Abstract
An appropriate inflammatory response plays critical roles in eliminating pathogens, whereas an excessive inflammatory response can cause tissue damage. Runt-related transcription factor 1 (RUNX1), a master regulator of hematopoiesis, plays critical roles in T cells; however, its roles in Toll-like receptor 4 (TLR4)-mediated inflammation in macrophages are unclear. Here, we demonstrated that upon TLR4 ligand stimulation by lipopolysaccharide (LPS), macrophages reduced the expression levels of RUNX1. Silencing of Runx1 attenuated the LPS-induced IL-1β and IL-6 production levels, but the TNF-α levels were not affected. Overexpression of RUNX1 promoted IL-1β and IL-6 production in response to LPS stimulation. Moreover, RUNX1 interacted with the NF-κB subunit p50, and coexpression of RUNX1 with p50 further enhanced the NF-κB luciferase activity. Importantly, treatment with the RUNX1 inhibitor, Ro 5-3335, protected mice from LPS-induced endotoxic shock and substantially reduced the IL-6 levels. These findings suggest that RUNX1 may be a new potential target for resolving TLR4-associated uncontrolled inflammation and preventing sepsis.
Keywords: inflammation, lipopolysaccharide (LPS), NF-κB (NF-KB), sepsis, Toll-like receptor 4 (TLR4), RUNX1, p50
Introduction
Toll-like receptor 4 (TLR4)4 signaling has critical roles in regulating the inflammation response against pathogens, especially Gram-negative bacteria (1). However, excessive inflammation can cause tissue damage, leading to sepsis and death. Sepsis often occurs in patients with infection and is the leading cause of death in hospitalized patients (2). It is accompanied by overly exuberant inflammatory responses (3). Previous studies have demonstrated that TLR4-deficient mice are resistant to lipopolysaccharides (LPS)-induced septic shock (4), and other surface receptors, including CD11b (5) and VEGFR-3 (6), also regulate TLR4 signaling-associated sepsis. Currently, exploring new effectors as potential targets to prevent sepsis are still under extensive investigation. Macrophages play critical roles in sepsis by regulating inflammation (7, 8). Macrophages can recognize pathogens through Toll-like receptors and produce various proinflammatory cytokines through transcription factor activation, such as nuclear factor κB (NF-κB) (9). The NF-κB family consists of five members: p50/p105, p52/p100, p65 (RelA), RelB, and c-Rel. They share an evolutionarily conserved Rel homology region and can form heterodimers or homodimers (10). The precursors of p50 subunit and p52 subunit are p105 and p100, respectively (11). However, both p50 and p52 lack a transcription activation domain, and they may inhibit transcription unless they interact with other NF-κB family members or other transcription factors containing a transcription activation domain (10). The p50-p65 heterodimer is the classical NF-κB family member (12).
The runx family, another group of key transcription factors, is composed of RUNX1 (also known as acute myeloid leukemia 1 (AML1)), RUNX2, and RUNX3 (13). In humans, RUNX1 is one of the genes most frequently altered by chromosome translocation and point mutations in acute myelogenous leukemia (14). In addition to participating in the regulation of cell cycle (15), proliferation (16), apoptosis (17), and ribosome biogenesis (18), RUNX1 is functionally related to the immune system. RUNX1 is critical in inducing the production of many genes in immune cells, such as IL-2 (19), IL-3 (20), macrophage colony-stimulating factor 1 receptor (CSF1R) (21), CSF2 (22), and CD4 (23). RUNX1 has been reported to be associated with autoimmune diseases, including rheumatoid arthritis (24) and systemic lupus erythematosus in humans (25). Previous studies suggest that RUNX1 mediates transactivation of Rorc to promote TH17 differentiation and interacts with RORγt to induce IL-17 production (26). Interestingly, RUNX1 is required for the optimal expression of Foxp3 in natural regulatory T cells and also interacts physically with FOXP3 (27). Moreover, RUNX1 deficiency in CD4+ T cells causes lethal lung inflammation (28). Although a role for RUNX1 has been suggested in T cells, it remains unclear about how RUNX1 regulates TLR4-mediated inflammation in macrophages.
Previous studies reported that RUNX1 could promote STAT3 activation and inhibit the expression of SOCS3/4 in epithelial cancer (29). The SOCS family, especially SOCS3/4, plays a negative regulatory role in TLR4-mediated inflammation (30). In this study, we asked whether RUNX1 played vital roles in TLR4-mediated inflammation. We found that RUNX1 interacted with the NF-κB subunit p50 to enhance the production of inflammatory cytokines in macrophages, such as IL-1β and IL-6, and a RUNX1 inhibitor, Ro 5-3335, could protect mice from LPS-induced shock. Our data identify RUNX1 as a positive regulator in macrophage inflammatory responses via the p50 pathway.
Results
Knockdown of Runx1 Expression Reduces the Production of Proinflammatory Cytokines in LPS-stimulated Macrophages
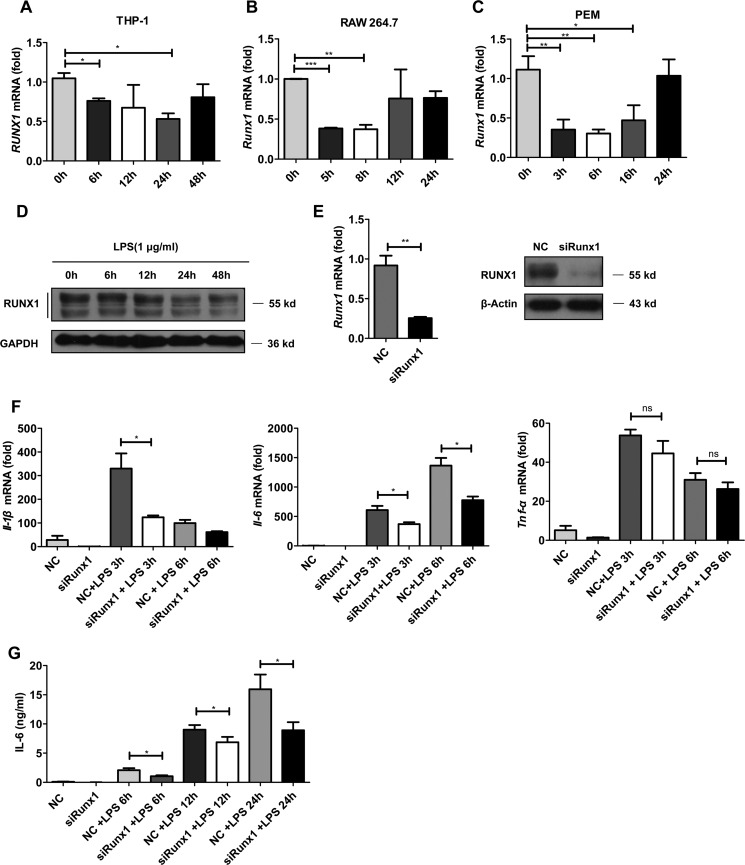
To explore the role of RUNX1 in macrophages, we first examined whether RUNX1 expression was regulated in response to TLR4 agonist (LPS) stimulation at the mRNA and protein levels. RUNX1 was expressed in the human monocytic cell line, THP-1, mouse macrophage-like cell line, RAW264.7, and primary mouse peritoneal macrophages (PEMs) (Fig. 1, A–C). The purity of CD11b+ F4/80+ PEMs was analyzed by FACS (supplemental Fig. S1). In response to LPS stimulation, RUNX1 expression at the mRNA level was significantly reduced in all these cells (Fig. 1, A–C, i.e. ∼6 h). We also detected a substantial decrease of RUNX1 at the protein level in these cells following LPS treatment (Fig. 1D, supplemental Fig. S2, A–B). To determine whether other stimuli affect the expression of Runx1 in macrophages, macrophages were stimulated with IL-1β, IL-6, or TNF-α. However, these cytokines did not modulate Runx1 mRNA and protein expression in PEMs (supplemental Fig. S3). These results demonstrate that RUNX1 is constitutively expressed in macrophages, and TLR4 activation decreases RUNX1 levels in macrophages.
