Abstract
Previous studies examining the reproductive health of alligators in Florida lakes indicate that a variety of developmental and health impacts can be attributed to a combination of environmental quality and exposures to environmental contaminants. The majority of these environmental contaminants have been shown to disrupt normal endocrine signaling. The potential that these environmental conditions and contaminants may influence epigenetic status and correlate to the health abnormalities was investigated in the current study. The red blood cell (RBC) (erythrocyte) in the alligator is nucleated so was used as an easily purified marker cell to investigate epigenetic programming. RBCs were collected from adult male alligators captured at three sites in Florida, each characterized by varying degrees of contamination. While Lake Woodruff (WO) has remained relatively pristine, Lake Apopka (AP) and Merritt Island (MI) convey exposures to different suites of contaminants. DNA was isolated and methylated DNA immuno-precipitation (MeDIP) was used to isolate methylated DNA that was then analyzed in a competitive hybridization using a genome-wide alligator tiling array for a MeDIP-Chip analysis. Pairwise comparisons of alligators from AP and MI to WO revealed alterations in the DNA methylome. The AP vs. WO comparison identified 85 differential DNA methylation regions (DMRs) with ⩾3 adjacent oligonucleotide tiling array probes and 15,451 DMRs with a single oligo probe analysis. The MI vs. WO comparison identified 75 DMRs with the ⩾3 oligo probe and 17,411 DMRs with the single oligo probe analysis. There was negligible overlap between the DMRs identified in AP vs. WO and MI vs. WO comparisons. In both comparisons DMRs were primarily associated with CpG deserts which are regions of low CpG density (1–2 CpG/100 bp). Although the alligator genome is not fully annotated, gene associations were identified and correlated to major gene class functional categories and pathways of endocrine relevance. Observations demonstrate that environmental quality may be associated with epigenetic programming and health status in the alligator. The epigenetic alterations may provide biomarkers to assess the environmental exposures and health impacts on these populations of alligators.
Keywords: Alligator, Environmental epigenetics, Endocrine disruptors, DNA methylation
1. Introduction
The ability of environmental factors and exposures to directly impact the development and health of an organism is mediated in large part through epigenetic mechanisms (Jirtle and Skinner, 2007; Skinner, 2014). Epigenetics is defined as “molecular factors and processes around DNA that regulate genome activity independent of DNA sequence and are mitotically stable” (Skinner, 2011). Biological processes depend on environmental exposures acting through epigenetic mechanisms, such as temperature induced sex determination in alligators and turtles or light induced seasonal breeding (Crews, 2003; Kohno et al., 2014; Parrott et al., 2014; Yoshimura, 2010). Abnormal exposures to factors such as environmental contaminants can also promote alterations in epigenetic programming that can lead to developmental and health impacts (Jirtle and Skinner, 2007; Skinner, 2014). Generally early life exposures have a greater impact on health and biology (Rinaudo and Wang, 2012). Although the specific environmental factors and epigenetic mechanisms may vary, all organisms from plants to humans are influenced by environmental epigenetics.
The American Alligator has been established as an ideal model for the assessment of health impacts resulting from environmental exposures (Guillette et al., 2007). A number of previous studies have demonstrated that alligators from lakes containing a variety of environmental toxicants have significant developmental and reproductive impairments when compared to those animals living in relatively pristine environments (Hamlin and Guillette, 2010; Orlando and Guillette, 2007). A well established example results from comparisons between two central Florida, USA lakes: Lake Apopka (AP) is characterized by moderate levels of agricultural and industrial contaminants, many of which are known endocrine disrupting chemicals (EDCs), and Lake Woodruff (WO) serves as reference site as it is geographically proximate but has remained relatively pristine and undeveloped (Guillette et al., 1994; Gunderson et al., 2001; Milnes et al., 2005, 2008; Moore et al., 2010; Rooney et al., 2003), Fig. 1. Alligators living in Lake Apopka display perturbations of the reproductive system including abnormal ovarian morphology, decreased robustness of sexually dimorphic gene expression within the gonad, and altered levels of circulating sex steroids (Horai et al., 2014). Exposures for a variety of trace elements, chemical toxicants, and known EDCs have been documented for those alligators living in Lake Apopka compared to Lake Woodruff (Guillette et al., 1994). The health of alligators living at Kennedy Space Center and Merritt Island National Wildlife Refuge (MI) is not as well characterized. However, high metal contaminations have been identified at MI and also shown to be present in the alligators liver (Horai et al., 2014). These populations reside in relatively close proximity and a previous study revealed no significantly different genetic structure at the population level (Davis et al., 2002). Clearly environmental factors such as photoperiod, temperature and nutrition can impact the development and health of these populations, however, previous studies have suggested these environmental differences appear negligible, but do need to be considered (Guillette et al., 1994; Horai et al., 2014). In contrast, the Lake Apopka and Merritt Island sites are registered Environmental Protection Agency (EPA) Superfund sites in regards to contamination levels and the Merritt Island site is near the Kennedy Space Center and Cape Canaveral Air Force base. Therefore, the alligator populations in these Florida lakes provide a good model to assess the impacts of environmental exposures.
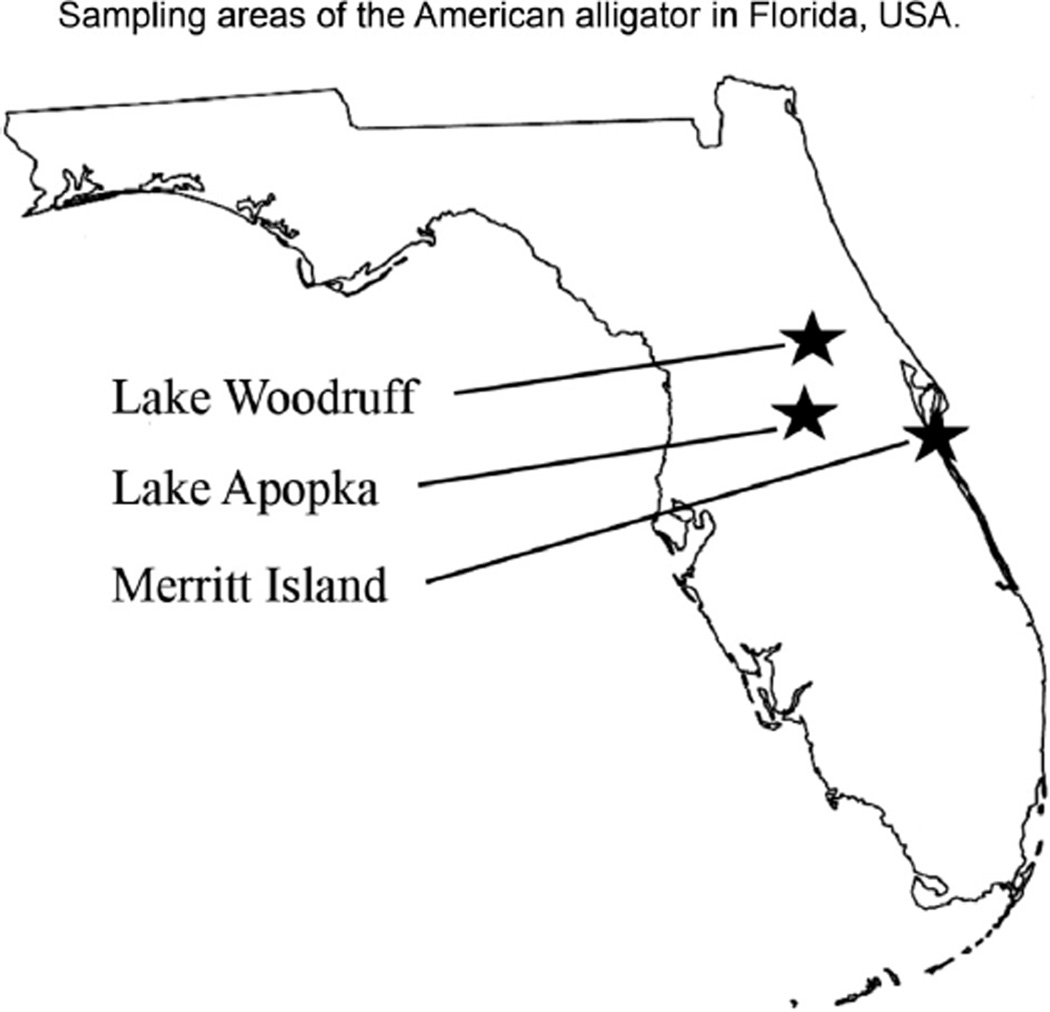
Fig. 1.
