analyses revealed that Tma108 targets were not enriched in mRNA encoding mitochondrial proteins. Instead, we observed that a large number of them encoded ATP-binding, RNA-binding or Zn-binding domains, suggesting that Tma108 could recognize specific nascent peptide features. This hypothesis was corroborated by the mass spectrometry analysis of Tma108 partners and the molecular dissection of the signals involved in Tma108 selective recognition of ATP2 and ASN1 mRNA. The latter also revealed the crucial role of the first translated amino acids of these targets in the recruitment of Tma108 to the translational machinery. Comparative genomic analysis and molecular modeling pointed to Tma108 as an original M1 metallopeptidase, making it the first example of a member of this protein family to be involved in nascent chain recognition during translation. In addition, a single amino acid substitution in the MAMEN motif (E296Q) demonstrated the involvement of the putative M1 metallopeptidase peptide-binding pocket in Tma108 substrate recognition. We put forward a model in which the Tma108 catalytic pocket has evolved to recognize specific nascent peptides and participate in the co-translational control of gene expression.
MATERIALS AND METHODS
Strain, plasmid, media used
All Saccharomyces cerevisiae strains used in this study are isogenic to BY4741: they are described in the supplementary methods (Supplementary Table S1). Plamids used for yeast transformation are also described in the supplementary methods (Supplementary Table S2).
Yeast were grown on rich medium (1% bactopeptone, 1% yeast extract) containing either 2% glucose (YPD) or 2% galactose (YPGal), as a carbon source. BY4743 and Tma108-PA strain were transformed with pRS416-derived plasmids (see supplementary methods). When required for plasmid selection, cells were grown on CSM medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% dextrose, 0.07% CSM mixture) depleted of the selective compound.
Antibodies
The following antibodies were used for western blotting or immunoprecipitation: rabbit IgG-HRP polyclonal antibody (PAP; code Z0113; Dako), mousse anti-rpl3 IgG (DSBH, clone 7.1.1), rabbit anti-Rpl1 igG (kind gift from F. Lacroute (17)).
FISH and mRNA localization quantitative analyses
The FISH experiments were performed as described previously (18). Diploid strains were used in order to better visualize the relative location of mRNA to the mitochondria. After overnight growth in YPGal medium, cells in the exponential phase were fixed with a final concentration of 4% paraformaldehyde, spheroplasted with lyticase, adhered to poly-l-lysine-treated coverslips and stored in 70% ethanol at −20°C. Five antisense oligonucleotide probes complementary to the targeted sequence were used to detect single mRNA molecules. Mitochondrial ribosomal RNA, ATP16 mRNA and ATP2 mRNA were detected using fluorescent probes as described previously (18). After overnight hybridization at 37°C, washing and 4,6-diamidino-2-phenylindole staining, coverslips were mounted with antifade solution. Three-dimensional imaging analysis was performed, by acquiring 41 images 200 nm apart along the z-axis on a Ti Nikon inverted microscope using a 60X numerical aperture objective.
To statistically analyze relative RNA localization in large cell populations (>100 cells), we used the CORSEN software (19). Quantification of distances between mRNA and mitochondria involved the following steps: (i) 3D segmentation and extraction of object features (coordinate and intensity); (ii) measurement of the distance between mRNA particles and the mitochondrial surface and (iii) statistical analysis of generated data, with distances weighted for intensity and using the median mRNA–mitochondrion distance for each cell.
Immunoprecipitation of ribonucleoprotein particles
For mass spectrometry and microarray analyses, ribonucleoparticle purifications from cells expressing proteinA-tagged baits (Tma108, Rpl16A, Scp160 or Tma46) were performed essentially as described in (20). Briefly, frozen grindates obtained from cells grown in rich medium were homogenized in nine volumes of extraction buffer (20 mM Hepes pH 7.5, 110 mM KOAc, 2 mM MgCl2, 0.1% Tween-20, 0,5% Triton X-100, 1 mM DTT, 1× protease inhibitors cocktail, complete EDTA-free, Roche and antifoam B, Sigma, 1:5000). When indicated, EDTA was added for a final concentration of 40 mM. The resulting extract was clarified by filtration through 1.6 μm GD/X Glass Microfiber syringe filters (25 mm, Whatman) and further incubated for 30 min at 4°C with IgG-conjugated magnetic beads. Beads were then washed three times with extraction buffer, once with washing buffer (0.1 M NH4OAc, 0.1 mM MgCl2) supplemented with 0.02% Tween-20 and four times with washing buffer without Tween-20. Beads were then split into two samples for protein and RNA analysis respectively. Immunoprecipitated proteins were eluted with 0.5 M NH4OH, 0.5 mM EDTA, lyophilized and resuspended either in SDS-sample buffer for SDS-PAGE or in 25 mM ammonium carbonate for mass spectrometry analysis. Immunoprecipitated RNA was eluted by treating the beads with proteinase K (0.2mg/ml, 30 min at 30°C) in the washing buffer supplemented with 0.1% SDS.
Small-scale immunoprecipitations (reporter construct studies) were performed using a similar procedure except that cells were lysed in the same buffer by bead beating using a Fastprep (Qbiogene). After a 10 000-g centrifugation at 4°C for 5 min, the soluble extract was processed for immunoprecipitation and RNA analysis as above.
Mass spectrometry analyses
The mass spectrometry proteomics data has been deposited on the ProteomeXchange Consortium (21) via the PRIDE partner repository with the dataset identifier PXD003174.
Detailed mass spectrometry analysis procedures are available in the supplementary methods.
Only proteins with a Mascot score higher than 150 were considered for further analysis. Protein score distributions of immunoprecipitations (IP) from the control (untagged BY4741) or proteinA-tagged strains were compared in order to distinguish specifically interacting proteins from the background (see Supplementary Figure S1 for an example). Based on the control distribution, a threshold score was chosen to keep <10% false positives. Proteins above this threshold score in the control or with a ratio of IP to control inferior to 2 were excluded.
Real time quantitative PCR analyses
Both total and immunoprecipitated RNA was purified using the Qiagen RNeasy mini kit and further treated with DNAse (Ambion TURBO DNA-Free kit). 500 ng of purified RNA was used for reverse transcription followed by real-time PCR analysis as described in (18). The oligonucleotides used are described in an additional file (Table S3 in supplementary methods).
Extraction of mitochondrion-associated RNA
Both mitochondrion-associated RNA and total cellular RNA were extracted as previously described (18) except that cycloheximide (200 μg/ml) was added on ice only in the fractionation buffer. RNA was extracted and purified using the RNeasy Mini Kit (Qiagen). The quality of the extraction was checked by quantitative PCR analysis as previously described (18).
Microarray hybridization and analysis
The microarray data and the related protocols are available at the GEO web site (www.ncbi.nlm.nih.gov/geo/) with the dataset identifiers E-MTAB-3922 and E-MTAB-3881.
Briefly, 1μg of RNA from each sample to be compared was reverse-transcribed and labeled with Cy3 or Cy5 dye using the indirect labeling procedure. We then hybridized labeled cDNAs with Agilent's ab 8×60K S. cerevisiae custom DNA chip (AMADID: 027945). Each hybridization was replicated using dye-swap. Arrays were read using an Agilent scanner at 2 μm resolution and the signal segmentation was done using the feature extraction software (Agilent). The data was normalized without background subtraction using the global Lowess method (22).
For the analysis of mitochondrion-associated RNAs, the enrichment ratios between cellular and mitochondrial extraction were calculated. A comparative analysis of BY4743 and tma108Δ/Δ mRNA localization was performed using LIMMA (23,24) to detect significant changes (P-value<0.05) in mRNA association to mitochondria.
For the analysis of ribosome-associated mRNA, we performed microarray hybridizations using cDNA reverse-transcribed from immunoprecipitation and the corresponding input sample. The LIMMA algorithm was used to determine the list of mRNAs specifically enriched in Tma108, Tma46 and Scp160 immunoprecipitations compared to Rpl16A. Only mRNAs with log2 ratios superior to 0.8 and an adjusted P-value of <0.05 were kept in the final list.
R software was used for category enrichment analysis (http://www.r-project.org/). GO (http://www.yeastgenome.org/download-data/curation) and Pfam (25) categories were assigned using the corresponding the databases. P-values to detect over-represented and under-represented GO or Pfam categories were calculated on the basis of a hypergeometric density distribution function as described in (26).
Molecular modeling of Tma108 structure
The tertiary structure of Tma108 was predicted using the Modeller (27) program interfaced with the HHpred server (28). The protein sequence of Tma108 from S. cerevisiae (UNIPROT code: P40462, 946 residues) was queried against the pdb70 database (Protein Data Bank (29) clustered at 70% sequence identity) using HHblits (30) to retrieve homologous sequences with known structures. Two of the best hits were selected as templates: human aminopeptidase A (PDB code: 4KX7, chain A) and porcine aminopeptidase N (PDB code: 4FKE, chain A). They both cover the entire protein (residues 4–944) with 20% of identities. The quality of the model was evaluated with Procheck (31). 85% of the residues were found in the most favored regions of the Ramachandran plot, indicating reasonable quality.