FIGURE 1.
Knockdown of Runx1 expression inhibits proinflammatory cytokine production in LPS-stimulated macrophages. THP-1 cells (A and D), RAW 264.7 cells (B), and PEMs (C) were stimulated with LPS at different doses for the indicated time points. The RUNX1 expression levels were detected by immunoblotting or RT-qPCR. The data are shown as the mean ± S.D. of a representative experiment. E–G, PEMs were transfected with Runx1 siRNA or nonspecific siRNA using the Lipofectamine RNAiMAX for 48 h, and the knockdown efficiency was measured by RT-qPCR and immunoblotting (E). These PEMs were then stimulated with 1 μg/ml of LPS for the indicated times. The Il-1β, Il-6, and Tnf-α mRNA levels were measured by RT-qPCR (F), and the supernatants IL-6 levels were determined by ELISA (G). The data are shown as the mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To study the role of RUNX1 in primary macrophages, we used specific siRNA to knockdown Runx1 in PEMs. The mRNA levels of Runx1 decreased by 70% in the siRunx1-transfected PEMs compared with the negative controls (Fig. 1E). The knockdown efficiency was also confirmed by Western blot analysis (Fig. 1E). Next, we analyzed the effects of Runx1 silencing on the production of inflammatory cytokines in macrophages. In response to LPS, the siRunx1-transfected PEMs produced diminished inflammatory cytokine expression at the mRNA level, including Il-1β and Il-6, but not Tnf-α, compared with the controls (Fig. 1F). We also examined IL-6 concentrations using an ELISA kit, and we confirmed that Runx1-silenced PEMs produced lower IL-6 levels than control cells at the indicated times (6, 12, and 24 h) (Fig. 1G). In addition to Il-1β or Il-6, the mRNA levels of other LPS-dependent genes including Il-12b, Csf2, and Ccl3 were also decreased in the siRunx1-transfected PEMs (supplemental Fig. S4, A–C).
Regulation of IL-6 and IL-1β Production by RUNX1 Overexpression or Treatment with a RUNX1 Inhibitor
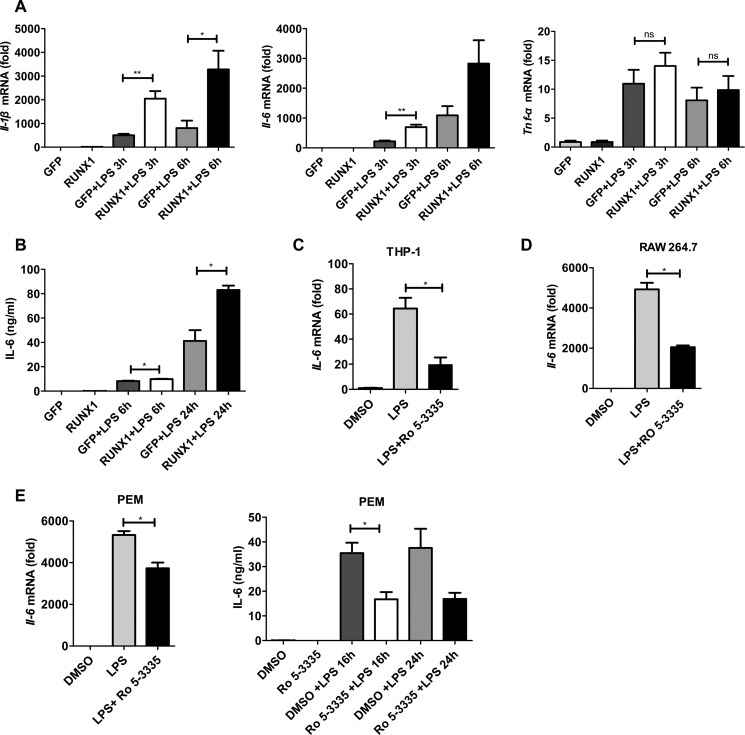
To further confirm the effect of RUNX1 on TLR4-induced production of proinflammatory cytokines, we overexpressed RUNX1 in RAW 264.7 cells. Upon LPS treatment, RAW 264.7 cells that overexpressed RUNX1 had profoundly enhanced Il-6 and Il-1β expression levels, but not Tnf-α, compared with control cells that overexpressed GFP (Fig. 2A). Additionally, RUNX1 overexpression also induced RAW 264.7 cells to secrete more IL-6 into the supernatant, as detected by ELISA (Fig. 2B).
FIGURE 2.
Regulation of IL-6 and IL-1β production by RUNX1 overexpression or treatment with a RUNX1 inhibitor. A and B, RUNX1-GFP or a GFP vector control (6 μg) were co-transfected with pCL-10A1 (6 μg) into 293T cells. The retroviral supernatants were harvested and used to infect RAW264.7 cells, which was followed by FACS sorting of the GFP+ cells. These stable RAW264.7 cells that overexpressed RUNX1 or GFP were stimulated with 100 ng/ml of LPS for the indicated times. The Il-1β, Il-6, and Tnf-α mRNA levels were measured with RT-qPCR (A), and the IL-6 levels in the supernatants were determined with ELISA (B). The data are shown as the mean ± S.E. of three independent experiments. THP-1 cells (C), RAW 264.7 cells (D), and PEMs (E) were stimulated with LPS (100 or 1000 ng/ml) for 4 h in the absence or presence of the RUNX1 inhibitor, Ro 5-3335 (50 μm). IL-6 levels were measured by RT-qPCR or ELISA. The data are shown as the mean ± S.D. of a representative of three independent experiments. *, p < 0.05; **, p < 0.01.
