Abstract
The transition from outcrossing to predominant self-fertilization is one of the most common evolutionary transitions in flowering plants. This shift is often accompanied by a suite of changes in floral and reproductive characters termed the selfing syndrome. Here, we characterize the genetic architecture and evolutionary forces underlying evolution of the selfing syndrome in Capsella rubella following its recent divergence from the outcrossing ancestor C. grandiflora. We conduct genotyping by multiplexed shotgun sequencing and map floral and reproductive traits in a large (N = 550) F2 population. Our results suggest that in contrast to previous studies of the selfing syndrome, changes at a few loci, some with major effects, have shaped the evolution of the selfing syndrome in Capsella. The directionality of QTL effects, as well as population genetic patterns of polymorphism and divergence at 318 loci, is consistent with a history of directional selection on the selfing syndrome. Our study is an important step toward characterizing the genetic basis and evolutionary forces underlying the evolution of the selfing syndrome in a genetically accessible model system.
Keywords: Floral evolution, mating system shift, next-generation sequencing, quantitative trait locus mapping, population genetics
The transition from outcrossing to predominant self-fertilization is one of the most common evolutionary transitions in flowering plants (Stebbins 1950). In association with this mating system shift, similar changes in a suite of floral and reproductive characters have evolved repeatedly (Barrett 2002). In general, selfers tend to have smaller, more inconspicuous flowers, a lower degree of separation between anthers and stigma, and lower pollen–ovule ratios than their outcrossing relatives. This combination of floral and reproductive characters is termed the selfing syndrome (Darwin 1876, Ornduff 1969).
Convergent evolution of the selfing syndrome strongly suggests that these floral and reproductive trait changes are adaptive. Indeed, there are several reasons to expect natural selection to favor the evolution of the selfing syndrome (reviewed in Sicard and Lenhard 2011). Reduced resource allocation to costly structures such as petals and nectaries should be favored in selfers, which are no longer dependent on pollinators for reproductive success (Sicard and Lenhard 2011). If there is a trade-off between resource allocation to pollen and ovule production, reduced pollen production is expected to be selected for in selfers (Charlesworth and Charlesworth 1981). Floral morphology could evolve to reduce the incidence of herbivory (Eckert et al. 2006), or simply as a result of selection for combinations of traits that improve autonomous selfing ability (Moeller and Geber 2005; Fishman and Willis 2008).
Quantitative genetic studies can aid in distinguishing between these different hypotheses for the evolution of the selfing syndrome. For instance, genetic correlations can point to the existence of intrinsic genetic trade-offs, and may suggest the presence of genetic constraints to floral evolution (Ashman and Majetic 2006). Quantitative trait locus (QTL) mapping can yield additional insights into the evolution of the selfing syndrome. A prevalence of QTL with allelic effects consistent with species differences is suggestive of a history of directional selection (Orr 1998) and overlapping QTL may suggest that underlying loci have pleiotropic effects, provided that resolution is sufficiently high. In addition, QTL mapping studies can yield information on the number and effect sizes of loci involved in the evolution of the selfing syndrome.
Dissecting the genetic architecture of the selfing syndrome through QTL mapping may thus allow for insights into adaptive evolution. However, despite the prevalence of the transition to selfing (Barrett 2002) and the vast literature on the causes and population genetic effects of this transition (reviewed in e.g., Barrett and Harder 1996; Charlesworth and Wright 2001; Charlesworth 2006), quantitative genetic studies of the selfing syndrome have only been conducted in a handful of systems. These studies have found that floral traits involved in the selfing syndrome mostly have a polygenic basis (e.g., in Mimulus, Lin and Ritland 1997; Fishman et al. 2002; Leptosiphon, Goodwillie et al. 2006; Oryza, Grillo et al. 2009; and Solanum, Bernacchi and Tanksley 1997) with little evidence for major genes. The directionality of QTL effects in the majority of these studies is consistent with a history of directional selection, although this is not always explicitly tested. Genetic correlations between floral traits are often positive (Ashman and Majetic 2006), and overlapping QTL for multiple floral traits (Bernacchi and Tanksley 1997; Fishman et al. 2002) suggest that genetic architecture may pose constraints for floral evolution, although distinguishing between close linkage and pleiotropy will require vastly improved mapping resolution.
Our knowledge of the molecular basis of selfing syndrome evolution is limited and only a single gene underlying a selfing syndrome trait has been cloned so far (Chen et al. 2007). Development of genomic resources for genetically accessible model systems is important to improve our understanding of the types of genetic changes underlying this evolutionary transition. Recent developments in sequencing technology hold the promise to facilitate such studies.
Here, we characterize the genetic architecture of the selfing syndrome in the crucifer genus Capsella, a promising model system for the study of mating system shifts. This genus harbors two diploid sister species that differ in their mating system, the highly self-fertilizing Capsella rubella, and the obligately outcrossing, self-incompatible C. grandiflora (Hurka and Neuffer 1997). The selfer C. rubella is thought to be derived from an outcrossing, C. grandiflora-like ancestor (Foxe et al. 2009; Guo et al. 2009). These two species differ not only in their geographical distribution and mating system, but also with respect to floral and reproductive traits. Capsella grandiflora is mainly found in the western Balkans and occasionally in northern Italy, whereas C. rubella has a wider circum-Mediterranean distribution (Hurka and Neuffer 1997). In C. rubella, there has been a derived loss of self-incompatibility and it exhibits the typical characteristics of the selfing syndrome (Hurka and Neuffer 1997). These changes have resulted in high rates of selfing, with effective selfing rates in natural populations of C. rubella estimated to be 0.90–0.97 (St. Onge et al. 2011).
Population genetic analysis of the S locus and multilocus data has suggested that the evolution of selfing in C. rubella was associated with a severe population bottleneck, suggestive of a major, rapid shift to high selfing rates from a small number of founding lineages (Foxe et al. 2009; Guo et al. 2009). Changes in floral and reproductive traits in C. rubella have probably evolved relatively rapidly, as the transition to selfing occurred recently, most likely within the last 50,000 years (Foxe et al. 2009; Guo et al. 2009). If floral evolution occurred subsequent to the founder event, adaptive morphological evolution in C. rubella would have proceeded with a limited amount of standing genetic variation.
Studies of the genetic basis of the selfing syndrome in Capsella are facilitated by the interfertility of the two diploid species, their close relationship to A. rabidopsis thaliana (Boivin et al. 2004), and the availability of a sequenced C. rubella genome. Here, we make full use of these advantages to map QTL for floral and reproductive traits in a large interspecific F2 population (N = 550). We generate a dense set of markers and genotype all individuals using a cost-effective technique based on massively parallel sequencing (MSG; Andolfatto et al. 2011). As a proof of concept, we map self-compatibility to a 255-kb region that encompasses the canonical Brassicaceae S locus. We assess the distribution and directionality of additive QTL effects, as well as the degree of overlap between QTL. Finally, we use population genomic data for 318 loci to explore whether regions harboring QTL for the selfing syndrome exhibit signs of directional selection. This study forms an important initial step in characterizing the genetic basis and evolutionary forces underlying recent evolution of the selfing syndrome in Capsella.
Materials and methods
PLANT MATERIAL
We generated an F2 mapping population from an interspecific cross between C. grandiflora and C. rubella. The F2 was generated by self-fertilizing a self-compatible F1 individual produced from a cross of an outbred C. grandiflora accession (2e-TS1) from Paleokastritsas, Greece, as seed parent and a C. rubella accession (1GR1) from Manolates, Samos, Greece, as pollen donor. Floral and reproductive traits (see below) were measured in both parents and the F1. In 2009, we grew a total of 700 F2 individuals alongside six selfed offspring of C. rubella 1GR1 and 16 accessions each of C. rubella and C. grandiflora sampled across the range of each species (Supporting information).
PLANT GROWTH CONDITIONS
Seeds were surface-sterilized, plated on half strength Murashige–Skoog medium and vernalized at 1.8°C for 18 days. Germination took place at room temperature over eight days, and F2 seeds had a consistently very high germination rate (>95%). Seedlings were transplanted to Pro-Mix BX (Premier Tech Horticulture, Riviére du Loup, Quebec, Canada) potting mix in 3.5-inch pots which were placed in a fully randomized design in the greenhouse at University of Toronto on June 15, 2009. Seedlings were acclimatized to natural light conditions for one week and were subsequently grown under long day conditions (22°C day/21°C night; 16 h light) with supplemental light from sodium high pressure lamps and biweekly fertilization with N:P:K (20:20:20) fertilizer.
PHENOTYPIC MEASUREMENTS
A total of 13 floral, vegetative, and reproductive characters were measured. We scored vegetative characters on all plants, whereas more labor-intensive floral and reproductive trait measurements were done on a subset of 550 F2s as well as on all C. rubella and C. grandiflora individuals.
