Abstract
A century ago, Alfred Nissle discovered that intentional intake of particular strains of Escherichia coli could treat patients suffering from infectious diseases. Since then, one of these strains became the most frequently used probiotic E. coli in research and was applied to a variety of human conditions. Here, properties of that E. coli Nissle 1917 strain are compared with other commercially available E. coli probiotic strains, with emphasis on their human applications. A literature search formed the basis of a summary of research findings reported for the probiotics Mutaflor, Symbioflor 2, and Colinfant. The closest relatives of the strains in these products are presented, and their genetic content, including the presence of virulence, genes is discussed. A similarity to pathogenic strains causing urinary tract infections is noticeable. Historic trends in research of probiotics treatment for particular human conditions are identified. The future of probiotic E. coli may lay in what Alfred Nissle originally discovered: to treat gastrointestinal infections, which nowadays are often caused by antibiotic-resistant pathogens.
Keywords: probiotics, Escherichia coli, Nissle, Symbioflor, human studies, applications
Introduction
Many probiotic products are based on particular strains of lactic acid bacteria, such as Lactobaccillus and Lactococcus species (both Firmicutes) or Bifidobacterium species (belonging to the Actinobacteria). Other genera of bacteria (and even yeast species) are also used for probiotic applications, including Escherichia coli, a member of the Gammaproteobacteria. This working horse of bacteriology is not only the most frequently studied bacterial species on the planet but also a rather complicated one, since it includes both commensal and pathogenic strains whose genomes can widely vary in size and gene content [1].
That E. coli is chosen as a probiotic would be in line with its presumed ubiquitous presence in the gut. But how often is E. coli actually present in a human gut, in what numbers, and is it a “major player” in that environment? Despite a plethora of data available on this species, these data are not easy to find. In his book on normal human microflora, Tannock describes that E. coli is typically found in the ileum (the last third of the small bowel) as well as in the colon, but not outnumbering other more numerous species [2]. Caugent and colleagues describe coexistence of transitory and persistent clones, with rapid changes in the genetic composition of the population, but quantitative data are not given [3]. The colon contains approximately 1.5 kg of wet-weight bacterial cells, while feces contains about 1012 bacteria per gram [4]. According to a publication in 1974 by Hill and Drasar, the human lower intestine contains, on average, 2·103 (in the terminal ileum) to 1.6·106 (in the cecum) colony-forming units (CFU) Enterobacteria per gram intestinal material; for feces (which reflects luminal flora of the recto-sigmoid region rather than the mucosal and villous crypts flora), the average is 2.5·107 CFU/g [5]. E. coli is only one of several Enterobacteria species typically present, and to put these numbers into perspective, these Enterobacteria are outnumbered by a factor of 100 to 1000 by Bacteroides and Gram-positive nonspore-forming anaerobes [2, 5]. In line with this, E. coli is not among the top 25 most prevalent bacterial species typically present in feces of human subjects consuming a Western diet [2]. The numbers quoted here were based on cultural findings, and the limitations of this procedure have long been recognized: a significant proportion of the bacteria in the gut are uncultivable. Nevertheless, since culturing of E. coli is well established, culture-dependent results should be sufficient for a quantitative estimate. It is, therefore, surprising how few quantitative data exist on colonization by E. coli in healthy individuals. In a recent publication comparing obese with normal-weight people, Zuo and coworkers reported around 108 CFU E. coli per gram feces for both groups [6]. Although, in recent times, metagenomics studies provide insights in the uncultivable fraction of the gut microbiome, those methods are rather insensitive and usually do not detect species present in fewer than 105 cells [7]. Moreover, findings are often reported as phyla (e.g., “Proteobacteria”) or taxonomic families (“Enterobacteriaceae”) rather than individual genera or species. It has been observed that there is only 15% overlap between metagenomics and culture-dependent methods [7]. In addition, sequence-dependent methods frequently over-estimate the diversity of species being present, for a number of reasons discussed elsewhere [8]. For these and maybe other reasons, metagenomic data hardly ever provide a quantitative estimate on the number of E. coli bacteria in the gut. In the outstanding, recently published catalogue of the human gut microbiome determined from 124 European individuals (based on fecal samples), Escherichia was not among the 56 most abundant species [9]. From browsing through a large amount of literature, it seems safe to say that E. coli is “often” present in a human gut, though in relatively low numbers, and whether it a “major team player” in that environment remains to be seen.
Despite this, E. coli bacteria are the basis of at least three commercially available probiotic products, known under the commercial names Mutaflor, Symbioflor 2, and Colinfant, respectively. These products have been used in multiple scientific investigations to unravel their presumed positive effects on human health. Mutaflor, produced by Ardeypharm GmbH (Herdecke, Germany, a pharmaceutical company founded in 1970), contains viable cells of a single E. coli strain called E. coli Nissle 1917. Symbioflor 2 (DSM 17252), produced by SymbioPharm GmbH (Herborn, Germany, founded in 1954), contains a concentrate of six E. coli genotypes. Colinfant is marketed by Dyntec (Terezín, Czech Republic) and contains a single E. coli strain; it is specifically marketed for use in newborns and infants and is mainly used in the Czech Republic.
An extensive scientific literature on E. coli Nissle 1917 (hereafter referred to as EcN) provided some remarkable insights in this probiotic strain, in particular when compared to the limited available literature on Symbioflor 2 and Colinfant, as reviewed here.
A brief comparison of basic facts about EcN, Symbioflor 2, and Colinfant
Some of the basic properties of EcN present in Mutaflor, of the six E. coli genotypes found in Symbioflor 2, and of the E. coli strain of Colinfant are summarized in Table 1. Of note is the 100-times higher recommended daily dose of Mutaflor compared to Symbioflor 2; the recommended daily dose of Colinfant is in between these two. Moreover, EcN is motile while the constituents of Symbioflor 2 lack flagella and are, thus, nonmotile. Plasmid content also varies and, as will be discussed in a next section, the closest known relatives of these commercial strains differ.
Table 1.
Trends in publication frequency over time for four keywords combined with “probiotics”
| EcN | Ref. | Symbioflor 2 | Ref. | Colinfant newborn | Ref. | |
|---|---|---|---|---|---|---|
| Information about the product | ||||||
| Description | Single E. coli strain | Six E. coli genotypes | Single E. coli strain | |||
| Product | Mutaflor capsulesa | Symbioflor 2 suspension | Powder for preparation of per oral solution | |||
| Contents | 2.5-25-109 CFU/capsule | b | 1.5–4.5 107 CFU/ml | c | 0.8–1.6 108 CFU/dosis | d |
| Recommended daily dose | 1-2 Capsules/day (2.5-50-109 CFU) | 2–4 ml (3.0–18 107 CFU) | 0.8–1.6 108 CFU three times/week | |||
| Maximum daily dose | 4 Capsules (10-1010 CFU) | 18 107 CFU | Not specified, maximum treatment 4 weeks | |||
| Information about the E. coli present in the product | ||||||
| Isolation date | “1917” (see main text) | [58] | 1954 | [88] | Data not available | |
| Serotype | 06:K5:H1 | [56] | Variable including 035,129, 0:169, rough, all H– | [27] | 083:K24:H31 | [105] |
| Plasmid content | 2 Cryptic plasmids | [56] | 12 Plasmids | [88] | No plasmids | [10] |
| Microcin production | Microcin M, H47 | [106] | Microcin S | [107] | Data not available | |
| Motility | Motile (flagella present) | [58] | Nonmotile (flagella absent) | [27] | Data not available | |
| No. of genes | 5324 Genes | [56] | 28,180 Genes belonging to 6486 gene families | [27] | Data not available | |
| Closest relatives | CFT073, ABU83972 (UPEC) | [91] | K12, ATCC8739 (commensals) | [27] | CFT073, 536 (UPEC) | [19] |
a Products with alternative delivery systems can be available, depending on geographic regions
b Product information leaflet Mutaflor, Ardeypharm
c Product information leaflet Symbioflor 2, SymbioPharm
d Product information leaflet Colinfant Newborn, Dyntec
Summary of publications related to EcN, Symbioflor 2, and Colinfant
A literature search was carried out to assess the current scientific knowledge on the three commercial products containing E. coli. Acknowledging that the PubMed database does not cover all available scientific literature, it was nevertheless chosen because it retrieves consistent findings, in contrast to the self-learning algorithms of web search engines. Thus, on 20 October 2015, PubMed searches were performed with the search terms “Nissle,” “Mutaflor”, “Symbioflor”, “DSM 17252”, “Colinfant”, or “E. coli strain A0 34/86” (for reasons explained below) to identify the then available relevant literature. Titles and abstracts (when available) of retrieved hits were screened to categorize the main subject of each study, as summarized in Table 2. The scientific literature on EcN is far more extensive than that of Symbioflor 2 or Colinfant, with 228, 21, and 11 relevant publications identified in PubMed, respectively.
Table 2.
