Abstract
Stat5a is a transcription factor utilized by several cytokine/hormone receptor signaling pathways that promotes transcription of genes associated with proliferation, differentiation, and survival of cancer cells. However, there are currently no clinically approved therapies that directly target Stat5a, despite ample evidence that it contributes to breast cancer pathogenesis. Here, deacetylation of the Stat5a coactivator and chromatin remodeling protein HMGN2 on lysine residue K2 by HDAC6 promotes Stat5a-mediated transcription and breast cancer growth. HDAC6 inhibition both in vitro and in vivo enhances HMGN2 acetylation with a concomitant reduction in Stat5a-mediated signaling, resulting in an inhibition of breast cancer growth. Furthermore, HMGN2 is highly acetylated at K2 in normal human breast tissue, but is deacetylated in primary breast tumors and lymph node metastases, suggesting that targeting HMGN2 deacetylation is a viable treatment for breast cancer. Together, these results reveal a novel mechanism by which HDAC6 activity promotes the transcription of Stat5a target genes and demonstrate utility of HDAC6 inhibition for breast cancer therapy.
Implications
HMGN2 deacetylation enhances Stat5a transcriptional activity, thereby regulating prolactin-induced gene transcription and breast cancer growth.
Keywords: Prolactin, Stat5a, HMGN2, HDAC6, breast cancer
Introduction
Signal transducer and activator of transcription 5a (Stat5a) is a transcription factor that promotes proliferation, differentiation, and survival of both the normal mammary gland and breast cancer cells. The significance of Stat5a in breast cancer pathogenesis is underscored in several genetic models that have identified Stat5a as an important factor for cancer initiation and progression. In the WAP-TAg mouse model of mammary cancer, hemizygous loss of Stat5a results in decreased tumor number, size, and delayed first tumor onset (1), while in conditional ERα in mammary epithelium (CERM) mice, loss of Stat5a significantly reduces ERα-induced hyperplastic alveolar nodules in both untreated and DMBA treated mice (2). Furthermore, in models where Stat5a is forcibly expressed, transgenic mice were more prone to mammary gland tumorigenesis (3, 4). Stat5a has also been shown to enhance proliferation of breast cancer cells by directly upregulating CISH and CyclinD1 transcription (5–7). Taken together, these data suggest that Stat5a is an important factor in the pathobiology of mammary cancer, and a deeper understanding of how Stat5a activity is regulated in breast cancer is critical for identifying therapeutic targets within this pathway.
Recent data from our laboratory indicate that the chromatin remodeling protein High Mobility Group Nucleosomal Binding Domain 2 (HMGN2) directly contributes to the activation of Stat5a target genes (8). Nuclear translocation of the prolactin receptor (PRLr) occurs after ligand-induced phosphorylation, where it functions as a coactivator through its interactions with Stat5a and HMGN2. The association of HMGN2 with Stat5a-responsive promoters via nuclear PRLr upregulates Stat5a-mediated transcription, presumably by allowing a more open chromatin conformation that enables engagement of transcriptional activators that elevate transcriptional output (8). The activity of HMGN proteins is affected by several post-translational modifications, including phosphorylation and acetylation. HMGN1 has been shown to be acetylated on several residues, including lysine residue 2 (K2), an event which reduces its ability to bind nucleosomes and prevents the unfolding of the higher order chromatin structure (9). Corroborating evidence has demonstrated that HMGN2 is acetylated on residue K2 and inhibits its association with histone cores (10).
While it has been shown that deacetylase activity is required for Stat5a-mediated transcriptional activation, neither the mechanism nor the target protein has been identified (11). Furthermore, it has been hypothesized that a non-histone protein is the target of histone deacetylase (HDAC) activity in Stat5a-mediated signaling, as histone-H3 and -H4 acetylation are not significantly changed at the Stat5a-regulated CISH promoter upon IL-3 stimulation (11). Using PRL-responsive genes as a model of Stat5a transcriptional activity, we sought to determine if HDACs could regulate HMGN2 acetylation levels and binding to Stat5a-responsive promoters, and therefore, Stat5a transcriptional activity.
Materials and Methods
Cell culture, transfection, and Prolactin
Human recombinant PRL was a gift from Dr. Anthony Kossiakoff, University of Chicago. PRL was used at a final concentration of 250ng/mL. Cell lines, culture conditions, and transfection have been previously described (8). The breast cancer cell lines MCF7, T47D, and MDA231 were obtained from ATCC. Re-authentication of these lines by short tandem repeat profiling was performed within the last 6 months by ATCC. The authenticity of the PDX line WHIM2 was confirmed by sequencing (12).
Antibodies
Rabbit polyclonal ac-K2-HMGN2 antibody was made by New England Peptides. The antibody was raised against an acetylated N-terminal HMGN2 peptide (P(KAc)RKAEGDAC), followed by affinity purification. To ensure specificity, a blocking peptide (P(KAc)RKAEGDAC) and a non-acetyl blocking peptide (PKRKAEGDAC) were used to ensure acetyl-specific interaction. Antibodies used for western blotting; Tubulin (Invitrogen, 322500, 1:2000); HDAC6 (Abcam, ab47181, 1:500); HMGN2 (Millipore, 07-252, 1:500; Cell Signaling, #9437, 1:2000) pan-acetyl lysine (Millipore, AB3879, 1:500); CISH (Santa Cruz, sc-15344, 1:500); CyclinD1 (Santa Cruz, sc-718, 1:500); CEBPβ (Santa Cruz, sc-150, 1:500); PARP/Cleaved PARP (Cell Signaling, #9542, 1:500); ac-K2-HMGN2 (1:50); acetyl-α-tubulin (Cell Signaling, #5335, 1:500). Secondary antibody was used at a dilution of 1:1000 for all antibodies except tubulin, which was 1:2000. Antibodies used for IHC: CyclinD1 (Santa Cruz, sc-718, 1:1500); acetyl-tubulin (Cell Signaling, D20G3, 1:100); ac-K2-HMGN2 (1:500).
Mouse xenograft model
All animal procedures were performed with approval from the Northwestern University Animal Care and Use Committee. Female nu/nu mice were obtained from Harlan Sprague-Dawley (Indianapolis, IN) at 4–6 weeks of age. Xengrafts were performed as described in (13). Mice were anesthetized using isofluorane and 1.5 million GFP-expressing MCF7 or 1 million GFP-luciferase MDA-MB-231 cells suspended in matrigel were injected into the 4th lactiferous duct. An estrogen pellet was also subcutaneously implanted in mice injected with MCF7 cells. Tumors were allowed to establish for 3 weeks before Bufexamac treatment. Bufexamac was dissolved in DMSO to a concentration of 300mg/mL and was added to a carrier consisting of PEG 400, propylene glycol, and Tween-80 in a ratio of 20:29:1. 50µL was injected intraperitoneally twice per day for a total daily dosage of 50mg/kg/d and 150mg/kg/d. Tumor size was measured weekly with a digital caliper. At the end of the study, mice were euthanized by CO2 asphyxiation and cervical dislocation. Tumors were formalin fixed and paraffin embedded for histology and immunohistochemistry.
Plasmids
HDAC6 pCMV-SPORT6 was purchased from Open Biosystems. HMGN2 pCMV was purchased from Origene. HMGN2 eGFP mutants were cloned into pEGFP-N1 (Clonetech) using standard techniques. All mutants were made using Quickchange Lightning mutagenesis (Stratagene) according to the manufacturer’s instructions along with the parental constructs listed above and the primers listed in Table S1.
siRNA
siHDAC6 (target sequence: 5’-GCAGTTAAATGAATTCCAT-3’) was used at a concentration of 40nM. Cells were transfected using RNAi max (Invitrogen) according to the manufacturer’s instructions. Non-targeting siRNA was used as a control. Experiments were performed after 48h of knockdown.
Luciferase assay
Luciferase assays have been previously described (8). The ratio of luciferase/renilla or luciferase/protein concentration was calculated and the results are reported as fold change compared to untreated empty vector control.
RNA extraction, cDNA synthesis, and RT-PCR
After indicated treatments, cells were washed with PBS and mRNA was isolated with an RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized using 1ug of total mRNA with qScript cDNA SuperMix (Quanta) according to the manufacturer’s specifications. qPCR was performed using Power SYBR-Green PCR Supermix (Applied Biosystems), 20ng cDNA (corresponding to mRNA concentration), and 1nM primers listed in Table S2 using an ABI 7900HT thermocycler (Applied Biosystems). Data were normalized to 18S or GAPDH and fold change was reperesented as 2−ΔΔCt (2−(Cttarget–Ct18SrRNA)PRL–(Cttarget–Ct18SrRNA)control) using untreated, empty vector cells as baseline.
Coimmunoprecipitation and chromatin immunoprecipitation
Coimmunoprecipitation was performed as previously described using TD buffer (8). ChIP was conducted as previously described until DNA isolation (14), and then the fast ChIP method was employed (15).
Proliferation
T47D, MCF7, or 231 cells were plated in 96-well plates and cultured in indicated media for 48–72 hours in the presence of growth factors or inhibitors. Each well was then pulsed with 0.5 µCi of 3H-thymidine (Amersham Pharmacia Biotech) for 4–6 hours. Cells were harvested onto a filtermat membrane with Filtermate Harvester (Perkin Elmer) before analysis with a MicroBeta TriLux Scintillation Counter (Perkin Elmer).
Migration
10,000 MCF7 cells were plated in an 8.0 micron pore size transwell insert (Costar). Serum free media was placed in the upper chamber and DMEM containing 10% FBS was placed in the lower chamber. Cells were treated with DMSO or Bufexamac in both the upper and lower chambers. Cells were allowed to migrate for 24h before cells from the lower chamber and bottom of the membrane were trypsinized and pelleted. Migrated cells were quantified using CyQUANT (Invitrogen) according to the manufacturer’s instructions.
Soft agar
Soft agar assays was carried out as previously described (16), with inhibitors added in each layer at indicated concentrations.
Trypan blue exclusion and cell viability
For the timecourse of cell growth, cells were incubated in indicated concentrations for the indicated timepoints. Media, along with trypsinized cells were spun down at 200g, then resuspended in PBS and trypan blue. Cells were counted on a Countess (Invitrogen) and the percentage of live cells was calculated. Cell viability was measured in the same manner on day 7 of treatment.
Tumor progression array
Breast tissues were obtained from the Tissue Core of the Northwestern University Breast Cancer Program. The tumor progression array consisted of patient-matched normal breast, primary tumor, and lymph node metastasis from 14 patients. For the array in this study, all tumors were ER+/PR+/Her2−. The presence of proper tissue sections were verified by H&E staining before IHC was performed.
