Summary
The Wnt pathway plays a crucial role in self‐renewal and differentiation of cells in the adult gut. In the present study, we revealed the functional consequences of inhibition of canonical Wnt signaling in the intestinal epithelium. The study was based on generation of a novel transgenic mouse strain enabling inducible expression of an N‐terminally truncated variant of nuclear Wnt effector T cell factor 4 (TCF4). The TCF4 variant acting as a dominant negative (dn) version of wild‐type (wt) TCF4 protein decreased transcription of β‐catenin‐TCF4‐responsive genes. Interestingly, suppression of Wnt/β‐catenin signaling affected asymmetric division of intestinal stem cells (ISCs) rather than proliferation. ISCs expressing the transgene underwent several rounds of division but lost their clonogenic potential and migrated out of the crypt. Expression profiling of crypt cells revealed that besides ISC‐specific markers, the dnTCF4 production downregulated expression levels of epithelial genes produced in other crypt cells including markers of Paneth cells. Additionally, in Apc conditional knockout mice, dnTCF activation efficiently suppressed growth of Apc‐deficient tumors. In summary, the generated mouse strain represents a convenient tool to study cell‐autonomous inhibition of β‐catenin‐Tcf‐mediated transcription. genesis 54:101–114, 2016. © 2016 The Authors genesis Published by Wiley Periodicals, Inc.
Keywords: β‐catenin, Cre/loxP, gene targeting, gut, Wnt pathway, TCF/LEF transcription factors
INTRODUCTION
The Wnt pathway represents one of the fundamental signaling mechanisms controlling cell‐fate decision during embryonic development and in adult tissues. Furthermore, aberrant activation of the pathway is associated with various disorders, including cancer or degenerative diseases [reviewed in (Clevers and Nusse, 2012)]. In the intestinal epithelium, the pathway regulates proliferation of ISCs [reviewed in (Krausova and Korinek, 2014)]. ISCs reside at the bottom of the crypts of Lieberkühn, submucosal invaginations of the single‐layer epithelium. ISCs divide approximately every 24 h, generating a pool of transit amplifying (TA) cells, rapidly proliferating cells that migrate upwards the crypt axis. At the crypt orifice these cells differentiate to several cell types that mainly include absorptive enterocytes, mucus‐producing goblet cells or hormone‐releasing enteroendocrine cells. In the small intestine, the differentiated cells cover finger‐like protrusions into the intestinal lumen called villi; the surface of the large intestine is flat. Within 3 to 7 days the differentiated cells fulfil their physiological role and are excluded from the epithelial layer. Bactericidal Paneth cells of the small intestine do not leave the crypt but stay at the crypt bottom and renew every 6 to 8 weeks (Garabedian et al., 1997).
The key molecular event of the best‐studied so‐called canonical Wnt signaling is regulation of the β‐catenin protein stability [reviewed in (Valenta et al., 2012)]. In the absence of signaling, the β‐catenin destruction complex that includes axis inhibition protein (Axin), adenomatous polyposis coli (APC), casein kinase 1 alpha (CK1α), and glycogen synthase kinase 3 (GSK3) mediates β‐catenin phosphorylation. The protein phosphorylated at the N‐terminal serine/threonine residues is subsequently degraded by the proteasome. Interaction of extracellular Wnt ligand with the Frizzled (Fz) receptor and lipoprotein‐related protein (LRP) co‐receptor inactivates the destruction complex and leads to β‐catenin accumulation in the cytoplasm and nucleus. Nuclear β‐catenin interacts with DNA‐binding proteins of the TCF/lymphoid enhancer factor (LEF) family. Since β‐catenin contains a strong transactivation domain, β‐catenin‐TCF/LEF complexes activate transcription of Wnt‐responsive genes such as Axin2, CD44, or Cyclin D1.
The involvement of Wnt/β‐catenin signaling in intestinal development and tissue maintenance was documented in numerous reports, including genetic ablation of β‐catenin or Tcf4 genes or production of diffusible extracellular Wnt signaling inhibitors. Different phenotypes have been documented, although loss of the proliferative capacity accompanied by the crypt demise appear to be the main type of damage observed in the majority of the studies. For example, elimination of the floxed β‐catenin alleles using β‐napthoflavone‐inducible Cre in Ah‐Cre transgenic mice resulted in increased epithelial cell apoptosis, reduced crypt and goblet cell number, and caused detachment of sheets of the differentiated cells from the villi (Ireland et al., 2004). In contrast, Fevr and colleagues observed loss of TA cells and crypt structures in hemizygous β‐catenin−/flox/Villin‐CreERT2 mice six days upon Cre activation. However, these authors conclude that the observed changes are caused solely by inhibition of Wnt/β‐catenin signaling. They also excluded the possibility that the phenotype is connected to reduced adhesion (Fevr et al., 2007). Transgenic expression of a secreted Wnt antagonist, Dickkopf‐1 (Dkk1), in the gut epithelium affected proliferation and crypt‐villus organization in the small intestine (Pinto et al., 2003). However, systemic expression of Dkk1 mediated by adenoviral delivery induced rapid degeneration of both the small intestine and colon (Kuhnert et al., 2004). Inactivation of Tcf4, a Tcf/Lef family member highly expressed in gut epithelia, resulted in neonatal lethality due to impaired proliferation of the small intestinal epithelium (Korinek et al., 1998). Proliferative defects were also observed upon conditional Tcf4 inactivation in all intestinal cell types (van Es et al., 2012a). In contrast, Angus‐Hill and colleagues documented cell hyperproliferation followed by necrotic death in the Tcf4‐deficient colon (Angus‐Hill et al., 2011). These seemingly contradictory results possibly reflect the dual, i.e. transcriptional repressive and activatory, role of Tcf4 and they are related to different targeting strategies of the Tcf4 locus.
To address the specific role of β‐catenin‐TCF/LEF‐mediated transcription we generated a mouse strain expressing N‐terminally truncated TCF4 protein from the Rosa26 locus. The TCF4 variant [(designated as dominant negative TCF4 (dnTCF4)] binds the regulatory regions in Wnt‐responsive genes. Nevertheless, due to the disruption of the β‐catenin interaction domain (Korinek et al., 1997; van de Wetering et al., 2002), it cannot associate with β‐catenin and acts as a nuclear blocker of canonical Wnt signaling. Additionally, a sequence encoding a tandem dimer (td) of red fluorescent protein Tomato flanked (i.e. “floxed”) by two loxP sites was inserted into the targeted locus. The floxed tdTomato was placed upstream of enhanced green fluorescent protein (EGFP)‐dnTCF4 cDNA and served as a transcription “roadblock” preventing expression of the downstream gene. Thus, although the Rosa26 locus was ubiquitously expressed, the EGFP‐dnTCF4 production was activated only in cells expressing Cre recombinase (Soriano, 1999). Such experimental design allowed cell‐autonomous suppression of β‐catenin‐TCF/LEF signaling that precluded any interference with other TCF/LEF‐independent β‐catenin functions (see Discussion for details). Importantly, knockin into the Rosa26 locus, which is dispensable during embryonic development or in adult individuals, did not modify (or damage) any other gene involved in Wnt signaling.
Here we show that selective expression of the EGFP‐dnTCF4 transgene affected the “stemness” of ISCs and led to their elimination from the small intestinal epithelium. In the healthy gut, stem cell dysfunction had no obvious impact on tissue homeostasis. However, in the genetic model of intestinal cancer based on conditional ablation of the Apc gene, the EGFP‐dnTCF4 production effectively attenuated neoplastic growth.
