Highlight
A novel CONSTANS-like gene in rice regulates developmental leaf senescence and confers drought sensitivity through the acceleration of senescence.
Key words: ARID, CO-like, drought, Ghd2, Oryza sativa, senescence
Abstract
CONSTANS (CO)-like genes have been intensively investigated for their roles in the regulation of photoperiodic flowering, but very limited information has been reported on their functions in other biological processes. Here, we found that a CO-like gene, Ghd2 (Grain number, plant height, and heading date2), which can increase the yield potential under normal growth condition just like its homologue Ghd7, is involved in the regulation of leaf senescence and drought resistance. Ghd2 is expressed mainly in the rice (Oryza sativa) leaf with the highest level detected at the grain-filling stage, and it is down-regulated by drought stress conditions. Overexpression of Ghd2 resulted in significantly reduced drought resistance, while its knockout mutant showed the opposite phenotype. The earlier senescence symptoms and the transcript up-regulation of many senescence-associated genes (SAGs) in Ghd2-overexpressing transgenic rice plants under drought stress conditions indicate that Ghd2 plays essential roles in accelerating drought-induced leaf senescence in rice. Moreover, developmental and dark-induced leaf senescence was accelerated in the Ghd2-overexpressing rice and delayed in the ghd2 mutant. Several SAGs were confirmed to be regulated by Ghd2 using a transient expression system in rice protoplasts. Ghd2 interacted with several regulatory proteins, including OsARID3, OsPURα, and three 14-3-3 proteins. OsARID3 and OsPURα showed expression patterns similar to Ghd2 in rice leaves, with the highest levels at the grain-filling stage, whereas OsARID3 and the 14-3-3 genes responded differently to drought stress conditions. These results indicate that Ghd2 functions as a regulator by integrating environmental signals with the senescence process into a developmental programme through interaction with different proteins.
Introduction
Leaf senescence is a precisely controlled process at the final stage of leaf growth and development, during which leaf cells are dismantled and recycled, ultimately leading to cell death (Lim et al., 2007). The initiation of leaf senescence is age dependent under optimal growth conditions, and macromolecules from source leaves are degraded and relocated to support the growth of actively growing tissues such as young leaves and reproducing seeds. Many unfavourable environmental conditions can cause premature senescence, including various abiotic and biotic stresses, among which drought stress constitutes one of the major abiotic factors threatening rice (Oryza sativa) yield (Zhang and Zhou, 2013; Albacete et al., 2014).
The most prominent visible change in senescent leaves is chlorophyll breakdown and degradation, and chloroplast dismantling is a major event during senescence (Ougham et al., 2008). At the metabolic level, catabolism takes the place of carbon assimilation. The generation of reactive oxygen species (ROS) is one of the earliest responses under stresses and senescence, and ROS, which primarily affect the chloroplast, are also signalling molecules which trigger senescence (Khanna-Chopra, 2012; Wang et al., 2013; Pinto-Marijuan and Munne-Bosch, 2014). During the senescence process, most of the free amino acid pools increase, and amino acids with a high ratio of nitrogen to carbon (N:C), including asparagine and arginine, can serve as nitrogen storage and transport compounds (Tetley and Thimann, 1974; Hildebrandt et al., 2015; Gaufichon et al., 2016). Sugar accumulation can also trigger leaf senescence (Wingler et al., 2006; Wingler and Roitsch, 2008).
A large number of senescence-related genes (SAGs) are up-regulated during senescence. They are involved in various aspects of the senescence process, including the onset of senescence, the breakdown and remobilization of biomolecules, regulation of the time course, and protection of senescent cells to ensure the accomplishment of the senescence event (Lee et al., 2001; Liu et al., 2011; Li et al., 2014). Among these SAGs, genes involved in the breakdown of chlorophylls such as SGR (stay green), NYC1 (non-yellow coloring1), PAO (pheide a oxygenase), and RCCR (chlorophyll catabolite reductase) are conserved in various species (Hortensteiner and Krautler, 2011; Balazadeh, 2014).
Many stress-responsive genes overlap with SAGs during senescence (Chen, 2002; Guo and Gan, 2012), and some regulatory genes are reported to control both stress and senescence processes. For instance, the Arabidopsis thaliana NAC transcription factor ANAC092/AtNAC2/ORE1 is involved in age-dependent senescence control, and disruption of ANAC092 increases the rate of seed germination under saline conditions (Balazadeh et al., 2010). NAC016 promotes both senescence and drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP (Kim et al., 2013; Sakuraba et al., 2015). The submergence-tolerance gene SUB1A enhances rice tolerance to drought and oxidative stress through a delay of leaf senescence (Fukao et al., 2012). A CCCH-type zinc finger family gene OsTZF1 delays leaf senescence, and improves rice tolerance to high-salt and drought stress conditions (Jan et al., 2013). An S-domain receptor-like kinase OsSIK2 enhances rice tolerance to salt and drought stress conditions, and delays dark-induced senescence (Chen et al., 2013a).
The CONSTANS (CO) gene identified in Arabidopsis plays an important role in the photoperiod pathway of flowering (Putterill et al., 1995). CO contains both CCT and B-box domains, and belongs to the CO-like or BBX gene family, defined with an emphasis on one of the two domains by different groups (Lagercrantz and Axelsson, 2000; Griffiths et al., 2003; Khanna et al., 2009; Crocco and Botto, 2013; Gangappa and Botto, 2014). Most CO-like genes containing a CCT domain are reported to be involved in the control of flowering time in various plant species. In rice, several such genes control the rice heading date, including Hd1, OsCOL3, OsCOL4, Ghd7, DTH2, OsCOL10, OsCCT01, OsCCT11, and OsCCT19 (Yano et al., 2000; Kim et al., 2008; Xue et al., 2008; Lee et al., 2010; Wu et al., 2013; Zhang et al., 2015; Tan et al., 2016). To date, only a few members of this family have been identified as regulators of abiotic stress responses. In Arabidopsis, overexpression of BBX24 (STO) and AtCOL4 improves salt tolerance (Nagaoka and Takano, 2003; Min et al., 2015). Suppression of BBX24 in Chrysanthemum decreases tolerance to freezing and drought stresses, whereas overexpression of this gene enhances tolerance to the stresses (Yang et al., 2014). Ghd7, which is a very important CO-like gene controlling grain number, plant height, and heading date in rice (Xue et al., 2008), also regulates stress tolerance. The expression of Ghd7 is up-regulated by cold treatment, but it is repressed by drought, abscisic acid (ABA), and high-temperature treatments (Weng et al., 2014). Overexpression of Ghd7HJ19 (an allele from rice variety Hejiang19) reduces the drought resistance of rice, and transformants containing an artificial microRNA construct of Ghd7 in the background of rice variety Zhonghua 11 show increased drought resistance (Weng et al., 2014). However, the regulation mechanism by which Ghd7 participates in stress tolerance is largely unknown.
In this work, we found that the novel CO-like gene Ghd2, which is a close homologue of Ghd7 and has similar roles in the control of grain number, heading date, and plant height, positively regulates drought stress-triggered early senescence in rice. Overexpression of Ghd2 accelerated developmental and dark-induced senescence. Ghd2 activated the expression of many SAGs. Furthermore, Ghd2 was shown to interact with several regulatory proteins, including OsARID3, OsPURα, and three 14-3-3 proteins. Our results suggest that Ghd2 is a multifunctional regulator involved in developmental and drought-induced leaf senescence.
Materials and methods
Vector construction and rice transformation
The cDNA sequence of Ghd2 was amplified from rice variety Zhonghua 11 (ZH11) and cloned into the T-vector (Promega). The cDNA was digested with Kpn I and Bam HI and cloned into the destination vectors pCAMBIA1301H-HPT (hygromycin resistance and driven by a rice LEA3 promoter) and pCAMBIA1301U-HPT-flag (hygromycin resistance, driven by a maize Ubiquitin promoter, and fused to 3×flag). To construct a CRISPR vector, a spacer sequence was cloned into the Bsa I-digested entry vector pOs-sgRNA, and then into the destination vector pH-Ubi-cas9-7 using the Gateway recombination reaction (Invitrogen). The overexpression and CRISPR vectors were introduced into the rice ZH11 cultivar by Agrobacterium-mediated transformation (Lin and Zhang, 2005).
Plant growth and stress treatments
To detect the diurnal expression pattern of Ghd2 under short day (SD) and long day (LD) conditions, wild type (WT) rice ZH11 were grown in a growth chamber (10h light/14h dark cycle for SD and 14h light/10h dark cycle for LD) and the top-second leaves were sampled at 33 days after germination (DAG).
To detect the transcript levels of Ghd2 under drought, dark, and ABA treatments, 2-week-old rice plants were subjected to drought stress (placing rice plants on facial tissues), dark (transferring the seedlings to a growth chamber without lighting), and ABA (transferring the seedlings to 100 µM ABA solution) treatments, and the shoots were sampled at the designated time points (0.5, 1, 3, and 6h). Rice plants kept in water were sampled at each time point to serve as controls.