Immunohistochemistry
FFPE tissue was cut into 4.0 micron sections baked overnight at 37°C. Slides were then deparaffinized and rehydrated through graded alcohols. Antigen retrieval was performed using 0.1mM EDTA pH 7.5 for ac-K2-HMGN2 antibody and citrate buffer, pH 6.0 for all other antibodies. Slides were cleaned in 1% Triton-X, rinsed, and incubated in 0.025% hydrogen peroxide to quench endogenous peroxidase activity. Slides were then blocked in a humidified chamber in 10% goat serum and 1% milk in PBS for 1h before incubation with appropriate antibodies in 5% goat serum and 1% milk overnight at room temp. The following day, slides were incubated for 1h with 1:1200 secondary antibody in 5% goat serum and 1% milk in PBS and a subsequent incubation in Vector ABC solution for 1h. Slides were then subjected to DAB staining for 10 min and then counterstained using Mayer’s hematoxylin. Bluing was performed in 0.4% ammonia water for 3 min before slides were dehydrated and mounted. Slides were then visually scored by a pathologist on a scale from 0–3, where 0 = no staining and 3 = intense staining.
Immunofluorescence
Immunofluorescence was performed as previously described (8) with cells preincubated with TSA for 2h before PRL treatment where indicated.
Microarray Procedures and Analysis
Cell Culture and RNA Isolation
Differential gene expression was assessed in MCF7 cells treated with DMSO (0.1%) control, prolactin (PRL; 250 ng/mL) and/or Bufexamac (100 µM, 0.1% DMSO). Prior to RNA isolation, MCF7 cells were plated at 60% confluency in 6 cm plates and incubated for 24 hours followed by an additional 24 hours of serum starvation in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY) and 1X ITS Liquid Media Supplement (Sigma Aldrich, St. Louis, MO). Cells were then treated for four hours with either DMSO or Bufexamac before stimulation with PRL. After two hours PRL stimulation, cells were washed with PBS and RNA was extracted using the MagMAX-96 for Microarrays Total RNA Isolation Kit (Invitrogen, Life Technologies, Carlsbad, CA) in an automated fashion using the magnetic particle processors MagMAX Express. RNA purity was judged by spectrophotometry at 260, 270, and 280 nm. RNA integrity was assessed by running 1 µL of every sample in RNA 6000 Nano LabChips on the 2100 Bioanalyzer (Agilent Technologies, Foster City, CA).
Microarray Hybridization and Data Acquisition
Each of the four cell treatment conditions (PRL− / DMSO, PRL+ / DMSO, PRL− / Bufexamac, PRL+ / Bufexamac) were assessed in three independently grown biological replicates. Each of the 12 RNA samples were hybridized in duplicate to two Human Genome U133A 2.0 Arrays (Affymetrix, Santa Barbara, CA) according to the Affymetrix protocol as previously described (17) with the following modifications: Starting with 500 ng of total RNA, we performed a single-strand cDNA synthesis primed with a T7-(dT24) oligonucleotide. Second strand cDNA synthesis was performed with E. coli DNA Polymerase I, and biotinylation of the cRNA was achieved by in vitro transcription (IVT) reaction using the GeneChip 3’ IVT Express Kit (Affymetrix, Santa Clara, CA). After a 37°C-incubation for 16 hours, the labeled cRNA was purified using the cRNA cleanup reagents from the GeneChip Sample Cleanup Module. As per the Affymetrix protocol, 10 µg of fragmented cRNA were hybridized on the GeneChip HG U133A 2.0 Arrays (Affymetrix Inc., Santa Clara, CA) for 16 hours at 60 rpm in a 45 °C hybridization oven. The arrays were washed and stained using the GeneChip Hybridization, Wash, and Stain Kit in the Affymetrix fluidics workstation. Every chip was scanned at a high resolution, on the Affymetrix GeneChip Scanner 3000 7G according to the GeneChip Expression Analysis Technical Manual procedures (Affymetrix, Santa Barbara, CA). After scanning, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip Operating Software v1.4 (GCOS) (Affymetrix, Santa Barbara, CA). Overall quality of each array was assessed by monitoring the 3′/5′ ratios for the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the percentage of “Present” genes (%P). Arrays exhibiting GAPDH and ACTIN 3′/5′ < 3.0 and %P > 40% were considered good quality arrays.
Differential Expression Analysis
The 24 resultant .CEL files were analyzed using the R statistical computing language and environment (18). The data quality of each microarray was assessed by examining the average background, percent of probe sets called present by the MAS5 detection call algorithm (19), and the 3’:5’ ratio for GAPDH and ACTIN. Additionally, to detect potential spatial artifacts resulting from sub-optimal hybridization conditions, probe level linear models were fit using the R Bioconductor package, “affyPLM”, and plots of the residuals were examined for each microarray (20, 21). Differential expression was assessed using the “affy” and “limma” Bioconductor packages (19, 22). Briefly, probesets were quantile normalized and processed by the Robust Multi-Array Average (RMA) algorithm (23) before the manufacturer’s control probesets and probesets considered “absent” in 20 or more arrays by the MAS5 algorithm were filtered from all arrays (24). Differential expression between treatment conditions was then assessed via moderated t-test adjusted for multiple hypotheses by the Benjamini & Hochberg method (25). The false discovery rate (FDR) was controlled so that only those probesets where q < 0.01 were deemed significant.
Hierarchical clustering was performed on the top 100 prolactin induced and top 100 prolactin inhibited genes (as defined by the expression fold change between the PRL− / DMSO and PRL+ / DMSO microarrays) utilizing the GenePattern public server (26–28). Microarrays were clustered using a pairwise average-linkage method and Pearson correlation as the similarity metric; genes were not subjected to clustering and are presented as originally ordered in the .GCT file supplied to the module.
Stat5 target genes were defined as previously reported (29) and include genes flanked by the classic Stat5 palindromic repeat binding motif as well as those identified by Kang et al. utilizing Stat5 ChIP-seq on murine mammary tissues at parturition. After converting Mus musculus gene annotations to human gene symbols, 847 unique genes comprised the Stat5 target gene list used in this study.
Statistical analysis
Statistical analysis was performed using appropriate statistical methods (student’s T-test, one- and two-way ANOVA) on GraphPad Prism 4. The results are shown as the mean with error bars depicting ± SEM. Statatistical significance is annotated with * p<0.05, ** p<0.01, *** p<0.001.
Results
HDAC6 regulates Stat5a transcriptional activity
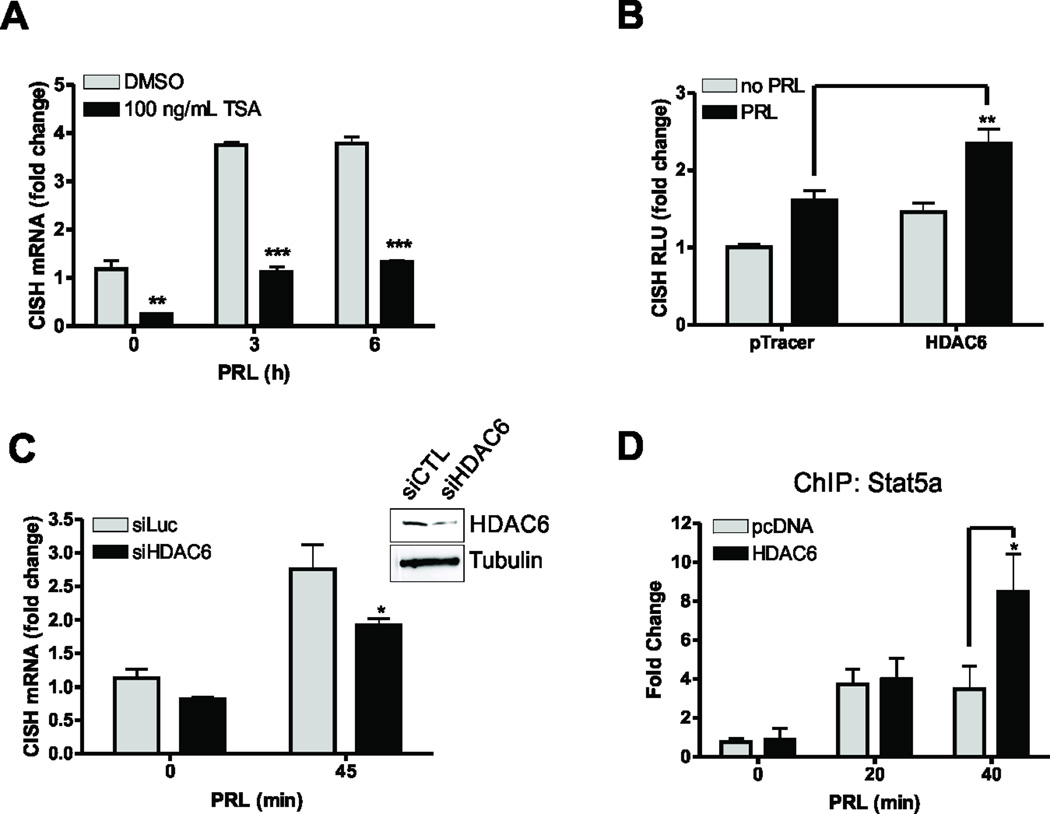
Jak2/Stat5a signaling has been shown to significantly contribute to mammary carcinogenesis (1, 2, 30), in part due to its effects on the pro-proliferative genes CISH and CCND1 (6, 7). Because HDAC activity has been shown to be required for IL-2- and IL-3-induced Stat5a-mediated transcription (11, 31), we reasoned that HDAC activity would be required for PRL-mediated activation of CISH transcription. Indeed, treatment of cells with the general HDAC inhibitor, trichostatin A (TSA), prevented PRL-mediated CISH mRNA transcription (Fig. 1A). HDAC6 has been shown to regulate other components of the PRL signaling cascade, namely by regulating PRLr dimerization (32). Thus, we next assessed whether HDAC6 could potentiate CISH luciferase reporter activity. Overexpression of HDAC6 increased CISH activity over control transfectants (Fig. 1B). As a complementary approach, siRNA knockdown of HDAC6 was employed. HDAC6 knockdown significantly decreased CISH mRNA expression as compared to control cells (Fig. 1C). Furthermore, overexpression of HDAC6 enhanced Stat5a binding to the CISH proximal promoter when analyzed by chromatin immunoprecipitation (Fig. 1D). Together, these data indicate that HDAC6 stabilizes the Stat5a transcriptional complex on the CISH proximal promoter to regulate Stat5a-mediated transcription.
