In tomatoes, formation of multiflowered inflorescences depends on a precisely timed process of meristem maturation mediated by the transcription factor gene TERMINATING FLOWER (TMF). Xu et al. show that TMF protein acts together with homologs of the Arabidopsis BLADE-ON-PETIOLE (BOP) transcriptional cofactors, defined by the conserved BTB/POZ domain.
Keywords: meristem, flowering, inflorescence, tomato
Abstract
Plant productivity depends on inflorescences, flower-bearing shoots that originate from the stem cell populations of shoot meristems. Inflorescence architecture determines flower production, which can vary dramatically both between and within species. In tomato plants, formation of multiflowered inflorescences depends on a precisely timed process of meristem maturation mediated by the transcription factor gene TERMINATING FLOWER (TMF), but the underlying mechanism is unknown. We show that TMF protein acts together with homologs of the Arabidopsis BLADE-ON-PETIOLE (BOP) transcriptional cofactors, defined by the conserved BTB (Broad complex, Tramtrack, and Bric-a-brac)/POZ (POX virus and zinc finger) domain. TMF and three tomato BOPs (SlBOPs) interact with themselves and each other, and TMF recruits SlBOPs to the nucleus, suggesting formation of a transcriptional complex. Like TMF, SlBOP gene expression is highest during vegetative and transitional stages of meristem maturation, and CRISPR/Cas9 elimination of SlBOP function causes pleiotropic defects, most notably simplification of inflorescences into single flowers, resembling tmf mutants. Flowering defects are enhanced in higher-order slbop tmf mutants, suggesting that SlBOPs function with additional factors. In support of this, SlBOPs interact with TMF homologs, mutations in which cause phenotypes like slbop mutants. Our findings reveal a new flowering module defined by SlBOP–TMF family interactions that ensures a progressive meristem maturation to promote inflorescence complexity.
Plants display remarkable diversity in when, where, and how many flowers are produced on inflorescences (Rickett 1944; Weberling 1989; Benlloch et al. 2007). Inflorescences originate from small groups of stem cells in shoot apical meristems (SAMs), which transition into inflorescence meristems (IMs) upon perceiving a combination of environmental and endogenous signals (Kobayashi and Weigel 2007; Andrés and Coupland 2012). Depending on the species, inflorescences can be simple, like the single flowers of tulips, or highly complex, like the extraordinarily branched inflorescences of hydrangeas (Weberling 1989; Prusinkiewicz et al. 2007). A quantitative range of complexity exists in between, and at the core of this variation are two major growth programs that influence meristem behavior in different ways following the transition to reproductive growth.
In monopodial plants such as Arabidopsis and maize, the SAM matures into a persistent reproductive state, and the IM generates flowers or inflorescence branches laterally, resulting in relatively narrow range of complexity (Hake 2008; Wang and Li 2008). In contrast, in sympodial plants, such as tomatoes and related nightshades (Solanaceae) (Knapp et al. 2004), meristems mature into terminal flowers, and new growth continuously arises from specialized axillary (sympodial) meristems that serve as the foundation for remarkable diversity in inflorescence architecture (Park et al. 2014).
Sympodial growth is most well understood in tomato, where hundreds of multiflowered inflorescences form from the coordinated activities of sequentially maturing vegetative and reproductive sympodial meristems (Pnueli et al. 1998; Lippman et al. 2008). In a typical plant, sympodial vegetative meristems (SYMs) develop in the axils of the last leaf formed from the primary shoot meristem (PSM) and canonical axillary meristems. Each SYM develops three leaves before terminating in the first flower of a multiflowered inflorescence, and this process continues indefinitely to form compound shoots, where each shoot unit is comprised of three leaves and an inflorescence (Pnueli et al. 1998). Similarly, inflorescences are condensed compound shoots that develop from sympodial IMs (SIMs), each of which initiates one new SIM before maturing into a flower. Several iterations of SIM termination and renewal result in a linear zigzag arrangement of multiple flowers (Lippman et al. 2008). Most tomato genotypes develop six to eight flowers on each inflorescence, but some cultivars and close wild relatives can exceed 20 flowers, and, in more distantly related wild species, inflorescences are even more floral due to branching. This diversity extends into the larger Solanaceae family, where inflorescence complexity ranges from single flowers in plants like peppers and petunias to the extremely branched inflorescences of many Solanaceous trees (Child 1979).
Our previous work on tomato inflorescence mutants and wild species have pointed to a prominent role for the process of meristem maturation in driving inflorescence diversity in sympodial plants (Park et al. 2012, 2014). Specifically, variation in the timing of maturation can modulate complexity such that a slower SIM maturation allows additional SIMs to form in each cycle, resulting in greater inflorescence complexity and vice versa. We previously described a mechanism that promotes meristem maturation in tomato, in which precise timing of activation of the homeobox gene COMPOUND INFLORESCENCE (S; homolog of Arabidopsis WOX9 and petunia EVG) followed by the F-box gene ANANTHA (AN; homolog of Arabidopsis UFO and petunia DOT) drives successive stages of SIM maturation to ensure that only one SIM develops in each sympodial cycle (Hepworth et al. 2006; Lippman et al. 2008; Rebocho et al. 2008; Souer et al. 2008). When S or AN is mutated, maturation is delayed (s mutants) or never achieved (an mutants), resulting in SIM overproliferation and highly branched inflorescences (Lippman et al. 2008; Park et al. 2012). Work in petunia revealed how AN completes the final stage of maturation (Souer et al. 2008). Upon its late expression in the floral meristem (FM), AN protein interacts with FALSIFLORA (FA; homolog of the Arabidopsis transcription factor LFY) to form a flower specification complex, which activates floral identity genes (Koes 2008; Souer et al. 2008).
We recently discovered a new genetic pathway that represses meristem maturation to maintain a vegetative state, defined by the tomato TERMINATING FLOWER (TMF) gene. TMF encodes a member of the conserved ALOG (Arabidopsis LSH1 and Oryza G1) protein family in plants, members of which contain a DNA-binding domain and have weak transcriptional activity (Iyer and Aravind 2012; MacAlister et al. 2012; Yoshida et al. 2013). We found that loss of TMF, one of 11 ALOG genes in tomato, causes much faster flowering and transformation of primary inflorescences into single flowers due to precocious activation of AN–FA during vegetative stages of PSM maturation (MacAlister et al. 2012). Surprisingly, inflorescences that develop from side shoots are unaffected, suggesting that TMF functions specifically to maintain a vegetative state during PSM maturation. Notably, mutations in the closest homolog of TMF in rice simplifies panicle architecture (Yoshida et al. 2013), but loss of the closest homologs in Arabidopsis (LIGHT-SENSITIVE HYPOCOTYL3 and LIGHT-SENSITIVE HYPOCOTYL4) has no effect on inflorescences, although overexpressing these genes suppresses lateral organ differentiation (Takeda et al. 2011). A handful of other ALOG genes in Arabidopsis and rice have reported roles in light signaling and floral organ development (Zhao et al. 2004; Yoshida et al. 2009; Cho and Zambryski 2011; Takeda et al. 2011; Sato et al. 2014). Thus, ALOG proteins represent a new, poorly understood family of growth regulators with prominent, species-specific roles in reproductive development. In this study, we explored the mechanism by which TMF represses meristem maturation to control inflorescence architecture and flower production in sympodial plants.
