Abstract
The advent of high-throughput genomics techniques, along with the completion of genome sequencing projects, identification of protein–protein interactions and reconstruction of genome-scale pathways, has accelerated the development of systems biology research in the yeast organism Saccharomyces cerevisiae. In particular, discovery of biological pathways in yeast has become an important forefront in systems biology, which aims to understand the interactions among molecules within a cell leading to certain cellular processes in response to a specific environment. While the existing theoretical and experimental approaches enable the investigation of well-known pathways involved in metabolism, gene regulation and signal transduction, bioinformatics methods offer new insights into computational modeling of biological pathways. A wide range of computational approaches has been proposed in the past for reconstructing biological pathways from high-throughput datasets. Here we review selected bioinformatics approaches for modeling biological pathways in S. cerevisiae, including metabolic pathways, gene-regulatory pathways and signaling pathways. We start with reviewing the research on biological pathways followed by discussing key biological databases. In addition, several representative computational approaches for modeling biological pathways in yeast are discussed.
Keywords: Saccharomyces cerevisiae, metabolic pathway, signaling regulation, gene regulatory network
Introduction
Biological pathways represent a series of molecular interactions within a cell at different conditions that lead to end-point biologicalfunctions. Signals from external environment trigger internal chemical reactions in biological pathways to tackle specific tasks. For example, the function of a MAPK-containing complex can be altered by the phosphorylation of components due to active MAPK in the MAPK signaling pathway[1]. Over the past decade, many large-scale experimental and computational approaches have been developed to decipher chemical reactions in metabolic pathways, gene regulation in regulatory network and the transmission of signals in signaling pathways.
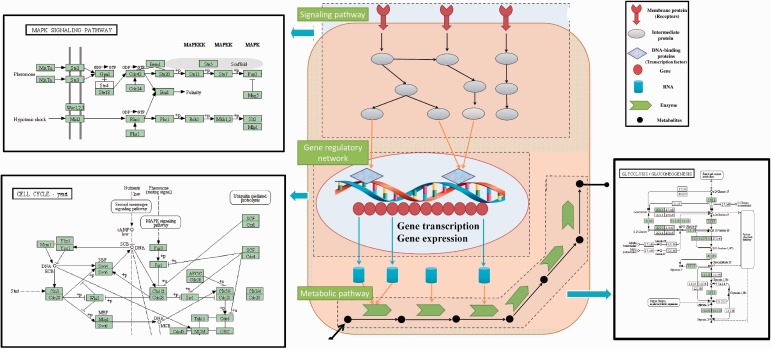
Signaling pathways process the chemical activities in response to the signals sent from the exterior of the cell to the internal receptor. Gene regulatory network represents the transcriptional regulation activities between genes and transcription factors. Metabolites in a metabolic pathway interact with each other inside the cell, with enzymes catalyzing the metabolic reactions. Different chemical signals stimulate specific proteins inside the signaling pathway to trigger specific reactions. Identification of relationship among genes, proteins or molecules in biological pathways is critical for understanding complex biological activities and biological functions. Figure 1 represents the interrelationship among signaling pathway, gene regulatory network and metabolic pathway within a cell accomplishing the biological activities. The three kinds of biological pathways can be represented by network graph in which the node represents the entity in the pathway and the edge represents the interaction between the entities.
Figure 1.
Interrelationship among signaling pathway, gene regulatory network and metabolic pathway in a cell. Cell starts to recognize and receive the signals by activating the membrane receptors, responding to the stimulus of the changes in the outside environment. The receptors can help transmit the signal outside the cell membrane into the signaling pathway inside the cell, activating a series of biochemical reactions. Take the MAPK signaling pathway in yeast (Top-right) from KEGG [2] as example, the signaling pathway is initialized by the stimuli of peptide mating pheromones, which are Mat-alpha and MatA, then the receptor Ste2 or Ste3 connects the peptide mating pheromone and activates the MAPK signal transduction cascades. The signaling cascade will finally produce the protein kinases that can enter the nucleus and activate the gene transcription by binding the transcription factors onto promoters of DNA. In the example of cell cycle yeast pathway (Bottom-left), the Cln3-Cdc28 protein kinases activate the transcription factors SBF and MNF, which regulate the Cln1/2 gene expression. The gene regulatory network can control the gene expression levels of mRNA by activating the transcription factors, and further translate the mRNA into proteins. Specific proteins, called enzymes, will participate in the metabolic pathway and catalyze a series of biochemical reactions by converting substrates into products, in which the product of one reaction becomes the substrate of next reactions. In the Glycolysis/Gluconeogenesis pathway (Bottom-right), experiments [3] showed the regulators identified in cell cycle also regulated the metabolic enzymes to catalyze the cellular metabolism. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Saccharomyces cerevisiae, as one of intensely studied single-cell eukaryotes, has been commonly used as a template organism to discover similar cellular processes and specific protein functions in other organisms. Many important functional pathways, such as lipid metabolism [4] and cell cycle [5], have been identified as similar cellular processes between yeast and human [6]. Through the historical development of systems biology research in yeast, S. cerevisiae has been widely studied from individual system components to complex module interactions in order to decipher the complete picture of the cellular processes.
The first complete genome sequencing of S. cerevisiae was achieved through the yeast genome project in 1996 [7]. With the completion of genome sequencing project, identification of protein–protein interactions became one of the key topics of focus on system-level molecular network study in S. cerevisiae, and was accomplished by different types of approaches, ranging from in vivo studies to in silico studies [8]. Yeast two-hybrid system was deemed as an effective in vivo technique to detect direct interactions between protein pairs based on the activation of functional transcription factors [9]. In the meantime, the in silico methods have proven their effectiveness of predicting potential interactions between proteins, for instance, based on the three-dimension structural similarities [10], and complemented the experimental approaches [8]. Interactions between proteins can also be indirectly identified through coexpressed genes using mRNA levels, which indicates that genes sharing similar expression patterns under a specific condition interact with each other [11]. Molecular interactions among cell activities also drove the study of gene coregulation in response to different conditions [12, 13]. Functionally correlated modules with sets of coregulated genes have been identified using S. cerevisiae expression datasets [14–16].
In addition to the detection of protein–protein interactions and gene regulatory network, there has also been a significant effort toward the reconstruction of metabolic pathways for understanding yeast genes in complex biological systems. The first genome-scale metabolic network has been manually curated for S. cerevisiae which contains 1175 metabolic reactions and 584 metabolites [17]. Several groups continued to reconstruct and expand the metabolic models by integrating the experimental and computational techniques [18–22].
At the same time, experimental approaches combined with computational methods have contributed toward the reconstruction of signaling pathways from microarray expression data and protein–protein interactions [23]. For example, NetSearch program [24] was proposed to determine the candidate pathways among protein interaction data and score each pathway by calculating the number of pathway members that were involved in the same cluster derived from the expression data. This method finally selected highest-ranking pathways and combined them into signaling pathway. Furthermore, through the study of signaling pathways in multiple species, most interactions between proteins in signaling pathways are directional, including activation, inhibition, phosphorylation, dephosphorylation and ubiquitination [25]. The authors proposed a signal-flow direction method to predict the potential upstream–downstream relationships between protein pairs in protein–protein interaction networks. This method was successfully used for accurate reconstruction of signaling pathways through protein interaction networks. Newer signal-flow approaches to signaling pathway reconstructions used the information on pathway components lying on the same signal transduction cascade to infer the order of the signal-flow using optimization techniques [26, 27]. Boolean modeling framework has also shown its good performance in analyzing signaling pathways [28, 29].