As the RUNX1 inhibitor, Ro 5-3335 has been previously well described (31), we next examined whether it could inhibit TLR4-triggered production of proinflammatory cytokines in macrophages. As shown in Fig. 2, C–E, Ro 5-3335 substantially blocked the expression of Il-6 at the mRNA and protein level in THP-1 cells, RAW 264.7 cells, and PEMs. We also found that Ro 5-3335 could inhibit the expression of Il-6 in PEMs stimulated with the TLR2 agonist, PGN, but not the TLR3 agonist, poly(I:C), or the TLR9 agonist, CpG (supplemental Fig. S5). These findings indicate that RUNX1 might positively regulate the TLR4 signaling pathway and proinflammatory cytokine production in macrophages.
RUNX1 Synergizes with the NF-κB Subunits to Enhance TLR4-triggered Inflammation
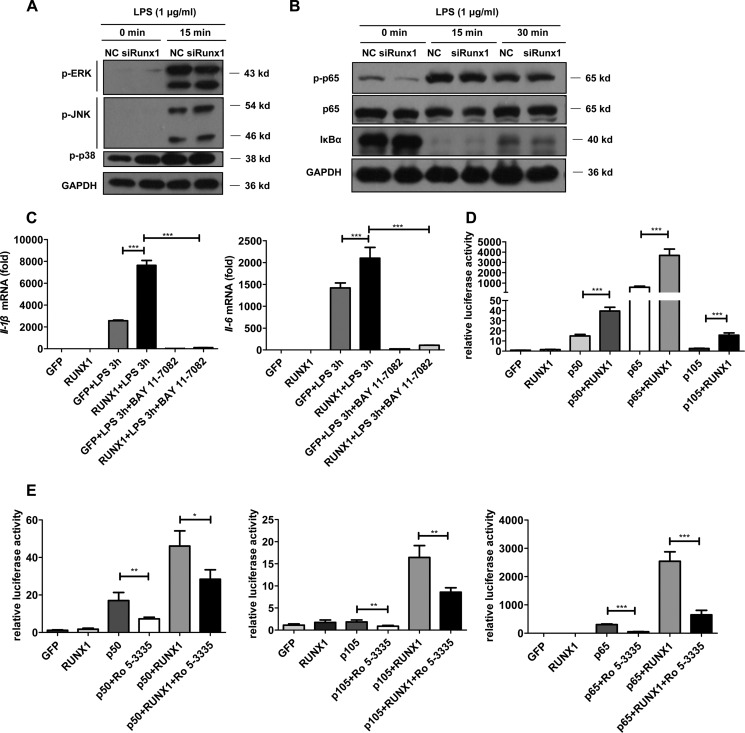
MAPKs and NF-κB signaling have critical roles in the induction of inflammatory cytokine production (32, 33). To reveal the function of RUNX1 in the TLR4 signaling pathway, we first analyzed the phosphorylation levels of MAPKs and NF-κB. Fig. 3, A–B, shows that Runx1 knockdown did not affect the ERK, JNK, p38, or p65 phosphorylation levels in PEMs. Additionally, we did not detect IκBα degradation differences between the Runx1-silenced and wild type PEMs (Fig. 3B). Moreover, the RUNX1 inhibitor, Ro 5-3335, did not affect p65 phosphorylation levels or IκBα degradation in PEMs (supplemental Fig. S6).
FIGURE 3.
RUNX1 synergizes with the NF-κB subunits to enhance TLR4-triggered inflammation. A and B, PEMs were transfected with Runx1 siRNA or nonspecific siRNA, then stimulated with 1 μg/ml of LPS for the indicated times. Cell lysates were extracted for immunoblotting to detect the phosphorylated ERK, P38, JNK, or p65 levels as well as the total IκBα and p65 levels. The data are shown as a representative of two independent experiments. C, the stable RAW 264.7 cell lines that overexpressed RUNX1 or GFP were stimulated with 100 ng/ml of LPS for 3 h in the presence of the NF-κB inhibitor, 10 μm BAY 11-7082, followed by detection of Il-1β and Il-6 by RT-qPCR. D and E, plasmids expressing RUNX1 or GFP were co-transfected with plasmids expressing p50, p65, or p105 into 293T cells in the presence of an NF-κB luciferase reporter plasmid and a Renilla luciferase plasmid. After 24 h, the cell lysates were assayed with a dual luciferase reporter assay system (D). Alternatively, after 8 h, Ro 5-3335 (50 μm) or DMSO was added for 18 h to examine the luciferase readings (E). The data are shown as the mean ± S.E. of at least two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Because RUNX1 is predominantly located in the nucleus (34), it is possible that RUNX1 might physically interact with or cooperate with other transcription factors in the nuclei to regulate inflammation. In agreement with this speculation, we observed that treatment with the NF-κB inhibitor, BAY 11-7082, strictly abrogated LPS-induced IL-6 and IL-1β production in the GFP control or RUNX1-overexpressing macrophages (Fig. 3C). Next, we examined the effect of RUNX1 on NF-κB activation using a dual-luciferase reporter assay. Although RUNX1 expression alone did not induce activation of the reporter gene in 293T cell, co-expression of RUNX1 and the NF-κB family members, p105, p50, or p65, significantly increased the NF-κB reporter activity compared with co-expression of the GFP control (Fig. 3D). The RUNX1 inhibitor, Ro 5-3335, partly reversed the synergized effect between RUNX1 and p50, p65, or p105 (Fig. 3E).