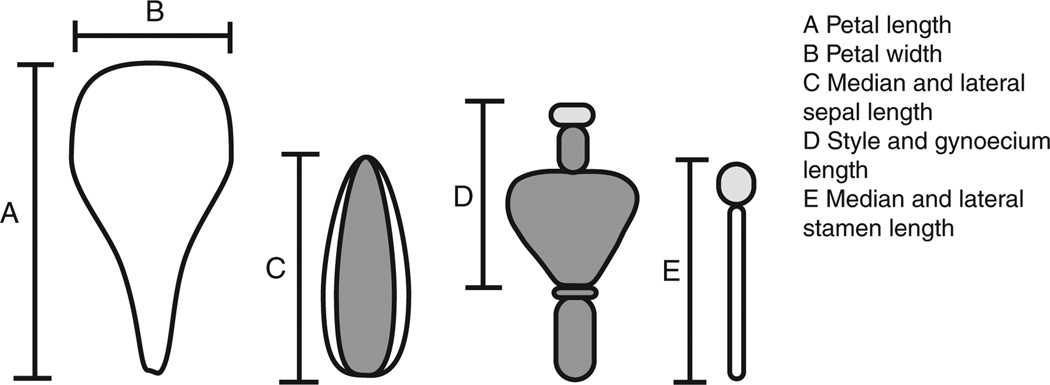
We measured seven floral traits: petal length and width, the length of lateral and median sepals, the length of lateral and median stamens, and the total length of the style and gynoecium (Fig 1). Floral measurements were done on three flowers from each individual. Measurements were based on digital images of dissected floral organs, taken with an Olympus SZTR1 dissecting microscope with an Infinity CCD camera (Olympus Canada, Markham, Ontario, Canada). Images were calibrated with a stage micrometer and measurements done using ImageJ 1.40 (Abramoff et al. 2004).
Figure 1.
Schematic showing floral measurements.
We assessed three reproductive traits: the number of pollen grains per flower, the number of ovules per flower, and self-incompatibility. Pollen and ovule counts were done on three flowers per plant, using a standard aniline blue-lactophenol staining protocol (Kearns and Inouye 1993) and a hemacytometer. As in Fishman et al. (2001), we tentatively classified those pollen grains that had a reduced diameter and did not stain strongly as inviable. Ovule counts were done under a dissecting microscope. We scored self-incompatibility as a binary trait at the end of the experiment, with plants producing any seeds by autonomous pollination classified as self-compatible and those that produced no seeds by autonomous pollination classified as self-incompatible.
For an overall assessment of plant size, we measured the length of the two longest leaves at the start of flowering, and to assess variation in phenology we scored the number of days to flowering and the number of rosette leaves at the start of flowering.
Some F2 individuals exhibited floral abnormalities, such as fusions between floral organs, on some of their inflorescences. To test whether this could be a result of inbreeding depression, we scored and mapped this as a binary trait.
DESCRIPTIVE STATISTICS
We assessed phenotypic differentiation between C. rubella and C. grandiflora with respect to floral and reproductive traits using the Wilcoxon rank sum test. To assess trait normality in the F2 population we used the Shapiro–Wilk normality test. Nonparametric measures of correlation (Spearman’s rho) were calculated for all pairwise combinations of traits in the F2.
MULTIPLEXED SHOTGUN SEQUENCING
To genotype our mapping population, we used multiplexed shotgun genotyping (MSG): a new approach based on shotgun sequencing of multiplexed Illumina libraries (Andolfatto et al. 2011). Briefly, MSG involves digesting genomic DNA with a restriction enzyme, followed by barcoding each sample with a unique adapter and pooling these samples for sequencing. Given a mapping population from a controlled cross and a reference genome, a Hidden Markov Model (HMM) is used to estimate ancestry probabilities for all markers in each individual from the MSG sequence data.
For genotyping of the 550 F2 individuals, we first extracted genomic DNA from frozen leaf tissue using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genomic DNA concentration was quantified using a Qubit BR kit (Invitrogen, Grand Island, NY) and a fluorometer and diluted to a standard concentration. Sequence library construction closely followed the MSG protocol described in Andolfatto et al. (2011). First, a total of 10 ng of genomic DNA was digested with Mse I (New England Biolabs, Ipswich, MA). We then ligated unique barcoded adapters to each sample and pooled samples to give six pools of 96 samples each (26 samples were included in several pools to increase coverage). Independent multiplexed sequencing libraries were constructed for each of these six pools. Ligated linker dimers were removed from each pool with Ampure beads (Beckman Coulter Genomics, Danvers, MA) and the cleaned ligation products were size-selected on an agarose gel to yield fragments of length 250–300 bp. FC2 flow-cell sequences were attached to ligation products using PCR. We sequenced each library on a separate lane of an Illumina Genome Analyzer IIX (Illumina, San Diego, CA) at Princeton Microarray Facility (http://www.genomics.princeton.edu/microarray/) using standard Illumina sequencing protocols.
SEQUENCE PARSING AND GENOTYPE CALLING
The MSG HMM algorithm (Andolfatto et al. 2011) requires reference genomes for both parents of the mapping population; however, the high heterozygosity of our C. grandiflora parent required us to take a two-step approach to generating reference parental genomes. First, the genome of our reference C. rubella mapping parent was constructed as part of our ongoing population genomics effort in Capsella (http://biology.mcgill.ca/vegi/index.html) and aided by a prerelease of the U.S. Department of Energy Joint Genome Institute C. rubella genome assembly (http://www.jgi.doe.gov/).
We then used all reads from the F2 mapping population MSG sequence to identify informative single-nucleotide polymorphisms (SNPs) (i.e., SNPs that segregate in the mapping population) and generate a synthetic C. grandiflora parental reference sequence. This procedure ensures that only informative SNPs that were identified in our F2 mapping population will be queried in the genotyping analysis. Details on construction of our C. rubella mapping parent genome and on identification of informative SNPs are given in Supplementary Information.
For all individuals in the mapping population, we assigned ancestry probabilities across chromosomes using MSG v0.3, a pipeline of scripts developed by Andolfatto et al. (2011) (available at http://genomics.princeton.edu/AndolfattoLab/MSG.html). Briefly, sequence reads were parsed by barcode into 550 groups corresponding to the 550 F2 individuals. Reads were mapped to the parental reference genomes using the Burrows–Wheeler algorithm (BWA; Li and Durbin 2009) with default settings, and a HMM was used to estimate a posterior probability of each possible genotype (in our case: homozygous C. rubella, heterozygous, or homozygous C. grandiflora) in a genomic region. Genotype probabilities for each informative marker position were obtained by imputing ancestry for all individuals at all positions that were typed in at least one individual. Using this pipeline we obtained genotype probabilities for a total of 121,979 SNPs. We set the HMM parameter γ which specifies the degree of uncertainty in the parental reference genomes to 0.03 for both parental genomes, the expected number of recombination events per genome per meiosis to 16 (i.e., at least one crossover per chromosome arm), and the model recombination rate parameter, rfac, to 1 × 10−6 (similar results were obtained with an rfac setting of 1). As we analyzed an F2 population, genotype priors were set to 0.5 for the heterozygote and 0.25 for each parental allele homozygote.
To simplify linkage map construction and facilitate QTL mapping, we assigned each individual a single genotype at each marker (i.e., a “hard ancestry call”) based on ancestry probabilities from the MSG HMM. A cutoff ancestry probability of 0.9 was used for genotype calling, and we filtered markers that had identical genotype configurations across the F2 so that only one of those markers was retained. If markers differed only in terms of missing ancestry calls, we kept the marker that had less missing data. The trimmed dataset was used for linkage map construction and QTL analyses using MQM mapping, which requires hard genotype calls. To test the use of MSG for fine-scale location of QTL, we also used a different filtering strategy and retained the HMM posterior probabilities for mapping. Specifically, we filtered genotype probabilities to retain markers that differed in posterior probability by more than 0.01 in at least one individual and genotype in our F2, and used those genotype probabilities (“soft ancestry calls”) instead of hard ancestry calls for QTL analyses. This procedure was used to map self-compatibility which was suggested to have a simple genetic basis in previous studies (Riley 1934; Nasrallah et al. 2007) and for which we could use a simpler interval mapping algorithm.
LINKAGE MAP CONSTRUCTION AND SEGREGATION DISTORTION
We constructed a linkage map in R/QTL (Broman et al. 2003) under default parameters, using a logarithm of odds (LOD) score cutoff of 6 to assign markers to linkage groups as suggested by Broman (2010). We tested for segregation distortion at each marker using a chi-square test and assessed significance after Bonferroni correction for multiple tests.
