Abstract
The importance of epistasis—or statistical interactions between genetic variants—to the development of complex disease in humans has been controversial. Genome-wide association studies of statistical interactions influencing human traits have recently become computationally feasible and have identified many putative interactions. However, statistical models used to detect interactions can be confounded, which makes it difficult to be certain that observed statistical interactions are evidence for true molecular epistasis. In this study, we investigate whether there is evidence for epistatic interactions between genetic variants within the cis-regulatory region that influence gene expression after accounting for technical, statistical, and biological confounding factors. We identified 1,119 (FDR = 5%) interactions that appear to regulate gene expression in human lymphoblastoid cell lines, a tightly controlled, largely genetically determined phenotype. Many of these interactions replicated in an independent dataset (90 of 803 tested, Bonferroni threshold). We then performed an exhaustive analysis of both known and novel confounders, including ceiling/floor effects, missing genotype combinations, haplotype effects, single variants tagged through linkage disequilibrium, and population stratification. Every interaction could be explained by at least one of these confounders, and replication in independent datasets did not protect against some confounders. Assuming that the confounding factors provide a more parsimonious explanation for each interaction, we find it unlikely that cis-regulatory interactions contribute strongly to human gene expression, which calls into question the relevance of cis-regulatory interactions for other human phenotypes. We additionally propose several best practices for epistasis testing to protect future studies from confounding.
Introduction
Epistasis, a phenomenon wherein the effect of a genetic variant on the phenotype is dependent on other genetic variants, was first identified more than a century ago; however, it has been highly contested whether or not epistasis plays an important role in the development of complex traits in humans. In model organisms, epistasis is commonly observed: variants associated with the trait of interest often interact with other variants, and more broadly, such interactions account for a notable proportion of variance in a multitude of phenotypes.1, 2, 3 Epistasis may play a similar role in humans—additive genetic effects are unable to account for the majority of heritability in most complex traits4, 5—but evidence for epistasis in human remains elusive. Most studies rely on the statistical association between genetic variants and phenotype to identify signs of epistasis, and the interactions identified are notoriously difficult to replicate.6, 7 This may be attributable to the inherent inability to tightly control a variety of factors when studying phenotypes in humans, or to the fact that most phenotypes studied are several steps removed from the underlying biological processes that influence them. These methodological limitations make it unclear whether the lack of observed epistasis in humans is a true feature of the genetic architecture, or whether epistasis is simply much more difficult to observe outside experimental systems.
Human-derived cell lines, as a proxy for primary tissue, provide a unique opportunity to investigate epistasis. Like model systems, the environment for cell lines can be tightly controlled, and moreover, comprehensive genetic and gene expression data can readily be collected. This enables the study of the genetic architecture underlying thousands of genes’ expression—a quantitative phenotype directly tied to the nucleotide sequence—through statistical association studies. Gene expression is an ideal phenotype to study epistasis, because molecular mechanisms that drive gene expression are known to involve complex molecular interactions among transcription factors and regulatory sequences, and experimental maps of chromatin looping and transcription factor binding enable biological interpretations for observed statistical interactions.8, 9 Moreover, the study of gene expression is also directly relevant to complex disease—although there are some striking examples of causal coding variants,10, 11 the vast majority of variants identified in genome-wide association studies are non-protein coding. Thus it is presumed that the disruption of gene regulation is causally involved in the development of many common diseases.12, 13 In several instances, it has been shown that single-nucleotide variants regulate gene expression by altering the function of regulatory elements and that these altered gene expression profiles result in clinical phenotypes.14, 15 By better understanding the genetic control of gene expression, we may therefore better understand the genetic architectures underlying complex disease.
Genetic variants associated with gene expression levels—termed expression quantitative trait loci (eQTL)—have been studied extensively in primary human tissue and in cell lines. In many eQTL analyses, a gene-based approached is taken wherein variants within the cis-regulatory region for a given gene are tested for association with its expression. Until recently, the number of association tests required to perform a similar genome-wide association test for interactions was not computationally feasible. However, advances in computational power are continually diminishing this barrier and two genome-wide studies of epistasis have identified replicating interactions.16, 17 The validity of these interactions, however, was questioned when it was demonstrated that through complex linkage disequilibrium (LD) patterns, these putative interactions could tag single-variant eQTL.18 Notably, all of the interactions identified in those studies were either no longer significant or were strongly attenuated when the effects of additional cis-eQTL were considered. This illustrates that, compared to single-locus analyses, the statistical models used to detect epistasis are subject to distinct confounding factors, which are rarely addressed in studies of epistasis.
In this study, we investigate whether evidence for epistasis within the cis-regulatory region in humans persists after systematically accounting for technical, statistical, and biological confounding factors. We performed a targeted investigation of interactions regulating gene expression levels in human lymphoblastoid cell lines (LCLs): the analysis was restricted to nominal eQTL within the target gene’s cis-regulatory region (p < 0.05) to drastically reduce the number of association tests performed1, 19 while retaining the genomic regions most likely to harbor pertinent regulatory elements. Few genes showed evidence of epistasis (165 of 11,465 genes tested), although multiple interactions were often detected for the same gene. A total of 1,119 interactions were identified, many of which replicated in an independent dataset (90 of 803 possible). We then investigated confounding factors—technical (variants within probe binding sites, ceiling/floor effect), statistical (missing genotype combinations, population stratification), and biological (haplotype effects, tagging cis-eQTL)—that provide alternative, more parsimonious explanations than biological epistasis. Ultimately, each of the interactions identified could be accounted for by an alternative mechanism, suggesting that the majority of statistical interactions identified without accounting for confounding factors are spurious associations. Many of these confounding factors are inherent to the statistical models used and will therefore generalize to other phenotypes; consequently, the analytic framework of this study will be of use to many future studies of statistical epistasis.
Subjects and Methods
Our code has been made freely available online (see Web Resources).
Genotyping and Gene Expression Data
The discovery dataset was comprised of individuals ascertained as part of the International HapMap Project, PhaseI+II,20 which consisted of 210 unrelated individuals with genome-wide genotyping data (Phase I+II, release 24). For each of these individuals, Stranger et al. collected and normalized gene expression levels from immortalized LCLs using the Sentrix Human-6 Expression Bead Chip, v.1.21 All probes with a HapMap SNP underlying the expression probe were removed from analysis.21 We applied a population normalization procedure, described by Veyrieras et al.,22 to the gene expression values such that the expression of each gene within each population followed a normal distribution. This removed population-level differences in gene expression, which enabled us to combine all ethnicities in our analysis. Our replication dataset consists of 232 unrelated individuals from the 1000 Genomes Project (1KG), for whom gene expression in LCLs was available. These individuals had been sequenced at low coverage as part of the 1KG Project;23 we used genetic data from phase I, version 3. Stranger et al. also collected and normalized gene expression levels in LCLs for these individuals using Illumina Sentrix Human-6 Expression BeadChip, v.2.24 We applied the same population normalization procedure22 to these data. Both the discovery and replication dataset are multiethnic; the sample composition by ethnicity is shown in Table 1.
Table 1.
Dataset Composition by Ethnicity
| Analysis | Total Sample Size |
Ethnicity |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CHB | CEU | GIH | JPT | LWK | MXL | MKK | YRI | ||
| Discovery | 210 | 45 | 60 | – | 45 | – | – | – | 60 |
| Replication | 232 | 34 | – | – | 35 | 80 | 38 | – | 45 |
The number of individuals of each ethnicity (1KG abbreviations) in the discovery and replication analyses.
