Abstract
The genetic determinants of osteoporosis remain poorly understood, and there is a large unmet need for new treatments in our ageing society. Thus, new approaches for gene discovery in skeletal disease are required to complement the current genome-wide association studies in human populations. The International Knockout Mouse Consortium (IKMC) and the International Mouse Phenotyping Consortium (IMPC) provide such an opportunity. The IKMC generates knockout mice representing each of the known protein-coding genes in C57BL/6 mice and, as part of the IMPC initiative, the Origins of Bone and Cartilage Disease project identifies mutants with significant outlier skeletal phenotypes. This initiative will add value to data from large human cohorts and provide a new understanding of bone and cartilage pathophysiology, ultimately leading to the identification of novel drug targets for the treatment of skeletal disease.
Keywords: osteoporosis, bone, genetics, gene discovery
Introduction
A novel strategy for osteoporosis gene discovery
Studies of human monogenic extreme phenotype disorders have been instrumental in discovering genetic and molecular mechanisms of common diseases including obesity and diabetes (Yamagata et al. 1996, Montague et al. 1997). However, collection of human extreme phenotype cohorts takes many years and requires significant effort and financial resource. A new approach to osteoporosis gene discovery involves systematic identification of extreme skeletal phenotypes in mutant mouse lines that carry single-gene knockouts representing all the known protein-coding genes. This approach has been made possible by the International Knockout Mouse Consortium (IKMC), whose aim is to disrupt each of the protein-coding genes in C57BL/6 mice, and the International Mouse Phenotyping Consortium (IMPC) that has established a multidisciplinary and broad primary phenotype screen to characterise these mutant mice. By using samples from mice that have undergone the IMPC phenotyping pipeline, a bespoke rapid-throughput multi-parameter skeletal phenotyping platform has been applied systematically to detect significant phenotypes by screening minimal number of samples. This phenotyping programme exploits the excellent replication of human skeletal disease in mice, and novel susceptibility genes can be validated by interrogating human osteoporosis cohorts.
‘Known unknowns’ in osteoporosis
Osteoporosis is a worldwide healthcare problem that causes up to 9 million fractures annually (Johnell & Kanis 2006). Within the EU, it is estimated that osteoporosis affects 30 million people and osteoporotic fractures cost €37 billion annually (Hernlund et al. 2013). These numbers are projected to rise with the increasing elderly population. Osteoporotic hip fractures are associated with a significant rise in mortality (Sattui & Saag 2014), especially during the year following fracture when it is estimated to be 8–36% (Abrahamsen et al. 2009).
The most important risk factors for osteoporotic fracture are low bone mineral density (BMD) (clinically assessed by dual-energy X-ray absorptiometry (DEXA or DXA)), increasing age and history of fracture (Johnell et al. 2005). There are two key determinants of adult BMD: the peak bone mass attained in early adulthood and the rate of bone loss during ageing. Variation in BMD has a large heritable genetic component. This is known from observations of familial clustering of osteoporosis (Seeman et al. 1989, Keen et al. 1999) and from twin studies that have calculated that the heritable contribution lies between 40 and 90% (Mitchell & Yerges-Armstrong 2011). The heritable contribution to variance in BMD is greatest in early adulthood (Gueguen et al. 1995), yet variation in the rate of bone loss per se is also genetically determined (Kelly et al. 1993). Thus, genetic mechanisms contribute significantly to the risk of osteoporosis. Nevertheless, the known BMD-associated genetic variants account for only 5.8% of the total variance (Estrada et al. 2012), indicating that the majority of susceptibility genes have yet to be identified.
Gene discovery from skeletal extreme phenotypes
Intrinsic and extrinsic factors, systemic hormones, neuronal innovation and mineral homeostasis can all affect bone mass. Significantly, many of the genes and signalling pathways involved in the intrinsic regulation of bone turnover and bone mass have been identified by the study of human monogenic disorders associated with extremes of BMD (Table 1). In traditional gene discovery, the loci of causative alleles would be identified by linkage analysis in the families of index cases, followed by positional cloning of the relevant genes (Alonso & Ralston 2014). Such studies have identified the two key regulatory pathways – canonical Wnt signalling and receptor activator of nuclear factor kappa-B ligand (RANKL)/RANK/osteoprotegerin (OPG) that respectively regulate the function of osteoblasts and osteoclasts. Both these pathways have subsequently been targeted by novel osteoporosis treatments.
Table 1.
Monogenic disorders that have identified key skeletal genes in bone remodelling.
| Disease | Clinical features | Gene | Mechanism | Reference | |
|---|---|---|---|---|---|
| Reduced bone mass | Osteoporosis- pseudoglioma syndrome (OPPGS) | Reduced bone mass and blindness | LRP5 | Loss-of-function mutations disrupt Wnt signalling and reduce osteoblastic bone formation. | (Gong et al. 2001) |
| Osteogenesis imperfecta | Increased bone fragility; blue sclerae in some types | COL1A1, COL1A2, CRTAP, LEPRE, PPIB | Loss-of-function mutations in collagen and collagen- processing proteins cause abnormal osteoid matrix, thereby impairing normal bone formation. | (Baldridge et al. 2008; Sykes et al. 1986; van Dijk et al. 2009) | |
| Juvenile-onset Paget disease | Short stature, fractures, skull enlargement, progressive deafness | TNFRSF11B (OPG) | Loss-of-function mutations disrupt inhibition of RANKL by osteoprotegerin, causing increased osteoclastic resorption of bone. | (Chong et al. 2003) | |
| X-linked osteoporosis | Juvenile-onset fractures in males | PLS3 | Loss-of-function mutations affect Plastin-3, an actin-binding protein. Mechanism osteoporosis is unknown. | (van Dijk et al. 2013) | |
| Increased bone mass | Osteopetrosis | Increased bone mass with fractures | TNFRSF11A (RANK), TNFSF11 (RANKL), CLCN7, TCIRG1, OSTM1 | Loss-of-function mutations affecting osteoclast differentiation and function cause reduced bone resorption. | (Frattini et al. 2000; Guerrini et al. 2008; Kornak et al. 2001; Pangrazio et al. 2006; Sobacchi et al. 2007) |
| Sclerosteosis and Van Buchem disease | Increased bone mass, syndactyly, entrapment neuropathies | SOST | Loss-of-function mutations affect inhibition of Wnt signalling by sclerostin, causing increased osteoblastic bone formation. | (Balemans et al. 2001) | |
| Autosomal dominant high bone mass | Increased bone density, entrapment neuropathies, square jaw and torus palatinus | LRP5 | Gain-of-function mutation in Wnt co-receptor causes increased osteoblastic bone formation. | (Boyden et al. 2002) |
The canonical Wnt/β-catenin signalling pathway is the key regulator of osteoblasts, which mediate bone formation (Balemans et al. 2001, Gong et al. 2001, Boyden et al. 2002, Little et al. 2002, Glass et al. 2005). The major Wnt antagonist sclerostin (SOST) was first identified by studying subjects with high bone mass due to sclerosteosis and Van Buchem disease (Balemans et al. 2001, Brunkow et al. 2001). The importance of Wnt signalling was further highlighted by the discovery of activating and inactivating mutations of the Wnt co-receptor LRP5, which results in high and low bone mass, respectively (Johnson et al. 1997, Little et al. 2002).
The RANK-RANKL-OPG pathway regulates osteoclasts, which mediate bone resorption. This signalling pathway was discovered in a functional screen of tumour necrosis factor (TNF)/TNF receptor superfamily members. OPG is an endogenous inhibiting decoy receptor of RANKL related to the TNF receptor. In vivo overexpression of OPG in mice was found to cause osteopetrosis due to impairment of the later stages of osteoclast differentiation (Simonet et al. 1997). Subsequently, human osteopetrosis phenotypes with increased BMD were found to be caused by mutations of RANK, RANKL and other related genes involved in osteoclast differentiation (Coudert et al. 2015).