RUNX1 Interacts with the p50 Subunit of NF-κB
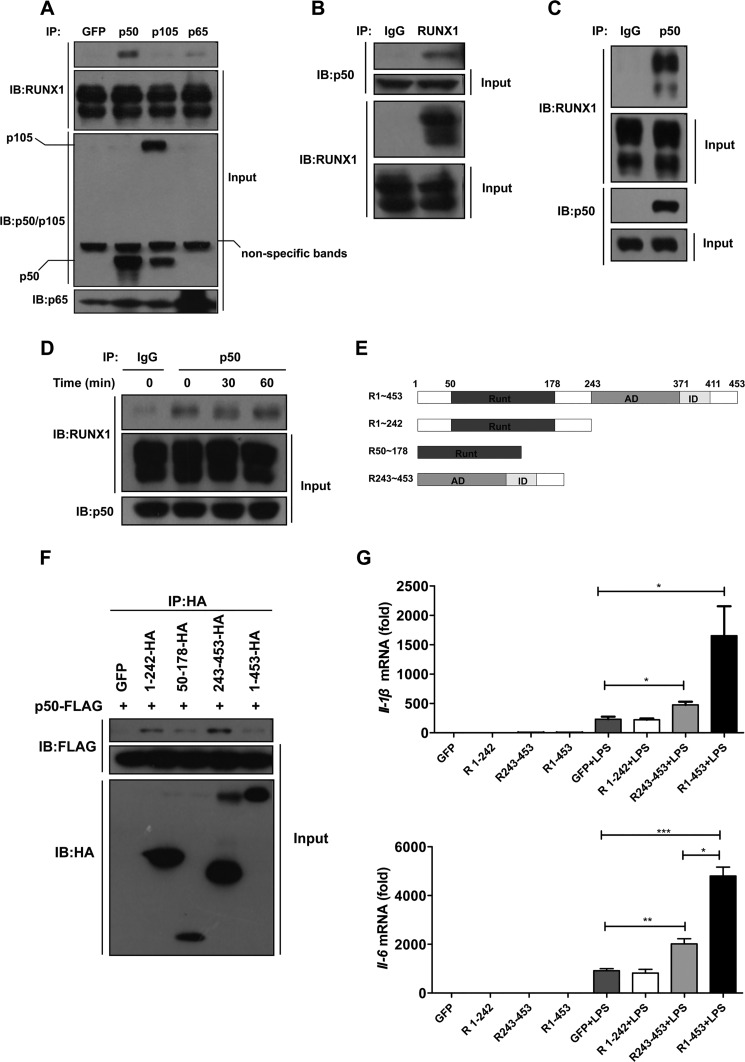
We next investigated whether RUNX1 could interact with p105, p50, or p65. 293T cells were transiently transfected with RUNX1 together with p105, p50, p65, or the GFP-myc vector control. Cell lysates were immunoprecipitated with anti-p50/p105, p65, or anti-Myc antibodies. We noticed that RUNX1 preferred to be coprecipitated with p50, compared with the other NF-κB family members including p105 and p65 (Fig. 4A). Immunoprecipitation using an anti-RUNX1 antibody was also performed, which indeed pulled down p50 (Fig. 4B). Next, we asked whether RUNX1 interacted with the endogenous p50, and confirmed the endogenous interaction between RUNX1 and p50 in THP-1 cells (Fig. 4C). The interaction between RUNX1 and p50 was not significantly changed by LPS treatment in RAW 264.7 at early time points (30 or 60 min), which overexpressed RUNX1 (Fig. 4D). These findings suggest that RUNX1 might regulate TLR4 signaling and proinflammatory cytokine production by binding to p50.
FIGURE 4.
RUNX1 interacts with the p50 subunit of NF-κB. A, RUNX1 was co-transfected with plasmids expressing GFP-myc, p50, p105, or p65 into 293T cells. The cell lysates were immunoprecipitated (IP) with the indicated antibodies followed by immunoblotting (IB) with an anti-RUNX1 antibody. B, 293T cells were co-transfected with p50 and RUNX1, and cell extracts were immunoprecipitated with an anti-RUNX1 antibody, followed by immunoblotting with the indicated antibodies. C, THP-1 cell lysates were immunoprecipitated with an anti-p50 antibody followed by immunoblotting with an anti-RUNX1 antibody to detect the endogenous interaction between RUNX1 and p50. D, RAW 264.7 cells that overexpressed RUNX1 were stimulated with LPS for 30 and 60 min, then cell lysates were immunoprecipitated with an anti-p50 antibody followed by immunoblotting with an anti-RUNX1 antibody. E, the RUNX1 truncations are listed (Runt, Runt domain; AD, activation domain; ID, inhibition domain). F, 293T cells were transiently co-transfected with p50-FLAG and HA-tagged RUNX1 truncations (R1∼242, R50–178, R243–453 or R1–453), then cell lysates were immunoprecipitated with an anti-HA antibody, followed by immunoblotting with an anti-FLAG antibody. The results are shown as a representative of three independent experiments. G, the stable RAW264.7 cells overexpressing GFP or the different RUNX1 truncations (R1∼242, R243–453, or R1–453) were stimulated with 100 ng/ml of LPS for 6 h. The Il-1β and Il-6 mRNA levels were measured with RT-qPCR. The data are shown as the mean ± S.E. of two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We further investigated whether RUNX1 could affect the recruitment of p50 to the Il-6 promoter using a ChIP analysis. Consistent with previous reports (35, 36), the levels of p50 on the Il-6 promoter were unchanged in macrophages in response to LPS treatment, and the RUNX1 inhibitor did not affect the recruitment of p50 to the Il-6 promoter (supplemental Fig. S7, left panel). However, the RUNX1 inhibitor greatly reduced the levels of RNA polymerase II binding to the Il-6 promoter (supplemental Fig. S7, right panel). These results support the role of RUNX1 in regulating IL-6 transcript in macrophages after LPS stimulation.
We then sought to determine which domain of RUNX1 could interact with p50. Different truncations of RUNX1 were constructed, including the N-terminal fragment (R1–242), the C-terminal fragment (R243–453), and the Runt domain (R50–178) (Fig. 4E). We noticed that all of these truncations could interact with p50 (Fig. 4F). This interaction is similar to the interaction between RUNX1 and Ets-1; both N-terminal and C-terminal fragments of RUNX1 can bind to Ets-1 (37). To further determine which domain of RUNX1 was responsible for NF-κB activation, we overexpressed different truncations of RUNX1 in RAW 264.7 cells. Following LPS stimulation, the C-terminal fragment and the full-length RUNX1 showed enhanced IL-6 expression, but the N-terminal fragment showed no significant effect (Fig. 4G). These results indicate that RUNX1 is dependent on its C-terminal region to augment TLR4-NF-κB activity.
A RUNX1 Inhibitor Protects against LPS-induced Septic Shock in Vivo
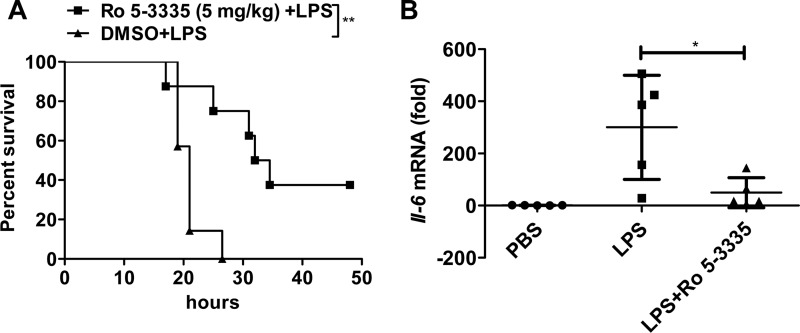
Excessive TLR4-NF-κB activation is critical for the induction of sepsis that is accompanied by exacerbated inflammatory responses (6, 7). To investigate the in vivo role of RUNX1 in inflammation, we utilized a murine endotoxic shock model via intraperitoneal injection of a lethal dose of LPS. LPS induced death in over 80% of mice at 20 h, whereas the RUNX1 inhibitor, Ro 5-3335, significantly improved the mouse survival rates at 20 h, 48 h, or longer time points when used at 5 mg/kg (Fig. 5A). We further examined the expression of proinflammatory cytokines in various organs and serum in these mice. Unexpectedly, we observed no significant differences in the Il-1β, Il-6, or Tnf-α mRNA levels in the liver, spleen, lungs, or serum IL-6 concentrations at 3 h after the Ro 5-3335 intervention when compared with the DMSO control mice.5 However, the Il-6 mRNA levels were selectively decreased in the kidneys of the treated mice at a later stage (i.e. 24 h) (Fig. 5B). These results indicate that the RUNX1 inhibitor may resolve IL-6 production in the kidneys and protect against LPS-induced septic shock.
