Abstract
Genome-wide association studies (GWASs) have revealed increased breast cancer risk associated with multiple genetic variants at 5p12. Here, we report the fine mapping of this locus using data from 104,660 subjects from 50 case-control studies in the Breast Cancer Association Consortium (BCAC). With data for 3,365 genotyped and imputed SNPs across a 1 Mb region (positions 44,394,495–45,364,167; NCBI build 37), we found evidence for at least three independent signals: the strongest signal, consisting of a single SNP rs10941679, was associated with risk of estrogen-receptor-positive (ER+) breast cancer (per-g allele OR ER+ = 1.15; 95% CI 1.13–1.18; p = 8.35 × 10−30). After adjustment for rs10941679, we detected signal 2, consisting of 38 SNPs more strongly associated with ER-negative (ER−) breast cancer (lead SNP rs6864776: per-a allele OR ER− = 1.10; 95% CI 1.05–1.14; p conditional = 1.44 × 10−12), and a single signal 3 SNP (rs200229088: per-t allele OR ER+ = 1.12; 95% CI 1.09–1.15; p conditional = 1.12 × 10−05). Expression quantitative trait locus analysis in normal breast tissues and breast tumors showed that the g (risk) allele of rs10941679 was associated with increased expression of FGF10 and MRPS30. Functional assays demonstrated that SNP rs10941679 maps to an enhancer element that physically interacts with the FGF10 and MRPS30 promoter regions in breast cancer cell lines. FGF10 is an oncogene that binds to FGFR2 and is overexpressed in ∼10% of human breast cancers, whereas MRPS30 plays a key role in apoptosis. These data suggest that the strongest signal of association at 5p12 is mediated through coordinated activation of FGF10 and MRPS30, two candidate genes for breast cancer pathogenesis.
Main Text
Strong evidence for the existence of a breast cancer (MIM: 114480) susceptibility locus at 5p12 has been observed through a GWAS in Iceland (SNP rs7703618),1 in the Breast Cancer Association Consortium (BCAC; SNP rs981782, 371 Kb centromeric),2 and in the Cancer GEnetic Markers of Susceptibility study (CGEMS; SNP rs4866929; 352 Kb centromeric; r2 = 0.18).3 A subsequent study, using 22 SNPs in ∼5,000 case subjects and ∼33,000 control subjects of European ancestry, reported that risk at this locus could be explained by two SNPs: rs4415084 and rs10941679.4 More recently, a BCAC study confirmed that rs10941679 was associated with risk of lower-grade, progesterone receptor (PGR [MIM: 607311])-positive breast cancer tumors.5
Here, we report the comprehensive fine-scale mapping of this locus in 104,660 subjects from 50 case-control studies participating in BCAC, including 41 studies from populations of European ancestry and nine of East Asian ancestry, and we explore the functional mechanisms underlying the associations in this region. Genotyping was conducted with the COGS array, a custom array comprising approximately 200,000 SNPs.6 After quality-control exclusions, we analyzed data from 48,155 case subjects and 43,612 control subjects of European ancestry and 6,269 case subjects and 6,624 control subjects of Asian ancestry. Estrogen receptor (ESR1 [MIM: 133430]) status of the primary tumor was available for 27,748 European and 4,997 Asian case subjects; of these, 7,646 (22%) European and 1,623 (32%) Asian case subjects were ER−.
We examined a 1 Mb region (positions 44,394,495–45,364,167; NCBI build 37 assembly) in which the 1000 Genomes Project cataloged 1,811 variants (March 2010 Pilot version 60 CEU project data). We aimed to genotype all 628 SNPs with minor allele frequency (MAF) > 2% and correlated with rs981782 and rs10941679 at r2 > 0.1 (n = 424), plus a set of SNPs designed to tag all remaining SNPs with r2 > 0.9 (n = 184), but we managed to include 563 SNPs with a designability score (DS) > 0.9 and which passed QC.6 IMPUTE v.2.0 was used to impute genotypes of all known SNPs in the region using the 1000 Genome Project data (March 2012 version) as a reference panel.