QTL ANALYSIS
For floral, vegetative, and reproductive traits that were scored on a continuous scale we mapped QTL by multiple QTL mapping (MQM) (Jansen 1993; Jansen and Stam 1994), which has benefits over interval mapping and composite interval mapping in terms of power and avoidance of false positives (Arends et al. 2010). After imputation of missing genotype data, significant cofactors were identified using an automated backward elimination procedure (Arends et al. 2010). We tested for QTL in 1 cM intervals and excluded cofactors in a window of 25 cM when testing for a QTL effect at a location. For traits that were measured on a binary scale, such as self-incompatibility and presence/absence of floral abnormalities, we used a binary interval mapping model instead (Xu and Atchley 1996). All these QTL analyses were conducted in R/QTL (Broman et al. 2003), using hard genotype calls (see section “Sequence Parsing and Genotype Calling” above).
We assessed significance of QTL using LOD scores, where LOD = log10(L0/L1), with L0 being the likelihood under the null hypothesis of no QTL in the interval and L1 the likelihood under the alternative hypothesis of a QTL in the interval. For each trait, genomewide significance thresholds (1% and 5%) were determined by 1000 permutations. For all QTL significant at P ≤ 0.01, we obtained 1.5-LOD and 2-LOD confidence intervals (CIs), as well as estimates of additive allelic effects and dominance deviations. We present additive effect sizes both standardized by the mean difference between the parental species, as in Fishman et al. (2002), and as the proportion of F2 phenotypic variance explained. We tested for pairwise interactions between all significant QTL and fit a model containing all main QTL effects and significant interactions. For each trait, we obtained an estimate of the total proportion of F2 phenotypic variation explained by this multiple QTL model. We tested the normality of residuals using a Shapiro–Wilk test.
To test whether our QTL effect size estimates could be biased due to variation in recombination rate and/or gene density (the “Noor effect”; Noor et al. 2001), we compared recombination rate and gene density within and outside of 1.5-LOD CIs for petal size traits using a Mann–Whitney test.
POPULATION GENETICS ANALYSIS
To test whether QTL regions show evidence for directional selection in C. rubella, we made use of our resequencing data from 354 exons in both C. rubella and C. grandiflora, analyzing only those loci with at least six samples sequenced in each species. These loci were sequenced as described in Slotte et al. (2010) and Qiu et al. (2011). Briefly, samples from eight Mediterranean populations of C. rubella and five populations of C. grandiflora were sampled for this study, and single large exons were amplified and sequenced on both strands. We analyzed polymorphism levels using a modified version of Polymorphorama (Andolfatto 2007; Haddrill et al. 2008), and used custom Perl scripts to calculate the number of fixed differences, and shared and unique polymorphisms. We determined the physical positions of these exons on the C. rubella genome using BLAST (Altschul et al. 1990), and identified exons that fell within the 2-LOD interval of QTL. To avoid very wide QTL intervals encompassing large fractions of chromosomes, we focused this analysis on QTL that had 2 LOD intervals that were 2 Mb or less. When multiple overlapping QTL fit these criteria, we used the narrower interval.
Directional selection is expected to lead to a reduction in the proportion of shared polymorphisms and an increase in the proportion of fixed differences between species (Foxe et al. 2009). We tested whether QTL regions exhibited such a signature using Fisher’s Exact test. In addition, we assessed whether the ratio of polymorphism in C. rubella and C. grandiflora differed between QTL regions and other genomic regions.
Results
PHENOTYPIC VARIATION BETWEEN SPECIES AND IN THE F2
We found significant phenotypic differentiation between C. rubella and C. grandiflora for all measured floral and reproductive traits, but not for leaf size and phenology traits (Table 1). The distribution of petal and reproductive traits in C. rubella did not overlap with that of C. grandiflora, whereas there was considerable overlap for other floral traits (Fig. 2).
Table 1.
Means and standard deviations for vegetative, floral and reproductive traits in Capsella rubella and C. grandiflora.
| Trait |
C. rubella (n=16) |
C. grandiflora (n=16) |
|---|---|---|
| Petal length (mm) | 2.45±0.21 | 4.24±0.47 |
| Petal width (mm) | 1.06±0.14 | 2.58±0.29 |
| Median sepal length (mm) |
1.95±0.20 | 2.38±0.22 |
| Lateral sepal length (mm) |
1.92±0.20 | 2.30±0.22 |
| Median stamen length (mm) |
2.08±0.15 | 2.42±0.20 |
| Lateral stamen length (mm) |
1.84±0.15 | 2.17±0.22 |
| Length of style + gynoecium (mm) |
1.89±0.20 | 2.19±0.29 |
| Ovules per flower | 21±2 | 14±2 |
| Pollen per flower | 5791±302 | 37453±4114 |
| Leaf number | 11±3 | 12±4 |
| Leaf length (cm) | 14.9±4.3 | 15.8±5.6 |
| Days to flowering | 24±3 | 24±2 |
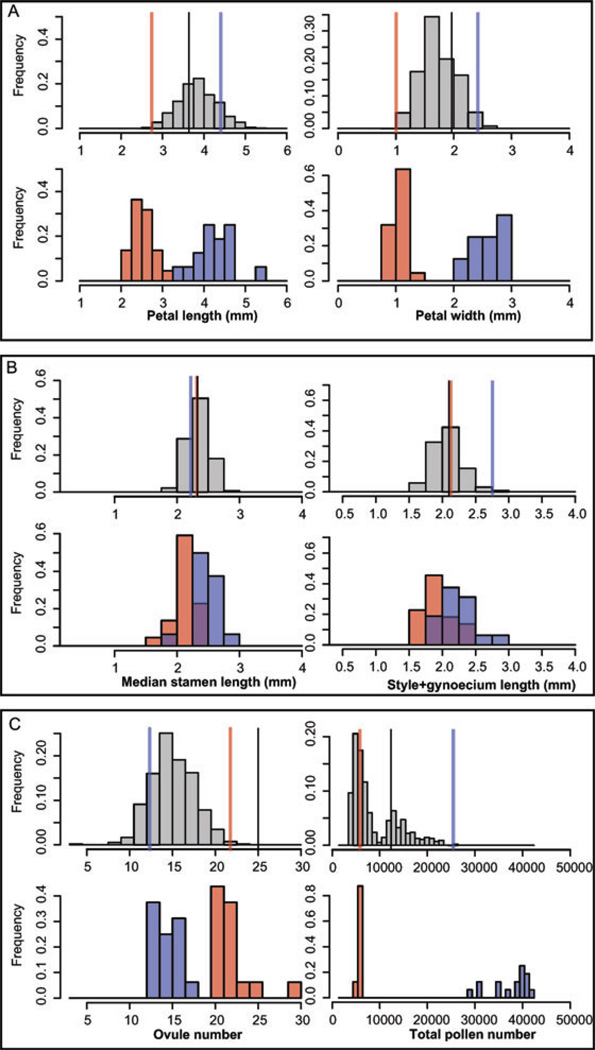
Figure 2.
Histograms of representative floral and reproductive traits in the F2 (grey) as well as in Capsella rubella (red) and C. grandiflora (blue). Phenotypic values of the parental accessions are shown as lines in the upper panel of each plot (C. rubella 1GR1 in red, C. grandiflora 2e-TS1 in blue, and the F1 value in black). The traits are A. Petal length (mm) (left), Petal width (mm) (right), B. Median stamen length (mm) (left), Style + gynoecium length (mm) (right), C. Ovule number per flower (left), and Pollen number per flower (right).
In the F2 population, all floral traits exhibited continuous variation with a unimodal distribution. Petal trait means in the F2 were intermediate between those of the parental accessions, with no clear evidence of transgressive segregation. For the other floral traits, as well as for ovule number and leaf size, more extreme values were often found in the F2 than in either parental accession. However, in most cases the range of variation in the F2 did not exceed that found in the parental species (Fig. 2).
All traits except petal and stamen length deviated significantly from normality (data not shown). However, only pollen number per flower had a clearly bimodal distribution. The lower mode of this distribution was close to the value for the C. rubella parental accession, with an additional peak intermediate between the parental values (Fig 2). The mean proportion of inviable pollen in the F2 was low (mean: 7.9 ± 10.7%) suggesting that hybrid incompatibilities affecting pollen viability are not rampant in this population. Phenotypic correlations were highest among floral traits (Fig. 3), whereas these traits exhibited a lower degree of correlation with vegetative and reproductive traits (Fig 3.).
Figure 3.
Character correlations in the F2 population. Scatterplots of all pairs of traits are given below the diagonal and Spearman’s correlation coefficients are given above the diagonal. Traits are color-coded with green for vegetative and phenology traits, yellow for floral traits and grey for reproductive traits.
