Abstract
Filamentous marine cyanobacteria produce bioactive natural products with both potential therapeutic value and capacity to be harmful to human health. Genome sequencing has revealed that cyanobacteria have the capacity to produce many more secondary metabolites than have been characterized. The biosynthetic pathways that encode cyanobacterial natural products are mostly uncharacterized, and lack of cyanobacterial genetic tools has largely prevented their heterologous expression. Hence, a combination of cutting edge and traditional techniques has been required to elucidate their secondary metabolite biosynthetic pathways. Here, we review the discovery and refined biochemical understanding of the olefin synthase and fatty acid ACP reductase/aldehyde deformylating oxygenase pathways to hydrocarbons, and the curacin A, jamaicamide A, lyngbyabellin, columbamide, and a trans-acyltransferase macrolactone pathway encoding phormidolide. We integrate into this discussion the use of genomics, mass spectrometric networking, biochemical characterization, and isolation and structure elucidation techniques.
Keywords: Cyanobacteria, Natural products, Biosynthesis, Mass spectrometry, Genomics
Introduction
Over the past three decades, natural products isolated from type III tropical filamentous cyanobacteria have provided numerous bioactive therapeutic lead compounds [51], as well as compounds with deleterious effects to human health [10]. In addition, a variety of methylated alkenes and alkanes are produced by several different clades of the phylum, including type III filamentous cyanobacteria [9]. The environmental role of these secondary metabolites is largely unknown, but potent activity in cytotoxicity assays suggests a potential ecological role as potential antagonistic or defense chemicals [42]. Along with interesting chemical functional groups and structural diversity, the cyanobacterial biosynthetic gene clusters described to date display a variety of novel biochemical features [22, 23]. In some cyanobacterial biosynthetic pathways, inter- and intra-species evolutionary adaptation is suggested by an apparent horizontal gene transfer within biosynthetic pathways, characterized by high gene homology but with a unique gene order and corresponding molecular structure [16].
Traditional isolation and structure elucidation techniques are robust and efficient when a secondary metabolite is produced in sufficient quantities. However, when they are produced in small quantities, several other newer approaches become helpful. For example, heterologous expression has been successfully accomplished to produce the cyanobacterial natural products O-demethylbarbamide [25] and lyngbyatoxin [41], and further development of a cyanobacterial “toolbox” is likely to facilitate these types of efforts in the future [54]. Online bioinformatics tools such as NCBI DELTA-BLAST, antiSMASH [3, 33], Nap-DoS [62] and NRPSpredictor [46] enable prediction of biosynthetic enzyme function from DNA sequence information. Advances in mass spectrometric data processing and visualization via molecular networking also enable rapid detection of new compounds that are available in only small quantities [56, 60], and in some cases novel structures can be reliably assigned using innovative algorithmic methods [40]. New cryoprobe designs for high-field NMR coupled with FAST data acquisition techniques are extending the reach of NMR-based structure elucidation to low nanomole quantities of natural products [5, 39].
This review summarizes a number of recent advances in the study of marine cyanobacterial secondary metabolite biosynthesis that have utilized an intriguing diversity of methodologies. For example, the OLS and FAAR/ADO pathways were initially discovered via a comparison of genes and hydrocarbon molecules between cyanobacterial species, and further probed biochemically to determine substrate preferences and mechanisms [32, 34, 47]. Biochemical studies of interacting polyketide synthase (PKS) modules in the curacin A pathway resulted in the characterization of type II docking domains, which mediate module association, thereby facilitating molecular chain elongation [57]. Overexpression of jamaicamide A genes jamA, jamB, and jamC from Moorea producens JHB, followed by in vitro biochemical analyses, has shed light on the mechanism of alkyne formation and has provided a potential mechanism for the creation of natural product derivatives possessing an alkyne for downstream synthetic modification via click chemistry [61]. In a related species, Moorea bouillonii PNG5-198, MS2-based Molecular Networking combined with genome mining was used to uncover the molecule columbamide A and its associated biosynthetic pathway, and revealed possible secondary metabolite pathway regulation features in three closely related Moorea species [26]. Bioinformatic analysis of genome sequences identified the phormidolide biosynthetic pathway in Leptolyngbya sp.; this pathway features trans-acting acyltransferases atypical of cyanobacterial biosynthesis (unpublished data). We discuss these recent biosynthetic findings, update previous reviews on this subject [22, 23], and highlight the potential for new genome comparison technologies in the context of their ability to aid in the discovery of novel cyanobacterial natural products and biosynthetic pathways.
Genome comparison and heterologous expression characterize cyanobacterial hydrocarbon pathways
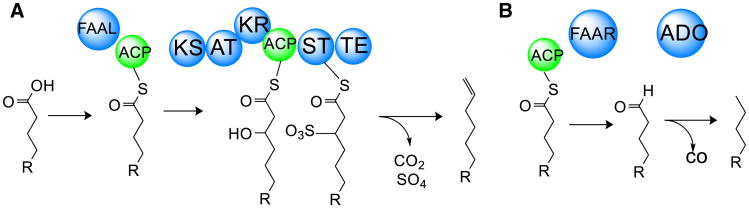
Cyanobacteria have been known to produce odd-chain length hydrocarbons for several decades [18, 59]. The curM gene, which generates the terminal olefin during the biosynthesis of curacin A in Moorea producens 3L, was used as a query sequence for mining the genome of Synechococcus sp. strain PCC 7002, which produces odd-chain length hydrocarbons with a terminal olefin (Fig. 1a) [34]. This research uncovered the olefin synthase (OLS) pathway, which is present in several different clades of cyanobacteria [9]. The biosynthetic portion of the pathway consists of (1) a fatty acyl-ACP ligase (FAAL) that uses ATP to activate a fatty acid of particular length, followed by linkage to the phosphopantetheine prosthetic group of an acyl carrier protein by way of an AMP-bound intermediate, and (2) a modular KS gene which shows significant homology to curM: a canonical KS domain with KS, AT, KR, and ACP domains, followed by a separate sulfotransferase and thioesterase module. The KS domain extends the preceding fatty acyl-ACP via an acetate unit and reduces the β-carbonyl to a hydroxy group. With the exception of Leptolyngbya sp. PCC 7376, all flamentous cyanobacteria which contain the OLS pathway split the FAAL-ACP and KS-AT-KR-ACP-ST-TE into two open reading frames, while in unicellular and baeocystous cyanobacteria, there is only one [9]. Sulfonation of the hydroxyl group via phosphoadenosine-phosphosulfate (PAPS), followed by concerted decarboxylation and desulfation, creates the terminal double bond [17]. Knockout and upregulation experiments in the native organism alternately eliminated or increased C19-hydrocarbon production, respectively, lending credence to the predicted role of the OLS pathway in terminal olefin production [34]. In later experiments, substrate feeding to both purified curM and the OLS KS/ST/TE gene cassette indicated that the OLS KS was unable to process 3-hydroxy 5-methoxy-dodecanoyl-CoA, in contrast to curM. However, both were able to process 3-hydroxy-dodecanoyl CoA [32].