Two additional replication datasets were used to investigate a promising interaction. The first consisted of 283 European-descent individuals from the Genotype-Tissue Expression (GTEx) Project, for whom gene expression in whole blood was assessed by RNA sequencing.25 Genotype data for these individuals were collected on both the HumanOmni5-Quad Array and the Infinium Exome Chip and then imputed to 1KG.25 The second dataset consisted of brain samples from autopsied European-descent individuals in the Mayo Late Onset Alzheimer’s Disease Consortium.26 These individuals were genotyped on the Illumina HumanHap300-Duo Genotyping Beadchip and gene expression was collected using the Illumina Whole-Genome DASL HT BeadChip.26 370 individuals had expression data available from cerebellum, and 385 had expression in the temporal cortex.
Generating SNP Pairs for Interaction Testing
To generate SNP pairs for each gene, we first identified all common SNPs within the gene’s cis-regulatory region. To be considered common, variants had to have a MAF > 5% when all ethnicities were combined. Based on cis-eQTL analyses,22 the cis-regulatory region was defined as starting 500 kb upstream of the gene’s start and ending 500 kb downstream of the gene’s stop (including the gene itself); gene boundaries were taken from ENSEMBL. Previously, these variants were individually tested for association with the gene’s expression level in the discovery dataset by Veyrieras et al.22 Based on this analysis, we filtered out SNPs whose marginal effects were not nominally associated with gene expression (excluded p > 0.05), under the hypothesis that nominally associated variants may represent weak marginal effects from a true underlying interaction. We then considered all possible SNP pairs among the remaining variants. Once this was done for each gene, more than 21 million SNP pairs were generated for interaction testing.
Identifying Significant Interactions
Each SNP pair was tested for interactions significantly associated with the expression of the gene for which it was generated. The following interaction model27 was used, which contains additive and dominant effects for each variant and all four possible interaction terms in order to ensure that variance is properly partitioned across the genetic terms:
| (Equation 1) |
where y represents gene expression, x1 and x2 use additive encoding to represent the genotype at SNP A and SNP B, respectively, z1 and z2 use Cordell’s27 dominant encoding to represent the genotype at SNP A and B, respectively, a1 and d1 are estimated coefficients representing the additive and dominant effects of SNP A, a2 and d2 are estimated coefficients representing the additive and dominant effects of SNP B, and iaa, iad, ida, and idd are estimated coefficients representing both additive and dominant interaction effects. The top three principal components were also included as covariates (PC1–3). To determine the significance of interactions, this model was compared to a reduced model lacking the four interaction terms using a likelihood ratio test (LRT):
| (Equation 2) |
This test was implemented with the program INTERSNP.28 We calculated an FDR of 5% using the qvalue package in R.29
Identification of Representative Interaction eQTL Models for Distinct Pairs of Interacting Genomic Loci
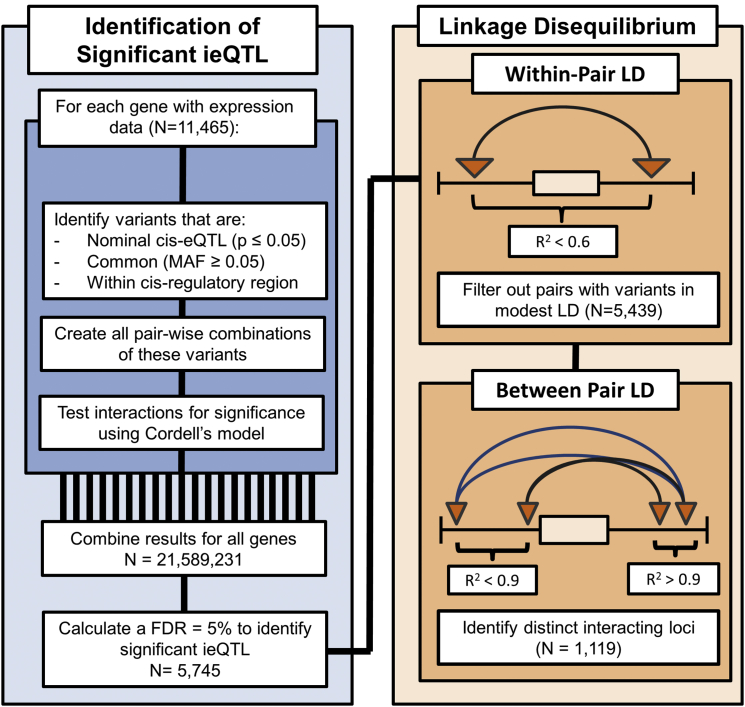
Some interaction eQTL (ieQTL) models identified in the discovery analysis were redundant due to LD. For two ieQTL models to be considered redundant, each SNP within one significant ieQTL model had to be in high LD (r2 ≥ 0.9) with a SNP within the second ieQTL model, and vice versa. By using this criterion, the pairs were effectively correlated at r2 ≥ 0.8, the threshold typically used for tag-SNP selection. The redundant SNP pairs have very similar βs for all parameters (Figure S2), indicating that they represent the same signal from a pair of interacting genomic loci. Redundant ieQTL models were grouped together. The model with the most significant LRT p value in the discovery analysis was used to represent the entire group in most analyses, so that each pair of interacting genomic loci was equally represented. A visual schematic of this process is provided in Figure 1.
Figure 1.
Workflow Used to Identify and Group ieQTL
In the discovery analysis, nominally significant cis-eQTL (denoted by triangles) were paired together and tested for interactions significantly associated with gene expression levels (denoted by arcs). The within-pair LD was then calculated (Figure S1), and interactions composed of variants in modest LD (r2 > 0.6) with one another were removed from the remainder of the analysis. Some of the remaining interactions represented the same pair of interacting genomic loci (Figure S2) and were partitioned into distinct groups (denoted by the arc color). For two interactions to be grouped together, each SNP within one significant ieQTL model had to be in high LD (r2 ≥ 0.9) with a SNP within the second ieQTL model, and vice versa.
Statistical Power Estimation
We performed simulation analyses to determine the power to identify interactions. We first randomly sampled a set of 20,000 SNP pairs having all nine genotype combinations present, and then we used the observed genetic data to simulate gene expression values. We simulated gene expression values based on the observed genotypes, the actual additive and dominant main effects for each of the two interacting variants, and an error term drawn from a standard normal distribution, and we embedded interaction terms of varying strength.
To properly represent the main effects of the variants, we used βs for the additive and dominant terms for each variant reflecting the actual effects within our dataset. We used
| (Equation 3) |
where y represents gene expression, x1 uses additive encoding to represent the genotype for the variant, z1 uses Cordell’s27 dominant encoding to represent the genotype, and the top three principal components were included as covariates (PC1–3).
We then determined the effect size for the interaction terms. There are four interaction terms in the model: additive by additive (iaa); additive by dominant; dominant by additive; and dominant by dominant. The iaa term is significant in all significant interaction models identified in the actual discovery analysis, whereas the other terms are not—these terms are included so that phenotypic variance is appropriately partitioned between genetic components. Consequently, these three interaction terms were treated as nuisance variables when simulating gene expression values; their βs were drawn from a normal distribution (mean = 0, standard deviation = 0.03). We used the effect sizes of cis-eQTL (p < 5.0 × 10−8) in our analysis to establish a “moderate” anticipated effect size (cis-eQTL median: β = 0.771) and a “high” anticipated effect size (cis-eQTL 75th percentile: β = 0.908). These βs are well within the range of observed effect sizes for significant interactions (iaa median: β = 0.65 and iaa max: β = 2.57). We then simulated gene expression data for each of the two effect sizes for each pair of SNPs.
Next, we performed the same LRT used in the discovery analysis to identify significant interactions. All interactions with p values below the FDR = 5% threshold (p ≤ 1.328 × 10−5) were considered significant. We then repeated this process 10 times using the same 20,000 pairs of variants. In each of these ten iterations, power was calculated as the total number of pairs found to have a significant interaction divided by the total number of simulated interactions tested. The mean and standard deviation across these ten iterations, broken out by variant MAF and LD, is reported in Table S1.