METHODS
Generation of Rosa26tdTomato Mice
Generation, housing of mice, and in vivo experiments were in compliance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and national and institutional guidelines. Animal care and experimental procedures were approved by the Animal Care Committee of the Institute of Molecular Genetics (Ref. 63/2013). The targeting construct was generated in the pEASY‐FLIRT vector (Pospichalova et al., 2011). The internal SacI restriction endonuclease site was used to clone human TCF4 cDNA (Genbank accession number Y11306, the cDNA encodes amino acids 31–597) into the pEGFP‐C1 vector (Clontech); tdTomato cDNA of tdTomato was kindly provided by Roger Tsian (UC San Diego, CA). The Simian virus 40 (SV40) early mRNA polyadenylation signal sequences (pAs) were derived from the pEGFP‐C1 vector (Clontech). ES R1 cells were grown on a feeder layer of MEF feeder cells (Stem Cell Technologies) treated with mitomycin C (for 2 h at final concentration 10 μg/mL; Sigma). ES cells were cultured in Glutamax Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 15% fetal bovine serum (FBS; ES cells tested; Hyclone), 2 mM l‐glutamine, 1 mM sodium pyruvate, 1× non‐essential amino acids, 0.1 mM β‐mercaptoethanol, 100 UI penicillin/streptomycin (all chemicals were purchased from Gibco). The complete medium was supplemented with conditioned media obtained from COS‐7 cells (kindly provided by Vladimir Divoky; Palacky University, Olomouc, Czech Republic) stably expressing mouse leukemia inhibitory factor (LIF). The targeting vector (25 μg) linearized by ClaI restriction endonuclease was electroporated (settings: 380 V, 25 μF, time constant ∼3.4 s) into 1 × 107 ES cells using the Gene Pulser II system (Bio‐Rad Laboratories). Cells harboring the integrated construct were selected using G418 (200 µg/mL Gibco); randomly targeted clones were counter‐selected by gancyclovir (0.2 µM; Sigma). After 10 days, colonies displaying red fluorescence were picked and expanded. Genomic DNA isolated from 24 clones was digested with EcoRI restriction enzyme and screened for homologous recombination at both arms of the targeted construct using Southern blotting. Correctly targeted ES cell clones were karyotyped and cells of one clone with the correct chromosomal count were injected into blastocysts of superovulated C57BL/6J females. The blastocyst injections were performed using the services of the Transgenic Unit (Institute of Molecular Genetics, Prague, Czech Republic). Of 14 chimeric males, one male was mated to C57BL/6J females to produce F1 generation Rosa26+/tdTomato heterozygous mice.
Mouse Strains, Genotyping, and Animal Manipulations
Animal genotyping was performed by PCR using genomic DNA isolated from tail biopsies as described previously (Pospichalova et al., 2011). The following primers were used to genotype the Rosa26 wild‐type (wt) and Rosa26tdTomato‐NeoR alleles (all targeted Rosa26 alleles are depicted in Fig. 1a): common forward primer P1: 5'‐TGTTTGTTCCAATATGGTAGCC‐3', wt reverse primer P2: 5'‐GTCTCTGCCTCCAGAGTGCT‐3', knock‐in reverse primer P3: 5'‐CGCGCATCTTCACCTTGTAG‐3'. PCR on the wt and Rosa26tdTomato‐NeoR allele generated the 401‐bp and 852‐bp DNA fragment, respectively. The Rosa26tdTomato allele generated by Flp‐mediated recombination of the pGK‐NeoR cassette was tested using primers P4: 5'‐GCGGATCCATGAAGTTCCTA‐3' and P5: 5'‐CCCCTCAGAGAAATGGAGTAGTTA‐3' (size of the PCR product: 171 bp). The recombined Rosa26dnTCF4 allele was detected with forward primer P6: 5'‐TGTAACT GTGGACAGAGGAG‐3' and reverse primer P7: 5'‐CG GACACGCTGAACTTGTGG‐3' (430 bp). Mouse strains ACTB‐Flp [strain nomenclature name: B6.Cg‐Tg(ACTFLPe)9205Dym/J], Lgr5‐EGFP‐CreERT2 [B6.129P2‐Lgr5tm1(cre/ERT2)Cle/J], Rosa26‐CreERT2 [B6.129‐Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J], Rosa26R [B6;129S4‐Gt(ROSA)26Sortm1Sor/J], and Vav1‐cre [B6.Cg‐Tg(Vav1‐cre)A2Kio/J] were purchased from The Jackson Laboratory (Bar Harbor, Maine) and genotyped as described in the genotyping protocols of the provider using tail biopsies. Apc‐CKO (B6.Cg‐Apctm2Rak) were obtained from Mouse Repository (NCI, Frederick) and were genotyped as described previously (Kuraguchi et al., 2006). Villin‐CreERT2 transgenic mice were kindly provided by Sylvie Robine (Institut Curie, Paris, France); these mice were genotyped using forward 5'‐AAAATTTG CCTGCATTACCG‐3' and reverse 5'‐ATTCTCCCACCG TCAGTACG‐3' primers (553 bp). Tamoxifen (Sigma, stock solution 100 mg/mL in ethanol) was administered by gavage (single dose: 5 mg of tamoxifen dissolved in sunflower oil). For tumor analysis, the mice were euthanized 16 days after a single tamoxifen dose and the intestines were dissected, washed in phosphate‐buffered saline (PBS) and fixed in 4% formaldehyde (v/v) in PBS for 1 day. Fixed intestines were embedded in paraffin, sectioned, and stained using hematoxylin and eosin (Sigma).
Figure 1.
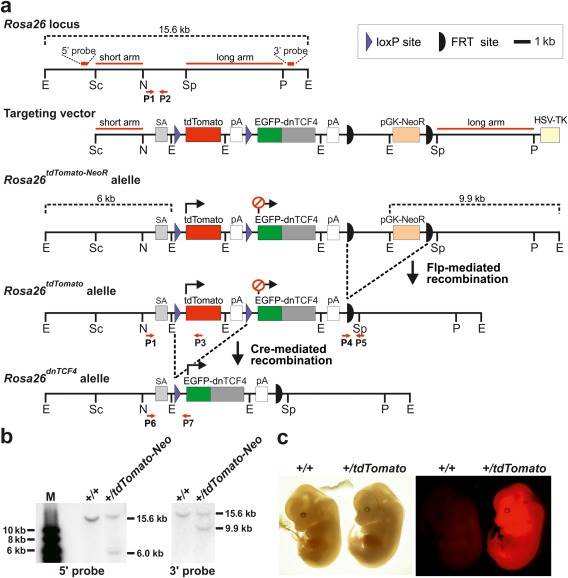
Generation of mice with inducible expression of N‐terminally truncated TCF4 protein. (a) The targeting strategy used for homologous recombination in ES cells. The targeting vector included the 5' “short” and 3' “long” Rosa26 homology arms and rabbit β‐globin splice acceptor site (SA) followed by transcription blocker [composed of tdTomato cDNA and the polyadenylation signal (pA)] flanked by two loxP sites (violet triangles). The sequence encoding dnTCF4 fused to the C‐terminus of EGFP was placed downstream of the blocker. The construct also contained the phosphoglycerate kinase promoter‐driven neomycin resistance gene (pGK‐NeoR) flanked by FRT sites (black semicircles) and the herpes simplex virus thymidine kinase (HSV‐TK) gene. HSV‐TK was used for selection against clones with randomly integrated vector. Recombination of the targeting vector with the Rosa26 locus generated the Rosa26tdTomato‐NeoR allele. The Rosa26tdTomato and Rosa26dnTCF4 alleles were produced using Flp‐ or Cre‐mediated recombination, respectively. Relevant restriction enzyme sites, primers for genotyping (P1‐P7), and positions of probes used for Southern blotting are indicated. E, EcoRI; N, NheI; P, PmlI; Sc, SacII; Sp, SspI. For clarity, the sizes of the genes and DNA elements do not completely correspond to the scale indicated in the upper right corner of the scheme. (b) Southern blot analysis of ES cell genomic DNA detecting the homologous recombination at the 5' or 3' end of the Rosa26 locus. EcoRI‐digested DNA was used for both blots. M, molecular weight markers. (c) Bright field (left) and fluorescent microscopy images (right) of wt and Rosa26+/tdTomato embryos isolated at E13.5. The presence of the tdTomato allele is manifested by whole‐body red fluorescence.