Multiple sequence alignments
Two multiple sequence alignments were generated using MAFFT (32) via the EMBL-EBI webserver (33) comprised of: (i) 14 sequences of M1 aminopeptidases and the sequence of Tma108 from S. cerevisiae, (ii) 18 sequences of Tma108 proteins from saccharomycetaceae (for details on the sequences used for the alignments see Table S4 in supplementary methods). We used Seaview to compute phylogenetic trees from the multiple sequence alignments with the pHyML program (parameters set to default). Name and accession numbers of the sequences used are available in the supplementary methods. The complete annotated alignment of Tma108 with M1 aminopeptidases is available in supplementary data 1. Tma108 orthologs used in the second alignment were obtained by reciprocal blast in the Saccharomycetaceae genome databases. We applied the same analysis to the closest paralogs of Tma108, Aap1 and Ape2, and observed that Tma108 orthologs could be clearly distinguished from those of Aap1 and Ape2 (see Supplementary Figures S2 and S3).
RESULTS
The Translation Machinery-Associated factor Tma108 impacts the localization of ATP2 mRNA to mitochondria
In a previous study (14), we demonstrated that translation was absolutely required for ATP2 mRNA to localize to the mitochondria. Our working model was that ATP2 mRNA localization was a co-translational event mediated by the targeting of its nascent amino-terminal peptide (Mitochondrial Targeting Sequence or MTS) to the mitochondrial surface (Supplementary Figure S4a).
We hypothesized that some ribosomal proteins could participate in the specific dynamics of translation required for ATP2 mRNA localization to the mitochondria. We performed FISH experiments to identify such proteins and analyzed the localization of ATP2 mRNA in several strains in which genes encoding structural ribosomal proteins and putative ribosomal partners (Tma proteins) were deleted (supplementary methods).
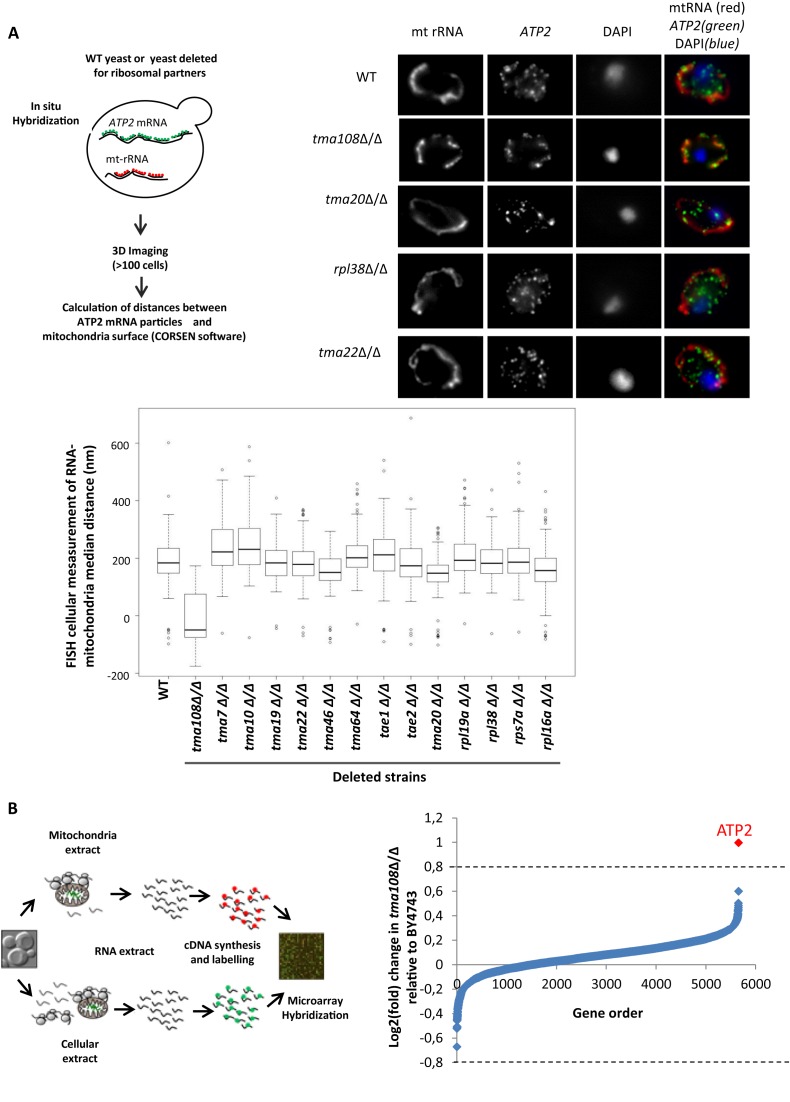
Fluorescent probes that allow for detection of single molecules of ATP2 mRNA were used in combination with probes directed against mitochondrial rRNA as markers for mitochondrial position (Figure 1A upper left). Three dimensional image acquisitions for more than 100 cells per strain were generated using Corsen which allowed us to calculate the median distances between ATP2 particles and mitochondrial surfaces (19). Distributions of median distances in cellular populations are represented in Figure 1A (bottom panel). Among the 14 tested strains, only the strain in which TMA108 was deleted showed a significant change in ATP2 mRNA localization compared to the wild type strain. In the absence of Tma108, most of the ATP2 mRNA particles co-localized with the mitochondria (Figure 1a upper right) and the median ATP2 mRNA-mitochondria distance shifted from 195 to −50 nm (Figure 1a bottom), indicating that Tma108 negatively impacted the localization of ATP2 mRNA in the vicinity of the mitochondria. This finding was then confirmed by a functional complementation experiment (Supplementary Figure S5) and by the observation that the over-expression of Tma108 led to a significant decrease in ATP2 mRNA mitochondrial localization (Supplementary Figure S6).
Figure 1.
The Translation Machinery-Associated factor Tma108 impacts ATP2 mRNA localization to mitochondria. (A) The impact of different translation machinery-associated factors on ATP2 mRNA localization. Experimental approach (top left). Fluorescent in situ experiments were performed in different mutants using probes to detect ATP2 mRNA and mitochondrial rRNA as a marker for mitochondria position. Typical FISH images (top right) show the impact of TMA108 deletion on ATP2 mRNA localization. In BY4743 strain, the ATP2 mRNA is partially localized to the mitochondria surface. TMA108 deletion led to a drastic increase in ATP2 mRNA localization to the mitochondria: most of the green spots co-localized with the mitochondrial red signal. ATP2 mRNA localization is not perturbed by TMA20, RPL38 or TMA22 deletions. The boxplots (bottom) represent the distributions of the median ATP2 mRNA-mitochondria distances in the analyzed cell populations. Similar FISH analyses were performed on a control mRNA, ATP16, and no significant change in mRNA localization was observed (data not shown). (B) Genome-wide analysis of the impact of Tma108 on the localization of mRNA to mitochondria. Experimental scheme (left) for the analysis of mRNA localization to mitochondria: total mRNA and mRNA associated with the mitochondrial fraction were isolated, reverse transcribed, and labeled for subsequent microarray analysis. Comparative analysis of the mRNA associated with the mitochondrial fraction in BY4743 and TMA108 deleted strain (right). The log2 fold changes in TMA108Δ/Δ relative to BY4743 are plotted: the values for each gene are the means of three biological replicates (see Supplemental Table S5). ATP2 is the single mRNA detected by LIMMA with a significant change in mRNA enrichment factor (P-value = 3.7 × 10−3).
ATP2 mRNA is one of the Mitochondria Localized nuclear-encoded mRNAs (MLRs), which were previously found to be preferentially localized at the mitochondrial surface (13,15). We then analyzed the impact of Tma108 activity on the spatial distribution of mRNA at a genome-wide level. In order to achieve this, we carried out comparative global analyses of mRNAs co-purified with mitochondrial fractions in the presence or absence of Tma108 using DNA microarrays (Figure 1B, left part). Consistent with our FISH analysis, we observed a 2-fold increase in ATP2 mRNA association with the mitochondrial fraction. Nevertheless, no other significant changes in mRNA enrichment factors were detected with this approach (Figure 1B, right part). This result showed that Tma108 specifically impacted ATP2 mRNA localization to the mitochondria without detectable effects on other localized RNA to mitochondria.
