Here, Burke et al. delineate a new pathway for mammalian small RNAs to enter the RNAi gene silencing machinery. They show that DUSP11 directly dephosphorylates viral triphosphate ncRNA transcripts and that this is required for efficient silencing by RISC, suggesting that mammalian viral pathogens can use DUSP11 to generate atypical microRNAs.
Keywords: BLV, adenovirus, RNAi, Alu, vault RNA, DUSP11
Abstract
RNA silencing is a conserved eukaryotic gene expression regulatory mechanism mediated by small RNAs. In Caenorhabditis elegans, the accumulation of a distinct class of siRNAs synthesized by an RNA-dependent RNA polymerase (RdRP) requires the PIR-1 phosphatase. However, the function of PIR-1 in RNAi has remained unclear. Since mammals lack an analogous siRNA biogenesis pathway, an RNA silencing role for the mammalian PIR-1 homolog (dual specificity phosphatase 11 [DUSP11]) was unexpected. Here, we show that the RNA triphosphatase activity of DUSP11 promotes the RNA silencing activity of viral microRNAs (miRNAs) derived from RNA polymerase III (RNAP III) transcribed precursors. Our results demonstrate that DUSP11 converts the 5′ triphosphate of miRNA precursors to a 5′ monophosphate, promoting loading of derivative 5p miRNAs into Argonaute proteins via a Dicer-coupled 5′ monophosphate-dependent strand selection mechanism. This mechanistic insight supports a likely shared function for PIR-1 in C. elegans. Furthermore, we show that DUSP11 modulates the 5′ end phosphate group and/or steady-state level of several host RNAP III transcripts, including vault RNAs and Alu transcripts. This study shows that steady-state levels of select noncoding RNAs are regulated by DUSP11 and defines a previously unknown portal for small RNA-mediated silencing in mammals, revealing that DUSP11-dependent RNA silencing activities are shared among diverse metazoans.
Transcripts generated by RNA polymerases initially contain a 5′ triphosphate. 5′ triphosphates on RNAs hinder access to exonucleases (Stevens 1980; Stevens and Poole 1995; Mathy et al. 2007) and are molecular patterns that can activate the antiviral response in mammalian cells (Hornung et al. 2006; Pichlmair et al. 2006; Nallagatla et al. 2007). Therefore, control of 5′ triphosphate status is likely important to maintain appropriate transcript levels and avoid inappropriate immune stimulation. Consistent with this, the 5′ ends of RNA polymerase II (RNAP II) transcripts are cotranscriptionally “capped,” where the 5′ triphosphate is removed and replaced with a methylated inverted 7-methyl guanosine. However, how the 5′ triphosphate status of RNAP I and RNAP III transcripts is regulated remains poorly understood. Recently, our work implicated a possible additional role of 5′ triphosphates in mammalian RNA silencing. We showed that a noncanonical viral precursor microRNA (pre-miRNA) that was transcribed by RNAP III was 5′ triphosphorylated but that its derivative miRNA contained a 5′ monophosphate due to a cellular phosphatase activity (Burke et al. 2014). Until now, the identity of this phosphatase and whether it is required for RNA silencing were unknown.
Many classes of small regulatory RNAs (∼18–30 nucleotide [nt]) have been identified. These include siRNAs, miRNAs, and Piwi-interacting RNAs (piRNAs), which are differentiated by their biogenesis pathway, structure, associated proteins, and cellular function. siRNAs and miRNAs mediate RNA silencing, a conserved mechanism of post-transcriptional gene regulation in eukaryotes (for review, see Carthew and Sontheimer 2009). During RNA silencing, siRNAs and miRNAs associate with an Argonaute (AGO) protein, a key component of the RNA-induced silencing complex (RISC), and guide RISC to target RNA transcripts via sequence complementarity. RISC association with target RNAs generally represses gene expression via cleavage or translational inhibition of the target messenger RNA (mRNA) transcript. Together, these small RNA effectors regulate diverse eukaryotic cellular processes, including the antiviral response, development, and homeostasis.
During RNAi in nematodes, RNA-dependent RNA polymerases (RdRPs) amplify siRNAs in response to exogenous RNAs (exo-RNAi) and endogenous RNAs (endo-RNAi) via distinct AGO pathways (Yigit et al. 2006; Gent et al. 2010; Vasale et al. 2010). During exo-RNAi, the RNase III enzyme Dicer processes foreign dsRNAs into primary siRNAs (Zamore et al. 2000; Bernstein et al. 2001; Hamilton et al. 2002), which are loaded into the primary AGO RDE-1 (Yigit et al. 2006). During endo-RNAi, an RdRP, in association with Dicer and the riboexonuclease ERI-1, synthesizes primary siRNAs ∼26 nt in length (26G-RNAs) from endogenous RNA transcripts. The 26G-RNAs are then loaded into the primary AGO ERGO-1 (Gent et al. 2010; Vasale et al. 2010). Upon RDE-1 and ERGO-1 association with target RNAs, RdRPs directly synthesize secondary siRNAs ∼22 nt in length (22G-RNAs) (Yigit et al. 2006; Gent et al. 2010; Vasale et al. 2010; Tsai et al. 2015). The 22G-RNAs primarily contain 5′ triphosphates and diphosphates (Pak and Fire 2007; Sijen et al. 2007) and associate with secondary worm-specific AGOs (WAGOs) to mediate repression of gene expression (Yigit et al. 2006). Robust accumulation of some 22G-RNAs and 26G-RNAs requires the Dicer-associated PIR-1 phosphatase (Duchaine et al. 2006; Chaves 2015). Since the human homolog of PIR-1, dual specificity phosphatase 11 (DUSP11), sequentially removes the γ and β phosphates from RNAs containing a 5′ triphosphate (Deshpande et al. 1999), the RNA triphosphatase activity of PIR-1 has been proposed to facilitate siRNA generation and function (Duchaine et al. 2006; Gent et al. 2010, Chaves 2015). However, how this promotes siRNA biogenesis has remained undetermined. Furthermore, because mammals do not encode RdRPs or amplify siRNAs (Schwarz et al. 2002), a similar role for DUSP11 in mammalian RNAi would not be expected.
miRNAs are a conserved class of small RNAs (∼22 nt) that post-transcriptionally regulate eukaryotic gene expression (Bartel 2004; Kim 2005). In contrast to siRNAs, miRNAs are initially embedded within stem–loop structures of longer primary miRNA (pri-miRNA) transcripts typically transcribed by RNAP II from defined genomic loci. The Microprocessor complex—composed of the RNase III enzyme Drosha and its RNA-binding partner, DGCR8—cleaves pri-miRNA stem–loops to liberate pre-miRNA hairpin structures (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004; Han et al. 2004; Landthaler et al. 2004). Upon export to the cytosol, Dicer processes pre-miRNAs (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001) into duplex miRNA strands (Lee et al. 2003) with 5′ monophosphate and 3′ hydroxyl termini (Hutvágner et al. 2001; Basyuk et al. 2003). The strands that map to the 5′ and 3′ ends of the pre-miRNA stem are denoted as the 5p strand and 3p strand, respectively. Generally, one of the strands, termed the guide strand, is preferentially stably loaded into an AGO protein. The other strand, termed the passenger strand, is rapidly turned over (Khvorova et al. 2003; Schwarz et al. 2003). Although the mechanisms of strand selection are still not fully understood, the thermodynamic stability at the ends of a miRNA duplex was shown to be a major factor (Khvorova et al. 2003; Schwarz et al. 2003). In addition, both the 5′ nucleotide of the miRNA and Dicer-associated proteins also influence strand selection (Frank et al. 2010; Noland and Doudna 2013). Upon association with AGO, the miRNA then guides RISC to target mRNAs via partial sequence complementarity, which results in translational repression and mRNA turnover (Bazzini et al. 2012; Djuranovic et al. 2012).
Of the >300 viral miRNAs identified to date, most are generated via the canonical miRNA biogenesis pathway described above (for review, see Grundhoff and Sullivan 2011; Kincaid and Sullivan 2012). However, a few viruses generate miRNAs using unconventional mechanisms and proteins, such as RNAP III, tRNase Z, and the integrator complex (Pfeffer et al. 2005; Bogerd et al. 2010; Diebel et al. 2010; Cazalla et al. 2011; Kincaid et al. 2012, 2014; Burke et al. 2014; Diebel et al. 2014; Feldman et al. 2014). Previous work from our laboratory and others has demonstrated that the miRNAs derived from the 5′ end of 5′ triphosphorylated miRNA precursors expressed by bovine leukemia virus (BLV) and human adenovirus (AdV) predominantly contain a 5′ monophosphate (Xu et al. 2009; Burke et al. 2014). This indicates the involvement of a phosphatase activity in the biogenesis of these miRNAs. Here, we identify the RNA triphosphatase responsible for this activity as DUSP11 and demonstrate that its RNA triphosphatase activity promotes 5′ monophosphate-dependent AGO association of the BLV and AdV 5p miRNAs. Furthermore, DUSP11 alters the phosphorylation status and steady-state levels of select host RNAP III RNAs, including the abundant vault RNAs (vtRNAs) and short interspersed nuclear element (SINE) Alu transcripts. This work reveals DUSP11 as a key modulator of the phosphate status, abundance, and RNA silencing activity of select RNAP III transcripts.
Results
Knockout of DUSP11 decreases the RISC activity of the BLV 5p miRNAs
The BLV miRNAs are encoded in compact, individual RNAP III type II genes (Kincaid et al. 2012; Burke et al. 2014). Similar to RNAP III-driven shRNAs, each BLV pre-miRNA is directly transcribed by RNAP III and, as a result, initially contains a 5′ triphosphate (Fig. 1A). However, the 5p miRNAs expressed in BLV-infected cells and cells transfected with plasmids encoding the BLV miRNA loci are predominantly 5′ monophosphorylated (Fig. 1A; Burke et al. 2014). This implicates an RNA triphosphatase in the BLV miRNA biogenesis pathway. To identify the putative phosphatase, we searched for phosphatases associated with RNAi machinery. Proteomics in Caenorhabditis elegans identified a Dicer-associated protein, PIR-1, that is required for the generation and/or RISC activity of 22G-RNAs (Duchaine et al. 2006). The human PIR-1 homolog DUSP11 associates with RNA (Yuan et al. 1998) and sequentially removes the γ phosphates and β phosphates from RNAs containing a 5′ triphosphate (Deshpande et al. 1999). Thus, we hypothesized that DUSP11 may be responsible for the dephosphorylation of the BLV 5p miRNAs.
Figure 1.
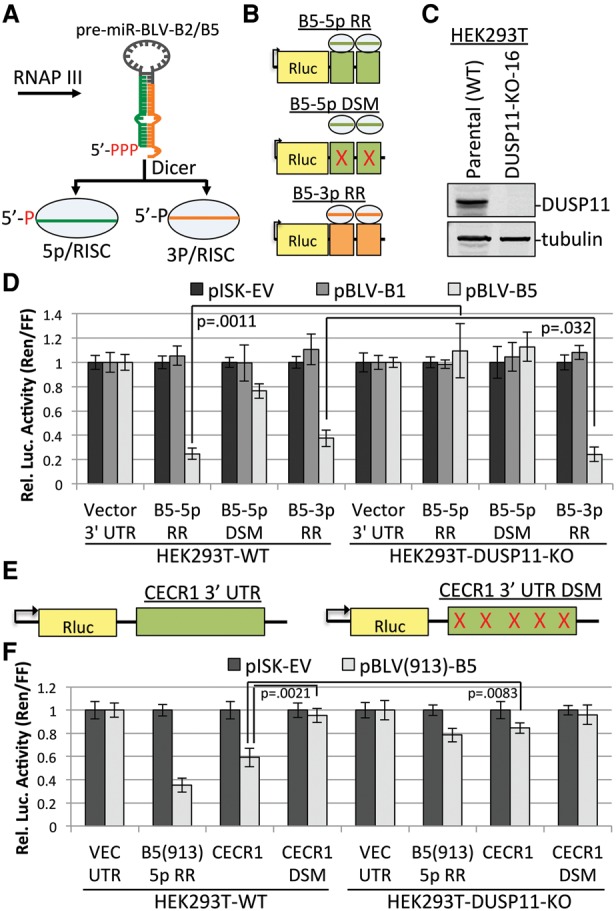
Knockout of DUSP11 decreases the RISC activity of the BLV 5p miRNAs. (A) Schematic diagram of the BLV miRNA biogenesis pathway. (B) Diagram of the BLV-B5 miRNA RISC reporters. Two sites complementary to either the 5p miRNA (green boxes) or the 3p miRNA (orange boxes) were inserted into the 3′ untranslated region (UTR) of Renilla luciferase. In the B5-5p docking site mutant (DSM), point mutations were made in the sequence complementary to the B5-5p seed to interfere with RISC-mediated silencing. (C) Immunoblot analysis to confirm CRISPR/Cas9-mediated knockout of DUSP11 in DUSP11 knockout clone 16 (DUSP11-KO-16) HEK293T cells as compared with the parental (wild-type [WT]) HEK293T cells. (D) Luciferase assay to measure the RISC activity of BLV-B5 miRNAs in parental (wild-type) and DUSP11 knockout clone 16 HEK293T cells. Bars represent the mean luciferase ratio (Renilla/firefly) ± SEM normalized to pISK-EV and the vector 3′ UTR reporter. Transfections were performed in triplicate in each experiment. Eight experiments were performed for vector 3′ UTR, B5-5p RR, and B5-3p RR cotransfected with pISK-EV and pBLV-B5. Four experiments were performed for vector 3′ UTR, B5-5p RR, and B5-3p RR cotransfected with pBLV-B1. Three experiments were performed for the B5-5p DSM with all expression vectors. (E) Diagram of the Rluc reporter containing the majority of the ovine CECR1 3′ UTR (green box), which has five potential target sites of BLV(913)-miR-B5-5p. Point mutations in all potential B5-5p target sites were made (red ×) to generate the Rluc CECR1 3′ UTR control vector (CECR1 DSM). (F) Luciferase assay measuring the repression of the CECR1 3′ UTR and B5-5p DSM by BLV-B5-5p in wild-type and DUSP11 knockout HEK293T cells. Bars represent the mean luciferase ratio (Renilla/firefly) ± SEM from four experiments. Values were normalized to pISK-EV and the vector 3′ UTR reporter.