The Ghd2-overexpression (Ghd2-OE) lines of the T1 generation with one copy of a T-DNA insertion (lines 7, 15, 16, 21, and 25) were planted for identification of the segregated non-transgenic (WT') and transgenic genotypes. Seeds from each genotype were harvested separately for further experiments.
For the drought and dark treatments, Ghd2-OE seeds were germinated on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 50mg/L hygromycin, while the WT' seeds were germinated on half-strength MS medium without hygromycin. At 10 DAG, seedlings were planted in pots with soil. At 30 DAG, drought (no watering until severe leaf rolling) and dark (no light for one week) stresses were applied. The top-second leaves from the WT' and Ghd2-OE plants were sampled when they became rolled during drought stress treatment for 3,3′-diaminobenzidine (DAB) staining, H2O2 quantification, transmission electron microscopy (TEM), scanning electron microscopy (SEM), and gene expression level detection. After 7 days of dark treatment, the leaves were sampled for TEM observation. The WT' and Ghd2-OE plants were grown in the field and progressive drought stress started at 60 DAG.
A DNA fragment harbouring the spacer sequence (−49 to +1019 from the start codon) in the segregated non-transgenic Ghd2-CRISPR plants was amplified and sequenced to determine the mutation. Seeds with a homozygous mutation near the protospacer adjacent motif were harvested separately. For phenotypic observation at the seedling stage, Ghd2-CRISPR and ZH11 seedlings were grown in pots and exposed to drought stress conditions the same as for the Ghd2-OE plants. For phenotypic observation at the reproductive stage, seeds of Ghd2-CRISPR and ZH11 were grown in the field and polyvinyl chloride (PVC) pots. Moderate drought stress was attained by stopping irrigation to the plants grown in the field at the booting stage until the leaves became fully rolled when observed at noon. Severe drought stress was attained by stopping the watering of the plants grown in PVC pots at the booting stage until the leaves began to turn yellow. The top-second leaves from the ZH11 and Ghd2-CRISPR plants during the drought stress treatment were sampled for gene expression level detection.
Gene expression quantification
The expression level of Ghd2 at the T0 generation was quantified by northern blot analysis using an α-32P-dCTP-labelled Ghd2-specific probe. The expression levels of other genes were detected by real-time quantitative reverse transcription (qRT)-PCR analysis (Livak and Schmittgen, 2001) using first-strand cDNA synthesized by Superscript III reverse transcriptase (Invitrogen) and performed with FastStart Universal SYBR Green Master (Rox) (Roche) on a real-time PCR system StepOnePlus or QuantStudio 6 Flex (Applied Biosystems). The rice Ubiquitin gene was used as the internal control.
DAB staining and H2O2 quantification
For DAB staining, leaves were immersed in 0.1mg/mL DAB with 50mM Tris-acetate buffer (pH 3.2), vacuum infiltrated, and placed in the light for 2 days at room temperature. The chlorophylls were removed by placing the samples in 100% ethanol at 37°C before photographing. The content of H2O2 was quantified using the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen).
TEM, SEM, and metabolite measurement
Leaves from Ghd2-OE and WT' plants at the seedling stage were sampled during drought stress treatment when the leaves became rolled. Normally grown Ghd2-OE and WT' plants were sampled at the same time as controls. TEM and SEM were conducted as described previously (Cao et al., 2013). The samples were freeze-dried and metabolites were measured according to the method previously described in detail (Chen et al., 2013b). Soluble sugars were determined from freeze-dried samples using the anthrone reagent (Yemm and Willis, 1954).
Microarray
Three Ghd2-OE lines (16, 21, and 25) were grown in pots alongside their respective WT' controls in a half-to-half manner, and the seedlings were harvested for microarray when the leaves became rolled during drought stress treatment. Microarray analysis was conducted by Affymetrix GeneChip service (CapitalBio) following the standard procedure.
Subcellular localization and bimolecular fluorescence complementation analysis
Ghd2 was fused in-frame with GFP in the vector HBT-sGFP, and Ghd2-GFP and Ghd7-CFP were co-transformed into rice protoplasts isolated from 2-week-old green seedlings using a polyethylene glycol (PEG)-mediated transformation assay. Protoplasts were incubated overnight at room temperature in the dark and the fluorescence signal was observed with a confocal microscope (Leica).
Full-length cDNAs of Ghd2 and the genes encoding Ghd2-interacting proteins (including GF14b, GF14c, and OsARID3) were amplified with the primers listed in Supplementary Table S1 at JXB online. The PCR products were digested and cloned into PvYNE and PvYCE (Waadt et al., 2008), respectively, to produce fusions with N- and C-terminal halves of YFPs, respectively. Combinations of bimolecular fluorescence complementation (BiFC) constructs were co-transformed into rice protoplasts isolated from 2-week-old green seedlings using a PEG-mediated transformation assay. After overnight incubation, the fluorescence was detected by confocal microscopy (Leica).
Transcriptional activation assay in rice protoplasts
Ghd2 was fused in-frame with the Gal4 DNA-binding domain in the effector vector Gal4BD, and co-transformed with the reporter vector GAL4-LUC. To test the transcriptional activation of the SAGs, Ghd2 was constructed in the effector vector (named ‘None’), while the promoters of the SAGs were constructed in the reporter vector (190LUC). The effector and reporter vectors were co-transformed together with the internal control vector Ubi-Rennila LUC in rice protoplasts isolated from 2-week-old green seedlings by PEG-mediated transformation. The two luciferases which lysed from the overnight incubated protoplasts were incubated with their substrates (Promega) and the luciferase activity was measured using the TECAN Infinite M200 System.
Transactivation assay in yeast and yeast two-hybrid screening
The full-length and truncated Ghd2 fragments were fused in-frame with the yeast Gal4 DNA-binding domain in the pDEST32 vector (Invitrogen). The fused pDEST32 vectors and the AD502 vector (Invitrogen) were co-transformed into the yeast strain Mav203 (Invitrogen), and the transactivation activity of Ghd2 was indicated by an X-gal assay according to the manufacturer’s manual (Invitrogen). For yeast two-hybrid (Y2H) screening, the Ghd2 deletions (N3 and C3) were used as baits. A cDNA library made from a mixture of rice tissues containing drought-treated leaves, seedlings, and callus was screened on synthetic complete-Leu-Trp-His medium containing 5mM (for N3) and 20mM (for C3) 3-amino-triazol (3-AT).
In vivo pull-down assay
Total proteins were extracted from leaves using extraction buffer [50mM Tris-HCl (pH 7.5), 150mM NaCl, 0.5% NP-40, 1mM phenylmethylsulfonyl fluoride]. Protein extracts were immunoprecipitated with ANTI-FLAG M2 Affinity Gel (A2220, Sigma) overnight at 4°C. Precipitated proteins were suspended with an equal volume of 2× sample buffer [125mM Tris HCl (pH 6.8), 4% SDS, 20% (v/v) glycerol, 0.004% bromophenol blue, 5% 2-mercaptoethanol], denatured at 98°C for 5min, and subjected to SDS-PAGE. The proteins were electroblotted onto polyvinylidene difluoride membrane and an antibody against OsARID3 (Xu et al., 2015) was used for the immunoblot analysis. The chemiluminescence detection kit ImmunStar WesternC (Bio-Rad) was used for the visualization of peroxidase activity of the secondary antibodies.
Results
Overexpression of OsK/Ghd2 in rice increased drought sensitivity
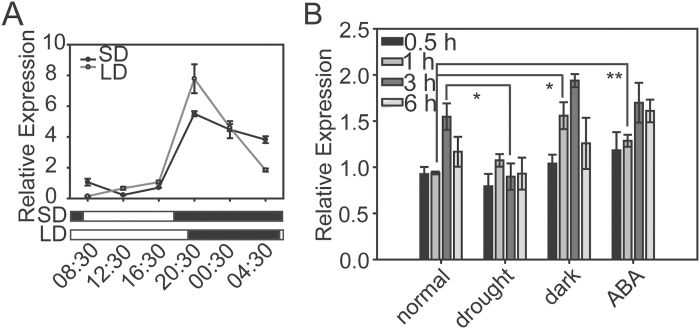
To investigate whether CCT domain-containing genes are involved in the regulation of the drought response in rice, first we checked the expression levels of this family under drought stress conditions. In the published microarray data GSE6901, which contains the gene expression profiles of rice seedlings under drought, salinity, and cold stresses (Jain et al., 2007), we found that three close homologues of CO-like genes in group II, originally named OsJ (OsCOL10), OsK, and OsL (Griffiths et al., 2003), were down-regulated under drought stress treatment (Supplementary Table S2). In our previous microarray results consisting of global genome expression analyses in shoots, flag leaves, and panicles under drought and salinity stresses (Zhou et al., 2007), we also found that OsK (LOC_Os02g49880) was significantly down-regulated under drought stress conditions (Supplementary Table S3). The expression of OsK was further investigated under drought, dark, and ABA treatments by performing qRT-PCR. The expression of OsK was analysed in the treated rice seedlings with their respective non-treated controls at each time point during the treatment, given that OsK exhibited a diurnal expression pattern under both the SD and LD growth conditions (Fig. 1A). Similar to the microarray results, the expression level of OsK was significantly down-regulated at 3h of drought stress treatment. However, OsK was slightly induced at 1h of dark and ABA treatments (Fig. 1B).