Figure 1. HDAC6 regulates Stat5a-mediated transcription.
(A) Representative qPCR analysis of CISH expression in MCF-7 cells that were serum starved for 24h before treatment with 200nM TSA. After 4h pretreatment with TSA, cells were treated with 250ng/mL PRL for indicated timepoints. Values are expressed as mean ± SEM and were normalized to GAPDH expression. (B) Representative CISH luciferase assay of MCF-7 cells that overexpress HDAC6. Values are expressed as RLU and are normalized to total protein. Data are presented as mean ± SEM. (C) qPCR analysis of CISH transcription in MCF-7 cells treated with non-silencing control or HDAC6 siRNA. Cells were serum starved for 24h before 250ng/mL PRL treatment. Inset shows efficiency of HDAC6 knockdown. Data are presented as mean ± SEM of CISH transcript normalized to GAPDH expression. (D) ChIP analysis of Stat5a on the endogenous CISH promoter in MCF-7 cells transfected with control vector or HDAC6. Cells were serum starved for 24h and treated with 250ng/mL PRL. Data are presented as mean ± SEM and were normalized to 18S RNA from 5% input. The fold change was determined using control transfected, no PRL as control. All experiments were performed at least 3 times, unless otherwise noted. Two-way ANOVA with Bonferroini multiple comparisons test was used for statistical analysis, * p<0.05; ** p<0.01; *** p<0.001.
Deacetylation of HMGN2 on residue K2 is critical for Stat5a-mediated CISH transcription
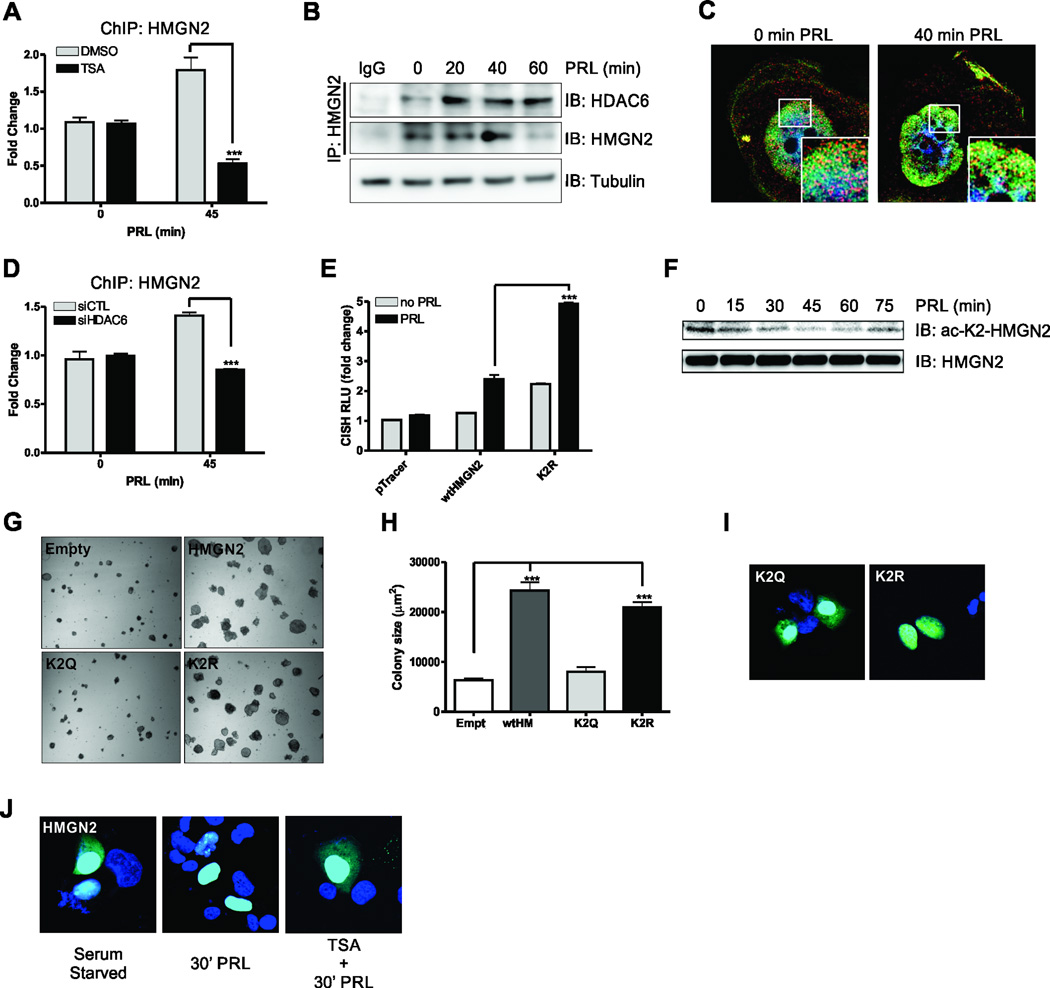
Recent data from our laboratory suggests that the interaction of nuclear PRLr with Stat5a and HMGN2 is required for optimal Stat5a-mediated gene transcription (8). Others have reported that when HMGN proteins are acetylated, their ability to bind nucleosomes is decreased (9), so we reasoned that HDAC6 might be responsible for HMGN2 deacetylation and subsequent binding to the CISH promoter. In order to determine if HDACs are important for HMGN2 binding to the CISH promoter, ChIP was performed on cells that had been treated with TSA. HMGN2 association with the CISH promoter was dramatically reduced with TSA treatment (Fig. 2A). Because HDAC6 potentiates Stat5a-mediated transcription, we next examined whether HDAC6 interacts with HMGN2. Coimmunoprecipitation confirmed its interaction and demonstrated its ligand dependence (Fig. 2B). Since HMGN2 is known to be primarily nuclear-localized and HDAC6 is recognized as a cytoplasmic protein, we performed structured illumination microscopy to identify the localization of the interaction. Under serum-starved conditions, we noted that HDAC6 was localized to the cytoplasm with nuclear localization readily apparent (Fig. 2C, left). While HMGN2 was primarily nuclear localized regardless of PRL treatment (Fig. 2C left, right), upon PRL stimulation, the HDAC6 apparent in the nucleus became strongly associated with HMGN2, particularly in the nuclear periphery (Fig. 2C, right inset). The appearance of HDAC6 in the nucleus is consistent with several reports that identify nuclear-localized HDAC6 under various stimuli (33–35). To determine if HDAC6 might regulate HMGN2 association with the CISH promoter, ChIP was performed in cells that had been treated with HDAC6 siRNA. HDAC6 knockdown significantly impaired HMGN2 association with the CISH proximal promoter after PRL stimulation (Fig. 2D).
Figure 2. HMGN2 K2 deacetylation is critical for PRL-mediated signaling and the oncogenic potential of breast cancer cells.
(A) ChIP analysis of HMGN2 on the endogenous CISH promoter in MCF-7 cells treated with DMSO or TSA. Cells were serum starved for 24h and treated with 200nM TSA for 4h before stimulation with 250ng/mL PRL. Data are presented as mean ± SEM and were normalized to 18S RNA from 5% input. The fold change was determined using control transfected, no PRL as control. (B) CoIP of endogenous HMGN2 with HDAC6. MCF-7 ells were serum starved for 24h before treatment with PRL for the indicated timepoints. Tubulin immunoblot on 5% input was used as a loading control. (C) Super resolution microscopy of HDAC6 (red) and HMGN2 (green) using the Nikon N-SIM. T47D cells were serum starved for 24h before treatment with PRL for indicated timepoints. (D) ChIP analysis of HMGN2 on the endogenous CISH promoter in control or HDAC6 siRNA transfected MCF-7 cells. Cells were serum starved for 24h and treated with 250ng/mL PRL. (E) CISH luciferase assay in MFC-7 cells transfected with the control vector, HMGN2, or the deacetylated-K2 HMGN2 mimic (K2R). Cells were serum starved for 24h and treated with 250ng/mL PRL for 24h. (F) MCF-7 cells were serum starved for 24h and treated with 250ng/mL PRL for the indicated timepoints and immunoblotted with an α-acetyl-K2-HMGN2 antibody. Total HMGN2 was used as a loading control. (G) Soft agar colony formation of MCF-7 cells that had been transfected with control vector, HMGN2, the acetylated K2 HMGN2 mimic (K2Q), or the K2R deacetylated-K2 HMGN2 mimic. Representative images of colony growth and quantification of colony size (H) are shown. (I) Confocal microscopy of eGFP K2Q and K2R HMGN2 fusion constructs. T47D cells were transfected with eGFP fusion constructs to assess the localization of the HMGN2 mutants. (J) Confocal microscopy of T47D cells that had been transfected with eGFP HMGN2 fusion construct. T47D cells were serum starved for 24h and were then pretreated for 4h with DMSO or TSA before stimulation with 250ng/mL PRL for 30 min. All experiments were performed at least 3 times, unless otherwise noted. One-way or Two-way ANOVA with Bonferroini multiple comparisons test was used for statistical analysis where appropriate, * p<0.05; ** p<0.01; *** p<0.001.
Because HDAC6 regulates HMGN2 association with the CISH proximal promoter, we reasoned that HMGN2 might become deacetylated upon PRL treatment. HMGN1 has been shown to be acetylated on several residues that are conserved in HMGN2: K2, located in the first nuclear localization signal (NLS); K35, K42, and K46, located in the nucleosome binding domain (NBD); and K50, K53, and K55, located in the second NLS (9). Mutation of K to R (deacetylated mimic) or K to Q (acetylated mimic) in either the NBD or the second NLS of HMGN2 had no effect on CISH luciferase activity (data not shown). However, mutation of the lysine residue in the first NLS had a dramatic effect on Stat5a activity. The K2R mutant robustly increased CISH luciferase over wtHMGN2 (Fig. 2E), suggesting that deacetylation of K2 is critical for the effects of HMGN2 on Stat5a-mediated transcription. We generated an acetyl-specific antibody, and as shown in Fig. S1A, the antibody is specific for acetylated-K2-HMGN2, as the acetyl-specific blocking peptide interfered with staining and the non-acetyl specific blocking peptide had no effect on staining. After treatment with PRL, the acetylation levels of HMGN2 K2 steadily decreased by approximately 50% over 45 min, returning to near normal levels of acetylation by 75 min post-stimulation (Fig. 2F), consistent with our hypothesis.
