Summary
The generation of neurons from neural stem cells requires large-scale changes in gene expression that are controlled to a large extent by proneural transcription factors, such as Ascl1. While recent studies have characterized the differentiation genes activated by proneural factors, less is known on the mechanisms that suppress progenitor cell identity. Here, we show that Ascl1 induces the transcription factor MyT1 while promoting neuronal differentiation. We combined functional studies of MyT1 during neurogenesis with the characterization of its transcriptional program. MyT1 binding is associated with repression of gene transcription in neural progenitor cells. It promotes neuronal differentiation by counteracting the inhibitory activity of Notch signaling at multiple levels, targeting the Notch1 receptor and many of its downstream targets. These include regulators of the neural progenitor program, such as Hes1, Sox2, Id3, and Olig1. Thus, Ascl1 suppresses Notch signaling cell-autonomously via MyT1, coupling neuronal differentiation with repression of the progenitor fate.
Keywords: MyT1, Ascl1, neurogenesis, Notch signaling, Hes1, transcription, Rbpj
Graphical Abstract
Highlights
-
•
MyT1 promotes neurogenesis downstream Ascl1
-
•
MyT1 represses Notch1 receptor and many of its downstream target genes
-
•
MyT1 represses Hes1 expression by direct DNA binding and competition with RBPJ
-
•
Ascl1 suppresses Notch signaling cell-autonomously while promoting differentiation
Vasconcelos et al. find that the transcription factor MyT1 promotes neuronal differentiation downstream the proneural factor Ascl1. MyT1 represses the transcription of Notch signaling components and targets genes, including important regulators of the neural progenitor program. Thus, Ascl1 activates differentiation genes while repressing the progenitor program via MyT1.
Introduction
The generation of neurons in the developing vertebrate nervous system requires the progression through a succession of distinct cellular states. These transitions have been particularly well characterized in the embryonic telencephalon, where radial glia (RG) cells located in the ventricular zone (VZ) have characteristics of neural stem cells and constitute the major progenitor type during the neurogenic period (Götz and Huttner, 2005). Upon division, an RG cell can give rise to another RG cell and either a neuronal committed intermediate progenitor (IP) or a post-mitotic neuron that migrate toward the subventricular zone (SVZ) and cortical plate (CP), respectively (Kriegstein and Alvarez-Buylla, 2009). These events are known to be coordinated, to large extent, by the opposing activities of proneural and Notch pathways.
Proneural transcription factors, such as Ascl1 (also known as Mash1), are both required and sufficient to induce a complete program of neuronal differentiation (Bertrand et al., 2002, Vasconcelos and Castro, 2014, Wilkinson et al., 2013). While activating neuronal differentiation, proneural proteins induce the transcription of Notch ligands, such as Dll1. Dll1 interacts with a transmembrane Notch receptor in neighboring cells, resulting in cleavage of the Notch intracellular domain (NICD) by gamma-secretase and its nuclear translocation, forming a complex with the DNA-binding transcription factor Rbpj and additional coactivators. Direct targets of this complex include the transcriptional repressors Hes1 and Hes5, which repress the expression of proneural genes and neuronal differentiation via a process called lateral inhibition (Kageyama et al., 2008, Louvi and Artavanis-Tsakonas, 2006). The induction of differentiation by proneural factors requires, therefore, the simultaneous repression of Notch receptor activity in differentiating progenitors by mechanisms that remain poorly understood.
Myelin transcription factor 1 (MyT1 or NZF2) is the founding member of a family of zinc-finger proteins comprising also MyT1Like (MyT1L or NZF1) and MyT3 (NZF3 or St18) (Jiang et al., 1996, Kim and Hudson, 1992, Yee and Yu, 1998). All factors are expressed with distinct patterns throughout the mouse developing nervous system. In situ hybridization studies have described Myt1 expression in differentiating progenitors and post-mitotic neuronal precursors, in both CNS and peripheral nervous system, starting at the beginning of the neurogenesis period (Matsushita et al., 2002, Matsushita et al., 2014). Evidence for a regulatory function of MyT1 in a neurogenic context was provided by functional studies in Xenopus embryos, where it counteracts lateral inhibition in synergy with the proneural factors X-Ngnr1, Xash3, or Xath5 (Bellefroid et al., 1996, Quan et al., 2004, Schneider et al., 2001). In mouse, the analysis of MyT1-null embryos has failed to provide insights into the function of MyT1 in the nervous system, presumably due to the observed ectopic upregulation of other family members in this mouse model (Hudson et al., 2011, Wang et al., 2007). More recently, the extensive use of MyT1L in neuronal reprogramming of mouse and human somatic cells (e.g., Pang et al., 2011 and Vierbuchen et al., 2010) has renewed the interest in understanding the function of MyT1 and its related factors in vertebrate neurogenesis.
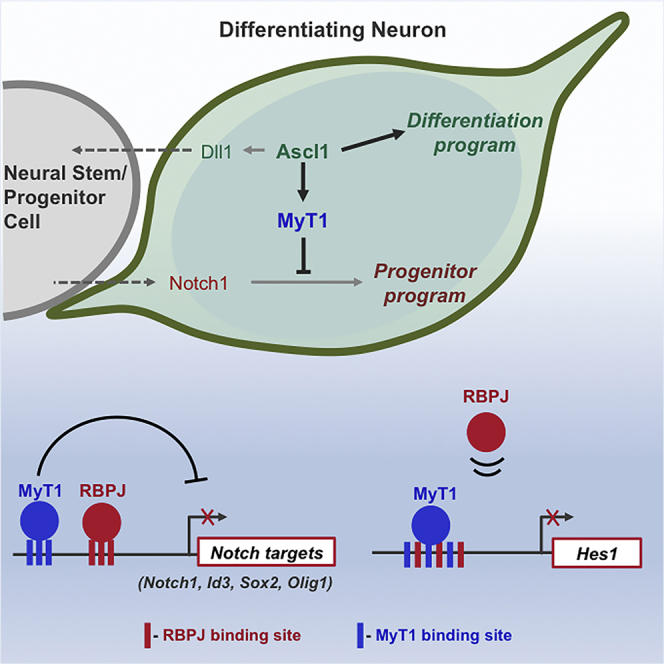
Here, we identify MyT1 as a direct target of the proneural factor Ascl1 at the onset of neuronal differentiation, and we investigate the function of MyT1 at this critical stage by combining acute functional experiments in the mouse telencephalon with the characterization of its transcriptional program. We found that MyT1 binding occurs mostly at active regulatory regions in undifferentiated neural stem/progenitor cells and is associated with transcriptional repression genome-wide. We further show that MyT1 acts at multiple levels to antagonize the inhibitory activity of Notch signaling, targeting both Notch pathway components and downstream targets. Notably, MyT1 promotes the downregulation of Hes1, a determinant step for the onset of neurogenesis, by competing with Rbpj for binding to the Hes1 promoter. Our results reveal a function of Ascl1 in inhibiting Notch signaling cell-autonomously, showing how activation of neuronal differentiation is tightly coordinated with repression of the progenitor program.
Results
Ascl1 Directly Activates the Transcription Factor MyT1
Several observations have suggested the zinc-finger transcription factor MyT1 may be under the regulation of Ascl1. Specifically, Myt1 expression is increased or decreased in expression profiling studies using DNA arrays upon Ascl1 gain and loss of function (GoF and LoF), respectively, both in mouse cultured neural stem/progenitor cells and in the embryonic telencephalon (Figure S1) (Castro et al., 2011, Gohlke et al., 2008, Raposo et al., 2015).
We started by analyzing the kinetics of MyT1 expression, using a cellular model of neurogenesis in which differentiation is triggered by the activation of an inducible version of Ascl1 protein (Ascl1-ERT2) in the neural stem cell line NS5 with 4-hydroxy-tamoxifen (Tam) (Raposo et al., 2015). Upon Ascl1 induction, MyT1 protein levels increased, as measured by immunocytochemistry and western blot (Figures 1A and 1B). Co-localization of MyT1 with the neuronal marker B-III-Tubulin (TuJ1) indicated that MyT1 expression occurred in differentiating neurons (Figure 1A). The increase in Myt1 expression occurred after the increase in Dll1 transcript, an early Ascl1 target gene, and preceded the increase in B-III-Tubulin transcript, an early neuronal marker that is also directly activated by Ascl1 (Castro et al., 2006, Castro et al., 2011) (Figure 1C). Thus, the timing of MyT1 induction is consistent with MyT1 being directly controlled by Ascl1.
