Abstract
We investigated mechanisms involved in the protection of zebrafish (Danio rerio) larvae by two probiotic candidate yeasts, Debaryomyces hansenii 97 (Dh97) and Yarrowia lypolitica 242 (Yl242), against a Vibrio anguillarum challenge. We determined the effect of different yeast concentrations (104–107 CFU/mL) to: (i) protect larvae from the challenge, (ii) reduce the in vivo pathogen concentration and (iii) modulate the innate immune response of the host. To evaluate the role of zebrafish microbiota in protection, the experiments were performed in conventionally raised and germ-free larvae. In vitro co-aggregation assays were performed to determine a direct yeast-pathogen interaction. Results showed that both yeasts significantly increased the survival rate of conventionally raised larvae challenged with V. anguillarum. The concentration of yeasts in larvae tended to increase with yeast inoculum, which was more pronounced for Dh97. Better protection was observed with Dh97 at a concentration of 106 CFU/mL compared to 104 CFU/mL. In germ-free conditions V. anguillarum reached higher concentrations in larvae and provoked significantly more mortality than in conventional conditions, revealing the protective role of the host microbiota. Interestingly, yeasts were equally (Dh97) or more effective (Yl242) in protecting germ-free than conventionally-raised larvae, showing that protection can be exerted only by yeasts and is not necessarily related to modulation of the host microbiota. Although none of the yeasts co-aggregated with V. anguillarum, they were able to reduce its proliferation in conventionally raised larvae, reduce initial pathogen concentration in germ-free larvae and prevent the upregulation of key components of the inflammatory/anti-inflammatory response (il1b, tnfa, c3, mpx, and il10, respectively). These results show that protection by yeasts of zebrafish larvae challenged with V. anguillarum relates to an in vivo anti-pathogen effect, the modulation of the innate immune system, and suggests that yeasts avoid the host-pathogen interaction through mechanisms independent of co-aggregation. This study shows, for the first time, the protective role of zebrafish microbiota against V. anguillarum infection, and reveals mechanisms involved in protection by two non-Saccharomyces yeasts against this pathogen.
Keywords: yeast probiotic, V. anguillarum, innate immune system, zebrafish, protective mechanisms
Introduction
A wide range of potentially probiotic bacteria have been tested in aquaculture to control infectious fish diseases (Hai, 2015). In contrast, few studies have addressed the protective effects of yeasts or the mechanisms involved in protection (Gatesoupe, 2007; Navarrete and Tovar-Ramírez, 2014). In those studies, modulation of the host immune system has been posited as a possible mechanism involved in the protection of fish against pathogens. An enhanced immune response, reflected by a higher IgM level, was observed in recovering juvenile leopard groupers (Mycteroperca rosacea) fed with Debaryomyces hansenii (CBS8339) and infected with the dinoflagellate Amyloodinium ocellatum (Reyes-Becerril et al., 2008). Olive flounder (Paralichthys olivaceus) infected with Uronema marinum and fed with the baker's yeast Saccharomyces cerevisiae (KCCM 11201) showed a significant increase in superoxide anion production and serum lysozyme activity compared to infected and non-yeast-fed fish (Harikrishnan et al., 2011). Similarly, Oreochromis niloticus treated with S. cerevisiae (BGY-25®) and infected with different fish pathogens revealed a significant increase in total protein, β and γ globulins compared to controls (Abu-Elala et al., 2013). All these studies have shown an effect on the immune system of the fish, probably due to immunostimulant compounds present in yeasts such as β-glucans, nucleic acids and/or mannanoligosaccharides (Li and Gatlin, 2006; Lokesh et al., 2012).
The control of an infectious disease can also be performed by limiting the growth of the pathogen in the host (Schneider, 2011). Although, there are few studies of antibacterial effects of yeasts compared to bacterial studies, several antagonistic properties against bacteria have been reported and reviewed (Hatoum et al., 2012). These include competition for nutrients, changes in pH, high production of ethanol, stimulation of immunoglobulins and antibacterial compounds by the host and inhibition of the attachment to intestinal cells (Hatoum et al., 2012). However, few in vivo studies using yeasts have shown the control of pathogen colonization and reduction of its concentration in broiler and mouse guts (Line et al., 1998; Correa França et al., 2015).
Recent studies demonstrated that a physical interaction (co-aggregation) between the yeast Saccharomyces boulardii and Salmonella enterica serovar Typhimurium could interfere with bacterial invasion, protecting mice against infection (Martins et al., 2013). Also, several structures of the yeast cell wall such as glucans, mannans, and chitin may play a role in co-aggregation with bacteria (Millsap et al., 1998; Hatoum et al., 2012). Therefore, we hypothesize that this mechanism could also be involved in fish protection by yeasts, interfering with host-pathogen interactions.
The initial contact of pathogens with host occurs in tissues colonized by microbiota such as the gut or skin. This microbiota protects the host from pathogens, in a process referred to as colonization resistance, involving direct and indirect mechanisms and impairing pathogen colonization and invasion (Belkaid and Hand, 2014; Pamer, 2016). The gut microbiota acts as a physical barrier to incoming pathogens by competitive exclusion such as competition for nutrients or attachment sites, production of antimicrobial molecules or stimulation of the host to produce antimicrobial compounds (Sekirov et al., 2010; Belkaid and Hand, 2014). The resistance capacity to colonization of the host microbiota against a pathogen can be studied using germ-free animals challenged with microorganisms. However, the few experiments performed in germ-free fish did not show protection against pathogens of the host microbiota (Rendueles et al., 2012; Oyarbide et al., 2015). In the context of fish protection by yeasts, the host microbiota, which plays crucial roles in important physiological processes such as the immune system maturation, has not been explored.
Zebrafish larvae have been used as a model to study interactions between a host and its microbiota or pathogens, and have multiple advantages which include small size, optical transparency of larvae, short generation times, and the possibility to perform in vivo analysis, which makes it a powerful platform to study the innate immune response to infection. The central immune molecules of the zebrafish immune system are similar with mammals (Rauta et al., 2012) and innate immunity can be studied in isolation from adaptive immunity, as the zebrafish lacks functional adaptive immunity until at least 3 weeks post-fertilization (Lam et al., 2004). Inflammation is the first biological response of the immune system to infection or irritation, where cytokines such as interleukin 1b and tumor necrosis factor a have an important role in initiating the pro-inflammatory responses once a microorganism enters the host (Bayne and Gerwick, 2001).
We recently reported on the protective effect of 13 different yeast strains isolated from the gut microbiota of healthy wild and reared fish against a Vibrio anguillarum challenge in the zebrafish (Danio rerio) model (Caruffo et al., 2015). Infected larvae pre-treated with yeasts showed significantly higher survival rate compared to non-treated larvae. In this study we selected two of those yeasts to explore some mechanisms involved in the observed protection. We determined yeast colonization capacity, the modulation of the innate immune response, the in vivo anti-V. anguillarum effects and co-aggregation with the pathogen. In addition we determined the role of the zebrafish microbiota in larval protection.
Materials and methods
Microorganisms and growing conditions
This study included 2 yeast strains previously isolated and identified from the gut of healthy fish (Raggi et al., 2014). Yarrowia lipolytica 242 (Yl242) was isolated from a wild yellowtail (Seriola lalandi) and D. hansenii 97 (Dh97) from a reared rainbow trout (Oncorhynchus mykiss). These two non-Saccharomyces species were selected due to their high abundance in commercial fish (Raggi et al., 2014), and the 2 yeast strains (Yl242 and Dh97) protected zebrafish larvae from a Vibrio anguillarum challenge, increasing its survival percentage (Caruffo et al., 2015). Yeasts were cultured according to Caruffo et al. (2015) in YPD broth (1% yeast extract, Difco, 1% peptone, Difco, 1% glucose, Merck) or YPD agar (YPD broth with 1.4% agar, Difco) supplemented with 0.05% chloramphenicol (Winkler), at 28°C under aerobic conditions. Inoculation of zebrafish larvae was performed with exponential growth cultures of yeast obtained in YPD broth at 28°C for 24 h.
Maintenance of conventionally raised (CONV-R) larvae
Tab5 embryos (wild type, WT) were maintained and raised according to Hedrera et al. (2013). All embryos were collected by natural spawning, staged according to Kimmel et al. (1995) and raised at 28°C in sterile E3 medium (1% NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 0.00003% methylene blue, Winkler, pH 7.0) in sterile Petri dishes (100 embryos/dish). 75% of the E3 volume was replaced daily with sterile E3 to avoid waste accumulation and oxygen limitation. At 3 dpf (days post-fertilization), larvae were transferred to six-well sterile tissue culture plates (20 larvae/well). Larvae were euthanized with an overdose of tricaine methanesulfonate (4%, MS-222, Sigma-Aldrich).
Germ-free larvae
Germ-free larvae were generated as previously described (Pham et al., 2008; Milligan-Myhre et al., 2011) with some modifications. Fertilized eggs, obtained by natural breeding, were collected and repeatedly washed in sterile E3 medium. In a UV treated hood, eggs were then washed 2 min with polyvinylpyrrolidone–iodine (PVP-I, 0.1%; MDK) and rinsed with sterile E3. Eggs were then immersed in sodium hypochlorite solution (0.003%) for 20 min, rinsed with sterile E3 and maintained for 4 h in E3 with antibiotics [kanamycin (Winkler) 5 μg/mL; ampicillin (Winkler) 200 μg/mL; amphotericin B (Calbiochem) 250 ng/mL; ceftazidime (Opko) 200 μg/mL and chloramphenicol (Winkler) 20 μg/mL]. The medium was replaced daily by fresh sterile E3 with antibiotics until 2 dpf. From 3 dpf on, larvae were maintained in sterile E3 without antibiotics.