Fig. 1.
Hydrocarbon producing pathways in cyanobacteria. a Organization of the Olefin Synthase (OLS) pathway—FAAL fatty acid-ACP ligase, ACP acyl carrier protein, KS ketosynthase, AT acyltransferase, KR ketoreductase, ST sulfotransferase, TE thioesterase. b Organization of the fatty acid ACP reductase (FAAR)/aldehyde deformylating oxygenase (ADO) pathway
Comparative genomics also helped identify the fatty acid ACP reductase/aldehyde deformylating oxygenase (FAAR/ADO) pathway. By comparing a set of similar genes in cyanobacteria that either did or did not produce odd-chain length alkanes, Schirmer et al. [47] were able to identify two genes in Synechococcus elongatus PCC 7942 that were common to all alkane-producing strains. Heterologous expression in E. coli identified the genes as forming alkanes, and knock-in and knock-out experiments confirmed their role in alkane production. In vitro biochemical experiments determined the first gene to be a fatty acid ACP reductase that catalyzes the reduction of a fatty acid to an aldehyde (Fig. 1b). X-ray crystallography along with expression of a homolog to the second gene lent evidence that it possessed aldehyde decarbonylation activity related to metal atom-catalyzed radical reactions [49]. Further biochemical and structural studies of the ADO enzyme provided evidence for an iron-catalyzed dioxygenase mechanism as the driver for alkane generation [21, 27]. The characterization of these two pathways to hydrocarbons in cyanobacteria was an important discovery that may have consequences to understanding cyanobacterial ecology and evolution [9], and provides an intriguing potential for expression of hydrocarbon-producing enzymes in industrial microbiology applications. In summary, a combination of genome mining and traditional pathway manipulation techniques enabled the identification and activity of these unique biochemical pathways.
Refinement of pathways via biochemical studies
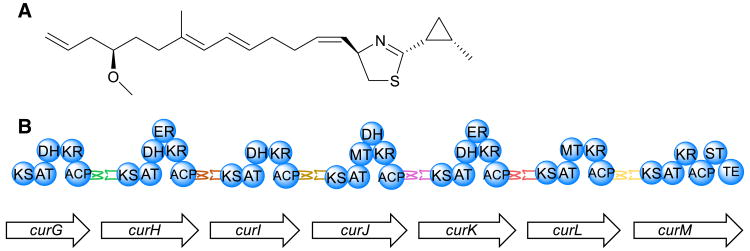
A combination of biochemistry, genome mining, and protein expression was employed to achieve a more fundamental understanding of the interaction between KS modules in the curacin A biosynthetic pathway. This pathway is remarkable in that it is in part composed of seven successive and separate KS modules containing the canonical KS-AT-ACP framework and associated tailoring enzymes, encoded by single genes for each KS module from curG-curM. Each module must interact consecutively with their downstream partner to ultimately produce curacin A (Fig. 2a) [8]. Docking domains, short 20–40 amino acid residue chains found on the C terminus of the upstream acyl carrier protein and downstream N-terminal ketosynthase enzyme in type-I PKS systems, foster selective and specific interaction between successive pathway modules, allowing the growing molecule to be passed from one module to the next with high fidelity (Fig. 2b) [14].
Fig. 2.
a Structure of curacin A, from Moorea producens 3L. b Representation of docking domains between successive PKS modules of curacin A pathway—DH dehydratase, ER enoyl reductase, MT methyltransferase
By comparing the amino acid sequences of several natural product pathways, a new type of docking domain, termed class 2, was identified in myxobacteria and cyanobacteria [57]. In contrast to previously elucidated class 1 docking domains that are found in actinobacteria, class 2 docking domains can employ a flexible structure and non-covalent interaction between ACP α-helix “coiled coils” that is distinct from class 1 docking domains, which employ helices responsible for both dimerization and interaction between ACP and KS, as determined by X-ray crystallography [6, 57]. Additionally, in class 2 docking domains, the ACP and KS can be brought close enough for the ACP to interact with the catalytic domains of the downstream KS module. This new structural interaction was demonstrated by the expression of chimeric enzymes wherein the class 1 docking domains found on two successive modules from the pikromycin pathway were replaced by cognate class 2 docking pairs identified from the curacin A pathway. This construct enabled the processing of synthetic pikromycin pathway intermediates, as measured by mass spectrometry. The insights gained from this multidisciplinary study revealed a broad potential use of class 2 docking domains in the design of expression systems for natural products.