RNA Isolation and quantitative polymerase chain reaction(qRT‐PCR)
Total RNAs were isolated from cells and tissues using RNABlue (TopBio) and reversely transcribed and analyzed by qRT‐PCR as described previously (Lukas et al., 2009). All primers (the first primer is in forward and the second primer in reverse orientation) were specific for mouse genes except for the primer set detecting the EGFP‐dnTCF4 transgene that was designed from human TCF4. The primers are listed in Supporting Information Table S1. The EGFP‐dnTCF4 transcript in cDNA was detected using forward 5'‐CGCCTAAAGAAGAGGCT GTG‐3' and reverse 5'‐CGGACACGCTGAACTTGTGG‐3' primers (356 bp).
Mouse EmbryonicFibroblasts (MEFs) and Recombinant Wnt3a Ligand
MEFs were isolated from E12 to E14 embryos. Embryos were washed in ice‐cold PBS; head and entrails (heart, liver) were excised and used for genotyping. The remaining tissues were incubated in Trypsin/EDTA (15 min at 37°C; Sigma), dissociated, and plated in Iscove's Modified Dulbecco's Medium [(IMDM); Sigma)] supplemented with 10% FBS, 100 UI penicillin/streptomycin, 2 mM l‐glutamine, 1× non‐essential amino acids. For Cre‐mediated recombination, cells were cultured for three days in 4‐OHT (final concentration 2 µM; Sigma); to prepare 1 mM stock solution, 4‐OHT was dissolved in ethanol. Mouse Wnt3a ligand was isolated from the culture medium of Wnt3a‐producing L cells according to the detailed protocol of Willert and colleagues (Willert et al., 2003).
Immunocytochemistry, Immunohistochemistry, and Immunoblotting
The techniques were performed as described previously (Doubravska et al., 2011; Waaler et al., 2012). For immunohistochemistry, dissected intestines were washed in ice‐cold PBS, fixed overnight in 4% (v/v) formaldehyde (Penta) and embedded in paraffin. Antibodies used for immunocytochemistry, immunohistochemistry, or immunoblotting: anti‐CtBP (mouse monoclonal, sc‐17759, Santa Cruz), anti‐EGFP (rabbit polyclonal, ab6556 and chicken polyclonal, ab13970, Abcam), anti‐Ki67 (mouse monoclonal, Mob 237, Diagnostic BioSystems). For fluorescent microscopy, Alexa 488 dye conjugated to a goat anti‐chicken and Alexa 594 dye conjugated to a goat anti‐rabbit antibody (Molecular Probes) and the DAPI nuclear stain (Molecular Probes) were used. For LacZ staining, mouse tissues were dissected and fixed for 2 h in fixative (1% formaldehyde, 0.2% glutaraldehyde, and 0.02% NP40 in PBS) at 4°C. The fixative was removed and tissues were washed three times for 5 min in PBS at RT. The LacZ substrate [5 mM K3FE(CN)6, 5 mM K4FE(CN)6 × 3H20, 2 mM MgCl2, 0.02% NP40, 0.1% sodium deoxycholate, and 1 mg/mL X‐gal (Amresco) in PBS] was added and tissues were incubated overnight at RT protected from the light. Next day, the substrate was removed and the tissue was washed three times for 5 min in PBS. The tissues were then fixed overnight in 4% formaldehyde, embedded in paraffin and sectioned. Specimens were counterstained using nuclear fast red (DiaPath). To quantify tissue hyperplasia induced by the loss of Apc, four ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ (control) and four Rosa26+/tdTomato/ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ (dnTCF4) 12‐week‐old mice were examined 16 days upon tamoxifen administration. The animals were obtained from two litters; from each litter two control and two dnTCF4 males were evaluated. Ten sections taken along the longitudinal axis of the small intestine were analyzed from each animal using fluorescent microscopy images and ImageJ software; (i.e., 40 images from each group). Sections were stained with anti‐Ki67 antibody and counterstained using DAPI. DAPI blue fluorescence was employed to quantify the total tissue area per one image. 4′,6‐diamidino‐2‐phenylindole dihydrochloride (DAPI) and anti‐Ki67 fluorescence (green) was utilized to quantify the proportion of proliferating cells in the tissue sections (estimated from the ratio between the green and blue fluorescent signal) (Tumova et al., 2014).
Statistical Analysis of Data
Results of the qRT‐PCR and proliferating cell analyses were evaluated by Student's t‐test.
RESULTS
Generation of a Mouse Strain with Inducible Expression of dnTCF4
To achieve inhibition of Wnt/β‐catenin signaling at the transcriptional level, a targeting construct based on the mouse ubiquitously expressed Rosa26 locus was generated (Fig. 1a). The construct contained 5' and 3' homology arms derived from the Rosa26 intron, splice acceptor (SA), and two cDNAs encoding either red fluorescent protein tdTomato floxed by two loxP sites or EGFP N‐terminally fused to human dnTCF4 lacking the first 31 amino acid residues. Both cDNAs were followed by SV40 pA. Finally, a neomycin expression cassette (pGK‐NeoR) flanked by two FRT sites was included in the construct to select targeted cell clones. Truncated dnTCF4 binds Wnt response elements in β‐catenin‐TCF/LEF target genes; however, due to the disruption of the β‐catenin‐interaction domain it does not associate with β‐catenin and functions as transcriptional repressor (Korinek et al., 1997). Since EGFP‐dnTCF4 was placed downstream of the loxP‐tdTomato‐pA‐loxP sequence, the EGFP‐dnTCF4 protein is not produced unless the floxed sequence is removed from genomic DNA by Cre‐mediated excision. The targeting construct was linearized using the unique ClaI restriction endonuclease recognition site and electroporated into mouse ES cells (Nagy et al., 1993). Genomic DNA isolated from ES cell clones that survived selection and constitutively produced tdTomato were subjected to Southern blotting to confirm correct homologous recombination at both 5' and 3' arms of the construct (Fig. 1b). Cells of one clone with appropriate integration and standard karyotype were injected into C57BL/6J blastocysts. Two females and 14 male chimeras were obtained. Rosa26+/tdTomato‐NeoR heterozygous mice were generated upon breeding of one highly chimeric male with wild‐type (wt) C57BL/6J females. Subsequently, the pGK‐NeoR selection cassette was removed from the targeted locus by crossing Rosa26+/tdTomato‐NeoR heterozygotes with ACTB‐flippase (Flp) transgenic mice expressing Flp from the promoter of the human β‐ACTIN gene (Rodriguez et al., 2000). Animals of both strains, i.e. Rosa26+/tdTomato‐NeoR and Rosa26+/tdTomato mice, were born in standard Mendelian ratio and exhibited full‐body red fluorescence (Fig. 1c and data not shown).