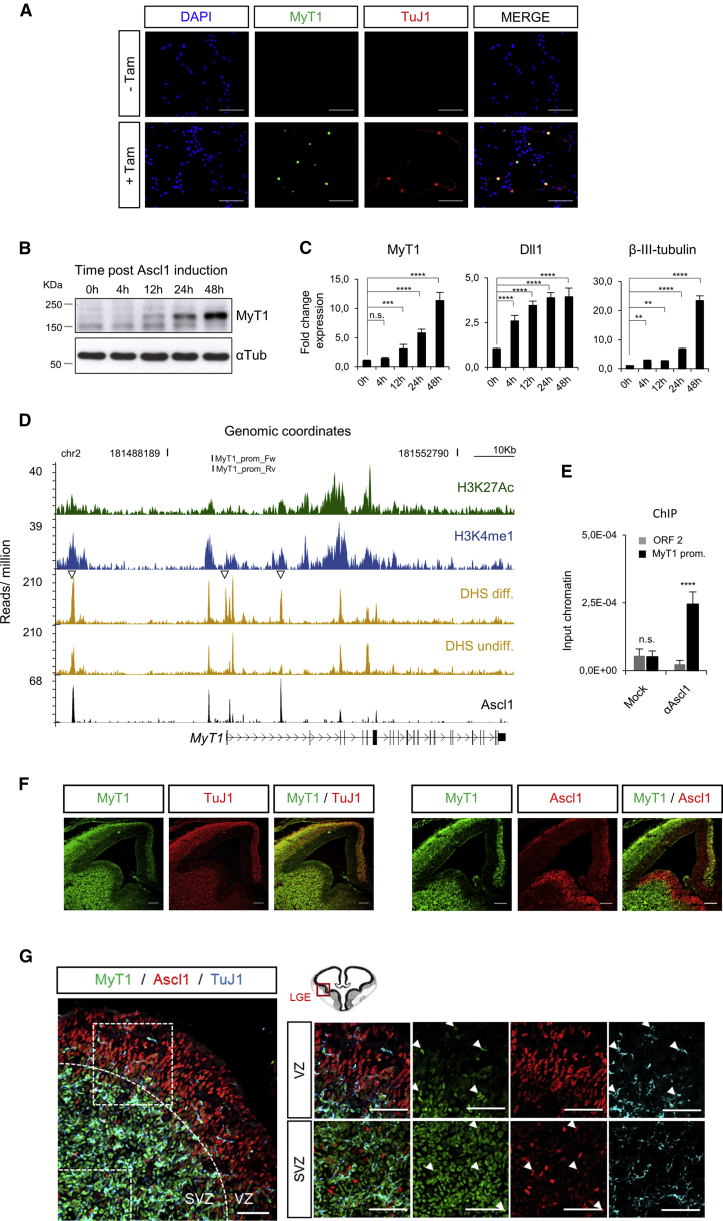
Figure 1.
MyT1 Is a Direct Target of Ascl1 during Neuronal Differentiation
(A) Immunocytochemical analysis of MyT1 (green) and TuJ1 (red) before (−Tam) and 48 hr after Tam induction (+Tam). Cell nuclei are labeled with DAPI (blue). Scale bar, 50 μm.
(B) Analysis of MyT1 protein levels by western blot post-Tam induction. α-tubulin was used as a loading control.
(C) RNA expression analysis of Myt1, Dll1, and B-III-Tubulin by qPCR post-Tam induction is shown.
(D) Ascl1 (black), H3K27ac (green), and H3K4me1 (blue) ChIP-seq and DNase-seq enrichment profiles (yellow) at Myt1 locus in undifferentiated and/or differentiating NS cells. MyT1 prom_Fw and MyT1 prom_Rv indicate genomic locations of primers used in (E).
(E) ChIP-qPCR of Ascl1 in chromatin extracted from E12.5 ventral telencephalon is shown. ORF1, negative control region; MyT1 prom., Myt1 promoter region amplified using the primers highlighted in (D).
(F) Immunohistochemical analysis for MyT1 (green) and neuronal marker B-III-Tubulin (TuJ1, red) (left panel) or MyT1 (green) and Ascl1 (red) (right panel) in frontal sections of E14.5 mouse telencephalon is shown. Scale bar, 100 μm.
(G) Immunohistochemical analysis for MyT1 (green), Ascl1 (red), and TuJ1 (cyan) in frontal sections of E14.5 mouse ventral telencephalon. Dashed squares indicate magnified regions of VZ and SVZ. White arrowheads indicate co-localization of MyT1 and TuJ1 in the VZ (upper panel) or MyT1 and Ascl1 in the SVZ (lower panel). VZ, ventricular zone; SVZ, subventricular zone. Scale bar, 100 μm.
Data are shown as mean ± SD; n.s., p > 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to one-way ANOVA with Bonferroni correction for multiple testing (C) and Student’s t test (E). See also Figure S1.
Effectively, visual inspection of the chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) enrichment profile of Ascl1-ERT2 in differentiating cells identified several peaks, corresponding to Ascl1 binding to active enhancer regions enriched for H3K4me1 and H3K27ac in the vicinity of the MyT1 gene (Figure 1D). Some Ascl1-binding events (BEs) occurred in closed chromatin regions in proliferating progenitors, which became opened during neuronal differentiation, as assessed by DNase sequencing (DNase-seq) (Figure 1D). This feature is associated with Ascl1 targets that are expressed de novo during differentiation (Raposo et al., 2015), and it may account for the late timing of MyT1 induction after Ascl1 expression. Ascl1 binding to Myt1 promoter was further validated by ChIP-qPCR in chromatin extracted from the developing ventral telencephalon (Figure 1E).
MyT1 Promotes Neurogenesis in the Mouse Telencephalon
Myt1 transcript is expressed in the germinal layers during the neurogenic period in scattered cells in the VZ, with highest expression in the SVZ (Matsushita et al., 2002, Matsushita et al., 2014). In the ventral telencephalon, MyT1 protein expression domain largely overlapped with that of the neuronal marker TuJ1 (Figure 1F). At the cellular level, many, but not all, MyT1-expressing cells in the SVZ co-expressed TuJ1 (Figure 1G), suggesting MyT1 was expressed both in IPs and differentiating neurons. In addition, few MyT1-expressing cells were interspersed in the VZ co-expressing TuJ1 (Figure 1G, arrowheads in upper panels). Although MyT1 and Ascl1 expression domains were mostly non-overlapping (Figure 1F), some cells co-expressing both factors could be found in the SVZ (Figure 1G, arrowheads in bottom panels), as expected from Ascl1-expressing IPs. Overall, the expression pattern was consistent with MyT1 starting to be expressed at the transition from RG cells to differentiating neurons, either directly (VZ) or indirectly through the generation of IPs (SVZ). The persistent expression of MyT1 in post-mitotic neuronal precursors suggests a function also at later stages of neurogenesis.
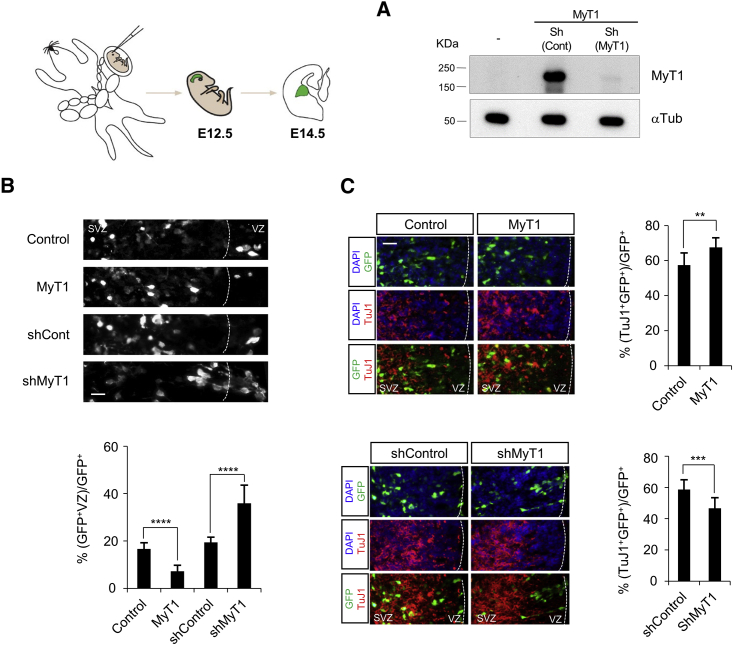
We next investigated MyT1 function in the telencephalon by performing acute GoF and LoF experiments by in utero electroporation. We targeted the lateral ganglionic eminence (LGE) at embryonic day (E)12.5, a stage at which MyT1 is the only member of its family to be significantly expressed in germinal layers (Matsushita et al., 2002, Matsushita et al., 2014). MyT1 overexpression resulted in more cells (∼56% increase) leaving the VZ, and this was associated with an increase in TuJ1-expressing cells (∼20% more) when compared to control conditions (Figures 2B and 2C). By contrast, knockdown of MyT1 with a small hairpin RNA (shRNA) expression vector (ShMyT1) (Figure 2A) resulted in more cells remaining in the VZ (∼85% increase) and fewer TuJ1-positive cells (∼17.4% decrease) (Figures 2B and 2C). We analyzed if the concomitant induction of other family members could be compensating the extent of the phenotype observed, by knocking down MyT1 together with MyT1L or MyT3. Indeed, a significant enhancement of the phenotype was observed when combining shMyT1 with shMyT1L, both in number of cells retained in the VZ and in the number of TuJ1-positive cells (Figure S2); the combination with ShMyT3 did not increase significantly the phenotype triggered by shMyT1 alone. Altogether, these experiments demonstrate that MyT1 promotes neurogenesis in the developing mouse embryo.
Figure 2.
MyT1 Promotes Neurogenesis in the Developing Telencephalon
(A) Analysis by western blot of MyT1 in P19 cells co-transfected with MyT1 expression vector, scramble shRNA (shControl), and/or MyT1 shRNA (shMyT1). α-tubulin was used as a loading control.