With the emerging growth of public databases by collecting the biological knowledge including ‘omics' data (genomic, proteomic, transcriptomic, metabolomics data etc.) and biochemical pathways, computational methods can be integrated with comprehensive experimental knowledge to improve the reconstruction of the biological pathways. Some widely used databases, such as KEGG [2, 30, 31] and Saccharomyces Genome Database (SGD) [32], have been discussed in detail in the next section.
In the remaining part of this review, we start with summarizing some key public data repositories used for biological pathway modeling followed by presenting selected bioinformatics approaches to pathway identification.
Data resource for S. cerevisiae
The current bioinformatics methods for pathway modeling mainly rely on known biological knowledge that has been experimentally validated through decades of study. This experimental knowledge can be used to evaluate the modeled pathways or integrate with data for pathway construction. Consequently, several repositories have been built and maintained by different research groups, which provide researchers with the access to biological information, such as mRNA expression data, protein–protein interactions or biochemical pathways. These integrated and comprehensive resources significantly facilitate various biological research and development. A list of selected databases which have been widely used for pathway modeling, is presented in Table 1.
Table 1.
Public repositories for S. cerevisiae
|
Saccharomyces cerevisiae Databases | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Database | Protein interaction | Biochemical pathway | Network dataset | Expression dataset | Metabolomics data (metabolite/reaction) | Year of publication | Lastest update | Accessibility | Reference |
| Biogrid | 342 878 | – | – | – | – | 2006 | 2015 | Download/Query | [33] |
| DIP | 24 618 | – | – | – | – | 2002 | 2014 | Download | [34] |
| MINT | 62 621 | – | – | – | – | 2002 | 2012 | Download | [35] |
| SGD | 340 493 | 187 | – | 339 | – | 1998 | 2015 | Download/Query | [32] |
| YeastNet V3 | 362 421 | – | 72 | – | – | 2004 | 2013 | Download | [36] |
| Yeast Interactome Database | – | – | 6 | – | – | 2008 | 2008 | Download/Query | [37] |
| MetaCyc | – | 268 | – | – | – | 2001 | 2015 | Download/Query | [38] |
| KEGG | – | 109 | – | – | – | 1999 | 2015 | Download/Query | [2] |
| ExpressDB | – | – | – | 46 | – | 2000 | 2006 | Download | [39] |
| yStrex | – | – | – | 82 | – | 2013 | 2014 | Download/Query | [40] |
| YMDB | – | – | – | – | 2027/916 | 2012 | 2012 | Download/Query | [41] |
| BIGG | – | – | – | – | iMM904: 1226/1557 | 2010 | 2015 | Query | [42] |
| iND750: 1056/1266 | |||||||||
Note. Twelve widely used databases are listed and the statistics of data in each database are presented. For example, SGD database provides 340 493 interaction records for yeast protein pairs from different types of experiments and 187 biochemical pathways for S. cerevisiae. SGD also provides 339 expression datasets from experiments. Besides the statistics, the website and related reference are also presented.
Databases, including Biogrid [33], Database of Interacting Proteins (DIP) [34], Molecular INTeraction database (MINT) [35] and Saccharomyces Genome Database (SGD) [32], store protein–protein interactions for S. cerevisiae with different scope and content, including the interactions observed from experiments or links predicted through computational methods. For example, latest version of BioGrid database contains 342 878 protein interactions that are directly extracted from publication using computational approaches [43]. MINT contains 62 621 experimentally validated protein–protein interactions that were manually collected from online publications. In addition to protein–protein interactions, SGD, ExpressDB [39] and yStrex database [40]contain yeast RNA expression datasets under different conditions and experiments. Compared to ExpressDB, SGD database maintains datasets most frequently and contains much more datasets including those generated in recent years. However, ExpressDB only stores expression datasets that were created prior to the year 2002.
There are three main databases that contain manually curated biological pathways representing the experimental knowledge from published literatures. MetaCyc [38] contains 268 pathways at present whereas KEGG [2, 30, 31]and SGD have 109 and 187 pathways, respectively, for S. cerevisiae. Pathways in the above mentioned databases are represented using different views. MetaCyc allows users to view individual pathway and the interconnections among pathways in a specific organism whereas KEGG combines several pathways from different species into one framework. Additionally, MetaCyc also provides the information whether a pathway has been experimentally validated [38].
The computationally integrated gene network for S. cerevisiae, including coexpression network, genetic interaction network and protein–protein network, can be downloaded from YeastNet database [36] and Yeast Interactome Database [37]. Metabolomics data for metabolic study are also maintained in YMDB [41] and BIGG [42].
Rapid growth in public databases covering a vast amount of biological knowledge is making the use of bioinformatics methods a more promising strategy for pathway modeling from both computational and biological point of view.
Bioinformatics approaches for biological pathway modeling
Computational approaches for modeling biological pathways can be developed using two types of modeling methods: network-based analysis and mathematical modeling. Network-based methods apply graph theory to discover relationships among nodes in the pathways, where each node represents a biological entity, such as gene or protein, and each edge represents the interaction type between node pairs. Such networks can be represented as directed or undirected graphs. Probabilistic graph model is a typical network-based approach that uses methods such as Bayesian networks to learn cellular networks from gene expression data.
Mathematical modeling learns and analyzes the underlying network by transforming the reactions and entities into matrix form. Several mathematical approaches have been developed to study biological pathways, in terms of different types of biochemical mechanism, and complexity of networks. Signaling pathways can be mathematically formalized through Boolean network [44] by representing large-scale networks. Ordinary differential equations can provide quantitative models describing the small-sized gene regulatory network [45]. Large-scale metabolic pathways are usually modeled using stoichiometric methods and flux balance analysis [46, 47]. Detailed review of various mathematical modeling approaches and their applications in yeast pathways have been presented in [48, 49].
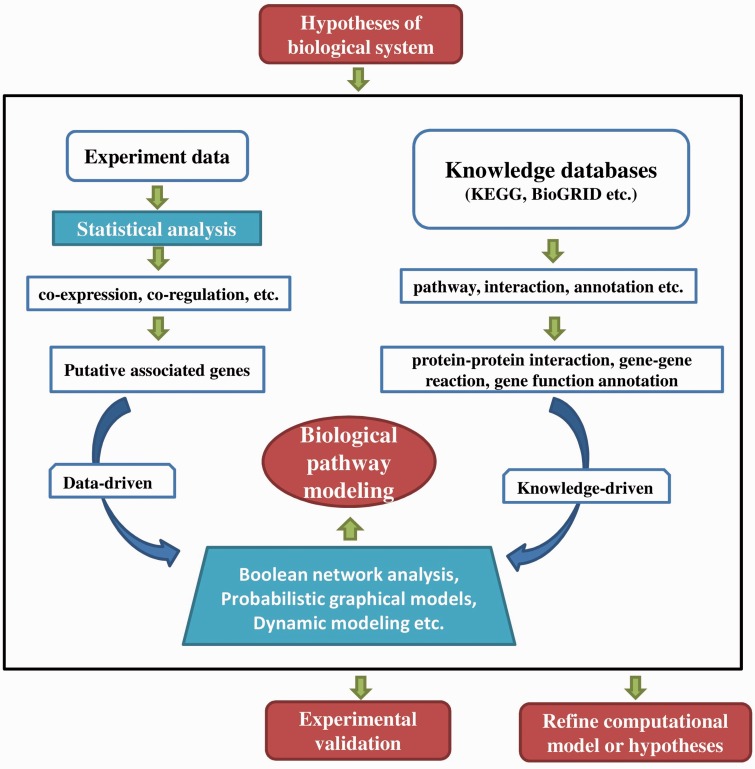
In this section, we mainly consider network-based bioinformatics approaches for modeling pathways. We start with discussing data-driven approaches to infer biological associations from different types of ‘omics' data, followed by describing knowledge-based methods that integrate prior knowledge with ‘omics' data to improve the pathway modeling. Figure 2 represents the flowchart for pathway modeling using computational approaches.