Fig. 1.
Diurnal expression pattern and the response of the Ghd2 or OsK gene to drought, dark, and ABA treatments. (A) Diurnal expression pattern of Ghd2 under LD and SD conditions. (B) Expression analysis of Ghd2 under drought, dark, and ABA treatments at 0.5, 1, 3, and 6h. Two-week-old seedlings kept in water were sampled simultaneously at each time point to serve as controls. Error bars indicate the SE based on three biological replicates. *P < 0.05, t-test; **P < 0.01, t-test.
We thereafter overexpressed OsK in the rice variety Zhonghua11 using a LEAP promoter, which exhibits a moderate expression level under normal growth conditions but it is strongly induced by drought stress and ABA treatment (Xiao et al., 2007). Northern blot was used to identify the overexpressing T0 seedlings, and six of them were found to be overexpressing OsK(Supplementary Fig. S1A). Southern blot results suggested that five seedling lines (7, 15, 16, 21, and 25) each had one copy of the T-DNA insertion (Supplementary Fig. S1B). Segregated non-transgenic lines (WT′) from the single-copy lines were used as negative controls in the following analyses. Under normal growth conditions, the OsK-overexpressing plants showed significantly increased grain number per panicle, plant height, and heading date (Supplementary Fig. S2), which is very similar to the phenotypes of the Ghd7-overexpressing rice (Xue et al., 2008). Taking into consideration the commonly adopted rule of gene nomenclature in rice (McCouch, 2008), we renamed OsK as Ghd2, because this gene confers functions similar to Ghd7 in the control of grain number, plant height, and heading date, and it is a close homologue of Ghd7.
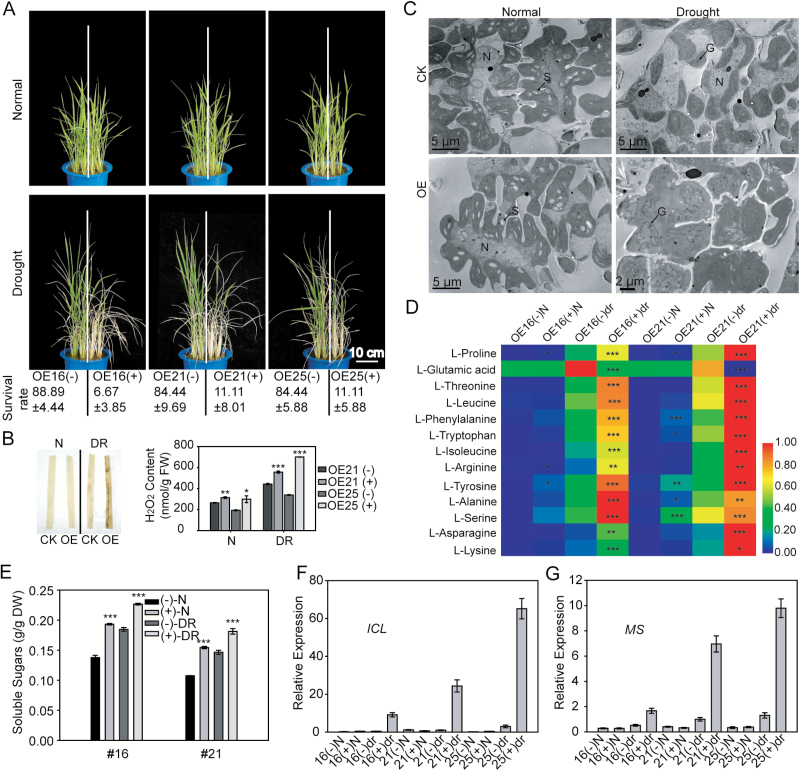
To check whether Ghd2 overexpression affected the drought resistance of rice, the Ghd2-OE and WT' plants were grown in the same pot in a half-to-half manner, and drought stress was applied at the seedling stage. To our surprise, the Ghd2-OE plants were more sensitive to drought stress than the WT'. During drought stress treatment, the Ghd2-OE plants showed an earlier leaf-rolling phenotype and less than 12% of them survived after recovery from a moderate drought stress treatment, whereas the survival rates for the WT' plants were above 84% (Fig. 2A). The Ghd2-OE plants also showed increased sensitivity to drought stress treatment when grown in the paddy field (Supplementary Fig. S3A). These results suggest that Ghd2 may negatively regulate drought resistance.
Fig. 2.
Overexpression of Ghd2 accelerated drought-induced leaf senescence at the seedling stage. (A) Phenotypes of the Ghd2-OE and WT' plants under drought stress treatment. Survival rates are shown below. Data represent the mean ± SE based on three replicates. Images presented above show five seedlings grown in one-half of each pot, whereas 15 seedlings grown in one-half of each pot were used for determining the survival rate. ‘+’ and ‘-’ refers to segregated transgenic and non-transgenic plants, respectively, from the Ghd2-OE lines. (B) H2O2 content in the Ghd2-OE and WT' plant leaves under normal and drought stress conditions. Left, DAB staining; right, H2O2 quantification. CK, the non-transgenic control (WT'). N, normal; DR, drought treatment; FW, fresh weight. Data represent the mean ± SE (n = 3). *P < 0.05, t-test; **P < 0.01, t-test; ***P < 0.005, t-test. (C) TEM images of the ultrastructure of Ghd2-OE and WT' plant leaves under normal and drought stress conditions. G, grana stack; N, nucleus; S, starch granule. (D) Contents of free amino acids in Ghd2-OE and WT’ plants at the seedling stage under normal and drought stress conditions. The data were analysed using the Heatmap Illustrator (http://hemi.biocuckoo.org/down.php). The data represent the mean ± SE (n = 3). *P < 0.05, t-test; **P < 0.01, t-test; ***P < 0.005, t-test. (E) Contents of soluble sugars in the Ghd2-OE and WT' plants. DW, dry weight. ***P < 0.005, t-test. (F and G) Validation of the expression changes of ICL (F) and MS (G) in a microarray analysis using qRT-PCR.
Drought-induced senescence was accelerated in Ghd2-OE plants
We first checked stomata of the Ghd2-OE and WT' plants because stomata affect the water-holding capacity of plants during drought stress. The results showed that there was no significant difference between the Ghd2-OE and WT' plants in stomatal density under both normal and drought conditions (Supplementary Fig. S4C). The percentage of opening stomata showed no significant difference either (Supplementary Fig. S4D). The water loss rates of detached rice leaves from the Ghd2-OE plants were similar to those of WT' plants (Supplementary Fig. S4E). Therefore, the role of Ghd2 in promoting drought sensitivity may not be related to stomata.
Careful observation of the phenotypic changes of the Ghd2-OE plants during the drought stress treatment revealed an early senescence symptom. As shown in Supplementary Fig. S3A (left), after being stressed by the progressive drought stress treatment (before leaf rolling), the leaves of the Ghd2-OE plants, especially in the leaf tips, began to turn yellow, but the leaves in the WT' plants remained green, similar to the non-treated rice plants. We performed DAB staining and found that H2O2 accumulated much more in the Ghd2-OE plants than in the WT' plants under drought stress treatment (Fig. 2B left panel). We also performed H2O2 quantification using an Amplex Red assay. A significant difference was detected under drought stress conditions, and the difference was detected even in leaves of normally grown plants, although the content was relatively low (Fig. 2B right panel).
Previous studies suggest that, as senescence proceeds, the ultrastructure of the leaf cells undergoes typical changes, as follows: chloroplasts become swelled and round, grana stacks and intergrana become disordered, and chloroplasts finally shrink and chloroplast components are completely decomposed (Kusaba et al., 2007; Park et al., 2007). We examined the ultrastructure of the Ghd2-OE and WT' leaves using TEM. As shown in Fig. 2C, there were no obvious differences under normal growth conditions, but under drought stress conditions, the chloroplasts in the Ghd2-OE plants were obviously swelled and the array of grana stacks were slightly disordered. However, the WT' chloroplasts maintained an appearance similar to the chloroplasts of normally grown plants, with the exception of the exhausted starch granules. These observations indicate that the drought-induced senescence was accelerated in the Ghd2-OE leaves compared to the WT'.
It is known that most of the free amino acid pools increase during the senescence process. We used gas chromatography-mass spectrometry to determine the contents of amino acids. The results showed that the contents of most of the free amino acids were significantly higher in the Ghd2-OE plants than in the WT' under drought stress conditions (Fig. 2D). We also measured soluble sugar content and the result showed that soluble sugars accumulated significantly more in the Ghd2-OE plants than in the WT' under both normal and drought stress conditions (Fig. 2E).