Sampling areas of American alligator in Florida, USA. The non-contaminated site Lake Woodruff (WO) and contaminated Superfund sites Lake Apopka (AP) and Merritt Island (MI) are indicated.
Epigenetic reprogramming is an essential aspect of normal development (Skinner, 2011). In most vertebrates examined, an erasure of DNA methylation takes place shortly after fertilization to create the embryonic stem cell that then undergoes cell specific de novo methylation of the genome as the fetus develops (Jirtle and Skinner, 2007; Morgan et al., 2005). All adult stem cells also have a reduced DNA methylation state that then is programmed to differentiate specific cell types (Jirtle and Skinner, 2007; Skinner, 2011, 2014). Early life exposures can alter these normal epigenetic programming events, and the alterations can be propagated in adult cell lineages resulting in distorted transcriptomes and increased disease susceptibility (Skinner, 2011). Exposure of the germ cells (i.e. sperm or egg) early in development can lead to generational impacts on health through epigenetic transgenerational inheritance mechanisms (Anway et al., 2005; Skinner, 2014). In addition to epigenetic programming during transgenerational inheritance and development, epigenetic states in adult animals are also directly influenced by nutrition, stress, and aging during the vertebrate lifespan. Although similar age and sex animals were used, and nutrition and temperature appear to be similar between the sites (Guillette et al., 1994; Horai et al., 2014), these environmental impacts need to be considered in data interpretation. Therefore, a resident population in a contaminated environment might harbor alterations to the epigenome resulting from environmental influences acting at various life history stages. These environmental epigenetic responses can lead to subsequent developmental and health effects.
The current study was designed to investigate potential alterations in the epigenome of alligators living in contaminated and non-contaminated lakes in Florida. Because environmental exposures are consistently present, it is not possible to determine the stage during the organism’s life history that these perturbations arise. However, early life exposures such as fetal or early postnatal have more dramatic effects on epigenetic programming and disease. The identification of an altered epigenome in a marker cell population provides an association with the environmental exposure, epigenetic alteration, and health impacts in the wildlife population. The study investigates the ability to identify epigenetic differences between the populations that are associated with different endocrine disruptor contamination exposures. Observations on a sensitive wildlife species exposed to relevant environmental contaminants can be used to suggest potential human population health impacts from similar exposures.
2. Methods
2.1. Sample collection
Blood and red blood cells were collected from 30 adult male alligators during a 10 day period in April of 2010. Samples were obtained 10 alligators from three research areas: Lake Apopka (AP), Lake Woodruff National Wildlife Refuge (WO), and one lagoon population located in Merritt Island National Wildlife Refuge (MI) (Fig. 1). Adult males of similar size and approximate age were sampled from each site, ranging from 223 cm to 384 cm across sites, Supplemental Table S1. The RBC samples were stored in RNA later and kept under −25 °C until DNA extractions were performed.
2.2. DNA Processing
Erythrocyte DNA was isolated with DNAeasy Blood and Tissue Kit (Qiagen, Valencia, CA) and then stored at −80 °C prior to analysis. DNA was sonicated following a previously described protocol (without protease inhibitors) (Tateno et al., 2000) and then purified using a series of washes and centrifugations (Ward et al., 1999) from a number of animals per collection analyzed. The same concentrations of DNA from individual RBC samples were then used to produce pools of DNA material. Two DNA pools were produced in total per site collection, each one containing the same amount of DNA from different animals. The number of individuals used per pool was 5. These DNA pools were fragmented by sonication and used for methylated DNA immunoprecipitation (MeDIP).
2.3. Differential DNA methylation region (DMR) analysis
MeDIP was performed as previously described (Guerrero-Bosagna et al., 2010) as follows: 6 µg of genomic DNA was subjected to a series of three 20 pulse sonications at 20% amplitude and the appropriate fragment size (200–1000 bp) was verified through 2% agarose gels; the sonicated genomic DNA was resuspended in 350 µl TE buffer and denatured for 10 min at 95 °C and then immediately placed on ice for 5 min; 100 µl of 5× IP buffer (50 mM Na-phosphate pH 7, 700 mM NaCl, 0.25% Triton X-100) was added to the sonicated and denatured DNA. An overnight incubation of the DNA was performed with 5 µg of antibody anti-5-methylCytidine monoclonal from Diagenode (Denville, NJ) at 4 °C on a rotating platform. Protein A/G beads from Santa Cruz were prewashed on PBS-BSA 0.1% and resuspended in 40 µl1 × IP buffer. Beads were then added to the DNA-antibody complex and incubated 2 h at 4 °C on a rotating platform. Beads bound to DNA-antibody complex were washed 3 times with 1 ml 1 × IP buffer; washes included incubation for 5 min at 4 °C on a rotating platform and then centrifugation at 6000 rpm for 2 min. Beads DNA-antibody complex were then resuspended in 250 µl digestion buffer (50 mM Tris HCl pH 8, 10 mM EDTA, 0.5% SDS) and 3.5 µl of proteinase K (20 mg/ml) was added to each sample and then incubated overnight at 55 °C on a rotating platform. DNA purification was performed first with phenol and then with chloroform: isoamyl alcohol. Two washes were then performed with 70% ethanol, 1 M NaCl and glycogen. MeDIP selected DNA was then resuspended in 30 µl TE buffer.
The array used for the differential methylation analysis was a DNA methylated custom array by Roche Nimblegen that consisted of a Whole Genome Tiling Array of the American Alligator (Alligator mississippiensis) made of two chip set arrays with a 200 bp resolution (121211-allmis-ms-meth-ux1). Probe sizes were 50–75 mer in length and median probe spacing was 200 bp. Two different comparative (MeDIP vs. MeDIP) hybridization experiments were performed (2 sub-arrays) for each contaminated lake collection (AP and MI) vs. the non-contaminated WO lake, with each sub-array including hybridizations from MeDIP DNA from DNA pools from these different collections. For one sub-array of each comparison, MeDIP DNA samples from the experimental groups (AP or MI) were labeled with Cy3 and MeDIP DNA samples from the non-contaminated WO were labeled with Cy5.
For each competitive hybridization experiment raw data from both the Cy3 and Cy5 channels were imported into R, checked for quality and converted into MA values. The normalization procedure is as previously described (Guerrero-Bosagna et al., 2010). Following normalization each adjacent ⩾3 probe set value represents the median intensity difference between AP or MI and control WO of a 600 bp window. Significance was assigned to probe differences between experimental comparisons AP or MI and reference WO samples by calculating the median value of the intensity differences as compared to a normal distribution scaled to the experimental mean and standard deviation of the normalized data. A Z-score and P-value were computed for each probe from that distribution. The statistically significant differential DNA methylation regions (DMR) were identified and p-values associated with each region represented, as previously described (Guerrero-Bosagna et al., 2010).
2.4. Additional bioinformatics and statistics
The December 2014 report (Green et al., 2014) of the A. mississippiensis genome produced by the Genome Sequencing Center was retrieved (http://crocgenome.hpc.msstate.edu/crocbase/gbrowsers.html, http://crocgenome.hpc.msstate.edu/gb2/gbrowse/gator/). A seed file was constructed and a BSgenome package was forged for using the Alligator DNA sequence in the R code (Herve Pages BSgenome: Infrastructure for Biostrings-based genome data packages. R package version 1.24.0). This sequence was used to design the custom tiling arrays and to perform the bioinformatics.