Results
Tomato BLADE-ON-PETIOLE (BOP) proteins interact with TMF
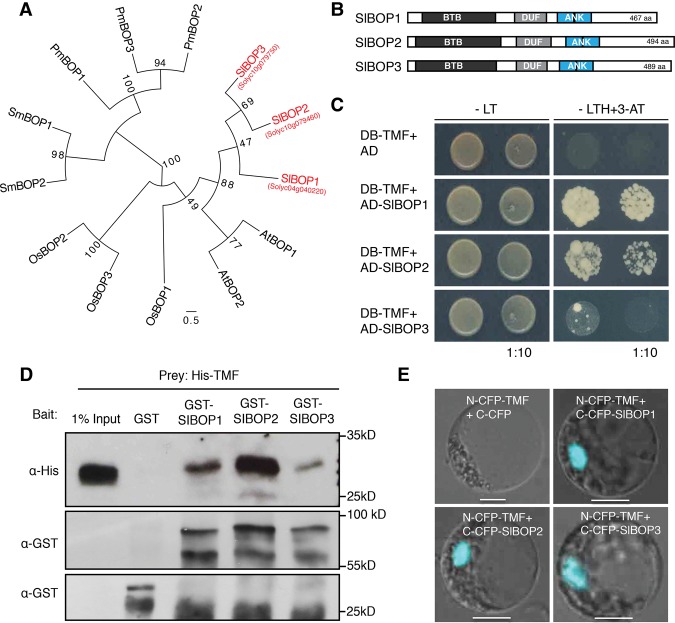
Using TMF as bait in a yeast two-hybrid screen, we previously identified 35 interacting proteins, several of which were annotated as transcription factors/cofactors (MacAlister et al. 2012). Among these were two homologs of the Arabidopsis BOP1 and BOP2 transcriptional coactivators, which have many reported functions in growth and development but are most recognized for their roles in leaf complexity and organ abscission (Ha 2003; Hepworth et al. 2005; Norberg et al. 2005; Ha et al. 2007; McKim et al. 2008). BOP proteins are members of the larger NPR1 (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1) protein family involved in plant defense, defined by two protein–protein interaction motifs found widely in plant and animal proteins: the BTB (Broad complex, Tramtrack, and Bric-a-brac)/POZ (POX virus and zinc finger) domain and ankyrin repeats (Fu and Dong 2013; Khan et al. 2014). Despite numerous genetic and molecular studies on the Arabidopsis BOP genes, little is known about the factors that cooperate with BOP proteins to control diverse aspects of development (Khan et al. 2014). For consistency, we designated the TMF-interacting tomato BOPs as SlBOP1 and SlBOP2. A homology search and phylogenetic analysis revealed SlBOP3 (Fig. 1A; Ichihashi et al. 2014) and a similar size and structure for all three SlBOP proteins (Fig. 1B). We confirmed TMF–SlBOP1/2 interactions in yeast with full-length coding sequences and found that SlBOP3 also interacted with TMF in yeast but less strongly than SlBOP1 and SlBOP2 (Fig. 1C). We validated these interactions using GST pull-down assays and bimolecular fluorescence complementation (BiFC) in tomato protoplasts, which further showed that all three SlBOPs interacted with TMF in the nucleus (Fig. 1D,E; Supplemental Fig. S1).
Figure 1.
SlBOP proteins interact physically with TMF. (A) Neighbor joining phylogenetic tree of full-length BOP family proteins from Solanum lycopersicum (Sl), Arabidopsis thaliana (At), Oryza sativa (Os), Selaginella moellendorffii (Sm), and Physcomitrella patens (Pm). Bootstrap values for 100 replicates are shown. (B) Schematic of protein structure shared by all SlBOP family members. (BTB) BTB/POZ domain (Pfam: 00651); (ANK) ankyrin repeats (Pfam: 00023); DUF3420 domain (Pfam:11900). (C) Yeast two-hybrid assays showing that SlBOP1, SlBOP2, and SlBOP3 interact with TMF. Bait (BD) and prey (AD) constructs were cotransformed into yeast cells as indicated. (BD) DNA-binding domain; (AD) activation domain; (−LT) nonselective medium minus Leu and Trp; (−LTH+3-AT) selective medium minus Leu, Trp, and His supplemented with 2 mM 3-amino-1, 2, 4-triazole (3-AT). (D) Pull-down assay showing interactions between recombinant expressed GST-SlBOP1, GST-SlBOP2, and GST-SlBOP3 and His-TMF. Anti-His and anti-GST antibodies were used to detect His-TMF, GST, and GST-SlBOP proteins, respectively. (E) BiFC assays in tomato protoplasts showing that SlBOP proteins interact with TMF in the nucleus. (N) N-terminal portion of CFP; (C) C-terminal portion of CFP. Bars, 10 µm.
SlBOPs are recruited to the nucleus by TMF to enhance its transcriptional activity
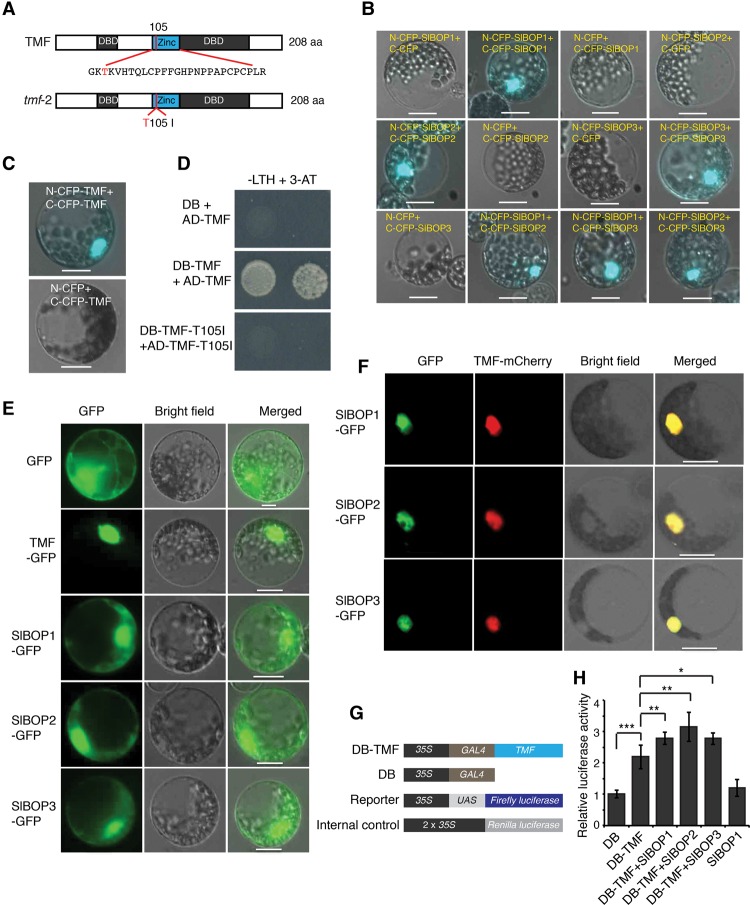
We showed previously that TMF, like other ALOG proteins, is a weak transcriptional activator (MacAlister et al. 2012), and, consistent with this, a recent comparative bioinformatics analysis of ALOG proteins revealed a DNA-binding domain with distant homology with retrotransposon integrase proteins (Fig. 2A; Iyer and Aravind 2012). We therefore reasoned that the role of TMF in flowering, meristem maturation, and inflorescence architecture is mediated by its transcriptional activity and that this activity might be influenced by SlBOP interactions. In Arabidopsis, accumulation of salicylic acid (SA) following pathogen infection induces deoligomerization of cytosolic NPR1 so that monomers can enter the nucleus and form a tetrameric “enhanceosome” with TGA family bZIP transcription factors (Boyle et al. 2009). Arabidopsis BOP1/2 also form homodimers and heterodimers (Jun et al. 2010), and alignment of the SlBOPs with NPR1 and BOP1/2 showed conservation of four cysteine intervals in the BTB/POZ domain that are essential for oligomerization (Supplemental Fig. S2). As expected, the SlBOPs also homodimerized and heterodimerized (Fig. 2B). Notably, TMF also formed homodimers, which we found is critical for function, as introduction of the same point mutation from a weak allele of tmf (tmf-2) (MacAlister et al. 2012) abolished dimerization (Fig. 2A,C,D).
Figure 2.
SlBOP proteins are recruited to the nucleus to form a transcriptional complex. (A) Schematic of protein structures showing putative DNA-binding domains and the tmf-2 amino acid change (T105I) in the zinc ribbon motif. (DBD) DNA binding domain; (Zinc) zinc ribbon motif (Iyer and Aravind 2012). (B) BiFC assays in tomato protoplasts showing homodimerization and hetrodimerization of SlBOP proteins. (C) BiFC assays in tomato protoplasts showing dimerization of TMF proteins. (D) Yeast two-hybrid assays showing failed dimerization of tmf-2. (E) Subcellular localization of transiently expressed SlBOP-GFP fusion proteins in tomato protoplasts showing cytoplasmic and nuclear localization. (F) Subcellular colocalization of transiently expressed TMF-mCherry with SlBOP-GFP fusion proteins in protoplasts. SlBOP proteins localize exclusively in the nucleus when TMF is coexpressed. (G) Schematics of constructs for dual-luciferase transcriptional activity assays in tomato protoplasts. The cauliflower mosaic virus (CaMV) 35S promoter driving five copies of yeast upstream activation sequence (UAS) fused with a firefly luciferase gene served as a reporter, and 2x35S-driven Renilla luciferase served as an internal control. (GAL4) Yeast transcription activator protein with a DNA-binding domain that specifically binds UAS. (H) Transcriptional activity of TMF–SlBOP protein complexes indicated by relative luciferase activity. Two independent experiments with three biological replicates for each experiment were performed. Data are means (±SD). n = 6. A two-tailed, two-sample Student's t-test was performed, and significant differences are represented by black asterisks. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Bars, 10 µm.