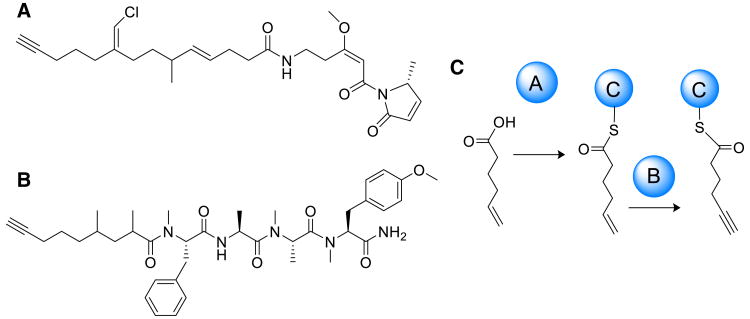
Alkynes present a unique molecular moiety found in bioactive bacterial and cyanobacterial natural products [37], including jamaicamide B [12] (Fig. 3a), viequeamide A [4], veraguamide [35], and carmabin A [19] (Fig. 3b). However, their biosynthetic mechanism of formation has remained uncharacterized. Comparison of the cyanobacterial biosynthetic gene clusters of carmabin A and jamaicamide A, both of which contain an alkyne in their predicted pathway starter units, pointed toward the protein products of a highly conserved set of three genes at the start of each biosynthetic pathway as being responsible for the formation of this functional group [61]. Further genome mining by Zhu et al. identified more than 80 pathways with a similar conserved set of genes [61]. Subsequent expression and purification of the JamA, B, C enzymes enabled mass spectrometry-based in vitro characterization of substrate specificity and cofactor requirements of the gene cassette. JamA was found to be an acyl-ACP synthetase with preference for 5-hexenoic acid, whereas JamB was characterized as a membrane-bound desaturase which generates the alkyne from hexenoic acid loaded onto the JamC acyl carrier protein (Fig. 3c). Additionally, creation of alkyne-tagged molecules was demonstrated in this study via heterologous expression of JamABC with a plant type III PKS, which could be useful in the creation of synthetic probes when integrated with click chemistry. Again, the combination of cyanobacterial genome mining with biochemical experiments and mass spectrometry was an effective approach to developing an understanding of this biosynthetic process, and has helped to develop a new biosynthetic tool for the creation of alkyne-based cellular probes.
Fig. 3.
a Structure of jamaicamide B. b Structure of carmabin A. c Enzymatic functions of JamABC: JamA acyl-ACP synthetase, JamB; membrane-bound desaturase, and JamC; acyl carrier protein
Genome mining: identifying trans-AT systems with intriguing biochemistry in cyanobacteria
Advances in sequencing technology have led to a rapid increase in the number of sequenced cyanobacterial genomes, which has in turn increased the number of identified modular biosynthetic gene clusters for secondary metabolites [23]. Our investigation of the secondary metabolite potential of a Leptolyngbya sp. collected from Sulawesi, Indonesia, led to the identification of a PKS pathway that was ultimately determined to encode for the toxic macrocyclic polyketide, phormidolide [2, 58]. This pathway possessed several unusual features from canonical PKS biosynthetic pathways and was annotated as a trans-AT system [2].
Trans-AT biosynthetic systems represent an emerging class of the biosynthetic pathway in heterotrophic bacteria with examples utilizing PKS [30] and hybrid PKS/NRPS modules [52]. In these systems, a separately encoded AT domain, typically upstream of the PKS megasynthase, loads substrates onto the modular ACPs. In some cases where a trans-AT is present, the modular AT domains, also known as “cis-”AT domains, may be absent [20] or in the case of leinamycin [52], be reduced (∼160 AA) when compared to full-length cis-AT proteins. In the latter case, these KS-AT adaptor regions (ATd) lack the conserved malonate-specific motif GHS[LVIFAM]G [11] which is present in the discrete trans-ATs, and show very low amino acid sequence identity to full-length cis- as well as trans-AT domains. It is possible that the KS-AT adaptor regions are non-functional and evolutionary artifacts [52]. In this latter case, exemplified by leinamycin, the discrete ATs act in trans and possess the acyltransferase activity. Trans-AT systems are hallmarked by PKS modules that are split between megasynthases [20, 30] and the possession of KS domains that are predicted to be inactive and are termed non-elongating KS0 domains [20, 30, 38]. These inactive or non-elongating KSs lack the catalytic His residue in the conserved motif HGTGT [13, 50] that is always present in elongating KS domains. Many trans-AT pathways encode hydroxymethylglutaryl coenzyme A (HMG-CoA) synthase (HCS) cassettes [13, 43, 50], which introduce methyl branches in growing polyketide chains at the C-3 position of the extended acetate unit. Enoyl-CoA hydratase pairs (ECH) are components of the aforementioned HCS cassette and have been shown to catalyze the successive dehydration and decarboxylation of an HMG intermediate during β-branch type methylation events. An interesting feature of the phormidolide pathway is the presence of ECH pairs embedded within the PKS megasynthases of the modular pathway in addition to the ECH pair in the HCS cassette of the tailoring enzyme suite. To date, we do not know the functional role of these “embedded” modular ECH pairs and a full biochemical analysis is needed to determine their role. Duplicated or tandem ACP domains are often observed at β-branch points within modules in the PKS megasynthase architecture [13, 20, 31]. These tandem domains appear to be effective in biosynthetic processes that require multienzyme events such as β-branching methylation [15].
Trans-AT pathways have been described in a diverse array of heterotrophic bacteria, but thus far have been rarely found in cyanobacteria. The first cyanobacterial trans-AT pathway was discovered from a lichen-associated Nostoc sp. encoding the polyketide natural product nosperin [24]. Interestingly, many of the reported trans-AT pathways have been described from bacteria that engage in symbiotic relationships with multicellular eukaryotes [43, 44], and this appears to be the case with this Nostoc sp. Intriguingly, our examination of genomic data from several filamentous marine cyanobacteria has led to the identification of the first trans-AT pathway described from this sub-group of cyanobacteria, and raises interesting questions about the distribution of these pathways as well as trends in the construction of stereochemically complex macrocyclic polyketides in these taxa. The pathway contains several of the deviations from canonical PKS systems described above, including discrete ATs, split modules, non-elongating KS0 domains, tandem ACP domains at β-branch points, and an HCS cassette [2] (Fig. 4). The organism in which the pathway was found, Leptolyngbya sp., is not known to engage in symbiotic relationships with eukaryotes, but rather has a strong tendency to form a biofilm-like substance. As more cyanobacterial genome sequence information becomes available, it will be interesting to observe if trans-AT pathways are of a more common occurrence and to further understand the evolution of these pathways and the biological role of the products they encode.