Variants within the Probe-Binding Site
To determine whether variants were within the probe binding locations, we first used BLAT to identify the probe binding location in hg19 coordinates. Some probes returned multiple hits; consequently, we filtered the binding sites (binding sites had to be on the same chromosome as the gene, have a length > 30 base pairs, and an identity score > 95%) to identify unique binding locations. We then exclusively looked within a subset of our discovery dataset with sequencing data in the 1KG Project (n = 174) to determine whether there were any variants within binding sites that might confound the interaction analysis.
Ceiling/Floor Effect
Microarrays have a limited dynamic range that is not able to capture the extremes of gene expression. If the combined additive effect of two variants exceeds the threshold of detection, their apparent combined effect will be less than the sum of their individual effects. Thus, they may be spuriously identified as interacting. If this occurs, there will be a characteristic pattern of βs: the main effects for variants will be in the same direction, and the interaction term β will be in the opposite direction. We looked for this characteristic pattern to determine an upper bound of the prevalence of the ceiling/floor effect within our results. First, we identified the significant variables (β ± SE could not contain zero) in the model. All interactions were then categorized as having 0, 1, or 2 SNPs with a significant main effect—either additive or dominant main effects counted; if both additive and dominant main effects were significant for the same variant, the one with the largest effect size was used to represent the main effect. For interactions where both variants had at least one significant main effect, we determined whether or not they had a concordant direction of effect. For those pairs with concordant directions of effect, we compared the significant interaction term with the largest absolute effect size to determine whether it was discordant with the main effects. If this was the case, the interaction had a pattern consistent with a ceiling/floor effect and was not considered clear evidence for epistasis.
Population Specific cis-eQTL
Population-specific cis-eQTL can confound the interaction analysis, even though gene expression values were population normalized and the top three PCs were included as covariates. To investigate this, we first stratified the discovery dataset by each of the three ethnicities (CEU, YRI, CHB+JPT) and tested each interaction for significance, using the same methodology. For interactions that were not significant (p < 0.05) in any of the populations, we determined whether the interacting variants were population-specific cis-eQTL using Equation 3. Variants with nominally significant (p < 0.05) main effects were considered cis-eQTL. If a variant was identified as a cis-eQTL in only a subset of populations, it was considered population specific.
Conditional cis-eQTL Analysis
To determine whether ieQTL pairs were tagging a cis-eQTL as suggested by Wood et al.,18 we first identified all nominal cis-eQTL (p < 0.05) for genes with significant ieQTL. To identify all nominal cis-eQTL, we used a subset of the discovery analysis individuals (n = 174) who were also sequenced as part of the 1KG Project.23 We used the called genotypes from Phase III, v.5. The same gene expression data previously described for the discovery set was used. Within this subset, we performed a single-marker cis-eQTL analysis for each common variant (MAF > 5%) within the cis-regulatory region,
| (Equation 4) |
where y represents gene expression, x1 uses additive encoding to represent the genotype for the variant, and the top three principal components were included as covariates (PC1–3). Variants with nominal significant (p < 0.05) main effects were considered cis-eQTL.
To determine whether any of these cis-eQTL could account for the interaction, we created all pairs of cis-eQTL and ieQTL for the same gene. We incorporated each cis-eQTL into each interaction model:
| (Equation 5) |
where y represents gene expression, x1 and x2 use additive encoding to represent the genotype at interacting SNPs A and B, respectively, z1 and z2 use Cordell’s dominant encoding to represent the genotype at interacting SNPs A and B, respectively, a1 and d1 are estimated coefficients representing the additive and dominant effects of SNP A, a2 and d2 are estimated coefficients representing the additive and dominant effects of SNP B, and iaa, iad, ida, and idd are estimated coefficients representing both additive and dominant interaction effects. The main effect of the cis-eQTL is represented with additive encoding by x3 and with dominant encoding by z3; the estimated coefficients corresponding to the main effects are a3 and d3, respectively. The top three principal components were also included as covariates (PC1–3). We then performed a LRT comparing this model to a reduced model lacking the interaction terms:
| (Equation 6) |
If the LRT p value of an interaction was nominally significant (p < 0.05) for all conditional analyses, we considered this evidence that the interaction and cis-eQTL represented independent signals.
Results
Discovery and Replication of Genetic Interactions that Impact Gene Expression Levels
We identified interactions between nominal cis-eQTL that were significantly associated with gene expression levels. Our analysis was conducted using 210 individuals from the HapMap Project, Phase I+II, on whom both genotyping20 and gene expression data within LCLs21 were available. A population normalization procedure was applied to the gene expression data, so that there were no systematic differences between populations.22 The overall workflow for the analysis is shown in Figure 1. For each gene with expression data (n = 11,465), we identified common SNPs (global MAF > 5%) within its cis-regulatory region, defined as 500 kb upstream to 500 kb downstream of the gene. To increase power, we considered only variants nominally associated with the gene’s expression (p < 0.05) in a single-marker analysis.22 We analyzed all pairwise combinations of these variants for each gene, resulting in more than 21 million SNP pairs. We then performed a likelihood ratio test (LRT) comparing a full model, which contains the top three PCs, main effects, and interaction terms, to a reduced model, containing only the covariates and main effects, to determine which interactions significantly improved model fit.27 Given the large number of correlated tests, we controlled the false discovery rate (FDR) at 5% (p ≤ 1.328 × 10−5) across p values from all LRT performed.29 Assuming moderate and large effect sizes, respectively, we had 21.6%–55.3% and 44.3%–8.9% power to detect interactions between high-frequency variants (MAF 0.2–0.5) in low LD with one another (Table S1).
LD between variants complicates the interpretation of the interaction models. We addressed two types of LD in significant interaction models: within-pair LD, defined as the LD between the variants in the same interaction model, and between-pair LD, defined as the LD between variants in different interaction models. Modest within-pair LD indicates that the variants may be identifying a haplotype, which could carry a single variant that is actually driving the association with gene expression. Wood et al. have demonstrated that even very stringent LD-pruning thresholds (r2 > 0.1 or D′ > 0.1) are insufficient to protect against confounding by cis-eQTL,18 so we adopted a two-stage strategy to address this concern. First, we removed all pairs with variants in modest LD with one another (r2 > 0.6) from the remainder of the analysis (median r2 between remaining pairs of interacting variants was 0.06, Figure S1). We then directly tested for confounding by cis-eQTL in a later analysis. Ultimately, 5,439 interaction models were both significant and passed the within-pair LD filtering criteria; they were significantly associated with the expression of 165 unique genes (Table S2). We then calculated between-pair LD, or the correlation of variants in different interaction models. Highly correlated interaction models were grouped together (Subjects and Methods, Figure 1) because they likely represent the same pair of interacting genomic loci, as evidenced by their very similar statistical models (Figure S2). The 5,439 interaction models represented 1,119 pairs of interacting genomic loci (Table S2). The interaction model with the most significant p value in the discovery analysis was selected to represent the entire group in all subsequent analyses, unless specifically stated otherwise, to ensure that each pair of interacting genomic loci was equally represented.
Next, we performed a replication analysis using an independent dataset of 232 unrelated individuals from the 1KG Project who had both whole-genome sequencing23 data and gene expression levels in LCLs24 available. All ieQTL composed of variants that were common (MAF > 5%) and had available genotyping data were tested for significant interactions with the same procedure used in the discovery analysis. Of the 803 ieQTL tested, 363 had p values < 0.05 and 90 passed a Bonferroni multiple testing correction for all tests performed in the replication analysis.
