Abstract
Virus-based nanomaterials are versatile materials that naturally self-assemble and have relevance for a broad range of applications including medicine, biotechnology, and energy.
This review provides an overview of recent developments in “chemical virology.” Viruses, as materials, provide unique nanoscale scaffolds that have relevance in chemical biology and nanotechnology, with diverse areas of applications. Some fundamental advantages of viruses, compared to synthetically programmed materials, include the highly precise spatial arrangement of their subunits into a diverse array of shapes and sizes and many available avenues for easy and reproducible modification. Here, we will first survey the broad distribution of viruses and various methods for producing virus-based nanoparticles, as well as engineering principles used to impart new functionalities. We will then examine the broad range of applications and implications of virus-based materials, focusing on the medical, biotechnology, and energy sectors. We anticipate that this field will continue to evolve and grow, with exciting new possibilities stemming from advancements in the rational design of virus-based nanomaterials.
1. Introduction
Nanoscale engineering is revolutionizing diverse disciplines in science and engineering. The use of viral scaffolds in particular has led to advancements of scientific knowledge in self-assembly and the development of novel materials with wide-ranging applications. Viruses have been studied for more than 100 years, and more than 5,000 types of viruses have been discovered and described. They come in a variety of shapes and sizes, and from a chemist's point of view they harbor many natural features that are uniquely relevant to nanotechnology and nanoscience. To date, it has not been feasible to synthetically create nanoparticles of comparable reproducibility, beauty, and utility. In a collaborative effort, research into “physical or chemical virology” is directed toward unraveling the processes of self-assembly and genome packaging, understanding and controlling self-assembly of virus-based materials into higher-order hierarchical structures, engineering and studying virus-based and virus-like materials for applications in the health and energy sectors, and scaled-up manufacturing of such materials for applications in clinics and in devices. In this review, we provide a general synopsis of the engineering of virus-based and virus-like materials and we will discuss the manifold and diverse applications of such. We start by introducing the use of viruses from a materials perspective and consider the methods for producing and modifying these particles. We then survey some recent developments in the expansion of their applications, with discussion focused on the utilization of virus-based materials for medicine (delivery systems and contrast agents), biotechnology (nanoreactors and sensing devices), and energy (battery electrodes and storage devices). Finally, we assess the opportunities and challenges for clinical or commercial application of virus-inspired materials.
2. Viruses in a materials world
Viruses usually bring to mind devastating disease and bear a negative connotation,1-3 especially with the recent outbreak of Ebola in 2014 that spread so quickly and proved difficult to control,4 as well as the current Zika virus outbreak that poses issues with microcephaly in newborns and may also possibly be linked to an increased risk of Guillain–Barré syndrome.5 Throughout history, infectious disease has plagued us, with the earliest recordings found from over 3000 years ago of smallpox in Egypt, India, and China.6 In fact, the mummy of Pharaoh Ramses V, who died around 1157 BC, possesses pustules and scarring reminiscent of smallpox infection. However, viruses also have positive qualities, and there have been many advances made in recent years in which nonpathogenic viruses and engineered virus-based nanomaterials have been utilized as three-dimensional scaffold materials for diagnostic and drug delivery systems as well as technological devices. Viruses were discovered to exist in 1892, and the first virus studied was the plant virus tobacco mosaic virus (TMV).7 It was not long after the discovery of viruses that they were considered for use in biotechnology and medicine. Early in the twentieth century, Frederick Twort and Felix d'Herelle independently reported the presence of bacterial viruses, or bacteriophages, and the idea of phage therapy to treat bacterial infections quickly took shape in the 1920's, although it was mainly practiced in the Soviet Union.8 The development of antibiotics largely overshadowed phage therapy, but there may be a comeback due to the increasing prevalence of antibiotics resistance,9 with benefits of phage therapy including greater specificity, lower toxicities, ability to disrupt bacterial biofilms, and ability to evolve to combat resistance.10
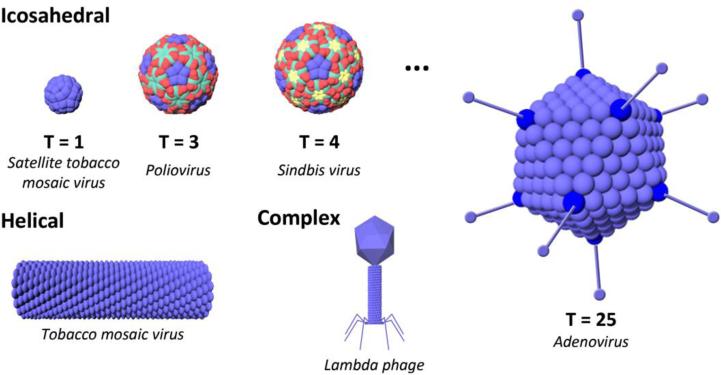
Aside from phage therapy, there are many other avenues for the use of viruses, and vaccines and gene therapy are likely the first applications that come to mind. However, the potential applications and current developments reach much farther. Around 2000, a group of researchers that included chemists, structural biologists, and virologists gave birth to a new field in which viruses are used for nanotechnology by demonstrating the ability to encapsulate materials within the capsid, address them chemically, and order them into crystal structures.11-14 In this manner, viruses can simply be used as well-ordered materials, separate from their normal role in infection. Most viruses are made up of coat protein subunits that naturally self-assemble into truly monodisperse particles. With more understanding of the coat protein building blocks and chemical biology, ever increasing complex assemblies can be programmed, including nanoboomerang- and tetrapod-shaped virus materials.15 Large-scale production of viruses can be easily achieved through propagation in their natural hosts or expression in a heterologous system (see Section 2.2). Additionally, these particles come in a variety of shapes and sizes16, 17 that can function as nanoscaffolds that can be easily and reproducibly modified18. As shown in Figure 1, the most common architectures are icosahedrons, filaments, and phage head-and-tail structures, but more diverse structures such as spindle-, zipper-, and bottle-shaped viruses also exist.17, 19
Figure 1. Some common viral architectures.
Viruses come in diverse shapes and sizes, with icosahedral and helical symmetries as well as more complex head-to-tail assemblies. For icosahedral viruses, examples of different triangulation numbers are shown (explained in Section 2.1), giving rise to different capsid sizes and structures. An example of a virus for each architecture is given in italics below the figures.
While there is the biotechnology arm where we seek to engineer particles for applications in medicine and energy, there is also a basic arm that investigates virus assembly and structure. These two arms of research are interconnected, with crosstalk between the two fields providing insights for advancement. For example, study of the physics of the packing signals of RNA viruses led to its application in the encapsulation of therapeutics for nanomedical applications (see Sections 4.1.4-4.1.5).14, 20 Additionally, fundamental understanding of the interactions involved in particle self-assembly informed the fabrication of novel imaging agents (see Section 4.1.2).21, 22 Through multidisciplinary collaboration, the use of viral scaffolds as unique materials for diverse applications can be realized.
2.1 Classification of viruses
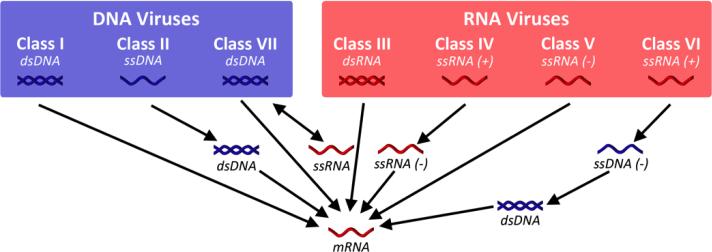
To differentiate between viruses containing their native nucleic acid, which are referred to as viral nanoparticles (VNPs), viruses devoid of their nucleic acid are considered virus-like particles (VLPs). Further classification of viruses can be based on a number of features, including the shape and structure of their capsids (as shown in Figure 1), the type of nucleic acid they contain (double-stranded (ds) or single-stranded (ss), RNA or DNA), and their host species. Classical virology taxonomy utilizes the Baltimore classification of viruses, in which the viruses are grouped both according to their genomes as well as their method of replication.23 Figure 2 illustrates the seven different classifications of viruses, demonstrating how they have evolved many different strategies for replication. However, for the most part, we will be considering plant viruses and bacteriophages (noninfectious particles) for use as materials, making the native cargo of the capsids less relevant. Mammalian viruses, such as adenovirus (class I – dsDNA virus) and adeno-associated virus (AAV, class II – ssDNA virus), do offer many advantages for applications in gene therapy, in which they can be administered to make modifications to the genetic sequence for therapeutic or prophylactic purposes (see Section 4.1.4).24 They also present opportunities in cancer immunotherapy, as seen in the recent approval of T-VEC for treatment of melanoma (see Section 4.1.3). Nevertheless, bacteriophage- and plant virus-derived materials may offer advantages, as their manufacture is scalable through fermentation and molecular farming. Additionally, these materials are not infectious toward mammals, adding another layer of safety. Both these factors are important considerations as we move toward clinical applications and commercialization.
Figure 2. Baltimore classification of viruses.
With the Baltimore classification, viruses are classified based on their genomic material as well as their method of replication.
Other than genomic content and host species, shape and size are important characteristics that should be considered for the choice of material used. Section 4.1.1 highlights some of the design guidelines for determining the desired properties for delivery vectors for nanomedical applications. For icosahedral viruses, the triangulation number, or T number, is one method of classification that gives an indication as to their size (see Figure 1). The T number was first described by Donald Caspar and Aaron Klug in 1962.25 By multiplying by 60, it can be used to determine the number of coat proteins in a capsid. For example, a T = 1 virus has 60 coat proteins, while a T = 4 virus has 240 coat proteins. The proteins are clustered into pentamers and hexamers, and a virus with icosahedral symmetry therefore consists of 12 pentamers and 10(T-1) hexamers. Size plays a factor in the transport and clearance behavior of a particle, as well as the amount of cargo that can be carried and delivered to a cell, a challenge with the smaller AAV (T = 1, ~20 nm in diameter). Additionally, the shape of the particles affects the possible modifications and functions that could be applied to the capsid. For example, icosahedrons have the advantage of possessing an interior cavity that can be used for the infusion and encapsulation of various payloads. On the other hand, high aspect ratio particles can be used to form wires, which can then be applied for energy applications. Overall, it is clear that there is a diverse library of virus particles to select from, no matter the application.
2.2 Production of viruses: fermentation, farming, and cell culture
A variety of methods have been developed for the production of virus particles, and we will discuss their manufacture in bacteria, yeast, insect cells, plants, and using cell-free systems, starting with one of the most widely utilized systems for the rapid production of proteins with ease of scale-up, Escherichia coli.26 Although other prokaryotic systems can also be considered, such as Pseudomonas fluorescens,27 the wealth of knowledge surrounding E. coli production makes it popular, and therefore it has also been widely applied for the production of VLPs. Viral coat proteins can be expressed and spontaneously self-assembled in the bacterial cells, and this has been demonstrated for bacteriophages, such as Qβ28 and MS2,29 as well as for heterologous expression of other viruses, such as plant virus-based TMV30 and mammalian virus-based hepatitis B virus (HBV) core particles.31
It should be noted that while VLPs formed in this way do not contain their own genomic content, they are also not “empty”, as they tend to package host nucleic acids. For example, about 25% of the mass of Qβ VLPs (a system that has undergone clinical testing) consists of E. coli RNA.32 For applications where the packaged nucleic acid is undersirable, several methods have been developed to remove the nucleic acid components after particle assembly.33-35 These methods include treatment with heavy metals such as lead acetate,33 incubation in alkaline conditions for RNA hydrolysis,34 and induction of osmotic shock using a high molarity sodium sulfate solution.35
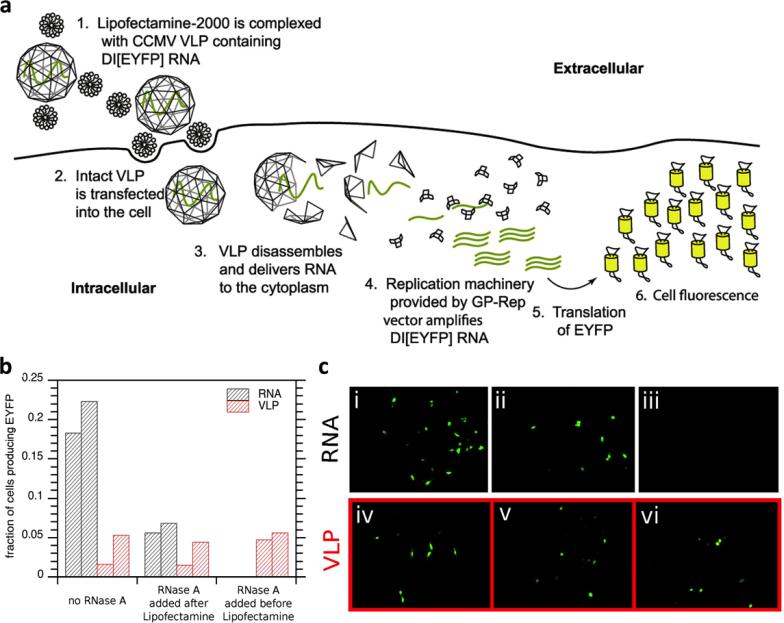
Another approach for production of empty VLPs is in vitro assembly of coat protein subunits after production in E. coli and purification. In vitro assembly is also a way to overcome challenges with insolubility of some eukaryotic capsid proteins in the bacteria cells that result in their accumulation in inclusion bodies.36 Digressing briefly, it is of significance to note that some headway has been made with producing soluble eukaryotic coat proteins with high yields and purity. This was demonstrated recently with the plant virus cowpea chlorotic mosaic virus (CCMV) through modulating several factors: an E. coli strain resistant to chloramphenicol was utilized, which helps inhibit protein transition to an insoluble state, and to give time for coat proteins to fold and maintain solubility, lower temperature, lower concentration of isopropyl-β-D-1-thiogalactopyranoside (IPTG) for induction of expression, and E. coli with a slower rate of protein synthesis were used.37 Returning to in vitro assembly, this method has been demonstrated for a wide range of VLPs, including those based on bacteriophages P2238 and PP7,39 plant viruses potato virus X (PVX)40 and CCMV,37 and mammalian viruses human papilloma virus (HPV)41 and human immunodeficiency virus (HIV).42 Some particles require a nucleic acid template in order to self-assemble,30, 39, 40 but others can be assembled to form empty capsids simply by altering conditions such as temperature, pH, and molarity.37, 38, 41, 42 For templated self-assembly, TMV for example has an origin of assembly that is thought to be required to drive its assembly,30 but other filamentous particles may not have such sequence specificity.40, 43 Knowledge of the self-assembly process of viruses can be important for determining the types of payloads that can be encapsulated as well as inform the design of novel architectures.15 Assembly of pure, empty particles in vivo in high yields is unique and has so far only been accomplished for the plant virus cowpea mosaic virus (CPMV), which will be discussed later with plant-based production systems.
In general, eukaryotic expression systems such as yeast, insect cells, and plants may be favored for production of assembled eukaryotic viruses as they are better able to secrete soluble eukaryotic proteins and can perform post-translational modifications, such as glycosylation, disulfide bond formation, and proteolytic processing.44-46 Yeast expression systems work similarly to bacterial systems and can also be scaled up using fermentation technology. Some common yeast species that have been developed for VLP production include Saccharomyces cerevisiae and Pichia pastoris,47 and VLPs that have successfully been produced in yeast include Qβ,47 CCMV,48 and HPV,49 which incidentally is how Merck produces the vaccine Gardasil.
In addition to bacteria and yeast cells, cultures of insect cells can also be utilized for VLP production. Baculovirus-based expression systems can be cultured in insect cell lines such as Spodoptera frugiperda (Sf) lines 9 and 21 and Trichoplusia ni moth cells.50 These viruses contain a large genome that is useful for incorporation of multiple genes of interest, but, due to the lack of unique restriction sites, also requires alternative strategies such as combining the use of recombination with shuttle vectors. This process tends to be a more time consuming and lower yielding method. The baculovirus-based expression system has been applied for the production of insect viruses such as flock house virus (FHV), plant viruses including CPMV, and mammalian viruses such as canine parvovirus (CPV) and HPV,49, 51-53 and it is GlaxoSmithKline's method of choice for producing its HPV vaccine Cervarix.
For production of plant VNPs and VLPs, plant-based expression systems are frequently used. Some common plant virus-based particles include red clover necrotic mottle virus (RCNMV), BMV, CCMV, CPMV, PVX, and TMV. To produce VNPs, plants can be infected by mechanical inoculation through applying purified virus solutions, infected leaf samples, cDNA of the virus genome, or even in vitro RNA transcripts to the leaves of the plant after gentle abrasion.54, 55 Agroinfiltration by injecting a suspension of Agrobacterium tumefaciens bacteria into leaves is also used for molecular farming in plants.56 These bacteria transfer part of their tumor inducing plasmid into the plant cell, which can be exploited for transient expression of genes of interest. Of note, Medicago Inc. uses this plant-based approach for the efficient production of VLP-based vaccines for influenza and rabies, among others. Other therapeutics such as the ZMAPP monoclonal antibody cocktail against Ebola virus (EBOV) from Mapp Biopharmaceutical also utilize plant production with agroinfiltration.57 Replication of intact VNPs such as BMV has been demonstrated.58 Additionally, viral capsids of CPMV completely devoid of RNA (either virus or host) can be produced in this way.59 Whereas BMV production utilized plasmids that transiently express BMV RNAs to systemically infect plants, for empty CPMV (eCPMV) VLP production, using plasmids encoding just two proteins was found to be sufficient: VP60, which is a precursor to CPMV's two coat proteins, and 24K proteinase for proteolytic processing of VP60. As mentioned previously, consistent empty VLP production in vivo has only been demonstrated for the eCPMV platform. Aside from farming in the plants themselves, some new technology that may be applied in the future for the production of VLPs is the use of plant cell packs for transient expression, where plant suspension cells are packed into a “cookie” through suctioning, then Agrobacterium containing the gene for the protein of interest applied.60 This approach has been proven to produce high yields of recombinant proteins and can be applied in a high throughput manner, making it an attractive option for VLP expression.
Finally, as an alternative to the above in vivo approaches, there has been some work involving VLP production using cell-free systems where cellular machinery for transcription and translation are used for protein expression in vitro. Some early work in this area utilized a eukaryotic system based on rabbit reticulocyte to study capsid assembly of hepatitis C virus (HCV), HBV, and three primate lentiviruses, but the yields were quite low (~10 ng/μL).61, 62 Since then, exploration with an E. coli-based system has achieved yields of around 400 ng/μL for MS2 and truncated HBV core antigen VLP production, with almost complete solubility, making it an excellent platform for rapid VLP production.63 Additionally, Qβ VLPs were able to be formed using this system through coproduction of its coat protein with a cytotoxic A2 protein that is normally naturally incorporated on the exterior of the capsid to facilitate infection, demonstrating the advantage of a cell-free system for cytotoxic protein production and regulating the relative expression of multiple proteins.64 As the cost of cell-free systems goes down, they may become more commonly applied for the production of VLPs.
3. Engineering virus-based scaffolds
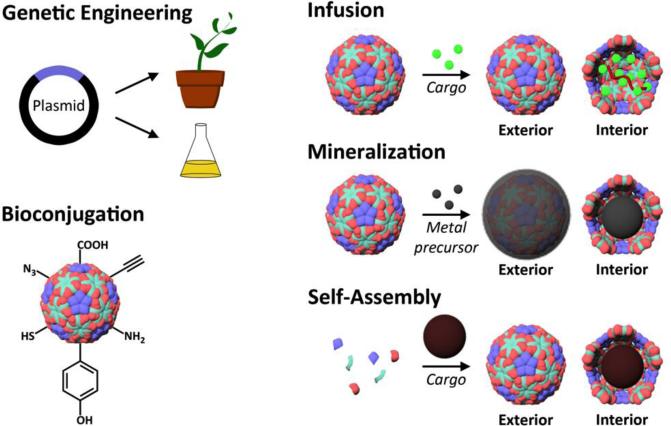
Since viruses have evolved to protect and efficiently deliver their nucleic acid cargo, they are able to withstand conditions required for chemical modification and retain a long shelf life. For modification, the interior cavity and exterior surface of the viral capsids can both be utilized, allowing for the encapsulation of sensitive compounds and the display of targeting moieties in precisely defined arrays, among other functions. The beauty and utility of these particles have been recognized, and efforts have been made toward mimicking these nanoscale architectures through self-assembly of protein nanomaterials.65 The unique genetically encoded protein shell architecture of virus-based scaffolds allows for a large range of techniques that can be used to tailor and modify these materials. Among the most frequently used of these that we will discuss are genetic engineering, bioconjugation, infusion, biomineralization, and self-assembly (Figure 3). As engineering capabilities improve, even greater diversities of virus-based and virus-like particles can be created, expanding the possible applications of these materials.
Figure 3. Techniques for modification of virus-based scaffolds.
Simplified illustrations show common methods for interior and exterior virus modification. To alter the composition of the protein capsid itself, genetic engineering can be used. With available exposed residues, bioconjugate chemistries can be performed. Through pores in the structure, small cargo can be infused into the capsid and then retained by reducing the pore size or electrostatic interactions. Interactions of metal precursors with the capsid can be used to selectively direct mineralization on the interior or exterior surface. Taking advantage of self-assembly of the viral scaffold, cargo introduced during assembly can be encapsulated.
3.1 Genetic engineering
The coat proteins of VNPs are determined by their genetic code. Nucleic acid sequences of viruses are relatively small, and therefore many of their genomes have been sequenced and are well characterized. Using genetic engineering, insertion or replacement of residues can be performed to add functional groups, with cysteine mutants being the most popular due to possible disulfide linkages, association with gold, and bioconjugation with thiol-selective chemistries.66-70 Insertion of unnatural amino acids is also possible, allowing for more diverse chemical modifications.71, 72 Additionally, removal of residues can be accomplished such that only a single unique reactive site remains on the coat protein.73 Aside from single residue modifications, larger changes such as insertion of purification tags can also be accomplished. For example, due to their affinity and coordination with nickel-nitrilotriacetic acid (Ni-NTA), polyhistidine tags have been expressed on viral capsids to serve as anchors for applications that include tethering them to surfaces, attaching other particles such as nanogold and iron oxide, and assembling higher-order structures.74-77 Display of other short peptide sequences have been demonstrated, including epitopes for vaccines78-80 and moieties for targeting receptors.81-83 Whole protein and protein domain insertions can also be achieved,84, 85 and even virus hybrids consisting of coat proteins expressing different proteins have been established through co-infection of plants, with verification by bimolecular fluorescence complementation.86 Aside from genetic engineering of the viral coat proteins, tags such as the antibody binding peptide Z33 can be genetically fused to fluorescent proteins, enzymes, and other proteins of interest.87 In this particular example, assembly of particles displaying the proteins can then be achieved by means of an intermediary antibody specific to the viral coat protein.
3.2 Bioconjugate chemistry
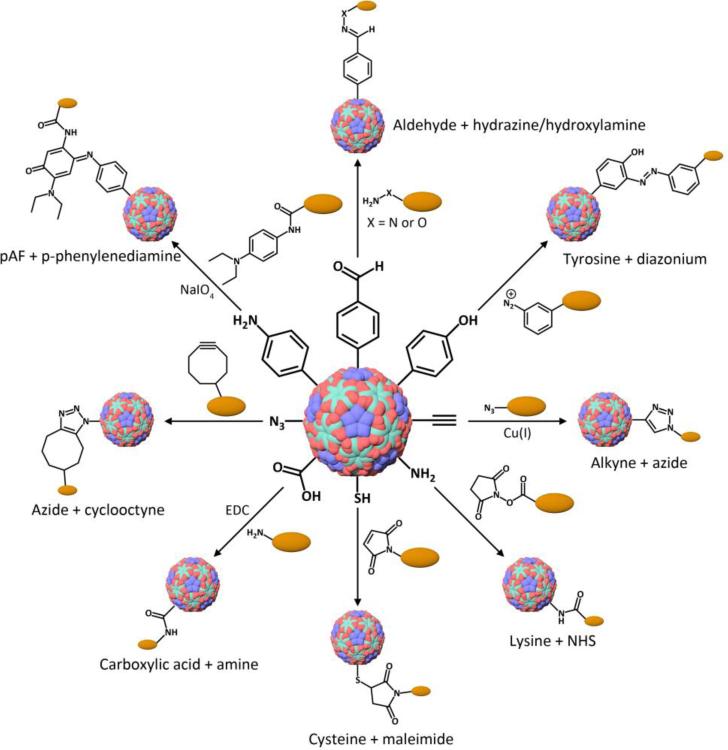
Conjugation strategies targeting both natural and unnatural amino acids on virus capsids allow for many possible modifications that may not be achievable through genetic engineering alone (Figure 4). Both the interior and exterior surfaces of many viruses have been shown to be amenable to chemical modifications.88-90 Some common groups that can be functionalized include lysine, aspartic/glutamic acid, cysteine, and tyrosine residues, which lend themselves to standard bioconjugation reactions involving N-hydroxysuccinimide (NHS) ester conjugation, carbodiimide activation, Michael addition, and azo coupling chemistries. Some alternatives to these natural amino acids include replacing methionine residues with homopropargyl glycine (HPG) or azidohomoalanine (AHA) residues to add alkyne or azide functionalities, respectively.71 Another interesting method utilizes mutant tRNA synthetases to attach unnatural amino acids to amber suppressor tRNAs for incorporation of these amino acids at amber stop codon sites.72, 91 Among the amino acids incorporated in this way are O-methyltyrosine, p-azidophenylalanine, p-acetylphenylalanine, p-benzoylphenylalanine, 3-(2-naphthyl)alanine, and p-aminophenylalanine (pAF). p-azidophenylalanine and pAF are particularly noteworthy due to providing azide and amine groups, respectively, for selective coupling reactions. As can be noted, incorporation of azide and alkyne groups is an especially widespread strategy. These conjugation handles allow for copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC), an efficient and biocompatible procedure that has found great versatility.92-96 Reaction without copper catalysis can also be achieved by utilizing cyclooctyne derivatives, which lower the activation barrier due to the ring strain.97
Figure 4. Bioconjugation reactions that can be used for virus modification.