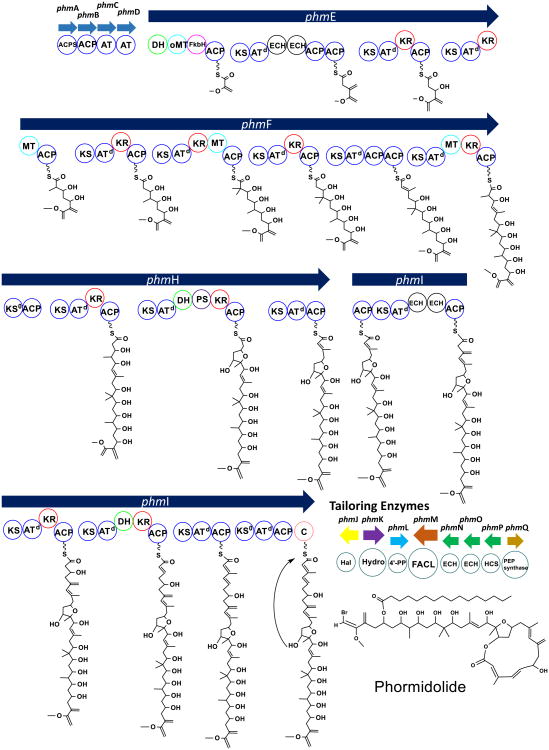
Fig. 4.
Organization of the phormidolide pathway. The pathway, identified from the genome of a Leptolyngbya sp., has many of the reported hallmarks of a trans-AT PKS system, including (1) discrete AT enzymes upstream of the PKS megasynthase architecture; (2) tandem ACP domains; (3) split modules; (4) non-elongating KS0 domains; (5) embedded ECH pairs and (6) HCS cassette. ACPS acyl carrier protein synthase, oMT O-methyltransferase, FkbH FkbH-phosphatase, ATd docking acyltransferase, KS0 non-elongating ketosynthase, PS pyran synthase, C NRPS-like condensation domain, Hal halogenase, Hydro hydrolase, 4′PP 4′-phosphopantetheine, FACL Fatty Acid CoA Ligase, HCS HMG-CoA synthase, PEP phosphoenolpyruvate
Discovery via genomic comparison and MS2-based molecular networking
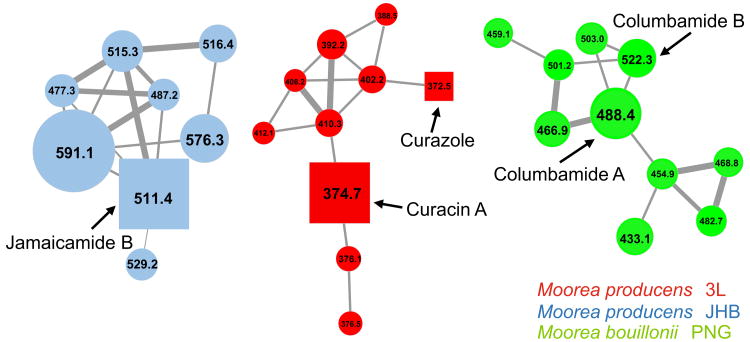
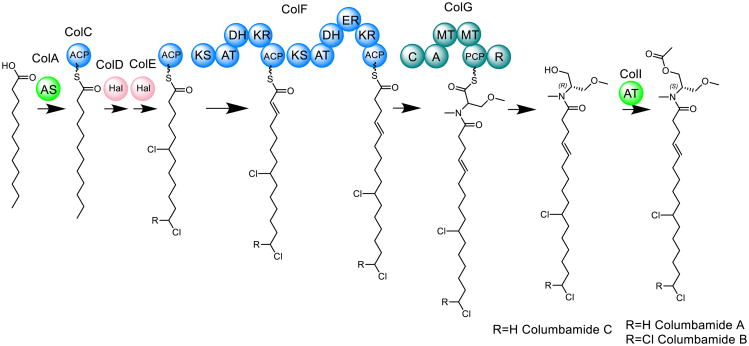
Combining new techniques like mass spectrometric profiling and bioinformatics can help identify interesting targets for chemical isolation and provide orthogonal means for natural product discovery. Mass spectrometric molecular networking [56, 60] is a useful tool for the identification of structural analogs within a chemical extract. Related families of natural products can be detected with algorithms that convert MS-profiled metabolomes to maps of structurally linked molecules on the basis of common fragment ions [55]. In addition, bioinformatic analyses of the biosynthetic gene clusters encoded in DNA sequence data can help to evaluate the uniqueness of compounds present within a genome [3, 33]. In a recent study, the mass spectrometric metabolic profiles of extracts from three marine cyanobacterial strains M. producens 3L, M. producens JHB and M. bouillonii PNG51905-8 were compared with respect to the biosynthetic pathways predicted from their sequenced genomes [26]. These same three extracts were subject to MS2-based molecular networking (Fig. 5). In addition to the well-studied cyanobacterial products curacin A, jamaicamide A and apratoxin A, a new class of chlorinated compounds, the columbamides, was discovered in relatively high yields in the extract of M. bouillonii PNG. Comparison of the known biosynthetic gene clusters from the Moorea strains led to the discovery of a putative regulatory gene, encoding a serine–histidine kinase, next to a biosynthetic gene cluster of unknown function in the M. bouillonii PNG genome. This same gene is adjacent to the curacin A and jamaicamide A biosynthetic gene clusters in M. producens 3L and JHB and, therefore, may be involved in the observed robust constitutive expression of these compounds. Subsequent isolation and structure elucidation primarily by NMR led to the discovery of the columbamides, alkyl amides with cannabinomimetic activity. Based on the bioinformatics analysis, nine enzymes are involved in the assembly of the columbamides. First, two novel halogenases are proposed to insert chlorine atoms at the terminal carbon and ω-7 positions of an acyl carrier protein-bound dodecanoic acid. This intermediate is then extended twice via the addition of acetate with two polyketide synthase modules, and then serine is added via an NRPS module. The molecule is ultimately released from the megasynthase through a reductase mechanism, and the resulting primary hydroxy group is acetylated to yield columbamide A. Figure 6 shows the proposed biosynthetic pathway and the chemical structures of the columbamides. The combined use of molecular networking and genomic comparison enabled the discovery of these unique molecules; integration of these techniques provides a powerful framework for novel bioactive metabolite discovery [26, 56].
Fig. 5.
Select molecular networks clusters derived from mass spectrometric analysis of extracts of M. producens 3L (red), M. producens JHB (blue), M. bouillonii PNG (green). In the nodes, squares indicate consensus MS/MS spectra to compounds in an in-house MS/MS-library of known molecules. The respective compound name of an identified molecule is given next to the square node. The node size is representative of the numbers of MS2-spectra obtained for that specific m/z, and is reflective of the relative abundance of the metabolite. Multiple clusters per compound derive from the fact that [M+H]+ and [M+Na]+ parent ions can fragment differently [60]
Fig. 6.
