ABSTRACT
Vector transmission is a critical stage in the viral life cycle, yet for most plant viruses how they interact with their vector is unknown or is explained by analogy with previously described relatives. Here we examined the mechanism underlying the transmission of citrus tristeza virus (CTV) by its aphid vector, Toxoptera citricida, with the objective of identifying what virus-encoded proteins it uses to interact with the vector. Using fluorescently labeled virions, we demonstrated that CTV binds specifically to the lining of the cibarium of the aphid. Through in vitro competitive binding assays between fluorescent virions and free viral proteins, we determined that the minor coat protein is involved in vector interaction. We also found that the presence of two heat shock-like proteins, p61 and p65, reduces virion binding in vitro. Additionally, treating the dissected mouthparts with proteases did not affect the binding of CTV virions. In contrast, chitinase treatment reduced CTV binding to the foregut. Finally, competition with glucose, N-acetyl-β-d-glucosamine, chitobiose, and chitotriose reduced the binding. These findings together suggest that CTV binds to the sugar moieties of the cuticular surface of the aphid cibarium, and the binding involves the concerted activity of three virus-encoded proteins.
IMPORTANCE Limited information is known about the specific interactions between citrus tristeza virus and its aphid vectors. These interactions are important for the process of successful transmission. In this study, we localized the CTV retention site as the cibarium of the aphid foregut. Moreover, we demonstrated that the nature of these interactions is protein-carbohydrate binding. The viral proteins, including the minor coat protein and two heat shock proteins, bind to sugar moieties on the surface of the foregut. These findings will help in understanding the transmission mechanism of CTV by the aphid vector and may help in developing control strategies which interfere with the CTV binding to its insect vector to block the transmission.
INTRODUCTION
The survival of a virus is dependent on its ability to move from host to host, which for many plant viruses requires an insect vector (1). The mechanisms by which viruses are transmitted by these insects have been classified into two general groups: circulative and noncirculative. More than half of the viruses with a described mode of transmission fall into the latter category and are defined by attachment to sites within the vector's stylet, cibarium, or foregut (2, 3). There are also differences in both the acquisition and retention of noncirculative viruses (4), previously described as nonpersistent versus semipersistent. While there is no clear demarcation between the two, nonpersistent viruses can be acquired and disseminated through probing and salivation within a matter of minutes, while most semipersistent viruses may only be acquired and subsequently transmitted through deep phloem feeding, which generally requires hours for acquisition, and vectors remain viruliferous for a few days (5).
For noncirculative viruses, transmission requires that virions bind or interact directly with receptors on the cuticular intima of the vector (3, 6). In some virus-vector systems, this involves direct interaction of the viral coat protein with the vector stylet or foregut organs (7, 8), whereas other systems use a nonvirion protein (variously termed helper components or transmission factors) to bridge the virion to the insect's mouthparts (9–12). For most plant viruses, the mechanism by which the virus and vector interact is unknown or is described by analogy from better studied virus-vector systems.
Within the Closteroviridae family, of which citrus tristeza virus (CTV) is a member, there are 38 species divided into four genera: the aphid-transmitted closteroviruses, the whitefly-transmitted criniviruses, the mealybug-transmitted ampeloviruses, and the velariviruses for which no vectors have yet been described (13). Despite this diversity of both species and vectors, only the mechanism of whitefly transmission of lettuce infectious yellows virus (LIYV) has been studied in detail, and it was found that the minor coat protein, which encapsidates one end of the virion, binds directly to the cibarium of the insect foregut (2, 8).
CTV is one of the most destructive diseases in citrus (14). This viral disease has killed more than 100 million trees that were propagated on the sour orange (Citrus aurantium) rootstock in Argentina, Brazil, United States, Spain, and Venezuela (15, 16). CTV symptoms depend on the virus strain, the citrus variety, and the scion-rootstock (17). Severe CTV strains cause one or a combination of the three main symptoms depending on the scion-rootstock combination (17). Infected plants grafted on sour orange show a quick decline and dieback. Sour orange, lemon, and grapefruit trees show stunting and yellowing; sweet orange, grapefruit, and mandarin trees show stem pitting (17). Citrus and a few close citrus relatives are the main hosts for CTV and its vector (18).
CTV is transmitted by seven aphid species, of which Aphis gossypii, Aphis spiraecola, Toxoptera aurantii, and Toxoptera citricida are considered to be major vectors (14). There is, however, considerable variation in the efficacy of transmission both between different vector species and between CTV isolates. T. citricida, the brown citrus aphid, is considered the most efficient and specialized of the vectors (19), for it has a host range limited primarily to rutaceous species and transmits most CTV isolates at higher rates than other aphids (20).
CTV is transmitted by brown citrus aphid in a semipersistent manner (21). Acquisition of CTV by brown citrus aphid ranges from a few seconds to 30 min, and the inoculation period ranges from 4 to 6 h (21). Once the aphid becomes infected with CTV, it remains infective for 24 h, and it loses infectivity after 48 h (21).
Here we examined the mechanism of the virus-vector interaction of the semipersistent aphid-transmitted virus CTV. We hypothesized that in addition to the minor coat protein, other viral proteins may also be implicated in the specific binding. We also hypothesized that CTV virions bind to the cuticular surface and not to proteins embedded in the mouthparts. As a first step in unraveling this complex interaction, we examined where virions bind to aphid mouthparts by using purified expressed CTV-encoded proteins to compete with fluorescent CTV virions in binding to dissected aphid mouthparts (foregut and stylet).
MATERIALS AND METHODS
Aphid colonies.
Citrus macrophylla (alemow) plants were used to rear aphids. The plants, produced from cuttings, were approximately 6 to 8 months old and 20 cm in height with multiple new shoots. All plants were kept in a U.S. Department of Agriculture Animal and Plant Health Inspection Service (USDA-APHIS)-approved secure greenhouse under a 16-h/8-h light/dark (L/D) photoperiod at 25°C temperature and 75% relative humidity until use. Brown citrus aphid colonies were reared on the above plants and kept inside mesh cages (width, 35.6 cm; length, 35.6 cm; height, 70 cm) in a growth chamber under controlled conditions similar to those of the greenhouse.
Development of green fluorescent protein-tagged CTV virion.