The DMR sequence homology BLAST search used the Gene NCBI database and correlated the epimutations associated (overlapped) with the genes. The three adjacent probes constituted approximately a 200 bp homology search. The KEGG pathway associations were identified as previously described (Skinner et al., 2012). All DMR genomic data obtained in the current study have been deposited in the NCBI public GEO database (GSE82318).
3. Results
The experimental design involved the collection of samples from adult male alligators from AP and MI as contaminated lakes and WO as a reference lake as previously described (Horai et al., 2014). The locations of these Florida lakes are shown in Fig. 1. Males of similar size and age were collected from all the sites during a 10-day period in April 2010, see Section 2 and Supplemental Table S1. Blood samples were collected and red blood cells (RBC), i.e. erythrocytes, purified by centrifugation as previously described (Skinner et al., 2014). The samples were frozen at −20 °C until use. The contamination of these lakes has previously been documented with a variety of toxicants identified in Table 1. Previously the health impacts on the alligators in the contaminated AP and MI lakes in comparison with the non-contaminated WO lake have been reported (Guillette et al., 1994; Gunderson et al., 2001; Milnes et al., 2005, 2008; Moore et al., 2010; Rooney et al., 2003), Table 1. The high contamination levels in the Superfund AP and MI sites have been reported (Guillette et al., 1994; Horai et al., 2014) in comparison to the negligible contamination in the WO site. Therefore, previous studies have documented the environmental toxicants, exposures and health impacts on the alligator populations used.
Table 1.
Representative examples of endocrine actions of various environmental contaminants on wildlife and documented health perturbations. American alligators from three sampling sites in Florida, USA: AP: Lake Apopka, MI: Merritt Island National Wildlife Refuge, and WO: Lake Woodruff National Wildlife Refuge. Endocrine activity associated with specific contaminants are presented. Health effects previously documented (Guillette, 2000; Horai et al., 2014) are listed.
| Site | Associated contaminants |
Endocrine activity | Health perturbations in alligators |
|---|---|---|---|
| Lake Apopka | p,p′-DDE | Xenoestrogen (Warembourg et al., 2016), | Altered sex steroid hormone concentrations |
| Antiandrogen (Wilson et al., 2008) | (Guillette et al., 1994, 1996; Moore et al., 2010) | ||
| p,p′-DDD | Xenoestrogen (Zhuang et al., 2012) | Reduced penis size (Guillette et al., 1996) | |
| trans- | Xenoestrogen (Rider et al., 2010) | Abated response to gonadotropin stimulation | |
| nonachlor | (Moore et al., 2010, 2012) | ||
| Dieldrin | Altered hormone metabolism (Fowler et al., 2007), | Increased presence of polyovular follicles | |
| Xenoestrogen (Soto et al., 1994) | (Guillette et al., 1994) | ||
| Mirex | Xenoestrogen (Freire et al., 2014; Warner, 1987) | Decreased robustness of sexually dimorphic gene expression (Milnes et al., 2008) |
|
| Toxaphene | Xenoestrogen (Hodges et al., 2000; Scippo et al., 2004) | Decreased hatch rate (Milnes et al., 2008; Woodward et al., 1993) |
|
| Merritt Island | Elevated Li, Fe, Ni, St, Sb, Pb, Bi, |
Thyroid hormone changes (Apostoli and Catalani, 2011; Lazarus, 2009) |
Reduced length (Trillanes et al., 2014) |
| V, As, S | HPG axis abnormalities (Rossi et al., 2016) Metaloestrogen (Apostoli and Catalani, 2011; Aquino et al., 2012; Darbre, 2006; Farooq, 2015; Gosse et al., 2014; Martin-Diaz et al., 2008) Decreased testosterone (Hutson, 2005; Kariyazono et al., 2015; Rotter et al., 2015) |
Decreased hatch rate (Lance et al., 2006) | |
| Lake Woodruff |
Reference site | None | None |
The epigenome analysis in the RBC involved the isolation of DNA followed by a methylated DNA immunoprecipitation (MeDIP). The MeDIP was then analyzed using a genome-wide alligator tiling array chip set (Chip) to identify differential DNA methylation regions (DMRs), as previously described (Guerrero-Bosagna et al., 2010; Manikkam et al., 2012). Comparative hybridizations between the AP vs. WO samples and MI vs. WO samples were made with two independent experiments with different sets of multiple animals from each site. The MeDIP-Chip analysis used single tiling array oligonucleotide probe resolution, two adjacent probe resolution and ⩾3 adjacent probe resolution, Fig. 2. For the AP vs. WO comparison 15,451 single probe DMR were identified and for ⩾3 adjacent probe 85 DMR were identified, Fig. 2 and Supplemental Table S2. For the MI vs. WO comparison 17,411 single probe DMR and for ⩾3 adjacent probe 75 DMR were identified, Fig. 2 and Supplemental Table S3. A mixture of hypermethylation or hypomethylation of individual DMR was observed for each data set. A statistical significance for individual probe differential methylation of p < 10−4 was used, with the ⩾3 adjacent probe DMR requiring each oligonucleotide probe to have statistical significance. Since the ⩾3 adjacent probe DMR is a much more stringent analysis that reduces false positives observed with the single oligonucleotide probe analysis, all subsequent data analysis used the ⩾3 probe data set.
Fig. 2.

DMR numbers and overlap. (A) The comparison AP vs. WO and MI vs. WO with p < 10−4 for single oligo probe (1+), two adjacent oligo probes (2+) and ⩾3 adjacent oligo probes (3+) detection. (B) Venn diagram overlap for single (1+) probe overlap. The DMR had to have overlapping sequence at the same genomic locations to be considered overlapped.
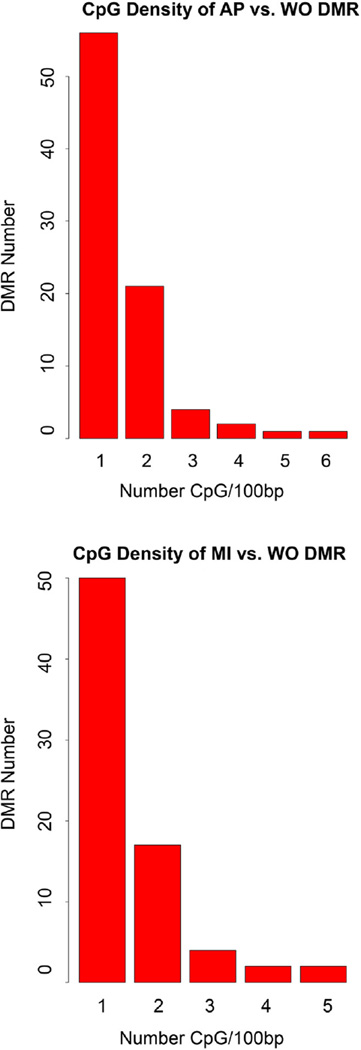
The overlap between the AP vs. WO 85 DMR and MI vs. WO 75 DMR using the ⩾3 oligo probe data set was found to be negligible. Only one genome contig/scaffold JH733431 had an AP DMR at 222669–224394 bp and MI DMR at 43066–44963. When the single oligo nucleotide DMR sets were compared there was approximately 10% of the DMR that were similar between the contaminated lake comparisons, Figure 2B. This overlap requires overlapping genomic locations of the DMR. Since the specific contaminants and doses of exposure are distinct between the AP and MI lakes, differences in the DMR are anticipated. A major genomic feature associated with the DMR involved CpG density. Previously we have observed that environmentally induced DMR primarily develop in CpG deserts with low CpG content (Skinner and Guerrero-Bosagna, 2014). The analysis of both the AP vs. WO and MI vs. WO DMR demonstrated a low density CpG content of 1 or 2 CpG per 100 bp, Fig. 3, which correlates with previously identified environmentally induced DMR.