Results of literature searches
| Retrieved with | “Nissle” or “Mutaflor” | “Symbioflor” or “DSM | “Colinfant” or “E. coli strain |
|---|---|---|---|
| Subject of publication | N = 228a | 17252” N = 21 | A0 34/86” N = 11b |
| In vitro studies | 49 | 2 | 0 |
| Mouse model studies | 40 | 2 | 0 |
| Rat model studies | 11 | 0 | 1 |
| Pig model studies | 14 | 0 | 2 |
| Human studies | 30 | 7 | 4 |
| Reviews and commentaries | 31 | 3 | 0 |
| Genome analysis and comparisons | 7 | 3 | 2 |
| Various | 29 | 3 | 2 |
| Other or unclear | 3 | 1 | 0 |
| Publications by Alfred Nissle | 14 | Not applicable | Not applicable |
Publications in Chinese and publications in which EcN was only used as a control were also excluded.
a The original search retrieved 335 articles, of which 107 were discarded as duplicates or not relevant
b Two publications in Czech were screened based on the translated abstract only
For EcN, 49 publications described results mainly obtained from in vitro work, performed to investigate its probiotic properties and mechanisms, and 51 publications presented animal studies using mice (40) or rats (11). Although the latter efforts have resulted in many valuable insights, they mostly inform us how the gut of these animals reacts to this E. coli strain. Compared to humans, these hosts have different diets, residual microbiota, lifestyles (nocturnal, nonsocial, etc.), and, to some extent, physiological properties, so that the insights gained from these models are not always relevant to the probiotic properties of EcN in a human host. Because this review concentrates on the question what E. coli-based probiotics do to the host(s) for which they are marketed, rodent studies are mostly ignored here, as are publications based on in vitro models. Thirty studies that describe human responses to EcN, which will be discussed in details in the next section, were identified. Of interest are also 14 studies of EcN in the porcine host, where the product is mainly used to combat infectious diseases. Multiple review and commentary articles that summarize and discuss studies with EcN were identified, but since these mostly reproduce results from primary publications, they are not in extenso included in this contribution. A number of publications described various properties and characteristics of EcN, or applications of derived ghost vesicles or culture supernatant, which for simplicity were lumped under the category “various” in Table 2. The complete genome of strain EcN has been sequenced twice, and based on this information, the strain has been compared with other E. coli genomes, which in all resulted in seven publications that were identified by the performed searches. Finally, PubMed retrieved 14 publications by Alfred Nissle, who originally isolated the strain that bears his name, which will be further discussed in a next section.
Of the 21 identified publications on Symbioflor 2 also summarized in Table 2, seven dealt with human studies. Three were review articles (including a systematic review), and three publications described characteristics of the genomes of the Symbioflor 2 genotypes. Only two in vitro studies and two mouse model studies could be identified. Further, three publications concentrated on various subjects (characterization of microcin S production, application in an artificial human organ culture model and transcriptome analysis), and the last one was an erratum.
After removal of a duplication, six publications that used Colinfant, of which four were studies with infants, newborns, or premature babies (Table 2), were identified. The absence of in vitro characterization studies for this product is surprising in the light of the published applications for these vulnerable hosts. One study described data obtained with a rat model, and one reported production of the pentasaccharide repeat unit that is characteristic for the O-antigen of Colinfant E. coli. In the first available reference on the name Colinfant [10], its content was described as a mutant of E. coli strain A0 34/86, so the search was extended with this term. This retrieved five more publications, two describing pig experiments, two genome comparison studies, and one characterization of a restriction–modification system.
Results obtained from human studies
Colinfant
Since the literature on Colinfant, which is presented here first, is limited, the literature was completed by following citations backwards in time, thus, extending the available publications that were summarized in Table 2. In a relatively recent publication, the strain used in this product was reported to be originally isolated from pig feces [11], although the original historical descriptions do not mention this (see below). By following the trace of publications on this strain backwards in time, the oldest publications that describe use of serotype O83 E. coli to colonize humans that could be identified (and are cited in more recent works describing Colinfant) are from 1967. In that year, a paper from Czech authors described how five breast-fed infants (3 days old) were given a dose of 5·108 CFU of E. coli strain O83, a strain described as “nonpathogenic for newborn germ-free piglets” without providing further information [12]. Colonization was determined by agglutination of isolates from feces. A second 1967 paper (not included in the original PubMed search summarized in the previous section) reported that the strain could still be detected in stools 25 weeks after dosage [13]. That publication described the strain as a gift from “Prof. Sedlák” (presumably, Prof. Jiří Sedlák of Vinohrady Hospital, Charles University in Prague), but its original source was not mentioned. That the feces of colonized infants contained mucosal antibodies against the strain was demonstrated a few years later [14]. A publication in 1991 described more details of what seems to be the same trial [15]; here, the strain is called E. coli O83 but described as O83:K24:H31, and the authors specifically mention that 1500 infants and newborns had been treated without causing any complications. The name Colinfant first appears in 1998 in a publication presenting more information on the strain; the product was compared to Mutaflor (both were described as “life vaccines”) for colonization properties in newborns [10]. It was reported that both strains colonized full-term newborns for 12 to 16 weeks (a nonspecified dose was administered for 4 days), though it was not described how the isolated strains were identified from stools. In the saliva of the challenged babies, IgA–IgM levels were elevated, and in their stools, IgA levels were elevated compared to nonchallenged controls. A randomized double-blind trial was also described in that 1998 publication, involving 230 colonized newborns and 204 controls; it reported reduced disease rates and mortality in the treatment group [10]. The cohort was followed up at the age of 10, when apparently, still, a lower incidence of infections could be demonstrated [16] (publication in Czech).
Prevention of infectious diseases and allergies was the target for a trial comparing 52 treated infants with 50 untreated infants as controls, both groups from allergic mothers, and 42 controls with nonallergic mothers [17]. Treated newborns received Colinfant from day 2 till 4 weeks of age. At month 3, both colonization and more frequent absence of pathogens in stools were observed; the incidence of allergies was also lower [17]. Finally, the product was given to 25 premature babies in intensive care, and these were compared to a control group of premature babies in a “conventional environment.” Again, the treated babies had fewer infections, resulting in lower antibiotic need [18] (publication in Czech). These studies were all performed by the same research group. No publications could be identified in which any of the reported effects were confirmed by others.
The exact nature of the E. coli strain present in Colinfant remains a bit obscure, as the 1998 publication in which the name Colinfant is introduced in the scientific literature is rather confusing about the exact nature of its component. According to that publication, the “parent strain” of the Colinfant content is E. coli strain A0 34/86, described as “enterotoxin negative” based on (nonpresented) experiments with 3-day old mice and rat intestinal loop experiments [10]. This parent strain was described as hemolytic due to HlyA production (presence of the hlyABCD operon), but deletion of hlyA produced a mutant strain called “O83,” which was no longer hemolytic. Unfortunately, from the literature, it remains unclear whether the experiments then described with 230 newborn babies were conducted with the hemolytic parental strain or with the nonhemolytic mutant O83 (the presented figures relate to strain “O83”). Either content of the probiotic Colinfant could be considered problematic for application in newborns. To address this question, recently, the commercial product was purchased and hemolytic activity was determined from the bacteria it contained (T. Wassenaar, A. Siegl, and C. Beifmohr, unpublished data). Indeed, the colonies produced weak but detectable hemolytic activity on sheep blood agar plates. This is a surprising phenotype and might raise safety concerns when administering the product to children, a concern that was not mentioned in the trial descriptions [10]. Whether it is ethical to provide a hemolytic strain to newborns and even premature babies would probably be questioned in some countries. In a 2006 paper, inactivation of the hlyA operon in E. coli A0 34/86 is described, to produce a product that is safe to use in pigs [11]. Another publication from 2006, investigating the genetic characteristics of strain A0 34/86, describes that the strain was purchased from Dyntec s.r.o., the company producing Colinfant, so it can be assumed that the findings relate to this hemolytic strain [19].
Symbioflor 2
The literature on Symbioflor 2 is also not very extensive. The earliest exposure study, published in 1998, investigated the humoral immune response in ten challenged healthy individuals. No effect on the composition of the gut microbiota could be observed, but a significant increase in circulating amounts of IgG, directed against the administered E. coli strains, was demonstrated [20]. In a study performed a decade later, 23 healthy volunteers were exposed to Symbioflor 2 and an increase in fecal inducible epithelial β-defensins, which remained elevated for at least 9 weeks after cessation of treatment [21], was reported. The study also included in vitro work to show that one genotype of Symbioflor 2, called G3/10, was responsible for the measured effect, which was comparable to the effects determined with EcN [21].
A randomized double-blind clinical trial was performed to compare Symbioflor 2 treatment of inflammatory bowel disease (IBD) patients with placebo, resulting in significantly more responders (27 out of 148) than in the control group (7/150) [22]. The probiotic was also tested in 203 children with IBD (age range of 4–18 years), who tolerated the treatment well and showed relieve of symptoms, although this was not a blinded or placebo-controlled study [23]. A systematic review and meta-analysis of clinical trials assessing probiotic treatment of IBD included two studies with live E. coli, comparing the Symbioflor 2 study mentioned above [22] with an EcN study [24]; in the meta-analysis, the effects were positive and comparable [25]. Recently, a human volunteer study was performed to investigate the colonization potential of Symbioflor 2, which resulted in long-term colonization in all five volunteers, for a period of at least 20 weeks; again, this prolonged colonization was due to one of the genotypes present in Symbioflor 2 only [26]. Interestingly, the strong colonizing strain (called G1/2) was not the same as the one that induced β-defensin production reported by Möndel and coworkers [21]. In one of the three publications on genomic characteristics of the Symbioflor 2 strains [27], the colonization capacity of this component G1/2 was correlated to presence of potential virulence genes, which will be addressed in more detail in a next section.