Sterility of larvae and E3 was monitored on day 3 dpf, and until day 9 dpf in non-inoculated larvae, as previously described (Pham et al., 2008; Milligan-Myhre et al., 2011). In brief, 3 larvae were homogenized in 150 μL of sterile phosphate buffer saline (PBS, Winkler; with a 25-gauge needle). One hundred microliter of the homogenate was plated in Trypticase Soy Agar (TSA, BBL), and 50 μL in Trypticase Soy Broth (TSB, BBL). Similarly, the sterility of the E3 medium was verified as previously described. We chose TSA according to a previous recommendation (Milligan-Myhre et al., 2011), and previous results showed that the microbiota of eggs and larvae reared in our facility were best described with this medium incubated aerobically at 28°C (data not shown).
Protection assays with different concentrations of yeasts
The protection experiments were performed as previously described (Caruffo et al., 2015) with different concentrations of each yeast. Yeast strains were grown at 28°C until the initial exponential phase, pelleted, re-suspended in E3 and transferred to 4 dpf zebrafish larvae at a final concentration ranging from 104 to 107 CFU/mL. Larvae were kept with yeast for 2 h at 28°C then transferred to E3. At 5 dpf, larvae were challenged by immersion with V. anguillarum at a concentration of 107 CFU/mL as previously described (Caruffo et al., 2015). The survival rate was recorded daily and monitored for 4 days post-challenge. Control groups were included: a group of larvae inoculated only with (i) yeasts, (ii) V. anguillarum, and (iii) non-inoculated larvae. Each group consisted of 60 larvae which were randomly distributed in three wells of a six-well sterile tissue culture plate (in triplicate, 20 larvae/well). Each experiment was independently performed 3 times. The experimental groups are described in Figure S1.
Yeast and V. anguillarum concentrations in zebrafish larvae
To determine the concentrations of yeast and V. anguillarum in larvae, 3 larvae of each group were individually homogenized in sterile PBS and serial dilutions were plated in YPD agar supplemented with 0.05% chloramphenicol (Winkler) for yeast count (CFU/larva) or CHROMagar™ Vibrio medium for V. anguillarum count.
Gene expression analysis (RT-qPCR) of innate immune genes
We evaluated the gene expression of some innate immune genes in larvae exposed to different treatments (Table 1). Three pools of 5 larvae per treatment were analyzed. Each pool of larvae was homogenized with a 25-gauge needle and RNA was obtained with the SV Total RNA Isolation System (Promega). cDNAs were synthesized using the ImProm-II™ Reverse Transcription System (Promega) according to the manufacturer's instructions in a TProfessional Thermocycler (Biometra). qPCR was performed in the LightCycler96 (Roche) using FastStart Essential DNA Green Master (Roche) in a 10 μL reaction with a final primer concentration of 500 nM. The primer sequences are detailed in Table 1. The thermal profile used was 95°C 10 min, 40 × (95°C × 30 s, 60°C × 30 s, 72°C × 30 s). Relative expression of RNAm was calculated using 2−ΔΔCT adjusted to primer efficiency (Pfaffl, 2001). beta actin 1 was used as housekeeping gene.
Table 1.
Primer sequences used for amplification of specific genes with the RT-qPCR technique.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon (pb) | References |
|---|---|---|---|---|
| b actin1 | TTCTGGTCGTACTACTGGTATTGTG | ATCTTCATCAGGTAGTCTGTCAGGT | 144 | Guan et al., 2011 |
| tnfa | GCGCTTTTCTGAATCCTACG | TGCCCAGTCTGTCTCCTTCT | 148 | Sepulcre et al., 2009 |
| il1b | TGGACTTCGCAGCACAAAATG | GTTCACTTCACGCTCTTGGATG | 150 | Kanther et al., 2014 |
| il10 | TCACGTCATGAACGAGATCC | CCTCTTGCATTTCACCATATCC | 151 | Zhang et al., 2012 |
| c3 | TGGGAGGCAATAGGCATGA | GCGTAGGATCCATCTGGTTTG | 100 | Rawls et al., 2004 |
| mpx | TCCAAAGCTATGTGGGATGTGA | GTCGTCCGGCAAAACTGAA | 90 | Rawls et al., 2007 |
Co-aggregation assays
Macroscopic and microscopic co-aggregation assays
Co-aggregation between the yeasts Dh97 or Yl242, and V. anguillarum was performed as previously described (Cisar et al., 1979; Stevens et al., 2015), with modifications. The cell suspensions were adjusted to an O.D. of 4 at 600 nm in co-aggregation buffer (TRIS 0.001M pH8, CaCl2 0.0001M, MgCl2 0.0001M, NaN3 0.02%, and NaCl 0.15 M; Winkler). Equal volumes (200 μL) of V. anguillarum and each yeast suspension were mixed in borosilicate tubes (12 × 75 mm, Schott) for at least 5 s in vortex. Visual co-aggregation was scored as previously described (Cisar et al., 1979). Control tubes containing 200 μL of each microorganism and 200 μL co-aggregation buffer were included to check potential auto-aggregation. All suspensions were observed in an optical microscope to observe any microscopic co-aggregation.
Spectrophotometric co-aggregation assays
Spectrometric co-aggregation experiments were performed using different media to suspend the microbial cells. Microbial pellets were suspended in 10 mL of PBS (Ogunremi et al., 2015), YPD (Furukawa et al., 2011) and E3, and adjusted to an O.D. of 1.0 at 600 nm. The suspensions of each yeast strain and V. anguillarum were mixed in equal volumes (5 mL) for 10 s in vortex. The upper suspension (1 mL) from each test was collected at 1 and 24 h, and O.D. was measured at 600 nm. Control tubes contained 10 mL of each microbial suspension. The percentage of co-aggregation was calculated using the following equation (Ogunremi et al., 2015):
Ax and Ay represent the O.D. of the two strains in the control tubes, and A(x + y) the O.D. of the mixture. A co-aggregation of >20% was considered positive.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 6 software (Graphpad Software, Inc). Survival data were analyzed using the Kaplan-Meier test and group differences were analyzed by the Wilcoxon test, using the Bonferroni correction for multiple comparisons. Differences in mean concentrations of yeasts and V. anguillarum were analyzed by Student's t-test. The correlation between yeast inoculum and colonization was evaluated by Spearman correlation. The analysis of the RT-qPCR results was calculated relative to the beta actin 1 transcript, and presented as relative expression (2−ΔΔCt); differences between groups were analyzed by ANOVA with the Dunnet multiple comparison corrected test. P ≤ 0.05 was considered significant.
Ethical statement
This study was carried out in strict accordance with the recommendations included in the “Guidelines for the care and use of fish in research” and the “Canadian Council on Animal Care's Guide to the Care and Use of Experimental Animals” (Canadian Council on Animal Care, 1989). The protocol was approved by the Committee on the Ethics of Animal Experiments of INTA, University of Chile and FONDECYT (FONDECYT 11110414).
Results
V. anguillarum challenge
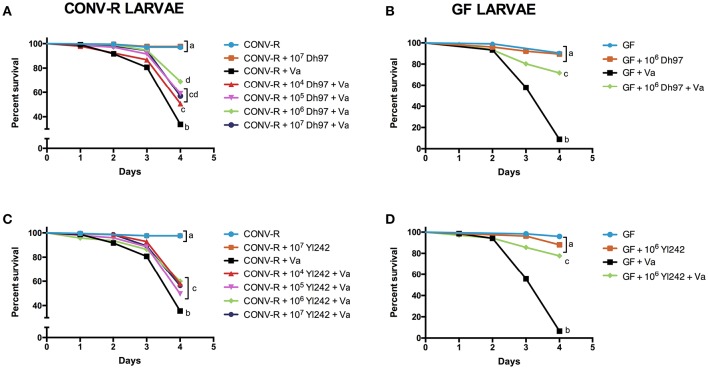
Figure 1 shows the survival rate (%) of zebrafish larvae exposed to different treatments (Figure S1). We observed a significant decrease in the survival rate of the conventionally raised (CONV-R) larvae exposed to the pathogen (Figures 1A,C). To evaluate the effect of zebrafish microbiota on the V. anguillarum challenge, we challenged germ-free (GF) larvae with the pathogen. A stronger lethal effect of the pathogen was observed in GF larvae than in CONV-R larvae (P < 0.001, unpaired t-test; Figures 1B,D).
Figure 1.
Protective effect of yeasts Debaryomyces hansenii (Dh97) and Yarrowia lipolytica (Yl242) against a V. anguillarum (Va) challenge in zebrafish larvae. Larvae were inoculated with different concentration (104–107 CFU/mL) of Dh97 (A,B) and Yl242 (C,D) on day 4 dpf, and challenged with V. anguillarum on day 5 dpf. Survival rate (%) of conventionally raised larvae (CONV-R) (A,C), and germ-free larvae (GF) (B,D) at 4 days post- V. anguillarum inoculation. The results show the mean ± SD of 3 independent experiments with three replicates each. Different letters indicate statistically significant differences among groups (Kaplan Meier, Wilcoxon P < 0.003).