EGFP‐dnTCF4 Decreased Expression of Wnt Signaling Target Genes in MEFs
Next, Rosa26+/tdTomato heterozygous mice were crossed to Rosa26‐CreERT2 animals expressing Cre recombinase estrogen receptor T2 fusion protein (CreERT2) from the Rosa26 locus (the corresponding allele was designated Rosa26CreERT2) (Ventura et al., 2007). The ERT2 partner immobilizes Cre enzyme in the cytoplasm until an antagonist of the estrogen receptor, tamoxifen [or its metabolite 4‐hydroxitamoxifen (4‐OHT)], is administered. Thus, the usage of CreERT2 allows excision of floxed DNA sequences in a timely manner (Indra et al., 1999). MEFs isolated from Rosa26CreERT2/tdTomato embryos at E13.5 were cultured with 4‐OHT or vehicle (ethanol) and genotyped by PCR to confirm the locus recombination and generation of the Rosa26dnTCF4 allele (Fig. 2a). Additionally, production of the EGFP‐dnTCF4 fusion protein was detected by immunoblotting (Fig. 2b). Furthermore, qRT‐PCR analysis was performed with RNA isolated from MEFs stimulated by recombinant Wnt3a. As shown in Figure 2b, mRNA levels of previously described Wnt signaling target genes Axin2, naked cuticle homolog 1 (Nkd1), and Troy were significantly decreased in recombined cells when compared to MEFs treated with vehicle only. Upon 4‐OHT‐induced recombination we noted a gradual decrease in red fluorescence; unfortunately, the fluorescent signal of EGFP‐dnTCF4 fusion protein was too “dim” for direct visualization by fluorescent microscopy. However, the nuclear EGFP‐dnTCF4 protein was clearly detected by immunocytochemical staining using anti‐EGPF antibody in MEFs growing in the presence of 4‐OHT (Fig. 2c). Moreover, EGFP‐dnTCF4 expression could be traced using fluorescence‐activated cell sorting (FACS) in the blood and bone marrow of Rosa26tdTomato/tdTomato/Vav1‐Cre+ and Rosa26+/tdTomato/Vav1‐Cre+ mice producing Cre enzyme in all cell types of the hematopoietic system (de Boer et al., 2003) (Supporting Information Fig. S1).
Figure 2.
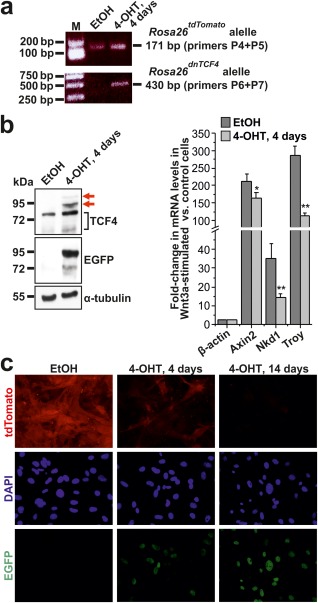
The Rosa26dnTCF4 allele suppresses Wnt signaling in MEFs. (a) PCR genotyping of MEFs isolated from Rosa26CreERT2/tdTomato embryos treated with solvent [ethanol (EtOH)] or 4‐OHT for 4 days to induce Cre‐mediated recombination of the transcription blocker. PCR of the Rosa26tdTomato and Rosa26dnTCF4 allele produced 171 bp or 430 bp DNA fragments, respectively. M, molecular weight markers. Notice that since CreERT2 is expressed from the Rosa26 locus, EGFP‐dnTCF4 is produced from a single Rosa26 allele. (b) Immunoblotting (left) and qRT‐PCR analysis (right) of MEFs isolated from Rosa26CreERT2/tdTomato embryo treated with EtOH or 4‐OHT for 4 days. EGFP‐dnTCF4 fusion protein (displaying lower mobility than endogenous Tcf4) was detected using anti‐TCF4 (red arrows) and anti‐EGFP antibodies. Notice that Tcf4 migrates in denaturing conditions as a double band. Immunoblotting with an anti‐α‐tubulin antibody was used as a loading control. Diagram shows results of the qRT‐PCR analysis of Wnt‐responsive genes Axin2, Nkd1, and Troy in MEFs stimulated overnight with recombinant Wnt3a. The “housekeeping” gene β‐actin (Actb) was also included in the test. The quantities of input cDNAs were normalized to Ubiquitin B (Ubb). The expression level of a given gene in unstimulated cells was arbitrarily set to 1. Error bars represent standard deviations (SDs); **P < 0.01 (Student's t‐test). (c) Fluorescent microscopy images of MEFs derived from Rosa26CreERT2/tdTomato embryo treated with EtOH (left panel) or 4‐OHT for 4 and 14 days. Nuclear EGFP‐dnTCF4 fusion protein was detected by immunocytochemical staining using an anti‐GFP antibody (green fluorescence); tdTomato protein was visualized by its native red fluorescence. Specimens were counterstained using the DAPI nuclear stain (blue). Original magnification: ×400.
EGFP‐dnTCF4 Affects Function of ISCs
To investigate the effect of EGFP‐dnTCF4 expression in the intestine, we analyzed intestinal epithelium of the Rosa26tdTomato/tdTomato/Villin‐CreERT2+ mice expressing tamoxifen‐regulated Cre in all intestinal cell lineages (el Marjou et al., 2004). The presence of EGFP‐dnTCF4 in the epithelium of duodenum, jejunum, ileum and colon was detected by immunoblotting using cellular lysates prepared from isolated tissue one day after tamoxifen administration. Both anti‐EGFP and TCF4‐specific antibodies detected the fusion protein in all investigated parts of the intestine (Fig. 3a). Moreover, immunohistochemical staining with anti‐EGFP antibody confirmed production of the nuclear EGFP‐dnTCF4 fusion protein in epithelial cells. However, no visible changes in the tissue architecture were observed at this time point and at days 7 and 21 upon tamoxifen administration. The absence of any phenotype was probably related to the fact that Cre‐mediated recombination of the transcription blocker leading to activation of the EGFP‐dnTCF4 transgene was mainly achieved in short‐living differentiated cells localized on the small intestinal villi or colonic surface. The conclusion was supported by the fact that virtually no EGFP‐dnTCF4‐positive cell was present in the small intestinal or colonic epithelium at day 21 upon tamoxifen administration. Strikingly, EGFP‐dnTCF4‐producing cells (including Paneth cells) were still noted at day 7 (Fig. 3b and data not shown). Such labeling was possibly caused by prolonged Cre‐mediated recombination in Villin‐CreERT2+ transgenic mice. Next, we intercrossed Rosa26tdTomato/tdTomato mice with Lgr5‐EGFP‐IRES‐CreERT2 animals. Lgr5‐EGFP‐IRES‐CreERT2 mice produce bicistronic mRNA encoding CreERT2 and EGFP from the leucine‐rich repeat‐containing G‐protein‐coupled receptor 5 (Lgr5) locus. Consequently, excision of floxed sequences can be achieved specifically in ISCs. Moreover, cytoplasmic EGFP (translated from the same mRNA as the CreERT2 enzyme) can be utilized as a surrogate ISC marker (Barker et al., 