Many Factors Confound Interaction Testing
Statistical interactions can be produced by a variety of factors other than biological epistasis, including technical artifacts, statistical artifacts, and LD artifacts driven by other biological processes. Technical artifacts are caused by the limitations of the data itself; for instance, limitations in the dynamic range of measureable gene expression can result in interactions being identified through the ceiling/floor effect. Statistical artifacts can result in an incorrect inference from a statistical model; for example, when there are population-level differences in the phenotype, analyzing multiple ethnicities together can produce spurious associations due to population stratification. Technical and statistical artifacts are especially troubling because they are unlikely to represent real biological association between the loci and phenotype. Other biological phenomena, namely haplotype effects and cis-eQTL effects, can be captured by interaction analyses due to LD patterns. We investigated whether the observed 1,119 significant ieQTL models from the discovery analysis could be explained by each of these phenomena.
Some Statistical Interactions Are Consistent with Confounding by Technical Limitations
The gene expression data used in this analysis was collected using microarrays. Microarray technology has a limited dynamic range, meaning that the upper and lower bound on the level of gene expression that microarrays can detect does not cover the full range observed in nature.
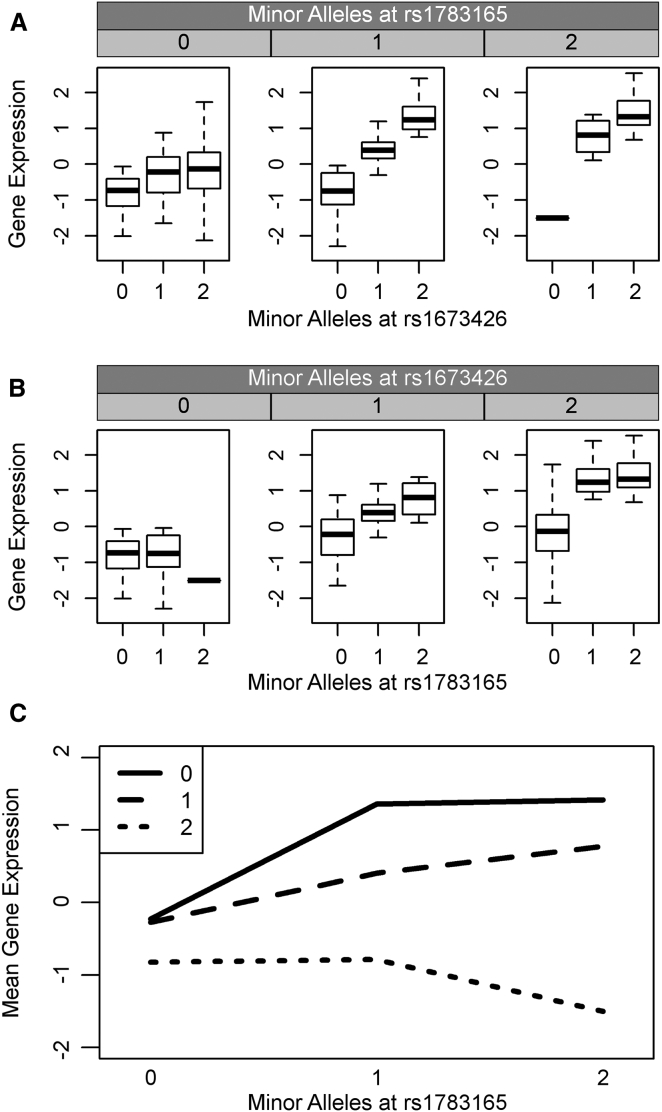
If the combined effect of two variants behaving additively exceeds the detectable limit, their individual effects will not be fully captured as they hit the maximum (i.e., ceiling) or minimum (i.e., floor) value detectable by microarrays. This phenomenon, known as the ceiling/floor effect, may result in such pairs of variants being spuriously identified as epistasis.30 Interactions caused by the ceiling/floor effect have a characteristic pattern of effects: the main effects of both variants have the same direction, and the interaction terms are in the opposite direction. For example, both main effects may increase gene expression, but the interactions will decrease gene expression. An example of an interaction putatively caused by the ceiling effect is shown in Figure 2. Of 1,119 locus pairs, 48 exhibited a pattern consistent with the ceiling/floor effect. It is possible that true genetic interactions could also produce this pattern; consequently, we consider this an upper bound of the influence of ceiling/floor artifacts within our analysis.
Figure 2.
The Interaction between rs1783165 and rs1673426 Associated with the Expression of PKHD1L1 May Be a Ceiling Effect
The ceiling effect, caused by limitations in the detectable range of gene expression, has a hallmark pattern—both variants have main effects with concordant direction of effect, and the interaction term has a discordant direction.
(A and B) The minor allele of rs1673426 (A) increases the expression of PKHD1L1. The minor allele of rs1783165 (B) also increases the expression of PKHD1L1, meaning both variants have a concordant direction of effect.
(C) The interaction plot depicts the mean gene expression for all individuals with the specified genotype combination, with each line representing the number of minor alleles at rs1673426. When there is only one minor allele at rs1673426, the mean gene expression increases for each minor allele at rs1783165; however, when there are two minor alleles at rs1673426, the increase in gene expression due to minor alleles at rs1783165 reaches a “maximum” at one minor allele. There is no additional increase in expression for having two minor alleles at rs1783165. This is denoted by the flat line connecting the two genotype combinations. Given that each minor allele at rs1783165 increases gene expression on the background of one minor allele at rs1673426, and that the maximum reached on the background of two minor alleles at rs1673426 is very close to the maximum gene expression levels possible to observe, we consider this an example of the ceiling effect.
The interpretation of microarray data is also complicated by genetic variants in the probe binding site, because different alleles may have different affinities for the probe. Probes containing any HapMap variant had previously been removed from the analysis;21, 22 however, HapMap does not provide comprehensive coverage of genetic variants. Consequently, we looked in a subset of individuals from the discovery analysis (n = 174) with low-coverage sequencing data through the 1KG Project to see whether genetic variants within the probe binding site may result in apparent interactions. The probes for 508 of 1,119 ieQTL contained a SNPs or indel in the 1KG Project. The probes for 255 ieQTL contained at least one common (MAF > 5%) variant. Although the conditional analysis (Subjects and Methods) performed later would likely account for the effect of these variants, we did not consider ieQTL with a common variant in the binding site evidence for biological epistasis. The probes for the remaining 253 ieQTL contained at least one rare variant, but no common variation. To determine whether these rare variants could result in the interaction, we performed the interaction analysis using only the 1KG individuals who did not have a rare variant in the probe binding site. The interactions for 200 ieQTL remained nominally significant (p < 0.05) when all individuals with rare variants were removed. Consequently, the interactions for 811 ieQTL are not attributable to variants within the probe binding sites.
Missing Genotype Combinations May Result in ieQTL
Linear regression models for epistasis may be unable to accurately decompose variance between genetic terms if there is either LD between the interacting variants or if there are missing genotype combinations. The issue of LD has previously been explored, and the Cordell model is robust to LD between variants when all genotype combinations are present.31 Consequently, we examined all interactions within the discovery dataset to see whether all of the nine possible two-locus genotype combinations were present. For 457 of the 1,119 ieQTL, at least one genotype combination was absent. Although failure to see certain two-locus genotypes may be due to lethal combinations, and thus perhaps is evidence for epistasis, it may also simply be a result of certain combinations being uncommon due to allele frequencies and the proximity between variants. Either way, the statistical model used cannot provide robust estimates unless all genotype combinations are present, and therefore, we do not consider these interactions as evidence for biological epistasis.