Given the intermolecular and intramolecular interactions between TMF and SlBOPs and considering elements of the NPR1-TGA2 mechanism as a framework, we sought evidence in support of TMF–SlBOP interactions resulting in the formation of transcriptional complexes. We first examined SlBOP subcellular localization in protoplasts. Unlike the exclusive nuclear localization of TMF, all three SlBOP-GFP fusion proteins were found in both the cytosol and the nucleus (Fig. 2E), similar to NPR1 and BOP1/2 (Kinkema et al. 2000; Hepworth et al. 2005). We next asked whether SlBOP localization is affected by TMF interactions or vice versa by coexpressing SlBOP-GFP with TMF-mCherry in protoplasts. Significantly, all SlBOP signals shifted exclusively to the nucleus in the presence of TMF (Fig. 2F), indicating that TMF recruits SlBOPs to the nucleus. To test the functional relevance of this recruitment, we assessed transcriptional activity of SlBOP–TMF by performing a dual-luciferase assay in protoplasts (Fig. 2G). Consistent with our findings in yeast (MacAlister et al. 2012), TMF showed weak transcriptional activity (Fig. 2H). Importantly, coexpressing each SlBOP with TMF resulted in a significant modest increase in transcriptional activity, particularly for SlBOP2 (Fig. 2H). Collectively, these results suggest that interactions between TMF and the SlBOPs are the foundation for SlBOP–TMF transcriptional complexes in vivo and that their formation depends on TMF recruitment of SlBOPs to the nucleus.
The SlBOP genes are expressed dynamically during meristem maturation
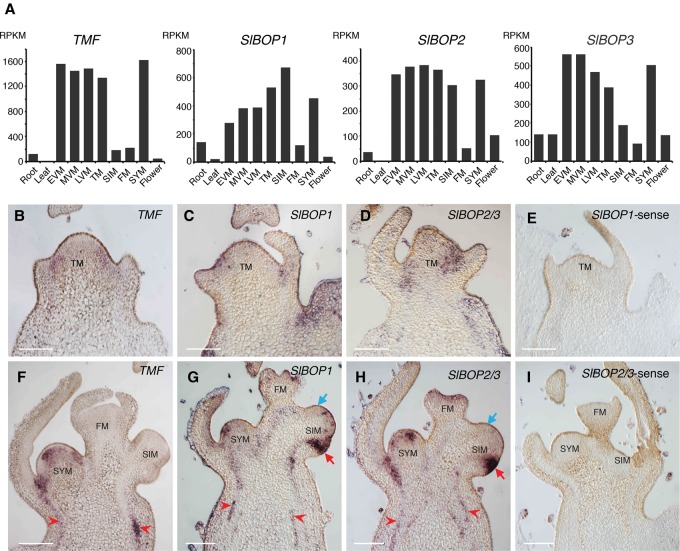
The likely existence of SlBOP–TMF complexes implicates SlBOP genes in meristem maturation and thus flowering and inflorescence development. In support of this, our tomato meristem maturation atlas (Park et al. 2012) showed that all three SlBOP genes are expressed to varying degrees in major tissues, including roots, leaves, and flowers, but are most prominently expressed in meristems (Fig. 3A). Notably, like TMF, the SlBOPs are highly expressed in vegetative and transition meristem (TM) stages of PSM maturation and then decrease in the FM. Expression levels in the SYM resemble the PSM (Fig. 3A). To substantiate these dynamics and compare SlBOP and TMF expression domains, we performed mRNA in situ hybridization. At the TM stage, all three SlBOP genes were expressed in domains overlapping with TMF, at the flanks of the meristem in presumptive boundary regions, and into initiating vasculature cells (Fig. 3B–E). These similarities extended to the SYM, but, unlike TMF, the SlBOPs maintained high levels of expression in SIMs (Fig. 3F–I).
Figure 3.
SlBOP genes are expressed dynamically during meristem maturation. (A) Normalized RNA sequencing (RNA-seq) read counts for TMF, SlBOP1, SlBOP2, and SlBOP3 in major tissues, including five stages of PSM maturation (Park et al. 2012). (EVM) Early vegetative meristem (fifth leaf initiated); (MVM) middle vegetative meristem (sixth leaf initiated); (LVM) late vegetative meristem (seventh leaf initiated); (TM) eighth (final) leaf initiated; (RPKM) reads per kilobase per million mapped reads. (B–I) In situ mRNA hybridization showing expressions of TMF, SlBOP1, and SlBOP2/3 in wild-type meristems. The top right denotes probe. TMF (B), SlBOP1 (C), and SlBOP2/3 (D) are expressed similarly at the periphery of the TM, marking lateral organ boundaries. Expressions overlap also in the SYM, but, unlike TMF (F), SlBOP1 (G) and SlBOP2/3 (H) are strongly expressed in the SIM. Vascular expression is also detected (red arrowheads). (E,I) SlBOP1 and SlBOP2/3 sense probe controls. Bars, 100 µm.
The SlBOP genes have pleiotropic roles in vegetative and reproductive development
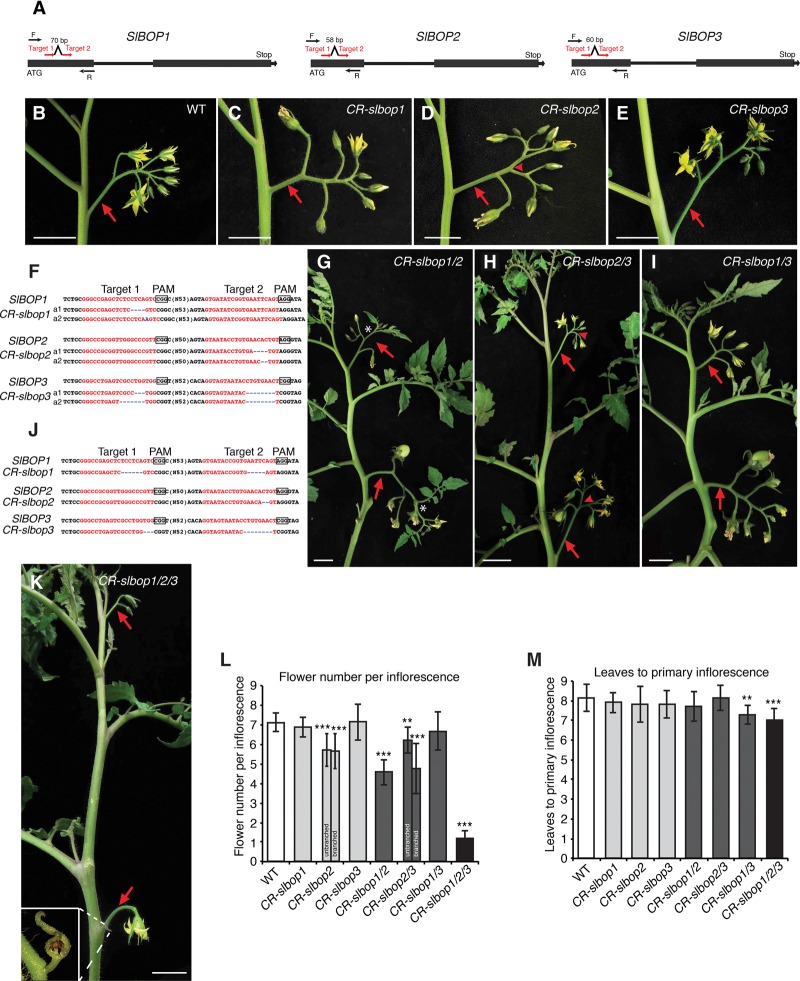
To dissect the individual and combined developmental roles of the SlBOP genes, we created loss-of-function null mutations using CRISPR/Cas9 gene editing. Following our standard protocol (Brooks et al. 2014), we targeted the first exon of each gene with two single-guide RNAs (sgRNAs) (Fig. 4A). Multiple independent first-generation (T0) transgenic lines screened by PCR and sequencing revealed chimeric CRISPR (CR)-slbop1, CR-slbop2, and CR-slbop3 mutants carrying insertion/deletion (indel) alleles of various sizes (Materials and Methods). To obtain homozygous stable mutants, we sequenced T1 progeny plants and isolated two out-of-frame deletion alleles for each gene that caused premature stop codons (Fig. 4B–F). One allele was chosen for further analyses.
