Abstract
The molecular machinery of the cyanobacterial circadian clock consists of three proteins: KaiA, KaiB, and KaiC. Through interactions among the three Kai proteins, the phosphorylation states of KaiC generate circadian oscillations in vitro in the presence of ATP. Here, we characterized the complex formation between KaiB and KaiC using a phospho-mimicking mutant of KaiC, which had an aspartate substitution at the Ser431 phosphorylation site and exhibited optimal binding to KaiB. Mass-spectrometric titration data showed that the proteins formed a complex exclusively in a 6:6 stoichiometry, indicating that KaiB bound to the KaiC hexamer with strong positive cooperativity. The inverse contrast-matching technique of small-angle neutron scattering enabled selective observation of KaiB in complex with the KaiC mutant with partial deuteration. It revealed a disk-shaped arrangement of the KaiB subunits on the outer surface of the KaiC C1 ring, which also serves as the interaction site for SasA, a histidine kinase that operates as a clock-output protein in the regulation of circadian transcription. These data suggest that cooperatively binding KaiB competes with SasA with respect to interaction with KaiC, thereby promoting the synergistic release of this clock-output protein from the circadian oscillator complex.
Organisms on this planet exhibit circadian rhythms to adapt to daily alterations in the environment. Cyanobacteria are photoautotrophic organisms capable of oxygen-producing photosynthesis, similar to that observed in eukaryotic algae and plants, which exhibit circadian rhythms and have become one of the most useful model organisms for circadian biology1,2,3. The central oscillator that generates the circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942 comprises only three proteins—KaiA, KaiB, and KaiC4. Through interactions among these proteins in the presence of ATP, KaiC undergoes phosphorylation and dephosphorylation cycles with the period of 24 h, which proceeds in vitro without daylight oscillation, indicating that the internal clock mechanism can be autonomous irrespective of transcriptional and translational feedback systems5,6,7.
KaiC forms a hexametric ring consisting of two rings—CI and CII2,3. Two specific residues positioned in the CII ring, S431 and T432, are phosphorylated and dephosphorylated in a 24-hour periodic manner5,6,8,9 as follows: KaiC-S/T → KaiC-S/pT → KaiC-pS/pT → KaiC-pS/T → KaiC-S/T, where S and T represent Ser431 and Thr432, respectively, and ‘p’ represents the phosphorylated residue. A series of phospho-mimicking KaiC mutants have generally been used for characterizing the functional roles of phosphorylation and the corresponding structural snapshots of the particular clock phase10,11,12. KaiA stimulates KaiC phosphorylation through the interaction of the A loops on the CII ring13, whereas KaiB accelerates the dephosphorylation process by interacting with the CI ring of phosphorylated KaiC14,15,16. During the latter process, KaiB undergoes a fold switch coupled with a transition from its homotetramerameric state to the KaiB–KaiC complex10. Formation of the KaiB–KaiC complex precludes the direct KaiA–KaiC interaction by promoting direct binding of KaiA to KaiB in the complex, thereby initiating a phase proceeding in the circadian cycle10,16. Furthermore, KaiB competes with SasA for binding to the KaiC hexamer. SasA is a histidine kinase that operates as a major clock-output protein that is released from the KaiABC clock oscillator during the regulation of circadian transcription10,13,17. Hence, the KaiB–KaiC interaction is a key event during oscillation of the cyanobacterial circadian protein system. Recent mass spectrometry (MS) analysis showed that KaiB can bind to phosphorylated KaiC at a stoichiometry of 6:618. To further understand the circadian clock mechanisms of KaiB–KaC assembly, we collected detailed structural information on the protein complexes using inverse contrast-matching small-angle neutron scattering (iCM-SANS) in conjunction with native MS analyses.
Results and Discussion
Analysis of the interaction between KaiB and KaiC mutants
In this study, KaiCAA, KaiCSE, KaiCDT, and KaiCDE were used as phospho-mimicking mutants to examine which phosphorylation form of KaiC exhibits the highest binding affinity to KaiB. KaiCDT and KaiCSE are mono-phosphorylated KaiC mimics at Ser431 and Thr432, respectively, whereas KaiCDE and KaiCAA are hyperphosphorylated and hypophosphorylated mutants regarding these sites.