About 28% of the F2 individuals were self-incompatible. Segregation of self-incompatibility did not deviate significantly from the 1:3 ratio expected under a single dominant locus with the C. rubella allele conferring self-compatibility (Chi-square test, χ2 = 1.08, df = 1, P = 0.30) as previously reported in crosses of C. grandiflora and C. rubella (Riley 1934; Nasrallah et al. 2007).
MSG
We used MSG to generate indexed 101 bp Illumina reads from each of our F2 individuals. We were able to map 67% of our approximately 188 million reads to the C. rubella reference nuclear genome assembly and identified a total of 121,979 informative SNPs among those reads. The median number of informative markers per individual was 6474, corresponding to a marker density of about 1 per 20 kb.
We used MSG v0.3 to assign ancestry probabilities for each F2 individual at each of these 121,979 markers. This resulted in a marker density of about 1 marker per kb. Ancestry probabilities were subsequently converted to hard ancestry calls (“genotypes”), of which we retained a total of 890 markers for linkage map construction and initial QTL analyses. For a test of the use of soft ancestry calls (genotype probabilities) from MSG for fine-scale location of QTL, we filtered soft ancestry calls and used those for QTL mapping.
LINKAGE MAP CONSTRUCTION
The resulting linkage map contained 890 markers and had eight linkage groups (Fig. 4), consistent with previous linkage mapping results (Boivin et al. 2004). The mean number of markers per linkage group was 111 (min: 93, max: 152) and the total map distance was 381.8 cM. The mean distance between markers was 0.4 cM, and the maximum distance between markers was 8.4 cM.
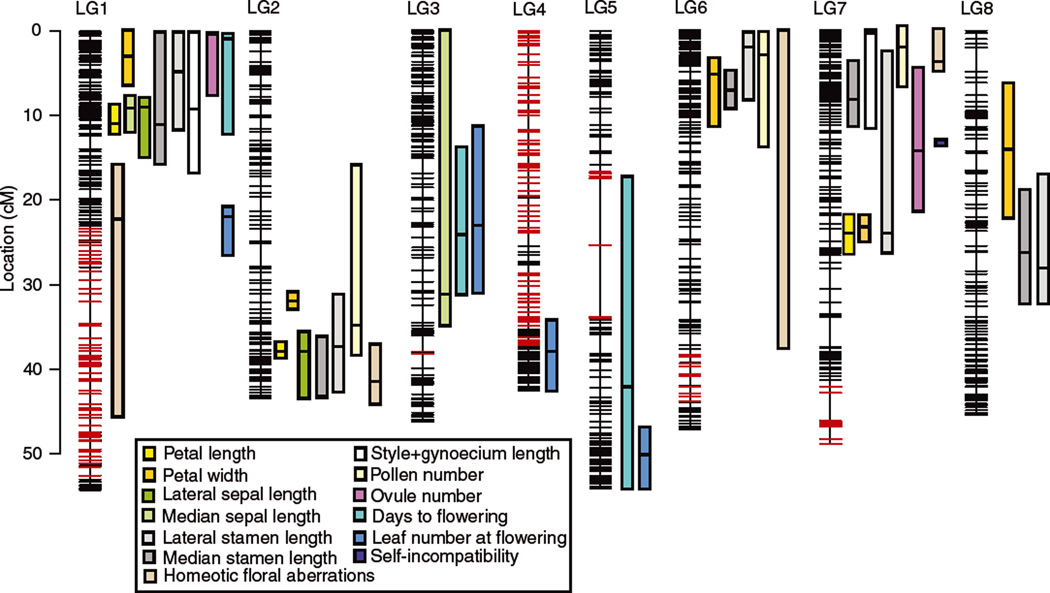
Figure 4.
Capsella linkage map and 1.5-LOD confidence intervals. Markers that exhibit significant segregation distorsion (at P < 0.05 after Bonferroni correction) are colored in red. All distances are in cM.
A total of 152 markers showed significant segregation distortion (Fig. 4). These markers mapped to five main regions: the lower part of LG1 and most of LG4 showed a consistent deficit of genotypes homozygous for the C. rubella allele, whereas regions on LG5, LG6, and the lower part of LG7 showed an excess of heterozygous genotypes.
QTL MAPPING
We identified a total of 41 QTL for the 13 phenotypic traits assessed in this study. For each floral trait we identified between two and five significant QTL, and there were a total of 24 significant QTL for the seven floral size traits measured (Fig. 4). These QTL co-localized to a great extent. Floral size QTL mainly mapped to five regions: the upper part of LG1, LG6 and LG7, the central part of LG8, and the lower part of LG2 (Fig. 4). QTL for reproductive traits (three QTL for pollen number and two for ovule number) also co-localized with floral trait QTL on LG1, LG2, LG6, and LG7 (Fig 4). In contrast, QTL for phenology traits did not overlap with floral size QTL, with the exception of one QTL for days to flowering that mapped to the upper part of LG1 (Fig. 4). We did not find any significant QTL for plant size at flowering.
The median width of 1.5-LOD CIs for floral and reproductive trait QTL was 7.6 cM or 4.4 Mb; however, 1.5-LOD CIs ranged from 1.8 to 34.6 cM (0.35 to 13.6 Mb). Petal size trait QTL on LG2 had the narrowest CIs of all continuous traits (1.8 cM or 0.7 Mb for petal length and 2.3 cM or 0.4 Mb for petal width; Table 2; Fig. 4).
Table 2.
List of significant QTL, including 1.5-LOD and 2-LOD confidence intervals and effect size estimates.
| Trait | QTL peak position (LG, cM) |
Peak LOD |
1.5-LOD CI (cM) |
2-LOD CI (cM) |
Additive effect (SE)1 |
Dominance deviation (SE) |
%Variance explained2 |
Relative homozygous effect3 |
|---|---|---|---|---|---|---|---|---|
| Petal length (mm) | 1, 11.1 | 77.91 | (8.5–12.1) | (8.1–12.1) | 0.37 (0.03) | −0.05 (0.03) | 26.90 | 0.41 |
| 2, 37.8 | 41.31 | (36.9–38.8) | (31.1–39) | 0.23 (0.03) | 0.24 (0.04) | 18.66 | 0.26 | |
| 7, 23.5 | 41.11 | (21.8–26.4) | (21.8–27.5) | 0.26 (0.03) | 0.02 (0.04) | 14.88 | 0.30 | |
| Petal width (mm) | 1, 2.8 | 15.87 | (0–6.5) | (0–10.3) | 0.12 (0.02) | −0.01 (0.03) | 6.11 | 0.16 |
| 2, 32.1 | 69.68 | (30.6–32.9) | (30.6–32.9) | 0.2 (0.02) | 0.16 (0.02) | 31.36 | 0.26 | |
| 6, 4.9 | 17.19 | (3.2–12) | (2.9–14.2) | 0.1 (0.02) | −0.03 (0.03) | 6.10 | 0.13 | |
| 7, 22.9 | 55.03 | (21.8–24.9) | (21.8–24.9) | 0.19 (0.02) | 0.01 (0.02) | 18.74 | 0.24 | |
| 8, 14.2 | 7.46 | (6.2–22) | (5.5–22) | 0.05 (0.02) | −0.01 (0.03) | 2.10 | 0.07 | |
| Median sepal length (mm) |
1, 9 | 31.37 | (7.7–12.1) | (7.4–14.2) | 0.19 (0.02) | 0 (0.02) | 22.40 | 0.90 |
| 3, 30.9 | 3.62 | (0–34.5) | (0–38) | 0.05 (0.02) | 0.01 (0.03) | 2.52 | 0.25 | |
| Lateral sepal length (mm) |
1, 9 | 25.95 | (8.1–15.3) | (7.7–16.4) | 0.18 (0.02) | −0.02 (0.02) | 15.94 | 0.95 |
| 2, 37.8 | 8.25 | (35.9–43.5) | (34.6–43.5) | 0.09 (0.02) | 0.1 (0.02) | 6.79 | 0.47 | |
| Median stamen length (mm) |
1, 11.1 | 5.99 | (0.1–15.8) | (0–16.4) | 0.05 (0.01) | −0.01 (0.01) | 4.58 | 0.29 |
| 2, 43.5 | 4.38 | (36.5–43.5) | (34.6–43.5) | 0.04 (0.01) | 0.04 (0.01) | 3.72 | 0.26 | |
| 6, 7 | 29.20 | (4.9–8.7) | (4.3–8.7) | −0.1 (0.01) | 0.01 (0.01) | 14.67 | −0.57 | |
| 7, 8 | 49.96 | (3.8–10.9) | (3–10.9) | 0.12 (0.01) | −0.01 (0.01) | 23.91 | 0.70 | |
| 8, 26 | 4.91 | (19–31.9) | (16.8–35.2) | 0.03 (0.01) | 0.01 (0.01) | 2.03 | 0.20 | |
| Lateral stamen length (mm) |
1, 4.9 | 5.36 | (0–11.5) | (0–12.1) | 0.05 (0.01) | −0.01 (0.02) | 3.48 | 0.30 |
| 2, 37.1 | 18.58 | (31.1–42.1) | (29.8–43) | 0.09 (0.01) | 0.06 (0.01) | 11.45 | 0.51 | |