We next assessed how mutation of the K2 residue affected soft agar colony formation of MCF7 cells. As shown in Fig. 2G–H, transfection of wtHMGN2 formed significantly larger colonies than empty vector control cells. However, when the acetylated mimic K2Q was transfected, mutant HMGN2 was unable to form colonies as efficiently as the wild-type protein. Furthermore, the deacetylated mimic K2R formed significantly larger colonies than both the control cells and K2Q transfectants, suggesting that deacetylation of HMGN2 is critical for its tumorigenic function.
It has previously been shown that selective pools of HMGN2 are acetylated by KAT2B on K2 and that this acetylation affects the interaction of HMGN2 with nucleosomes (9, 10). Because the deacetylated residue on HMGN2 is in the first NLS and deacetylated HMGN proteins have been shown to be localized to the nucleus, we reasoned that the acetylated HMGN2 mimic K2Q would be localized in part to the cytoplasm. To this end, eGFP fusion proteins of the K2Q acetylated mimic and the K2R deacetylated mimic were constructed. Mutants were transfected into MCF7 cells and subjected to confocal microscopy. As shown in Fig. 2I, the K2R deacetylated mimic was exclusively localized to the nucleus whereas the K2Q acetylated mimic was partially localized to the cytoplasm. The partial localization to the cytoplasm was expected, as several alternate mechanisms also regulate the nuclear internalization of HMGN2, that include the phosphorylation of residues S20 and S28 (36). Since acetylation affects nuclear localization of HMGN2, and HDAC6 recruitment is PRL-dependent, we next assessed the effects of PRL treatment on wtHMGN2 localization. Serum starved cells had appreciable amounts of cytoplasmically localized HMGN2 when subjected to confocal microscopy (Fig. 2J left). Conversely, when cells were treated with PRL, virtually all HMGN2 became localized to the nucleus (Fig. 2J middle). To show that this effect was due to HMGN2 deacetylation, cells were pretreated for 2h with TSA before PRL treatment. As anticipated, there were again appreciable amounts of HMGN2 localized to the cytoplasm (Fig. 2J right). Together, these data suggest that K2 deacetylation is a partial mechanism by which PRL stimulation either induces or maintains HMGN2 nuclear localization to promote Stat5a-mediated gene transcription.
HDAC6 inhibition impedes Stat5a-mediated signaling
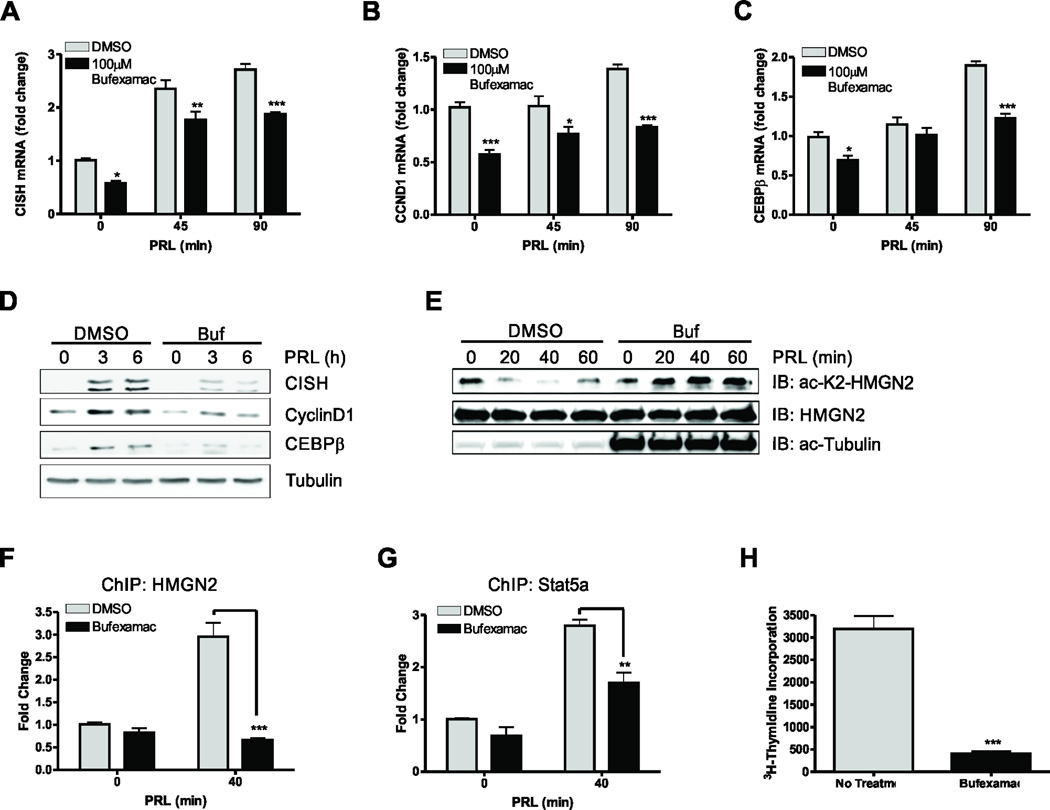
Because Stat5a-mediated signaling is regulated by the deacetylation of HMGN2, we reasoned that treatment with an HDAC6 inhibitor would prevent transcription of Stat5a target genes. Bufexamac, a non-steroidal anti-inflammatory drug (NSAID), was recently shown to also be a specific class IIb HDAC inhibitor having greatest efficacy for HDAC6 (37). Indeed, treatment with Bufexamac inhibited PRL-induced transcription of CISH (Fig. 3A), CCND1 (Fig. 3B), and CEBPβ (Fig. 3C). Furthermore, treatment of MCF7 cells with Bufexamac inhibited CISH, CyclinD1, and CEBPβ protein expression (Fig. 3D). To complement the data obtained using Bufexamac and to exclude off-target effects, the highly specific HDAC6 inhibitors tubacin (38) and tubastatin A were also utilized to determine how PRL-mediated transcription was affected by more specific HDAC6 inhibition. Treatment of cells with tubacin significantly inhibited transcription of CISH (Fig. S2A), CCND1 (Fig. S2B), and CEBPβ (Fig. S2C). Furthermore, treatment of MCF7 cells with tubacin inhibited CISH, CyclinD1, and CEBPβ protein expression, while the inactive analog of tubacin, niltubacin, had no effects (Fig. S2D). Treatment of MCF7 cells with tubastatin A also impaired PRL-induced transcription of CISH (Fig. S2E), CCDN1 (Fig. S2F), and CEBPβ (Fig. S2G), suggesting that HDAC6 directly regulates Stat5a-mediated transcriptions.
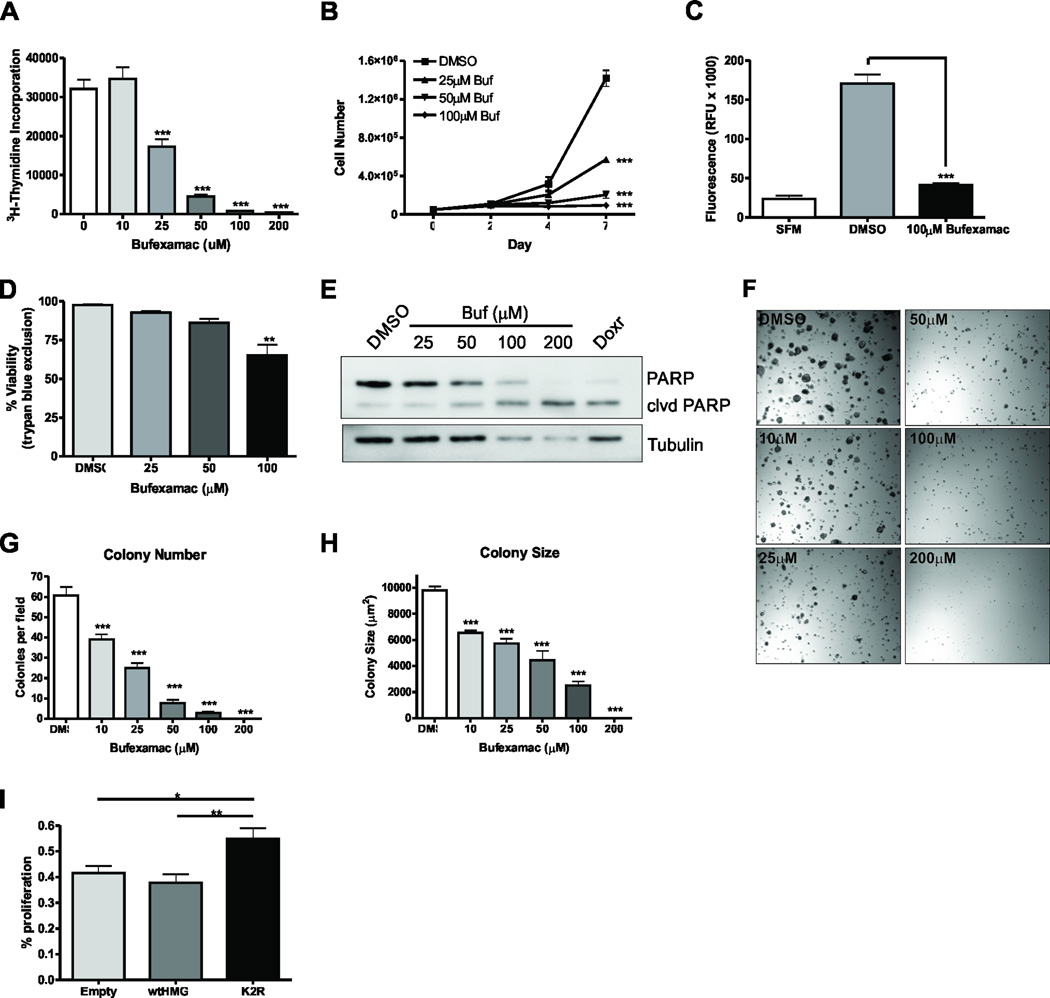
Figure 3. Bufexamac inhibits PRL-mediated signaling.