Previous studies have indicated that KaiB is apt to bind phosphorylated KaiC mutants in which glutamate or aspartate residue occupies position 431 (e.g., KaiCEE, KaiCDE, KaiCEA, or KaiCDT)9,19,20. Prior to our detailed structural analyses, we used Blue native-polyacrylamide gel electrophoresis (BN-PAGE) to investigate the binding abilities of KaiB to a series of KaiC phospho-mimic mutants, in which the KaiC hexamer formed two conformational states: ground-state (gs)-KaiC, which was stable, and competent-state (cs)-KaiC (Fig. 1), which was labile and degraded into monomers upon binding of Coomassie Brilliant Blue)21. KaiB forms stable complexes with wild-type KaiC (KaiCWT) as well as KaiCDE and KaiCDT mutants. KaiCDT mimicked an optimally phosphorylated form of KaiC in terms of KaiB binding in comparison with KaiCWT and the other KaiC mutants, KaiCDE, KaiCAA, and KaiCSE (Fig. 1). Therefore, the interaction between KaiB and KaiCDT was further characterized.
Figure 1. KaiCDT predominantly binds to KaiB.
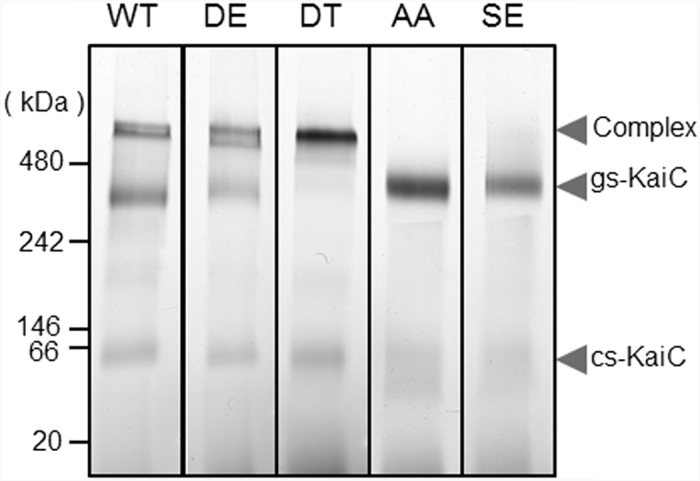
KaiCWT, KaiCDE, KaiCDT, KaiCAA, and KaiCSE were incubated with KaiB at 30 °C for 12 h and were subjected to BN-PAGE. The upper and lower bands with respect to the 480-kDa position corresponded to KaiB–KaiC complex and ground-state (gs)-KaiC, respectively. The 66-kDa protein bands corresponded to the competent state (cs)-KaiC. The gs-KaiC and cs-KaiC were donated in the previous work21. The full-length gels of BN-PAGE are presented in Supplemental Fig. S1.
Oligomeric state of the KaiB–KaiC complex
To characterize the oligomeric state of the KaiB–KaiC complex in solution, we performed native MS and sedimentation velocity analytical ultracentrifugation (SV-AUC) analyses. Our native MS data indicated that upon titration with KaiB, the KaiCDT hexamer formed a uniform complex with a molecular mass of 428,600 ± 410 Da, which corresponded to a 6:6 stoichiometry (Fig. 2 and Supplemental Fig. S2), consistent with previous native MS data obtained using hyperphosphorylated KaiCWT18. Our titration results underscore the fact that the complex formed between KaiB and KaiCDT only existed in a 6:6 stoichiometry, indicating that the six KaiB protomers bind to the KaiC hexamer with strong positive cooperativity. The SV-AUC data confirmed that KaiCDT forms a homogeneous hexamer and that KaiB and KaiCDT form a 6:6 complex with a sedimentation coefficient of 13.7 S (Supplemental Fig. S3).
Figure 2. Characterization of the KaiB–KaiCDT complex.