Fig. 3.
DMR CpG density. The CpG density (CpG/100 bp) and associated DMR (3+ probe) number is presented for (A) AP vs. WO and (B) MI vs. WO. The DMR with a specific CpG density were added to give the total number of DMR for the DMR identified with ⩾3 adjacent oligonucleotide probes.
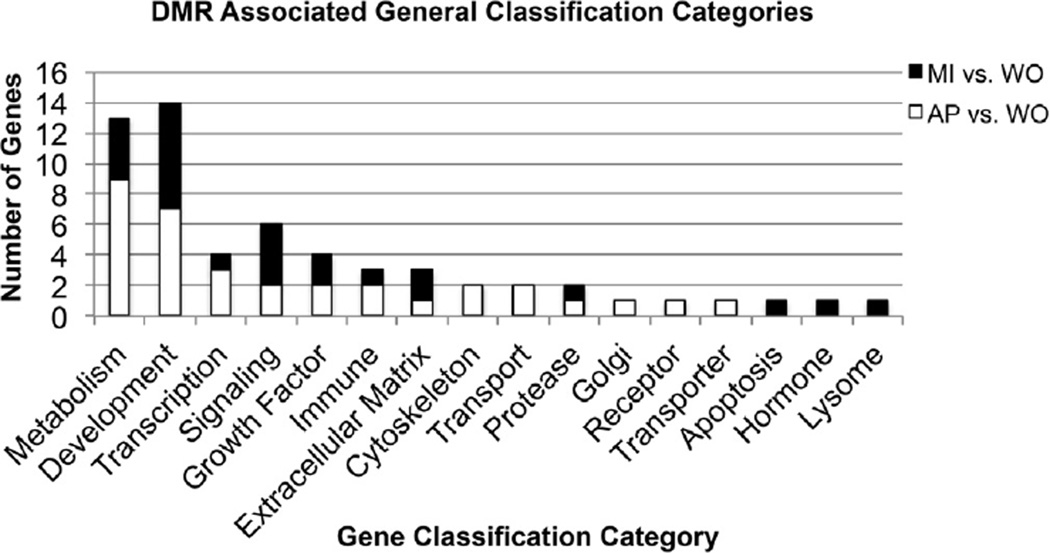
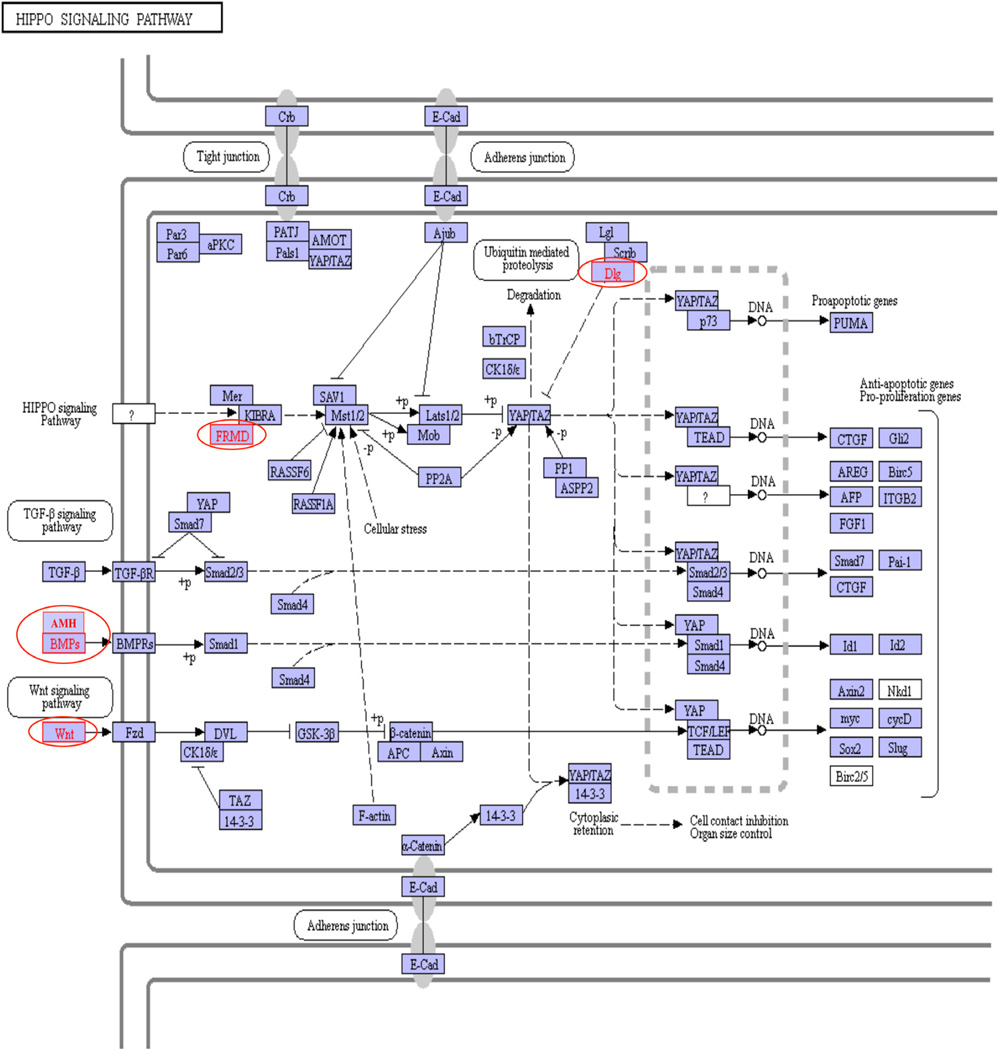
The majority of the alligator genome is not fully annotated such that many of the sites are simple locations on the genome, Supplemental Tables S2 and Supplemental Tables S3 (Green et al., 2014). However, some annotation has occurred and we did sequence homology blast searches for DMR sequences within 5 Kb of a gene to identify the homologies of those genes. The genes currently known to be associated with the AP vs. WO DMR are presented in Table 2 and MI vs. WO DMR are presented in Table 3. The gene classification categories for the genes associated with the DMR are also identified. The major gene classification categories of metabolism, signaling and transcription are present in both DMR data sets, Fig. 4. A KEGG pathway analysis of these DMR gene sets demonstrated an endocrine relevant pathway Hippo signaling pathway was the top pathway with 4 genes (AMH, DLG1, FRMD6, WNT11) involving hormone and growth factor signaling, Fig. 5. Additional pathways for the AP vs. WO DMR associated genes included the metabolic pathway (ALDH5A1, MCEE, SORD and SPR) the calcium signaling pathway (GNAL, IGH), the phospholipase D pathway (EGF, IGH) and PI3 K-Akt pathway (EGF, IGH). The MI vs. WO DMR associated gene pathways included the hippo signaling pathway, viral myocarditis (CXADR, IGH) and HTLV-1 infection pathway (DCG1, WNT11). Therefore a number of endocrine related pathways are present in both DMR data sets, Tables 2 and 3.
Table 2.