E. coli Nissle
Most studies in which EcN was applied to treat human illness relate to inflammatory bowel disease (IBD). The oldest published study describing human exposure, from 1989, collected data from 1074 patients with either functional enteric disorders or IBD, who had taken EcN as recorded by their physician [28]. The study reported a tolerance in more than 90% of the patients, with initial side effects not requiring termination in 2.8%, and termination due to adverse reactions in 1.5% of the patients. Of those with functional intestinal disorders, 84% subjectively judged the treatment to be good to very good; for IBD, this was 78%. As already mentioned above, a double-blind placebo-controlled clinical trial confirmed that IBD patients indeed responded to EcN treatment, though significant improvement was only apparent after 10 to 11 weeks of treatment [24]. In this study, patients with altered enteric microbiota (e.g., after gastroenterocolitis or following the intake of antibiotics) responded best. The abovementioned observation that human β-defensins were induced in vitro and detected at 10 to 15-fold increase following EcN treatment [21] could provide a mechanistic explanation of this positive effect.
Ulcerative colitis (UC), a subcategory of IBD in which the lining of the colon is specifically affected, is typically treated with mesalazine. Already in 1997, a randomized double-blind clinical trial involving UC patients compared 12 weeks of EcN treatment with the gold standard of mesalazine treatment, showing similar start and end scores of the clinical activity index as well as similar relapse rates for both treatments [29]. These results were confirmed 2 years later by another research group [30] and again in a larger study by the research group of the 1997 study [31]. UC can lead to colorectal cancer, for which microsatellite instability in the genome is a possible driver. Rather disappointingly, a follow-up study showed that neither EcN nor mesalazine treatment could prevent the formation of such microsatellite instability in UC patients [32]. Effectiveness of EcN similar to mesalazine was also demonstrated in children between 11 and 18 years suffering from UC [33]. When acute distal UC was treated by rectal administration of EcN, it produced mixed results with no effectiveness for the intention-to-treatment population, but efficacy for the per-protocol population, resulting in dose-dependent remission rates [34]. A volunteer study with healthy individuals was performed to assess if mesalazine could be taken together with EcN, which turned out to have no effect on survival of the probiotic in stools [35]. Whether concomitant treatment with both would increase effectiveness for UC treatment has not yet been described.
In contrast to these positive findings, the effect of EcN on UC was questioned by a recent study comparing pretreatment with ciprofloxacin before EcN administration [36]: 78% of patients receiving ciprofloxacin followed by placebo reached remission, compared to 66% of those receiving EcN after ciprofloxacin. Similarly, for the group receiving placebo instead of ciprofloxacin, again those taking EcN showed lower remission than those receiving twice the placebo (54% and 86%, respectively) [36].
Crohn’s disease, another variant of IBD, is related to adherent-invasive E. coli (AIEC) colonization. It is, therefore, not surprising that treatment for this disorder is rarely tested with EcN, as probiotic products not containing E. coli are usually preferred. A meta-analysis compared all available data on probiotic treatment of CD [37] included only one study that compared EcN treatment with placebo. That study showed no statistically significant difference [38]. More recently, in vitro experiments with biopsies from CD patients and healthy controls showed that EcN lacked the capacity to compete with AIEC [39], which makes it unlikely that EcN can be effective to treat CD.
Since bloating is sometimes reported as a side effect of EcN intake, this was investigated in a randomized double-blind study with healthy volunteers [40]. EcN was well tolerated and did not significantly affect abdominal symptoms, stool frequency, or stool consistency; it had no effect on intestinal gas dynamics.
Other enteric disorders were also challenged with EcN treatment. For instance, a randomized double-blind clinical trial involved 70 patients with constipation [41]. After 4 weeks of therapy, their stool frequency had increased significantly compared with controls, confirmed by crossover patients, when verum patients were changed to placebo and vice versa. When liver cirrhosis patients were treated with EcN, they showed improved liver functions (as by Child–Pugh classification) and reduced endotoxin levels though there were no other significant improvements [42, 43] (publication in Czech). These two publications, describing data obtained from the same study, are thus far the only described application of EcN to treat liver disease.
In a prospective open trial, uncomplicated diverticular disease of the colon was treated with antimicrobials and absorbents, followed by EcN for 5 weeks, which prolonged remission time [44]. Another open trial, this time treating collagenous colitis, showed therapeutic benefit when EcN treatment lasted for at least 4 weeks [45].
Babies were also treated with EcN. In a randomized double-blind clinical trial, healthy newborns were given the bacteria during their first 5 days of life [46]. The stools were EcN-positive in >90% of infants for as long as 6 months. A variety of pathogens was shown to be absent or reduced in numbers during the study. The same group performed a randomized double-blind trial on premature infants where an increase in EcN-specific IgA antibodies could be demonstrated [47]. EcN was further shown in a double-blind randomized trial to shorten acute (viral) diarrhea in infants and toddlers with 2.3 days [48]. Finally, to complete the positive findings for various applications, a case was described of Clostridium difficile-negative pseudomembranous colitis following antimicrobial treatment in an adult, who was successfully treated with multiple intestinal lavages, followed by EcN administration [49].
Less successful was an attempt to treat subjects with hay fever (grass pollen-allergic rhinoconjunctivitis). A randomized double-blind clinical trial with 6 months of treatment covering the full exposure season did not show effects on symptoms, pollen-specific humoral IgE or IgA levels, or a skin prick test [50]. Mutaflor was also not able to reduce carriage of multidrug-resistant E. coli in elderly residents of long-term care facilities [51], though, in this study, only two out of 12 treated subjects had detectable EcN in their feces.
This latter observation brings up an interesting question: how well is EcN capable to colonize the human gut? The applied daily dose of Mutaflor is relatively high (see Table 1), and in several of the abovementioned trials, the product was taken for weeks at a time. Colonization of newborn babies was demonstrated, but in those hosts, the gut is still relatively unoccupied [46]. When seven adult volunteers were orally challenged for 1 week, EcN could only be detected in the stools of four of these [52]. The study combining EcN with mesalazine mentioned above reported that, for the control group (not receiving mesalazine), within 2 weeks after secession of EcN administration, the strain could be detected in the stools of only 40% of individuals, dropping to 20% after 9 weeks [35]. These findings demonstrate that EcN is not very efficient to colonize the human gut long-term. Attempts to improve this colonization potential were not identified. In one animal study, the colonization properties of EcN in mice could be enhanced by introduction of a missense mutation in a histidine kinase gene, but this did not improve its capacity to outcompete enterohemorhagic E. coli challenge [53].
It has been argued that some of the effects of probiotics are caused by cellular components, so that the organisms would not be required to be alive and establish colonization in the gut [54]. Nevertheless, according to Caselli and coworkers, the definitions of probiotics used by the World Health Organization and the Food and Health Organization include that they have to “remain viable and stable after culture, manipulation, and storage before consumption (and) have to survive to gastric acid and biliary and pancreatic digestion” [55]. These authors argue that, since the host’s pattern recognition receptors (PRRs) play a pivoting role in probiotic applications, it would be irrelevant whether the bacteria were dead or alive [55]. If this argument were accepted, one can only wonder why probiotic products are based on living organisms, which are more difficult to produce, store, and quality control than preparations of cellular components. My gut feeling (pun intended) is that living organisms have more effect than dead ones, though there are few data to support one or the other view.
The origin of E. coli Nissle 1917
The story of the origin of the Nissle 1917 strain has been told many times and even featured in the publications announcing the release of the sequenced EcN genome [56, 57]. According to this story, Alfred Nissle had isolated the strain from the stool of a First World War soldier, who was the only one of his unit not suffering from dysentery. Some citations state that the lucky soldier remained free from “diarrhea” [58] or “shigellosis” [59] while yet others mention his resistance was against typhoid fever. The original publications cited for this information vary, as Nissle was rather productive and many of his papers are still being cited. Some of the citations refer to his earliest publication, from 1916 [60] cited by, among others, Pöhlmann et al. [61], or a work from 1918 [62], cited by authors who elaborately mention that the soldier had been stationed on the Balkan peninsula [63] although this information was never described by Nissle himself. Others cite a 1925 paper [64], cited by Zyrek et al. [65] which is the oldest record in PubMed mentioning the term “Mutaflor” (though, in fact, Nissle introduced the commercial name in a footnote in a publication from 1919 [66], but that publication is not represented in PubMed). His late works are also frequently cited, e.g., a publication from 1959 [67], cited by Verna and Lucak [59] where it is reported that the soldier was protected from shigellosis, or from 1961 [68] (cited in the review article [58]), or even the posthumous publication that appeared in 1966, a year after Nissle’s death, in which the author, aged 90, had looked back at his long career [69] (cited on page 213 by Tannock [70]).
It is heart-warming to see that so many international authors still read original literature in the German language, but the various citations for this information, and the variation in description of what should be a historical consistent story, is a bit suspicious. Moreover, some of these original publications are hard to come by. I was able to retrieve five original publications written by Alfred Nissle to check what exactly he had published on Mutaflor.
