ABSTRACT
Neural stem cells (NSCs) within the adult hippocampal dentate gyrus reside in the subgranular zone (SGZ). A dynamic network of signaling mechanisms controls the balance between the maintenance of NSC identity, and their subsequent differentiation into dentate granule neurons. Recently, the ubiquitin-specific protease 9 X-linked (USP9X) was shown to be important for hippocampal morphogenesis, as mice lacking this gene exhibited a higher proportion of proliferating NSCs, yet a decrease in neuronal numbers, within the postnatal dentate gyrus. Here we reveal that Usp9x-deficiency results in the upregulation of numerous oligodendrocytic and myelin-associated genes within the postnatal hippocampus. Moreover, cell counts reveal a significant increase in oligodendrocyte precursor cells and mature oligodendrocytes per unit volume of the mutant dentate gyrus. Collectively, these findings indicate that USP9X may regulate NSC lineage determination within the postnatal SGZ.
KEYWORDS: hippocampal neurogenesis, neural stem cell, oligodendrocyte, subgranular zone, USP9X
NSCs within the adult hippocampus maintain the capacity to undergo neurogenesis throughout life.1 The continual supply of new neurons arising from the neurogenic niche of the SGZ is crucial for learning,2 memory,3 and spatial navigation.4 To ensure the stem cell pool is maintained throughout adulthood, NSCs predominantly remain in a state of quiescence and self-renew when they undergo proliferation.5,6 Under normal conditions in the adult brain, NSCs within the SGZ predominantly undergo neurogenic differentiation to generate dentate granule neurons.1 However, these NSCs are multipotent,7 as they retain the ability to produce astrocytes,8 and can be induced to generate oligodendrocytes via virally-mediated reprogramming.9,10 Multiple intrinsic and extrinsic signaling mechanisms act in cohesion to determine the fate of NSCs within the SGZ and ensure proper hippocampal neurogenesis. Critically, disruption at any stage of these processes can have profound consequences on the development and function of the hippocampus, leading to cognitive deficits11 and psychiatric disorders.12 Therefore, it is imperative to understand the underlying mechanisms governing the maintenance of the NSC pool and their path to neurogenic differentiation.
The deubiquitylating enzyme USP9X has previously been shown to play a pivotal role with relation to NSC biology. USP9X promotes the self-renewal of embryonic stem cell-derived NSCs in vitro.13 In vivo, USP9X is highly expressed throughout the developing central nervous system,14 the postnatal hippocampus,15 and within the adult neurogenic niches.13 The conditional ablation of Usp9x from NSCs within the embryonic forebrain (via Emx1-Cre-mediated deletion) results in a number of cortical abnormalities,14 most notably the severe reduction in the size of the postnatal hippocampus and dentate gyrus, evident as early as one week after birth.15 We recently analyzed cellular populations within the postnatal dentate gyrus of these Usp9x-deficient mice and showed a significant decrease in the total number of NSCs, neuroblasts and mature neurons.15 Interestingly, when we examined the proportion of NSCs that were quiescent versus those that were proliferating, we found a significantly higher proportion of proliferating NSCs in the SGZ of Usp9x-deficient mice, and a concomitant decrease in the proportion of quiescent NSCs, compared to controls.15 Together, these findings indicate that Usp9x-deficiency culminates in abnormal neurogenesis within the postnatal SGZ, which contributes to, at least in part, the reduced size of the dentate gyrus. However, it remains unclear as to why the lack of Usp9x leads to an increase in the relative proportion of proliferating NSCs within the SGZ but a reduced number of neuronal cells in the dentate granule cell layer. In this paper, we posited that the lack of Usp9x might result in the premature differentiation of NSCs toward the astrocytic or oligodendrocytic lineage.