| 6, 2 | 26.85 | (0.5–7.9) | (0.3–8.7) | −0.1 (0.01) | −0.01 (0.01) | 14.14 | −0.61 | |
| 7, 23.5 | 22.13 | (2.1–26.6) | (1.9–27.5) | 0.09 (0.01) | −0.01 (0.01) | 12.45 | 0.52 | |
| 8, 28.4 | 4.52 | (16.8–31.9) | (11.7–34.8) | 0.04 (0.01) | 0.01 (0.02) | 3.04 | 0.26 | |
| Style + gynoecium length (mm) |
1, 9 | 12.88 | (0–16.4) | (0–16.6) | 0.1 (0.01) | 0.01 (0.02) | 9.98 | 0.65 |
| 7, 0 | 3.82 | (0–8.8) | (0–10.6) | −0.05 (0.01) | −0.05 (0.02) | 4.01 | −0.34 | |
| Ovule number | 1, 0 | 4.83 | (0–7.4) | (0–9.1) | 0.73 (0.16) | 0.1 (0.22) | 3.52 | −0.19 |
| 7, 14 | 8.44 | (4.3–20.9) | (2.4–21.6) | −0.92 (0.16) | 0.24 (0.21) | 5.98 | 0.24 | |
| Pollen number | 2, 34.6 | 7.25 | (15.9–38.2) | (8.6–41.4) | 1479 (297) | 159 (415) | 6.08 | 0.09 |
| 6, 2.9 | 5.98 | (0–13.4) | (0–15.5) | 1374 (309) | 403 (415) | 4.12 | 0.09 | |
| 7, 1.9 | 13.31 | (0–6.5) | (0–8.2) | 2193 (293) | −213.8 (401.44) | 9.36 | 0.14 | |
| Leaf number at flowering |
1, 22.2 | 21.84 | (20.8–26.4) | (19.8–27.4) | −2.55 (0.29) | −0.1 (0.39) | 18.57 | |
| 3, 23 | 3.48 | (11.1–30.8) | (8.5–45.4) | −0.86 (0.31) | −0.21 (0.41) | 4.44 | ||
| 4, 38.1 | 3.28 | (34.3–42.5) | (33.1–42.5) | −0.85 (0.31) | −0.4 (0.41) | 3.84 | ||
| 5, 50 | 23.22 | (46.9–54.1) | (46.1–54.1) | −2.59 (0.28) | 0.19 (0.38) | 12.38 | ||
| Days to flowering | 1, 1.1 | 3.77 | (0–12.1) | (0–13.9) | −0.98 (0.25) | −0.23 (0.34) | 3.19 | |
| 3, 24.5 | 13.90 | (13.7–30.8) | (13.1–30.8) | −1.97 (0.25) | −0.17 (0.33) | 10.38 | ||
| 5, 42.1 | 3.57 | (17.5–54.1) | (17.5–54.1) | −1.08 (0.26) | −0.04 (0.34) | 3.03 | ||
| Homeotic floral aberrations |
1, 22.2 | 4.23 | (16.6–41.3) | (15.8–45.3) | 0.10 (0.03) | −0.10 (0.04) | 4.12 | |
| 2, 41.4 | 5.50 | (37.8–41.4) | (37.1–43.5) | 0.15 (0.03) | −0.09 (0.04) | 2.47 | ||
| 6, 0 | 3.49 | (0–7.2) | (0–37.2) | −0.13(0.03) | −0.02 (0.04) | 1.03 | ||
| 7, 3.8 | 28.43 | (0–4.6) | (0–4.6) | −0.34(0.03) | −0.14 (0.04) | 20.35 |
Additive effect: half the mean phenotypic difference between homozygotes for the C. grandiflora allele and homozygotes for the C. rubella allele.
Per cent F2 phenotypic variance explained by QTL.
Homozygous additive effect (2a) relative to mean phenotypic difference between C. grandiflora and C. rubella for traits that differ significantly between C. grandiflora and C. rubella.
For 25 of the 29 floral size and reproductive trait QTL, the direction of allelic effects was consistent with phenotypic differences between species. Floral size QTL with allelic effects opposite to expectation were only found for traits whose phenotypic distributions overlap between C. rubella and C. grandiflora (e.g., stamen length and the total length of style and gynoecium; Table 2).
Altogether, a model with significant QTL explained between 10 and 64% of the phenotypic variation for floral and reproductive traits in the F2 population. Petal size QTL explained the highest proportion of F2 variance (petal width: 64%; petal length: 63%), whereas QTL for stamen length explained an intermediate proportion (median stamen length: 50%, lateral stamen length: 46%) and QTL for reproductive traits had the lowest explanatory power (pollen number: 19%, ovule number: 10%) (Table S2).
Individual QTL effects were also greatest for petal size traits; the leading QTL for petal width and length explained 31% and 27% of F2 phenotypic variation, respectively (Table 2). Homozygous additive effects at leading QTL for these traits accounted for a large fraction of the phenotypic difference between C. grandiflora and C. rubella (26% for petal width and 41% for petal length; Table 2). Overall, dominance deviations were small in relation to additive effects of QTL, with the exception of floral size QTL mapping to the lower part of LG2, where the C. rubella QTL allele was largely recessive for both petal length, petal width, lateral sepal length, and stamen length (Table 2). Pairwise epistatic effects were found for petal length (QTL on LG1 and LG7), stamen length (lateral stamen length: between QTL on LG1 and LG6, LG6 and LG8; median stamen length: between QTL on LG1 and LG2), and days to flowering (between QTL on LG1 and LG3; LG1 and LG4), but explained a low proportion of F2 variation (0.4–1.4%).
With the exception of pollen number, the distributions of residuals after accounting for QTL effects were unimodal and reasonably symmetric (Fig. S2). There were no significant deviations from normality of residuals for petal length or width, or for median and lateral stamen lengths. However, we did find slight but significant departures from residual normality for some traits (e.g., sepal and style length, pollen/ovule number, and phenology traits; Fig. S2). As standard QTL mapping methods are robust to deviations from normality when significance is determined by permutations and with dense genotyping information (Broman and Sen 2009), we did not transform the data.
We did not find evidence for a “Noor effect” causing overestimates of effect sizes for petal traits, as there were no significant differences between QTL regions and the remainder of the genome in either recombination rate or gene density.
About 40% of F2 individuals exhibited floral abnormalities on some inflorescences (Table S2). Partly recessive C. rubella alleles on LG7 explain most of the F2 variation for this trait; however, two minor QTL on LG1 and LG2 with fully or partly recessive C. grandiflora alleles also contribute to some degree. We did not find evidence for significant interaction between QTL for floral abnormalities in a two-locus QTL scan.