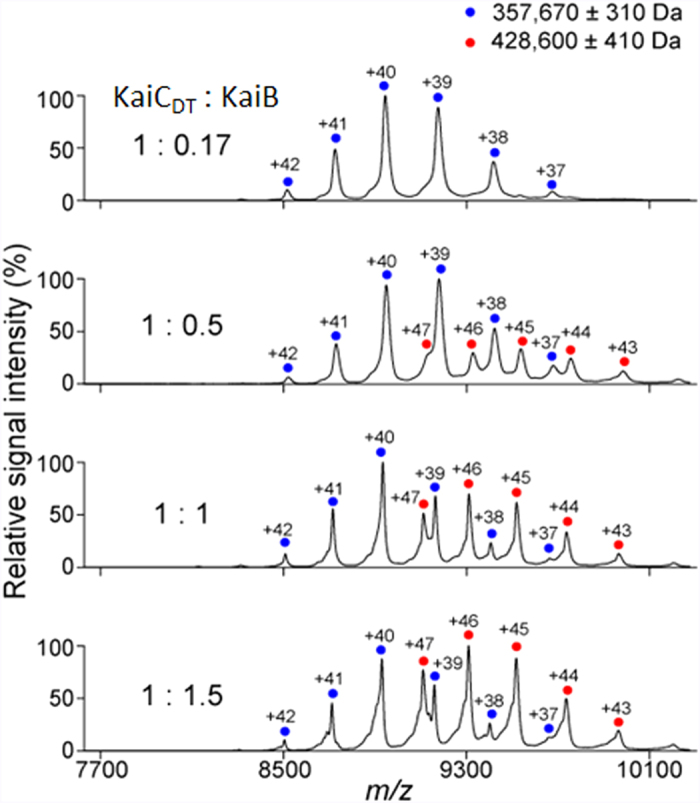
Mass spectra of mixtures of KaiCDT and KaiB at 1:0.17, 1:0.5, 1:1, and 1:1.5 molar ratios (KaiCDT to KaiB). Blue and red circles show the ion series of the KaiCDT homo-hexamer and the 6:6 hetero-dodecamer complexes of KaiCDT and KaiB, respectively.
Spatial arrangement of the KaiB subunits in the KaiB–KaiC complex
We analyzed the spatial arrangement of the KaiB subunits in the 6:6 KaiB–KaiCDT complex in an aqueous solution by small-angle scattering. In this approach, the scattering intensity arising from each domain is proportional to the square of its molecular mass. This means that in conventional small-angle scattering such as small-angle X-ray scattering, scattering from the much larger KaiC (with a molecular mass of 65 kDa) dominates that of KaiB (with a molecular mass of 14 kDa). Therefore, it is difficult to collect structural information on KaiB from the scattering profile of the complex. Our strategy for overcoming this difficulty was to weaken or erase scattering from the KaiC subunits in the complex using the iCM-SANS method22. In neutron scattering, an isotope effect is most remarkable between hydrogen and deuterium: the scattering lengths of hydrogen and deuterium are −3.74 fm and +6.67 fm, respectively. This enables control of the scattering length densities of the solvent and the properly deuterated protein components in the complex23,24, facilitating elimination of the scattering originating from those protein components22.