The above results indicate that Ghd2 overexpression may accelerate drought-induced leaf senescence. To further confirm this at the gene expression level, we compared the whole genome expression profiles to identify differentially expressed genes (DEGs) between the Ghd2-OE and WT' plants under drought stress treatment. Leaves from three Ghd2-OE lines (line numbers 16, 21, and 25) and their corresponding WT's were sampled for microarray analysis when the leaves became rolled during the drought stress treatment. Gene Ontology (GO) analysis revealed that ‘amino acid transmembrane transport’ and ‘anion transmembrane transport’ were the main overrepresented terms under the ‘biological processes’ category. The GO terms with the highest proportion of DEGs under the ‘molecular function’ category were ‘transmembrane transporter activity’ and ‘symporter activity’ (Supplementary Table S4). Many SAGs showed higher expression levels in the Ghd2-OE plants than in the WT' (Table 1). Of these up-regulated SAGs, genes involved in chlorophyll degradation, including OsNAP, OsSGR, OsNYC3, OsPAO, OsRCCR1, and OsRCCR3, showed more than 2-fold higher expression in the Ghd2-OE plants. Three genes (OsPPDKB, OsAS1, and OsGS1;2), which function in asparagine synthesis for the long-distance transport of nitrogen (Funayama et al., 2013; Hartmann et al., 2015; Ohashi et al., 2015) were also up-regulated in the Ghd2-OE plants. Notably, the expression of two key genes in the glyoxylate cycle, Isocitrate lyase (ICL) and Malate synthase (MS), were elevated to very high levels in the Ghd2-OE plants under drought stress conditions. The expression levels of ICL and MS were also increased in the WT' plants during drought stress treatment, but the induction was significantly stronger in the Ghd2-OE plants (Fig. 2F, G). The glyoxylate cycle is involved in the oil reserve mobilization in the germination of oil crops, and the activation of the glyoxysome during leaf senescence is proposed to mobilize thylakoid membrane lipids (Eastmond and Graham, 2001; Graham, 2008). Therefore, the up-regulation of key genes of the glyoxylate cycle in the Ghd2-OE plants may imply a faster mobilization of the thylakoid membrane lipids.
Table 1.
Microarray results of SAGs in the Ghd2-OE and WT’ plants during drought stress treatment.
| ID | Name | #16 (+/-) | #21 (+/-) | #25 (+/-) |
|---|---|---|---|---|
| LOC_Os07g34520 | ICL | 40.61 | 22.73 | 23.10 |
| LOC_Os04g40990 | MS | 14.42 | 8.55 | 8.35 |
| LOC_Os09g36200 | SGR | 2.11 | 3.08 | 1.78 |
| LOC_Os03g18130 | AS1 | 1.82 | 2.10 | 2.78 |
| LOC_Os03g12290 | GS1;2 | 1.48 | 3.02 | 2.27 |
| LOC_Os10g25030 | RCCR1 | 2.66 | 2.85 | 2.04 |
| LOC_Os03g21060 | NAP | 1.45 | 2.79 | 2.02 |
| LOC_Os10g25030 | RCCR3 | 2.66 | 2.78 | 1.95 |
| LOC_Os05g33570 | PPDKB | 1.30 | 2.72 | 1.40 |
| LOC_Os06g24730 | NYC3 | 2.05 | 2.67 | 2.18 |
| LOC_Os03g05310 | PAO | 1.52 | 2.09 | 1.46 |
| LOC_Os10g25040 | RCCR2 | 1.36 | 1.77 | 1.92 |
| LOC_Os07g37250 | NYC4 | 1.22 | 1.58 | 1.30 |
| LOC_Os02g57260 | l57 | 1.31 | 1.45 | 1.38 |
| LOC_Os01g12710 | NYC1 | 1.35 | 1.44 | 1.71 |
‘+/-’, ratios of expression levels in transgenic positive plants versus expression levels in transgenic negative plants.
Our findings with regards the ultrastructure of the chloroplast; H2O2, free amino acid, and soluble sugar content; and the gene expression of SAGs together suggest that the senescence process is accelerated by Ghd2 overexpression.
Disruption of Ghd2 enhanced drought resistance and delayed leaf senescence
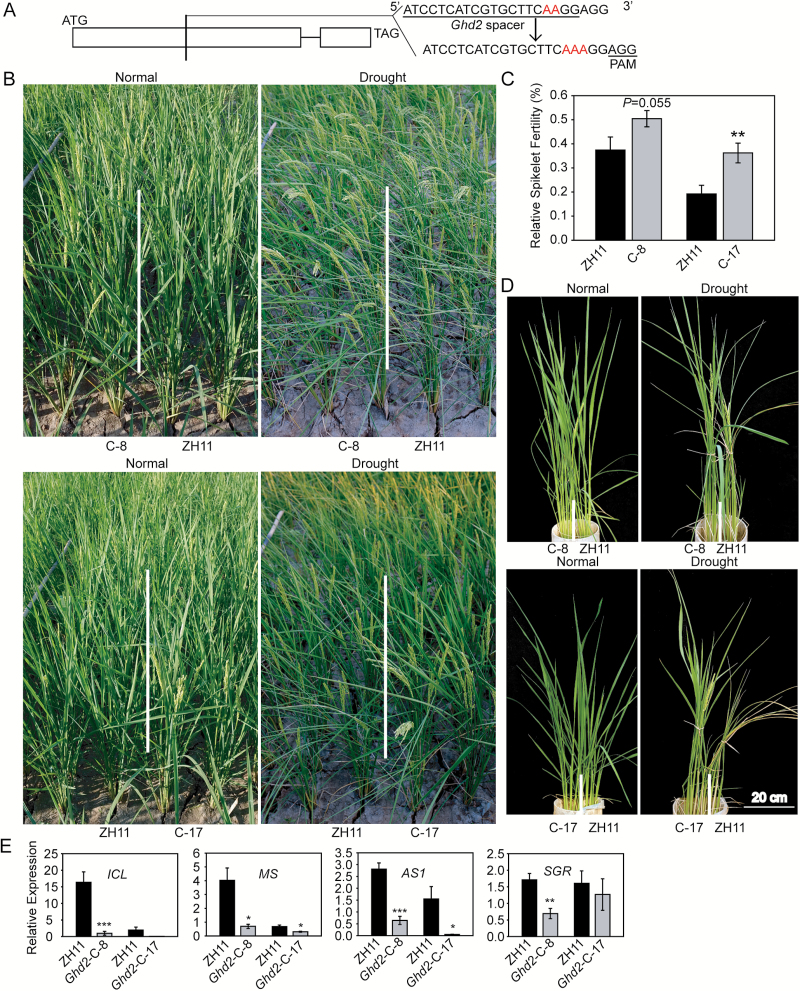
We further checked whether knockout mutation of Ghd2 could affect drought resistance. CRISPR/CAS9 technology (Miao et al., 2013) was used to produce a mutant of Ghd2 because a T-DNA insertion mutant for this gene was unavailable. We designed sgRNA targeted to the middle of the first exon of the Ghd2 gene (Fig. 3A), and the CRISPR construct was transformed into ZH11. Single-copy transgenic lines were identified by Southern blot analysis (Supplementary Fig. S1C), and progenies without CAS9 and the hygromycin-resistance gene (Hpt) insertion were selected by PCR using primers for both the CAS9 and Hpt sequences. Sequencing of the PCR amplifications covering the target site suggested that the homozygous lines (line numbers 8, 17, and 20) were mutated by a single nucleotide (A) insertion.
Fig. 3.
Ghd2-CRISPR plants showed delayed drought-induced leaf senescence. (A) Diagram showing the structure of Ghd2 for CRISPR. (B) Phenotypes of the Ghd2-CRISPR (C-x) and WT (ZH11) plants during drought stress treatment in the field facilitated with a moveable rain-off shelter. (C) Relative spikelet fertility of the Ghd2-CRISPR and WT plants after moderate drought stress treatment at the panicle development stage in the field. Data represent the mean ± SE (n = 12). **P < 0.01, t-test. (D) Phenotypes of the Ghd2-CRISPR and WT plants during severe drought stress treatment at the reproductive stage in PVC pots. (E) Real-time qRT-PCR results showing the expression levels of the SAGs in the Ghd2-CRISPR and WT plants during drought stress treatment. Data represent the mean ± SE (n = 4). *P < 0.05, t-test; **P < 0.01, t-test; ***P < 0.005, t-test.
At the seedling stage, the Ghd2-CRISPR lines showed improved drought resistance (Supplementary Fig. S3B). During a moderate drought stress treatment at the panicle development stage, the Ghd2-CRISPR lines showed an obvious delay of leaf-rolling compared with the WT (Fig. 3B). The relative spikelet fertility was higher in the Ghd2-CRISPR plants (Fig. 3C). We also tested leaf senescence of the Ghd2-CRISPR plants grown in PVC pots with severe drought stress treatment at the reproductive stage. With the extended period of drought stress, the older leaves of the WT plants began to turn yellow, whereas all of the Ghd2-CRISPR leaves remained green, indicating that the drought-induced senescence was delayed in the Ghd2-CRISPR plants (Fig. 3D). The H2O2 content was also significantly lower in the Ghd2-CRISPR plants under the drought stress conditions (Supplementary Fig. S5). qRT-PCR showed that the SAGs, including ICL, MS, OsAS1, and OsSGR, were down-regulated in the Ghd2-CRISPR plants during drought stress treatment (Fig. 3E). These results further suggest that Ghd2 positively affects drought-induced leaf senescence.