(B and C) In utero electroporation of control or MyT1-, control shRNA- (shControl), or MyT1 shRNA- (shMyT1) expressing vectors in E12.5 mouse ventral telencephalon. Immunofluorescence analysis on a coronal section of the telencephalon for GFP (green or gray) and TuJ1 (red) 2 days post-electroporation (E14.5) is shown. Cell nuclei are labeled with DAPI (blue). Histograms represent the quantification of VZ to SVZ transition based on the fraction of GFP+ cells that are retained in the VZ (GFP+VZ/GFP+) (B) and of neuronal differentiation based on the fraction of GFP+ cells that express TuJ1 ((TuJ1+GFP+)/GFP+) (C). VZ, ventricular zone; SVZ, subventricular zone. Scale bars, 20 and 50 μm (B and C).
Data are shown as mean ± SD; ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to Student’s t test (B and C). See also Figure S2.
MyT1 Functions as a Transcriptional Repressor during Neurogenesis
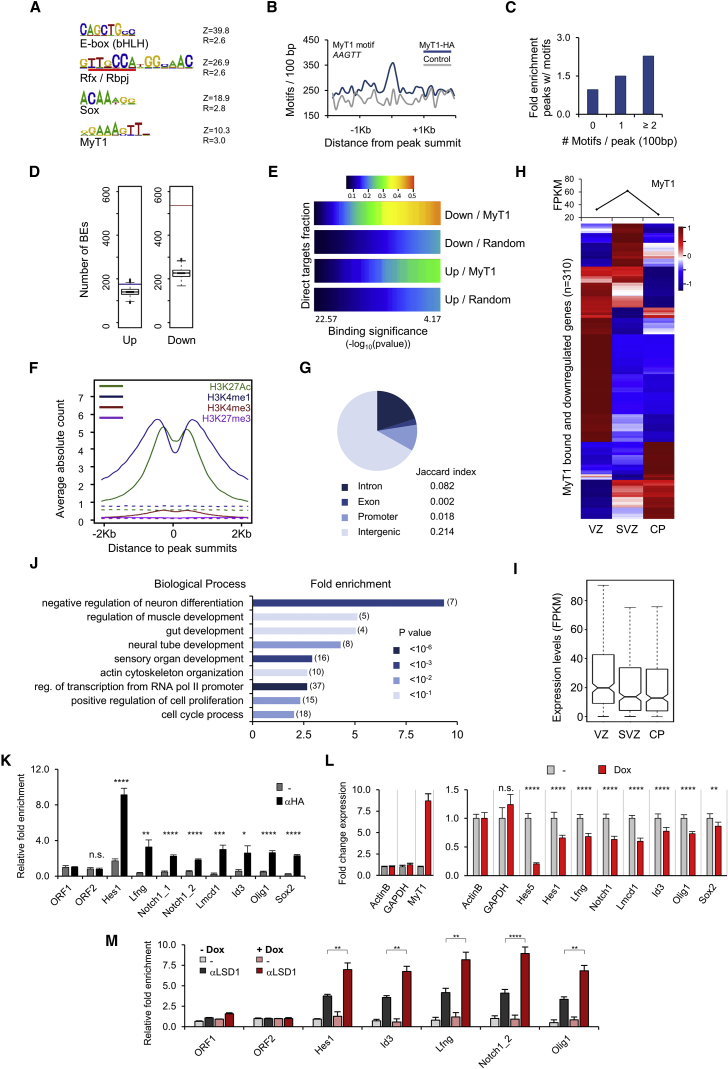
To understand the molecular basis of MyT1 function at the onset of differentiation, we characterized its target genes upon acute GoF in neural stem/progenitor cells. For this, we infected NS5 cells with a lentivirus expressing a doxycycline (Dox)-inducible HA-tagged version of MyT1, and we characterized the gene expression changes 4 hr post MyT1 induction using DNA arrays (Figure S3). This identified 1,764 deregulated genes (p < 0.05), of which 57% were downregulated and 43% were upregulated (Table S1). We performed genome-wide mapping of MyT1 binding by ChIP-seq using an antibody against the HA-tag of MyT1, and we identified 7,615 BEs (q < 10−2) associated with 4,448 unique genes following a nearest gene annotation (Table S2). A de novo search found the MyT1-binding motif (Gamsjaeger et al., 2008, Kim and Hudson, 1992) to be enriched at peak summits, with peaks with one or multiple motifs significantly enriched when compared to a control dataset (Figures 3A–3C). In addition, a compound Rfx/Rbpj motif, E box (bHLH) and Sox DNA-binding motifs were also overrepresented. Hierarchical clustering of MyT1 peaks according to the presence of these motifs revealed their occurrence was largely mutually exclusive (Figure S4A). Although the largest group bore the MyT1 motif, the data suggest that many MyT1 peaks result from indirect recruitment of MyT1 by other TFs.
Figure 3.
MyT1 Binding Represses Neural Progenitor Genes
(A) Top overrepresented motifs in 50-bp regions centered at MyT1 peak summits are shown. Z, Z score; R, enrichment ratio.
(B) Frequency distribution of MyT1 motif centered at MyT1 peak summits (blue) or 2 kb upstream (control, gray). y axis represents the number of motifs in bins of 50 bp.
(C) Fold enrichment of MyT1 peaks with distinct number of MyT1 motifs as compared to control regions is shown.
(D) Number of MyT1 BEs associated with up- (blue bar) or downregulated (red bar) genes following a nearest gene annotation in MyT1 GoF DNA arrays. Statistical significance was assessed by comparing the association with 1,000 randomized datasets of equal size and assuming a normal distribution. Test data are represented as box with median of test and first and third quartiles. Whiskers, ±1.5× interquartile range (IQR).
(E) Heatmap displaying the cumulative fraction of deregulated genes in MyT1 GoF that are directly regulated by MyT1. Number of transcripts with expression FC > 1.2 are plotted against MyT1 BEs with increasing p value (bin = 94 BEs). Statistical significance was assessed by comparing the association with 100 randomly generated ChIP-seq datasets of equal size.
(F) ChIP-seq enrichment profiles of histone marks in undifferentiated NS5 cells, in 4-kb genomic regions centered at MyT1 peak summits. Dashed lines show the enrichment profile of histone marks at random genomic regions.
(G) Pie chart represents the fraction of MyT1 BE overlapping gene features.
(H) Heatmap representing the expression of MyT1 direct targets (bound and downregulated) in distinct layers of the E14.5 mouse embryonic telencephalon. Color code refers to relative gene expression levels. Hierarchical clustering was done using Pearson correlation. Absolute levels of expression of MyT1 are represented in the graph above. VZ, ventricular zone; SVZ, subventricular zone; CP, cortical plate.
(I) Absolute expression levels (fragments per kilobase per million mapped reads [FPKM]) of MyT1 direct targets in distinct layers of the E14.5 mouse embryonic telencephalon. Data distribution is represented as box with median and first and third quartiles (whiskers, ±1.5× IQR; notches, ±1.5× IQR/n1/2).
(J) Enrichment of representative GO biological process terms associated with MyT1 targets (genes bound and downregulated by MyT1) is shown.
(K) Validation of MyT1 binding to selected genes by ChIP-qPCR with anti-HA antibody in NS5 cells upon MyT1 GoF is shown. ORF1 and ORF2, negative control regions. Mean ± SD of triplicate assays.
(L) Validation of gene expression changes of selected genes in NS5 cells 4 hr upon MyT1 GoF by expression qPCR is shown. Mean ± SD of triplicate assays.
(M) ChIP-qPCR using anti-LSD1 antibody in NS5 cells before (−Dox) and 4 hr after (+Dox) MyT1 GoF is shown. ORF1 and ORF2, negative control regions.
Data are shown as mean ± SD; n.s., p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to Student’s t test (K–M). See also Figures S3 and S4.
Previous reports have implicated MyT1 both in transcriptional activation and repression (Bellefroid et al., 1996, Hu et al., 2013, Romm et al., 2005, Wang et al., 2008, Yokoyama et al., 2014). To gain an insight into the global transcriptional response mediated by MyT1, we integrated the location analysis of MyT1 with the expression profiling to find that 14% of sites are associated with a deregulated gene. Strikingly, MyT1 BEs were statistically significantly associated with downregulated genes (p < 3.45 × 10−51), whereas such association with upregulated genes was not found (Figure 3D). Notably, this differential association with downregulated genes was maintained when considering only peaks containing the MyT1 motif, or those without it (Figure S4B), suggesting that all types of MyT1 BEs are associated with repression of gene expression.
We next determined the fractions of up- and downregulated genes associated with at least one MyT1 BE (direct targets), considering increasing p value cutoffs for MyT1 binding (Figure 3E). We found that downregulated genes were strongly enriched for MyT1 direct targets, in contrast to upregulated genes (Figure 3E, compare Down/MyT1 with Up/MyT1), and that such enrichment was statistically significant (Figure 3E, compare Down/MyT1 with Down/Random), confirming that MyT1 binding is associated with gene repression genome-wide.
Genome-wide maps of histone modifications in proliferating NS5 cells (Mikkelsen et al., 2007, Raposo et al., 2015) showed significant enrichment of H3K27ac, H3K4me1, and H3K4me3 at MyT1 BEs, as compared to random genomic regions (while no enrichment of the repressive mark H3K27me3 was found) (Figure 3F). In addition, most MyT1 BEs were found within intergenic or intronic regions, with a smaller but significant fraction occurring at gene promoters (Figure 3G). These results suggest that MyT1 binds to distal enhancer and promoter regions bearing active marks in neural stem/progenitor cells, and they are in line with MyT1’s role as a transcriptional repressor at onset of differentiation.
