Highlight
The mitochondrial pentatricopeptide repeat protein AtGRS1 edits RNA at four sites and is critical for development. The abscisic acid response gene ABI5 participates in the short root phenotype of grs1.
Key words: ABI5, mitochondria, pentatricopeptide repeat proteins, RNA editing, root.
Abstract
Most pentatricopeptide repeat (PPR) proteins are involved in organelle post-transcriptional processes, including RNA editing. The PPR proteins include the PLS subfamily, containing characteristic triplets of P, L, and S motifs; however, their editing mechanisms and roles in developmental processes are not fully understood. In this study, we isolated the Arabidopsis thaliana Growing slowly 1 (AtGRS1) gene and showed that it functions in RNA editing and plant development. Arabidopsis null mutants of grs1 exhibit slow growth and sterility. Further analysis showed that cell division activity was reduced dramatically in the roots of grs1 plants. We determined that GRS1 is a nuclear-encoded mitochondria-localized PPR protein, and is a member of the PLS subfamily. GRS1 is responsible for the RNA editing at four specific sites of four mitochondrial mRNAs: nad1-265, nad4L-55, nad6-103, and rps4-377. The first three of these mRNAs encode for the subunits of complex I of the electron transport chain in mitochondria. Thus, the activity of complex I is strongly reduced in grs1. Changes in RPS4 editing in grs1 plants affect mitochondrial ribosome biogenesis. Expression of the alternative respiratory pathway and the abscisic acid response gene ABI5 were up-regulated in grs1 mutant plants. Genetic analysis revealed that ABI5 is involved in the short root phenotype of grs1. Taken together, our results indicate that AtGRS1 regulates plant development by controlling RNA editing in Arabidopsis.
Introduction
Pentatricopeptide repeat (PPR) proteins are a class of RNA binding proteins characterized by the presence of a degenerate 35-amino-acid repeat, the PPR motif, which is arranged in tandem 2–50 times (Small and Peeters, 2000). The PPR motif (P motif) has another two variants, namely the S (short) motif with a length of 31 amino acids and the L (long) motif with a length of 35–36 amino acids. Based on their motifs, PPR proteins are divided into two subfamilies: the P subfamily has only P motifs, and the PLS subfamily contains characteristic triplets of P, L, and S motifs. Most members of the PLS subfamily contain extra conserved domains at their C-terminus, and these are designated the E, E+, and DYW domains (Lurin et al., 2004; Cheng et al., 2016).
PPR proteins are involved in many aspects of RNA processing in mitochondria and chloroplasts, including RNA cleavage, splicing, editing, and translation, and play crucial roles in plant developmental processes and responses to environmental stresses (Andrés et al., 2007; Zehrmann et al., 2009; Liu et al., 2010; Murayama et al., 2012; Zhu et al., 2012; Haili et al., 2013; Mei et al., 2014; Yang et al., 2014; Hsieh et al., 2015). RNA editing is an important step in the post-transcriptional control of organelle gene expression. Most RNA editing in plants results in the conversion of cytidine (C) to uridine (U) (Covello and Gray, 1989; Gualberto et al., 1989; Hiesel et al., 1989; Shikanai, 2006; Chateigner-Boutin and Small, 2010). In the mitochondria of Arabidopsis, approximately 500 C-to-U editing sites had been uncovered (Giegé and Brennicke, 1999; Bentolila et al., 2005, 2008). The mechanism of the editing reaction puzzled researchers for many years, until the first PPR protein, CHLORORESPIRATORY REDUCTION 4, was found to be involved in chloroplast RNA editing (Kotera et al., 2005). Since then, PPR proteins have been found to be involved in RNA editing and all the discovered trans-factors involved in RNA editing in plants belong to the PLS subfamily (Takenaka et al., 2013; Shikanai, 2015). Although several PPR proteins target individual sites, some are found to recognize more than two and even as many as eight sites (Kim et al., 2009; Zehrmann et al., 2009, 2012; Zhu et al., 2012; Glass et al., 2015). Although recently bioinformatics, biochemical, and structural analyses have shown that PPR proteins recognize RNA in one-motif to one-nucleotide binding mode (Yagi et al., 2013; Yin et al., 2013; Barkan and Small, 2014), the mechanism of how a single PPR protein recognizes multiple target sequences still needs further investigation.
Mutations in many RNA-editing PPR proteins do not result in any evident developmental defect (Zehrmann et al., 2009; Verbitskiy et al., 2010; Härtel et al., 2013), although some PPRs are important in development (Yu et al., 2009; Koprivova et al., 2010; Liu et al., 2010; Murayama et al., 2012; Haili et al., 2013; Yang et al., 2014). The relationship between mutant phenotype and RNA editing has not received much attention until recently. Mutations in PPR proteins involved in chloroplast RNA editing have been shown to impair chloroplast biogenesis (Yu et al., 2009). Several reports have shown that an increase in reactive oxygen species (ROS) is responsible for the developmental defects observed in the mitochondrial RNA editing by those mutant PPRs (Liu et al., 2010; Yang et al., 2014). The nature of other signaling pathways linking PPRs involved in mitochondrial RNA editing and plant development remains largely unknown.
In this study, we analyzed the Arabidopsis T-DNA knockout mutant grs1-1, which displays a phenotype of slow growth and sterility. Genetic and molecular analysis indicates that the GRS1 gene encodes a PPR protein. Further studies showed that GRS1 is required for the RNA editing of four mitochondrial transcripts. The upstream sequences of these editing sites share some conserved nucleotides. The lack of RNA editing at these sites leads to reduced levels of functional mitochondrial complex I and affects mitochondrial ribosome biogenesis. Abscisic acid (ABA) response gene ABI5 but not ROS is involved in the short root phenotype in grs1.
Materials and methods
Mutant library construction and selection of grs1-1
We generated an Arabidopsis mutant library with T-DNA encoding LAT52::EGFP, a cell-autonomous pollen-specific reporter (Twell et al., 1989; Sessions et al., 2002), and a hygromycin-resistance gene. T-DNA mutagenesis was carried out on qrt1 plants (Preuss et al., 1994), where mature pollen grains maintain male meiotic products in tetrads (Supplementary Fig. S1A, B at JXB online). Hygromycin-resistant plants, heterozygous for a single locus T-DNA insertion, produced tetrads with two mutant pollen grains emitting green fluorescent protein (GFP) fluorescence, and two wild-type grains that did not display any GFP activity (Supplementary Fig. S1C, D). This simplified the process of determining whether a T2 plant was heterozygous (tetrads are two GFP+ to two GFP−, HYG resistant), homozygous (all four tetrad members are GFP+, HYG resistant) (Supplementary Fig. S1E, F) or wild-type (all four tetrads members are GFP−) for a T-DNA induced mutation.
For grs1-1 selection, T1 seeds were obtained by self-pollination of hygromycin-resistant grs1-1 plant and sown on 1/2 MS plates with hygromycin to select grs1-1 seedlings. Thirty-two hygromycin-resistant seedlings were grown on soil and the pollen grains of each plant were visualized under a fluorescence microscope to determining whether a T2 plant was heterozygotes, homozygotes, or wild-type. T1 seeds were sown on 1/2 MS plates for germination.