Tma108 interacts with ribosomes bound to the ATP2 mRNA
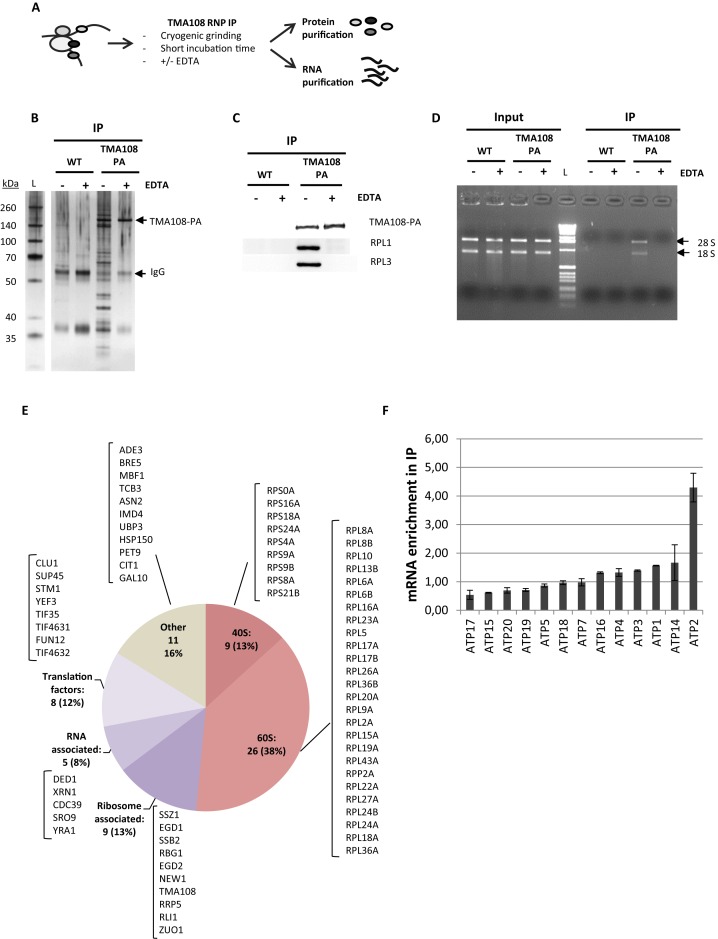
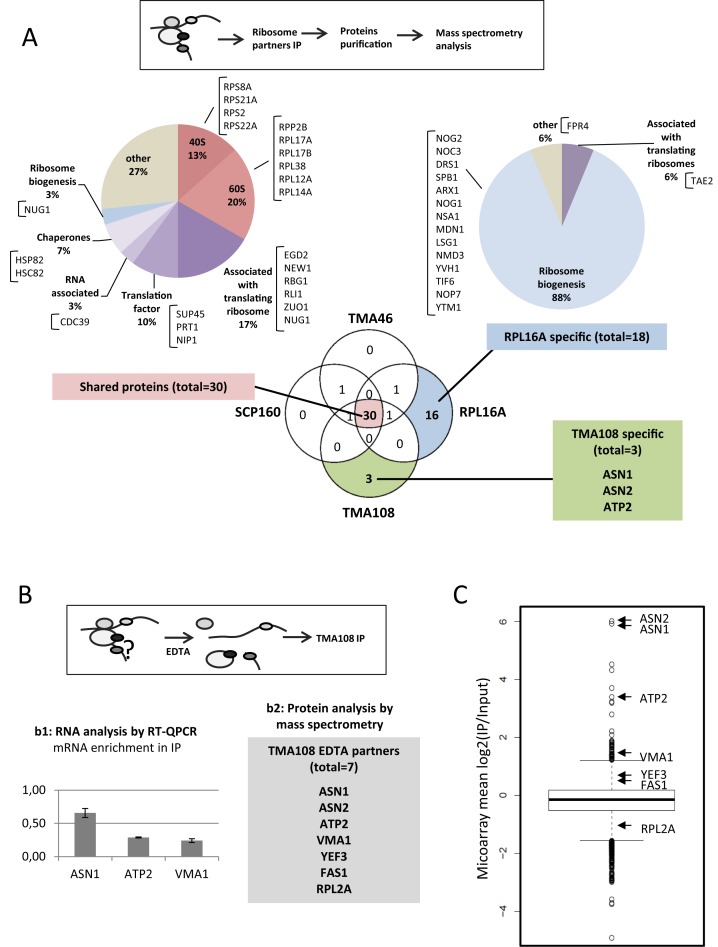
In their pioneering work to identify new ribosomal partners, Fleischer and colleagues demonstrated that Tma108 was present in salt washes of ribosome purifications and cosedimented in sucrose gradients with polysomes in an EDTA-dependent manner (16). Nevertheless, they were not able to confirm this interaction by direct immunoprecipitation of Tma108. In order to clarify the potential association between Tma108 and the ATP2 mRNA translation machinery, we performed immunoprecipitations of a proteinA-tagged version of Tma108 in conditions that maintained ribonucleoprotein (RNP) structures and were previously used to analyze ribosomal particles (20) (Figure 2A). This technique allowed us to purify both protein (Figure 2B, C and E) and RNA components (Figure 2D and F) of the RNP complexes which were immunoprecipitated together with Tma108.
Figure 2.
Tma108 interacts with translating ribosomes that are associated with ATP2 mRNA. (A) Experimental approach. Immunoprecipitations were performed on cells grown on rich medium, expressing Tma108-PA and untagged cells (WT) in presence (+) or in absence (−) of 40 mM EDTA. Protein (B, C and E) and RNA (D,F) contents were analyzed from total extract (Input) and immunoprecipitated samples (IP). (B) Input and immunoprecipitated fractions obtained from untagged or Tma108-PA expressing-cells were analyzed by SDS-PAGE followed by silver staining. The position of the Tma108-PA fusion and IgG are indicated. (C) Input and immunoprecipitated fractions obtained from untagged or Tma108-PA expressing-cells were analyzed by SDS-PAGE followed by western-blots using antibodies directed to ribosomal proteins (Rpl1, Rpl3). (D) Following RNA isolation, input and immunoprecipitated fractions obtained from untagged or Tma108-PA expressing-cells were analyzed by agarose gel electrophoresis with ethidium bromide staining (L: 1kb DNA ladder biolabs). The bands corresponding to 28S and 18S rRNA are indicated. (E) Diagram showing the functional distribution of proteins detected by mass spectrometry analysis of Tma108-PA IP in the absence of EDTA. The names of the proteins included in the different sectors are indicated (see Supplementary Table S6). Only proteins reproducibly detected with a MS score superior to the threshold determined with the untagged sample were selected (see Supplementary Figure S1). Most of these proteins are lost when IP are performed in the presence of EDTA (see Figure 5B). (F) Real-time PCR analysis of mRNA immunoprecipitated with Tma108-PA IP. For each analyzed mRNA, the enrichment factor corresponds to the ratio IP/Input normalized using a set of control mRNA (Act1,Trx2,Tpm2,Jen1,Flr1). After normalization, the mean enrichment factor for the control mRNA set is equal to 1. When EDTA is added in the IP, ATP2 mRNA is no longer enriched compared to the other mRNA (see Figure 5B). No specific enrichment of ATP2 mRNA is detected when analyzing IP performed with the untagged strain (data not shown).
Analyses of the protein fractions using mass spectrometry showed that >80% of the proteins co-immunoprecipitated with Tma108 (Figure 2B) were known components of the translation machinery (Figure 2E). They included core members of the 60S and 40S subunits, general translation factors (Clu1, Fun12, Stm1, Sup45, Tif35, Tif4631, Tif4632, Yef3), as well as proteins associated with the ribosome (Rbg1, Rli1, Rrp5, New1), the nascent peptide (Egd1, Egd2, Ssb2, Ssz1, Zuo1) or the translated mRNA (Ded1, Cdc39, Sro9, Xrn1, Yra1). These results were confirmed by immunoblotting of Rpl1 and Rpl3 (Figure 2C). The association between Tma108 and the ribosome was also supported by the detection of rRNA in the Tma108 IP and not in the mock IP (Figure 2D). Strikingly, all these interactions were lost upon EDTA treatment (Figure 2B, C and D), which dissociated the 40S and the 60S subunits of the translating ribosome, indicating that Tma108 might interact with the mature mRNA-ribosome complex.
Next, we used real-time quantitative PCR to analyze the mRNA present in the ribosomal particles which had immunoprecipitated with Tma108. We observed a clear and reproducible enrichment of ATP2 mRNAs in the Tma108 IP, compared to a control set of mRNA (Figure 2F). We further analyzed the presence within the Tma108 IP of diverse mRNAs encoding the other subunits of the mitochondrial ATP synthase complex. Among these, some are similarly nuclear-encoded and localized to the mitochondria (ATP1, ATP3 and ATP4) (34). No enrichment was measured for any of the 12 other ATP synthase-encoding mRNAs. This last observation is consistent with our previous finding that only ATP2 mRNA localization is disturbed in the absence of Tma108 (Figure 1B).
Our results show that Tma108 may interact with ribosomes engaged in translation. However, this interaction would be restricted to ribosome translating specific mRNAs, such as ATP2.
Tma108 targets ribosomes that translate particular sets of mRNAs.