To test the hypothesis that Usp9x-deficiency leads to precocious differentiation of NSCs into glia, we performed microarray-based transcriptomic profiling on hippocampi from mice in which Usp9x had been conditionally ablated from neural progenitors within the dorsal telencephalon using an Emx1-Cre allele. These mice will be referred to as Usp9x−/Y; Emx1-Cre mice from here onwards, while littermate controls mice will be referred to as Usp9xloxP/Y mice. Microarray analysis of postnatal day (P) 14 mutant and control hippocampi revealed that over 600 genes were differentially regulated in the mutant hippocampus. Significance of differentially expressed transcripts were identified at p < 0.05 and a log2 ratio fold change of <−0.5 or >0.5. Notably, many of the genes that were upregulated in the mutant hippocampi were related to the oligodendrocytic lineage, including myelin oligodendrocyte glycoprotein (Mog),16 myelin-associated oligodendrocytic basic protein (Mobp),17 myelin associated glycoprotein (Mag),18 CNPase (Cnp),18 and claudin-1119 (Cldn11; Fig. 1A, B). In contrast, a number of factors related to NSC self-renewal and neurogenesis were significantly downregulated, including Tenascin C (Tnc),20 fatty acid binding protein 7, brain (Fabp7),21 and hairy and enhancer of split 522 (Hes5; Fig. 1B). This is consistent with the previously reported role for USP9X in neural progenitor cell self-renewal,13 and the reduction in dentate granule neurons within the postnatal Usp9x−/Y; Emx1-Cre dentate gyrus.15 Validation of these microarray data was performed using quantitative polymerase chain reaction (qPCR) on cDNA generated from mRNA isolated from independent hippocampal samples from P14 mice. This analysis confirmed that the relative expression of these oligodendrocyte-associated genes was significantly elevated in mutant samples compared to controls (Fig. 1C). Interestingly, expression of caspase 3 (Casp3) was significantly reduced (Fig. 1B, C). As this factor is expressed in apoptotic cells,23 this suggests that the smaller dentate gyrus phenotype of Usp9x-deficient mice is unlikely to be related to abnormal cell death. This finding was supported by immunocytochemical analysis of cleaved caspase 3 expression, which revealed fewer apoptotic cells in the mutant dentate gyrus in comparison to controls at P14 (data not shown). It should be noted, however, that a decrease in cell death may simply be a reflection of the fact that there are reduced numbers of new-born neurons present in the mutant hippocampus. Finally, the qPCR analysis also confirmed that the expression of Tnc, Fabp7, Hes5 and the dentate granule neuron marker prospero homeobox protein-1 (Prox1)24 was significantly reduced in the mutant in comparison to the controls (Fig. 1C). Collectively, these data are consistent with previous reports into the phenotype of Usp9x-deficient mice,15 suggesting that in the absence of Usp9x, NSC self-renewal and neurogenesis are impaired. Moreover, these data further suggest that Usp9x-deficient NSCs in the SGZ, which normally produce neurons in vivo,1 may potentially be biased to produce myelinating oligodendrocytes in the absence of this factor.
Figure 1.

Lack of Usp9x results in the upregulated mRNA expression levels of oligodendrocyte and myelin associated genes within the hippocampus at P14. (A) Signal intensity plot showing the differentially expressed genes for comparison between Usp9xloxP/Y control mice vs. Usp9x−/Y; Emx1-Cre mutant mice. Only the genes that were statistically (p < 0.05) and magnitudually (log2 ratio fold change <−0.5 or > 0.5) differentially expressed were colored. Upregulated genes are illustrated in red and downregulated genes are illustrated in green. All other genes are shown in gray. The dotted diagonal line shows equal intensity across the experimental conditions while the solid lines correspond to a log2 ratio fold change of <−1 or > 1. (B) Summary of the microarray results, showing the gene symbol, gene name, log2 ratio fold change, and p value of key significantly misregulated genes in the hippocampus of mutant mice at P14. (C) qPCR validation of the microarray results, demonstrating increased relative levels of oligodendrocyte and myelin associated mRNAs, as well as reduced levels of neurogenic mRNAs, in the hippocampus of mutant mice at P14. *p < 0.05; **p < 0.01; ***p < 0.001, t-test.