Plant materials and growth conditions
Arabadopsis thaliana qrt1 (Preuss et al., 1994) was used as a wild-type strain. The grs1-1 allele was isolated from our mutant library with hygromycin resistance (Wu et al., 2012, Supplementary data). The grs1-2 (CS428796) and gin1-3 lines were obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio, USA). The mutant abi5-1 (Liu et al., 2012) was provided by Dr Lei Zhang (College of Life Sciences, Wuhan University). The transgenic line pCyclinB1;1:Dbox-GUS (Colon-Carmona et al., 1999) was provided by Dr Jian Xu (Temasek Life Sciences Laboratory, Singapore). Seeds were surface-sterilized with 20% bleach for 10min, and washed three times with sterile distilled water. Seeds were stratified for 3 d at 4 °C and then sown on 1/2 MS plates with 1.0% (w/v) sucrose. To decrease the ROS level in seedlings, diphenyleneiodonium (DPI, 100 μM, Sigma) or reduced glutathione (GSH, 300 μM, Sigma) was added to the culture media. Agar plates were placed in a growth room with a photoperiod of 16h light/8h dark. For kanamycin selection, 50mg l–1 of kanamycin (Sigma) was supplemented to the media. Similarly, 50mg l–1 of hygromycin (Roche) and 10mg l–1L of sulfadiazin (Sigma) were added for hygromycin selection and sulfadiazin selection, respectively. Plants were grown on soil in a greenhouse under long-day conditions (16h light/8h dark) at 22 °C.
Cloning of the T-DNA flanking sequence and characterization of the grs1-1 and grs1-2 alleles
The T-DNA flanking sequence in the grs1-1 mutant was cloned by TAIL-PCR (Liu et al., 1995). The authenticity of the cloned sequence was confirmed by PCR using two pairs primers located around the T-DNA left border (GRS1-T1, TGGAACAAGTTCATCACGGTTTC; LB-S, CCAAAATCCAGTACTAAAATCCAG) and right border (GRS1-T2, ATTCATGGTTTGTGCATAAAAAGAG; RB-S, CGCGCGGTGTCATCTATG). For the grs1-2 allele, the T-DNA site was confirmed by PCR using the following primers: GRS1-RP, GTGAAAATGGGAGCAAAAGTG; and LB3, TAGCATCTGAATTTCATAACCAATCTCGATACAC.
Vector construction and plant transformation
Plasmids P092, P093, and P094 were produced as described previously (Wu et al., 2012; Yan et al., 2016). To generate the pGRS1::GRS1 complementation construct, a 3876-bp wild-type genomic sequence containing the AT4G32430 gene, 1078-bp upstream of the ATG codon and 506-bp downstream of the TAG codon sequences, was PCR-amplified (primers: GRS1-F1, NNNNGGTACCTGATGTTTTGGGAGCGACTTC; and GRS1-R1, NNNNCTCGAGACCAAACTCATACCTTAAAGCCATC) from genomic DNA and was then cloned into the P092 plasmid with T-DNA encoding pLAT52::DsRED and a kanamycin-resistance gene (Supplementary Fig. S2C). To examine the subcellular location of GRS1, we amplified and cloned the 35S promoters into P094 to generate the 35S::EGFP construct. Then the GRS1 ORF was amplified (primers: GRS1-CDS1, NNNNGGTACCATGACCCTTCTGAACTATCTACACTGT; and GRS1-CDS2, NNNNCTCGAGAACTGCAACTTTCCCC TCCAAATTCATC) from genomic DNA and cloned into the 35S::EGFP plasmid to generate a 35S::GRS1-EGFP construct. To produce the mitochondrial marker line, we amplified the TagRFP-T (Shibata et al., 2010) and put it under the control of 35S to generate 35S::RFP. Then we amplified and cloned the 129-bp DNA fragment containing the mitochondria-targeted pre-sequence of the located F1-ATPase gene At5g13450 (Robison et al., 2009) (using primers MITO-1, NNNNGGTACCGCCACCATGGCTAATCGTTTCAGATCAGG; and MITO-2, NNNNCTGCAGTGTTTGAGCAGAAGCA GTTGCATAAG) into 35S::RFP to generate the 35S::Mito-RFP construct. To investigate the expression pattern of GRS1, the GRS1 promoter was amplified (primers: GRS1-F1 as above, and GRS1-R2: NNNNCTCGAGAGAAGCAAACTAGTCGGATTCTAATTC) and put upstream of GUS (β-glucuronidase) in P093 to generate pGRS1::GUS. All the gene constructs were transferred into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants by the floral dip method (Clough and Bent, 1998).
Genotype analysis of the genomic complemented lines
To identify the genotype of the genomic complemented lines, the DNA of these plants was extracted and PCR analysis was conducted using three pairs of primers (S1+A1, S2+A1, S1+A2) (Supplementary Fig. S2B, C): Primer S1, CATCTGTAGGCAACAGTTTCATCAC located upstream of the T-DNA insertion site; Primer S2, CCAAAATCCAGTACTAAAATCCAG located around the T-DNA left border; Primer A1, CTCTTCTCTCGCTTTTTAAGTTGC located downstream of the AT4G32430 gene and beyond the genomic fragment used for complementation; and Primer A2, TGACTTAGTTGATTTGGAGGGTG located downstream of the genomic fragment used for complementation.
Histochemical analysis of GUS activity
For pCYCB1;1:Dbox-GUS staining, we crossed the pCYCB1;1:Dbox-GUS stable lines with grs1-1 mutant plants. F2 seeds were obtained by self-pollination of F1 and sown on 1/2 MS plates with hygromycin to select seedlings with the grs1-1 background. Individual F3 seeds were obtained by self-pollination of these seedlings and sown on 1/2 MS plates for germination. GUS activity analysis was performed with 8-d-old seedlings (with normal roots and short roots), and the lines with all normal roots with GUS activity were regarded as homozygous for pCYCB1;1:Dbox-GUS. The seedlings with short roots were regarded as homozygoous for both pCYCB1;1:Dbox-GUS and grs1-1.
The histochemical analysis of GUS activity was performed according to Vielle-Calzada et al. (2000). Plant tissues were incubated at 37 °C in GUS-staining solution [2mM 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) in 50mM sodium phosphate buffer, pH 7.0] containing 0.1% Triton X-100, 2mM K4Fe(CN)6 and 2mM K3Fe(CN)6. The stained tissues were then transferred to 70% (v/v) ethanol solution. Samples were mounted with traditional clearing solution and placed under a microscope (Olympus) fitted with differential interference contrast optics for imaging.
Analysis of subcellular localization of GRS1
The iPSORT Prediction program (Bannai et al., 2002) predicted that GRS1 is targeted to the mitochondria. To confirm its mitochondrial localization, transgenic plants containing the 35S::GRS1-EGFP construct were crossed with a transgenic mitochondrial marker line expressing 35S::mito-RFP. The petal cells of the F1 progeny were visualized using a FV1000 confocal laser-scanning microscope (CLSM; Olympus). GFP fluorescence was detected with excitation at 488nm and emission at 510–530nm; red fluorescent protein (RFP) fluorescence was detected with excitation at 568nm and emission at 590–620nm.
Analysis of RNA editing
The status of Arabidopsis mitochondrial RNA editing in grs1 plants was examined as described by Zehrmann et al. (2008). Total RNA was extracted from 20-d-old grs1 and wild-type seedlings. Complementary DNA fragments of all mitochondrial transcripts containing RNA editing sites were amplified by RT-PCR. The primers used in this experiment are given in Supplementary Table S3. The amplified PCR products were directly sequenced and the results were compared to the corresponding DNA sequence for each transcript.