Presented in the figure are some of the more common reactions for functionalization of viruses. Other methods discussed in the text include atom-transfer radical polymerization, ring-opening metathesis polymerization, and supramolecular interactions.
Additional reaction handles that have been utilized include aldehydes and ketones for hydrazone or oxime condensation reactions.88, 98-100 Selective formation of aldehydes or ketones is possible, where pyridoxal 5'-phosphate (PLP)-mediated transamination specific for the N-terminus leads to in situ oxidation of the N-terminal amine.101 The formed ketone or an aldehyde group can then be used to form stable oxime linkages with alkoxyamines. With the availability of a wide range of chemically modifiable natural and unnatural amino acids, multiple functional groups can be simultaneously incorporated within a single virus-based particle to result in the formation of a versatile, multifunctional platform.
One area of particle modification that is of great interest is the formation of protein/polymer hybrid conjugates. Polyethylene glycol (PEG) is a polymer frequently used for shielding biological interactions, and attachment of PEG through aforementioned bioconjugation techniques is fairly standard.89, 102, 103 More sophisticated polymerization chemistry techniques have also been shown to be applicable for polymer attachment to virus-based particles, and more widespread application of these could confer advantages of better efficiency and more control over the polydispersity. Atom-transfer radical polymerization (ATRP) is one such method, where small initiators can be added to the particles first then polymerization from the capsid carried out through the introduction of monomers, resulting in easier purification of the smaller reagents as well as overcoming challenges with steric hindrance of large bulky polymers.33, 104, 105 Incorporation of polymers using this method has proven to useful for the attachment or complexation of large payloads of MR contrast agents, chemotherapeutics, and siRNA and for both interior33, 104 and exterior105 modification. Polymers could also be synthesized first with ATRP before attachment to the viral capsid, such as for the display of glycoproteins.106 (ROMP) is another method for biocompatible polymer synthesis, and it was utilized to prepare water-soluble polynorbornene-based polymers with strict size and architecture control, which had a good safety profile when attached to Qβ and delivered to fibroblast cells.107
Aside from chemistries that rely on covalent bonds, supramolecular chemistry strategies can also be utilized for virus modification. For example, by taking advantage of the hydrophobic interior of β-cyclodextrin that allows it to accommodate a range of guest molecules, virus particles first modified with β-cyclodextrin moieties can then be functionalized with derivatives of such guest molecules. This approach has been demonstrated using derivatives of adamantine for the display of an imaging agent, chemotherapeutic drug, targeting ligand, and PEG polymer.108 In a similar manner, charge-transfer interactions between electron donors and acceptors can also be used for derivatization of viral scaffolds. As an illustration, attachment of pyrene molecules allowed for particle functionalization through interaction with electron-deficient dinitrophenyl and pyridinium motifs.109
3.3 Infusion
The interior of viral capsids can be used as a cage for encapsulation of foreign cargo. Viruses are generally flexible and contain pores that allow for diffusion of small molecules, such as drugs and contrast agents, into and out of the capsid. Retention of the molecules inside the capsid can then be achieved through electrostatic and/or affinity interactions with the nucleic acid within the shell93, 110, 111 or interactions with polymers conjugated internally.33, 104 Encapsulation of molecules can also be accomplished by gating using pH or metal ion concentration to trigger structural transitions. Using the gating process, molecules are allowed to diffuse into the particle under an environment where the capsid is in a swollen, open conformation, and then the molecules are trapped within the capsid as the pores are closed off through change in buffer conditions.112-114 Depending on the desired application, the molecules of interest can either remain encapsulated within the particles or released over time. Examples of infusion for imaging and drug delivery are described in Sections 4.1.2 and 4.1.5. Another application of infusion is for introducing metal precursors into the capsid for interior mineralization, which is discussed next.
3.4 Mineralization
Viral particles can also serve as templates in the biomineralization process with unique size and shape control. Through tuning electrostatics or the use of mineralization-directing peptides, nucleation of precursor metal ions and subsequent shape-constrained mineralization can be realized. Peptide nucleators and binders were identified by screening using phage display techniques against various substrates, such as GaAs and ZnS,115, 116 and shown to be highly specific. Mineralization has been demonstrated for the interior11, 117-119 and the exterior120-122 surfaces of particles, as well as for both icosahedral11, 117, 118, 122 and rod-shaped119-121 viruses. These resulting hybrid inorganic-organic materials find use in a variety of functions, ranging from applications in energy as semiconductors (see Section 4.3) to medicine as contrast agents (see Section 4.1).
3.5 Self-assembly
While we have mostly considered these virus-based particles as intact scaffolds we can build from, they can also be taken apart and reassembled, either with their natural genome or with foreign cargo. There is a great breadth in the types of cargo that can be encapsulated through self-assembly, including gold nanoparticles, quantum dots, and photosensitizer drugs.21, 123-126 Since coat proteins naturally self-assemble around negatively charged nucleic acids, in general a more negative surface charge results in more efficient encapsulation of the foreign cargo.123 Native packaging mechanisms can also be utilized, which was demonstrated with adding oligonucleotides mimicking the origin of assembly for RCNMV's packaging of its RNA on various nanoparticles to induce particle formation around the different cargo.127 Size plays a factor in assembly due to its effect on the radius of curvature, and differently sized cargo could result in alterations in the morphology and physical characteristics of the capsid.123-125
As seen in Section 3.1, coat proteins can be genetically modified for incorporation of foreign protein cargo during particle assembly. One study fused the coat proteins of CCMV to elastin-like polypeptides (ELPs), which exhibit lower critical solution temperature (LCST) behavior, and the investigators found two different self-assembly pathways from the resultant hybrid.128 While VLPs consisting of 90 coat protein dimers were formed under normal pH-induced self-assembly conditions, when the NaCl concentration was increased to lower the ELP transition temperature, the ELP-induced assembly resulted in the formation of smaller particles consisting of 30 coat protein dimers. Enzyme facilitation is another method for coat protein modification, which was demonstrated through the use of sortase A to covalently attach a protein with a C-terminal LPETG tag to glycines at the N-terminus of CCMV coat proteins before assembly for protein encapsulation.129 Specific binding interactions with the coat protein can also be exploited for self-assembly. Engineering of coiled-coil protein interactions was established by introducing a lysine coil at the N-terminus of CCMV that can associate with a glutamic acid coil at the C-terminus of a fluorescent protein, which resulted in encapsulation of the protein when the two modified proteins were combined.130 Introducing histidine tags that have affinity for Ni-NTA is another method.77 Additionally, some interactions that naturally exist for some particles include the association of scaffold proteins with bacteriophage P22 that aids in viral assembly131, 132 and the binding of translational repression operator proteins to RNA stem-loops within MS2 bacteriophages.14 Fusions to these proteins can then be utilized for encapsulation of materials of interest, such as enzyme cascades and therapeutic molecules.
4. Applications of virus-based particles
4.1 Medicine
Viruses have been applied broadly in medicine for diagnostic and therapeutic purposes, and many are in the pipeline undergoing clinical trials for oncotherapy and as gene therapy vectors.133, 134 In fact, there is currently much excitement over the recent approval of the oncotherapy talimogene laherparepvec (T-VEC) manufactured by Amgen,135 and T-VEC will be discussed in more detail in Section 4.1.3. Bacteriophages and plant viruses are particularly attractive tools for biomedical applications because they do not replicate within mammalian cells, and therefore the platforms may add another layer of safety. In this section, we will explore the use of virus-based particles as delivery vehicles targeted toward imaging and treatment of diseases and as scaffolds that interact with the local environment, which can be utilized for vaccines, immunotherapy, and tissue engineering.
4.1.1 Nanomedical viral engineering design rules
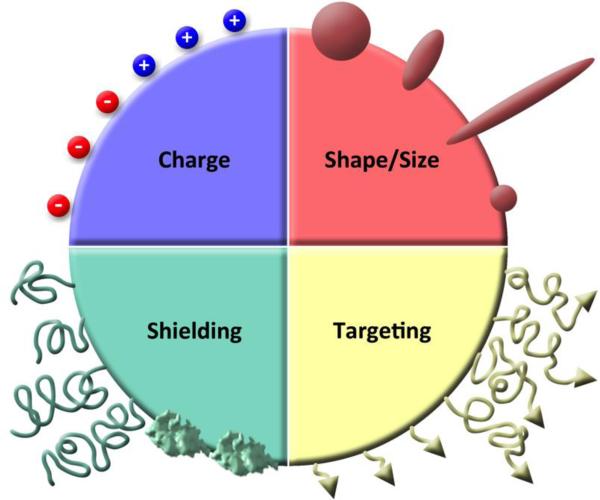
Some important considerations for the design of viruses for applications in vivo include charge, shape, and surface ligand presentation (Figure 5). These design parameters affect their circulation in the body as well as their cellular interactions and tissue specificity. Some general principles have been established specifically for virus-based particles,136 and we will highlight some of the lessons here. It is important to note that although these principles provide a good guideline, in vivo studies are crucial for ascertaining how new proposed particles will behave due to the intricacy and complexity of biological interactions, which cannot be fully predicted through in vitro testing or modeling.
Figure 5. Design parameters to consider for nanoparticle engineering.
Parameters include charge (positive or negative), shape and size (different aspect ratio filaments and diameter spheres), shielding (self proteins/peptides and polymers of various sizes and densities), and targeting (ligands for receptors or environmental factors displayed on different linkers and at various densities).
In terms of charge, there appears to be a trend where virus-based particles with negative surface charge tend to have shorter circulation times. This was observed with negatively charged CCMV, CPMV, and TMV, which have circulation half-lives of less than 10 minutes.137-139 In comparison, the half-lives for positively charged Qβ and M13 are on the order of 4-5 hours.140, 141 The effect of charge on plasma clearance was made more evident when much quicker clearance for both bacteriophages was observed with the neutralization of their positive lysine residues.140, 141 Additionally, the reverse study with a single amino acid substitution of glutamic acid residues with lysines using bacteriophage λ resulted in over a 1000-fold higher circulation time.142 A notable exception to this trend is PVX, which is expected to be longer circulating based on its positive charge and abundance of surface lysines but in fact has a quick clearance half-life of around 10 minutes.143 A more recent study reported a negative zeta potential for PVX,144 likely due to different buffer conditions used, which indicates further investigation into the charge of PVX under in vivo conditions is crucial for confirming whether or not it defies convention.
Additional influences based on charge include altering how particles interact with mammalian cells and tumor transport rates. Due to the abundant presence of proteoglycan in the cell membrane conferring a negative charge and collagen within the tumor interstitial space conferring a positive charge, positively charged particles are more likely to have enhanced binding to mammalian cells145 and are better able to avoid aggregation and penetrate tumor tissue.143, 146 Some examples demonstrating these charge-based effects include polyarginine-decorated CPMV found to be taken up eight times more efficiently than native CPMV in a human cervical cancer cell line145 and positively charged PVX shown to be able to penetrate to the tumor core unlike negatively charged CPMV.143 In the latter case, PVX's filamentous nature also allowed it to better avoid the macrophagocytic system, leading to greater tumor homing.
It is likely that the shape and flexibility of PVX plays an additional role in its ability to diffuse throughout the tumor. A comparison between the diffusion profiles of a spherical and rod-shaped particle was performed with CPMV and TMV using a spheroid model, and it was shown that whereas CPMV experienced a steady diffusion profile, TMV exhibited a two-phase diffusion behavior that entailed an extremely rapid early loading phase, which could be attributed to its movement axially, acting like a needle.147 Some other advantageous properties that are conferred by elongated particles include better margination toward the vessel wall and stronger adherence due to greater surface area for interaction, which not only have implications for tumor homing but also for enhanced targeting of cardiovascular disease.148, 149
Shape is a difficult parameter to account for due to the challenge of producing monodisperse particles that can be precisely and reproducibly tailored at the nanoscale, but this challenge can be surmounted using VNPs and VLPs due to the specificity of their self-assembly process. Some bottom-up assembly approaches have been investigated recently to elucidate the role of aspect ratio in cell uptake, biodistribution, and tumor homing. In one approach, CPMV particles were linked together to form chains in order to maintain charge and surface properties while modifying the aspect ratio, and dimers with an aspect ratio of 2 were found to target cancer cells more efficiently than single particles.150 Higher aspect ratios were investigated in another study that utilized in vitro assembly of TMV around synthetic RNA to form rods of various lengths (300, 130, and 60 nm, corresponding to aspect ratios of 16.5, 7, and 3.5).151 For receptor-targeted particles modified with the RGD ligand, rods with an intermediate aspect ratio of 7 were found to be more efficient at tumor targeting due to a combination of better macrophage avoidance and greater adhesion to target integrins compared to the short rods and better diffusion within the tumor compared to the long rods. Based on the aforementioned spheroid study with TMV,147 it is likely that the three aspect ratios experience similar axial diffusion during the initial rapid phase, but then longer particles are impeded during the slower distribution phase. In contrast, PVX particles with an aspect ratio of 40 appear not be hindered by their length and in fact experience better penetration in relation to icosahedral CPMV.143 Some possible reasons to account for this include a thinner cross-section and greater flexibility for PVX compared to TMV. To further expand understanding of the role of shape, other factors such as density and flexibility should be considered in future studies.
Surface presentation of shielding polymers also plays a role in the in vivo behavior of virus-based particles. PEG is the standard polymer used to reduce immunogenicity and nonspecific cell interactions. The importance of polymer coatings is particularly apparent in the study highlighted above with different aspect ratios of TMV. Although it was found that targeted particles fared better when intermediate in size, coating the TMV with PEG allowed the shorter rods to be better able to avoid clearance and, paired with their superior diffusion properties within the tumor, resulted in increased passive tumor targeting of the short 60 nm PEGylated rods.151
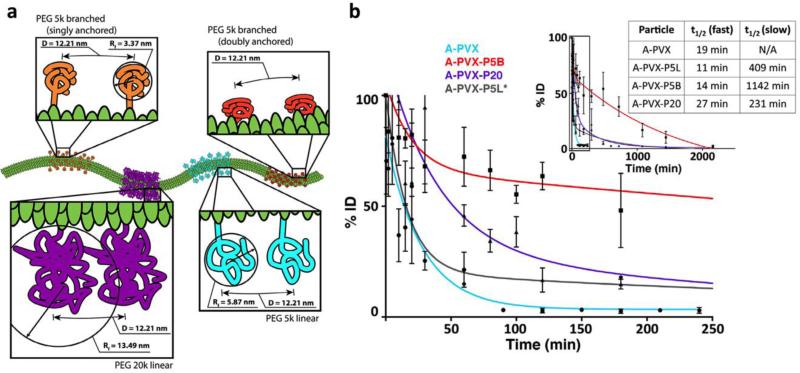
Surface PEGylation has been applied to many VNPs and has been established as a broadly applicable method for extending circulation time.102, 138, 152, 153 Additionally, differences in the route of clearance was also observed, with non-PEGylated TMV and PVX filaments getting filtered through the kidneys, while PEGylated particles do not, likely due to the increase in the width of the particles after conjugation preventing renal filtration.102, 138, 154 The conformation of the PEG coating can be predicted computationally through estimating its surface coverage on the particle and its hydrodynamic radius to determine the packing density of the polymer. The use of higher molecular weight PEG generally results in a higher hydrodynamic radius and thus better shielding, but hydrodynamic radius is only an average and polymers can extend and collapse in solution. Despite a smaller predicted radius, branched PEG with multiple sites of attachment to the particle has been shown to be more effective at shielding than linear PEG four times its molecular weight, likely because simultaneous tethering of the ends of the PEG traps it closer to the particle, reducing its movement and the possibility of nonspecific protein adsorption. We hypothesize that the branched PEG leads to a more efficient shield, preventing the formation of a protein corona that may tag the virus-based nanoparticles for recognition by the innate immune system and lead to removal from circulation (Figure 6).102 Therefore, the dynamics of PEG in solution should also be considered when determining its conformation for optimization of particle shielding.
Figure 6. Effect of PEG shielding on PVX clearance.
a) Diagram of conformations of PEGs of different lengths and geometries displayed on PVX based on calculations of grafting density and Flory dimension. b) Pharmacokinetics of the various PEGylated particles when injected in Balb/C mice show better shielding of the 5k branched polymer. Reproduced with permission from ref. 102. Copyright 2015 Elsevier.
Other polymer coatings are also being studied, with poly(N-(2-hydroxypropyl)methacrylamide) (pHPMA) being another hydrophilic polymer used, particularly with AAV and adenovirus in order to eliminate normal infection pathways and allow redirection of the viruses through other pathways.155, 156 Some cationic polymers, such as poly(amidoamine) (PAMAM) dendrimers157 and polyethyleneimine,158 have also been explored for shielding from infection as well as for improving transfection efficiency. For more control and diversity of polymerization, chemistries have been established for grafting polymers to and from viral scaffolds using ATRP and ROMP, as discussed in Section 3.2, but their properties in vivo have not yet been established. The use of serum albumin has been recently investigated for coating of TMV and shown to be more effective than medium-length PEG (5000 Da), with circulation times up to 10-fold greater in comparison.159 Self peptides based on human CD47 could also be considered for inhibiting phagocytic clearance of the nanoparticles.160
Along with surface modifications that allow them to avoid undesirable cell interactions, particles can be enhanced for specific cell targeting through the display of receptor-specific or disease environment-specific ligands. Some examples of targets that have been used for specific uptake of virus-based nanoparticles include epidermal growth factor receptor (EGFR)161, 162 and folate receptor (FR).163, 164 In such a manner, overexpressed receptors or environmental cues can be tracked for diagnostic or drug delivery purposes, which will be discussed in the following sections. To obtain the most favorable ligand display density, there is a balance between increasing avidity and reducing cellular receptor depletion that arises from increased ligand density.165 While multivalency and a higher degree of labeling with targeting ligands is beneficial for stronger cellular interactions, too many ligands may reduce the extent of endocytosis through exhaustion of cellular receptors. Another design parameter for the inclusion of targeting ligands is the linker used for attachment. For example, the inclusion of PEG can assist in increasing circulation time and avoiding nonspecificity as discussed above. Additionally, PEG linkers can improve cell targeting by adding flexibility and enhancing presentation of targeting peptides.166 By altering the characteristics of the linker, the interaction of the ligand with its target and the overall behavior of the particle in vivo can be tuned.
4.1.2 Imaging
Viruses have been used for tissue-specific imaging and delivery of contrast agents in applications of optical imaging, magnetic resonance imaging (MRI), and positron emission tomography (PET). The utility of using viruses as imaging probes comes from the d iversity of approaches for modification of the particles as well as the ease of precise assembly. In addition, clearance and removal from the body are critical for preventing toxicity from tissue retention of contrast agents, and many VNP platforms tend to be cleared quickly from the body (half-life of minutes)137-139, 154 compared to some synthetic materials that require months for clearance, such as carbon nanotubes, gold, and silica.167-169 Imaging is an important tool in medicine for diagnostics and for visualization of disease localization and progression, as well as treatment success. With improvements in imaging technology, earlier disease detection and better prognosis can be realized. The ability to track particles further aids in the evaluation of drug delivery platforms, as it can be used for confirmation of cell-specific uptake and investigation of interactions of particles within the body, such as their clearance, biodistribution, and immunogenicity.
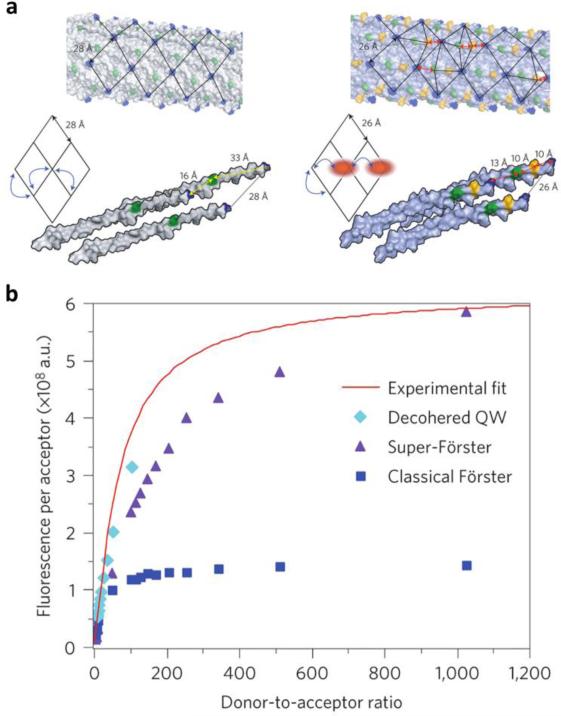
Fluorescence imaging is the main modality for preclinical evaluation and was used to aid in the establishment of the design rules in Section 4.1.1. Fluorescent agents can be incorporated into viral capsids through bioconjugation,170, 171 genetic engineering,84, 86 infusion,110, 114 and self-assembly.21, 130 Fluorescence is useful for quantification of particle uptake using flow cytometry, visualization of particle localization through confocal microscopy, and determination of biodistribution using in vivo imaging. Although high dye densities can easily be achieved through efficient capsid modification strategies, sensitivity decreases after a certain threshold due to fluorophores experiencing quenching when placed at distances less than approximately 10 nm. Therefore, a fairly low density of around 10% is more ideal for achieving optimal fluorescence intensity.172 Encapsulation of indocyanine green can be utilized as a method for near infrared (NIR) photoacoustic imaging, and it has shown greater photostability compared to the chromophore alone.173 As advancements are made, another aspect that could be explored is the integration of gold nanoparticles with fluorophores for metal-enhanced fluorescence with improved quantum yields and decreased photobleaching.174
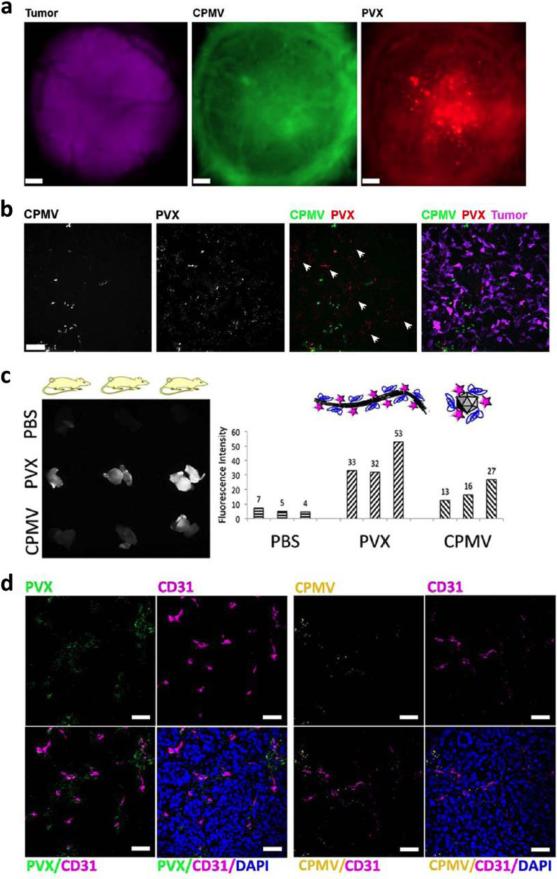
First iteration native and PEGylated particles can be directly evaluated for their biodistribution, pharmacokinetics, and tumor homing behavior through fluorescence imaging.84, 90, 102, 138, 143, 175 Overall, particles are cleared mainly through the liver and spleen, with filamentous particles having a higher rate of spleen clearance compared to icosahedra l particles,138, 143 localizing with B cells within the white pulp over time.102, 138 Due to leaky vasculature and poor lymphatic drainage, the enhanced permeability and retention (EPR) effect is found in solid tumors and can be utilized for tumor imaging through nanoparticle deposition. Using both mouse and chicken chorioallantoic membrane (CAM) models with tumor xenografts, the passive partitioning of particles to the tumor can be observed (Figure 7).84, 143 As discussed in the previous section, evaluation of localization of particles inside the tumor revealed enhanced accumulation and penetration of rod-shaped particles.143
Figure 7. Imaging of tumor uptake and distribution of CPMV and PVX.
a) Comparison of icosahedral CPMV (green) and filamentous PVX (red) distribution when coinjected in a CAM model of chick embryos prepared with vascularized GFP-expressing human fibrosarcoma HT1080 or human epithelial carcinoma HEp3 tumors (magenta), with PVX better able to penetrate to the tumor core. Scale bar = 190 μm. b) Fluorescence microscopy of 8 μm tumor sections showing CPMV having limited distribution, while PVX is spread throughout the tumor and found in areas devoid of CPMV (white arrowheads). c) Image of tumors from an HT-29 colon cancer mouse xenograft model after intravenous injection of CPMV and PVX particles (left) and quantitation of fluorescence intensity (right). d) Immunofluorescence staining of 10 μm tumor sections showing CPMV (pseudocolored in yellow) remaining close to the endothelium (stained with FITC-labeled CD31 antibody pseudocolored in pink) and PVX (pseudocolored in green) having better tissue penetration properties. Nuclei were stained with DAPI (blue). Scale bars are 30 μm. Reproduced with permission from ref. 143. Copyright 2012 American Chemical Society.