To determine whether DUSP11 functions in the biogenesis of the BLV miRNAs, we first made RISC reporters for the BLV-B5 miRNAs (Fig. 1A) by inserting two sites complementary to either BLV-miR-B5-5p (B5-5p RR) or BLV-miR-B5-3p (B5-3p RR) into the 3′ untranslated region (UTR) of Renilla luciferase (Rluc) (Fig. 1B). To control for B5-5p specificity, we made a BLV-miR-B5-5p RISC reporter with three point mutations in the B5-5p seed docking site mutant (DSM) (Fig. 1B). HEK293T DUSP11 knockout cell lines were then generated using the CRISPR/Cas9 system. Immunoblot analysis confirmed the absence of DUSP11 in clonal cell lines (Fig. 1C). We then performed RISC reporter assays for the BLV-B5 miRNAs in the parental (wild-type) and DUSP11 knockout HEK293T cells. In wild-type cells, cotransfection of pBLV-B5 but not the control vectors (pISK-EV or pBLV-B1) markedly decreased the luciferase ratio (Renilla/firefly) of the B5-5p RR (75%) and the B5-3p RR (60%), whereas fourfold less repression was observed with the B5-5p DSM reporter (Fig. 1D). These findings confirm the efficacy of the BLV-B5 miRNA RISC reporters and indicate that the BLV-B5-5p and BLV-B5-3p miRNAs are active in RISC.
In contrast to the wild-type cells, the luciferase ratio (Renilla/firefly) of B5-5p RR did not appreciably decrease with BLV-B5 coexpression in DUSP11 knockout cells, whereas the repression of B5-3p RR increased ∼1.5-fold (Fig. 1D). Thus, knockout of DUSP11 significantly decreased (approximately fivefold) BLV-miR-B5-5p activity in RISC but slightly increased BLV-miR-B5-3p activity in RISC. Similar results were obtained by siRNA-mediated knockdown of DUSP11, which resulted in a twofold derepression of the BLV-B5-5p RR, while the repression of B5-3p RR increased slightly (Supplemental Fig. S1A,B). These siRNA results rule out off-target effects of the CRISPR/Cas9 as accounting for the BLV B5-5p phenotype that we observed in DUSP11 knockout cells. The RISC activity of BLV-miR-B2-5p also decreased in the DUSP11 knockout cells, although the magnitude of derepression of B2-5p RR was less than what we observed for B5-5p RR (Supplemental Fig. S1C). We observed a decrease in RISC activity of the BLV-B2-5p and BLV-B5-5p miRNAs in DUSP11 knockout cell lines in both the HEK293 and A549 parental backgrounds, demonstrating conserved activity of DUSP11 in different cell types (Supplemental Fig. S1D–F). In contrast to the BLV miRNAs, the RISC activity of the SV40-RI257 miRNAs (Chen et al. 2013a), which contain 5′ monophosphates as a result of canonical Drosha- and Dicer-mediated endonucleolytic cleavage (Hutvágner et al. 2001; Basyuk et al. 2003), remained unaffected by knockout of DUSP11 (Supplemental Fig. S2A). These data demonstrate that DUSP11 specifically promotes the RISC activity of 5p miRNAs derived from the 5′ triphosphorylated BLV pre-miRNAs.
To determine whether the lack of DUSP11 affects miRNA-mediated down-regulation of a more biologically relevant target mRNA, we searched for possible target sites of all known isoforms of BLV-miR-B5-5p in bovine transcripts that are at least indirectly targeted by BLV miRNAs during infection of primary peripheral blood mononuclear cells (PBMCs) (Gillet et al. 2016). We identified one transcript, CECR1, with four predicted miRNA docking sites for BLV(913)-miR-B5-5p in its 3′ UTR. In sheep, the common cancer model for BLV infection, five possible target sites of BLV(913)-miR-B5-5p (four of five shared with bovine) were identified in the CECR1 3′ UTR. To determine whether the CERC1 transcript could be directly regulated by BLV(913)-miR-B5-5p, we generated chimeric luciferase reporter plasmids that contain the majority of the ovine CECR1 3′ UTR as well as a control reporter in which all five putative BLV-miR-B5-5p docking sites were mutated (Fig. 1E). In wild-type cells, cotransfection of pBLV(913)-miR-B5 resulted in significant repression of CECR1 but not the CECR1 DSM (Fig. 1F). These data indicate that BLV(913)-miR-B5-5p can target the CECR1 3′ UTR, resulting in repression of gene expression. However, in the DUSP11 knockout cells, cotransfection of pBLV(913)-miR-B5 did not result in substantial repression of CECR1 3′ UTR reporter, indicating that DUSP11 is required for BLV-miR-B5-5p-mediated regulation of the CECR1 3′ UTR. These data show that DUSP11 can affect the regulatory capacity of a BLV 5p miRNA on a transcript previously implicated as being affected by BLV infection.
Knockout of DUSP11 decreases the steady-state levels of BLV 5p miRNAs
To gain insight into how DUSP11 promotes BLV 5p miRNA RISC activity, we examined the effect of DUSP11 on the steady-state levels of the BLV pre-miRNAs and miRNAs. We cotransfected parental, DUSP11 knockout, or Dicer knockout (Bogerd et al. 2014) HEK293T cells with a transfection control (pHSUR4) and the BLV-B2 or the BLV-B5 miRNA expression vectors and performed Northern blot analysis. Consistent with previous studies (Burke et al. 2014), the levels and RISC activity of BLV- B2 and BLV- B5 miRNAs were substantially reduced in Dicer knockout cells (Fig. 2A; Supplemental Fig. S2C–E), indicating that BLV miRNA biogenesis is largely dependent on Dicer. Similar to knockout of Dicer, the levels of BLV-miR-B2-5p and BLV-miR-B5-5p were markedly reduced in the DUSP11 knockout cells as compared with the wild-type cells (Fig. 2A). However, the corresponding 3p miRNAs were elevated in the DUSP11 knockout cells, indicating that DUSP11 is not required for Dicer-mediated processing of the BLV pre-miRNAs to generate the BLV 3p miRNAs. Knockout of DUSP11 did not affect the canonical SV40-RI257 miRNAs (Supplemental Fig. S2B), suggesting that the DUSP11-dependent biogenesis of the BLV 5p miRNAs is specific to 5′ triphosphorylated precursors. Combined, these results show that DUSP11 is not required for Dicer processing of the BLV pre-miRNAs but is required for accumulation of BLV 5p miRNAs.
Figure 2.
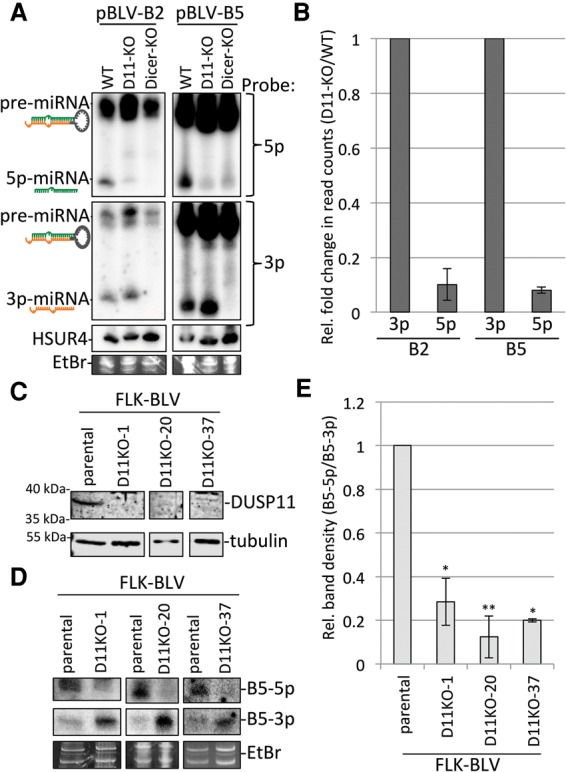
Knockout of DUSP11 decreases the accumulation of BLV 5p miRNAs. (A) Northern blot analysis of BLV-B2 and BLV-B5 miRNAs in parental (wild-type [WT]), DUSP11 knockout (D11-KO), and NoDice-2-20 (Dicer-KO) HEK293T cells. Cells were cotransfected with transfection control vector, pHSUR4, and each BLV miRNA expression vector. The membrane was first blotted with probes specific for the 5p miRNAs, stripped, reprobed for the 3p miRNAs, stripped, and reprobed for HSUR4 RNA. (B) Next-generation small RNA sequencing in parental (wild-type) and DUSP11 knockout HEK293T cells transiently expressing the BLV-B2 and BLV-B5 miRNAs. Bars represent the average fold change (DUSP11 knockout/wild-type) in a number of small RNA reads of the most abundant isoform of each indicated miRNA (Supplemental Fig. S3A) from two independent experiments ±the standard deviation. n = 2. The fold change of the 5p miRNAs was normalized to the fold change of the corresponding 3p miRNAs. (C) Western blot analysis of DUSP11 in parental FLK-BLV cells (fetal lamb kidney cells persistently infected with BLV) and various FLK-BLV-DUSP11 knockout cell lines (D11KO-1, D11KO-20, and D11KO-37). (D) Northern analysis of the BLV-B5 miRNAs in the parental, D11KO-1, D11-KO-20, and D11KO-37 FLK-BLV cell lines. The membrane was first probed for the 3p miRNA, stripped, and reprobed for the 5p miRNA. (E) Quantitation of the relative band density (5p/3p) of the BLV-B5 miRNAs by Northern blot analysis. Bars represent the average ± the standard deviation from three experiments. Values were normalized to the parental cells. The P-values were calculated using one sample t-test. (*) P < 0.05; (**) P < 0.005.
To independently confirm the results of our Northern blot analysis, we performed high-throughput small RNA sequencing in the parental (wild-type) and DUSP11 knockout HEK293T cells transfected with pBLV-B2 and pBLV-B5 vectors. Consistent with our Northern blot analysis (Fig. 2A), the read counts for BLVmiR-B2-5p and BLV-miR-B5-5p were substantially reduced (∼10-fold relative to the corresponding 3p miRNAs) in the DUSP11 knockout cells as compared with the wild-type cells (Fig. 2B). Because T4 RNA ligase catalyzes the ligation of 5′ monophosphorylated RNAs to RNAs with a 3′ hydroxyl group, 5′ triphosphorylated RNAs are excluded from small RNA libraries unless converted to a 5′ monophosphate (Lau et al. 2001). Since this could account for the decrease in the read counts of the BLV 5p miRNAs in the DUSP11 knockout cells, we treated the small RNAs with RNA 5′ polyphosphatase (which removes the γ and β phosphates from 5′ triphosphorylated RNA) prior to preparing the small RNA libraries. As expected, the read counts of abundant host miRNAs were largely unaffected by RNA 5′ polyphosphatase (Supplemental Fig. S3A), whereas the read counts of some small RNAs (18–32 nt) derived from the 5′ end, but not the 3′ end, of host RNAP III transcribed RNAs (including the 5S rRNA and Y-RNAs) increased with RNA 5′ polyphosphatase treatment (Supplemental Fig. S3B; Supplemental Data 1). This confirmed that RNA 5′ polyphosphatase treatment increased incorporation of 5′ triphosphorylated RNAs into our small RNA libraries. Pretreatment with RNA 5′ polyphosphatase did not appreciably or consistently increase (less than twofold relative to the 3p miRNAs) the read counts of the BLV 5p miRNAs (Supplemental Fig. S3A). This result suggests that the BLV 5p miRNAs that remain 5′ triphosphorylated are rapidly turned over. We note that all of the above trends were observed in two independent small RNA libraries, providing confidence in the reproducibility of these findings. Thus, both the Northern blot analysis and RNA sequencing (RNA-seq) results indicate that knockout of DUSP11 decreases the steady-state levels of the BLV 5p miRNAs.