Ghd2 regulates developmental and dark-induced senescence
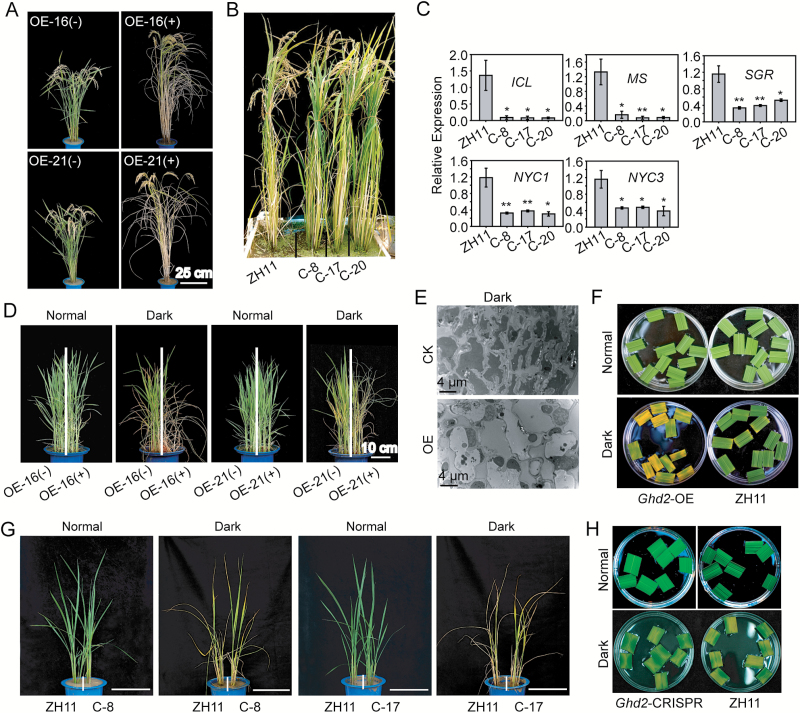
When grown in the field under non-stressed conditions, Ghd2-OE plants showed mild early senescence symptoms before flowering (Supplementary Fig. S2A), and the symptoms became increasingly obvious during the grain-filling process. At 35 days after flowering (DAF), the Ghd2-OE plants exhibited a significantly accelerated leaf-yellowing phenotype (Fig. 4A). In contrast, leaf senescence was delayed in the Ghd2-CRISPR plants at the late grain-filling stage, and the Ghd2-CRISPR plants exhibited a significantly delayed leaf-yellowing phenotype at 50 DAF (Fig. 4B). We also examined the expression of the SAGs at the late grain-filling stage, and found that ICL, MS, OsSGR, OsNYC1, and OsNYC3 had relatively lower expression levels in the Ghd2-CRISPR plants than in the WT (Fig. 4C). Of note, Ghd2 was strongly expressed in the matured leaves approaching senescence, especially at the grain-filling stage (Supplementary Fig. S6). These results suggest that Ghd2 may also be involved in the regulation of developmental leaf senescence, especially at the grain-filling stage.
Fig. 4.
Phenotypes of the Ghd2-OE and Ghd2-CRISPR plants during developmental and dark-induced leaf senescence. (A) Phenotypes of the Ghd2-OE and WT’ rice at 35 DAF. (B) Phenotypes of the Ghd2-CRISPR and WT rice at 50 DAF. (C) Expression of SAGs in the leaves of Ghd2-CRISPR and WT plants at the late grain-filling stage. Data represent the mean ± SE (n = 4). *P < 0.05, t-test; **P < 0.01, t-test. (D) Phenotypes of the Ghd2-OE(+) and WT' (Ghd2-OE(-)) rice seedlings after dark treatment. ‘+’ and ‘-’ refers to segregated transgenic and non-transgenic plants, respectively, from the Ghd2-OE lines. (E) TEM images of the ultrastructure of Ghd2-OE and CK (the non-transgenic control, WT') plant leaves after dark treatment. (F) Phenotypes of the Ghd2-OE and WT plant leaves after dark treatment. Segments of top-second leaves were incubated in water and dark-treated for 3 days before photographing. (G) Phenotypes of the Ghd2-CRISPR and WT rice seedlings after dark treatment. Bar = 20cm. (H) Phenotypes of the Ghd2-CRISPR and WT plant leaves after dark treatment. Leaf segments were dark-treated for 1 week.
We further examined whether dark treatment could accelerate the senescence of Ghd2-OE plants because dark treatment has frequently been used to simulate synchronous senescence (Cha et al., 2002; Kusaba et al., 2007; Morita et al., 2009). Ghd2-OE and WT' seedlings grown in the same pots were exposed to dark conditions for 1 week and then recovered with normal light conditions. As expected, the Ghd2-OE plants showed accelerated leaf senescence with dark treatment, with leaves yellowing faster and more seedlings dying after recovery (Fig. 4D). After dark treatment, the number of thylakoids was significantly lower in the Ghd2-OE leaf blade tips than in WT', as observed by TEM (Fig. 4E). The leaf segments of Ghd2-OE showed faster yellowing than the WT' after the dark treatment (Fig. 4F). In contrast to the Ghd2-OE plants, both the intact (Fig. 4G) and detached (Fig. 4H) leaves from the Ghd2-CRISPR plants showed delayed senescence compared to WT under dark treatment. The above results together suggest that Ghd2 promotes both developmental and dark-induced senescence in addition to drought-induced senescence.
Ghd2 activated the expression of senescence-related genes
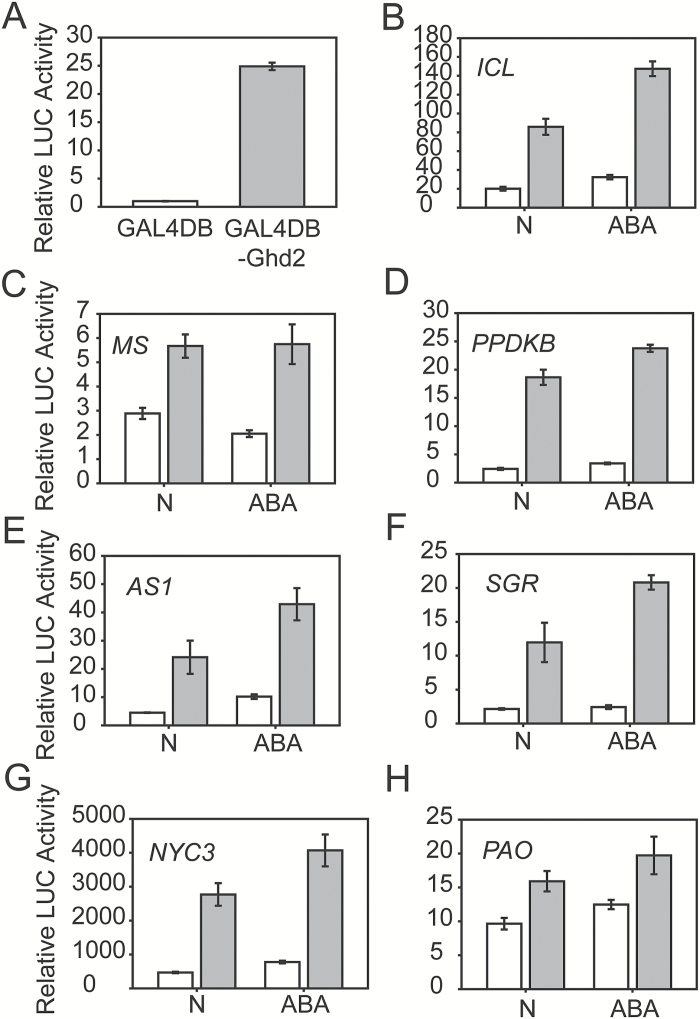
Similar to other reported CO-like proteins, a Ghd2-GFP fusion was localized in the nuclei of rice protoplasts (Supplementary Fig. S7A). It has been reported that CO-like proteins possess trans-activation activity in yeast, and the activation domain is located in the middle region between the CCT domain and the B-box domain (Ben-Naim et al., 2006; Min et al., 2015). Ghd2 also possessed trans-activation activity, and the activation assay of a series of deletion mutants of Ghd2 in yeast showed that the activation domain was also located in the middle region of the Ghd2 protein (Supplementary Fig. S7). We also confirmed that Ghd2 has high trans-activation activity in protoplasts (Fig. 5A).
Fig. 5.
Ghd2 activated the expression of SAGs in protoplasts. Effector (Ghd2) and reporter (promoter) constructs were co-transformed into rice protoplasts using a dual firefly-Renilla luciferase system. (A) Trans-activation activity of Ghd2 in rice protoplasts. 35S::Gal4BD:Ghd2 was co-transformed with Gal4::LUC and strongly induced LUC expression. (B–H) 35S::Ghd2 induced the expression of LUC driven by the promoters of SAGs under normal and ABA treatment conditions. The white bar shows the LUC activity of the empty effector vector and the grey bar the LUC activity of 35S::Ghd2 co-transformed with the reporter constructs.