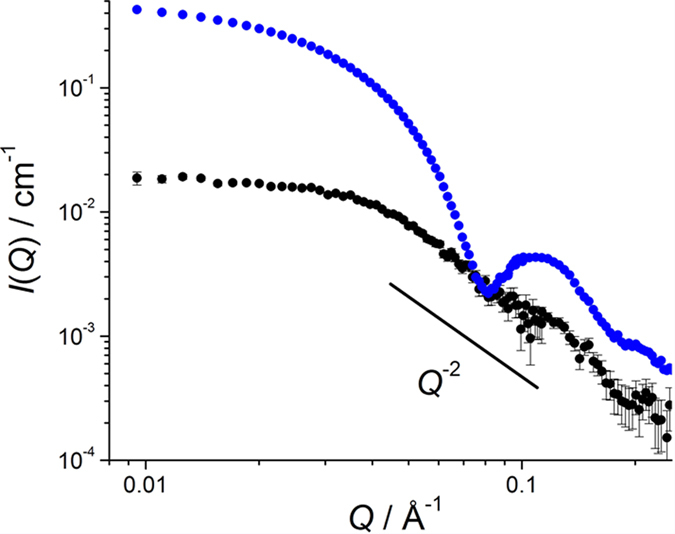
In this study, we prepared the 6:6 KaiB–KaiCDT complex consisting of the partially deuterated KaiCDT subunits and the nondeuterated KaiB (h-KaiB) subunits. The solvent D2O ratio at the contrast matching point was determined to eliminate scattering originating from the partially deuterated KaiCDT subunits. The observed SANS intensity at 0.029 Å−1, IO(0.029), of the partially deuterated KaiCDT protein alone dissolved in 0% D2O at a concentration of 1 mg/mL, was found to be 0.575 ± 0.003 cm−1 (Supplemental Fig. S4). In addition, IC(0.029) was calculated as a function of the protein deuteration ratio based on the amino-acid sequence of KaiCDT. From the square roots of IO(0.029) and IC(0.029), the deuteration ratio of the partially deuterated KaiCDT preparation was found to be 72.2% (Supplemental Fig. S5). Hereafter, this KaiCDT preparation will be referred to as 72d-KaiCDT. In 97% D2O solvent, 72d-KaiCDT was virtually invisible in terms of scattering because the scattering length density of KaiCDT was matched to that of the solvent (Supplemental Figs S4 and S6). As a result, we could clearly exclusively observe the neutron scattering of KaiB subunits in the complex. Figure 3 compares the SANS profiles of 72d-KaiCDT+h-KaiB and h-KaiCDT+h-KaiB complexes in 97% D2O. The scattering of 72d-KaiCDT+h-KaiB complex showed marked differences from that of the h-KaiCDT+h-KaiB complex in which both KaiB and KaiC fully contributed to scattering. In addition, the scattering profile of the 72d-KaiCDT+h-KaiB complex approximately followed the Q−2 power law indicating that the six KaiB subunits were arranged into a disk-like shape25.
Figure 3. SANS profiles of 72d-KaiCDT+h-KaiB (black) and h-KaiCDT+h-KaiB (blue) complexes in 97% D2O.
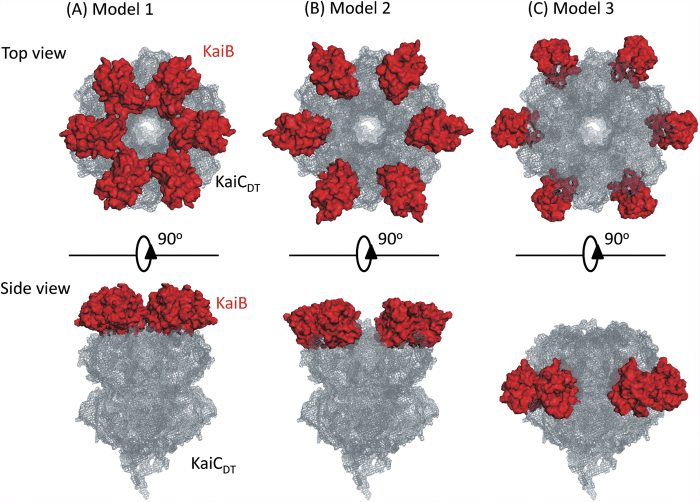
With the assumption of six-fold symmetry of arrangement of the six KaiB subunits along with their KaiC-binding surfaces consistent with the previously reported deuterium exchange mass spectrometric data10, we examined the following three distinct structural models of the KaiB–KaiCDT complex: Model 1: The KaiB subunits form a hexameric ring on the top of the KaiC C1-ring. The distance between the center of mass of each KaiB component and the six-fold axis of KaiCDT is set to be 33 Å, which is the minimum distance with avoidance of steric hindrance between the KaiB subunits (Fig. 4A). Model 2: The KaiB subunits are located on the edge of the KaiC C1-ring. The distance between the center of mass of KaiB and the six-fold axis of KaiCDT is set to be 45 Å, which is the maximum distance with direct interactions between the KaiB and KaiCDT components (Fig. 4B). Model 3: The KaiB subunits are located on the side of the KaiC C1-ring with 60 Å of the distance between center of mass of KaiB and the six-fold axis of KaiCDT (Fig. 4C).