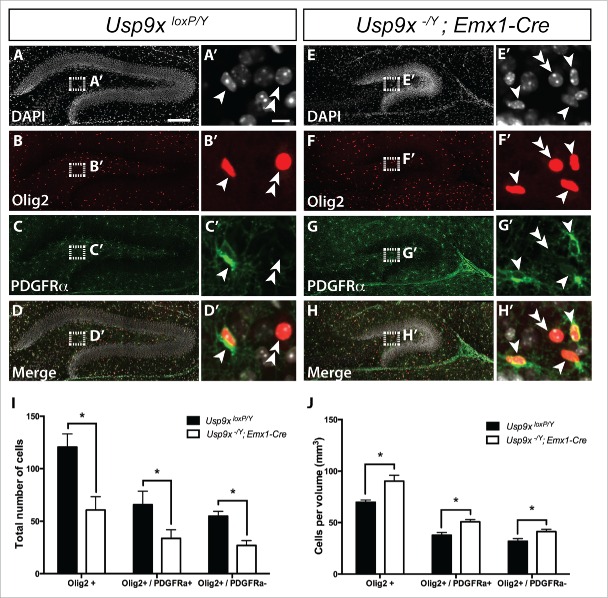
To investigate whether the elevated expression of oligodendrocyte-associated genes was reflected at a cellular level, we next performed immunocytochemical analyses of P14 Usp9x−/Y; Emx1-Cre and Usp9xloxP/Y mice using antibodies against oligodendrocyte transcription factor 2 (Olig2), which identifies all cells within the oligodendrocytic lineage25 and platelet-derived growth factor receptor α (PDGFRα), which, in conjunction with Olig2, labels oligodendrocyte precursor cells.26,27 We performed co-immunofluorescence labeling with these 2 antibodies on hippocampal tissue from P14 mutant and control brains, followed by confocal microscopy, and counted cells of the oligodendrocyte lineage (Olig2+), oligodendrocyte precursor cells (Olig2+/PDGFRα+) and mature oligodendrocytes (Olig2+/PDGFRα−) (Fig. 2A-H). As oligodendrocytes differentiating from NSCs within the SGZ migrate into the hilus of the dentate gyrus,9 we counted both the total number of immuno-positive cells within the hilus, as well as normalizing cell counts relative to the volume of the respective hilar regions. This analysis revealed that the total number of cells within the oligodendrocytic lineage, including oligodendrocyte precursor cells and mature oligodendrocytes, was reduced in Usp9x-deficient mice in comparison to controls (Fig. 2I). This is consistent with the markedly reduced size of the dentate gyrus within Usp9x−/Y; Emx1-Cre mice at P14 (Fig. 2A, E).15 Interestingly, however, normalized counts relative to the volume of the hilar region revealed that there were significantly elevated numbers of Olig2+ cells per mm3 in P14 Usp9x−/Y; Emx1-Cre mice in comparison to Usp9xloxP/Y controls, a finding reflected in the relative increase of oligodendrocyte precursor cells and mature oligodendrocytes per unit volume (Fig. 2J). Furthermore, although the absence of Usp9x leads to a higher density of oligodendrocytic cells in the dentate gyrus, there seems to be no effect on the ability of oligodendrocyte precursor cells to differentiate and mature. A similar analysis of astrocytes was performed using co-labeling of astrocytic markers glial fibrillary acidic protein (GFAP) and s100 calcium-binding protein β (s100β; Fig. 3A-H).28 We found that the total number of astrocytes in the mutant was reduced in Usp9x-deficient mice in comparison to controls (Fig. 3I). However, there was no significant change in the number of astrocytes per unit volume of the mutant hilar region in comparison to wild-type controls at P14 (Fig. 3J). When considered in light of the elevated proportion of proliferating NSCs and the reduced number of neuronal cells in the dentate gyrus of P14 Usp9X-deficient mice,15 these findings indicate that the absence of Usp9x may result in the abnormal production of oligodendrocytic cells instead of neurons within the postnatal hippocampus.
Figure 2.
Increased density of oligodendrocytes in the dentate gyrus of Usp9x−/Y; Emx1-Cre mice. Co-immunofluorescence labeling and confocal microscopy was performed on hippocampal sections of Usp9xloxP/Y (A–D) and Usp9x−/Y; Emx1-Cre (E–H) at P14. Cell nuclei were labeled with DAPI (A, E). Oligodendrocyte precursors were defined as cells expressing both Olig2 (red in (B)and F) and PDGFRα (green in C and G). Mature oligodendrocytes were defined as cells that only expressed Olig2. The merged panels are shown in (D, H). The insets reveal a higher magnification view of the boxed region showing oligodendrocyte precursor cells (arrowheads) and mature oligodendrocytes (double arrowheads). Quantification of labeled cells was performed within the hilar region of the dentate gyrus. Total numbers of Olig2+ oligodendrocytes, including Olig2+/PDGRFα+ precursors and Olig2+/PDGRFα− mature oligodendrocytes in the mutant mice were significantly reduced compared to controls (I). Normalized cell counts relative to the volume of the hilar region revealed a significant increase in oligodendrocytic cells per mm3 within the mutant compared to controls, including elevated numbers of oligodendrocyte precursors and mature oligodendrocytes (J). *p < 0.05, t-test. Scale bar in (A): (A-H) – 150 µm; (A’): (A’-H’) −10 µm.
Figure 3.