Thereafter, we examined whether Ghd2 could activate the expression of some SAGs by using a dual firefly-Renilla luciferase system in rice protoplasts. The results showed that overexpression of Ghd2 in rice protoplasts resulted in the up-regulated expression of several SAGs, including ICL and MS in the glyoxylate cycle, OsPPDKB and OsAS1 in amino acid remobilization, and OsSGR, OsNYC3, and OsPAO in chloroplast degradation (Fig. 5B–H). Because ABA is an important phytohormone participating in leaf senescence (Liang et al., 2014), and Ghd2 was induced by ABA (Fig. 1B), we treated protoplasts with ABA and observed that ABA treatment enhanced the induction of these SAGs, with the exception of MS (Fig. 5B–H). These results indicate that Ghd2 can activate the expression of some SAGs, which partially explains the accelerated senescence by Ghd2 overexpression.
Identification of Ghd2-interacting proteins
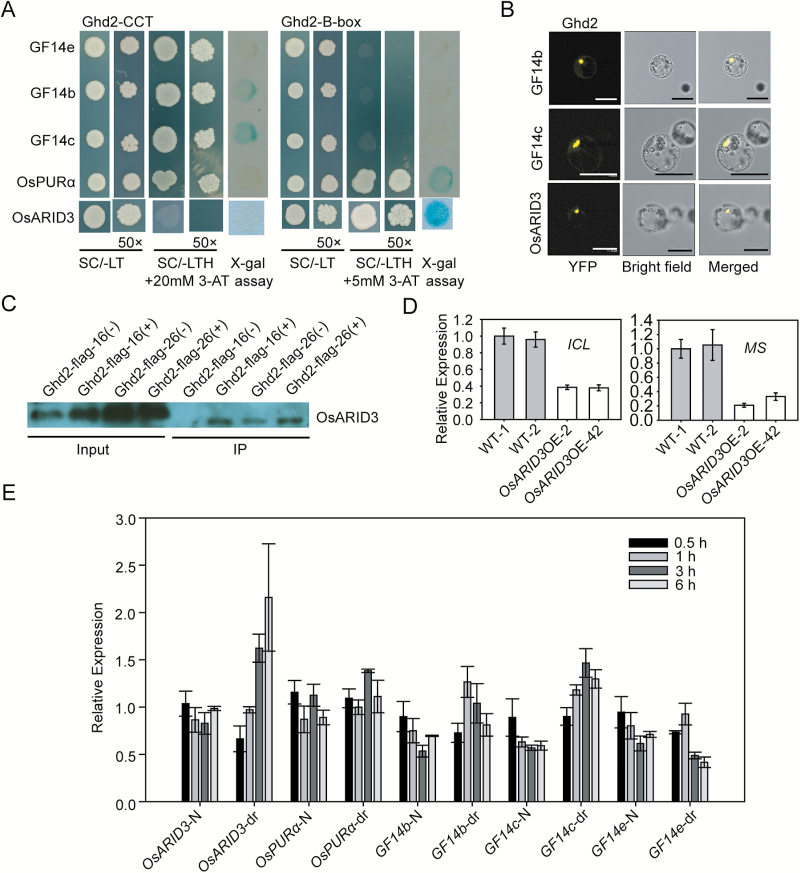
Both the CCT domain and B-box domain are able to mediate protein–protein interactions (Ben-Naim et al., 2006). We performed Y2H screening to identify the interacting proteins of Ghd2. Because the complete Ghd2 protein has strong activation activity, we used N3 and C3, two deletions without self-activation activity and containing N-terminal B-box and C-terminal CCT domains, respectively (Supplementary Fig. S7B), as baits in the screening. Sequencing of the positive clones from the Y2H screening resulted in two candidates (OsARID3 and OsPURα) for N3-interacting proteins and three candidates (OsGF14 b, c, and e) for C3-interacting proteins (Fig. 6A). These candidate proteins were validated to interact with Ghd2 by both the HIS3 and lacZ reporters, except OsGF14e, which could only be validated by the HIS3 reporter. We further confirmed the interactions for OsGF14b, OsGF14c, and OsARID3 by BiFC (Fig. 6B). The interaction between Ghd2 and OsARID3 was further confirmed using an in vivo pull-down assay (Fig. 6C).
Fig. 6.
Identification of Ghd2-interacting proteins. (A) 14-3-3 proteins (GF14b, GF14c, and GF14e), OsPURα, and OsARID3 interact with C3 or N3 deletions of Ghd2 in Y2H assays. Colonies grown on Synthetic complete-Leu-Trp medium (SC/-LT) were tested by both X-gal assay and selection on Synthetic complete-Leu-Trp-His medium (SC/-LTH) containing 5 and 20mM 3-AT for N3 and C3, respectively. (B) 14-3-3 proteins (GF14b and GF14c) and OsARID3 were confirmed to interact with Ghd2 by BiFC. YFP, yellow fluorescent protein. Bar = 20 μm. (C) Confirmation of the interaction between Ghd2 and OsARID3 by an in vivo pull-down assay. (D) Expression of ICL and MS in the OsARID3-OE and WT plants. (E) Expression analysis of OsPURα, OsARID3, and 14-3-3 proteins under drought stress conditions. This figure is available in colour at JXB online.
We checked the microarray results for OsARID3-overexpression plants (OsARID3-OE) generated previously in our group (Xu et al., 2015). Interestingly, we found that ICL and MS were down-regulated in the OsARID3-OE plants, which is in contrast to the up-regulation of ICL and MS in the Ghd2-OE plants. qRT-PCR analysis confirmed this result (Fig. 6D), suggesting that Ghd2 and OsARID3 may work antagonistically in regulating the two SAGs.
The transcript levels of these Ghd2-interacting proteins were examined in response to drought stress treatment. The results showed that OsARID3 was induced by drought stress treatment (Fig. 6E), which is the opposite to the drought-suppressed expression of Ghd2 (Fig. 1B). OsPURα showed no significant change in response to drought stress treatment, while the three 14-3-3 genes responded differently, with GF14b and GF14c being induced and GF14e being repressed by drought stress treatment (Fig. 6E). We also checked the diurnal expression patterns of these genes at different developmental stages. We found that the expression patterns of OsARID3 and OsPURα were very similar to Ghd2, with the highest levels occurring during the night at the grain-filling stage (Supplementary Fig. S6). These results indicate that Ghd2-interacting proteins may be involved in different biological processes, including developmental and drought-induced leaf senescence regulated by Ghd2.
Discussion
Different roles of Ghd2 under optimal growth and stress conditions
CO-like genes have been intensively studied with a focus on their roles in photoperiodic flowering, but very limited information related to abiotic stresses has been reported for this gene family. We found that Ghd2 not only controls grain number, heading date, and plant height (Supplementary Fig. S2A), but also confers drought sensitivity by accelerating drought-induced premature senescence. To date, the molecular link between yield potential and stress tolerance has seldom been addressed. A previous study reported that Ghd7, a close homologue of Ghd2, plays a negative role in drought resistance (Weng et al., 2014), but the mechanism was not completely revealed. Both Ghd2 and Ghd7 are down-regulated by drought stress conditions. Therefore, these CO-like genes, including Ghd2 and Ghd7, may be good candidates to investigate the regulation mechanisms of how plants achieve high yield under favourable growth conditions, and how plants avoid severe yield loss or failure of reproduction under unfavourable environmental conditions. Ghd2 is rhythmically expressed in rice leaves (Fig. 1A) and delays the rice heading date (Supplementary Fig. S2A). The longer vegetative growth period enables rice plants to produce more leaves, and the relatively high expression of Ghd2 at the grain-filling stage could promote the translocation of nutrients from senescent leaves synthesized during the vegetative phase to developing seeds. On the other hand, down-regulation of Ghd2 during drought stress conditions alleviates drought-induced leaf senescence, and thus more photosynthetic leaves remain for seed development.
Ghd2 confers drought sensitivity by accelerating drought-induced leaf senescence
Senescence is a tightly regulated process accompanied by co-ordinated expression of SAGs. To date, several genes have been characterized to play key roles in the regulation of leaf senescence in rice, especially in processes of chlorophyll breakdown and degradation, including OsSGR, OsNYC1, OsNYC3, OsPAO, and OsRCCR (Cha et al., 2002; Kusaba et al., 2007; Park et al., 2007; Morita et al., 2009; Tang et al., 2011), which have been used as marker genes for leaf senescence. OsNAP has been identified as an important regulator in senescence that directly or indirectly regulates SAGs, and it has been suggested as an ideal marker for the onset of leaf senescence in rice (Liang et al., 2014). Frequently used marker genes for leaf senescence also include Osl57 and Osl85, which are involved in fatty acid degradation and remobilization and encode 3-ketoacyl CoA thiolase and ICL respectively (Lee et al., 2001). In this study, we found most of these known marker genes were up-regulated in the Ghd2-OE plants during drought stress (Table 1), suggesting that Ghd2 may regulate leaf senescence by altering the expression of these genes.