(A–C) qPCR analysis of CISH (A), CCND1 (B), and CEBPB (C) in MCF-7 cells treated with Bufexamac. MCF7 cells were serum starved for 24h and were then pretreated for 4h with 100µM Bufexamac prior to 250ng/mL PRL treatment for the indicated timepoints. Data are presented as mean ± SEM of CISH transcript normalized to GAPDH expression. (D) Immunoblot of CISH, Cyclin D1, and CEBPβ on MCF-7 cells that had been treated with Bufexamac. Cells were treated as in (A) for the indicated timepoints. (E) Immunoblot of acetyl-K2-HMGN2 in the presence or absence of the HDAC6 inhibitor Bufexamac. MCF-7 cells were serum starved for 24h and then pretreated for 4h with Bufexamac or DMSO before stimulation with 250ng/mL PRL for the indicated timepoints. Total HMGN2 was used as a loading control and acetyl-tubulin was used as a positive control for inhibition of HDAC6 with Bufexamac (F) ChIP analysis of HMGN2 on the endogenous CISH promoter in MCF-7 cells treated with DMSO or Bufexamac. Cells were serum starved for 24h and treated with 200nM Bufexamac for 4h before stimulation with 250ng/mL PRL. Data are presented as mean ± SEM and were normalized to 18S RNA from 5% input. The fold change was determined using control transfected, no PRL as control. (G) ChIP analysis of Stat5a on the CISH promoter on MCF-7 cells treated as in (F). (H)3H-thymidine incorporation was assessed in MCF-7 cells treated with DMSO or Bufexamac. Cells were grown for 48h in serum free media for 48h supplemented with 250ng/mL PRL before addition of 3H-thymidine for 4h. Student’s T test or Two-way ANOVA with Bonferroini multiple comparisons test was used for statistical analysis, * p<0.05; ** p<0.01; *** p<0.001.
We next assessed whether the reduction in Stat5a-mediated gene transcription was associated with an impairment of HMGN2 deacetylation in Bufexamac treated cells. We treated cells with PRL in the absence or presence of Bufexamac and revealed that HMGN2 was deacetylated after 20–40 minutes of PRL stimulation in cells treated with DMSO, but remained acetylated upon PRL stimulation in the presence of Bufexamac, suggesting that HDAC6 is responsible for the deacetylation of HMGN2 upon PRL stimulation (Fig 3E). In order to determine whether this effect correlated to a decrease in HMGN2 binding to the CISH promoter, chromatin immunoprecipitation was performed on cells that had been treated with Bufexamac. Indeed, there was a significant decrease in HGMN2 binding to the CISH promoter when cells were pretreated with Bufexamac (Fig. 3F). Furthermore, there was a significant decrease in Stat5a association with the CISH promoter, suggesting that treatment with Bufexamac prevents association and/or stabilization of the Stat5a transcriptional complex on the CISH proximal promoter (Fig. 3G). Since Stat5a-mediated signaling was inhibited by Bufexamac treatment, we next assessed how Bufexamac affected PRL-induced proliferation of MCF7 cells. As shown in Fig. 3H, 3H-thymidine incorporation was inhibited by Bufexamac in cells incubated in the presence of PRL.
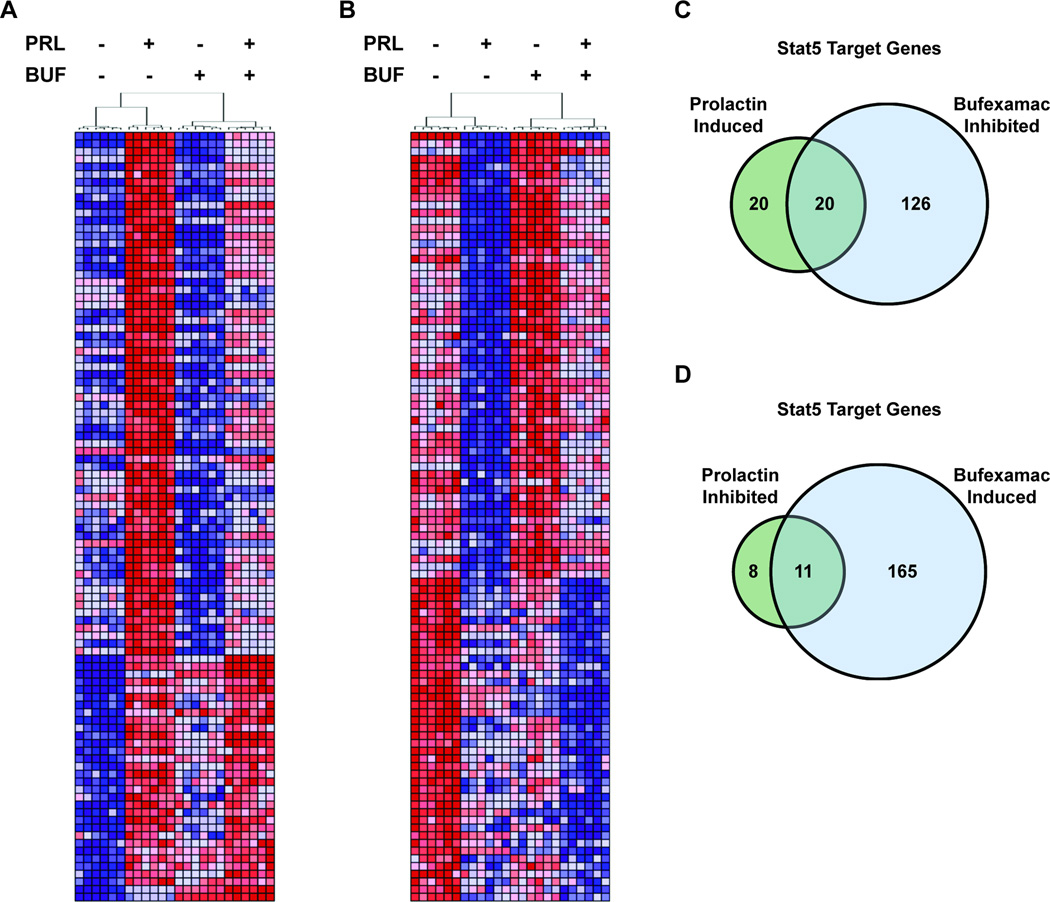
To determine the extent to which prolactin and Bufexamac altered global gene expression in human breast cancer cells, we utilized a series of expression microarrays and RNA derived from MCF7 cells treated with prolactin and/or Bufexamac. Analysis of the resultant data revealed 1,112 and 1,282 genes whose expression was significantly (FDR<0.01) induced or inhibited, respectively, by stimulation with prolactin. 2,896 and 3,048 genes were significantly (FDR<0.01) induced or inhibited, respectively, following Bufexamac treatment. After applying a fold change cutoff of 1.2 to the data, prolactin induced the expression of 504 genes and inhibited the expression of 376 genes, while Bufexamac inhibited 1,894 genes and induced 1,795 genes. The original microarray .CEL files and normalized expression values (as described in Material and Methods) were published online and are accessible via the NCBI Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81217. There was high concordance between the prolactin stimulated differential expression reported here and that previously reported by our group (39). Regarding the prolactin induced, Bufexamac inhibited expression of the prolactin receptor target genes CISH, CCND1, and CEBPB, the microarray data was in agreement with the qPCR and immunoblot data reported elsewhere in this manuscript (Fig. 3). Bufexamac also opposed the expression changes stimulated by prolactin on a global scale. Bufexamac significantly inhibited 68% of the top 100 prolactin induced genes (Fig. 4A) and significantly induced 58% of the top 100 prolactin inhibited genes (Fig. 4B). Of the 504 and 376 genes induced and inhibited, respectively, by prolactin with a fold change greater than 1.2, Bufexamac significantly inhibited 59% (295/504) and induced 44% (164/376). Interestingly, Bufexamac treatment also significantly inhibited the expression of the prolactin receptor gene itself (fold change = 1.6), an effect observed with and without prolactin stimulation.
Figure 4. Bufexamac opposes prolactin induced gene expression globally and at STAT5 target genes.
(A, B) Heat maps depict expression values of the top 100 prolactin induced (A) and prolactin inhibited (B) genes (represented by rows) identified by microarray analysis. Samples are represented by columns and are ordered as determined by hierarchical clustering. Samples clustered with technical and biological replicates according to the treatment conditions indicated above each dendrogram (PRL; prolactin, BUF; Bufexamac). Red and blue represent high and low gene expression, respectively. (C, D) Venn diagrams quantify the STAT5 target genes* from the microarray analysis that are significantly prolactin induced (C) or prolactin inhibited (D) with fold change >1.2 that are also significantly inhibited (C) or induced (D) by Bufexamac. *Genes identified as Stat5 target genes are defined as those previously reported by Kang et al. (see Materials and Methods).
We also sought to determine if, like its global effects, Bufexamac opposed the actions of prolactin at Stat5 target genes. Indeed, utilizing a previously reported set of Stat5 target genes (29), we found that of the 40 Stat5 target genes significantly induced by prolactin with a fold change greater than 1.2, 50% (20/40) were also significantly inhibited by Bufexamac (Fig. 4C). Of the 19 Stat5 target genes significantly inhibited by prolactin with a fold change greater than 1.2, 58% (11/19) were significantly induced by Bufexamac (Fig. 4D).
HDAC6 inhibition impedes breast cancer growth and tumorigenic phenotype in vitro
Because Bufexamac inhibited Stat5a-mediated signaling and proliferation, the effect of Bufexamac treatment on breast cancer growth in vitro under normal growth conditions was examined next. Proliferation of MCF7 cells treated with Bufexamac was inhibited in a dose-dependent manner as measured by both 3H-thymidine incorporation (Fig. 5A) and trypan blue exclusion (Fig. 5B). Correspondingly, tubacin treatment (Fig. S3A) and tubastatin A (Fig S3B) inhibited proliferation of MCF7 cells in a dose-dependent manner. We also sought to determine how treatment of the triple negative basal-type MDA-MB-231 cell line was affected by Bufexamac treatment. As shown in Fig. S3C, Bufexamac treatment also inhibited proliferation of MDA-MB-231 cells, but to a lesser extent than MCF7 cells. These results may be partially explained by the fact that there are reduced levels of PRLr in MDA-MB-231 cells compared to MCF7 cells. Because Bufexamac is also an NSAID, the effect of a comparable NSAID that does not inhibit HDAC activity was examined on MCF7 cells. Treatment of breast cancer cells with similar doses of Ibuprofen had no effect on 3H-thymidine incorporation (Fig. S3D), suggesting that HDAC6 inhibition is the primary cause of the anti-proliferative effects of Bufexamac.
Figure 5. Bufexamac inhibits the tumorigenic properties of breast cancer cell in vitro.