Regulatory mechanisms in bone turnover
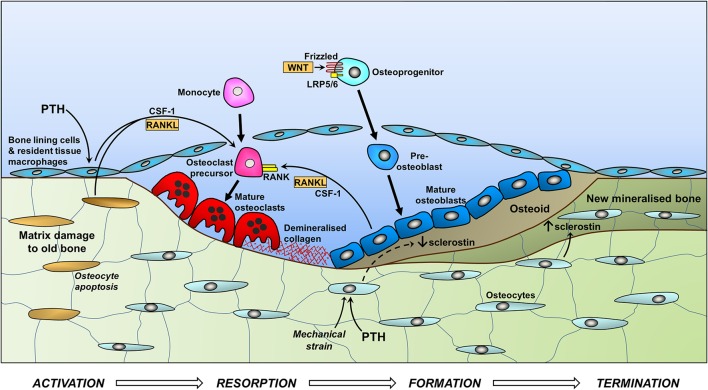
Knowledge of the signalling pathways that regulate bone turnover is essential for understanding the pathophysiology of osteoporosis. The manifest complexity of the signalling pathways and networks that regulate the cellular processes involved in dynamic bone turnover is significant as small differences in function of individual components, including those not yet discovered, may have a combined effect on heritable risk of osteoporosis. The opposing processes of bone resorption and formation are tightly regulated by critical mechanisms including the Wnt signalling and RANKL/RANK/OPG pathways (Fig. 1). At the cellular level, bone remodelling takes place in multicellular units, which comprise co-located osteoclasts and osteoblasts within a bone remodelling cavity (Raggatt & Partridge 2010). The bone remodelling cycle is initiated by osteocytes in response to altered mechanical loading (Nakashima et al. 2011), local microdamage and systemic factors such as parathyroid hormone (Goldring 2015). Unloading stimulates expression of RANKL and the Wnt inhibitors sclerostin and Dickkopf-related protein 1 (DKK-1) in osteocytes, thus increasing osteoclastic bone resorption and decreasing osteoblastic bone formation. Repetitive loading can cause fatigue-induced microdamage and focal tissue injury that leads to osteocyte apoptosis, following which pro-osteoclastogenic signalling is initiated by a discrete population of adjacent osteocytes (Kennedy et al. 2012). By contrast, increased mechanical loading decreases expression of sclerostin and DKK-1 in osteocytes and leads to increased osteoblastic bone formation (Robling et al. 2008). In their basal state, osteocytes maintain quiescence by inhibiting osteoclastogenesis by secreting transforming growth factor β (TGFβ) and inhibiting Wnt-activated osteoblastic bone formation by secreting sclerostin (Heino et al. 2002, Tu et al. 2012). If TGFβ levels fall, bone-lining cells become activated and join osteocytes in secreting the cytokines monocyte/macrophage colony-stimulating factor (M-CSF) and RANKL that stimulate recruitment and differentiation of circulating osteoclast monocyte progenitors (Nakashima et al. 2011). Mature multi-nucleated bone-resorbing osteoclasts adhere to the cell surface and create resorption pits in the middle of the multicellular unit. The osteoclasts secrete acid and proteases including cathepsin-K that degrade the bone matrix, leaving demineralised collagen that is resorbed by macrophages. Wnt-activated osteoblasts subsequently secrete and mineralise new bone matrix (osteoid) to fill the resorption cavity. When the repair is complete, the bone surface returns to its quiescent state and sclerostin stimulates mineralisation of the new osteoid (Tu et al. 2012).
Figure 1.
Schematic representing the bone remodelling processes of the ‘basic multicellular unit’ in the endosteal surface of trabecular bone. Activation: microdamage to the bone causes osteocyte apoptosis, reducing local basal inhibition of osteoclastogenesis. Resorption: in response to PTH signalling, RANKL and CSF-1 (colony-stimulating factor-1) increase recruitment, proliferation and differentiation of osteoclasts, which demineralise the bone matrix and then digest the collagen matrix, the remnants being removed by macrophages. Formation: PTH and mechanical activation of osteocytes reduce sclerostin expression, removing the potent inhibition of Wnt-mediated osteoblast differentiation (via cell surface receptor Frizzled and co-receptors LRP5 and LRP6) and bone formation. Termination: in response to increasing levels of sclerostin, bone formation ceases and newly deposited osteoid is mineralised.
Gene discovery drives therapeutic innovation
Identification of the genes and regulatory networks that determine normal bone formation, maintenance and strength has facilitated the recent development of new targeted treatments to increase BMD and reduce fracture risk. There is a pressing need for new treatment options in osteoporosis as current treatments achieve at best partial relative risk reduction of fracture. Bisphosphonates remain the mainstay of treatment for primary and secondary prevention of osteoporotic fracture, despite the recognition uncommon but significant side effects (Kennel & Drake 2009). Other treatments including strontium ranelate and raloxifene are no longer advocated as first line because of associated cardiovascular risks (Barrett-Connor et al. 2006, Abrahamsen et al. 2014).
New treatments for osteoporosis include small-molecule inhibitors and biological therapies have been developed based on this new understanding of bone turnover. Denosumab is a fully humanised monoclonal antibody to RANKL that mimics the endogenous inhibiting activity of OPG (Lacey et al. 2012). Denosumab is administered by 6-month injection; yet, its use is restricted by high cost and its effects are rapidly reversible. Another new class of osteoporosis drugs are those that target the osteoclast-secreted protease cathepsin-K (Makras et al. 2015). However, most of the cathepsin-K inhibitors have adverse off-target effects, with the exception of odanacatib, which has now successfully completed phase III trials (Bone et al. 2015). Humanised monoclonal antibodies to the Wnt antagonist sclerostin (romosozumab and blosozumab) are in development as new anabolic agents (Shah et al. 2015, Sim & Ebeling 2015). However, since romosozumab is currently in phase III trials and blosozumab has only completed phase II trials, effect on fracture risk is still unknown.
Despite some recent advances, there remains a large unmet need for new therapeutic targets in osteoporosis. Of all the newer targeted treatments, only the small-molecule odanacatib is administered orally. The requirement for parenteral subcutaneous administration is an impediment to use of teriparatide and all monoclonal antibody treatments. It is hoped that further advances in knowledge of the complex mechanisms of bone turnover regulation and the genetic basis of the heritability of osteoporosis will enable the accelerated development of new osteoporosis treatments. The current goals in osteoporosis gene discovery must be to identify novel targets with a systematic methodology that may replicate the successes of past studies in Mendelian extreme phenotype cohorts.
Systematic osteoporosis gene discovery in human studies
The Human Genome Project and association studies
In order to identify novel genes that relate to heritability of osteoporosis systematically, new population-based methodologies were required and have been used extensively. Linkage analysis and positional cloning techniques, used to identify gene mutations in Mendelian disorders, were less successful when applied to population cohorts with low BMD. Several genome-wide linkage studies were performed on high and low BMD cohorts to detect quantitative trait loci (QTLs) that regulate BMD. However, these studies produced heterogeneous results, and when combined in a meta-analysis, none of the QTLs reached genome-wide significance (Ioannidis et al. 2007).
It was the subsequent development of GWAS analysis that made it possible to search for genetic influences on complex polygenic traits such as osteoporosis. After the first sequence of the human genome was completed, the International HapMap Project was established to catalogue the common genetic variation between the genomes of 249 individuals in four different populations, leading to the mapping of over 1 million single-nucleotide polymorphisms (SNPs) within the human genome (International HapMap Consortium 2005). A major aim of the HapMap Project was to facilitate the study of the contribution of normal genetic variation to the inheritance of complex polygenic phenotypic traits such as osteoporosis (Frazer et al. 2009). In GWAS analyses, large population-based cohorts containing many thousands of subjects are genotyped by chip-based microarrays for the presence of millions of SNPs, or alternatively copy-number variants (CNVs). These data are used to perform hypothesis-free significance testing for association of the inheritance of the SNPs or CNVs with disease phenotypes (Manolio 2010).
To date, GWAS have identified more than 60 loci associated with BMD, confirming the polygenic nature of this variable (Richards et al. 2012). Less data are, however, available for fracture risk than BMD (Mitchell & Streeten 2013). Although estimation of areal BMD by DEXA scanning is a good predictor of fracture susceptibility, it does not necessarily reflect bone quality as DEXA does not account for bone geometry, size and strength (Marshall et al. 1996).
Population-based genetic cohort studies have significant limitations in relation to osteoporosis gene discovery. When GWAS data are combined in meta-analyses, variability in the characterisation of phenotype can be an important confounder, and population studies are also limited by the quality and reproducibility of the phenotyping (Visscher et al. 2012). Many of the genomic loci identified by GWAS map to genes in pathways already known to be related to bone biology, such as the Wnt/β-catenin, RANK-RANKL-OPG, mesenchymal stem cell differentiation and SOX9-regulated endochondral ossification pathways (Mitchell & Streeten 2013). Overall, GWAS analysis has not yet led to the discovery of major new mechanisms that regulate bone turnover regulation and predispose to osteoporosis. Indeed, contrary to initial expectations, GWAS analyses failed to discover sufficient determinants of polygenic heritable traits to enable individual disease risk prediction. An osteoporosis risk prediction model using the combined weighted effects of 63 BMD-decreasing alleles in a population-based study containing 2836 women explained only 5.8% of the total variance in femoral neck BMD (Estrada et al. 2012).
A further difficulty in identifying susceptibility genes by GWAS analysis arises from the fact that identified SNPs are rarely functionally significant, although their loci may identify adjacent pathogenic pathways and mechanisms. The co-association of the SNP with the disease phenotype indicates that it is in linkage disequilibrium with a gene (or a non-coding regulatory element) that has a constituent functional role that is contributory to the phenotype (Cantor et al. 2010). The size of the detected effect is itself of little importance, and it is typical that the effect size of individual identified loci is inversely proportional to the population sample size in a GWAS. The mean effect size of SNPs identified in the largest osteoporosis GWAS meta-analysis to date was 0.048 standard deviations (s.d.s) and the largest effect size was 0.1 s.d. (Estrada et al. 2012, Zheng et al. 2015).