Effect of yeast strain inoculum on survival of V. anguillarum-challenged larvae
The effect of the yeast inoculum (CFU/mL) on the survival rate (%) of V. anguillarum-challenged larvae is shown in Figure 1. We observed that both yeasts, Dh97 and Yl242, significantly protected CONV-R larvae from the V. anguillarum challenge (Figures 1A,C). In Dh97 a tendency of dose-dependent protection until 106 CFU/mL was observed, with higher protection of CONV-R larvae pre-treated with 106 CFU/mL compared to 104 CFU/mL (Figure 1A). On the contrary, yeast Yl242, displayed a similar protective effect independent of the concentration used (Figure 1C). Likewise, at 105 and 106 CFU/mL, Dh97 was more effective in protecting CONV-R larvae compared to Yl242 (P < 0.05 and P < 0.01, respectively; Kaplan-Meier, log-rank post-test).
In GF larvae challenged with V. anguillarum, both yeasts were able to increase survival rate significantly at 106 CFU/mL (Figures 1B,D). The effectiveness of yeast Dh97 was similar in GF and CONV-R larvae (P = 0.7209, unpaired t-test; Figures 1A,B); whereas yeast Yl242 was more effective in protecting GF than CONV-R larvae (P < 0.001, unpaired t-test; Figures 1C,D).
Concentration and persistence of yeasts and V. anguillarum in CONV-R and GF larvae
To evaluate if the protection of larvae was related to the concentration and persistence of yeast or V. anguillarum in larvae, we determined the cultivable count of these microorganisms in larvae. The initial concentration of yeast reached in CONV-R larvae (4 dpf), after 2 h immersion depended on the yeast species and dose (Table 2 and Figure S2). At the same inoculum in E3 medium (CFU/mL), Dh97 reached significantly higher concentrations in CONV-R larvae than Yl242 (P < 0.05, unpaired t-test). For both yeasts we observed a positive correlation between yeast inoculum and yeast concentration in CONV-R larvae (CFU/larva; r = 0.9203, P < 0.0001 for Dh97; and r = 0.7778, P < 0.001 for Yl242, Spearman correlation test; Figure S2). In GF larvae (4 dpf) Dh97 reached similar concentrations as in CONV-R larvae (P > 0.05, unpaired t-test); whereas Yl242 reached higher concentrations (P < 0.05, unpaired t-test; Table 2). For all experimental groups, both yeasts persisted in larvae and at 9 dpf reached similar or higher concentrations compared to 4 dpf (Table 2).
Table 2.
Initial counts (4 dpf) and persistence (9 dpf) of yeasts in CONV-R and germ-free (GF) larvae.
| Yeast concentration (log10CFU/larva) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Yeast dose | CONV-R + Dh97 | CONV-R + Dh97 + Va | GF + Dh97 | GF + Dh97 + Va | ||||
| log10CFU/mL | 4 dpf | 9 dpf | 4 dpf | 9 dpf | 4 dpf | 9 dpf | 4 dpf | 9 dpf |
| ni | < | < | < | < | < | < | < | < |
| 4 | 2.4 ± 0.2 | 2.5 ± 0.5 | 2.4 ± 0.2 | 3.8 ± 0.0** | − | − | − | − |
| 5 | 3.3 ± 0.1 | 3.4 ± 0.2 | 3.3 ± 0.1 | 3.2 ± 0.3 | − | − | − | − |
| 6 | 3.2 ± 0.1 | 3.6 ± 0.3 | 3.2 ± 0.1 | 2.8 ± 0.0* | 3.0 ± 0.1 | 4,2 ± 0.2*** | 3,0 ± 0.1 | 3.4 ± 0.4 |
| 7 | 3.5 ± 0.1 | 3.8 ± 0.3 | 3.5 ± 0.1 | 3.8 ± 0.1 | − | − | − | − |
| CONV-R + Yl242 | CONV-R + Yl242 + Va | GF + Yl242 | GF + Yl242 + Va | |||||
| 4 dpf | 9 dpf | 4 dpf | 9 dpf | 4 dpf | 9 dpf | 4 dpf | 9 dpf | |
| ni | < | < | < | < | < | < | < | < |
| 4 | 1.9 ± 0.1 | 3.0 ± 0.0** | 1.9 ± 0.1 | 2.8 ± 0.2* | − | − | − | − |
| 5 | 2.1 ± 0.0 | 3.3 ± 0.0** | 2.1 ± 0.0 | 3.0 ± 0.0** | − | − | − | − |
| 6 | 2.3 ± 0.1 | 3.4 ± 0.1** | 2.3 ± 0.1 | 3.2 ± 0.2* | 3.1 ± 0.4 | 3.5 ± 0.2 | 3.5 ± 0.1 | 3.8 ± 0.0** |
| 7 | 2.3 ± 0.0 | 3.3 ± 0.0** | 2.3 ± 0.0 | 3.3 ± 0.3* | − | − | − | − |
Unpaired t-test
P ≤ 0.05;
P ≤ 0.005;
P ≤ 0.001, indicates significant differences between the yeast concentration at 4 and 9 dpf.
ni, larvae not inoculated with yeasts.
<, < 1 (log10CFU/larva).
−, Not determined.
The initial concentration of V. anguillarum in CONV-R larvae at 5 dpf not treated with yeasts reached on average log10 2.8 CFU/larva, and persisted with the same concentration at 9 dpf (Table 3). The pre-treatment with yeast Dh97 generally did not significantly affect the initial concentration of the pathogen for CONV-R larvae (ANOVA P > 0.05); however, unexpectedly the pre-treatment with Yl242 significantly enhanced the initial pathogen concentration compared to larvae not inoculated with yeasts (ANOVA P < 0.05; Table 3).
Table 3.
Initial counts (5 dpf) and persistence (9 dpf) of V. anguillarum in CONV-R and germ-free (GF) larvae.
| V. anguillarum concentration (log10CFU/larva) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Yeast dose | CONV-R + Dh97 + Va | GF + Dh97 + Va | CONV-R + Yl242 + Va | GF + Yl242 + Va | ||||
| log10CFU/mL | 5 dpf | 9 dpf | 5 dpf | 9 dpf | 5 dpf | 9 dpf | 5 dpf | 9 dpf |
| ni | a2.8 ± 0.2 | a2.9 ± 0.1 | 4.1 ± 0.1 | 4.3 ± 0.5 | a2.8 ± 0.2 | a2.9 ± 0.1 | 4.1 ± 0.1 | 4.3 ± 0.5 |
| 4 | a2.9 ± 0.1 | b2.1 ± 0.1** | − | − | b3.3 ± 0.3 | a1.8 ± 0.0** | − | − |
| 5 | a2.6 ± 0.2 | b2.0 ± 0.1* | − | − | b3.2 ± 0.1 | a1.7 ± 0.1*** | − | − |
| 6 | b3.2 ± 0.1 | b2.0 ± 0.0** | 2.3 ± 0.0 | 4.3 ± 0.1*** | b3.2 ± 0.1 | a3.4 ± 0.1 | 3.2 ± 0.0 | 4.1 ± 0.1** |
| 7 | a2.6 ± 0.0 | b2.0 ± 0.4 | − | − | b3.2 ± 0.4 | a3.0 ± 0.0 | − | − |
Unpaired t-test
P ≤ 0.05;
P ≤ 0.005;
P ≤ 0.001, indicates significant differences between the V. anguillarum concentration at 4 and 9 dpf. Letters indicate differences between treated and not inoculated larvae at the respective day, ANOVA with Dunnet multiple comparison corrected test.
ni, larvae not inoculated with yeasts.
−, Not determined.
Comparing the pathogen load in CONV-R larvae at the end of the challenge (9 dpf) with the initial concentration (5 dpf), we observed that all concentrations of Dh97 significantly reduced the pathogen load (P < 0.05, unpaired t-test), except for the higher yeast doses (log10 7 CFU/larva). However, comparing with non-yeast inoculated larvae all Dh97 doses were equally effective in reducing pathogen concentration at 9 dpf (Table 3; ANOVA P < 0.005). Similarly, pre-treatment with Yl242 reduced the pathogen load at 9 dpf compared to 5 dpf and compared to larvae not inoculated with yeast (P < 0.005, unpaired t-test). However, only yeast doses of log10 4 and log10 5 CFU/larva were effective.
When GF larvae were challenged with V. anguillarum, the initial pathogen concentration at 5 dpf reached significantly higher levels than in CONV-R larvae (P < 0.05, unpaired t-test). The pre-treatment of GF larvae with both yeasts significantly reduced the initial pathogen concentration (at 5 dpf), compared to GF-challenged larvae (P < 0.05, unpaired t-test). However, neither yeast avoided V. anguillarum growth in GF larvae; counts of the pathogen at 9 dpf reached a similar level to larvae not inoculated with yeasts. These results suggest that larval protection by yeasts against a V. anguillarum challenge is not only due to a reduction in the host pathogen load, and other mechanisms such as immune modulation may be involved.