To determine whether DUSP11 is important for BLV 5p miRNA accumulation during infection, we used the CRISPR/Cas9 system to generate DUSP11 knockout cell lines in FLK-BLV cells, which are fetal lamb kidney cells persistently infected with BLV. Multiple individual FLK-BLV-DUSP11 knockout clonal cell lines were confirmed by both immunoblot analysis (Fig. 2C) and genomic sequencing. Consistent with our above results, Northern blot analysis revealed that the ratio of the BLV-B5 miRNAs (5p to 3p) was decreased (approximately fivefold) in the FLK-BLV-DUSP11 knockout cell lines as compared with the parental FLK-BLV cells (Fig. 2D,E). These findings demonstrate that DUSP11 modulates the 5p to 3p ratio of the BLV miRNAs in infected cells.
DUSP11 catalytic activity promotes BLV 5p miRNA accumulation and RISC activity
To determine whether depletion of DUSP11 enzymatic activity is responsible for the decrease in BLV 5p miRNA steady-state levels and RISC activity in DUSP11 knockout cells, we rescued DUSP11 expression in the DUSP11 knockout cells and analyzed BLV miRNA expression and RISC activity. We cloned the DUSP11 ORF with a N-terminal 3xFlag epitope tag into the pLenti vector (pLenti-D11). We also generated a DUSP11 catalytic mutant (pLenti-D11-CM) by mutating Cys152, which is required for RNA triphosphatase activity (Deshpande et al. 1999), to a serine. We generated lentivirus particles and stably transduced HEK293T DUSP11 knockout cells with pLenti-EV (DUSP11-KO-EV), pLenti-DUSP11 (DUSP11-KO-D11), and pLenti-DUSP11-CM (DUSP11-KO-D11-CM). Immunoblot analysis confirmed that both DUSP11-KO-D11 and DUSP11-KO-D11-CM express DUSP11 at levels comparable with the parental 293T cells (Fig. 3A). Note that, as expected, the 3xFlag tag slightly reduced electrophoretic mobility.
Figure 3.
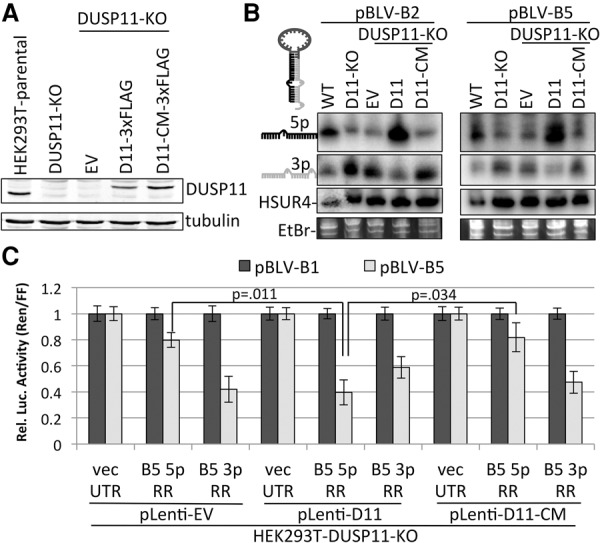
DUSP11 catalytic activity is required for BLV 5p miRNA accumulation and RISC activity. (A) Immunoblot analysis of parental HEK293T cells (wild-type), parental DUSP11 knockout clone 16 (DUSP11-KO) cells, and DUSP11 knockout clone 16 cells transduced with pLenti-EV (EV), pLenti-DUSP11-3xFlag (D11-3xFlag), or the pLenti-DUSP11-CM-3xFlag catalytic mutant expression vector (D11-CM-3xFlag). (B) Northern blot analysis of BLV-B2 and BLV-B5 miRNAs in the indicated cell lines. The membrane was first blotted with probes specific for the 5p miRNAs, stripped, reprobed for the 3p miRNAs, stripped, and reprobed for HSUR4 RNA. (C) Luciferase assay to measure the RISC activity of BLV-B5 miRNAs in the DUSP11 knockout cells transduced with the indicated vectors. Bars represent the mean luciferase ratio (Renilla/firefly) ± SEM from four experiments in which transfections were performed in triplicate.
As observed above, Northern blot analysis revealed decreased levels of BLV-miR-B2-5p and BLV-miR-B5-5p and increased corresponding 3p miRNA levels in the DUSP11 knockout cells (Fig. 3B). Importantly, we observed a substantial increase in BLV-miR-B2-5p and BLV-miR-B5-5p levels relative to the respective 3p miRNAs in the DUSP11-KO-D11 cells but not in the control DUSP11-KO-EV cells or DUSP11-KO-D11-CM cells (Fig. 3B). In contrast, the 5p to 3p ratio of the control SV40-RI257 canonical miRNAs remained constant in all of the cell lines (Supplemental Fig. S2F). Consistent with the Northern blot analysis, RISC reporter assays revealed that BLV-miR-B5-5p RISC activity was restored in the DUSP11-rescued cell line but not the DUSP11 catalytic mutant-rescued cell line (Fig. 3C). Similar results for both the Northern blot analysis and RISC reporter assays were obtained by transiently expressing DUSP11 and the DUSP11 catalytic mutant in DUSP11 knockout HEK293T cells (Supplemental Fig. S4A–C). We note that overexpression of DUSP11 in the parental HEK293T cells increased BLV 5p miRNA accumulation, which suggests that DUSP11 is a limiting factor in BLV 5p miRNA accumulation (Supplemental Fig. S4B). These results demonstrate that DUSP11 catalytic activity is required to rescue the defects in accumulation and RISC activity of the BLV 5p miRNAs observed in the DUSP11 knockout cells. Thus, DUSP11 RNA triphosphatase activity promotes both the accumulation and RISC activity of the BLV 5p miRNAs.
DUSP11 directly dephosphorylates the BLV pre-miRNAs and miRNAs
DUSP11 has been shown to sequentially remove the γ and β phosphates from 5′ triphosphorylated trinucleotide RNAs in vitro (Deshpande et al. 1999). To determine whether DUSP11 can directly dephosphorylate the BLV pre-miRNAs and/or 5p miRNA, we tested whether DUSP11 can dephosphorylate the BLV pre-miRNAs and miRNAs in vitro. We generated luciferase (negative control), DUSP11, and DUSP11 catalytic mutant proteins via in vitro transcription/translation, which was confirmed by immunoblot analysis (Fig. 4A). We then incubated radiolabeled [γ-32P]-BLV-pre-miR-B5 and [γ-32P]-BLV-miR-B5-5p mimics with buffer only, the positive control calf intestinal phosphatase (CIP), luciferase, DUSP11, or DUSP11 catalytic mutant. Ethidium bromide (EtBr) staining of fractionated RNAs showed that the levels of both the pre-miRNA and miRNA mimics remained constant in each reaction (Fig. 4B). However, as compared with buffer only and luciferase negative controls, positive control CIP treatment dramatically reduced the amount of radiolabeled [γ-32P]-BLV-pre-miR-B5 and [γ-32P]-BLV-miR-B5-5p. Similar to CIP, DUSP11 reduced the amount of radiolabeled [γ-32P]-BLV-pre-miR-B5 and [γ-32P]-BLV-miR-B5-5p, whereas the DUSP11 catalytic mutant did not (Fig. 4B). These results demonstrate that DUSP11 catalytic activity can promote removal of the γ phosphate from both the BLV pre-miRNAs and 5p miRNAs.
Figure 4.
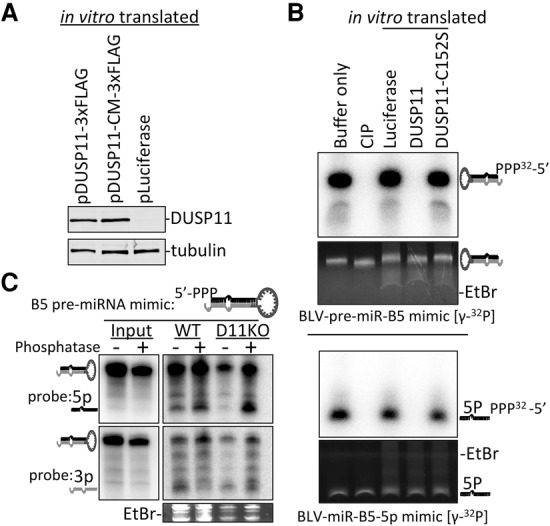
DUSP11 directly dephosphorylates the BLV pre-miRNAs and 5p miRNAs. (A) Immunoblot analysis to confirm expression of DUSP11 and DUSP11 catalytic mutant proteins generated using in vitro transcription/translation. The membrane was probed using anti-DUSP11 and anti-tubulin antibodies. (B) In vitro phosphatase reactions on the [γ-32P]-BLV-pre-miR-B5 mimic and [γ-32P]-BLV-miR-B5-5p miRNA mimic using CIP (positive control) or the in vitro translated DUSP11, DUSP11 catalytic mutant, or luciferase (negative control) from A. Reactions were fractionated on 15% PAGE/8 M urea, and RNAs were stained with EtBr. RNAs were then transferred to a membrane, exposed to a storage phosphor screen, and imaged on a Typhoon bimolecular imager. (C) Northern blot analysis from wild-type and DUSP11 knockout HEK293T cells transfected with a 5′ triphosphorylated BLV-B5 pre-miRNA mimic pretreated with (+) or without (−) RNA 5′ polyphosphatase. The blot was first probed for the 5p miRNA arm (green), stripped, and reprobed for the 3p arm (orange). Note that a lighter exposure for the input RNA is shown as compared with the RNA recovered from cells.
To test whether DUSP11 modifies the 5′ triphosphate of the BLV pre-miRNA and/or 5p miRNAs in cells, we analyzed miRNA accumulation from a BLV-pre-miR-B5 mimic containing either 5′ triphosphate (5′-ppp-pre-miR-B5) or a 5′ monophosphate (5′-p-pre-miR-B5). The 5′-ppp-pre-miR-B5 mimic was generated in vitro using T7 polymerase-mediated transcription and then treated with or without RNA 5′ polyphosphatase to generate the 5′ monophosphorylated mimic. We then transfected the BLV-pre-miR-B5 mimics into wild-type and DUSP11 knockout HEK293T cells and performed Northern blot analysis. Pretreatment with RNA 5′ polyphosphatase increased levels of the 5p derivative and decreased levels of the 3p derivative in both wild-type and DUSP11 knockout cells (Fig. 4C), indicating that the 5′ phosphate group affects the 5p to 3p ratio. The defect in 5p accumulation from the 5′-ppp-pre-miR-B5 mimic was more pronounced in the DUSP11 knockout cells as compared with the wild-type cells, suggesting that DUSP11 dephosphorylates a portion of the 5′-ppp-pre-miR-B5 mimic in the wild-type cells. Similar results were obtained using a BLV-pre-miR-B2 mimic (Supplemental Fig. S5A). Importantly, the defect in 5p miRNA accumulation from the 5′-ppp-BLV-pre-miR-B5 mimic (as compared with the 5′-p-B5-pre-miR mimic) phenocopied the defect in 5p miRNA accumulation from RNAP III transcribed pre-miRNAs observed in the DUSP11 knockout cells. Together, these results suggest that DUSP11-meditated conversion of the 5′ triphosphate on either the BLV pre-miRNAs and/or 5p miRNAs to a 5′ monophosphate promotes accumulation of the BLV 5p miRNAs in cells.
DUSP11 promotes AGO association with the BLV 5p miRNAs through 5′ monophosphate-dependent strand selection
Our above data demonstrate that BLV 5p miRNA accumulation and RISC activity are dependent on DUSP11. As AGO association is necessary for RISC activity and increases the steady-state levels of miRNAs through strand selection (Khvorova et al. 2003; Schwarz et al. 2003), we hypothesized that knockout of DUSP11 decreases AGO association with the BLV 5p miRNAs. To test this, we performed RNA immunoprecipitation (RIP) using a pan-AGO antibody to enrich for AGO-associated RNAs from wild-type and DUSP11 knockout HEK293T cells expressing BLV-miR-B2 and BLV-miR-B5. Immunoblot analysis confirmed that AGO RIP enriched for AGO proteins (Fig. 5A). Input and AGO RIP-recovered RNAs were subjected to Northern blot analysis. The BLV pre-miRNAs and longer noncoding RNAs (ncRNAs; as observed by EtBr staining) were largely depleted in the AGO RIP samples. However, the BLV miRNAs were efficiently recovered, indicating that AGO RIP enriched for AGO-associated BLV miRNAs (Fig. 5B). AGO association with both BLV-miR-B2-5p and BLV-miR-B5-5p was higher (approximately threefold to fivefold) in the wild-type cells than in the DUSP11 knockout cells. Conversely, AGO association with BLV-miR-B2-3p and BLV-miR-B5-3p miRNAs was higher (approximately twofold to fourfold) in the DUSP11 knockout cells as compared with the wild-type cells (Fig. 5B). Similar results were obtained by performing RIP using an anti-Flag antibody from wild-type and DUSP11 knockout cells expressing individual Flag-tagged AGOs 1–4 (Fig. 5C). These results demonstrate that knockout of DUSP11 results in decreased AGO-associated BLV 5p miRNAs and increased AGO-associated BLV 3p miRNAs.