To address the limitation that standard GWAS microarrays only test common genetic variants (minor allele frequency (MAF) > 5%), larger reference panels have been created from the recent UK10K and 1000 Genomes projects. ‘Next-generation sequencing’ techniques have enabled direct imputation of low-frequency coding and non-coding variants found by whole-genome sequencing of large osteoporosis cohorts. Thus far, only one low-frequency variant with genome-wide significance for skeletal disease has been identified by these techniques (Zheng et al. 2015). The variant is near a novel locus, engrailed homeobox-1 (EN1) and has an estimated effect size on lumbar spine BMD of 0.2 s.d.s, which is four-fold larger than the mean effect size of previously reported common variants.
Transcriptomics and osteoporosis
One limitation of the GWAS data is that the majority of identified SNPs may be located within poorly annotated regulatory elements in intronic or intergenic non-coding regions (Cooper 2010). Indeed, it is increasingly appreciated that heritable variation of osteoporosis risk is manifest in more ways than just variation in the coding sequence of genes. Expression levels of RNA and proteins, as regulated by gene regulation networks and epigenetic control, are also of great importance. Consequently, study of variations in levels of RNA transcripts has also been applied to identify new molecular mechanisms and signalling pathways involved in regulation of BMD. Furthermore, it is anticipated that analysis of non-coding RNAs will lead to a better understanding of the significance of intergenic loci identified by GWAS (Hangauer et al. 2013).
Transcriptomic studies have used high-throughput microarray analysis to quantify mRNA expression in osteoporotic and non-osteoporotic human bone, precursor mesenchymal stem cells and primary osteoblast cultures (Wu et al. 2013). In a microarray study of gene expression in human osteoblasts, 1606 genes were found to be differentially expressed between osteoporotic and non-osteoporotic subjects (Trost et al. 2010). Many of these genes had been identified previously by GWAS analysis, but several were new and could not have been predicted. Thus, osteoporosis transcriptomics studies have confirmed that numerous distinct signalling pathways are involved in the regulation of bone turnover and bone mass. However, replication of these studies has been difficult and confounded by limited sample size, poor signal to noise ratio and technical limitations of commercially available microarrays (Wu et al. 2013). Future work will increasingly use next-generation sequencing methods, which do not require a priori knowledge of the transcriptome sequence and have a superior sensitivity, specificity and dynamic range in comparison with current microarrays (Vikman et al. 2014). With use of advanced statistical techniques, it is possible to discover new splice variants and non-coding sequences. Despite these advances, RNA-sequencing approaches require preparation of high-quality RNA from sufficient numbers of physiologically relevant samples. Accordingly, it is not yet possible to study gene expression in osteoporosis using large population-based sample sizes, and RNA sequencing is likely to be most useful in the analysis of animal models.
Proteomics
Proteomics allows the simultaneous analysis of all proteins in a sample of cells by antibody-based purification and mass spectrometry techniques. A number of studies have demonstrated differential regulation of cellular protein expression in osteoporosis (Wu et al. 2013). Most of the studies have been performed in vitro using osteoblast and osteoclast cell cultures. Human proteomic studies have predominantly used peripheral circulating monocytes as precursors to osteoclasts (Deng et al. 2011a,b), or bone marrow-derived mesenchymal stem cells (Choi et al. 2010). However, reproducibility of such results has been limited.
Epigenomics
The heritability of osteoporosis may also be mediated by epigenetic gene regulation, in which gene or allele expression is ‘imprinted’ by DNA methylation and histone modifications (Holroyd et al. 2012). Epigenome-wide studies have only recently been performed in relation to osteoporosis. Transcriptome gene expression microarray, epigenomic miRNA microarray and methylome sequencing were simultaneously performed using circulating monocytes from five subjects with low hip BMD and five with high BMD, in order to integrate transcriptomic and epigenomic data (Zhang et al. 2015). The aim is to reveal the higher regulatory mechanisms (‘interaction network modules’) underlying the genetic control of osteoporosis heritability. However, the problems of low sample number and difficulty of accessing human skeletal tissues are important limitations in such approaches.
The mouse as an essential tool for osteoporosis gene discovery
Human population studies have been limited in their ability to identify novel genetic determinants of osteoporosis, although they have confirmed that bone-regulating genes identified in the study of rare extreme phenotypes contribute to the complex heritability of secondary osteoporosis. Across the breadth of human biology, there are huge knowledge gaps as the function of most genes and the heritability of many complex diseases remain unknown (Schofield et al. 2012). In order to meet the challenge of discovering the functional relationship between human genes and their phenotypic effects, powerful new experimental tools are required. Model organisms provide the ability to draw genetic and physiological parallels to human genetic systems, and their use has complemented many advances in human molecular genetics. In particular, an extensive range of techniques for experimental genetic manipulation have been developed in mice.
The genome of the C57BL/6J mouse strain was sequenced in 2002 (Waterston et al. 2002), shortly after the human genome. Thus, laboratory mice provide a unique resource that can be used to generate genetically modified models of human diseases (Schofield et al. 2012). Currently, 17,055 mouse–human homologs have been annotated in the Mouse Genome Database (Dolan et al. 2015). Critically, the selective mutation or deletion (‘knockout’) of individual genes in mice can be used to identify and characterise gene function in vivo.
International knockout mouse programmes
Such is the utility of knockout mice that a number of mutagenesis programmes have collaborated to cryo-preserve mouse lines for unrestricted use by the research community. The IKMC was established with the goal of creating a complete resource of reporter-tagged null mutations for all protein-coding genes in C57BL/6 mouse ES cells by 2021 (Collins et al. 2007).
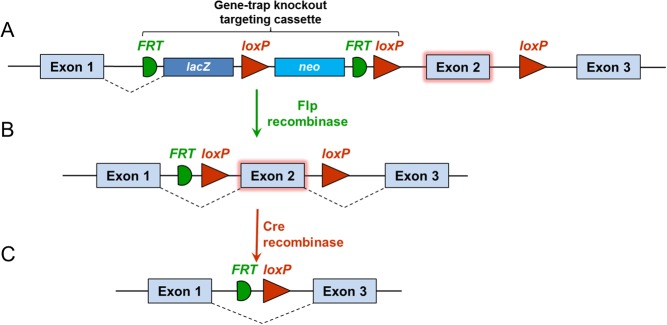
The IKMC-targeted mutagenesis techniques involve use of bacterial artificial chromosome (BAC)-based ‘Knockout-First’ conditional ready gene targeting vectors that use homologous recombination to incorporate a lacZ selection cassette in the relevant targeted sequence (Fig. 2) (Testa et al. 2004). This enables the generated mouse lines to be versatile and powerful tools for research. The Knockout-First vectors combine two functions by creating either a reporter-tagged knockout or a conditional mutation if the gene-trap cassette is removed by FLP recombinase, thereby reverting the knockout mutation to wild type, although with addition of loxP sites that flank a functionally critical exon (Skarnes et al. 2011). The consequence of this is that temporal or tissue-specific analysis of gene function can be performed by crossing mice bearing the allele containing the LoxP-flanked (‘floxed’) critical exon with transgenic mice that express Cre recombinase under control of a constitutive or inducible cell type-specific promoter.
Figure 2.
Knockout-first strategy for creating dual-purpose knockout/conditional alleles. Bacterial artificial chromosome (BAC)-based targeting vectors are inserted by homologous recombination into mouse ES cells. Recombination steps with Cre or Flp recombinase are illustrated. (A) Knockout-first allele (reporter-tagged insertion allele). Gene-trap knockout is generated using a targeting cassette containing the marker genes lacZ and neomycin. A separate loxP site is inserted on the other side of a critical exon (Exon 2). (B) Conditional allele (post-Flp). By crossing mice with a Flp deleter strain, the gene-trap knockout is reversed and a floxed allele is created, enabling conditional Cre recombinase-mediated gene inactivation. (C) Deletion allele (post-Flp and Cre with no reporter).
Pipeline phenotyping of knockout mice
The IKMC provides readily available knockout mice that can be used for functional investigation of candidate genes, for example in loci identified by GWAS analysis (Cox & Church 2011). The singular ambition to deduce the function of all the genes discovered in the Human Genome Project, however, has led to the development of ambitious large-scale systematic phenotyping programmes of all knockout mouse lines with deletions of homologs to human genes (Bradley et al. 2012). These projects enable the study of the extreme phenotype potential of every known coding gene, thereby providing the potential for systematic discovery of novel critical skeletal genes. What would otherwise have been individually impossible in terms of resources and complexity has been achieved by the coordination and the standardisation of a series of international programmes to form a major worldwide project The IMPC (Koscielny et al. 2014).