Figure 4. Structural models of the KaiB–KaiCDT complex.
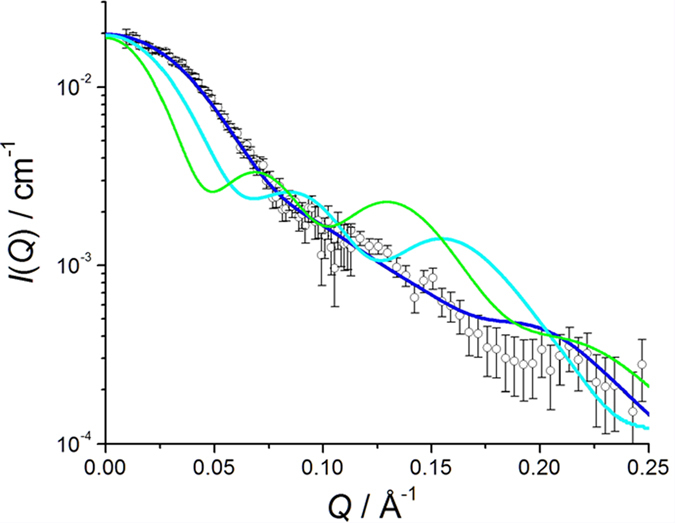
The scattering profiles with 72d-KaiC and h-KaiB in 97% D2O were calculated for each model and compared with the experimental data (Fig. 5). The results clearly showed that Model 1 illustrated the best structural arrangement of the KaiB subunits in the complex. Furthermore, the experimentally determined gyration radius, 33.6 Å, was consistent with that computed from Model 1, 33.2 Å, in comparison with those from Model 2 (44.2 Å) and Model 3 (59.4 Å). The hexameric KaiB ring forming on the KaiC hexamer was consistent with C6-symmetrized particle-image-based cryoEM structure26. In this interaction mode, the hexameric KaiB subunits cover the top surface of the KaiC C1 ring including the interaction site of SasA10. Therefore, it is conceivable that the cooperative binding of KaiB as competitive inhibitor against SasA promotes synergistic release of this clock-output protein from the KaiC hexamer.
Figure 5. SANS profiles calculated from Model 1 (blue), Model 2 (cyan), and Model 3 (green) along with the experimentally obtained profile (open circle).
In summary, we obtained structural information on the KaiB–KaiC complexes using iCM-SANS in conjunction with native MS analyses, providing insights into the working mechanisms of the circadian clock comprised of Kai proteins.
Methods
Protein expression and purification
KaiA, KaiB, and KaiC originating from S. elongatus PCC 7942 were expressed in Escherichia coli as Strep-tagged recombinant proteins and purified as previously described21. The expression plasmids of the KaiC mutants (KaiCDE, KaiCDT, KaiCAA, and KaiCSE) were also constructed according to a previous study21. KaiCDE is a mutant of KaiC with aspartate and glutamate residues at positions 431 and 432, respectively, that mimics hyperphosphorylated KaiC, whereas KaiCAA is a mutant with alanine residues at these positions that mimics hypophosphorylated KaiC. For preparation of the deuterated proteins, the bacterial cells were grown in M9 minimal media containing glucose as a mixture with varying ratios of isotopically natural and fully deuterated glucose (1,2,3,4,5,6,6-D7, 98%, Cambridge Isotope Laboratories, Inc.), along with varying ratios of H2O and D2O as previously described23,24.
BN-PAGE
BN-PAGE was performed using a NativePAGETM Novex Bis-Tris Gel System (Invitrogen) according to the manufacturer’s protocols with some modifications as previously described21.