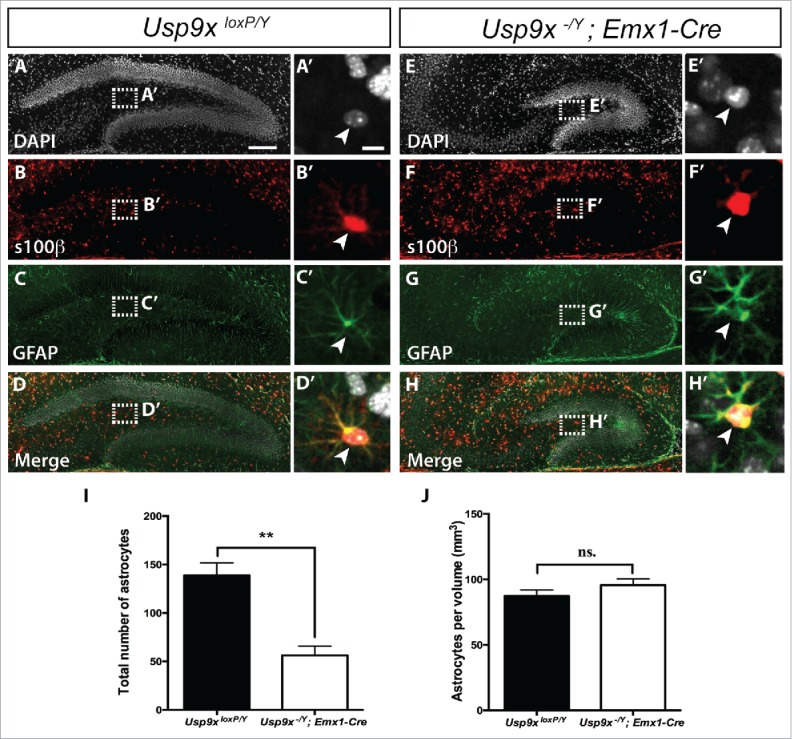
Astrocytic cell density did not change in the dentate gyrus of Usp9x−/Y; Emx1-Cre mice. Co-immunofluorescence labeling and confocal microscopy was performed on hippocampal sections of Usp9xloxP/Y (A–D) and Usp9x−/Y; Emx1-Cre (E–H) at P14. Cell nuclei were labeled with DAPI (A, E). Astrocytes were defined as cells expressing both s100β (red in B and F) and GFAP (green in C and G). The merged panels are shown in (D, H). The insets reveal a higher magnification view of the boxed region showing astrocytes (arrowheads). Quantification of labeled cells was performed within the hilar region of the dentate gyrus. There were significantly fewer total numbers of GFAP+/s100β+ astrocytes in the mutant mice compared to controls (I). Normalized cell counts relative to the volume of the hilar region revealed no significant changes in astrocytes per mm3 within the mutant compared to controls (J). (ns) Not significant, **p < 0.001, t-test. Scale bar in (A): (A-H) – 150 µm; (A’): (A’-H’) −10 µm.
NSCs residing within the SGZ of the postnatal and adult dentate gyrus do not normally produce oligodendrocytes, instead these cells predominantly generate Prox1-expressing dentate granule neurons,1,16 as well as a small proportion of astrocytes.8 However, the SGZ NSCs can be directed to differentiate into oligodendrocytes, both in vitro and in vivo.9,10 Indeed, a recent report revealed that retrovirally-driven expression of key oligodendrocytic genes, including Olig2, Sox10 and Ascl1, within the dentate gyrus of adult mice was sufficient to induce the differentiation of hippocampal NSCs into oligodendrocyte precursor cells and myelinating mature oligodendrocytes.10 How could the loss of Usp9x culminate in oligodendrogenesis? At this stage this is unclear, however previous reports of interactions between USP9X and members of the Notch signaling pathway,29 coupled with the reduced expression of Notch pathway members revealed here (Hes5, Fabp7 and Tnc) suggests that a primary role for USP9X, in the context of NSCs, is the maintenance of their identity and self-renewal capacity. Identification of why the loss of Usp9x potentially biases NSCs toward oligodendrocytic differentiation will require a more comprehensive analysis of the USP9X interactome, coupled with proteomic analysis of NSCs lacking this enzyme. The importance of such future studies is emphasized by the fact that aberrant USP9X expression has been associated with a number of human neurological disorders that exhibit abnormalities in hippocampal function, including X-linked intellectual disability,30 epilepsy,31 and Parkinson's disease.32
Although our findings implicate USP9X in the inhibition of oligodendrocytic differentiation of NSCs within the SGZ, there are caveats to this interpretation of the data. Firstly, although we noted a correlation between Usp9x-deficiency and the production of oligodendrocytes, the data presented here does not confirm that the absence of USP9X in NSCs directly leads to their differentiation into oligodendrocytes. To address this, lineage tracing experiments using the Usp9x conditional allele crossed with an inducible NSC-specific Cre driver (e.g. Nestin Cre ERT2),33 coupled with ethynyl deoxyuridine (EdU) labeling to identify proliferating cells, could be used to directly demonstrate if Usp9x-deficient cells generate oligodendrocytes within the postnatal dentate gyrus hilus. Moreover, proliferating oligodendrocyte precursor cells are also found distributed throughout non-neurogenic areas of the hippocampus.34 Thus, although we counted cells only within the hilar region of the dentate gyrus, it is similarly unclear as to whether the generation of oligodendrocytes in the Usp9x-deficient hippocampus was a result of NSCs in the SGZ abnormally