The international phenotyping pipeline incorporates standardised and validated tests with protocols shared by contributing mouse clinics across the world (de Angelis et al. 2015). The pipelines incorporate 20 phenotyping tests that capture 413 parameters and cover all systems divided between the categories of morphology, metabolism, cardiovascular, bone, neurobehavioural and sensory, haematology, clinical chemistry and allergy/immune. The phenotyping tests are performed at defined times in the first 16 postnatal weeks and require a minimum cohort of seven male and seven female mice (Brown & Moore 2012). New statistical models have had to be developed, in order to ensure the reproducibility of the multivariate data generated (Karp et al. 2015). In an initial report of phenotype data on mutant lines representing 320 unique genes, 83% of mutant lines were outliers (de Angelis et al. 2015). Of the 250 lines reported in an early phase of the phenotyping pipeline contributed by the IMPC member Wellcome Trust Sanger Institute (WTSI), 104 were lethal or sub-viable and were phenotyped as heterozygotes; nonetheless, haploinsufficient phenotypes were detected in 38 of these lines (White et al. 2013). Ongoing results from the international multicentre phenotyping projects are released at http://www.Mousephenotype.org/.
Skeletal phenotyping screen of knockout mouse lines
Ancillary IMPC projects
The systematic phenotyping of knockout mice by the IMPC has already begun to reveal genes whose deletion has an effect on BMD (estimated by whole-body DEXA), implying a functional role in bone development and skeletal physiology. To date, 79 out of 1820 lines have an outlier phenotype associated with decreased BMD (IMPC phenotype MP:0000063) and 41 lines have increased BMD (IMPC phenotype MP:0000062). However, a major limitation is the poor sensitivity and specificity of DXA for the analysis of the mouse skeleton (Holmen et al. 2004). Other skeletal parameters included in the IMPC phenotyping are body length and X-ray skeletal survey to detect gross anatomical variation. As critical determinants of healthy bone include bone mineral content (BMC), bone strength and other three-dimensional morphological parameters, the IMPC phenotype screen lacks the precision required for comprehensive detection of bone structure and strength abnormalities.
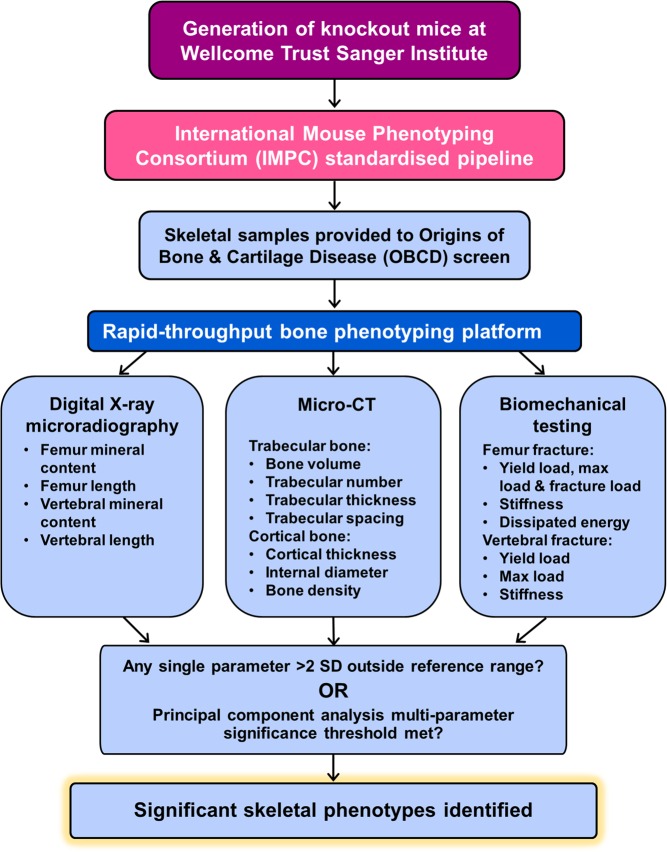
To address these limitations, a number of ancillary specialist screens have been established. The Origins of Bone and Cartilage Disease (OBCD) project (http://www.boneandcartilage.com/) is performing a multi-parameter skeletal phenotyping screen of mouse lines generated by the WTSI, an IMPC partner institution, using methods that give critical functional and structural information regarding skeletal phenotype (Fig. 3). The goals of the OBCD project are to (i) identify novel pathways regulating normal bone development, maintenance and resilience; (ii) uncover new genetic determinants of osteoporosis; and (iii) provide in vivo models to elucidate their molecular basis and investigate novel treatments.
Figure 3.
Flow chart showing how the OBCD bone phenotyping platform leads to identification of significant abnormal skeletal phenotypes, in conjunction with the IMPC standardised phenotyping project.
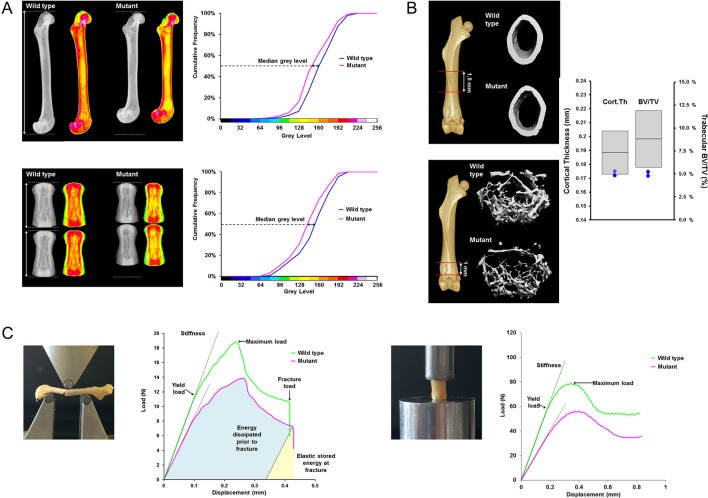
New imaging and biomechanical techniques have been developed to detect abnormalities of bone structure and strength that parallel those occurring in human disease. Cross-disciplinary collaboration with the fields of biophysics, microimaging and statistics has enabled development of a bespoke rapid-throughput multi-parameter bone phenotyping platform (Fig. 4) (Bassett et al. 2012a, Esapa et al. 2012).
Figure 4.
OBCD skeletal phenotyping methods. (A) X-ray microradiography images of femur and fifth to sixth tail vertebrae from wild-type and mutant mice. Low bone mineral content is represented in green/yellow colour and high bone mineral content is represented in red/pink colour in pseudo-coloured images. Cumulative frequency graphs showing difference in bone mineral content between wild-type and mutant mice. (B) Micro-CT images of cortical and trabecular bone from wild-type and mutant mice. Cortical thickness and trabecular bone volume/total volume (BV/TV) parameters in the mutant are shown in comparison with reference mean ± 2 standard deviations. (C) Femur three-point bend and vertebral compression analysis with load–displacement curves illustrating biomechanical parameters.
OBCD phenotyping techniques
Limbs and caudal vertebrae from knockout mice are analysed at 16 weeks of age after completion of the IMPC phenotyping pipeline at WTSI. No additional animals are required for the OBCD screen, and transportation of samples is logistically simple. Each batch of 75 samples, which contains both mutants and wild-type controls, is analysed blind with genotypes only being assigned on completion of the batch’s phenotyping.
Digital X-ray microradiography is performed on femurs and tail vertebrae, and the images are analysed to determine bone length and BMC (Bassett et al. 2012b). X-ray images are recorded at 10 µm pixel resolution using a Faxitron MX20 specimen radiography system. Bone length is determined using ImageJ 1.41 software (http://rsb.info.nih.gov/ij/). The relative mineral content of the calcified tissues is quantified after calibrating each image to three internal standards. Measurement of the median grey level is used to identify outliers with increased or decreased BMC relative to reference data obtained for more than 100 wild-type controls of the same genetic background.
Micro-CT analysis has been optimised to determine femoral trabecular bone parameters including trabecular bone volume as proportion of tissue volume (BV/TV), trabecular thickness and trabecular number. In addition, cortical bone parameters of cortical thickness, cortical diameter and cortical volumetric BMD are calculated (Bouxsein et al. 2010, Esapa et al. 2012). A Scanco micro-CT 50 system allows automated rapid-throughput high-resolution imaging. Three-dimensional quantitative image analysis is performed using clearly defined regions of interest and compared with reference data.
Biomechanical variables of bone strength and toughness are derived from femur three-point bend test load–displacement curves and tail vertebrae compression testing (Esapa et al. 2012). Load–displacement curves are plotted so that yield load, maximum load and fracture load can be determined. Stiffness is determined from the slope of the linear (elastic) part of the load–displacement curve.