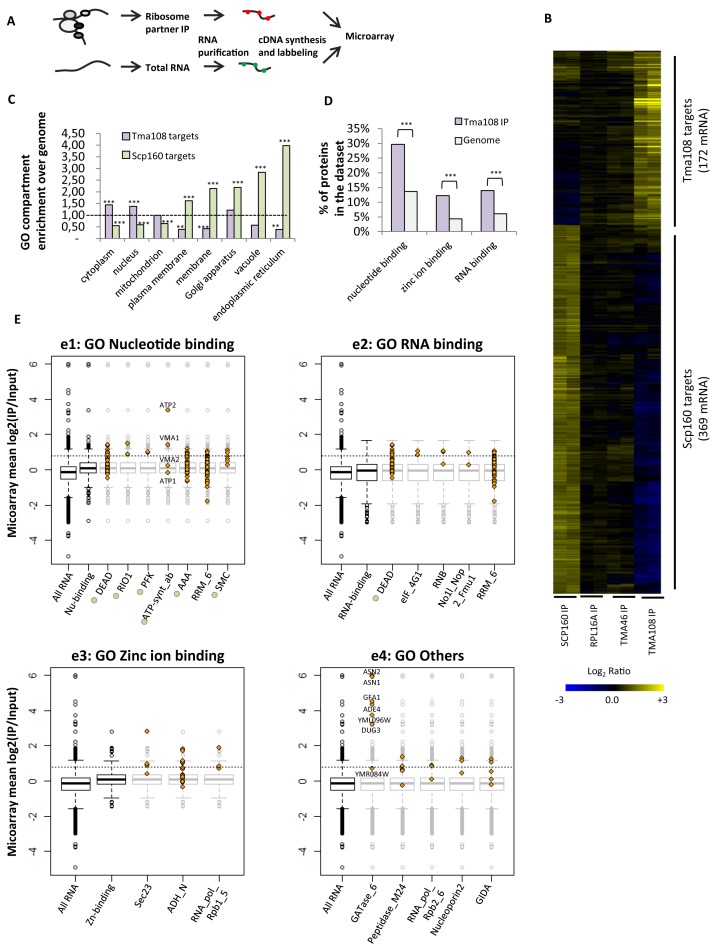
Subsequently, we used microarray technology to identify the whole set of mRNAs which were enriched in the Tma108 IP (Figure 3A). In order to determine the selectivity of Tma108-mRNA association, we also analyzed the mRNAs obtained from the immunoprecipitation of Rpl16a, a core component of the ribosome, and two other ribosomal partners, Scp160 and Tma46 (16,35). In each case, the isolation of ribosomal particles was confirmed by the detection of rRNA in the immunopurified extracts (data not shown). Two independent IP experiments were used for microarray hybridization and the replicates were highly consistent, as shown in the hierarchical clustering analysis in Figure 3B. Statistical analysis defined 174 mRNA specifically enriched in the Tma108 IP dataset compared to the Rpl16a dataset, which was used as a proxy for general translational activity (36,37). We also identified 369 mRNAs enriched in the Scp160 IP, a result consistent with previous publications, demonstrating that Scp160 associates with specific subsets of mRNAs (2,38). However, we did not detect any specific subset of mRNA associated with Tma46 as compared to Rpl16a, suggesting that Tma46 interacts with the entire ribosomal population without selectivity for any translated mRNAs. Interestingly, Tma108 and Scp160 targets show almost no overlap (only 6 mRNAs are shared in the two datasets). Gene ontology (GO) analysis revealed that Scp160 targets mRNA encoding proteins expressed in the secretory pathway compartments (membranes, vacuole, Golgi, endoplasmic reticulum) (Figure 3C), in agreement with an earlier study (2). Conversely, mRNAs encoding cytoplasmic and nuclear proteins were over-represented among Tma108 targets (Figure 3C). These observations support the fact that Tma108 and Scp160 define different subsets of ribosomal particles associated with distinct mRNAs. In addition, they reveal that Tma108 targets only a very limited fraction of the cellular mRNA (3%) indicating high selectivity for this translation machinery-associated factor.
Figure 3.
Tma108 targets ribosome translating a subset of the cellular mRNA. (A) Strategy developed to identify mRNA specifically enriched with factors associated with translating ribosomes. Immunoprecipitations were performed from cells expressing proteinA tagged versions of Tma108, Scp160 or Tma46. As a control, the whole set of ribosomes was immunoprecipitated using cells expressing a tagged version of Rpl16a. (B) Hierarchically clustered heat map showed IP/input log2(ratios) for mRNA enriched with at least one of the tested ribosomal partners in two independent experiments (Supplementary Tables S7 and S8). Each line represents an experiment, each column a gene. (C) Analysis of over-enrichment and under-enrichment of GO terms (cellular component) among Tma108 and Scp160 mRNA targets (Supplementary Table S9). The extent of the enrichment for each GO term is expressed as a fold enrichment over genome. Significant over- and under-enrichments are indicated (***P-value < 10−3, **P-value < 5 × 10−2). (D) Tma108 targets are highly enriched in defined molecular functions. Analysis of enrichment of the GO terms revealed three main categories of over-represented functions: Nucleotide binding (P-value: 2.3 × 10−8), Zinc ion binding (P-value: 1.5 × 10−5) and RNA binding (P-value: 9.7 × 10−5). Over-represented functional categories included in these three GO terms are not represented (Supplementary Table S10). (E) Analysis of Pfam domains over-represented in proteins encoded by Tma108 targets. Eighteen Pfam domains that showed a significant enrichment (P-value < 0.05) in Tma108 dataset were selected (identifiers and P-value in Supplementary Table S11). They were classified according to their belonging to the different GO terms (function) enriched in Tma108 list (e1: Nucleotide binding, e2: RNA binding, e3:Zinc ion binding, e4: Others). The boxplots represent the distribution of mean log2(IP/Input) values obtained in microarray analyses of Tma108 immunoprecipitation for the whole cellular mRNA and for the analyzed GO categories. For each Pfam domain, the values of log2(IP/Input) measured for each individual gene included in the Pfam dataset are plotted (yellow diamond) overlapped on the boxplot (light gray) of the corresponding GO term (e1–3) or of the total RNA dataset (e4). Pfam domains indicated with green circles correspond to domains with ATPase activity. Dashed lines correspond to the 0.8 threshold that was chosen to determine Tma108 targets.
Tma108-associated ribosomes preferentially target mRNAs encoding proteins bearing ATP-binding, RNA-binding or Zinc-binding domains
In order to further understand Tma108 selectivity, we analyzed the representation of the diverse Gene Ontology categories (process and function) within the Tma108 mRNA target dataset. Two cellular processes were significantly over-represented amongst Tma108 targets: Glutamine metabolic process (P-value: 7.8 × 10−7) and Stress granule assembly (P-value: 5.2 × 10−5). However, the mRNAs linked to these processes only represent 6% of the mRNA enriched in the Tma108 dataset. In contrast, 45% of the mRNAs co-immunoprecipitated with Tma108 belong to at least one of the following three "biochemical function" categories: Nucleotide binding (P-value: 2.3 × 10−8), Zinc ion binding (P-value: 1.5 × 10−5) and RNA binding (P-value: 9.7 × 10−5) (Figure 3D). These observations indicate that Tma108 mRNA targets do not encode proteins involved in a common cellular pathway, but rather proteins that share common biochemical properties. In particular, 24% of the mRNAs translated by Tma108-associated ribosomes encode ATP-binding proteins.
We then questioned whether or not this over-representation of defined biochemical functions amongst the Tma108 targets was due to its selectivity for specific protein domains encoded by the target mRNAs. Enrichment analysis identified 18 over-represented Pfam domains, among which seven were nucleotide-binding domains (Figure 3E, panel 1), five were RNA-binding domains (Figure 3E, panel 2), three were Zinc ion binding domains (Figure 3E, panel 3) and five corresponded to other protein motifs (Figure 3E, panel 4). These domains were found in 51% of the nucleotide-binding proteins, 67% of the RNA-binding proteins and 38% of Zinc-binding proteins identified in the Tma108 dataset respectively. Interestingly, six out of the seven identified nucleotide-binding domains had ATPase activity (Figure 3E, panel 1). In several cases, we observed that all the members of the Pfam family corresponded to mRNAs targeted by Tma108 (Figure 3E). The most striking result was obtained for the Glutamine amidotransferase domain category (GATase), whose members (Asn1, Asn2, Ade4, Gfa1, Yml096w, Dug3) had the highest enrichment values in the microarray experiments (Figure 3E, panel 4). This selectivity for the GATase domain accounts for the over-representation of proteins involved in glutamine metabolism as mentioned above. To a lesser extent, the same kinds of results were observed for several other domains, notably RIO1, PFK, eIF-4G1, Sec 23, RNA_pol_Rpb1_5 and nucleoporin2. However, for certain domains, only a subset of the mRNA was enriched compared to the global distribution. This was the case for the ATP synthase α/β domain family which includes two subunits of the mitochondrial ATP synthase (Atp1, Atp2) and two subunits of the vacuolar H+-ATPase (Vma1, Vma2): in this case, only ATP2 and VMA1 mRNAs co-purified with Tma108 (Figure 3E, panel 1). For large families of proteins, like DEAD-box proteins or AAA-ATPases, only a subset of the family members (14% of the DEAD box proteins, 13% of the AAA ATPase) was enriched in Tma108 immunoprecipitation.
Our analysis thus reveals that Tma108 preferentially targets ribosomes translating ATP-binding, RNA-binding or Zn-binding proteins. This suggests that Tma108 selectivity could be based on the presence of specific functional domains in the proteins encoded by its mRNA targets. However, other parameters are involved in Tma108 selectivity, as attested by the fact that some proteins containing one of the aforementioned domains do not have their mRNA enriched in the Tma108 IP.