Case-control analyses were conducted on 3,365 SNPs (563 genotyped and 2,776 imputed at r2 > 0.3). In European-ancestry women, 461 of these SNPs were associated with overall breast cancer risk, 489 with ER+ and 38 with ER− breast cancer risk (p < 10−4; Table S1). SNP rs10941679 showed the strongest overall association (MAF = 0.27, per-minor (g) allele: OR = 1.12; 95% CI 1.10–1.14; p = 2.55 × 10−26; Figure 1, Tables 1 and S1). To identify additional association signals at this region, we conducted a forward stepwise logistic regression examining SNPs with univariate p < 0.1 (n = 1,040).6 The most parsimonious model included three variants: SNP1 rs10941679 (signal 1), SNP2 rs6864776 (signal 2; conditional p = 6.22 × 10−11), and SNP3 rs200229088 (signal 3; conditional p = 1.12 × 10−5, borderline significance; Table S2). SNP1 and SNP3 are weakly correlated (r2 = 0.15) but SNP2 was uncorrelated with the other two (r2 = 0.07 and 0.05).
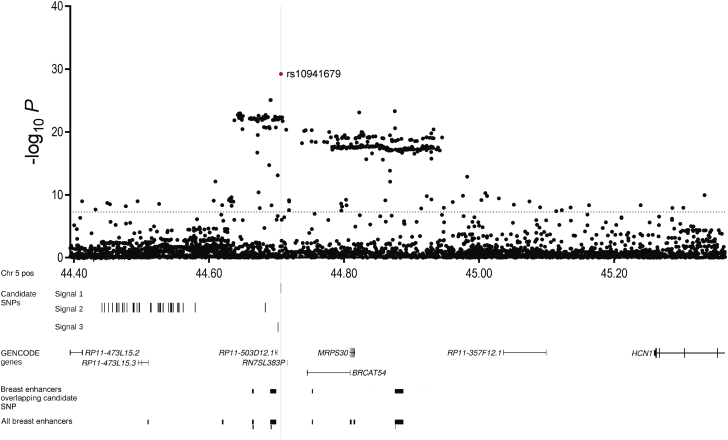
Figure 1.
Manhattan Plot of the 5p12 Breast Cancer Susceptibility Locus
SNPs are plotted according to their chromosomal position on the x axis and their overall p values (log10 values, likelihood ratio test) from the European BCAC studies (48,155 case and 43,612 control subjects) on the y axis. The purple dotted line intersects the y axis at p = 10−8 and indicates genome-wide significance. Candidate SNPs in signal 1 (rs10941679), signal 2 (38 SNPs), and signal 3 (rs200229088) are shown as short vertical lines. The locations of annotated genes and putative lncRNA transcripts from GENCODE and enhancers predicted in Corradin et al.13 and Hnisz et al.12 from breast cancer cell lines are shown in the bottom panels.
Table 1.
Associations of the Top SNPs from Each Signal with Overall Breast Cancer Risk and Breast Cancer Stratified by ER Status
| Sig | SNP | Com | Min | MAF∗ | OR Overall 95% CI | p Overall | Conditional p Value | OR ER− | p ER− | OR ER+ | p ER+ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Europeans | |||||||||||
| 1 | rs10941679 | A | G | 0.27 | 1.12 (1.10–1.14) | 2.55 × 10−26 | 6.55 × 10−24 | 1.04 (1–1.08) | 0.059 | 1.15 (1.13–1.18) | 8.35 × 10−30 |
| 2 | rs6864776 | G | A | 0.23 | 1.04 (1.02–1.06) | 7.84 × 10−4 | 1.44 × 10−12 | 1.10 (1.05–1.14) | 2.5 × 10−5 | 1.02 (0.99–1.05) | 0.08 |
| 3 | rs200229088 | TTG | T | 0.31 | 1.09 (1.07–1.12) | 2.28 × 10−12 | 1.12 × 10−5 | 1.03 (0.99–1.09) | 0.11 | 1.12 (1.09–1.15) | 7.51 × 10−14 |
| Asians | |||||||||||
| 1 | rs10941679 | A | G | 0.50 | 1.09 (1.04–1.15) | 9.12 × 10−4 | 0.0859 | 1.03 (0.95–1.11) | 0.53 | 1.11 (1.04–1.18) | 1.32 × 10−3 |
| 2 | rs6864776 | G | A | 0.32 | 0.94 (0.89–1.00) | 3.47 × 10−2 | 0.8901 | 0.95 (0.87–1.04) | 0.28 | 0.94 (0.89–1.00) | 6.24 × 10−2 |
| 3 | rs200229088 | TTG | T | 0.37 | 1.09 (1.02–1.15) | 6.52 × 10−3 | 0.9149 | 1.04 (0.95–1.14) | 0.43 | 1.08 (1.00–1.16) | 3.65 × 10−2 |
Abbreviations are as follows: Com, common alleles; Min, minor alleles; MAF, minor allele frequency; OR, per-allele odds ratios (OR); 95% CI, 95% confidence intervals and 1 degree of freedom; p, significance levels for overall breast cancer are indicated in European and Asian case-control studies, and separately for ER+ and ER− disease.