Phenotypic characterization
For the determination of the root meristem size, root tips were excised from seedlings 8 d after germination, and examined with a differential interference contrast (DIC) microscope (Olympus).
Measurement of ROS in roots
For nitrobluetetrazolium (NBT) staining to detect superoxides, seedlings were incubated in a reaction buffer containing 1mM NBT (Sigma-Aldrich) and 20mM K-phosphate at pH 6.0 for 20min. The seedlings stained by NBT were washed three times with water and then transferred to acetic acid:ethanol (1:3, v/v) solution. To enable 3, 3- diaminobenzidine (DAB) staining to detect H2O2, the seedlings were incubated in 0.3mg ml–1 DAB (Sigma-Aldrich) dissolved in 50mM Tris-HCl (pH 5.0) for 12h. The seedlings stained by DAB were washed three times with water, and were then examined in 10% glycerol with an Olympus microscope.
Quantitative RT-PCR
Total RNAs of seeds before germination and 7-d-old seedlings were extracted using the RNAqueous® Phenol-free total RNA Isolation kit (Ambion)according to the manufacturer’s protocol. After digestion with RNase-free DNase I (Promega), the first strand of cDNA was synthesized using oligo-dT and M-MLV reverse transcriptase (Invitrogen). Quantitative PCR analysis was performed using FastStart Essential DNA Green Master (Roche) on a CFX ConnectTM Real-Time System (BioRad). Each experiment was repeated three times and samples were normalized using UBQ10 expression. Data acquisition and analyses used Bio-Rad CFX Manager software; the relative expression levels were measured using the 2(–∆∆Ct) analysis method and the error bars in the figures represent the variance of three replicates. The genes and the primers used for detection of the mRNA expression are listed in Supplementary Table S4.
Detection of enzyme activity of complex I
Analysis of the NADP dehydrogenase activity of mitochondrion complex I was performed according to Wu et al. (015). Proteins of crude organelle extract from young seedlings were solubilized with 1% (v/v) digitonin and resolved by Blue Native-PAGE. After PAGE, the NADH dehydrogenase activity of complex I was visualized by incubation of the gel in the presence of 1mM nitroblue tetrazolium (NBT) and 0.2mM NADH in 0.05M MOPS (pH 7.6).
Results
GRS1 plays an essential role in vegetative and reproductive development
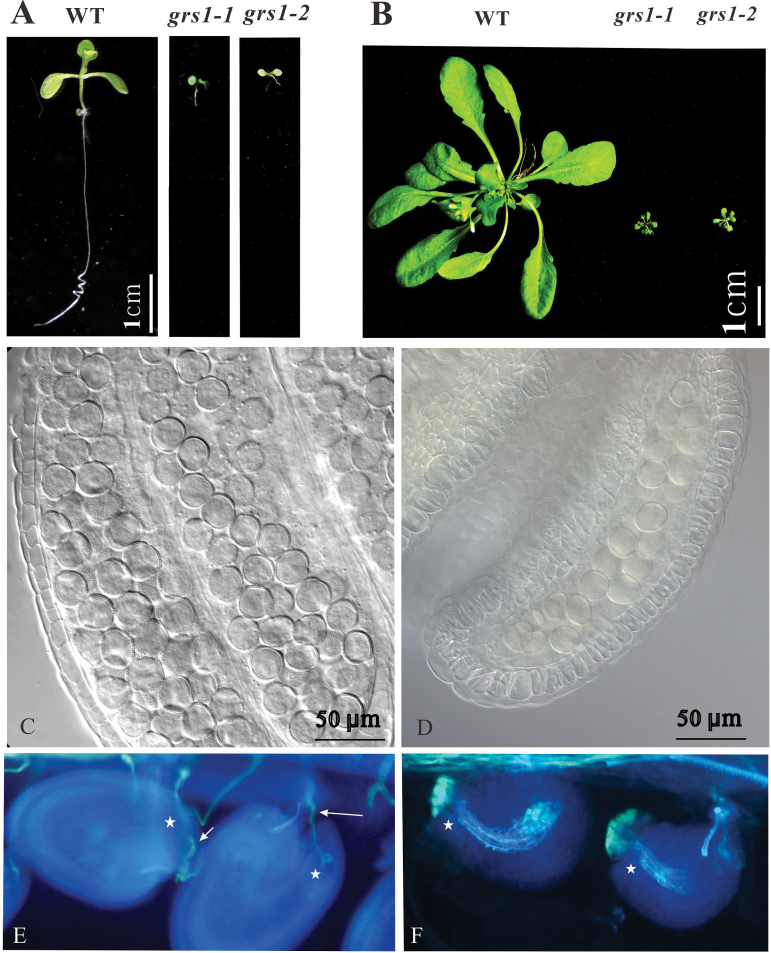
We generated an Arabidopsis mutant library to simplify the process of screening mutants whose homozygotes were lethal or exhibited growth retardation (Supplementary Fig. S1). One mutant displaying an extremely slow growth phenotype was isolated and named growing slowly1 (grs1-1). When we analyzed the effect of grs1-1 on plant development, we found that grs1-1/+ heterozygous plants had no visible morphological abnormalities in vegetative and reproductive organs compared with wild-type plants. grs1-1 homozygous plants, however, exhibited multiple phenotypes as shown in Fig. 1. Thirty-two hygromycin-resistant T2 plant were heterozygous grs1-1, suggesting that the grs1-1 homozygotes are either lethal or exhibited growth retardation. T1 seeds of grs1-1 were sown on 1/2 MS plates for germination and about 25% of 11-d-old seedlings showed an extremely slow growth phenotype (Supplementary Fig. S2A). The DNA of these slow-growth seedlings was extracted and PCR analysis confirmed that they were homozygous for grs1-1 (Supplementary Fig. S2D). grs1-1 homozygous seedlings only survived on MS medium plates, and their vegetative growth was strongly affected (Fig. 1A, B). Opening the siliques of grs1-1 two days after flowering revealed the absence of developed seeds. To determine which parent was responsible for the aborted phenotype, we performed reciprocal crosses of grs1-1 and wild-type plants. Both females and males were found to be sterile in grsl-1 mutant plants. Further analysis showed that the number of pollen grains in grs1-1 was much lower than in the wild-type; female gametophyte development in grs1-1 was also found to be retarded and did not appear to be able to attract wild-type pollen tubes into the ovules (Fig. 1C–F).
Fig. 1.
Several developmental processes are impaired in grs1. (A) Root growth of 8-d-old seedlings in wild-type (WT), grs1-1, and grs1-2 plants. (B) Appearance of 35-d-old plants in wild-type, grs1-1, and grs1-2. (C, D) Anther of wild-type (C) and grs1-1 (D) plants. The amount of pollen grains in grs1-1 is much lower compared to the wild-type. (E, F) Aniline blue staining of pollen tube guidance in ovules. Ovules attract pollen tubes (indicated by arrows) in the wild-type (E), but no pollen tubes are observed in the ovules of grs1-1 homozygous plants (F). The stars indicate the micropylar end of the ovules.