Pilot study
Before commencing the OBCD skeletal phenotyping screen, a prospective pilot study of 100 unselected knockout mouse lines was undertaken (Bassett et al. 2012a). As part of the pilot project, it was necessary to determine reference ranges and coefficients of variation for each of the study parameters for female C57BL/6 wild-type mice. Principal component analysis was used to optimise the detection of significant skeletal phenotypes in multivariate outliers, as significant abnormal phenotypes may only be detected when variances in all parameters are considered (Rousseeuw & Van Zomeren 1990). As reference data were established in a large number of genetically identical wild-type mice, power calculations determined that samples from only two animals were required to detect a significant abnormality that represents an outlier phenotype.
In the pilot study of 100 knockout mouse lines, nine new genetic determinants of bone mass and strength were identified, none of which had been identified previously in GWAS analysis of osteoporosis cohorts or could have been predicted a priori. Analysis of the contrasting patterns of abnormality amongst the different phenotypic parameters enabled phenotypes to be categorised in a way that could be mapped directly to human skeletal disease. Bones were classified as either (i) weak but flexible with low BMC (as in osteoporosis), (ii) weak and brittle with low BMC (typical of matrix disorders including osteogenesis imperfecta) or (iii) strong but brittle with high BMC (high bone mass disorders such as osteopetrosis). The nine new determinants of bone mass and strength identified included five genes whose deletion results in low bone mass (Bbx, Cadm1, Fam73b, Prpsap2 and Slc38a10) and four whose deletion results in high bone mass (Asxl1, Setdb1, Spns2 and Trim45). The study also confirmed the low bone mass phenotype identified previously in Sparc knockout mice (Delany et al. 2000). Three of the knockout lines carried heterozygous mutations (Asxl1, Setdb1 and Trim45), whereas the rest were homozygotes.
Other skeletal phenotyping programmes
Although the OBCD pilot study was the first approach to be published, similar phenotype screening methods have been undertaken by others. Lexicon Pharmaceuticals, Inc. recently published selected results from a screen of knockout mouse lines to search for potential osteoporosis drug targets (Brommage et al. 2014). This phenotyping screen included three techniques (skeletal DEXA of live mice, micro-CT of dissected bones and histological examination of decalcified bones). Ten novel genes were named, and three further unnamed novel genes coding for apparent potential osteoporosis drug targets were alluded to.
The IMPC-constituent knockout mouse programme (KOMP) of the Jackson Laboratory has recently commenced its own skeletal phenotyping project that involves rapid micro-CT and automated bone and joint cartilage histology (http://bonebase.org/">http://bonebase.org/">http://bonebase.org/). This screen focuses on detecting evidence of variations in skeletal cellular function. Histomorphometry is performed by a recently innovated high-throughput process that involves computer-automated signal detection for the particular cell type-specific stains. Data are accrued by automated analysis that calculates the percentage of the bone surface containing the light signal from each stain, thereby suggesting the pattern of disruption of cellular activity in the trabecular bone of the femur and vertebra that may account for the architectural observations seen in micro-CT (Hong et al. 2012). Besides phenotyping inbred lines from the IKMC mutant mouse repository (Yoshiki & Moriwaki 2006), the Bonebase phenotyping project is phenotyping mouse lines from the ‘Collaborative Cross’ project, which has created hybrids from eight founder inbred strains in order to perform genetic mapping studies to identify the QTLs that contribute to complex traits and diseases (Bogue et al. 2015). Similarly, Bonebase is also studying ‘diversity outbred’ lines created to produce a genetic resource to facilitate high-resolution mapping of the effects of allelic heterozygosity that replicates the complexity of the human population (Svenson et al. 2012).
Current OBCD project goals
The OBCD project is currently funded by a Wellcome Trust Strategic Award to undertake skeletal phenotyping of all knockout mouse lines generated at the Sanger Institute. Results are available at the OBCD website and also uploaded to the IMPC mouse portal. Although the IMPC parent project is powered robustly to assess and catalogue the unknown pleiotropic effects of gene deletion, the OBCD screen is designed for rapid-throughput hypothesis generation. Once extreme phenotypes are detected, they can be selected for additional in-depth analysis.
Detailed analysis of extreme phenotypes
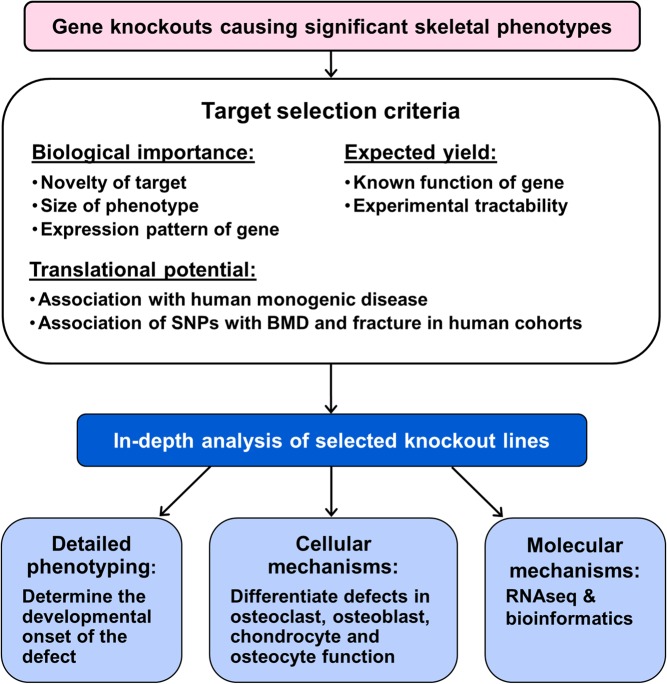
Knockout mice with extreme skeletal phenotypes are considered for additional detailed analysis and the selection procedure follows a specific algorithm (Fig. 5). Although novelty is a key criterion, phenotype severity, biological plausibility, human disease association and experimental tractability are also critical considerations (Duncan et al. 2011, van Dijk et al. 2014).
Figure 5.
Flow chart outlining selection of knockout mouse lines for further study and analysis.
Detailed phenotyping includes skeletal analysis during prenatal and postnatal development, at peak bone mass and in adulthood. Juvenile analysis establishes the role of gene in skeletal development and growth, whereas adult analysis establishes its role in skeletal maintenance and repair. Gene expression pattern of the deleted gene can be investigated by LacZ staining, taking advantage of the reporter gene included in the knockout-first gene targeting cassette. Temporal expression can be established at different time points in embryonic and adult mice.
To determine the cellular basis of the abnormal phenotype, static and dynamic histomorphometry can be performed to identify abnormalities of osteoclastic bone resorption and osteoblastic bone formation (Bassett et al. 2010). Primary chondrocytes, osteoblasts and osteoclasts, cultures from wild-type and mutant mice can be undertaken to determine the consequences of gene deletion on cell proliferation, differentiation and function (Bassett et al. 2008). If these studies suggest that the phenotype is a consequence of a defect in a specific bone cell lineage, conditional deletion of the gene in the specific cell type may be used to determine if the phenotype of the global knockout is recapitulated. By crossing the knockout mouse line with an Flp deleter strain, the gene-trap knockout is reversed and a loxP-flanked (‘floxed’) allele is generated (Fig. 2). Subsequently, floxed mice can be crossed with bone cell lineage-specific Cre strains (Murray et al. 2012) (Table 2).
Table 2.
Examples of cell-specific promoter-driven Cre recombinases in available transgenic mouse lines
| Cell type | Mouse lines expressing Cre-recombinase with cell-specific promoter | Equivalent inducible Cre-recombinase | References |
|---|---|---|---|
| Osteoblasts | OC-Cre, Col1a1-Cre | Col1-CreERT2 | (Kim et al. 2004; Nakanishi et al. 2008; Zhang et al. 2002) |
| Osteoclasts | Ctsk-Cre, LysM-Cre | Ctsk-CreERT2 | (Chiu et al. 2004; Sanchez-Fernandez et al. 2012) |
| Osteocytes | Dmp1-Cre | Dmp1-CreERT2 | (Lu et al. 2007; Powell et al. 2011) |
| Chondrocytes | Col2-Cre | Agc1-CreERT2 | (Henry et al. 2009; Yoon et al. 2005) |
To determine the molecular basis of the skeletal phenotype, the effect of the gene knockout on global gene expression can be examined by whole-genome microarray analysis of RNA extracted from bones of the knockout mice or from cell cultures. Cluster analysis is performed to identify patterns of gene expression that suggest involvement of specific signalling pathways. In addition, whole transcriptome analysis can be performed by RNA sequencing with next-generation sequencing techniques. Bioinformatics analysis can determine the effect of the gene deletion on complex gene networks including alternative splice variants and regulatory elements such as non-coding RNAs (Vikman et al. 2014).