Innate immune response induced in larvae
To determine the role of yeast in immune modulation of the host we evaluated the relative expression of innate immune response marker genes in CONV-R and GF larvae challenged with V. anguillarum, including interleukin 1 beta (il1b), tumor necrosis factor a (tnfa), interleukin 10 (il10), complement component 3 (c3) and myeloid-specific peroxidase (mpx).
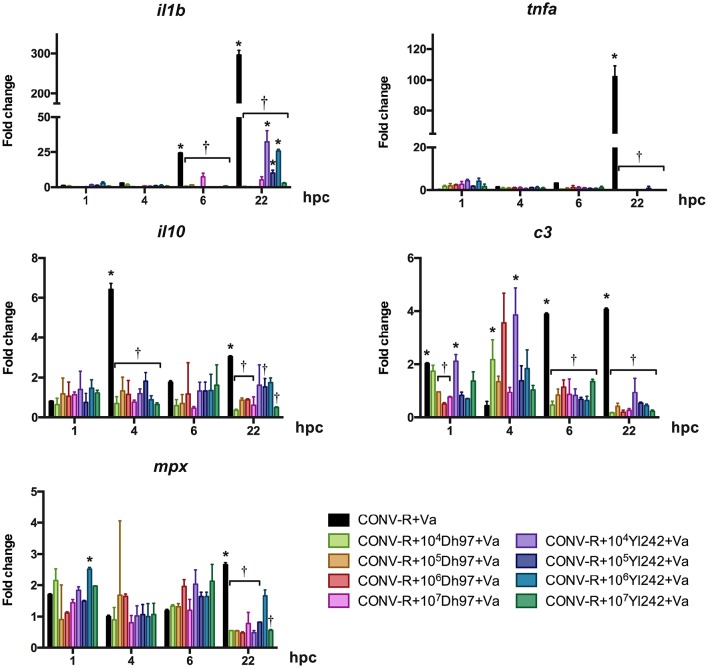
CONV-R larvae challenged with V. anguillarum showed a significant upregulation of il1b at 6 and 22 h post-challenge (hpc), c3 at 1, 6, and 22 h post-challenge (hpc) and tnfa and mpx at 22 hpc compared to un-challenged CONV-R larvae (Figure 2, Supplementary Table 1). The transcription level of the anti-inflammatory cytokine il-10 was upregulated at 4 and 22 hpc. Interestingly, pre-treatment with yeast Dh97 or Yl242 significantly prevented the upregulation of all these genes (Figure 2, Supplementary Table 1). In general, all yeast doses were equally effective to prevent the upregulation of these genes (Figure 2, Supplementary Table 1).
Figure 2.
Expression of innate immune genes analyzed by qPCR in conventionally raised (CONV-R) larvae challenged with V. anguillarum at 5 dpf, pre-treated at 4 dpf with different concentrations of each yeast, relative to non-challenged CONV-R larvae. Dh97, Debaryomyces hansenii 97; Yl242, Yarrowia lipolytica 242; il1b, interleukin 1 beta; tnfa, tumor necrosis factor a; c3, complement component 3; mpx, myeloid-specific peroxidase; il10, interleukin 10; hpc, hours post V. anguillarum-challenge. Data were normalized to beta actin 1. The results show the mean ± SD of 3 independent experiments with three replicates each. *Indicates statistically significant differences of the experimental groups with non-challenged CONV-R larvae, †Indicates statistically significant differences of the V. anguillarum-challenged larvae treated with yeasts (CONV-R + yeast + Va) with V. anguillarum-challenged CONV-R larvae (CONV-R + Va).
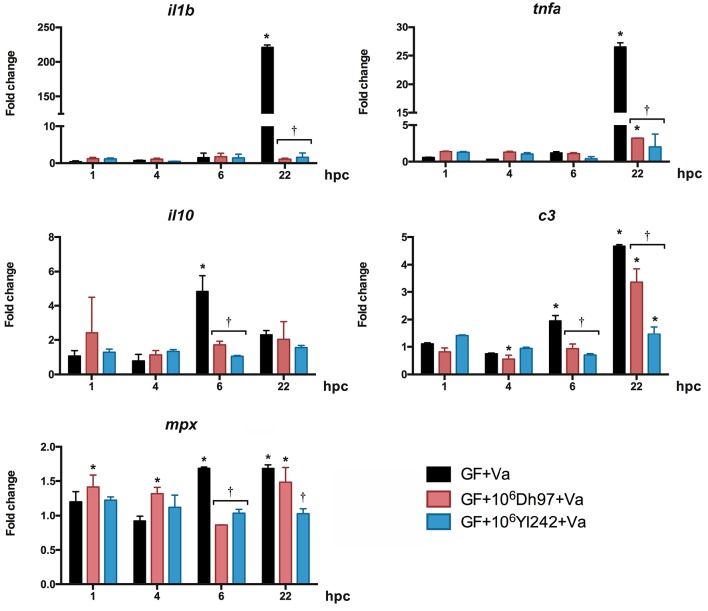
In GF larvae challenged with V. anguillarum, il1b, tnfa, and c3 were significantly upregulated at 22 hpc, as in CONV-R larvae (Figure 3, Supplementary Table 2). il10 was significantly upregulated at 6 hpc, and mpx was significantly upregulated at 6 and 22 hpc. Both yeasts, Dh97 and Yl242, significantly prevented the upregulation of il1b, tnfa, and c3 at 22 hpc, and il10 at 6 hpc (Figure 3, Supplementary Table 2).
Figure 3.
Expression of innate-immune genes analyzed by qPCR in germ-free (GF) larvae challenged with V. anguillarum (Va) at 5 dpf, pre-treated at 4 dpf with 106 CFU/mL of each yeast, relative to non-challenged germ-free larvae. Dh97, Debaryomyces hansenii 97; Yl242, Yarrowia lipolytica 242; il1b, interleukin 1 beta; tnfa, tumor necrosis factor a; c3, complement component 3; mpx, myeloid-specific peroxidase; il10, interleukin 10; hpc, hours post V. anguillarum challenge. Data were normalized to beta actin 1. The results show the mean ± SD of 3 independent experiments with three replicates each. *Indicates statistically significant differences of the experimental groups with non-challenged GF larvae, †Indicates statistically significant differences between the V. anguillarum-challenged GF larvae treated with yeasts (GF + yeast + Va) with V. anguillarum-challenged GF larvae (GF + Va).
To evaluate if yeasts alone could stimulate the innate immune system of larvae we measured the expression of the same genes in CONV-R and GF larvae treated with each yeast (Figures S3, S4, respectively). In CONV-R larvae (Figure S3 and Supplementary Table 3). il1b was upregulated by the two yeasts at 6 and 30 hpt (hour post-treatment), tnfa was upregulated by yeast Yl242 at concentrations of 104 and 105 (CFU/mL) at 46 hpt, il10 was upregulated at 1, 6, 30, and 46 hpt by both yeasts, c3 was upregulated only by some doses of yeast Dh97 at 6 and 24 hpt. Finally, mpx was only upregulated by Yl242 at 24 hpt at a dose of 106 CFU/mL. None of the genes evaluated showed a dose-effect response. The overall gene expression induced by yeasts in GF larvae showed less change than in CONV-R (Figure S4 and Supplementary Table 4). Dh97 significantly downregulated the expression of c3 at 1, 6, 22, 28, and 30 hpt and upregulated the expression of the gene at 46 hpt. On the other hand, yeast Yl242 upregulated il1b at 30 hpt, tnfa at 24 and 30 hpt, il10 at 28 and 30 hpt, c3 at 4, 24, 28, and 46 hpt, and mpx at 28, and 30 hpt, and downregulated mpx at 1 and 24 hpt.
Co-aggregation studies
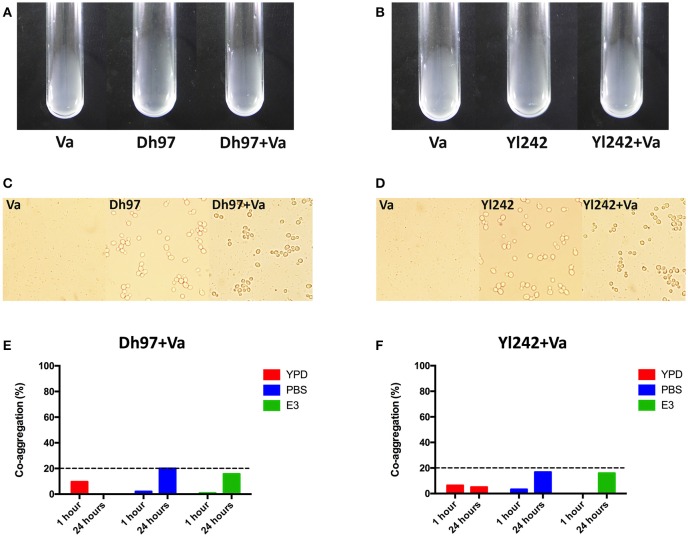
We determined if Dh97 and Yl242 yeasts could bind V. anguillarum through co-aggregation analysis. We did not detect any visual (Figures 4A,B) or microscopic auto- or co-aggregation (Figures 4C,D). We also performed a quantitative spectrophotometric co-aggregation assay using different media (YPD, PBS, or E3), since it has been reported that co-aggregation depends greatly on the conditions used (Millsap et al., 1998). All co-aggregation percentages were less than 20% in all media tested, showing no co-aggregation between the yeasts and V. anguillarum (Figures 4E,F).