(A)3H-thymidine incorporation was assessed in cells treated with DMSO or Bufexamac. MCF7 cells were grown under normal growth conditions with the indicated concentrations of Bufexamac for 48h. 3H-thymidine was added 4h prior to harvesting. (B) MCF-7 cell growth was assessed by trypan blue exclusion. 50,000 cells were seeded and allowed to grow for the indicated timepoints with the indicated concentrations of Bufexamac. Data are presented as mean cell number ± SEM of total live cells. (C) MCF7 cells were allowed to migrate towards 10% FBS for 24h in an 8µm pore size transwell migration assay. (D) Trypan blue exclusion assay of MCF-7 cells treated as in (B) after day 7 of treatment. (E) Immunoblot of PARP cleavage on Bufexamac treated cells. MCF-7 cells were grown in the presence of the indicated treatment for 3 days. Doxorubicin is shown as a positive control and tubulin is used as a loading control. (F) Soft agar colony formation assay of MCF-7 cells assessing a dose response to Bufexamac treatement. Colony number (G) and colony size (H) were assessed after 14 days in the presence of Bufexamac at the indicated concentrations. (I)3H-thymidine incorporation assay assessing the ability of Bufexamac to inhibit proliferation of MCF-7 cells in the presence of the constitutively deacetylated K2 HMGN2 mimic. Cells were transfected with empty vector, HMGN2, or K2R and treated with 25µM Bufexamac for 48h. 3H-thymidine was added 4h prior to harvesting and percent proliferation was calculated comparing Bufexamac treated cells to DMSO treated cells for each transfectant. All experiments were performed at least 3 times, unless otherwise noted. One-way or Two-way ANOVA with Bonferroini multiple comparisons test was used for statistical analysis where appropriate, * p<0.05; ** p<0.01; *** p<0.001.
The effects of Bufexamac treatment on breast cancer cell phenotype were next examined. MCF7 cell migration across a transwell membrane towards 10% FBS was significantly reduced in cells treated with Bufexamac (Fig. 5C). Breast cancer cell viability was also decreased as a function of Bufexamac dose (Fig. 5D). Furthermore, there was a significant and dose-dependent increase in PARP cleavage (Fig. 5E), suggesting that Bufexamac induces apoptosis in breast cancer cells, particularly at higher doses. Anchorage-independent growth was next assessed in control versus Bufexamac treated cells. As shown in (Fig. 5F–H), there was a dose dependent decrease in both colony number (Fig. 5G) and colony size (Fig. 5H) as measured by a soft agar colony formation assay. We next reasoned that the effects of Bufexamac might be diminished in cells that express the deacetylated K2 HMGN2 mimic because HDAC6 inhibition cannot deacetylate this mutant protein on residue K2. In cells overexpressing the constitutively deacetylated HMGN2 K2R mutant, the effects of Bufexamac treatment on proliferation were mitigated compared to the wild-type transfected control as measured by 3H-thymidine incorporation (Fig. 5I).
Bufexamac inhibits breast cancer growth in vivo
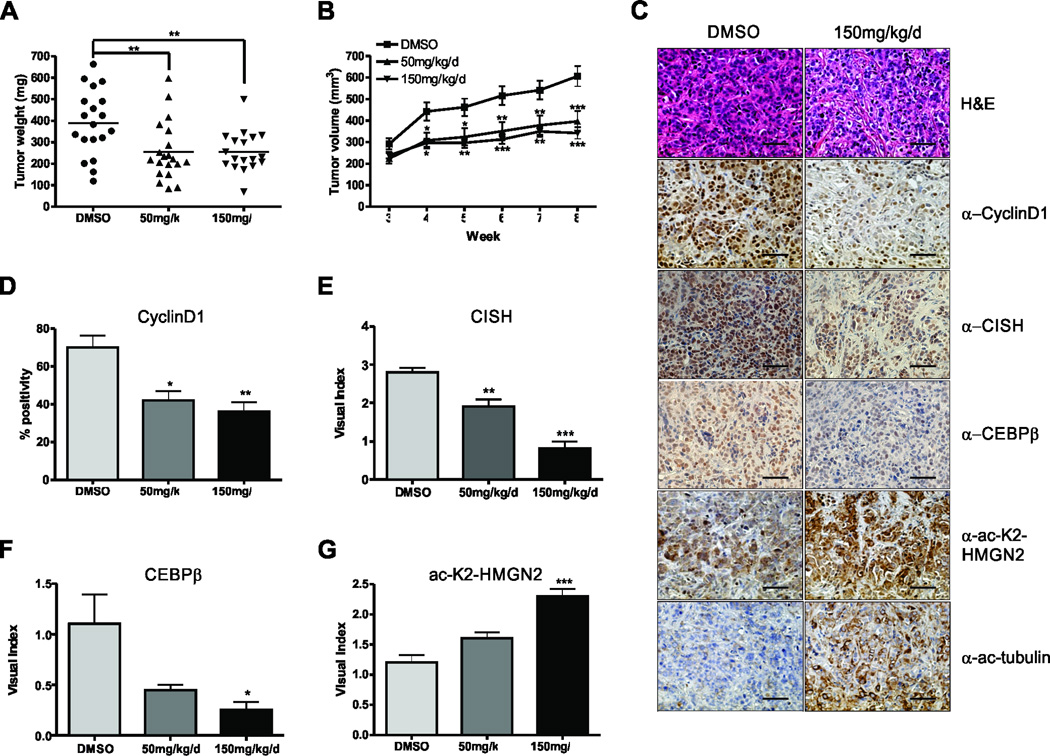
Since Bufexamac had dramatic effects on breast cancer cell proliferation, migration, and soft agar colony formation, the effects of Bufexamac treatment in a mouse xenograft model of breast cancer was examined. ER+/PR+ MCF7 and triple negative MDA-MB-231 cell lines were injected into the 4th lactiferous ducts of nude mice (13). Tumors were allowed to establish for 3 weeks before twice daily intraperitoneal injections of Bufexamac were administered. Tumor volume was significantly decreased in both MCF7 (Fig. 6A–B) and MDA-MB-231 tumor xenografts (Fig. S4A and S4B). Formalin-fixed paraffin embedded tumors were sectioned and stained for the Stat5a target genes cyclinD1, CISH, and CEBPβ. As shown in Fig. 6C, Bufexamac treatment decreased their expression as a function of dose (quantified in Fig. 6D–F). In order to assess whether Bufexamac was affecting the levels of acetyl-K2-HMGN2 in vivo, we utilized the acetyl-K2-HMGN2-specific antibody. In mice treated with Bufexamac, there was a notable increase in acetyl-K2-HMGN2 (Fig. 6C and 6G) as well as acetyl-tubulin, as a positive control (Fig. 6C).
Figure 6. Bufexamac inhibits tumor growth in vivo.
(A) MCF7 cells suspended in matrigel were xenografted into the 4th lactiferous ducts of nu/nu mice. Tumor growth was monitored for 3 weeks before twice daily intraperitoneal injections of Bufexamac. Final tumor weight is shown after sacrifice. (B) Xenografted tumors were measured weekly by calipers and volume was calculated using the formula V = (D*d2)/2, where V = tumor volume, D = tumor length (longer), and d = tumor width (shorter). (C) Representative images of formalin fixed, paraffin embedded tumor sections that were stained with indicated antibodies for IHC. Scale bar = 20µm. (D–G) Quantification of IHC is shown for α-CyclinD1 (D), CISH (E), CEBPβ (F), and α-ac-K2-HMGN2 (G). Visual scoring was determined on a scale from 0–3, where 0 = no staining and 3 - intense staining. One-way or Two-way ANOVA with Bonferroini multiple comparisons test was used for statistical analysis where appropriate, * p<0.05; ** p<0.01; *** p<0.001.
Ac-K2-HGMN2 is decreased as a function of neoplastic progression in human breast cancer
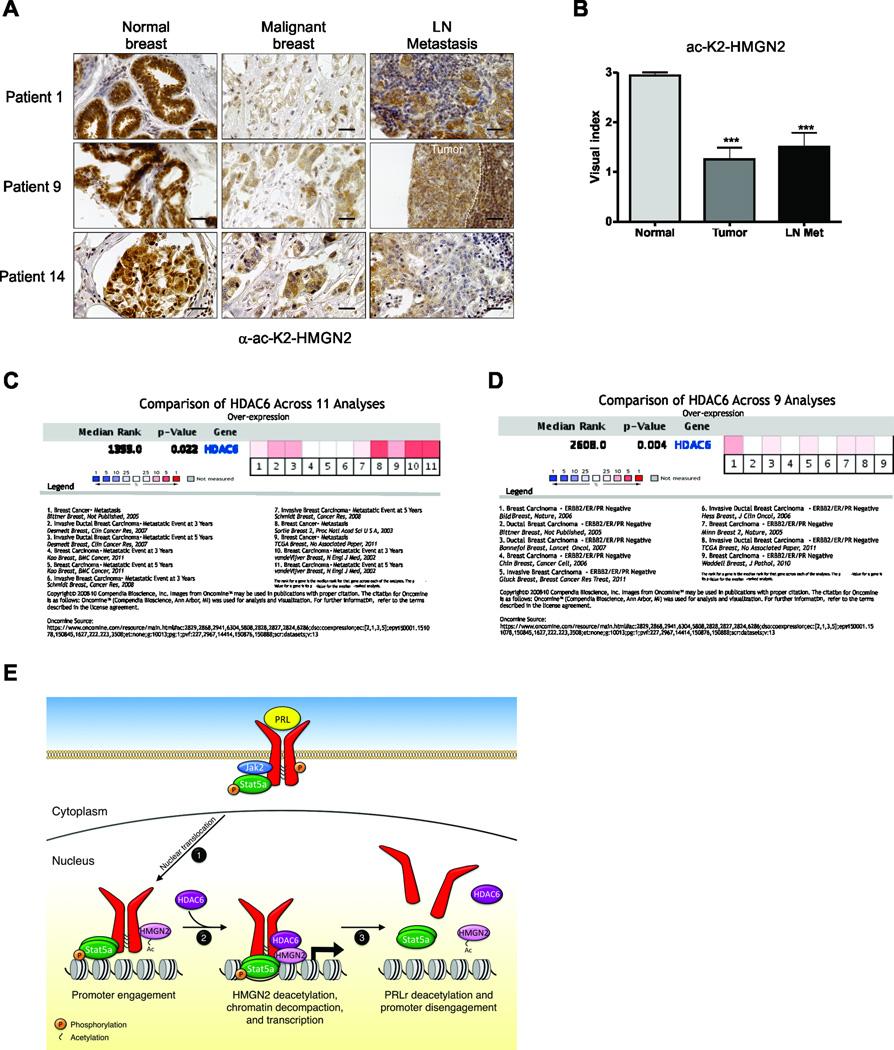
We next sought to determine whether acetyl-K2-HMGN2 levels change with malignant progression of human breast cancer. A tumor progression array consisting of 14 patient-matched samples of normal breast, primary tumor, and lymph node metastasis was stained with α-ac-K2-HMGN2. Fig. 7A demonstrates 3 representative matched patients in which HMGN2 is highly acetylated in normal breast epithelium, but is significantly reduced in primary tumors and lymph node metastases. The entire population of the progression array is quantified in Fig. 7B. We next sought to determine whether HDAC6 expression correlated with prognosis in Oncomine datasets. As shown in Fig. 7C, a meta-analysis of HDAC6 expression in Oncomine (http://www.oncomine.org) revealed a correlation with metastatic progression within 3–5 years. Furthermore, HDAC6 overexpression was associated with triple negative breast cancer (TNBC) (Fig. 7D). Given this overexpression, and the inhibitory effect of Befuxemac on the growth of the ER- cell line MDA-MB-231, additional confirmatory studies with a triple negative patient derived xenograft (PDX) with Bufexemac were performed. To that end, The TNBC PDX WHIM2 line was obtained (12), and treated with Bufexamac. As shown in Fig. S5, we noted a dose-dependent decrease in growth of this PDX line, similar to results observed in Fig. S3. Together, these data indicate that HDAC6 expression correlates with breast cancer progression and aggressive biology, and suggests that targeting HMGN2 K2 deacetylation with HDAC6-specific inhibitors may be an effective breast cancer therapeutic.