MyT1 Represses Notch Pathway Components and Regulators of the Neural Progenitor Program
We next assessed the expression of the identified MyT1 targets (bound and downregulated) at distinct stages of the neuronal lineage, using expression profiling datasets representative of various layers of the E14.5 mouse embryonic cortex (Fietz et al., 2012). Notably, most MyT1 direct targets (69.0%) were downregulated during neuronal differentiation in vivo, as indicated by a decrease in expression from VZ to SVZ or CP (Figure 3H). In addition, their absolute levels of expression were highest at VZ, when compared to SVZ and CP (Figure 3I).
To investigate the function of MyT1 direct targets, we performed gene ontology (GO) analysis on a high-confidence list of 402 MyT1 targets (bound and repressed by MyT1) (Table S3). Enriched terms (GO biological processes) suggested a role in maintenance of a progenitor state (negative regulation of neuron differentiation, positive regulation of cell proliferation, and cell-cycle process), and they were associated with Notch pathway genes (e.g., Notch1, Lfng, Dtx4, and Hes1) (Figure 3J; Table S4). In addition, MyT1 targets encoded important transcriptional regulators (regulation of transcription from RNA polymerase II [Pol II] promoter) (Figure 3J) of the neural progenitor program, such as Sox2, Id3, and Olig1 (Bai et al., 2007, Englund et al., 2005, Gómez-López et al., 2011) (Figures 3K and 3L; Table S4).
MyT1 has been shown to physically interact with the histone demethylase LSD1 in a complex that contains CoREST and promotes transcriptional repression (Yokoyama et al., 2014). To address if LSD1 is recruited by MyT1 to the abovementioned targets associated with a MyT1 motif, we performed anti-LSD1 ChIP-qPCR. We observed a substantial increase of LSD1 recruitment to MyT1 target sites upon MyT1 GoF, supporting a role for this histone demethylase in MyT1-mediated transcriptional repression in neural progenitor cells (Figure 3M).
MyT1 Promotes Neurogenesis by Counteracting Notch-Signaling Activity
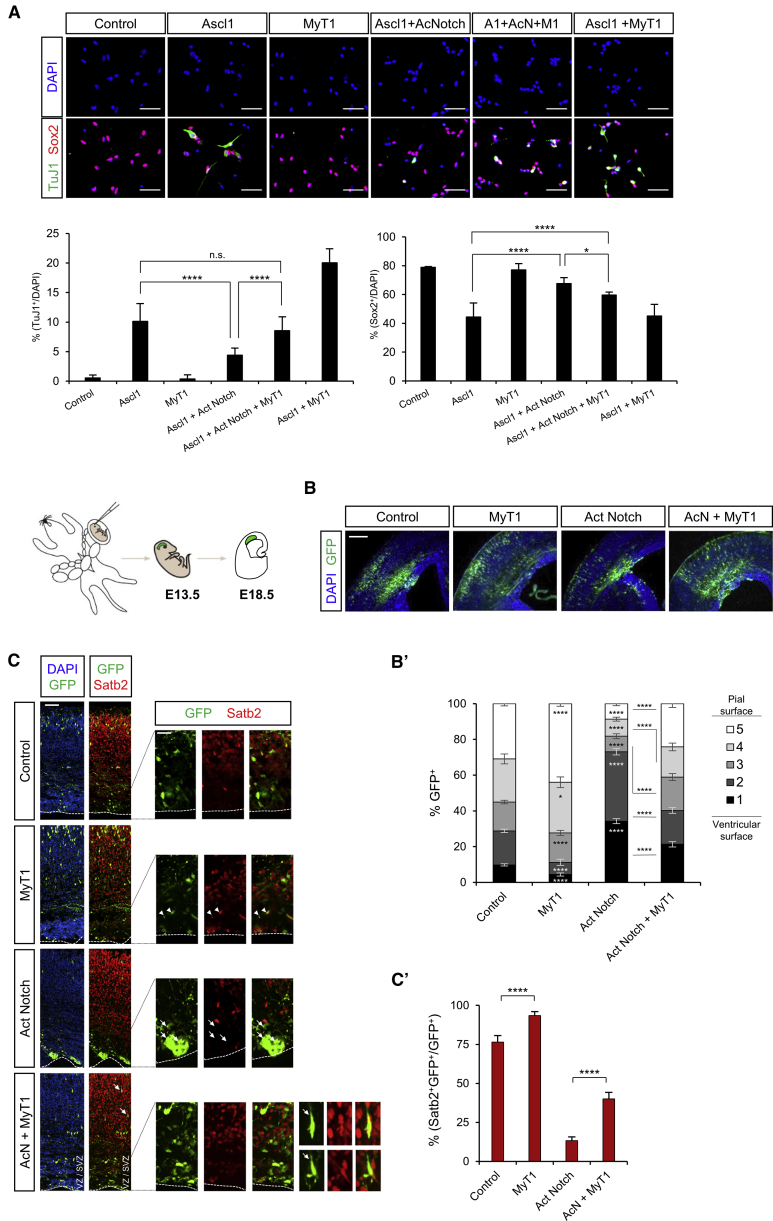
Given the prominence of Notch-related genes among MyT1 targets and in line with previous findings (Bellefroid et al., 1996, Hu et al., 2013, Romm et al., 2005, Wang et al., 2008, Yokoyama et al., 2014), we tested if MyT1 can counteract the inhibitory effect of Notch signaling in neurogenesis. We infected NS5 cells with lentiviruses expressing Ascl1, MyT1, and a membrane-bound dominant active version of the Notch1 receptor, whose cleavage is independent of a ligand/receptor interaction (Act Notch). As expected, Ascl1 expression induced the generation of TuJ1-positive neurons with a concomitant decrease of Sox2 expression (Berninger et al., 2007, Farah et al., 2000, Nakada et al., 2004), and this effect was inhibited by the co-expression of Act Notch (Gaiano et al., 2000) (Figure 4A). Importantly, MyT1 alone did not trigger neuronal differentiation, but its co-expression with Ascl1 overcame the inhibition by Act Notch, as observed by a rescue of the number of TuJ1-positive cells to levels similar to those obtained with Ascl1 alone (with Sox2 expression following a similar trend). In addition, a strong increase in the differentiation activity of Ascl1 alone was observed when both factors (Ascl1 + MyT1) were co-expressed (Figures 4A and 4B, respectively). Altogether, our results suggest that MyT1 alone is not able to promote neuronal differentiation but that it enhances the activity of the proneural factor by counteracting the inhibitory activity of the Notch pathway.
Figure 4.
MyT1 Counteracts Inhibition of Neuronal Differentiation by Notch Signaling
(A) Immunocytochemical analysis of Sox2 (red) and TuJ1 (green) upon infection with control or Ascl1-, Act Notch-, and/or MyT1-inducible lentiviruses 48 hr post-Dox. Nuclei were labeled with DAPI (blue). Histograms represent the percentage of Sox2+/DAPI or TuJ1+/DAPI cells in each condition. Scale bar, 40 μm.
(B and C) In utero electroporation of control or MyT1, Act Notch, or Act Notch + MyT1 expression vectors in E13.5 mouse dorsal telencephalon. Immunofluorescence analysis of coronal sections of GFP (green) and the neuronal marker Satb2 (red) 5 days after electroporation (E18.5) is shown. Cell nuclei are labeled with DAPI (blue). Scale bars, 500 μm (B) and 50 and 20 μm (C, left and right). (B’) Histogram represents the percentage of electroporated cells present in five bins (1 to 5 from ventricular to pial surface, respectively) of equal length spanning the cortical thickness. (C’) Histogram represents the percentage of (Satb2+GFP+)/GFP+ cells in each condition. Arrowheads indicate examples of Satb2+GFP+ cells. Arrows indicate examples of Satb2−GFP+ cells.
Data are shown as mean ± SD; n.s., p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to one-way ANOVA with Bonferroni correction for multiple testing (A) and Student’s t test (B’–C’).
We next tested whether MyT1 could similarly counteract Notch signaling in vivo by co-electroporating MyT1 and Act Notch expression vectors in the dorsal telencephalon. As expected, expression of Act Notch inhibited endogenous neurogenesis, resulting in the retention of electroporated cells closer to the ventricular surface and in a decreased number of cells expressing the neuronal marker Satb2 (Figures 4B and 4C), whereas MyT1 overexpression had the opposite effect (Figures 4B and 4C). Notably, co-expression of Act Notch and MyT1 rescued significantly the Act Notch phenotype (Figures 4B and 4C). Overall, these experiments support the conclusion that MyT1 counteracts Notch signaling during neurogenesis.
MyT1 Counteracts Notch Activation of Hes1 Expression
Considering the well-established role of Hes1 as a downstream effector of Notch signaling in neurogenesis (Artavanis-Tsakonas et al., 1999, Shimojo et al., 2008), we next focused on the regulation of this MyT1 target gene. Analysis of the MyT1 ChIP-seq profile showed strong enrichment at the Hes1 proximal promoter region, centered on three evolutionarily conserved MyT1 motifs (Figure 5A). Strikingly, these motifs were found in tandem, and partially overlapping, with three Rbpj-binding motifs (TC box) previously shown to mediate the regulation of Hes1 by Notch/Rbpj (Arnett et al., 2010, Jarriault et al., 1995, Nam et al., 2007) (Figure 5B). To test the ability of MyT1 to bind to its cognate motifs, we performed an electrophoretic mobility shift assay (EMSA) using a Hes1 oligonucleotide probe spanning the MyT1 motifs or mutated versions. MyT1 was able to directly bind to this Hes1 promoter probe, as shown by the formation of a large DNA/protein complex (Figure 5C). The formation of this complex was strongly reduced or abolished when each of the MyT1 motifs was disrupted separately or simultaneously, respectively, suggesting cooperative binding of MyT1 to the three sites.