His 1916 paper [60] is very informative and deserves to be summarized here in some detail. He described experiments he performed with E. coli (“Koli” as he called them) isolated from stool samples. He cultured these on agar plates (“Endoagarplatten”) and spiked them with a fixed ratio of 2:3 of “Typhusbacillen” (in later experiments, he cultured the mixture in a broth for 6 or 7 h before plating). On the plates, he observed that some, but not all, of the tested E. coli isolates could inhibit or reduce the growth of what we now call Salmonella enterica serovar Typhi. He called this phenomenon “antagonism” and expressed the number of “Typhus” colonies per 100 “Koli”-colonies as the “antagonistic index” of the assessed E. coli isolate. He then compared this index between strains, describing in detail the difficulty in standardizing the test, performed with inclusion of control strains with a proven high or low antagonistic index. At the extremes, he observed that the strongest antagonistic strains were 1350 times better able to inhibit S. Typhi growth than the weakest tested isolates.
Nissle tried to explain the observed differences in antagonism by differences in lactic acid production, but observed there was no correlation, nor was indole production related. He further reported that strongly antagonistic strains not only inhibited S. Typhi but also “Paratyphus-Dysenterie”, “Kruse-Shiga”, “Dysenterie Flexner”, and “Proteus”. These old names refer to Salmonella enterica serovar Paratyphi, Shigella dysenteriae, Shigella flexneri, and either Proteus vulgaris or P. mirabilis (neither of which are strongly pathogenic but that was not known at that time). He was even able to show antagonism between different E. coli strains, by clever use of a strain that was unable to produce gas on a medium containing glucose (“Traubenzucker”, his strain was an atypical non-gas-former, as have subsequently been characterized and described [71]). This indicator strain was defeated by strong antagonistic E. coli strains but not by weakly antagonistic ones. Nissle further showed that the antagonistic property was lost after heating at 60 °C, which we now know is indicative of proteins or peptides. The phenotype he determined was most probably the result of microcins; it is now known that EcN produces microcins MccM and MccH47 [72].
When he compared E. coli strains obtained from 15 fecal samples of healthy individuals, three were very strong, five were strong, four were medium, and three were weakly antagonistic (it is unclear whether these experiments were performed with the indicator E. coli or with Typhus). In contrast, 25 strains from “pathological stools” produced two reasonably strong, five medium, and 18 weak antagonists. From these observations, Nissle concluded that a strong antagonistic E. coli in the gut might protect against enteric diseases.
The three “very strong” antagonistic strains from healthy stool were described in a bit more detail. One was found by chance, but the other two came from selected individuals: Nissle had visited an army hospital (“Verwundeten-Lazarett”) and had specifically selected patients who reported to have never suffered from enteric diseases, even though they had been exposed to acutely infected patients. He identified two such persons (it was not described why they were hospitalized or where they had served) who produced E. coli with an antagonistic index of 100:10 and 100:3, respectively.
Nissle had no means to conserve his strains other than by subculture. He describes that one of his precious strong antagonizers lost its phenotype after 2 years. He was able to protect the other two, one isolated in March 1915 and the other in September of that year (unclear is, whether both of these were the isolates from the soldiers), from that fate by storing them in capsules made of paraffin or wax. To test their positive effect against infections, he first gave these capsules to healthy volunteers and, after proven safety, to diseased patients. The rest of the 1916 publication describes successful treatments of 11 cases. Case 11 suffered from Typhus infection from 25 July till 8 August 1915 and shed high numbers of “Typhusbacillen” since (from 12 September till 17 April 1916) when treatment was started. The person shed low numbers till 28 April. after which Thypus bacteria could no longer be detected in his stool. The treatment consisted of 52 capsules of “Kolireinkultur” administered in 37 days. Nissle described that the person produced the administered high-antagonistic E. coli in his stool 3 weeks after termination of treatmen (at least he was able to isolate a strain with similar antagonistic properties from the stool). Nissle suggested the capsules as treatment for acute intestinal infections and speculated it might even be effective against other infections, giving the example of diphtheria. The 1916 publication demonstrates the extraordinary scientific skills of Nissle. Not only was he a sharp observer, he also designed his experiments under standardized conditions with inclusion of the necessary controls, tested explanatory hypotheses and discarded these based on experimental evidence, developed means to store his biological material, and performed clinical trials avant la lettre.
In his 1918 publication [62], Nissle continued his investigations with the capsules that were from then on commercially produced under the name “Mutaflor,” though it remains unclear whether the E. coli strain used in the product was his isolate from March or September 1915. Who the individual was from whose stool it was isolated, whether it was one of the soldiers, and if so, where he had been stationed is not disclosed by Nissle. What we can deduc from the literature is that the strain was originally isolated in 1915, possibly from a soldier of unknown nationality, who at that time was treated in a German army hospital. The year “1917” that has become part of the name of this strain present in Mutaflor neither refers to the year of isolation, nor to the year the strain was first publicly described.
Pig studies
Apart from a high number of animal studies involving rodents, mostly performed as a model to study human colonization, quite a few publications were identified that studied performance of probiotic E. coli in the porcine host, and these deserve a bit more attention here.
The EcN strain is quite capable to colonize pigs persistently [73]. Even in piglet populations not deliberately fed with EcN, the strain could be identified, suggesting that the strain has established itself persistently in some swine populations [74]. In line with the original in vitro antagonistic activity of EcN, the product is used to combat infections in pigs. For instance, a 2006 publication showed that the strain could protect the porcine host against pathogenic E. coli, in particular against toxigenic E. coli, which frequently causes post-weaning diarrhea in piglets [75]. Likewise, EcN (but not Bifidobacterium choerinum) could protect pigs from S. enterica serovar Typhimurium, though this was only tested in a gnotobiotic pig mode [76]. In vitro results suggested that this protection was due to inhibition of adherence, for which F1C fimbriae and the flagella of EcN were considered responsible [77]. A few years later, this was confirmed by in vivo studies using atypical enteropathogenic E. coli as the challenge; the most important protective step of EcN in the porcine gut was inhibition of adhesion and formation of microcolonies [78].
Production of bacteriocins was demonstrated in approximately 30% of naturally occurring porcine E. coli strains that belonged to all four major phylotypes [79] These researchers investigated if EcN in mature animals (4–5 month old sows) with and without administration of indomethacin (which in presence of bacteria induces entero- and colopathy) affected the presence of bacteriocin-producing E. coli. The hypothesis was that EcN would colonize, thereby alter the residual microbiota and decrease the toxicity of indomethacin. However, EcN did not colonize very well and there was no effect on overall bacteriocin production, though some shifts in presence of particular bacteriocin-types were observed [79]. Whereas the actions of indomethacin and EcN on the colonic mucosa are opposite, when tested in combination, this did not result in a positive effect but actually resulted in the worst impact on stomach, and small and large bowel (the height of cryptal mucosa and widening of the basement size of colonocytes were taken as measures for deteriorating impact on the epithelium), as demonstrated in a second publication by the same group [80]. In a follow-up, it was reported that EcN could indeed lower the incidence of bacteriocin-producing E. coli, though it still did not result in stable colonization [81]. One publication studied the effect of combined colonization with Lactobacillus rhamnosus GG and EcN in gnotobiotic pigs that were fed rice bran and then challenged with human rotavirus diarrhea. According to these authors, rice bran on its own protects against this virus (an explanatory mechanism was not given), while, given in combination with EcN, the protection was 100% and resulted in a thousand-fold drop of virus titre [82].
The immune response to colonization in young pigs was investigated by determination of quantitative immune cell responses and mucosal transcriptional responses for production of cytokines [83]. Disappointedly, neither was influenced by presence of EcN, with the exception of an increase in mucosal CD8+ cells in the ascending colon. Lastly, calprotectin production was studied in germ-free and gnotobiotic pigs primary colonized with EcN, as a model for treatment of IBD in humans [84].
Compared to the considerable literature on EcN, the other two E. coli probiotic products have hardly been studied in the porcine host. For Colinfant, this is a bit surprising, as the parental strain A0 34/86 was apparently a porcine commensal [11]. The publication describing its porcine origin presents evidence that deletion of the HlyA operon in this parental strain did not affect colonization potential in new-born piglets, but did improve viability of colostrum-deprived gnotobiotic animals [11]. Only one other publication on Colinfant in pigs could be identified, which compared the immune response of germfree pigs exposed to strain A0 34/86 with a commensal O86 strain; both resulted in recruitment of dendritic cells [85].
No published information is available on Symbioflor 2 in pigs, but preliminary studies showed that none of the components of this product was able to colonize 8-week-old pigs (I. Hennig-Pauka and C. Beimfohr, personal communication).
Genome comparisons, closest relatives, and virulence genes
Assuming that Colinfant contains strain A0 34/86, its constituent is rich in fimbrial adhesins, iron-uptake systems, toxins, microcins, proteases, and autotransporters, according to Hejnova et al. [86]. When the strain was compared to four other E. coli strains based on limited genome sequences, it most resembled uropathogenic E. coli strain CFT073, despite the differences in serotype, while it was most distantly related to commensal E. coli K12 strain MG1655 [86]. These findings were confirmed in a follow-up study, which further reported presence and inducible expression of yersiniabactin (an iron-uptake gene cluster) which strain A0 34/86 shares with uropathogenic E. coli strains CFT073 and 536 [19]. The close relatedness to strain CFT073 was also observed when 20 kb sequences flanking the HlyA operon of A0 34/86 were analyzed [11].