Haplotype Effects Captured through Complex LD Patterns May Produce ieQTL
In some LD architectures, a combination of two variants can identify haplotypes. Although there is evidence to suggest haplotypes form in response to biological interactions between variants,32, 33 haplotypes may simply carry a single variant that additively regulates gene expression. Thus, interactions between two variants in LD with one another may simply be tagging a cis-eQTL. Wood et al. demonstrated that this could occur even when strict LD-pruning thresholds (r2 > 0.1 or D′ > 0.1) were used; therefore, we consider it unlikely that any LD-pruning threshold would be sufficient to eliminate confounding by cis-eQTL.18 Consequently, we adopted a two-stage strategy to address haplotype effects, wherein we first use a lenient LD threshold to filter out interactions and then directly tested whether the interaction can be accounted for by cis-eQTL.
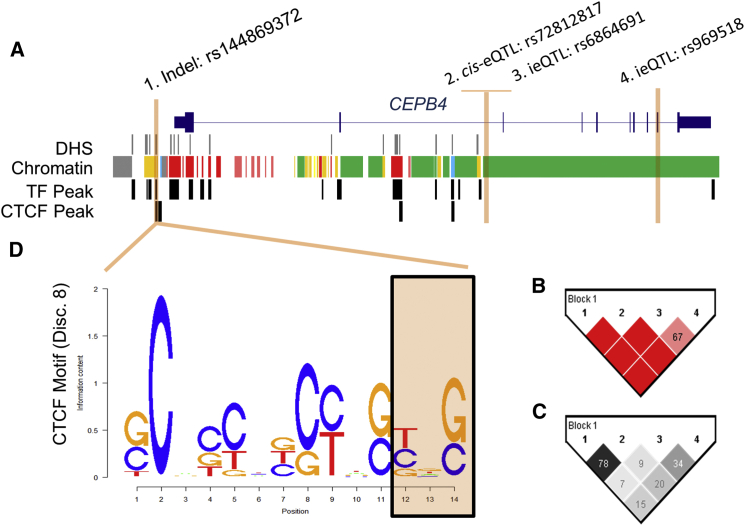
In the first stage, we used LD patterns to filter out variants in moderate LD with one another, because they probably represent a haplotype. We did this by first removing all interaction models composed of variants in modest LD with one another (r2 > 0.6) from all portions of the study, as previously mentioned. We then investigated whether or not variants within the same interaction model were in modest LD with one another as assessed by D′; of the 1,119 interacting loci, 806 had D′ values < 0.6. We did not consider any of the variants with D′ thresholds exceeding this threshold as evidence for epistasis, because they probably carry a single variant driving the effect. An example of this phenomenon observed in our data is illustrated in Figure 3. The distribution of LD statistics, both r2 and D′, for interaction models is shown in Figure S1.
Figure 3.
Interactions Impacting the Expression of CPEB4 May Represent Haplotype Effects
(A) A significant interaction between rs6864691 and rs969518 regulating the expression of CPEB4 was identified. The cis-eQTL rs72812817 mediated this interaction in the conditional analysis; however, none of these variants were within putative regulatory elements in GM12878 assayed by the ENCODE Project.
(B) However, a D′ heatmap of the region (the numbers correspond to SNP labels in A) illustrated that an indel, rs144869372, always occurred on the background of the cis-eQTL (D′ = 1).
(C) This occurs despite modest r2 values, as shown in the r2 heatmap of the region. There is evidence from ENCODE (A) suggesting the indel may be functional, as it occurs within both a ChromHMM strong enhancer (yellow) and a CTCF binding peak in GM12878.
(D) Notably, the indel is predicted to alter the binding of CTCF by HaploReg, by altering the last three nucleotides in the binding motif. Given the functional genomics evidence, the indel may be the causal variant and is detected by interactions that tag the haplotype carrying the indel.
In the second stage of the analysis, we directly tested whether or not the interaction could be accounted for by cis-eQTL by conditioning the interaction on each of the target gene’s cis-eQTL in turn. We first identified all nominal, common cis-eQTL (p < 0.05) for the interaction’s regulated gene using a subset of individuals from our discovery dataset (n = 174) with sequencing data available through the 1KG Project so that we would have a comprehensive list of genetic variation. Although the 1KG sequencing data is low coverage, it is extremely unlikely we would fail to detect the effect of a common cis-eQTL—1KG estimates they had 99.3% power to detect variants of 1% frequency.23 Even if a common cis-eQTL was missed, all variants that could tag it through LD would additionally have to be absent for its effect to not be captured in the conditional analysis. We then created all pairs of cis-eQTL and ieQTL for the same gene. For each of these combinations, we performed a conditional analysis in which the additive and dominant main effect for the cis-eQTL were incorporated into both the full and reduced model used in the LRT to determine the significance of the interaction. The majority of interactions appeared to be mediated by cis-eQTL (Figure 4); however, 139 of the 965 testable ieQTL remained significant (p < 0.05) in all conditional analyses performed, indicating that these interactions are not explained by cis-eQTL.
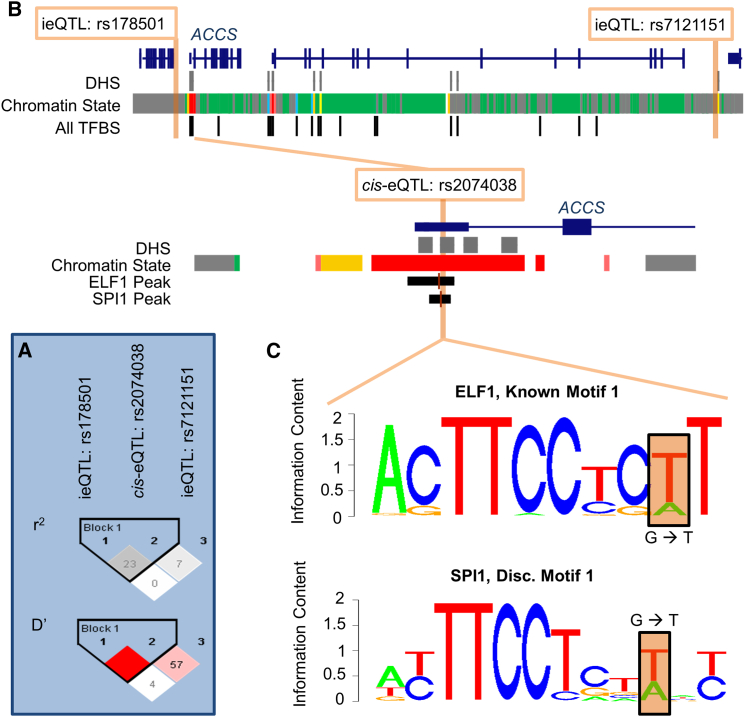
Figure 4.
The Interacting SNPs Regulating ACCS Are Probably Tagging a Single-Variant cis-eQTL through Linkage Disequilibrium
The interaction between rs178501 and rs7121151 is mediated by the cis-eQTL rs2074038 in the conditional analysis (interaction p value > 0.05).
(A) While the interacting variants are in low LD with the cis-eQTL based on r2, their high D′ indicates they often occur on the same haplotype.
(B) The interacting variants are not located within DNase hypersensitivity sites, predicted chromatin states with a regulatory function (GM12878 Combined), or any of the uniform binding peaks identified for all transcription factors tested in GM12878 by ENCODE; however, the cis-eQTL is located within the canonical promoter for ACCS, a DNase hypersensitivity site, and numerous transcription factor binding peaks identified in GM12878 by ENCODE.
(C) Notably, the cis-eQTL occurs within a binding peak for both ELF1 and SPI1 in GM12878 and also alters the binding motifs of these transcription factors at the position highlighted in orange. Thus, the cis-eQTL rs2074038 is probably the causal variant, and the interaction is simply capturing its effect through LD.