Analytical ultracentrifugation
The sedimentation velocity method was used to characterize KaiB, KaiCDT, and their complex. Concentrations of KaiB, KaiCDT, and their complex were 40 μM, 30 μM, and 60 μM, respectively, in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM ATP, 5 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA. The samples were placed in analytical cells with double-sector centerpieces with sapphire windows. The experiments were performed using an Optima XL-I analytical ultracentrifuge (Beckman-Coulter) at 20 °C and an angular velocity of 50,000 rpm for KaiB, 30,000 rpm for KaiCDT and 20,000 rpm for their complex. Data were recorded with Rayleigh interference optical system, followed by the analysis with a c(s) distribution of the Lamm equation solutions calculated by the Sedfit v 14.427. The partial specific volume of the KaiB, 0.7556 ml/g, KaiCDT, 0.7361 ml/g, and their complex, 0.7458 ml/g were calculated from their amino acid compositions using the program SEDNTERP ver.1.09, buffer density (1.00540 g/cm3), and buffer viscosity (1.0245 cP) were estimated using the program SEDNTERP ver.1.09.
MS under non-denaturing conditions
The purified KaiCDT and KaiB proteins (20 μM and 3.3, 10, 20, and 30 μM monomer, respectively) were mixed, incubated at 30 °C for 4 h, and buffer-exchanged into 150 mM ammonium acetate, pH 7.5, by passing the proteins through a Bio-Spin 6 column (Bio-Rad). The buffer-exchanged KaiCDT–KaiB complexes were immediately analyzed by nanoflow electrospray ionization MS using gold-coated glass capillaries made in house (approximately 2–5 μL sample loaded per analysis). Spectra were recorded on a SYNAPT G2-Si HDMS mass spectrometer (Waters, Milford, MA, USA) in positive ionization mode at 1.33 kV with a 150 V sampling cone voltage and source offset voltage, 0 V trap and transfer collision energy, and 5 mL/min trap gas flow. The spectra were calibrated using 1 mg/mL cesium iodide and analyzed using MassLynx software (Waters).
SANS measurements
SANS experiments were performed using the D22 instrument installed at the Institut Laue-Langevin (ILL), Grenoble, France. The SANS intensities were observed with 6.0 Å neutrons and two sample-to-detector distances of 5.6 m and 2.0 m: the covered q-ranges are 0.0095 to 0.25 Å−1. The temperature was maintained at 20 °C in the irradiation. The observed SANS intensity was corrected for background, empty cell and buffer scatterings, and transmission factors and subsequently converted to the absolute scale by GRASP software using incident beam flux (http://www.ill.eu/instruments-support/instruments-groups/groups/lss/grasp/home/). For SANS measurements, 1–2 mg/mL KaiC was dissolved in buffer containing 50 mM NaH2PO4 (pH 7.8), 150 mM NaCl, 1 mM ATP, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 50 mM L-arginine, 50 mM L-glutamic acid, and various concentration of D2O (ISOTECH) in presence and absence of KaiB. All SANS samples are listed in Table S1.
3D structure modeling
For simulation of the SANS profile of KaiB, the 3D-structural model of a fold switch KaiB with strept tag was developed on the basis of SWISS-MODEL28,29,30. Furthermore, 3D models of the KaiB–KaiC complex were built by arranging six KaiB subunits on hexametric KaiC ring with a pseudo-sixfold symmetry. In all three models, the interaction surface of KaiB is facing KaiCDT (Supplemental Fig. S7).
Additional Information
How to cite this article: Sugiyama, M. et al. Structural characterization of the circadian clock protein complex composed of KaiB and KaiC by inverse contrast-matching small-angle neutron scattering. Sci. Rep. 6, 35567; doi: 10.1038/srep35567 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants (24310068, 26102524, and 15H02042 to M.S., 15K18515 to R.I., 15K18492 to R.M., 26102543 and 16H00784 to K.T., 16J08362 to K.O., 25102001, 25102008, and 15H02491 to K.K. and 25121722, 26102530, 15K14457 and 16H00770 to S.U.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, by the Okazaki ORION project, and by the Joint Studies Program in the Okazaki BIO-NEXT project of the Okazaki Institute for Integrative Bioscience. The SANS experiments at ILL were performed under proposal No. 8-03-838.