Figure 5.
MyT1 Counteracts the Activity of Rbpj/Notch at the Hes1 Promoter
(A) MyT1 (blue) and RBPJ (red) ChIP-seq enrichment profiles at the vicinity of Hes1 gene. Regions contained in Hes1::Luc and EMSA probe are indicated below figure.
(B) Species alignment of Hes1 proximal promoter region spanning MyT1 (blue) and Rbpj (red) binding motifs is shown.
(C) EMSA shows MyT1 binding to a Hes1 promoter probe with wild-type (WT) sequence or mutations in MyT1 binding sites. n.s., non-specific band.
(D–G) Transcriptional assay in P19 cells co-transfected with control or MyT1 and/or Act Notch expression vectors and a reporter construct expressing luciferase under the control of Hes1 proximal promoter region (pHes1::Luc) or mutated versions in which MyT1-binding sites were individually or simultaneously disrupted. Equal amounts of expression vectors were used, except in (G) in which the various MyT1:ActNotch ratios used are indicated. Mean ± SD of quadruplicate assays.
(H) Transcriptional assay in P19 cells co-transfected with a control or MyT1 and/or RBPJ-VP16 (Blanpain et al., 2006) expression vectors together with pHes1::Luc is shown.
Data are shown as mean ± SD; n.s., p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to Student’s t test (F and G) or one-way ANOVA test with Bonferroni correction for multiple testing (D, E, and H). See also Figure S5.
We next investigated whether MyT1 could counteract the activity of Notch signaling on a luciferase reporter construct containing the Hes1 proximal promoter region spanning all MyT1 and Rbpj sites (pHes1::Luc) (Nishimura et al., 1998). A transcriptional assay in transfected P19 cells showed Act Notch strongly activates the Hes1 promoter, while co-expression of MyT1 completely abrogated this activity (Figure 5D). To test if the observed effect of MyT1 is dependent on DNA binding, we performed mutagenesis on the MyT1-binding sites. Mutations were designed so as to minimize any effect on Notch activation, resulting in promoter constructs that still responded, albeit less strongly, to Act Notch (Figure 5E). Disruption of individual binding sites partially impaired the activity of MyT1 (Figure 5F), while simultaneous disruption of the three sites (BS1 + 2 + 3) had a stronger effect readily observed when lower ratios of MyT1/Act Notch were used (Figure 5G). Similar results were obtained using a dominant active version of Rbpj fused to the VP16 activation domain (RBPJ-VP16) (Figure 5H), further supporting that MyT1 regulates Hes1 at the promoter level. Importantly, no other transcription factor tested was able to counteract Act Notch activity on Hes1 promoter (Figure S5). Altogether, we concluded that MyT1 counteracts the activity of the Notch/Rbpj complex on the Hes1 promoter via a mechanism that depends on binding to the three MyT1-binding sites.
Next, we analyzed MyT1 protein and Hes1 transcript expression patterns in the embryonic mouse telencephalon by double immuno/in situ hybridization, and we concluded these are largely non-overlapping (Figure 6A). To further investigate this with single-cell resolution, we re-examined the molecular signatures of 96 E11.5 dorsal telencephalic neural stem/progenitor cells obtained by single-cell RNA sequencing (RNA-seq) (Hagey and Muhr, 2014). Profiling using principal component 2 (PC2), which orders cells through differentiation according to markers that reflect the progression of cortical neurogenesis (Hagey and Muhr, 2014), showed an inverse correlation between Myt1 and Hes1 expression (Figure 6B, upper graph), with a Spearman correlation coefficient of reads per kilobase per million mapped reads (RPKM) levels of −0.407. In spite of this, examination of absolute RPKM levels showed that co-expression of both genes was detected in a small but significant fraction of cells, suggesting the onset of Myt1 expression occurs in cells that still express Hes1 (Figure 6B, bottom panel). Thus, MyT1 expression is inversely correlated, but partially overlapping, with that of Hes1, which is consistent with MyT1 playing a determinant role in switching off Hes1 transcription at the onset of neurogenesis.
Figure 6.
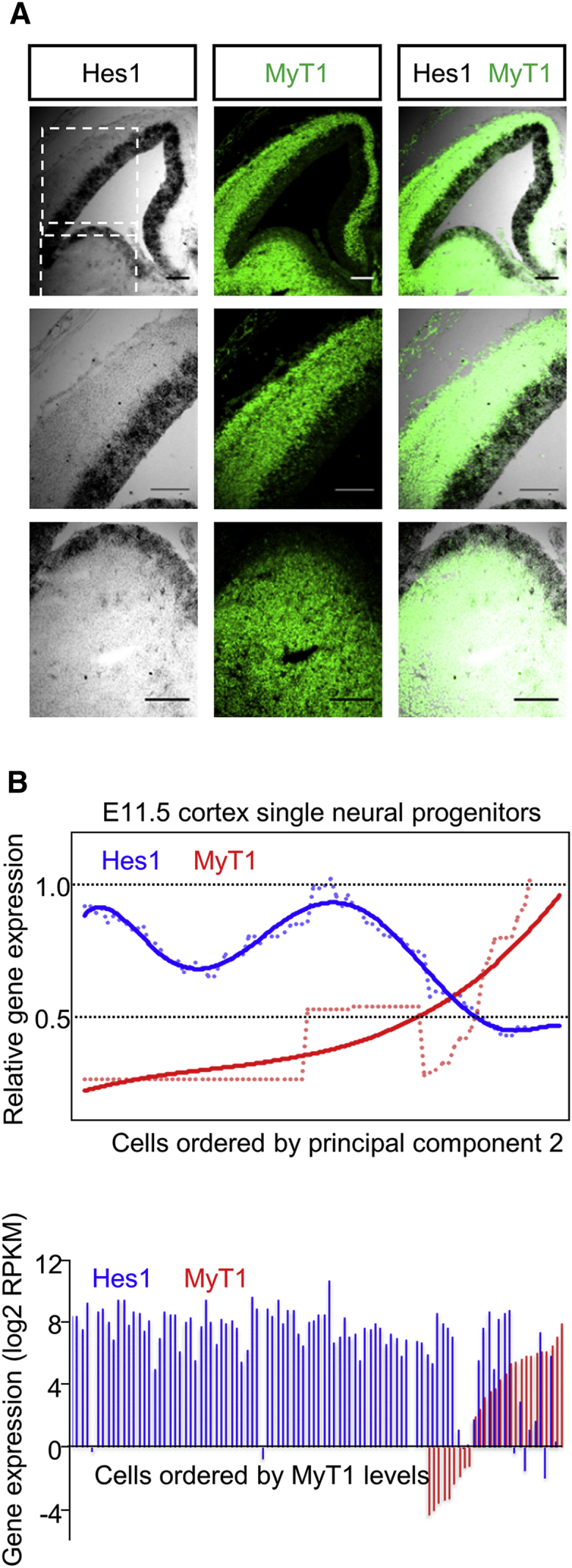
Comparative Analysis of MyT1 and Hes1 Expression in Neural Progenitor Cells
(A) Immunofluorescence of MyT1 (green) and in situ hybridization for Hes1 (gray) in frontal sections of E14.5 mouse embryonic telencephalon. Dashed squares indicate magnified regions of dorsal (middle) or ventral (bottom panels) domains. Scale bar, 140 μm.
(B) Top panel: relative expression of Myt1 and Hes1 in 96 E11.5 dorsal telencephalic neural progenitor cells, determined by single-cell RNA-seq. Dots represent rolling averages, ordered by principal component 2 (PC2), with polynomial trend lines overlapping. Bottom panel: absolute expression levels of Myt1 and Hes1 in neural progenitor cells are shown as above, ordered by MyT1 expression levels.