Cell division is impaired in grs1-1
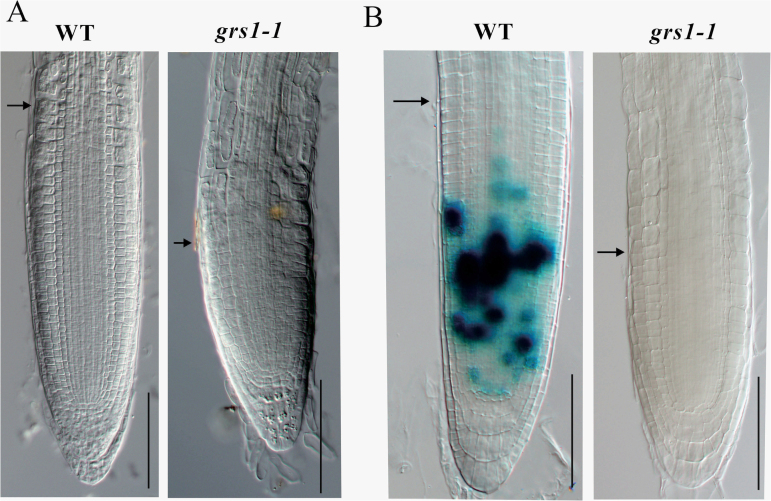
After germination, the growth rate of the primary root was dramatically reduced in grs1-1 plants compared to the wild-type. To determine the cellular basis for the observed defects in the root development of grs1-1 plants, we examined the size of the root meristem in seedlings 8 d after germination. It was observed that the size of root meristem in grs1-1 was much shorter than that of the wild-type (Fig. 2A). To further substantiate the role of GRS1 in controlling root cell division, we crossed pCyclin B1;1:Dbox-GUS stable lines (Colon-Carmona et al., 1999) with grs1-1 mutant plants. The pCyclin B1;1:Dbox-GUS reporter allows the visualization of cells at the G2-M phase of the cell cycle, and thus to monitor mitotic activity in the root meristem (Colon-Carmona et al., 1999). In contrast to the wild-type, we found that there was almost no GUS signal in grs1-1 roots (Fig. 2B). The results indicate that the number of dividing cells was reduced dramatically in grs1-1compared to wild-type roots.
Fig. 2.
The activity of the root meristem division is reduced in grs1-1. (A) The root meristematic zone of 8-d-old wild-type (WT) plants is much longer than that of grs1-1 plants. (B) Expression of pCyclinB1;1:Dbox-GUS in the meristematic zone of 8-d-old WT and grs1-1 seedlings. Arrows indicate the boundary between the root meristematic and elongation zone. Scale bars = 100 µm.
Molecular characterization of grs1-1
Arabidopsis grs1-1 plants were generated by T-DNA insertion with resistance to hygromycin. All the grs1-1/+ heterozygous plants were resistant to hygromycin, suggesting that the mutant phenotype was caused by a T-DNA insertion. We cloned the T-DNA flanking sequence by using the thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) technique (Liu et al., 1995). The grs1-1 mutant was shown to carry a T-DNA insertion in the gene AT4G32430 located 1325bp downstream of the ATG start codon (Fig. 3A, Supplementary Fig. S2B). Another allele containing a T-DNA insertion in the GRS1 gene, CS428796, was obtained from the Arabidopsis Biological Resource Center. We verified that the CS428796 mutant carries a T-DNA insertion in the AT4G32430 gene at 850bp downstream of the ATG start codon (Fig. 3A). We then renamed the CS428796 allele grs1-2. Homozygous grs1-2 plants were found to phenocopy grs1-1 homozygous plants (Fig. 1A, B).
Fig. 3.
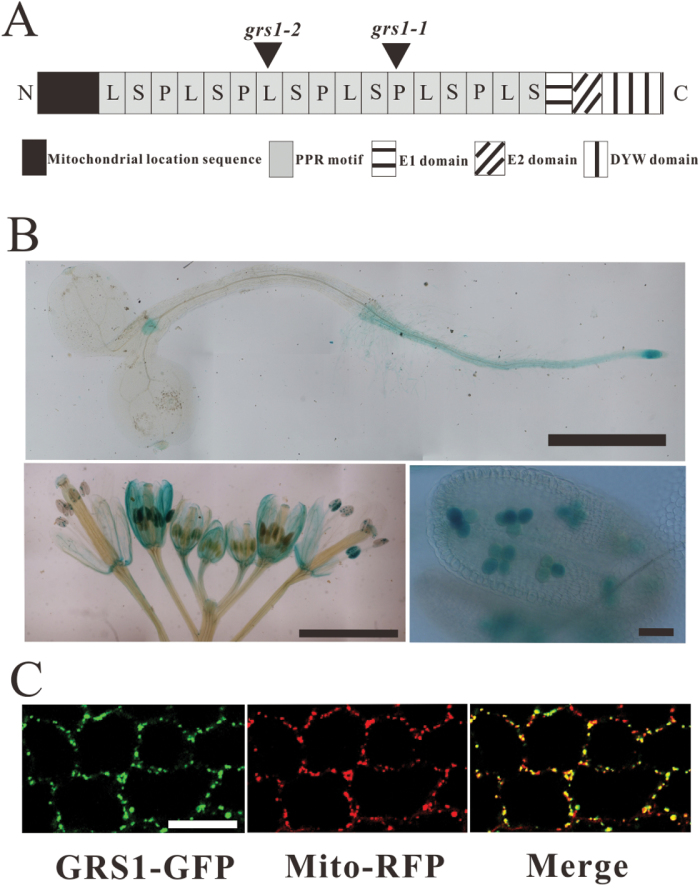
Structural features, expression patterns, and subcellular localization of GRS1. (A) Diagram showing the relative position of the T-DNA insertion in the GRS1 gene and the structural features of the GRS1 protein. Various protein domains are indicated below the diagram. (B) GUS expression patterns in different plant parts of transgenic proGRS1::GUS lines. Top row: 7-d-old seedling. GUS signal is observed in root and shoot meristems. Scale bar = 5mm. Bottom row, left: an inflorescence, scale bar = 3mm. right: an anther with pollen grains, scale bar = 50 µm. (C) Localization of GRS1-GFP protein in the mitochondria. Petal cells of plant co-expressing GRS1-GFP and the mitochondrial marker Mito-RFP were examined with confocal laser scanning microscopy. From left to right: green fluorescent signal from GRS1-GFP; red fluorescent signal from the mitochondrial marker Mito-RFP; merged picture with green and red signals showing co-localization. Scale bar = 20µm.
To confirm that the grs1-1 mutant phenotypes were indeed caused by knockout of the AT4G32430 gene, we performed a complementation test with the genomic sequence of AT4G32430. Fifty-nine T1 transgenic plants were screened on double-resistance plates with hygromycin and kanamycin (for the transformed genomic sequence). Among them, eleven plants were homozygous for grs1-1. All these grs1-1 homozygous plants carrying the fragments of the exogenous genomic sequence (resistance to kanamycin) showed no obvious differences compared to the wild-type, and were named the genomic complemented lines (homozygous for grs1-1, heterozygous for exogenous genomic fragment) (Supplementary Fig. S2A). Genotype analysis confirmed the genomic complemented lines contained both the mutated grs1-1 version and expression of the wild-type version (Supplementary Fig. S2D). These results indicate that the AT4G32430 gene can successfully complement the grs1-1 phenotype. The AT4G32430 gene was therefore renamed as GRS1.