The top signal, SNP1 rs10941679, is markedly more significant than any other SNP in the locus (likelihood ratio > 10,000:1). Hence, the most parsimonious explanation is that this SNP is causally related to risk. The next most strongly associated SNP, after adjustment for signal 1 SNP rs10941679, was rs6864776, representing signal 2 (OR per minor allele = 1.04; 95% CI 1.02–1.06; p = 7.84 × 10−4; conditional p = 1.44 × 10−12). Within signal 2, a further 37 SNPs correlated with rs6864776 at r2 > 0.6, had likelihood ratios of <100:1 relative to rs6864776, and hence could not be excluded from being causative statistically (Table S2). After adjustment for both signal 1 SNP rs10941679 and signal 2 top SNP rs6864776, a single SNP remained: rs200229088 (OR overall = 1.09, 95%; CI 1.07–1.12; p = 2.28 × 10−12; conditional p = 1.12 × 10−5). There are no other SNPs correlated with rs200229088 that could explain this association. All other SNPs were excluded from causality (likelihood ratio > 10,000:1; Table S2). Two of the excluded variants had been previously postulated as likely causative variants4, 7 and so we investigated these in more depth. We found both SNPs to be partially correlated with all three signals and consequently display initially inflated effects, which are adjusted by the conditional analyses. Thus, SNP rs44150844 (r2 with signal 1 SNP rs10941679 = 0.51, with signal 2 SNP rs6864776 = 0.11, and with signal 3 SNP rs200229088 = 0.37) has odds against causality > 10 million:1 versus signal 1 candidate rs10941679. Similarly, SNP rs7716600, which is an eQTL for MRPS30 expression7 (r2 with SNP rs10941679 = 0.77, with SNP rs6864776 = 0.05, and with SNP rs200229088 = 0.12) has odds against causality >160,000:1 versus signal 1 candidate rs10941679. These exclusions of former causal candidates highlight the need for fine-mapping studies before conducting functional analyses.
Haplotype analyses were conducted using the above three signal-representative variants, which generated eight haplotypes (Table 2). Haplotypes carrying the rare allele of signal 3 SNP rs200229088 conferred higher risks than corresponding haplotypes carrying the common allele, consistent with this allele having an independent effect. Haplotype G, carrying the minor alleles of both the signal 1 and 2 representative SNPs, is very rare and reveals that their risk alleles are negatively correlated, which is also consistent with our finding that signal 2 top SNP rs6864776 increases in significance after conditioning on signal 1 SNP rs10941679 (Table 1).
Table 2.
Haplotype Analysis across the BCAC Studies
| Haplotypes | rs10941679 Signal 1 | rs6864776 Signal 2 | rs200229088 Signal 3 | Haplotype Frequency | OR | p Value |
|---|---|---|---|---|---|---|
| A | 1 | 1 | 1 | 0.395440 | – | – |
| B | 1 | 1 | 2 | 0.120099 | 1.06 (1.02–1.10) | 1.49 × 10−3 |
| C | 1 | 2 | 1 | 0.199599 | 1.10 (1.06–1.13) | 7.76 × 10−11 |
| D | 1 | 2 | 2 | 0.018665 | 1.15 (1.04–1.27) | 5.03 × 10−3 |
| E | 2 | 1 | 1 | 0.098169 | 1.14 (1.09–1.19) | 1.45 × 10−11 |
| F | 2 | 1 | 2 | 0.154525 | 1.20 (1.16–1.24) | 2.72 × 10−30 |
| G | 2 | 2 | 1 | 0.004248 | 0.91 (0.72–1.15) | 4.15 × 10−1 |
| H | 2 | 2 | 2 | 0.009253 | 1.28 (1.10–1.48) | 1.14 × 10−3 |
Each haplotype was compared to the ancestral haplotype carrying the common alleles of signal 1 SNP rs10941679, signal 2 SNP rs6864776, and signal 3 SNP rs200229088 (haplotype A).