Figure 2.
Flowchart for pathway modeling using computational approaches. The bioinformatics methods for pathway modeling starts with the hypothesis of pathway construction which can be derived from experiments or theory. Then computational methods can be performed on the experiment data (e.g. Microarray data, RNASeq data) and knowledge information (e.g. Pathway information, functional annotation) to model the biological pathways. The predicted pathways can be refined by evaluating each model with experiments and hypothesis. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Data-driven analysis
With the rapid development of high-throughput techniques, different types of ‘omics' data become available for a deeper understanding of systems biology in yeast. Different computational approaches were proposed to model the biological pathways through the data of transcriptomes, proteomics, thermodynamics and metabolomics. In this section, we briefly review several popular bioinformatics approaches, which were developed to the biological networks in yeast from different types of data.
Transcriptome study
Transcriptomic profiling experiments generated microarray data or RNASeq data to represent the gene activity, by measuring the change of expression levels within a cell under different conditions. Many efficient algorithms were designed to infer the interaction and relationship among the cellular entities from gene expression data, by following the assumption: proteins encoded by coexpressed genes are more likely to interact in the same pathway and similar gene expression patterns tend to share similar biological function [50, 51]. Similarity of expression profiles can be formalized by several measures, such as Pearson correlation coefficient (PCC) and mutual information [52]. Pearson correlation coefficient is a standard way to represent coexpression measurement, which calculates the degree of linear relationship between the expression values of gene pairs across the multiple conditions. Studies have emphasized on the construction of gene coexpression networks to infer cellular function from S. cerevisiae datasets [13, 16, 53–61].
R package GTOM [55] was developed to infer the coexpression networks from yeast cell cycle datasets, where a novel measurement ‘topological overlap' together with Pearson correlation coefficients was designed for network analysis. PCC was also applied on yeast expression data to confirm that gene pairs with highly correlated expression profiles were regulated by pair of interacting loci on chromosomes [56]. In the another work, the PCC was applied in the first step as similarity measurement to score each protein pair [57]; this proposed method assessed and ranked the reliability of protein interactions to improve the prediction accuracy.
To overcome the shortcoming of missing gene–gene relationship information when transforming a pair of gene expression data across all samples into correlation coefficients [59], identification of gene expression patterns based on local similarity [58] has been proposed. This method measures how significantly data fluctuates across experimental samples and how the regulation pattern represents each gene expression (up- or down-regulated).
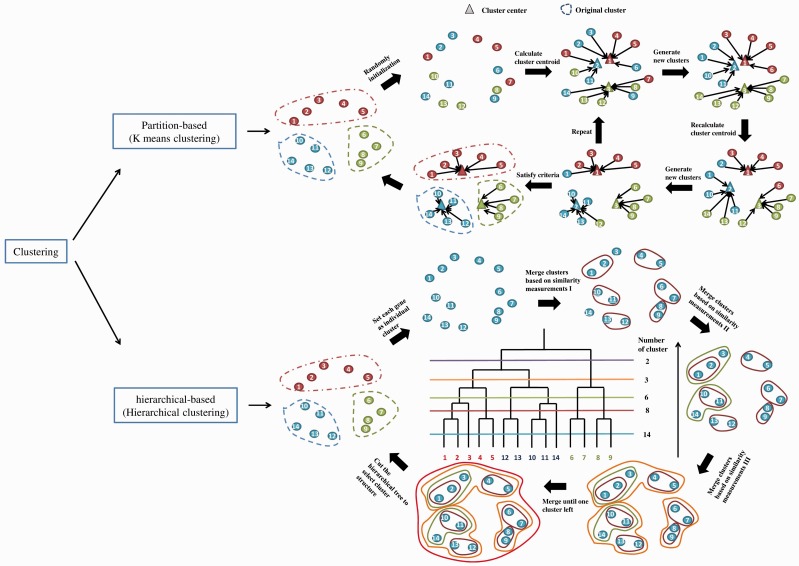
Clustering algorithms have been successfully applied on yeast gene expression data to discover similar patterns among genes and identify coexpressed or coregulated genes involved in the same biological processes [62, 63]. K-means clustering and hierarchical clustering are two basic types of clustering algorithms. K-means clustering is a widely used algorithm to partition gene expression matrix into multiple subsets based on squared error criterion, however, hierarchical clustering groups genes with similar expression profiles to build a hierarchical tree structure from gene expression data, a graphically visualization of clusters through dendrogram [64]. Figure 3 represents how the two types of methods are applied to form the clusters.
Figure 3.
Clustering algorithms. We discussed two typical clustering algorithms applied in yeast pathway modeling. The clustering methods are classified into two categories: partition-based clustering (Top) and hierarchical-based clustering (Bottom). (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
NetSearch [24] used K-means clustering to reconstruct the yeast MAPK signaling pathways. K-means clustering was also applied for protein function prediction by generating the coexpression groups from yeast gene expression data [65].
However, both the two clustering methods have disadvantages, suffering from sensitive to outlier or limited to cluster size. Other novel approaches with substantial improvements have been developed to discover relationships among genes using expression data based on similarity or prior knowledge. Zhu et al. [66] proposed a network constrained clustering method where the similarity between gene expression profiles was measured by the shortest-path distance in a gene coexpression network. Learning the number of the clusters automatically without specifying it in the initial step [67] was also applied. Detailed reviews of approaches to perform clustering analysis on gene expression data have been presented in [68–70].
Coexpression analysis was used as starting point to explore biological pathways. This analysis allows learning the similarity of expression profiles and interactions between genes. Applications of Gaussian graphical model (GGMs) and Bayesian networks on gene expression data have shown robust performances in constructing biological pathways [29, 71], by expanding the coexpression network into gene regulatory network [72, 73], metabolic pathway [74] and signaling pathway [75]. In these probabilistic models, regulation between each pair of genes depended on their conditional independence given the expression levels of the rest of genes.
The basic procedure of fitting a Gaussian graphical model (GGM) follows several steps. For any gene expression data , where n is the number of genes and m is the number of conditions, under the assumption of multivariate normal distribution, the variance matrix for all gene pairs is firstly calculated and inversely transformed into precision matrix . The conditional independence network can be inferred from the precision matrix where genes Xa and Xb are connected in the graph if. The Gaussian graphical network is inferred by the maximum likelihood estimation of the precision matrix. Several GMM-based approaches have been proposed to represent the yeast gene network. Authors [76] applied partial correlation derived from standard correlation in the precision matrix to discover regulatory genetic links involved in yeast metabolic pathways among a small set of genes. Also -regularized methods were integrated into Gaussian graphical models to solve the singularity problem of precision matrix in the case of high-dimension data by generating sparse covariance matrices. Gaussian graphical models combined with Graphical lasso algorithm, constrained -minimization and covariate-adjusted precision matrix estimation method were recently applied to construct yeast mitogen-activated protein kinase signaling pathway [77–79].
Gaussian graphical models aim to construct undirected graphs to represent coregulated gene networks while Bayesian networks discover the causal relationship between gene pairs and are represented as directed acyclic graph. The effectiveness of Bayesian network methods using S. cerevisiae cell cycle data has been discussed in [80, 81].