Although the rapid-throughput skeletal phenotyping screen is a powerful technique to identify new gene that regulates bone mineralisation and strength, it has limitations related to (i) the use of mice as a model system, (ii) analysis of only knockout animals and (iii) the specific skeletal phenotyping methods selected.
There are well-described differences in bone structure, bone remodelling and hormonal changes between humans and mice. Despite these differences, key molecules that regulate bone and cartilage have been shown to have the same functions in mice and humans and many inherited skeletal disorders are recapitulated in genetically modified mice. In addition, although there is no mouse menopause, the consequences of oestrogen deficiency can still be studied in mice using the ovariectomy provocation model. Furthermore, if extreme skeletal phenotypes are identified in mouse knockout lines, genetic variation in the homologous human genes can be investigated in large well-characterised human populations such as those included in the GEnetic Factors for OSteoporosis Consortium (GeFOS) (http://www.gefos.org/).
The IKMC/IMPC knockout strategy aims to determine the physiological role of genes by identifying the pathophysiological consequences of loss-of-function but will not identify phenotypes associated with gain-of-function mutations, epigenetic modifications or other environmental risk factors. Nevertheless, if a significant skeletal phenotype is detected in knockout animals, the consequences of gain-of-function mutation could then be studied by using CRISPR-Cas-based targeted gene-editing techniques to generate mouse models (Gaj et al. 2013, Sander & Joung 2014).
Robust, sensitive and specific skeletal phenotyping screening requires a combination of complementary, rapid-throughput methodologies but can never be exhaustive. However, once an extreme phenotype has been identified and prioritised for further detailed analysis, other important determinants of bone strength such as tissue mineralisation and bone geometry can be determined by quantitative backscattered electron scanning electron microscopy (Bassett et al. 2010) and statistical shape modelling (Yang et al. 2006), respectively.
In conclusion, systematic, rapid-throughput skeletal phenotyping of genetically modified mice is an exciting new approach that complements human population studies of complex polygenic disorders and has the potential to identify important new signalling pathways involved in the pathogenesis of skeletal disease.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
This work was supported by a Wellcome Trust Strategic Award (grant number 101123) and a Wellcome Trust Project Grant (grant number 094134).
References
- Abrahamsen B, van Staa T, Ariely R, Olson M, Cooper C. 2009. Excess mortality following hip fracture: a systematic epidemiological review. Osteoporosis International 20 1633–1650. ( 10.1007/s00198-009-0920-3) [DOI] [PubMed] [Google Scholar]
- Abrahamsen B, Grove EL, Vestergaard P. 2014. Nationwide registry-based analysis of cardiovascular risk factors and adverse outcomes in patients treated with strontium ranelate. Osteoporosis International 25 757–762. ( 10.1007/s00198-013-2469-4) [DOI] [PubMed] [Google Scholar]
- Alonso N, Ralston SH. 2014. Unveiling the mysteries of the genetics of osteoporosis. Journal of Endocrinological Investigation 37 925–934. ( 10.1007/s40618-014-0149-7) [DOI] [PubMed] [Google Scholar]
- Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, Pepin MG, Weis M, Eyre DR, Walsh J, et al. 2008. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Human Mutation 29 1435–1442. ( 10.1002/humu.20799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, et al. 2001. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Human Molecular Genetics 10 537–543. ( 10.1093/hmg/10.5.537) [DOI] [PubMed] [Google Scholar]
- Barrett-Connor E, Mosca L, Collins P, Geiger MJ, Grady D, Kornitzer M, McNabb MA, Wenger NK. 2006. Effects of raloxifene on cardiovascular events and breast cancer in postmenopausal women. New England Journal of Medicine 355 125–137. ( 10.1056/NEJMoa062462) [DOI] [PubMed] [Google Scholar]
- Bassett JH, Williams AJ, Murphy E, Boyde A, Howell PG, Swinhoe R, Archanco M, Flamant F, Samarut J, Costagliola S, et al. 2008. A lack of thyroid hormones rather than excess thyrotropin causes abnormal skeletal development in hypothyroidism. Molecular Endocrinology 22 501–512. ( 10.1210/me.2007-0221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett JH, Boyde A, Howell PG, Bassett RH, Galliford TM, Archanco M, Evans H, Lawson MA, Croucher P, St Germain DL, et al. 2010. Optimal bone strength and mineralization requires the type 2 iodothyronine deiodinase in osteoblasts. PNAS 107 7604–7609. ( 10.1073/pnas.0911346107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett JH, Gogakos A, White JK, Evans H, Jacques RM, van der Spek AH, Ramirez-Solis R, Ryder E, Sunter D, Boyde A, et al. 2012a. Rapid-throughput skeletal phenotyping of 100 knockout mice identifies 9 new genes that determine bone strength. PLoS Genetics 8 e1002858 ( 10.1371/journal.pgen.1002858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett JH, van der Spek A, Gogakos A, Williams GR. 2012b. Quantitative X-ray imaging of rodent bone by Faxitron. Methods in Molecular Biology 816 499–506. ( 10.1007/978-1-61779-415-5_29) [DOI] [PubMed] [Google Scholar]
- Bogue MA, Churchill GA, Chesler EJ. 2015. Collaborative cross and diversity outbred data resources in the mouse phenome database. Mammalian Genome 26 511–520. ( 10.1007/s00335-015-9595-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone HG, Dempster DW, Eisman JA, Greenspan SL, McClung MR, Nakamura T, Papapoulos S, Shih WJ, Rybak-Feiglin A, Santora AC, et al. 2015. Odanacatib for the treatment of postmenopausal osteoporosis: development history and design and participant characteristics of LOFT, the Long-Term Odanacatib Fracture Trial. Osteoporosis International 26 699–712. ( 10.1007/s00198-014-2944-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of Bone and Mineral Research 25 1468–1486. ( 10.1002/jbmr.141) [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. 2002. High bone density due to a mutation in LDL-receptor-related protein 5. New England Journal of Medicine 346 1513–1521. ( 10.1056/NEJMoa013444) [DOI] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Bürger A, Bult CJ, et al. 2012. The mammalian gene function resource: the international knockout mouse consortium. Mammalian Genome 23 580–586. ( 10.1007/s00335-012-9422-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brommage R, Liu J, Hansen GM, Kirkpatrick LL, Potter DG, Sands AT, Zambrowicz B, Powell DR, Vogel P. 2014. High-throughput screening of mouse gene knockouts identifies established and novel skeletal phenotypes. Bone Research 2 14034 ( 10.1038/boneres.2014.34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW. 2012. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mammalian Genome 23 632–640. ( 10.1007/s00335-012-9427-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, et al. 2001. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. American Journal of Human Genetics 68 577–589. ( 10.1086/318811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. 2010. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. American Journal of Human Genetics 86 6–22. ( 10.1016/j.ajhg.2009.11.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WS, McManus JF, Notini AJ, Cassady AI, Zajac JD, Davey RA. 2004. Transgenic mice that express Cre recombinase in osteoclasts. Genesis 39 178–185. ( 10.1002/gene.20041) [DOI] [PubMed] [Google Scholar]
- Choi YA, Lim J, Kim KM, Acharya B, Cho JY, Bae YC, Shin HI, Kim SY, Park EK. 2010. Secretome analysis of human BMSCs and identification of SMOC1 as an important ECM protein in osteoblast differentiation. Journal of Proteome Research 9 2946–2956. ( 10.1021/pr901110q) [DOI] [PubMed] [Google Scholar]
- Chong B, Hegde M, Fawkner M, Simonet S, Cassinelli H, Coker M, Kanis J, Seidel J, Tau C, Tuysuz B, et al. 2003. Idiopathic hyperphosphatasia and TNFRSF11B mutations: relationships between phenotype and genotype. Journal of Bone and Mineral Research 18 2095–2104. ( 10.1359/jbmr.2003.18.12.2095) [DOI] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. 2007. A mouse for all reasons. Cell 128 9–13. ( 10.1016/j.cell.2006.12.018) [DOI] [PubMed] [Google Scholar]