MyT1 Represses Many Notch/Rbpj Transcriptional Targets
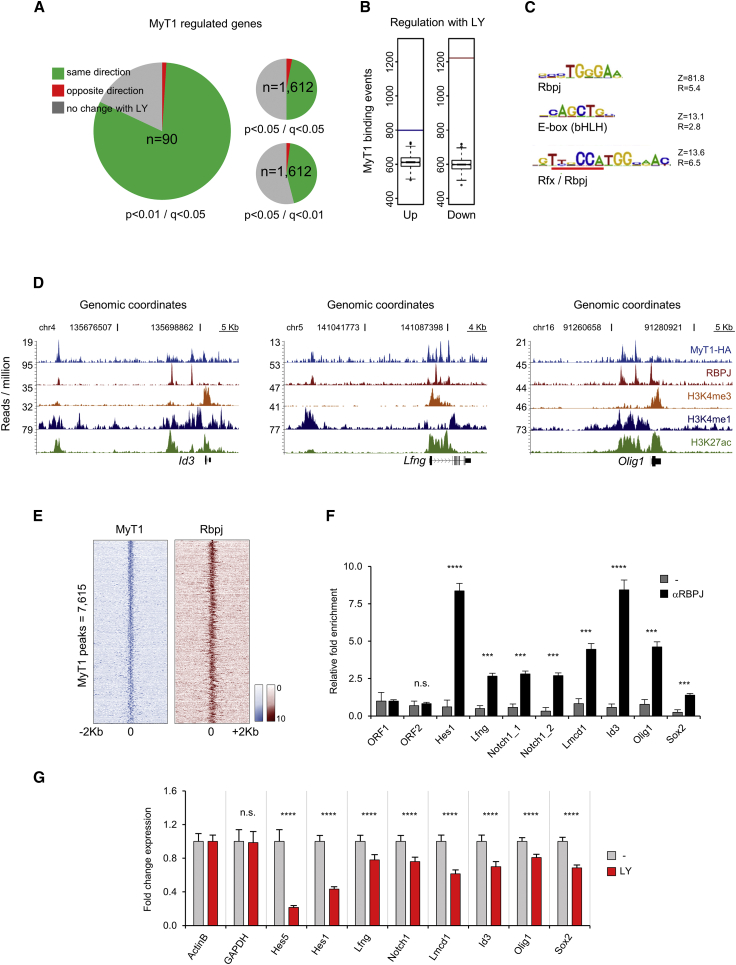
The overrepresentation of a putative Rbpj-binding motif at MyT1 target sites (Figure 3A) suggests the direct control of Notch target genes by MyT1 may extend beyond the regulation of Hes1. To investigate this, we compared the transcriptional changes observed in our MyT1 GoF model with those that take place upon exposure of NS5 cells to the pharmacological inhibitor of gamma-secretase LY411575 (LY), (Lanz et al., 2004), as assessed by RNA sequencing (RNA-seq) (Table S5). When considering a high-confidence list of genes deregulated upon MyT1 induction (p < 0.01, n = 90), the vast majority (81%) was also deregulated upon LY treatment, with all these but two genes changing in the same direction (Figure 7A). Notably, this same trend (expression of common deregulated genes changes in same direction) was maintained when decreasing the stringency cutoff for deregulation by MyT1 (p < 0.05, n = 1,612), or simultaneously increasing the stringency cutoff for deregulation upon LY treatment (q < 0.01, n = 1,612) (Figure 7A). Thus, MyT1 expression induces a global change in gene expression that resembles that triggered by the inhibition of Notch signaling.
Figure 7.
MyT1 and Notch Pathways Oppositely Regulate Many Transcriptional Targets
(A) Pie charts representing the percentage of genes deregulated in NS5 cells 4 hr upon MyT1 GoF and deregulated in the same direction (green), opposite direction (red), or unchanged/data unavailable (gray) 4 hr upon LY treatment. Cutoff values for analysis upon MyT1 GoF (p value) and exposure to LY (q value) are indicated in figure next to each pie chart. n, total number of genes.
(B) Number of MyT1 BEs associated with genes that are up- (blue bar) or downregulated (red bar) upon LY treatment. Boxplot distributions of MyT1-binding associations, which can be found testing against 1,000 different random sets of genes, are shown. Test data are represented as box with median of test and first and third quartiles (whiskers, ±1.5× IQR).
(C) Top overrepresented motifs in 50-bp regions centered at MyT1 peak summits are shown. Z, Z score; R, enrichment ratio.
(D) Examples of MyT1 (blue) and RBPJ (red) ChIP-seq enrichment profiles at vicinity of common bound genes and associated H3K4me3 (orange), H3K4me1 (dark blue), and H3K27ac (green) ChIP-seq enrichment profiles are shown.
(E) Density plot of MyT1 (blue) and RBPJ (red) ChIP-seq reads at 4-kb genomic regions centered at MyT1 peak summits (signal intensity represents normalized tag count) is shown.
(F) Validation of RBPJ binding to selected genes by ChIP-qPCR in NS5 cells is shown. ORF1 and ORF2, negative control regions. Mean ± SD of triplicate assays.
(G) Validation of gene expression changes of selected genes in NS5 cells 4 hr after LY treatment by expression qPCR is shown.
Data are shown as mean ± SD; n.s., p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to Student’s t test (F and G). See also Figure S6.
Considering the short time points (4 hr) at which both expression profiling experiments were performed, it is likely that many genes downstream Notch are direct MyT1 targets. In agreement with this, we found a much stronger statistical association of MyT1 BEs with genes downregulated by LY than with upregulated ones (p < 1.6 × 10−70 versus p < 1.1 × 10−8) (Figure 7B). To evaluate the extent to which MyT1 represses genes directly activated by the Notch/Rbpj complex, we performed genome-wide mapping of Rbpj in NS5 cells by ChIP-seq, resulting in the identification of 15,281 BEs (q < 10−2) (Table S6; Figure 7D) associated with 9,215 unique genes. As expected, a de novo search for DNA motifs enriched at Rbpj BEs found the TC box to be the most enriched motif, in addition to an E box (bHLH) and the compound Rfx/Rbpj motif (Figure 7C). A strong Rbpj-sequencing signal was found at genomic regions centered at MyT1 peak summits, confirming that MyT1 often binds in close vicinity to Rbpj (Figure 7E), as illustrated for several targets (Figures 5A and 7D). Notably, most (80.4%) of MyT1 direct targets identified were also bound by Rbpj. MyT1 and RBPJ binding and their opposite effects in target gene expression were validated for several genes (Figures 3K, 3L, 7F, and 7G). Most significantly was the ability of both pathways to oppositely regulate the expression of Notch1, further validated by the ability of MyT1 to counteract the activation of Notch1 gene promoter by Act Notch in a transcriptional assay (Figures S6A–S6C). In conclusion, MyT1 can counteract the Notch pathway at distinct levels, repressing Notch-signaling components and many of its downstream targets.
Discussion
In spite of its identification two decades ago, little progress has been made toward understanding the function of the zinc-finger factor MyT1 in vertebrate neurogenesis. After initial studies in Xenopus embryos suggested X-MyT1 promotes neurogenesis by counteracting lateral inhibition (Bellefroid et al., 1996), a confirmation of such model using mouse genetics has been hampered by functional redundancy with other family members (Wang et al., 2007). In addition, no molecular basis has been ever established for MyT1 function, to large extent due to the lack of identified target genes. Here we provide evidence of a neurogenic role for MyT1 in the developing mouse embryo, and we characterize its transcriptional program.
Activation of neuronal differentiation requires the concomitant repression of the gene expression program that operates in neural stem/progenitor cells, including the downregulation of the Notch transcriptional program (Imayoshi and Kageyama, 2014, Kageyama et al., 2008). This is often attributed to decreased Notch input signaling from neighboring cells (Kawaguchi et al., 2008, Nelson et al., 2013), which results in default repression of Notch target genes via the recruitment of corepressors by Rbpj. However, recent studies have reported that IPs exhibit attenuated Rbpj signaling as compared to RG cells, and they are unable to induce the expression of Hes genes upon activation of the Notch receptor (Kawaguchi et al., 2008, Mizutani et al., 2007, Nelson et al., 2013). This suggests the existence of cell-autonomous mechanisms to inhibit the Notch program at the onset of differentiation, which could be explained by the regulation of Notch target genes by transcription factors such as MyT1. In addition, MyT1 interacts with the Notch pathway also by regulating various pathway components. Most notably, the repression of Notch1 receptor gene may lead to an overall decrease of Notch-signaling levels by MyT1. In agreement with this, the expression of Hes5, a well-established readout of Notch signaling, was strongly downregulated upon MyT1 expression with no apparent binding of MyT1 to its promoter region (Figures 3L, S6D, and S6E).
Among the targets identified, Hes1 is likely to be a major effector of MyT1 in neurogenesis. Hes1 expression intrinsically oscillates in proliferating neural stem/progenitor cells, by a mechanism that relies on its activation by Notch signaling. It is believed that the onset of differentiation depends critically on the sustained downregulation of Hes1 expression; however, the mechanisms that repress Hes1 in this context are poorly understood (Imayoshi and Kageyama, 2014, Imayoshi et al., 2013). The cis architecture of the Hes1 promoter, where MyT1 and Rbpj sites are found interspersed and partially overlapping, was revealed to be a very efficient mechanism to counteract Notch activity. This feature may be unique to Hes1, as we did not find a similar cis architecture at other regulatory regions targeted by both pathways.
It has been shown that MyT1L cooperates with Ascl1 in reprogramming mouse embryonic fibroblasts (MEFs) into induced neurons (Vierbuchen et al., 2010). Although it is not clear what the levels of canonical Notch signaling are in MEFs, Hes1 expression is well documented in this cellular context. MyT1 family members have high sequence homology within the zinc-finger domains, and all have been shown to recognize the MyT1 DNA-binding motif (Bellefroid et al., 1996, Jiang et al., 1996, Yee and Yu, 1998). Therefore, it is possible that MyT1L functions, at least in part, by repressing Hes1 during reprogramming. In support of this, we found that MyT1L counteracts the Notch activation of Hes1 promoter with the same efficiency as MyT1 (Figure S7A). In addition, ChIP-qPCR analysis of an HA-tagged version of MyT1L revealed a strong enrichment at Hes1 proximal promoter soon after ectopic expression in MEFs (Figure S7B).
The broad expression of MyT1 in the embryonic nervous system (Matsushita et al., 2014), together with the importance of Notch signaling in neurogenesis, suggests that the regulatory events we identified in the telencephalon may occur pan-neuronally throughout the embryonic nervous system. In addition, while our study focuses on the function of MyT1 at the onset of neuronal differentiation, further research should also address the function of MyT1 at later stages along the neuronal lineage.