2007). Since the most efficient recombination between loxP sites mainly occurs in the small intestine (Fafilek et al., 2013; Powell et al., 2012), we focused on this part of the gastrointestinal tract. Rosa26tdTomato/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ mice were injected with three consecutive doses of tamoxifen and analyzed 1 and 21 days later. Control animals (of the same genotype) were injected with solvent only. Interestingly, at the first time point the EGFP‐specific signal was absent; however, EGFP‐producing cells re‐appeared at the second time point. In contrast, lysozyme‐positive Paneth cells were detected in the crypts at both time points (Fig. 4a). Moreover, anti‐Ki67 staining did not reveal any remarkably differences in the amounts or localization of proliferating cells (Fig. 4b). Our attempt to visualize EGFP‐dnTCF protein in the crypt compartment using fluorescent or immunohistochemical detection was unsuccessful, probably due to the low sensitivity of staining in proliferating cells; nevertheless, EGFP‐dnTCF mRNA could be readily detected in total RNA isolated from freshly isolated crypts at day 4 upon (first) tamoxifen injection (Supporting Information Fig. S2). To analyze transcriptional changes in the crypts related to inhibition of Wnt/β‐catenin signaling, qRT‐PCR analysis was performed using total RNA isolated from small intestinal crypts. RNA samples were obtained from Rosa26tdTomato/tdTomato/Lgr5‐EGFP‐CreERT2+ and control Rosa26tdTomato/tdTomato mice injected with a single dose of tamoxifen and sacrificed one and 21 days later. In agreement with the fluorescent microscopy images, we observed temporal downregulation in mRNA levels of genes specifically expressed in cells located at the bottom of the crypt. The genes included Paneth cell markers ephrin receptor B3 (EphB3) and lysozyme 1 (Lyz1) (van Es et al., 2005) or genes with broader expression in various crypt cells such as division cycle associated 7 (Cdca7), ephrin receptor B2 (EphB2), naked cuticle homolog 1 (Nkd1), and SRY (sex determining region Y)‐box 9 (Sox9) (Batlle et al., 2002; Schuijers et al., 2015; Stancikova et al., 2015). Remarkably, the most robust downregulation was recorded for three genes that encode Wnt signaling‐activated markers of ISCs achaete‐scute complex homolog 2 (Ascl2), Lgr5, and Troy (Fafilek et al., 2013; van der Flier et al., 2009). Interestingly, olfactomedin 4 (Olfm4), which represents another ISC marker but whose expression is not regulated by Wnt signaling (van der Flier et al., 2009), was decreased only moderately. Finally, expression of keratin 20 (Krt20), which marks terminally differentiated cells, was at the first time point substantially increased, but at the later time point returned to its original level (Fig. 4c). To follow the fate of EGFP‐dnTCF4‐expressing ISCs, lineage tracing was performed using reporter mouse strain Rosa26R generated by “knock‐in” of the bacterial β‐galactosidase (LacZ) gene into the Rosa26 locus (the corresponding allele was designated as Rosa26R) (Soriano, 1999). Rosa26R/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ and control Rosa26R/+/Lgr5‐EGFP‐IRES‐CreERT2+ mice were injected with a single dose of tamoxifen and analyzed at several later time points. One day after tamoxifen administration, LacZ‐expressing cells were located at the bottom of the small intestinal crypts in both Rosa26R/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ and Rosa26R/+/Lgr5‐EGFP‐IRES‐CreERT2+ animals. In control mice, at day 4 (upon tamoxifen administration) LacZ‐positive cells emanated from the crypts, reaching the top of the villi at day 7. The streams of “blue” cells persisted in the tissue for at least 3 weeks, confirming the stem cell origin of the labeled clones. In contrast, in Rosa26R/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ animals, the situation was markedly different. At day 4 the labeled cells still persisted in the crypts; however, at day 7 the majority of LacZ‐positive cells were not localized to the crypts, but the cells moved (separately or as short “ribbons”) toward the top of the villi. Finally, 3 weeks upon tamoxifen administration, the small intestinal epithelium was practically devoid of blue cells except for some occasional clones that probably escaped recombination of the Rosa26tdTomato allele (Fig. 4d; detailed images of the crypts are given in Supporting Information Fig. S3).
Figure 3.
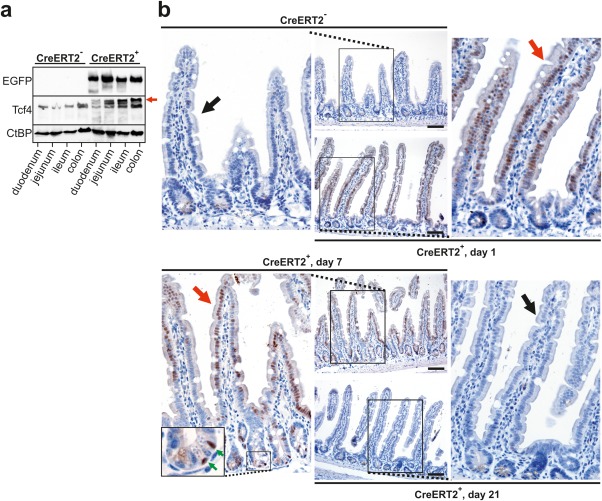
Transitory production of EGFP‐dnTCF protein in the intestine of Rosa26tdTomato/tdTomato/Villin‐CreERT2+ mice. Notice that in these mice EGFP‐dnTCF4 can be produced from both Rosa26 alleles. (a) Immunoblotting analysis using cell lysates prepared from the indicated parts of the intestinal epithelium of Rosa26tdTomato/tdTomato (CreERT2‐) and Rosa26tdTomato/tdTomato/Villin‐CreERT2+ (CreERT2+) mice one day after a single dose of tamoxifen. Notice that whereas an anti‐TCF4 antibody detected both endogenous Tcf4 and transgenic EGFP‐dnTCF4 protein (red arrow), the EGFP‐specific signal was visible in CreERT2+ animals only. Immunoblotting with an antibody recognizing C‐terminal binding protein (CtBP) was used as a loading control. (b) Immunohistochemical detection of EGFP‐dnTCF4 protein in the jejunum of CreERT2‐ and CreERT2+ mice using an EGFP‐specific antibody and 3,3'‐diaminobenzidine (DAB) staining (brownish precipitate); specimens were counterstained with hematoxylin (blue nuclei). CreERT2+ mice were sacrificed at days 1, 7, and 21 upon tamoxifen administration. Notice the positive nuclei of villus epithelial cells at days 1 and 7 (red arrows) and diminished nuclear EGFP‐dnTCF4 positivity in the intestinal epithelium of CreERT2‐ mouse or in CreERT2+ animal sacrificed at the last time point (black arrows). Green arrows in the inset placed in the bottom left image point to labeled Paneth cells. Scale bar: 0.15 mm.
Figure 4.