GRS1 encodes a mitochondria-targeted pentatricopeptide repeat protein
To investigate the expression pattern of GRS1, we fused the GRS1 promoter sequence to a GUS reporter gene, and transformed this construct into the wild-type. In seedlings, GRS1::GUS was preferentially expressed in the meristematic region of both roots and stems. In flowers, GUS activity was detected in the sepal, stigma, stamen, and pollen grains (Fig. 3B).
BLAST analysis identified GRS1 as a member of the PPR family, more specifically belonging to the PLS subfamily. Thus, GRS1 encodes a PLS-type pentatricopeptide repeat protein, as proposed by Lurin et al. (2004). It consists of six PPR-like S, six PPR-like L, and five P motifs with E1, E2, and DYW C-terminal extensions (Lurin et al., 2004; Barkan and Small, 2014; Cheng et al., 2016) (Fig. 3A, Supplementary Fig. S3, and Table S1). The iPSORT Prediction program (Bannai et al., 2002) predicted that GRS1 is targeted to mitochondria and, indeed, GRS1-GFP was found to co-localize with the mitochondria-localized Mito-RFP (Fig. 3C), indicating that GRS1 is a nuclear-encoded mitochondrial protein.
GRS1 is required for mitochondrial RNA editing
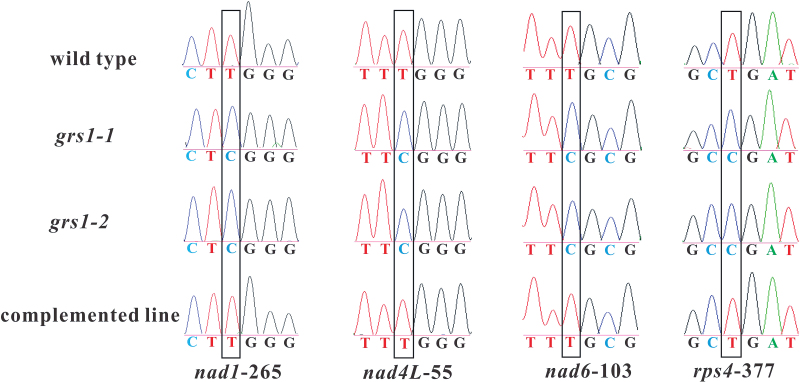
Since GRS1 encodes a DYW-type PPR protein, we tested its involvement in mitochondrial RNA editing. We identified several unedited sites in the mitochondrial RNA in the grs1-1 mutants. Our results revealed that C-to-U editing at the positions of nad1-265, nad4L-55, nad6-103, and rps4-377 was specifically blocked in the grs1-1 plants. Editing of these four sites is also inhibited in grs1-2 mutants (Fig. 4). The C-to-U editing in the nad1 mRNA results in an arginine-to-tryptophane amino acid change (R89W) in the NAD1 protein. The C-to-U editing in the nad4L mRNA results in an arginine-to-tryptophane amino acid change (R19W) in the NAD4L protein. The C-to-U editing in the nad6 mRNA results in an arginine-to-cystine amino acid change (R35C) in the NAD6 protein. The C-to-U editing in the rps4 mRNA results in a proline-to-leucine amino acid change (P126L) in the RPS4 protein. Editing of the four mRNAs at these four editing sites was highly efficient in the wild-type, as shown by the detection of a single peak equivalent to the T nucleotide at these positions, whereas editing of these sites was totally abolished in grs1-1 and grs1-2 mutants (Fig. 4). Editing deficiencies of the mutant alleles were restored in the grs1-1 complemented lines (Fig. 4). These results confirmed that mutation in the GRS1 gene was responsible for the defect of mitochondrial RNA editing in the grs1-1 mutants.
Fig. 4.
GRS1 is responsible for RNA editing of four sites in Arabidopsis mitochondria. Wild-type plants show that RNA editing of the mitochondrial editing sites nad1-265, nad4L-55, nad6-103, and rps4-377 is efficient, while in grs1-1 and grs1-2 these sites are not edited. Editing deficiencies were restored in the grs1-1 complemented lines.
Complex I function and mitoribosomal biogenesis are impaired in grs1-1 mutants
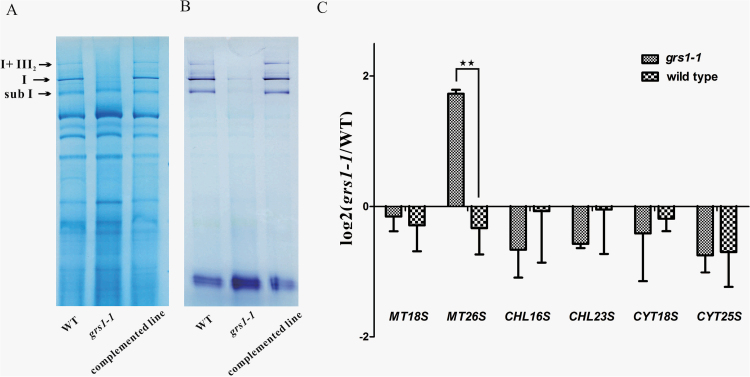
The proteins NAD1, NAD4L, and NAD6 are components of the mitochondrial electron transport chain complex I (NADH dehydrogenase). Having observed that RNA editing of these genes was altered in grs1-1 mutants and resulted in amino acid changes, we hypothesized that RNA editing defects of these transcripts may lead to complex I malfunction in grs1-1 mutants. To test this hypothesis, we isolated crude mitochondria from seedlings of wild-type, grs1-1 mutants, and grs1-1 complemented lines. Separation of mitochondrial complexes by blue-native PAGE and NADH dehydrogenase activity staining showed that both protein levels and activity of complex I could barely be detected in grs1-1 mutants (Fig. 5A, B).
Fig. 5.
Complex I activity and mitoribosomal biogenesis are affected in grs1-1 mutants. (A) Proteins of crude organelle extractions from young seedlings of wild-type (WT), grs1-1, and grs1-1 complemented lines were stained by Coomassie blue. (B) In-gel assay of NADH dehydrogenase activity in WT, grs1-1, and grs1-1 complemented lines. Activity of complex I could hardly be detected in grs1-1. The activity staining bands on the lower part of the gel correspond to the activity of the dehydrolipoamide dehydrogenase, which can serve as a loading control. I+ III2, mitochondrial complex I and complex III super-complex; I, mitochondrial complex I; sub I, mitochondrial sub-complex I. (C) Accumulation of rRNAs as a proxy for corresponding ribosomal subunits in grs1-1 compared with wild-type plants. Levels of rRNA transcripts of large subunits and small subunits in mitochondrial, chloroplast, and cytosolic ribosomes are shown. The values obtained were averaged for three biological replicates, with error bars representing SD. Statistically significant differences between grs1-1 and the wild-type are indicated: **P<0.01 (Student’s t-test).