differentiating toward the oligodendrocytic lineage instead of a neurogenic path, or whether the lack of Usp9x influenced the proliferation of these randomly distributed oligodendrocyte precursors throughout the hippocampus. Again, the use of an inducible NSC-specific Cre driver in future studies will clarify this question. Overall, given the expression of USP9X by NSCs within the adult neurogenic niches, as well as the reported role for this enzyme in maintaining NSC self-renewal in vitro,13 our findings support the hypothesis that USP9X plays an important role in regulating NSC biology within the postnatal dentate gyrus.
Methods
Animal breeding
Mice were bred as we described previously.15 Male mice lacking the Usp9x allele inherited the Emx1-Cre allele (referred to as Usp9x−/Y; Emx1-Cre), while Cre-negative males were used as controls (referred to as Usp9xloxP/Y). All mouse breeding was performed under the ethical clearance approved by Griffith University Animal Ethics Committee. All experiments were performed in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, and were carried out in accordance with The University of Queensland Institutional Biosafety committee.
Microarray
Four Usp9x−/Y; Emx1-Cre and 4 Usp9xloxP/Y animals were used for microarray analyses. RNA was isolated from dissected hippocampus using an RNase kit (Qiagen). Prior to microarray, the integrity of the RNA was verified on an Agilent Bioanalyzer RNA Nano 6000. The Affymetrix Mouse Gene 2.0 ST microarray was performed at the Ramaciotti Center for Genomics, The University of New South Wales, Australia. Data was processed, quantile normalized and differentially expressed transcripts were identified at p < 0.05 and log2 ratio fold change of <−0.5 or >0.5 using R/BioConductor limma package,35 at the QFAB Bioinformatics, The University of Queensland, Australia. False discovery rate (FDR) correction was performed on the p-values.36
Quantitative polymerase chain reaction
Quantitative polymerase chain reaction (qPCR) was performed on hippocampal tissue from P14 animals (6 Usp9x−/Y; Emx1-Cre and 4 Usp9xloxP/Y) using standard protocol as we described previously.15 Gene expression was calculated using −ΔΔ Ct-method relative to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (Gapdh). All the samples were tested in triplicate, and each experiment was repeated a technical triplicate. Primer sequences used were:
Mog_Forward:5′GACCTGCAGGAGGATCGTAG3′
Mog_Reverse: 5′ACCAAGAAGAGGCAGCAATG3′
Mobp_Forward: 5′AATGAGAGCAAGACAAGCGG3′
Mobp_Reverse: 5′TCCTTGGCCATTTTCTGACT3′
Mag_Forward: 5′CGGGTTGGATTTTACCACAC3′
Mag_Reverse: 5′CTGCCTTCAACCTGTCTGTG3′
Cldn11_Forward: 5′GCTGGGGTGCTCCTTATTCT3′
Cldn11_Reverse: 5′CAACCTGCGTACAGCGAGTA3′
Cnp_Forward: 5′GTTCTGAGACCCTCCGAAAA3′
Cnp_Reverse: 5′CCTTGGGTTCATCTCCAGAA3′
Casp3_Forward: 5′TGCTGGTGGGATCAAAGC3′
Casp3_Reverse: 5′TGAATCCACTGAGGTTTTGTTG3′
Tnc_Forward: 5′AGTCCAGGACAGACGGAAAC3′
Tnc_Reverse: 5′AAAACCATCAGTACCACGGC3′
Fabp7_Forward: 5′ CGGACAATGCACATTCAAG3′
Fabp7_Reverse: 5′TCTTTGCCATCCCACTTCTG3′
Hes5_Forward: 5′CCAGGAAAACCGACTG3′
Hes5_Reverse: 5′AACTCCTGCTCCAGCAGCA3′
Prox1_Forward: 5′GGCATTGAAAAACTCCCGTA3′
Prox1_Reverse: 5′GCTATACCGAGCCCTCAACA3′
Gapdh_Forward: 5′ GCACAGTCAAGGCCGAGAAT3′
Gapdh_Reverse: 5′ GCCTTCTCCATGGTGGTGAA3′
Immunocytochemistry labeling and image analysis