Besides passive tumor homing properties, natural interactions of viruses with certain cells can also be exploited. CPMV in particular exhibits unique specificity in interacting with surface vimentin, which is found on endothelial, cancer, and inflammatory cells.176-179 The native affinity of CPMV for surface vimentin allows for high-resolution imaging of microvasculature up to 500 μm in depth, which cannot be achieved through the use of other nanoparticles, as they tend to aggregate and block the vasculature.180 This interaction can be utilized for a range of applications, such as delivery to a panel of cancer cells including cervical, breast, and colon cancer cell lines,110 delineation of atherosclerotic lesions,177 and intravital imaging of tumor vasculature and angiogenesis.180 Another example of an existing endogenous association is CPV with transferrin receptor (TfR), an important receptor for iron transport into cells and highly upregulated by numerous cancer cell lines.52 Even after dye labeling, CPV retains its specificity for TfR and was shown to bind to receptors found on HeLa cervical cancer cells, HT-29 colon cancer cells, and MDA-MB-231 breast cancer cells. As a quick side note, in vivo imaging of bacterial infections and differentiation between F-positive and F-negative E. coli strains is also possible through specificity of binding of M13 phage.181
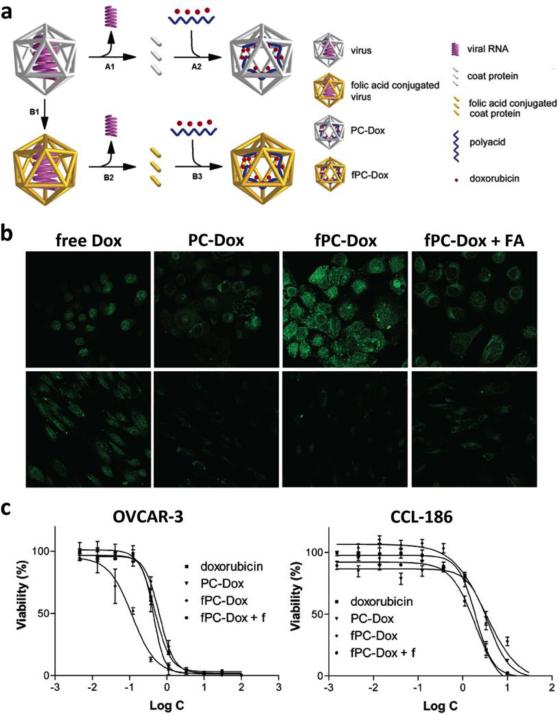
Specificity can also be introduced through the incorporation of targeting ligands for molecular imaging. RGD is a targeting peptide that is frequently used due to its high affinity for αvβ3 and αvβ5 integrins, which are involved in angiogenesis and associated with cancer proliferation.81, 151, 182, 183 The association of RGD-targeted particles with tumor vascular endothelium has been demonstrated in mice, although the study also indicated that better tumor localization would be achieved with greater circulation time imparted through incorporation of a better shielding linker (see Section 4.1.1).151 CPMV displaying peptide F56, which was discovered through phage display, has been used to target vascular endothelial growth fa ctor receptor-1 (VEGFR-1), with accumulation throughout the tumor observed compared to no detectable uptake of non-targeted particles.99 Other options that have been explored include FR targeting with folic acid (FA),164, 184, 185 TfR with transferrin,186 and prostate specific membrane antigen (PSMA) with a PSMA antibody.187 In recent years, a target that has been approached from many angles is epidermal growth factor receptor (EGFR), an important biomarker overexpressed on many malignant cell types. Strategies range from display of EGF on Qβ through genetic engineering,162 using phage antibody libraries to select for fd phages with single-chain antibody variable fragments (scFvs) specific for EGFR as well as its related receptor human epidermal growth factor receptor 2 (HER2),101 conjugation of EGFR antibodies on MS2,188 and also chemical attachment of GE11 peptide on PVX.161 These studies all evaluated cell binding in vitro and there are some promising results indicating partitioning of targeted particles to tumor cells compared to macrophages in co-cultures,161 and it would be of interest to see their development in mouse models.
Outside of membrane proteins of cancer cells, proteins highly expressed by activated endothelial cells, such as vascular cell adhesion molecule (VCAM)-1, can be utilized for targeted imaging of cardiovascular disease and atherosclerotic plaques.148 Beyond such strategies, matrix and secreted proteins are also advantageous targets. For example, collagen189 and secreted protein acidic and rich in cysteine (SPARC),190, 191 an extracellular matrix glycoprotein, can be detected for tumor imaging through target-specific peptides displayed on M13. SPARC in particular has been successfully targeted for deep tissue imaging of lung cancer,191 and it has even been used for guided resection of ovarian cancer through the pairing of M13 with fluorescent single-walled carbon nanotubes (SWNTs),190 which allows for non-photobleached fluorescence with less background in the second NIR window ranging from 950-1400 nm. The polymerized fibrin found in thrombi has also been investigated for the delineation of blood clots using MS2, CPMV, and TMV equipped with GPRPP149, 192 and CREKA149 pentapeptide amino acid sequences.
The above studies investigating imaging of thrombosis149 and atherosclerosis148 established target-specific imaging not only with optical but also with MR imaging. MRI is a clinically relevant method for noninvasive disease characterization with good soft tissue contrast, and the use of contrast agents in conjunction with MR can improve the signal-to-noise ratio to highlight differences between diseased and normal tissues. Gadolinium is one such paramagnetic contrast agent that can be used to achieve brighter signal in T1-weighted imaging. Molecular imaging of atherosclerotic plaques was achieved at dosages 400 times lower than clinically used for angiography with the encapsulation Gd chelated with 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid, or Gd(DOTA), within TMV targeted to VCAM-1.148 The high contrast can be attributed to the high payload of 1200 Gd/TMV, the slower molecular tumbling rate resulting from attachment of Gd(DOTA) to the macromolecule, inclusion of the targeting peptide, and the advantage imparted by the shape of TMV for drifting laterally to the vessel wall.
This latest result is the culmination of numerous studies by several groups that formed stepping stones along the way. While early work looked at the direct binding of Gd to natural metal binding sites in the capsid of CCMV,193 the use of chelation and bioconjugation was quickly introduced to mitigate concerns of free Gd leading to toxicity in patients with underlying kidney disease, with explorations using both DOTA93, 194 and diethylenetriaminepentaacetic acid (DTPA).195 These approaches resulted in per particle T1 relaxivities on the order of 1,000 to 8,000 mM−1 s−1 measured at 64 MHz. While fairly high and much greater than 20 mM−1 s−1 for Gd alone, these values do not approach the measurement of around 28,000 mM−1 s−1 per particle from the initial study with direct attachment of Gd.193 To improve contrast, interior and exterior labeling196, 197 as well as rigidity of linkers198 was explored with MS2 using chelators based on hydroxypyridinonate (HOPO) due to its 3-fold relaxivity enhancement compared to clinically used Magnevist, or Gd(DTPA). Internally modified capsids and more rigid linkers, in particular the S,S enantiomer of 1,2-cyclohexyldiamine, each demonstrated approximately 30 to 40% higher relaxivities. Enhancing Gd loading was another method explored to increase per particle relaxivity, either using ATRP to amplify density of groups with which to attach the contrast agents104, 105 or using branched oligomers with multiple Gd(DTPA) complexes attached.199, 200 The greatest success with this approach led to the incorporation of over 9,000 Gd(DTPA) per P22 particle, with per particle relaxivities exceeding 200,000 mM−1 s−1.104 In vivo imaging was first demonstrated using P22 conjugated to Gd(DTPA) in order to visualize blood vessels in a mouse, with clear depiction of the carotid artery, mammary arteries, the jugular vein, and veins in the head at a resolution of 250 μm.200 More recently, relaxivities approaching 1,000,000 mM−1 s−1 per particle were reported by utilizing TMV's greater surface area conferred by its shape to introduce a large payload of Gd(DOTA), accompanied by thermal transition of the rods using conditions that result in 200 nm spheres.201 Coating interiorly labeled TMV particles with silica could potentially increase the relaxivity 3-fold as well as lead to greater macrophage uptake and hence contrast.202 It is expected that a combination of these research directions investigating chelators, linkers, conjugation, shape shifting, and coating will result in particles with even greater contrast for better visualization of disease.
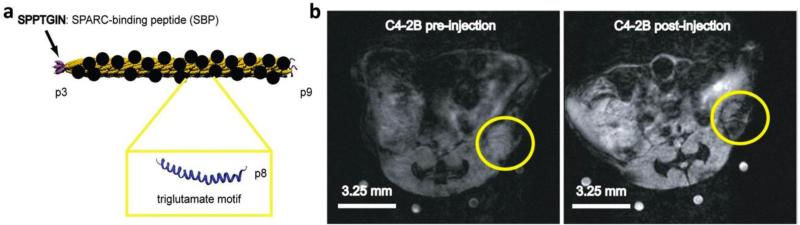
Apart from Gd-based contrast enhancement, manganese and iron oxide are other contrast agents that have been investigated. Manganese research is relatively new and labeling of P22 with Mn porphyrins was shown to have a per particle relaxivity of 7,000 mM−1 s−1 at 90 MHz, and while this is low compared to advancements in Gd imaging, it is a promising avenue to pursue due to the reduced toxicity of Mn.203 Unlike Gd and Mn, iron oxide is a contrast agent for T2-weighted imaging and is observed from a resultant darker image. Interestingly, the first demonstration of MR imaging with iron oxide was in plants, where cubic iron oxide nanoparticles were encapsulated within BMV, Nicotiana benthamiana leaves were infiltrated with the modified virus, and imaging was performed with cell-to-cell trafficking of the encapsulated iron oxide observed.204 Encapsulation of iron oxide nanoparticles and phantom imaging has been demonstrated with hepatitis B core VLPs.77 Moving toward translation, attachment of iron oxide nanoparticles along with SPARC binding peptides to the surface of M13 was effective for the imaging of prostate cancer (Figure 8).205
Figure 8. Targeted MR imaging of prostate cancer with M13.
a) Diagram of M13 structure with the major p8 proteins displaying a triglutamate motif for the multivalent display of iron oxide nanocrystals (black circles) and the p3 proteins at the end of the virus displaying SPARC binding peptide (pink) for targeting. b) MR scans of mice using a 7 T small animal MR scanner with subcutaneous C4-2B tumors (encircled) before (left) and 24 hours after (right) M13 injection displayed dark contrast from the targeted particles against the bright image of the tumor. Reproduced with permission from ref. 205. Copyright 2012 Nature Publishing Group.
Another MR contrast approach that is quite new is chemical exchange saturation transfer (CEST) and hyperCEST imaging. Xenon-based agents in particular have been explored for viruses. After selective saturation of these nuclei, an enhanced water signal is observed due to saturation transfer to surrounding bulk water. This technique has found success with MS2,206 M13,207 and fd,208 with sensitivities as low as 230 fM.207 By additionally incorporating scFVs that recognize EGFR, molecular imaging and contrast specificity were demonstrated with MDA-MB-231 cancer cells, with essentially no contrast in Jurkat negative control cells.208 Due to the more than 10,000-fold increase in sensitivity, there is a lot of potential in this new technology.
PET imaging is another sensitive imaging modality and relies on the detection of radiotracers. It has been utilized for ascertaining the biodistribution of non-PEGylated209 and PEGylated210 MS2 capsids through incorporation of [18F]fluorobenzaldehyde and 64Cu chelated with DOTA, respectively. Taking it a step further, biodistribution of encapsulated or non-encapsulated superparamagnetic iron oxide nanoparticles, 18F, and poly-L-lysine (PLL) cation (for packaging 18F) within hemagglutinating virus of Japan envelopes (HVJ-Es) was studied to determine whether magnetic stimulus can be used to redirect the viruses to the head, and it was clear that the application of the magnets altered the fate of the viruses, with increased signal in the head.211 Targeting ligands were also explored in conjunction with PET imaging, with RGD for targeting human sarcoma212 and glioma213 xenografts and GE11 for targeting an EGFR positive liver cancer xenograft model.157 In the context of virus-based particles, there has been less work with PET compared to the other imaging modalities. While PET has its advantages of high sensitivity and ability to image more deeply, radiotoxicity is an issue. For the purposes of simply detecting particle localization for diagnostics, MRI may be more ideal to pursue as technology improves.
4.1.3 Vaccines and immunotherapy
We will begin our foray into viral vectors for combatting diseases starting with vaccines, which has an extensive history and is likely the first medical application of viruses. Its popularization had an illustrious beginning in 1796 with Edward Jenner's experiment inoculating his gardener's eight-year-old son with cowpox, which resulted in protecting the boy from subsequent challenges with the more serious smallpox virus.214 While knowledge of viruses would not come until 100 years later, with the work of Dmitry Ivanovsky and Martinus Beijerinck filtering TMV from plant sap and demonstrating its infectivity and replication,7 the medical application of viruses had its roots here.
Instead of live viruses, safer alternatives for vaccines have since been established, including attenuated viruses, inactivated or subunit viruses, non-infectious VLPs, nanoparticle delivery, and nucleic acid vaccines.215, 216 Vaccines have been researched for a wide range of diseases, with great success for some diseases such as polio217 and measles,218 but some important vaccines such as for HIV and EBOV are still lacking, which are discussed below. Eliciting effective and long-term immune response is one challenge for vaccines, and the use of virus capsids offers the advantages of multivalent antigen presentation, incorporation of multiple epitopes, and particle stability. Since the field is enormous, we would like to feature just some of the research on vaccination, focusing on a few studies in the areas of infectious disease, brain disorders, and cancer. For a more comprehensive overview of virus-based particles for vaccines, the reader is invited to consult further reviews.219, 220
In the realm of infectious diseases, HIV is particularly challenging to address due to sequence diversity and difficulty in generating broadly neutralizing antibodies. This is likely to be partially due to its structural characteristics, consisting of a low number of envelope spikes that allows it to escape recognition as foreign and increases the likelihood of evolving envelope determinants that mimic self.221 High density display of HIV antigens is one method to combat this, with trimeric glycoproteins gp41 and gp120, as well as their precursor gp160 being highly targeted. As an example, a recent investigation studied the effect of presentation of the particularly conserved membrane-proximal external region (MPER) of HIV-1 gp41 on VLPs, and the approach produced anti-MPER antibodies that showed neutralizing activity in a rabbit model.222 Albeit moderate, the production of neutralizing antibodies is a valuable therapeutic response that could be improved through modifying MPER presentation. Another potential target for vaccines is CCR5, a cellular self-protein found to be involved as a co-receptor for HIV replication and pathogenesis. High-density display of CCR5-based peptides on Qβ resulted in high IgG antibody titers, which was able to protect 25% of rhesus macaques against intravaginal challenge with the highly virulent SIVmac251 strain.223 In terms of potential vaccines undergoing clinical trials, some success has been seen for the use of a treatment combining ALVAC, a canarypox vector vaccine, with boosters of AIDSVAX, a gp120 subunit vaccine, where vaccine efficacy of 31.2% was observed in a study consisting of 16,395 subjects in Thailand.224 While work still remains to be done to improve the efficacy of HIV vaccines, great strides have been made in recent years toward its realization.
Another area where vaccine production is of great interest is for protection from the highly virulent and deadly EBOV. Ebola VLPs have been generated consisting of glycoprotein, nucleoprotein, and VP40 matrix protein from the virus using a baculovirus expression system, and cynomolgus macaques vaccinated with the VLPs were completely protected against lethal EBOV challenge, with strong T cell responses likely contributing.225 Further investigation of Ebola VLPs consisting of glycoprotein and VP40 also produced in insect cells demonstrated the potential of delivery without adjuvants and revealed a strong immune response that protected against lethal challenge in mice when high doses (50 μg) were utilized.226 Optimization is still needed to enhance immunogenicity, and some prospects include improving glycoprotein incorporation during VLP assembly and including immunostimulatory molecules within the particles. A potential safer alternative to Ebola VLPs is the use of other viruses to display EBOV antigens instead. Vesicular stomatitis virus (VSV) is one such virus that has been studied, and using highly attenuated forms of VSV that have been genetically engineered to incorporate EBOV glycoprotein in place of its own has been an effective strategy, with a single dose being sufficient to protect both guinea pigs and macaques from challenge.227 These results are highly encouraging, and it will be interesting to see if there is effective protection against other strains of EBOV.
Aside from vaccines for human viral infections, it is of great interest to investigate animal vaccines as well for the protection of pets and livestock. An early study inserted a short epitope from the VP2 capsid protein of mink enteris virus (MEV) within the capsid of CPMV and found that it imparted protective immunity against clinical disease in mink, with a dose of 1 mg not only offering complete protection but also reducing shedding of the virus.79 Since the epitope occurs in canine parvovirus and feline panleukopenia virus as well, the same platform could be used for protection of minks, dogs, and cats. As another example, foot-and-mouth disease virus (FMDV) is a highly infectious virus that affects cloven-hoofed animals such as cattle and sheep, which are important in farming. Empty FMDV capsids produced using a baculovirus system and tested in guinea pigs were able to generate neutralizing antibodies against FMDV, but at a lower level than the commercial inactivated vaccine.228 There was still good antigenicity and immunogenicity, and use of crude protein extracts may have resulted in lower particle quantities in the experimental setup. Thus, the results are exciting for the use of noninfectious empty capsids to treat FMDV. Bluetongue virus (BTV) is another problematic disease that has been detrimental to the agricultural industry due to its high morbidity and mortality, affecting ruminants such as sheep and cattle. VLPs of BTV produced using a baculovirus expression system were found to be protective against infection when tested in sheep, with effective delivery of both single-serotype and multi-serotype cocktails and no interference observed from the presence of antibodies against other serotypes.229 BTV VLPs can also be assembled in plants using a CPMV-based HyperTrans vector system, and vaccination with these VLPs provided protection against BTV challenge.230 The particles were found to elicit a strong antibody response in sheep after a booster dose, comparable to live, attenuated virus used in a commercial vaccine. The development of vaccines in plant-based systems could result in cheap, easily scalable production without the danger of animal pathogen contamination.231
Interestingly, vaccines can also be applied to brain disorders such as addiction and Alzheimer's disease. Nicotine from tobacco use is the most common drug addiction worldwide, and reduction of nicotine transport to the brain has been found to decrease dependency on the drug due to reducing stimulation of the mesolimbic reward system.232 Nicotine covalently coupled to Qβ resulted in high drug-specific IgG antibody production in vaccinated mice, and the binding of the antibodies to nicotine caused a decrease in nicotine levels in the brain of up to 90% in individual mice.233 Further, Phase I trials found that this approach was safe and well tolerated, with high antibody production against nicotine in all individuals. Phase II trials have demonstrated that nicotine vaccines can be effective for patients quitting smoking, but there is still room to improve antibody titers for greater efficacy.234
Turning to Alzheimer's disease, amyloid-β (Aβ) peptide deposits are associated with the development of the neurodegenerative disorder and can be targeted with vaccines to reduce aggregation and ameliorate symptoms. In one study, hepatitis B virus core proteins displaying two 15-amino acid Aβ fragments taken from the N-terminus were assembled into chimeric VLPs and used in the immunization of an Alzheimer's transgenic mouse model without inclusion of additional adjuvant.235 The VLPs elicited a potent humoral response that reduced Aβ deposition and microgliosis, and there was a resultant improvement in learning and memory, with immunized mice more readily learning and remembering the location of the hidden platform in a Morris water maze. Another therapeutic that has potential for Alzheimer's disease is CAD106, a Qβ-based vaccine displaying the first six amino acids of Aβ that is undergoing Phase II testing for long-term treatment of patients with mild Alzheimer's disease.236 Overall, multiple exposures to CAD106 resulted in a prolonged time to antibody titer decline, and the treatment had favorable safety and tolerability profiles, with no occurrences of the severe adverse responses found in Aβ antibody therapies, such as meningoencephalitis and autoimmune disease. More time is needed to observe the long-term treatment effects of the vaccine, but so far the results are promising.
Finally, we look at the treatment of cancer through a few different immunotherapy approaches. Recently, Merck brought Gardasil 9 to market, an HPV vaccine upgrade from Gardasil that protects against 9 serotypes of HPV that account for 90% of HPV-related cancers.237 While this is fantastic news, alternatives are necessary due to issues with cost and distribution in the developing world. Commercial vaccines are VLPs derived from the L1 major capsid protein of HPV, which is not conserved across serotypes. Thus, a simpler approach that can be more broadly protective would be more ideal. As an example, efforts have been made to produce highly immunogenic L2 VLPs that are stable over time without refrigeration.238 Although the L2 minor capsid protein of HPV is less exposed and less immunogenic, it is highly conserved across serotypes, and MS2 displaying a short L2 peptide from HPV type 16 worked particularly well in preclinical mouse models. Reconstituted virus after spray drying remained highly immunogenic without use of an adjuvant even after 7 months of storage at room temperature, and mice vaccinated with the 16L2-MS2 VLPs were additionally protected from heterologous HPV pseudovirions of types 31 and 45, while Gardasil only protected against type 31.
Vaccines can also be used for protection by stimulating the immune system against the cancer cells themselves. HER2 is a receptor overexpressed on breast cancers that tend to be more aggressive and is one potential target for cancer immunotherapy. Presentation of P4378-394, a B-cell epitope from the extracellular domain of HER2, on PVX led to higher antibody titers that were specific to HER2 compared to soluble P4 peptide alone.239 PVX-based carriers are promising for vaccines due to their tropism toward B cells154 and large surface area imparted by their filamentous nature leading to high multivalency. A different approach against HER2 cancer cells resulted in T cell mediated tumor prevention by utilizing the association between the minor capsid protein VP2 of murine polyomavirus (MPyV) with the internal surface of major capsid protein VP1.240 The VP2 coat proteins were fused to HER21-683 and the particles were assembled in such a way that immunization of mice with the VLPs resulted in antibodies to VP1 but not HER2. On the other hand, the VLPs induced the production of HER2-specific T cells, and in both an autochthonous HER2 breast cancer mouse model and a model in which mice were challenged with HER2-positive D2F2/E2 cells (but not in a model with HER2-negative D2F2 cells), complete protection against tumor growth was observed for over 80% of the mice. Therefore, this is a potential approach for a potent prophylactic vaccine against HER2 cancer. Beside cell surface receptors, Tn antigen, a tumor-associated carbohydrate antigen, has been widely explored for presentation on viral capsids, with investigations into utilizing particles such as CPMV, TMV, and Qβ.241-243 Although Tn is one of the weakest antigens, strategic patterned display of the antigen was able to induce a potent humoral immune response that recognized human tumor cells. Evaluation of Tn presentation for in vivo cancer protection will be an essential next step for carrying this forward as a cancer vaccine therapeutic.
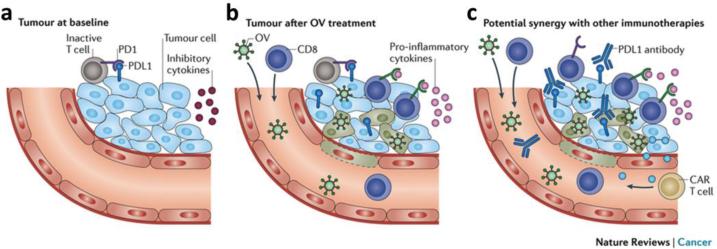
It is clear that by taking advantage of the body's natural immune system, myriad applications for virus-based vaccines can be realized. We would like to briefly highlight the application of viruses for immunotherapy as well, especially with the recent groundbreaking approval of the first oncolytic virus (OV) for cancer therapy by the FDA in October 2015.135 T-VEC was approved for the treatment of melanoma patients and is a viral vector based on HSV type 1. The mechanism of action for OVs is not yet well understood but appears to involve both lysis from more rapid replication within tumor cells as well as promotion of systemic anti-tumor immune response.244 T-VEC encodes for granulocyte–macrophage colony-stimulating factor (GM-CSF), a cytokine involved in dendritic cell recruitment maturation that aids in additional stimulation of anti-tumor immunity. In phase III clinical trials, treatment with T-VEC led to durable responses, even for patients with advanced stage IV disease. The approval of T-VEC is a significant step forward, and future investigations will be important for enhancing efficacy through combination approaches (Figure 9) as well as for the establishment of safety profiles and regulatory guidelines. For more information on the action of OVs and other recent developments in this field, please refer to the following reviews.244, 245
Figure 9. Oncolytic virus therapy action and potential synergy.
a) Immune clearance of tumors at baseline is inhibited by inactivation of T cells through binding of their programmed cell death protein 1 (PD1) receptor to programmed death ligand 1 (PDL1) expressed on tumor cells as well as by secretion of inhibitory cytokines. b) OV treatment triggers local expression of pro-inflammatory cytokines and/or overrides immune checkpoint inhibition, resulting in immune stimulation and recruitment of immune cells. c) Combination of OV therapy with other immunotherapies such as PDL1 antibodies and chimeric antigen receptor-expressing T cells can be used to enhance immune responses. Reproduced with permission from ref. 245. Copyright 2015 Nature Publishing Group.