Tma108 is a non-canonical member of the M1 aminopeptidase family, harboring a peptide-binding domain that might have evolved to reach specific substrates
Our observation regarding the selectivity of Tma108 targeting raised questions about the nature of the physical interactions driving this process and the specificity of the underlying molecular mechanism. We used phylogenic analyses and homology-based modeling to define the domains of the protein that could contribute to such specific interactions. Tma108 has been shown to be an essential gene in some genetic background (39), we assumed that selective pressures could have shaped the conservation of Tma108 residues according to their functional relevance.
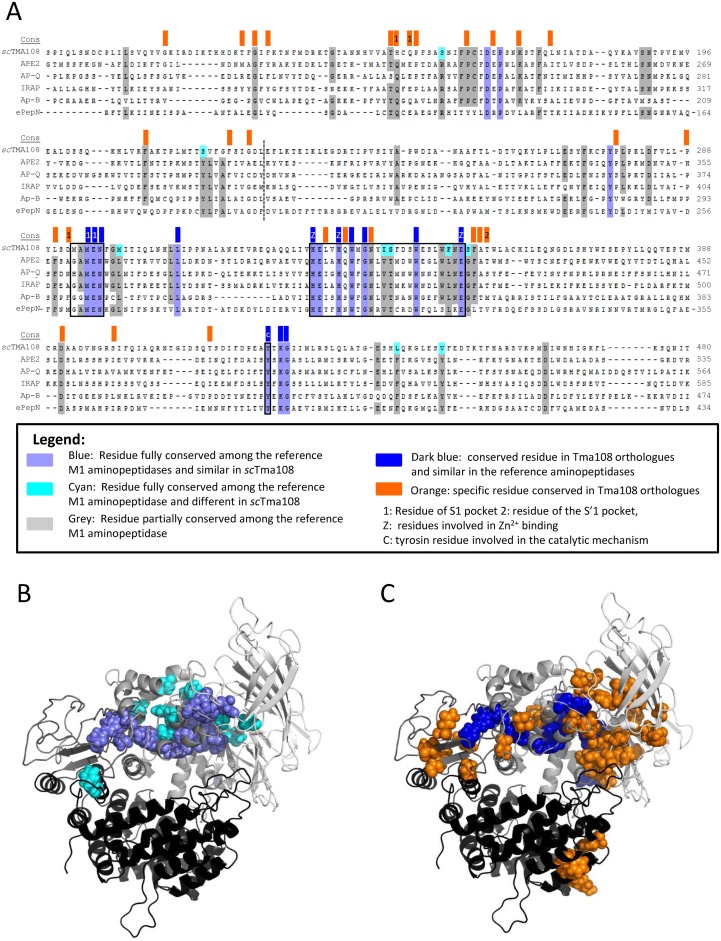
As Tma108 has been identified as a member of the M1 aminopeptidase family, we could compare it to 14 well-characterized M1 aminopeptidases (11 Homo sapiens, 1 E. coli, 2 S. cerevisiae sequences) using multiple alignments of the primary structures. The phylogenic tree obtained from this analysis (Supplementary Figure S3) showed that Tma108 was not closely related to previously known aminopeptidases and defined an independent branch of the tree (as opposed to its closest yeast relatives, Ape2 and Aap1). Nevertheless, the alignment (Figure 4A) confirmed the good conservation of the characteristic Zn-binding motif of M1 metallopeptidases, HEXXHX18E (40). We also observed that the tyrosine residue involved in the catalytic mechanism and generally located 80–90 residues after the HEXXHX18E motif in M1 aminopeptidases was conserved in Tma108 (40). The (G/A/H/V)(G/A)MEN motif, found in 77% of the M1 family proteins and involved in substrate binding and recognition (41), was also present in Tma108. Only the 293M in the first position of the motif (MAMEN) is not congruent with the defined signature. Finally, most of the residues common to all reference M1 aminopeptidases (blue in Figure 4A and B) were also observed in Tma108, as displayed in the 3D model (Figure 4B). The residues specific to Tma108 (cyan in Figure 4A and B) are mostly located in the periphery of the catalytic pocket.
Figure 4.
Tma108, a new protein of the M1 aminopeptidase family. (A) S. cerevisiae Tma108 (scTma108) was compared to 14 well-characterized M1 aminopeptidases using multiple alignment of primary structures, followed by residue conservation analysis. The alignment shown corresponds to the most conserved region and is restricted to five M1 aminopeptidases. Residues in blue are fully conserved among the 15 M1 aminopeptidases (including Tma108), those in grey are partially conserved (90% of the sequences shown). Residues that are conserved in all the M1 aminopeptidases except in Tma108 are indicated in cyan on Tma108 sequence. Furthermore, Tma108 conservation was analyzed after retrieval of Tma108 orthologs in the diverse sequenced yeast genomes. Eighteen sequences from species (including S. cerevisiae) distributed along the phylogenic tree of the Saccharomycetaceae, were selected to detect residues that are fully conserved. These residues are indicated by color boxes at the top of the alignment (line labeled cons): dark blue boxes indicate residues that are conserved among Tma108 orthologs and similar in the reference aminopeptidases, orange boxes indicate residues that are specific to Tma108 orthologs. Numbers in the boxes indicate residues that were previously shown to be implicated in M1 aminopeptidase activities (based on the crystal structure of aminopeptidase N(42,43)): binding of the first amino acid of the substrate in S1 pocket (1), binding of the second amino acid of the substrate in S'1 pocket (2), binding of the Zn2+ cofactor (z), catalysis (c). Frames on the alignment indicate the two sequence signatures of the M1 family, (G/A/H/V)(G/A)MEN and HEXXHX18E, and the tyrosine involved in the catalytic mechanism. A dashed line indicates the separation between domains 1 and 2 of Tma108 inferred by homology based structure modeling (see below). (B and C) 3D molecular model of Tma108 of S. cerevisiae. Four domains are shown in different colors from light gray to black: NH2-terminus (1–227), central (228–490), hinge (491–578) and COOH terminal (579–946). Residues underscored by the two conservation analyses (b: conserved residues in all the M1 aminopeptidases, c: conserved residues in all the Tma108 orthologs) described in (a) are colored in the structure (same color code as in a).
We analyzed the sequence conservation of Tma108 amongst the potential orthologs of Tma108 found in the Saccharomycetaceae familly. We identified 57 fully conserved residues, among which 15 were identical to the residues found in all the reference aminopeptidases (residues in dark blue in Figure 4A and C). According to their position in the 3D model (Figure 4C), these residues make up the central part of the M1 aminopeptidase catalytic pocket. Interestingly, we also observed 42 residues that are specific and conserved in Tma108 orthologs (orange in Figure 4A and C). Although they are not contiguous in the Tma108 sequence, these residues are clustered along the catalytic pocket. Some of them (153H, 155Q, 292D and 357T) correspond to positions that were demonstrated to be part of the S1 or the S'1 pocket involved in the substrate recognition of aminopeptidase N in E. coli (42,43).
M1 aminopeptidases are specialized in the hydrolysis of N-terminal residues of peptides, although some of them have evolved to recognize other substrates (this is notably the case of the Leukotriene A4 hydroxylases). Our analysis highlighted the conservation of the catalytic pocket but also, the existence of residues specific to the Tma108 protein. These results suggest that the selectivity of Tma108 is linked to the recognition of specific peptides by this region.
Tma108 directly interacts with the proteins translated by the Tma108-associated ribosomes
In order to gain insight into Tma108 interactions, we tried to identify proteins that specifically associate with Tma108 ribosomes. For this purpose, a comparative proteomic analysis of the proteins co-immunoprecipitated with Tma108, Scp160, Tma46 and Rpl16a was performed (Figure 5A). Proteins found in the four immunoprecipitation datasets mainly corresponded to ribosomal proteins and translation factors. In addition, ribosomal biogenesis factors were exclusively detected in Rpl16a immunoprecipitation, demonstrating that the ribosomes associated with Tma108, Scp160 and Tma46 were mature translating ribosomes. Strikingly, the only proteins which were specific to the Tma108 IPs were Asn1, Asn2 and Atp2, whose mRNAs were amongst those most enriched in the Tma108 IPs (Figure 3E, panels 1 and 4). Since our immunoprecipitations were performed in conditions which maintained the integrity of ribonucleoprotein complexes, our analysis was likely to detect the nascent chains of Asn1, Asn2 and Atp2 being translated by the Tma108-specific ribosomes. These results prompted us to investigate whether the specificity of Tma108 for proteins harboring particular Pfam domains could be due to its direct interaction with these proteins during their translation.
Figure 5.