Preparation of tissue sections and immunocytochemical labeling was performed as described.15 Briefly, P14 brains were embedded in noble agar and sectioned on a coronal plane at 50 µm using a vibratome. Primary antibodies used include: Olig2 (polyclonal rabbit, 1:400, EMD Millipore), PDGFRα (polyclonal goat, 1:100, R&D Systems), cleaved caspase 3 (polyclonal rabbit, 1:200, Cell Signaling) and GFAP (monoclonal mouse, 1:400, EMD Millipore). Corresponding secondaries antibodies included donkey 488, Cy3, 555 (Jackson), and s100β (rabbit conjugated AlexaFluor 647) before being counter-stained with 4′, 6-diamidino-2-phenylindole (DAPI). All image acquisition and analysis was performed as previously described.15 For all analyses we had 6 rostro-caudal sequential hippocampal sections, with each section containing the left and right hippocampi, per animal to image and count. Cell counts for Olig2+/PDGFRα− cells, Olig2+/PDGFRα+, and GFAP+/S100b+ cells were performed within the hilar region of the hippocampal dentate gyrus in a 10 µm z-stack, which consisted of 10 consecutive 1 µm-thick optical sections. The hilus was defined by the area below the SGZ and between the superior and inferior blades of the dentate gyrus. The normalized cell counts were calculated by dividing the number of cells by the volume of the respective hilar region. Student's t-tests were used to compare all quantification datasets, where we used n = 3 animals per genotype at the age of P14. Statistical significance was established at a p-value of < 0.05. Error bars represent standard error of the mean (SEM). Data analysis was performed blind to the genotype of the sample.
Abbreviations
- GFAP
glial fibrillary acidic protein
- NSC
neural stem cell
- Olig2
oligodendrocyte transcription factor 2
- PDGFRα
platelet-derived growth factor α
- qPCR
quantitative polymerase chain reactions
- s100β
s100 calcium-binding protein β
- SGZ
subgranular zone
- USP9X
ubiquitin-specific protease 9 X-linked
Acknowledgments
We gratefully acknowledge the Queensland Brain Institute's Advanced Microscopy Facility for their assistance with microscopy. Many thanks to Stephen Rudd and Xin-Yi Chuang from QFAB Bioinformatics, The University of Queensland for technical analyses on the microarray data.
Funding
This work was supported by National Health and Medical Research Council project grants (1057751 to MP and 1009248 to SAW) and an Australian Research Council Discovery Grant (DP160100368 to MP). MP was supported by a fellowship (Australian Research Council Future Fellowship; FT120100170). SO was supported by an Australian Postgraduate Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- [1].Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med 1998; 4:1313-7; PMID:9809557; http://dx.doi.org/ 10.1038/3305 [DOI] [PubMed] [Google Scholar]
- [2].Gould E, Beylin A, Tanapat P, Reeves A, Shors TJ. Learning enhances adult neurogenesis in the hippocampal formation. Nat Neurosci 1999; 2:260-5; PMID:10195219; http://dx.doi.org/ 10.1038/6365 [DOI] [PubMed] [Google Scholar]
- [3].Saxe MD, Battaglia F, Wang JW, Malleret G, David DJ, Monckton JE, Garcia AD, Sofroniew MV, Kandel ER, Santarelli L, et al.. Ablation of hippocampal neurogenesis impairs contextual fear conditioning and synaptic plasticity in the dentate gyrus. Proc Natl Acad Sci USA 2006; 103:17501-6; PMID:17088541; http://dx.doi.org/ 10.1073/pnas.0607207103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jessberger S, Clark RE, Broadbent NJ, Clemenson GD Jr, Consiglio A, Lie DC, Squire LR, Gage FH. Dentate gyrus-specific knockdown of adult neurogenesis impairs spatial and object recognition memory in adult rats. Learn Mem 2009; 16:147-54; PMID:19181621; http://dx.doi.org/ 10.1101/lm.1172609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Götz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 2010; 6:445-56; PMID:20452319; http://dx.doi.org/ 10.1016/j.stem.2010.03.017 [DOI] [PubMed] [Google Scholar]