Since the RPS4 protein is a component of the small subunit (SSU) of the mitoribosome, we tested whether the change in RPS4 editing in the grs1 mutants affects mitochondrial ribosome biogenesis. As rRNAs are unstable when unassembled, rRNA levels can serve as a marker for the accumulation of ribosomal subunits (Walter et al., 2010; Kwasniak et al., 2013). We determined the abundance of mitochondrial (mt 18S and mt 26S), chloroplast (chl 16S and chl 23S) and cytosolic (cyt 18S and cyt 25S) rRNAs. The mt 18S showed no evident difference between grs1-1 and the wild-type, while a significant increase was observed for mt 26S rRNA in grs1-1 plants compared to the wild-type (Fig. 5C), with the increased ratio of mt 26S to mt 18S indicating an imbalance between mitoribosomal subunits. The chl 16S, chl 23S, cyt 18S, and cyt 25S showed no obvious differences between grs1-1 and the wild-type (Fig. 5C), suggesting the grs1 mutation only affects mitochondrial ribosome biogenesis.
An alternative respiratory pathway is activated in grs1-1 mutants
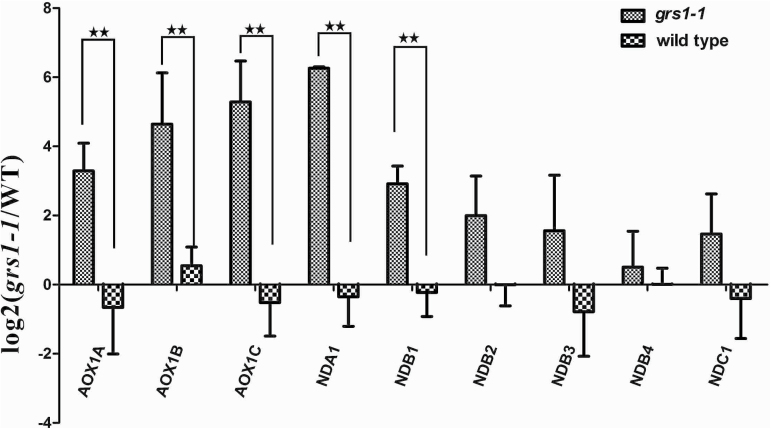
Lack of complex I activities is known to result in elevated levels of an alternative respiratory pathway in Arabidopsis (Yuan and Liu, 2012). The components of this alternative respiratory pathway include several alternative NAD(P)H dehydrogenases (NDs) and alternative oxidases (AOXs). To determine whether grs1-1 mutants had the same phenotype, we performed quantitative RT-PCR assays for the transcripts levels of six ND genes and three AOX genes in wild-type and grs1-1 plants. As shown in Fig. 6, the expression levels of the nine examined genes in grs1-1 increased significantly relative to the wild-type. These results indicate that the alternative respiratory pathway is activated in grs1-1. grs1-2 mutants had a similar phenotype with up-regulation of transcripts for alternative respiration compared with the wild-type. (Supplementary Fig. S4).
Fig. 6.
The alternative respiratory pathway is activated in grs1-1. The expression levels of alternative respiratory pathway genes in grs1-1 increased significantly relative to the wild-type. These genes include three alternative oxidases (AOXs) and six alternative NAD(P)H dehydrogenases (NDs). The values obtained were averaged for three independent experiments, with error bars representing SD. Statistically significant differences between grs1-1 and the wild-type are indicated: **P<0.01 (Student’s t-test;).
The grs1-1 mutant does not accumulate higher amounts of ROS than the wild-type
Reports have shown that impaired activity of the mitochondrial electron transport chain of complex I can cause a redox imbalance and increases in ROS accumulation, leading to the accumulation of more ROS in mutants than in the wild-type (Liu et al., 2010; Yang et al., 2014). We analyzed the ROS levels in grs1-1 mutants and wild-type plants and showed that grs1-1 mutants do not accumulate higher amounts of ROS than the wild-type (Fig. 7A, B). Consistent with these results, addition of the reducing agent glutathione (GSH) or diphenyleneiodonium chloride (DPI) was not able to complement the root growth defects of grs1-1 mutant plants (Fig. 7C).
Fig. 7.
grs1-1 mutants do not accumulate more ROS than the wild-type. (A) Nitroblue tetrazolium (NBT) staining for superoxide in primary root tips of wild-type and grs1-1 plants. (B) 3, 3-diaminobenzidine (DAB) staining for H2O2 in primary root tips of wild-type and grs1-1 plants. Scale bars = 100 µm. (C) Root meristem cell number in wild-type, grs1-1, and grs1-1 with addition of reducing agents glutathione (GSH) or diphenyleneiodonium chloride (DPI). The values obtained were averaged for n>20, with error bars representing SD. Statistically significant differences are indicated: **P<0.01 (Student’s t-test;).
abi5 partially rescues the post-germination growth arrest of grs1-1
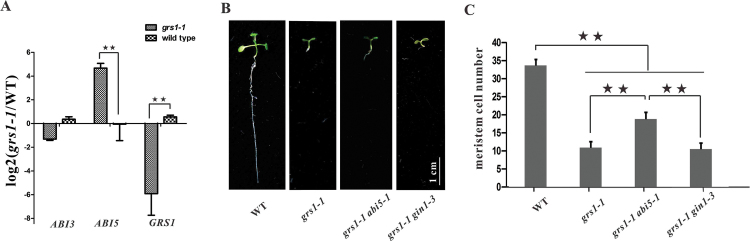
Since grs1 mutant display defects in seed germination and post-germination growth, it is possible that the ABA signaling pathway is activated in these mutants. Given that the transcription factors ABI3 and ABI5 are key proteins in the ABA signaling pathway (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001), expression of ABI3 and ABI5 was analyzed in grs1-1 mutant and wild-type seedling plants 8 d after germination. Expression of ABI5 was found to be significantly up-regulated in grs1-1 mutants, whereas expression levels of ABI3 were not significantly altered (Fig. 8A), implying that ABI5, but not ABI3, is activated in grs1-1 mutants and is involved in the short-root phenotype. To test this hypothesis, the grs1-1 abi5-1 double-mutant was generated, and it showed longer roots than those of the grs1-1 mutants (Fig. 8B, Supplementary Table S2). While only about 10 cells could be observed in the meristems of in grs1-1 mutants, approximately 20 cells were established in the meristem of grs1-1 abi5-1 double-mutant plants (Fig. 8C). These results indicate that abi5-1 partially rescues the post-germination growth arrest of the grs1-1 mutants.
Fig. 8.
The abi5-1 mutant partially rescues the post-germination growth arrest of grs1-1 mutants. (A) Relative expression of ABI3, ABI5, and GRS1 in wild-type and grs1-1 plants. (B) Root growth of 8-d-old seedlings of wild-type, grs1-1, and grs1-1 abi5-1 and grs1-1 gin1-3 double-mutants. Scale bar = 1cm. (C) Root meristem cell number in wild-type, grs1-1, and grs1-1 abi5-1 and grs1-1 gin1-3 double-mutants. The values obtained were averaged for n>20, with error bars representing SD. Statistically significant differences are indicated: **P<0.01 (Student’s t-test;).
We then tested whether a decrease in the ABA content in grs1-1 mutants can rescue the post-germination growth arrest of these plants. The gin1-3 mutant line is a knockout allele of the ABA2 gene, one of the key genes involved in ABA synthesis However, the grs1-1 gin1-3 double-mutant did not show any evident differences compared with the grs1-1 single-mutant plants in post-germination growth (Fig. 8B, C).