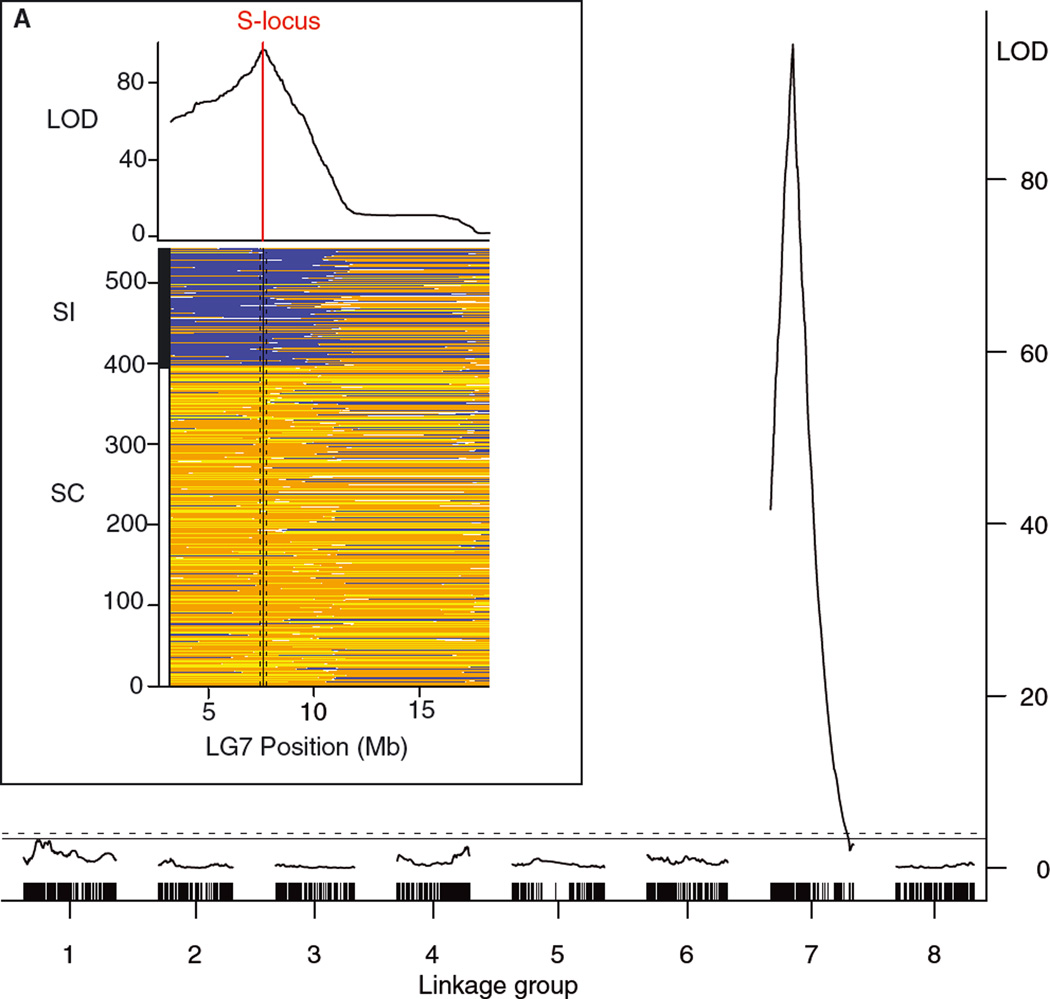
In agreement with the 1:3 segregation of self-compatibility in our cross, SC mapped to a single, strongly significant QTL at ~7.6 Mb on LG7, with a dominant C. rubella allele conferring self-compatibility (Fig. 5). We mapped self-compatibility using 5361 markers on LG7 with soft ancestry calls from MSG. Using this method, the peak of the QTL for self-compatibility was located 50 kb downstream of the S locus and the 255 kb wide 1.5-LOD interval included both key S locus genes SRK and SCR. This is consistent with a previous report that self-compatibility maps to the S locus in crosses between C. rubella and C. grandiflora (Nasrallah et al. 2007). The 1.5-LOD CI for self-compatibility did not overlap with those for floral traits, but overlapped with a very wide CI for a QTL for ovule number (Fig. 4). Consistent with this, self-incompatible plants also produced slightly fewer ovules per flower than self-compatible plants on average (14.2 vs 15.4 ovules/flower; Mann–Whitney test, W = 38,374.5; P = 4.4 × 10−7).
Figure 5.
Fine-mapping of QTL for self-incompatibility on LG7. The large figure shows the LOD profile resulting from interval mapping of SI with hard genotype calls, and the inset shows the fine-scale location of the QTL to a region of LG7 containing the S locus, as well as the ancestry probabilities for each individual across LG7, sorted by self-incompatibility status (SI: self-incompatible, SC: self-compatible). For each individual, a blue bar indicates high probability (>0.9) that the region is homozygous for the Capsella grandiflora allele, a yellow bar indicates high probability that the region is homozygous for the C. rubella allele and an orange bar indicates high probability of heterozygosity.
POPULATION GENETICS OF QTL
We identified four nonoverlapping QTL that had 2 LOD intervals of 2MB or less in the C. rubella genome. Out of 318 loci that were resequenced in at least six samples from each species, seven of our resequenced exons fall within three of these four QTL regions (Table 3). Of these seven loci, only a single segregating site was observed in C. rubella, compared with 85 segregating sites in C. grandiflora. This contrasts with the relative diversity overall, where C. rubella has 766 segregating sites compared with C. grandiflora’s 3395 (two-tailed Fisher’s exact test P < 0.01). QTL regions therefore exhibit a more extreme reduction in diversity than other genomic regions in C. rubella. When we compare the proportion of shared polymorphisms and fixed differences, our seven loci falling under QTL similarly show an excess of fixed differences relative to shared polymorphism (two-tailed Fisher’s exact test, P < 0.01).
Table 3.
Population genetics of narrow (2 LOD interval <2MB) QTL regions compared to the rest of the genome
| QTL | Locus | Segregating sites C. grandiflora |
Segregating sites C. rubella |
Fixed differences |
Shared polymorphism |
|---|---|---|---|---|---|
| LG1, Petal length | At1g18880 | 3 | 1 | 3 | 0 |
| At1g18900 | 12 | 0 | 1 | 0 | |
| At1g20510 | 10 | 0 | 1 | 0 | |
| At1g20960 | 5 | 0 | 0 | 0 | |
| LG2, Petal width | At1g68530 | 10 | 0 | 0 | 0 |
| LG7, Self-compatibility | At4g21390 | 25 | 0 | 3 | 0 |
| At4g21380 | 20 | 0 | 1 | 0 | |
| All QTL | 85 | 1 | 9 | 0 | |
| Total genes | 3395 | 766 | 111 | 327 |
Discussion
In this study, we have begun to characterize the genetic architecture of the selfing syndrome in Capsella by QTL mapping of floral and reproductive traits that differ between the self-fertilizing species C. rubella and the obligate outcrosser C. grandiflora. In addition, we use population genetic data to test for selection on genomic regions that affect the selfing syndrome.
LINKAGE MAP AND SEGREGATION DISTORTION
Our study highlights the strength of the methods based on massively parallel sequencing for marker discovery and genotyping. Using multiplexed shotgun sequencing (MSG; Andolfatto et al. 2011), we identified a total of 121,979 markers or about one marker every kb. Given the rapid development of sequencing and genotyping technology, mapping resolution will increasingly be limited by the amount of recombination in the analyzed pedigree rather than by marker availability. Indeed, this is the case in our cross, where 890 markers remain after filtering those with identical genotype configurations across F2 individuals.
Our linkage map contained eight linkage groups, as expected from previous studies (e.g., Boivin et al. 2004). As is common for interspecific crosses (Rieseberg et al. 2000), a substantial proportion of markers (17%) showed significant segregation distortion. Inbreeding depression could be expected to cause segregation distortion in our study, as we used an outbred, highly heterozygous C. grandiflora individual as a mapping parent, and deleterious recessive C. grandiflora alleles would be rendered homozygous in the F2 mapping population. However, none of the regions that exhibited segregation distortion showed a deficit of homozygotes for the C. grandiflora allele, and the germination and survival rate in the F2 population was very high (>95%). Thus, inbreeding depression does not appear to be a major factor underlying segregation distortion in our cross. Other possible explanations include hybrid incompatibilities (Moyle et al. 2006) or loci that affect pollen performance and/or pollen-style interactions (Fishman et al. 2008).
NUMBER OF LOCI, EFFECT SIZES, AND PLEIOTROPY
We identified a total of 41 QTL for all the 13 traits examined. The number of QTL per trait is modest, between two and five, and additive effects of leading QTL for floral size traits explain a considerable proportion of F2 variance as well as divergence between species (e.g., 32% of F2 variance; 26% of interspecific divergence for petal width). Furthermore, homozygous additive effects at the three significant QTL for petal length and width can account for the majority of the divergence in petal length between C. grandiflora and C. rubella. Thus, changes at a few genomic regions appear to be sufficient to cause the severe reduction in petal size seen in C. rubella.
As we used a large mapping population (N = 550), our effect size estimates should not be greatly overestimated due to the Beavis effect (Beavis 1994). Variation in recombination rates also seems unlikely to bias our estimates (“the Noor effect”; Noor et al. 2001), as petal size QTL regions did not have significantly lower recombination rates or higher gene densities than the rest of the genome. There was also no evidence for large-effect QTL being located in regions of unusually high gene densities or low recombination rates, as expected under the Noor effect (Fig. S3). It is possible that there are additional minor QTL of small effect that we did not have the power to detect in this study. However, we note that our study has high power to detect QTL with even very small effect (e.g., explaining about 3% of the F2 variance in petal size traits) assuming environmental and genetic variances as estimated for Capsella in recombinant inbred lines by Sicard et al. (2011). This conclusion was not sensitive to the exact values of environmental and genetic variance, as a similar estimated minimum detectable QTL effect was obtained with an environmental variance twice as large.