Experimental Procedures
Immunohistochemistry and In Situ RNA Hybridization
Embryonic brains were processed for in situ hybridization or immunostaining as previously described (Castro et al., 2011). For double in situ hybridization/immunohistochemistry, frozen sections were treated as for in situ hybridization until the last post-fixation step, before immunostaining. The Hes1 probe was produced from the plasmid pBluescriptII SK-Hes1. NS5 cells were grown on glass coverslips coated with poly-L-Lysine (Sigma-Aldrich) and fixed with 4% formaldehyde for 10 min. See the Supplemental Experimental Procedures for antibody dilutions used. Bright-field and fluorescent images of fixed sections and coverslips were acquired using the microscope Leica DMRA2 or the laser-scanning confocal microscope Zeiss LSM510 Meta.
In Utero Electroporation
Mouse MyT1 cDNA (Nielsen et al., 2004) and AcNotch coding sequences were cloned upstream of an internal ribosomal entry site (IRES) and an nuclear localization signal (NLS)-tagged GFP in the pCAG expression vector (Niwa et al., 1991). Control and shRNA plasmids used (Sigma-Aldrich) were pLKO.1 scramble shRNA (Addgene 1864), pLKO MyT1 shRNA (TRCN0000081610), pLKO MyT1L shRNA (TRCN0000012109), and pLKO MyT3 shRNA (TRCN0000042479). Electroporation in utero was employed at E12.5 or E13.5 to deliver expression vectors (1 μl DNA plasmid corresponding to 3 μg mixed with 0.03% fast-green dye in PBS) to the ventricular RG cells of mouse embryos, as previously described (Saito, 2006, Sessa et al., 2008). All experiments were conducted upon the approval and following the guidelines of animal care and use committees from San Raffaele Scientific Institute (SRSI) and Instituto Gulbenkian de Ciência (IGC).
Culture and Infection of NS5 Cells
NS5 (Conti et al., 2005) and NS5-Ascl1-ERT2 cells (Raposo et al., 2015) were cultured in mouse Neurocult NSC basal medium supplemented with mouse Neurocult NSC proliferation supplement (STEMCELL Technologies), penicillin-streptomycin (100 U/mL) (Gibco), epidermal growth factor (EGF, 10 ng/mL) (PeproTech), basic FGF (10 ng/mL) (PeproTech), and laminin (1 μg/mL) (Sigma-Aldrich). For expression profiling and ChIP, NS5-MyT1-HA/GFP TetON cells were generated upon infection with MyT1-HA TetON-FUW, eGFP TetON-FUW, and M2rtTA TetON-FUW lentiviruses and were further expanded. When indicated, cells were treated with 50 nM Tam (Sigma-Aldrich), 2 μg/mL Dox hyclate (Sigma-Aldrich), or 10 nM LY (Lanz et al., 2004). Replication-incompetent lentiviruses were produced by transient transfection of HEK293T cells with TetON-FUW vectors co-transfected with psPAX2 and pCMV-VSVG. Lentiviral particles were concentrated from supernatant by ultracentrifugation at 9,000 × g for 4 hr and resuspended in 0.1% BSA PBS.
Gene Expression, DNA Microarrays, and RNA-Seq
Gene expression analysis of cultured NS5 cells by real-time qPCR with PerfeCTa SYBR Green FastMix, ROX (Quanta Biosciences) was carried out according to the manufacturer’s instructions on an ABI7900 HT machine (Applied Biosystems), using cDNA produced with High Capacity RNA-to-cDNA Master Mix (Applied Biosystems) after Trizol RNA extraction (see the Supplemental Experimental Procedures for primers). Microarray analysis was performed on biological triplicates of NS5 MyT1-HA TetON cells 0 and 4 hr post-Dox induction. RNA concentration and purity were confirmed using Agilent 2100 Bioanalyzer with RNA Nano Kit (Agilent Technologies). RNA (100 ng) was processed with Ambion WT Expression Kit (Life Technologies) and hybridized to the Affymetrix Mouse Gene 1.0 ST Array. CEL files were analyzed using Chispter software (version 3.1.0, Kallio et al., 2011) using RNA normalization and empirical Bayes two-group test with Benjamini-Hochberg post hoc for p value correction. Samples used for RNA-seq analysis were obtained from biological triplicates of NS5 0 and 4 hr post-LY treatment, and total RNA was extracted as described above. The sequencing library was prepared according to the TruSeq RNA sample preparation version 2 protocol (Illumina) and sequenced on an Illumina HiSeq 2000. Raw reads were aligned to NCBI37 (mm9) mouse genome using Tophat2 version 2.0.3 (Kim et al., 2013). Pairwise analysis of differential gene expression was performed with cuffdiff version 2.1.1 using fragment bias and multi-read corrections (Trapnell et al., 2013). Genomic sequence and gene annotation were obtained from ENSEMBL (May 2012, version 67).
ChIP and ChIP-Seq
Ascl1 ChIP from embryonic telencephalon was performed as previously described (Castro et al., 2011). NS5 cells were fixed sequentially with 2 mM di(N-succimidyl) glutarate and 1% formaldehyde in PBS and lysed, sonicated, and immunoprecipitated as described previously (Castro et al., 2011), using anti-HA (ab1424, Abcam), anti-Rbpj (D10A4, Cell Signaling Technology), or anti-LSD1 (ab17721, Abcam) antibodies. DNA sequences were quantified by real-time PCR (see the Supplemental Experimental Procedures for primers). For sequencing, DNA libraries were prepared from 10 ng immunoprecipitated DNA according to the manufacturer’s protocol. Paired-end sequencing was performed using MySeq. Raw reads were mapped to the mouse genome (NCBI37/mm9) with Bowtie version 0.12.7 (Langmead et al., 2009). Data were processed further with MACS version 1.4.1 (Zhang et al., 2008) to define the locations of BEs. Further details of data processing are described in the Supplemental Experimental Procedures.
In Vitro Binding and Transcriptional Assays
EMSAs were performed as described previously (Castro et al., 2006), using probes (Supplemental Experimental Procedures) labeled with [γ-32P] ATP (PerkinElmer) using T4 polynucleotyde kinase (New England Biolabs). Control, MyT1, and RBPJ proteins produced by coupled in vitro transcription and translation in rabbit reticulocyte lysates (TNT, Promega) were incubated with probe in 20-μL binding reactions (15% glycerol, 20 mM HEPES [pH 7.9], 5 mM MgCl2, 50 mM KCl, 0.1 mM ZnSO4, 0.01% Triton X-100, 10 mM DTT, 5 mM PMSF, and 0.2 μg/uL herring sperm DNA; Sigma-Aldrich D7290) for 20 min at room temperature (RT). Reporter gene assays in transfected P19 cells were performed as previously described (Castro et al., 2011). Oligonucleotides used to mutate Hes1::Luc are described in the Supplemental Experimental Procedures.
Statistical Procedures
Where indicated, data from at least three (qPCR assays) or four (luciferase assays) biological replicates are presented as mean ± SD. For in utero electroporations, >500 cells from at least three sections were counted, using three embryos per condition. Differentiation in culture was scored counting >1,500 cells from three coverslips per condition. In all cases, statistical significance was calculated by performing a two-tailed, unpaired Student’s t test or one-way ANOVA test with Bonferroni correction (see figure legends).
Author Contributions
F.F.V. performed most experimental work with contributions from C.L. and D.M.T. V.T. performed the in vivo gene expression analysis and A.S. the in utero electroporation experiments. A.A.S.F.R. did most of the bioinformatics analysis with contributions from F.F.V. D.W.H. and J.M. performed the single-cell transcriptomics analysis. D.S.C., F.F.V., A.S., and V.B. designed experimental work. D.S.C. and F.F.V. wrote the manuscript, which was read and approved by all others.
Acknowledgments
We thank the IGC Bioinformatics Unit for expert assistance in data analysis, the European Molecular Biology Laboratory (EMBL) Gene Core high-throughput sequencing Unit for ChIP-seq library preparation and sequencing, and the IGC Gene Expression Unit for DNA array processing and hybridization. We thank Guoqiang Gu for MyT1 and MyT1L antibodies; Brad G. Hoffman for the MyT3 antibody; and Ryoichiro Kageyama, Tasuku Honjo, Warren S. Pear, and Tohru Marunouchi for plasmids. This study was funded by Fundação para a Ciência e Tecnologia (FCT) grants UID/Multi/04555/2013 and PTDC/NEU-NMC/031572012 and Marie Curie CIG (303644) to D.S.C., the Telethon grant (GGP15096) and the Italian Ministry of Health Young investigator grant (GR-2013-02355540) to A.S., and an FCT doctoral fellowship (SFRH/BD/51178/2010) to F.F.V. D.S.C. is supported by the FCT Investigator program (IF/00413/2012).
Published: October 4, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and twelve tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.09.024.
Accession Numbers
The accession numbers for all genomic datasets reported in this paper are Array Express (http://www.ebi.ac.uk/arrayexpress): E-MTAB-4330 (MyT1 ChIP-seq), E-MTAB-4494 (Rbpj ChIP-seq in NS5 cells), E-MTAB-4335 (MyT1 GoF in NS5 cell DNA arrays), and E-MTAB-4331 (RNA-seq from NS5 cells treated with LY).