Figure 4.
Co-aggregation assays between the yeasts Debaryomyces hansenii (Dh97) or Yarrowia lipolytica (Yl242) with V. anguillarum (Va). (A,B) Macroscopic co-aggregation assays. Equal volumes (200 μL) of V. anguillarum and (A) Dh97 or (B) Yl242 suspensions in co-aggregation buffer were mixed in borosilicate tubes (12 × 75 mm, Schott) for at least 5 s on vortex (Dh97 or Yl242 + Va tubes). Control tubes containing 200 μL of each microorganism and 200 μL of co-aggregation buffer (Va, Dh97, and Yl242 tubes) were included to check potential auto-aggregation. (C,D) Microscopic co-aggregation assays: all suspensions (from A,B) were observed in an optical microscope to observe any microscopic co-aggregation. (E,F) Spectrophotometric co-aggregation assays. A suspension of each yeast strain and V. anguillarum in YPD, PBS, and E3 (O.D. of 1.0 at 600 nm) were mixed in equal volumes (5 mL) for 10 s in vortex. Control tubes contained 10 mL of each microbial suspension. The O.D. (600 nm) of the upper suspension (1 mL) from each tubes was measured at 1 and 24 h. The percentage of co-aggregation was calculated as previously described (Ogunremi et al., 2015).
Discussion
The development of new probiotics requires not only in vivo demonstration of their benefits, but also an understanding of the mechanisms involved in their effects. In this study we explored some mechanisms involved in the protection of zebrafish larvae against a V. anguillarum challenge by two probiotic yeasts, D. hansenii 97 (Dh97) and Y. lipolytica 242 (Yl242), isolated from the intestine of healthy fish (Raggi et al., 2014). We analyzed the effect of both yeasts on in vivo pathogen concentration, modulation of the host innate immune system and co-aggregation with the pathogen. In addition, the effect of zebrafish microbiota on the survival of larvae was determined using germ-free (GF) larvae.
As previously reported (Caruffo et al., 2015), challenging CONV-R zebrafish larvae with V. anguillarum provokes high mortality (>60%) at 4 days post-challenge (dpc). Studies performed in mouse models have shown that an intact microbiota protects the host against pathogen attack (Endt et al., 2010). To determine the potential protective role of the zebrafish microbiota against V. anguillarum challenge, GF larvae were exposed to the pathogen. Our results show, for the first time in zebrafish, that the resident microbiota can protect the host from V. anguillarum infection, since GF animals showed significantly more mortality (92%) than CONV-R larvae. In opposition to our results, no protective role of the resident zebrafish microbiota was detected in larvae exposed to the same pathogen (Oyarbide et al., 2015) or Edwarsiella ictaluri (Rendueles et al., 2012). Similarly, no protective effect of the host microbiota was observed in a novel infection model of gnotobiotic Nile tilapia with E. ictaluri (Situmorang et al., 2014). These results suggest that the specific composition of the resident microbiota of zebrafish in our facilities may be more effective in protecting larvae against V. anguillarum infection. Future studies should characterize the composition of this microbiota to elucidate the specific microorganisms involved in protection.
We then explored the capacity of two yeast strains isolated from healthy fish to protect zebrafish larvae from V. anguillarum challenge. In conventionalized conditions at a dose of 106 CFU/mL, yeast Dh97 was more effective than Yl242 in protecting the larvae. The different colonization capacity of the yeasts in CONV-R larvae may explain this difference. In germ-free conditions the yeasts showed similar colonization capacities and were equally effective against the pathogen, and surprisingly, more effective than the host microbiota to protect larvae. Overall, the results suggest that yeast concentrations inside larvae were more determinant than yeast proliferation to protect them from a unique dose of the pathogen. In addition, these results showed that protection exerted by yeasts is not necessarily related to the modulation of the host microbiota.
The concentration of the pathogen in CONV-R larvae at the moment of the challenge and at the end of the experiment were similar (P > 0.05), and reached about log10 2.8 CFU/larva. In a similar study (Oyarbide et al., 2015) with larvae exposed to V. anguillarum, the pathogen reached higher concentrations (log10 5.9 and 5.8 per larvae at 5 and 6 dpf, respectively) and produced higher mortality (100% after 3 days post-V. anguillarum challenge at 8 dpf) than in our study. This result could be due to differences in the virulence of the strains, differences in the susceptibility of the hosts, higher concentration of V. anguillarum inoculated or to the design of the experiment, in which larvae were constantly exposed to the pathogen (Oyarbide et al., 2015), in contrast with our study. The reduced survival observed in challenged GF larvae during our experiments could be related to higher concentrations of the pathogen in larvae (>1 log) than those observed for challenged CONV-R larvae and/or due to the lack of host microbiota protection.
We then evaluated the capacity of the yeasts to reduce the pathogen concentration in V. anguillarum-challenged larvae in CONV-R and GF conditions. The two yeasts modified pathogen concentrations during the challenge. The initial concentration of the pathogen reached in CONV-R larvae was not modified, except at the higher doses of Yl242, which increased the initial concentration of the pathogen, suggesting that this yeast could stimulate V. anguillarum entrance to the host. The possible mechanisms explaining this point could involve greater habitat availability inside the gut by eventual modification of the gut microbiota by Yl242, or modification of intestinal mucus layer, enhancing the chemotactic swimming of V. anguillarum toward the intestinal mucus (O'Toole et al., 1999), although these hypotheses merit more study. At the end of the challenge both yeasts tended to reduce the pathogen concentration in CONV-R larvae. In germ-free larvae, this anti-bacterial effect was only observed at the beginning of the challenge, but yeasts were unable to control pathogen growth; the pathogen reached the same concentration at 9 dpf as in germ-free larvae not treated with yeasts. These results contrast with those obtained in yeast-treated CONV-R larvae, where yeasts tended to reduce the bacterial load at 9 dpf. This difference is probably due to an indirect effect of the yeast on the host microbiota, because reduction of pathogen growth was not observed in germ-free larvae. Surprisingly, in spite of the higher V. anguillarum concentration, in germ-free larvae the survival of yeast-treated larvae was equivalent to yeast-treated CONV-R larvae. We hypothesize that yeasts could exert other mechanisms to reduce the virulence of V. anguillarum that could explain lower mortality observed. These findings are in accordance with a previous study showing that protection of S. boulardii against Salmonella infection in mice is not related to in vivo antagonism (Martins et al., 2013). Overall, these results showed that larval protection by yeasts is not always associated with an in vivo anti-pathogen effect, as previously described (Schneider, 2011). These results suggest that other mechanisms besides the control of pathogen replication may be involved in protection, such as modulation of the immune response of the host.
The inflammatory signaling cascade is triggered when the host receptors involved are capable of binding to the bacteria or their products. This process results in the production of several pro-inflammatory cytokines such as il1b and tnfa (van der Vaart et al., 2012). A strong inflammatory response in larvae was observed after the V. anguillarum challenge, reflected by a robust upregulation at the transcriptional level of tnfa and il1b, as previously described in zebrafish larvae infected with V. anguillarum (Oyarbide et al., 2015) and E. ictaluri (Rendueles et al., 2012). The challenged larvae also exhibited an up-regulation of the mRNA level of c3, mpx and il10. c3 is the best characterized component of the complement system; it plays a central role in all activation pathways (Lee et al., 2013) and it is crucial in the early immune response of fish larvae (Løvoll et al., 2007). Its expression is induced by LPS, and in zebrafish it plays a role in inflammatory processes and regeneration (Forn-Cuní et al., 2014). mpx is one of the most specific markers for neutrophil and its precursors. Its expression is related to myelopoiesis (Bennett et al., 2001; Glenn et al., 2014). The upregulation of this gene in challenged larvae could reflect active neutrophil proliferation derived from the inflammatory response induced by the pathogen. il10 targets various leukocytes and mainly represses or modulates excessive inflammatory responses (Ouyang et al., 2011). The induction of this cytokine reveals a modulatory response of the host to the induced inflammation triggered by the pathogen. Importantly, in our study the analysis of cytokine expression was performed only until 22 h post-V. anguillarum challenge, since previous reports have shown that most of the transcripts are modulated in the first 24 h after V. anguillarum infection (Rojo et al., 2007; Zhang et al., 2013; Liu et al., 2014; Oyarbide et al., 2015). Previous results showed a significant increase of tnfa, il1b, and il10 over time in zebrafish larvae infected by E. ictaluri up to 3 days post-infection (Rendueles et al., 2012). In our study, it would be important to evaluate the immune modulation exerted by V. anguillarum until the end of the trial (4 dpc), to determine its correlation with larval mortality.
The pre-treatment of CONV-R and germ-free larvae with yeasts completely prevented upregulation of all immune relevant genes evaluated at 22 hpc. It has been previously described that yeasts can also show anti-inflammatory effects. The yeast S. cerevisiae var. boulardii can modulate the immune system response during bacterial infection (Czerucka et al., 2007; Moslehi-Jenabian et al., 2010). This yeast can exert anti-inflammatory effects related to the suppression of NF-κB activation, inhibition of the pro-inflammatory cytokine gene expression and stimulation of PPAR-γ expression, reducing enterocyte responses to pro-inflammatory cytokines. Whether, these mechanisms could be involved in larval protection merits further analysis.