Figure 5.
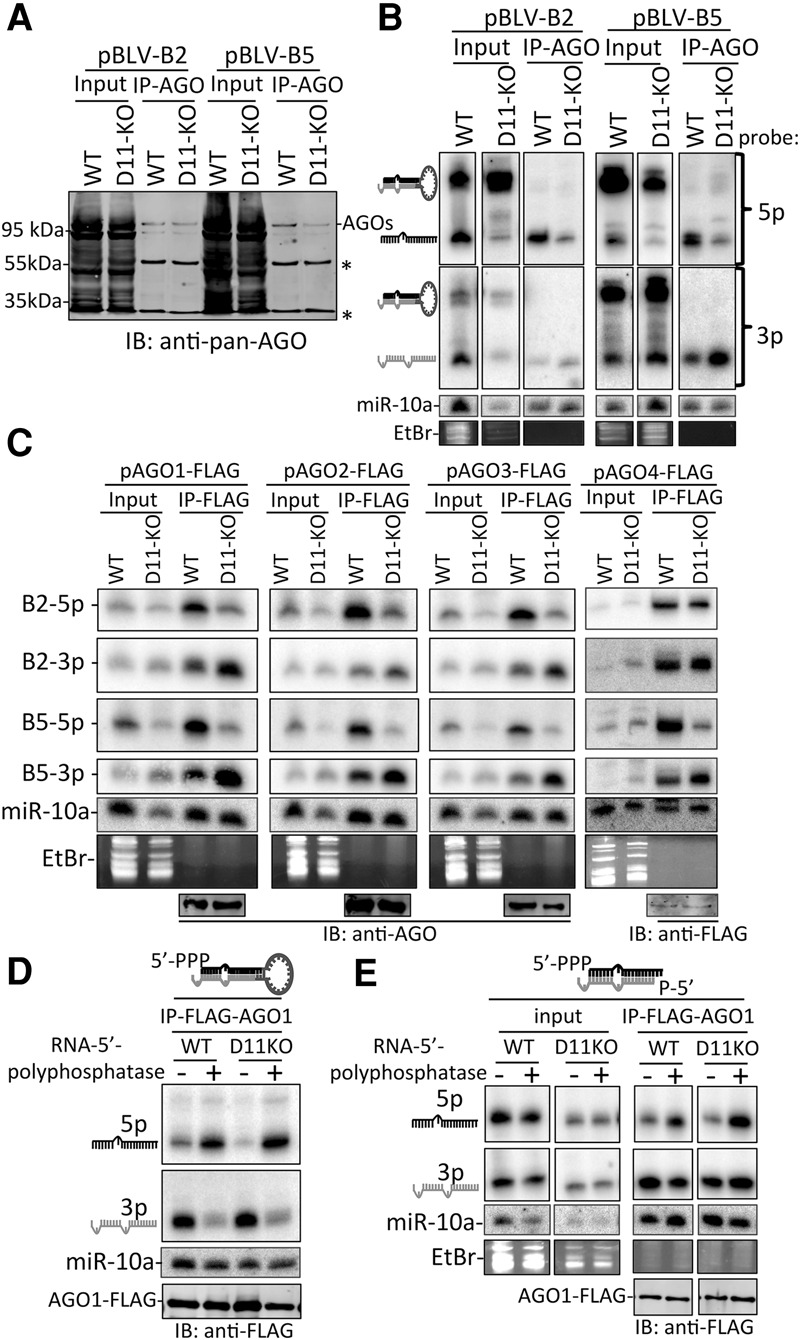
Knockout of DUSP11 decreases AGO association with 5′ triphosphorylated BLV 5p miRNAs. (A) Immunoblot analysis of input lysate (0.1%) and RIP (5%) of endogenous AGO proteins (∼95 kDa) using pan-AGO antibody (2A8) in parental (wild-type [WT]) and DUSP11 knockout (D11-KO) HEK293T cells transfected with either pBLV-B2 or pBLV-B5 expression vectors. Asterisks indicate IgG light and heavy chains. (B) Northern blot analysis of input RNA (2.5%) and RNA recovered from RIP samples (50%) in A. The membrane was first probed with 5p probes, stripped, and reprobed with the 3p probes. (C) RIP–Northern blot analysis of BLV B2 and B5 miRNAs associated with individual AGO proteins. Parental (wild-type) and DUSP11 knockout HEK293T cells were cotransfected with pBLV-B2 and pBLV-B5 and each of the indicated AGO-Flag expression vectors. The individual AGOs were precipitated using the anti-Flag M2 antibody. Total input RNA (5% for AGOs1–3 and 10% for AGO4) and RIP samples (95%) were subjected to Northern blot analysis. Blots were first probed for BLV-miR-B5-5p, sequentially stripped, and reprobed for BLV-miR-B5-3p, BLV-miR-B2-5p, and BLV-miR-B2-3p. (D) RIP–Northern blot analysis of parental and DUSP11 knockout HEK293T cells cotransfected with pAGO1-Flag and the BLV-B5 pre-miRNA mimic pretreated with (+; 5′-p-B5) or without (−; 5′-ppp-B5) RNA 5′ polyphosphatase. RIP was performed using the anti-Flag M2 antibody 48 h after transfection of mimics and pAGO1-Flag. RNA recovered from RIP-Flag was subjected to Northern blot analysis. The membrane was first probed for the 5p miRNA arm, stripped, and reprobed for the 3p arm. Immunoblot analysis of the RIP-Flag samples is shown below the Northern blot to show the immunoprecipitation efficiency of AGO1-Flag between the RNA 5′ polyphosphatase-treated (+) and untreated (−) samples. (E) Similar to D except using the BLV-B5 duplexed miRNA mimics in which the 5p arm was pretreated with (+) or without (−) RNA 5′ polyphosphatase. All of the duplexes contained a 3p miRNA mimic that was treated with RNA 5′ polyphosphatase to mimic the 5′ monophosphate generated by Dicer processing.
Our above data strongly suggest that DUSP11-mediated dephosphorylation of the BLV pre-miRNAs and/or 5p miRNAs promotes 5p miRNA accumulation in AGO proteins. Because the 5′ monophosphate group of miRNAs is known to interact with the MID domain of AGO (Schirle and Macrae 2012), we hypothesized that a 5′ triphosphate on the BLV pre-miRNAs may reduce loading and/or association of the derivative 5p miRNA with AGO. To directly test this, we analyzed AGO association of miRNAs derived from the 5′-ppp-BLV-pre-miR-B5 mimic treated with or without RNA 5′ polyphosphatase as described above. We cotransfected wild-type and DUSP11 knockout cells with the BLV-pre-miR-B5 mimics and pAGO1-Flag and performed RIP analysis. Northern blot analysis revealed that treatment of the 5′-ppp-BLV-pre-miR-B5 mimic with RNA 5′ polyphosphatase markedly increased AGO1 association with the 5p derivative while sharply decreasing AGO1 association with the 3p derivative (Fig. 5D). As the magnitude of the increase of the 5p derivative was equal to the decrease in the 3p derivative upon RNA 5′ polyphosphatase treatment, this suggests that the 5′ phosphate group alters strand selection into AGO1. This effect was observed in both the wild-type and DUSP11 knockout cells, suggesting that the pre-miRNAs were not efficiently dephosphorylated by DUSP11 when transfected into wild-type cells under these experimental conditions, perhaps due to saturation of cytosolic DUSP11, which has been shown to localize to the nucleus (Yuan et al. 1998). Nevertheless, AGO1 association with the 5p derivative was slightly less in the DUSP11 knockout cells in comparison with the wild-type cells. This indicates that DUSP11 dephosphorylated a small fraction of either the BLV pre-miRNA mimic or the derivative 5p miRNA in the wild-type cells. Similar results were obtained by performing AGO2 RIP using both BLV-pre-miR-B2 and BLV-pre-miR-B5 mimics (Supplemental Fig. S5B). These data demonstrate that 5p miRNAs generated from 5′ triphosphorylated BLV pre-miRNAs inefficiently associate with AGO.
To determine whether the 5′ monophosphate-dependent AGO strand bias of the B5 miRNAs is downstream from Dicer processing, we performed AGO1 RIP using B5 duplex miRNA mimics in which the 5p arm contained either a 5′ triphosphate or 5′ monophosphate. The BLV-miR-B5-5p and BLV-miR-B5-3p mimics were generated in vitro using T7 polymerase. The BLV-miR-B5-5p miRNA mimic was treated with or without RNA 5′ polyphosphatase and annealed to the BLV-miR-B5-3p mimic, which was pretreated with RNA 5′ polyphosphatase to mimic the 5′ monophosphate generated by Dicer-mediated endonucleolytic cleavage of the pre-miRNA. We cotransfected wild-type and DUSP11 knockout HEK293T cells with pAGO1-Flag and the B5 mimics and then immunoprecipitated AGO1. Northern blot analysis revealed that RNA 5′ polyphosphatase treatment increased 5p miRNA association with AGO1 in both wild-type and DUSP11 knockout cells (Fig. 5E). Similar results were obtained by performing RIP for AGO2-Flag (Supplemental Fig. S5C). These data indicate that a 5′ triphosphate group on a miRNA reduces association with AGO. However, unlike what we observed for the derivative of the pre-miRNA mimics, we did not observe a reciprocal decrease in AGO association of the 3p miRNA when the 5′ triphosphorylated 5p miRNA was treated with RNA 5′ polyphosphatase. As RNA 5′ polyphosphatase treatment of the 5′-ppp-BLV-pre-miR-B5 mimic significantly reduced AGO association with the 3p derivative (Fig. 5D), this suggests that 5′ monophosphate-dependent AGO strand bias is coupled to or enhanced by Dicer processing of the pre-miRNAs.
Combined, these data demonstrate that DUSP11 converts the 5′ triphosphate on the BLV-B2 and BLV-B5 pre-miRNAs to a 5′ monophosphate, which promotes stable incorporation of BLV-miR-B2-5p and BLV-miR-B5-5p into AGO through a Dicer-coupled 5′ monophosphate-dependent strand selection mechanism.
DUSP11 promotes accumulation, AGO association, and RISC activity of the 5p miRNA derived from human AdV viral-associated (VA) RNAI
Group C AdVs express two VA RNAs, VA RNAI (VAI) and VA RNAII (VAII), which are ∼160-nt highly structured ncRNAs. In addition to its role in inhibiting the antiviral response (O'Malley et al. 1986, 1989), VAI serves as a noncanonical pre-miRNA, giving rise to derivative miRNAs (mivaRNAs) via Dicer processing (Lu and Cullen 2004; Aparicio et al. 2006; Xu et al. 2007; Furuse et al. 2013). Interestingly, VAI is a primary RNAP III transcript that predominantly contains a 5′ triphosphate (Vennström et al. 1978). However, similar to the BLV 5p miRNAs, the RISC-active mivaRNAI-5p isoform from human AdV type 5 (AdV5) was shown to predominantly contain a 5′ monophosphate (Xu et al. 2009). Thus, we wanted to determine whether DUSP11 is responsible for the dephosphorylation of mivaRNAI-5p.
To test whether DUSP11 may be responsible for the dephosphorylation of mivaRNAI-5p, we analyzed the steady-state levels of VAI miRNAs in parental (wild-type) and DUSP11 knockout HEK293T cells transfected with VAI expression vector (pVAI). Northern blot analysis revealed that mivaRNAI-5p levels, but not the mivaRNAI-3p levels, were lower in the DUSP11 knockout cells as compared with the wild-type cells (Fig. 6A). We observed a similar decrease in mivaRNAI-5p levels in DUSP11-KO-D11-CM cells as compared with the DUSP11-KO-D11 cells, indicating that DUSP11 catalytic activity increases accumulation of mivaRNAI-5p. Next, we transfected wild-type and DUSP11 knockout cells with a negative control vector (pcDNA3.1), pAGO1-Flag, or pAGO2-Flag. The cells were then either transfected with pVAI or infected with AdV. RIP using the anti-Flag antibody revealed that mivaRNAI-5p was more associated (approximately threefold relative to mivaRNAI-3p) with AGOs in the wild-type cells as compared with the DUSP11 knockout cells from both the pVAI transfected and AdV5-infected cells (Fig. 6B). Furthermore, analysis of mivaRNAI-5p RISC activity revealed a decrease (approximately twofold) in mivaRNAI-5p RISC activity in DUSP11 knockout cells as compared with activity in the wild-type cells (Fig. 6C). This decrease in RISC activity was largely rescued in the DUSP11-KO-D11 cells but not in the negative control DUSP11-KO-EV or DUSP11-KO-D11-CM cell lines. These data demonstrate that DUSP11 catalytic activity increases mivaRNAI-5p accumulation, AGO association, and RISC activity.