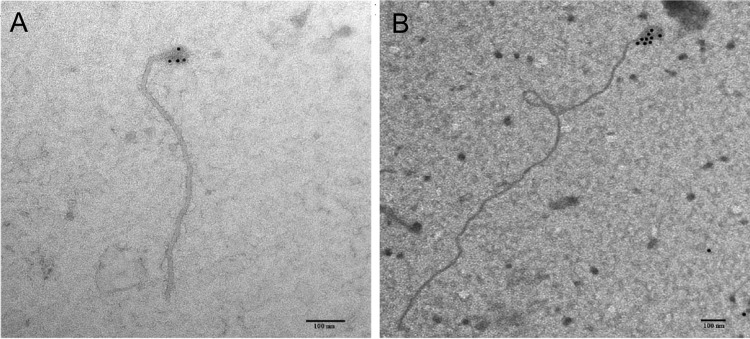
A full-length green fluorescent protein (GFP)-tagged CTV virion was developed through the insertion of a read-through element and a GFP open reading frame (ORF) at the end of the p27 ORF. The read-through domain was assembled from three fragments: (i) from base 11558 of the T36 genome (NCBI GenBank accession number U16304) to the 3′ end of the p27 (base 16058), with a silent point mutation of the codon prior to the stop codon and insertion of three additional codons after the stop to encourage read-through of the stop codon (22) and the first 23 bases of GFP; (ii) from the latter to the 5′ end of GFP, with a duplication of the last 43 nucleotides (nt) of the p27 ORF to increase the length of the p25 controller element; and (iii) from the latter to base 17294 of the T36 genome (Table 1). These amplicons were assembled into a contiguous fragment by overlap PCR and ligated into the PstI-PmeI-digested T36-based infectious clone as above. Clones were confirmed by sequencing and then were used to transform Agrobacterium tumefaciens EHA105 (23) for infiltration into Nicotiana benthamiana, propagated for 5 days, and then purified using sucrose gradient ultracentrifugation as described previously (24). Integration of the GFP read-through p27 protein into the CTV virion was confirmed using a rabbit anti-GFP antibody (Clontech, Palo Alto, CA), with secondary detection by 10-nm colloidal gold-labeled goat anti-rabbit antibodies (Sigma-Aldrich, St. Louis, MO) on a Morgagni 268 (FEI Company, Hillsboro, OR) transmission electron microscope (Fig. 1).
TABLE 1.
Primers used for the amplification and assembly of the infectious clone and protein expression constructsa
| Construct and gene/region | Sense | Sequence (5′ to 3′) | Binding site (KC517488) |
|---|---|---|---|
| CTV GFP-p27 fusionb | |||
| Outer primers | + | GCAATCTCGAGACTAGTTAGTGCTGTCTCTCCCGTATATC | 11661–11688 |
| − | CGTGTCTAAGTCACGCTAAACAAAGTGAC | 17272–17300 | |
| P27 to GFP read-through | + | TACGCGATTTGGGTAAGTACCTATAGCAATTACAGATGGCTAGCAAAGGAGAAGAACT | 16025–16044 |
| − | AGTTCTTCTCCTTTGCTAGCCATCTGTAATTGCTATAGGTACTTACCCAAATCGCGTA | 16025–16044 | |
| GFP CP controller element | + | TACAAATAACTCGAGGGGTAGTTGGTTTGGGGACGGTAACATTATAC | 16051–16076 |
| − | GTATAATGTTACCGTCCCCAAACCAACTACCCCTCGAGTTATTTGTA | 16051–16076 | |
| E. coli/pRSETc | |||
| L1 | + | ATGCGGATCCATGAGTGGGCGGCGAGTTTGTTAC | 1122–1142 |
| − | CGATCTCGAGCTAACCAACCAAATGGTGGTTAGG | 1542–1562 | |
| L2 | + | ATGCGGATCCATGCGGTTTGTGGTGTGTGTTGAAG | 2580–2601 |
| − | CGATCTCGAGGCCCATCTTATGATACTTATTGAG | 2985–3008 | |
| p6 | + | TCGATGGGGATCCATGGACTGTGTGATTCAAGG | 11874–11893 |
| − | CGATAAGCTTTTAGATAGTGGCGTGAGTGC | 12010–12029 | |
| p27 | + | TCGATGGGGATCCATGGCGGGTTACACGATG | 15325–15342 |
| − | CGATAAGCTTCTACAAATACTTTCCCAAATC | 16027–16047 | |
| p25 | + | TCGATGGGGATCCATGGACGACGAGACAAAGAA | 16141–16160 |
| − | CGATAAGCTTTCAACGTGTGTTAAATTTCCCAAG | 16789–16812 | |
| p18 | + | TCGATGGGGATCCATGTCAGGCAGCTTGG | 16778–16793 |
| − | CGATAAGCTTCTAAGTCGCGCTAAACAAAGC | 17224–17244 | |
| p13 | + | TCGATGGGGATCCATGGGTATTCGACGCGTG | 17315–17332 |
| − | CGATAAGCTTCTAGTTATCGCAAGGTAAGA | 17655–17674 | |
| p20 | + | TCGATGGGGATCCATGCGAGCTTACTTTAGTG | 17750–17768 |
| − | CGATAAGCTTCTACACGCATGAAGGAGAAAC | 18278–18298 | |
| p23 | + | TCGATGGGGATCCATGAACGATACTAGCGGAC | 18380–18398 |
| − | CGATAAGCTTTCAGATGAAGTGGTGTTCACG | 18989–19009 | |
| A. tumefaciens/pCAMBIA 1380d | |||
| 35S promoter start | + | ATCGGGCGCGCCGATCTCCTTTGCCCCAGAGA | NAe |
| 35S–6×His-p61 overlap | + | CATTTCATTTGGAGAGGATGCATCATCATCATCATCATTCGTCTCATCACGTATG | 13746–13762 |
| − | CATACGTGATGAGACGAATGATGATGATGATGATGCATCCTCTCCAAATGAAATG | 13746–13762 | |
| p61-Nos overlap | + | GGTTACACGATGCTTCCTAGCGTTCAAACATTTGGCAAT | 15331–15346 |
| − | ATTGCCAAATGTTTGAACGCTAGGAAGCATCGTGTAACC | 15331–15346 | |
| 35S–6×His-p65 overlap | + | CATTTCATTTGGAGAGGATGCATCATCATCATCATCATGTGCTTCTGGGTCTAGAC | 12038–12055 |
| − | GTCTAGACCCAGAAGCACATGATGATGATGATGATGCATCCTCTCCAAATGAAATG | 12038–12055 | |
| p65-Nos overlap | + | TGGAAAGAGTACCTCTCTAGCGTTCAAACATTTGGCAAT | 15300–15316 |
| − | ATTGCCAAATGTTTGAACGCTAGAGAGGTACTCTTTCCA | 15300–15316 | |
| Nos terminator end | − | CGATCTGCAGCCCGATCTAGTAACATAGATG | NA |
Primer binding sites are given according to the sequence of isolate FS577 (NCBI GenBank accession no. KC517488) and restriction enzyme recognition sites are underlined, where appropriate.