Comparison: AP vs. WO DMR gene associations.
| Gene symbol | Full name | Classification | Accession # |
|---|---|---|---|
| LOC102569753 | Gamma-1-syntrophin-like | Development | XM 006258375.1 |
| IGH | Immunglobulin heavy chain region genomic sequence | Immune | JQ479335.1 |
| LMAN1 | Hodgsonii lectin, mannose-binding, 1 | Development | XM 005972817.1 |
| STX17 | Syntaxin 17 | Cytoskeleton | XM 006260382.1 |
| RNPEPL1 | Arginyl aminopeptidase (aminopeptidase B)-like 1 | Metabolism | XM 006260799.1 |
| LOC104386233 | Testis-expressed sequence 2 protein-like | Development | XM 009994809.1 |
| SPR | Sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | Metabolism | XM 006361301.1 |
| IQCK | IQ motif containing K | Development | XM 006259008.1 |
| LOC102387914 | Nucleotide-binding oligomerization domain-containing protein 2-like | Transcription | XM 006039339.1 |
| ITPKA | Inositol-trisphosphate 3-kinase A | Signaling | XM 006265889.1 |
| LOC102574832 | Myosin-9-like | Cytoskeleton | XM 006266873.1 |
| TMEM65 | Transmembrane protein 65 | Extracellular Matrix | XM 006028471.1 |
| LOC102562113 | Lysosomal acid lipase/cholesteryl ester hydrolase-like | Metabolism | XM 006272247.1 |
| B4GALNT3 | Beta-1,4-N-acetyl-galactosaminyl transferase 3 | Golgi | XM 006265693.1 |
| LOC102381513 | Zinc finger protein 135-like | Transcription | XM 006036439.1 |
| SLC16A7 | Solute carrier family 16 (monocarboxylate transporter), member 7 | Transport | XM 005297971.2 |
| V2R | Vomeronasal type 2 receptor | Receptor | KJ847402.1 |
| LOC102382251 | Fatty acid desaturase 1-like | Metabolism | XM 006033326.1 |
| GNAL | Guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type | Signaling | XM 006036342.1 |
| LOC101951603 | Ovochymase-2-like | Development | XM 008170299.1 |
| LOC101941680 | Cytochrome P450 1B1 | Metabolism | XM_008174397.1 |
| SLC44A2 | Solute carrier family 44 (choline transporter), member 2 | Transport | XM_006265591.1 |
| EGF | Epidermal growth factor | Growth factor | XM_006264080.1 |
| LOC102383552 | Magnesium transporter NIPA2-like | Transporter | XM_006026533.1 |
| SORD | Sorbitol dehydrogenase | Metabolism | XM_006020303.1 |
| DNAH9 | Dynein, axonemal, heavy chain 9 | Development | XM_006022043.1 |
| LOC102385461 | General transcription factor IIE subunit 2-like | Transcription | XM_006033990.1 |
| ALDH5A1 | Aldehyde dehydrogenase 5 family, member A1 | Metabolism | XM_006260546.1 |
| MMP23B | Matrix metallopeptidase 23B | Protease | XM_006016178.1 |
| FRMD6 | FERM domain containing 6 | Development | XM_006276033.1 |
| GALNT18 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 | Metabolism | XM_006276188.1 |
| LOC102380209 | Class I histocompatibility antigen, F10 alpha chain-like | Immune | XM_006034603.1 |
| LTBP2 | Latent transforming growth factor beta binding protein 2 | Growth factor | XM_006015345.1 |
| MCEE | Methylmalonyl CoA epimerase | Metabolism | XM_006020025.1 |
Table 3.
Comparison: MI vs. WO DMR gene associations.
| Gene symbol | Full name | Classification | Accession # |
|---|---|---|---|
| KIF6 | Kinesin family member 6 | Extracellular Matrix |
XM_006257878.1 |
| LOC102382038 | C-type lectin domain family 2 member d-like | Signaling | XM_006038095.1 |
| IGH | Immunglobulin heavy chain region | Immune | JQ479336.1 |
| DLG1 | Large homolog 1 (Drosophila) | Development | XM_006258584.1 |
| WIPF3 | WAS/WASL interacting protein family, member 3 | Development | XM_006019590.1 |
| LOC102368067 | Inhibitor of apoptosis protein-like | Apoptosis | XM_006032933.1 |
| LOC102371633 | Multiple epidermal growth factor-like domains protein 6-like | Growth factor | XM_006018743.1 |
| LOC102448388 | G-protein coupled receptor 183-like | Signaling | XM_006115385.1 |
| LOC101941259 | Collagen alpha-1(III) chain-like | Extracellular matrix |
XM_005310545.1 |
| LOC102388048 | Cytochrome P450 26B1-like | Metabolism | XM_006034634.1 |
| LOC102382571 | ADP-ribosylation factor-like protein 13B–like | Metabolism | XM_006026530.1 |
| NRTN | Neurturin | Growth factor | XM_006017519.1 |
| GRPEL1 | GrpE-like 1, mitochondrial (E. coli) | Development | XM_006017017.1 |
| OTC | Ornithine carbamoyltransferase | Metabolism | NM_001287307.1 |
| DUSP3 | Dual specificity phosphatase 3 | Signaling | XM_006268590.1 |
| CXADR | Coxsackie virus and adenovirus receptor | Development | XM_006022916.1 |
| OAF | OAF homolog (Drosophila) | Development | XM_006259692.1 |
| SMCR8 | Smith-Magenis syndrome chromosome region, candidate 8 | Development | XM_006024036.1 |
| AMH | Anti-mullerian hormone | Hormone | AB546782.1 |
| WNT11 | Wingless-type MMTV integration site family, member 11 | Signaling | XM_006021962.1 |
| SRD5A1 | Steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
Metabolism | XM_006271131.1 |
| LOC102371443 | Zinc finger protein 436-like | Transcription | XM_006037782.1 |
| BLOC1S6 | Biogenesis of lysosomal organelles complex-1, subunit 6, pallidin | Lysosome | XM_006030425.1 |
| PHYPADRAFT_164024 | Patens predicted protein | Development | XM_001763795.1 |
| LOC102565785 | Desumoylating isopeptidase 2-like | Protease | XM_006277696.1 |
Fig. 4.
DMR associated gene classifications. AP vs. WO (hatched bar) and MI vs. WO (open bar) gene numbers with specific gene classification categories are presented.
Fig. 5.
Endocrine related Hippo Signaling Pathway. The genes involved in the pathway are identified and those DMR associated genes are circled and lettered in red.
4. Discussion
The alligator is an ideal wildlife model to assess environmental impacts on health (Guillette et al., 2007). The ability to have specific populations in contaminated vs. non-contaminated lakes allows a natural population to be assessed. Previous studies have documented the contamination and health in the Florida lakes studied (Guillette et al., 1994; Gunderson et al., 2001; Milnes et al., 2005, 2008; Moore et al., 2010; Rooney et al., 2003). The natural condition of having a wide variety of contaminants and exposures for multiple generations and exposures on various developmental periods is established in these alligator populations. Therefore, while the specific actions of an individual toxicant at a specific developmental period exposure cannot be determined, the model more accurately portrays the effect of environmental quality on the epigenome in wildlife and human populations. Previously environmental factors such as photoperiod, temperature and nutrition have been suggested to be minimal between the sites (Guillette et al., 1994; Horai et al., 2014), however, they are factors that need to be considered in data interpretation. In contrast, the EPA Superfund sites of AP and MI have extensive documentation of contaminant exposures and dose information to suggest one of the major environmental differences between the AP and MI sites with the WO site are contaminants. The range of exposures from agricultural toxicants, such as DDT and atrazine, to metals, such as mercury, is what would be expected in these lakes (Guillette, 2000; Guillette et al., 1994; Gunderson et al., 2001; Hewitt et al., 2002; Horai et al., 2014) and other exposure sites. The major contaminants and endocrine activities are presented in Table 1. The current study was designed to simply assess if alterations in epigenetic programming correlated with the animal exposures.
In these Florida lakes the impact of contamination on alligator development and health in the AP and MI lakes has been established (Guillette et al., 1994; Gunderson et al., 2001; Milnes et al., 2005, 2008; Moore et al., 2010; Rooney et al., 2003). This includes endocrine abnormalities such as altered levels of circulating sex steroids and aberrant hormone metabolism (Milnes et al., 2005; Moore et al., 2010). Abnormal gonadal development and reproductive function have also been confirmed (Guillette et al., 1994; Gunderson et al., 2001; Milnes et al., 2008). Therefore, developmental effects and pathologies have been observed in the contaminated AP and MI lakes in comparison to the non-contaminated WO lake, Table 1.