Proteome study
In the past decades, genome-scale protein interaction networks have been experimentally or computationally generated for S. cerevisiae [82, 83]. Interaction between proteins reflects the function association within the cellular system and thus the identification of protein–protein interactions is an important step toward the reconstruction of biological pathways. A comprehensive list of computational methods for identifying protein–protein interaction has been presented in [8]. Several methods have been developed to construct biological pathways by integrating protein interactions with transcriptomics data for S. cerevisiae. For example, the authors [84] curated an initial pathway model by gathering the galactose-related genes from existing knowledge and measured global cellular response by perturbing each gene in pathway. The paths between perturbed genes and regulated genes involved in the same metabolic pathway were identified based on known physical interactions. Steffen et al. [24] proposed a method to identify interaction subnetworks in regulatory and signaling pathways from protein interaction data and gene expression data across different conditions. They ranked each subnetwork based on the significant change in the expression profiles of the genes in the network. Tornow et al. [85] explored function modules through protein interaction networks based on the strength of correlation between gene pairs derived from expression data. They proposed a framework to discover a group of genes that were biologically correlated in genetic or cellular networks. Liu et al. [25] proposed novel score functions for protein–protein interaction data and gene expression data, and applied them on each subnetwork of yeast MAPK pathways with different permutation of protein sets. Scott et al. [86] improved the path searching algorithm by integrating the color coding approach and used a probabilistic approach to assign weights to each interaction of genes based on logistic regression and searched the paths of given lengths with the highest scores, where the score was defined as the product of weights of the edges in path. They built a logistic model from three random variables: (i) the number of times each interaction was identified in multiple experiments, (ii) the Pearson correlation between expression profiles for each pair of genes, and (iii) small world clustering coefficients. They developed two algorithms for finding paths and pathway structures in several yeast signaling pathways with high accuracy. This was further improved by heuristic search for pathway construction from protein–protein interaction and gene expression data [87]. To further address the edge orientation problems, integer programming approaches and genetic algorithms were proposed to search the optimal paths between sources and targets from global protein interactions for signaling pathway construction [88–92]. All these methods followed similar modeling framework in which path searching among the pool of interactions combining with path scoring strategy or constraints-based algorithms. Other network analyses for modeling biological pathways on yeast, such as Boolean networks, have been reviewed in [93].
These bioinformatics methods integrating proteomic data with transcriptomic data provide an alternative approach to understand biological pathways.
Metabolome/Fluxome study
As we have described, the transcriptomic data and proteomic data represent a series of cellular functions in the top-bottom biological process, traversing from signaling pathway to gene regulatory network. Metabolites, as the end products of cellular process, participate in the metabolic pathways connected by biochemical reactions. Integrated analysis of gene, protein and metabolites with biochemical reactions can facilitate the genome-scale network reconstruction within an organism. Metabolomics data represent quantitative profiles of metabolites over a series of metabolic processes under different conditions, which can facilitate to understand the enzyme regulation in the metabolism through the changes in the level of metabolites. The metabolomics data of S. cerevisiae are provided in the databases [41, 42], which were intensively applied to model the metabolic pathways that were perturbed underlying different conditions. Generally, metabolomics profiling data were applied to infer the intracellular fluxes in yeast metabolic system, which contributes to the fluxomics study to discover the intracellular pathway activities [94, 95]. Due to the property of steady state in the metabolic system, flux balance analysis [46, 47] has been widely used for analyzing the fluxes space and studying biochemical networks.
Knowledge-based analysis
A growing number of public databases, such as KEGG [2, 30, 31] and Saccharomyces Genome Database (SGD) [32], have been created to provide information about function annotation, protein interactions and experimentally validated biological pathways (Table 1). These databases serve as excellent resources to facilitate pathway predictions and models. Most researches have focused on integrating function annotation and protein–protein networks with expression data to improve the accuracy and precision of pathway construction. In the last section, we have reviewed computational approaches to construct biological pathways based on different kind of ‘omics' data. To improve the accuracy of inferred pathways, data integration, which combines proteomic data, validated pathways, functional annotation and transcriptomic data, has been proposed. In this section, we reviewed several key methods for biological pathway modeling that utilize existing prior knowledge together with ‘omics' data.
Integration of biological pathway and ‘omic’ data
Protein–protein interaction networks play key role in understanding the biochemical processes and construct the biological pathways within cells. However, protein–protein interaction networks are almost undirected, which only indicate the presence of interactions between proteins. This shortcoming presents a substantial challenge for pathway modeling with high accuracy since most biological pathways, such as signaling pathways and metabolic pathways, contain different types of directional interactions and reactions: (i) activation or inhibition of the transcription of gene for a protein under specific signal transductions response and (ii) gene regulation by phosphorylation or dephosphorylation [96, 97]. In order to reproduce biological processes with higher accuracy during the construction of biological pathways, several approaches have been proposed to extract prior biological knowledge from the known pathways, provided by KEGG, SGD and MetaCyc, and incorporate them into pathway modeling. Several statistical approaches [98, 99] have utilized pathways information from KEGG and combined them with microarray dataset to identify genes and subnetworks in several KEGG transcriptional pathways associated with diseases, by applying hidden Markov-random field model. Authors [100] applied regression analysis with network-constrained method where they added gene pairs in the same pathway as penalty in the network-constrained regularization criterion for estimating the parameter in the regression model. Qi et al. [73] utilized prior biological knowledge of gene–gene interactions extracted from the KEGG database and applied Bayesian probabilistic graphical model to enlarge the metabolic pathway of yeast on the basis of initial pathways in KEGG database by sampling coexpressed genes from gene clusters derived from gene expression data. This integrated method improved the prediction accuracy compared to pure data-driven methods [70, 101]. Motivated by the successful applications of applying machine learning and data mining approaches in bioinformatics problems, Li et al. [102] developed a novel method to transform each metabolic pathway into a list of number by representing the features of graph property, chemical functional group and chemical structure set. The author finally constructed the vector matrix with 16 features for all pathways from yeast species, and applied nearest neighbor algorithm to identify the metabolic pathway.
Integration of functional annotation and ‘omics' data
Functional similarity serves as the basis of coexpression networks and protein–protein interactions, and it is the key assumption to model the biological pathways from the gene expression data or protein interactions. Computational approaches combining gene ontology or chemical functional modules with gene expression data for modeling biological pathway have become a promising research direction in recent years. Gene ontology (GO) [103] represents the gene function and relationships with hierarchical structure in terms of three ontology categories: biological process, molecular function and cellular components. Semantic similarity measures have been applied to relate the genes in terms of GO terms and function annotation to discover the interactions between genes [104, 105]. Integration of function annotation and mRNA expression data on S. cerevisiae was proposed in [106] by applying Bayesian network method on the multiple modules consisting of highly correlated genes based on GO annotations and expression data. In this work, significantly affected genes under given experimental conditions were extracted initially and assigned a similarity score for each pair of genes based on the similarity of GO terms. Then different functional groups were generated based on the degree of dependency between genes derived from gene expression data. Modules were learned through Bayesian network method and were combined to form final genetic interaction network. Authors [107] generated the functional annotation relationship for proteins in the pathways from the KEGG database (template) and proteins in protein–protein interaction networks (target), and built a functional template-target mining strategy to search the signaling pathway segments from protein interaction networks [108]. This method also improved the accuracy and precision for yeast S. cerevisiae compared to earlier methods and had the ability to recover some missing links in the signaling pathway. To incorporate more biological information, authors [109] integrated functional similarity with pathways information, protein domain annotation and protein domain interactions to construct a probabilistic structure prior for Bayesian network inference. Independent of these methods, several other functional annotation-based methods have been proposed to infer biological pathway and obtain better insights into the cellular functions and regulation machinery [60, 110, 111].