Supplemental Information
Table S1. Genes Deregulated 4 hr after MyT1 GoF in NS5 Cells, Related to Figure 3
Table S2. Genomic Coordinates of MyT1-HA BEs Determined by ChIP-Seq, Related to Figure 3
Table S3. Genes Bound and Repressed by MyT1 GoF, Related to Figure 3
Table S4. Identity of MyT1 Target Genes Associated with Enriched GO Terms, Related to Figure 3
Table S5. Expression Profiling Dataset upon Notch Inhibition, Related to Figure 5
Table S6. Genomic Coordinates of RBPJ BEs Determined by ChIP-Seq, Related to Figure 7
References
- Arnett K.L., Hass M., McArthur D.G., Ilagan M.X.G., Aster J.C., Kopan R., Blacklow S.C. Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat. Struct. Mol. Biol. 2010;17:1312–1317. doi: 10.1038/nsmb.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F., Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Bellefroid E.J., Bourguignon C., Hollemann T., Ma Q., Anderson D.J., Kintner C., Pieler T. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87:1191–1202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- Berninger B., Costa M.R., Koch U., Schroeder T., Sutor B., Grothe B., Götz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Castro D.S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Lowry W.E., Pasolli H.A., Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro D.S., Skowronska-Krawczyk D., Armant O., Donaldson I.J., Parras C., Hunt C., Critchley J.A., Nguyen L., Gossler A., Göttgens B. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Castro D.S., Martynoga B., Parras C., Ramesh V., Pacary E., Johnston C., Drechsel D., Lebel-Potter M., Garcia L.G., Hunt C. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.-L., Cattaneo E., Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R.A., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M.H., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Fietz S.A., Lachmann R., Brandl H., Kircher M., Samusik N., Schröder R., Lakshmanaperumal N., Henry I., Vogt J., Riehn A. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. USA. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N., Nye J.S., Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R., Swanton M.K., Kobus F.J., Lehtomaki E., Lowry J.A., Kwan A.H., Matthews J.M., Mackay J.P. Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1. J. Biol. Chem. 2008;283:5158–5167. doi: 10.1074/jbc.M703772200. [DOI] [PubMed] [Google Scholar]
- Gohlke J.M., Armant O., Parham F.M., Smith M.V., Zimmer C., Castro D.S., Nguyen L., Parker J.S., Gradwohl G., Portier C.J., Guillemot F. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-López S., Wiskow O., Favaro R., Nicolis S.K., Price D.J., Pollard S.M., Smith A. Sox2 and Pax6 maintain the proliferative and developmental potential of gliogenic neural stem cells In vitro. Glia. 2011;59:1588–1599. doi: 10.1002/glia.21201. [DOI] [PubMed] [Google Scholar]
- Götz M., Huttner W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hagey D.W., Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9:1908–1920. doi: 10.1016/j.celrep.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Hu J., Ho A.L., Yuan L., Hu B., Hua S., Hwang S.S., Zhang J., Hu T., Zheng H., Gan B. From the Cover: Neutralization of terminal differentiation in gliomagenesis. Proc. Natl. Acad. Sci. USA. 2013;110:14520–14527. doi: 10.1073/pnas.1308610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L.D., Romm E., Berndt J.A., Nielsen J.A. A tool for examining the role of the zinc finger myelin transcription factor 1 (Myt1) in neural development: Myt1 knock-in mice. Transgenic Res. 2011;20:951–961. doi: 10.1007/s11248-010-9470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Kageyama R. Oscillatory control of bHLH factors in neural progenitors. Trends Neurosci. 2014;37:531–538. doi: 10.1016/j.tins.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E.H., Kopan R., Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yu V.C., Buchholz F., O’Connell S., Rhodes S.J., Candeloro C., Xia Y.R., Lusis A.J., Rosenfeld M.G. A novel family of Cys-Cys, His-Cys zinc finger transcription factors expressed in developing nervous system and pituitary gland. J. Biol. Chem. 1996;271:10723–10730. doi: 10.1074/jbc.271.18.10723. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Shimojo H., Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- Kallio M.A., Tuimala J.T., Hupponen T., Klemelä P., Gentile M., Scheinin I., Koski M., Käki J., Korpelainen E.I. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A., Ikawa T., Kasukawa T., Ueda H.R., Kurimoto K., Saitou M., Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kim J.G., Hudson L.D. Novel member of the zinc finger superfamily: A C2-HC finger that recognizes a glia-specific gene. Mol. Cell. Biol. 1992;12:5632–5639. doi: 10.1128/mcb.12.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz T.A., Hosley J.D., Adams W.J., Merchant K.M. Studies of Abeta pharmacodynamics in the brain, cerebrospinal fluid, and plasma in young (plaque-free) Tg2576 mice using the gamma-secretase inhibitor N2-[(2S)-2-(3,5-difluorophenyl)-2-hydroxyethanoyl]-N1-[(7S)-5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl]-L-alaninamide (LY-411575) J. Pharmacol. Exp. Ther. 2004;309:49–55. doi: 10.1124/jpet.103.060715. [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Matsushita F., Kameyama T., Marunouchi T. NZF-2b is a novel predominant form of mouse NZF-2/MyT1, expressed in differentiated neurons especially at higher levels in newly generated ones. Mech. Dev. 2002;118:209–213. doi: 10.1016/s0925-4773(02)00250-2. [DOI] [PubMed] [Google Scholar]
- Matsushita F., Kameyama T., Kadokawa Y., Marunouchi T. Spatiotemporal expression pattern of Myt/NZF family zinc finger transcription factors during mouse nervous system development. Dev. Dyn. 2014;243:588–600. doi: 10.1002/dvdy.24091. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Nakada Y., Hunsaker T.L., Henke R.M., Johnson J.E. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Nam Y., Sliz P., Pear W.S., Aster J.C., Blacklow S.C. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. USA. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.R., Hodge R.D., Bedogni F., Hevner R.F. Dynamic interactions between intermediate neurogenic progenitors and radial glia in embryonic mouse neocortex: potential role in Dll1-Notch signaling. J. Neurosci. 2013;33:9122–9139. doi: 10.1523/JNEUROSCI.0791-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.A., Berndt J.A., Hudson L.D., Armstrong R.C. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol. Cell. Neurosci. 2004;25:111–123. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Nishimura M., Isaka F., Ishibashi M., Tomita K., Tsuda H., Nakanishi S., Kageyama R. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics. 1998;49:69–75. doi: 10.1006/geno.1998.5213. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Pang Z.P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D.R., Yang T.Q., Citri A., Sebastiano V., Marro S., Südhof T.C., Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan X.-J., Denayer T., Yan J., Jafar-Nejad H., Philippi A., Lichtarge O., Vleminckx K., Hassan B.A. Evolution of neural precursor selection: functional divergence of proneural proteins. Development. 2004;131:1679–1689. doi: 10.1242/dev.01055. [DOI] [PubMed] [Google Scholar]
- Raposo A.A.S.F., Vasconcelos F.F., Drechsel D., Marie C., Johnston C., Dolle D., Bithell A., Gillotin S., van den Berg D.L.C., Ettwiller L. Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 2015;10:1544–1556. doi: 10.1016/j.celrep.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romm E., Nielsen J.A., Kim J.G., Hudson L.D. Myt1 family recruits histone deacetylase to regulate neural transcription. J. Neurochem. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Schneider M.L., Turner D.L., Vetter M.L. Notch signaling can inhibit Xath5 function in the neural plate and developing retina. Mol. Cell. Neurosci. 2001;18:458–472. doi: 10.1006/mcne.2001.1040. [DOI] [PubMed] [Google Scholar]
- Sessa A., Mao C.A., Hadjantonakis A.K., Klein W.H., Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H., Ohtsuka T., Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos F.F., Castro D.S. Transcriptional control of vertebrate neurogenesis by the proneural factor Ascl1. Front. Cell. Neurosci. 2014;8:412. doi: 10.3389/fncel.2014.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhang J., Zhao A., Hipkens S., Magnuson M.A., Gu G. Loss of Myt1 function partially compromises endocrine islet cell differentiation and pancreatic physiological function in the mouse. Mech. Dev. 2007;124:898–910. doi: 10.1016/j.mod.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hecksher-Sorensen J., Xu Y., Zhao A., Dor Y., Rosenberg L., Serup P., Gu G. Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev. Biol. 2008;317:531–540. doi: 10.1016/j.ydbio.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G., Dennis D., Schuurmans C. Proneural genes in neocortical development. Neuroscience. 2013;253:256–273. doi: 10.1016/j.neuroscience.2013.08.029. [DOI] [PubMed] [Google Scholar]
- Yee K.S., Yu V.C. Isolation and characterization of a novel member of the neural zinc finger factor/myelin transcription factor family with transcriptional repression activity. J. Biol. Chem. 1998;273:5366–5374. doi: 10.1074/jbc.273.9.5366. [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Igarashi K., Sato T., Takagi K., Otsuka I M., Shishido Y., Baba T., Ito R., Kanno J., Ohkawa Y. Identification of myelin transcription factor 1 (MyT1) as a subunit of the neural cell type-specific lysine-specific demethylase 1 (LSD1) complex. J. Biol. Chem. 2014;289:18152–18162. doi: 10.1074/jbc.M114.566448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes Deregulated 4 hr after MyT1 GoF in NS5 Cells, Related to Figure 3
Table S2. Genomic Coordinates of MyT1-HA BEs Determined by ChIP-Seq, Related to Figure 3
Table S3. Genes Bound and Repressed by MyT1 GoF, Related to Figure 3
Table S4. Identity of MyT1 Target Genes Associated with Enriched GO Terms, Related to Figure 3
Table S5. Expression Profiling Dataset upon Notch Inhibition, Related to Figure 5
Table S6. Genomic Coordinates of RBPJ BEs Determined by ChIP-Seq, Related to Figure 7