Previously a large number of environmental factors and toxicants have been shown to alter epigenetic programming and impact normal physiology and disease (Gomes and Pelosi, 2013; Mirbahai and Chipman, 2014; Skinner, 2014). Epigenetic mechanisms such as DNA methylation, histone modifications, chromatin structure and non-coding RNA all have the ability to dramatically influence genome activity and gene expression (Jirtle and Skinner, 2007; Skinner, 2014). In contrast to DNA sequence that cannot be modified by the vast majority of environmental factors, the epigenome can be dramatically modified by the environment, particularly in early development periods (Jirtle and Skinner, 2007; Skinner, 2011, 2014). This is a normal biological process in allowing environment to mediate its actions on biology through the epigenome. Therefore, when abnormal environmental factors such as toxicants are present the epigenome can be modified to impact development and health. The current study was designed to investigate this using the alligator as a model.
Epigenetic alterations were identified in the RBCs of adult male alligators in the contaminated lakes compared to the non-contaminated lake. The MeDIP-Chip analysis on RBCs as a marker cell identified 85 DMR in the AP vs. WO comparison and 75 DMR in the MI vs. WO comparison. This was a genome-wide analysis on the newly developed alligator genome (Green et al., 2014). This genome has not been completely assembled or annotated yet, but the genome-wide analysis was possible since the entire sequence is known. A custom alligator genome-wide tiling array was used in the current study. A less stringent analysis identified a large number of single oligonucleotide probe DMR, but we focused on the more stringent and reproducible DMR involving ⩾ 3 adjacent oligonucleotide probes in the DMR. The overlap between the AP vs. WO and the MI vs. WO DMR sets identified negligible DMR in common in the alligator RBC between the contaminated lakes. Although approximately 10% overlap was present in the single oligo DMR comparison, Fig. 2. The majority of DMR were unique between the two comparisons. Therefore, the RBC had altered epigenetic programming that was site specific and associated with the different environmental exposures in the contaminated lakes, Table 1.
A genomic feature found in previous environmentally responsive DMR is a low density CpG desert (Skinner and Guerrero-Bosagna, 2014). Interestingly, the alligator DMR identified also occur in these same low density CpG regions of 1 or 2 CpG per 100 bp. Similar observations have been made in multiple species (Skinner and Guerrero-Bosagna, 2014) such that the CpG deserts are thought to be important targets for environmentally induced reprogramming.
Due to the incomplete nature of the alligator genome annotation we investigated sequence homology in a blast search to identify gene associations with the DMR. Less than half of the DMR identified have known gene associations, but those genes identified were further investigated, Supplemental Tables S2 and Supplemental Tables S3. The gene classification categories identified have a variety of categories with the endocrine signaling classification being predominant, Fig. 4. A DMR associated gene pathway analysis identified a number of pathways for both the AP vs. WO and MI vs. WO data sets. Interestingly, one of the top pathways was the endocrine related Hippo signaling pathway, Fig. 5, showing hormone and growth factor signaling. Additional signaling pathways that can be influenced by endocrine agents were also identified such as the PI3 K-AKT and calcium signaling pathways. Therefore, the DMR associated genes identified in both the AP vs. WO and MI vs. WO datasets have the capacity to alter endocrine signaling. Further annotation of the alligator genome will allow this DMR data set to be investigated more extensively. Several interesting regulatory factors such as EGF, V2R and TGFβ2 in the AP vs. WO data set and AMH, WNT11 and neurtorin in the MI vs. WO data set suggests potential regulatory impacts on tissue physiology and development. This supports the concept that the altered epigenetic states identified may impact genome activity and endocrine physiology. Although the health impacts cannot be directly assessed, the potential altered genome activity has the capacity to promote alligator health abnormalities, as previously identified, Table 1.
The study of natural populations and the impacts of environment is a common area of study in population genetics. Environmental exposures generally do not have the capacity to directly alter DNA sequence such that genetic alterations are minimal. In contrast, these exposures can dramatically alter the epigenome as observed in the current study. The potential to do population epigenetics to study the effects of environmental exposures and variable ecologies on wildlife phenotypic variation is supported by the observations of the current study. However, the comparison of the three lakes needs to consider natural exposures such as temperature, nutrition and human development factors, as well as the contamination exposure differences. Although the natural exposure factors have been suggested to be relatively similar between the sites (Guillette et al., 1994), they need to be considered in the data interpretation. The two contaminated lake sites of AP and MI are EPA designated Superfund sites with high exposure contaminant levels documented, such that the contaminants are a significant difference between the AP and MI sites, and very different to the non-contaminated WO site. The interpretation is that the contaminant levels are a major exposure driving the differences in epigenetics observed. Future studies involving additional reference sites and contaminated sites are needed to investigate population variation within the sites and between sites. The more advanced molecular technology of next generation sequencing will facilitate this type of analysis. Therefore, the current study provides the initial report to demonstrate epigenetic differences can be observed between the sites. Future studies are now needed to further investigate the effects of specific exposures and determine the population variation of the sites studied.
The environment has the ability to directly alter epigenetic programming to influence the development and health of an individual or population. The current study identified altered epigenetic programming in the alligator populations that are in contaminated lakes vs. a non-contaminated lake and associated this with altered health impacts on the populations. Although the study of natural populations does not allow direct causal relationships to be established, the correlations observed are supported by laboratory animal experiments as previously reported (Jirtle and Skinner, 2007; Skinner, 2014). The alligator and Florida lake populations were found to be an excellent model to study such associations in wildlife species that mimic potential human exposures in comparable environments. The identification of epigenetic reprogramming suggests the epigenetic changes and DMR may be used as molecular biomarkers for exposures and correlated disease (Manikkam et al., 2012). In the event such environmentally induced epigenetic alterations are present in the germline the potential transgenerational inheritance phenotypes may be observed. Although not possible to be concluded from the current study, such information would suggest potential generational impacts on the alligator populations due to the environmental exposures.
Supplementary Material
Acknowledgments
We thank Dr. Ingrid Sadler-Riggleman, Dr. Daniel Beck and Ms. Jayleana Barton for technical assistance and Ms. Heather Johnson for assistance in preparation of the manuscript. We would also like to thank Ashley Boggs, Russ Lowers, Allan Woodward and Florida Fish and Wildlife Commission employees for assistance with field collections and permitting. The research was supported by South Carolina Centers of Economic Excellence grant to LJG and NIH grant to MKS.
Finally, we would like to dedicate this manuscript to our late friend, mentor, colleague, and co-author, Louis J. Guillette, Jr.
Abbreviations
- RBC
red blood cell
- WO
Lake Woodruff
- AP
Lake Apopka
- MI
Merritt Island
- MeDIP
methylated DNA immunoprecipitation
- DMRs
differential DNA methylation regions
- EDCs
endocrine disrupting compounds
Footnotes
Author contributions
Study conception and design: LJG, MKS.
Acquisition of data: BBP, EN, MMH.
Analysis and interpretation of data: LJG, MKS, BBP, EN, MMH.
Drafting of manuscript: MKS, LJG.
Critical revision: BBP, EN, MMH, MKS.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ygcen.2016.04.012.