Figure 4.
CRISPR/Cas9-engineered mutations in SlBOP genes cause strong inflorescence defects. (A) Schematics illustrating two sgRNAs (red arrows) targeting each SlBOP gene. Black arrows represent PCR genotyping primers. (B–E) A multiflowered inflorescence typical of a wild-type plant (B) and representative inflorescences from single mutants of CR-slbop1 (C), CR-slbop2 (D), and CR-slbop3 (E). The red arrowhead in D indicates a single branching event often observed in CR-slbop2 mutants. (F) Sequences from homozygous T1 transgenic progeny plants lacking the Cas9 transgene showing CRISPR/Cas9-induced out-of-frame deletions (blue dashed lines) for CR-slbop1, CR-slbop2, and CR-slbop3, resulting in two independent null loss-of-function alleles (a1 and a2). The red font highlights sgRNA targets, and black boxes indicate protospacer-adjacent motif (PAM) sequences. (G–I) A representative shoot with two successive inflorescences from CR-slbop1/2 (G), CR-slbop2/3 (H), and CR-slbop1/3 (I) double mutants. The white star in G shows a typical CR-slbop1/2 inflorescence terminating with a leaf or reverting to a vegetative shoot. The red arrowhead in H indicates a single branching event in CR-slbop2/3 double-mutant inflorescences. (J) Sequences showing out-of-frame deletion alleles for each SlBOP gene in the CR-slbop1/2/3 triple mutant. (K) Representative CR-slbop1/2/3 triple-mutant plant showing that all multiflowered inflorescences are transformed into single- or two-flowered inflorescences (arrows). The inset shows an aborted second flower bud with bracts. (L,M) Quantification and statistical comparison of flowers per inflorescence (L) and flowering time (M) in wild-type and CR-slbop single, double, and triple mutants. Note that flower number for CR-bop1/2 plants was quantified as the flowers on each inflorescence before vegetative reversion, and flower number of CR-bop2/3 was quantified as the flowers on each inflorescence branch from branched inflorescences and also unbranched inflorescences separately. Data are means (±SD). n = 12–18 in L; n = 10–15 in M. A two-tailed, two-sample Student's t-test was performed, and significant differences are represented by black asterisks. (**) P < 0.01; (***) P < 0.001. Red arrows indicate inflorescences. Bars: B–K, 2 cm.
Similar to Arabidopsis bop1/2 single and double mutants, all three CR-slbop mutants displayed altered leaf complexity and loss of floral organ abscission (Supplemental Fig. S3A–D). For example, CR-slbop2 mutants showed increased leaf complexity compared with wild-type plants, primarily reflected in the formation of additional intercalary leaflets on the central rachis, perhaps due to increased meristem activity along the marginal blastozone, as in Arabidopsis bop mutants (Ha 2003; Khan et al. 2014). In support of this, in some conditions, we observed fusion between neighboring intercalaries and shoots forming along the rachis of CR-slbop2 leaves (Supplemental Fig. S3C,O). These results suggest that the SlBOPs have overlapping roles in leaf development.
In contrast to the pronounced changes in leaf development and organ abscission, the CR-slbop mutants displayed only subtle inflorescence defects. The most obvious changes were observed in CR-slbop2, where more than half of the inflorescences underwent a single branching event and developed fewer flowers on each inflorescence compared with wild type (Fig. 4D,L). In addition, the arrangement of flowers in both CR-slbop2 and CR-slbop3 inflorescences was frequently disrupted due to defects in pedicel orientation and increased internode length between flowers compared with wild type (Fig. 4B–E). Reproductive defects extended to flower and fruit development; floral organs were often fused between whorls (e.g., stamens fused to carpels) in CR-slbop2 and CR-slbop3, and, consequently, fruits were misshapen and displayed scarring (Supplemental Fig. S3J–N). Several of these reproductive defects, particularly changes in pedicel orientation and internode elongation, were reminiscent of bop mutants in Arabidopsis and barley (Xu et al. 2010; Khan et al. 2012; Jost et al. 2016). Thus, similar to BOP genes in other species, the tomato SlBOP family functions in multiple developmental contexts (Couzigou et al. 2012; Wu et al. 2012; Ichihashi et al. 2014; Khan et al. 2014; Tavakol et al. 2015; Jost et al. 2016).
Higher-order CR-slbop mutants show enhanced leaf and inflorescence defects
Given that all three SlBOP proteins interact with TMF and that their genes share overlapping meristem expression patterns, we created higher-order mutants to determine whether the SlBOPs have redundant roles in vegetative and reproductive development. To generate all mutant combinations, we crossed T0 plants and screened the progeny of F1 plants. Notably, crossing T0 chimeric CR-slbop lines allowed us to exploit CRISPR/Cas9 to reciprocally induce mutations in the wild-type SlBOP alleles introduced in the F1 cross (Materials and Methods). Even more, this approach allowed us to efficiently generate mutations in the closely linked SlBOP2 and SlBOP3 genes (231 kb apart on chromosome 10).
All three CR-slbop double-mutant combinations showed pleiotropic changes similar to those in the single mutants, including changes in leaf complexity and loss of organ abscission (Supplemental Fig. S3E–G). Notably, CR-slbop1/2 and CR-slbop2/3 leaves produced even more intercalary leaflets than the single mutants. Surprisingly, CR-slbop1/3 leaves developed fewer intercalary leaflets and therefore appeared simpler. These findings suggest a more complex relationship among the SlBOPs in determining leaf complexity (Ichihashi et al. 2014).
In addition to enhanced leaf phenotypes, the CR-slbop double mutants displayed a range of modifications to inflorescence architecture. Most dramatically, CR-slbop1/2 inflorescences developed fewer flowers than wild type, reverted to a vegetative shoot on the primary inflorescence, and later formed inflorescences terminated in leaves (Fig. 4G,L). In contrast, CR-slbop2/3 inflorescences showed no vegetative characteristics but branched more frequently than CR-slbop2 mutants alone and also developed fewer flowers (Fig. 4H,L). CR-slbop1/3 inflorescences were more similar to wild type, but, interestingly, we found that these double mutants flowered slightly faster than wild type by one leaf (Fig. 4I,M).
The SlBOP genes act together to control inflorescence architecture and flower production
To test whether all three SlBOP genes act together to control flowering and inflorescence development, we generated triple mutants by crossing homozygous CR-slbop1/2 and CR-slbop2/3 double mutants and screening progeny from F1 plants (Materials and Methods). Similar to CR-slbop1/3 double mutants, the triple mutants also flowered faster than the wild type (Fig. 4M). Most strikingly, we found that all inflorescences were transformed into one or two flowers (Fig. 4J–L; Supplemental Fig. S3H). To understand the developmental basis for this dramatic simplification of inflorescence complexity, we compared reproductive stages of meristem development from the double and triple mutants and found that CR-slbop1/2 plants initiated fewer SIMs than wild type, CR-slbop2/3 plants initiated two SIMs on the sides of the first FM, and triple mutants nearly always failed to initiate SIMs (Fig. 5). Interestingly, loss of SIMs was frequently accompanied by the formation of small bracts. These findings, based on a complete series of CR-slbop mutants, show that all three SlBOP genes are required to initiate and perpetuate the formation and maturation of SIMs.
Figure 5.