Discussion
Organism model for computational modeling approaches
As we have described, genome of S. cerevisiae has been intensively studied throughout cell levels with integrated analysis (e.g., genome, proteome, interactome, transcriptome, metabolome). In the past decades, substantial amount of omic data were generated to elucidate the yeast biological system, including gene expression data, protein–gene and protein–protein interactions, protein levels, and fluxes measurements of metabolite level [112]. The large-scale omics data provided a powerful test ground for computational modeling approaches to construct the biological pathways in yeast. Furthermore, due to the large part of yeast genes sharing the similar functions with homolog in other species, and the simplest cell structure in S. cerevisiae, we think applying the computational approaches on pathway modeling for S. cerevisiae is valuable and flexible because of several advantages. Firstly, large known biological data provided a significant prior knowledgebase for most computational approaches to learn the parameters and fit the models. And relatively simpler network structure with substantial prior knowledge may generate networks with high accuracy and less false-positive interactions, which also provides better interpretation of biological process in other species. More importantly, most computational methods will experience the limitation of network size. For example, in the Boolean network and Bayesian network, the possible subnetworks is super-exponential to the number of genes, which is most suitable to the small networks with no more than hundreds genes [113]. In addition, the availability of time series gene expression data in yeast also made the differential equations suitable to analyze the flux changes and gene regulation over time. Overall, yeast can be served as ideal organism model for the evaluation of computational methods that are useful to study other organisms.
From yeast to human: application of biological pathways modeling
Yeast was widely considered as preferred organism model in both experimental and computational research, not just because it is simplest unicellular eukaryote that is easy to manipulate, but also because of the similar characteristics in cellular system between yeast and human cells. In other words, the study of modeling biological pathways in yeast will facilitate the understanding of biological processes in humans. Many important biological processes in human cell pathways can be studied in yeast, such as lipid metabolism [4], and cell cycle [5]. Furthermore, compared to higher eukaryotes, genome of S. cerevisiae has the relatively small number of genes (∼6000) so that yeast has been widely studied under different conditions (e.g., cell types, temperatures) and single-cell levels (e.g., genome, proteome, transcriptome, metabolome). And the fact that a large part of yeast genes have the human orthologues made researchers easier to understand the biological activities in human cells. Generally, the knowledge of biological pathways modeling in yeast can be applied to human cells by the following protocol. The candidate pathways can be reconstructed by applying computational or experimental approaches on high-throughput experimental data from yeast models, and the identified networks in yeast can be served as the basis for reconstruction of human cell pathways. R. Usaite et al. [114] applied subnetwork searching algorithms on the integrated omic data to study the regulation of human AMP-activated kinases based on the analysis of regulatory network of the yeast orthologue Snf1 protein kinase. This work connected the similar function between the yeast Snf1 and human orthologue for better understanding the protein function and gene regulation in human cells. Recent study [115] detected potential cancer-related human signaling network which is orthologous to the yeast NaCI subnetwork detected by integer linear programming, which provided insights for understanding the human disease biology.
Integration of biological pathways
As discussed above, we mainly discussed about the computational modeling methods for three types of biological pathways, by utilizing different kind of data. However, the availability of methods considering all levels of biological pathways will also largely facilitate the genome-scale network reconstruction. Generally, each approach was designed to model the specific type of biological network because of its unique features and properties. For example, flux balance analysis (FBA) [47] is not suitable for signaling pathway because signaling pathway acts different functions responding to environmental changes, which often fail to reach steady state for FBA to simulate. However, graph-based approaches, such as Boolean network or Bayesian network have been developed to model the signaling pathways and gene regulatory network. Some efforts have been put to propose the methods connecting two of the three pathways [116–119]. Gonçalves et al. [49] provides an overview on the scope and limit of current methods for integrating multiple pathways. Especially, for yeast model, Chandrasekaran et al. [120] has proposed their method to build genome-scale integrated model and showed the ability to integrate metabolic and regulatory network model. Lee et al. [121] also proposed a strategy by using the flux balance analysis to integrate signaling, metabolic and regulatory processes. However, a more comprehensive analysis is still in need to understand the whole cellular system and to better address the complexity of integrated signaling-regulatory-metabolic networks at the genome scale; more experimental data representing the interactions among the three pathways can be also incorporated to model the pathways.
Conclusion
We reviewed a number of bioinformatics approaches for yeast pathway modeling based on data-driven methods as well as knowledge-driven methods. Data-driven methods were developed to discover the biological pathways using solely the gene expression data or protein–protein interactions data. Knowledge-driven methods can integrate multiple source of information to effectively predict the biological networks. In particular, we focused on bioinformatics methods for modeling biological pathways through ‘omics' data and applying network-based analysis to construct the pathways.
Clustering algorithms are applied to identify coexpression groups for function prediction and biological network analysis. Probabilistic graphical model provides a statistical means to infer the network structures from gene expression data, where edges represents regulation between gene pairs relying on the conditional independence given the rest genes in network. However, pathways inferred from solely ‘omics' data do not guarantee their biological interpretations. By utilizing information stored in public databases, such as the knowledge of protein–protein interactions, pathway information and functional annotation, integrated analyses can be performed to improve biological interpretations of the inferred pathways. Physical protein–protein interactions can be used to further refine the constructed model and discover true cellular reactions. The directionality information in the interactions and reactions in curated pathways can help improve the prediction accuracy and expand the biological pathways. Functional annotation provides a functional template to recognize the coregulation network with the ability to recover links in the signaling pathways. Table 2 listed several web servers providing the analysis of biological pathways in terms of different input data types and objectives. Both data-driven and knowledge-driven approaches cover a wide range of the statistical regression models to network-based probabilistic models, and even though modeling biological pathways from different kinds of data information is still challenging, computational methods with integrated knowledge are expected to improve the automation of the reconstruction process for biological pathways.
Table 2.
Web servers providing the analyses of biological pathways
| Number | Web server | Objective | Data Input | URL | Reference |
|---|---|---|---|---|---|
| 1 | GeneNetwork | Inferring genetic network architecture from microarray data | Gene expression data | http://genenetwork.sbl.bc.sinica | [122] |
| 2 | MetaReg | Modeling and analysis of a biological network from high throughput data | Gene expression data | http://acgt.cs.tau.ac.il/metareg/application.html | [123] |
| 3 | WGCNA | Identifying clusters (modules) of highly correlated genes | Gene expression data | http://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/Rpackages/WGCNA/ | [124] |
| 4 | YEASTRACT-DISCOVERER | Identifying and visualizing transcription regulatory networks and associations between TF and target genes | Gene expression data | http://www.yeastract.com/ | [125] |
| 5 | GeneNT | Network constrained (NC) clustering | Gene expression data | http://crantastic.org/packages/GeneNT | [122] |
| 6 | NetworkAnalyst | Network analysis and visualization by mining gene expression data | Gene expression data | http://www.networkanalyst.ca/NetworkAnalyst/ | [126] |
| 7 | KEGGanim | Visualize the gene expression data in KEGG pathways | Gene expression data | http://biit.cs.ut.ee/kegganim/ | [127] |
| 8 | ASIAN | Infer a framework of regulatory networks from gene expression data. | Gene expression data | http://www.mrc-lmb.cam.ac.uk/genomes/madanm/blang/methods/AburataniS.ASIAN.awebserver forinferringa.html | [128] |
| 9 | ArrayXPath | Visualize gene-expression data in integrated biological pathway | Gene expression data | http://www.snubi.org/software/ArrayXPath/ | [129] |
| 10 | VisHiC | Cluster and interpret gene expression microarray data | Gene expression data | http://www.hsls.pitt.edu/obrc/index.php?page=gene_expression_tools | [130] |
| 11 | PANA | Integrate the functional annotation with gene expression data to discover functional relationship among pathways | Functional annotation data, Gene expression data | http://cs.uns.edu.ar/∼ip/PANA/ | [60] |
| 12 | NetSearch | Identifying signaling pathways from microarray expression data and protein interactions | Gene expression data, Protein–protein interaction | http://arep.med.harvard.edu/NetSearch/runprog.html | [24] |
| 13 | Struct2net | Protein–protein interaction detection based on structural information with functional annotation | Protein–protein interaction data, Protein sequence | http://groups.csail.mit.edu/cb/struct2net/webserver/ | [131, 132] |
| 14 | GraphWeb | Mine large biological networks for smaller modules, discover novel candidates and connections for known pathways | Protein–protein interaction data, Directed regulatory data | http://biit.cs.ut.ee/graphweb/ | [133] |
| 15 | NeAT | Analysis of biological networks/pathway, including path finding, network clustering, etc. | Functional annotation data, Gene expression data, protein interaction data | http://rsat.ulb.ac.be/rsat/index_neat.html | [134] |
| 16 | KOBAS server | Annotate protein sequences with KEGG Orthology terms and identify significantly enriched pathways | Protein sequence data | http://kobas.cbi.pku.edu.cn/home.do | [135] |
| 17 | PHT-Pathway Hunter Tool | Perform shortest path analysis in the metabolic pathway | Substrate and product metabolite of a reaction in the pathway | http://pht.tu-bs.de/ | [136] |
Key Points
Substantial bioinformatics approaches have been applied to construct the biological pathways in yeast from high throughput data and protein–protein interactions.