Figure 6.
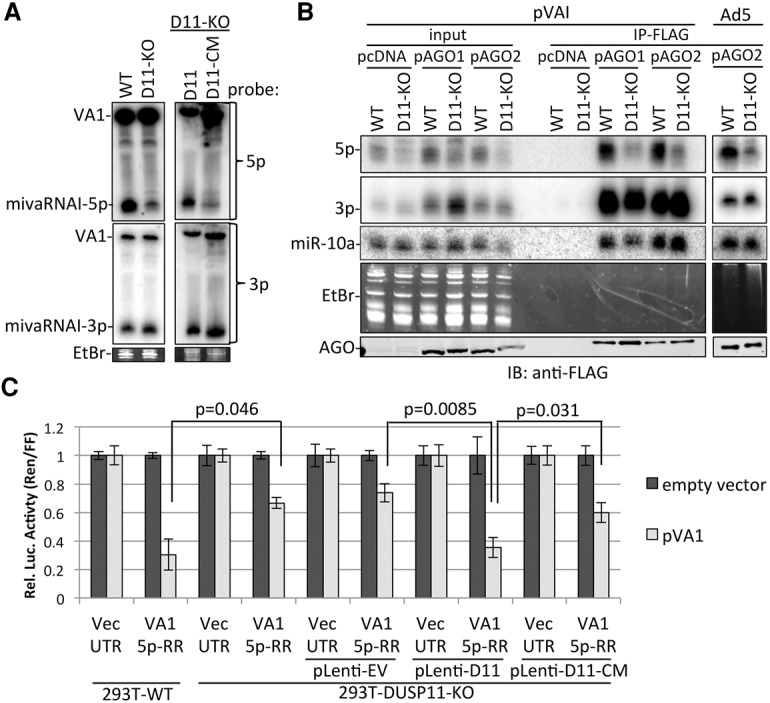
DUSP11 promotes accumulation, AGO association, and RISC activity of AdV5 mivaRNAI-5p. (A) Northern blot analysis of the 5p and 3p miRNAs derived from AdV5 VA RNAI from parental (wild-type [WT]), DUSP11 knockout (D11-KO), DUSP11-KO-pLenti-D11, and DUSP11-KO-pLenti-DUSP11-CM HEK293T cells transfected with pVAI. The membrane was first probed for the 5p miRNA, stripped, and reprobed for the 3p miRNA. (B) AGO RIP analysis of VAI-derived miRNAs. Parental (wild-type) and DUSP11 knockout HEK293T cells were cotransfected with pcDNA-EV, pAGO1-Flag, or pAGO2-Flag and either cotransfected with pVAI or infected with AdV5. RIP was performed 48 h after transfection using anti-Flag M2 antibody. One percent of the input RNA and 95% of RIP-recovered RNA were subjected to Northern blot analysis. The membrane was first probed for the 5p miRNA, stripped, and reprobed for the 3p miRNA. Immunoblot analysis was performed on 0.1% of input lysate and 5% of the Flag RIP. (C) Luciferase assay to measure RISC activity of the 5p miRNA derived from VAI RNA in parental (wild-type), DUSP11 knockout, DUSP11-KO-pLenti-D11, and DUSP11-KO-pLenti-DUSP11-CM HEK293T cells. Bars represent the mean luciferase ratio (Renilla/firefly) ± SEM from three experiments (wild-type and DUSP11 knockout) or four experiments (stable cell lines) in which transfections were performed in triplicates.
DUSP11 alters the phosphorylation state and steady-state levels of select RNAP III RNAs
We examined the role of DUSP11 on cellular RNAs. As expected, based on our above data, the levels of individual host miRNAs, which contain a 5′ monophosphate, are largely unaffected by knockout of DUSP11 in HEK293T cells (Supplemental Fig. S6A). To test whether knockout of DUSP11 affects the non-miRNA transcriptome, we used thermostable group II intron reverse transcriptase high-throughput sequencing (TGIRT-seq), which enables the analysis of full-length, highly structured RNAs (Mohr et al. 2013; Katibah et al. 2014; Nottingham et al. 2016; Qin et al. 2016). We used two different RNA-seq library preparation protocols in order to test for differences in the greatest range of host RNAs. In the first protocol, ribosomal-depleted (ribo-depleted) total RNA was first fragmented as is typical for RNA-seq experiments to assess mRNA abundances. In the second protocol, intact ribo-depleted total RNA was used as input in order to analyze full-length shorter ncRNA transcripts. We observed no overlap between the set of protein-coding genes up-regulated by knockout of DUSP11 in the HEK293T versus A549 backgrounds and only a single gene down-regulated in common (ABCG2) (Supplemental Fig. S6B; Supplemental Data 1). Since DUSP11 has been implicated previously as a potential splicing factor (Yuan et al. 1998), we quantitated isoform abundance using the Cufflinks package. We did not observe substantial differences in isoform abundances between the parental and corresponding DUSP11 knockout cell lines (Supplemental Fig. S6C; Supplemental Data 1). Thus, overall, DUSP11 does not appear to alter protein-coding gene expression to a high degree in the cellular contexts that we examined.
Next, we assayed short (<500-nt) noncoding gene expression in the unfragmented RNA-seq libraries (Supplemental Fig. S6D; Supplemental Data 1). To assay for differences in phosphate status, we developed a TGIRT-seq-based method in which we incorporated Terminator treatment for inferring the 5′ triphosphate status of short RNAs en masse. To assess the effectiveness of the Terminator treatment, we first examined the effects on small nucleolar RNA (snoRNAs), which typically possess a 5′ monophosphate (Kiss and Filipowicz 1993; Nag et al. 1993). Terminator treatment reduced the abundance of snoRNAs in all four treated libraries, confirming that Terminator activity can identify 5′ monophosphate transcripts in our libraries (Fig. 7A; Supplemental Fig. S6E). We then tested whether DUSP11 knockout altered the susceptibility of host genes to Terminator treatment. We focused our analysis on the classic RNAP III transcribed host RNAs, since they do not undergo 7-methylguanylate capping like mRNAs. We observed that a subset of RNAP III transcribed ncRNAs (vtRNAs, Y-RNAs, and RMRP RNA) exhibited increased resistance to Terminator treatment in the DUSP11 knockout cell lines in both parental cell backgrounds (Fig. 7B). These results are consistent with DUSP11 altering the phosphate status of select full-length RNAP III transcripts.
Figure 7.
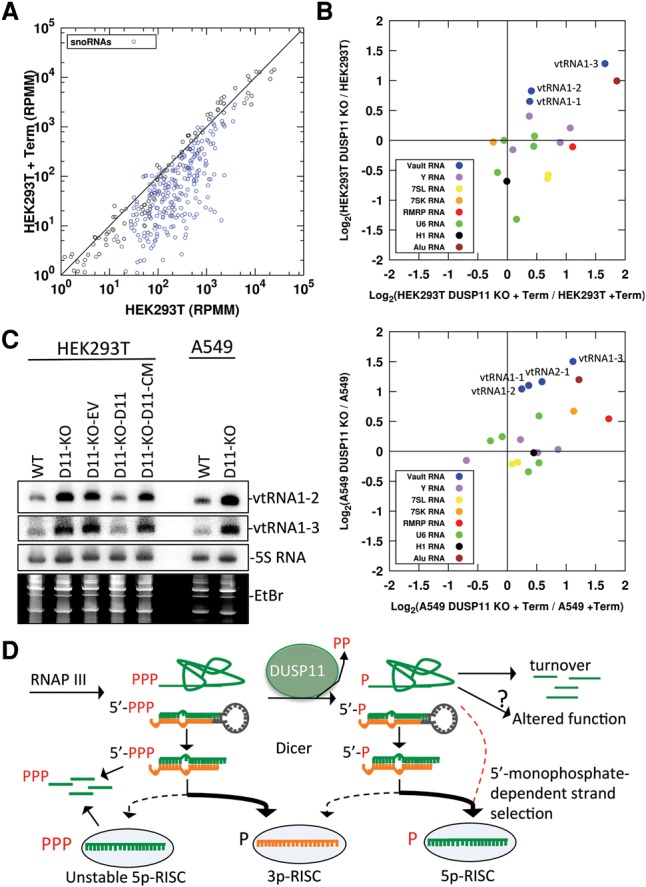
Analysis of cellular RNAs in DUSP11 knockout cell lines. (A) Host gene expression (RefSeq genes ≤500 nt with no annotated coding sequence) in HEK293T cells with or without Terminator treatment assayed by unfragmented TGIRT-seq. Read counts from the indicated cell line library mapping to annotated host genes in reads per million mapped (RPMM) are plotted on each axis. snoRNAs are indicated with blue circles. (B) Expression analysis of select host RNAP III transcribed genes in HEK293T, HEK293T DUSP11 knockout, A549, and A549 DUSP11 knockout cell lines assayed by fragmented TGIRT-seq with and without Terminator treatment. The log base 2 ratio of gene counts from the indicated libraries is plotted on each axis. (C) Northern blot analysis of candidate ncRNA DUSP11 targets from the indicated cell lines. EtBr-stained low-molecular-weight RNA is provided as an additional loading control. The membrane was first probed for vtRNA1-2, stripped, and reprobed for the indicated RNAs. (D) Model for the role of DUSP11 in RNA silencing and modulation of RNAP III transcripts in mammalian cells. RNAP III transcribed RNAs initially contain a 5′ triphosphate. DUSP11 dephosphorylates a fraction of these RNAs. This reduces the steady-state level and alters the activity/function of some RNAs. For RNAP III transcribed miRNA precursors, the 5p arm of the resulting 5′ monophosphorylated miRNA precursors is predominantly loaded into AGO proteins to generate stable/functional 5p RISCs. The miRNA precursors that remain 5′ triphosphorylated predominantly load the 3p miRNA in AGO, while the 5′ triphosphorylated 5p miRNAs are rapidly degraded. Furthermore, the 5′ triphosphorylated 5p miRNAs that are incorporated into AGO to generate an unstable 5p RISC, promoting degradation of the complex/5p miRNA. The red dashed arrow indicates the DUSP11-dependent, Dicer-independent route for the BLV 5p miRNAs and other noncanonical interfering RNAs.
To determine whether DUSP11 affects the steady-state levels of short ncRNAs, we compared the DUSP11 knockout to the relevant parental line and a minimum abundance of at least 10 reads per million mapped (RPMM) as cutoff in our unfragmented TGIRT libraries. This analysis revealed that the vtRNAs (vtRNA1-1, vtRNA1-2, and vtRNA1-3) increased approximately twofold in the absence of DUSP11 in both the HEK293T and A549 parental cell backgrounds (Fig. 7B; Supplemental Data 1). To independently verify this, we performed Northern blot analysis on total RNA harvested from HEK293T and HEK293T DUSP11 knockout-derived cell lines. As compared with parental HEK293T cells, the steady-state levels of vtRNA1-2 and vtRNA1-3 were increased in the HEK293T DUSP11 knockout cells (Fig. 7C). A similar increase in vtRNA1-2 and vtRNA1-3 was observed in the A549 DUSP11 knockout cells as compared with the parental A549 cells (Fig. 7C). Expression of DUSP11 but not the DUSP11 catalytic mutant in the DUSP11 knockout HEK293T cells reduced vtRNA1-2 and vtRNA1-3 to levels comparable with the parental HEK293T cells (Fig. 7C). These results validate the TGIRT RNA-seq results and demonstrate that DUSP11 catalytic activity modulates the steady-state levels of vtRNAs.