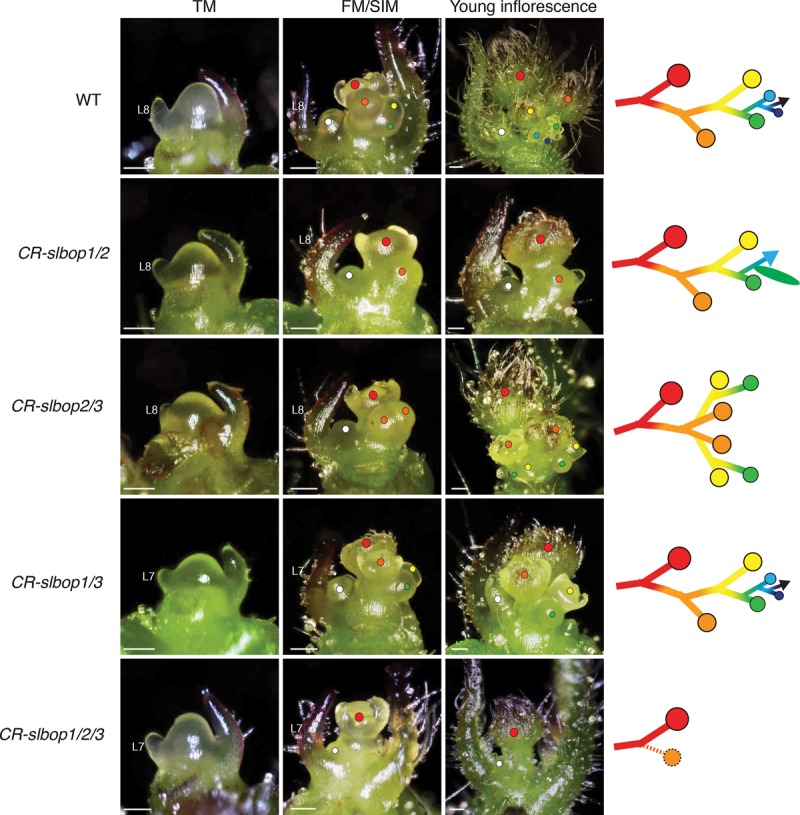
Meristem maturation of slbop mutants. Stereoscope images comparing PSM maturation from wild type and CR-slbop double and triple mutants. Note that the TM stage is indistinguishable, with defects arising at the FM stage when the first SIM is initiated. The wild type shows the typical zigzag reiteration of SIMs that gives rise to multiflowered inflorescences. Varying defects of SIM reiteration occur in slbop double mutants, including reduced SIM production and vegetative reversion in CR-slbop1/2, two SIMs initiated in CR-slbop2/3, and normal SIM reiteration in slbop1/3. The FM stage from CR-slbop1/2/3 shows a failure to initiate SIM reiteration, resulting in one to two flowers per inflorescence. Colored dots on stereoscope images reflect successive SIM initiation (yellow, green, blue, etc.) and FM termination (red and orange dots). The white dot indicates the first SYM in the axil of the last formed leaf on the PSM. Diagrams at the right reflect the resulting inflorescences in each genotype. The black arrow in wild type indicates continued SIM reiteration, the blue arrow and green oval in CR-slbop1/2 indicate vegetative reversion, and the dotted orange line and circle in CR-slbop1/2/3 indicate infrequent formation of a second SIM. Bars, 100 µm.
TMF and SlBOPs act synergistically to prevent precocious flowering and promote inflorescence complexity
Our combined molecular and genetic dissection of SlBOP function suggested a close relationship with TMF in controlling flowering, meristem maturation, and inflorescence architecture. However, there are several notable distinctions between the reproductive phenotypes of tmf and CR-slbop mutants. Specifically, tmf mutants flower much faster than CR-slbop triple mutants, after only four leaves compared with seven leaves in CR-slbop triple mutants and eight leaves in wild type (MacAlister et al. 2012). Furthermore, only the primary inflorescence of tmf is single-flowered, and this flower often develops leaf-like sepals. In contrast, all inflorescences on CR-slbop1/2/3 triple-mutant plants develop only one to two flowers, and these flowers are morphologically much more similar to wild type than to tmf single flowers. These similarities and distinctions suggested that TMF and SlBOP functions do not completely overlap.
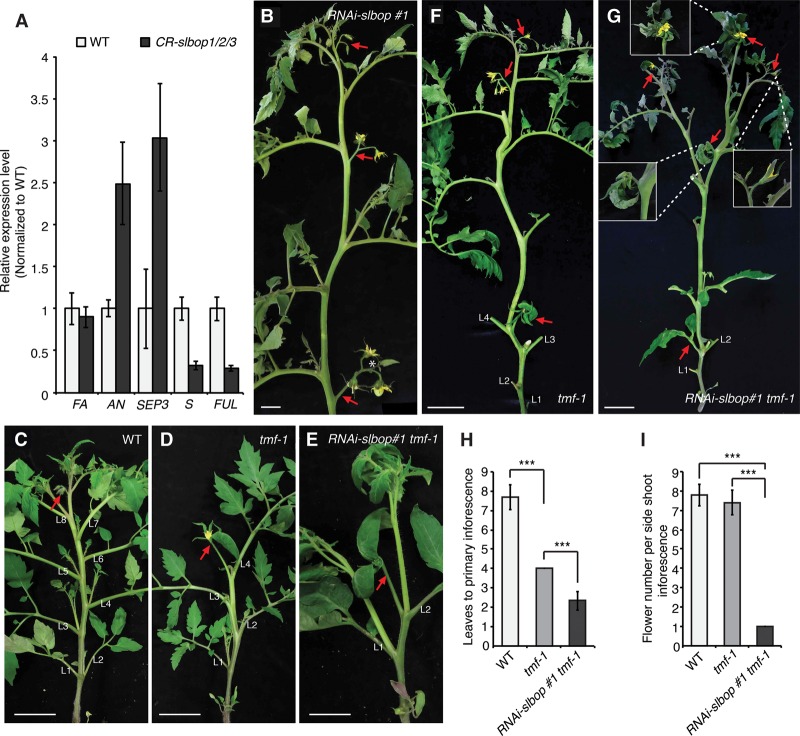
To explore this possibility, we performed several genetic analyses. Given that tmf phenotypes are due to precocious activation of the AN–FA complex and that driving AN expression one stage earlier than its normal activation using the promoter of the S gene closely mimics CR-slbop1/2/3 triple-mutant inflorescences (MacAlister et al. 2012), we asked whether the CR-slbop1/2/3 reproductive phenotypes depend on AN or FA. In support of this, the FM identity genes AN and SEPALLATA3 (SEP3) were both up-regulated more than twofold in the TM stage of CR-slbop triple mutants (Fig. 6A). This subtle change is consistent with a slightly precocious activation of AN–FA and thus adoption of floral fate during meristem maturation, explaining CR-slbop early flowering and simplified inflorescences. We tested this genetically by combining slbop with an and fa loss-of-function mutations. To reduce the complexity of this analysis, we took advantage of RNAi to simultaneously knock down the activity of all three SlBOP genes in a dominant manner (Supplemental Fig. S4). We recovered two lines showing down-regulation of all three SlBOPs with a phenotypic severity between CR-slbop1/2 double and CR-slbop1/2/3 triple mutants (Materials and Methods; Fig. 6B; Supplemental Fig. S5). We used one of these lines to generate RNAi-slbop an and RNAi-slbop fa double mutants, and both double mutants developed branched inflorescences lacking flowers due to floral organ identity defects typical of an and fa (Lippman et al. 2008). However, branching was not as severe as in an or fa alone, and the inflorescences of RNAi-slbop an plants often developed leaves (Supplemental Fig. S6).
Figure 6.
Loss of SlBOP activity enhances tmf mutant phenotypes. (A) Quantitative RT–PCR (qRT–PCR) on the TM stage of wild-type and CR-slbop1/2/3 primary shoots for the FM identity genes FA, AN, and SEP3 compared with the flowering transition gene S and a tomato homolog of Arabidopsis FRUITFUL (FUL). Values are the averages of two biological and six technical replicates. UBIQUITIN (UBI) served as an internal control, and values were normalized relative to wild type. (B) Representative main shoot from an RNAi-slbop plant showing two- or three-flowered inflorescences. (C–E) Representative primary shoots showing typical multiflowered primary inflorescence after eight leaves in wild type (C) compared with tmf-1, which flowers after four leaves and produces a single-flowered primary inflorescence with leaf-like sepals (D). (E) RNAi-slbop tmf-1 plants flower after only two leaves, and the sepals of the single-flowered primary inflorescence are even more leaf-like. (F,G) A whole-plant view of tmf-1 and RNAi-slbop tmf-1 plants showing that the typical multiflowered inflorescences from tmf-1 side shoots (F) are transformed into single flowers with leaf-like sepals in RNAi-slbop tmf-1 plants (G). (H,I) Quantification and comparison of flowering time (as measured by leaves to primary inflorescence) and flower number of the primary inflorescence from the indicated genotypes. (L) Leaf. Red arrows indicate inflorescences. Data are means (±SD). n = 6 in A; n = 15 in H and I. A two-tailed, two-sample Student's t-test was performed, and significant differences are represented by black asterisks. (***) P < 0.001. Bars: B, 2 cm; C–G, 5 cm.