It has been widely described that neutrophil migration in zebrafish larvae, considered a key hallmark in an inflammatory process, is correlated with the expression of some inflammatory cytokines such as tnfa and il1b (Barros-Becker et al., 2012; Hedrera et al., 2013; de Oliveira et al., 2016). In a previous study (Caruffo et al., 2015) we observed an increase in neutrophil migration outside the hematopoietic region at 3 hpc in CONV-R larvae challenged with V. anguillarum, showing an inflammatory response of the host. Although, in the present study we did not evaluate neutrophil migration, we would expect an increase in neutrophil migration outside the hematopoietic tissue during all the infection period with V. anguillarum in CONV-R and germ-free larvae, concomitant with a reduced number of inflammatory cells in larvae pre-treated with yeasts.
Yeasts contain β-glucans, mannoproteins, and chitin in their cell walls, and also nucleotides which can stimulate the immune system by binding to specific receptors (Reyes-Becerril et al., 2008; Oyarbide et al., 2012; Barreto-Bergter and Figueiredo, 2014). The immuno-stimulatory effect of yeast β-glucans, which are part of the pathogen-associated molecular patterns (PAMPS), is well-known and has proven to be efficient in different fish species (Bricknell and Dalmo, 2005; Magnadottir, 2010) including zebrafish (Rodríguez et al., 2009). β-glucans are located on the inner cell wall layer of yeasts, protected by an outer layer of mannoproteins (Erwing and Gow, 2016). It has been described that the immune effect of β-glucans depends on their structure and the level of exposure of these molecules to the host immune cells (Navarrete and Tovar-Ramírez, 2014; Erwing and Gow, 2016). For example, juvenile rainbow trout (O. mykiss) fed with a beta-mercaptoethanol-treated S. cerevisiae-supplemented diet (with an expected more open structure of the yeast cell wall due to the breaks of the disulfide bonds between mannoproteins) showed higher stimulation of the immune system and an enhanced survival rate against Yersinia ruckeri compared to fish fed with whole-cell yeast (Tukmechi et al., 2011). It is noteworthy that all studies have been performed with β-glucans derived from S. cerevisiae, and little is known about the immunomodulatory effect of β-glucans derived from non-S. cerevisiae yeasts. It is known that yeast species have different cell wall composition, with different proportions of glucans (Nguyen et al., 1998), suggesting that they can differentially modulate the host immune system. This difference could explain, in part, the different immune modulations observed with Dh97 and Yl242, or the different protection magnitude by different yeast species in a V. anguillarum challenge model (Caruffo et al., 2015).
Previous work showed that immune stimulation by yeast in gilthead seabream (Sparus aurata L.) displays increased or decreased expression of the immune genes according to the organ evaluated (intestine, head kidney, and liver; Reyes-Becerril et al., 2008). In this study we tested the stimulation of the innate immune system in larvae treated with yeasts. Although we did not observe a time-, dose-, or yeast-specific response, yeasts were able to modulate some of the genes evaluated. The lack of a clear tendency in cytokine expression could be explained because we evaluated the transcripts in the entire larvae and not in each organ. The magnitude of cytokine expression induced by both yeasts in non-V. anguillarum challenged larvae was lower than in those stimulated by the pathogen. This could be related to the point discussed above, i.e., to the grade of exposure of immune-stimulating molecules in the cell wall of these two yeasts, or because the interaction of V. anguillarum with the host is greater due to the invasive nature of this pathogen. Related to the last point, it has been reported that larvae challenged with GFP-labeled V. anguillarum harbor the pathogen in the gastrointestinal tract at 3 hpc (O'Toole et al., 2004), as we previously observed (Caruffo et al., 2015), and in the head and tail after 6 hpc (O'Toole et al., 2004). By contrast, five probiotic yeast candidates including Yl242 were only observed in the gastrointestinal tract 5 days after yeast treatment (Caruffo et al., 2015).
In our study both yeasts were able to remain viable in larvae until the end of the V. anguillarum challenge. However, we do not know if protection against this pathogen or immune modulation needs viable yeast. This point is essential to a better understanding of the mechanisms involved in yeast protection. One would expect that protection mechanisms by dead yeasts could include competition for the physical space (in the gut), stimulation of the immune system by their cell wall components, adhesion to the pathogen impeding its invasion of the host (Moslehi-Jenabian et al., 2010) or promoting its elimination by feces (Pontier-Bres et al., 2014). On the other hand, live yeasts could also contribute with secreted factors (Ran et al., 2016). The importance of yeast viability on the probiotic effect has been recently studied and shows that the effect is influenced by fish density (Ran et al., 2015, 2016). Under high stocking density, supplementation of live S. cerevisiae in the feed of Nile tilapia significantly enhanced resistance of fish against Aeromonas hydrophila compared to heat-inactivated yeast (Ran et al., 2016). However, under normal fish density both live and inactivated yeast protected the host against infection by A. hydrophila. In addition, live yeast, but not inactivated yeast, reduced intestinal expression of tnfα, tgfβ, and il1β (Ran et al., 2015), showing the importance of secreted factors in the host immune modulation. In our study, it seems that multiple species-specific mechanisms are involved in protection against V. anguillarum. Future studies including protection experiments using dead yeast (i.e., heat-inactivated yeast) will help to elucidate this issue.
It has been reported that S. cerevisiae var. boulardii can prevent the adherence and translocation of bacteria to enterocytes, which can be explained in part by their ability to bind bacteria (Moslehi-Jenabian et al., 2010). Because yeast treatments completely abolish the inflammatory response induced by V. anguillarum, we tested the hypothesis that yeasts could adhere to V. anguillarum, impeding or reducing its contact with the host, which would explain in part the protective role of yeasts. These experiments were performed with in vitro co-aggregation assays. Co-aggregation has been defined as a specific recognition and adhesion of genetically distinct bacteria when they are in suspension, which is mediated by adhesins and polysaccharide receptors on the cell surface of co-aggregating cells (Kolenbrander, 2000; Rickard et al., 2003; Vornhagen et al., 2013). This specific interaction has been observed in human intestinal bacteria (Ledder et al., 2008), and recently between yeast and bacteria (Martins et al., 2013; Stevens et al., 2015). We did not observe any in vitro co-aggregation between yeasts and V. anguillarum in any of the assays performed. A previous study showed in vivo binding of the yeast S. cerevisiae (UFMG 905) with S. enterica serovar Typhimurium, reducing its translocation and invasion in mice (Martins et al., 2013). Whether this yeast-V. anguillarum interaction could occur in vivo requires further investigation.
In addition to the mechanisms evaluated in this study, yeasts can protect the host from pathogens via other pathways, mainly in the gut. For example, yeasts can improve the intestinal barrier function, stabilizing tight junctions and reducing pathogen translocation (Moslehi-Jenabian et al., 2010). The trophic effect of yeasts due to the production of polyamines (mainly spermine and spermidine) has been well described in humans, rodents and fish (Tovar-Ramírez et al., 2004; Buts and De Keyser, 2006). Although, this trophic effect has not been evaluated in the protection against a pathogen challenge, it could be postulated that this mechanism may also improve host survival. Recently, yeasts have also been shown to affect the intestinal traffic of the pathogen Salmonella Typhimurium. The adhesion of the pathogen and the yeast modifies pathogen distribution in the lumen, increasing its elimination in feces (Pontier-Bres et al., 2014). Further studies are necessary to elucidate if these mechanisms are also involved in the protection of zebrafish larvae against the V. anguillarum challenge by yeasts Dh97 and Yl242.
In conclusion, our results revealed that protection of zebrafish larvae against a V. anguillarum challenge with two non-Saccharomyces yeasts involves strain-specific mechanisms. Yeasts were able to modulate the innate immune system of the host and showed an in vivo anti-pathogen effect; however, the lower mortality with yeast pretreatment does not always correlate with lower pathogen burden. This suggests that other protection mechanisms may be involved. In addition, using GF larvae we highlighted the importance of the normal resident microbiota to enhance the host response to a bacterial infection, and showed the utility of using probiotic yeasts to restore or even improve the beneficial effect exerted by the host microbiota. Whether the beneficial effects of yeasts include other mechanisms will be explored in future investigations. Thus, our results provide new insight into the complex microbial interaction between a beneficial and pathogenic microorganisms and the host in the context of health and disease.
Author contributions
MC, NN, and PN conceived and designed the experiments. MC, NN, OS performed the experiments. MG, NF performed the co-aggregation experiments. MC, NN, KG, CF, AR, and PN analyzed the data. MC, PN wrote the paper.