Population-Specific eQTLs May Produce Statistical Interactions
In our discovery and replication analyses, we analyzed multiple ethnicities together. When there are population differences in the distribution of both genotypes and phenotypes, analyzing multiple populations together can lead to spurious results, due to a phenomenon known as population stratification. The population normalization procedure applied to the gene expression data removes systematic population differences in the phenotype, thereby enabling multiple ethnicities to be combined for analysis without risk of known complications from population stratification. Although this approach has been used in other studies, we also controlled for the top three PCs in our analysis to adjust for residual ethnicity-dependent effects.22, 34 Furthermore, we performed a stratified analysis, wherein we tested each of the 1,119 ieQTL in each of the three discovery ethnicities (CEU, YRI, and CHB+JPT) separately. Although the Cordell model was not robust in the stratified analysis in many cases (due to the reduced sample size, all nine possible two-locus genotype combinations were often not observed in all populations), 859 of 1,119 ieQTL were at least nominally significant (p < 0.05) in at least one population, suggesting that population stratification is unlikely to account for their significance.
However, the interaction for 260 ieQTL was completely attenuated in the stratified analysis. In some cases, this may be attributed to reduced power to detect effects because the sample size is smaller; however, it could also suggest that interaction testing was subject to an unknown form of population stratification. Upon further investigation, we found that 234 of 260 ieQTL attenuated in the stratified analysis involved at least one population-specific cis-eQTL, meaning that a variant was a significant cis-eQTL in only a subset of populations. Population-specific cis-eQTL may be a product of reduced power to detect effects when allele frequencies are different between populations; however, there were also instances in which variants with very similar allele frequencies had different marginal effects across populations (Figure S3).24 Such variants might be a product of population-dependent ability to tag causal cis-eQTL due to differential LD patterns. In relation to interaction testing, systematic differences in both the main effect of each variant and the frequency of two-locus genotype combinations between populations resulted in a spurious interaction signature; an example is provided in Figure 5. To investigate whether population-specific effects may impact the 859 ieQTL that were nominally significant in at least one population, we calculated the within-population LD between each pair of interacting variants. 689 of 859 ieQTL were significant in at least one population where the variants were not in LD with one another (r2 and D′ < 0.6) (Table S3). We did not consider the 170 ieQTL that were exclusively significant in populations with population-specific haplotypes as clear evidence for biological epistasis. Ultimately, 689 of the 1,119 ieQTL were inconsistent with population-specific effects.
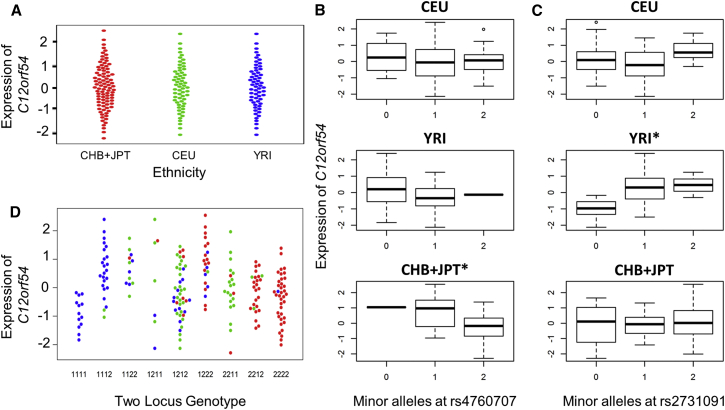
Figure 5.
Population-Specific eQTLs May Underlie ieQTL Regulating C12orf54
The interaction between rs2731091 and rs4760707 regulating C12orf54 replicated but was not nominally significant (p < 0.05) in any population in the stratified analysis.
(A) Due to the population normalization procedure, there are not systematic differences in the expression of C12orf54 between populations; however, we found that each variant was a population-specific cis-eQTL.
(B) rs4760707 was a cis-eQTL in CHB+JPT (p = 7.25 × 10−6) but not in YRI (p = 0.17) or CEU (p = 0.96).
(C) rs2731091 significantly regulated gene expression as a cis-eQTL in YRI (p = 7.28 × 10−6) but not CEU (p = 0.14) or CHB+JPT (p = 0.84).
(D) There were clear population differences in the frequency of two-locus genotypes between populations; in combination, it appears the population differences in two-locus genotypes and population-specific cis-eQTL produced a nuanced form of population stratification.
ieQTL Can Be Entirely Accounted for by Alternative Mechanisms
Ultimately, we investigated whether confounding factors could cumulatively account for all the interactions identified in this analysis (Tables 2 and S3). Of the 1,119 interacting genomic loci identified, 90 significantly replicated using a Bonferroni multiple testing correction threshold. Of these, 26 ieQTL could be explained by technical artifacts (i.e., the ceiling/floor effect and/or variants within the probe binding sites). 50 of the remaining 64 ieQTL could be explained by statistical artifacts (i.e., population stratification and/or missing genotypes). Biological explanations other than epistasis—namely haplotype effects or the tagging of cis-eQTL—could account for all remaining ieQTL that replicated at the most stringent Bonferroni level.
Table 2.
Proportion of Interactions Consistent with Confounding Factors
| Confounder |
All Interactions (n = 1,119) |
Bonferroni Replicating Interactions (n = 90) |
||
|---|---|---|---|---|
| Total | (%) | Total | (%) | |
| Ceiling/floor Effect | 48 | (4.30) | 11 | (12.22) |
| Variants in probe | 308 | (27.52) | 15 | (16.68) |
| cis-eQTL | 980 | (87.58) | 78 | (86.68) |
| D′ haplotype | 313 | (27.97) | 43 | (47.78) |
| Population-specific effects | 430 | (38.43) | 58 | (64.44) |
| Missing genotypes | 457 | (40.84) | 37 | (41.11) |
We counted the number of interactions consistent with each alternative explanation; interactions can be consistent with multiple confounders. We considered two categories of interactions: all interactions identified (n = 1,119) and the subset of those that replicated with p values exceeding the Bonferroni multiple testing correction threshold for the entire replication analysis (n = 90).
We additionally investigated the impact of filtering out interactions consistent with confounding prior to the replication analysis. Removing these interactions prior to replication testing had a considerable influence on the multiple testing correction threshold: only 86 of the 1,119 interactions identified in the discovery analysis were not consistent with the ceiling/floor effect, population stratification, variants within the probe binding site, missing genotype combinations, haplotype effects, or the tagging of cis-eQTL (Table S4). 37 of the 86 ieQTL had sufficient data to be tested in the replication analysis, and although none replicated at the adjusted Bonferroni multiple testing correction threshold, two interactions did replicate with nominal significance (p < 0.05). One of these, the interaction between rs1549791 and rs7115749 to regulate APIP, did not have a consistent direction of effect between the discovery and replication datasets (Figure S4) and thus was not considered evidence for epistasis. The remaining interaction, between rs1262808 and rs11615099 regulating the expression of MYRFL, had concordant effects in both the discovery and replication datasets (Figure 6). It did not pass the multiple testing correction threshold in the initial replication analysis (p = 2.03 × 10−3) though, so we further examined additional datasets.