Compared with the complete dependence of tmf phenotypes on AN and FA (MacAlister et al. 2012), this partial suppression suggested that the early flowering and simplified inflorescences of slbop triple mutants rely on additional factors and further points to a complex interplay between the SlBOPs and TMF in reproductive development. In support of this, we found that combining RNAi-slbop with tmf dramatically enhanced flowering and inflorescence defects. Compared with our null tmf-1 allele, which flowers after four leaves, RNAi-slbop tmf-1 mutants flowered after only two leaves and developed single-flowered inflorescences throughout the entire plant, and the sepals of these flowers were even more leaf-like (Fig. 6C–I). We validated this synergistic interaction by combining RNAi-slbop with the weaker tmf-2 allele that is less early flowering than tmf-1 and shows weak penetrance (9%) of single-flower primary inflorescences (Supplemental Fig. S7A,D; MacAlister et al. 2012). Significantly, RNAi-slbop tmf-2 plants also flowered after two leaves, the penetrance of single-flower primary inflorescences exceeded 90%, and all inflorescences from side shoots were transformed to one to two flowers with leaf-like sepals (Supplemental Fig. S7B–D). Taken together, these genetic interactions demonstrate that the roles of the SlBOPs in flowering and inflorescence development do not depend exclusively on TMF and suggest that other factors are involved. In this regard, it is notable that three TMF family members (TFAMs) are expressed predominantly in meristems, with TFAM1 and TFAM2 showing expression dynamics closely resembling TMF and SlBOP1 and SlBOP2 (Supplemental Fig. S8A; Park et al. 2012). Even more, we found that both TFAM1 and TFAM2 proteins interact with all three SlBOPs in yeast, and CRISPR/Cas9-generated null mutations in each gene result in phenotypes that match multiple defects of slbop single mutants, most significantly reduced flower production in CR-tfam1 and a high frequency of inflorescences with single branching events in CR-tfam2 (Supplemental Fig. S8B–H). In addition, CR-tfam1 mutants showed altered leaf complexity, fused floral organs, loss of floral organ abscission, and defects in fruit shape (Supplemental Fig. S8E,I–K). These results suggest that, like the SlBOPs, TMF family members have independent and overlapping redundant functions and further point to the existence of multiple SlBOP–ALOG transcriptional complexes.
Discussion
Despite many studies over the last decade, surprisingly little is known about the molecular mechanisms by which the conserved BOP protein family regulates diverse aspects of plant development (Khan et al. 2014). Indeed, only one BOP protein partner is known in Arabidopsis, a TGA family bZIP transcription factor, PERIANTHIA (PAN), which shares overlapping roles with BOP1/2 in regulating floral patterning (Hepworth et al. 2005). Here, we showed that the tomato SlBOP family, in part through interactions with the ALOG transcriptional regulator TMF, plays a major role in flowering and inflorescence architecture by modulating meristem maturation. All BOP proteins share ankryin repeats and the BTB/POZ domains with the NPR1 family, including four cysteine residues in the BTB/POZ domain that are important for oligomerization (Jun et al. 2010). However, BOP proteins lack the NPR1 domains required for SA-induced translocation and transactivation activity, raising the questions of whether there is regulated transport of cytoplasmically localized BOP proteins to the nucleus and what factors might be involved (Boyle et al. 2009; Spoel et al. 2009; Jun et al. 2010; Fu and Dong 2013). Our findings from interaction and expression assays, including evidence that TMF recruits SlBOPs to the nucleus, strongly suggest that SlBOP–TMF complexes exist in vivo, providing the first insights into a new molecular mechanism underlying one of the many developmental roles of the BOP family (Hepworth et al. 2005; Jun et al. 2010; Khan et al. 2014).
Like BOP genes in other species, the SlBOPs seem to have unequal redundant functions in vegetative and reproductive development. For example, whereas Arabidopsis BOP1 seems to have a more prominent role in leaf development, BOP2 functions more during reproductive growth (Khan et al. 2014). Similarly, recent work in barley has shown that two BOP homologs regulate tillering, ligule development, proximal–distal leaf patterning, and several aspects of inflorescence development, including internode length and floral organ development (Tavakol et al. 2015; Jost et al. 2016). Our data suggest that SlBOP2 is the dominant family member in tomato. However, the earlier flowering and extreme simplification of inflorescences in CR-slbop triple mutants contrasts the comparatively weak bop flowering and inflorescence phenotypes in Arabidopsis and barley (Hepworth et al. 2005; Norberg et al. 2005; Xu et al. 2010; Andres et al. 2015; Khan et al. 2015; Jost et al. 2016). Although some inflorescence defects are shared among the three species (e.g., elongated internodes and abnormal pedicel orientation), it is striking that eliminating BOP activity in Arabidopsis results in a weak loss of IM determinacy, whereas the few-flowered inflorescences of tomato CR-slbop mutants are based on precocious meristem maturation that leads to enhanced SIM termination. This difference could reflect the evolution of distinct growth habits where the SlBOP family may have been co-opted in sympodial plants to serve as major regulators of meristem maturation, mediated in part through interactions with TMF.
However, it is telling that, unlike tmf mutants, mutations in the floral specification genes AN and FA do not completely suppress slbop flowering defects, suggesting that the SlBOPs and TMF act in partially overlapping pathways and that other factors function with SlBOP proteins to control flowering, meristem maturation, and inflorescence architecture. Indeed, we show that at least two TMF homologs interact with the SlBOPs, and CR-tfam mutants display a range of phenotypes similar to CR-slbops. These additional interactions could explain the dramatically enhanced flowering defects in RNAi-slbop tmf plants. In this scenario, when TMF is lost, SlBOPs still interact with TMF family members, and, in slbop triple mutants, TMF and family members still function on their own and/or with other transcription factors/cofactors (MacAlister et al. 2012). Thus, the stronger phenotypes of plants lacking TMF and the SlBOPs could be due to the disruption of parallel pathways (and transcriptional complexes) that regulate flowering and meristem maturation by converging on the same downstream targets. Following this logic, the enhanced early flowering in RNAi-slbop tmf plants compared with CR-slbop triple mutants and tmf might be due to even earlier activation of AN. In support of this, expressing AN in the earliest stages of meristem maturation using the TMF promoter results in plants that flower after only two leaves and develop single-flower primary inflorescences, mimicking RNAi-slbop tmf plants (Supplemental Fig. S9). Further combined biochemical and genetic studies, including in vivo identification and functional characterization of additional partners of SlBOP and ALOG family members, along with revealing their transcriptional targets in diverse tissues, will help resolve the molecular interplay between these deeply conserved protein families in plant development.
Materials and methods
Plant materials and growth conditions
The tomato (Solanum lycopersicum) cultivar M82 was used in this study. Greenhouse plants were grown under natural light with supplementation from high-pressure sodium bulbs (50 mM/m2/sec) on a 16-h light/8-h dark photoperiod. Daytime and nighttime temperatures were 26°C–28°C and 18°C–20°C, respectively.
Agrobacterium tumefaciens-mediated transformations were performed using a standard protocol (Van Eck et al. 2006), with transplanting and acclimation of transgenic plants performed as described (Brooks et al. 2014).
To introduce RNAi-slbop into tmf mutants, we crossed RNAi-slbop #1 to tmf-1 and tmf-2, respectively, and self-pollinated the F1 plants. We then genotyped tmf-1 and tmf-2 mutant plants harboring RNAi-slbop transgene by PCR from F2 progeny plants. To generate RNAi-slbop an and RNAi-slbop fa mutant plants, we crossed RNAi-slbop #1 into plants heterozygous for strong alleles of an (an-e1546) and fa (LA0854) and genotyped F1 plants to obtain an and fa heterozygous plants that also carried the RNAi-slbop transgene. The RNAi-slbop an and RNAi-slbop fa mutant plants were then obtained by genotyping F2 progeny plants for an and fa mutations and the RNAi-slbop transgene.
To express the AN gene under the promoter of TMF, we used the transactivation system (Ornitz et al. 1991; Liang et al. 2006), as done previously (MacAlister et al. 2012). Briefly, a 3.8-kb DNA fragment immediately upstream of TMF was PCR-amplified and subcloned in front of LhG4. AN cDNA was subcloned behind a 6× tandem OP array. Both pTMF::LhG4 and OP::AN were cloned into the pART27 binary vector for transformation. LhG4 lines were crossed to the appropriate OP lines, and F1 plants (pTMF≫AN) were phenotyped.