Funding
This work was supported by FONDECYT N°11110414. MC acknowledges a scholarship from CONICYT N°21110848 and Dr. Stekel fellowship, INTA-Nestlé.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Estela Blanco, Diego Merino, and Lafayette Eaton for editing and revising the English grammar.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00127
References
- Abu-Elala N., Marzouk M., Moustafa M. (2013). Use of different Saccharomyces cerevisiae biotic forms as immune-modulator and growth promoter for Oreochromis niloticus challenged with some fish pathogens. Int. J. Vet. Sci. Med. 1, 21–29. 10.1016/j.ijvsm.2013.05.001 [DOI] [Google Scholar]
- Barreto-Bergter E., Figueiredo R. (2014). Fungal glycans and the innate immune recognition. Front. Cell. Infect. Microbiol. 4:145. 10.3389/fcimb.2014.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros-Becker F., Romero J., Pulgar A., Feijóo C. G. (2012). Persistent oxytetracycline exposure induces an inflammatory process that improves regenerative capacity in zebrafish larvae. PLoS ONE 7:e36827. 10.1371/journal.pone.0036827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne C. J., Gerwick L. (2001). The acute phase response and innate immunity of fish. Dev. Comp. Immunol. 25, 725–743. 10.1016/S0145-305X(01)00033-7 [DOI] [PubMed] [Google Scholar]
- Belkaid Y., Hand T. W. (2014). Role of the Microbiota in immunity and inflammation. Cell 157, 121–141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C. M., Kanki J. P., Rhodes J., Liu T. X., Paw B. H., Kieran M. W., et al. (2001). Myelopoiesis in the zebrafish, Danio rerio. Blood 98, 643–651. 10.1182/blood.V98.3.643 [DOI] [PubMed] [Google Scholar]
- Bricknell I., Dalmo R. (2005). The use of immunostimulants in fish larval aquaculture. Fish Shellfish Immunol. 19, 457–472. 10.1016/j.fsi.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Buts J. P., De Keyser N. (2006). Effects of Saccharomyces boulardii on intestinal mucosa. Dig. Dis. Sci. 51, 1485–1492. 10.1007/s10620-005-9016-x [DOI] [PubMed] [Google Scholar]
- Canadian Council on Animal Care (1989). Guide to the Care and Use of Experimental Animals, Vol. 2. Ottawa, ON: Canadian Council on Animal Care. [Google Scholar]
- Caruffo M., Navarrete N., Salgado O., Díaz A., López P., García K., et al. (2015). Potential probiotic yeasts isolated from the fish gut protect zebrafish (Danio rerio) from a Vibrio anguillarum challenge. Front. Microbiol. 6:1093. 10.3389/fmicb.2015.01093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar J., Kolenbrander P., McIntire F. (1979). Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infection Immun. 24, 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa França R., Rochedo Conceiçã F., Mendonça M., Haubert L., Sabadin G., Diaz de Oliveira P., et al. (2015). Pichia pastoris X-33 has probiotic properties with remarkable antibacterial activity against Salmonella Typhimurium. Appl. Microbiol. Biotechnol. 99, 7953–7961. 10.1007/s00253-015-6696-9 [DOI] [PubMed] [Google Scholar]
- Czerucka D., Piche T., Rampal P. (2007). Review article: yeast as probiotics – Saccharomyces boulardii. Aliment. Pharmacol. Ther. 26, 767–778. 10.1111/j.1365-2036.2007.03442.x [DOI] [PubMed] [Google Scholar]
- de Oliveira S., Rosowski E. E., Huttenlocher A. (2016). Neutrophil migration in infection and wound repair: going forward in reverse. Nat. Rev. Immunol. 16, 378–391. 10.1038/nri.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endt K., Stecher B., Chaffron S., Slack E., Tchitchek N., Benecke A., et al. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6:e1001097. 10.1371/journal.ppat.1001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwing L. P., Gow N. A. R. (2016). Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 14, 163–176. 10.1038/nrmicro.2015.21 [DOI] [PubMed] [Google Scholar]
- Forn-Cuní G., Reis E. S., Dios S., Posada D., Lambris J. D., Figueras A., et al. (2014). The evolution and appearance of C3 duplications in fish originate an exclusive teleost c3 gene form with anti-inflammatory activity. PLoS ONE 9:e99673. 10.1371/journal.pone.0099673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa S., Nojima N., Yoshida K., Hirayama S., Ogihara H., Morinaga Y. (2011). The importance of inter-species cell-cell co-aggregation between Lactobacillus plantarum ML11-11 and Saccharomyces cerevisiae BY4741 in mixed-species biofilm formation. Biosci. Biotechnol. Biochem. 75, 1430–1434. 10.1271/bbb.100817 [DOI] [PubMed] [Google Scholar]
- Gatesoupe J. (2007). Live yeasts in the gut: natural occurrence, dietary introduction, and their effects on fish health and development. Aquaculture 267, 20–30. 10.1016/j.aquaculture.2007.01.005 [DOI] [Google Scholar]
- Glenn N. O., Schumacher J. A., Kim H. J., Zhao E. J., Skerniskyte J., Sumanas S. (2014). Distinct regulation of the anterior and posterior myeloperoxidase expression by Etv2 and Gata1 during primitive Granulopoiesis in zebrafish. Dev Biol. 393, 149–159. 10.1016/j.ydbio.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B., Ma H., Wang Y., Hu Y., Lin Z., Zhu Z., et al. (2011). Vitreoscilla hemoglobin (VHb) overexpression increases hypoxia tolerance in zebrafish (Danio rerio). Mar. Biotechnol. 13, 336–344. 10.1007/s10126-010-9305-z [DOI] [PubMed] [Google Scholar]
- Hai N. (2015). The use of probiotics in aquaculture. J. Appl. Microbiol. 119, 917–935. 10.1111/jam.12886 [DOI] [PubMed] [Google Scholar]
- Harikrishnan R., Kim M. C., Kim J. S., Balasundaram C., Heo M. S. (2011). Immunomodulatory effect of probiotics enriched diets on Uronema marinum infected olive flounder. Fish Shellfish Immunol. 30, 964–971. 10.1016/j.fsi.2011.01.030 [DOI] [PubMed] [Google Scholar]
- Hatoum R., Labrie S., Fliss I. (2012). Antimicrobial and probiotic properties of yeasts: from fundamental to novel applications. Front. Microbiol. 3:421. 10.3389/fmicb.2012.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrera M. I., Galdames J. A., Jimenez-Reyes M. F., Reyes A. E., Avendaño-Herrera R., Romero J., et al. (2013). Soybean meal induces intestinal inflammation in zebrafish larvae. PLoS ONE 8:e69983. 10.1371/journal.pone.0069983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M., Tomkovich S., Xiaolun S., Grosser M. R., Koo J., Flynn E. J., III., et al. (2014). Commensal microbiota stimulate systemic neutrophil migration through induction of serum amyloid A. Cell. Microbiol. 16, 1053–1067. 10.1111/cmi.12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. (2000). Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54, 413–437. 10.1146/annurev.micro.54.1.413 [DOI] [PubMed] [Google Scholar]
- Lam S. H., Chua H. L., Gong Z., Lam T. J., Sin Y. M. (2004). Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 28, 9–28. 10.1016/S0145-305X(03)00103-4 [DOI] [PubMed] [Google Scholar]
- Ledder R. G., Timperley A. S., Friswell M. K., Macfarlane S., McBain A. J. (2008). Coaggregation between and among human intestinal and oral bacteria. FEMS Microbiol. Ecol. 66, 630–636. 10.1111/j.1574-6941.2008.00525.x [DOI] [PubMed] [Google Scholar]
- Lee J. W., Lee Y. M., Lee J. H., Noh J. K., Kim H. C., Park C. J., et al. (2013). The expression analysis of complement component C3 during early developmental stages in Olive Flounder (Paralichthys olivaceus). Dev Reprod. 17, 311–319. 10.12717/DR.2013.17.4.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Gatlin D. (2006). Nucleotide nutrition in fish: current knowledge and future applications. Aquaculture 251, 141–152. 10.1016/j.aquaculture.2005.01.009 [DOI] [Google Scholar]
- Line J. E., Bailey J. S., Cox N. A., Stern N. J., Tompkins T. (1998). Effect of yeast-supplemented feed on Salmonella and Campylobacter populations in broilers. Poultry Sci. 77, 405–410. 10.1093/ps/77.3.405 [DOI] [PubMed] [Google Scholar]
- Liu X., Wu H., Chang X., Tang Y., Liu Q., Zhang Y. (2014). Notable mucosal immune responses induced in the intestine of zebrafish (Danio rerio) bath-vaccinated with a live attenuated Vibrio anguillarum vaccine. Fish Shellfish Immunol. 40, 99–108. 10.1016/j.fsi.2014.06.030 [DOI] [PubMed] [Google Scholar]
- Lokesh J., Fernandes J. M. O., Korsnes K., Bergh Ø., Brinchmann M. F., Kiron V. (2012). Transcriptional regulation of cytokines in the intestine of Atlantic cod fed yeast derived mannan oligosaccharide or β-Glucan and challenged with Vibrio anguillarum. Fish Shellfish Immunol. 33, 626–631. 10.1016/j.fsi.2012.06.017 [DOI] [PubMed] [Google Scholar]
- Løvoll M., Johnsen H., Boshra H., Bøgwald J., Sunyer J. O., Dalmo R. A. (2007). The ontogeny and extrahepatic expression of complement factor C3 in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 23, 542–552. 10.1016/j.fsi.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Magnadottir B. (2010). Immunological control of fish diseases. Mar. Biotechnol. 12, 361–379. 10.1007/s10126-010-9279-x [DOI] [PubMed] [Google Scholar]