FIGURE 5.
A RUNX1 inhibitor protects against LPS-induced septic shock in vivo. LPS (20 mg/kg) was used to induce septic shock in C57BL/6 mice in the presence or absence of the RUNX1 inhibitor, Ro 5-3335. The survival rates of these mice were recorded (A), and the Il-6 mRNA levels in the kidneys were detected by RT-qPCR (B). The data are shown as the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01.
Discussion
Excessive TLR4-NF-κB signaling is closely related to uncontrolled inflammation, tissue damage, and septic shock (38, 39) and is critical to identifying new key regulators in the TLR4-NF-κB pathway, as they could be potential targets that prevent sepsis. RUNX1 is a master transcription factor in hematopoiesis (40) and plays indispensable roles in T cells and autoimmune diseases (28, 41, 42). However, its role is still unknown in the regulation of TLR4-NF-κB signaling in macrophages.
In the current study, we found that RUNX1 mRNA and protein expression levels decreased following LPS stimulation in macrophages. Silencing of Runx1 and treatment with the RUNX1 inhibitor, Ro 5-3335, attenuated the expression of Il-6 and Il-1β, but not Tnf-α, in mouse peritoneal macrophages, RAW 264.7 cells, and human THP-1 cells. In agreement with this, RUNX1 overexpression enhanced the production of TLR4-induced IL-6 and IL-1β. These findings indicate that RUNX1 is a positive regulator of the TLR4 pathway for the induction of the inflammatory response. Our study is consistent with previous findings that RUNX1 could promote STAT3 phosphorylation by inhibiting SOCS3/4 expression in epithelial cancer cells (29), and STAT3 is a major signal component in IL-6 signaling (43). Although another recent study reported that RUNX1 inhibits NF-κB signaling by blocking the activity of the IκB kinase in leukemia (44), we detected no differences in IκBα degradation and p65 phosphorylation after silencing Runx1 or inhibition of RUNX1 activity in LPS-treated murine primary macrophages. These contradictory observations might be due to the different cell types that were used and treated with different experimental methods.
In our study, we also determined whether RUNX1 affected expression of the MyD88-independent genes, such as Ifn-β and Ccl5. No significant difference of their expression was detected at the mRNA levels in RAW 264.7 cells overexpressed by either GFP or RUNX1 (supplemental Fig. S8, A and B). In agreement with this, RUNX1 did not affect the IRF3 phosphorylation levels (supplemental Fig. S8, C and D). These findings suggest that RUNX1 may not affect type I interferon production.
In agreement with the finding that RUNX1 is located predominantly in the nucleus (34), coexpression of RUNX1 with NF-κB family members p50, p105, or p65, enhanced the NF-κB luciferase activity. However, we observed that RUNX1 specifically interacted with p50, but not p65, and LPS did not affect the interaction between RUNX1 and p50 at early time points. This might explain why RUNX1 did not affect the Tnf-α expression levels in macrophages, since p50-deficient macrophages induce normal Tnf-α expression in response to LPS stimulation (45). Previous studies have demonstrated that p50 lacks a transcriptional activation domain (10, 46), and p50 needs to interact with other transcription factors or transcription coactivators, including p65/RelA, RelB, C-Rel, BCL3, and IκBζ (10), for the induction of its downstream target genes. Our study provides the first evidence that RUNX1 binds to p50 and synergizes as a transcriptional coactivator for the production of IL-6 and IL-1β in macrophages. As noticed, the C-terminal fragment of RUNX1 was responsible for NF-κB activation. Since the C-terminal fragment of RUNX1 contains the activation domain, it suggests that p50 might coordinate with the RUNX1 activation domain to activate gene expression. Interestingly, RUNX1a, an isoform of RUNX1, contains a highly conserved N-terminal fragment of RUNX1 (i.e. 1–242 amino acids) (13). RUNX1a promotes hematopoietic lineage commitment from human pluripotent stem cell (47) and was overexpressed in acute leukemia (48). Because NF-κB plays critical roles in hematopoietic differentiation and leukemia (49, 50), it is interesting to ask whether RUNX1a might bind to p50 to regulate hematopoietic differentiation and acute leukemia. Nevertheless, no ortholog of human RUNX1a exists in mice (51). Additionally, our data indicates that the N-terminal fragment of RUNX1 was not responsible for NF-κB activation in macrophages. This suggests that RUNX1a cannot compromise the role of RUNX1 in macrophages for the interaction with p50.
Although RUNX1 did not affect the recruitment of p50 to the Il-6 promoter, it is indispensable for the recruitment of RNA polymerase II to the Il-6 promoter. This indicates that RUNX1 might act as a transcriptional coactivator of p50 for the production of IL-6 in macrophages.
Another implication of our findings is the in vivo function of the RUNX1 inhibitor, Ro 5-3335, in preventing LPS-induced septic shock. The RUNX1 inhibitor, Ro 5-3335, substantially improved the survival rates in the LPS-induced sepsis model. Indeed, we observed a significant reduction of Il-6 levels at the later stage in mouse kidneys after the Ro 5-3335 treatment. This may be due to the facts that IκBζ binds to p50 and that this is dependent on p50 to selectively promote Il-6 transcription, not Tnf-α, in macrophages (35). As septic shock is the leading cause of acute kidney injury and IL-6 is a vital mediator of acute kidney injury (52, 53), we propose that the RUNX1 inhibitor may protect against LPS-induced septic shock by attenuating IL-6 production and blocking acute kidney injury. RUNX1 might be a new potential therapeutic target for resolving TLR4 signaling that is related to acute excessive inflammation or septic shock.
Experimental Procedures
Reagents and Mice
Antibodies against RUNX1, p50/p105, p65, and IκBα were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against p-p65, p-p38, p-ERK, p-JNK, and p-IRF3 were obtained from Cell Signaling Technology (Danvers, MA). Antibodies against GAPDH were obtained from Beyotime Institute of Biotechnology (Shanghai, China). Anti-HA, anti-Myc, anti-β-Actin-HRP, and anti-FLAG antibodies were purchased from Sigma. Anti-CD11b-FITC and anti-F4/80-APC antibodies, as well as ELISA kits for IL-6 were from eBioscience (San Diego, CA). ChIP-grade antibody against p50 was purchased from Abcam (Cambridge, UK). The EZ-ChIPTM Chromatin Immunoprecipitation Kit was obtained from Merck Millipore (Billerica, MA). Murine IL-1β, IL-6, and TNF-α were obtained from PeproTech (Rocky Hill, NJ). PGN, CpG, and poly(I:C) were purchased from InvivoGen (San Diego, CA). The RUNX1 inhibitor, Ro 5-3335, was purchased from Tocris Bioscience (Bristol, UK). The NF-κB inhibitor, BAY 11-7082, was purchased from Beyotime Institute of Biotechnology. Highly purified LPS from Escherichia coli O55:B5 were obtained from Sigma. Male C57BL/6 wild type mice (8–10 weeks old) were purchased from the Model Animal Research Center of Nanjing University, Nanjing, China.