We examined the associations of these three SNPs in the Asian case-control studies within BCAC. SNP1 and SNP3 both replicated in the Asian studies and the relative risk estimates with overall breast cancer were consistent with those seen in the European population: per g-allele OR (rs10941679) = 1.09; 95% CI 1.04–1.15; p = 0.0009, conditional p = 0.0859 and per t-allele OR (rs200229088) = 1.09; 95% CI 1.02–1.15; p = 0.0065, conditional p = 0.9149 (Table 1). SNP2 was not replicated in Asians (per a-allele OR = 0.94; 95% CI 0.89–1.00; p = 0.034, conditional p = 0.8901) (Table 1).
We investigated the associations of these three signals with tumor subtypes based on ER status. SNP1 rs10941679 was largely associated with ER+ breast cancer (OR ER+ = 1.15; 95% CI 1.13–1.18; p = 8.35 × 10−30 versus OR ER− disease = 1.04; 95% CI 1.00–1.08; p = 0.059; p heterogeneity = 1.5 × 10−5; Table 1) as was SNP3 rs200229088 (OR ER+ = 1.12; 95% CI 1.09–1.15; p = 7.51 × 10−14 versus OR ER− = 1.03; 95% CI 0.99–1.09; p = 0.11, p heterogeneity = 0.02). By contrast, SNP2 rs6864776 was moderately associated with ER− but not ER+ tumors (OR ER− = 1.10; 95% CI 1.05–1.14; p = 2.55 × 10−5 versus OR ER+ = 1.02; 95% CI 0.99–1.05; p = 0.08; p heterogeneity = 0.01; Table 1).
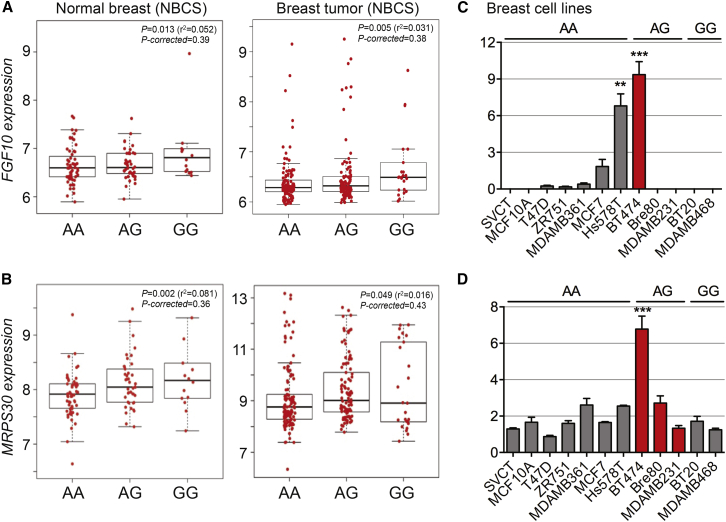
Candidate SNPs 1–3 span a 1.7 Mb region on 5p12 that includes three annotated genes—FGF10 (MIM: 602115), MRPS30 (MIM: 611991), and HCN1 (MIM: 602780)—and several putative long noncoding RNAs (lncRNAs; Figure 1). To identify potential target gene(s), we examined the associations of the three lead SNPs with expression levels of genes located within 1 Mb in three different studies: (1) 116 normal breast samples and 241 breast tumors from the Norwegian Breast Cancer Study (NBCS),8 (2) 93 normal and 765 breast cancer tissues from the TCGA study (germline genotype data from Affymetrix SNP 6 array were obtained from TCGA dbGAP data portal9), and (3) 183 normal breast samples from the Genotype-Tissue Expression (GTEx) project.10 The SNP1 rs10941679 risk-associated g-allele was moderately associated with increased FGF10 mRNA expression in NBCS normal breast (p = 0.013, p corrected = 0.39) and breast tumors (p = 0.005, p corrected = 0.38) as well as in GTEx normal breast (p corrected = 0.02; Figures 2A and S1A). The effect in TCGA was in the same direction, though not significant (normal breast p = 0.353, p corrected = 0.95 and breast tumors p = 0.057, p corrected = 0.41; Figure S1B). The g-allele was also associated with increased expression of MRPS30 in the NBCS normal (p = 0.002, p corrected = 0.36) and breast tumors (p = 0.049, p corrected = 0.43), in GTEx normal breast (p corrected = 0.002), and in TCGA (normal breast p = 6.86 × 10−5, p corrected = 5.31 × 10−3 and breast tumors p = 7.21 × 10−6, p corrected = 9.35 × 10−4; Figures 2B, S1A, and S1C). No associations were observed with SNP2 rs6864776 or SNP3 variant rs200229088. We also measured endogenous levels of FGF10, MRPS30, and nearby lncRNAs FGF10-AS1, BRCAT54, RP11-503D12.1, and RP11-473L15.3 mRNA in breast cell lines homozygous (A/A or G/G) or heterozygous (A/G) for the common allele of SNP1 (Table S3, Figures 2C, 2D, S2, and S3). Total RNA from cell lines was extracted using Trizol and complementary DNA synthesized using random primers as per manufacturers’ instructions. Quantitative PCR (qPCR) were performed using TaqMan assays for FGF10 and MRPS30 normalized against beta-glucuronidase (GUSB [MIM: 611499]) or with SYTO9 for lncRNAs normalized against TATA box-binding protein (TBP [MIM: 600075]; primers are listed in Table S4). Although