Proposed biosynthetic pathway of the columbamides in M. bouillonii PNG–AS acyl-ACP synthetase, A adenylation domain, PCP peptidyl carrier protein, R reductase
Development of a cyanobacterial reference genome to foster more accurate genome assembly
Despite the enormous biosynthetic potential of tropical filamentous cyanobacteria, extensive genome mining has remained elusive due to a lack of sequences, which themselves are sparse due to difficulties in culturing these organisms. Currently available molecular biology tools and heterologous expression systems are typically designed for unicellular, heterotrophic bacteria. However, the few available genome sequences for tropical filamentous cyanobacteria are rich with biosynthetic gene clusters. For example, in a comparison of 126 different cyanobacterial genomes, Moorea producens 3L contained 14 PKS, NRPS, or PKS-NRPS biosynthetic gene clusters, the third highest total in the study [7, 48]. However, only four of these clusters encode known products, one cluster is shared between several different genomes but is not elucidated, and nine cannot be connected by bioinformatics to any known metabolite. The genome of M. producens 3L currently contains 161 scaffolds and cannot be completed with the available data due to the lack of a closely related reference sequence; this situation is mirrored for several unpublished genome sequences as well. The closest completed genome to serve as a potential reference to Moorea species is from Microcoleus sp. PCC 7113. This latter genome contains one chromosome and 8 plasmids comprising 7.95 megabases (unpublished data, GenBank access number CP003630.1). In silico predictions of phylogenetic homology using the Genome-to-Genome Distance Calculator [1] indicate that Microcoleus sp. PCC 7113 has only a 12.7–20.7 % similarity index to M. producens 3L. This value is very low compared to the similarity of M. producens 3L to M. producens JHB (58.5–66.8 %), and Moorea sp. PAL 15AUG08-1 (48–57.7 %).
Development of a completed reference genome of a single Moorea species is crucial to future genome comparisons, genome-driven discovery of novel natural products, and a more complete understanding of the evolutionary aspects of this interesting genus. In this regard, the genome of a Moorea sp. PAL15AUG08-1 was sequenced with both Pacific Biosciences (PacBio) and Illumina MiSeq platforms, and is thus the most suitable candidate to be a reference as it is the most complete marine filamentous cyanobacterial genome to date (unpublished). Additionally, Moorea sp. PAL15Aug08-1 is considered a “super-producer”, as it is estimated that 19.8 % of its genome is devoted to natural products biosynthesis; this is nearly four times the average cyanobacterial genome of 5 % encoding for natural products [48]. A total of 43 biosynthetic gene clusters have been identified, including 9 NRPS, 5 PKS and 12 hybrid NRPS/PKS clusters; this is 5 times the average number of NRPS/PKS clusters in cyanobacterial genomes (Fig. 7) [7]. However, only two known compounds have been isolated from this strain to date, palmyramide A [53] and curacin D [29], indicating that there is considerable potential for discovery of new compounds from this strain.
Fig. 7.
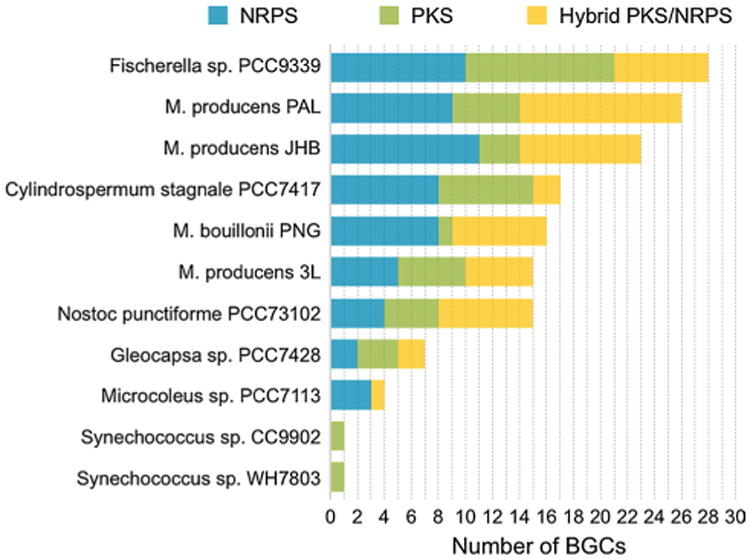
Comparison between PKS/NRPS clusters of 4 Moorea strains with 7 (out of 126) additional cyanobacteria reviewed by Calteau et al., 2014 [7]. The 7 genomes were selected to compare two low secondary metabolite producers (Synechococcus genomes), two average secondary metabolite producers producers (Microcoleus sp. and Gleocapsa sp.) and three high secondary metabolite producers producers (Nostoc punctiforme, C. stagnela and Fischerella sp.). With the exception of Fischerella sp. PCC9339, only final scaffolds were considered, as incomplete genomes tend to present fragmented biosynthetic gene
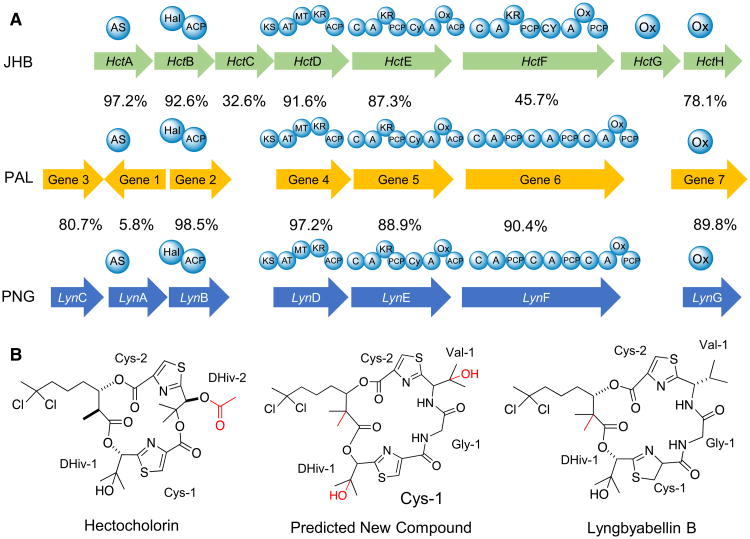
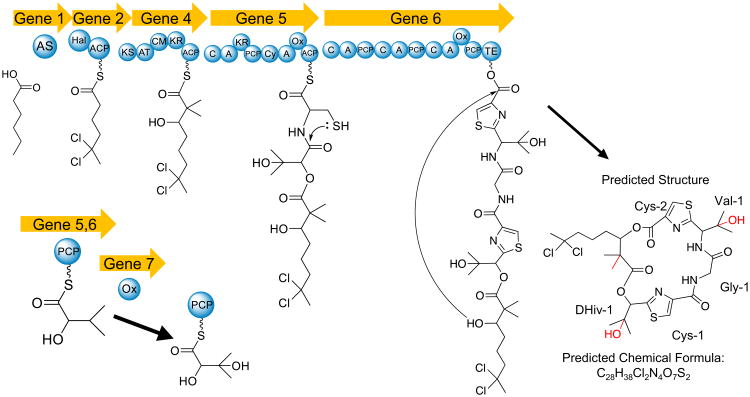
One example of a 50 Kb hybrid NRPS/PKS cluster that is under current investigation encodes for a natural product with significant homology to the biosynthetic pathway for the antifungal lipopeptide hectochlorin A [45] and cytotoxic metabolite lyngbyabellin A [28] (Fig. 8a, b). The first predicted biosynthetic gene of this cluster has 97 % identity with hctA and encodes for a fatty acyl-ACP ligase. However, it is translated in the opposite direction of the remainder of the cluster, possibly indicating an inversion in this ORF relative to the other clusters. The homologous gene in the lyngbyabellin pathway is smaller, which explains the lower identity with the first gene in the PAL cluster, but still shows significant conservation in the regions that overlap. The second gene in this new cluster is highly homologous to the second genes in the hectochlorin and lyngbyabellin gene clusters, and is presumably responsible for the formation