Finally, we performed an analysis of Alu-derived elements, which are also short ncRNAs transcribed by RNAP III (Chu et al. 1995). Due to the low abundance and repetitive nature of Alu transcripts, we used a strategy that pooled Alu transcript reads that met the following criteria. First, we used RepeatMasker annotations to count all reads mapping to within ±3 nt of the 5′ end of annotated Alu derived loci in the unfragmented TGIRT-seq libraries. This was to enrich for Alu reads derived from Alu loci versus degradation of Alu sequences embedded in other transcripts. We summed the 5′ Alu reads across all loci with one or more RPMM in either library for quantitation (Fig. 7B). The sum of these Alu reads showed greater abundance and Terminator resistance in the DUSP11 knockout lines, consistent with DUSP11 controlling the phosphate status and nuclease susceptibility of the 5′ ends of Alu transcripts. We then selected the most abundant quantifiable loci (≥10 RPMM) for further inspection (Supplemental Fig. S6F). This identified four loci in the HEK293T background. Notably, all four displayed higher steady-state levels in the DUSP11 knockout line (Supplemental Fig. S6F). Similar results were observed for seven out of the eight Alu loci identified in the A549 background (Supplemental Fig. S6F). Taken together, these data demonstrate that DUSP11 affects the phosphate status and/or steady-state levels of a diverse group of RNAP III RNAs.
Discussion
Here, we identify the first biologically relevant RNA substrates of DUSP11 and demonstrate that DUSP11 can function to alter the phosphorylation status, steady-state levels, and RNAi activity of select RNAP III transcripts. We show that DUSP11 RNA triphosphatase activity promotes accumulation, AGO association, and RISC activity of noncanonical miRNAs expressed by a bovine retrovirus (BLV) and an unrelated pathogenic human DNA virus (AdV). Based on our data, we propose that DUSP11 converts the 5′ triphosphate of diverse noncanonical miRNA precursors to a 5′ monophosphate, which is required for efficient association of the derivative 5p miRNAs with AGO proteins (Fig. 7D).
Knockout of DUSP11 RNA triphosphatase activity substantially reduced the RISC activity and steady-state levels of BLV-miR-B2-5p and BLV-miR-B5-5p (Figs. 1–3). Interestingly, we observed low amounts of RISC-active BLV-miR-B5-5p in Dicer knockout cells (Fig. 2A; Supplemental Fig. S2C–E), indicating that a portion of BLV 5p miRNAs can be generated via a Dicer-independent mechanism. Notably, a recent study showed that a fraction of human 5p miRNAs are also generated via a Dicer-independent mechanism (Kim et al. 2016). We note that low levels of the BLV 5p miRNAs were observed in DUSP11 knockout cells (Fig. 2A; Supplemental Fig. S4B). These results are consistent with the 5′ monophosphate-dependent small RNA-seq results (Fig. 2B; Supplemental Fig. S3A) and suggest that a small fraction (∼5%–10%) of BLV pre-miRNAs/miRNAs becomes 5′ monophosphorylated via an alternative mechanism, such as nonenzymatic hydrolysis, a redundant phosphatase, or endonucleolytic cleavage. Nevertheless, our data indicate that DUSP11 converts a substantial fraction of the BLV pre-miRNAs and/or 5p miRNAs from a 5′ triphosphate to a 5′ monophosphate, thereby promoting accumulation and RISC activity of the BLV 5p miRNAs (Figs. 1–4).
The reduction (∼10-fold) in total BLV 5p miRNA levels in DUSP11 knockout cells without a corresponding increase in BLV-5′-ppp–5p miRNAs (less than twofold) (Supplemental Fig. S3A) suggests that the BLV-5′-ppp–5p miRNAs are not loaded into AGO and, as a result, are rapidly turned over, similar to miRNA passenger strands (Khvorova et al. 2003; Schwarz et al. 2003). Consistent with this, AGO association with the BLV 5p miRNAs was reduced in DUSP11 knockout cells (Fig. 5A–C). Furthermore, the steady-state levels, AGO association, and RISC activity of the BLV 3p miRNAs were increased in the DUSP11 knockout cells (Figs. 1–5), indicating that the 3p miRNAs are preferentially loaded into AGO. These results suggest that DUSP11-mediated dephosphorylation of BLV pre-miRNAs and/or miRNAs promotes AGO association with the 5p miRNA through the process of strand selection. In support of this, we observed a strong inversion in AGO association with the 5p and 3p derivatives from 5′-p-pre-miRNAs as compared with 5′-ppp-pre-miRNAs. The 5′-ppp-pre-miRNAs generated much fewer AGO-associated 5p miRNAs than the 5′-p-pre-miRNAs, whereas 5′-ppp-pre-miRNAs generated much more AGO-associated 3p miRNAs than the 5′-p-pre-miRNAs (Fig. 5D; Supplemental Fig. S5B). We did not observe a similar robust inversion in strand selection using miRNA duplexes (Fig. 5E; Supplemental Fig. S5C), although AGO association with the 5′-ppp-5p-miRNA was reduced in comparison with the 5′-p-5p-miRNA. These findings suggest that 5′-ppp-5p-miRNA may unstably associate with AGO. Combined, these data demonstrate that the 5′-phosphate group can strongly influence AGO association with small RNAs via a 5′ monophosphate-dependent strand selection mechanism coupled to or enhanced by Dicer and/or Dicer-associated proteins during pre-miRNA processing. Since 5′ triphosphorylated shRNAs and siRNAs have technological applications and are potential therapeutic agents (Paddison et al. 2002; ter Brake et al. 2006; Lin et al. 2012; Chen et al. 2013b; Baek et al. 2015), DUSP11 activity may be a major factor affecting the utility of these approaches.
During endo-RNAi in C. elegans, the 5′ monophosphorylated 26G-RNAs are thought to initially contain a 5′ triphosphate due to direct synthesis by RRF-3 (Gent et al. 2010; Vasale et al. 2010). This observation implicated PIR-1 in 26G-RNA dephosphorylation (Gent et al. 2010), which is consistent with recent findings showing that PIR-1 is required for 26G-RNA accumulation (Chaves 2015). However, the relevant function of 26G-RNA dephosphorylation remains unclear. Two alternative models have been proposed (Chaves 2015). In the first, PIR-1-mediated dephosphorylation of a precursor RNA substrate is required to generate the 26G-RNAs via Dicer processing. In the second, PIR-1-mediated dephosphorylation of 26G-RNAs promotes productive loading of 26G-RNAs with the primary ERGO-1 AGO. Our data with DUSP11 support the latter model.
While the activity of DUSP11 in controlling the phosphorylation status and abundance of select RNAP III transcripts is clear, the extent of DUSP11-dependent RNAi involving endogenous mammalian RNAs remains unclear. We did not observe significant global changes in the level of host miRNAs or mRNAs in DUSP11 knockout cells (Supplemental Fig. S6A–D; Supplemental Data 1). These results are consistent with DUSP11 not being required for normal housekeeping processes, including host miRNA biogenesis and mRNA splicing, at least in the contexts that we examined. However, we did observe increases in abundance and/or Terminator resistance for some RNAP III transcribed host ncRNAs (Fig. 7B,C). These findings are consistent with an increase in the RNAP I transcribed 5′ETS spacer fragment during 45S rRNA processing that was observed upon DUSP11 knockdown (Tafforeau et al. 2013). Combined, these results suggest that DUSP11 is involved in the regulation, maturation, and/or turnover of some 5′ triphosphorylated host-encoded ncRNAs transcribed by RNAP I and RNAP III.
Our small RNA-seq data indicate that a higher portion of RNAs that map to the loci of multiple RNAP III transcribed RNAs, including small RNA derivatives (18–32 nt) from the 5′ end of the 5S rRNA and Y-RNAs, is 5′ triphosphorylated in DUSP11 knockout cells (Supplemental Fig. S3B,C; Supplemental Data 1). Meta-analysis of previously published AGO-associated RNAs (Hafner et al. 2010) revealed that some of the 5′ end derivatives of the 5S rRNA associate with AGO proteins (Supplemental Fig. S3D). Although it is unclear whether their association with AGO is direct, we note that nucleotide transitions, a hallmark of direct protein association in PAR-CLIP (photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation) studies, are observed in RNAs mapping to the 5′ end of 5S rRNA (Supplemental Fig. S3D). These RNAs are slightly longer than canonical miRNAs and are too low in abundance to have canonical silencing function. However, it is plausible that they possess noncanonical functions or function as silencing RNAs when more abundant in other cellular contexts. In support of this, AGO has been shown to associate with the genomic loci of 5S RNA genes via RNA intermediates (Woolnough et al. 2015). Therefore, it is conceivable that some DUSP11-dependent RISC-loaded RNAs play a role in recruiting RISC to RNAP III loci, which will be the subject of future studies.
DUSP11 homologs are observed in diverse metazoans. Our findings suggest that DUSP11/PIR-1-mediated dephosphorylation is an anciently conserved mechanism to permit loading of small RNAs that initially contain a 5′ triphosphate into AGO proteins. Furthermore, we show that DUSP11 is a previously unappreciated modulator of select RNAP III transcript phosphate status and abundance in mammalian cells. Because mammals have evolved a complex protein-based response to viruses that can be triggered by 5′ triphosphorylated RNAs, it will be important to investigate the activity of DUSP11 during pathogen defense response. In this regard, it is interesting to note that Alu transcripts, which, in the appropriate cellular context, are known to induce the inflammasome innate immune response (Kaneko et al. 2011; Tarallo et al. 2012; Kerur et al. 2013), are higher in DUSP11−/− cells (Fig. 7B; Supplemental Fig. S6F).
In conclusion, our findings demonstrate that DUSP11 functions to control the 5′ phosphate status of select RNAP III transcripts. The biological outcomes of this regulation include altering the steady-state levels of abundant and known immunogenic transcripts as well as the propensity for stable association of diverse viral miRNAs in RISC. It will therefore be interesting to learn whether DUSP11 more broadly participates in the inflammatory response and RNA silencing of host transcripts.
Materials and methods
Plasmids
The BLV miRNA expression vectors and corresponding RISC reporters are described in Burke et al. (2014). The B2-5p and B5-5p DSM RISC reporters were made by fill-in PCR using Phusion polymerase (New England Biolabs) and the B5-5p and B2-5p DSM primers (Supplemental Table S1). The amplicons were then cloned into the Xho1/Xba1 sites of pcDNA3.1dsRluc (Seo et al. 2008). The DUSP11 expression vectors were generated via PCR amplification of the DUSP11 ORF from pDONR221-DUSP11 vector (DNASU, clone HsCD00043373), which had a G-to-T point mutation at nucleotide 494. This was corrected by PCR-directed mutagenesis using DUSP11 491 sense and DUSP11 506 antisense primers (Supplemental Table S1). A 3x-Flag encoding sequence was then inserted after the start codon by PCR using DUSP11-3xNT-Flag primers (Supplemental Table S1). The C152S mutation was generated by PCR-directed mutagenesis using DUSP11 C152S primers (Supplemental Table S1). The DUSP11-3xFlag and DUSP11-CM-3xFlag amplicons were cloned into the Kpn1/Xba1 sites of pcDNA3.1+ and the Xho1/Xba1 sites of pLenti CMV Blast empty (Addgene, plasmid 17486). The pISK-HSUR4 vector was described previously (Kincaid et al. 2014). For the DUSP11 CRISPR/CAS9 guide RNAs (gRNAs), the D11_gRNA_1, D11_gRNA_2, and D11_FLK_gRNA gene blocks (Supplemental Table S1) were cloned into pIDT-SMART-Kan (pISK). The VAI-5p RISC reporter was made by fill-in PCR of VAI-5p-RR sense and antisense primers (Supplemental Table S1) and cloned into the Xho1/Xba1 sites of pcDNA3.1dsRluc. pVAI was made by amplifying the VAI locus from the pAdvantage plasmid using the VAI primers (Supplemental Table S1) and was cloned into the Kpn1/Xba1 site of puc19. The mivaRNAs-5p RISC reporters were made as described above using the VAI 5p RR oligos (Supplemental Table S1). The pIRESneo-Flag/HA Ago1, pIRESneo-Flag/HA Ago2, pIRESneo-Flag/HA Ago3, and pIRESneo-Flag/HA Ago4 plasmids (Meister et al. 2004) were obtained from Addgene. The SV40-RI257 miRNA expression vector and RISC reporters are described in Chen et al. (2013a). Potential BLV 5p miRNA docking sites in 3′ UTR sequences were identified using the miRanda algorithm with the strict seed alignment option (Enright et al. 2004). To make the CECR1 and CECR1 DSM reporters, the CECR1_gBlock or CECR1_DSM_gBlock (Supplemental Table S1)—which contain the majority of the ovine CECR1 3′ UTR (XM_012175609.2) or the CECR1 DSM control, which contains three to four point mutations in nucleotides complementary to the seed of BLV(913)-miR-B5-5p at each predicted docking site—were inserted into the Xho1/Xba1 sites of pcDNA3.1dsRluc.
Cell culture
HEK293T and HEK293T Dicer knockout (NoDice 2-20) cells were provided by Dr. Bryan Cullen (Bogerd et al. 2014). HEK293 cells and A549 cells were obtained from American Type Culture Collection. Cells were maintained in DMEM supplemented with 10% (v/v) FBS (Cellgro). Transfections were performed using Lipofectamine2000 (Invitrogen). FLK-BLV cells were maintained in DMEM supplemented with 10% (v/v) FBS.