Of the six genotypes present in Symbioflor 2, three also contain the HlyA operon as described by Willenbrock et al. [87], though its expression is weaker than that of Colinfant and very weak compared to pathogenic hemolytic strains. The genetic content of the Symbioflor 2 strains was initially compared to other E. coli strains by microarray analysis, revealing similarity to nonpathogenic K12 strain MG1655 [87]. A more thorough comparison was possible when the genome sequences of the genotypes became available [88]. This revealed the presence of a number of putative virulence and fitness genes, not only HlyA but also Type 1 and Type 3 fimbriae, aerobactin, enterobactin, and serine protease autotransporters, as well as various colicins [27]. In silico MLST analysis with 28 other strains revealed that, with the exception of G3/10, five Symbioflor 2 genotypes were of the same sequence type as commensal E. coli K12 MG1655, while, in a genome-wide comparison, all genotypes clustered together, mixed with the three commensal strains K12 MG1655, ATCC8739, and BL21 DE3 [27].
The close similarity of EcN to uropathogenic E. coli CFT073 and 536, all of serotype O6, was already known ten years ago [89, 90], and this similarity was confirmed when the genome of EcN was unravelled [57, 56]. Vejborg and colleagues concluded, based on comparison of sequence mutations, that EcN, CFT073, and closely related strain 83972 (a urine isolate that had not caused symptoms) have a common virulent ancestor [91]. The similarity between these three strains is extended to their transcriptome [92] illustrating, as those authors commented, that “the distance from a pathogen to a probiotic strain can be surprisingly short.”
In conclusion, two of the three commercial products contain E. coli with weak hemolytic activity, two of the three have close similarity to uropathogenic E. coli (UPEC) strains, and all three contain a number of genes that improves fitness or, when present in a pathogenic strain, increases virulence. This may not be a coincidence. In view of the fact that there is not one publication describing probiotic effects of a truly commensal K-12 strain, it is tempting to hypothesize that a reminiscence of virulence is required to produce beneficial effects to the host. In line with this hypothesis, it was reported that EcN’s genotoxic effect due to hybrid nonribosomal peptide–polyketides (which induce DNA double strand breaks in eukaryotic cells) is required for the beneficial effect on IBD, at least in a rat model [93].
Two key observations from previous sections presented above, namely, the similarity of probiotic E. coli strains to UPEC isolates and their colonization capacity in pigs, are of interest in light of the suggestion that UPEC infections may have a zoonotic origin. Most studies investigating this relationship concentrate on avian pathogenic E. coli (e.g., refs. [94, 95]), but another potential source might be porcine strains. Possibly, such strains have opposite effects in humans: although they have the potential to cause urinary tract infections, when their pathogenic potential is reduced, this could result in a strain with probiotic characteristics. This hypothesis can be tested by a detailed genomic comparison of porcine, UPEC, and probiotic strains.
Probiotics as health elixir?
As discussed in a previous section, a century ago, the first probiotic E. coli was applied to combat enteric infections, which in pre-antibiotic times were a serious health threat. But it was also described above that the products have subsequently been applied to treat a variety of diseases and conditions, while infectious diseases are no longer the target. Meanwhile, the products are commercialized using rather vague targets such as a “healthy immune system.” These trends are not specific for E. coli products.
Using PubMed as a reference source, on 7 December 2015, the number of publications over the years that could be retrieved with the search term “probiotics”, in combination with either the term “infection”, “allergy”, “ulcerative colitis” or “irritable bowel” was recorded to determine how frequent these research subjects have been investigated over the years. The results are plotted in Fig. 1, which shows in grey the absolute counts of the publications retrieved with “probiotics” (right-hand scale) and in colors the relative counts (as percentages) for searches where “probiotics” was combined with a second term. As can be seen, the total number of publications on “probiotics” increases steadily per year, and of these, approximately 14% are somehow related to infection (blue curve), a percentage that has been relatively stable over the past decade. Literature on probiotics and ulcerative colitis (green) clearly peaked in 2003 and then declined, while literature on probiotics and irritable bowel disease (red) followed a few years later, with peaks in 2006 and 2009, after which the relative attention also decreased. Lastly, publications on probiotics in combination with allergy (purple) have peaked around 2005–2009 and, since then, declined. By and large, these general findings for all publications on probiotics are reflected by the literature on E. coli-based products.
Fig. 1.
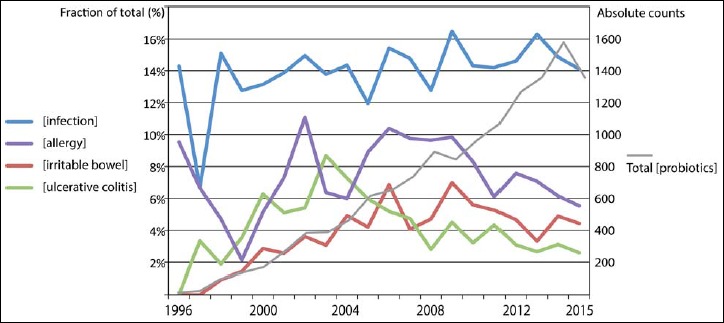
Trends in publication frequency over time for four keywords combined with “probiotics”
It is interesting to note that the probiotic strains tested over the years have remained largely the same, and this not only applies to E. coli but also to the strains of Lactobacillus, Lactococcus, and Bifidobacterium species that are frequently used in probiotics. However, as pointed out by others [55], it is doubtful that only a handful of bacterial strains and species could benefit patients suffering from all these conditions, not to mention the less frequently investigated applications such as obesity, constipation, liver diseases, eczema, and type 2 diabetes, among others.
Probiotics seem to be a panacea to treat a wide variety of conditions, as if it were a modern health elixir, enriched with a scientific scent. This is all the more surprising in light of the knowledge that not all probiotic strains are good colonizers, for which EcN would be an example, as discussed above. It seems that some of the published research is driven by fashion, while treatment is tried against a moving target. The mode of action against whatever is tested is often investigated in animal studies that may be inadequate to assess what truly happens in humans.
Possible future applications of probiotic E. coli
Despite the scepticism expressed above, some recent publications direct to future applications of probiotic E. coli that are worth mentioning here, acknowledging that these application have as yet only been tested in animal models.
It was already mentioned that the antagonistic effects against pathogens originally described by Nissle are nowadays only employed in the porcine host. The historic application of probiotic E. coli to combat gastro-intestinal infections in humans was replaced by subsequently discovered antibiotics, which provided a more effective treatment. However, that picture is changing, as ever more pathogens are becoming resistant against commonly used antibiotics. This has resulted in a rediscovery of previously dismissed remedies, for instance, bacteriophage therapy, once popular against infectious diseases, in particular in Eastern Europe [96]. Maybe it is time to reconsider the effectiveness of probiotics to treat (mild) infections caused by enteropathogens, resistant or not. In line with Nissle’s original observations, the in vitro inhibitory effect of EcN and a novel E. coli isolate (strain 1307) on growth of Shiga-toxin-producing STEC strains (otherwise known as EHEC) was described in a 2009 publication [97], and similar antagonistic effects were recently described against EHEC serotypes O104:H4 and O157:H7 [98]. This is particularly interesting, as antibiotic treatment of EHEC infections is contraindicated. As yet, human clinical trials for this old and rediscovered application have not been published.
The antagonistic effects of EcN are not only due to bacteriocins: a highly efficient uptake of iron helps EcN outcompeting pathogenic Salmonella Typhimurium, at least in the murine gut [99]. In addition, novel insights in the metabolism of E. coli in the gut have demonstrated that bacterial competition in the gut is also driven by nutrient dependency [100]. Such insights could explain how EcN, together with an E. coli strain HS, could prevent colonization of EHEC in mice, as the two strains together metabolized the five sugars on which EHEC mainly depends for growth [100].
Probiotic E. coli is not particularly suitable for use as vehicles to deliver drugs or other bioactive molecules in the human gut, as such heterologous proteins must pass two membranes in order to be secreted, while their three-dimensional structure must be maintained. For delivery of interleukin-10, which furthermore requires dimerization in order to be biologically active, the yeast Saccharomyces boulardii turned out to be a more suitable vehicle than E. coli [61]. E. coli is capable, however, to alter the pharmacokinetics of particular drugs. At least in rats, it has been demonstrated that the biological half-life of the anti-arrhythmic drug amiodarone (used for treatment of ventricular tachycardia and ventricular fibrillation) was extended by presence of EcN in the rat’s gut, though E. coli strain ATCC 25922 did not have an effect [101]. Such findings point out potential unintended side effects of probiotics, and when confirmed in humans, such pharmacokinetic effects can also be deliberately applied.
Ever since the discovery of tumor-necrotizing effects of E. coli extracts, such as lipid A, the possibility to treat cancer with bacteria has been considered. It has been shown that human mammary tumors established in mice collect bacteria such as E. coli or Salmonella Typhimurium following intravenous injection [102]. Expression of apoptosis-inducing toxins (e.g., azurin) can enhance the effectivity of bacterial cancer treatment [103], while ferritin-overexpression provides a practical marker for tumor detection by magnetic resonance imaging (MRI) [104]. All these findings and applications are as yet based on murine studies only, but may point towards future directions of research.