Discussion
Putative cis-acting elements recognized by GRS1
Recently bioinformatics, biochemical, and structural analyses have shown that PPR proteins recognize RNA in one-motif to one-nucleotide binding mode (Kim et al., 2009; Yagi et al., 2013; Yin et al., 2013; Barkan and Small, 2014). The major determinant is the amino acid at position 5 of the motif (Yagi et al., 2013; Yin et al., 2013; Barkan and Small, 2014; Cheng et al., 2016). The second major determinant is at position 2 of the motif and position 35 of the following motif (Yagi et al., 2013; Yin et al., 2013; Barkan and Small, 2014; Cheng et al., 2016). The site-specific RNA editing factors PPR and the RNA target sequences show optimal correlations when the PPR domains are aligned with the nucleotide sequences upstream of RNA editing sites up to the fourth nucleotide (nucleotide −4). The last S motif of GRS1 is accordingly positioned at the −4 nucleotides site of all the editing sites (Supplementary Fig. S3 and Table S1). In this way, the conserved A nucleotide at position −12 and G nucleotide at position −6 are consistent with the predictions of bioinformatics (Kim et al., 2009; Yagi et al., 2013; Yin et al., 2013; Barkan and Small, 2014).
Cis-elements located between 20 to 25 nucleotides upstream and one to three nucleotides downstream of the edited C are known to be important in the context of RNA editing in mitochondria and plastids (Zehrmann et al., 2009; Barkan and Small, 2014). When comparing the context of the four RNA sites edited by GRS1, five nucleotides are identical in addition to the edited C (Supplementary Fig. S3), suggesting that these positions are important for guiding editing through GRS1 in the mitochondria. These five nucleotides, however, are not sufficient to specify a unique site in the plant mitochondrial transcriptome. An in silico screen identified NAD4-403, another editing site with the same RNA context in the mitochondrial genome (Supplementary Fig. S3). NAD4-403 is edited normally in the wild-type and in the grs1 mutant, confirming that the five shared nucleotide positions are not sufficient to guide editing through GRS1. More information may be provided by other nucleotides inside the context of RNA editing of the four sites to ensure GRS1 specifically binds to them. It was reported that PPR proteins distinguish purines from pyrimidines much better than they distinguish between C/U or A/G (Yagi et al., 2014; Kindgren et al., 2015). The conservation between these four sequences is better than shown when this is taken into account, with several other nucleotide positions, such as −4, −7, −9, −14, and −15, showing expected matches to the protein sequence in addition to the ones that have been indicated. The correlations of the amino acid codes in GRS1 and the diversity of its targeted RNA bases can offer more information for predicting whether a PPR protein can bind a particular RNA.
Comparison of grs1-1 plants with other Arabidopsis complex I mutant lines
Loss of GRS1 directly affects the editing of three components of complex I: nad1-265, nad4L-55, and nad6-103, which in turn impair the function of complex I. Most complex I mutants show a retarded growth phenotype, such as ahg11 (Murayama et al., 2012), abo5 (Liu et al., 2010), abo8 (Yang et al., 2014), bir6 (Koprivova et al., 2010), css1 (Nakagawa and Sakurai, 2006), indh (Wydro et al., 2013), mtsf1 (Haili et al., 2013), nMat1 (Keren et al., 2012), nMat2 (Keren et al., 2009), nMat4 (Cohen et al., 2014), otp43 (de Longevialle et al., 2007), otp439 and tang2 (Colas des Francs-Small et al., 2014), slg1 (Sung et al., 2010), slo2 (Zhu et al., 2012), slo3 (Hsieh et al., 2015), and also assmk1 (small kernel 1), which has been shown to be responsible for loss of editing of NAD7-448 transcripts in maize and rice (Li et al., 2014).
The phenotype of grs1-1 plants, however, cannot be fully explained by the loss of function of complex I. The defects observed in grs1-1 plants are much stronger than those of mutants defective in complex I activity such as the slo2 (Zhu et al., 2012), opt43 (de Longevialle et al., 2007), nMat1 (Keren et al., 2012) and ndufs4 mutants (Meyer et al., 2009). Impaired activity of the mitochondrial electron transport chain of complex I can cause a redox imbalance and increases in ROS accumulation, leading to the accumulation of more ROS in mutants than in the wild-type (Liu et al., 2010; Yang et al., 2014); however, the grs1-1 mutants do not accumulate more ROS than the wild-type. Consistent with these results, addition of GSH or DPI could not restore the root growth defects of grs1-1 mutant plants. The results indicate that other signals must be responsible for the retarded growth phenotype observed in grs1-1 plants.
ABA is a well-established key player in seed germination and post-germination growth. Furthermore, some reports have shown that mutations of PPR proteins result in mutant plants that are more sensitive to ABA than wild-type plants (Liu et al., 2010; Murayama et al., 2012; Yang et al., 2014). Expression of ABI5 was found to be significantly up-regulated in the grs1-1 mutants compared to the wild-type plants, while expression of ABI3 was not up-regulated in grs1-1 mutant plants compared to the wild-type. The results indicate that the up-regulated expression of ABI5 is independent of the ABA signal. The grs1-1 abi5-1 double-mutant displayed higher root meristem cell numbers than the grs1-1single-mutant plants. The results indicate that abi5-1 partially rescued the post-germination growth arrest of grs1-1 mutant plants. The grs1-1 gin1-3 double-mutant, however, could not partially rescue the post-germination growth arrest of grs1-1 mutant plants. These findings suggest that ABI5, but not ABA, is involved in the post-germination growth arrest of grs1-1 mutant plants. The mechanism through which grs1-1 mutant plants activate ABI5 remains an interesting question for future investigation.
Other factors must be involved in the root growth defects of the grs1-1 mutant plants, since abi5-1 only partially rescued their post-germination growth arrest. One possibility is that the mutation of GRS1 also impairs the function of mitoribosomes, leading to a dysfunction of mitochondria in addition to the loss of function of complex I. This scenario is found in mcsf1 mutants, where the activity of complexes I and IV are both reduced, leading to severe defects in embryo development, which is arrested at the early globular stage (Zmudjak et al., 2013).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Rapid identification of heterozygous and homozygous mutants through pollen fluorescence.
Figure S2. Genomic complement fragment of At4g32430 rescues the phenotype of grs1-1.
Figure S3. Putative coordination of PPR motifs of GRS1 and RNA nucleotides around the editing sites targeted by GRS1.
Figure S4. Relative expression of alternative pathway genes in wild-type and grs1-2.
Table S1. PLS repeat structure of At4g32430.
Table S2. The root length of 8-d-old seedlings.
Table S3. Primers used for RNA editing analysis.
Table S4. Primers used for quantitative RT-PCR.
Acknowledgements
We thank Dr Jian Xu for providing the transgenic line pCYCB1;1:Dbox-GUS and Dr Lei Zhang for providing abi5-1 mutant seeds. This work was supported by project number 2013CB945100 of the National Natural Science Foundation (grant No. 31570317, 31270362).