Cell Culture
Primary mouse PEMs from C57BL/6 wild type mice were prepared as described previously (6). PEMs, RAW 264.7 cells (kind gifts from Dr. B. Sun, SIBCB, CAS, Shanghai, China), and 293T cells (kind gifts from Dr. J. F. Chen, SIBCB, CAS, Shanghai, China) were cultured with complete DMEM containing 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml). Human THP-1 cells (kind gifts from Dr. B. Ge, Tongji University, Shanghai, China) were maintained in complete 1640 medium supplemented with 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml). For macrophage induction, THP-1 cells were stimulated with 100 ng/ml of phorbol 12-myristate 13-acetate for 24 h and allowed to rest for 8–12 h. PEMs were obtained from mice that received an intraperitoneal injection of 2.5–3 ml of 3% Brewer thioglycollate medium for 4 days, and the purity of CD11b+F4/80+ macrophages was analyzed by FACS (supplemental Fig. S1).
Plasmids
The MigR1-RUNX1 plasmid was a gift from Dr. Shi Jingyi (Shanghai Institute of Hematology). HA-tagged RUNX1 and various RUNX1 truncations (54) (R1∼242, R50–178, and R243–453) were inserted into the XhoI-EcoRI site of the mammalian expression vector, pcDNA3.1 or MigR1. Human p65 (a kind gift from Dr. Z Zhou, SIBCB, China), p50, and p105 were also inserted into the XhoI-EcoRI site of the vector, pcDNA3.1. The primers are listed in the supplemental information (supplemental Table S1).
Retroviral Transduction
To generate retroviral particles, MigR1-RUNX1/RUNX1 truncations (R1∼242, R243–453)-GFP (6 μg) were co-transfected with pCL-10A1 (6 μg) into 293T cells by the calcium phosphate method. After 24 h, the retroviral supernatants were collected to infect RAW264.7 cells, and the cells were passaged three times, followed by flow cytometry sorting of GFP+ cells.
siRNA Transfection and RT-qPCR
PEMs (5 × 105) were transfected with 20 nm siGENOME SMARTpool mouse Runx1 siRNA (Dharmacon, Lafayette, CO) or with 20 nm nonspecific siRNA using the Lipofectamine RNAiMAX kit (Invitrogen) according to the manufacturer's instructions. The siRNA sequences are listed in the supplemental information (supplemental Tables S2 and S3). Total RNA was extracted with RNAiso reagent (TaKaRa Ltd, Kyoto, Japan). cDNA was generated from 1 μg of RNA using Moloney MLV transcriptase (Promega) and examined with quantitative RT-qPCR with SYBR Green Master Mix on a CFX-96 machine (Bio-Rad). siRNA sequences or the primers for detecting Il-1β, Il-6, and Tnf-α were used as described previously (6), and are listed in supplemental Table S4. To determine the specificity of siRNA, expression of the unrelated proteins, including Wdr7, Asap1, Naip2, and Vps13b, were detected by RT-qPCR (supplemental Fig. S9).
Cell Stimulation and Cytokine ELISA
PEMs, RAW 264.7, and phorbol 12-myristate 13-acetate-differentiated THP-1 cells were seeded in 12-well plates and stimulated with LPS (100 or 1000 ng/ml) for 3, 6, 12, or 24 h in the absence or presence of 50 μm Ro 5-3335 or 10 μm BAY 11–7082, which was added as a pretreatment for 1 h. The supernatants were collected for cytokine analysis using a commercial ELISA kit according to the manufacturer's instructions.
Luciferase Assay
293T (5 × 105) cells were co-transfected with 0.1 μg of expression plasmid (p50, p65, or p105) and 0.25 μg of NF-κB luciferase reporter plasmid containing 1/20 Renilla luciferase plasmid with 0.8 μg of pcDNA3.1-RUNX1 or pcDNA3.1-GFP plasmid using the Lipofectamine 2000 reagent. The cells were recovered for 24 h and measured with a dual luciferase reporter assay system (Promega). For the Ro 5-3335 intervention experiment (31), briefly, 293T cells were transiently co-transfected with 0.1 μg of expression plasmids encoding p50, p105, or p65 plus 0.8 μg of pcDNA3.1-RUNX1. After 6–8 h, the medium was removed and 50 μm Ro 5-3335 or DMSO was added into the new medium. Then, 18 h later, the cell lysates were assayed with an NF-κB luciferase assay.
Coimmunoprecipitation and Western Blot Analysis
293T cells (5 × 106) were co-transfected with 6 μg of pcDNA3.1-p50/p105/p65 plasmid and 6 μg of pcDNA3.1-RUNX1-HA or HA-tagged various RUNX1 truncations, and the whole cell lysates were collected to perform co-immunoprecipitation experiments with the indicated antibodies. The endogenous interactions between RUNX1 and p50 were examined by co-immunoprecipitation in RAW 264.7 or THP-1 cells. PEMs, THP-1, or RAW 264.7 cells were seeded in 12-well plates and stimulated with LPS (1000 ng/ml) for 3, 6, 12, or 24 h, and the whole cell lysates were subjected to immunoblotting with anti-RUNX1 antibodies.
Chromatin Immunoprecipitation
RAW 264.7 cells were stimulated with 100 ng/ml of LPS for 5 h in the absence or presence of 50 μm Ro 5-3335. ChIP experiments were performed with the EZ-ChIPTM kit according to the manufacturer's instructions. To measure enrichment, the purified DNA was quantified by qPCR with the primers covering NF-κB binding site of the murine Il-6 promoter used as described previously (55). The primers are listed in the supplemental information (supplemental Table S5).
LPS Shock Model and Ro 5-3335 Treatment
Male C57BL/6 mice (8–10 weeks old) received an intraperitoneal injection of the RUNX1 inhibitor, Ro 5-3335 (5 mg/kg), with 5% DMSO in PBS or 5% DMSO in PBS for 3 h, following by an intraperitoneal injection with a lethal (20 mg/kg) or sublethal dose (10 mg/kg) of LPS to induce LPS shock as described previously (56). All the mice were observed and the survival rates were calculated. Serum, liver, lung, kidney, and spleen samples were collected at the indicated time points for cytokine measurements.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 5, and statistically significant differences were determined by a two-tailed Student's t test with 95% confidence intervals.
Author Contributions
M.-C. L. and S.-Y. Z. performed the experiments and statistical analysis. D.-Y. F., J. X., C.-D. X., and W.-Y. L. helped with the experiments. H.-Y. W., T. Z., and M.-C. L. designed the study. H.-Y. W., M.-C. L., T. Z., and S.-Y. Z. prepared the manuscript.