the number of ER+ breast cell lines carrying the risk allele was limited, FGF10 and MRPS30 mRNA levels were significantly higher in the BT474 heterozygous cell line (Figures 2C and 2D). BRCAT54 was detected in the majority of cell lines but its expression appears to be genotype independent (Figure S3A). FGF10-AS1, RP11-503D12.1, and RP11-473L15.3 transcripts were either expressed at very low levels or not detected in the cell lines analyzed (Figures S3B–S3D). Therefore, although we cannot rule out the possibility that the risk SNPs may influence local lncRNA expression, the low or absent transcript levels precluded any further evaluation.
Figure 2.
Association of rs10941679 with FGF10 and MRPS30 Expression in Normal Breast Tissues, Breast Tumors, and Breast Cancer Cell Lines
(A and B) FGF10 (A) or MRPS30 (B) expression in normal breast (n = 116) or breast tumors from NBCS dataset (n = 241). SNP genotypes are shown on the x axis and log2-normalized gene expression values on the y axis. p values are presented before and after correction for multiple testing using FDR as implemented in p.adjust function in R. Each box plot shows the median rank normalized gene expression (horizontal line), the first through third quartiles (box), and 1.5× the interquartile range (whiskers).
(C and D) Endogenous FGF10 (Hs00610298_m1) (C) or MRPS30 (Hs00169612_m1) (D) expression measured by qPCR in untreated breast cell lines and normalized to GUSB (4326320E). Error bars denote SEM (n = 3). p values were determined with a two-tailed t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
Candidate causal SNPs were then explored using publicly available datasets from ENCODE,11 which includes information such as the location of promoter and enhancer histone marks, open chromatin, bound proteins, and altered motifs for the MCF7 breast cancer cell line, and from Hnisz et al.12 and Corradin et al.13 to identify the location of likely enhancers and their gene targets in a cell-specific context. Analysis of cis enhancer-gene interactions via PreSTIGE13 showed evidence of putative regulatory elements (PREs) surrounding the top risk-associated SNPs in MCF7 breast cancer cells, but no histone-marked elements harboring a risk SNP in this cell line or in a range of cell lines and tissues analyzed in Roadmap (Figures 1 and S4). However, it is possible that certain epigenetic marks may be detected only in a specific cell subtype such as breast stem cells or in response to an external stimulus.
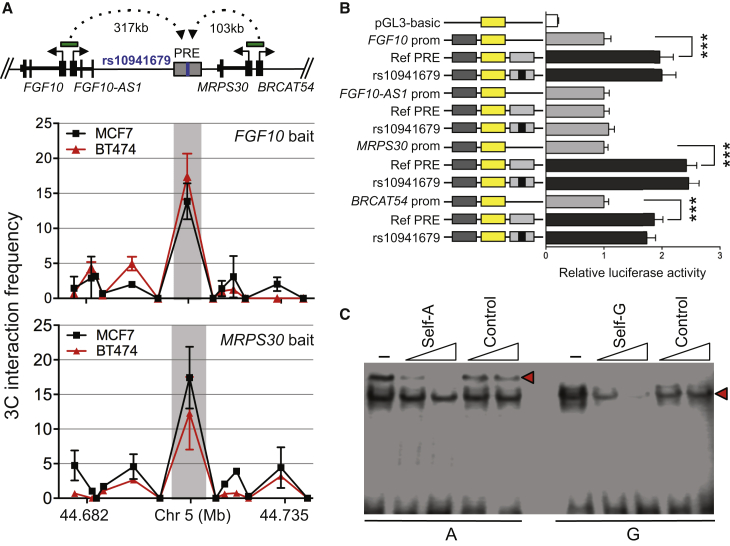
To identify target gene(s), we performed chromatin conformation capture (3C) assays in ER+ MCF7, BT474, and MDA-MB-361 and ER− MDA-MB-231 breast cancer cell lines and Bre80 normal breast cells (Table S5).8 3C libraries were created by cross-linking the chromatin from cell lines; DNA was then digested with EcoRI, which flanks 12 contiguous fragments that cover the PRE, and the FGF10, MRPS30, and HCN1 promoters (Table S6); DNA was religated and decrosslinked; and qPCR with primers for the bait (gene promoters) and interactors (12 PRE fragments) was performed to detect the presence of ligation products, representing gene loops. BAC clones covering the regions of interest were used to normalize for PCR efficiency. These assays showed that the PRE containing SNP1 frequently interacted with the FGF10 and MRPS30 promoter regions in MCF7 and BT474 breast cancer cell lines, but only with MRPS30 in the MDA-MB-361, MDA-MB-231, and Bre80 cell lines. This latter result was expected because FGF10 is not expressed or expressed at very low levels in these cell lines (Figures 