Generation of DUSP11 knockout cell lines
Cells (T25-flask; 50% confluency) were cotransfected with 4.2 µg of hCas9 vector, 1.2 µg of pISK-D11_gRNA_1, 1.2 µg of pISK-D11_gRNA_2, and 600 ng of pLX304 using Lipofectamine2000 (Invitrogen). Twenty-four hours after transfection, the medium was replaced with medium containing 5 µg/mL blasticidin. Selective medium was replaced 3 d after transfection. Five days after transfection, selective medium was replaced with normal growth medium. The medium was replaced every 2–3 d until 13 d after transfection. Cells were then replated by serial dilution to generate individual clones. Individual clones were propagated and screened by immunoblot analysis. The FLK DUSP11 knockout cells were made similarly as described above using 4.2 µg of hCas9 vector, 2.4 µg of the D11_FLK_gRNA (Supplemental Table S1), 600 ng of pcDNA3.1-puro, and 5 µg/mL puromycin for selection. Individual clones were screened by Western blot analysis and confirmed by Sanger sequencing of the target region, which was amplified using the D11_FLK primers (Supplemental Table S1) and cloned into the Xho1/Xba1 sites of pcDNA3.1dsRluc or pISK. The following clones had mutations in both alleles (nucleotides are relative to start of the ovine DUSP11 ORF; XM_004006079.3): FLK-BLV-D11-KO-1 had a 1-nt deletion (nucleotide 296; allele 1) and a 1-nt insertion between nucleotides 295 and 296 (allele 2). FLK-BLV-D11-KO-20 had an 8-nt deletion (nucleotides 295–305; allele 1) and 2-nt deletion (nucleotides 297 and 298; allele 2). FLK-BLV-D11-KO-37 had a 14-nt deletion (nucleotides 291–304; allele 1) and a 1-nt deletion (nucleotide 297; allele 2).
Knockdown of DUSP11
HEK293T cells (six-well format, 70% confluency) were transfected with 50 pmol of either silencer select negative control siRNA (scrambled; Ambion) or DUSP11-specific siRNAs (Ambion, AM16708; siRNA ID 105842). Forty-eight hours later, cells were reseeded into a 12-well format (1:3 ratio) and cotransfected with 1 µg of either pBLV-B1 or pBLV-B5 per well, 5 ng of Rluc-RISC reporters per well, and 10 pmol of either DUSP11-specific siRNAs or negative control siRNA per well. Transfections were performed in triplicate. Dual-luciferase assay was performed 24–48 h after transfection of reporters and miRNA expression vectors using GloMax 96 Microplate Luminometer (Promega). Data were normalized to negative control siRNAs, pBLV-B1, and Rluc vector 3′ UTR.
RISC reporter assays in the DUSP11 knockout cell lines
For analysis of the RISC activity of the B5 miRNAs in the parental (wild-type), DUSP11 knockout, DUSP11-KO-EV, DUSP11-KO-D11, and DUSP11-KO-D11-CM HEK293T cells, the cells (24-well format, 70% confluent) were cotransfected with 500 ng of pISK-EV, pBLV-B1, or pBLV-B5 per well; 2.5 ng of the firefly luciferase expression vector (pcDNA3.1dsLuc2CP) per well; and 2.5 ng of each individual B5 RISC reporter per well using 2-µL of Lipofectamine2000 per well. A pooled transfection reaction for each miRNA expression vector/reporter combination was made and equally distributed to each cell line. Forty-eight hours after transfection, lysates were harvested, and dual-luciferase assay was performed. Raw Renilla/firefly ratios were normalized to pISK-EV and Rluc vector 3′ UTR for each data set. To assay B2 miRNAs and B5 miRNAs in parental and DUSP11-KO-5 HEK293 cells, cells (24-well format, 70% confluent) were cotransfected with 50 ng of pISK-EV, pBLV-B2, or pBLV-B5 per well; 2.5 ng of the firefly luciferase expression vector (pcDNA3.1dsLuc2CP) per well; and 2.5 ng of each individual RISC reporter per well using 1 µL of Lipofectamine2000 per well. Dual luciferase was performed 48 h after transfection. To measure the RISC activity of the B2 and B5 miRNA in parental and DUSP11-KO-45 A549 cells, cells were cotransfected with 500 ng of pISK-EV, pBLV-B2, or pBLV-B5 per well; 10 ng of the firefly luciferase expression vector (pcDNA3.1dsLuc2CP) per well; and 10 ng of each individual of the B5 RISC reporter per well using 2-µL of Lipofectamine2000 per well. Transfection was noticeably toxic to the DUSP11 knockout A549 cells, so the luciferase assay was performed 24 h after transfection. Transfections were performed in triplicate for each experiment. The bars in the graphs represent the average of all of the normalized ratios. The SEM was calculated from the standard deviation of all of the data points, with each experiment performed in triplicate representing one sample (n).
Immunoblot analysis for DUSP11
Cells were lysed in SDS solution (1% SDS, 2% β-mercaptoethanol) by boiling for 10 min followed by 1 min of vortexing (Hume et al. 2008). Equal volumes of lysates were fractionated on 12.5% SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad). Anti-DUSP11 polyclonal rabbit antibody (1:2000 dilution; Proteintech, catalog no. 10204-2-AP) and anti-α-Tubulin monoclonal mouse antibody (1:10,000 dilution; Sigma-Aldrich, catalog no. T6199) in phosphate-buffered saline (PBS) with 0.1% Tween20 (PBST) and 5% BSA was used to blot for DUSP11 and α-Tubulin. After three washes with PBST, membranes were blotted with IRDye 800CW and IRDye 680LT secondary antibodies (1:10,000 dilution; LI-COR) in PBST with 5% BSA. Blots were washed four times with PBST and then scanned on an Odyssey CLx infrared imaging system (LI-COR).
Generation of stable cell lines expressing DUSP11 and DUSP11 catalytic mutant
Generation of the lentiviral particles, transduction, and selection with blasticidin were performed as described previously (Kincaid et al. 2013).
Northern blot analysis
Northern blots were performed as described in McClure et al. (2011). Briefly, RNA was isolated using PIG-B (Weber et al. 1998), fractionated on 15% PAGE-urea gel, and transferred to Amersham Hybond N+ membrane (GE Healthcare). The membrane was then probed in ExpressHyb hybridization solution (Clontech) with the indicated DNA oligos (Integrated DNA Technologies) (Supplemental Table S1) radiolabeled with 6000 Ci/mmol [γ-32P]-ATP by T4 polynucleotide kinase (T4PNK; New England Biolabs). Membranes were exposed to a storage phosphor screen (GE Healthcare) and visualized with a Typhoon biomolecular imager (GE Healthcare). Membranes were stripped by incubating the membrane in boiling 0.1% SDS with agitation for 5 min, which was repeated three times.
Generation of the BLV pre-miRNA and miRNA mimics
The B2 and B5 pre-miRNA mimics and B5 5p and 3p miRNA mimics were generated using the AmpliScribe T7-Flash transcription kit (Epicentre) and T7 B5 pre-miRNA 5G oligos, T7 B5 G5p strand oligos, T7 B5 G3p strand oligos, T7 B2 pre-miRNA 5′-G oligos (Supplemental Table S1) according to the manufacturer's instructions. The 5′ adenosine of the pre-miRNAs and miRNA was changed to a 5′ guanosine in the mimics for efficient T7-mediated transcription. The RNAs were extracted with PIG-B and fractionated on nondenaturing 20% PAGE. The pre-miRNA bands (∼55 nt) and miRNA bands (∼22 nt) were then gel-excised and purified as described in Burke et al. (2015). To convert the 5′ triphosphate to a 5′ monophosphate, 10 µg of the RNAs was then treated with or without 1 µL of RNA 5′ polyphosphatase (Epicentre) in a 20-µL reaction and incubated for 1 h at 37°C. RNA was then purified via ammonium acetate precipitation in 75% EtOH. The BLV-B2 and BLV-B5 pre-miRNA mimics were flash-annealed in 100 mM Tris (pH 7.5), 30 mM NaCl, and 3 mM MgCl2. The BLV-B5 miRNA mimics were annealed in siRNA annealing buffer (Dharmacon). For the BLV pre-miRNA mimic experiments, wild-type and DUSP11 knockout HEK293T cells (24-well format; 70% confluent) were transfected with 2 ng of the pre-miRNA mimics. Total RNA was harvested 24–48 h after transfection and subjected to Northern blot analysis. For AGO RIP analysis of the BLV pre-miRNA mimics, cells (10-cm dish, 70% confluent) were cotransfected with 100 ng of pre-miRNA mimic and 7.5 µg of either pIRESneo-Flag/HA Ago1 or pIRESneo-Flag/HA Ago2. For the B5 miRNA duplex mimics, HEK293T and HEK293T DUSP11 knockout cells (10-cm dish; 70% confluent) were cotransfected with 20 pmol of B5 miRNA duplex mimics and either pIRESneo-Flag/HA Ago1 or pIRESneo-Flag/HA Ago2. RIP–Northern blot analysis was then performed as described below.
In vitro phosphatase reactions
In vitro translated DUSP11 and DUSP11 catalytic mutant proteins were generated using the TnT quick-coupled transcription/translation system (Promega) and the pcDNA3.1-DUSP11 and pcDNA3.1-DUSP11-CM plasmids as templates. The radiolabeled [γ-32P]-B5-pre-miRNA and [γ-32P]-B5-miRNA mimics were generated by standard T7 polymerase reactions containing 8 µL of 6000 Ci/mmol [γ-32P]ATP and T7 B5 pre-miRNA and T7 B5 5p strand oligos (Supplemental Table S2), which encode for a 5′ adeonsine. The mimics were then purified using MicroSpin G-25 columns (GE Healthcare). Ten picomoles of each mimic was incubated in a 20-µL reaction (50 mM Tris, 10 mM KCl, 5 mM DTT, protease inhibitor [Roche], 1 U of SUPERase In RNase inhibitor [ThermoFisher]) that contained 3.3 µL of in vitro translated products (DUSP11-3xFlag, DUSP11-CM-3x-Flag, or the negative control luciferase) or 1 µg of CIP (New England Biolabs) for 1 h at 37°C. The reactions were fractionated on 0.5× 8 M Tris–borate–EDTA–urea–15% polyacrylamide denaturing gel. RNAs were visualized by EtBr staining. The RNAs were then transferred to Amersham Hybond N+ membranes (GE Healthcare). Membranes were exposed to a storage phosphor screen (GE Healthcare) and visualized with a Typhoon biomolecular imager (GE Healthcare).
RIP of AGO proteins
RIP was performed as described in Flores et al. (2014). For RIP of the BLV miRNAs with endogenous AGO proteins, HEK293T and HEK293T DUSP11 knockout cells were seeded in a 15-cm dish. When 70% confluent, cells were transfected with 15 µg of pBLV-B2 or pBLV-B5. Forty-eight hours after transfection, cells were collected, washed in PBS, and pelleted. One percent of cells were removed for total input RNA isolation. The cells were then lysed in 3 vol (∼1 mL) of NP40 lysis buffer (50 mM HEPES at pH 7.5, 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% [v/v] NP-40, 0.5 mM DTT, protease inhibitors [Roche]) for 10 min on ice. The lysate was centrifuged at 5000g for 15 min at 4°C. The supernatant was collected (∼1 m), and 0.1% was saved for immunoblot analysis. The supernatant was then added to 50 µL of Protein G Dynabeads (Life Technologies) preloaded with 5 µg of anti-pan-AGO 2A8 monoclonal mouse antibody (Diagenode) and incubated for 2 h at 4°C with rotation. The lysate was removed, and the beads were washed 10 times with NT2 buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40) for 5 min at 4°C with rotation. After the last wash was removed, the beads were resuspended in 100 µL of NT2 buffer, and 5 µL (5%) was removed for immunoblot analysis. Four units of proteinase K (New England Biolabs) were then added to the remaining 95 µL of beads resuspended in NT2 buffer and incubated for 30 min at 55°C with agitation. The RNA was then extracted with PIG-B followed by precipitation in 1 vol of isopropanol, 0.3 M sodium acetate, and 1 µg of glycogen. The recovered RNA was then subjected to Northern blot analysis and next-generation small RNA-seq.
For RIP analysis of the individual AGO proteins, HEK293T and HEK293T DUSP11 knockout cells (10-cm dish, 70% confluent) were cotransfected with 3 µg of pBLV-B2, 3 µg of pBLV-B5, and 3 µg of individual AGO-Flag expression vectors (pIRESneo-Flag/HA Ago1, pIRESneo-Flag/HA Ago2, or pIRESneo-Flag/HA Ago3) (Meister et al. 2004). Forty-eight hours after transfection, RIP–Northern blot analysis was performed using the anti-Flag M2 mouse monoclonal antibody (Sigma-Aldrich).