Figure 7. HMGN2 acetylation on residue K2 is lost during malignant progression of human breast cancer.
(A) A breast cancer progression array consisting of 14 patient-matched samples of normal human breast, primary tumors, and lymph node metastases were subjected to IHC analysis of ac-K2-HMGN2. Representative images of α-ac-K2-HMGN2 IHC of 3 separate patients is shown. Scale bar = 40µm. (B) Quantification of the entire breast cancer progression array shown in (A). (C) Meta-analysis of HDAC6 expression in 11 breast cancer datasets for patients who have a metastatic event within 3 or 5 years. (D) Meta-analysis of HDAC6 expression in 9 breast cancer datasets for triple negative disease. (E) Schematic of Stat5-mediated signaling: 1) Ligand binding induces phosphorylation and nuclear translocation of the Stat5a complex to PRL-induced target genes; 2) HDAC6 deacetylates PRLr-bound HMGN2 and allows chromatin decompaction and efficient Stat5a-mediated transcription of target genes; 3) Acetylation of HMGN2 and dissociation of the PRLr/Stat5a/HMGN2 complex from DNA. One-way ANOVA with Bonferroini multiple comparisons test was used for statistical analysis, *** p<0.001.
Discussion
In this report, we identify HDAC6 as a novel potentiator of Stat5a-mediated signaling and demonstrated that its effects on HMGN2 deacetylation are critical for Stat5a function. Treatment of breast cancer cells with the pan-HDAC inhibitor TSA, as well as the HDAC6-specific inhibitors tubacin, tubastatin A, and Bufexamac, disrupted the stability of the Stat5a chromatin complex and prevented efficient Stat5a-mediated signaling. Furthermore, treatment of murine xenograft models of both ER+/PR+ and ER−/PR− breast cancers with the HDAC6 inhibitor Bufexamac resulted in restoration of acetylated HMGN2 levels and decreased CISH, CEBPβ, and CyclinD1 expression, thereby preventing cellular proliferation and reducing tumor burden. Additionally, we demonstrated that acetylation levels of HMGN2 on residue K2 decrease during human malignant progression, suggesting that targeting this pathway is highly relevant to human pathology.
There are several potential mechanisms by which HMGN proteins can alter transcriptional activity: 1) HMGN proteins have been shown to compete with histone H1 binding to chromatin, which results in decreased compactness in the higher-order chromatin structure (40); 2) HMGN proteins have been shown to modulate histone modifications. HMGN1 has been shown to increase the acetylation levels of H3K14 (41), decrease the levels of H3S10 phosphorylation (42), and inhibit H2AS1 phosphorylation (43); and 3) HMGN proteins have been shown to inhibit ATP-dependent chromatin remodeling, suggesting that HMGN proteins may act to maintain either an active or a repressive state of a gene (44). Together, these studies suggest that HMGN proteins can affect the post-translational modifications of the core histones in response to various stimuli, thus altering chromatin compaction and DNA accessibility.
While it has previously been shown that HMGN2 localization changes upon phosphorylation (9, 36), the effects of HMGN2 acetylation remain less clear. It has been established that acetylated HMGN proteins possess reduced binding affinity to chromatin (9, 10). It is interesting to note that the deacetylated HMGN2 lysine residue is located in the first NLS. Histone H3 and H4, which also contain dual NLS, have been shown to be acetylated in the N-terminal NLS, which acted as an NLS mask, thereby reducing their binding to importin α/β(45). HMGN1 and HMGN2 have also been shown to utilize importin α/β heterodimers to enter the nucleus (46). This, along with our data showing that HMGN2 acetylation on residue K2 results in cytoplasmic accumulation, suggests that acetylation may mask the NLS and impair nuclear import of HMGN2. However, there remained a significant pool of nuclear-localized mutant HMGN2, suggesting that there are other factors important for nuclear import or retention, such as additional acetylated lysine residues, other posttranslational modifications, or the second NLS.
While the presence of an NES and the SE14 domain would suggest that HDAC6 is exclusively cytoplasmic, there have been several reports of nuclear-localized HDAC6. Upon differentiation and growth arrest, a fraction of HDAC6 was shown to become nuclear localized (34). A recent study demonstrated that estrogen treatment of breast cancer cells resulted in the relocalization of HDAC6 from the cytoplasm to the nuclear periphery (35). Additionally, it was shown that HDAC6 is recruited to estrogen-responsive promoters and acts as an LCoR cofactor to regulate estrogen responsiveness of breast cancer cells (33). These data, in addition to our data in showing that HDAC6 interacts with HMGN2 in the nuclear periphery, supports evidence that HDAC6 also functions in the nucleus as a regulator of gene transcription.
Precise balance of acetylation and deacetylation appears to be critical for regulating the PRL signaling cascade. It has been shown that both PRLr and Stat5b acetylation is required for their dimerization (32). Treatment of cells with PRL results in CBP-mediated acetylation of the PRLr within the cytoplasmic loop and mutation of these acetylation sites results in a decrease in LHRE luficerase expression. Acetylation is reversed when SIRT2 or to a lesser extent HDAC6 is cotransfected. Similar results were obtained with Stat5b (whose sites of acetylation are conserved in Stat5a); dimerization, and presumably nuclear import of Stat5b, requires acetylation (32). While these findings may appear at odds with our results demonstrating that HDAC6 deacetylates HMGN2 on the lysine at position 2 thereby enhancing HMGN2 binding to chromatin, deacetylation of multiple proteins by the same deacetylase within the nucleus may ensure that signaling is appropriately turned off after a stimulus. HDAC6 may bind and deacetylate HMGN2 initially, thereby enhancing chromatin accessibility and stabilizing the basal transcriptional machinery, and then subsequently deacetylate nuclear PRLr and/or Stat5, resulting in monomerization and release from the promoter to stop signal transduction. Our results with HMGN2 clearly demonstrate that deacetylation of HMGN2 occurs between 15 and 45 minutes of signal transduction with acetylation returning by 60–75 minutes post-stimulus, suggesting that there is a counterregulatory mechanism that ensures HMGN2 disassociates from chromatin to halt PRL-mediated signaling (Fig 7E).
HDAC6 plays an important role in cellular homeostasis in normal tissues and its role in the pathogenesis breast cancer is becoming increasingly appreciated. HDAC6 has been correlated with increased motility and invasion in breast cancer cells, in part due to its effects on tubulin and cortactin deacetylation (47–50). Acetylated tubulin is found in stable microtubules, but is absent in dynamic cellular structures. HDAC6 deacetylates tubulin to regulate microtubule turnover and therefore, motility (51). Cortactin, an F-actin binding protein that has reduced binding affinity towards F-actin when acetylated, is deacetylated by HDAC6 which enhances its binding to F-actin to regulate migration (48). Additionally, estrogen treatment of MCF7 cells in vitro upregulates HDAC6 expression, which results in increased motility in part due to its effects on tubulin deacetylation (52). While these reports implicate HDAC6 in breast cancer migration and metastasis, there are also studies that show HDAC6 plays a role in tumor initiation and progression. In vivo, mice lacking HDAC6 were shown to be resistant to mammary tumor formation in the oncogene-induced ErbB2 and Ras models and also in chemically induced models of mammary cancer (53). Furthermore, mouse embryonic fibroblasts (MEFs) from HDAC6−/− mice were shown to form fewer colonies when transduced with SV40 early region and RasG12V than wild-type MEFs, suggesting that HDAC6 plays a role in oncogene-induced transformation (53). Additionally, reduced levels of HDAC6 in SKOV ovarian cancer cells, and SKBR3 and MCF7 breast cancer cells resulted in a 5–30-fold reduction in soft agar colony formation (53). Together, these results suggest that HDAC6 plays an essential role in in the migratory phenotype of breast cancer cells, in addition to being required for the oncogenic potential of breast cancer cells.
Much of the data described above were performed in ER+ breast cancer cells, but data presented in this manuscript and elsewhere indicate that HDAC6 treatment may be effective regardless of breast cancer subtype. In our study, we identified that Stat5a signaling is attenuated in both ER+ and TNBC following Bufexemac treatment, though the efficacy appeared to be higher in the ER+ subtypes. Part of this may be due to reduced reliance on PRLr signaling in TNBCs, but several others have shown additional mechanisms of HDACi efficacy in TNBC aside from cortactin deacetylation. Phthalates, an environmental toxin associated with increased risk of breast cancer and resistance to endocrine therapy, are agonists of the aryl hydrocarbon receptor (AhR). It was revealed that phthalates activate the AhR/HDAC6/c-Myc signaling pathway in TNBC cells and promoted their tumorigenicity (54). Others have shown that HDAC6 (and HDAC3) promotes survivin expression in TNBC, thereby enhancing survival of cells, a phenotype reversed by treatment with voronistat (55). Interestingly, a phase I–II study of 55 patients with stage IIA-IIIC breast cancer receiving vorinostat plus paclitaxel and doxorubicin-cyclophosphamide revealed that 54% of Her2+ patients and 27% of TNBC patients responded to treatment, while none of the 12 ER+ patients responded (56). The small sample sizes and the pan-HDACi activity of vorinostat in this trial make the data difficult to draw concrete conclusions on subsets of patients likely to benefit from HDACi therapy, although it appears that additional biomarkers will be needed to stratify patients most likely to respond to HDACi.