Assembly and integration of a GFP read-through domain at the C terminus of the minor coat protein (CP) into the full-length T36 infectious clone.
Cloning of CTV proteins into the pRSET binary vector for expression in E. coli BL21(DE3).
Cloning of CTV proteins into the pCAMBIA 1380 vector for agrobacterium-mediated transient expression in N. benthamiana.
NA, not applicable.
FIG 1.
Transmission electron micrographs of the citrus tristeza virus (CTV) virion after immunodetection using gold-labeled antibodies (bar = 100 nm). (A) Immunodetection using antibodies produced against the wild-type minor coat protein. (B) Immunodetection using antibodies produced against the modified minor coat protein, including a C terminus GFP read-through at the 5′ end of the virion. Note that the minor coat protein encapsidates the 5′ end.
Expression of CTV proteins.
Isolate T68-1, which has a high transmission efficacy, was selected as the source for the proteins; cDNA was synthesized from this isolate using SuperScript III reverse transcriptase with random hexamers (Invitrogen) according to the manufacturer's instructions. The CTV ORFs for genes p6, p27, p25, p13, p18, p20, and p23 and those for the L1 and L2 protease domains were amplified (Table 1) using T68-1 cDNA under standard PCR conditions. Amplicons were inserted into the pRSET-A vector (Invitrogen, Waltham, MA), which adds a 6×His tag and enterokinase cleavage site to the N terminus of the expressed protein, using appropriate restriction sites (Table 1). Clones were confirmed by sequencing and then were used to transform BL21(DE3) pLysS Escherichia coli (Promega Corp., Madison, WI) for protein expression. The His tag was not cleaved by enterokinase in the following experiments.
The p61 and p65 proteins, which are lethal to E. coli, were expressed in N. benthamiana under a 35S promoter. Fragments containing the 35S promoter, p61 or p65 genes with a 6×His tag at the N terminus, or the NOS terminator were amplified separately (Table 1) and assembled by overlap PCR. These fragments were digested and inserted into the pCAMBIA 1380 binary vector (Cambia, Canberra, Australia) and used to transform A. tumefaciens EHA105, according to published protocols (25). Proteins, expressed by either method, were purified using HisTrap columns in an ÄKTA Prime Plus protein purification system (GE Healthcare, Piscataway, NJ), according to the manufacturer's instructions. Eluted proteins were checked by SDS-PAGE, quantitated, and diluted to 25 ng/ml for competition assays.
CTV localization in aphids.
To localize the virus binding sites, five apterous T. citricida adults and late-instar nymphs, starved for 5 h prior to the assay, were fed on 100 μl of the purified GFP-labeled CTV virion preparation sandwiched between thin Parafilm membranes (Bemis NA, Neenah, WI). The aphids were permitted to feed on the preparation for 3 h and were then transferred to virus-free membranes containing 100 μl of water to passage unbound virions through their gut. Aphids were then decapitated with a razor blade, and the heads were mounted on glass slides and examined for fluorescence using a Bio-Rad ZOE fluorescent cell imager (Bio-Rad Laboratories, Hercules, CA).
Enzymatic treatments of aphid mouthparts.
To determine the nature of the virion-binding site within the aphid's cibarium, individual T. citricida were placed on filter paper under an ×2 to ×225 trinocular extreme wide-field microscope (AmScope, Irvine, CA), and the head was dissected with a razor blade. The anterior gut assembly, from the stylet to midgut, was removed and placed in 0.1 M sodium cacodylate buffer (11). Batches of five were treated with 25, 50, and 100 nano-international units (nIU)/ml of (i) trypsin (Sigma-Aldrich), (ii) pronase E (Sigma-Aldrich), or (iii) proteinase K (Sigma-Aldrich), in 50 mM phosphate-buffered saline (PBS) with 0.05% SDS for 2 h at 37°C, or with 25, 50, and 100 nIU/ml of (iv) chitinase in 50 mM PBS for 1 h at room temperature. After incubation, the stylet-to-foregut organs were then washed three times with 50 mM PBS. They were then incubated with 10 μl of the purified GFP-labeled CTV virion preparation in 100 μl of PBS for a further 4 h, washed again, and then mounted onto glass slides and examined for fluorescence using a Zeiss A-1 fluorescence microscope equipped with a AxioCam ICc1 and GFP longpass filter (Carl Zeiss, Thornwood, NY). GFP-labeled CTV virion fluorescence intensities were determined using ImageJ software (NIH) and compared to that of the untreated GFP-labeled CTV virion control.
Competitive binding of CTV proteins and virions to aphids.
To examine the role of individual CTV proteins on the transmission process, we competitively bound individually expressed CTV proteins and fluorescently labeled CTV virions to excised aphid stylet-to-foregut assemblies. To conduct the competitive binding experiments, the aphid stylet through to the foregut organs were excised as described for the enzymatic digests. Batches of 10 were incubated with 25 ng of individual purified CTV protein for 4 h at room temperature and washed three times with 50 mM PBS buffer. Foreguts were then incubated with 10 μl of the GFP-labeled CTV virion preparation in 100 μl of PBS for a further 4 h. The positive-control samples were incubated with the virion preparation alone, while the negative (minimum fluorescence) control was incubated with buffer only. After incubation, samples were washed three times with PBS, and fluorescence intensities were determined as described earlier. Five replicates of each protein-virion combination were examined, and statistical analysis was performed according to the enzymatic treatment assays above.
Competitive binding of lectins and virions to aphids.
To examine whether CTV binds to the cuticular layer of foregut or embedded proteins, we competitively bound lectins and fluorescently labeled CTV virions to excised aphid stylet-to-foregut assemblies. Experiments were conducted as described above with CTV proteins. Lectins and other proteins used in this experiment are listed in Table 2.