Conclusions and outlook
Over the past few decades, probiotic applications of E. coli have gained scientific credibility, though a lot of research has been conducted with in vitro or animal models only, while relatively few human studies support the claimed or predicted effects. Of the three products discussed here, most is known about EcN, which is a surprisingly poor long-term colonizer of the human gut. This strain was originally isolated based on its antagonistic capacity in order to combat infections, but this treatment application became out of fashion as antibiotics entered the field. Now that these threaten to backfire, with pathogenic bacteria sooner or later developing resistance against the antimicrobials they are exposed to, interest has increased in alternative methods that have been applied in the past, including phage therapy and probiotics. This could introduce a revival of the original concept introduced exactly a century ago by Alfred Nissle.
Moreover, the safety of probiotic E. coli no longer needs to be questioned, despite the presence of recognized virulence-associated characteristics. This opens new avenues, in particular in combination with the capacity to specifically colonize cancerous tissue. If the results obtained with mice can eventually be reproduced in humans, probiotic E. coli could one day serve as an internal vehicle to transport chemotherapeutic drugs directly into the tumors where they should exclusively be active.
Future studies to unravel the role of specific genes in bacterial fitness and capacity for colonization, infection, and pathogenicity require a more holistic approach, whereby conditions may determine whether a strain is a commensal, probiotic, or pathogen. Even so-called UPEC strains are often apathogenic colonizers of the gut and only cause infections when they happen to reach sufficient numbers in the urogenital tract. Such observations, combined with the fact that probiotic strains resemble UPEC strains in many ways (though they never cause urinary tract infections), illustrates that there is no strict division between pathogenic and nonpathogenic E. coli strains. As with so many other biological features, it seems the E. coli world is not black and white but consists of many shades of grey.
Acknowledgements
The author thanks Prof. Manfred Kist (Freiburg) for useful comments and suggestions.
Funding Statement
Founding sources SymbioPharm GmbH provided financial support to write this review; however, it had no influence on the content of the article.
References
- 1.Kaas RS, Friis C, Ussery DW, Aarestrup FM: Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13, 577 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tannock GW. (1995). Normal Microflora. An Introduction to Microbes Inhabiting the Human Body, Chapman and Hall, London, UK [Google Scholar]
- 3.Caugant DA, Levin BR, Selander RK: Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98, 467–490 (1981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sender R, Fuchs S, Milo R: Revised estimates for the number of human and bacteria cells in the body. Plos Biol 14(8), e1002533 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill MJ, Drasar BS: The normal colonic bacterial flora. Gut 16, 318–323 (1975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuo HJ, Xie ZM, Zhang WW, Li YR, Wang W, Ding XB, Pei XF: Gut bacteria alteration in obese people and its relationship with gene polymorphism. World J Gastroenterol 17, 1076–1081 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D: The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev 28, 237–264 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avershina E, Rudi K: Confusion about the species richness of human gut microbiota. Benef Microbes 6, 657–659 (2015) [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renaul P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium. Bork P, Ehrlich SD, Wang J: A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 59–65 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodinová-Zádníková R, Sonnenborn U, Tlaskalová H: Probiotics and E. coli infections in man. Vet Q 20 Suppl 3, S78–81 (1998) [PubMed] [Google Scholar]
- 11.Sheshko V, Hejnova J, Rehakova Z, Sinkora J, Faldyna M, Alexa P, Felsberg J, Nemcova R, Bomba A, Sebo P: HlyA knock out yields a safer Escherichia coli A0 34/86 varian with unaffected colonization capacity in piglets. FEMS Immunol Med Microbiol 48, 257–266 (2006) [DOI] [PubMed] [Google Scholar]
- 12.Lodinová R, Jouja V, Lanc A: Influence of the intestinal flora on the development of immune reactions in infants. J Bacteriol 93, 797–800 (1967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodinová R, Jouja V, Lanc A: Experimentelle Besiedlung des Darmtrakts von Neugeborenen mit dem E. coli 083 und Untersuchungen über die Bildung von Antikorpern. Z Immunitaetsforsch 133, 229–237 (1967) [PubMed] [Google Scholar]
- 14.Lodinová R, Wagner V: Development of faecal immunoglobulins and coproantibodies in infants after artificial oral colonization with E. coli 083. Experientia 26, 188 (1970) [DOI] [PubMed] [Google Scholar]
- 15.Lodinová-Zádníková R, Slavíková M, Tlaskalová-Hogenová H, Adlerberth I, Hanson LA, Wold A, Carlsson B, Svanborg C, Mellander L: The antibody response in breastfed and non-breast-fed infants after artificial colonization of the intestine with Escherichia coli O83. Pediatr Res 29, 396–399 (1991) [DOI] [PubMed] [Google Scholar]
- 16.Lodinová-Zádníková R, Prokesová L, Tlaskalová H, Kocourková I, Zizka J, Stranák Z: Influence of oral colonization with probiotic E. coli strain after birth on frequency of recurrent infections, allergy and development of some immunologic parameters. Long-term studies. Ceska Gynekol 69 Suppl 1, 91–97 (2004) [PubMed] [Google Scholar]
- 17.Kocourková I, Žádníková R, Žižka J, Rosová V: Effect of oral application of a probiotic E. coli strain on the intestinal microbiota of children of allergic mothers during the first year of life. Folia Microbiol (Praha) 52, 189–193 (2007) [DOI] [PubMed] [Google Scholar]
- 18.Lodinová-Zádníková R, Cukrovská B, Stranák Z: Effect of care in a protected environment on the occurrence of nosocomial infections, mucosal colonization of pathogenic microbiota and development of indicators of immunity in premature infants. Ceska Gynekol 67 Suppl 1, 23–28 (2002) [PubMed] [Google Scholar]
- 19.Hejnova J, Pages D, Rusniok C, Glaser P, Sebo P, Buchrieser C: Specific regions of genome plasticity and genetic diversity of the commensal Escherichia coli A0 34/86. Int J Med Microbiol 296, 541–546 (2006) [DOI] [PubMed] [Google Scholar]
- 20.Jansen GJ, Wildeboer-Veloo AC, van der Waaij D, Degener JE: Escherichia coli as a probiotic? Infection 26, 232–233 (1998) [DOI] [PubMed] [Google Scholar]
- 21.Möndel M, Schroeder BO, Zimmermann K, Huber H, Nuding S, Beisner J, Fellermann K, Stange EF, Wehkamp J: Probiotic E. coli treatment mediates antimicrobial human beta-defensin synthesis and fecal excretion in humans. Mucosal Immunol 2, 166–172 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enck P, Zimmermann K, Menke G, Klosterhalfen S: Randomized controlled treatment trial of irritable bowel syndrome with a probiotic E. coli preparation (DSM17252) compared to placebo. Z Gastroenterol 47, 209–214. (2009) Erratum in Z Gastroenterol 52, 64 (2014) [DOI] [PubMed] [Google Scholar]
- 23.Martens U, Enck P, Zieseniss E: Probiotic treatment of irritable bowel syndrome in children. Ger Med Sci 8, Doc07 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruis W, Chrubasik S, Boehm S, Stange C, Schulze J: A double-blind placebo-controlled trial to study therapeutic effects of probiotic Escherichia coli Nissle 1917 in subgroups of patients with irritable bowel syndrome. Int J Colorectal Dis 27, 467–474 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford AC, Quigley EM, Lacy BE, Lembo AJ, Saito YA, Schiller LR, Soffer EE, Spiegel BM, Moayyedi P: Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am J Gastroenterol 109, 1547–1561 (2014) [DOI] [PubMed] [Google Scholar]
- 26.Wassenaar TM, Beimfohr C, Geske T, Zimmermann K: Voluntarily exposure to a single, high dose of probiotic Escherichia coli results in prolonged colonisation. Benef Microbes 5, 367–375 (2014) [DOI] [PubMed] [Google Scholar]
- 27.Wassenaar TM, Zschüttig A, Beimfohr C, Geske T, Auerbach C, Cook H, Zimmermann K, Gunzer F: Virulence genes in a probiotic E. coli product with a recorded long history of safe use. Eur J Microbiol Immunol (Bp) 5, 81–93 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schütz E: Behandlung von Darmerkrankungen mit Mutaflor. Fortschr Med 107, 599–602 (1989) [PubMed] [Google Scholar]
- 29.Kruis W, Schütz E, Fric P, Fixa B, Judmaier G, Stolte M: Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther 11, 853–858 (1997) [DOI] [PubMed] [Google Scholar]
- 30.Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT: Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet, 354, 635–639 (1999) [DOI] [PubMed] [Google Scholar]