Our results thus differ from those of previous studies of the selfing syndrome that found a large number of QTL, each of small effect (e.g., Fishman et al. 2002; Goodwillie et al. 2006). For instance, in an interspecific Mimulus guttatus–M. nasutus mapping population, Fishman et al. (2002) found at least 11 QTL for each trait examined; although our study has similar power, we detect only two to five QTL per trait. The distribution of effect sizes also appears to differ between Mimulus and Capsella, as none of the relative homozygous effects in Mimulus estimated by Fishman et al. (2002) are as great as the largest that we find in Capsella.
Another way of assessing effect sizes is to relate them to levels of standing variation in the ancestral population or species. Scaled this way, leading QTL for petal size traits have homozygous additive effects of about 1.6 times the C. grandiflora standard deviation. Although it is difficult to compare directly, this is about twice as large as the leading corolla width effect size in Mimulus when standardized by the variation seen in the M. guttatus Iron Mountain population (Fishman et al. 2002).
Thus, regardless of the exact measure of effect size used, the evolution of the selfing syndrome in Capsella seems to have involved fewer genes, potentially of larger effect than in Mimulus. This could in part be due to differences in the demographics of the transition to selfing in these two genera. In Capsella, there was a severe reduction in effective population size in association with the transition to selfing (Foxe et al. 2009; Guo et al. 2009). If floral evolution occurred subsequent to the bottleneck, such population size reductions may have rendered selection on alleles of small effect inefficient, resulting in the preferential fixation of alleles of larger effect (Hamblin et al. 2011). Additionally, because outcrossing Capsella flowers are not as large and showy as those of Mimulus, fewer mutational steps may be required to achieve the selfing syndrome phenotype.
Previous studies have found high correlations between floral size traits, but lower correlations between floral and vegetative traits (e.g., Bernacchi and Tanksley 1997; Lin and Ritland 1997; Fishman et al. 2002; Georgiady et al. 2002; Goodwillie et al. 2006). Our results agree with this pattern, as phenotypic correlations were high between floral size traits, but lower between floral traits and both vegetative/phenology-related and reproductive traits. Consistent with this, we also found that floral size QTL co-localized to a great extent, but less so with QTL for phenology traits. These QTL map to five main genomic regions on linkage groups 1, 2, 6, 7, and 8. Distinguishing between pleiotropy and close linkage as a cause for co-localization of QTL will require additional fine-mapping; however, at present our results suggest the selfing syndrome could have evolved through mostly major-effect changes at a modest number of loci.
INBREEDING DEPRESSION
Our F2 mapping population allowed us to assess the degree of dominance at individual QTL. Most QTL had additive effects that were considerably greater than dominance effects, the most prominent exception being a QTL on LG2 affecting several floral size traits, for which the C. rubella allele was largely recessive. Overall, there was no tendency for alleles from either species to be mostly recessive, and the majority of allelic effects were in the direction expected from the phenotypic divergence between species. If inbreeding depression affected our QTL mapping of floral and reproductive traits, we would expect to see recessive C. grandiflora alleles causing reduced flower size or pollen/ovule number. As this was not the case, we conclude that our QTL mapping results are unlikely to be severely affected by inbreeding depression.
We observed abnormal flowers on some inflorescences in about 40% of all F2 individuals. To test whether this could be a result of inbreeding depression, we mapped QTL for this trait. If floral abnormalities were a result of inbreeding depression, we would expect them to be a result of partly or completely recessive C. grandiflora alleles. There were indeed two QTL that showed this pattern, however, together they only accounted for about 7% of the F2 variation, and the major QTL for this trait instead featured a recessive C. rubella allele. As we have not observed this phenotype in C. rubella, and did not find any evidence for two-locus epistatic interactions, we hypothesize that higher order interactions or nuclear-cytoplasmic incompatibilities may be involved.
SELECTION ON THE SELFING SYNDROME
As mentioned above, the majority (86%) of the QTL effects for floral and reproductive traits were in the direction expected from phenotypic differences between the two species. This is consistent with directional selection favoring the evolution of the selfing syndrome of C. rubella. However, we did not conduct a formal test for directional selection such as Orr’s sign test (Orr 1998), due to the relatively small number of QTL per trait and the possible nonindependence of QTL for different floral traits due to pleiotropy.
Population genetic analysis of 318 loci sequenced in both C. rubella and C. grandiflora yielded some additional evidence for selection on the selfing syndrome. Narrow QTL regions show an excess of fixed differences relative to polymorphism in C. rubella, as expected if selective sweeps in C. rubella have affected these regions (Foxe et al. 2009). Although these results are consistent with QTL regions being the target of recent selective sweeps, it will be important to integrate these results with genome-wide patterns of polymorphism, because the severe population bottleneck in C. rubella could generate large-scale heterogeneity in polymorphism that cannot be accounted for here, with short genomic fragments sequenced across the genome.
EVOLUTION OF SELFING
Previous population genetic analysis in this species pair has suggested very recent origins of C. rubella in the context of a severe population bottleneck leading to a substantial loss of genetic diversity (Foxe et al. 2009; Guo et al. 2009). Evidence for a severe genome-wide population bottleneck is consistent with a self-compatible lineage experiencing a rapid shift to high selfing, rather than a protracted spread of selfing modifiers through a previously outcrossing population. Models for the evolution of selfing suggest that a major modifier to high selfing can evolve even in the context of high inbreeding depression (Lande and Schemske 1985), and selection for reproductive assurance associated with colonization of new postglacial habitats may have enhanced this spread (Foxe et al. 2009).
Our results agree with Riley’s (1934) conclusion that self-compatibility is caused by a dominant C. rubella allele, and confirms a previous report that self-compatibility maps to the canonical Brassicaceae S-locus in Capsella (Nasrallah et al. 2007). The switch to self-compatibility might alone have resulted in high rates of selfing, as introgression of the self-compatibility allele from C. rubella into C. grandiflora yields a mean autonomous selfing efficiency of ~0.4–0.5, about half that of present-day C. rubella (Sicard et al. 2011). Plants that have high selfing efficiency under greenhouse settings may still outcross to a large extent if pollinators and mates are available. Thus, although additional field-based experiments would be important to test the effect of self-compatibility per se on the realized selfing rate, it is possible that the mutation conferring self-compatibility itself comprised a major mutation to high selfing.
If there is extensive pleiotropy, this may have facilitated subsequent evolution of the selfing syndrome in the new self-compatible lineage. An attractive hypothesis is that following the loss of SI, selection for improved efficacy of autonomous self-pollination resulted in correlated changes in floral and reproductive traits (Sicard et al., 2011). In any case, changes in petal size likely occurred prior to the geographical spread of C. rubella, as our study finds petal size QTL in similar genomic locations as Sicard et al. (2011), despite the fact that our C. rubella mapping parents are from widely separated geographical locations (Greece vs. Canary Islands).
Conclusions
In this study, we have conducted QTL mapping of floral and reproductive traits that differ between the outcrosser C. grandiflora and the predominantly selfing C. rubella. We find a modest number of QTL for each floral and reproductive trait examined. In contrast to other systems, evolution of the selfing syndrome in Capsella seems to have involved fewer loci, potentially of larger effect. The directionality of QTL effects and patterns of polymorphism and divergence in QTL regions suggest that the selfing syndrome has been subject to directional selection. This study therefore provides an important basis for further studies of the evolutionary forces and genetic changes that underlie evolution of the selfing syndrome.
Supplementary Material
Acknowledgments
We thank S. Cai, C. Olteanu, L. Mastropaolo, and G. Raczkowski for technical assistance with plant care and phenotyping, and A. Lu for assistance with Perl scripts for trimming and parsing genotype data from MSG output. We thank Y.-W. Lee, H. Schielzeth, A. Sicard and M. Lenhard for insightful comments and discussion on this manuscript. We thank T. Hu for assistance in implementing MSG, A. Platts for assistance with analysis of the interim C. rubella genome assembly, D. Weigel, J. Schmutz, D. Rohksar, K. Barrie, S. Prochnik and the U.S. Department of Energy Joint Genome Institute for allowing us to use a pre-release version of the genome. This study was funded by an Ontario Government Early Researcher Award and an NSERC Discovery Grant to SIW, by research grants from the Royal Swedish Academy of Sciences and the Lars Hierta foundation to TS. This research was partly supported by funding from a NIGMS Center of Excellence grant P50 GM071508 to PA and funding from the National Science Foundation Grant DEB-0946398 to PA. KMH was supported by a Canadian Institute of Health (CIHR) fellowship. Illumina sequencing of the Cr1Gr1 genome was funded by a Genome Canada/Genome Quebec grant to T. Bureau, S. Wright, M. Blanchette, D. Schoen, P. Harrison, and J. Stinchcombe. C. rubella genome sequence data were produced by the U.S. Department of Energy Joint Genome Institute, supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting Information
The following supporting information is available for this article:
Table S1. Accession information.