TABLE 2.
Proteins, sugars, and enzymes used in competition assays for binding of citrus tristeza virus proteins with excised mouthparts of the brown citrus aphid, Toxoptera citricia
| Protein, sugar, or enzyme | Competitora | Characterization, activity, and/or affinity |
|---|---|---|
| Viral proteins | L1 | Protease |
| L2 | Protease | |
| P13 | Host range determinant | |
| P18 | Host range determinant | |
| P20 | Silencing suppressor | |
| P23 | Silencing suppressor | |
| P25 | Coat protein and silencing suppressor | |
| P27 | Minor coat protein | |
| P61 | Heat shock protein 90 h | |
| P65 | Heat shock protein 70 h | |
| Proteins | ConA | Lectin with affinity to carbohydrates with terminal mannose or glucose (αMan > αGlc > GlcNAc) |
| LCA | Lectin with affinity to branched mannose with fucose linked α(1,6) to the N-acetylglucosamine, (αMan > αGlc > GlcNAc) | |
| WGA | Lectin with affinity to N-linked oligosaccharides, [GlcNAc(β1,4GlcNAc) > βGlcNAc] | |
| PNA | Lectin with affinity to terminal β-galactose, (Galβ1,3GalNAc > α and βGal) | |
| BSA | Neutral protein (no sugar affinity) | |
| OV | Glycoprotein with affinity to lectins | |
| Sugars | d-Galactose | Reducing monosaccharide (Fisher projection) |
| d-Mannose | Reducing monosaccharide (Fisher projection) | |
| d-Glucose | Reducing monosaccharide (Fisher projection) | |
| Methyl α-d-galactopyranoside | Pyranose form of galactose (Haworth projection) | |
| Methyl α-d-mannopyranoside | Pyranose form of mannose (Haworth projection) | |
| Methyl α-d-glucopyranoside | Pyranose form of glucose (Haworth projection) | |
| N-Acetylglucosamine (GlcNAc) | Monomer of chitin (the insect cytoskeleton) | |
| Chitobiose (GlcNAc)2 | Dimer of N-acetylglucosamine | |
| Chitotriose (GlcNAc)3 | Trimer of N-acetylglucosamine | |
| Enzymes | Pronase E | Protease with activity against denatured and native proteins leading to complete or nearly complete digestion into individual amino acids |
| Protease K | Protease cleaves native proteins between amino acids X and Y (X ↓ Y), when X is an aliphatic, aromatic, or hydrophobic amino acid and Y is any amino acid | |
| Trypsin | Trypsin cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, except when either is followed by proline | |
| Chitinase | Chitinase is an extracellular enzyme complex that degrades chitin into chitobioses |
ConA, concanavalin A; LCA, lens culinaris agglutinin; WGA, wheat germ agglutinin; PNA, peanut agglutinin; BSA, bovine serum albumin; OV, ovalbumin.
Competitive binding of sugars and CTV virions to aphids.
Batches of 10 aphid stylets were incubated with 10 μl of the GFP-labeled CTV virion preparation in 100 μl of PBS, including gradient concentrations (50 mM, 100 mM, 200 mM, and 500 mM) of different sugars as described in Table 2. The positive-control samples were incubated with virion preparation alone, while the negative (minimum fluorescence) control was incubated with buffer only. After incubation, samples were washed three times with PBS, and fluorescence intensities were determined as described earlier. Five replicates of each protein-virion combination were examined, and statistical analysis was performed according to the enzymatic treatment assays above.
Statistical analysis.
In order to study the effect of enzymatic activity and the presence of competitors, analysis of variance (ANOVA) and post hoc Tukey honestly significant difference (HSD) tests were performed among treatments using SPSS v 19.0 (SPSS, Chicago, IL).
RESULTS
Retention site of CTV virion is located on the cibarium of T. citricida.
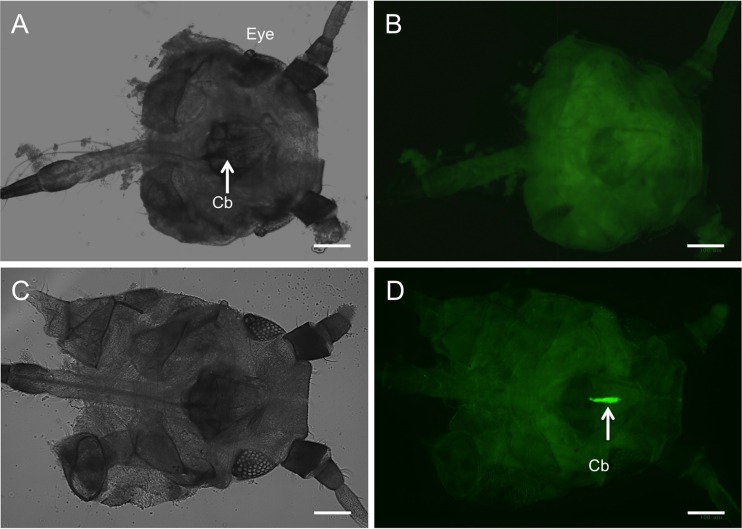
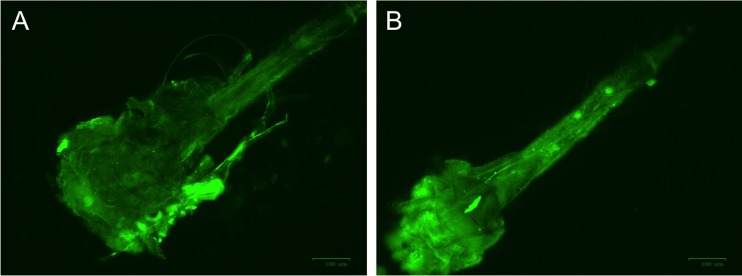
To examine the interaction between virus and vector, we first needed to localize where the virion binds to the aphid. Here we used a fluorescently labeled virion, containing a fusion of GFP to the C terminus of the minor coat protein via a read-through domain. These modified virions, when centrifuged through a sucrose gradient, retained the fluorescence with the virion band; integration of p27 GFP into the end of the virions was confirmed by electron microscopy using antibodies specific to GFP (Fig. 1). When these labeled virions were fed to aphids through a Parafilm membrane and subsequently examined by fluorescence microscopy, we observed fluorescence only in the cibarium of T. citricida (Fig. 2). No fluorescence was observed in the stylet, salivary glands, or other structures in the head. Similar localization of the virion-binding site was observed after incubation of virions with dissected stylet-to-foregut assemblies (Fig. 3).