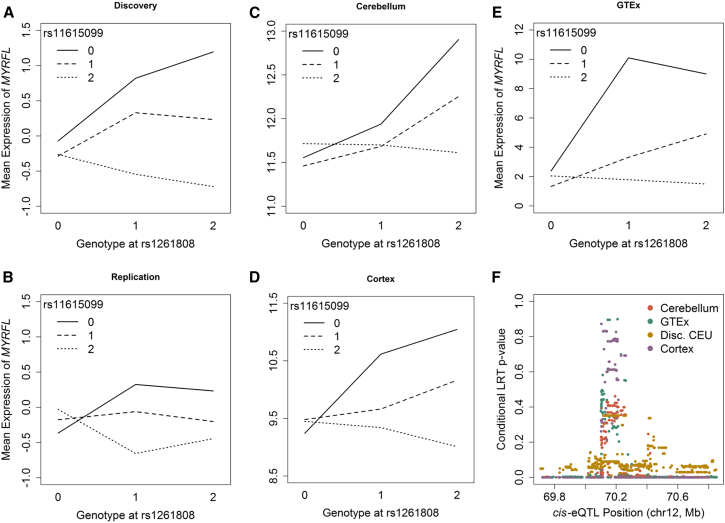
Figure 6.
Despite Consistent Replication, the Interaction Regulating MYRFL Is Attributable to cis-eQTL
(A–E) In each interaction plot, all individuals are categorized according to their two-locus genotype at rs1262808 and rs11615099. The mean expression of MYRFL for all individuals with each of the nine possible two-locus genotypes is shown here for the (A) discovery, (B) replication, (C) Mayo, cerebellum, (D) Mayo, cortex, (E) GTEx, whole blood datasets. The interaction plot illustrates a consistent trend across all datasets, this interaction is mediated by cis-eQTL.
(F) Conditional cis-eQTL analyses were conducted in the discovery (CEU only, yellow); GTEx (purple); Mayo, cerebellum (teal); and Mayo, temporal cortex (orange). For each conditional analysis, the conditional LRT p value is plotted by the genomic position of the cis-eQTL conditioned on. The p value peak observed in this region illustrates that cis-eQTL completely attenuate the interaction when they are conditioned on.
Replication Does Not Protect against Confounding
To determine whether the interaction between rs1262808 and rs11615099 regulating the expression of MYRFL was robust, we examined it in several additional datasets, some of which leveraged different technologies to assess gene expression levels, used primary tissues rather than cell lines, and were collected in different cellular contexts. First, we examined the interaction in 283 individuals from the Genotype-Tissue Expression (GTEx) Project with RNA sequencing of gene expression in whole blood and found a significant (p = 6.96 × 10−4) and similar pattern of effect (Figure 6). The same trend was also observed in 370 European-descent individuals with gene expression in both cerebellum (p = 7.50 × 10−4) and temporal cortex (p = 1.23 × 10−11), illustrating that the interaction was found in very different cellular conditions and was robust in four completely independent datasets (Figure 6). Despite significant and consistent replication, however, there was still the possibility that this interaction was attributable to confounding factors: the conditional cis-eQTL analysis was conducted in the multi-ethnic discovery dataset, meaning that population-specific LD patterns could have obfuscated the signal from a single variant enough to result in the residual significance of the interaction term. To account for this, we performed conditional cis-eQTL analyses in the additional replication datasets composed only of European-descent individuals and found cis-eQTL that completely attenuated the significance of the interaction signal in all cases. Although the cis-eQTL that most attenuates the signal varies between datasets, all tag the same locus (Figure 6). The same locus also attenuates the interaction completely in a conditional analysis on the CEU subset of the discovery dataset (Figure 6). Thus, despite consistent replication in numerous datasets, this interaction can be explained by confounding by cis-eQTL.
Discussion
In this study, we analyzed more than 21 million pairs of cis-regulatory variants for epistatic interactions influencing gene expression and found limited evidence for epistasis within the cis-regulatory region of genes. Fewer than 2% of genes tested (165 of 11,465) had significant interactions between regulatory genetic variants that appeared to influence their expression in the tightly controlled context of LCLs. Nonetheless, 90 of the 1,119 significant interactions replicated in independent datasets. We then performed a comprehensive investigation of known and novel potential confounding factors on the identified interactions (haplotype effects, ceiling/floor effect, single variant eQTL tagged through LD, missing genotype combinations, population stratification, and others) and found that all the interactions—even those that replicated—could be explained by at least one technical, statistical, or biological confounder. Thus, our findings do not support a major role for large effect interactions between common variants within the cis-regulatory region influencing the regulation of gene expression in LCLs.
Additionally, this study provides a trait-independent framework for protecting future interaction studies from confounding. Prior to performing any association testing, there are two levels of quality control required for statistical studies of epistasis: those adopted in GWAS best practices,35, 36, 37, 38 which are aimed at ensuring that individual genetic variants are called with high accuracy, and those that check whether a given pair of genetic variants is appropriate for interaction testing (i.e., missing genotype and the within-pair LD filters). Even when these quality-control measures are applied prior to the discovery analysis, significant interactions need to be further examined for evidence of confounding by single variants tagged through LD and for population-specific effects. We advise removing interactions consistent with these confounders prior to replication, as this reduced the number of putative interactions carried forward substantially, and consequently, the multiple testing penalty. The ceiling/floor effect is a more complicated confounder, as it is difficult to statistically disambiguate whether consistent interactions are caused by technical limitations or by biological epistasis. Consequently, we recommend interactions consistent with the ceiling/floor effect be flagged, rather than filtered out, and validated with an alternative technology if possible. It is still critical to replicate interactions to ensure they have robust, consistent effects, despite replication being insufficient to protect against confounding. Given how pervasive confounding factors are, it is critical to explicitly account for them through additional quality-control procedures and post hoc analyses in future studies to reduce spurious results.
To strike a balance between maximizing the power to detect effects and thoroughly investigating potentially interacting loci, we performed a focused analysis of common variants with significant marginal effects in the cis-regulatory region, which harbors the majority of known regulatory elements. We were moderately powered to detect interactions between common variants in low LD with one another with effects commensurate with the single-locus eQTL found in this dataset. Although additional statistical interactions with either smaller effect sizes or between less frequent genotype combinations would likely be identified with increased power, every example of a significant interaction we did identify was consistent with at least one confounding factor. Thus, we did not find compelling evidence that cis-regulatory interactions contribute strongly to the genetic architecture of gene expression; however, there are several additional limitations to our study. First, cell lines are a model system, and thus are not perfectly representative of primary tissue. Second, we analyzed multiple ethnicities simultaneously in an effort to increase sample size; however, doing so also increased the heterogeneity of our sample, which may have obfuscated some interactions. Therefore, our findings do not preclude the existence of epistasis within the cis-regulatory region, and we recommend that future studies of regulatory epistasis consider potential interactions that (1) occur within haplotypes (consistent with reports from Corradin et al.33 and Lappalainen et al.32), (2) have smaller effect sizes than those detected in similarly powered single-locus eQTL studies, (3) occur among less frequent genotype combinations, including rare variants, (4) involve variants without marginal eQTL effects (though evidence in model organisms suggests that these are rare1), and/or (5) are context dependent (e.g., inducible eQTL effects). Observing statistical interactions in these contexts could reconcile our findings with molecular studies, many of which use mutagenesis to generate genetic variation that would not be observed in population-based studies, that illustrate that transcription factors (TF) interact with each other to influence promoter and enhancer activity.39, 40, 41
Genetic interactions involving distant variants could also be a mechanism through which epistasis influences complex traits. However, we did not investigate interactions involving variants outside of the cis-regulatory region because evidence from eQTL studies in humans suggests that trans-eQTL effects are less robust, less common, and have smaller effect sizes.42, 43 This, coupled with the substantial increases in the number of association tests required to investigate trans interactions, would have resulted in reduced power to detect such effects. Nonetheless, interactions between distant variants (i.e., gene-by-gene interactions) may still be important to the biology of disease in humans. Increases in the sample size of eQTL datasets and the corresponding increases in statistical power will enable future in-depth studies of trans interactions that may help to illuminate the biological mechanisms through which genetic variants are associated with disease. However, trans interactions are not protected from many of the confounders influencing the study of cis interactions,18 and thus studies of trans interactions will need to explicitly account for these issues as well.