References
- Andrés C, Lurin C, Small ID. 2007. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiologia Plantarum 129, 14–22. [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18, 298–305. [DOI] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65, 415–442. [DOI] [PubMed] [Google Scholar]
- Bentolila S, Chateigner-Boutin A-L, Hanson MR. 2005. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiology 139, 2006–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Elliott LE, Hanson MR. 2008. Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics 178, 1693–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A-L, Small I. 2010. Plant RNA editing. RNA Biology 7, 213–219. [DOI] [PubMed] [Google Scholar]
- Cheng S, Gutmann B, Zhong X, et al. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. The Plant Journal 85, 532–547. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen S, Zmudjak M, Colas des Francs-Small C, et al. 2014. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. The Plant Journal 78, 253–268. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Falcon de Longevialle A, Li Y, Lowe E, Tanz SK, Smith C, Bevan MW, Small I. 2014. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiology 165, 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P. 1999. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. The Plant Journal 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. 1989. RNA editing in plant mitochondria. FASEB Journal 7, 64–71. [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID. 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. The Plant Cell 19, 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P, Brennicke A. 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proceedings of the National Academy of Sciences, USA 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass F, Härtel B, Zehrmann A, Verbitskiy D, Takenaka M. 2015. MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Molecular Plant 8, 1466–1477. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Lamattina L, Bonnard G, Weil J-H, Grienenberger J-M. 1989. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341, 660–662. [DOI] [PubMed] [Google Scholar]
- Haili N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H. 2013. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Research 41, 6650–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel B, Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. 2013. MEF10 is required for RNA editing at nad2-842 in mitochondria of Arabidopsis thaliana and interacts with MORF8. Plant Molecular Biology 81, 337–346. [DOI] [PubMed] [Google Scholar]
- Hiesel R, Wissinger B, Schuster W, Brennicke A. 1989. RNA editing in plant mitochondria. Science 246, 1632–1634. [DOI] [PubMed] [Google Scholar]
- Hsieh WY, Liao JC, Chang C, Harrison T, Boucher C, Hsieh MH. 2015. The SLOW GROWTH 3 pentatricopeptide repeat protein is required for the splicing of mitochondrial nad7 intron 2 in Arabidopsis. Plant Physiology 168, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O. 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araujo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. The Plant Journal 71, 413–426. [DOI] [PubMed] [Google Scholar]
- Kim SR, Yang JI, Moon S, Ryu CH, An K, Kim KM, Yim J, An G. 2009. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. The Plant Journal 59, 738–749. [DOI] [PubMed] [Google Scholar]
- Kindgren P, Yap A, Bond CS, Small I. 2015. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. The Plant Cell 27, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, des Francs-Small CC, Calder G, Mugford ST, Tanz S, Lee BR, Zechmann B, Small I, Kopriva S. 2010. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. The Journal of Biological Chemistry 285, 32192–32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. 2005. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. [DOI] [PubMed] [Google Scholar]
- Kwasniak M, Majewski P, Skibior R, Adamowicz A, Czarna M, Sliwinska E, Janska H. 2013. Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in arabidopsis mitochondria. The Plant Cell 25, 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Zhang YF, Hou M, et al. 2014. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). The Plant Journal 79, 797–809. [DOI] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. The Plant Journal 63, 749–765. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP. 2012. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. Journal of Experimental Botany 63, 6371–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences, USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Jiang SC, Lu YF, et al. 2014. Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling. Journal of Experimental Botany 65, 5317–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Tomaz T, Carroll A, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH. 2009. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiology 151, 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama M, Hayashi S, Nishimura N, Ishide M, Kobayashi K, Yagi Y, Asami T, Nakamura T, Shinozaki K, Hirayama T. 2012. Isolation of Arabidopsis ahg11, a weak ABA hypersensitive mutant defective in nad4 RNA editing. Journal of Experimental Botany 63, 5301–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Sakurai N. 2006. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant & Cell Physiology 47, 772–783. [DOI] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW. 1994. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264, 1458–1459. [DOI] [PubMed] [Google Scholar]
- Robison MM, Ling X, Smid MP, Zarei A, Wolyn DJ. 2009. Antisense expression of mitochondrial ATP synthase subunits OSCP (ATP5) and γ (ATP3) alters leaf morphology, metabolism and gene expression in Arabidopsis. Plant & Cell Physiology 50, 1840–1850. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, et al. 2002. A high-throughput Arabidopsis reverse genetics system. The Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Inuzuka T, Yoshida H, Sugiura H, Wada I, Maki M. 2010. The ALG-2 binding site in Sec31A influences the retention kinetics of Sec31A at the endoplasmic reticulum exit sites as revealed by live-cell time-lapse imaging. Bioscience, Biotechnology, & Biochemistry 74, 1819–1826. [DOI] [PubMed] [Google Scholar]
- Shikanai T. 2006. RNA editing in plant organelles: machinery, physiological function and evolution. Cellular and Molecular Life Sciences 63, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. 2015. RNA editing in plants: machinery and flexibility of site recognition. Biochimica et Biophysica Acta 1847, 779–785. [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. 2000. The PPR motif – a TPR-related motif prevalent in plant organellar proteins. Trends in Biochemical Sciences 25, 45–47. [DOI] [PubMed] [Google Scholar]
- Sung TY, Tseng CC, Hsieh MH. 2010. The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. The Plant Journal 63, 499–511. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A. 2013. RNA editing in plants and its evolution. Annual Review of Genetics 47, 335–352. [DOI] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S. 1989. Isolation and expression of an anther-specific gene from tomato. Molecular and General Genetics 217, 240–245. [DOI] [PubMed] [Google Scholar]
- Verbitskiy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M. 2010. The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. The Plant Journal 61, 446–455. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U. 2000. Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Walter M, Piepenburg K, Schottler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R. 2010. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. The Plant Journal 64, 851–863. [DOI] [PubMed] [Google Scholar]
- Wu J, Sun Y, Zhao Y, et al. 2015. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Research 25, 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Peng XB, Li WW, He R, Xin HP, Sun MX. 2012. Mitochondrial GCD1 dysfunction reveals reciprocal cell-to-cell signaling during the maturation of Arabidopsis female gametes. Developmental Cell 23, 1043–1058. [DOI] [PubMed] [Google Scholar]
- Wydro MM, Sharma P, Foster JM, Bych K, Meyer EH, Balk J. 2013. The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis [corrected]. The Plant Cell 25, 4014–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. 2013. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 8, e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Nakamura T, Small I. 2014. The potential for manipulating RNA with pentatricopeptide repeat proteins. The Plant Journal 78, 772–782. [DOI] [PubMed] [Google Scholar]
- Yan HL, Chen D, Wang YF, Huang J, Sun MX, Peng XB. 2016. Ribosomal protein L18aB is required for both male gametophyte function and embryo development in Arabidopsis. Scientific Reports 6, 31195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang J, He J, Qin Y, Hua D, Duan Y, Chen Z, Gong Z. 2014. ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genetics 10, e1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, et al. 2013. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. 2009. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. The Plant Journal 59, 1011–1023. [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu D. 2012. Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. The Plant Journal 70, 432–444. [DOI] [PubMed] [Google Scholar]
- Zehrmann A, van der Merwe J, Verbitskiy D, Härtel B, Brennicke A, Takenaka M. 2012. The DYW-class PPR protein MEF7 is required for RNA editing at four sites in mitochondria of Arabidopsis thaliana. RNA Biology 9, 155–161. [DOI] [PubMed] [Google Scholar]
- Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. 2008. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion 8, 319–327. [DOI] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. 2009. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. The Plant Cell 21, 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Dugardeyn J, Zhang C, et al. 2012. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. The Plant Journal 71, 836–849. [DOI] [PubMed] [Google Scholar]
- Zmudjak M, Colas des Francs-Small C, Keren I, Shaya F, Belausov E, Small I, Ostersetzer-Biran O. 2013. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. The New Phytologist 199, 379–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.