of the geminal-dichloro group in these molecules. One disparity is that hctC, a transposase of unknown function, is not present in the new cluster; however, this gene is most likely not related to compound biosynthesis. However, a similar putative transposase gene is found in the lyngbyabellin A gene cluster, and suggests that the new cluster is more distant in its evolutionary history. The third biosynthetic gene in the new cluster is 97.2 % identical to lynD, a PKS module responsible for a single acetate extension, along with a C-methylation from S-adenosyl methionine (sAM). Curiously, in the formation of lyngbyabellin A, a double methylation occurs at this juncture instead of the single methylation observed in hectochlorin. The biochemical basis for this double methylation from a single cMT domain is currently not understood. However, due to the significant conservation between amino acid residues in the lyngbyabellin A cMT domain and the PAL cluster cMT domain, it is reasonable to predict that a double methylation occurs at this step in the new cluster. The fourth gene in the cluster is 94 % identical to hctE gene and is composed of one NRPS/PKS module and one NRPS module (C-A-KR-PCP and Heterocyclization(Cy)-A-Oxidase(Ox)-ACP). In the first amino acid module, online A-domain prediction software (NRPSPredictor2) is unable to discern which amino acid (or keto acid) is activated; however, given its 93 % identity with the first A domain in hctE, it is reasonable to predict that it may also activate 2-hydroxyisovaleric acid. In this regard, a KR domain is observed between the A and PCP domains, similar to those found in hctE and lynE genes, suggesting the initial activation of an α-keto acid with subsequent in situ reduction to an α-hydroxy acid. The second module in this ORF has the same structure of the second module of hctE, and is thus predicted to activate and then cyclize a cysteine residue after its incorporation. The fifth gene in this cluster does not share significant homology to any part of the hectochlorin or lyngbyabellin A gene clusters. This gene has 3 NRPS modules that are predicted to incorporate glycine, valine and cysteine; the latter cysteine is further predicted to be heterocyclized. This is followed by a thioesterase that may catalyze the hydrolysis from the biosynthetic assembly line. Lastly, the sixth gene encodes for a putative P450 monooxygenase that may be responsible for oxidation events during the biosynthesis of this compound, analogous to similar motifs involved in the biosynthesis of hectochlorin and lyngbyabellin A. The final structure of this metabolite is predicted to be similar to lyngbyabellin B (Fig. 9) which is produced by M. producens; however, the biosynthesis of lyngbyabellin B remains to be elucidated [36]. Analysis of LCMS data and molecular networks generated from M. producens PAL 15AUG08-1 chemical extracts has yet to uncover any lyngbyabellin analogs and, hence, the compound appears not to be produced under its current conditions of laboratory culture.
Fig. 8.
a Comparison between hectochlorin and lyngbyabellin A biosynthetic genes versus the new gene cluster identified in M. producens PAL 15AUG08-1. The percentage values represent the amino acid identities of MAFFT alignments between PAL genes and their equivalents above and below in JHB and PNG pathways. b Comparison between structures and residues for the predicted new compound, hectochlorin and lyngbyabellin B. In red, post-translational modifications that are not clearly predicted by biosynthetic gene clusters. Cy heterocyclization domain, Ox oxidase
Fig. 9.
Predicted biosynthesis of M. producens PAL 15AUG08-1 macrolide of structural similarity to hectochlorin and lyngbyabellin B. Red sections represent uncertain positions of P450 oxidation
Conclusion
Recent progress in understanding marine cyanobacterial natural product pathways has required a broad range of methods to explore questions concerning their biosynthesis. While the development of more robust heterologous expression tools would usher a transformative approach to studying cyanobacterial natural products, it is clear that the combination of genome analysis, mass spectrometry, and biochemical studies has provided effective tools for understanding their fundamental pathways. This combination of techniques will most certainly be used, along with improvements in expression tools, to probe the molecules and biosynthetic pathways in cultured cyanobacterial specimens as well as metagenomic and environmental samples. Advances in mass spectrometry visualization have enabled efficient identification of molecular analogs, in particular natural product families. Similarly, development of a complete reference genome for a filamentous marine cyanobacterium will facilitate the more rapid mapping of genomic data and hence the discovery of new biosynthetic gene clusters.