Our findings (along with prior reports)18 illustrate that significant interaction effects can be due to a variety of confounding factors. This demonstrates that significant statistical interactions do not necessarily imply a biological relationship with either the phenotype or between the variants themselves. To account for this, some confounders can be addressed as part of quality-control procedures prior to performing any association tests (i.e., missing genotype check, removing variants in probe binding sites, and LD filtering), whereas others—such as confounding by single variants with strong effects—will probably require specific post hoc analyses after the initial association is identified. Furthermore, replication—long held as the gold standard for genetic association studies—does not safeguard against these confounders, because they can be due to artifacts that are consistent across multiple datasets. Given the pervasive nature of confounding, it must be considered in all future studies of epistasis. The analytic approach used in this study provides a trait-independent framework for explicitly examining confounding factors in interaction studies and avoiding reporting spurious results.
Acknowledgments
We thank Laura Wiley for normalizing gene expression values within the replication dataset. We also thank Jacob Hall, Corinne Simonti, and R. Michael Sivley for their help and advice on this project. This work was supported by a human genetics training grant, T32GM080178, from the NIH.
Published: September 15, 2016
Footnotes
Supplemental Data include four figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.022.
Web Resources
Interaction eQTL Analysis, http://www.icompbio.net/?page_id=1654
Supplemental Data
This file provides all 5,439 interactions identified in the discovery analysis. When these interactions appeared to represent the same signal, due to LD, they were placed into groups (n = 1,119) and a representative interaction was chosen. We provide the group identifier for each of the interactions, and the group’s representative interaction.
We examined whether or not the 1,119 interactions could be explained by confounding factors. Here, we present which alternative explanations could account for each interaction.
References
- 1.Bloom J.S., Kotenko I., Sadhu M.J., Treusch S., Albert F.W., Kruglyak L. Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat. Commun. 2015;6:8712. doi: 10.1038/ncomms9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang W., Richards S., Carbone M.A., Zhu D., Anholt R.R.H., Ayroles J.F., Duncan L., Jordan K.W., Lawrence F., Magwire M.M. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. USA. 2012;109:15553–15559. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyler A.L., Donahue L.R., Churchill G.A., Carter G.W. Weak epistasis generally stabilizes phenotypes in a mouse intercross. PLoS Genet. 2016;12:e1005805. doi: 10.1371/journal.pgen.1005805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuk O., Hechter E., Sunyaev S.R., Lander E.S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei W.-H., Hemani G., Haley C.S. Detecting epistasis in human complex traits. Nat. Rev. Genet. 2014;15:722–733. doi: 10.1038/nrg3747. [DOI] [PubMed] [Google Scholar]
- 7.Murk W., Bracken M.B., DeWan A.T. Confronting the missing epistasis problem: on the reproducibility of gene-gene interactions. Hum. Genet. 2015;134:837–849. doi: 10.1007/s00439-015-1564-3. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 11.Klein R.J., Zeiss C., Chew E.Y., Tsai J.-Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaub M.A., Boyle A.P., Kundaje A., Batzoglou S., Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smemo S., Tena J.J., Kim K.-H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown A.A., Buil A., Viñuela A., Lappalainen T., Zheng H.F., Richards J.B., Small K.S., Spector T.D., Dermitzakis E.T., Durbin R. Genetic interactions affecting human gene expression identified by variance association mapping. eLife. 2014;3:e01381. doi: 10.7554/eLife.01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemani G., Shakhbazov K., Westra H.-J., Esko T., Henders A.K., McRae A.F., Yang J., Gibson G., Martin N.G., Metspalu A. Detection and replication of epistasis influencing transcription in humans. Nature. 2014;508:249–253. doi: 10.1038/nature13005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Wood A.R., Tuke M.A., Nalls M.A., Hernandez D.G., Bandinelli S., Singleton A.B., Melzer D., Ferrucci L., Frayling T.M., Weedon M.N. Another explanation for apparent epistasis. Nature. 2014;514:E3–E5. doi: 10.1038/nature13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kooperberg C., Leblanc M. Increasing the power of identifying gene x gene interactions in genome-wide association studies. Genet. Epidemiol. 2008;32:255–263. doi: 10.1002/gepi.20300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veyrieras J.-B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith J., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou F., Chai H.S., Younkin C.S., Allen M., Crook J., Pankratz V.S., Carrasquillo M.M., Rowley C.N., Nair A.A., Middha S., Alzheimer’s Disease Genetics Consortium Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordell H.J. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 28.Herold C., Steffens M., Brockschmidt F.F., Baur M.P., Becker T. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics. 2009;25:3275–3281. doi: 10.1093/bioinformatics/btp596. [DOI] [PubMed] [Google Scholar]
- 29.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis-Beck M.S., Bryman A., Liao T.F., editors. The SAGE Encyclopedia of Social Science Research Methods. SAGE Publications; 2003. [Google Scholar]
- 31.Zeng Z.-B., Wang T., Zou W. Modeling quantitative trait loci and interpretation of models. Genetics. 2005;169:1711–1725. doi: 10.1534/genetics.104.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lappalainen T., Montgomery S.B., Nica A.C., Dermitzakis E.T. Epistatic selection between coding and regulatory variation in human evolution and disease. Am. J. Hum. Genet. 2011;89:459–463. doi: 10.1016/j.ajhg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sal lari R., Lupien M., Markowitz S., Scacheri P.C. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker J., Wendland J.R., Haenisch B., Nöthen M.M., Schumacher J. A systematic eQTL study of cis-trans epistasis in 210 HapMap individuals. Eur. J. Hum. Genet. 2012;20:97–101. doi: 10.1038/ejhg.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pluzhnikov A., Below J.E., Konkashbaev A., Tikhomirov A., Kistner-Griffin E., Roe C.A., Nicolae D.L., Cox N.J. Spoiling the whole bunch: quality control aimed at preserving the integrity of high-throughput genotyping. Am. J. Hum. Genet. 2010;87:123–128. doi: 10.1016/j.ajhg.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J., GENEVA Investigators Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuvich R.L., Armstrong L.L., Bielinski S.J., Bradford Y., Carlson C.S., Crawford D.C., Crenshaw A.T., de Andrade M., Doheny K.F., Haines J.L. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet. Epidemiol. 2011;35:887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner S., Armstrong L.L., Bradford Y., Carlson C.S., Crawford D.C., Crenshaw A.T., de Andrade M., Doheny K.F., Haines J.L., Hayes G. Chapter 1. Curr. Protoc. Hum. Genet; 2011. pp. 1–18. (Quality control procedures for genome-wide association studies). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gertz J., Siggia E.D., Cohen B.A. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiore C., Cohen B.A. Interactions between pluripotency factors specify cis-regulation in embryonic stem cells. Genome Res. 2016;26:778–786. doi: 10.1101/gr.200733.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwasnieski J.C., Mogno I., Myers C.A., Corbo J.C., Cohen B.A. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl. Acad. Sci. USA. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 43.Grundberg E., Kwan T., Ge B., Lam K.C.L., Koka V., Kindmark A., Mallmin H., Dias J., Verlaan D.J., Ouimet M. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19:1942–1952. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file provides all 5,439 interactions identified in the discovery analysis. When these interactions appeared to represent the same signal, due to LD, they were placed into groups (n = 1,119) and a representative interaction was chosen. We provide the group identifier for each of the interactions, and the group’s representative interaction.
We examined whether or not the 1,119 interactions could be explained by confounding factors. Here, we present which alternative explanations could account for each interaction.