- 31.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, Kamm MA, Weismueller J, Beglinger C, Stolte M, Wolff C, Schulze J: Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53, 1617–1623 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goel A, Mittal A, Evstatiev R, Nemeth M, Kruis W, Stolte M, Boland CR, Gasche C: In vivo effects of mesalazine or E. coli Nissle 1917 on microsatellite in stability in ulcerative colitis. Aliment Pharmacol Ther 30, 634–642 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henker J, Müller S, Laass MW, Schreiner A, Schulze J: Probiotic Escherichia coli Nissle 1917 (EcN) for successful remission maintenance of ulcerative colitis in children and adolescents: an open-label pilot study. Z Gastroenterol 46, 874–875 (2008) [DOI] [PubMed] [Google Scholar]
- 34.Matthes H, Krummenerl T, Giensch M, Wolff C, Schulze J: Clinical trial: probiotic treatment of acute distal ulcerative colitis with rectally administered Escherichia coli Nissle 1917 (EcN). BMC Complement Altern Med 10, 13 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joeres-Nguyen-Xuan TH, Boehm SK, Joeres L, Schulze J, Kruis W: Survival of the probiotic Escherichia coli Nissle 1917 (EcN) in the gastrointestinal tract given in combination with oral mesalamine to healthy volunteers. Inflamm Bowel Dis 16, 256–262 (2010) [DOI] [PubMed] [Google Scholar]
- 36.Petersen AM, Mirsepasi H, Halkjær SI, Mortensen EM, Nordgaard-Lassen I, Krogfelt KA: Ciprofloxacin and probiotic Escherichia coli Nissle add-on treatment in active ulcerative colitis: a double-blind randomized placebo controlled clinical trial. J Crohns Colitis 8, 1498–1505 (2014) [DOI] [PubMed] [Google Scholar]
- 37.Rolfe VE, Fortun PJ, Hawkey CJ, Bath-Hextall F: Probiotics for maintenance of remission in Crohn’s disease. Cochrane Database Syst Rev 4, CD004826 (2006) [DOI] [PubMed] [Google Scholar]
- 38.Malchow HA: Crohn’s disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn’s disease? J Clin Gastroenterol 25, 653–658 (1997) [DOI] [PubMed] [Google Scholar]
- 39.Jensen SR, Fink LN, Nielsen OH, Brynskov J, Brix S: Ex vivo intestinal adhesion of Escherichia coli LF82 in Crohn’s disease. Microb Pathog 51, 426–431 (2011) [DOI] [PubMed] [Google Scholar]
- 40.Hernando-Harder AC, von Bünau R, Nadarajah M, Singer MV, Harder H: Influence of E. coli strain Nissle 1917 (EcN) on intestinal gas dynamics and abdominal sensation. Dig Dis Sci 53, 443–450 (2008) [DOI] [PubMed] [Google Scholar]
- 41.Möllenbrink M, Bruckschen E: Behandlung der chronischen Obstipation mit physiologischen Escherichia coli-Bakterien. Ergebnisse einer klinischen Studie die Wirksamkeit und Verträglichkeit von mikrobiologischen Therapie mit dem E. coli Stamm Nissle 1917 (Mutaflor). Med Klin (Munich) 89, 587–593 (1994) [PubMed] [Google Scholar]
- 42.Lata J, Juránková J, Príbramská V, Fric P, Senkyrík M, Díte P, Kroupa R: Effect of administration of Escherichia coli Nissle (Mutaflor) on intestinal colonisation, endo-toxemia, liver function and minimal hepatic encephalopathy in patients with liver cirrhosis. Vnitr Lek 52, 215–219 (2006) [PubMed] [Google Scholar]
- 43.Lata J, Novotný I, Príbramská V, Juránková J, Fric P, Kroupa R, Stibůrek O: The effect of probiotics on gut flora, level of endotoxin and Child–Pugh score in cirrhotic patients: results of a double-blind randomized study. Eur J Gastroenterol Hepatol 19, 1111–1113 (2007) [DOI] [PubMed] [Google Scholar]
- 44.Fric P, Zavoral M: The effect of non-pathogenic Escherichia coli in symptomatic uncomplicated diverticular disease of the colon. Eur J Gastroenterol Hepatol 15, 313–315 (2003) [DOI] [PubMed] [Google Scholar]
- 45.Tromm A, Niewerth U, Khoury M, Baestlein E, Wilhelms G, Schulze J, Stolte M: The probiotic E. coli strain Nissle 1917 for the treatment of collagenous colitis: first results of an open-label trial. Z Gastroenterol 42, 365–369 (2004) [DOI] [PubMed] [Google Scholar]
- 46.Lodinová-Zádniková R, Sonnenborn U: Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol Neonate 71, 224–232 (1997) [DOI] [PubMed] [Google Scholar]
- 47.Cukrowska B, LodInová-ZádnIková R, Enders C, Sonnenborn U, Schulze J, Tlaskalová-Hogenová H: Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand J Immunol 55, 204–209 (2002) [DOI] [PubMed] [Google Scholar]
- 48.Henker J, Laass M, Blokhin BM, Bolbot YK, Maydannik VG, Elze M, Wolff C, Schulze J: The probiotic Escherichia coli strain Nissle 1917 (EcN) stops acute diarrhoea in infants and toddlers. Eur J Pediatr 166, 311–318 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goerg KJ, Schlörer E: Probiotische Therapie einer pseudomembranösen Kolitis: Kombination aus intestinaler Lavage und oraler Gabe von Escherichia coli. Dtsch Med Wochenschr 123, 1274–1278 (1998) [DOI] [PubMed] [Google Scholar]
- 50.Dölle S, Berg J, Rasche C, Worm M: Tolerability and clinical outcome of coseasonal treatment with Escherichia coli strain Nissle 1917 in grass pollen-allergic subjects. Int Arch Allergy Immunol 163, 29–35 (2014) [DOI] [PubMed] [Google Scholar]
- 51.Tannock GW, Tiong IS, Priest P, Munro K, Taylor C, Richardson A, Schultz M: Testing probiotic strain Escherichia coli Nissle 1917 (Mutaflor) for its ability to reduce carriage of multidrug-resistant E. coli by elderly residents in long-term care facilities. J Med Microbiol 60, 366–370 (2011) [DOI] [PubMed] [Google Scholar]
- 52.Prilassnig M, Wenisch C, Daxboeck F, Feierl G: Are probiotics detectable in human feces after oral uptake by healthy volunteers? Wien Klin Wochenschr 119, 456–462 (2007) [DOI] [PubMed] [Google Scholar]
- 53.Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Møller J, Krogfelt KA, Kazmierczak K, Kenney LJ, Con-way T, Cohen PS. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect Immun 82, 670–682 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lochs H: A question of survival? Interaction between probiotics and the gastrointestinal tract. Wien Klin Wochenschr 119, 441–443 (2007) [DOI] [PubMed] [Google Scholar]
- 55.Caselli M, Cassol F, Calò G, Holton J, Zuliani G, Gasbarrini A: Actual concept of “probiotics”: is it more functional to science or business? World J Gastroenterol 19, 1527–1540 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reister M, Hoffmeier K, Krezdorn N, Rotter B, Liang C, Rund S, Dandekar T, Sonnenborn U, Oelschlaeger TA: Complete genome sequence of the gram-negative probiotic Escherichia coli strain Nissle 1917. J Biotechnol 187, 106–107 (2014) [DOI] [PubMed] [Google Scholar]
- 57.Cress BF, Linhardt RJ, Koffas MA: Draft genome sequence of Escherichia coli strain Nissle 1917 (Serovar O6:K5:H1). Genome Announc, 28, 1(2), e0004713 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobi CA, Malfertheiner P: Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig Dis 29, 600–607 (2011) [DOI] [PubMed] [Google Scholar]
- 59.Verna EC, Lucak S: Use of probiotics in gastrointestinal disorders: what to recommend? Therap Adv Gastroenterol 3, 307–319 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nissle A: Über die Grundlagen einer neuen ursächlichen Bekämpfung der pathologischen Darmflora. Deutsche Medizinische Wochenschrift 42, 1181–1184 (1916) [Google Scholar]
- 61.Pöhlmann C, Thomas M, Förster S, Brandt M, Hartmann M, Bleich A, Gunzer F: Improving health from the inside: use of engineered intestinal microorganisms as in situ cytokine delivery system. Bioengineered 4, 172–179 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nissle A: Die antagonistische Behandlung chronischer Darmstörungen mit Colibakterien. Med Klin 2, 29–33 (1918) [Google Scholar]
- 63.Ortuño Sahagún D, Márquez-Aguirre AL, Quintero-Fabián S, López-Roa RI, Rojas-Mayorquín AE: Modulation of PPAR-γ by nutraceutics as complementary treatment for obesity-related disorders and inflammatory diseases. PPAR Res 2012, 318613 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nissle A: Weiteres über Grundlagen und Praxis der Mutaflor-Behandlung. Deutsche Medizinische Wochenschrift 44, 1809–1813 (1925) [Google Scholar]
- 65.Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA: Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol 9, 804–816 (2007) [DOI] [PubMed] [Google Scholar]
- 66.Nissle A: Weiteres über die Mutaflorbehandlung unter besonderer Berü cksichtigung der chronischen Ruhr. Münchener Medizinische Wochenschrift 25, 678–681 (1919) [Google Scholar]
- 67.Nissle A: Erläuterung über die Bedeutung der Kolondysbakterie und den Wirkungsmechanismus der Colitherapie (“Mutaflor”). Die Medizinische 4, 1017–1022 (1959) [PubMed] [Google Scholar]
- 68.Nissle A: Alte und neue Erfahrungen über Heilerfolge durch Sanierung der Dickdarmflora mit Mutaflor bei Magen-Darmkrankheiten. Medizinische Welt 29–30, 1519–1523 (1961) [Google Scholar]
- 69.Nissle A: Zur Klärung der Beziehungen zwischen Krankheitsursache, Symptomen und rationeller Therapie. Medizinische Welt 23, 1290–1294 (1966) [PubMed] [Google Scholar]
- 70.Tannock GW. (2002): Probiotics and Prebiotics: Where are we going? Caister Academic Press, Norfolk, England [Google Scholar]