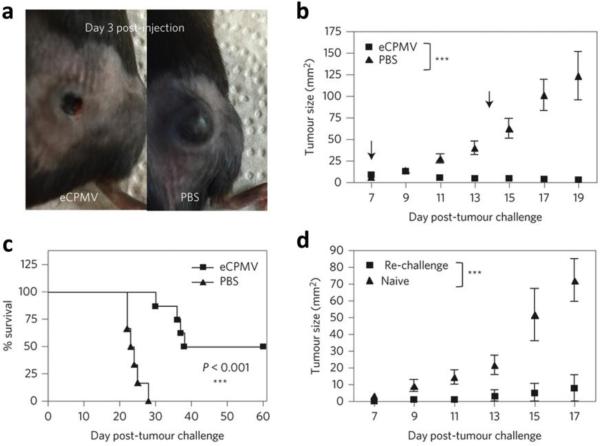
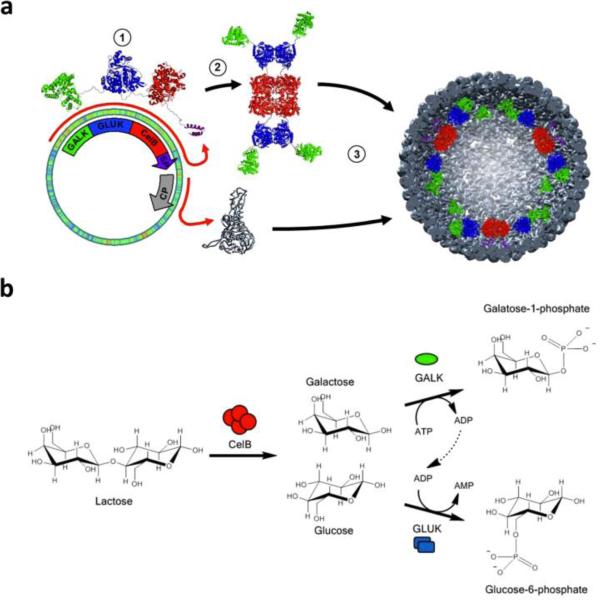
As a final example of the effectiveness of virus particles for immunostimulation, a recent study, in which we collaborated with the Fiering Lab at Darmouth University, demonstrated that in situ vaccination of tumors with eCPMV, just the capsid without any nucleic acid or modification, could help overcome the immunosuppressive tumor microenvironment.246 Treatment with eCPMV led to reduction and even regression of tumor growth and metastasis in a variety of mouse models, including melanoma, ovarian carcinoma, colon cancer, and breast carcinoma. eCPMV was found to specifically target and activate neutrophils in the tumor microenvironment, leading to a strong and rapid anti-tumor response. The response was found to be systemic and durable, with mice that eliminated primary B16F10 melanoma tumors through eCPMV-mediated immunity resistant to re-challenge and three out of four mice completely rejecting the tumor (Figure 10). Systemic protection is likely the result of immune memory against tumor antigens and mediated by T cells. These results were established for unmodified eCPMV, therefore opening the opportunity for further enhancement of efficacy through the display of antigens or the inclusion of adjuvants and chemotherapeutics.
Figure 10. Systemic anti-tumor immunity after in situ vaccination with eCPMV.
a) Images of mice with flank B16F10 melanoma tumors three days after intradermal injection of either eCPMV or PBS demonstrate slower growth with eCPMV. b) Tumor measurements of the mice after treatment (arrows indicate treatment days), with significant decrease in tumor progression rate for eCPMV (n = 8 for eCPMV, n = 6 for PBS). c) Kaplan-Meier curves illustrate survival of half the mice treated with eCPMV, with complete elimination of primary tumors observed for those mice. d) Rechallenge on the opposite flank 4 weeks later (n = 4/group) also saw delayed growth for eCPMV, and 3 out of 4 mice did not develop new tumors. *p < 0.05; **p < 0.01; ***p < 0.001. Reproduced with permission from ref. 246. Copyright 2015 Nature Publishing Group.
Overall, while there are still difficulties that exist for the development of vaccines targeting chronic diseases, cancer, and infectious diseases, progress is being made and recent results show great promise in the efficacy of some novel vaccines. The potential of VLPs for vaccines and immunotherapies is quite evident, with opportunities for the treatment of not only infectious diseases but also addiction, brain disorders, cancer, and more.
4.1.4 Gene delivery
In addition to vaccines, viruses have been applied for the delivery of full-length DNA, small interfering RNAs, and machinery for genome editing, such as nucleases, for treatment of a wide range of disorders. Gene delivery has had a long history, beginning with its initial conceptualization over 40 years ago.247 Since then, there has been an extensive body of work dedicated to the realization of gene therapy for treatment of diseases, with Parkinson's disease,248 cystic fibrosis,249 hemophilia,250 and cancer251 being just a few examples. The extension of gene therapy for the general public is starting to become more realistic, with the approval of UniQure's Glybera for clinical use in Europe in 2012 opening the door for bringing gene therapies into the market.252 The AAV therapy delivers the lipoprotein lipase gene to make up for deficiency in patients who cannot process triglycerides. Other gene therapies are poised to push toward commercialization in the coming year, among which includes an AAV gene therapy from Spark Therapeutics for the treatment of Leber congenital amaurosis visual impairment, which is caused by defects in the RPE65 gene.253 Based on lack of serious adverse events from initial studies and positive phase III trial results, they are expected to advance toward filing a Biologics License Application with the U.S. Food and Drug Administration.
The road to this point has certainly not been easy, with tragedy striking in 1999 when Jesse Gelsinger died during a clinical trial from a massive immune response instigated by the adenoviral vector meant to correct for his ornithine transcarbamoylase metabolic deficiency.254 Only a few years later, despite great success for many children, retrovirus-based gene treatment for X-linked severe combined immunodeficiency resulted in the development of leukemia in several young children as a result of gene insertion near oncogenes.255 Despite these misfortunate and discouraging results in the past, more research has led to a better understanding of gene delivery, its potential pitfalls, and how to overcome them, leading to a lot more control over accomplishing the purpose of gene delivery with less severe and fewer adverse effects. Due to the plethora of work done in this area, we can by no means be comprehensive in our review of gene delivery, and there are many excellent reviews that we invite the reader to consult.256-258 However, we will take a glimpse at a few studies to demonstrate the breadth of the field and the exciting studies being performed. We will then focus more on some recent developments in improving the safety profile of adenovirus and retargeting it for cell-specific delivery as well as the progress in utilizing non-mammalian viruses for gene delivery.
Adenovirus is an icosahedral non-enveloped virus with a core diameter of 90 nm and fibers that extend from its penton bases, which allow attachment to the host cell through the coxsackie-adenovirus receptor (CAR) (see Figure 1).259 Following binding, adenovirus can further bind to integrin receptors through RGD displayed at the base of the fibers. The specificity of adenovirus for mammalian cells can be utilized for gene therapy for cases such as glioblastoma, a highly aggressive brain cancer, with the ability to prolong time to death or reintervention.260 This tactic works by injecting adenovirus containing cDNA for herpes simplex virus thymidine kinase (HSV-tk) around the lesion after surgical resection of the tumor. Following systemic delivery of the prodrug ganciclovir, cells that are transduced with HSV-tk can then phosphorylate the prodrug to ganciclovir triphosphate, a cytotoxic nucleotide analogue that becomes incorporated into the DNA of actively proliferating cells. Adenovirus gene delivery can also be utilized as an antiviral therapeutic through RNA interference (RNAi), and this has been demonstrated for HBV using delivery of anti-HBV primary microRNA (pri-miR) cassettes driven by a murine transthyretin (MTTR) promoter.261 The promoter is a liver-specific transcription regulatory element and imparted high specificity, with barely detectable expression in non-liver cells. Furthermore, expression of anti-HBV pri-miRs resulted in knockdown of HBV replication up to 94% in mice.
As an alternative to adenovirus delivery, AAV is particularly popular due to its very low immunogenicity, inability to self-replicate, and capacity to target non-dividing cells.262 Stereotactic delivery of nerve growth factor (NGF) into nucleus basalis neurons in the basal forebrain has been shown to be a well-tolerated method for long-term NGF expression for the purposes of protecting patients with Alzheimer's disease from neural degeneration.263 AAV vectors can additionally provide protection from simian/human immunodeficiency virus (SHIV).264 Through delivery of a transgene encoding for eCD4-Ig, which binds to the CD4 envelope glycoprotein of HIV-1 as well as co-receptor CCR5 with high avidity, protection from multiple infectious doses of SHIV was conferred in rhesus macaques. AAV/phage hybrids (AAVP) have also been created in order to combine the targeting potential of phages with the transgene expression efficiency of AAV by inserting cis-regulatory elements from AAV into the M13 phage vector genome flanking the transgene cassette.212 Using the HSV-tk and ganciclovir approach described above for adenovirus, AAVP displaying RGD peptides was able to home to sarcoma cells and instigate transgene expression and tumor regression in a rat SKLMS1 human soft-tissue sarcoma xenograft model.
A similar outcome for combining targeting and transfection can also be achieved through covalent coating of adenovirus with a hydrophilic polymer to result in retargeting of the virus as well as protection from antibody neutralization.155 High immunogenicity is one of the downfalls for adenovirus delivery, resulting mainly from intravascular injection,265 and antibodies produced are primarily against the hexon capsid protein.266 While adenovirus can still be utilized for local administration, by shielding the hexons and incorporating targeting ligands, systemic delivery with target selectivity independent of CAR is made possible. Amine-reactive pHPMA-based polymers (mentioned previously in Section 4.1.1) have been used for adenovirus coating, and further modification of the virus with targeting ligands such as basic fibroblast growth factor (bFGF) and vascular endothelial growth factor (VEGF)155 or antibodies against E-selectin and P-selectin glycoprotein ligand-1 (PSGL-1) fused to IgG1 Fc267 have been demonstrated for retargeting of adenovirus to cancer cells and tumor-associated vasculature. Additionally, pHPMA-coated adenovirus with activatable cell penetrating peptides attached enabled cytoplasmic delivery of the virus to metalloproteinase-overexpressing tumor cells.268 Transduction with pHPMA copolymer coatings was shown to be maintained not only in cell cultures but also in vivo in mouse models.267, 269 PEG has also been shown to be an effective polymer for shielding270 and retargeting.271 As an improvement to simple PEG coatings, complexation with copolymers of PEG and PEI resulted in the ability to transduce CAR-negative NIH 3T3 cells, with the added benefit of less toxicity compared to PEI alone.272 Aside from PEI, cationic poly(amidoamine) (PAMAM) dendrimers can also be used for enhancing gene delivery. Through the addition of EGFR-specific ligand GE11, dendrimer-coated adenovirus carrying the sodium iodide symporter gene (NIS) was successfully applied for radiovirotherapy in a liver cancer xenograft mouse model, in addition taking advantage of iodide for 124I PET imaging of viral distribution.157
While mammalian viruses have the machinery for gene transduction, phages and plant viruses also hold potential for gene delivery and gene silencing. Early phage gene delivery was reported through targeting M13 to EGF with a mammalian gene cassette inserted into the vector backbone.273 Transduction was low but could be improved through the use of multivalent phagemid-based vectors (discussed in Section 4.2.1) and genotoxic treatment such as heat shock, UV irradiation, and camptothecin treatment. As a more practical method to overcome cellular barriers of mammalian cells, inspiration from HBV was taken and the PreS1 region of the HBV envelope protein involved in virus attachment during infection was displayed on bacteriophage T7, which resulted in more efficient gene transfer when tested in HepG2 human hepatocellular carcinoma cells.274 Another hybrid phage complex, this time with cationic polymers poly-D-lysine (PDL) and diethylaminoethyl-dextran (DEAE-DEX), was also able to improve transgene expression, and cell type specificity was retained from the display of RGD.275 Gene delivery with plant viruses is only beginning to take shape and has recently been demonstrated through assembly of CCMV coat proteins around heterologous RNA derived from Sindbis virus (SINV), which was shown to be released into the cytoplasm of mammalian cells through co-delivery of Lipofectamine-2000 (Figure 11).276
Figure 11. Gene delivery to mammalian cells using CCMV plant virus.
a) Strategy for delivering DI[EYFP], defective interfering RNA for enhanced yellow fluorescent protein (EYFP) derived from SINV, through cotransfection of CCMV containing the gene with Lipofectamine-2000. b) Flow cytometry analysis of transduction efficiency showing lower efficiency for VLP transduction than naked RNA but the cargo is protected from RNase A. c) Corresponding fluorescence microscopy images (columns are in same order as bar graph) showing EYFP signal due to transduction for all conditions except for naked RNA incubated with RNase A. Reproduced with permission from ref. 276. Copyright 2013 Elsevier.
For gene silencing, MS2 phage has been especially popular for packaging of RNA. Encapsulation of a cocktail of anti-cyclin small interfering RNAs (siRNAs) to silence expression of cyclin A2, cyclin B1, cyclin D1, and cyclin E1 in MS2 was shown to protect the RNA from degradation for over 3 months when stored in the fridge.277 Targeting these VLPs to hepatocellular carcinoma cells resulted induced apoptosis in over 90% of the cells at 150 pM siRNA concentration, with no substantial effect on the viability of normal hepatocytes. siRNA delivery with MS2 has been corroborated, with delivery of Bcl2 siRNA packaged within MS2 targeted to TfR of HeLa cells causing enhanced gene knockdown and apoptosis compared to non-targeted particles and having effectiveness similar to a commercial lipid transfection reagent.20 miRNA-mediated RNA interference (RNAi) has also been explored, with MS2 conjugated with HIV-1 Tat47–57 peptides shown to be able to effectively transfer encapsulated pre-miRNA into a range of cell lines and tissue types, which was processed into mature miRNA and subsequently suppressed expression of specific target gene.278 The same group also showed packaging of antisense RNA delivered with the same Tat peptide for inhibition of hepatitis C virus (HCV) RNA translation.279 While so far these studies have mainly been proof-of-concept, they suggest VLPs have much potential as gene delivery systems, with many directions for gene therapy applications.
4.1.5 Drug delivery
In addition to gene delivery, viruses can also be used for the delivery of drugs, where viruses are applied as carriers of therapeutic cargo for photothermal therapy, photodynamic therapy (PDT), and chemotherapy. Photothermal therapy is an area whose potential has barely been tapped in the virus realm. In general, heat is produced by metallic nanostructures through the absorption of NIR or infrared light to induce hyperthermia, which can be utilized for killing susceptible tumor cells. Using such an approach, treatment can be applied to a specific area and toxicity elsewhere is reduced. So far, there has been one study, in which 1.3 nm gold was covalently attached to adenovirus and delivered to HeLa cells, where the possibility of using virus-based particles for a combination of photothermal and gene cancer therapy was discussed.280 Other studies since then have also been successful in attaching gold to VNPs,281, 282 but photothermal therapy using these formulations has yet to be explored.
Photodynamic therapy, like photothermal therapy, utilizes light as a trigger, except with the effect of creating localized cytotoxic reactive oxygen species (ROS) for therapy. Initially, PDT was applied as an antimicrobial approach, using CCMV functionalized with a ruthenium-based photosensitizer and directed to Staphylococcus aureus using both an electrostatic approach with PLL and a targeted approach with an antibody for protein A, which is present in the cell wall.283 PDT was demonstrated at standard antimicrobial fluence rates of up to 55.2 J cm−2, with cell reduction of about 3 orders of magnitude observed and more selectivity imparted through the use of anti-protein A for targeting. Since then, PDT has been applied as a therapeutic against cancer cells. The buckyball C60 has been attached to CPMV and Qβ to improve solubilization of the photosensitizer,94, 284 and to aid in delivery to and treatment of PC-3 prostate cancer cells.284 Internal conjugation of porphyrins to MS2 combined with external display of aptamers for protein tyrosine kinase 7 receptors on Jurkat leukemia T cells has been established for selective killing of Jurkat cells when cultured with erythrocytes after 20 min of illumination at 415 nm.285 Additionally, as a proof-of-concept study, glycan decoration of Qβ was shown to be an effective strategy for targeting cells bearing CD22 receptors that are involved in the regulation of B cells, and this was illustrated with the delivery of metalloporphyrins for specific elimination of CD22-positive cells.286 To move toward clinical application of PDT, use of photosensizers that can be excited in the NIR range would be more ideal for better tissue penetration. Phthalocyanine dyes are one such class of photosensitizers, and self-assembly of CCMV with phthalocyanine encapsulated has been explored recently.125, 287 CCMV coencapsulated with Gd(DOTA) micelles and phthalocyanine dyes resulted in better capsid stability and further imparts the possibility to perform combination therapy and MR imaging,287 and thus it only remains to investigate their potential value in PDT.
Virus-based and other nanoparticles have also been developed for the delivery of chemotherapies. Chemotherapy is associated with dose-limiting toxicities, and specific delivery to cancer cells using carrier systems increases safety as well as targeted payload delivery.288, 289 In particular, doxorubicin (Dox) delivery has been studied extensively, likely popular in part due to the clinical success of Doxil, a liposomal formulation of Dox.290 Dox works through intercalating into DNA and causing oxidative DNA damage.291 Conjugation of Dox can be achieved by pH-cleavable hydrazone linkages, as demonstrated in one study where an alkyne-functionalized hydrazone linker was used for attachment to azide-functionalized polymers grown from the capsid of Qβ by ATRP.105 In such a manner, the Dox could then be subsequently released by low pH cleavage of the linker after particle uptake, which was shown to maintain efficacy in the killing of HeLa cells. Methods for conjugation have also been recently established for spheres made from thermal transitioning of TMV and utilized for the loading of doxorubicin, which was found to be effective for chemotherapeutic delivery to breast cancer cells.292 Besides chemical conjugation, Dox can be loaded by infusion into RCNMV and incorporated into a fibrous matrix made up of polylactic acid (PLA) and polyethylene oxide (PEO) nanofibers.113 The combination of the two systems could be tailored to result in either a two-phase release profile or a first order release profile, depending on whether the virus was co-spun with the fibers or the matrix immersed in the virus solution after electrospinning, respectively.
Specific delivery of Dox to cells has been achieved through conjugation of FA to HCRSV and CMV,163, 184 SPARC binding peptides to M13,293 and peptides for CD46 receptor and N-cadherin targeting to RCNMV,294 as well as by utilizing CPMV's natural interactions with surface vimentin.295 For instance, with Dox encapsulated within Hibiscus chlorotic ringspot virus (HCRSV) further conjugated with FA, more efficient inhibition of OVCAR-3 ovarian cancer cell growth was achieved, while no difference in drug efficacy was observed when CCL-186 human diploid fibroblast cells were tested as a control for normal cells (Figure 12).184 As another example, CPMV was used for treatment of HeLa cells through the display of Dox attached either by a direct covalent bond or through a disulfide linker.295 With the disulfide linkage, the cell killing was similar to free Dox, likely due to release of the drug in cell culture media before particle uptake, but covalent attachment of Dox to CPMV resulted in more efficient cell killing than free Dox, with almost complete elimination of the cells at a concentration of 1.45 μM for the CPMV formulation whereas cells treated with free Dox were still completely viable.
Figure 12. FA targeting for specific cell killing with Dox.
a) Schematic of formation of HCRSV-based protein cages without (PC-Dox) and with FA conjugation (fPC-Dox) where Dox is encapsulated during capsid reassembly with the inclusion of polyacid. b) Confocal microscopy of Dox uptake for OVCAR-3 ovarian cancer cells and CCL-186 fibroblast cells incubated with free doxorubicin, PC-Dox, fPC-Dox, and fPC-Dox in the presence of FA. c) Cell viability curves of cells after treatment with varying concentrations of the different formulations showing fPC-Dox had greater inhibition of OVCAR-3 cells without affecting pattern of CCL-186 inhibition. Reproduced with permission from ref. 184. Copyright 2007 American Chemical Society.
Virus-based platforms have also been explored for the co-delivery of therapies. In one study, M13 was investigated for its feasibility in delivering hygromycin and Dox specifically to either SKBR3 human breast adenocarcinoma cells that overexpress HER2 or A431 human epidermoid carcinoma cells that overexpress EGFR.296 Dox delivery was successful and was shown to require a cathepsin-B cleavable peptide linker for efficacy, but delivery of hygromycin was particularly impressive, with over a 1000-fold improvement in potency compared to free drug administration. However, different targeting strategies were used, making it difficult to draw conclusions as to the relative efficacy of the two drugs. Another study assessed the range of therapeutic cargo that could be encapsulated within MS2, including doxorubicin, cisplatin, and 5-fluorouracil (5-FU), along with ricin toxin A-chain (RTA).277 Using SP94-targeted formulations that bind to Hep3B hepatocellular carcinoma (HCC) cells with 104-fold higher avidity compared to other cell types, the encapsulation of Dox individually was shown to be more effective than free Dox, while encapsulation of a cocktail comprised of all three chemotherapeutics was even more effective, with an IC50 concentration below 1 nM. Delivery of the toxin RTA was also remarkable, with almost complete elimination of Hep3B cells at a concentration of 100 fM without affecting the viability of control cells.
While the targeted chemotherapeutic cocktail above was highly effective, other approaches using prodrugs have also been investigated to further reduce the risk of toxicity. For example, instead of direct delivery of 5-FU as a treatment option, encapsulation of the enzyme yeast cytosine deaminase (yCD) is an alternative method that results in the presence of 5-FU in the target cell by conversion of 5-fluorocytosine and as a consequence causes target cell death.297 In another study that utilized conversion of a prodrug, delivery of an exogenous protein horseradish peroxidase (HRP) was demonstrated with M13 displaying Ypep2 peptides, which have selectivity for PC-3 prostate cancer cells.298 After delivery, HRP was able to oxidize indole-3-acetic acid to produce a peroxyl radical that led to cytotoxicity.
Other anticancer drugs that have been investigated for virus-based nanoparticle delivery include taxol,299 bortezomib (or BTZ),300 and trastuzumab (or Herceptin).144 Cardiovascular disease is another route for virus-based therapeutic intervention, and CPMV delivery of chromium has shown promise for protecting against diabetic atherosclerosis in vascular smooth muscle cells.301 Additionally, filamentous phages have been applied for the delivery of antibacterial agents, including neomycin and chloramphenicol for growth inhibition of E. coli, S. aureus, and Streptococcus pyogenes.302, 303 Antiviral delivery with viral vectors is a new development, and the similar tropism of the plant virus CPMV to antigen presenting cells that are commonly subverted by pathogenic viruses was exploited for combatting chronic infectious disease caused by the prototypic arenavirus lymphocytic choriomenigitis virus.111
Only very recently was drug delivery with VNPs or VLPs demonstrated in vivo.304 Since TMV is a hollow nanotube with a negatively charged interior channel, it could be taken advantage of for the loading of cationic drugs. The highly potent platinum DNA-binding drug phenanthriplatin was introduced within the carrier and shown to be released under acidic conditions. When TMV delivery of phenanthriplatin was applied in a triple negative breast cancer mouse xenograft model, much greater efficacy was observed for TMV-phenanthriplatin compared to free drug or clinically used cisplatin controls, with 4-fold smaller tumor growth. This is likely due to better transport of the drug to the tumor cells with the nanocarrier. The combination of TMV and phenanthriplatin shows potential for bringing an effective new chemotherapy into the clinic, but virus-based nanoparticle drug delivery is a quickly growing field, and it is expected that progression toward other clinical applications is also fast approaching.
4.1.6 Tissue engineering
Viruses have been incorporated into biocompatible tissue engineering scaffolds for directing cell growth, alignment, and differentiation. The neighboring environment affects the behavior of cells, both in terms biological cues such as presence of chemokines and ligand as well as physical cues such as topology and mechanical moduli. Using this knowledge, scaffolds can be designed to regulate cells in a manner suitable for applications in the repair or replacement of damaged tissue.
As a first step, cell adhesion to viral scaffolds was investigated. Pioneering work utilized layer-by-layer (LbL) assembly of CPMV and polymer poly(diallyldimethylammonium chloride) (PDDA) to form thin films that were found to aid in the adhesion and proliferation of NIH-3T3 fibroblasts, with more layers leading to more CPMV adsorption and greater cell adhesion.305 Research then moved toward the use of filamentous viruses, as they better mimic the structure of the extracellular matrix (ECM). Coating of TMV with different cell binding motifs derived from integrin binding matrix proteins collagen and fibronectin onto a high binding plate demonstrated that the peptide sequence displayed plays a role in cell adhesion and morphology.306 While cells cultured on TMV with RGD motifs formed filopodial extensions, they adhered more weakly compared to cells that remained rounded when cultured on TMV displaying P15, a collagen I mimetic sequence. For screening the influence of various biochemical cues on cell proliferation and morphology, phage-chips have been constructed such that arrays of M13 nanofibers labeled with various peptides or growth factors and self-assembled on gold chips can be monitored using surface plasmon resonance (SPR) spectroscopy for their effect on cultured cells.307 Further progression toward mimicking the ECM looked at synthesizing fibrous matrices through electrospinning of RGD-modified viruses with polymers.308, 309 Nanofibers made up of TMV with polyvinyl alcohol (PVA)308 and M13 with poly(lactic-coglycolic acid) (PLGA)309 formed biodegradable fibrous matrices that enhanced cell adhesion, proliferation, and spreading of baby hamster kidney (BHK) and fibroblast cells, respectively, compared to scaffolds of the polymers alone.