FIG 2.
In vivo localization of the GFP-labeled CTV virion acquired by feeding of brown citrus aphids, T. citricida, through Parafilm. (A) Negative control aphid head and cibarium (Cb) under visible light. (B) The same negative-control aphid cibarium imaged using a 515-nm longpass fluorescein isothiocyanate (FITC)-GFP filter. (C) Aphid head with localized GFP-labeled CTV virion in the cibarium under visible light microscopy. (D) Same CTV-positive cibarium under 515-nm longpass FITC-GFP filter showing the retention site of CTV. Images were taken using the ZOE fluorescent cell imager (bar = 100 μm).
FIG 3.
In vitro localization of the GFP-labeled CTV virion in the dissected foregut of brown citrus aphid, T. citricida after incubation with GFP-labeled CTV virion, (A) Dissected mouthpart (stylet-to-foregut assembly) without incubation (negative control). (B) Dissected mouthpart incubated with GFP-labeled CTV virion showing the retention site of CTV. Note that most of light green fluorescence localized in the cibarium. Images were taken using the ZOE fluorescent cell imager (bar = 100 μm).
Although we noticed some fluorescence in the cibarium area, the majority of the fluorescence was concentrated and localized within the cibarium. Given the specific localization of the virion to the cibarium, we wished to investigate the composition of the putative target of the virion.
Minor coat protein (p27) and heat shock proteins (p61 and p65) implicated in binding to cibarium.
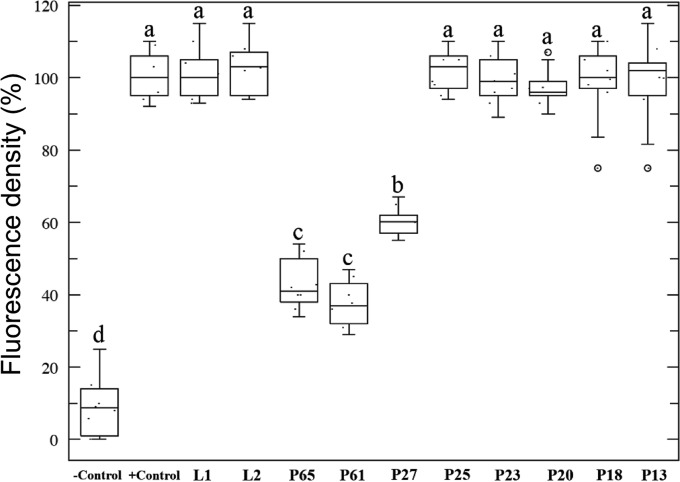
To examine whether CTV proteins specifically interact with the cibarium, we conducted a series of in vitro competitive binding assays between individually expressed CTV proteins, developed from the highly transmissible T68 isolate, and the GFP-labeled CTV virion to excised T. citricida stylet-to-foregut organs, containing the cibarium, and compared the changes in fluorescence intensity to that of the GFP-labeled virion alone. We found that the majority of CTV proteins tested, including leader proteases (L1 and L2), p23, p20, p18, p13, and the major coat protein (p25), did not show a significant change in GFP-labeled virion fluorescence in the cibarium relative to the virion-alone positive control (Fig. 4). This indicated that their presence did not interfere with or complement the binding of the labeled virion and that these proteins do not apparently interact with the aphid. However, three CTV proteins, p27, p65, and p61, each reduced fluorescence and, by extension, the binding efficacy of the intact virion through competition (Fig. 4).
FIG 4.
Competition assay between expressed CTV proteins and the GFP-labeled CTV virion for in vitro binding to dissected T. citricida foreguts and stylets. The fluorescence intensity of the competitive binding assay was assessed against the binding efficacy of the GFP-labeled CTV virion-only positive control. Horizontal thick lines indicate the medians, boxes show the interquartile ranges, including 25 to 75% of the values, and whiskers show the highest and the lowest values in each set. The circles represent outliers. Letters on the bars indicate significant differences (P < 0.05). To calculate the percentage of fluorescence density, the mean optical density of the positive control (mouthpart incubated with GFP-labeled virion without any competitor) was normalized to 100 (maximum fluorescence); then data points from all treatments were calculated relative to 100.
CTV virions bind to the sugar surface of aphid cibarium.
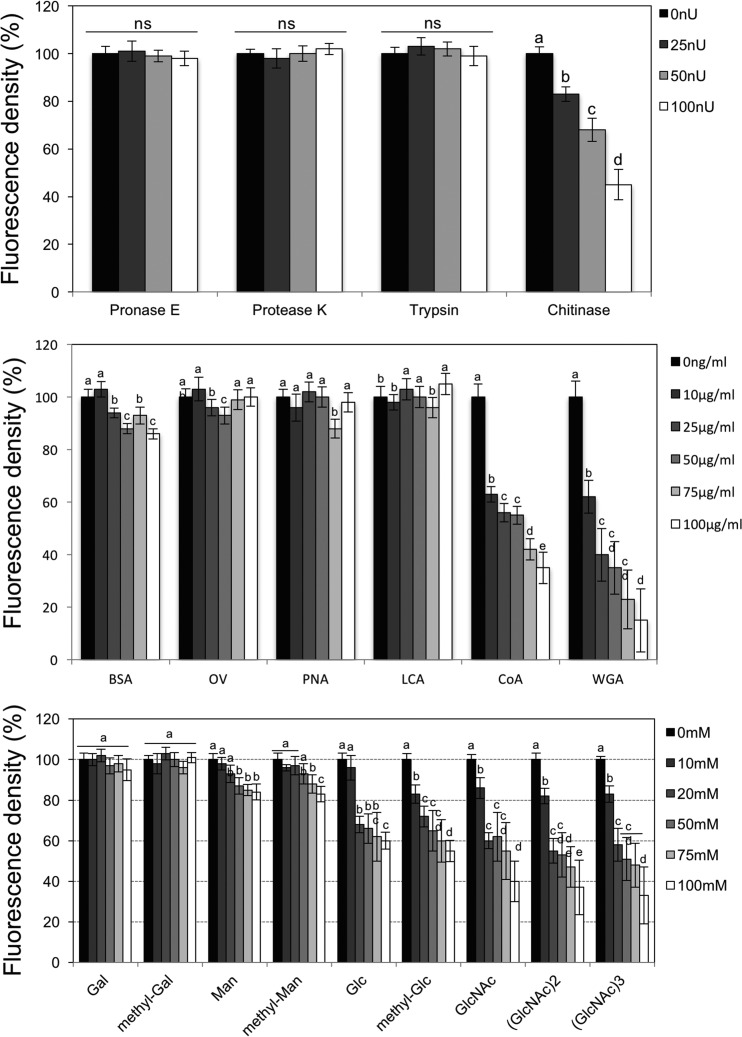
We treated dissected aphid stylet-cibarium-foregut structures with enzymatic digestion using 25, 50, and 100 nIU concentrations of pronase E, proteinase K, trypsin, and chitinase, followed by incubation with the GFP-labeled CTV virion. Digestion with 100 nIU/ml chitinase gave the greatest reduction in GFP fluorescence and, thus likely, virion binding (Fig. 5A). Lower concentrations of chitinase produced no significant effect, nor did any concentration of pronase E, proteinase K, or trypsin. This suggests that the target is chitin but might also be a protein embedded in a chitin matrix.