Table S2. Quantitative trait locus (QTL) information.
Figure S1. LOD profiles.
Figure S2. Distributions of residuals.
Figure S3. Gene densities and recombination rates.
Supporting Information may be found in the online version of this article.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
LITERATURE CITED
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andolfatto PD. Hitchhiking effects of recurrent beneficial amino acid substitutions in Drosophila melanogaster. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P, Davison D, Erezyilmaz D, Hu TT, Mast J, Sunayama-Morita T, Stern DL. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 2011;21:610–617. doi: 10.1101/gr.115402.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, Broman KW. R/qtl: high-throughput multiple QTL mapping. Bioinformatics. 2010;26:2990–2992. doi: 10.1093/bioinformatics/btq565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman T, Majetic C. Genetic constraints on floral evolution: a review and evaluation of patterns. Heredity. 2006;96:343–352. doi: 10.1038/sj.hdy.6800815. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. The evolution of plant sexual diversity. Nat. Rev. Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- Barrett SCH, Harder LD. Ecology and evolution of plant mating. Trends Ecol. Evol. 1996;11:73–79. doi: 10.1016/0169-5347(96)81046-9. [DOI] [PubMed] [Google Scholar]
- Beavis WD. The power and deceit of QTL experiments: lessons from comparative QTL studies. In: Wilkinson DB, editor. Proceedings of the 49th Annual Corn and Sorghum Research Conference. Washington, DC: American Seed Trade Organization; 1994. pp. 250–266. [Google Scholar]
- Bernacchi D, Tanksley SD. An interspecific backcross of Lycopersicon esculentum x L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics. 1997;147:861–877. doi: 10.1093/genetics/147.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin K, Acarkan A, Mbulu R, Clarenz O, Schmidt R. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 2004;135:735–744. doi: 10.1104/pp.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. Technical Report #214. University of Wisconsin-Madison, Department of Biostatistics and Medical Informatics; 2010. Genetic map construction with R/qtl. [Google Scholar]
- Broman KW, Sen S. A guide to QTL mapping with R/qtl. New York, NY: Springer; 2009. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Evolution of plant breeding systems. Curr. Biol. 2006;16:R726–R735. doi: 10.1016/j.cub.2006.07.068. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Allocation of resources to male and female functions in hermaphrodites. Biol. J. Linn. Soc. 1981;15:57–74. [Google Scholar]
- Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- Chen K-Y, Cong B, Wing R, Vrebalov J, Tanksley SD. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science. 2007;318:643–645. doi: 10.1126/science.1148428. [DOI] [PubMed] [Google Scholar]
- Darwin C. The effects of cross and self fertilization in the vegetable kingdom. London, U.K.: John Murray; 1876. [Google Scholar]
- Eckert CG, Samis KE, Dart S. Reproductive assurance and the evolution of uniparental reproduction in flowering plants. In: Harder LD, Barrett SCH, editors. Ecology and evolution of flowers. Oxford, U.K.: Oxford Univ. Press; 2006. pp. 183–203. [Google Scholar]
- Fishman L, Willis JH. Pollen limitation and natural selection on floral characters in the yellow monkeyflower, Mimulus guttatus. New Phytol. 2008;177:802–810. doi: 10.1111/j.1469-8137.2007.02265.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Morgan E, Willis JH. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics. 2001;159:1701–1716. doi: 10.1093/genetics/159.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Kelly A, Willis J. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution. 2002;56:2138–2155. doi: 10.1111/j.0014-3820.2002.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Aagaard J, Tuthill JC. Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution. 2008;62:2958–2970. doi: 10.1111/j.1558-5646.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Foxe J, Slotte T, Stahl E, Neuffer B, Hurka H, Wright S. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl. Acad. Sci. USA. 2009;106:5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiady MS, Whitkus RW, Lord EM. Genetic analysis of traits distinguishing outcrossing and self-pollinating forms of currant tomato, Lycopersicon pimpinellifolium (Jusl.) Mill. Genetics. 2002;161:333–344. doi: 10.1093/genetics/161.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwillie C, Ritland C, Ritland K. The genetic basis of floral traits associated with mating system evolution in Leptosiphon (Polemoniaceae): an analysis of quantitative trait loci. Evolution. 2006;60:491–504. [PubMed] [Google Scholar]
- Grillo M, Li C, Fowlkes A, Briggeman T, Zhou A, Schemske D, Sang T. Genetic architecture for the adaptive origin of annual wild rice, Oryza nivara. Evolution. 2009;63:870–883. doi: 10.1111/j.1558-5646.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- Guo Y, Bechsgaard J, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup M. Recent speciation of Capsella rubella from Capsella grandiflora associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl. Acad. Sci. USA. 2009;106:5246–5251. doi: 10.1073/pnas.0808012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P, Bachtrog D, Andolfatto P. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol. Biol. Evol. 2008;25:1825–1834. doi: 10.1093/molbev/msn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT, Buckler ES, Jannink J-L. Population genetics of genomics-based crop improvement methods. Trends Genet. 2011;27:98–106. doi: 10.1016/j.tig.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Hurka H, Neuffer B. Evolutionary processes in the genus Capsella (Brassicaceae) Plant Syst. Evol. 1997;206:295–316. [Google Scholar]
- Jansen RC. Interval mapping of multiple quantitative trait loci. Genetics. 1993;135:205–211. doi: 10.1093/genetics/135.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Stam P. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 1994;136:1447–1455. doi: 10.1093/genetics/136.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns CA, Inouye DW. Techniques for pollination biologists. Niwot, CO: Univ. of Colorado Press; 1993. [Google Scholar]
- Lande R, Schemske DW. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Ritland K. Quantitative trait loci differentiating the out-breeding Mimulus guttatus from the inbreeding M. platycalyx. Genetics. 1997;146:1115–1121. doi: 10.1093/genetics/146.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller D, Geber M. Ecological context of the evolution of self-pollination in Clarkia xantiana: Population size, plant communities, and reproductive assurance. Evolution. 2005;59:786–799. doi: 10.1554/04-656. [DOI] [PubMed] [Google Scholar]
- Moyle LC, Graham EB. Proceedings of the SMBE Tri-National Young Investigators’ Workshop 2005. Genome-wide associations between hybrid sterility QTL and marker transmission ratio distortion. Mol. Biol. Evol. 2006;23:973–980. doi: 10.1093/molbev/msj112. [DOI] [PubMed] [Google Scholar]
- Nasrallah JB, Liu P, Sherman-Broyles S, Schmidt R, Nasrallah ME. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics. 2007;175:1965–1973. doi: 10.1534/genetics.106.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Cunningham AL, Larkin JC. Consequences of recombination rate variation on quantitative trait locus mapping studies. Simulations based on the Drosophila melanogaster genome. Genetics. 2001;159:581–588. doi: 10.1093/genetics/159.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornduff R. Reproductive biology in relation to systematics. Taxon. 1969;18:121–133. [Google Scholar]
- Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Zeng K, Slotte T, Wright SI, Charlesworth D. Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species. Genome Biol. Evol. 2011;3:868–880. doi: 10.1093/gbe/evr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Baird SJ, Gardner KA. Hybridization, introgression, and linkage evolution. Plant Mol. Biol. 2000;42:205–224. [PubMed] [Google Scholar]
- Riley HP. A further test showing the dominance of self-fertility to self-sterility in shepherd’s purse. Am. Nat. 1934;68:60–64. [Google Scholar]
- Sicard A, Lenhard M. The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann. Bot. 2011;107:1433–1443. doi: 10.1093/aob/mcr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A, Stacey N, Hermann K, Dessoly J, Neuffer B, Lenhard M. Genetics, evolution, and adaptive significance of the selfing syndrome in the genus Capsella. Plant Cell. 2011;23:3156–3171. doi: 10.1105/tpc.111.088237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora a plant species with a large effective population size. Mol. Biol. Evol. 2010;27:1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Variation and evolution in plants. New York: Columbia Univ. Press; 1950. [Google Scholar]
- St. Onge KR, Källman T, Slotte T, Lascoux M, Palmé AE. Contrasting demographic history and population structure in Capsella rubella and Capsella grandiflora two closely related species with different mating systems. Mol. Ecol. 2011;20:3306–3320. doi: 10.1111/j.1365-294X.2011.05189.x. [DOI] [PubMed] [Google Scholar]
- Xu S, Atchley WR. Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics. 1996;143:1417–1424. doi: 10.1093/genetics/143.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.