Coexpression analysis are mainly the starting point of discovering protein–protein interactions and cofunctional modules, which can further drive the study of signaling pathway modeling, gene coregulation exploration and construction of metabolic pathway.
Data-driven methods, such as clustering approaches, probabilistic graphical models and Bayesian network, help discover the biological pathways using solely the gene expression eta or protein–protein interactions data.
Knowledge-driven methods, by incorporating the protein–protein interactions, pathway information and functional annotations provided by available public database, can significantly improve the performance of biological pathway modeling.
Funding
This work is partially supported by the National Science Foundation Computing and Communication Foundations (NSF CCF) under Grant No. 1451316 to D.Z. And this work is also supported by an National Institutes of Health (NIH) under Grant No. R01GM093123 to J.C. This work is also supported by the National Science Foundation (NSF CAREER) under Grant No. DBI1149224 to J.C.
Biographies
Jie Hou is a graduate student in the department of Computer Science Ph.D. program at University of Missouri, Columbia. His research interests are computational biology and bioinformatics.
Lipi Acharya works as Associate Scientist at Dow AgroSciences. Her research focuses on the development and application of computational methods to address problems in computational biology and bioinformatics.
Dongxiao Zhu is an Associate Professor in the Department of Computer Science at Wayne State University. His research is in the areas of computational biology, bioinformatics, health informatics and the interface with data mining and machine learning.
Jianlin Cheng is an Associate Professor in the Department of Computer Science at University of Missouri, Columbia. His research is focused on bioinformatics, computational biology, data mining and machine learning.
References
- 1.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 2007;1773:1311–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2011:40(Database issue):D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estévez-García IO, Cordoba-Gonzalez V, Lara-Padilla E, et al. Glucose and glutamine metabolism control by APC and SCF during the G1-to-S phase transition of the cell cycle. J Physiol Biochem 2014;70:569–81. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen J. Systems biology of lipid metabolism: from yeast to human. FEBS Lett 2009;583:3905–13. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell LH. Nobel lecture: yeast and cancer. Biosci Rep 2002;22:373–94. [DOI] [PubMed] [Google Scholar]
- 6.Botstein D, Chervitz SA, Cherry JM. Yeast as a model organism. Science (New York, NY) 1997;277:1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goffeau A, Barrell B, Bussey H, et al. Life with 6000 genes. Science 1996;274:546–67. [DOI] [PubMed] [Google Scholar]
- 8.Rao VS, Srinivas K, Sujini G, et al. Protein-protein interaction detection: methods and analysis. Int J Proteomics 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamdi A, Colas P. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol Sci 2012;33:109–18. [DOI] [PubMed] [Google Scholar]
- 10.Zhang QC, Petrey D, Deng L, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 2012;490:556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge H, Liu Z, Church GM, et al. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet 2001;29:482–6. [DOI] [PubMed] [Google Scholar]
- 12.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 2008;91:243–8. [DOI] [PubMed] [Google Scholar]
- 13.Lee W-P, Tzou W-S. Computational methods for discovering gene networks from expression data. Brief Bioinform 2009;10:408–23. [DOI] [PubMed] [Google Scholar]
- 14.Ihmels J, Friedlander G, Bergmann S, et al. Revealing modular organization in the yeast transcriptional network. Nat Genet 2002;31:370–7. [DOI] [PubMed] [Google Scholar]
- 15.Segal E, Shapira M, Regev A, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 2003;34:166–76. [DOI] [PubMed] [Google Scholar]
- 16.Brohée S, Janky Rs, Abdel-Sater F, et al. Unraveling networks of co-regulated genes on the sole basis of genome sequences. Nucleic Acids Res 2011;39(15):6340–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Förster J, Famili I, Fu P, et al. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 2003;13:244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte NC, Herrgård MJ, Palsson BØ. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 2004;14:1298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobson PD, Smallbone K, Jameson D, et al. Further developments towards a genome-scale metabolic model of yeast. BMC Syst Biol 2010;4:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heavner BD, Smallbone K, Barker B, et al. Yeast 5–an expanded reconstruction of the Saccharomyces cerevisiae metabolic network. BMC Syst Biol 2012;6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrgård MJ, Swainston N, Dobson P, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol 2008;26:1155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuepfer L, Sauer U, Blank LM. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 2005;15:1421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samaga R, Klamt S. Modeling approaches for qualitative and semi-quantitative analysis of cellular signaling networks. Cell Commun Signal 2013;11:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steffen M, Petti A, Aach J, et al. Automated modelling of signal transduction networks. BMC Bioinformatics 2002;3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Zhao H. A computational approach for ordering signal transduction pathway components from genomics and proteomics Data. BMC Bioinformatics 2004;5:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acharya L, Judeh T, Duan Z, et al. GSGS: A computational framework to reconstruct signaling pathways from gene sets. IEEE/ACM Trans Comput Biol Bioinform 2012;9:438–50. [DOI] [PubMed] [Google Scholar]
- 27.Acharya LR, Judeh T, Wang G, et al. Optimal structural inference of signaling pathways from unordered and overlapping gene sets. Bioinformatics 2012;28:546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen TS, Oliveira AP, Nielsen J. Reconstruction and logical modeling of glucose repression signaling pathways in Saccharomyces cerevisiae. BMC Syst Biol 2009;3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pe’er D. Bayesian network analysis of signaling networks: a primer. Sci STKE 2005;281:l4. [DOI] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014;42:D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherry JM, Hong EL, Amundsen C, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 2012;40(Database issue):D700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatr-Aryamontri A, Breitkreutz B-J, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res 2015;43(Database issue):D470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xenarios I, Salwinski L, Duan XJ, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 2002;30:303–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licata L, Briganti L, Peluso D, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 2012;40:D857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Shin J, Kim E, et al. YeastNet v3: a public database of data-specific and integrated functional gene networks for Saccharomyces cerevisiae. Nucleic Acids Res 2014;42(Database issue):D731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Braun P, Yıldırım MA, et al. High-quality binary protein interaction map of the yeast interactome network. Science 2008;322:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger CJ, Zhang P, Mueller LA, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 2004;32:D438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aach J, Rindone W, Church GM. Systematic management and analysis of yeast gene expression data. Genome Res 2000;10:431–45. [DOI] [PubMed] [Google Scholar]