Multiple sequence alignments and phylogenetic analysis
Full-length SlBOP1/2 gene and protein sequences were identified by BLASTN using the sequences obtained from yeast two-hybrid screening as queries against the coding sequences of ITAG 2.4 (https://blast.ncbi.nlm.nih.gov/Blast.cgi). SlBOP3 was obtained by BLAST searches using the sequences of SlBOP1/2 and Arabidopsis BOP1/2 as queries against the coding sequences of ITAG 2.4 (https://blast.ncbi.nlm.nih.gov/Blast.cgi). We identified the Oryza sativa, Selaginella moellendorffii, and Physcomitrella patens BOP family members using BLASTP through the NCBI BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were generated using SeaView 4 (Gouy et al. 2010), and a phylogenetic tree was generated from protein sequences using MAFFT (version 7.221) (Katoh and Standley 2013).
Yeast two-hybrid
Yeast two-hybrid screening was performed by Hybrigenics Services (MacAlister et al. 2012). To confirm interactions between TMF and SlBOPs by yeast two-hybrid, the full-length coding sequence of each protein was cloned to pGBD and pGAD vectors, respectively (James et al. 1996). The BD-TMF, BD-TFAM1, BD-TFAM2, and AD-SlBOP constructs were cotransformed into yeast strain PJ69-4A, and the yeast two-hybrid assay was carried out using the protocol as described (James et al. 1996).
Recombinant protein expression and pull-down assay
To construct the plasmids that express His-TMF and GST-SlBOP proteins, the coding sequences of TMF and SlBOP1/2/3 were subcloned into pDONR221 by BP reaction (Invitrogen) and then recombined into pET-61-DEST and pET-60-DEST (Novagen) by LR reaction (Invitrogen). To produce recombinant proteins, each expression construct was transformed into Escherichia coli BL21 (DE3). In brief, bacteria were cultured in LB medium at 37°C to reach 1.2 (OD600) and cooled for 30 min on ice. The cells were then continuously cultured for 15 h at 16°C after adding 0.8 mM isopropyl-b-D-thiogalactopyranoside (IPTG). Cells were then harvested and lysed by sonication. The cell lysates were centrifuged at 16,000g for 30 min, and the resulting supernatants were incubated with His Mag Sepharose Ni (GE Healthcare) for purifying His-TMF proteins and MagneGST protein purification beads (Promega) for GST alone and GST-SlBOP proteins, respectively. The proteins were then purified according to the manufacturer's instructions.
For an in vitro GST-SlBOP pull-down assay, 0.5 mg of recombinant GST and GST-SlBOP proteins bound to MagneGST beads was blocked by 5% BSA for 2 h at 4°C followed by incubation with 0.3 mg of His-TMF for an additional 2 h at 4°C. The magnetic beads were then washed five times with ice-cold 1× PBS buffer. The bound proteins were eluted by 1× SDS-PAGE buffer with heating for 10 min at 95°C. His-TMF and GST-SlBOP proteins were detected by a monoclonal anti-His antibody (MBL Life Science) and a monoclonal anti-GST antibody (Santa Cruz Biotechnology), respectively.
Subcellular localization and BiFC assays in tomato protoplasts
To generate the transient expression constructs for subcellular localization, eGFP was fused to the C terminus of SlBOP1, SlBOP2, SlBOP3, and TMF, respectively. Fusion protein expression was driven by the cauliflower mosaic virus (CaMV) 35S promoter in transient expression vectors as described previously (Xu et al. 2012). To produce constructs for the BiFC assays, the coding regions of TMF and SlBOPs were ligated into split CFP vectors pSCYNE and pSCYCE to generate N-CFP-TMF, C-CFP-SlBO1, C-CFP-SlBO2, and C-CFP-SlBO3, respectively (Waadt et al. 2008). The constructs for subcellular localization and BiFC assay were transfected into tomato protoplasts as described previously (Xu et al. 2015). Cells were incubated for 12 h at 28°C before harvesting for microscopy. Imaging was performed on a Zeiss Axioplan 2 fluorescence microscope with an AxioCam camera.
Dual-luciferase assays in tomato protoplasts
For the Gal4-DNA-binding domain fusions, the TMF, SlBOP1, SlBOP2, and SlBOP3 coding sequences were cloned into the pDB vector by BamHI and KpnI restriction sites to generate the effectors (Ohta et al. 2001). pDB empty vector served as a negative control. A Renilla luciferase (LUC) gene under the control of a CaMV 35S promoter was used as an internal control, and a firefly LUC gene under the control of four copies of the Gal4 upstream activating sequence fused to a minimal 35S promoter served as a reporter (Ohta et al. 2001). For the transient expression in tomato protoplasts, the plasmids for effector, reporter, and internal control were cotransfected into tomato protoplasts using a standard PEG-mediated transfection protocol (Xu et al. 2015) at a plasmid ratio of 6:6:1 (effector:reporter:internal control). After culturing for 16 h, dual-luciferase assays were performed by following the instructions of the Promega dual-luciferase reporter assay system, and the luciferase activity was measured by the GloMaxTM20-20 luminometer (Wei et al. 2009). For data collection and statistical analyses, two independent experiments with three biological replicates per experiment were performed.
Meristem imaging, tissue collection, and quantitative RT–PCR (qRT–PCR)
Hand-dissected tomato meristems and stereomicroscope imaging of meristems were obtained according to our standard protocols (Park et al. 2012). TM stage collection, RNA extraction, and cDNA preparation of wild type and RNAi-slbop were performed according to our previously published protocols (Park et al. 2012). Briefly, total RNA was extracted from 20–30 meristems per biological replicate using a PicoPure RNA extraction kit (Arcturus). Total RNA (0.5–1 μg) was treated with DNase I and used for cDNA synthesis with a SuperScript III RT kit (Invitrogen). qRT–PCR was performed using gene-specific primers in the reaction system of iQ SYBR Green Supermix (Bio-Rad) on the CFX96 real-time system (Bio-Rad) following the manufacturer's instructions (Supplemental Table 2). UBIQUITIN was used as a control.
The expression patterns of SlBOP1, SlBOP2, SlBOP3, and TMF were acquired from the tomato tissue RNA sequencing (RNA-seq) database (http://tomatolab.cshl.edu/~lippmanlab2/allexp_query.html). RNA-seq data from different tissues (e.g., Fig. 3A) were mined from the tomato genome project transcriptome profiling data sets deposited in the NCBI Sequence Read Archive (SRA) under accession number SRP010775 (Tomato_Genome_Consortium 2012) and from our meristem maturation atlas (Park et al. 2012).
mRNA in situ hybridization
mRNA in situ hybridization was performed using standard protocols (Jackson 1992) with slight modifications. Briefly, to generate probes of SlBOP1 and SlBOP2/3, full-length coding sequences of SlBOP1 and SlBOP2, whose coding sequences are largely identical to SlBOP3 (∼86%), were amplified from cDNA using KOD Xtreme hot start DNA polymerase (Novagen), and the resulting products were ligated into StrataClone pSC-A-amp/kan vector (Agilent Technologies). Plasmids were linearized, and, depending on insert orientation, T7 or T3 RNA polymerase was used for in vitro transcription (Roche). Full-length probes were used for the hybridization. Meristem stages were as described previously (Park et al. 2012). For fixation, meristems were hand-dissected and fixed in 4% paraformaldehyde with 0.3% Triton-X under vacuum.
CRISPR/Cas9 gene editing and genotyping and phenotyping of resulting mutants
CRISPR/Cas9 mutagenesis, plant regeneration, and greenhouse care were performed according to our standard protocols (Brooks et al. 2014). Briefly, constructs were designed using two sgRNAs targeting the first exon of each target gene to generate various indels within coding sequences (see Supplemental Table 1 for a list of sgRNAs). For genotyping each first-generation (T0) transgenic line, three different leaf samples were collected to capture all possible induced mutant alleles due to sectoring (chimeric), and genomic DNA was extracted using a standard protocol. Each plant was genotyped by PCR for the presence of the Cas9/sgRNA1/sgRNA2 construct with primers designed to amplify a region spanning the 3′ end of the 35S promoter and the 5′ end of Cas9. The CRISPR/Cas9 T-DNA-positive lines were further genotyped for indel mutations using a forward primer to the left of the sgRNA1 target sequence and a reverse primer to the right of the sgRNA2 target sequence (Supplemental Table 1). PCR products from selected plants were purified for cloning into the pSC-A-amp/kan vector (Stratagene). A minimum of eight clones per PCR product was sequenced. To generate higher-order CR-slbop double mutants, crosses were conducted between each T0 single mutant, and F1 plants were self-pollinated. The F2 progenies were then screened by PCR genotyping and sequencing as described above. To generate all combinations of CR-slbop double mutants, we took advantage of T0 chimeric plants of CR-slbop single mutants in which CRISPR/Cas9 can reciprocally induce mutations in the F1 cross, and this strategy also allowed us to efficiently introduce mutations in the closely linked SlBOP2 and SlBOP3 genes. The CR-slbop double mutants were obtained by genotyping the progeny of F1 plants. To create triple mutants, crosses were made between genotyping confirmed F1 double-mutant plants for CR-slbop1/2 and CR-slbop2/3, which allowed immediate creation of homozygous or biallelic mutations in SlBOP2. The CR-slbop1/2/3 triple mutants were then obtained by screening the progeny of the F1 plants.