- Martins F. S., Vieira A. T., Elian S. D., Arantes R. M., Tiago F. C., Sousa L. P., et al. (2013). Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect. 15, 270–279. 10.1016/j.micinf.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Milligan-Myhre K., Charette J., Phennicie R., Stephens W., Rawls J., Guillemin K., et al. (2011). Study of host–microbe interactions in zebrafish, in The Zebrafish: Disease Models and Chemicals Screens, eds Detrich W., Westerfield M., Zon L. (Cambridge, MA: Academic Press; ), 87–116. [Google Scholar]
- Millsap K. W., van der Mei H. C., Bos R., Busscher H. J. (1998). Adhesive interactions between medically important yeasts and bacteria. FEMS Microbiol. Ecol. 21, 321–336. 10.1111/j.1574-6976.1998.tb00356.x [DOI] [PubMed] [Google Scholar]
- Moslehi-Jenabian S., Pedersen L. L., Jespersen L. (2010). Beneficial effects of probiotic and food borne yeasts on human health. Nutrients 2, 449–473. 10.3390/nu2040449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete P., Tovar-Ramírez D. (2014). Use of yeasts as probiotics in fish aquaculture, in Sustainable Aquaculture Techniques, eds Hernandez-Vergara M., Pérez-Rostro C. (Rijeka: InTech; ), 135–172. [Google Scholar]
- Nguyen T. H., Fleet G. H., Rogers P. L. (1998). Composition of the cell walls of several yeast species. Appl. Microbiol. Biotechnol. 50, 206–212. 10.1007/s002530051278 [DOI] [PubMed] [Google Scholar]
- Ogunremi O. R., Sanni A. I., Agrawal R. (2015). Probiotic potentials of yeasts isolated from some cereal-based Nigerian traditional fermented food products. J. Appl. Microbiol. 119, 797–808. 10.1111/jam.12875 [DOI] [PubMed] [Google Scholar]
- O'Toole R., Lundberg S., Fredriksson S. A., Jansson A., Nilsson B., Wolf-Watz H. (1999). The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J. Bacteriol. 181, 4308–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole R., Von Hofsten J., Rosqvist R., Olsson P. E., Wolf-Watz H. (2004). Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microb. Pathog. 37, 41–46. 10.1016/j.micpath.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Rutz S., Crellin N. K., Valdez P. A., Hymowitz S. G. (2011). Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 29, 71–109. 10.1146/annurev-immunol-031210-101312 [DOI] [PubMed] [Google Scholar]
- Oyarbide U., Iturria I., Rainieri S., Pardo M. (2015). Use of gnotobiotic zebrafish to study Vibrio anguillarum pathogenicity. Zebrafish 12, 71–80. 10.1089/zeb.2014.0972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarbide U., Rainieri S., Pardo M. A. (2012). Zebrafish (Danio rerio) larvae as a system to test the efficacy of polysaccharides as immunostimulants. Zebrafish 9, 74–84. 10.1089/zeb.2011.0724 [DOI] [PubMed] [Google Scholar]
- Pamer E. (2016). Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352, 535–538. 10.1126/science.aad9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham L. N., Kanther M., Semova I., Rawls J. F. (2008). Methods for generating and colonizing gnotobiotic zebrafish. Nat. Protoc. 3, 1862–1875. 10.1038/nprot.2008.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier-Bres R., Munro P., Boyer L., Anty R., Imbert V., Terciolo C., et al. (2014). Saccharomyces boulardii modifies Salmonella typhimurium traffic and host immune responses along the intestinal tract. PLoS ONE 9:e103069. 10.1371/journal.pone.0103069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggi P., Lopez P., Diaz A., Carrasco D., Silva A., Velez A., et al. (2014). Debaryomyces hansenii and Rhodotorula mucilaginosa comprised the yeast core gut microbiota of wild and reared carnivorous salmonids, croaker and yellowtail. Environ. Microbiol. 16, 2791–2803. 10.1111/1462-2920.12397 [DOI] [PubMed] [Google Scholar]
- Ran C., Huang L., Hu J., Tacon P., He S., Li Z., et al. (2016). Effects of dietary live and heat-inactive baker's yeast on growth, gut health, and disease resistance of Nile tilapia under high rearing density. Fish Shellfish Immunol. 56, 263–271. 10.1016/j.fsi.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Ran C., Huang L., Liu Z., Xu L., Yang Y., Tacon P., et al. (2015). A Comparison of the beneficial effects of live and heat-inactivated baker's yeast on Nile Tilapia: suggestions on the role and function of the secretory metabolites released from the yeast. PLoS ONE 10:e0145448. 10.1371/journal.pone.0145448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauta P. R., Nayak B., Das S. (2012). Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol. Lett. 148, 23–33. 10.1016/j.imlet.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Rawls J. F., Mahowald M. A., Goodman A. L., Trent C. M., Gordon J. I. (2007). In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. U.S.A. 104, 7622–7627. 10.1073/pnas.0702386104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J. F., Samuel B. S., Gordon J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 101, 4596–4601. 10.1073/pnas.0400706101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendueles O., Ferrières L., Frétaud M., Bégaud E., Herbomel P., Levraud J. P., et al. (2012). A new zebrafish model of oro-intestinal pathogen colonization reveals a key role for adhesion in protection by probiotic bacteria. PLoS Pathogen. 8:e1002815. 10.1371/journal.ppat.1002815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Becerril M., Tovar-Ramírez D., Ascencia-Valle F., Civera-Cerecedo R., Gracia-López V., Barbosa-Solomieu V. (2008). Effects of dietary live yeast Debaryomyces hansenii on the immune and antioxidant system in juvenile leopard grouper Mycteroperca rosacea exposed to stress. Aquaculture 280, 39–44. 10.1016/j.aquaculture.2008.03.056 [DOI] [Google Scholar]
- Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E., Handley P. S. (2003). Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11, 94–100. 10.1016/S0966-842X(02)00034-3 [DOI] [PubMed] [Google Scholar]
- Rodríguez I., Chamorro R., Novoa B., Figueras A. (2009). β-Glucan administration enhances disease resistance and some innate immune responses in zebrafish (Danio rerio). Fish Shellfish Immunol. 27, 369–373. 10.1016/j.fsi.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Rojo O. M., de Ilarduya A., Estonba Pardo, M. A. (2007). Innate immune gene expression in individual zebrafish after Vibrio anguillarum inoculation. Fish Shellfish Immunol. 23, 1285–1293. 10.1016/j.fsi.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Schneider D. (2011). Tracing personalized health curves during infections. PLoS Biol. 9:e1001158. 10.1371/journal.pbio.1001158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S. L., Antunes L. C., Finlay B. B. (2010). Gut Microbiota in Health and Disease. Physiol. Rev. 90, 859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- Sepulcre M., Alcaraz-Pérez F., López-Muñoz A., Roca F., Meseguer J., Cayuela M., et al. (2009). Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-κB activation. J. Immunol. 182, 1836–1845. 10.4049/jimmunol.0801755 [DOI] [PubMed] [Google Scholar]
- Situmorang M. L., Dierckens K., Mlingi F. T., Van Delsen B., Bossier P. (2014). Development of a bacterial challenge test for gnotobiotic Nile tilapia Oreochromis niloticus larvae. Dis. Aquat. Organ. 109, 23–33. 10.3354/dao02721 [DOI] [PubMed] [Google Scholar]
- Stevens M. R., Luo T. L., Vornhagen J., Jakubovics N. S., Gilsdorf J. R., Marrs C. F., et al. (2015). Coaggregation occurs between microorganisms isolated from different environments. FEMS Microbiol. Ecol. 91:pii: fiv123. 10.1093/femsec/fiv123 [DOI] [PubMed] [Google Scholar]
- Tovar-Ramírez D., Zambonino-Infante J., Cahu C., Gatesoupe F. J., Vázquez-Juárez R. (2004). Influence of dietary live yeast on European sea bass (Dicentrarchus labrax) larval development. Aquaculture 234, 415–427. 10.1016/j.aquaculture.2004.01.028 [DOI] [Google Scholar]
- Tukmechi A., Rahmati Andani H. R., Manaffar R., Sheikhzadeh N. (2011). Dietary administration of β-mercapto-ethanol treated Saccharomyces cerevisiae enhanced the growth, innate immune response and disease resistance of the rainbow trout, Oncorhynchus mykiss. Fish Shellfish Immunol. 30, 923–928. 10.1016/j.fsi.2011.01.016 [DOI] [PubMed] [Google Scholar]
- van der Vaart M., Spaink H. P., Meijer A. H. (2012). Pathogen recognition and activation of the innate immune response in zebrafish. Adv. Hematol. 2012:159807. 10.1155/2012/159807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornhagen J., Stevens M., McCormick D. W., Dowd S. E., Eisenberg J. N., Boles B. R., et al. (2013). Coaggregation occurs amongst bacteria within and between biofilms in domestic showerheads. Biofouling 29, 53–68. 10.1080/08927014.2012.744395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Fei C., Wu H., Yang M., Liu Q., Wang Q., et al. (2013). Transcriptome profiling reveals Th17-like immune responses induced in zebrafish bath-vaccinated with a live attenuated Vibrio anguillarum. PLoS ONE 8:e73871. 10.1371/journal.pone.0073871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu H., Xiao J., Wang Q., Liu Q., Zhang Y. (2012). Immune responses of zebrafish (Danio rerio) induced by bath-vaccination with a live attenuated Vibrio anguillarum vaccine candidate. Fish Shellfish Immunol. 33, 36–41. 10.1016/j.fsi.2012.03.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.