In the current study, we have identified a novel mechanism by which Stat5a is regulated and provide insight into how nuclear HDAC6 can contribute to transcriptional regulation and breast cancer pathogenesis. In vivo use of the class II HDAC inhibitor Bufexamac prevented HMGN2 deacetylation, Stat5a-mediated transcription, and breast cancer growth. Overall, these data suggest that targeted HDAC6 inhibition may be a viable option as a breast cancer therapeutic.
Supplementary Material
Acknowledgments
We would like to thank Dr. Debu Chakravarti (Northwestern University) for the use of lentiviral reagents, Dr. Anthony Kossiakoff (University of Chicago) for providing human recombinant PRL, Dr. Stuart L. Schreiber (Harvard University) for the HDAC6 constructs, and Manop Buranapramest for critical reading of the manuscript.
Financial Support: This work was supported in part by grants from the Avon and Lynn Sage Foundations (CVC). TRM acknowledges support from the NIH/NCI (T32CA106195), the American Cancer Society – Friends of Rob Kinas, the Medical Research Foundation, and the Cathy and Jim Rudd Career Development Award for Cancer Research. This work was also supported in part by Komen grants awarded to Dr. Lisa M. Coussens.
Footnotes
Conflict of Interest Statement: The authors disclose no potential conflicts of interest.
References and Notes
- 1.Ren S, Cai HR, Li M, Furth PA. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- 2.Miermont AM, Parrish AR, Furth PA. Role of ERalpha in the differential response of Stat5a loss in susceptibility to mammary preneoplasia and DMBA-induced carcinogenesis. Carcinogenesis. 2010;31:1124–1131. doi: 10.1093/carcin/bgq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iavnilovitch E, Groner B, Barash I. Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Mol Cancer Res. 2002;1:32–47. [PubMed] [Google Scholar]
- 4.Humphreys RC, Hennighausen L. Signal transducer and activator of transcription 5a influences mammary epithelial cell survival and tumorigenesis. Cell Growth Differ. 1999;10:685–694. [PubMed] [Google Scholar]
- 5.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 6.Borges S, Moudilou E, Vouyovitch C, Chiesa J, Lobie P, Mertani H, et al. Involvement of a JAK/STAT pathway inhibitor: cytokine inducible SH2 containing protein in breast cancer. Adv Exp Med Biol. 2008;617:321–329. doi: 10.1007/978-0-387-69080-3_30. [DOI] [PubMed] [Google Scholar]
- 7.Brockman JL, Schroeder MD, Schuler LA. Molecular endocrinology. Vol. 16. Baltimore, Md: 2002. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway; pp. 774–784. [DOI] [PubMed] [Google Scholar]
- 8.Fiorillo AA, Medler TR, Feeney YB, Liu Y, Tommerdahl KL, Clevenger CV. HMGN2 inducibly binds a novel transactivation domain in nuclear PRLr to coordinate Stat5a-mediated transcription. Mol Endocrinol. 2011;25:1550–1564. doi: 10.1210/me.2011-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergel M, Herrera JE, Thatcher BJ, Prymakowska-Bosak M, Vassilev A, Nakatani Y, et al. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. The Journal of biological chemistry. 2000;275:11514–11520. doi: 10.1074/jbc.275.15.11514. [DOI] [PubMed] [Google Scholar]
- 10.Herrera JE, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Molecular and cellular biology. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rascle A, Lees E. Chromatin acetylation and remodeling at the Cis promoter during STAT5-induced transcription. Nucleic Acids Res. 2003;31:6882–6890. doi: 10.1093/nar/gkg907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrell JC, Dye WW, Allred DC, Jedlicka P, Spoelstra NS, Sartorius CA, et al. Estrogen receptor positive breast cancer metastasis: altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer research. 2006;66:9308–9315. doi: 10.1158/0008-5472.CAN-06-1769. [DOI] [PubMed] [Google Scholar]
- 14.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson J, Denisenko O, Bomsztyk K. The fast chromatin immunoprecipitation method. Methods Mol Biol. 2009;567:45–57. doi: 10.1007/978-1-60327-414-2_3. [DOI] [PubMed] [Google Scholar]
- 16.Galbaugh T, Feeney YB, Clevenger CV. Prolactin receptor-integrin cross-talk mediated by SIRPalpha in breast cancer cells. Mol Cancer Res. 2010;8:1413–1424. doi: 10.1158/1541-7786.MCR-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumur CI, Nasim S, Best AM, Archer KJ, Ladd AC, Mas VR, et al. Evaluation of quality-control criteria for microarray gene expression analysis. Clin Chem. 2004;50:1994–2002. doi: 10.1373/clinchem.2004.033225. [DOI] [PubMed] [Google Scholar]
- 18.RDevelopmentCoreTeam. R: A Language and Environment for Statistical Computing. 2011 Available from: http://www.R-project.org/
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. Experimental design and low-level analysis of microarray data. Int Rev Neurobiol. 2004;60:25–58. doi: 10.1016/S0074-7742(04)60002-X. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archer KJ, Reese SE. Detection call algorithms for high-throughput gene expression microarray data. Brief Bioinform. 2010;11:244–252. doi: 10.1093/bib/bbp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 26.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 27.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 29.Kang K, Yamaji D, Yoo KH, Robinson GW, Hennighausen L. Mammary-Specific Gene Activation Is Defined by Progressive Recruitment of STAT5 during Pregnancy and the Establishment of H3K4me3 Marks. Mol Cell Biol. 2014;34:464–473. doi: 10.1128/MCB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakamoto K, Triplett AA, Schuler LA, Wagner KU. Janus kinase 2 is required for the initiation but not maintenance of prolactin-induced mammary cancer. Oncogene. 2010;29:5359–5369. doi: 10.1038/onc.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Molecular and cellular biology. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, et al. Acetylation modulates prolactin receptor dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19314–19319. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palijan A, Fernandes I, Bastien Y, Tang L, Verway M, Kourelis M, et al. Function of histone deacetylase 6 as a cofactor of nuclear receptor coregulator LCoR. The Journal of biological chemistry. 2009;284:30264–30274. doi: 10.1074/jbc.M109.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verdel A, Curtet S, Brocard MP, Rousseaux S, Lemercier C, Yoshida M, et al. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 35.Riolo MT, Cooper ZA, Holloway MP, Cheng Y, Bianchi C, Yakirevich E, et al. Histone Deacetylase 6 (HDAC6) Deacetylates Survivin for Its Nuclear Export in Breast Cancer. The Journal of biological chemistry. 2012;287:10885–10893. doi: 10.1074/jbc.M111.308791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louie DF, Gloor KK, Galasinski SC, Resing KA, Ahn NG. Phosphorylation and subcellular redistribution of high mobility group proteins 14 and 17, analyzed by mass spectrometry. Protein Sci. 2000;9:170–179. doi: 10.1110/ps.9.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 38.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiorillo AA, Medler TR, Feeney YB, Wetz SM, Tommerdahl KL, Clevenger CV. The Prolactin Receptor Transactivation Domain Is Associated with Steroid Hormone Receptor Expression and Malignant Progression of Breast Cancer. Am J Pathol. 2013;182:217–233. doi: 10.1016/j.ajpath.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. The EMBO journal. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, et al. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Molecular cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson-Saliba M, Siddon NA, Clarkson MJ, Tremethick DJ, Jans DA. Distinct importin recognition properties of histones and chromatin assembly factors. FEBS letters. 2000;467:169–174. doi: 10.1016/s0014-5793(00)01142-x. [DOI] [PubMed] [Google Scholar]
- 46.Johnson-Saliba M, Siddon NA, Clarkson MJ, Tremethick DJ, Jans DA. Distinct importin recognition properties of histones and chromatin assembly factors. FEBS Lett. 2000;467:169–174. doi: 10.1016/s0014-5793(00)01142-x. [DOI] [PubMed] [Google Scholar]
- 47.Rey M, Irondelle M, Waharte F, Lizarraga F, Chavrier P. HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur J Cell Biol. 2011;90:128–135. doi: 10.1016/j.ejcb.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Molecular cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azuma K, Urano T, Horie-Inoue K, Hayashi S, Sakai R, Ouchi Y, et al. Association of estrogen receptor alpha and histone deacetylase 6 causes rapid deacetylation of tubulin in breast cancer cells. Cancer research. 2009;69:2935–2940. doi: 10.1158/0008-5472.CAN-08-3458. [DOI] [PubMed] [Google Scholar]
- 50.Williams KA, Zhang M, Xiang S, Hu C, Wu JY, Zhang S, et al. Extracellular signal-regulated kinase (ERK) phosphorylates histone deacetylase 6 (HDAC6) at serine 1035 to stimulate cell migration. The Journal of biological chemistry. 2013;288:33156–33170. doi: 10.1074/jbc.M113.472506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 52.Saji S, Kawakami M, Hayashi S, Yoshida N, Hirose M, Horiguchi S, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene. 2005;24:4531–4539. doi: 10.1038/sj.onc.1208646. [DOI] [PubMed] [Google Scholar]
- 53.Lee YS, Lim KH, Guo X, Kawaguchi Y, Gao Y, Barrientos T, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer research. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh TH, Tsai CF, Hsu CY, Kuo PL, Lee JN, Chai CY, et al. Phthalates induce proliferation and invasiveness of estrogen receptor-negative breast cancer through the AhR/HDAC6/c-Myc signaling pathway. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:778–787. doi: 10.1096/fj.11-191742. [DOI] [PubMed] [Google Scholar]
- 55.Lee JY, Kuo CW, Tsai SL, Cheng SM, Chen SH, Chan HH, et al. Inhibition of HDAC3- and HDAC6-Promoted Survivin Expression Plays an Important Role in SAHA-Induced Autophagy and Viability Reduction in Breast Cancer Cells. Front Pharmacol. 2016;7:81. doi: 10.3389/fphar.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu Y, Hershman DL, Bhalla K, Fiskus W, Pellegrino CM, Andreopoulou E, et al. A phase I–II study of the histone deacetylase inhibitor vorinostat plus sequential weekly paclitaxel and doxorubicin-cyclophosphamide in locally advanced breast cancer. Breast Cancer Res Treat. 2014;146:145–152. doi: 10.1007/s10549-014-3008-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.