Tma108 directly interacts with proteins encoded by its associated translating complexes. (A) Comparison of the composition of Tma108, Scp160 and Tma46-associated ribosomes with the composition of canonical (Rpl16A-associated) ribosomes. Experimental strategy (top panel). Immunoprecipitations of proteinA-tagged versions of Tma108, Scp160, Tma46 or Rpl16a were followed by mass spectrometry analysis of their protein partners. Venn Diagramm represents the overlap of the four different sets of co-immunoprecipitated proteins: in each sector the number of interactants reproducibly detected in two independent experiments is indicated (Supplementary Table S12). Chart pies show the functional distribution of proteins for shared and Rpl16a specific dataset. The names of the proteins included in the different sectors are indicated. The 3 proteins specifically detected in Tma108 IP are also indicated. (B) Analysis of Tma108 partners following ribosome dissociation with EDTA. Experimental strategy (top panel) used to identify Tma108 interacting partners. Immunoprecipitations (n = 2) were performed on cells expressing Tma108-PA in the presence of 40 mM EDTA (see Figure 2B–D). RNAs (b1) were analyzed by real-time PCR. For each analyzed mRNA, the enrichment factor corresponds to the ratio IP/Input normalized using a set of control mRNA (Act1, Trx2, Jen1, Flr1). Mass spectrometry analysis (b2) of proteins co-immunoprecipitated with Tma108 in the presence of EDTA: only seven proteins were reproducibly detected in that condition. (C) Most of the Tma108-specific proteins partners are translated by Tma108-associated ribosomes. The boxplot represents the distribution of mean log2(IP/Input) values obtained following microarray analysis of Tma108-associated mRNAs (in the absence of EDTA). Arrows point the values for the mRNAs corresponding to the proteins that were detected as specific Tma108 partners (Figure 5B, panel 2).
To test this hypothesis, we took advantage of the well-characterized EDTA-mediated dissociation of translating ribosomes (16). The cell extracts were treated with 40 mM EDTA before performing Tma108 IPs, in order to identify which components of the ribosomal RNPs were interacting Tma108 partners (Figure 5B). Under these conditions, we could not detect any enrichment for ATP2, ASN1 or VMA1 mRNAs (Figure 5B, panel 1). Similarly, mass spectrometry analyses revealed that ribosomal proteins no longer co-immunoprecipitated with Tma108 after EDTA treatment (see also Figure 2C and D). Still, seven proteins were reproducibly detected in EDTA-treated Tma108 IPs (Figure 5B, panel 2). Four of them (Asn1, Asn2, Atp2 and Vma1) were translated by Tma108-associated ribosomes as determined by our previous microarray analyses (Figure 5C). Thus, although the EDTA treatment had successfully dissociated the Tma108 ribonucleoparticles, the detection of these four proteins suggests a direct interaction between Tma108 and their nascent polypeptides.
Tma108 recognizes the N-terminus of the nascent peptides during translation, a process that involves its putative peptide-binding pocket
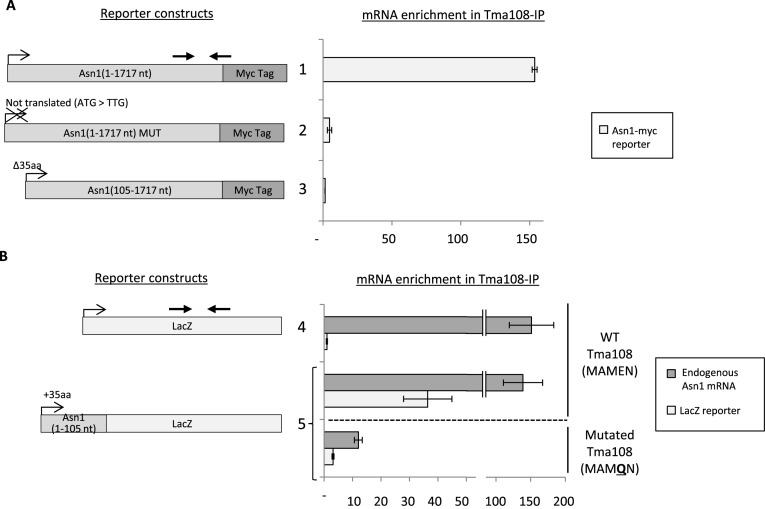
Our results strongly suggest that Tma108 is a specific nascent chain-associated factor. According to this model, several predictions can be made: (i) the coding sequence (without the untranslated regions) might be sufficient to recruit Tma108, (ii) impairment of the translatability of the mRNA might prevent the recruitment of Tma108 and (iii) the N-terminal part of the target might be involved in the selective recruitment of Tma108. To test these predictions, we assessed the ability of Tma108 to recognize different mutated versions of Asn1 (Figure 6A) and Atp2 (Supplementary Figure S7a) fused to reporters, by measuring the co-purification of the fusion-encoding mRNAs with Tma108-PA. As expected, we observed a high enrichment of an mRNA corresponding to the coding sequence of Asn1 fused to 13 repeats of the myc epitope and whose expression was controlled by the ZWF1 promoter (Figure 6A, construct #1), in the Tma108 IP. The mRNA enrichment factors for this construct were similar to those measured for the endogenous ASN1 mRNA (data not shown, for an example see Figure 6B) suggesting that the untranslated regions of the ASN1 mRNA are not involved in the recruitment of Tma108. In the absence of translation (ATG→TTG mutant, Figure 6A, construct #2), ASN1-myc mRNA was no longer enriched in the IP, while endogenous ASN1 mRNAs were still co-immunoprecipitated with Tma108 (data not shown). mRNA quantification indicated that this effect was not caused by mRNA degradation in the absence of translation, since the start codon mutation had little effect on the level of Asn1-myc mRNA (Supplementary Figure S7b). This result demonstrates that translation is absolutely necessary for Tma108 to interact with Asn1-myc mRNA, and supports the model of an interaction with the nascent chain. Furthermore, we observed that an mRNA encoding a truncated version of ASN1-myc, whose first 35 amino acids had been deleted, was no longer associated to Tma108 (Figure 6A, construct #3). Additionally, these first 35 amino acids were sufficient to recruit Tma108 to a LacZ reporter mRNA (Figure 6B constructs #4 and 5). Since our genomic analysis pointed to Tma108 as an original M1 metallopeptidase (Figure 4), we performed a mutational analysis to test if the region corresponding to the putative catalytic peptide-binding pocket was involved in the recognition of these 35 amino acids. The M1 aminopeptidase family differs from other gluzincins (defined by the HEXXHX18E motif) by the (G/A/H/V)(G/A)MEN motif, an anionic coordination motif that recognizes the free N terminus of substrates (40). Previous studies focusing on various M1 metallopeptidases have demonstrated that mutations in this motif, notably the replacement of the Glu residue by a Gln, decreased or abolished aminopeptidase activity, while maintaining the general structure of the catalytic pocket (44–46). We constructed a mutant strain with a single amino acid substitution in the genomic MAMEN motif (E296Q) of Tma108-PA and verified that this mutated version of Tma108 was correctly expressed and immunoprecipitated (Supplementary Figure S7c). Strikingly, we observed that Tma108-E296Q lost its ability to interact with both the endogenous ASN1 and the LacZ reporter mRNA fused to the 35 first amino acids of Asn1 (Figure 6B, construct #5). Furthermore, the ability of this mutant to interact with ribosomes was decreased, as indicated by a strong reduction in the amounts of the ribosomal proteins Rpl1 and Rpl3 in the Tma108 immunoprecipitation (Supplementary Figure S7c). This result supports the hypothesis that the M1 metallopeptidase peptide-binding pocket of Tma108 is involved in the recruitment of Tma108 on ribosomal particles, most likely by the recognition of the N-terminal residues of the targeted protein.
Figure 6.
The translation of the N-terminal region of Asn1 is required to recruit Tma108 on the ribosome. Real-time quantitative RT-PCR analyses of the enrichment in Tma108-PA IPs of various ASN1 versions fused to a 13-myc Tag (A, construct 1–3) or a LacZ gene (B, construct 4–5). For construct 5, the experiment was also performed with a mutated version of the Tma108-PA (E296Q) in which the glutamate of the MAMEN motif was replaced by a glutamine. The arrows in bold indicate the positions of the primers used for the Q-PCR. The mean enrichment factors were obtained from two independent immunoprecipitation experiments by calculating the ratio IP/Input normalized using ACT1 and ATP1 as control mRNAs. In each experiment, the enrichment of the endogenous ATP2 mRNA was measured and confirmed that Tma108-PA ribonucleoparticles were efficiently immunoprecipitated (data not shown).
A specific signal present in the first 35 Asn1 amino acids is thereby necessary and sufficient to recruit Tma108 early during translation. However, the enrichment factors obtained for the N-terminal residues of Asn1 fused to the LacZ protein were not as high as those obtained for its full sequence. This suggests that an additional sequence within the Asn1 polypeptide might be involved in anchoring Tma108 to the nascent protein. Additionally, we observed that the full-length Asn1-myc protein is specifically detected in Tma108 IPs (Supplementary Figure S7d). This observation indicates that Tma108 remains associated with its target until translation is complete and may even be involved in post-translational events. Similar results showing the essential role of active translation and of the N-terminal residues of the protein for the recruitment of Tma108 were obtained with Atp2 reporter constructs (Supplementary Figure S7a).