As an advancement from cell adhesion and proliferation, external stimulus can also direct the orientation of cell growth, which is important for the function of many cell types such as cardiac and skeletal myocytes. Taking advantage of available viruses with elongated geometries, M13 and TMV have been utilized for cell alignment. In an early study, M13 with RGD motifs was self-assembled into parallel arrangements through slow drying or dragging methods to form thin films on which oriented growth of NIH-3T3 and Chinese hamster ovary (CHO) cells were achieved, with the shear method producing the most consistent results.310 Investigation of the ECM deposited by cells cultured on these films revealed correlation between cellular alignment and the orientation of fibronectin and collagen I deposition.311 NIH-3T3 fibroblasts produced more ECM proteins, which resulted in more of a tendency to deviate from the original patterning, and thus the fibroblasts displayed reduced alignment over time as well as when compared with less ECM-producing BHK cells. Other methods for the alignment of viral nanorods, and subsequently cells, include shearing force from fluid flow through glass capillaries312, 313 and microcontact printing combined with dip-coating.314 Myogenic differentiation of myoblast cells can be achieved through exposure to differentiation media after oriented cell growth.313
In addition to forming 2D films, fabrication of 3D aligned fibers is possible based on interfacial polyionic complexation, which was demonstrated by injecting negatively charged, RGD-labeled M13 phage into a solution of cationic polymers PEI, PLL, and chitosan.315 NIH-3T3 fibroblast cells encapsulated through co-injection with the phage solution grew well within the fibers and after seven days began to spread along the matrix within the phage fibers, demonstrating the potential of virus scaffolds for cell growth and remodeling. 3D tissue cultures have also been formed through treatment of cells with hydrogels that can then be magnetically levitated, and cell clustering could be controlled by the magnetic field profiles of the magnets used.316 Hydrogels were fabricated through simple combination of solutions of gold nanoparticles with M13 displaying RGD,317, 318 with the additional inclusion of magnetic iron oxide nanoparticles for levitation. The 3D cell cultures were able to recapitulate in vivo behavior, as observed by similar protein expression patterns as well as infiltration of highly invasive glioblastoma cells when co-cultured with astrocytes.
An application of phage nanofiber formation is the growth and differentiation of neural cells. Fibers containing M13 displaying RGD or IKVAV, a laminin motif that plays a role in neural cell adhesion and neurite extension, were shown to be advantageous for neural progenitor cell (NPC) proliferation and differentiation, with extension of neurites parallel the fibers observed.82 The cell binding motifs are crucial for inducing neural cell growth, and it has been demonstrated that using a control RGE peptide results in a drastic decrease in cell adhesion and neurite outgrowth.319 There is a range of cell binding peptides derived from fibronectin and collagen that have been investigated and found to be effective for supporting neural growth and enhancing neurite extension,320 and it would be of interest to further study the specific interactions between the cells and the scaffold to better understand the roles of the peptides in cell differentiation. In addition to cell binding motifs, immobilization of growth factors to phages has been demonstrated to retain bioactivity, with bFGF shown to promote NPC proliferation and NGF leading to greater neural differentiation.321 Furthermore, a recent study demonstrated that electroactivity could also be used to augment neural tissue regeneration.322 TMV that was modified with polyaniline and further doped with poly(styrenesulfonate) (PSS) was aligned by flow through capillary tubes and shown to increase the percentage of cells with neurite outgrowths as well as the percentage of cells with bipolar morphology.
Virus scaffolds are not limited to neural differentiation, and by far the most extensive research has been performed in osteogenic differentiation. An early study looked at osteoblast differentiation of bone marrow stromal cells on icosahedral turnip yellow mosaic virus (TYMV).323 Osteocalcin gene expression and onset of mineralization were found 7 days earlier for cells grown on TYMV-coated substrates compared to tissue culture plastic when cultured in osteogenic media, indicating the nanotopology imparted by the TYMV supports osteogenic differentiation. Similar data was found for substrates coated with rod-shaped TMV, with further DNA microarray data showing differential expression in a large panel of genes and bone morphogenetic protein 2 (BMP2) in particular found to be especially important in osteogenic differentiation in this manner.324 There was a rapid onset in BMP2 gene and protein expression, and this enhancement was only found when TMV was coated on the substrate and not when added to the cell media, verifying the role of nanotopology.325 Additional conjugation of phosphate groups to the exterior of TMV further boosted differentiation by aiding in the incorporation of calcium and consequently highly enriched mineralization of the ECM.326 A related strategy for mineralization utilized genetic engineering to display highly negatively charged E8 peptides on M13 phage, which could then be self-assembled into nanofibers in the presence of calcium ions and led to hydroxyapatite formation with the addition of phosphate ions.327
The effect of multivalent presentation of various ligands has also been studied, and rapid differentiation and nodule formation was observed for substrates with TMV coated with RGD after only 2 days in serum-free osteogenic media.328 While this was found for TMV genetically engineered to display RGD, TMV chemically conjugated to RGD also enhanced bone differentiation.329 In the absence of osteogenic supplements, presentation of RGD with the addition of synergy peptide PHSRN on M13 was actually found to be sufficient to induce osteoblastic differentiation of mesenchymal stem cells (MSCs).330 Display of DGEA peptide derived from collagen.331 along with PDPLEPRREVCE derived from osteocalcin and YGFGG derived from osteogenic growth peptide332 on M13 have also been shown to accelerate proliferation and differentiation of MSCs into osteoblasts. M13 is capable of self-assembly into long-range ordered morphologies using dip-coating methods, forming nematic orthogonal twists, cholesteric helical ribbons, and smectic helicolidal nanofilaments based on conditions such as phage concentration and pulling speed.333 Films formed using RGD and EEEE peptide-labeled phages could be used to control both soft and hard tissue formation, with smectic hilcolidal nanofilament surfaces in particular demonstrated to form enamel-like composites when treated with calcium and phosphate ions. Moving toward 3D scaffolds, TMV-RGD was incorporated within porous alginate hydrogels and resultantly produced greater cell attachment and enhanced osteogenic differentiation.83 3D printed scaffolds composed of hydroxyapatite and β-tricalcium phosphate is another approach, and introduction of M13-RGD combined with chitosan within the scaffold pore not only led to osteogenesis but also angiogenesis, with inclusion of VEGF further enhancing the effect (Figure 13).334
Figure 13. 3D printed virus-activated bone scaffold with angiogenesis.
a) Schematic of 3D printed bioceramic bone scaffold incorporating negatively charged RGD-labeled phage nanofibers using positively charged chitosan for new bone and blood formation when seeded with MSCs. b) Images of scaffold architecture. Scanning electron microscopy (SEM) of bone scaffold showed macro-scale (1) and micro-scale (2) pores, as well as pores filled with chitosan and phage matrix (5). Atomic force microscopy (AFM) (3) and transmission electron microscopy (TEM) (4) demonstrated morphology of phage nanofibers. 3D confocal fluorescence imaging showed presence of dye-labeled phage (red) within matrix-filled pores (6), and brightfield imaging revealed support of MSC adhesion for both the scaffold pores (7) and columns (8). c) Immunofluorescence staining for endothelial CD31 (1, 3, 5) and hematoxylin and eosin (H&E) staining (2, 4, 6) of implants of negative control (wild-type phage), virus-activated matrix (VAM), and positive control (RGD-phage with VEGF) scaffolds, respectively, as well as quantitative analysis (7) showed VAM promotes angiogenesis at an intermediate level. (**p<0.01). Reproduced with permission from ref. 334. Copyright 2014 John Wiley & Sons.
A new direction in this area is the incorporation of gene delivery within tissue engineering constructs. Mutant FLAG-tagged AAV was tethered to scaffolds made up of PLGA and gelatin sponge through anti-FLAG antibodies, and virus transduction was observed when HeLa cells were seeded onto the scaffold, both in vitro and when implanted in vivo into nude mice.335 Furthermore, transduction was observed for cells cultured on drop-cast films consisting of hybrid phages constructed from the combination of M13-RGD phage with an AAV-derived gene cassette.336 While still in its early stages, with further developments, tissue regeneration and reprogramming of cellular defects could be made possible through the combination of tissue engineering and gene delivery.
For future translation of virus-based tissue scaffolds, the immunogenicity and long-term effects of the viruses must be considered. Investigation of the in vivo behavior of implanted porous alginate hydrogels containing TMV and TMV-RGD revealed good biocompatibility, as evidenced by normal wound healing, hydrogel biodegradation over time, no pathological inflammation, and very little immune response triggered as opposed to intramuscular injection of native TMV.337 In addition to degradation of the hydrogel, it is expected that the protein-based viruses will also be degraded over time by cell proteinases, thus mitigating concerns of the implications of long-term deposition of the material. Since previous work has shown that viral substrates can be utilized for rapid differentiation in serum-free media, thus without the presence of xenogeneic proteins and growth factors,328 implantation of tissue scaffolds after ex vivo culturing of cells is a tangible reality.
4.2 Biotechnology
In the realm of biotechnology, viruses have found use for a variety of applications including peptide display technologies, confined synthesis, multiplexed sensors, diagnostics, nanoreactors, catalysts, as well as agriculture, several examples of which are discussed in the following sections.
4.2.1 Phage display technologies
The first of these technologies we will cover is phage display, which is routinely applied for a myriad of applications. The display of foreign sequences on filamentous phages was first described by George Smith in 1985.338 Since then, it has become a prominent method for the selection of peptides and antibodies that have affinity for specific targets, encompassing both organic and inorganic matter. Many reviews have been written that delve into the depths of phage display technology.339-343 Our aim in this section is to discuss the characteristics of phage display and highlight a few of the interesting studies and some of the latest applications.
Based on the original research in which f1 phage was studied,338 E. coli filamentous bacteriophages, which include f1, fd, and M13, are some of the more common platforms utilized for phage display. The general structure of filamentous phages is shown in Figure 14,344 which illustrates how the phages are comprised of a number of minor and major coat proteins. Out of these, the pIII and pVIII proteins are what are usually utilized for the display of foreign proteins,345 likely due to greater accessibility at the tip and sides, respectively, but display with pVI,346 pVII,347, 348 and pIX348 have also been successfully implemented. In brief, phage display utilizes the genetic programming of the phage coat proteins; insertion of random sequences of DNA within the protein genes can be used to form a library of phages with billions of different foreign peptides or proteins presented.342 After several rounds of panning and amplification to isolate the specific phages that bind to a target, the identity of the binding peptides or proteins displayed can be determined through sequencing. Whereas insertions at pVIII are limited to around 9-mers,349 cyclic phage libraries350 and libraries displaying sequences of up to 38 random residues on pIII are possible.351 Phage display is not only limited to filamentous phages, as libraries based on other phages such as lambda and T7 are also possible.352, 353
Figure 14. Filamentous phage structure.
a) Schematic of phage structure showing how the five structural proteins are arranged around its ssDNA genome. b) Legend labeling the structural proteins with approximate values for size, weight, and copies/virion. Reproduced with permission from ref. 344. Copyright 2011 Løset et al.
Another alternative that has proven useful in phage display is the use of phagemids.354 Phagemids typically contain traditional plasmid aspects, with a plasmid replication origin, restriction enzyme recognition sites, and an antibiotic selection marker, as well as phage aspects, including a phage origin of replication and a gene for pIII fused to any protein of interest (Figure 15).355 By co-infection with a helper phage to supply other structural proteins required for phage formation, complete virions assembled around phagemid DNA can be recovered. The advantages of using phagemids include ease of cloning and recombination, more efficient transformation, a smaller genome for incorporation of larger foreign genes, and greater genetic stability over multiple rounds of propagation.354
Figure 15. Phage display cycle with phagemid.
A library of DNA sequences with random variations of the protein of interest (POI) displayed on the pIII coat protein is cloned into a phagemid vector. After transformation of E. coli cells and subsequent infection with helper phages, the phage library is created. Using an immobilized target molecule, rounds of selection and amplification are performed until phages with the highest affinity are isolated. DNA sequencing can be used to identify the phages, and/or directed evolution can be used to create new libraries for panning. Reproduced with permission from ref. 355. Copyright 2011 Biochemical Society.
Phage and phagemid display technology have given rise to numerous opportunities for the isolation of protein-based ligands for a range of applications. In its simplest form, screening can be performed in vitro, where the target of interest is immobilized on a solid support. Using this technique, peptides have been identified that are specific for targets that include inorganic materials such as hydroxyapatite,356 silver,357 and quantum dots,13 as well as organic materials such as microtubules,358 fibrin,359 and integrins.360 As some examples, these binding peptides can be used for biomineralization for formation of hard tissue356 as well as for nucleation, growth, and patterning of metals.357 It should be noted that many of the examples found in other sections that involve mineralization rely on the use of metal binding peptides for nucleation. Additionally, peptides can be selected that not only bind but also obstruct the function of its targets, such as demonstrated with peptides identified for inhibition of proteases such as human neutrophil elastase,361 as well as cancer-associated matrix metalloproteinases.362
The versatility of the technology can be expanded beyond peptide identification. For example, phage libraries can be designed for the display Fab antigen-binding fragments and scFvs,339 which has implications for identification of antibodies for bioassays as well as immunotherapy. Some examples include the construction of a Fab phage display library for the isolation of antibodies specific to human prostate cancer cells363 and the generation and identification of scFv antibodies that recognize a marker of angiogenesis, VEGFR-3.364 Besides Fabs and scFvs, an alternative method of display where the antibody heavy- and light-chain variable regions were separately displayed on pVII and pIX, respectively, was demonstrated to effectively drive the formation of functional Fv heterodimers, which may have greater affinity and stability compared to scFvs.365 Enzymes and their substrates can also be displayed on phages, either independently or together on a single particle, in order to screen for functional enzyme catalysts.366 Co-presentation of the enzyme and substrate can be achieved using an intervening linker between the two, and then active enzymes can be selected for by panning for product formation.367
Aside from immobilization of isolated targets on a surface, selection methods have also been demonstrated with cultured cells, as well as ex vivo and in vivo, to select for cell binding peptides.343 Using panning against cell lines of various types, such as fibroblasts and myoblasts,368 peptides with high cellular specificity can be identified. In this manner, peptides have been isolated that have specificity for endothelial cells associated with atherosclerosis,369 breast cancer cells,370 hepatocellular carcinoma cells,371 melanoma cells,372 and ovarian cells,373 among others. Ex vivo panning works similarly, except the library is screened against cells or tumor masses that have been isolated, and binding peptides have been identified for a range of cell types that include neuroblastoma cells,374 islet cells,375 and colonic adenoma cells.376
In vivo phage display is particularly noteworthy and lends itself to many different applications. For example, peptides targeting specific organs can be examined using in vivo screening of random peptides in a mouse model.377 After multiple rounds of phage administration, isolation, and amplification, peptides that localize to the brain and kidney were identified. Furthermore, attachment of brain targeting peptides to fixed red blood cells resulted in the accumulation of the cells in the brain at a greater extent than the kidneys. Another investigation that also studied kidney targeting in vivo found that peptide specificity could be used for the tailoring of pharmacokinetics.378 By directing clearance toward the kidneys and away from the reticuloendothelial system, more rapid clearance could be achieved. In vivo screening has also been applied in humans for the mapping of unique zip codes lining the vasculature.379 Unique tripeptide motifs specific for various regions around the body were identified after biopsies of the bone marrow, fat tissue, skeletal muscle, prostate, skin, and liver were performed, which could be utilized toward the creation of a map of molecular signatures along the human vasculature.
As the above examples indicate, the reach of phage display applications is extensive and encompasses mineralization, in vitro assays, targeted delivery, molecular imaging, vaccines, and tissue engineering.
4.2.2 Sensing and multiplexed systems
The multivalency of viruses can be applied for sensing and multiplexed systems; the high degree of multivalency has been shown to improve the detection limit in a number of settings. Phages identified from a library against the desired target sequence can be directly applied as sensors. Genetic engineering simplifies the manufacture of large quantities of phages displaying specific targeting moieties, and several reviews cover the wide range of sensing applications available for these materials.380, 381 Besides phages, many other virus-based platform technologies have also been developed for signal detection and amplification. In our examples, we will cover the broad range of virus-based materials incorporated in manifold sensing technologies, which include antibody-based, electrochemical, and optical techniques.
Enzyme-linked immunosorbent assay (ELISA) is a traditional technique for the detection of target antigens.382 Although several variations exist, it generally relies on three components: (1) a probe specific for the target, (2) an antibody tagged with an enzyme to detect the probe, and (3) a substrate that is converted in the presence of the enzyme. The first probe tends to also be an antibody with specificity for the target, but phage display has introduced the opportunity for using peptides for target recognition. For example, phages decorated with peptides isolated from phage display have been used in ELISAs for the detection of anthrax spores383 and the surface antigen of HBV.384 For an alternative to the presentation of peptides on viruses, antibodies can also be presented on the viral scaffold for multivalent detection, and the previous section discussed how phage display and genetic engineering could be used for the display of antibodies and fragments thereof. As another approach, it was recently demonstrated that functionalization of PVX-based nanoparticles with protein A fragments can be used to display whole antibody molecules by using protein A's property of binding to the Fc region of the heavy chain of IgGs.85 The PVX particles could then be used as a plug-and-play system for the display of a variety of antibodies, with a level of orientational control not achievable with chemical conjugation.
Instead of sample immobilization followed by probing for antigens in the sample with a virus-based detector, viruses displaying antigens could first be immobilized before incubation with the sample to detect for the presence of certain antibodies.385 This was demonstrated as a successful approach for the diagnosis of primary Sjögren's syndrome (pSjS), a chronic systemic autoimmune disease whose heterogeneity often delays diagnosis. Lipo peptide derived from the human autoantigen lipocalin was displayed on PVX, and the nanoparticle platform was shown to be specific to pSjS patient sera and had greater reactivity than the peptide alone. On the other hand, eCPMV-based display of lipo was not reactive, likely due to a smaller density of peptide display. Regardless, the PVX ELISA has promise for future implementation, and an additional benefit found was the stability of the coating, with no loss of specificity or sensitivity observed even after two months of storage at 4°C.
Another aspect of ELISAs is the enzyme used for quantification via substrate conversion. HRP is a popular enzyme for ELISAs due to its ability to convert chromogenic substrates into colored products and chemiluminescent substrates into fluorescent products.382 Immobilization of HRP, as well as glucose oxidase (GOX), has been explored using CPMV386 and TMV387 platforms. While the addition of sensing molecules to the virions will be required for their use in ELISAs, the display of the enzymes themselves on the particles can be directly used for sensing. GOX catalyzes the oxidation of glucose and concurrently generates hydrogen peroxide, which can then be reduced to water by HRP in the presence of a substrate. Therefore, the two enzymes can be coupled to form a glucose sensing system, where HRP substrate conversion can be used for detection. In fact, the TMV nanorods resulted in up to 45-fold higher substrate conversion rates than control samples with the same input of enzymes, which could be due to a combination of a greater surface area and better steric accessibility of the presented enzymes. This is only one example of the potential of enzyme-based sensors, and the immobilization of enzymes that catalyze other reactions could also be considered for the creation of sensitive biosensors.
The output of immunosorbent assays need not be enzyme-linked, and an alternative readout options include fluorescence from the particles. For example, phages isolated after panning against staphylococcal enterotoxin B (SEB), an agent that can cause food poisoning, were labeled with the fluorophore Cy5 and used to probe for SEB.388 Based on fluorescence readings, SEB was detectable down to a concentration of 1.4 ng/well. As another example, dye-labeled CPMV conjugated with antibodies was used for the detection of SEB, botulinum toxin, and the bacterium Campylobacter jejuni.389 The detection of SEB was specifically quantified, and the limit of detection was improved for the CPMV formulation when compared with a mole equivalent of dye-labeled antibody.
Fluorescence can be used for other sensing functions, such as found with the application of fluorescently labeled CPMV to DNA microarray sensors.390 By additionally coupling NeutrAvidin to the capsid, the CPMV could be used as a detection reagent. One result of the investigation demonstrated that the delivery of multiple dyes using CPMV resulted in signal amplification and led to the detection of 14% more genes compared to the control in a rat expression array (Figure 16). Another fluorescence sensing application utilized LbL assembly of M13 labeled with quantum dots along with quenchers that can be displayed by the explosive 2,4,6-trinitrotoluene (TNT).391 The design of the M13 film allowed for highly selective detection of TNT at a sub ppb level through the evolution of fluorescence signal when the quenchers were displaced.
Figure 16. Microarrays hybridized with cDNA made from rat total RNA.
a) Result from cDNA labeled with Cy5-dCTP control. b) Result from cDNA labeled with biotin-functionalized dCTP and dUTP followed by binding with NeutrAvidin-functionalized CPMV-Cy5. Both strategies resulted in a density of labeling of about one every 50 bases, but the CPMV-based probe resulted in greater sensitivity, detecting 71% of the features compared to 57% for the control. Reproduced with permission from ref. 390. Copyright 2009 Elsevier.
Optical sensors also encompass those that utilize SPR for detection. For example, LbL assembly of cationic M13 with anionic gold nanoparticles resulted in the development of an SPR spectrum that was sensitive to humidity.392 It is therefore possible to utilize electrostatic assembly to integrate viruses in humidity sensing devices. SPR has also been applied as sensors for immunoassays, such as exemplified by the detection of the food-borne bacterium Listeria monocytogenes using a gold SPR sensor chip.393 M13 displaying an scFv antibody recognizing L. monocytogenes cells was immobilized on the sensor chip before injection of samples for measurement. The change in resonance due to the binding of cells allowed for specific detection down to levels of around 2 × 106 cfu/mL.
Surface enhanced Raman spectroscopy (SERS) has been another approach that has been considered for sensing.394 It relies on signal enhancement of the Raman signal when in close proximity to the surface of a noble metal. Due to the sensitivity of the technique and the unique signature of various reporters, SERS is a promising technique for multiplexed analysis. M13 phage, selected by phage display for binding to the model rabbit anti-goat IgG antigen, was labeled with Cy3 Raman reporters that were conjugated to Au@Ag core–shell nanoparticles. Due to a high surface area for reporter presentation, the resultant M13 construct imparted an exponential increase in the Raman intensity observed when compared to similarly labeled antibodies against the model antigen. M13 has also been applied in a colorimetric sensor that was based on changes in the modulation of its self-assembled structure, but this application is discussed in more detail in Section 4.3.1.395
Besides optical sensing, viruses have also found application in electrochemical sensing. The three different examples we will highlight involve electrochemical impedance spectroscopy396 and amperometry397, 398 for measurements. In the first example, a gold electrode surface on which a M13 monolayer was attached was used for the detection of an antibody as well as prostate-specific membrane antigen (PSMA), a marker of prostate cancer.396 The resistive component of the impedance, ZRe, measured from 2 to 500 kHz, increased upon analyte binding, and measurement of this characteristic could be used for highly specific detection of down to concentrations of around 100 nM. In the next example, a thin film of TMV conjugated with electroactive oligoaniline was used for the detection of volatile organic compounds, specifically methanol and ethanol.397 The response current was measured from the thin film sensor, and there was a high response observed in the presence of ethanol and methanol over what was observed for acetonitrile, isobutyl alcohol, tetrahydrofuran, toluene, and acetone. Moreover, the current measurements exhibited reproducibility as well as a quick response time for both absorption and desorption of the compounds. Finally, the last example involves the application of a solution of TMV displaying binding peptides for TNT for in-solution sensing of the molecule.398 As a result of binding of TNT to TMV, the diffusion coefficient of the TNT was reduced and a differential Faradaic current signature of the electroactive compound was caused. The differential current was proportional to TNT concentration and therefore could be used for TNT sensing. Detection using this method is not limited to just TNT and can be expanded to other relatively small electroactive species.
4.2.3 Diagnostic Controls
Another sensing approach, which is widely applied for disease diagnostics, involves using assays that detect and amplify the nucleic acid content of infectious agents such as bacteria and viruses.399-401 Quality control is an important consideration to ensure results from these assays are accurate. For example, failures with the well-known polymerase chain reaction (PCR) assay could occur due to nucleic acid degradation or the presence of inhibitors such as bile salts and polysaccharides in clinical samples, which could potentially lead to reduction in polymerase binding or activity.402 False negatives due to failures during processing may be monitored by incorporating known RNA or DNA into samples to serve as a positive internal control to verify that nucleic acid degradation did not occur during processing.
To this end, viral capsids have found utility due to their ability to shield nucleic acids from nuclease digestion, serving as a better mimic for use in viral assays and allowing for long-term storage of the controls. The design of so-called “Armored RNA” (developed and patented by Asuragen and Cenetron Diagnostics) was the first such development and functions as a well-characterized control that is resistant to RNase.403 Armored RNA is comprised of an RNA standard sequence encapsulated within an MS2 capsid, which can be co-produced using an expression vector in E. coli (Figure 17). If so desired, the RNA within these particles can be subsequently released through heating at 70°C for 5 minutes. In the pioneering study, the control was tested in an HIV reverse transcription-PCR (RT-PCR) assay using a non-infectious consensus sequence taken from the HIV-1 gag gene. The Armored RNA particles were found to be stable in anticoagulated plasma with no loss in signal after storage in a variety of conditions: 4°C for 2 months, −20°C for 6 months, and even after five freeze-thaw cycles.
Figure 17. Example of Armored RNA packaging.
Transcribed recombinant RNA, in this case an exogenous HCV-2b consensus sequence, can be packaged within self-assembled MS2 coat proteins. Reproduced with permission from ref. 404. Copyright 2009 American Association for Clinical Chemistry.