- [6].Jones KM, Sarić N, Russell JP, Andoniadou CL, Scambler PJ, Basson MA. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 2015; 33:196-210; PMID:25183173; http://dx.doi.org/ 10.1002/stem.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 2011; 145:1142-55; PMID:21664664; http://dx.doi.org/ 10.1016/j.cell.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, Fishell G, Koulakov A, Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 2011; 8:566-79; PMID:21549330; http://dx.doi.org/ 10.1016/j.stem.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jessberger S, Toni N, Clemenson GD Jr, Ray J, Gage FH. Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci 2008; 11:888-93; PMID:18587391; http://dx.doi.org/ 10.1038/nn.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Braun SM, Pilz GA, Machado RA, Moss J, Becher B, Toni N, Jessberger S. Programming hippocampal neural stem/progenitor cells into oligodendrocytes enhances remyelination in the adult brain after injury. Cell Rep 2015; 11:1679-85; PMID:26074082; http://dx.doi.org/ 10.1016/j.celrep.2015.05.024 [DOI] [PubMed] [Google Scholar]
- [11].Gargaro AC, Sakamoto AC, Bianchin MM, Geraldi Cde V, Scorsi-Rosset S, Coimbra ER, Carlotti CG Jr, Assirati JA, Velasco TR. Atypical neuropsychological profiles and cognitive outcome in mesial temporal lobe epilepsy. Epilepsy Behav 2013; 27:461-9; PMID:23611738; http://dx.doi.org/ 10.1016/j.yebeh.2013.03.002 [DOI] [PubMed] [Google Scholar]
- [12].Heuser K, Taubøll E, Nagelhus EA, Cvancarova M, Petter Ottersen O, Gjerstad L. Phenotypic characteristics of temporal lobe epilepsy: the impact of hippocampal sclerosis. Acta Neurol Scand Suppl 2009; 189:8-13; PMID:19566491; http://dx.doi.org/ 10.1111/j.1600-0404.2009.01205.x [DOI] [PubMed] [Google Scholar]
- [13].Jolly LA, Taylor V, Wood SA. USP9X enhances the polarity and self- renewal of embryonic stem cell-derived neural progenitors. Mol Biol Cell 2009; 20:2015-29; PMID:19176755; http://dx.doi.org/ 10.1091/mbc.E08-06-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stegeman S, Jolly LA, Premarathne S, Gecz J, Richards LJ, Mackay-Sim A, Wood SA. Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFβ-mediated axonogenesis. PLoS One 2013; 8:e68287; PMID:23861879; http://dx.doi.org/ 10.1371/journal.pone.0068287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Oishi S, Premarathne S, Harvey TJ, Iyer S, Dixon C, Alexander S, Burne THJ, Wood SA, Piper M. Usp9x-deficiency disrupts the morphological development of the postnatal hippocampal dentate gyrus. Sci Rep 2016; 6:25783; PMID:27181636; http://dx.doi.org/ 10.1038/srep25783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gil V, Bichler Z, Lee JK, Seira O, Llorens F, Bribian A, Morales R, Claverol-Tinture E, Soriano E, Sumoy L, et al.. Developmental expression of the oligodendrocyte myelin glycoprotein in the mouse telencephalon. Cereb Cortex 2010; 20:1769-79; PMID:19892785; http://dx.doi.org/ 10.1093/cercor/bhp246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Montague P, McCallion AS, Davies RW, Griffiths IR. Myelin-associated oligodendrocytic basic protein: a family of abundant CNS myelin proteins in search of a function. Dev Neurosci 2006; 28:479-87; PMID:17028425; http://dx.doi.org/ 10.1159/000095110 [DOI] [PubMed] [Google Scholar]
- [18].Campagnoni AT. Molecular biology of myelin proteins from the central nervous system. molecular biology of myelin proteins from the central nervous system. J Neurochem 1988; 51:1-14; PMID:2454292 [DOI] [PubMed] [Google Scholar]
- [19].Tiwari-Woodruff SK, Buznikov AG, Vu TQ, Micevych PE, Chen K, Kornblum HI, Bronstein JM. OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes. J Cell Biol 2001; 153:295-305; PMID:11309411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Garcion E, Halilagic A, Faissner A, ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development 2004; 131:3423-32; PMID:15226258; http://dx.doi.org/ 10.1242/dev.01202 [DOI] [PubMed] [Google Scholar]