Small RNA-seq
For small RNA-seq of total small RNAs, small RNAs (<70 nt) were isolated as described in Burke et al. (2015). The small RNAs were treated with or without RNA 5′ polyphosphatase. RNA was then recovered via ammonium acetate precipitation in 75% EtOH. Small RNA libraries were prepared for Illumina small RNA-seq using the multiplex small RNA library preparation set (New England Biolabs) and sequenced on Illumina HiSeq 2500 and NextSeq 500 platforms (Genomic Sequencing and Analysis Facility, University of Texas at Austin). Adapter sequences were trimmed from the reads using FastX clipper from the FastX toolkit software (http://hannonlab.cshl.edu/fastx_toolkit). The preprocessed reads were then mapped to reference sequences using the SHRiMP2 software package (Rumble et al. 2009). Small RNA reads for the BLV miRNAs and RNAP III-derived small RNAs were quantitated using custom scripts. Host miRNAs were quantitated using mirUtils, a custom script provided by Anna Battenhouse, Nathan Abell, and Vishy Iyer (http://mirutils.sourceforge.net).
RNA-seq using TGIRTs
TGIRT-seq libraries were constructed essentially as described (Nottingham et al. 2016; Qin et al. 2016). Briefly, total RNA from HEK293T cells or A549 cells was first ribo-depleted by using a RiboZero Gold (human/mouse/rat) kit (Illumina). The resulting ribo− RNA was then either mock-treated or digested with 1 U of Terminator exonuclease (Epicentre) according to the manufacturer's instructions. Pretreated RNAs were then incubated with T4PNK (Epicentre) to remove 3′ phosphates or 2′,3′ cyclic phosphates, which inhibit TGIRT template switching (Mohr et al. 2013). Additionally, a subset of pretreated RNAs was fragmented prior to T4PNK treatment with an NEBNext Magnesium RNA fragmentation module (New England Biolabs) by incubating RNAs for 7 min at 94°C. RNA reaction cleanup used a modified version of the Zymo RNA Clean and Concentrator (Zymo Research) protocol (i.e., addition of eight sample volumes of ethanol to increase retention of very small RNA species).
cDNAs were synthesized via TGIRT template switching with 1 µM purified TeI4c-ΔEn-MRF reverse transcriptase for 15 min at 60°C as described (Mohr et al. 2013; Qin et al. 2016). During synthesis, a DNA oligonucleotide containing the complement of an Illumina Read 2 sequencing primer-binding site was seamlessly linked to the 5′ cDNA end. After reaction cleanup, a DNA oligonucleotide containing the complement of an Illumina Read 1 sequencing primer-binding site was then ligated to the cDNA 3′ end by using thermostable 5′ AppDNA/RNA ligase (New England Biolabs). Properly ligated cDNAs were then amplified for 12 cycles of PCR in order to synthesize the second strand and add Illumina capture and index sequences. Libraries were size-selected with Ampure XP beads (Beckman-Coulter) and evaluated on an Agilent 2100 Bioanalyzer. TGIRT-seq libraries were sequenced on the Illumina NextSeq 500 platform (150-nt single-end reads) at the Genome Sequencing and Analysis Facility at the University of Texas at Austin.
Reads were mapped to the University of California at Santa Cruz hg19 human genome reference sequence using Bowtie2 in local alignment mode (Langmead and Salzberg 2012). Read counts per RefSeq annotated gene were generated using the featureCounts program in the Subread package (Liao et al. 2014). To identify Alu-derived loci, mapped reads with an alignment score ≥200 and beginning ±3 nt from the 5′ end of RepeatMasker annotated Alu-derived sequences were counted, and only loci with one or more RPMM were included in the analysis (http:// www.repeatmasker.org). For isoform analysis, adaptor sequences were trimmed using CutAdapt (Martin 2011). Trimmed reads were mapped using HISAT2 (Pertea et al. 2016). The Cufflinks package was used to assemble and estimate transcript abundance using Gencode 24lift37 annotations (Trapnell et al. 2010). Plots were generated using the gnuplot package. All RNA-seq data can be accessed through NCBI BioProject PRJNA342202.
Supplementary Material
Acknowledgments
We thank Dr. Bryan Cullen for the parental HEK293T and HEK293T NoDice cell lines, and members of C.S.S.'s laboratory for comments regarding this manuscript. Funding was provided by National Institutes of Health (R01AI077746 to C.S.S., and R01GM37949 and GM37951 to A.M.L.), a Burroughs Wellcome Investigators in Pathogenesis Award, Cancer Prevention and Research Institute of Texas (RP110098), and a University of Texas at Austin Institute for Cellular and Molecular Biology fellowship. J.M.B. initiated the project. J.M.B., R.P.K., A.M.L., and C.S.S. designed the experiments. J.M.B., R.P.K., and R.M.N. performed the experiments. J.M.B., R.P.K., A.M.L., R.M.N., and C.S.S. interpreted the data. J.M.B. and C.S.S. wrote the manuscript with help from the other authors.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.282616.116.
References
- Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. 2006. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 80: 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SE, Kim H, Kim KB, Yoon S, Choe J, Suh W, Jeong YJ, Cho YH, Kim DE. 2015. Dual effects of duplex RNA harboring 5′-terminal triphosphate on gene silencing and RIG-I mediated innate immune response. Biochem Biophys Res Commun 456: 591–597. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Basyuk E, Suavet F, Doglio A, Bordonne R, Bertrand E. 2003. Human let-7 stem–loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res 31: 6593–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. 2010. A mammalian herpesvirus uses non-canonical expression and processing mechanisms to generate viral microRNAs. Mol Cell 37: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. 2014. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20: 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Bass CR, Kincaid RP, Sullivan CS. 2014. Identification of tri-phosphatase activity in the biogenesis of retroviral microRNAs and RNAP III-generated shRNAs. Nucleic Acids Res 42: 13949–13962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Kuny CV, Kincaid RP, Sullivan CS. 2015. Identification, validation, and characterization of noncanonical miRNAs. Methods 91: 57–68. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Xie M, Steitz JA. 2011. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell 43: 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves D. 2015. The RNA 5′ phosphatase PIR-1 cooperates with Dicer to produce endogenous small RNAs and suppress viral replication in C. elegans. PhD thesis University of Lisbon, Lisbon, Portugal. [Google Scholar]
- Chen CJ, Cox JE, Kincaid RP, Martinez A, Sullivan CS. 2013a. Divergent microRNA targetomes of closely related circulating strains of polyomavirus. J Virol 87: 11135–11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qian Y, Yan F, Tu J, Yang X, Xing Y, Chen Z. 2013b. 5′-triphosphate-siRNA activates RIG-I-dependent type I interferon production and enhances inhibition of hepatitis B virus replication in HepG2.2.15 cells. Eur J Pharmacol 721: 86–95. [DOI] [PubMed] [Google Scholar]
- Chu WM, Liu WM, Schmid CW. 1995. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res 23: 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Deshpande T, Takagi T, Hao L, Buratowski S, Charbonneau H. 1999. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J Biol Chem 274: 16590–16594. [DOI] [PubMed] [Google Scholar]
- Diebel KW, Smith AL, van Dyk LF. 2010. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16: 170–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebel KW, Claypool DJ, van Dyk LF. 2014. A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing. Gene 544: 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2004. MicroRNA targets in Drosophila. Genome Biol 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ER, Kara M, Coleman CB, Katrina R, Grau KR, Oko LM, Krueger BJ, Renne R, van Dyk LF, Tibbetts SA. 2014. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. mBio 5: e00981–e001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Kennedy EM, Skalsky RL, Cullen BR. 2014. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res 42: 4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. 2010. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822. [DOI] [PubMed] [Google Scholar]
- Furuse Y, Ornelles DA, Cullen BR. 2013. Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA. Virology 447: 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet NA, Hamaidia M, de Brogniez A, Gutiérrez G, Renotte N, Reichert M, Trono K, Willems L. 2016. Bovine leukemia virus small noncoding RNAs are functional elements that regulate replication and contribute to oncogenesis in vivo. PLoS Pathog 12: e1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411: 325–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Martin S, et al. 2006. Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997. [DOI] [PubMed] [Google Scholar]
- Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. 2008. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 320: 797–799. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katibah GE, Qin Y, Sidote DJ, Yao J, Lambowitz AM, Collins K. 2014. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc Natl Acad Sci 111: 12025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Hirano Y, Tarallo V, Fowler BJ, Bastos-Carvalho A, Yasuma T, Yasuma R, Kim Y, Hinton DR, Kirschning CJ, et al. 2013. TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Invest Ophthalmol Vis Sci 54: 7395–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Kim VN. 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim B, Kim VM. 2016. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci 133: E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8: e1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci 109: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RP, Burke JM, Cox JC, de Villiers EM, Sullivan CS. 2013. A human torque teno virus encodes a microRNA that inhibits interferon signaling. PLoS Pathog 9: e1003818. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kincaid RP, Chen Y, Cox JE, Rethwilm A, Sullivan CS. 2014. Non-canonical miRNA biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. mBio 5: e00074–e00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W. 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J 12: 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Lin L, Liu Q, Berube N, Detmer S, Zhoua Y. 2012. 5′-triphosphate-short interfering RNA: potent inhibition of influenza A virus infection by gene silencing and RIG-I activation. J Virol 86: 10359–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 78: 12868–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. [Google Scholar]
- Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 2007. 5′ to 3′-exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′-stability of mRNA. Cell 129: 681–692. [DOI] [PubMed] [Google Scholar]
- McClure LV, Lin YT, Sullivan CS. 2011. Detection of viral microRNAs by Northern blot analysis. Methods Mol Biol 72: 153–171. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al. 2013. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19: 958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag MK, Thai TT, Ruff EA, Selvamurugan N, Kunnimalaiyaan M, Eliceiri GL. 1993. Genes for E1, E2, and E3 small nucleolar RNAs. Proc Natl Acad Sci 90: 9001–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 2007. 5′-triphosphate-dependent activation of PKR by RNAs with short stem–loops. Science 318: 1455–1458. [DOI] [PubMed] [Google Scholar]
- Noland CL, Doudna JA. 2013. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 19: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham RM, Wu DC, Qin Y, Yao J, Hunicke-Smith S, Lambowitz AM. 2016. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22: 597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RP, Mariano TM, Siekierka J, Mathews MB. 1986. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 44: 391–400. [DOI] [PubMed] [Google Scholar]
- O'Malley RP, Duncan RF, Hershey JW, Mathews MB. 1989. Modification of protein synthesis initiation factors and the shut-off of host protein synthesis in adenovirus-infected cells. Virology 168: 112–118. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 16: 948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244. [DOI] [PubMed] [Google Scholar]
- Pertea M, Daehwan K, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′ phosphates. Science 314: 997–1001. [DOI] [PubMed] [Google Scholar]
- Qin Y, Yao J, Wu DC, Nottingham RM, Mohr S, Hunicke-Smith S, Lambowitz AM. 2016. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22: 111–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. 2009. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol 5: e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Macrae IJ. 2012. The crystal structure of human Argonaute2. Science 336: 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Haley B, Zamore PD. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell 10: 537–548. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS. 2008. Evolutionarily conserved function of a viral microRNA. J Virol 82: 9823–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247. [DOI] [PubMed] [Google Scholar]
- Stevens A. 1980. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J Biol Chem 255: 3080–3085. [PubMed] [Google Scholar]
- Stevens A, Poole TL. 1995. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J Biol Chem 270: 16063–16069. [DOI] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51: 539–551. [DOI] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, et al. 2012. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M, Berkhout B. 2006. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 14: 883–892. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Chen CC, Conte D Jr, Moresco JJ, Chaves DA, Mitani S, Yates JR III, Tsai MD, Mello CC. 2015. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D Jr. 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci 107: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennström B, Pettersson U, Philipson L. 1978. Two initiation sites for adenovirus 5.5S RNA. Nucleic Acids Res 5: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Bolander ME, Sarkar G. 1998. PIG-B: a homemade monophasic cocktail for the extraction of RNA. Mol Biotechnol 9: 73–77. [DOI] [PubMed] [Google Scholar]
- Woolnough JL, Atwood BL, Giles KE. 2015. Argonaute 2 binds directly to tRNA genes and promotes gene repression in cis. Mol Cell Biol 35: 2278–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Segerman B, Zhou X, Akusjärvi G. 2007. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol 81: 10540–10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Gkountela S, Saeed K, Akusjärvi G. 2009. The 5′-end heterogeneity of adenovirus virus-associated RNAI contributes to the asymmetric guide strand incorporation into the RNA-induced silencing complex. Nucleic Acids Res 37: 6950–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Li DM, Sun H. 1998. PIR1, a novel phosphatase that exhibits high affinity to RNA. ribonucleoprotein complexes. J Biol Chem 273: 20347–20353. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