Footnotes
Author Contributions Conceived and designed experiments: M.S., H.Y. and K.K. Prepared recombinant proteins and the complex: Y.Y., R.M., K.T., H.Y., K.O. and K.T. Performed native PAGE: K.O. and K.T. Performed analytical ultracentrifugation and mass spectrometry experiments: M.N., K.I. and S.U. Performed SANS experiments and analysis: M.S., H.Y., Y.Y., L.P., A.M., R.I., N.S. and Y.O. Wrote the paper: M.S., H.Y. and K.K.
References
- The Cyanobacteria: Molecular Biology, Genomics and Evolution, (Caister Academic Press, Norfolk, UK, 2008). [Google Scholar]
- Pattanayek R. et al. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell 15, 375–388 (2004). [DOI] [PubMed] [Google Scholar]
- Xu Y. et al. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA 101, 13933–13938 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M. et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 (1998). [DOI] [PubMed] [Google Scholar]
- Rust M. J., Markson J. S., Lane W. S., Fisher D. S. & O’Shea E. K. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318, 809–812 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M. et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005). [DOI] [PubMed] [Google Scholar]
- Tomita J., Nakajima M., Kondo T. & Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254 (2005). [DOI] [PubMed] [Google Scholar]
- Nishiwaki T. et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA 101, 13927–13932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T. et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J 26, 4029–4037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. G. et al. Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349, 324–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe J. et al. Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316 (2015). [DOI] [PubMed] [Google Scholar]
- Murayama Y. et al. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J 30, 68–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R. et al. Combined SAXS/EM based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS One 6, e23697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama H. et al. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell 23, 161–171 (2006). [DOI] [PubMed] [Google Scholar]
- Kitayama Y., Iwasaki H., Nishiwaki T. & Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J 22, 2127–2134 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh R., Nishimura A., Yasui S., Onai K. & Ishiura M. The ATP-mediated regulation of KaiB-KaiC interaction in the cyanobacterial circadian clock. PLoS One 8, e80200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R. et al. The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J Biol Chem 287, 29506–29515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J. et al. Insight into cyanobacterial circadian timing from structural details of the KaiB-KaiC interaction. Proc Natl Acad Sci USA 111, 1379–1384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA 107, 14805–14810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Chew J., Chockanathan U. & Rust M. J. Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc Natl Acad Sci USA 111, E3937–E3945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama K., Azai C., Nakamura K., Tanaka S. & Terauchi K. Conversion between two conformational states of KaiC is induced by ATP hydrolysis as a trigger for cyanobacterial circadian oscillation. Sci Rep 6, 32443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M. et al. Conformational characterization of a protein complex involving intrinsically disordered protein by small-angle neutron scattering using the inverse contrast matching method: a case study of interaction between α-synuclein and PbaB tetramer as a model chaperone. J. Appl. Crystallography 47, 430–435 (2014). [Google Scholar]
- Sugiyama M. et al. Kinetic asymmetry of subunit exchange of homooligomeric protein as revealed by deuteration-assisted small-angle neutron scattering. Biophys J 101, 2037–2042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M. et al. Spatial arrangement and functional role of a subunits of proteasome activator PA28 in hetero-oligomeric form. Biochem Biophys Res Commun 432, 141–145 (2013). [DOI] [PubMed] [Google Scholar]
- Schmidt W. P. Some fundamental concepts and techniques useful in small-angle scattering studies of disordered solids. In Modern aspects of small-angle scattering (ed. Brumberger H.) xv, 463 p. (Kluwer Academic Publishers, Dordrecht; Boston, 1995). [Google Scholar]
- Villarreal S. A. et al. CryoEM and molecular dynamics of the circadian KaiB-KaiC complex indicates that KaiB monomers interact with KaiC and block ATP binding clefts. J Mol Biol 425, 3311–3324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F., Arnold K., Kunzli M., Bordoli L. & Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37, D387–D392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J. & Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.