- [21].Gerstner JR, Bremer QZ, Vander Heyden WM, Lavaute TM, Yin JC, Landry CF. Brain fatty acid binding protein (Fabp7) is diurnally regulated in astrocytes and hippocampal granule cell precursors in adult rodent brain. PLoS One 2008; 3:e1631; PMID:18286188; http://dx.doi.org/ 10.1371/journal.pone.0001631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ 2008; 50(Suppl 1):S97-103; PMID:18430159; http://dx.doi.org/ 10.1111/j.1440-169X.2008.00993.x [DOI] [PubMed] [Google Scholar]
- [23].Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999; 6:99-104; PMID:10200555; http://dx.doi.org/ 10.1038/sj.cdd.4400476 [DOI] [PubMed] [Google Scholar]
- [24].Lavado A, Lagutin OV, Chow LM, Baker SJ, Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol 2010; 8:e1000460; PMID:20808958; http://dx.doi.org/ 10.1371/journal.pbio.1000460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mei F, Wang H, Liu S, Niu J, Wang L, He Y, Etxeberria A, Chan JR, Xiao L. Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes. J Neurosci 2013; 33:8454-62; PMID:23658182; http://dx.doi.org/ 10.1523/JNEUROSCI.2453-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hall A, Giese NA, Richardson WD. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development 1996; 122:4085-94; PMID:9012528 [DOI] [PubMed] [Google Scholar]
- [27].Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 2002; 109:61-73; PMID:11955447 [DOI] [PubMed] [Google Scholar]
- [28].von Bohlen und Halbach O. Immunohistological markers for proliferative events, gliogenesis, and neurogenesis within the adult hippocampus. Cell Tissue Res 2011; 345:1-19; PMID:21647561; http://dx.doi.org/ 10.1007/s00441-011-1196-4 [DOI] [PubMed] [Google Scholar]
- [29].Murtaza M, Jolly LA, Gecz J, Wood SA. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci 2015; 72:2075-89; PMID:25672900; http://dx.doi.org/ 10.1007/s00018-015-1851-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Homan CC, Kumar R, Nguyen LS, Haan E, Raymond FL, Abidi F, Raynaud M, Schwartz CE, Wood SA, Gecz J, et al.. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet 2014; 94:470-8; PMID:24607389; http://dx.doi.org/ 10.1016/j.ajhg.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Paemka L, Mahajan VB, Ehaideb SN, Skeie JM, Tan MC, Wu S, Cox AJ, Sowers LP, Gecz J, Jolly L, et al.. Seizures are regulated by ubiquitin-specific peptidase 9 X-linked (USP9X), a de-ubiquitinase. PLoS Genet 2015; 11:e1005022; PMID:25763846; http://dx.doi.org/ 10.1371/journal.pgen.1005022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rott R, Szargel R, Haskin J, Bandopadhyay R, Lees AJ, Shani V, Engelender S. α-Synuclein fate is determined by USP9Xregulated monoubiquitination. Proc Natl Acad Sci USA 2011; 108(46):18666-71; PMID:22065755; http://dx.doi.org/ 10.1073/pnas.1105725108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sun MY, Yetman MJ, Lee TC, Chen Y, Jankowsky JL. Specificity and efficiency of reporter expression in adult neural progenitors vary substantially among nestin-CreER (T2) lines. J Comp Neurol. 2014; 522:1191-208; PMID:24519019; http://dx.doi.org/ 10.1002/cne.23497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Matsumoto Y1, Tsunekawa Y, Nomura T, Suto F, Matsumata M, Tsuchiya S, Osumi N. Differential proliferation rhythm of neural progenitor and oligodendrocyte precursor cells in the young adult hippocampus. PLoS One 2011; 6:e27628; PMID:22110700; http://dx.doi.org/ 10.1371/journal.pone.0027628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smyth GK. 2005, ‘limma: Linear Models for Microarray Data’, in Gentleman R., Carey V., Huber W., Irizarry R. & Dudoit S. (eds), Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Springer: New York, pp. 397-420. [Google Scholar]
- [36].Yoav B, Daniel Y. Quantitative Trait Loci Analysis Using the False Discovery Rate. Genetics 2005; 171:783-90; PMID:15956674; http://dx.doi.org/ 10.1534/genetics.104.036699 [DOI] [PMC free article] [PubMed] [Google Scholar]