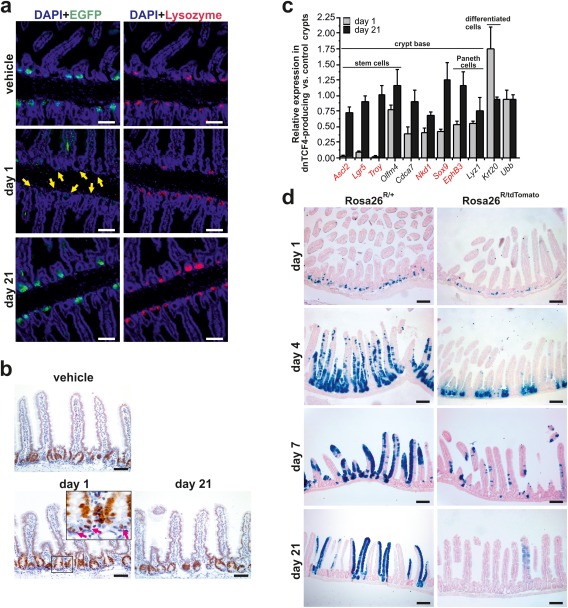
EGFP‐dnTCF4‐expressing ISCs do not contribute to tissue homeostasis. (a) Confocal microscopy images of immunohistochemical staining performed in the proximal part of the small intestine of Rosa26tdTomato/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ mice. Mice were injected by vehicle (mineral oil) only or by three consecutive doses of tamoxifen and analyzed next day or 21 days upon the first tamoxifen injection (indicated as “day 1” or “day 21,” respectively). Paraffin‐embedded sections were stained using an anti‐EGFP or anti‐lysozyme antibody to visualize ISCs (and/or EGFP‐dnTCF4‐producing cells; green fluorescence) and Paneth cells (red fluorescence). The specimens were co‐stained with DAPI nuclear stain (blue fluorescence), and merged images are shown. Notice the absence of (any) EGFP‐positive cells at the base of the crypts (yellow arrows) of “day 1” mice. (b) Distribution of proliferating Ki67‐positive cells in the small intestine of Rosa26tdTomato/tdTomato/Lgr5‐EGFP‐CreERT2+ mice. The animals were treated as in panel (a). Pink arrows in the inset indicate anti‐Ki67‐stained cells located at the base of the crypts. The specimens were counterstained with hematoxylin. (c) EGFP‐dnTCF4 alters gene expression signature in cells localized in the small intestinal crypts. Expression profiling of the small intestinal crypts isolated from Rosa26tdTomato/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ (CreERT2+) and control Rosa26tdTomato/tdTomato (CreERT2‐) mice. Animals in both groups were injected with a single dose of tamoxifen and analyzed 1 and 21 days later (in the diagram indicated as the “day 1” and “day 21” dataset, respectively). The relative expression levels (at given time points) of the indicated gene in CreERT2+ vs. CreERT‐ crypts are shown. Corresponding crossing point (Cp) values are given in Supporting Information Table S2. Genes directly activated by Wnt/β‐catenin signaling are in red. The quantities of input cDNAs were normalized to Actb mRNA. The relative expression of another housekeeping gene, Ubb, is also shown. The analysis was performed in technical triplicates; error bars represent SDs from three analyzed mice (for each genotype). (d) Lineage tracing experiment performed in the middle part of the small intestine of Rosa26R/+/Lgr5‐EGFP‐IRES‐CreERT2+ (designated as Rosa26R/+) and Rosa26R/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ (Rosa26R/tdTomato) mice. Specimens were obtained 1, 4, 7, and 21 days after a single tamoxifen injection. Three animals of each genotype were analyzed for the given time point; representative images are shown. Scale bar: 0.15 mm (a, b), 0.3 mm (d).
EGFP‐dnTCF4 Inhibits Tumor Growth in Conditional Apc Knockout Mice
Targeted inactivation of Apc in the mouse intestine using conditional (CKO) allele of the gene results in tissue hyperplasia accompanied by expansion of the stem cell compartment and tumor formation (Sansom et al., 2004). Deletion of Apc has to occur in ISCs; otherwise, the transformed cells fail to progress to malignancy (Barker et al., 2009). To test the ability of EGFP‐dnTCF4 to suppress tumor growth, the Apc gene was inactivated in Rosa26+/tdTomato/ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ and ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ mice. The animals were injected by a single dose of tamoxifen to induce Cre nuclear activity and their small intestine was analyzed 16 days later. Histological analysis of ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ mice revealed a continuous “zone” of transformed small intestinal epithelium as described previously (Barker et al., 2009). The intestinal tumors in Rosa26+/tdTomato/ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ animals were substantially smaller. Additionally, the lesions contained lower amounts of EGFP‐positive cells (Supporting Information Fig. S4). Similarly to the small intestine of the Rosa26R/tdTomato/Lgr5‐EGFP‐IRES‐CreERT2+ mice, we never detected a prominent nuclear signal specific for EGFP‐dnTCF4 protein. This indicated that the detected EGFP signal originated from the Lgr5‐EGFP‐IRES‐CreERT2 allele. Subsequently, we quantified the difference in tissue growth and proportion of proliferating cells in Rosa26+/tdTomato/ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ and ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ animals. Since distinct borders between individual neoplastic lesions could not be distinguished, the extent of tissue hyperplasia was determined by assessment of DAPI signal in fluorescent microscopy images. Additionally, by quantifying the ratio between DAPI (blue) and Ki67‐specific (green) fluorescent signal, cell proliferation was estimated using the same image series (Fig. 5b). As shown in Figure 5c, dnTCF4 production significantly reduced the average tissue area recorded in one microscopy image (0.23 mm2 vs. 0.53 mm2). Correspondingly, the proportion of Ki67‐positive cells was substantially lower in Rosa26+/tdTomato/ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ mice than in control ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ animals (7.88% vs. 24.97%). Taken together, our results indicate that suppression of the β‐catenin‐TCF/LEF signaling inhibited the clonogenic capacity of ISCs, which were unable to contribute to tissue homeostasis or—upon transformation—to initiate tumor growth.
Figure 5.
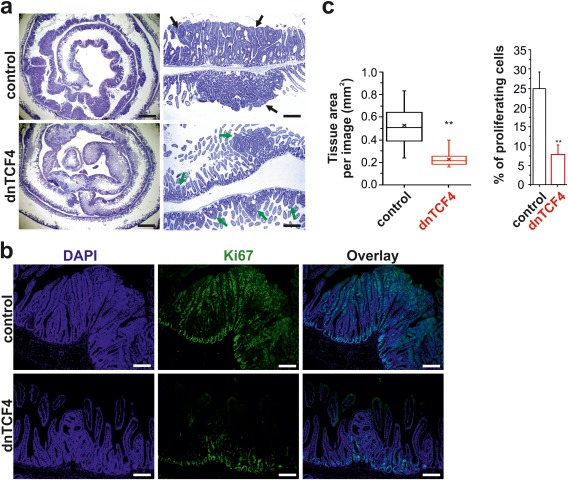
Decreased tumor formation in Apc‐deficient intestine of mice harboring the Rosa26dnTCF4 allele. (a) Left, representative microscopy images showing hematoxylin and eosin‐stained specimens of the rolled small intestine (the duodenum is centered) obtained from ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ (control) and Rosa26+/tdTomato/ApcCKO/CKO/Lgr5‐EGFP‐IRES‐CreERT2+ (dnTCF4) mice 16 days after a single dose of tamoxifen. Right, hematoxylin‐stained immunohistochemical sections prepared from the proximal part of the small intestine. Notice the differences in the size of the neoplastic adenomatous lesions formed in control (black arrows) and dnTCF (green arrows) mice. Scale bar: 3 mm (left image); 0.75 mm (right image). (b) Representative fluorescent microscopy images of the small intestinal sections stained with an anti‐Ki67 antibody to visualize proliferating cells; the specimens were counterstained with DAPI nuclear stain (corresponding DAB‐based staining is shown in Supporting Information Fig. S5). Scale bar: 0.3 mm. (c) Quantification of tissue hyperplasia induced by the loss of Apc. Ten sections taken along the longitudinal axis of the small intestine were analyzed from each animal; two control and two dnTCF4 mice were examined. Left, DAPI fluorescence was employed to quantify the total tissue area per one image. The boxed areas correspond to the second and third quartiles; the spread of the values is also shown. Median and average value is indicated by the transverse line and cross, respectively. Right, both DAPI and anti‐Ki67 fluorescence was utilized to quantify the proportion of proliferating cells in the tissue sections. **P < 0.01; error bars indicate SDs.