Supplementary Material
Acknowledgments
We thank members of the Lippman laboratory, especially C. Brooks, J. Dalrymple, and A. Krainer, for technical assistance, and R.J. Ontiveros for contributing to the luciferase assays. We thank W. Wang for assistance with tomato transformation, Z. Lemmon for bioinformatics assistance, Y. Eshed (Weizmann) for discussions, and T. Mulligan and S. Vermylen from Cold Spring Harbor Laboratory Uplands Farm and staff from the Long Island Horticultural Research and Extension Center for assistance with plant care. This research was supported by the Next-Generation BioGreen 21 Program (System and Synthetic Agro-biotech Center, PJ0118832016) to S.J.P, a National Science Foundation Plant Genome Research Program grant to J.V.E. and Z.B.L. (IOS-1237880), and a National Science Foundation grant (IOS-1556171) to Z.B.L.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.288415.116.
References
- Andrés F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Andres F, Romera-Branchat M, Martínez-Gallegos R, Patel V, Schneeberger K, Jang S, Altmüller J, Nürnberg P, Coupland G. 2015. Floral induction in Arabidopsis thaliana by FLOWERING LOCUS T requires direct repression of BLADE-ON-PETIOLE genes by homeodomain protein PENNYWISE. Plant Physiol 169: 2187–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueno F. 2007. Floral initiation and inflorescence architecture: a comparative view. Ann Bot 100: 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Després C. 2009. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21: 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J. 2014. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child A. 1979. A review of branching patterns in the Solanaceae In The biology and taxonomy of the Solanaceae (ed. Hawkes JG, et al. ), pp. 345–356. Academic Press, London. [Google Scholar]
- Cho E, Zambryski PC. 2011. ORGAN BOUNDARY1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc Natl Acad Sci 108: 2154–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou J-M, Zhukov V, Mondy S, Abu el Heba G, Cosson V, Ellis THN, Ambrose M, Wen J, Tadege M, Tikhonovich I, et al. 2012. NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE Genes. Plant Cell 24: 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. 2013. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Ha CM. 2003. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130: 161–172. [DOI] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC. 2007. BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19: 1809–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S. 2008. Inflorescence architecture: the transition from branches to flowers. Curr Biol 18: R1106–R1108. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. 2005. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Klenz JE, Haughn GW. 2006. UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223: 769–778. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Aguilar-Martínez JA, Farhi M, Chitwood DH, Kumar R, Millon LV, Peng J, Maloof JN, Sinha NR. 2014. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci 111: E2616–E2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L. 2012. ALOG domains: provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol Direct 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. 1992. In-situ hybridization in plants In Molecular plant pathology: a practical approach, Vol. I. (ed. Gurr MJ, et al. ), pp. 164–173. Oxford University Press, Oxford, England. [Google Scholar]
- James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Taketa S, Mascher M, Himmelbach A, Yuo T, Shahinnia F, Rutten T, Druka A, Schmutzer T, Steuernagel B, et al. 2016. A homolog of Blade-On-Petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol 171: 1113–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Ha CM, Fletcher JC. 2010. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22: 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Xu M, Murmu J, Tabb P, Liu Y, Storey K, McKim SM, Douglas CJ, Hepworth SR. 2012. Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol 158: 946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Xu H, Hepworth SR. 2014. BLADE-ON-PETIOLE genes: setting boundaries in development and defense. Plant Sci 215-216: 157–171. [DOI] [PubMed] [Google Scholar]
- Khan M, Ragni L, Tabb P, Salasini BC, Chatfield S, Datla R, Lock J, Kuai X, Despres C, Proveniers M, et al. 2015. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis. Plant Physiol 169: 2166–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. 2000. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Bohs L, Nee M, Spooner DM. 2004. Solanaceae—a model for linking genomics with biodiversity. Comp Funct Genomics 5: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D. 2007. Move on up, it's time for change mobile signals controlling photoperiod-dependent flowering. Genes Dev 21: 2371–2384. [DOI] [PubMed] [Google Scholar]
- Koes R. 2008. Evolution and development of virtual inflorescences. Trends Plant Sci 13: 1–3. [DOI] [PubMed] [Google Scholar]
- Liang D, Wu C, Li C, Xu C, Zhang J, Kilian A, Li X, Zhang Q, Xiong L. 2006. Establishment of a patterned GAL4-VP16 transactivation system for discovering gene function in rice. Plant J 46: 1059–1072. [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D. 2008. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6: 2424–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C, Park SJ, Jiang K, Marcel F, Bendahmane A, Izkovich Y, Eshed Y, Lippman ZB. 2012. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat Genet 44: 1393–1398. [DOI] [PubMed] [Google Scholar]
- McKim SM, Stenvik G-E, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, Aalen RB, Haughn GW. 2008. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135: 1537–1546. [DOI] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O. 2005. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132: 2203–2213. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P. 1991. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci 88: 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Schatz MC, Lippman ZB. 2012. Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci 109: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Eshed Y, Lippman ZB. 2014. Meristem maturation and inflorescence architecture-lessons from the Solanaceae. Curr Opin Plant Biol 17: 70–71. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. 1998. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. 2007. Evolution and development of inflorescence architectures. Science 316: 1452–1456. [DOI] [PubMed] [Google Scholar]
- Rebocho AB, Bliek M, Kusters E, Castel R, Procissi A, Roobeek I, Souer E, Koes R. 2008. Role of EVERGREEN in the development of the cymose petunia Inflorescence. Dev Cell 15: 437–447. [DOI] [PubMed] [Google Scholar]
- Rickett HW. 1944. The classification of inflorescences. Bot Rev 10: 187–231. [Google Scholar]
- Sato D-S, Ohmori Y, Nagashima H, Toriba T, Hirano H-Y. 2014. A role for TRIANGULAR HULL1 in fine-tuning spikelet morphogenesis in rice. Genes Genet Syst 89: 61–69. [DOI] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RAM, Koes R. 2008. Patterning of inflorescences and flowers by the F-Box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell 20: 2033–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. 2009. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Hanano K, Kariya A, Shimizu S, Zhao L, Matsui M, Tasaka M, Aida M. 2011. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J 66: 1066–1077. [DOI] [PubMed] [Google Scholar]
- Tavakol E, Okagaki R, Verderio G, Shariati JV, Hussien A, Bilgic H, Scanlon MJ, Todt NR, Close TJ, Druka A, et al. 2015. The Barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol 168: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato_Genome_Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck J, Kirk DD, Walmsley AM. 2006. Tomato (Lycopersicum esculentum). Methods Mol Biol 343: 459–473. [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. 2008. Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Weberling F. 1989. Morphology of flowers and inflorescences. Cambridge University Press, Cambridge. [Google Scholar]
- Wei W, Huang J, Hao YJ, Zou HF, Wang HW, Zhao JY, Liu XY, Zhang WK, Ma B, Zhang JS, et al. 2009. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS One 4: e7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X-M, Yu Y, Han L-B, Li C-L, Wang H-Y, Zhong N-Q, Yao Y, Xia G-X. 2012. The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol 159: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, McKim SM, Murmu J, Haughn GW, Hepworth SR. 2010. Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J 63: 974–989. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang Y, Yu Y, Duan J, Liao Z, Xiong G, Meng X, Liu G, Qian Q, Li J. 2012. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat Commun 3: 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu Y-H, Jiang K, Brooks C, Ogawa-Ohnishi M, Xiong G, Pauly M, et al. 2015. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet 47: 784–792. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Suzaki T, Tanaka W, Hirano H-Y. 2009. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci 106: 20103–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, et al. 2013. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci 110: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Nakazawa M, Takase T, Manabe K, Kobayashi M, Seki M, Shinozaki K, Matsui M. 2004. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J 37: 694–706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.