2C, 3A, S5, and S6). Notably, both genes share a bidirectional promoter with the lncRNAs FGF10-AS1 and BRCAT54, raising the possibility that these transcripts are also targets of the PRE (Figure 3A). No additional interactions were detected between the PRE and other annotated genes within 1 Mb of the PRE, including HCN1 (Figure S5). To assess the potential impact of SNP1 on the identified chromatin interactions, allele-specific 3C was performed in heterozygous BT474 cell lines.8 However, the sequence profiles revealed that SNP1 had no significant effect on chromatin looping (Figure S7).
Figure 3.
Distal Regulation of FGF10 and MRPS30 at the 5p12 Risk Region
(A) 3C interaction profiles between the FGF10/FGF10AS-1 or MRPS30/BRCAT54 bidirectional promoters and the putative regulatory element (PRE; gray bar) containing SNP rs10941679. Anchor points are set at the promoters. Graphs represent one of three independent experiments (see Figure S5B). Error bars denote SD.
(B) Luciferase reporter assays after transient transfection of ER+ BT474 breast cancer cell lines. The PRE containing the major SNP allele was cloned downstream of target gene promoter-driven luciferase constructs (Ref PRE). The risk g-allele was engineered into the constructs and designated by the rs ID. Primers are listed in Table S7. Error bars denote 95% confidence intervals from three independent experiments. p values were determined by 2-way ANOVA followed by Dunnett’s multiple comparisons test (∗∗∗p < 0.001).
(C) EMSA for oligonucleotides containing SNP rs1094617 with the A = common allele and G = minor allele as indicated below the panel, assayed using BT474 nuclear extracts. Primers are listed in Table S8. Labels above each lane indicate inclusion of competitor oligonucleotides at 30- and 100-fold molar excess, respectively: (-) no competitor and control denotes a non-specific competitor. A red arrowhead shows a band of different mobility detected between the common and minor alleles.
The regulatory capability of the PRE, combined with the effect of SNP1, was further examined in reporter assays. Promoter-driven luciferase reporter constructs were generated by the insertion of PCR-amplified fragments containing FGF10, FGF10-AS1, MRPS30, or BRCAT54 promoters into pGL3-Basic.14 A 1,736-bp PRE fragment (containing either the common or minor allele of rs10941679) was then generated by PCR and cloned downstream of the modified pGL3-promoter constructs (Table S7). MCF7 and BT474 breast cancer cell lines plus Bre80 normal breast cells were transfected with the reporter plasmids and luciferase activity was measured 24 hr after transfection. To correct for any differences in transfection efficiency or cell lysate preparation, Firefly luciferase activity was normalized to Renilla. Notably, the “Ref PRE” acted as a transcriptional enhancer, leading to a 2- to 3-fold increase in FGF10, MRPS30, and BRCAT54 promoter activity, but had no effect on the FGF10-AS1 promoter in MCF7 and BT474 cells (Figures 3B and S8). The enhancer activity was also observed for the MRPS30 and BRCAT54 promoters in Bre80 cells (Figure S8). In all cell lines, inclusion of the SNP1 risk (g) allele had no significant effect on the PRE enhancer activity. Although this appears to rule out an effect of this SNP on transactivation, it is possible that SNP1 affects the recruitment of key proteins required for the epigenetic modification of the enhancer, which would not be observed in a reporter assay. Another possibility is that the SNP effect may be observed only under certain biological conditions such as growth factor stimulation.