Since these initial results, Armored RNA has been applied as a control for a vast array of assays, which include the detection of HCV,404-406 respiratory viruses such as influenza and severe acute respiratory syndrome (SARS) viruses,407-409 enteroviruses,410, 411 and West Nile virus.412 It has even been utilized as a surrogate virus for the detection of animal pathogens, including classical swine fever virus (CSFV), FMDV, and vesicular stomatitis virus (VSV).413 Several developments in the technology have been made to enhance the utility of Armored RNA. In the original area of HIV detection, Armored RNA was expanded beyond RT-PCR assays to the branched DNA (bDNA) assay, which provides a reliable method for quantification of HIV-1 RNA but requires a longer 3 kb RNA control.414 The standard strategy for RNA encapsulation within MS2 is limited to only around 2 kb, but increasing the packaging efficiency to overcome this limit can be achieved through incorporating more translational repressor stem-loops within the RNA, which specifically interact with the MS2 capsid and trigger the self-assembly of the viral shell around the cargo.415 In such a manner, Armored RNA containing the longer HIV pol gene was successfully synthesized and performed reliably as a standard for the bDNA assay.414 HCV Armored RNA has also been developed for RT-PCR and bDNA assays, and additional work demonstrated its applicability in genotyping assays as well, allowing the distinction of a specific subtype of HCV.404 For real-time RT-PCR reactions, false negatives could occur when a singleplex primer/probe assay is used since mismatches with a set of primers could exist in a number of samples, thus dual-specific RNA controls in duplex assays were tested, and resultant enhancement in the sensitivity of detection was found compared to monospecific assays.405, 416
Beyond controls for the detection of single virus types, Armored RNA chimeras have also been created.407, 417 In one study, a chimeric RNA sequence was derived from a mix of gene fragments from HCV, HIV-1, SARS coronavirus 1, and SARS coronavirus 2.417 They were able to package the fairly large 1.2 kb RNA sequence within MS2 and demonstrate its versatility as a control for the multiple different RT-PCR assays for the detection of each individual virus. In another study, RNA fragments from influenza A, influenza B, and SARS viruses were spliced together into one fragment in order to create a single control for the simultaneous testing of these common respiratory viruses that may result in similar clinical symptoms.407 Using multiplex RT-PCR for the three viruses with different reporter dyes, simultaneous amplification and detection could be achieved with a highly sensitive detection limit of 101 copies/μl of the Armored RNA for all the viruses.
A natural extension of Armored RNA technology is the encapsulation of DNA within a bacteriophage capsid for quality control of DNA viruses. Early work in this area utilized lambda and filamentous fd phages for DNA packaging, and greater stability of the DNA was observed.418-420 Containment of control dsDNA within lambda phage conferred resistance to DNase digestion, with reliable amplification in PCR assays for HBV418, 419 and cytomegalovirus.418 When stored in SM buffer, the protected DNA was stable for at least a few months, but stability for more than 5 days in plasma at room temperature could not be achieved, which may be a result of greater susceptibility of lambda to plasma proteases compared to MS2.418, 419 For ssDNA, encapsulation in fd phage was demonstrated with DNA taken from parvovirus B19.420 The resultant controls were resistant to nuclease degradation and performed similarly to the native virus in PCR assays, with very similar growth curves observed. Furthermore, the constructs remained stable at both 37°C and 45°C when diluted in human plasma for periods of at least 4 weeks. More recently, a return to the Armored RNA roots was taken with the demonstration of encapsulation of dsDNA in MS2, which resulted in extra stability over lambda as was found for fd phage but without being limited to ssDNA.421 By conjugating sulfhydryl-modified DNA sequences of interest to an amine-modified stem-loop DNA structure, assembly of dissociated MS2 CPs could then be triggered around the stem-loops to form Armored DNA. Using this strategy, the formation of MS2 capsids packaging HBV and HPV DNA sequences with lengths ranging from 1.3 to 6.5 kb was accomplished, which is astonishing given that the genome of MS2 is only 3.5 kb. The Armored DNA controls performed well in PCR and genotyping assays, and storage in newborn calf serum even after 6 months at 4 °C was shown to not affect performance.
Encapsulated controls are not only limited to bacteriophages. For example, recombinant RNA particles based on CPMV can also be utilized as internal controls for RT-PCR assays.422 To produce these particles, a cDNA clone was engineered to contain sequences for the desired control RNA alongside RNA-2 of CPMV, which codes for its coat and movement proteins. In the proof-of-concept study, two sequences from FMDV and one from swine vesicular disease virus (SVDV) were cloned together into the cassette. Using agroinfiltration of cowpea plants with the combination of this plasmid along with another plasmid for RNA-1 of CPMV, which is responsible for its replication and proteolytic processing, recombinant CPMV particles were propagated in and recovered from their host plant. The CPMV component containing RNA-2 was separated from the RNA-1 component using a Nycodenz density gradient, resulting in a non-infectious construct due to its reliance on RNA-1 for replication. The particles performed well as positive controls for the detection of both FMDV and SVDV. Additionally, they were resistant to RNase and performed reliably even after 33 days storage in a 10% suspension of bovine epithelium at room temperature. This method may provide a low cost alternative to Armored RNA, while maintaining the advantages of stability and rapid production.
Another alternative utilizes TMV coat proteins for assembly of a positive control, with the helical symmetry of the resultant particle more realistically mimicking the stability of filamentous viruses. This approach was recently demonstrated for RT-PCR detection of EBOV.423 To construct the EBOV-TMV mimic, purified CP from TMV was reassembled around an RNA transcript containing an EBOV sequence fragment and a shortened TMV sequence containing the origin of assembly (OAS) necessary for TMV assembly (Figure 18). The EBOV sequence was taken from a region in the RNA-dependent RNA polymerase gene (L-gene) that showed homology between all published EBOV sequences. Detection of both the EBOV and TMV sequences was accomplished using multiplex RT-PCR with the EBOV-TMV particle. Aside from the EBOV primer binding sites, the EBOV sequence was scrambled, therefore posing no threat of infection. Overall, EBOV-TMV is a scalable construct that could be easily adapted as a control for EBOV diagnostics, and the technology could be further applied for mimicking other filamentous viruses.
Figure 18. Manufacture of EBOV-TMV.
a) EBOV-TMV is manufactured by disassembly of TMV propagated in N. benthamiana plants into individual coat proteins that are then reassembled around synthetic RNA transcripts containing EBOV and TMV gene sequences. b) TEM of negatively-stained wild-type TMV rods. c) TEM of shorter EBOV-TMV rods demonstrating successful reconstitution. Scale bar = 100 nm. Reproduced with permission from ref. 423. Copyright 2016 Nature Publishing Group.
4.2.4 Nanoreactors
One of the first demonstrations of the utility of using viruses for the display of biocatalysts was based on genetic engineering of PVX to display a lipase enzyme.424 Although the catalytic activity of the bound enzyme was lower than the free enzyme, the study demonstrated the potential for generating catalytically active nanoparticles that can self-assemble and be easily propagated. Since then, interest in the functionalization of virus particles with enzymes has grown. Coupled with the ability of the viral capsid to self-assemble around a diverse range of cargo rather than simply nucleic acids, encapsulation of enzymes to form nanoreactors has become of increasing relevance to biotechnology.
Initial work with the formation of such nanoreactors looked at the incorporation of a single enzyme within the capsid. Through disassembly and reassembly of the capsid, HRP was encapsulated within CCMV, resulting in an estimated one in every 130 capsids containing the enzyme and allowing study of the enzyme at a single-molecule level.425 Comparison of the activity of encapsulated and non-encapsulated enzyme showed different signatures due to the time necessary for the dihydrorhodamine 6G substrate to diffuse in and the fluorescent rhodamine 6G product to diffuse out of the capsid. Resultantly, product accumulation and delayed loss of fluorescence was observed with the spatially constrained HRP. Further investigation with the confinement of the enzyme Pseudozyma antartica lipase B (PalB) within CCMV utilized a heterodimeric coiled-coil linker to first attach PalB to CP subunits before assembly.426 Through varying the ratio of CP with and without PalB during assembly, the average number of encapsulated enzymes could be controlled. Encapsulated PalB had a higher activity compared to non-encapsulated PalB, which was hypothesized to be due to a higher enzyme concentration when considering just the capsid alone. Additionally, there was no effect on the reaction velocity when varying the number of encapsulated enzymes between one and four PalB per capsid, which is likely due the presence of generally only one substrate molecule per capsid, making more than one enzyme unnecessary for substrate conversion.
Tethered encapsulation was also explored using RNA aptamers.427 The technique utilized co-expression of Qβ CP, Rev-tagged Peptidase E (PepE) or firefly luciferase, and a bifunctional mRNA containing an α-Rev aptamer and a Qβ genome packaging hairpin on its two ends for the encapsulation of the enzyme during expression and assembly. This strategy resulted in up to 18 enzymes encapsulated within the particle. The enzymes remained active after encapsulation, and the capsid was found to offer some protection against thermal degradation, protease digestion, as well as hydrophobic adsorption. A one-pot expression-assembly approach has also been utilized for enzyme tethering within P22, with initial studies investigating programmed encapsulation of alcohol dehydrogenase D (AdhD)428 and homotetrameric β-glycosidase enzyme CelB429 using plasmids harboring both genes for the P22 CP and for its scaffold protein (SP) fused to the enzyme of interest. SPs associate with the interior of the P22 capsid in such a way that the enzymes are consequently encapsulated during expression and assembly of the proteins in E. coli. AdhD encapsulated in P22 was less active due to enzyme crowding effects but overall had a similar catalytic efficiency and did not exhibit substrate inhibition, unlike free AdhD.428 With encapsulation of CelB, which is only active in its tetrameric form, incorporation of multimeric enzymes in P22 was demonstrated.429 Surprisingly, unlike AdhD encapsulated CelB did not result in loss in activity or change in substrate affinity. Additionally, embedding the encapsulated CelB in an acrylamide gel and dehydrating and rehydrating the gel resulted in over 60% retention in activity, which could be attributed to substrate diffusion limitations, therefore presenting the utility of P22 nanoreactors for enzyme immobilization applications.
A consideration for the use of capsids as a nanoreactor is the influence of electrostatics on substrate diffusion into the carrier.430 MS2 CPs were assembled around negatively charged alkaline phosphatase (PhoA-neg), and the effect of capsid mutation adding either negative or positive charges around its pores was explored. Not unexpectedly, while introduction of negative charge had an inhibitory effect, additional positive charge resulted in greater catalysis of the negatively charged phosphatase substrate. Therefore, engineering of nanoreactors can be used to control the flux and extent of reaction. Another benefit of nanoreactors, also seen above with PepE, is the bestowment of stabilization. As an additional example, encapsulation within P22 of phosphotriesterase, which has low heat tolerance and is prone to hydrolysis, yielded protection from proteases and desiccation as well as thermal stability with activity even at 60°C, making it a more practical option as an enzyme for combatting various harmful insecticides and nerve agents.431 The strategy for sequestration of enzymes within P22 during expression is also valuable for the recovery of otherwise insoluble proteins.432 Some recombinant proteins are trafficked to inclusion bodies during production in E. coli, making recovery difficult. α-galactosidase (GalA) is one such protein that was studied, and encapsulation within P22 was shown to allow for successful rescue of properly folded GalA. Encapsulation resulted in very highly active enzyme, which was hypothesized to be most likely due to more correctly folded and active enzymes when produced in this manner.
There is a range of other applications to consider for nanoreactors, many of which have been demonstrated using P22 viral scaffolds. For example, it was determined that the immobilization of cytochrome P450 (CYP450) inside P22 could be used for its stabilization and delivery to human cervix carcinoma cells with retention of substrate conversion activity, which could be further exploited for enzymatic prodrug therapies.132 In addition, encapsulation of NADH oxidase, which predominantly reduces oxygen to hydrogen peroxide, was demonstrated as a method for bacterial growth inhibition by triggering oxidative damage in E. coli cultures.433 Notably, a recent breakthrough in nanoreactor technology is its use for the catalysis of hydrogen production.434 Sequestration of oxygen-tolerant nickel-iron (NiFe)-hydrogenase in P22 provided greater stability for the hydrogenase and resultantly, a greater than 100-fold increase in proton reduction activity over the free enzyme was observed. With the scalable production of encapsulated hydrogenase through simple fermentation, cheap and sustainable clean fuel production can be realized.
Further development of viral nanoreactors also explored the introduction of multiple enzymes tethered to P22.131 By the close proximity of enzymes involved in a cascade, the product of one enzyme can be efficiently taken as the input of the next. Assessment of enzyme cascade encapsulation was evaluated with the tetrameric CelB, the monomeric ATP-dependent galactokinase (GALK), and the dimeric ADP-dependent glucokinase (GLUK), which processes lactose into galactose and glucose, phosphorylates galactose to form galactose-1-phosphate, and phosphorylates glucose to glucose-6-phosphate, respectively (Figure 19). Using multienzyme GLUK-CelBSP and GALK-GLUK-CelB-SP fusions, co-encapsulation of two- and three-enzyme cascades within P22 was achieved. When compared to a 1:1 mixture of individually encapsulated CelB-P22 and GLUK-P22 controls, GLUK-CelB-P22 showed greater enzymatic conversion under inhibitory conditions for CelB. GALK-GLUK-CelB-P22 additionally showed a greater than 2-fold faster turnover rate than GLUK-CelB-P22, suggesting all three proteins assembled property into their active forms and were successfully encapsulated. Therefore, enzyme assemblies are of import in facilitating the construction of complex metabolic systems.
Figure 19. Encapsulation of enzyme cascade in P22.
a) Schematic of P22 nanoreactor assembly where a multienzyme GALK, GLUK, and CelB fusion gene with an additional SP scaffolding domain is encapsulated in the capsid. The expression of the fusion protein allows the enzymes to form the oligomers required for activity (tetramer for CelB, dimer for GLUK). The enzymes are colored green, blue, and red, respectively, for the GALK, GLUK, and CelB fusion, and the CP is shown in gray and SP in purple. b) Illustration of the metabolic pathways of the enzymes and how they are coupled. Reproduced with permission from ref. 131. Copyright 2014 American Chemical Society.
4.2.5 Agricultural applications
An interesting area that has just recently been considered is the application of plant viruses in agriculture. Since their natural hosts for infection are plants, at first glance it appears counterintuitive to apply such nanoparticles for this particular application. However, the pioneering study utilized RCNMV for combatting parasitic root nematode infections and demonstrated that such particles have better mobility in the soil, therefore improving the bioavailability of the abamectin pesticide for nematode control.112 Using ligand gating for infusion (see Section 3.3), the neutrally charged pesticide could be loaded within RCNMV. In such a manner, the abamectin cargo is protected against oxidation and can be released over time. Encapsulated and free abamectin showed similar efficacies in liquid nematode cultures, but when tested on infected tomato seedlings, the viral delivery of abamectin resulted in healthier root growth and reduced root galling compared to abamectin alone. These results have important implications for the agricultural industry, where parasitic nematodes have resulted in astronomical costs on the order of $118 billion worldwide due to crop damage,435 and future considerations with noninfectious VLPs could further refine this strategy of using the naturally evolved mechanisms of such viruses for cargo delivery to plants.
4.2.6 Plant-based pharmaceutical production
In Section 2.2, we discussed how GlaxoSmithKline, Merck, Medicago Inc., and Mapp Biopharmaceutical adopted expression systems derived from viruses for the production of VLP vaccines and antibody cocktails. Plants, as used by Medicago Inc. and Mapp Biopharmaceutical, are not as well known for the expression of recombinant proteins as the more widely used E. coli and yeast expression systems, so we will focus on some examples of plant-based production systems in this section to illustrate some of their advantages. The utilization of plants as a production platform has undergone rapid growth, and cost-effective, highly scalable, and safe production of protein pharmaceuticals with post-translational modifications can now be achieved using plant viral vectors.436, 437 Due to these advantages in cost and production, plant systems offer the potential for rapid pharmaceutical development, especially for more impoverished areas.
One of the first examples of transfecting plants with viral vectors that demonstrated rapid production of a protein relevant to pharmaceutics utilized an expression vector based on TMV.438 High level heterologous expression of biologically active α-trichosanthin, which can inhibit HIV replication in vitro, was achieved through insertion of the gene into a TMV plasmid. By controlling its transcription using a subgenomic promoter, regulation of the expression of the specific gene could be achieved. Further research of such N. benthamiana-based expression systems found that suppression of post-transcriptional gene silencing (PTGS), the plant's adaptive immune system, resulted in enhanced efficiency and was possible through co-expression of the P19 protein of tomato bushy stunt virus (TBSV).439
In parallel, the development of the magnICON “deconstructed” TMV vector system440 has resulted in advancements in the production capacity without need for PTGS suppression.441 The magnICON system involves engineering DNA modules for replication, the gene of interest, and recombination, with the advantage of being easily modifiable using Gateway technology, a universal system for cloning. The modules are then delivered to plants by Agrobacterium and result in transient gene expression and production of viral RNA replicons that can replicate autonomously.440 Furthermore, through “magnifection,” or weak vacuum infiltration of the plants immersed in the Agrobacterium suspension, transfection at a large scale can be quickly achieved without the need of the CP gene or the wait for systemic plant movement.442 A large variety of biologically relevant pharmaceutics have been produced using magnICON, including VLP vaccines,443, 444 antibodies,445, 446 plague antigens,447 cytokines,448 and growth hormones.449
The TMV RNA-based overexpression (TRBO) vector is another replicon system that has been developed and has shown promise for high-level protein expression, with greater yields than demonstrated with the aforementioned P19-enhanced transient expression (Figure 20).450 Essentially, the TRBO vector is a 35S promoter-driven TMV expression vector with the CP gene removed. The CP deletion resulted in a higher efficiency of recombinant protein expression, which was demonstrated for a range of proteins, including GFP, adenosine kinase, the 10th domain from human fibronectin, and some proteinases. Another study also demonstrated the usefulness of the TRBO vector for production of R8, a chimeric allergen derived from dust mites that could be applied for asthma diagnosis or immunotherapy.451
Figure 20. Quantitative analysis of GFP expression with TRBO vector.
Fluorescence (in μg GFP/g infiltrated tissue) was measured from N. benthamiana leaves after agrobacterium infiltration (top). Leaves were also imaged under UV light (bottom). The labels in the figure indicate the plasmids used for transformation, and the results indicate the superior expression of protein with TRBO vector compared to previous expression vectors, even with P19 enhancement. pJL-24 is a previous iteration of a 35S promoter-driven TMV-based expression vector that included the expression of all the TMV genes in addition to the gene insert, pJL3:P19 is a plasmid for the expression of the RNA-silencing suppressor protein P19, pJL TRBO-G is a GFP-expressing TRBO vector, and p35S:GFP is a plasmid with GFP expression under the control of a 35S promoter. Reproduced with permission from ref. 450. Copyright 2007 American Society of Plant Biologists.
Similar expression systems have been derived from plasmids based on PVX and other potexviruses. Complete PVX constructs can be used for the expression of recombinant proteins alongside PVX CPs through insertion of an internal ribosome entry site, which allows for initiation of translation in the middle of an mRNA, between the two genes.452 Similar constructs that instead have an intervening FMDV 2A cleavage sequence can be used to create proteins expressed as a fusion to the CP.453 While larger inserts tend to result in genetic instability and loss of the gene of interest,454 the combination of introducing a heterologous subgenomic promoter from bamboo mosaic virus and deleting a portion at the N-terminus of the CP was found to be a successful strategy to increase transgene expression stability.455
Deconstructed vectors have also been used for PVX-based expression systems. For example, through replacement of the CP gene and the triple gene block (TGB) that encodes movement proteins, efficient production of the gene insert can be achieved.456 Suppression of PTGS helped to improve yields, with transient co-expression of P19 from TBSV or HC-Pro from tobacco etch virus (TEV) resulting in a 44% and 83% increase in gene expression, respectively.457 Amplicon-plus Targeting Technology utilizes this benefit of gene silencing suppression to enhance production.458 This technology involves using a transgenic TEV-B tobacco line that expresses the suppressor gene that is then infected with a PVX amplicon containing the gene of interest. Successful production of a highly labile L1 vaccine protein from canine oral papillomavirus was achieved when this system was used for the expression of the recombinant protein fused to a chloroplast targeting peptide. Coupling of PVX and TMV viral vector systems has also been evaluated and was demonstrated to be a useful approach for resolving complications from competition between multiple replicons in the same cells.446 With the noncompetitive viral systems, expression of assembled oligomeric proteins can be accomplished, such as for construction of full-length IgG with its heavy and light chains.445, 446
Some other examples of potexvirus vectors include vectors based on plantago asiatica mosaic virus (PlAMV)459 and foxtail mosaic virus (FoMV).460 The recombinant PlAMV vector was found to have greater genetic stability and longer retention of the inserted gene compared to PVX, likely due to stronger RNA silencing suppression activity found from the first movement protein in its TGB.459 On the other hand, the FoMV-based FECT vector series utilized deconstructed plasmids in which the CP and TGB were deleted and required co-expression of the P19 suppressor of PTGS.460
Moving on to other systems, CPMV-based expression systems have also shown great versatility for the enhanced expression of a large range of proteins, including antibodies,461 human gastric lipase,462 and VLP vaccines (such as BTV VLPs discussed in Section 4.1.3).230 CPMV is a bipartite virus with RNA-1 providing replication and protein processing capabilities and RNA-2 coding for movement and coat proteins. By altering the shorter RNA-2 through removal of the movement and coat proteins and appending the gene of interest, expression of foreign genes can be achieved through inoculation of plants with constructs of both RNA-1 and the modified RNA-2.463, 464 Agroinfiltration of just the RNA-2 construct with the TBSV P19 suppressor of silencing was found to overcome the necessity for RNA-1, and additional elimination of the second start codon in the 5’ untranslated region (UTR) of RNA-2, located at position 161, resulted in hypertranslation of the downstream protein, likely due to AUG 161 being inhibitory for overall translation.465 With this discovery, high protein expression could be achieved without restrictions on insert size and without the need for RNA-dependent RNA polymerases.
The hypertranslatable CPMV or CPMV-HT system described above was packaged into pEAQ expression vectors for easy recombination, where the gene of interest can be inserted between the modified 5’ UTR and the 3’ UTR of RNA-2, and with P19 expressed from the same plasmid.466 The pEAQ vectors have broad applications, but perhaps one of its greatest advantages is the production of VLPs for vaccines. Medicago Inc. uses CPMV-HT for its large-scale production of enveloped influenza H5 VLPs, with observed budding from the plasma membrane resulting in envelopment and similar structural characteristics to influenza viral particles.467 eCPMV can also be produced using the pEAQ system, where the necessary proteins for CPMV formation, namely its VP60 CP precursor and a 24K proteinase, could either be expressed from the same vector or from two CPMV-HT vectors to form the mature eCPMV particle devoid of any nucleic acid encapsulated.468 Since the pEAQ vector is nonreplicative, levels of expression of multiple proteins can be controlled through co-infiltration of appropriate concentrations of the expression vectors. In such a manner more complex VLPs such as BTV VLPs consisting of up to four different proteins can be assembled.230
Our last examples of plant viral expression vectors come from geminiviruses, which are small ssDNA viruses with twinned capsid morphology. The in-plant activation (INPACT) expression platform is notable for its use of a split-gene cassette in stably transformed plants.469, 470 The INPACT cassette is based on a deconstructed tobacco yellow dwarf virus (TYDV) genome and consists of two components: (1) the gene of interest split into two exons with flanking large intergenic regions (LIRs) and a small intergenic region (SIR) in between, and (2) Rep and RepA activator genes required for replication and activation of recombinant protein production that are inducible by ethanol. Binding of Rep to a site within the LIR initiates rolling circle replication of the replicon system. With ethanol induction, protein production is separate from plant growth, allowing high levels of protein expression that could be cytotoxic or inhibit plant development.
Another method uses a single-vector DNA replicon system based on bean yellow dwarf virus (BeYDV) and is exemplified by the production of oligomeric monoclonal antibodies protective for EBOV.471 The plasmid contains two tandemly linked replicons for the heavy and light chains and only requires the SIR, LIR, and Rep/RepA viral components. The system resulted in noncompeting replicon amplification and protein expression, with efficient assembly of the IgG tetramer. As a final example, pRIC is a similar BeYDV-derived autonomously replicating vector472 that presented an advancement to a previous high expressing but non-replicating pTRAc cassette.473 The pRIC replicon gene vector was created by the addition of the SIR, LIR, and Rep/RepA genes and resulted in gene amplification up to 2 orders of magnitude and up to 7-fold greater production of HPV-16 major CP L1 and HIV-1C p24 subunit vaccine antigens compared to pTRAc.472
There are clearly many options for quick and high yielding recombinant protein production in plants. Although MagnICON has been used widely in the past, its utilization of multiple modules likely detracts from its efficiency, and the development of other systems using single constructs may lead to greater popularity. The main applications for plant-based production have been for antibody and vaccine production, but pharmaceutical proteins of all types can also be produced with the above techniques.
4.3 Energy and nanostructured materials
The design and development of devices with nanoscale features open the door for novel and more efficient ways to capture, store, and transfer energy. Since viruses are self-assembled from coat proteins into nanoscale structures, and the protein-based building blocks also show an inherent propensity to self-assemble into higher-order hierarchical assemblies,124, 474-479 they provide an ideal building scaffold for the design of nanostructured materials. The versatility of hybrid virus-based materials in energy sciences and applications has already been recognized. Examples are highlighted in the following sections of the functionalization of virus-based materials to yield energy-relevant materials such as light harvesting systems, plasmonic metamaterials, and energy and data storage systems.