References
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostoli P, Catalani S. Metal ions affecting reproduction and development. Met. Ions Life Sci. 2011;8:263–303. [PubMed] [Google Scholar]
- Aquino NB, Sevigny MB, Sabangan J, Louie MC. The role of cadmium and nickel in estrogen receptor signaling and breast cancer: metalloestrogens or not? J. Environ. Sci. Health, C Environ. Carcinog. Ecotoxicol. Rev. 2012;30:189–224. doi: 10.1080/10590501.2012.705159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews D. Sex determination: where environment and genetics meet. Evol. Dev. 2003;5:50–55. doi: 10.1046/j.1525-142x.2003.03008.x. [DOI] [PubMed] [Google Scholar]
- Darbre PD. Metalloestrogens: an emerging class of inorganic xenoestrogens with potential to add to the oestrogenic burden of the human breast. J. Appl. Toxicol. 2006;26:191–197. doi: 10.1002/jat.1135. [DOI] [PubMed] [Google Scholar]
- Davis LM, Glenn TC, Strickland DC, Guillette LJ, Jr, Elsey RM, Rhodes WE, Dessauer HC, Sawyer RH. Microsatellite DNA analyses support an east-west phylogeographic split of American alligator populations. J. Exp. Zool. 2002;294:352–372. doi: 10.1002/jez.10189. [DOI] [PubMed] [Google Scholar]
- Farooq A. Structural and functional diversity of estrogen receptor ligands. Curr. Top. Med. Chem. 2015;15:1372–1384. doi: 10.2174/1568026615666150413154841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler PA, Abramovich DR, Haites NE, Cash P, Groome NP, Al-Qahtani A, Murray TJ, Lea RG. Human fetal testis Leydig cell disruption by exposure to the pesticide dieldrin at low concentrations. Hum. Reprod. 2007;22:2919–2927. doi: 10.1093/humrep/dem256. [DOI] [PubMed] [Google Scholar]
- Freire C, Koifman RJ, Sarcinelli PN, Rosa AC, Clapauch R, Koifman S. Association between serum levels of organochlorine pesticides and sex hormones in adults living in a heavily contaminated area in Brazil. Int. J. Hyg. Environ. Health. 2014;217:370–378. doi: 10.1016/j.ijheh.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Gomes MV, Pelosi GG. Epigenetic vulnerability and the environmental influence on health. Exp. Biol. Med. 2013;238:859–865. doi: 10.1177/1535370213490630. [DOI] [PubMed] [Google Scholar]
- Gosse JA, Taylor VF, Jackson BP, Hamilton JW, Bodwell JE. Monomethylated trivalent arsenic species disrupt steroid receptor interactions with their DNA response elements at non-cytotoxic cellular concentrations. J. Appl. Toxicol. 2014;34:498–505. doi: 10.1002/jat.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St John JA, Capella-Gutierrez S, Castoe TA, Kern C, Fujita MK, Opazo JC, Jurka J, Kojima KK, Caballero J, Hubley RM, Smit AF, Platt RN, Lavoie CA, Ramakodi MP, Finger JW, Jr, Suh A, Isberg SR, Miles L, Chong AY, Jaratlerdsiri W, Gongora J, Moran C, Iriarte A, McCormack J, Burgess SC, Edwards SV, Lyons E, Williams C, Breen M, Howard JT, Gresham CR, Peterson DG, Schmitz J, Pollock DD, Haussler D, Triplett EW, Zhang G, Irie N, Jarvis ED, Brochu CA, Schmidt CJ, McCarthy FM, Faircloth BC, Hoffmann FG, Glenn TC, Gabaldon T, Paten B, Ray DA. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science. 2014;346:1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner M. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillette LJ., Jr Contaminant-induced endocrine disruption in wildlife. IGF Res.: Official Journal of the Growth Hormone Research Society and the International IGF Research Society. 2000;10(Suppl. B):S45–S50. doi: 10.1016/s1096-6374(00)80009-x. [DOI] [PubMed] [Google Scholar]
- Guillette LJ, Jr, Gross TS, Masson GR, Matter JM, Percival HF, Woodward AR. Developmental abnormalities of the gonad and abnormal sex hormone concentrations in juvenile alligators from contaminated and control lakes in Florida. Environ. Health Perspect. 1994;102:680–688. doi: 10.1289/ehp.94102680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillette LJ, Jr, Pickford DB, Crain DA, Rooney AA, Percival HF. Reduction in penis size and plasma testosterone concentrations in juvenile alligators living in a contaminated environment. Gen. Comp. Endocrinol. 1996;101:32–42. doi: 10.1006/gcen.1996.0005. [DOI] [PubMed] [Google Scholar]
- Guillette LJ, Jr, Edwards TM, Moore BC. Alligators, contaminants and steroid hormones. Environ. Sci.: An International Journal of Environmental Physiology and Toxicology. 2007;14:331–347. [PubMed] [Google Scholar]
- Gunderson MP, LeBlanc GA, Guillette LJ., Jr Alterations in sexually dimorphic biotransformation of testosterone in juvenile American alligators (Alligator mississippiensis) from contaminated lakes. Environ. Health Perspect. 2001;109:1257–1264. doi: 10.1289/ehp.011091257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin HJ, Guillette LJ., Jr Birth defects in wildlife: the role of environmental contaminants as inducers of reproductive and developmental dysfunction. Syst. Biol. Reprod. Med. 2010;56:113–121. doi: 10.3109/19396360903244598. [DOI] [PubMed] [Google Scholar]
- Hewitt EA, Crain DA, Gunderson MP, Guillette LJ., Jr Thyroid status in juvenile alligators (Alligator mississippiensis) from contaminated and reference sites on Lake Okeechobee, Florida, USA. Chemosphere. 2002;47:1129–1135. doi: 10.1016/s0045-6535(02)00090-5. [DOI] [PubMed] [Google Scholar]
- Hodges LC, Bergerson JS, Hunter DS, Walker CL. Estrogenic effects of organochlorine pesticides on uterine leiomyoma cells in vitro. Toxicol. Sci.: An Official Journal of the Society of Toxicology. 2000;54:355–364. doi: 10.1093/toxsci/54.2.355. [DOI] [PubMed] [Google Scholar]
- Horai S, Itai T, Noguchi T, Yasuda Y, Adachi H, Hyobu Y, Riyadi AS, Boggs AS, Lowers R, Guillette LJ, Jr, Tanabe S. Concentrations of trace elements in American alligators (Alligator mississippiensis) from Florida, USA. Chemosphere. 2014;108:159–167. doi: 10.1016/j.chemosphere.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Hutson JC. Effects of bismuth citrate on the viability and function of Leydig cells and testicular macrophages. J. Appl. Toxicol. 2005;25:234–238. doi: 10.1002/jat.1060. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariyazono Y, Taura J, Hattori Y, Ishii Y, Narimatsu S, Fujimura M, Takeda T, Yamada H. Effect of in utero exposure to endocrine disruptors on fetal steroidogenesis governed by the pituitary-gonad axis: a study in rats using different ways of administration. J. Toxicol. Sci. 2015;40:909–916. doi: 10.2131/jts.40.909. [DOI] [PubMed] [Google Scholar]
- Kohno S, Parrott BB, Yatsu R, Miyagawa S, Moore BC, Iguchi T, Guillette L., Jr Gonadal differentiation in reptiles exhibiting environmental sex determination. Sexual Dev.: Genetics, Molecular Biology, Evolution, Endocrinology, Embryology, and Pathology of Sex Determination and Differentiation. 2014;8:208–226. doi: 10.1159/000358892. [DOI] [PubMed] [Google Scholar]
- Lance VA, Horn TR, Elsey RM, de Peyster A. Chronic incidental lead ingestion in a group of captive-reared alligators (Alligator mississippiensis): possible contribution to reproductive failure. Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 2006;142:30–35. doi: 10.1016/j.cbpc.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lazarus JH. Lithium and thyroid. Best Pract. Res. Clin. Endocrinol. Metab. 2009;23:723–733. doi: 10.1016/j.beem.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS ONE. 2012;7:e31901. doi: 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Diaz ML, Sales D, DelValls A. Toxicokinetic approach for the assessment of endocrine disruption effects of contaminated dredged material using female Carcinus maenas. Ecotoxicology. 2008;17:495–503. doi: 10.1007/s10646-008-0203-3. [DOI] [PubMed] [Google Scholar]