- Cooper DN. 2010. Functional intronic polymorphisms: buried treasure awaiting discovery within our genes. Human Genomics 5 284–288. ( 10.1186/1479-7364-4-5-284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert AE, de Vernejoul MC, Muraca M, Del Fattore A. 2015. Osteopetrosis and its relevance for the discovery of new functions associated with the skeleton. International Journal of Endocrinology 2015 372156 ( 10.1155/2015/265151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RD, Church CD. 2011. Mouse models and the interpretation of human GWAS in type 2 diabetes and obesity. Disease Models & Mechanisms 4 155–164. ( 10.1242/dmm.000414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Angelis MH, Nicholson G, Selloum M, White JK, Morgan H, Ramirez-Solis R, Sorg T, Wells S, Fuchs H, Fray M, et al. 2015. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nature Genetics 47 969–978. ( 10.1038/ng.3360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany AM, Amling M, Priemel M, Howe C, Baron R, Canalis E. 2000. Osteopenia and decreased bone formation in osteonectin-deficient mice. Journal of Clinical Investigation 105 915–923. ( 10.1172/JCI7039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng FY, Lei SF, Chen XD, Tan LJ, Zhu XZ, Deng HW. 2011a. An integrative study ascertained SOD2 as a susceptibility gene for osteoporosis in Chinese. Journal of Bone and Mineral Research 26 2695–2701. ( 10.1002/jbmr.471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng FY, Lei SF, Zhang Y, Zhang YL, Zheng YP, Zhang LS, Pan R, Wang L, Tian Q, Shen H, et al. 2011b. Peripheral blood monocyte-expressed ANXA2 gene is involved in pathogenesis of osteoporosis in humans. Molecular & Cellular Proteomics 10 M111.011700 ( 10.1074/mcp.m111.011700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan ME, Baldarelli RM, Bello SM, Ni L, McAndrews MS, Bult CJ, Kadin JA, Richardson JE, Ringwald M, Eppig JT, et al. 2015. Orthology for comparative genomics in the mouse genome database. Mammalian Genome 26 305–313. ( 10.1007/s00335-015-9588-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC, Eastell R, Prince RL, Eisman JA, Jones G, et al. 2011. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genetics 7 e1001372 ( 10.1371/journal.pgen.1001372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esapa CT, Bassett JH, Evans H, Croucher PI, Williams GR, Thakker RV. 2012. Bone mineral content and density. Current Protocols in Mouse Biology 2 365–400. ( 10.1002/9780470942390.mo120124) [DOI] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, et al. 2012. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nature Genetics 44 491–501. ( 10.1038/ng.2249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, et al. 2000. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nature Genetics 25 343–346. ( 10.1038/77131) [DOI] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. 2009. Human genetic variation and its contribution to complex traits. Nature Reviews Genetics 10 241–251. ( 10.1038/nrg2554) [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology 31 397–405. ( 10.1016/j.tibtech.2013.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DA, 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G. 2005. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Developmental Cell 8 751–764. ( 10.1016/j.devcel.2005.02.017) [DOI] [PubMed] [Google Scholar]
- Goldring SR. 2015. The osteocyte: key player in regulating bone turnover. RMD Open 1 e000049 ( 10.1136/rmdopen-2015-000049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. 2001. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107 513–523. ( 10.1016/S0092-8674(01)00571-2) [DOI] [PubMed] [Google Scholar]
- Gueguen R, Jouanny P, Guillemin F, Kuntz C, Pourel J, Siest G. 1995. Segregation analysis and variance components analysis of bone mineral density in healthy families. Journal of Bone and Mineral Research 10 2017–2022. ( 10.1002/jbmr.5650101223) [DOI] [PubMed] [Google Scholar]
- Guerrini MM, Sobacchi C, Cassani B, Abinun M, Kilic SS, Pangrazio A, Moratto D, Mazzolari E, Clayton-Smith J, Orchard P, et al. 2008. Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. American Journal of Human Genetics 83 64–76. ( 10.1016/j.ajhg.2008.06.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genetics 9 e1003569 ( 10.1371/journal.pgen.1003569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino TJ, Hentunen TA, Vaananen HK. 2002. Osteocytes inhibit osteoclastic bone resorption through transforming growth factor-beta: enhancement by estrogen. Journal of Cellular Biochemistry 85 185–197. ( 10.1002/jcb.10109) [DOI] [PubMed] [Google Scholar]
- Henry SP, Jang CW, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. 2009. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis 47 805–814. ( 10.1002/dvg.20564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernlund E, Svedbom A, Ivergard M, Compston J, Cooper C, Stenmark J, McCloskey EV, Jonsson B, Kanis JA. 2013. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Archives of Osteoporosis 8 136 ( 10.1007/s11657-013-0136-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF, Glatt V, Bouxsein ML, Ai M, Warman ML, et al. 2004. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. Journal of Bone and Mineral Research 19 2033–2040. ( 10.1359/jbmr.040907) [DOI] [PubMed] [Google Scholar]
- Holroyd C, Harvey N, Dennison E, Cooper C. 2012. Epigenetic influences in the developmental origins of osteoporosis. Osteoporosis International 23 401–410. ( 10.1007/s00198-011-1671-5) [DOI] [PubMed] [Google Scholar]
- Hong SH, Jiang X, Chen L, Josh P, Shin DG, Rowe D. 2012. Computer-automated static, dynamic and cellular bone histomorphometry. Journal of Tissue Science & Engineering 5 (Supplement 1) 004 ( 10.4172/2157-7552.S1-004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium 2005. A haplotype map of the human genome. Nature 437 1299–1320. ( 10.1038/nature04226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Ng MY, Sham PC, Zintzaras E, Lewis CM, Deng HW, Econs MJ, Karasik D, Devoto M, Kammerer CM, et al. 2007. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. Journal of Bone and Mineral Research 22 173–183. ( 10.1359/jbmr.060806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnell O, Kanis JA. 2006. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporosis International 17 1726–1733. ( 10.1007/s00198-006-0172-4) [DOI] [PubMed] [Google Scholar]
- Johnell O, Kanis JA, Oden A, Johansson H, De Laet C, Delmas P, Eisman JA, Fujiwara S, Kroger H, Mellstrom D, et al. 2005. Predictive value of BMD for hip and other fractures. Journal of Bone and Mineral Research 20 1185–1194. ( 10.1359/JBMR.050304) [DOI] [PubMed] [Google Scholar]
- Johnson ML, Gong G, Kimberling W, Reckér SM, Kimmel DB, Recker RB. 1997. Linkage of a gene causing high bone mass to human chromosome 11 (11q12-13). American Journal of Human Genetics 60 1326–1332. ( 10.1086/515470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Meehan TF, Morgan H, Mason JC, Blake A, Kurbatova N, Smedley D, Jacobsen J, Mott RF, Iyer V, et al. 2015. Applying the ARRIVE guidelines to an in vivo database. PLoS Biology 13 e1002151 ( 10.1371/journal.pbio.1002151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen RW, Hart DJ, Arden NK, Doyle DV, Spector TD. 1999. Family history of appendicular fracture and risk of osteoporosis: a population-based study. Osteoporosis International 10 161–166. ( 10.1007/s001980050211) [DOI] [PubMed] [Google Scholar]
- Kelly PJ, Nguyen T, Hopper J, Pocock N, Sambrook P, Eisman J. 1993. Changes in axial bone density with age: a twin study. Journal of Bone and Mineral Research 8 11–17. ( 10.1002/jbmr.5650080103) [DOI] [PubMed] [Google Scholar]
- Kennedy OD, Herman BC, Laudier DM, Majeska RJ, Sun HB, Schaffler MB. 2012. Activation of resorption in fatigue-loaded bone involves both apoptosis and active pro-osteoclastogenic signaling by distinct osteocyte populations. Bone 50 1115–1122. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennel KA, Drake MT. 2009. Adverse effects of bisphosphonates: implications for osteoporosis management. Mayo Clinic Proceedings 84 632–637. ( 10.1016/S0025-6196(11)60752-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Nakashima K, de Crombrugghe B. 2004. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth. American Journal of Pathology 165 1875–1882. ( 10.1016/S0002-9440(10)63240-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornak U, Kasper D, Bosl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. 2001. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 104 205–215. ( 10.1016/S0092-8674(01)00206-9) [DOI] [PubMed] [Google Scholar]
- Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, Blake A, Chen CK, Easty R, Di Fenza A, et al. 2014. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Research 42 D802–D809. ( 10.1093/nar/gkt977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DL, Boyle WJ, Simonet WS, Kostenuik PJ, Dougall WC, Sullivan JK, San Martin J, Dansey R. 2012. Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nature Reviews. Drug Discovery 11 401–419. ( 10.1038/nrd3705) [DOI] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, et al. 2002. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. American Journal of Human Genetics 2002. 70 11–19. ( 10.1086/338450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. 2007. DMP1-targeted Cre expression in odontoblasts and osteocytes. Journal of Dental Research 86 320–325. ( 10.1177/154405910708600404) [DOI] [PubMed] [Google Scholar]
- Makras P, Delaroudis S, Anastasilakis AD. 2015. Novel therapies for osteoporosis. Metabolism 64 1199–1214. () [DOI] [PubMed] [Google Scholar]
- Manolio TA. 2010. Genomewide association studies and assessment of the risk of disease. New England Journal of Medicine 363 166–176. ( 10.1056/NEJMra0905980) [DOI] [PubMed] [Google Scholar]