4.3.1 Principles of self-assembly: wires, sheets, and arrays
A hurdle to the production of mesoscale nanostructured materials is the availability of high-precision manufacturing technologies that facilitate large-scale assembly while also providing spatial control at the 1-100 nanometer level.480 Top-down approaches, derived from technology implemented by the computer industry, have progressed to provide tighter control of feature dimensions with impeccable reproducibility. To program feature components, they rely on lithographic fabrication, such as photolithography,481 microcontact printing (or soft lithography),482, 483 block copolymer nanolithography,484, 485 nanoimprint lithography,486 and scanning-probe or dip-pen lithography.487, 488 Although top-down approaches facilitate extraordinary reproducibility in the writing of nanoscale features at the centimeter size scale, the technology is highly specialized and feature sizes are still limited.
On the other hand, bottom-up approaches seek to achieve directed and controlled assembly of individual components into hierarchical architectures, and they more closely mimic biological systems, cells, and organisms, which can orchestrate complex energy conversion functionalities. Developments in the art and science of self-assembly have made tremendous contributions to the 3D organization of composite materials, rendering high precision manufacturing of energy-relevant biomolecular and inorganic materials possible. For example, DNA-based “programming” exploits the sequence-specificity of base pairing to precisely position materials in 2D and 3D space.489-499 Chemical programming of hierarchical structures has also been devised, as seen in the synthesis of branched dendrimer systems.500-505 Other methods include the use of synthetic block copolymers506 or the application of orthogonal pairs of coiled-coil peptides507, 508 to induce self-assembly of nanoparticles. It is clear that self-assembly holds great potential for the nanomanufacturing of mesoscale materials through simple solution-based bottom-up synthesis, and virus-based self-assembly is one such approach that presents several unique advantages.
For example, high aspect ratio virus particles, such as the plant virus TMV and bacteriophage M13, form excellent biology-derived scaffolds for the templating and synthesis of inorganic matter to produce nanowires at the mesoscale.282, 509-511 In the case of TMV, mineralization can be achieved both in its interior channel119 and around its exterior surface,282 leading to explicit control of the width of the wires. By coupling viral particles with mineralization techniques, semiconducting, superconducting, and insulating nanowires can be formed, resulting in hybrid materials with properties of interest to energy sciences and the electronic industry.
High-order assemblies bridging the nano-to-mesoscale can also be achieved with self-assembly. For instance, TMV building blocks can be specifically directed to assemble end-to-end (or head-to-tail) or side-to-side when exposed to appropriate bathing conditions.512, 513 At acidic pH, TMV rods tend to align head-to-tail and form long wires due to hydrophobic interactions between the dipolar ends of the TMV rod.514 The 1D TMV wires can be further stabilized and condensed to form bundles with the assistance of aniline through in situ polymerization of polyaniline.478 By additionally incorporating DNA hybridization, progress has also been made toward greater control in the specific ordering of viral particles when assembled end-to-end.515 To accomplish this, the ends of M13 phage were functionalized with different DNA oligonucleotides in such a manner that introduction of the appropriate hybridizing oligonucleotides led to the assembly of ordered phage trimers (Figure 21). Although this work investigated controlling the sequence of phages labeled with different fluorescent dyes, in the future it can be further extended for the formation of heterofunctional multiphage structures with distinct moieties that impart more complex functionalities.
Figure 21. Ordered assembly of phage trimer by DNA hybridization.
a) Diagram of design of multiphage structure using DNA hybridization. pIX protein displaying DNA sequence A and pIII protein displaying sequence B are linked by complementary sequence C. Similiarly, pIX and pIII proteins displaying sequences D and E, respectively, are linked by complementary sequence F. b) Fluorescence microscopy image of phages after assembly illustrated the specific arrangement of the phages. Reproduced with permission from ref. 515. Copyright 2013 American Chemical Society.
The high aspect ratio structures formed by TMV and phages M13 and fd have also long been used to produce and study liquid crystalline arrangements, which may find applications in next-generation electronic displays. To yield liquid crystalline assemblies, in-solution mixing protocols have been developed to drive the alignment and spatial organization of the proteinaceous building blocks. A classical approach makes use of concentration gradients such that a nematic liquid crystalline phase is generated at a critical concentration.516 The onset of ordering can be explained by the Onsager theory for isotropic-nematic phase transition, which states that there is competition between translational and orientational entropy, leading to higher densities favoring the nematic phase in order to confer greater translational entropy.517, 518
Filamentous viruses are able to form the same mesophases exhibited by other rod-shaped liquid crystal materials: nematic, cholesteric, smectic A, and smectic C, and they can exist interchangeably.477, 519 For example, M13 phage-based liquid crystals present a nematic phase at low concentrations. As the concentration increases, a cholesteric liquid crystalline phase is observed. Finally, at high concentrations, the system exhibits a smectic liquid crystalline phase. Furthermore, based on the anisotropic nature of the filamentous viruses, external stimuli such as electric and magnetic fields can be applied to tune the ordering, therefore allowing alignment of the whole sample.13, 520, 521
Ordering can also be achieved at liquid/liquid and liquid/solid interfaces. For instance, when the oil/water interface between perfluorodecalin and water was explored, it was found that TMV rods aligned parallel to the interface at low concentrations but aligned normal to the interface at high concentrations, which could be attributed to a compromise between maximizing interfacial interactions and minimizing electrostatic repulsion between the rods.475 Parallel alignment of TMV can also be achieved through assembly at interfaces between buffer and a lipid monolayer supported on a submerged hydrophobic substrate, with the inclusion of Ca2+ ions helping to screen Coulomb repulsion between the particles.522, 523 Another possibility for the formation of assemblies of high aspect ratio viruses is through mixtures with spheres; the increased free energy at the interface favors the assembly of uniform structures.524, 525 This was observed in a study where isotropic, nematic, lamellar, and crystalline phases were obta ined through the modulation of the concentration of TMV, the concentration of BSA or PEG spheres, and the ionic strength.
Addition of divalent metal cations has also been shown to induce the formation of ordered aggregates of TMV. Precipitates of TMV with nematic liquid crystal properties were formed with the addition of ions such as Cd2+, Zn2+, Pb2+, Cu2+, and Ni2+, an effect that did not extend to the alkaline earth metals Ca2+ and Mg2+.526 Furthermore, highly ordered, optically birefringent films were formed when the aggregates were dried on a glass surface. It is hypothesized that TMV contains low-affinity metal binding sites that induce its crosslinking in the presence of the metal ions. While beneficial for liquid crystal applications, this behavior poses an important consideration when formation of well-dispersed biomineralized TMV is desired in the formation of hybrid materials. For example, achieving conditions for dense copper deposition without aggregation of the TMV templates is a challenge.527 Nevertheless, exciting new properties may be possible as a result of particle assembly. Assembly of TMV in the presence of Ba2+ is of particular interest, as instead of the liquid crystalline ordering found for other divalent metal ions studied, the TMV crystallized into a hexagonal superlattice that may have interesting electronic properties.528
In addition to in-solution techniques, electrospinning, wet spinning, and dip coating have been applied to generate long-range ordered filamentous virus-based films.333, 529 For example, varied chiral liquid crystalline M13 phage films were obtained with the use of dip coating methods.333 By carefully adjusting phage concentration, pulling speed, bathing conditions, M13 surface chemistry, and surface chemistry of the solid support, supramolecular M13 films with nematic orthogonal twists, cholesteric helical ribbons, or smectic helicolidal nanofilaments could be produced. These M13 arrays exhibited unique optical and photonic properties such that the films showed iridescence when illuminated, which has potential applications as reflectors or displays. Another potential area of application lies in colorimetric sensing. For example, matrices formed using dip coating of M13 displaying binding peptides for the explosive TNT showed significant enough structure changes in the presence of TNT vapors that levels down 300 ppb could be selectively distinguished (Figure 22).395
Figure 22. Phage litmus for TNT detection.
a) Phages genetically engineered with binding peptide for TNT were self-assembled through dip coating with varying pulling speeds to form bundled structures that resulted in colored matrices. b) Structural changes upon TNT binding resulted in color changes that can be detected using an iPhone-based analysis system down to a level of 300 ppb, with dashed red line showing the sensitivity limit. c) Images and processed fingerprints from the colorimetric sensor after exposure to MNT, DNT, and TNT demonstrated selective sensing of TNT over the similar molecules. d) Principal component analysis of the color changes further verified selectivity. Reproduced with permission from ref. 395. Copyright 2014 Nature Publishing Group.
Another area of interest is the use of viral building blocks for the patterning of surfaces, which can be accomplished using a variety of strategies, including conjugation chemistries530, 531 and electrostatic interactions.479, 532-536 The patterning of virus-based nanobuilding blocks has also been demonstrated using photolithographic and microcontact printing strategies, where a hard elastomeric pattern is “inked” with virions and stamped onto a functionalized surface primed to interact with the virus-based nanoparticles.537 Toward free-standing films, we have recently demonstrated the development of detachable mesoporous films, using a combination of nanosphere lithography and electrodeposition to form nanopatterned, conducting virus–polymer arrays (Figure 23).538 To accomplish this, a hexagonally close-packed array of polystyrene (PS) latex microspheres was created using colloidal or nanosphere lithography, yielding a mesoporous architecture. A conducting poly(pyrrole-co-pyrrole-3-carboxylic acid) film was then electrochemically polymerized in the interstitial voids between the PS beads by cyclic voltammetry. Following PS template removal, CPMV particles were deposited atop the conducting polymer film through electrostatic adsorption and hydrogen bonding. The resultant Janus polymer–virus film was found to be robust and stable, allowing it to be electrochemically delaminated from the substrate.
Figure 23. Creation of free-standing Janus mesoporous virus film.
Topographical tapping mode AFM images illustrate the initial formation of close-packed PS microspheres (a), partial removal of PS spheres (b) after patterned poly(pyrrole-co-pyrrole-3-carboxylic acid) electropolymerization (c), and a non-patterned film (d). e) Optical microscope image (480 μm by 360 μm) after overlaying CPMV on the patterned polymer through electrostatics and delaminating the film to create a free-standing film. Insets show topographical AFM images at indicated points in the film. Reproduced with permission from ref. 538.
Other methods for VNP immobilization onto a surface have utilized DNA hybridization, a powerful technique proven for effective guided assembly of materials in 3D space.489-499 DNA hybridization facilitates controlled and reversible assembly through adjustment of sequence specificity, temperature, or chemical cues (e.g. access of free nucleotides). The TMV building block is one example where nucleic acid hybridization was applied for the programming of materials.539, 540 Here, TMV underwent a mild disassembly protocol to expose the 5’ end of its genome, which could then be used as a code to guide the assembly of vertical TMV arrays, either directly with the corresponding complementary oligonucleotides displayed on the surface,539 or through an intervening DNA linker with sequences specific for both the TMV and the surface540. As an alternative nucleic acid-based method, RNA templates containing the TMV OAS can be patterned onto surfaces, giving a cue for the in situ formation of TMV arrays from the surface up when purified TMV coat proteins are added.541, 542
Icosahedrons also make excellent building blocks for self-programmed assembly of crystalline arrays and lattices. Two-dimensional virus arrays of CPMV have been formed through a drop-and-dry method on a mica substrate, with packing symmetry controlled by modulating the PEG surfactant concentration.543 The organization of the rhombic and hexagonal closed packed structures formed can be attributed to both steric requirements as well as the charge distribution around the capsid surface. Another approach utilizes interfacial adsorption of viral particles under a cationic lipid monolayer that is formed at the surface of an aqueous solution.544, 545 Based on studies with CPMV and TYMV, varying 2D array architectures such as rectangular, hexagonal, and rhombic lattices were achieved using this method.
For 3D crystals, a standard technique applied is depletion condensation, in which PEG is a common condensing agent used for the induction of virus crystallization in bulk.546 Toward the programming of binary materials and photonic crystals, the organization of compact structures of Qβ phage and gold nanoparticles through DNA interconnectors was reported, and the binary lattice was shown to form a NaTl lattice structure that contained interpenetrating organic and inorganic diamond lattices.547 Assembly of viruses can also be mediated by electrostatic self-assembly,548 and organization of particles into crystalline lattices using this method has been demonstrated through the introduction of dendrons and dendrimers.549-551 More recently, binary AB nanoparticle superlattices were achieved using icosahedral CCMV and either gold nanoparticles552 or avidin553. The patchy, but symmetrical, charge distribution on the virus capsid surface enables the formation of binary cubic structures. The authors have also recently extended these concepts to the non-viral protein cage ferritin, yielding photoactive biohybrid crystals with phthalocyanine dyes.554 More specifically, face-centered cubic crystals up to 100 microns in size were obtained in which the phthalocyanines maintained their native properties, suggesting that such materials could find applications in photodynamic therapy, water treatment, diagnostic arrays, and as an oxidant in organic synthesis.
4.3.2 Data storage and battery electrodes
The programmability of the viral scaffold, combined with its propensity to self-assemble into 3D hierarchical architectures, has led to its application toward devices such as battery electrodes, digital memory devices, and energy storage devices. As discussed and illustrated in earlier sections, viruses offer a framework for ligand and peptide display. As a result, they can facilitate the precise deposition of inorganic and organic materials such as metals,67, 533, 555-557 metal oxides,511 semiconductors,558 graphene,559 and carbon nanotubes,560 all of which are energy-relevant materials.
To achieve digital memory, virus hybrids can be created such that they exhibit conductance switching behavior.561, 562 One method to fabricate such a device involved decorating TMV with Pt nanoparticles and using them to form a composite layer in a PVA matrix sandwiched between two electrodes.561 As a result of charge trapping in the nanoparticles, the device exhibited bistable low conductance OFF and high conductance ON states, which were observed with the application of a reverse bias below −2.4 V or a forward bias above 3 V, respectively (Figure 24). In a similar manner, CPMV decorated with semiconducting quantum dots also demonstrated bistable ON/OFF electrical behavior.562 Additionally, for both systems, it was shown that process was reversible and could undergo repeated cycles of reading and writing. Although the maximum limit was found to be around 400 cycles, suggesting the need for further refinement, these investigations have established the use of biomaterial hybrid systems for memory storage and motivate the exploration of future possibilities.
Figure 24. TMV-based digital memory device.
a) TEM image of TMV with approximately 10 nm-sized Pt nanoparticles uniformly attached. b) I-V curves of device created with an active layer derived from the TMV-Pt nanowires (illustrated in inset). The curves demonstrate the conductance switching behavior of the device, with a switch to the ON state during the first bias scan (blue filled circles) at 3.1 V, stability in the ON state with the second scan (blue open circles), and a return to the OFF state during a reverse scan (blue squares) at −2.4 V. On the other hand, devices made from only TMV (red triangles) and only Pt nanoparticles (red diamonds) showed no conductance switching behavior. Reproduced with permission from ref. 561. Copyright 2006 Nature Publishing Group.
Some early work with scaffolds formed by CPMV investigated the positioning of redox-active species, such as ferrocene and viologen, on the viral template.563, 564 The multivalent redox-active nanoparticles exhibited simultaneous multielectron transfer, indicating that the multiple redox centers are independent and behave as essentially non-interacting redox units. Such materials may be envisioned as multielectron reservoirs and as nanoscale electron transfer mediators in redox catalysis or amperometric biosensors.
Another electrical application for viruses involves their utilization as battery electrodes. Research has been performed for the creation of both cathodes and anodes based on M13 as well as TMV templates.510, 556, 557, 565-568 Using self-assembly of the viruses, fabrication of high performing small battery electrodes was made possible. Early studies utilized M13 to grow cobalt oxide nanowires that served as the active anode material, and they demonstrated that virus network formation is a versatile method that could be applied for the formation of electrodes on both rigid and flexible substrates with full electrochemical functionality and greater capacity.510, 565 Additional hybridization of the nanowires with Ni nanoparticles was found to enhance efficiency and cycle life.568 When M13 was instead used for biotemplating of manganese oxide to serve as a cathode for lithium-oxygen batteries, improved capacity of porous networks formed by the virus was also observed.566 On the other hand, TMV electrode development utilized its ability to assemble vertically on a substrate to form “nanoforests” with high surface area and room to accommodate for volume expansion.556, 557 Engineering of the TMV anode involved Sn nanoparticle deposition on Ni-coated TMV, which was then further coated with carbon. In such a way, Sn aggregation that usually occurs when it expands during performance in sodium ion batteries was suppressed.556 Alternatively, the TMV-based cathode was designed for integrated lithium ion batteries, and it was formed by coating TMV-templated Ni nanorods with Ti, LiFePO4, then carbon.557 Aside from excellent electrochemical performance, both TMV electrodes exhibited high mechanical and electronic integrity.
An additional M13-templated electrode example that is of note is one that utilized iridium oxide as the anodic material.511 IrO2 is exciting because it is an electrochromic material, which experiences color change through electrical potential application, a useful property for paper-like display devices. By chemically attaching gold nanoparticles to M13 before mineralization of IrO2, IrO2–Au hybrid nanowires were formed to enhance electron mobility. Porous electrodes made of these nanowires exhibited remarkable switching times of 35 ms for oxidation/coloration and 25 ms for reduction/bleaching, which is promising for applications requiring fast electrochromics.
Further energy materials that can be engineered with viral scaffolds for energy storage and generation devices include graphene sheet conducting frameworks and Au–Pt core–shell catalytic nanowires.559, 569 In the first case, M13 was used to stabilize graphene sheets in aqueous media stabilized to prevent the spontaneous aggregation between individual graphene sheets.559 M13 also served as a viral template to facilitated the nucleation of bismuth oxyfluoride, a conversion reaction cathode material, to form graphene/bismuth oxyfluoride nanocomposites that could be used as a conducting framework for energy storage with improved electron transfer kinetics. In the case of the catalytic nanowires mentioned above, M13 was used as a template for mineralization of Au followed by Pt, a design with fuel cell applications.569 With the Au core as a co-catalyst to oxidize and remove carbon monoxide, the Pt nanowires could be used to oxidize ethanol with less vulnerability to any such carbonaceous species that are formed. This strategy demonstrated excellent catalytic activity, and it additionally has advantages of simple scale-up synthesis, greater durability of the Pt catalyst, and lower cost due to the presence of M13 reducing dead volume of Au in the core.
As a final example of the energy generation potential of phages, liquid crystals formed from M13 phage have been shown to exhibit piezoelectric properties.570 Glutamic acid residues were introduced at the N-terminus of the pVIII coat proteins in order to create a greater net dipole moment directed from the N-terminus to the positively charged C-terminus, which resulted in an enhancement in the effective piezoelectric coefficient of the phage films. Self-assembled phage monolayers with nematic structure and phage multilayers with smectic structure all demonstrated piezoelectric behavior, with the response increasing in accordance to increasing film thickness until saturation occurred at around a thickness of 100 nm. The investigation further demonstrated easily fabrication of multilayer phage films through spontaneous ordering after dropcasting, and the energy output from mechanical pressure of the film was sufficient to turn on a liquid crystal display. Overall, this phage-based technology presents an opportunity for environmentally friendly energy generation.
4.3.3 Light harvesting and catalysis
Solar energy is another source of green energy. Light harvesting systems, such as found in the photosynthesis machinery of plants, enable the conversion of light into energy.571 Chromophores transport energy using Förster resonance energy transfer (FRET), whereby electrons are excited upon photon absorption by one chromophore then transfer their energy to another chromophore as they undergo relaxation through further photon emission. The excitation energy gets transferred in a cascade from donors to acceptors when the energy levels of absorption and emission match. The photosynthesis system of plants contains precisely spaced arrays of chromophores that facilitate light harvesting and conversion into energy and is one o f nature's most sophisticated energy conversion complexes.571
Synthetic light harvesting systems are of great interest in basic energy science and technology development for implementation in solar panels and other photovoltaic devices. Like photosynthetic machinery, viral capsids provide a means to precisely position chromophores with spatial control at the sub-nanometer level. Recognizing this engineering design space, a few examples have been published using the self-assembling TMV scaffold.68, 572, 573 In brief, TMV coat proteins were covalently labeled with either a donor or an acceptor from a FRET pair and then assembled into disk or rod structures at a controlled ratio of donors to acceptors. Through assembly, the chromophores could be positioned in close proximity at tunable distances in such a way to achieve the most efficient FRET. Similarly, M13 displaying zinc porphyrins has also shown promise as a light harvesting antenna, and FRET was observed in which the porphyrin served as both a donor and an acceptor for exciton migration along the virus.574
A very recent study has revealed groundbreaking work that further enhances the efficiency of energy transport in light harvesting devices.575 For the first time, experimental evidence was found that demonstrated a super-Förster regime where quantum coherence and classical incoherent mechanisms interact to enhance exciton transfer efficiency compared to what can be found with classical FRET. Genetic engineering of M13 was utilized to regulate the distances between chromophore binding sites to be either around 33 Å for the “Classical Förster” clone or 10 Å for the “Super-Förster” clone, a distance short enough for increased coupling strength but not too short to experience complete quenching. Fluorescence measurements from the virions modified with a range of donor-to-acceptor ratios of chromophores demonstrated the superior behavior of the Super-Förster clone (Figure 25). Additionally, there was a 68% enhancement in exciton diffusion length, an exciting outcome demonstrating enhanced exciton transport that has previously only been theorized.576
Figure 25. M13 Super-Förster clone for enhanced exciton transport.
a) M13 Classical Förster (left) and Super-Förster clones (right) showing engineered binding sites for chromophores (N-terminus in blue, pre-existing lysine residue in green, and inserted lysine residue in orange) and the networks for energy transport between the residues. Blue arrows show classical incoherent exciton hopping, while red ellipses indicate exciton delocalization. b) Experimental data of fluorescence per acceptor to donor-to-acceptor ratio of the Super-Förster clone is best matched by numerical simulations based on Super-Förster theory and decohered quantum walk (QW) rather than based on classical Förster. Reproduced with permission from ref. 575. Copyright 2016 Nature Publishing Group.
As another part of the light harvesting process, filamentous viruses have also been employed for electron harvesting as nanowire-based photoanodes due to their ability to form porous networks with high surface area. Previous work has investigated the implementation of M13 in dye-sensitized solar cells (DSSCs).560, 577-579 The design of DSSCs involves the presence of a dye or photosensitizer for light absorption that, when excited, transfers the energy into the conduction band of a TiO2 photoanode, where it then diffuses to a current collector such as fluorine-doped tin oxide. By using M13 as a sacrificial template for photoanode formation, it was shown that a highly interconnected porous TiO2 structure could be achieved for greater charge transport efficiency. Strategies to create a porous TiO2 network included using electrostatic interactions for complexation of TiO2 nanoparticles and M13 viruses,577 layer-by-layer phage deposition before TiO2 nucleation,578 and crosslinking of the phages by glutaraldehyde to form a hydrogen before TiO2 nucleation.579 After removal of the sacrificial viral scaffold, the photoanode was found to possess improved electron diffusio n properties. With additional incorporation of gold nanoparticles, the dye molecules were found to also experience greater light absorption through localized surface plasmon resonance, therefore further improving light harvesting.579 As another step toward improvement of DSSCs and other photovoltaics, stabilization of SWNTs by M13 through non-covalent binding before biomineralization of TiO2 was used for successful incorporation of the SWNTs in the photoanode.560 Due to the high electron mobility of the SWNTs, a high power conversion of 10.6% was observed with this strategy.
Another example of M13-based photovoltaic cells involves the mineralization of perovskite nanocrystals, which have unique catalytic, electric, and magnetic properties.558 In particular, M13 was used as a template for strontium titanate (STO) and bismuth ferrite (BFO) mineralization. The study presented the first report of the photovoltaic properties of BFO nanoparticles, which showed effective absorption of visible light but a solar power conversion efficiency of only 0.17%. On the other hand, the STO nanowires exhibited photocatalytic water reduction properties, and they could be used for the efficient hydrogen production under both UV and visible light. Additional research in this area investigated the improving the performance of STO nanowires under visible light.580 Whereas dye-sensitization was required in the previous study,558 a new fabrication method was developed that utilized ammonia gas treatment to tune the optical absorption of the STO nanowires, which was successful for the manufacture of visible-light active photocatalysts.
Light conversion has also been explored with TMV.555 In the study, TMV was arranged vertically on a gold-coated indium tin oxide (ITO) surface, and the particles were then coated with nickel, ITO, followed by CuO; the ITO served as a current collector, while the CuO served as a photocathode. At a high enough TMV density, the film was found to form an antireflective surface due to its high surface area and surface roughness, which is beneficial for greater photoelectrochemical cell efficiency. Various CuO thicknesses were investigated, and a thickness of 520 nm was found to produce one of the highest photocurrent densities reported for CuO systems its size. Overall, the properties of this virus-templated surface bring much promise for future photoelectrochemical applications, such as catalytic water conversion.