FIG 5.
The binding of CTV particles and foregut surface is similar to the lectin-sugar interaction. (A) Effect of the enzymatic treatment of dissected T. citricida foreguts and stylets on the in vitro binding of the GFP-labeled CTV virion. Five replicates of the each of four enzymatic treatments were tested at three different concentrations, and the fluorescence intensity was compared to that of the untreated GFP-labeled CTV virion-only positive control. (B) Competition between lectins and the GFP-labeled CTV virion for in vitro binding to dissected T. citricida foreguts and stylets. (C) Competition between sugars and the GFP-labeled CTV virion for in vitro binding to dissected T. citricida foreguts and stylets. Letters on the bars indicate significant differences (P < 0.05). To calculate the percentage of fluorescence density, the mean optical density of the positive control (mouthpart incubated with GFP-labeled virion without any competitor) was normalized to 100 (maximum fluorescence); then data points from all treatments were calculated relative to 100. ns, not significant; nU, nano-international unit.
In order to test whether the virions bind to sugars rather than proteins, we carried out competition assays with several lectins and sugars. Competition assays using lectins showed that wheat germ agglutinin (WGA) and concanavalin A (ConA) decreased the fluorescence intensity, indicating a reduction in virion binding to the cibarium (Fig. 5B). Interestingly, only the presence of sugars, including Glc, GlcNAc, (GlcNAc)2, (GlcNAc)3, Man, and methyl-Man reduced the binding of virions to the cibarium (Fig. 5C). On the other hand, Gal and methyl-Gal did not interfere with the binding (Fig. 5C). The decrease in binding was correlated with concentration increases in Glc, GlcNAc, (GlcNAc)2, (GlcNAc)3 ConA, and WGA, which indicates specific reduction. Some concentrations of bovine serum albumin (BSA) and lens culinaris agglutinin (LCA) treatment reduced the binding, but there was no trend (Fig. 5B and C).
These results together suggested that CTV virions bind to the N-acetylglucosamine (NAG) moieties of the cuticular surface of the aphid cibarium.
DISCUSSION
Noncirculative plant viruses have evolved close and specific interactions with their insect vectors (1, 6) and have been observed to bind or localize to multiple locations of their hemipteran vectors, from the stylet to the foregut (2, 11, 26). These interactions involve binding of virions, either directly or via helper proteins, to specific protein receptors on the cuticular intima of the vector (12, 27). Mutation of these viral vector-binding proteins has been shown to reduce or abolish transmission (28, 29), which, in the case of potyviruses at least, correlates with a lack of virus retention in the stylet (30). This suggests that viral vector-binding proteins interact with their vectors in a state of equilibrium, tight enough to be temporarily retained, but loose enough to be released into the next host.
To examine the interaction between CTV and its aphid vector, we produced a fluorescently labeled CTV virion by creating a read-through GFP fusion to the minor coat protein. These labeled virions were found to bind specifically to the cibarium of the aphid, much as LIYV virions do in their whitefly vector (26). What was most intriguing was that virions of this strain (T36) bound to the cibarium after membrane feeding, suggesting that for CTV at least, there is no correlation with lack of retention and poor transmissibility. This binding was abolished only by predigestion of the cuticular intima with chitinase and was not affected by protease treatments, suggesting that virions bind to the sugar moieties and not to proteins embedded in the cuticle. To confirm this result, competition assays with monosaccharides and lectins were performed. Only Glu, GlcNAc, (GlcNAc)2, and (GlcNAc)3, wheat germ agglutinin, and concanavalin A reduced the binding of GFP-labeled virions. These findings together strongly suggest the CTV virion binds to the sugar moieties on the cuticular surface of cibarium and more specifically, chitin. Other vector-borne plant pathogens behave similarly. For instance, Xylella fastidiosa, a Gram-negative bacterium, multiplies and forms a biofilm on the surface of the foregut of its insect vectors (31). Interestingly, although this bacterium propagates in its vector, it is localized on the cibarium of the foregut and does not circulate within the body (31). It has been shown that N-acetylglucosamine inhibits bacterial adhesion to vector foregut extracts and intact wings and that attachment to leafhopper surfaces is affected in the presence of specific polysaccharides (32). On the other hand, X. fastidiosa binds to different polysaccharides using cell surface carbohydrate-binding proteins, including the afimbrial adhesins, hemagglutinin-like proteins (32).
This led us to ask what viral genes or proteins of CTV are involved in this virus-vector interaction (binding to the cuticular surface of the foregut) and why T36 differs from other extant CTV isolates. Using competitive binding of expressed CTV proteins with fluorescently tagged virions, we identified three CTV proteins, p27, p61, and p65, as being involved in the virus binding to chitin in the brown citrus aphid. p25 and p27 have been shown to be part of the virion, and p61 and p65 are suspected but not proven. We tested nearly all of the proteins that CTV expresses (except the replicase and p33) in the competition assays.
The minor coat protein p27 was a likely candidate for vector interaction because the minor coat protein of a related member of the Closteroviridae, LIYV, previously had been shown to bind to the cibarium of its whitefly vector (2). The involvement of p61 and p65 homologues, which are present in all extant members of the Closteroviridae (33), was unexpected. In CTV, these two proteins are essential for virion assembly and act to restrict encapsidation by the minor coat protein to the 5′ end of the virion (34). Furthermore, it was shown that neither protein functioned alone during assembly; both worked only in combination (34). Here, we found that both p61 and p65 also have a role in aphid transmission of CTV.