To seek further evidence that SNP1 lies within an enhancer element, we performed electrophoretic mobility shift assays (EMSAs) for both the protective (a) and risk (g) alleles.15 Nuclear lysates were prepared from ER+ BT474, MCF7, and MDA-MB-361 or ER− MDA-MB-231 and Hs578T cells using the NE-PER nuclear and cytoplasmic extraction reagents. Biotinylated oligonucleotide duplexes were prepared by combining sense and antisense oligonucleotides, heat annealing, and slow cooling. Duplex-bound complexes were transferred onto Zeta-Probe positively charged nylon membranes by semi-dry transfer then cross-linked onto the membranes. Membranes were processed with the LightShift Chemiluminescent EMSA kit as per the manufacturer’s instructions, and signals were visualized with the C-DiGit blot scanner. For SNP1, we observed allele-specific binding by nuclear proteins only in the ER+ BT474, MCF7, and MDA-MB-361 extracts (Figures 3C and S9). The protein-DNA complexes were shown to be specific, as demonstrated by increasing amounts of cold self-competitor (Figures 3C and S9 and Table S8).
Further EMSAs using competitor DNA or antibody supershifts against predicted transcription factors (TFs) suggested four proteins bound to the SNP site including FOXA1, FOXA2, CEBPB, and OCT1 (Figure S10 and Table S9). To confirm TF binding in vivo, we performed chromatin immunoprecipitation (ChIP) in heterozygous BT474 cells as previously described (Table S10).15 When compared to an IgG control antibody, we observed a moderate enrichment in FOXA1 and OCT1 binding to DNA overlapping SNP rs10941679, but no difference between alleles in this cell line (Figure S11). In addition, western blot analysis indicated that FOXA1 protein expression was restricted to the ER+ breast cancer cell lines analyzed, whereas OCT1 was more widely expressed (Figure S12). FOXA1 is a pioneer factor and master regulator of ER activity due to its ability to open local chromatin and recruit ER to target gene promoters.16 Notably, breast cancer-associated SNPs are enriched for FOXA1 binding17 and several studies have linked cooperative binding of FOXA1, ER, and OCT1 to increased gene transcription.18, 19 Consistent with our eQTL data, it is tempting to speculate that in specific ER+ cell subtypes and/or conditions, rs10941679 alters FOXA1 affinity and OCT1 recruitment leading to target gene activation.
In conclusion, we have provided evidence for at least three independent causal SNPs with effects on the risk of breast cancer at this locus. The minor g-allele of signal 1 SNP rs10941679 conferred a 15% increased risk of ER+ breast cancer and higher expression levels of the MRPS30 and FGF10 genes and was the most strongly associated SNP with MRPS30 expression in this 1 Mb region. MRPS30—also called PDCD9 (Programmed Cell Death protein 9)—encodes a mitochondrial ribosomal protein involved in apoptosis.20 Although the role of mitochondria in apoptosis remains unclear, it is well established that cytochrome c and other pro-apoptotic proteins are released during cell death initiation.20 Clearly, further investigation of the function of this protein is now merited. By contrast, FGF10 is an extensively studied gene with compelling data suggesting its involvement in breast tumorigenesis. FGF10 is a member of the fibroblast growth factor (FGF) family and encodes a glycoprotein that specifically binds to FGFR2 (splice FGFR2IIIb) to control signaling pathways including cell differentiation, proliferation, and apoptosis.21 Variants regulating FGFR2 (MIM: 176943) have the strongest association with ER+ breast cancer susceptibility identified to date.22 FGF10 is overexpressed in ∼10% of human breast cancers23 and increased levels of FGF10 are highly correlated with proliferation rate of breast cancer cell lines and cancer cell invasion.24, 25 It signals through multiple downstream pathways including MAPK and WNT and genes such as FGFR2, CCND1 (MIM: 168461), and TGFB1 (MIM: 190180),21, 24 all known to play key roles in breast cancer. Therapeutic targeting of FGFs and their receptors (FGFRs) is currently a major area of drug development research, and the identification of a subgroup of individuals diagnosed with breast cancer with alterations in these pathways may open new avenues for personalized medicine and pathway-targeted treatments.
Published: September 15, 2016
Footnotes
Supplemental Data include Supplemental Acknowledgments, 12 figures, 10 tables, and consortia information and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.017.
Contributor Information
Alison M. Dunning, Email: amd24@medschl.cam.ac.uk.