- 40.Wanichthanarak K, Nookaew I, Petranovic D. yStreX: yeast stress expression database. Database 2014;2014:bau068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jewison T, Knox C, Neveu V, et al. YMDB: the yeast metabolome database. Nucleic Acids Res 2012;40(Database issue):D815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schellenberger J, Park JO, Conrad TM, et al. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 2010;11:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirschman L, Burns GAC, Krallinger M, et al. Text mining for the biocuration workflow. Database 2012;2012:bas020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R-S, Saadatpour A, Albert R. Boolean modeling in systems biology: an overview of methodology and applications. Phys Biol 2012;9:055001. [DOI] [PubMed] [Google Scholar]
- 45.Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol 2008;9:770–80. [DOI] [PubMed] [Google Scholar]
- 46.Blazier AS, Papin JA. Integration of expression data in genome-scale metabolic network reconstructions. Front Physiol 2012;3:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niklas J, Schneider K, Heinzle E. Metabolic flux analysis in eukaryotes. Curr Opin Biotechnol 2010;21:63–9. [DOI] [PubMed] [Google Scholar]
- 48.Österlund T, Nookaew I, Nielsen J. Fifteen years of large scale metabolic modeling of yeast: developments and impacts. Biotechnol Adv 2012;30:979–88. [DOI] [PubMed] [Google Scholar]
- 49.Gonçalves E, Bucher J, Ryll A, et al. Bridging the layers: towards integration of signal transduction, regulation and metabolism into mathematical models. Mol BioSyst 2013;9:1576–83. [DOI] [PubMed] [Google Scholar]
- 50.Holter NS, Mitra M, Maritan A, et al. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci 2000;97:8409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan J, Dean AK, Zhang W. A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst Biol 2010;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics 2012;13:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markowetz F, Spang R. Inferring cellular networks–a review. BMC Bioinformatics 2007;8:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Penfold CA, Wild DL. How to infer gene networks from expression profiles, revisited. Interface Focus 2011;1:857–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 2007;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Zheng W, Zhao H, et al. Statistical analysis reveals co-expression patterns of many pairs of genes in yeast are jointly regulated by interacting loci. PLoS Genet 2013;9:e1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karagoz K, Arga KY. Assessment of high-confidence protein–protein interactome in yeast. Comput Biol chem 2013;45:1–8. [DOI] [PubMed] [Google Scholar]
- 58.Roy S, Bhattacharyya DK, Kalita JK. Reconstruction of gene co-expression network from microarray data using local expression patterns. BMC Bioinformatics 2014;15:S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Priness I, Maimon O, Ben-Gal I. Evaluation of gene-expression clustering via mutual information distance measure. BMC Bioinformatics 2007;8:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ponzoni I, Nueda MJ, Tarazona S, et al. Pathway network inference from gene expression data. BMC Syst Biol 2014;8:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu D, Hero III AO. Bayesian hierarchical model for large-scale covariance matrix estimation. Journal of Computational Biology 2007;14:1311–26. [DOI] [PubMed] [Google Scholar]
- 62.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allocco DJ, Kohane IS, Butte AJ. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 2004;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang D, Tang C, Zhang A. Cluster analysis for gene expression data: A survey. IEEE Trans Knowl Data Eng 2004;16:1370–86. [Google Scholar]
- 65.Tran LH, Tran LH. Hypergraph and protein function prediction with gene expression data. J Automation and Control Engineering 2015;3:164–70. [Google Scholar]
- 66.Zhu D, Hero AO, Cheng H, et al. Network constrained clustering for gene microarray data. Bioinformatics 2005;21:4014–20. [DOI] [PubMed] [Google Scholar]
- 67.Al-Shboul B, Myaeng S. Initializing k-means using genetic algorithms. World Academy of Science, Engineering and Technology 2009;54:114–8. [Google Scholar]
- 68.Kerr G, Ruskin HJ, Crane M, et al. Techniques for clustering gene expression data. Comput Biol Med 2008;38:283–93. [DOI] [PubMed] [Google Scholar]
- 69.Pirim H, Ekşioğlu B, Perkins AD, et al. Clustering of high throughput gene expression data. Comput Oper Res 2012;39:3046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaskowiak PA, Campello RJ, Costa Filho IG. Proximity measures for clustering gene expression microarray data: a validation methodology and a comparative analysis. IEEE/ACM Trans Comput Biol Bioinform 2013;10:845–57. [DOI] [PubMed] [Google Scholar]
- 71.Friedman N. Inferring cellular networks using probabilistic graphical models. Science 2004;303:799–805. [DOI] [PubMed] [Google Scholar]
- 72.Zheng J, Chaturvedi I, Rajapakse JC. Integration of epigenetic data in bayesian network modeling of gene regulatory network. In: Pvattern Recognition in Bioinformatics. Springer, 2011;7036:87–96. [Google Scholar]
- 73.Chen H, Maduranga D, Mundra P, et al. Integrating epigenetic prior in dynamic bayesian network for gene regulatory network inference. In: IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Singapore 2013, pp. 76–82. IEEE. http://ieeexplore.ieee.org/xpl/articleDetails.jsp?tp=&arnumber=6595391&url=http%3A%2F%2Fieeexplore.ieee.org%2Fxpls%2Ficp.jsp%3Farnumber%3D6595391. [Google Scholar]
- 74.Qi Q, Li J, Cheng J. Reconstruction of metabolic pathways by combining probabilistic graphical model-based and knowledge-based methods. In: Proceedings of BMC, Cincinnati, OH, USA 2014, S5 BioMed Central Ltd; http://www.biomedcentral.com/1753-6561/8/S6/S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gat-Viks I, Shamir R. Refinement and expansion of signaling pathways: the osmotic response network in yeast. Genome Res 2007;17:358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu X, Ye Y, Subramanian K. Interactive analysis of gene interactions using graphical gaussian model. In: Proceedings of the 3nd ACM SIGKDD Workshop on Data Mining in Bioinformatics (BIOKDD 2003), Washington, DC, USA. ACM 2003, Vol. 3, pp. 63–9. http://dl.acm.org/citation.cfm?id=980984. [Google Scholar]
- 77.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 2008;9:432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai T, Liu W, Luo X. A constrained ℓ 1 minimization approach to sparse precision matrix estimation. J Am Stat Assoc 2011;106:594–607. [Google Scholar]
- 79.Cai TT, Li H, Liu W, et al. Covariate-adjusted precision matrix estimation with an application in genetical genomics. Biometrika 2013;100:139–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nariai N, Kim S, Imoto S, et al. Using protein-protein interactions for refining gene networks estimated from microarray data by Bayesian networks. In: Pacific Symposium on Biocomputing, World Scientific, 2004, 336–47. [DOI] [PubMed] [Google Scholar]
- 81.Bulashevska S, Eils R. Inferring genetic regulatory logic from expression data. Bioinformatics 2005;21:2706–13. [DOI] [PubMed] [Google Scholar]
- 82.Ito T, Chiba T, Ozawa R, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 2001;98:4569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 2006;440:637–43. [DOI] [PubMed] [Google Scholar]
- 84.Ideker T, Thorsson V, Ranish JA, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 2001;292:929–34. [DOI] [PubMed] [Google Scholar]
- 85.Tornow S, Mewes H. Functional modules by relating protein interaction networks and gene expression. Nucleic Acids Res 2003;31:6283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scott J, Ideker T, Karp RM, et al. Efficient algorithms for detecting signaling pathways in protein interaction networks. J Comput Biol 2006;13:133–44. [DOI] [PubMed] [Google Scholar]
- 87.Yeh C-Y, Yeh H-Y, Arias CR, et al. Pathway detection from protein interaction networks and gene expression data using color-coding methods and A* search algorithms. ScientificWorldJournal 2012;2012:315797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gitter A, Klein-Seetharaman J, Gupta A, et al. Discovering pathways by orienting edges in protein interaction networks. Nucleic Acids Res 2011;39:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nguyen HA, Vu CL, Tu MP, et al. Discovery of pathways in protein–protein interaction networks using a genetic algorithm. Data Knowl Eng 2015;96:19–31. [Google Scholar]