Acknowledgments
This work was supported by NIH Grants CA108874 and GM107550-01. Karin Kleigrewe was supported by a fellowship within the Postdoc-Programme of the German Academic Exchange Service (DAAD). Tiago Ferreira Leão is supported by the Ministry of Education of Brazil, D.F., CAPES fellowship, no. 13425137.
Contributor Information
Nathan A. Moss, Email: namoss@ucsd.edu.
William H. Gerwick, Email: wgerwick@ucsd.edu.
References
- 1.Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertin M, Vulpanovici A, Monroe EA, Korobeynikov A, Sherman DH, Gerwick L, Gerwick WH. The phormidolide biosynthetic gene cluster: a trans-AT PKS pathway encoding a toxic macrocyclic polyketide. ChemBioChem. 2015 doi: 10.1002/cbic.201500467. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucl Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boudreau PD, Byrum T, Liu WT, Dorrestein PC, Gerwick WH. Viequeamide A, a cytotoxic member of the kulolide superfamily of cyclic depsipeptides from a marine button cyanobacterium. J Nat Prod. 2012;75:1560–1570. doi: 10.1021/np300321b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breton RC, Reynolds WF. Using NMR to identify and characterize natural products. Nat Prod Rep. 2013;30:501–524. doi: 10.1039/c2np20104f. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz TJ, Geders TW, Bartley FE, III, Reynolds KA, Smith JL, Sherman DH. Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains. ACS Chem Biol. 2009;4:41–52. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calteau A, Fewer DP, Latifi A, Coursin T, Laurent T, Jokela J, Kerfeld CA, Sivonen K, et al. Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genom. 2014;15:977. doi: 10.1186/1471-2164-15-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 9.Coates RC, Podell S, Korobeynikov A, Lapidus A, Pevzner P, Sherman DH, Gerwick L, et al. Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PloS One. 2014;9(1):e85140. doi: 10.1371/journal.pone.0085140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codd GA, Morrison LF, Metcalf JS. Cyanobacterial toxins: risk management for health protection. Toxicol Appl Pharmacol. 2005;203:264–272. doi: 10.1016/j.taap.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Davies C, Heath RJ, White SW, Rock CO. The 1.8 Å crystal structure and active-site architecture of β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure. 2000;8:185–195. doi: 10.1016/s0969-2126(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 12.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 13.El-Sayed AK, Hothersall J, Cooper SM, Stephens E, Simpson TJ, Thomas CM. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem Biol. 2003;10:419–430. doi: 10.1016/s1074-5521(03)00091-7. [DOI] [PubMed] [Google Scholar]
- 14.Gokhale RS, Tsuji S, Cane DE, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 15.Gu L, Eisman EB, Dutta S, Franzmann TM, Walter S, Gerwick WH, Georgios S, Sherman DH. Tandem acyl carrier proteins in the curacin biosynthetic pathway promote consecutive multienzyme reactions with a synergistic effect. Angew Chem Int Ed. 2011;50:2795–2798. doi: 10.1002/anie.201005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Håkansson K, Wipf P, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu L, Wang B, Kulkarni A, Gehret JJ, Lloyd KR, Gerwick L, Gerwick WH, Wipf P, et al. Polyketide decarboxylative chain termination preceded by O-sulfonation in curacin A biosynthesis. J Am Chem Soc. 2009;131:16033–16035. doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han J, Calvin M. Hydrocarbon distribution of algae and bacteria, and microbiological activity in sediments. Proc Natl Acad Sci. 1969;64:436–443. doi: 10.1073/pnas.64.2.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper GJ, Orjala J, Schatzman RC, Gerwick WH. Carmabins A and B, new lipopeptides from the Caribbean cyanobacterium Lyngbya majuscula. J Nat Prod. 1998;61:529–533. doi: 10.1021/np970443p. [DOI] [PubMed] [Google Scholar]
- 20.Irschik H, Kopp M, Weissman KJ, Buntin K, Piel J, Müller R. Analysis of the sorangicin gene cluster reinforces the utility of a combined phylogenetic/retrobiosynthetic analysis for deciphering natural product assembly by trans-AT PKS. ChemBioChem. 2010;11:1840–1849. doi: 10.1002/cbic.201000313. [DOI] [PubMed] [Google Scholar]
- 21.Jia C, Li M, Li J, Zhang J, Zhang H, Cao P, Lu X, et al. Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases. Protein Cell. 2015;6:55–67. doi: 10.1007/s13238-014-0108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones AC, Gu L, Sorrels CM, Sherman DH, Gerwick WH. New tricks from ancient algae: natural products biosynthesis in marine cyanobacteria. Curr Opin Chem Biol. 2009;13:216–223. doi: 10.1016/j.cbpa.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones AC, Monroe EA, Eisman EB, Gerwick L, Sherman DH, Gerwick WH. The unique mechanistic transformations involved in the biosynthesis of modular natural products from marine cyanobacteria. Nat Prod Rep. 2010;27:1048–1065. doi: 10.1039/c000535e. [DOI] [PubMed] [Google Scholar]
- 24.Kampa A, Gagunashvili AN, Gulder TA, Morinaka BI, Daolio C, Godejohann M, Miao V, Piel Jörn, et al. Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses. Proc Nat Acad Sci. 2013;110:E3129–E3137. doi: 10.1073/pnas.1305867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim EJ, Lee JH, Choi H, Pereira AR, Ban YH, Yoo YJ, Kim E, Park JW, et al. Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product. Org Lett. 2012;14:5824–5827. doi: 10.1021/ol302575h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleigrewe K, Almaliti J, Tian IY, Kinnel RB, Korobeynikov A, Monroe EA, Duggan BM, di Marzo V, et al. Combining mass spectrometric metabolic profiling with genomic analysis: a powerful approach for discovering natural products from cyanobacteria. J Nat Prod. 2015;78:1671–1682. doi: 10.1021/acs.jnatprod.5b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li N, Nørgaard H, Warui DM, Booker SJ, Krebs C, Bollinger JM., Jr Conversion of fatty aldehydes to alka(e)nes and formate by a cyanobacterial aldehyde decarbonylase: cryptic redox by an unusual dimetal oxygenase. J Am Chem Soc. 2011;133:6158–6161. doi: 10.1021/ja2013517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luesch H, Yoshida WY, Moore RE, Paul VJ, Mooberry SL. Isolation, structure determination, and biological activity of lyngbyabellin A from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:611–615. doi: 10.1021/np990543q. [DOI] [PubMed] [Google Scholar]