- 71.Fishbein M: The aerogenic response of Escherichia coli and strains of Aerobacter in EC broth and selected sugar broths at elevated temperatures. Appl Microbiol 10, 79–85 (1962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vassiliadis G, Destoumieux-Garzón D, Lombard C, Rebuffat S, Peduzzi J: Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob Agents Chemother 54, 288–297 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barth S, Duncker S, Hempe J, Breves G, Baljer G, Bauerfeind R: Escherichia coli Nissle 1917 for probiotic use in piglets: evidence for intestinal colonization. J Appl Microbiol 107, 1697–1710 (2009) [DOI] [PubMed] [Google Scholar]
- 74.Kleta S, Steinrück H, Breves G, Duncker S, Laturnus C, Wieler LH, Schierack P: Detection and distribution of probiotic Escherichia coli Nissle 1917 clones in swine herds in Germany. J Appl Microbiol 101, 1357–1366 (2006) [DOI] [PubMed] [Google Scholar]
- 75.Schroeder B, Duncker S, Barth S, Bauerfeind R, Gruber AD, Deppenmeier S, Breves G: Preventive effects of the probiotic Escherichia coli strain Nissle 1917 on acute secretory diarrhea in a pig model of intestinal infection. Dig Dis Sci 51, 724–731 (2006) [DOI] [PubMed] [Google Scholar]
- 76.Splichalova A, Trebichavsky I, Rada V, Vlkova E, Sonnenborn U, Splichal I: Interference of Bifidobacterium choerinum or Escherichia coli Nissle 1917 with Salmonella Typhimurium in gnotobiotic piglets correlates with cytokine patterns in blood and intestine. Clin Exp Immunol 163, 242–249 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schierack P, Kleta S, Tedin K, Babila JT, Oswald S, Oelschlaeger TA, Hiemann R, Paetzold S, Wieler LH: E. coli Nissle 1917 Affects Salmonella adhesion to porcine intestinal epithelial cells. PLoS One 6(2), e14712 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kleta S, Nordhoff M, Tedin K, Wieler LH, Kolenda R, Oswald S, Oelschlaeger TA, Bleiss W, Schierack P: Role of F1C fimbriae, flagella, and secreted bacterial components in the inhibitory effect of probiotic Escherichia coli Nissle 1917 on atypical enteropathogenic E. coli infection. Infect Immun 82, 1801–1802 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bureš J, Smajs D, Květina J, Förstl M, Smarda J, Kohoutová D, Kuneš M, Cyrany J, Tacheci I, Rejchrt S, Lesná J, Vorisek V, Kopáčová M: Bacteriocinogeny in experimental pigs treated with indomethacin and Escherichia coli Nissle. World J Gastroenterol 17, 609–617 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bureš J, Pejchal J, Květina J, Tichý A, Rejchrt S, Kuneš M, Kopáčová M: Morphometric analysis of the porcine gastrointestinal tract in a 10-day high-dose indomethacin administration with or without probiotic bacteria Escherichia coli Nissle 1917. Hum Exp Toxicol 30, 1955–1962 (2011) [DOI] [PubMed] [Google Scholar]
- 81.Smajs D, Bureš J, Smarda J, Chaloupková E, Květina J, Förstl M, Kohoutová D, Kuneš M, Rejchrt S, Lesná J, Kopáčová M: Experimental administration of the probiotic Escherichia coli strain Nissle 1917 results in decreased diversity of E. coli strains in pigs. Curr Microbiol 64, 205–210 (2012) [DOI] [PubMed] [Google Scholar]
- 82.Yang X, Twitchell E, Li G, Wen K, Weiss M, Kocher J, Lei S, Ramesh A, Ryan EP, Yuan L: High protective efficacy of rice bran against human rotavirus diarrhea via enhancing probiotic growth, gut barrier function, and innate immunity. Sci Rep 5, 15004 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duncker SC, Lorentz A, Schroeder B, Breves G, Bischoff SC: Effect of orally administered probiotic E. coli strain Nissle 1917 on intestinal mucosal immune cells of healthy young pigs. Vet Immunol Immunopathol, 111, 239–250 (2006) [DOI] [PubMed] [Google Scholar]
- 84.Splíchal I, Fagerhol MK, Trebichavský I, Splíchalová A, Schulze J: The effect of intestinal colonization of germ-free pigs with Escherichia coli on calprotect in levels in plasma, intestinal and bronchoalveolar lavages. Immunobiology 209, 681–687 (2005) [DOI] [PubMed] [Google Scholar]
- 85.Haverson K, Rehakova Z, Sinkora J, Sver L, Bailey M: Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol 119, 243–253 (2007) [DOI] [PubMed] [Google Scholar]
- 86.Hejnova J, Dobrindt U, Nemcova R, Rusniok C, Bomba A, Frangeul L, Hacker J, Glaser P, Sebo P, Buchrieser C: Characterization of the flexible genome complement of the commensal Escherichia coli strain A0 34/86 (O83:K24:H31). Microbiology 151, 385–398 (2005) [DOI] [PubMed] [Google Scholar]
- 87.Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW: Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol 8(12), R267 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zschüttig A, Auerbach C, Meltke S, Eichhorn C, Brandt M, Blom J, Goesmann A, Jarek M, Scharfe M, Zimmermann K, Wassenaar TM, Gunzer F: Complete sequence of probiotic Symbioflor 2 Escherichia coli strain G3/10 and draft sequences of Symbioflor 2 E. coli strains G1/2, G4/9, G5, G6/7, and G8. Genome Announc 3(2), pii: e01330–14 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U: Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186, 5432–5441 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun J, Gunzer F, Westendorf AM, Buer J, Scharfe M, Jarek M, Gössling F, Blöcker H, Zeng AP: Genomic peculiarity of coding sequences and metabolic potential of probiotic Escherichia coli strain Nissle 1917 inferred from raw genome data. J Biotechnol 117, 147–161 (2005) [DOI] [PubMed] [Google Scholar]
- 91.Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P: A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics 283, 469–484 (2010) [DOI] [PubMed] [Google Scholar]
- 92.Hancock V, Vejborg RM, Klemm P: Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073, comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics 284, 437–454 (2010) [DOI] [PubMed] [Google Scholar]
- 93.Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, Bacquié V, Pénary M, Gaultier E, Nougayrède JP, Fioramonti J, Oswald E: Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3, 501–609 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antáo EM, Laturnus C, Diehl I, Glodde S, Homeier T, Böhnke U, Steinrück H, Philipp HC, Wieler LH: Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol 297, 163–176 (2007) [DOI] [PubMed] [Google Scholar]
- 95.Mitchell NM, Johnson JR, Johnston B, Curtiss R, 3rd, Mellata M: Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl Environ Microbiol 81, 1177–1187 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakonieczna A, Cooper CJ, Gryko R: Bacteriophages and bacteriophage-derived endolysins as potential therapeutics to combat Gram-positive spore forming bacteria. J Appl Microbiol 119, 620–631 (2015) [DOI] [PubMed] [Google Scholar]
- 97.Reissbrodt R, Hammes WP, dal Bello F, Prager R, Fruth A, Hantke K, Rakin A, Starcic-Erjavec M, Williams PH: Inhibition of growth of Shiga toxin-producing Escherichia coli by nonpathogenic Escherichia coli. FEMS Microbiol Lett 290, 62–69 (2009) [DOI] [PubMed] [Google Scholar]
- 98.Rund SA, Rohde H, Sonnenborn U, Oelschlaeger TA: Antagonistic effects of probiotic Escherichia coli Nissle 1917 on EHEC strains of serotype O104:H4 and O157:H7. Int J Med Microbiol 303, 1–8 (2013) [DOI] [PubMed] [Google Scholar]
- 99.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M: Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conway T, Cohen PS: Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol Spectr 3(3) (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matuskova Z, Anzenbacherova E, Vecera R, Tlaskalova-Hogenova H, Kolar M, Anzenbacher P: Administration of a probiotic can change drug pharmacokinetics: effect of E. coli Nissle 1917 on amidarone absorption in rats. PLoS One 9(2), e87150 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stritzker J, Weibel S, Seubert C, Götz A, Tresch A, van Rooijen N, Oelschlaeger TA, Hill PJ, Gentschev I, Szalay AA: Enterobacterial tumor colonization in mice depends on bacterial metabolism and macrophages but is independent of chemotaxis and motility. Int J Med Microbiol 300, 449–456 (2010) [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y, Zhang Y, Xia L, Zhang X, Ding X, Yan F, Wu F: Escherichia coli Nissle 1917 targets and restrains mouse B16 melanoma and 4T1 breast tumors through expression of azurin protein. Appl Environ Microbiol 78, 7603–7610 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hill PJ, Stritzker J, Scadeng M, Geissinger U, Haddad D, Basse-Lüsebrink TC, Gbureck U, Jakob P, Szalay AA: Magnetic resonance imaging of tumors colonized with bacterial ferritin-expressing Escherichia coli. PLoS One 6(10), e25409 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mandal PK, Misra AK: Concise synthesis of the pentasaccharide O-antigen of Escherichia coli O83:K24:H31 present in the Colinfant vaccine. Glycoconj J 25, 713–722 (2008) [DOI] [PubMed] [Google Scholar]
- 106.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K: The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149, 2557–2570 (2003) [DOI] [PubMed] [Google Scholar]
- 107.Zschüttig A, Zimmermann K, Blom J, Goesmann A, Pöhlmann C, Gunzer F: Identification and characterization of microcin S, a new antibacterial peptide produced by probiotic Escherichia coli G3/10. PLoS One 7(3), e33351 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]