Supplementary Material
This work was supported in part by Ministry of Science and Technology of China Grants 2016YFD05002007, 2016YFC0902200, and 2012CB910800, Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB19000000, National Natural Science Foundation of China Grants 81270801, 81470941, 31422018, 81571617, and 81570508, and Instrument Developing Project of the Chinese Academy of Sciences Grant YZ201339. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S9 and Tables S1–S5.
M. Luo, S. Zhou, D. Feng, J. Xiao, W. Li, C. Xu, H. Wang and T. Zhou, unpublished data.
- TLR4
- Toll-like receptor 4
- BCL3
- B-cell CLL/lymphoma 3
- C-Rel
- v-rel avian reticuloendotheliosis viral oncogene homolog
- IκBζ
- nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor ζ
- IRF3
- interferon regulatory factor 3
- NF-κB
- nuclear factor-light-chain enhancer of activated B cell
- PEMs
- primary mouse peritoneal macrophages
- RelB
- v-rel avian reticuloendotheliosis viral oncogene homolog B
- RUNX1
- Runt-related transcription factor 1
- SOCS
- suppressor of cytokine signaling
- qPCR
- quantitative PCR.
References
- 1. Brubaker S. W., Bonham K. S., Zanoni I., and Kagan J. C. (2015) Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deutschman C. S., and Tracey K. J. (2014) Sepsis: current dogma and new perspectives. Immunity 40, 463–475 [DOI] [PubMed] [Google Scholar]
- 3. Angus D. C., and van der Poll T. (2013) Severe sepsis and septic shock. N. Engl. J. Med. 369, 840–851 [DOI] [PubMed] [Google Scholar]
- 4. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., and Akira S. (1999) Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 5. Han C., Jin J., Xu S., Liu H., Li N., and Cao X. (2010) Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol 11, 734–742 [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y., Lu Y., Ma L., Cao X., Xiao J., Chen J., Jiao S., Gao Y., Liu C., Duan Z., Li D., He Y., Wei B., and Wang H. (2014) Activation of vascular endothelial growth factor receptor-3 in macrophages restrains TLR4-NF-κB signaling and protects against endotoxin shock. Immunity 40, 501–514 [DOI] [PubMed] [Google Scholar]
- 7. Shalova I. N., Lim J. Y., Chittezhath M., Zinkernagel A. S., Beasley F., Hernández-Jiménez E., Toledano V., Cubillos-Zapata C., Rapisarda A., Chen J., Duan K., Yang H., Poidinger M., Melillo G., Nizet V., et al. (2015) Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity 42, 484–498 [DOI] [PubMed] [Google Scholar]
- 8. Xia M., Liu J., Wu X., Liu S., Li G., Han C., Song L., Li Z., Wang Q., Wang J., Xu T., and Cao X. (2013) Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 39, 470–481 [DOI] [PubMed] [Google Scholar]
- 9. Aderem A. (2001) Role of Toll-like receptors in inflammatory response in macrophages. Crit. Care Med. 29, S16–S18 [DOI] [PubMed] [Google Scholar]
- 10. Vallabhapurapu S., and Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 11. Xiao G., Harhaj E. W., and Sun S. C. (2001) NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 12. Urban M. B., Schreck R., and Baeuerle P. A. (1991) NF-kappa B contacts DNA by a heterodimer of the p50 and p65 subunit. EMBO J. 10, 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ito Y., Bae S. C., and Chuang L. S. (2015) The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer 15, 81–95 [DOI] [PubMed] [Google Scholar]
- 14. Osato M. (2004) Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 23, 4284–4296 [DOI] [PubMed] [Google Scholar]
- 15. Bernardin-Fried F., Kummalue T., Leijen S., Collector M. I., Ravid K., and Friedman A. D. (2004) AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J. Biol. Chem. 279, 15678–15687 [DOI] [PubMed] [Google Scholar]
- 16. Zhang L., Fried F. B., Guo H., and Friedman A. D. (2008) Cyclin-dependent kinase phosphorylation of RUNX1/AML1 on 3 sites increases transactivation potency and stimulates cell proliferation. Blood 111, 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wildey G. M., and Howe P. H. (2009) Runx1 is a co-activator with FOXO3 to mediate transforming growth factor β (TGFβ)-induced Bim transcription in hepatic cells. J. Biol. Chem. 284, 20227–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai X., Gao L., Teng L., Ge J., Oo Z. M., Kumar A. R., Gilliland D. G., Mason P. J., Tan K., and Speck N. A. (2015) Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17, 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong W. F., Kurokawa M., Satake M., and Kohu K. (2011) Down-regulation of Runx1 expression by TCR signal involves an autoregulatory mechanism and contributes to IL-2 production. J. Biol. Chem. 286, 11110–11118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uchida H., Zhang J., and Nimer S. D. (1997) AML1A and AML1B can transactivate the human IL-3 promoter. J. Immunol. 158, 2251–2258 [PubMed] [Google Scholar]
- 21. Zhang D. E., Hetherington C. J., Meyers S., Rhoades K. L., Larson C. J., Chen H. M., Hiebert S. W., and Tenen D. G. (1996) CCAAT enhancer binding protein (C/EBP) and AML1 (CBFA2) synergistically activate the M-CSF receptor promoter. Mol. Cell Biol. 16, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frank R., Zhang J., Uchida H., Meyers S., Hiebert S. W., and Nimer S. D. (1995) The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene 11, 2667–2674 [PubMed] [Google Scholar]
- 23. Taniuchi I., Osato M., Egawa T., Sunshine M. J., Bae S. C., Komori T., Ito Y., and Littman D. R. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 [DOI] [PubMed] [Google Scholar]
- 24. Tokuhiro S., Yamada R., Chang X., Suzuki A., Kochi Y., Sawada T., Suzuki M., Nagasaki M., Ohtsuki M., Ono M., Furukawa H., Nagashima M., Yoshino S., Mabuchi A., Sekine A., et al. (2003) An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 35, 341–348 [DOI] [PubMed] [Google Scholar]
- 25. Prokunina L., Castillejo-López C., Oberg F., Gunnarsson I., Berg L., Magnusson V., Brookes A. J., Tentler D., Kristjansdóttir H., Gröndal G., Bolstad A. I., Svenungsson E., Lundberg I., Sturfelt G., Jönssen A., et al. (2002) A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat. Genet. 32, 666–669 [DOI] [PubMed] [Google Scholar]
- 26. Lazarevic V., Chen X., Shim J. H., Hwang E. S., Jang E., Bolm A. N., Oukka M., Kuchroo V. K., and Glimcher L. H. (2011) T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12, 96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rudra D., Egawa T., Chong M. M., Treuting P., Littman D. R., and Rudensky A. Y. (2009) Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10, 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong W. F., Kohu K., Nakamura A., Ebina M., Kikuchi T., Tazawa R., Tanaka K., Kon S., Funaki T., Sugahara-Tobinai A., Looi C. Y., Endo S., Funayama R., Kurokawa M., Habu S., et al. (2012) Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J. Immunol. 188, 5408–5420 [DOI] [PubMed] [Google Scholar]