- 29.Márquez B, Verdier-Pinard P, Hamel E, Gerwick WH. Curacin D, an antimitotic agent from the marine cyanobacterium Lyngbya majuscula. Phytochemistry. 1998;49:2387–2389. doi: 10.1016/s0031-9422(98)00365-3. [DOI] [PubMed] [Google Scholar]
- 30.Matilla MA, Stöckmann H, Leeper FJ, Salmond GP. Bacterial biosynthetic gene clusters encoding the anti-cancer haterumalide class of molecules: biogenesis of the broad spectrum antifungal and anti-oomycete compound oocydin A. J Biol Chem. 2012;287:39125–39138. doi: 10.1074/jbc.M112.401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattheus W, Gao LJ, Herdewijn P, Landuyt B, Verhaegen J, Masschelein J, Volckaert G, Lavigne R. Isolation and purification of a new kalimantacin/batumin-related polyketide antibiotic and elucidation of its biosynthesis gene cluster. Chem Biol. 2010;17:149–159. doi: 10.1016/j.chembiol.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy JG, Eisman EB, Kulkarni S, Gerwick L, Gerwick WH, Wipf P, Sherman DH, Smith JL. Structural basis of functional group activation by sulfotransferases in complex metabolic pathways. ACS Chem Biol. 2012;7:1994–2003. doi: 10.1021/cb300385m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucl Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendez-Perez D, Begemann MB, Pfleger BF. Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl Env Microbiol. 2011;77:4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mevers E, Liu WT, Engene N, Mohimani H, Byrum T, Pevzner PA, Dorrestein PC, Spadafora C, et al. Cytotoxic veraguamides, alkynyl bromide-containing cyclic depsipeptides from the marine cyanobacterium cf Oscillatoria margaritifera. J Nat Prod. 2011;74:928–936. doi: 10.1021/np200077f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milligan KE, Marquez BL, Williamson RT, Gerwick WH. Lyngbyabellin B, a toxic and antifungal secondary metabolite from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1440–1443. doi: 10.1021/np000133y. [DOI] [PubMed] [Google Scholar]
- 37.Minto RE, Blacklock BJ. Biosynthesis and function of polyacetylenes and allied natural products. Prog Lipid Res. 2008;47:233–306. doi: 10.1016/j.plipres.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moldenhauer J, Chen XH, Borriss R, Piel J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angew Chem. 2007;119:8343–8345. doi: 10.1002/anie.200703386. [DOI] [PubMed] [Google Scholar]
- 39.Molinski TF. NMR of natural products at the ‘nanomole-scale’. Nat Prod Rep. 2010;27:321–329. doi: 10.1039/b920545b. [DOI] [PubMed] [Google Scholar]
- 40.Ng J, Bandeira N, Liu WT, Ghassemian M, Simmons TL, Gerwick WH, Linington R, Dorrestein PC, et al. Dereplication and de novo sequencing of nonribosomal peptides. Nat Method. 2009;6:596–599. doi: 10.1038/nmeth.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ongley SE, Bian X, Zhang Y, Chau R, Gerwick WH, Müller R, Neilan BA. High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chem Biol. 2013;8:1888–1893. doi: 10.1021/cb400189j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul VJ, Arthur KE, Ritson-Williams R, Ross C, Sharp K. Chemical defenses: from compounds to communities. Biol Bull. 2007;213:226–251. doi: 10.2307/25066642. [DOI] [PubMed] [Google Scholar]
- 43.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Nat Acad Sci. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Nat Acad Sci. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramaswamy AV, Sorrels CM, Gerwick WH. Cloning and biochemical characterization of the hectochlorin biosynthetic gene cluster from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2007;70:1977–1986. doi: 10.1021/np0704250. [DOI] [PubMed] [Google Scholar]
- 46.Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucl Acids Res. 2011;39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schirmer A, Rude MA, Li X, Popova E, Cardayre D. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 48.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Kerfeld CA. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Nat Acad Sci. 2013;110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stubbe J, van der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 50.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, et al. Identification of the putative bryostatin polyketide synthase gene cluster from Candidatus Endobugula sertula, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 51.Tan LT. Marine cyanobacteria: a prolific source of bioactive natural products as drug leads. In: Kim SK, editor. Marine microbiology: bioactive compounds and biotechnological applications. Wiley; New York: 2013. pp. 59–81. online edition. [Google Scholar]
- 52.Tang GL, Cheng YQ, Shen B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 53.Taniguchi M, Nunnery JK, Engene N, Esquenazi E, Byrum T, Dorrestein PC, Gerwick WH. Palmyramide A, a cyclic depsipeptide from a Palmyra Atoll collection of the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2009;73:393–398. doi: 10.1021/np900428h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taton A, Unglaub F, Wright NE, Zeng WY, Paz-Yepez J, Brahamsha B, Palenik B, Peterson TC, et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucl Acids Res. 2014;17:e136. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whicher JR, Smaga SS, Hansen DA, Brown WC, Gerwick WH, Sherman DH, Smith JL. Cyanobacterial polyketide synthase docking domains: a tool for engineering natural product biosynthesis. Chem Biol. 2013;20:1340–1351. doi: 10.1016/j.chembiol.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williamson RT, Boulanger A, Vulpanovici A, Roberts MA, Gerwick WH. Structure and absolute stereochemistry of phormidolide, a new toxic metabolite from the marine cyanobacterium Phormidium sp. J Org Chem. 2002;67:7927–7936. doi: 10.1021/jo020240s. [DOI] [PubMed] [Google Scholar]
- 59.Winters K, Parker PL, Van Baalen C. Hydrocarbons of blue-green algae: geochemical significance. Science. 1969;163:467–468. doi: 10.1126/science.163.3866.467. [DOI] [PubMed] [Google Scholar]
- 60.Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, et al. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu X, Liu J, Zhang W. De novo biosynthesis of terminal alkyne-labeled natural products. Nat Chem Biol. 2015;11:115–120. doi: 10.1038/nchembio.1718. [DOI] [PubMed] [Google Scholar]
- 62.Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7(3):e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]