Leaf senescence is associated with nutrient remobilization, especially the recycling of nitrogen from the photosynthetic apparatus. Amino acid metabolism is reprogrammed during the senescence process and Asn in particular can serve as the major shunt for storage and long-distance transport of nitrogen from sources to sinks because of its high N:C ratio and relatively inert feature (Lam et al., 1994; Lam et al., 2003). The levels of most free amino acids including Asn were significantly higher in the Ghd2-OE plants than in the WT′ under drought stress conditions, further supporting the roles of Ghd2 in accelerating senescence. Expression of key genes in the glyoxylate cycle, ICL and MS, are increased during leaf senescence (Pastori and Rio, 1997), and PPDK was suggested to salvage carbon from glyoxysome- mediated lipid degradation, thus providing the precursor for the synthesis of Asn (Lin and Wu, 2004; Hartmann et al., 2015). Rice OsAS1 is responsible for the final step in Asn biosynthesis and OsGS1;2 is involved in the pathway (Ohashi et al., 2015). The up-regulation of ICL, MS, OsPPDKB, and OsAS1 in the Ghd2-OE plants during drought stress (Table 1) and the activation of these genes by Ghd2 in rice protoplast (Fig. 5) are in accordance with the higher Asn content (Fig. 2D). These results suggest that Ghd2 may reprogram amino acid metabolism through regulation of genes such as ICL, MS, OsPPDKB, and OsAS1 to redirect C and N transport from sources to sinks. Given that the glyoxylate cycle can convert membrane lipids to soluble carbohydrates or substrates for energy metabolism (Eastmond and Graham, 2001), we speculate that the carbohydrates salvaged from the glyoxylate cycle can also partly contribute to the higher soluble sugar content in the Ghd2-OE plants (Fig. 2E). Furthermore, the term ‘transmembrane transport’ was overrepresented in the up-regulated genes in the Ghd2-OE plants (Supplementary Table S4), implying a possible relevance to accelerated nutrient transport. This evidence together suggests that Ghd2 positively regulates the expression of SAGs during drought stress to accelerate senescence-related processes such as chloroplast breakdown and degradation, metabolism reprogramming, and nutrient mobilization, which finally leads to drought sensitivity.
Ghd2 promotes developmental and dark-induced senescence
Under optimal growth conditions, leaf senescence is initiated in an age-dependent manner, and crops including rice undergo leaf senescence at the grain-filling and maturation stages in the field (Lim et al., 2007; Schippers, 2015). Analysis of OsNAP, which regulates leaf senescence in an age-dependent manner and indicates the onset of senescence, suggests that the age-dependent leaf senescence process in rice is initiated at the tillering stage but not before the four-leaf stage (Liang et al., 2014). In line with this result, the Ghd2-OE plants showed slightly accelerated leaf senescence at the tillering stage in the field (Supplementary Fig. S2A), and senescence was significantly accelerated during the grain-filling and maturation stages (Fig. 4A), whereas senescence was delayed in the Ghd2-CRISPR plants (Fig. 4B), suggesting that Ghd2 can also promote developmental senescence in addition to drought-induced senescence. Because developmental leaf senescence at the grain-filling and maturation stages involves nutrient translocation from senescent leaves to developing seeds (Liang et al., 2014), the relatively high expression of Ghd2 in matured leaves especially at the grain-filling stage (Supplementary Fig. S6) could be related to nutrient translocation in the developmental senescence process. The acceleration of leaf senescence at the grain-filling stage by Ghd2 is reasonable because Ghd2 overexpression could delay flowering time (Supplementary Fig. S2) and there is a great demand for the late flowering plants to quickly finish the grain-filling process before encountering unfavourable low temperature conditions. In addition, accelerated dark-induced leaf senescence in Ghd2-OE plants and delayed dark-induced leaf senescence in Ghd2-CRISPR plants (Fig. 4D–H) further confirms the involvement of Ghd2 in senescence regulation.
Ghd2-interacting proteins and their diverse roles
It has been reported that co-factors such as HAP3 and HAP5 could interact with CO-like proteins to form a complex and bind target genes (Ben-Naim et al., 2006; Wenkel et al., 2006). In addition to HAPs, other CO-like interacting proteins could also play important roles in the regulation of CO-like genes. 14-3-3 proteins μ and ν interact with CO and influence various biological processes, such as transition to flowering, early phytochrome response, red light signalling, and chloroplast development (Folta et al., 2008; Mayfield et al., 2007; Mayfield et al., 2012). Specifically, 14-3-3 proteins form a florigen activation complex with Hd3a and OsFD1 to control flowering time (Taoka et al., 2011). Some 14-3-3 genes have been reported to be responsive to various stresses, such as drought, heat, salt, and cold (Chen et al., 2006; Denison et al., 2011). In this study, we have identified three close homologues of the 14-3-3 protein, including GF14b, GF14c, and GF14e, which specifically interact with the CCT domain of Ghd2. We found that the three rice 14-3-3 genes responded differently to drought stress conditions, with GF14b and GF14c being induced and GF14e being repressed by drought stress treatment. The wide spread participation of 14-3-3 proteins in various biological processes could be associated with the diverse functions of Ghd2, including the regulation of flowering time and/or drought resistance.
In addition to the CCT domain-interacting 14-3-3 proteins, OsPURα and OsARID3 specifically interact with the B-box domain of Ghd2. OsPURα is a DNA/RNA-binding protein (Chang et al., 2011), and it may facilitate the transcriptional regulation of Ghd2 via its DNA-binding activity. Interestingly, we also identified OsARID3 as a Ghd2-interacting protein. OsARID3 is a member of the rice AT-rich interaction domain (ARID) family, which was identified as being required for shoot apical meristem development (Xu et al., 2015). ICL and MS, which were found to be up-regulated in the Ghd2-OE plants, were down-regulated in the OsARID3-OE plants, suggesting that Ghd2 and OsARID3 may work antagonistically in regulating the genes associated with senescence.
We noticed that the expression profiles of OsPURα and OsARID3 were different from that of Ghd2. OsPURα showed no obvious transcript level change in the drought stress treatment, but its expression pattern was similar to Ghd2, with the highest level at the grain-filling stage, indicating that OsPURα may participate in the regulation of developmental senescence along with Ghd2. However, OsARID3 was induced by drought stress treatment, and it was also expressed in rice leaves with the highest level at the grain-filling stage, indicating that OsARID3 may be involved in both developmental and drought-induced leaf senescence mediated by Ghd2.
In conclusion, Ghd2 is a CO-like transcription factor which affects drought sensitivity by accelerating drought-induced leaf senescence. Ghd2 is also involved in the regulation of developmental and dark-induced leaf senescence. The accelerated leaf senescence exhibited by Ghd2 overexpression can be partially explained by the regulation of several SAGs, including ICL/Osl85, MS, OsPPDKB, OsAS1, OsSGR, OsNYC3, and OsPAO. The interaction proteins of Ghd2, including OsARID3, OsPURα, and 14-3-3 proteins, could participate in the Ghd2-mediated regulation network in developmental and stress-induced leaf senescence.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Table S2. Public microarray results of OsK, OsL, and OsJ in response to drought stress treatment.
Table S3. Public microarray results of OsK in response to drought stress treatment in three rice organs.
Table S4. GO analysis of up-regulated DEGs in the Ghd2-OE plants.
Figure S1. Expression level and copy number detection.
Figure S2. Phenotypes of the Ghd2-OE plants.
Figure S3. Phenotypes of the Ghd2-OE and Ghd2-CRISPR plants under drought stress treatment.
Figure S4. Stomata and water loss rate in the Ghd2-OE and WT' plants.
Figure S5. H2O2 content in the Ghd2-CRISPR and WT plants under drought stress treatment.
Figure S6. Diurnal expression levels of Ghd2, OsPURα, OsARID3, and 14-3-3 proteins at different developmental stages.
Figure S7. Ghd2 is located in the nucleus and functions as a transcriptional activator.
Acknowledgements
We acknowledge Lei Wang for providing the 35S:Ghd7-CFP plasmid; Dr Masaru Ohme-Takagi (National Institute of Advanced Industrial Science and Technology, Japan) for providing the 35S-GAL4-LUC, GAL4-LUC, and 190LUC plasmids; and Dr Shouyi Chen (Institute of Genetics and Developmental Biology, China) for providing the GAL4BD, None, and Ubi-Rennila LUC plasmids. We also thank Dong Li and Jian Wan for measuring the metabolites, and Jianbo Cao for TEM observation. This work was supported by grants from the National Program for Basic Research of China (2012CB114305) and the National Program on High Technology Development (2014AA10A600, 2016YFD0100604).