Our analysis of the signals involved in Tma108 recruitment to both ATP2 and ASN1 mRNA emphasize the crucial role of translation and of the N-terminal amino acids of Tma108 targets. These results strongly support the assumption that Tma108 is a specific nascent chain-associated factor that is selectively loaded onto N-terminal amino acids during translation (Figure 7). This selective recognition of targets involves the catalytic peptide-binding pocket of Tma108, which is enriched in conserved residues specific to the Tma108 family (Figure 4C).
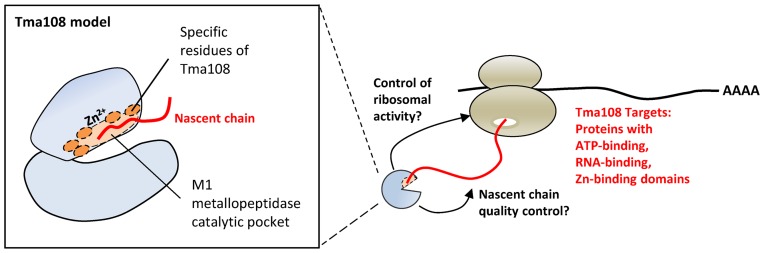
Figure 7.
Model highlighting the co-translational capture of selected nascent chain by Tma108. This study has revealed that Tma108, a putative M1 metallopeptidase, interacts with a subset of ribosomal particles translating mRNA encoding proteins with ATP-binding, RNA-binding and Zinc-binding domains (right). We propose that Tma108 is recruited on selected nascent-chain through interactions involving specific residues located in its catalytic pocket (left). The observation that Tma108 is essential in some cellular contexts (39) strongly suggests that Tma108 could be involved in specific co-translational events.
DISCUSSION
While studying candidate genes potentially involved in ATP2 mRNA mitochondrial localization, we identified Tma108 as a a specific ribosome-associated factor. Indeed, we observed that Tma108 has a high selectivity for mRNA encoding proteins of diverse subcellular compartments with ATP-binding, RNA-binding or Zinc-binding domains (Figure 3). Tma108 represents the first example of M1 metallopeptidase family involved in ribosomal complexes. Our results support the fact that Tma108-ribosome specificity relies on the recognition of the first amino acids of the nascent chain during translation, possibly through its conserved catalytic pocket. Strikingly, the deletion of Tma108 was previously shown to be lethal depending on the yeast's genetic background (39). This latter observation strongly supports the hypothesis that, in certain situations, specific co-translational events involving Tma108 could be essential for cellular homeostasis.
Tma108, a novel nascent chain-associated factor, interacts co-translationally with a subset of the nascent proteome
Tma108 was identified in 2006 by Fleischer et al. as a new translation machinery-associated factor of unknown function (16). In this study, we used affinity purification experiments coupled with mass spectrometry and microarray analyses, under conditions that preserve the structure of translating complexes, in order to identify the mRNAs translated by Tma108-associated ribosomes. Using this approach, we demonstrated that Tma108 was specifically associated with a sub-population of ribosomes translating 174 mRNA encoding proteins localized to various sub-cellular compartments. Moreover, immunoprecipitations in the presence of EDTA, which dissociates the different components of the polysome complexe, revealed that Tma108 might specifically interact with the nascent peptide. This was attested by the exclusive recovery of several proteins encoded by the target mRNAs, while the translated mRNA and the ribosomal subunits were lost. Only 7 of the 174 potential targets of Tma108 were detected using this approach. This could be caused by the unstable and transient nature of interactions with the nascent chains, as highlighted by previous studies which exhibit the difficulty of capturing the targets of factors acting co-translationally on nascent chains by proteomic approaches (37). Still, the existence of a direct RNA-independent interaction between Tma108 and the nascent chains of Asn1 and Atp2 was strongly supported by the observation that the translation of the first N-terminal amino acids of these proteins was required for Tma108 to associate with ASN1/ATP2 mRNAs (Figure 6 and Supplementary Figure S7).
Since we observed that most of Tma108 targets have ATP-binding, RNA-binding or Zinc-binding domains we can infer that Tma108 specifically selects ribosomes translating proteins that share common biochemical properties (Figure 3). Similar results were obtained by the Frydman group, which demonstrated that various chaperones take over subsets of nascent chains with common fates or folding constraints in yeast (36,37). For instance, the HSP70 SSB chaperone interacts with longer, more slowly translated and aggregation-prone nascent polypeptides. Likewise, our results indicate that Tma108 could recognize specific features within the nascent chain, as indicated by its apparent selectivity for some Pfam domains. However key features distinguish Tma108 from these Hsp proteins. First, the pool of Tma108 targets (<200) is much more restricted than that of the chaperones studied to date (typically 2000 mRNA targets for HSP70 SSB). Then, Tma108 recognizes a specific signal within the first amino acids of the nascent chain (Figure 6 and Supplementary Figure S7). Finally, Tma108 belongs to the M1 aminopeptidase family and does not share any structural features with Hsp proteins (as shown below).
In 2007, after the discovery of multiple RNA binding proteins and non-coding regulatory RNAs (mi-RNA, si-RNA), Keene proposed the post-transcriptional RNA regulon model. In this model, functionally related mRNAs are co-regulated at different steps from transcription to translation by diverse trans-acting factors (1). Similarly, the observation that nascent chain-associated factors, like Tma108, define different subsets of ribosomal particles, could indicate that specific quality controls are involved in the translation of proteins with different biochemical properties (36,37,9).
Tma108, a member of the M1 metallopeptidase family, interacts with specific nascent chains
A myriad of factors were shown to be involved in welcoming the nascent chain, but their selectivity was poorly explored (47–49). In this context, Tma108 would be a unique example of a nascent-chain associated factor with such narrow selectivity, which raises the question of the molecular basis of this selectivity. Tma108 belongs to the M1 metallopeptidase family characterized by a Zn-binding catalytic pocket in which peptides are docked and their N-terminal residue hydrolyzed (40). Although these enzymes are found in organisms ranging from bacteria and plants to vertebrates (50) and have been involved in a variety of important biological functions (51), until now none of them have been shown to interact with nascent peptides during translation.
Comparative analyses revealed that the catalytic pocket is well conserved in Tma108, underscoring its importance for Tma108 function. Since peptides are the natural substrates of M1 metalloproteases, it was consistent to speculate that Tma108 evolved to recognize the nascent chain during translation. This hypothesis was corroborated by data from immunoprecipitation experiments in conditions that dissociate the translating ribosomal complexes (Figure 5) and by the observation that the translatability of the targeted mRNAs is crucial to recruit Tma108 (Figure 6 and Supplementary Figure S7). Finally, a single amino acid substitution (E296Q), in the MAMEN peptide-binding motif destroyed the ability of Tma108 to recognize the Asn1 nascent chain, further demonstrating the importance of this domain for Tma108 function.
It is unknown whether or not Tma108 displays peptidase activity that could contribute to the specific turnover of improperly folded proteins. Further studies, including mutagenesis and in vitro assays, will be necessary to fully elucidate the structure-function relationships within Tma108. How M1 metalloproteases select their substrates among the diverse cellular proteins is still unclear and a systematic exploration of Tma108 residues involved in that process would be necessary to answer this question. Indeed, we observed the existence of conserved residues surrounding the putative catalytic region of Tma108. These residues were specific to the Tma108 orthogroup and could be implicated in the selectivity of Tma108 for its substrates. We demonstrated that interaction between Tma108's putative peptide-binding pocket and Asn1's first 35 residues was sufficient to recruit Tma108 on a LacZ reporter gene (Figure 6). Interestingly, according to the Pfam database (25), the GATase domain of Asn1 corresponds to amino acids 3–165, demonstrating that the signal responsible for the recruitment of Tma108 overlaps with this domain. Nevertheless, we were not able to isolate a common motif within the first 50 amino acids of the Tma108 targets (data not shown) thus, it would be interesting to investigate whether a specific structure adopted by the nascent chain is involved in the recruitment of Tma108.
The impact of Tma108 on protein synthesis: lessons from the ATP2 model mRNA
Our observation that Tma108 targets a ribosome subpopulation raises the question of its impact on the translational process. Recent works, including our study, demonstrate that ribosomes serve as docking sites for a multitude of enzymes, targeting factors and chaperones acting on the nascent polypeptides emerging from its exit tunnel (47–49). It is likely that nascent-chain associated factors couple ribosome activity with proper folding and quality control of the nascent proteome, thereby allowing for fine tuning of protein synthesis.