- Marshall D, Johnell O, Wedel H. 1996. Meta-analysis of how well measures of bone mineral density predict occurrence of osteoporotic fractures. BMJ 312 1254–1259. ( 10.1136/bmj.312.7041.1254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Streeten EA. 2013. Clinical impact of recent genetic discoveries in osteoporosis. Application of Clinical Genetics 6 75–85. ( 10.2147/TACG.S52047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Yerges-Armstrong LM. 2011. The genetics of bone loss: challenges and prospects. Journal of Clinical Endocrinology and Metabolism 96 1258–1268. ( 10.1210/jc.2010-2865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, Sewter CP, Digby JE, Mohammed SN, Hurst JA, et al. 1997. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature 387 903–908. ( 10.1038/43185) [DOI] [PubMed] [Google Scholar]
- Murray SA, Eppig JT, Smedley D, Simpson EM, Rosenthal N. 2012. Beyond knockouts: cre resources for conditional mutagenesis. Mammalian Genome 23 587–599. ( 10.1007/s00335-012-9430-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi R, Akiyama H, Kimura H, Otsuki B, Shimizu M, Tsuboyama T, Nakamura T. 2008. Osteoblast-targeted expression of Sfrp4 in mice results in low bone mass. Journal of Bone and Mineral Research 23 271–277. ( 10.1359/jbmr.071007) [DOI] [PubMed] [Google Scholar]
- Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, Bonewald LF, Kodama T, Wutz A, Wagner EF, et al. 2011. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nature Medicine 17 1231–1234. ( 10.1038/nm.2452) [DOI] [PubMed] [Google Scholar]
- Pangrazio A, Poliani PL, Megarbane A, Lefranc G, Lanino E, Di Rocco M, Rucci F, Lucchini F, Ravanini M, Facchetti F, et al. 2006. Mutations in OSTM1 (grey lethal) define a particularly severe form of autosomal recessive osteopetrosis with neural involvement. Journal of Bone and Mineral Research 21 1098–1105. ( 10.1359/jbmr.060403) [DOI] [PubMed] [Google Scholar]
- Powell WF, Jr, Barry KJ, Tulum I, Kobayashi T, Harris SE, Bringhurst FR, Pajevic PD. 2011. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. Journal of Endocrinology 209 21–32. ( 10.1530/joe-10-0308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggatt LJ, Partridge NC. 2010. Cellular and molecular mechanisms of bone remodeling. Journal of Biological Chemistry 285 25103–25108. ( 10.1074/jbc.R109.041087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JB, Zheng HF, Spector TD. 2012. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nature Reviews Genetics 13 576–588. ( 10.1038/nrg3228) [DOI] [PubMed] [Google Scholar]
- Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE, et al. 2008. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. Journal of Biological Chemistry 283 5866–5875. ( 10.1074/jbc.M705092200) [DOI] [PubMed] [Google Scholar]
- Rousseeuw P, Van Zomeren B. 1990. Unmasking multivariate outliers and leverage points. Journal of the American Statistical Association 85 633–639. ( 10.1080/01621459.1990.10474920) [DOI] [Google Scholar]
- Sanchez-Fernandez MA, Sbacchi S, Correa-Tapia M, Naumann R, Klemm J, Chambon P, Al-Robaiy S, Blessing M, Hoflack B. 2012. Transgenic mice for a tamoxifen-induced, conditional expression of the Cre recombinase in osteoclasts. PLoS ONE 7 e37592 ( 10.1371/journal.pone.0037592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology 32 347–355. ( 10.1038/nbt.2842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattui SE, Saag KG. 2014. Fracture mortality: associations with epidemiology and osteoporosis treatment. Nature Reviews Endocrinology 10 592–602. ( 10.1038/nrendo.2014.125) [DOI] [PubMed] [Google Scholar]
- Schofield PN, Hoehndorf R, Gkoutos GV. 2012. Mouse genetic and phenotypic resources for human genetics. Human Mutation 33 826–836. ( 10.1002/humu.22077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman E, Hopper JL, Bach LA, Cooper ME, Parkinson E, McKay J, Jerums G. 1989. Reduced bone mass in daughters of women with osteoporosis. New England Journal of Medicine 320 554–558. ( 10.1056/NEJM198903023200903) [DOI] [PubMed] [Google Scholar]
- Shah AD, Shoback D, Lewiecki EM. 2015. Sclerostin inhibition: a novel therapeutic approach in the treatment of osteoporosis. International Journal of Women’s Health 7 565–580. ( 10.2147/ijwh.s73244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim I-W, Ebeling PR. 2015. Romosozumab/CDP7851 for the treatment of osteoporosis. Expert Review of Endocrinology & Metabolism 10 471–481. ( 10.1586/17446651.2015.1079482) [DOI] [PubMed] [Google Scholar]
- Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Luthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, et al. 1997. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89 309–319. ( 10.1016/S0092-8674(00)80209-3) [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474 337–342. ( 10.1038/nature10163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobacchi C, Frattini A, Guerrini MM, Abinun M, Pangrazio A, Susani L, Bredius R, Mancini G, Cant A, Bishop N, et al. 2007. Osteoclast-poor human osteopetrosis due to mutations in the gene encoding RANKL. Nature Genetics 39 960–962. ( 10.1038/ng2076) [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. 2012. High-resolution genetic mapping using the mouse diversity outbred population. Genetics 190 437–447. ( 10.1534/genetics.111.132597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes B, Ogilvie D, Wordsworth P, Jones N. 1986. Osteogenesis imperfecta is linked to both type I collagen structural genes. Lancet 2 69–72. ( 10.1016/S0140-6736(86)91609-0) [DOI] [PubMed] [Google Scholar]
- Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. 2004. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38 151–158. ( 10.1002/gene.20012) [DOI] [PubMed] [Google Scholar]
- Trost Z, Trebse R, Prezelj J, Komadina R, Logar DB, Marc J. 2010. A microarray based identification of osteoporosis-related genes in primary culture of human osteoblasts. Bone 46 72–80. () [DOI] [PubMed] [Google Scholar]
- Tu X, Rhee Y, Condon KW, Bivi N, Allen MR, Dwyer D, Stolina M, Turner CH, Robling AG, Plotkin LI, et al. 2012. Sost downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading. Bone 50 209–217. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk FS, Nesbitt IM, Zwikstra EH, Nikkels PG, Piersma SR, Fratantoni SA, Jimenez CR, Huizer M, Morsman AC, Cobben JM, et al. 2009. PPIB mutations cause severe osteogenesis imperfecta. American Journal of Human Genetics 85 521–527. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk FS, Zillikens MC, Micha D, Riessland M, Marcelis CL, de Die-Smulders CE, Milbradt J, Franken AA, Harsevoort AJ, Lichtenbelt KD, et al. 2013. PLS3 mutations in X-linked osteoporosis with fractures. New England Journal of Medicine 369 1529–1536. ( 10.1056/NEJMoa1308223) [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. 2014. Ten years of next-generation sequencing technology. Trends in Genetics 30 418–426. ( 10.1016/j.tig.2014.07.001) [DOI] [PubMed] [Google Scholar]
- Vikman P, Fadista J, Oskolkov N. 2014. RNA sequencing: current and prospective uses in metabolic research. Journal of Molecular Endocrinology 53 R93–R101. ( 10.1530/JME-14-0170) [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. 2012. Five years of GWAS discovery. American Journal of Human Genetics 90 7–24. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420 520–562. ( 10.1038/nature01262) [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154 452–464. ( 10.1016/j.cell.2013.06.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zhang L, Han Y, Lin Y, Deng HW. 2013. Genome-wide approaches for identifying genetic risk factors for osteoporosis. Genome Medicine 5 44 ( 10.1186/gm448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox RD, Lathrop GM, Boriraj VV, et al. 1996. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384 455–458. ( 10.1038/384455a0) [DOI] [PubMed] [Google Scholar]
- Yang Y, Bull A, Rueckert D, Hill A. 2006. 3D statistical shape modeling of long bones. In Biomedical Image Registration, pp 306–314. Eds Pluim JPW, Likar B. eFA Gerritsen. Berlin, Germany: Springer-Verlag; ( 10.1007/11784012_37) [DOI] [Google Scholar]
- Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. 2005. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. PNAS 102 5062–5067. ( 10.1073/pnas.0500031102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiki A, Moriwaki K. 2006. Mouse phenome research: implications of genetic background. ILAR Journal 47 94–102. ( 10.1093/ilar.47.2.94) [DOI] [PubMed] [Google Scholar]
- Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, et al. 2002. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. Journal of Biological Chemistry 277 44005–44012. ( 10.1074/jbc.M208265200) [DOI] [PubMed] [Google Scholar]
- Zhang JG, Tan LJ, Xu C, He H, Tian Q, Zhou Y, Qiu C, Chen XD, Deng HW. 2015. Integrative analysis of transcriptomic and epigenomic data to reveal regulation patterns for BMD variation. PLoS ONE 10 e0138524 ( 10.1371/journal.pone.0138524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, Dahia CL, Park-Min KH, Tobias JH, Kooperberg C, et al. 2015. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 526 112–117. ( 10.1038/nature14878) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a