DISCUSSION
During the last several years, a wealth of results uncovered novel β‐catenin interactors and additional components and molecular mechanisms of the Wnt pathway activation. For example, besides the TCF/LEF proteins, β‐catenin binds and modulates activity of several other transcription factors including androgen receptor (AR), liver receptor homolog 1 (LRH1), hypoxia induced factor 1α (HIF1α), and Sox17 (SRY (sex determining region Y)‐box 17; reviewed in Valenta et al. (2012). Recently, Yes‐associated protein 1 (YAP1) and related transcriptional coactivator TAZ [alternative name for WW domain‐containing transcription regulator 1 (WWTR1)] were discovered as critical components of the β‐catenin destruction complex (Azzolin et al., 2014). Wnt signaling stabilizes TAZ and promotes TAZ‐dependent transcriptional activation of genes regulated by the TEA domain/Transcription Enhancer Factor (TEAD) family of transcription factors, the main binding partners of both TAZ and YAP1 (Azzolin et al., 2012). Moreover, β‐catenin and Yes‐associated protein (YAP) act as coactivators of the T‐BOX 5 (TBX5) nuclear factor (Rosenbluh et al., 2012). Although recent results of Park et al. questioned a direct participation of the β‐catenin destruction complex in regulation of the YAP/TAZ activity (Park et al., 2015), it is evident that Wnt signaling activates not only the β‐catenin‐TCF/LEF‐dependent program, but also responses mediated by additional DNA‐binding proteins and transcriptional mediators. The matter is even more complicated due to the fact that besides its engagement in transcriptional regulation, β‐catenin plays a structural role in E‐cadherin‐based adherens junctions. It is therefore difficult to relate the exact contribution of distinct, i.e. signaling or structural, β‐catenin functions to the various effects observed.
In the present study we utilized Cre/loxP‐mediated expression of dnTCF4, a truncated variant of the TCF4 nuclear factor, to study the effects of the canonical (i.e. β‐catenin‐TCF/LEF‐dependent) Wnt signaling pathway inhibition in the intestine. Importantly, our approach based on Cre/loxP‐mediated induction of EGFP‐dnTCF4 precluded any interference with additional β‐catenin functions. In addition, targeting the Rosa26 locus retained other genes encoding the Wnt pathway components or regulators intact. Thus, the experimental design was principally different from the methods used in previous reports. Consequently, differences in the observed phenotypes might be related to the particular methodological setup. Several lines of transgenic mice were used to trigger EGFP‐dnTCF4 expression in different cellular types and tissue. In agreement with previously published results, suppression of Wnt/β‐catenin signaling in the hematopoietic system did not affect lineage differentiation (Zhao et al., 2007); however, we observed clear interference between the EGFP‐dnTCF4 production and the process of leukemic transformation (Alberich‐Jorda and Korinek, unpublished results). Similarly, triggering EGFP‐dnTCF4 expression throughout the epithelia of the small and large intestine using Villin‐CreERT2 transgenic mice did not have any impact on the tissue. Nevertheless, since the EGFP‐dnTCF4 production was mainly detected in short‐lived differentiated cells localized outside of the crypts (Fig. 3b), i.e. in epithelial compartments that are not dependent on Wnt signaling, the absence of any defect was not surprising. Activation of the EGFP‐dnTCF4 transgene specifically in ISCs markedly changed the gene transcription profiles in the small intestinal crypts. As expected, ISC‐specific Wnt signaling target genes were amongst the most robustly downregulated genes. Of note, Lgr5‐EGFP‐IRES‐CreERT2 animals contain only one functional allele of the Lgr5 gene; however, similar results were obtained using Troy‐CreERT2 transgenic mice producing CreERT2 enzyme specifically in ISCs (Fafilek et al., 2013) (data not shown). In contrast, the Olfm4 gene, which represents an additional but Wnt signaling‐independent ISCs marker, was downregulated only moderately. The possible fate of ISCs could be deduced from the lineage tracing experiment in Rosa26R mice. The setup precluded production of EGFP‐dnTCF4 from both Rosa26 alleles; however, production of the Wnt inhibitor was sufficient to alter the ISC behavior. Strikingly, suppression of Wnt/β‐catenin did not impact proliferation but rather asymmetric division and/or positional information of ISCs. Modified ISCs underwent several rounds of division and migrated out of the crypt. Besides ISC markers, the EGFP‐dnTCF4 production modulated expression of other epithelial genes including Paneth cell‐specific genes EphB3 and Lyz1. The result implies that phenotypic changes (or demise) of ISCs impacts on transcriptional profiles of other crypts cells. The fact that (partial) loss of ISCs did not affect the tissue architecture is in accord with previous studies describing the effects of diphtheria toxin‐mediated elimination of ISCs (Buczacki et al., 2013; Tian et al., 2011). These studies showed that epithelial cells retain remarkable plasticity and upon ISC loss, crypt cells positioned above the stem and Paneth cell compartment can revert their phenotype and functionally substitute for the lost ISCs. These ISC‐substituting cells might be precursors of Paneth cells or other secretory lineages (Buczacki et al., 2013; Roth et al., 2012; van Es et al., 2012b) or TA cells (Asfaha et al., 2015).
The majority of human adenocarcinomas of the colon and rectum (colorectal carcinoma) display hyperactive Wnt signaling that is mainly related to the loss of the APC tumor suppressor. A minor fraction of colorectal carcinomas contains mutations inactivating additional components of the β‐catenin destruction complex AXIN1 or AXIN2, or harbors genetic changes leading to production of an oncogenic (stable) variant of β‐catenin (CancerGenomeAtlasNetwork, 2012). In either case, inappropriate transcription of the β‐catenin‐TCF/LEF target genes represents a hallmark of cancer cells transformed by aberrant Wnt signaling. This concept was recently questioned by a study showing that APC deficiency promotes signaling mediated by YAP. Intact APC protein decreases YAP activity through the Hippo pathway, i.e. independently of the β‐catenin destruction complex. Consequently, in APC‐deficient colorectal cancer cells, YAP activates a significant portion of genes related to tumor initiation and/or progression. Additionally, targeted inactivation of the Yap gene in the mouse intestine prevented formation of neoplasia related to Apc loss, supporting importance of the APC‐Hippo‐YAP axis in intestinal tumorigenesis (Cai et al., 2015). Using Apc conditional knockout mice we observed suppression of tumor growth upon EGFP‐dnTCF activation. Thus, it is evident that besides YAP signaling, the β‐catenin‐TCF/LEF‐mediated transcriptional program is indispensable for epithelial cell transformation.
Supporting information
Supporting Information
Supporting Information Figure S1
Supporting Information Figure S2
Supporting Information Figure S3
Supporting Information Figure S4
Supporting Information Figure S5
Supporting Information Table S1
Supporting Information Table S2
ACKNOWLEDGMENTS
The authors thank R. Tsien for tdTomato cDNA, S. Robine for Villin‐Cre mice, T. Stopka for Vav1‐cre mice, V. Divoky for LIF‐producing COS7 cells, and S. Takacova for critically reading the manuscript. The authors also thank Z. Kozmik for the idea to modulate the Wnt pathway output by gene knock‐in into the Rosa26 locus. The authors express their special thanks to the members of the Transgenic and Archiving Module, the Czech Centre for Phenogenomics, for generation of transgenic mice. L.J., B.F., and V.K. conceived and designed the experiments. L.J., B.F., M.H., M.K., M.V., and M.A‐J. performed the experiments. L.J., B.F., M.K., and V.K. analyzed the data. E.S., K.G., M.A., and R.S. provided administrative and technical support. L.J. and V.K. wrote the manuscript.