Stacey L. Edwards, Email: stacey.edwards@qimrberghofer.edu.au.
Web Resources
1000 Genomes, http://www.1000genomes.org
Cancer Cell Line Encyclopedia (CCLE), https://portals.broadinstitute.org/ccle/home
ENCODE, https://www.encodeproject.org/
GTEx Portal, http://www.gtexportal.org/home/
OMIM, http://www.omim.org/
PreSTIGE, http://genetics.case.edu/prestige/
The Cancer Genome Atlas, http://cancergenome.nih.gov/
Supplemental Data
References
- 1.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 2.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., SEARCH collaborators. kConFab. AOCS Management Group Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 5.Milne R.L., Goode E.L., García-Closas M., Couch F.J., Severi G., Hein R., Fredericksen Z., Malats N., Zamora M.P., Arias Pérez J.I., GENICA Network. kConFab Investigators. AOCS Group Confirmation of 5p12 as a susceptibility locus for progesterone-receptor-positive, lower grade breast cancer. Cancer Epidemiol. Biomarkers Prev. 2011;20:2222–2231. doi: 10.1158/1055-9965.EPI-11-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., Breast and Ovarian Cancer Susceptibility Collaboration. Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) kConFab Investigators. Australian Ovarian Cancer Study Group. GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361, e1–e2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quigley D.A., Fiorito E., Nord S., Van Loo P., Alnæs G.G., Fleischer T., Tost J., Moen Vollan H.K., Tramm T., Overgaard J. The 5p12 breast cancer susceptibility locus affects MRPS30 expression in estrogen-receptor positive tumors. Mol. Oncol. 2014;8:273–284. doi: 10.1016/j.molonc.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghoussaini M., Edwards S.L., Michailidou K., Nord S., Cowper-Sal Lari R., Desai K., Kar S., Hillman K.M., Kaufmann S., Glubb D.M., Australian Ovarian Cancer Management Group. Australian Ovarian Cancer Management Group Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q., Seo J.H., Stranger B., McKenna A., Pe’er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sal lari R., Lupien M., Markowitz S., Scacheri P.C. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glubb D.M., Maranian M.J., Michailidou K., Pooley K.A., Meyer K.B., Kar S., Carlebur S., O’Reilly M., Betts J.A., Hillman K.M., GENICA Network. kConFab Investigators. Norwegian Breast Cancer Study Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am. J. Hum. Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunning A.M., Michailidou K., Kuchenbaecker K.B., Thompson D., French J.D., Beesley J., Healey C.S., Kar S., Pooley K.A., Lopez-Knowles E., EMBRACE. GEMO Study Collaborators. HEBON. kConFab Investigators Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowper-Sal lari R., Zhang X., Wright J.B., Bailey S.D., Cole M.D., Eeckhoute J., Moore J.H., Lupien M. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer K.B., Maia A.T., O’Reilly M., Teschendorff A.E., Chin S.F., Caldas C., Ponder B.A. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belikov S., Astrand C., Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol. Cell. Biol. 2009;29:5413–5425. doi: 10.1128/MCB.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavdar Koc E., Ranasinghe A., Burkhart W., Blackburn K., Koc H., Moseley A., Spremulli L.L. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 21.Turner N., Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 22.Meyer K.B., O’Reilly M., Michailidou K., Carlebur S., Edwards S.L., French J.D., Prathalingham R., Dennis J., Bolla M.K., Wang Q., GENICA Network. kConFab Investigators. Australian Ovarian Cancer Study Group Fine-scale mapping of the FGFR2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1. Am. J. Hum. Genet. 2013;93:1046–1060. doi: 10.1016/j.ajhg.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theodorou V., Boer M., Weigelt B., Jonkers J., van der Valk M., Hilkens J. Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene. 2004;23:6047–6055. doi: 10.1038/sj.onc.1207816. [DOI] [PubMed] [Google Scholar]
- 24.Abolhassani A., Riazi G.H., Azizi E., Amanpour S., Muhammadnejad S., Haddadi M., Zekri A., Shirkoohi R. FGF10: type III epithelial mesenchymal transition and invasion in breast cancer cell lines. J. Cancer. 2014;5:537–547. doi: 10.7150/jca.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chioni A.M., Grose R. Negative regulation of fibroblast growth factor 10 (FGF-10) by polyoma enhancer activator 3 (PEA3) Eur. J. Cell Biol. 2009;88:371–384. doi: 10.1016/j.ejcb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.