The T68 isolate was used as a source for the individual expressed proteins as it is the most highly transmissible CTV isolate we possess, being transmitted at a rate of approximately 40% versus <1% for T36 (S. J. Harper, unpublished data). The individual proteins of T68 share approximately 95% amino acid identity with those of T36, but those differences are responsible for a significant change in the aphid transmission phenotype. The fluorescently labeled particles were derived from isolate T36, the parental isolate of the infectious clone used (34); at the time of writing, this is the only infectious clone available. Complementation has been shown between isolates from different CTV strains, including T36 and T68, affecting systemic infection of citrus (35), as well as aphid transmission (Harper, unpublished), which suggests that the proteins of different strains can interact.
Our results indicated that three CTV proteins, p27, p61, and p65, may be involved in the aphid transmission process. We can hypothesize that the minor coat protein binds directly to the aphid's cibarium, as has been shown for the related crinivirus LIYV to its whitefly vector (2). Given that p27 encapsidates the 5′ end of the virion (36), the reduction in GFP-labeled virion binding to the cibarium of dissected aphids in vitro in the presence of extraneous p27 might be explained as competition between virion-bound and free p27 for binding sites, supporting a direct virus-vector interaction mechanism for this protein. A similar reduction in binding of fluorescent virions to the aphid foregut was observed in the presence of extraneous p61 or p65. Their similar levels of competition with virion binding suggest that all three proteins might be functioning in transmission as components of the virion. In fact, homologues of p61 and p65 have been reported to be components of virions in other members of the Closteroviridae: beet yellows virus and LIYV (8, 37).
Viruses with a phloem-limited tropism, such as CTV, have developed close and specific interactions with their vectors (6), but for many of these viruses, the mechanism of interaction and the viral proteins involved remain unknown and may only be explained by analogy. While it was likely from the outset that the minor coat protein of CTV is involved in virus-vector interaction, as it is in other members of the Closteroviridae (2), further research will be required to reveal the function of the molecular chaperones p61 and p65 in aphid transmission of CTV, and our research suggests that they play a role.
ACKNOWLEDGMENTS
We acknowledge C. L. Davis for assistance with the electron microscopy, C. T. Bierman for performing enzyme-linked immunosorbent assays (ELISAs), and L. Lindsey for assistance with the aphid dissection and assays.
This research was supported by grants from Southern Gardens Citrus, by an endowment from the J. R. and Addie S. Graves family, and by the UF Agricultural Experiment Station.
Funding Statement
Southern Gardens Citrus, by an endowment from the J. R. and Addie S. Graves family, and the UF Agricultural Experiment Station.
REFERENCES
- 1.Ng JCK, Falk BW. 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu Rev Phytopathol 44:183–212. doi: 10.1146/annurev.phyto.44.070505.143325. [DOI] [PubMed] [Google Scholar]
- 2.Chen AY, Walker GP, Carter D, Ng JC. 2011. A virus capsid component mediates virion retention and transmission by its insect vector. Proc Natl Acad Sci U S A 108:16777–16782. doi: 10.1073/pnas.1109384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc S, Uzest M, Drucker M. 2011. New research horizons in vector-transmission of plant viruses. Curr Opin Microbiol 14:483– 491. doi: 10.1016/j.mib.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Nault LR. 1997. Arthropod transmission of plant viruses: a new synthesis. Ann Entomol Soc Am 90:521– 541. doi: 10.1093/aesa/90.5.521. [DOI] [Google Scholar]
- 5.Moreno A, Tjallingii WF, Fernandez-Mata G, Fereres A. 2012. Differences in the mechanism of inoculation between a semi-persistent and a non-persistent aphid-transmitted plant virus. J Gen Virol 93:662–667. doi: 10.1099/vir.0.037887-0. [DOI] [PubMed] [Google Scholar]
- 6.Ng JC, Zhou JS. 2015. Insect vector-plant virus interactions associated with non-circulative, semi-persistent transmission: current perspectives and future challenges. Curr Opin Virol 15:48–55. doi: 10.1016/j.coviro.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Pirone TP, Blanc S. 1996. Helper-dependent transmission of plant viruses. Annu Rev Phytopathol 34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 8.Tian T, Rubio L, Yeh HH, Crawford B, Falk BW. 1999. Lettuce infectious yellows virus: in vitro acquisition analysis using partially purified virions and the whitefly Bemisia tabaci. J Gen Virol 80:1111–1117. doi: 10.1099/0022-1317-80-5-1111. [DOI] [PubMed] [Google Scholar]
- 9.Drucker M, Froissart R, Hebrard E, Uzest M, Ravallec M, Esperandieu P, Mani JC, Pugniere M, Roquet F, Fereres A, Blanc S. 2002. Intracellular distribution of viral gene products regulates a complex mechanism of cauliflower mosaic virus acquisition by its aphid vector. Proc Natl Acad Sci U S A 99:2422–2427. doi: 10.1073/pnas.042587799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froissart R, Michalakis Y, Blanc S. 2002. Helper component-transcomplementation in the vector transmission of plant viruses. Phytopathology 92:576–579. doi: 10.1094/PHYTO.2002.92.6.576. [DOI] [PubMed] [Google Scholar]
- 11.Uzest M, Gargani D, Drucker M, Hebrard E, Garzo E, Candresse T, Fereres A, Blanc S. 2007. A protein key to plant virus transmission at the tip of the insect vector stylet. Proc Natl Acad Sci U S A 104:17959–17964. doi: 10.1073/pnas.0706608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzest M, Gargani D, Dombrovsky A, Cazevieille C, Cot D, Blanc S. 2010. The “acrostyle”: a newly described anatomical structure in aphid stylets. Arthropod Struct Dev 39:221–229. doi: 10.1016/j.asd.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Martelli GP, Abou Ghanem-Sabanadzovic N, Agranovsky AA, Al Rwahnih M, Dolja VV, Dovas CI, Fuchs M, Gugerli P, Hu JS, Jelkmann W, Katis N. 2012. Taxonomic revision of the family Closteroviridae with special reference to the grapevine leafroll-associated members of the genus Ampelovirus and the putative species unassigned to the family. J Plant Pathol 94:7–19. [Google Scholar]