LITERATURE CITED
- Angus‐Hill ML, Elbert KM, Hidalgo J, Capecchi MR. 2011. T‐cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci USA 108:4914–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, Westphalen CB, von Burstin J, Mastracci TL, Worthley DL, Guha C, Quante M, Rustgi AK, Wang TC. 2015. Krt19(+)/Lgr5(‐) cells are radioresistant cancer‐initiating stem cells in the colon and intestine. Cell Stem Cell 16:627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, Fassina A, Cordenonsi M, Piccolo S. 2014. YAP/TAZ incorporation in the beta‐catenin destruction complex orchestrates the Wnt response. Cell 158:157–170. [DOI] [PubMed] [Google Scholar]
- Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. 2012. Role of TAZ as mediator of Wnt signaling. Cell 151:1443–1456. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. 2009. Crypt stem cells as the cells‐of‐origin of intestinal cancer. Nature 457:608–611. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007. [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. 2002. Beta‐catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111:251–263. [DOI] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. 2013. Intestinal label‐retaining cells are secretory precursors expressing Lgr5. Nature 495:65–69. [DOI] [PubMed] [Google Scholar]
- Cai J, Maitra A, Anders RA, Taketo MM, Pan D. 2015. beta‐Catenin destruction complex‐independent regulation of Hippo‐YAP signaling by APC in intestinal tumorigenesis. Genes Dev 29:1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CancerGenomeAtlasNetwork. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. 2012. Wnt/beta‐catenin signaling and disease. Cell 149:1192–1205. [DOI] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol 33:314–325. [DOI] [PubMed] [Google Scholar]
- Doubravska L, Krausova M, Gradl D, Vojtechova M, Tumova L, Lukas J, Valenta T, Pospichalova V, Fafilek B, Plachy J, Sebesta O, Korinek V. 2011. Fatty acid modification of Wnt1 and Wnt3a at serine is prerequisite for lipidation at cysteine and is essential for Wnt signalling. Cell Signal 23:837–848. [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. 2004. Tissue‐specific and inducible Cre‐mediated recombination in the gut epithelium. Genesis 39:186–193. [DOI] [PubMed] [Google Scholar]
- Fafilek B, Krausova M, Vojtechova M, Pospichalova V, Tumova L, Sloncova E, Huranova M, Stancikova J, Hlavata A, Svec J, Sedlacek R, Luksan O, Oliverius M, Voska L, Jirsa M, Paces J, Kolar M, Krivjanska M, Klimesova K, Tlaskalova‐Hogenova H, Korinek V. 2013. Troy, a tumor necrosis factor receptor family member, interacts with lgr5 to inhibit wnt signaling in intestinal stem cells. Gastroenterology 144:381–391. [DOI] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, Huelsken J. 2007. Wnt/beta‐catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol 27:7551–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. 1997. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J Biol Chem 272:23729–23740. [DOI] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. 1999. Temporally‐controlled site‐specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen‐inducible Cre‐ER(T) and Cre‐ER(T2) recombinases. Nucleic Acids Res 27:4324–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ. 2004. Inducible Cre‐mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta‐catenin. Gastroenterology 126:1236–1246. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. 1998. Depletion of epithelial stem‐cell compartments in the small intestine of mice lacking Tcf‐4. Nat Genet 19:379–383. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. 1997. Constitutive transcriptional activation by a beta‐catenin‐Tcf complex in APC‐/‐ colon carcinoma. Science 275:1784–1787. [DOI] [PubMed] [Google Scholar]
- Krausova M, Korinek V. 2014. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal 26:570–579. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf‐1. Proc Natl Acad Sci USA 101:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraguchi M, Wang XP, Bronson RT, Rothenberg R, Ohene‐Baah NY, Lund JJ, Kucherlapati M, Maas RL, Kucherlapati R. 2006. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet 2:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Mazna P, Valenta T, Doubravska L, Pospichalova V, Vojtechova M, Fafilek B, Ivanek R, Plachy J, Novak J, Korinek V. 2009. Dazap2 modulates transcription driven by the Wnt effector TCF‐4. Nucleic Acids Res 37:3007–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow‐Newerly W, Roder JC. 1993. Derivation of completely cell culture‐derived mice from early‐passage embryonic stem cells. Proc Natl Acad Sci USA 90:8424–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Kim YC, Yu B, Moroishi T, Mo JS, Plouffe SW, Meng Z, Lin KC, Yu FX, Alexander CM, Wang CY, Guan KL. 2015. Alternative Wnt Signaling Activates YAP/TAZ. Cell 162:780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 17:1709–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospichalova V, Tureckova J, Fafilek B, Vojtechova M, Krausova M, Lukas J, Sloncova E, Takacova S, Divoky V, Leprince D, Plachy J, Korinek V. 2011. Generation of two modified mouse alleles of the Hic1 tumor suppressor gene. Genesis 49:142–151. [DOI] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, Coffey RJ. 2012. The Pan‐ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. 2000. High‐efficiency deleter mice show that FLPe is an alternative to Cre‐loxP. Nat Genet 25:139–140. [DOI] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, Shao DD, Schumacher SE, Weir BA, Vazquez F, Cowley GS, Root DE, Mesirov JP, Beroukhim R, Kuo CJ, Goessling W, Hahn WC. 2012. beta‐Catenin‐driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151:1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Franken P, Sacchetti A, Kremer A, Anderson K, Sansom O, Fodde R. 2012. Paneth cells in intestinal homeostasis and tissue injury. PLoS One 7:e38965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon‐Assmann P, Clevers H, Nathke IS, Clarke AR, Winton DJ. 2004. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 18:1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J, Junker JP, Mokry M, Hatzis P, Koo BK, Sasselli V, van der Flier LG, Cuppen E, van Oudenaarden A, Clevers H. 2015. Ascl2 acts as an R‐spondin/Wnt‐responsive switch to control stemness in intestinal crypts. Cell Stem Cell 16:158–170. [DOI] [PubMed] [Google Scholar]
- Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21:70–71. [DOI] [PubMed] [Google Scholar]
- Stancikova J, Krausova M, Kolar M, Fafilek B, Svec J, Sedlacek R, Neroldova M, Dobes J, Horazna M, Janeckova L, Vojtechova M, Oliverius M, Jirsa M, Korinek V. 2015. NKD1 marks intestinal and liver tumors linked to aberrant Wnt signaling. Cell Signal 27:245–256. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. 2011. A reserve stem cell population in small intestine renders Lgr5‐positive cells dispensable. Nature 478:255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumova L, Pombinho AR, Vojtechova M, Stancikova J, Gradl D, Krausova M, Sloncova E, Horazna M, Kriz V, Machonova O, Jindrich J, Zdrahal Z, Bartunek P, Korinek V. 2014. Monensin inhibits canonical Wnt signaling in human colorectal cancer cells and suppresses tumor growth in multiple intestinal neoplasia mice. Mol Cancer Ther 13:812–822. [DOI] [PubMed] [Google Scholar]
- Valenta T, Hausmann G, Basler K. 2012. The many faces and functions of beta‐catenin. Embo J 31:2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon‐Pon‐Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. 2002. The beta‐catenin/TCF‐4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241–250. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. 2009. Transcription factor achaete scute‐like 2 controls intestinal stem cell fate. Cell 136:903–912. [DOI] [PubMed] [Google Scholar]
- van Es JH, Haegebarth A, Kujala P, Itzkovitz S, Koo BK, Boj SF, Korving J, van den Born M, van Oudenaarden A, Robine S, Clevers H. 2012a. A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self‐renewal. Mol Cell Biol. 32:1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, Taketo MM, Clevers H. 2005. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol 7:381–386. [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens AC, Barker N, van Oudenaarden A, Clevers H. 2012b. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14:1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445:661–665. [DOI] [PubMed] [Google Scholar]
- Waaler J, Machon O, Tumova L, Dinh H, Korinek V, Wilson SR, Paulsen JE, Pedersen NM, Eide TJ, Machonova O, Gradl D, Voronkov A, von Kries JP, Krauss S. 2012. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res 72:2822–2832. [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR 3rd, Nusse R. 2003. Wnt proteins are lipid‐modified and can act as stem cell growth factors. Nature 423:448–452. [DOI] [PubMed] [Google Scholar]
- Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. 2007. Loss of beta‐catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell 12:528–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information Figure S1
Supporting Information Figure S2
Supporting Information Figure S3
Supporting Information Figure S4
Supporting Information Figure S5
Supporting Information Table S1
Supporting Information Table S2