- Milnes MR, Bermudez DS, Bryan TA, Gunderson MP, Guillette LJ., Jr Altered neonatal development and endocrine function in Alligator mississippiensis associated with a contaminated environment. Biol. Reprod. 2005;73:1004–1010. doi: 10.1095/biolreprod.105.041012. [DOI] [PubMed] [Google Scholar]
- Milnes MR, Bryan TA, Katsu Y, Kohno S, Moore BC, Iguchi T, Guillette LJ., Jr Increased posthatching mortality and loss of sexually dimorphic gene expression in alligators (Alligator mississippiensis) from a contaminated environment. Biol. Reprod. 2008;78:932–938. doi: 10.1095/biolreprod.107.064915. [DOI] [PubMed] [Google Scholar]
- Mirbahai L, Chipman JK. Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposuresMutation research. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2014;764–765:10–17. doi: 10.1016/j.mrgentox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Moore BC, Kohno S, Cook RW, Alvers AL, Hamlin HJ, Woodruff TK, Guillette LJ. Altered sex hormone concentrations and gonadal mRNA expression levels of activin signaling factors in hatchling alligators from a contaminated Florida lake. J. Exp. Zool. Part A, Ecol. Genet. Physiol. 2010;313:218–230. doi: 10.1002/jez.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BC, Roark AM, Kohno S, Hamlin HJ, Guillette LJ., Jr Geneenvironment interactions: the potential role of contaminants in somatic growth and the development of the reproductive system of the American alligator. Mol. Cell. Endocrinol. 2012;354:111–120. doi: 10.1016/j.mce.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Orlando EF, Guillette LJ., Jr Sexual dimorphic responses in wildlife exposed to endocrine disrupting chemicals. Environ. Res. 2007;104:163–173. doi: 10.1016/j.envres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Parrott BB, Kohno S, Cloy-McCoy JA, Guillette LJ., Jr Differential incubation temperatures result in dimorphic DNA methylation patterning of the SOX9 and aromatase promoters in gonads of alligator (Alligator mississippiensis) embryos. Biol. Reprod. 2014;90:2. doi: 10.1095/biolreprod.113.111468. [DOI] [PubMed] [Google Scholar]
- Rider CV, Hartig PC, Cardon MC, Lambright CR, Bobseine KL, Guillette LJ, Jr, Gray LE, Jr, Wilson VS. Differences in sensitivity but not selectivity of xenoestrogen binding to alligator versus human estrogen receptor alpha. Environ. Toxicol. Chem./SETAC. 2010;29:2064–2071. doi: 10.1002/etc.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo P, Wang E. Fetal programming and metabolic syndrome. Annu. Rev. Physiol. 2012;74:107–130. doi: 10.1146/annurev-physiol-020911-153245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney AA, Bermudez DS, Guillette LJ., Jr Altered histology of the thymus and spleen in contaminant-exposed juvenile American alligators. J. Morphol. 2003;256:349–359. doi: 10.1002/jmor.10090. [DOI] [PubMed] [Google Scholar]
- Rossi EM, Marques VB, Nunes Dde O, Carneiro MT, Podratz PL, Merlo E, dos Santos L, Graceli JB. Acute iron overload leads to hypothalamic-pituitary-gonadal axis abnormalities in female rats. Toxicol. Lett. 2016;240:196–213. doi: 10.1016/j.toxlet.2015.10.027. [DOI] [PubMed] [Google Scholar]
- Rotter I, Kosik-Bogacka DI, Dolegowska B, Safranow K, Kuczynska M, Laszczynska M. Analysis of the relationship between the blood concentration of several metals, macro- and micronutrients and endocrine disorders associated with male aging. Environ. Geochem. Health. 2015 doi: 10.1007/s10653-015-9758-0. [DOI] [PubMed] [Google Scholar]
- Scippo ML, Argiris C, Van WeerdtDe Weerdt C, Muller M, Willemsen P, Martial J, Maghuin-Rogister G. Recombinant human estrogen, androgen and progesterone receptors for detection of potential endocrine disruptors. Anal. Bioanal. Chem. 2004;378:664–669. doi: 10.1007/s00216-003-2251-0. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenet.: Official Journal of the DNA Methylation Society. 2011;6:838–842. doi: 10.4161/epi.6.7.16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol. Cell. Endocrinol. 2014;398:4–12. doi: 10.1016/j.mce.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Guerrero-Bosagna C. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genom. 2014;15:692. doi: 10.1186/1471-2164-15-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Haque MM, Zhang B, Savenkova M. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol. 2012;13:R91. doi: 10.1186/gb-2012-13-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Guerrero-Bosagna C, Haque MM, Nilsson EE, Koop JAH, Knutie SA, Clayton DH. Epigenetics and the evolution of Darwin’s Finches. Genome Biol. Evol. 2014;6:1972–1989. doi: 10.1093/gbe/evu158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto AM, Chung KL, Sonnenschein C. The pesticides endosulfan, toxaphene, and dieldrin have estrogenic effects on human estrogen-sensitive cells. Environ. Health Perspect. 1994;102:380–383. doi: 10.1289/ehp.94102380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H, Kimura Y, Yanagimachi R. Sonication per se is not as deleterious to sperm chromosomes as previously inferred. Biol. Reprod. 2000;63:341–346. doi: 10.1095/biolreprod63.1.341. [DOI] [PubMed] [Google Scholar]
- Trillanes CE, Perez-Jimenez JC, Rosiles-Martinez R, Gonzalez-Jauregui M. Metals in the caudal scutes of Morelet’s crocodile (Crocodylus moreletii) from the southern Gulf of Mexico. Bull. Environ. Contam. Toxicol. 2014;93:423–428. doi: 10.1007/s00128-014-1349-8. [DOI] [PubMed] [Google Scholar]
- Ward WS, Kimura Y, Yanagimachi R. An intact sperm nuclear matrix may be necessary for the mouse paternal genome to participate in embryonic development. Biol. Reprod. 1999;60:702–706. doi: 10.1095/biolreprod60.3.702. [DOI] [PubMed] [Google Scholar]
- Warembourg C, Debost-Legrand A, Bonvallot N, Massart C, Garlantezec R, Monfort C, Gaudreau E, Chevrier C, Cordier S. Exposure of pregnant women to persistent organic pollutants and cord sex hormone levels. Hum. Reprod. 2016;31:190–198. doi: 10.1093/humrep/dev260. [DOI] [PubMed] [Google Scholar]
- Warner W. Inhibitory effect of chlordecone and mirex on steroid synthesis in Y-1 cells. J. Environ. Pathol. Toxicol. Oncol. 1987;7:47–54. [PubMed] [Google Scholar]
- Wilson VS, Blystone CR, Hotchkiss AK, Rider CV, Gray LE., Jr Diverse mechanisms of anti-androgen action: impact on male rat reproductive tract development. Int. J. Androl. 2008;31:178–187. doi: 10.1111/j.1365-2605.2007.00861.x. [DOI] [PubMed] [Google Scholar]
- Woodward AR, Percival HF, Jennings ML, Moore CT. Low clutch viability of American alligators on Lake Apopka. Florida Scientist. 1993;56:52–63. [Google Scholar]
- Yoshimura T. Neuroendocrine mechanism of seasonal reproduction in birds and mammals. Anim. Sci. J. (Nihon Chikusan Gakkaiho) 2010;81:403–410. doi: 10.1111/j.1740-0929.2010.00777.x. [DOI] [PubMed] [Google Scholar]
- Zhuang S, Zhang J, Wen Y, Zhang C, Liu W. Distinct mechanisms of endocrine disruption of DDT-related pesticides toward estrogen receptor alpha and estrogen-related receptor gamma. Environ. Toxicol. Chem./SETAC. 2012;31:2597–2605. doi: 10.1002/etc.1986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.