References
- Albacete AA, Martinez-Andujar C, Perez-Alfocea F. 2014. Hormonal and metabolic regulation of source-sink relations under salinity and drought: from plant survival to crop yield stability. Biotechnology Advances 32, 12–30. [DOI] [PubMed] [Google Scholar]
- Balazadeh S. 2014. Stay-green not always stays green. Molecular Plant 7, 1264–1266. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Kohler B, Mueller-Roeber B. 2010. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. The Plant Journal 62, 250–264. [DOI] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. 2006. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. The Plant Journal 46, 462–476. [DOI] [PubMed] [Google Scholar]
- Cao J, Song Y, Wu H, Qin L, Hu L, Hao R. 2013. Ultrastructural studies on the natural leaf senescence of Cinnamomum camphora. Scanning 35, 336–343. [DOI] [PubMed] [Google Scholar]
- Cha K-W, Lee Y-J, Koh H-J, Lee B-M, Nam Y-W, Paek N-C. 2002. Isolation, characterization, and mapping of the stay green mutant in rice. Theoretical and Applied Genetics 104, 526–532. [DOI] [PubMed] [Google Scholar]
- Chang JC, Liao YC, Yang CC, Wang AY. 2011. The purine-rich DNA-binding protein OsPuralpha participates in the regulation of the rice sucrose synthase 1 gene expression. Physiologia Plantarum 143, 219–234. [DOI] [PubMed] [Google Scholar]
- Chen F, Li Q, Sun L, He Z. 2006. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Research 13, 53–63. [DOI] [PubMed] [Google Scholar]
- Chen LJ, Wuriyanghan H, Zhang YQ, Duan KX, Chen HW, Li QT, Lu X, He SJ, Ma B, Zhang WK, Lin Q, Chen SY, Zhang JS. 2013. a. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiology 163, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. 2002. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. The Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J. 2013. b. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Molecular Plant 6, 1769–1780. [DOI] [PubMed] [Google Scholar]
- Crocco CD, Botto JF. 2013. BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531, 44–52. [DOI] [PubMed] [Google Scholar]
- Denison FC, Paul AL, Zupanska AK, Ferl RJ. 2011. 14-3-3 proteins in plant physiology. Seminars in Cell & Development Biology 22, 720–727. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Graham IA. 2001. Re-examining the role of the glyoxylate cycle in oilseeds. Trends in Plant Science 6, 72–77. [DOI] [PubMed] [Google Scholar]
- Folta KM, Paul AL, Mayfield JD, Ferl RJ. 2008. 14-3-3 isoforms participate in red light signaling and photoperiodic flowering. Plant Signaling & Behavior 3, 304–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J. 2012. The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiology 160, 1795–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama K, Kojima S, Tabuchi-Kobayashi M, Sawa Y, Nakayama Y, Hayakawa T, Yamaya T. 2013. Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant & Cell Physiology 54, 934–943. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF. 2014. The BBX family of plant transcription factors. Trends in Plant Science 19, 460–470. [DOI] [PubMed] [Google Scholar]
- Gaufichon L, Rothstein SJ, Suzuki A. 2016. Asparagine metabolic pathways in Arabidopsis. Plant & Cell Physiology 57, 675–689. [DOI] [PubMed] [Google Scholar]
- Graham IA. 2008. Seed storage oil mobilization. Annual Review of Plant Biology 59, 115–142. [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. 2003. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiology 131, 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan SS. 2012. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant, Cell & Environment 35, 644–655. [DOI] [PubMed] [Google Scholar]
- Hartmann L, Pedrotti L, Weiste C, et al. 2015. Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. The Plant Cell 27, 2244–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araujo WL, Braun HP. 2015. Amino acid catabolism in plants. Molecular Plant 8, 1563–1579. [DOI] [PubMed] [Google Scholar]
- Hortensteiner S, Krautler B. 2011. Chlorophyll breakdown in higher plants. Biochimca et Biophysica Acta 1807, 977–988. [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. 2007. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology 143, 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K. 2013. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiology 161, 1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Chopra R. 2012. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481. [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. 2009. The Arabidopsis B-box zinc finger family. The Plant Cell 21, 3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Yun CH, Lee JH, Jang YH, Park HY, Kim JK. 2008. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228, 355–365. [DOI] [PubMed] [Google Scholar]
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC. 2013. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant & Cell Physiology 54, 1660–1672. [DOI] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, Tanaka A. 2007. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. The Plant Cell 19, 1362–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U, Axelsson T. 2000. Rapid evolution of the family of CONSTANS LIKE genes in plants. Molecular Biology and Evolution 17, 1499–1507. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Peng SS-Y, Coruzzi GM. 1994. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiology 106, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Wong P, Chan HK, Yam KM, Chen L, Chow CM, Coruzzi GM. 2003. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiology 132, 926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R-H, Wang C-H, Huang L-T, Chen S-CG. 2001. Leaf senescence in rice plants cloning and characterization of senescence up-regulated genes. Journal of Experimental Botany 52, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Lee YS, Jeong DH, Lee DY, et al. 2010. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. The Plant Journal 63, 18–30. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao Y, Liu X, Peng J, Guo H, Luo J. 2014. LSD 2.0: an update of the leaf senescence database. Nucleic Acids Research 42, D1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, et al. 2014. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proceedings of National Academy of Sciences, USA 111, 10013–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Lin JF, Wu SH. 2004. Molecular events in senescing Arabidopsis leaves. The Plant Journal 39, 612–628. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Liu X, Li Z, Jiang Z, Zhao Y, Peng J, Jin J, Guo H, Luo J. 2011. LSD: a leaf senescence database. Nucleic Acids Research 39, D1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mayfield JD, Folta KM, Paul AL, Ferl RJ. 2007. The 14-3-3 proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiology 145, 1692–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JD, Paul AL, Ferl RJ. 2012. The 14-3-3 proteins of Arabidopsis regulate root growth and chloroplast development as components of the photosensory system. Journal of Experimental Botany 63, 3061–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch SR. 2008. Gene nomenclature system for rice. Rice 1, 72–84. [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. 2013. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Research 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JH, Chung JS, Lee KH, Kim CS. 2015. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. Journal of Integrative Plant Biology 57, 313–324. [DOI] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M. 2009. Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. The Plant Journal 59, 940–952. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nagaoka S, Takano T. 2003. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. Journal of Experimental Botany 54, 2231–2237. [DOI] [PubMed] [Google Scholar]
- Ohashi M, Ishiyama K, Kojima S, Konishi N, Nakano K, Kanno K, Hayakawa T, Yamaya T. 2015. Asparagine synthetase1, but not asparagine synthetase2, is responsible for the biosynthesis of asparagine following the supply of ammonium to rice roots. Plant & Cell Physiology 56, 769–778. [DOI] [PubMed] [Google Scholar]
- Ougham H, Hortensteiner S, Armstead I, Donnison I, King I, Thomas H, Mur L. 2008. The control of chlorophyll catabolism and the status of yellowing as a biomarker of leaf senescence. Plant Biology 10 Suppl 1, 4–14. [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, et al. 2007. The senescence-induced staygreen protein regulates chlorophyll degradation. The Plant Cell 19, 1649–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori CM, Rio LAd. 1997. Natural senescence of pea leaves. Plant Physiology 113, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Marijuan M, Munne-Bosch S. 2014. Photo-oxidative stress markers as a measure of abiotic stress-induced leaf senescence: advantages and limitations. Journal of Experimental Botany 65, 3845–3857. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC. 2015. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. The Plant Cell 27, 1771–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers JH. 2015. Transcriptional networks in leaf senescence. Current Opinion in Plant Biology 27, 77–83. [DOI] [PubMed] [Google Scholar]
- Tan J, Jin M, Wang J, et al. 2016. OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant & Cell Physiology 57, 798–812. [DOI] [PubMed] [Google Scholar]
- Tang Y, Li M, Chen Y, Wu P, Wu G, Jiang H. 2011. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. Journal of Plant Physiology 168, 1952–1959. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, et al. 2011. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. [DOI] [PubMed] [Google Scholar]
- Tetley RM, Thimann KV. 1974. The metabolism of oat leaves during senescence I. Respiration, carbohydrate metabolism, and the action of cytokinins. Plant Physiology 54, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. The Plant Journal 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lin A, Loake GJ, Chu C. 2013. H2O2-induced leaf cell death and the crosstalk of reactive nitric/oxygen species. Journal of Integrative Plant Biology 55, 202–208. [DOI] [PubMed] [Google Scholar]
- Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang Q. 2014. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiology 164, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. 2006. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell 18, 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Purdy S, MacLean JA, Pourtau N. 2006. The role of sugars in integrating environmental signals during the regulation of leaf senescence. Journal of Experimental Botany 57, 391–399. [DOI] [PubMed] [Google Scholar]
- Wingler A, Roitsch T. 2008. Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biology 10 Suppl 1, 50–62. [DOI] [PubMed] [Google Scholar]
- Wu W, Zheng XM, Lu G, et al. 2013. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proceedings of the National Academy of Sciences, USA 110, 2775–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. 2007. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theoretical and Applied Genetics 115, 35–46. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zong W, Hou X, Yao J, Liu H, Li X, Zhao Y, Xiong L. 2015. OsARID3, an AT-rich interaction domain-containing protein, is required for shoot meristem development in rice. The Plant Journal 83, 806–817. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ma C, Xu Y, et al. 2014. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. The Plant Cell 26, 2038–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell 12, 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. 1954. The estimation of carbohydrates in plant extracts by anthrone. The Biochemical Journal 57, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou C. 2013. Signal transduction in leaf senescence. Plant Molecular Biology 82, 539–545. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Q, Dong H, et al. 2015. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Scientific Reports 5, 7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, Jiao Y, et al. 2007. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Molecular Biology 63, 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.