- 29. Scheitz C. J., Lee T. S., McDermitt D. J., and Tumbar T. (2012) Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 31, 4124–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elliott J., and Johnston J. A. (2004) SOCS: role in inflammation, allergy and homeostasis. Trends Immunol. 25, 434–440 [DOI] [PubMed] [Google Scholar]
- 31. Cunningham L., Finckbeiner S., Hyde R. K., Southall N., Marugan J., Yedavalli V. R., Dehdashti S. J., Reinhold W. C., Alemu L., Zhao L., Yeh J. R., Sood R., Pommier Y., Austin C. P., Jeang K. T., et al. (2012) Identification of benzodiazepine Ro5–3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFβ interaction. Proc. Natl. Acad. Sci. U.S.A. 109, 14592–14597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hommes D. W., Peppelenbosch M. P., and van Deventer S. J. (2003) Mitogen activated protein (MAP) kinase signal transduction pathways and novel anti-inflammatory targets. Gut 52, 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoffmann A., Natoli G., and Ghosh G. (2006) Transcriptional regulation via the NF-κB signaling module. Oncogene 25, 6706–6716 [DOI] [PubMed] [Google Scholar]
- 34. Yoshida N., Ogata T., Tanabe K., Li S., Nakazato M., Kohu K., Takafuta T., Shapiro S., Ohta Y., Satake M., and Watanabe T. (2005) Filamin A-bound PEBP2β/CBFβ is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Mol. Cell Biol. 25, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., et al. (2004) Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430, 218–222 [DOI] [PubMed] [Google Scholar]
- 36. Zhong H., May M. J., Jimi E., and Ghosh S. (2002) The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636 [DOI] [PubMed] [Google Scholar]
- 37. Kim W. Y., Sieweke M., Ogawa E., Wee H. J., Englmeier U., Graf T., and Ito Y. (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 18, 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu H., Chen G., Wyburn K. R., Yin J., Bertolino P., Eris J. M., Alexander S. I., Sharland A. F., and Chadban S. J. (2007) TLR4 activation mediates kidney ischemia/reperfusion injury. J. Clin. Invest. 117, 2847–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tauseef M., Knezevic N., Chava K. R., Smith M., Sukriti S., Gianaris N., Obukhov A. G., Vogel S. M., Schraufnagel D. E., Dietrich A., Birnbaumer L., Malik A. B., and Mehta D. (2012) TLR4 activation of TRPC6-dependent calcium signaling mediates endotoxin-induced lung vascular permeability and inflammation. J. Exp. Med. 209, 1953–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Growney J. D., Shigematsu H., Li Z., Lee B. H., Adelsperger J., Rowan R., Curley D. P., Kutok J. L., Akashi K., Williams I. R., Speck N. A., and Gilliland D. G. (2005) Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kitoh A., Ono M., Naoe Y., Ohkura N., Yamaguchi T., Yaguchi H., Kitabayashi I., Tsukada T., Nomura T., Miyachi Y., Taniuchi I., and Sakaguchi S. (2009) Indispensable role of the Runx1-Cbfβ transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 31, 609–620 [DOI] [PubMed] [Google Scholar]
- 42. Wang Y., Godec J., Ben-Aissa K., Cui K. R., Zhao K., Pucsek A. B., Lee Y. K., Weaver C. T., Yagi R., and Lazarevic V. (2014) The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-γ-producing T helper 17 cells. Immunity 40, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bromberg J., and Wang T. C. (2009) Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 15, 79–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakagawa M., Shimabe M., Watanabe-Okochi N., Arai S., Yoshimi A., Shinohara A., Nishimoto N., Kataoka K., Sato T., Kumano K., Nannya Y., Ichikawa M., Imai Y., and Kurokawa M. (2011) AML1/RUNX1 functions as a cytoplasmic attenuator of NF-κB signaling in the repression of myeloid tumors. Blood 118, 6626–6637 [DOI] [PubMed] [Google Scholar]
- 45. Sha W. C., Liou H. C., Tuomanen E. I., and Baltimore D. (1995) Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80, 321–330 [DOI] [PubMed] [Google Scholar]
- 46. Collins P. E., Grassia G., Colleran A., Kiely P. A., Ialenti A., Maffia P., and Carmody R. J. (2015) Mapping the interaction of B cell leukemia 3 (BCL-3) and nuclear factor κB (NF-κB) p50 identifies a BCL-3-mimetic anti-inflammatory peptide. J. Biol. Chem. 290, 15687–15696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ran D., Shia W. J., Lo M. C., Fan J. B., Knorr D. A., Ferrell P. I., Ye Z., Yan M., Cheng L., Kaufman D. S., and Zhang D. E. (2013) RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood 121, 2882–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu X., Zhang Q., Zhang D. E., Zhou C., Xing H., Tian Z., Rao Q., Wang M., and Wang J. (2009) Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia 23, 739–745 [DOI] [PubMed] [Google Scholar]
- 49. Bottero V., Withoff S., and Verma I. M. (2006) NF-κB and the regulation of hematopoiesis. Cell Death Differ. 13, 785–797 [DOI] [PubMed] [Google Scholar]
- 50. Guzman M. L., Neering S. J., Upchurch D., Grimes B., Howard D. S., Rizzieri D. A., Luger S. M., and Jordan C. T. (2001) Nuclear factor-κB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 98, 2301–2307 [DOI] [PubMed] [Google Scholar]
- 51. Komeno Y., Yan M., Matsuura S., Lam K., Lo M. C., Huang Y. J., Tenen D. G., Downing J. R., and Zhang D. E. (2014) Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood 123, 3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zarjou A., and Agarwal A. (2011) Sepsis and acute kidney injury. J. Am. Soc. Nephrol. 22, 999–1006 [DOI] [PubMed] [Google Scholar]
- 53. Nechemia-Arbely Y., Barkan D., Pizov G., Shriki A., Rose-John S., Galun E., and Axelrod J. H. (2008) IL-6/IL-6R axis plays a critical role in acute kidney injury. J. Am. Soc. Nephrol. 19, 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fujimoto T., Anderson K., Jacobsen S. E., Nishikawa S. I., and Nerlov C. (2007) Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPα interaction. EMBO J. 26, 2361–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee K., Na W., Lee J. Y., Na J., Cho H., Wu H., Yune T. Y., Kim W. S., and Ju B. G. (2012) Molecular mechanism of Jmjd3-mediated interleukin-6 gene regulation in endothelial cells underlying spinal cord injury. J. Neurochem. 122, 272–282 [DOI] [PubMed] [Google Scholar]
- 56. Li W., Xiao J., Zhou X., Xu M., Hu C., Xu X., Lu Y., Liu C., Xue S., Nie L., Zhang H., Li Z., Zhang Y., Ji F., Hui L., Tao W., Wei B., and Wang H. (2015) STK4 regulates TLR pathways and protects against chronic inflammation-related hepatocellular carcinoma. J. Clin. Invest. 125, 4239–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.