Strikingly, our single molecule FISH and subcellular fractionation data clearly demonstrated that the absence of Tma108 disturbed the spatio-temporal translation of ATP2 mRNA (Figure 1). Based on previous studies, we put forward the model that ATP2 mRNA localization to the mitochondria is mediated by the ability of its nascent amino-terminal peptide (the mitochondrial targeting peptide or MTS) to interact co-translationally with the mitochondria itself (14,52) (Supplementary Figure S4a). We also demonstrated that ATP2 mRNA localization to the mitochondria was the result of a narrow equilibrium between targeting and elongation velocities of the nascent chain. Indeed, the blocking of elongation with cycloheximide drastically increased ATP2 mRNA localization ((13) and unpublished data). On the contrary, the uncoupling of the mitochondrial proton gradient, a prerequisite for MTS interaction with the mitochondria, delocalized ATP2 mRNA (14). We propose that Tma108 may participate in regulating the dynamics of ATP2 translation and that the impairment of these dynamics upon Tma108 inactivation indirectly alters its delivery to the mitochondria. Several non-exclusive hypotheses may explain the negative impact of Tma108 on ATP2 mRNA localization (Supplementary Figure S4b). First, Tma108 could stimulate the elongation of ATP2 mRNA and thus accelerate the release of the translation machinery. This would disrupt the mRNA targeting process, which requires a constant interaction between the mRNA and the ribosome until their association with the mitochondria. Alternatively, Tma108 could participate in a specific co-translational quality control and its interaction with the Atp2 nascent chain could mask the MTS and reduce its ability to interact with the mitochondria.
Tma108 targets about 200 protein-coding mRNAs involved in diverse cellular pathways but sharing common biochemical properties. Quantitative proteomic analysis indicates that Tma108 does not significantly affect the steady state levels of its targets (Supplementary Figure S8). This result indicates that Tma108 function is not related to translation initiation, the rate-determining step of translation (53). This result is consistent with the role of Tma108 as a nascent chain-associated factor, recruited once the translation process is engaged. Our results also demonstrate that the absence of Tma108 does not impact the stability of the targeted proteins. However, it should be noted that these quantitative proteomic experiments have been conducted in a strain background in which Tma108 is dispensable. The observation that Tma108 is essential in other genetic backgrounds (39) strongly suggests that specific co-translational events involving Tma108 could be essential for cellular homeostasis.
Supplementary Material
Acknowledgments
We are grateful to Thierry Foulon, Sandrine Cadel, and Micheline Fromont-Racine for their useful advice and critical readings of the manuscript. We thank the “Plateforme de protéomique structurale et fonctionnelle de l'Institut Jacques Monod" for our mass spectrometry analyses. We thank Stéphane Le Crom and Jean-Charles Cadoret for granting us access to their microarray facilities. We also thank Alice Lambert and Baptiste Saudemont for the careful proofreading of the manuscript.
Footnotes
Present address: Mathilde Garcia, UMR7238, UPMC/CNRS, Paris, 75006, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IBPS (Institut de Biologie Paris Seine); STRUDYEV project (ANR-10-JCJC-1603) of the Agence Nationale pour la Recherche. Funding for open access charge: IBPS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 2.Hogan D.J., Riordan D.P., Gerber A.P., Herschlag D., Brown P.O. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue S., Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipovska A., Rackham O. Specialization from synthesis: how ribosome diversity can customize protein function. FEBS Lett. 2013;587:1189–1197. doi: 10.1016/j.febslet.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 5.Shi Z., Barna M. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 2015;31:31–54. doi: 10.1146/annurev-cellbio-100814-125346. [DOI] [PubMed] [Google Scholar]
- 6.Adams D.R., Ron D., Kiely P.A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal.: CCS. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao S., Brown A., Santhanam B., Hegde R.S. Structure and assembly pathway of the ribosome quality control complex. Mol. cell. 2015;57:433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao S., Hegde R.S. Target selection during protein quality control. Trends Biochem. Sci. 2016;41:124–137. doi: 10.1016/j.tibs.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Pausch P., Singh U., Ahmed Y.L., Pillet B., Murat G., Altegoer F., Stier G., Thoms M., Hurt E., Sinning I., et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 2015;6:7494. doi: 10.1038/ncomms8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung H., Gkogkas C.G., Sonenberg N., Holt C.E. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margeot A., Blugeon C., Sylvestre J., Vialette S., Jacq C., Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21:6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saint-Georges Y., Garcia M., Delaveau T., Jourdren L., Le Crom S., Lemoine S., Tanty V., Devaux F., Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia M., Delaveau T., Goussard S., Jacq C. Mitochondrial presequence and open reading frame mediate asymmetric localization of messenger RNA. EMBO Rep. 2010;11:285–291. doi: 10.1038/embor.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams C.C., Jan C.H., Weissman J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischer T.C., Weaver C.M., McAfee K.J., Jennings J.L., Link A.J. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petitjean A., Bonneaud N., Lacroute F. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal proteins. Mol. Cell. Biol. 1995;15:5071–5081. doi: 10.1128/mcb.15.9.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia M., Darzacq X., Devaux F., Singer R.H., Jacq C. Yeast mitochondrial transcriptomics. Methods Mol. Biol. 2007;372:505–528. doi: 10.1007/978-1-59745-365-3_35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jourdren L., Delaveau T., Marquenet E., Jacq C., Garcia M. CORSEN, a new software dedicated to microscope-based 3D distance measurements: mRNA-mitochondria distance, from single-cell to population analyses. RNA. 2010;16:1301–1307. doi: 10.1261/rna.1996810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oeffinger M., Wei K.E., Rogers R., DeGrasse J.A., Chait B.T., Aitchison J.D., Rout M.P. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 21.Vizcaino J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J.A., Sun Z., Farrah T., Bandeira N., et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemoine S., Combes F., Servant N., Le Crom S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics. 2006;7:467. doi: 10.1186/1471-2105-7-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 28.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 29.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remmert M., Biegert A., Hauser A., Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. Procheck – a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 32.Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 33.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia M., Darzacq X., Delaveau T., Jourdren L., Singer R.H., Jacq C. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell. 2007;18:362–368. doi: 10.1091/mbc.E06-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang B.D., Fridovich-Keil J.L. Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res. 2000;28:1576–1584. doi: 10.1093/nar/28.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willmund F., del Alamo M., Pechmann S., Chen T., Albanese V., Dammer E.B., Peng J., Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.del Alamo M., Hogan D.J., Pechmann S., Albanese V., Brown P.O., Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li A.M., Watson A., Fridovich-Keil J.L. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830–1837. doi: 10.1093/nar/gkg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowell R.D., Ryan O., Jansen A., Cheung D., Agarwala S., Danford T., Bernstein D.A., Rolfe P.A., Heisler L.E., Chin B., et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hooper N.M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 41.Cadel S., Darmon C., Pernier J., Herve G., Foulon T. The M1 family of vertebrate aminopeptidases: role of evolutionarily conserved tyrosines in the enzymatic mechanism of aminopeptidase B. Biochimie. 2015;109:67–77. doi: 10.1016/j.biochi.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Ito K., Nakajima Y., Onohara Y., Takeo M., Nakashima K., Matsubara F., Ito T., Yoshimoto T. Crystal structure of aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli and conformational change of methionine 260 involved in substrate recognition. J. Biol. Chem. 2006;281:33664–33676. doi: 10.1074/jbc.M605203200. [DOI] [PubMed] [Google Scholar]
- 43.Addlagatta A., Gay L., Matthews B.W. Structural basis for the unusual specificity of Escherichia coli aminopeptidase N. Biochemistry. 2008;47:5303–5311. doi: 10.1021/bi7022333. [DOI] [PubMed] [Google Scholar]
- 44.Luciani N., Marie-Claire C., Ruffet E., Beaumont A., Roques B.P., Fournie-Zaluski M.C. Characterization of Glu350 as a critical residue involved in the N-terminal amine binding site of aminopeptidase N (EC 3.4.11.2): insights into its mechanism of action. Biochemistry. 1998;37:686–692. doi: 10.1021/bi971705p. [DOI] [PubMed] [Google Scholar]
- 45.Laustsen P.G., Vang S., Kristensen T. Mutational analysis of the active site of human insulin-regulated aminopeptidase. Eur. J. Biochem./FEBS. 2001;268:98–104. doi: 10.1046/j.1432-1327.2001.01848.x. [DOI] [PubMed] [Google Scholar]
- 46.Vazeux G., Iturrioz X., Corvol P., Llorens-Cortes C. A glutamate residue contributes to the exopeptidase specificity in aminopeptidase A. Biochem. J. 1998;334:407–413. doi: 10.1042/bj3340407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramer G., Boehringer D., Ban N., Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 48.Gloge F., Becker A.H., Kramer G., Bukau B. Co-translational mechanisms of protein maturation. Curr. Opin. Struct. Biol. 2014;24:24–33. doi: 10.1016/j.sbi.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Pechmann S., Willmund F., Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page M.J., Di Cera E. Evolution of peptidase diversity. J. Biol. Chem. 2008;283:30010–30014. doi: 10.1074/jbc.M804650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tholander F., Muroya A., Roques B.P., Fournie-Zaluski M.C., Thunnissen M.M., Haeggstrom J.Z. Structure-based dissection of the active site chemistry of leukotriene A4 hydrolase: implications for M1 aminopeptidases and inhibitor design. Chem. Biol. 2008;15:920–929. doi: 10.1016/j.chembiol.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 52.Eliyahu E., Pnueli L., Melamed D., Scherrer T., Gerber A.P., Pines O., Rapaport D., Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell. Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.