- 90.Silverbush D, Elberfeld M, Sharan R. Optimally orienting physical networks. J Comput Biol 2011;18:1437–48. [DOI] [PubMed] [Google Scholar]
- 91.Silverbush D, Sharan R. Network orientation via shortest paths. Bioinformatics 2014;30(10):1449–55. [DOI] [PubMed] [Google Scholar]
- 92.Anh NH, Long VC, Phuong TM, et al. A genetic-based approach for discovering pathways in protein-protein interaction networks. In: International Conference of Soft Computing and Pattern Recognition (SoCPaR), Hanoi, Vietnam. 2013, pp. 79–85. IEEE; http://ieeexplore.ieee.org/xpl/articleDetails.jsp?arnumber=7054106&filter%3DAND(p_IS_Number%3A7054094). [Google Scholar]
- 93.Bornholdt S. Boolean network models of cellular regulation: prospects and limitations. J R Soc Interface 2008;5:S85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mo ML, Palsson BØ, Herrgård MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol 2009;3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martínez JL, Bordel S, Hong KK, et al. Gcn4p and the Crabtree effect of yeast: drawing the causal model of the Crabtree effect in Saccharomyces cerevisiae and explaining evolutionary trade ‐ offs of adaptation to galactose through systems biology. FEMS Yeast Res 2014;14:654–62. [Google Scholar]
- 96.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009;43:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oliveira AP, Ludwig C, Picotti P, et al. Regulation of yeast central metabolism by enzyme phosphorylation. Mol Syst Biol 2012;8:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics 2007;23:1537–44. [DOI] [PubMed] [Google Scholar]
- 99.Wei Z, Li H. A hidden spatial-temporal Markov random field model for network-based analysis of time course gene expression data. Ann Appl Stat 2008:408–29. [Google Scholar]
- 100.Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 2008;24:1175–82. [DOI] [PubMed] [Google Scholar]
- 101.Green ML, Karp PD. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics 2004;5:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen L, Zeng W-M, Cai Y-D, et al. Prediction of metabolic pathway using graph property, chemical functional group and chemical structural set. Curr Bioinform 2013;8:200–7. [Google Scholar]
- 103.Consortium GO. The Gene Ontology [GO] databaseandinformaticsresource. NucleicAcidsRes 2004;32:D258–61. [Google Scholar]
- 104.Pesquita C, Faria D, Bastos H, et al. Evaluating GO-based semantic similarity measures. In: Proceedings of the 10th Annual Bio-Ontologies Meeting, Bienna, Austria 2007, p. 38. [Google Scholar]
- 105.Pesquita C, Faria D, Falcao AO, et al. Semantic similarity in biomedical ontologies. PLoS Comput Biol 2009;5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee PH, Lee D. Modularized learning of genetic interaction networks from biological annotations and mRNA expression data. Bioinformatics 2005;21:2739–47. [DOI] [PubMed] [Google Scholar]
- 107.Bebek G, Yang J. PathFinder: mining signal transduction pathway segments from protein-protein interaction networks. BMC Bioinformatics 2007;8:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Agrawal R, Imieliński T, Swami AN. Mining association rules between sets of items in large databases. In: Proceedings of ACM SIGMOD International Conference on Management of Data, Washington, DC. 1993, New York: ACM Press, pp. 207–16. [Google Scholar]
- 109.Praveen P, Fröhlich H. Boosting probabilistic graphical model inference by incorporating prior knowledge from multiple sources. PloS ONE 2013;8:e67410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu X, Zhu L, Guo J, et al. Prediction of yeast protein–protein interaction network: insights from the Gene Ontology and annotations. Nucleic Acids Res 2006;34:2137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.James K, Wipat A, Hallinan J. Integration of full-coverage probabilistic functional networks with relevance to specific biological processes. In: Data Integration in the Life Sciences, Manchester, UK. 2009, pp. 31–46. Springer; http://link.springer.com/chapter/10.1007%2F978-3-642-02879-3_4. [Google Scholar]
- 112.Joyce AR, Palsson BØ. The model organism as a system: integrating'omics' data sets. Nat Rev Mol Cell Biol 2006;7:198–210. [DOI] [PubMed] [Google Scholar]
- 113.Ristevski B. A survey of models for inference of gene regulatory networks. Nonlinear Anal 2013;18:444–65. [Google Scholar]
- 114.Usaite R, Jewett MC, Oliveira AP, et al. Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol Syst Biol 2009;5:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chasman D, Ho YH, Berry DB, et al. Pathway connectivity and signaling coordination in the yeast stress ‐ activated signaling network. Mol Syst Biol 2014;10:759. [Google Scholar]
- 116.Shlomi T, Eisenberg Y, Sharan R, et al. A genome ‐ scale computational study of the interplay between transcriptional regulation and metabolism. Mol Syst Biol 2007;3:101. [Google Scholar]
- 117.Chandrasekaran S, Price ND. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci USA 2010;107:17845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yizhak K, Benyamini T, Liebermeister W, et al. Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics 2010;26:i255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mosca E, Alfieri R, Maj C, et al. Computational modeling of the metabolic States regulated by the kinase akt. Front Physiol 2011;3:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chandrasekaran S, Price ND. Metabolic constraint-based refinement of transcriptional regulatory networks. PLoS Comput Biol 2013;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Min Lee J, Gianchandani EP, Eddy JA, et al. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput Biol 2008;4:e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu C-C, Huang H-C, Juan H-F, et al. GeneNetwork: an interactive tool for reconstruction of genetic networks using microarray data. Bioinformatics 2004;20:3691–3. [DOI] [PubMed] [Google Scholar]
- 123.Ulitsky I, Gat-Viks I, Shamir R. MetaReg: a platform for modeling, analysis and visualization of biological systems using large-scale experimental data. Genome Biol 2008;9:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Teixeira MC, Monteiro PT, Guerreiro JF, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res 2014;42(Database issue):D161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015;10:823–44. [DOI] [PubMed] [Google Scholar]
- 127.Adler P, Reimand J, Jänes J, et al. KEGGanim: pathway animations for high-throughput data. Bioinformatics 2008;24:588–90. [DOI] [PubMed] [Google Scholar]
- 128.Aburatani S, Goto K, Saito S, et al. ASIAN: a web server for inferring a regulatory network framework from gene expression profiles. Nucleic Acids Res 2005;33:W659–W664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chung H-J, Park CH, Han MR, et al. ArrayXPath II: mapping and visualizing microarray gene expression data with biomedical ontologies and integrated pathway resources using Scalable Vector Graphics. Nucleic Acids Res 2005;33(Web Server issue):W621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Krushevskaya D, Peterson H, Reimand J, et al. VisHiC—hierarchical functional enrichment analysis of microarray data. Nucleic Acids Res 2009;37:W587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Singh R, Park D, Xu J, et al. Struct2Net: a web service to predict protein–protein interactions using a structure-based approach. Nucleic Acids Res 2010;38(Web Server issue):W508–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Singh R, Xu J, Berger B. Struct2net: integrating structure into protein-protein interaction prediction. In: Pacific Symposium on Biocomputing, Grand Wailea, Maui, Hawaii. 2006, pp. 403–14. World Scientific. [PubMed] [Google Scholar]
- 133.Reimand J, Tooming L, Peterson H, et al. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res 2008;36:W452–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brohée S, Faust K, Lima-Mendez G, et al. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res 2008;36:W444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu J, Mao X, Cai T, et al. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res 2006;34:W720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rahman SA, Advani P, Schunk R, et al. Metabolic pathway analysis web service (Pathway Hunter Tool at CUBIC). Bioinformatics 2005;21:1189–93. [DOI] [PubMed] [Google Scholar]