- 14.Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L. 2008. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol 9:251–268. doi: 10.1111/j.1364-3703.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocha-Peña MA, Lee RF, Lastra R, Niblett CL, OchoaCorona F, Garnsey SM, Yokomi RK. 1995. Citrus tristeza virus and its aphid vector Toxoptera citricida, threats to citrus production in the Caribbean and central and North America. Plant Dis 79:437–445. doi: 10.1094/PD-79-0437. [DOI] [Google Scholar]
- 16.Atta S, Zhou C, Zhou Y, Cao M, Wang XF. 2012. Distribution and research advances of Citrus tristeza virus. J Integr Agr 11:346–358. doi: 10.1016/S2095-3119(12)60019-7. [DOI] [Google Scholar]
- 17.Cĕrni S, Rusčĭc J, Nolasco G, Gatin Z, Krajacĭć M, Šoric D. 2008. Stem pitting and seedling yellows symptoms of Citrus tristeza virus infection may be determined by minor sequence variants. Virus Genes 36:241–249. doi: 10.1007/s11262-007-0183-z. [DOI] [PubMed] [Google Scholar]
- 18.Roistacher CN, Bar-Joseph M. 1987. Transmission of citrus tristeza virus (CTV) by Aphis gossypii and by graft inoculation to and from Passiflora species. Phytophylactica 19:179–182. [Google Scholar]
- 19.Biswas KK, Tarafdar A, Diwedi S, Lee RF. 2012. Distribution, genetic diversity and recombination analysis of citrus tristeza virus of India. Virus Genes 45:139–148. doi: 10.1007/s11262-012-0748-3. [DOI] [PubMed] [Google Scholar]
- 20.Yokomi RK, Lastra R, Stoetzel MB, Damsteegt VD, Lee RF, Garnsey SM, Gottwald TR, Rocha-Peña MA, Niblett CL. 1994. Establishment of the brown citrus aphid (Homoptera, Aphididae) in Central-America and the Caribbean Basin and transmission of citrus tristeza virus. J Econ Entomol 87:1078–1085. doi: 10.1093/jee/87.4.1078. [DOI] [Google Scholar]
- 21.Bar-Joseph M, Marcus R, Lee RF. 1989. The continuous challenge of citrus tristeza virus control. Annu Rev Phytopathol 27:291–316. doi: 10.1146/annurev.py.27.090189.001451. [DOI] [Google Scholar]
- 22.Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. 1991. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 218:365–373. doi: 10.1016/0022-2836(91)90718-L. [DOI] [PubMed] [Google Scholar]
- 23.Hood EE, Gelvin SB, Melchers S, Hoekema A. 1993. New Agrobacterium helper plasmids for gene transfer to plants (EHA105). Trans Res 2:208–218. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- 24.Gowda S, Satyanarayana T, Robertson CJ, Garnsey SM, Dawson WO. 2005. Infection of citrus plants with virions generated in Nicotiana benthamiana plants agroinfiltrated with a binary vector based citrus tristeza virus, p 23–33. Abstr 16th Conf Int Org Citrus Virologists, International Organization of Citrus Virologists, Riverside, CA. [Google Scholar]
- 25.English J, Davenport G, Elmayan T, Vaucheret H, Baulcombe D. 1997. Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J 12:597–603. doi: 10.1046/j.1365-313X.1997.d01-13.x. [DOI] [Google Scholar]
- 26.Ng JC. 2013. A quantum dot-immunofluorescent labeling method to investigate the interactions between a crinivirus and its whitefly vector. Front Microbiol 4:77. doi: 10.3389/fmicb.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Calvino L, Goytia E, López-Abella D, Giner A, Urizarna M, Vilaplana L, López-Moya JJ. 2010. The helper-component protease transmission factor of tobacco etch potyvirus binds specifically to an aphid ribosomal protein homologous to the laminin receptor precursor. J Gen Virol 91:2862–2873. doi: 10.1099/vir.0.022335-0. [DOI] [PubMed] [Google Scholar]
- 28.Blanc S, Ammar ED, Garcia-Lampasona S, Dolja VV, Llave C, Baker J, Pirone TP. 1998. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J Gen Virol 79:3119–3122. doi: 10.1099/0022-1317-79-12-3119. [DOI] [PubMed] [Google Scholar]
- 29.Stewart LR, Medina Tian VT, Turina M, Falk BW, Ng JC. 2010. A mutation in the Lettuce infectious yellows virus minor coat protein disrupts whitefly transmission but not in planta systemic movement. J Virol 84:12165–12173. doi: 10.1128/JVI.01192-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang RY, Ammar ED, Thornbury DW, Lopez-Moya JJ, Pirone TP. 1996. Loss of potyvirus transmissibility and helper-component activity correlate with non-retention of virions in aphid stylets. J Gen Virol 77:861–867. doi: 10.1099/0022-1317-77-5-861. [DOI] [PubMed] [Google Scholar]
- 31.Purcell AH, Finlay A. 1979. Evidence for non-circulative transmission of Pierce's disease bacterium by sharpshooter leafhoppers. Phytopathology 69(4):393–395. doi: 10.1094/Phyto-69-393. [DOI] [Google Scholar]
- 32.Killiny N, Almeida RPP. 2009. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl Environ Microbiol 75:521–528. doi: 10.1128/AEM.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolja VV, Kreuze JF, Valkonen JPT. 2006. Comparative and functional genomics of closteroviruses. Virus Res 117:38–51. doi: 10.1016/j.virusres.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satyanarayana T, Gowda S, Ayllón MA, Dawson WO. 2004. Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc Natl Acad Sci U S A 101:799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper SJ, Cowell SJ, Dawson WO. 2015. With a little help from my friends: complementation as a survival strategy for viruses in a long-lived host system. Virology 478:123–128. doi: 10.1016/j.virol.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 36.Febres VJ, Ashoulin L, Mawassi M, Frank A, Bar-Joseph M, Manjunath KL, Lee RF, Niblett CL. 1996. The p27 protein is present at one end of citrus tristeza virus particles. Phytopathology 86:1331–1335. [Google Scholar]
- 37.Alzhanova DV, Prokhnevsky AI, Peremyslov VV, Dolja VV. 2007. Virion tails of beet yellows virus: coordinated assembly by three structural proteins. Virology 359:220–226. doi: 10.1016/j.virol.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]