4.3.4 Plasmonic metamaterials
Plasmonic nanostructures have already led to breakthroughs in optics and nanophotonics,581-584 as well as in biotechnology and biomedicine.585, 586 For nanometer-sized noble metal particles, the de Broglie wavelength of the valence electrons is of the same order of magnitude as the size of the particle, and as a result quantum size effects may be observed. The characteristic plasmon resonance bands that lead to extraordinary metal enhancement effects arise due to the oscillation of the valence electrons at a collective oscillation frequency.586, 587 Knowing this, nanostructures can be designed and organized such that their plasmonic bands are finely tuned. For applications in the body, a plasmonic range in the NIR imaging window where light can penetrate several centimeters into the tissue is desirable. The substantial absorption and scattering properties of plasmonic nanostructures can then be used as contrast agents for optical imaging, such as optical coherence tomography588, 589 and photoacoustic imaging.590 As a demonstration of a potential application of plasmonics, sensing using viral plasmonics has been achieved by using M13 phage loaded with gold nanocubes for SERS.591 Alternatively, the plasmonic nanostructures also have applications in a therapeutic context due to the transformation of the absorbed light into heat,592, 593 which could be applied for the thermal ablation of tumors or the controlled release of drugs.594, 595 Use as “theranostic” reagents, where diagnostic imaging and therapeutic functions are merged, presents another option.596, 597 Beyond healthcare, it should be noted that plasmonics can also be applied to many disciplines, including nonlinear plasmonics, electronic transport, local heating, biosensors, quantum optics, and metamaterials.598
Virus-based plasmonic metamaterial synthesis was first demonstrated with BMV shells assembled around gold nanoparticle cores, where 3D crystals formed by the particles exhibited an optical transmission spectrum indicative of multipolar plasmonic coupling between adjacent gold cores.124 More recently, MS2 phages were functionalized with internal gold nanoparticles and external fluorophores, with the intervening distance controlled using oligonucleotide hairpins (Figure 26).174 Enhancements in fluorescence intensity and corresponding decreases in lifetime were observed, and the results correlated well with expectations of the effect of the separation between the plasmon and gain materials.599-601 In the reverse configuration (internal fluorophores and gold nanoparticle antennas), fluorescence enhancement was also observed, and it was found to be a function of nanoparticle size and the separation between the plasmon and gain material.602
Figure 26. Controlled display of gold nanoparticles and fluorophores for enhanced fluorescence.
a) Schematic for the assembly of MS2 around gold nanoparticles followed by attachment of DNA hairpins to place fluorophores a fixed distance away from the capsid. b) Images from total internal reflection fluorescence microscopy of MS2 labeled with fluorophores set 3 bp away from the capsid, with (left) and without (right) gold encapsulated, demonstrating metal-enhanced fluorescence. Reproduced with permission from ref. 174. Copyright 2013 American Chemical Society.
Some interesting electromagnetic effects can only be generated through well-structured ordering of plasmonic materials, a property that viruses can provide. A recent example exploring this potential involved the precise placement of plasmonic gold nanoparticles with defined spacing and symmetry using cysteine-functionalized CPMV.281 The CPMV was engineered to allow gold to be placed at the 12 vertices around the capsid. The resultant plasmonic nanocluster demonstrated resonances at visible wavelengths due to interactions of neighboring gold nanoparticles, and finite-element simulations suggest the structure is likely to give rise to a 10-fold enhancement of the local electromagnetic field through near-field coupling.
5. Summary and future directions
Overall, viruses have been exploited in the development of a dizzying array of applications that fall under the broad scope of medicine, biotechnology, and energy. While some areas in medicine, such as vaccines and gene delivery, have been around for a while, the application of viruses in other areas, such as drug delivery and tissue engineering, have only recently been conceptualized and developed. For many diseases, there is still much to tackle to improve the effectiveness of their diagnosis and treatment. One challenge of great interest for the transition of new virus-based nanomaterials into the clinic will be to attain more exquisite control over the ability to regulate the immune response and clearance behavior for specific applications. The immunogenicity of the proteinaceous coat of viral particles can be advantageous for immunotherapy but is undesirable when the particles are used as nanocarriers for imaging and drug delivery. It has been shown that the elimination of particles by antibodies can tuned with PEGylation, and alternative coatings such as serum albumin can further reduce an immune response. By conferring stealth properties, repeat administration of virus-based nanomaterials for applications in diagnostics and therapeutics will become achievable.
In biotechnology, there have been many advances in the field of virus-based devices for sensing, diagnostics, catalysis, agriculture, and pharmaceutical production. To highlight some achievements, phage display has become a familiar technique, Armored RNA as a molecular diagnostic control is commercially available, and scaled-up manufacture of pharmaceutics in plants has been shown to be a cheap and effective alternative. Exciting developments just underway include the efficient catalysis of hydrogen for clean fuel production and the use of plant viruses for combatting diseases in plants. It will not be long before the widespread adoption of newer technology such as these follows.
Finally, it should be abundantly clear that viruses present a great opportunity in the manufacture of energy-relevant materials. Along with their ability to self-assemble into ordered arrays, viruses offer the possibility for precise positioning at the nanoscale, a distinct advantage that can reveal novel material properties such as enhancements in plasmonic behavior. The recent breakthrough in enhancing light harvesting efficiency exemplifies the advantage of the well-ordered virus scaffold. Deeper understanding of virus structure and assembly, in combination with new approaches in synthetic virology, will yield ever more complex formulations that will enable new functions. It remains exciting at the virus-chemistry interface!
Acknowledgements
This work was supported in parts by grants from the National Science Foundation (CAREER DMR-1452257 to NFS for sensing and CHE-1306447 to NFS for virus:polymer chemistry, CMMI-1333651 for virus nanomanufacturing), the National Institutes of Health (F31 HL129703 to AMW, R03EB020602 to NFS for shape engineering of VNPs, R01CA202814 and R21EB020946 to NFS for imaging, R21HL121130 to NFS for cardiovascular targeting), the Department of Energy (BES DE-SC0008068 to NSF for basic energy science), the American Cancer Society (128319-RSG-15-144-01-CDD to NFS for drug delivery), and the Susan G. Komen Foundation (CCR14298962 to NFS for vaccines).
Biography
 Amy M. Wen
Amy M. Wen
Amy M. Wen received her BSE from Duke University in 2010. She then pursued her PhD at Case Western Reserve University under the supervision of Prof. Nicole F. Steinmetz in the Department of Biomedical Engineering. Her current research interests focus on using bioinspired approaches to develop design guidelines and strategies for therapeutic intervention.
 Nicole F. Steinmetz
Nicole F. Steinmetz
Nicole F. Steinmetz is an Assistant Professor of Biomedical Engineering at Case Western Reserve University School of Medicine. Her research interests are in applying synthetic virology approaches toward next-generation biomedicines and materials. She trained at RWTH-Aachen University in Germany (Masters in Molecular Biotechnology) and received her PhD in Bionanotechnology from John Innes Centre, UK. She then moved to the Scripps Research Institute for her postdoctoral research under a NIH K99/R00 award. Dr. Steinmetz was named a 2015 Young Innovator of Cellular and Biomolecular Engineering (BMES), and she is a 2014 Crain's Cleveland Business 40 under 40 honoree and a 2011 Mt. Sinai Scholar. She has won many prestigious awards, including a 2016 American Cancer Society Research Scholar Award, a 2015 NSF CAREER Award, and a 2014 Susan G. Komen Career Catalyst Grant. Dr. Steinmetz has authored more than 100 peer-reviewed journal articles, reviews, book chapters, and patent applications; she has authored and edited books on virus-based nanotechnology.
References
- 1.Averhoff FM, Glass N, Holtzman D. Clin. Infect. Dis. 2012;55:S10–S15. doi: 10.1093/cid/cis361. [DOI] [PubMed] [Google Scholar]
- 2.De Cock KM, Jaffe HW, Curran JW. Aids. 2012;26:1205–1213. doi: 10.1097/QAD.0b013e328354622a. [DOI] [PubMed] [Google Scholar]
- 3.Graham-Rowe D. Nature. 2011;480:S2–S3. doi: 10.1038/480S2a. [DOI] [PubMed] [Google Scholar]
- 4.Ebola Response Team WHO. N. Engl. J. Med. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauci AS, Morens DM. N. Engl. J. Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 6.Behbehani AM. Microbiol. Rev. 1983;47:455–509. doi: 10.1128/mr.47.4.455-509.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lustig A, Levine AJ. J. Virol. 1992;66:4629–4631. doi: 10.1128/jvi.66.8.4629-4631.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanishvili N. Adv. Virus Res. 2012;83:3–40. doi: 10.1016/B978-0-12-394438-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 9.Reardon S. Nature. 2014;510:15–16. doi: 10.1038/510015a. [DOI] [PubMed] [Google Scholar]
- 10.Loc-Carrillo C, Abedon ST. Bacteriophage. 2011;1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas T, Young M. Nature. 1998;393:152–155. [Google Scholar]
- 12.Wang Q, Lin T, Tang L, Johnson JE, Finn MG. Angew. Chem. Int. Ed. 2002;41:459–462. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 13.Lee SW, Mao C, Flynn CE, Belcher AM. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 14.Brown WL, Mastico RA, Wu M, Heal KG, Adams CJ, Murray JB, Simpson JC, Lord JM, Taylor-Robinson AW, Stockley PG. Intervirology. 2002;45:371–380. doi: 10.1159/000067930. [DOI] [PubMed] [Google Scholar]
- 15.Eber FJ, Eiben S, Jeske H, Wege C. Nanoscale. 2015;7:344–355. doi: 10.1039/c4nr05434b. [DOI] [PubMed] [Google Scholar]
- 16.Natarajan P, Lander GC, Shepherd CM, Reddy VS, Brooks CL, 3rd, Johnson JE. Nat. Rev. Microbiol. 2005;3:809–817. doi: 10.1038/nrmicro1283. [DOI] [PubMed] [Google Scholar]
- 17.Prangishvili D, Garrett RA. Trends Microbiol. 2005;13:535–542. doi: 10.1016/j.tim.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Douglas T, Young M. Science. 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 19.Prangishvili D, Forterre P, Garrett RA. Nat. Rev. Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 20.Galaway FA, Stockley PG. Mol. Pharm. 2013;10:59–68. doi: 10.1021/mp3003368. [DOI] [PubMed] [Google Scholar]
- 21.Dixit SK, Goicochea NL, Daniel MC, Murali A, Bronstein L, De M, Stein B, Rotello VM, Kao CC, Dragnea B. Nano Lett. 2006;6:1993–1999. doi: 10.1021/nl061165u. [DOI] [PubMed] [Google Scholar]
- 22.Malyutin AG, Easterday R, Lozovyy Y, Spilotros A, Cheng H, Sanchez-Felix OR, Stein BD, Morgan DG, Svergun DI, Dragnea B, Bronstein LM. Chem. Mater. 2015;27:327–335. [Google Scholar]
- 23.Baltimore D. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirth T, Parker N, Yla-Herttuala S. Gene. 2013;525:162–169. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- 25.Caspar DL, Klug A. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 26.Rosano GL, Ceccarelli EA. Front. Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelps JP, Dao P, Jin H, Rasochova L. J. Biotechnol. 2007;128:290–296. doi: 10.1016/j.jbiotec.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Bessa J, Schmitz N, Hinton HJ, Schwarz K, Jegerlehner A, Bachmann MF. Eur. J. Immunol. 2008;38:114–126. doi: 10.1002/eji.200636959. [DOI] [PubMed] [Google Scholar]
- 29.Pickett GG, Peabody DS. Nucleic Acids Res. 1993;21:4621–4626. doi: 10.1093/nar/21.19.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang DJ, Roberts IM, Wilson TM. Proc. Natl. Acad. Sci. USA. 1994;91:9067–9071. doi: 10.1073/pnas.91.19.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birnbaum F, Nassal M. J. Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jennings GT, Bachmann MF. Annu. Rev. Pharmacol. Toxicol. 2009;49:303–326. doi: 10.1146/annurev-pharmtox-061008-103129. [DOI] [PubMed] [Google Scholar]
- 33.Hovlid ML, Lau JL, Breitenkamp K, Higginson CJ, Laufer B, Manchester M, Finn MG. ACS Nano. 2014;8:8003–8014. doi: 10.1021/nn502043d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochoa WF, Chatterji A, Lin T, Johnson JE. Chem. Biol. 2006;13:771–778. doi: 10.1016/j.chembiol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Liu CM, Jin Q, Sutton A, Chen L. Bioconjug. Chem. 2005;16:1054–1057. doi: 10.1021/bc050108+. [DOI] [PubMed] [Google Scholar]
- 36.Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. Virology. 1995;207:486–494. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- 37.Diaz-Valle A, Garcia-Salcedo YM, Chavez-Calvillo G, Silva-Rosales L, Carrillo-Tripp M. J. Virol. Methods. 2015;225:23–29. doi: 10.1016/j.jviromet.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 38.Fuller MT, King J. J. Mol. Biol. 1982;156:633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- 39.Caldeira JC, Peabody DS. J. Nanobiotechnology. 2007;5:10. doi: 10.1186/1477-3155-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arkhipenko MV, Petrova EK, Nikitin NA, Protopopova AD, Dubrovin EV, Yaminskii IV, Rodionova NP, Karpova OV, Atabekov JG. Acta Naturae. 2011;3:40–46. [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Cripe TP, Estes PA, Lyon MK, Rose RC, Garcea RL. J. Virol. 1997;71:2988–2995. doi: 10.1128/jvi.71.4.2988-2995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morikawa Y, Goto T, Momose F. J. Biol. Chem. 2004;279:31964–31972. doi: 10.1074/jbc.M313432200. [DOI] [PubMed] [Google Scholar]
- 43.Bragard C, Duncan GH, Wesley SV, Naidu RA, Mayo MA. J. Gen. Virol. 2000;81:267–272. doi: 10.1099/0022-1317-81-1-267. [DOI] [PubMed] [Google Scholar]
- 44.Celik E, Calik P. Biotechnol. Adv. 2012;30:1108–1118. doi: 10.1016/j.biotechadv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Murphy CI, Piwnica-Worms H, Grunwald S, Romanow WG, Francis N, Fan HY. Curr. Protoc. Mol. Biol. 2004 doi: 10.1002/0471142727.mb1610s65. Chapter 16, Unit 16.19. [DOI] [PubMed] [Google Scholar]
- 46.Webster DE, Thomas MC. Biotechnol. Adv. 2012;30:410–418. doi: 10.1016/j.biotechadv.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 47.Freivalds J, Dislers A, Ose V, Skrastina D, Cielens I, Pumpens P, Sasnauskas K, Kazaks A. J. Biotechnol. 2006;123:297–303. doi: 10.1016/j.jbiotec.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Brumfield S, Willits D, Tang L, Johnson JE, Douglas T, Young M. J. Gen. Virol. 2004;85:1049–1053. doi: 10.1099/vir.0.19688-0. [DOI] [PubMed] [Google Scholar]
- 49.Wang JW, Roden RB. Expert Rev. Vaccines. 2013;12:129–141. doi: 10.1586/erv.12.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Felberbaum RS. Biotechnol. J. 2015;10:702–714. doi: 10.1002/biot.201400438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneemann A, Dasgupta R, Johnson JE, Rueckert RR. J. Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh P, Destito G, Schneemann A, Manchester M. J. Nanobiotechnology. 2006;4:2. doi: 10.1186/1477-3155-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shanks M, Lomonossoff GP. J. Gen. Virol. 2000;81:3093–3097. doi: 10.1099/0022-1317-81-12-3093. [DOI] [PubMed] [Google Scholar]
- 54.Chapman S, Kavanagh T, Baulcombe D. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 55.Ahlquist P, French R, Janda M, Loesch-Fries LS. Proc. Natl. Acad. Sci. USA. 1984;81:7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischer R, Drossard J, Commandeur U, Schillberg S, Emans N. Biotechnol. Appl. Biochem. 1999;30(Pt 2):101–108. [PubMed] [Google Scholar]
- 57.Yao J, Weng Y, Dickey A, Wang KY. Int. J. Mol. Sci. 2015;16:28549–28565. doi: 10.3390/ijms161226122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gopinath K, Dragnea B, Kao C. J. Virol. 2005;79:14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders K, Sainsbury F, Lomonossoff GP. Virology. 2009;393:329–337. doi: 10.1016/j.virol.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 60.Rademacher T. 2013 Int. Pat., PCT/EP2013/000296.
- 61.Klein KC, Polyak SJ, Lingappa JR. J. Virol. 2004;78:9257–9269. doi: 10.1128/JVI.78.17.9257-9269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lingappa JR, Newman MA, Klein KC, Dooher JE. Virology. 2005;333:114–123. doi: 10.1016/j.virol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 63.Bundy BC, Franciszkowicz MJ, Swartz JR. Biotechnol. Bioeng. 2008;100:28–37. doi: 10.1002/bit.21716. [DOI] [PubMed] [Google Scholar]
- 64.Smith MT, Varner CT, Bush DB, Bundy BC. Biotechnol. Prog. 2012;28:549–555. doi: 10.1002/btpr.744. [DOI] [PubMed] [Google Scholar]
- 65.King NP, Bale JB, Sheffler W, McNamara DE, Gonen S, Gonen T, Yeates TO, Baker D. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, Wege C. Nanoscale. 2013;5:3808–3816. doi: 10.1039/c3nr33724c. [DOI] [PubMed] [Google Scholar]
- 67.Klem MT, Willits D, Young M, Douglas T. J. Am. Chem. Soc. 2003;125:10806–10807. doi: 10.1021/ja0363718. [DOI] [PubMed] [Google Scholar]
- 68.Miller RA, Presley AD, Francis MB. J. Am. Chem. Soc. 2007;129:3104–3109. doi: 10.1021/ja063887t. [DOI] [PubMed] [Google Scholar]
- 69.Peabody DS. J. Nanobiotechnology. 2003;1:5. doi: 10.1186/1477-3155-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q, Lin T, Johnson JE, Finn MG. Chem. Biol. 2002;9:813–819. doi: 10.1016/s1074-5521(02)00166-7. [DOI] [PubMed] [Google Scholar]
- 71.Strable E, Prasuhn DE, Jr., Udit AK, Brown S, Link AJ, Ngo JT, Lander G, Quispe J, Potter CS, Carragher B, Tirrell DA, Finn MG. Bioconjug. Chem. 2008;19:866–875. doi: 10.1021/bc700390r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carrico ZM, Romanini DW, Mehl RA, Francis MB. Chem. Commun. (Camb) 2008 doi: 10.1039/b717826c. DOI: 10.1039/b717826c, 1205-1207. [DOI] [PubMed] [Google Scholar]
- 73.Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, Lin T. Chem. Biol. 2004;11:855–863. doi: 10.1016/j.chembiol.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 74.Chatterji A, Ochoa WF, Ueno T, Lin T, Johnson JE. Nano Lett. 2005;5:597–602. doi: 10.1021/nl048007s. [DOI] [PubMed] [Google Scholar]
- 75.Medintz IL, Sapsford KE, Konnert JH, Chatterji A, Lin T, Johnson JE, Mattoussi H. Langmuir. 2005;21:5501–5510. doi: 10.1021/la0468287. [DOI] [PubMed] [Google Scholar]
- 76.Udit AK, Brown S, Baksh MM, Finn MG. J. Inorg. Biochem. 2008;102:2142–2146. doi: 10.1016/j.jinorgbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Shen L, Zhou J, Wang Y, Kang N, Ke X, Bi S, Ren L. Small. 2015;11:1190–1196. doi: 10.1002/smll.201401952. [DOI] [PubMed] [Google Scholar]
- 78.Staczek J, Bendahmane M, Gilleland LB, Beachy RN, Gilleland HE., Jr. Vaccine. 2000;18:2266–2274. doi: 10.1016/s0264-410x(99)00571-x. [DOI] [PubMed] [Google Scholar]
- 79.Dalsgaard K, Uttenthal A, Jones TD, Xu F, Merryweather A, Hamilton WD, Langeveld JP, Boshuizen RS, Kamstrup S, Lomonossoff GP, Porta C, Vela C, Casal JI, Meloen RH, Rodgers PB. Nat. Biotechnol. 1997;15:248–252. doi: 10.1038/nbt0397-248. [DOI] [PubMed] [Google Scholar]
- 80.Koo M, Bendahmane M, Lettieri GA, Paoletti AD, Lane TE, Fitchen JH, Buchmeier MJ, Beachy RN. Proc. Natl. Acad. Sci. USA. 1999;96:7774–7779. doi: 10.1073/pnas.96.14.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi RU, Kanesashi SN, Inoue T, Enomoto T, Kawano MA, Tsukamoto H, Takeshita F, Imai T, Ochiya T, Kataoka K, Yamaguchi Y, Handa H. J. Biotechnol. 2008;135:385–392. doi: 10.1016/j.jbiotec.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 82.Merzlyak A, Indrakanti S, Lee SW. Nano Lett. 2009;9:846–852. doi: 10.1021/nl8036728. [DOI] [PubMed] [Google Scholar]
- 83.Luckanagul J, Lee LA, Nguyen QL, Sitasuwan P, Yang X, Shazly T, Wang Q. Biomacromolecules. 2012;13:3949–3958. doi: 10.1021/bm301180c. [DOI] [PubMed] [Google Scholar]
- 84.Shukla S, Dickmeis C, Nagarajan AS, Fischer R, Commandeur U, Steinmetz NF. Biomater. Sci. 2014;2:784–797. doi: 10.1039/c3bm60277j. [DOI] [PubMed] [Google Scholar]
- 85.Uhde-Holzem K, McBurney M, Tiu BD, Advincula RC, Fischer R, Commandeur U, Steinmetz NF. Macromol. Biosci. 2015 doi: 10.1002/mabi.201500280. DOI: 10.1002/mabi.201500280. [DOI] [PubMed] [Google Scholar]
- 86.Dickmeis C, Honickel MMA, Fischer R, Commandeur U. Front. Bioeng. Biotechnol. 2015:3. doi: 10.3389/fbioe.2015.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pille J, Cardinale D, Carette N, Di Primo C, Besong-Ndika J, Walter J, Lecoq H, van Eldijk MB, Smits FC, Schoffelen S, van Hest JC, Makinen K, Michon T. Biomacromolecules. 2013;14:4351–4359. doi: 10.1021/bm401291u. [DOI] [PubMed] [Google Scholar]
- 88.Schlick TL, Ding Z, Kovacs EW, Francis MB. J. Am. Chem. Soc. 2005;127:3718–3723. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]
- 89.Kovacs EW, Hooker JM, Romanini DW, Holder PG, Berry KE, Francis MB. Bioconjug. Chem. 2007;18:1140–1147. doi: 10.1021/bc070006e. [DOI] [PubMed] [Google Scholar]
- 90.Wen AM, Shukla S, Saxena P, Aljabali AA, Yildiz I, Dey S, Mealy JE, Yang AC, Evans DJ, Lomonossoff GP, Steinmetz NF. Biomacromolecules. 2012;13:3990–4001. doi: 10.1021/bm301278f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tian F, Tsao ML, Schultz PG. J. Am. Chem. Soc. 2004;126:15962–15963. doi: 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
- 92.Bruckman MA, Kaur G, Lee LA, Xie F, Sepulveda J, Breitenkamp R, Zhang X, Joralemon M, Russell TP, Emrick T, Wang Q. Chembiochem. 2008;9:519–523. doi: 10.1002/cbic.200700559. [DOI] [PubMed] [Google Scholar]
- 93.Prasuhn DE, Jr., Yeh RM, Obenaus A, Manchester M, Finn MG. Chem. Commun. (Camb) 2007 doi: 10.1039/b615084e. DOI: 10.1039/b615084e, 1269-1271. [DOI] [PubMed] [Google Scholar]
- 94.Steinmetz NF, Hong V, Spoerke ED, Lu P, Breitenkamp K, Finn MG, Manchester M. J. Am. Chem. Soc. 2009;131:17093–17095. doi: 10.1021/ja902293w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. Int. Ed. Engl. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uchida M, Morris DS, Kang S, Jolley CC, Lucon J, Liepold LO, LaFrance B, Prevelige PE, Jr., Douglas T. Langmuir. 2012;28:1998–2006. doi: 10.1021/la203866c. [DOI] [PubMed] [Google Scholar]
- 97.Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 98.Dirksen A, Dawson PE. Bioconjug. Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brunel FM, Lewis JD, Destito G, Steinmetz NF, Manchester M, Stuhlmann H, Dawson PE. Nano Lett. 2010;10:1093–1097. doi: 10.1021/nl1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Venter PA, Dirksen A, Thomas D, Manchester M, Dawson PE, Schneemann A. Biomacromolecules. 2011;12:2293–2301. doi: 10.1021/bm200369e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carrico ZM, Farkas ME, Zhou Y, Hsiao SC, Marks JD, Chokhawala H, Clark DS, Francis MB. ACS Nano. 2012;6:6675–6680. doi: 10.1021/nn301134z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee KL, Shukla S, Wu M, Ayat NR, El Sanadi CE, Wen AM, Edelbrock JF, Pokorski JK, Commandeur U, Dubyak GR, Steinmetz NF. Acta Biomater. 2015;19:166–179. doi: 10.1016/j.actbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Nat. Protoc. 2010;5:1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lucon J, Qazi S, Uchida M, Bedwell GJ, LaFrance B, Prevelige PE, Jr., Douglas T. Nat. Chem. 2012;4:781–788. doi: 10.1038/nchem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pokorski JK, Breitenkamp K, Liepold LO, Qazi S, Finn MG. J. Am. Chem. Soc. 2011;133:9242–9245. doi: 10.1021/ja203286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sen Gupta S, Raja KS, Kaltgrad E, Strable E, Finn MG. Chem. Commun. (Camb) 2005 doi: 10.1039/b502444g. DOI: 10.1039/b502444g, 4315-4317. [DOI] [PubMed] [Google Scholar]
- 107.Isarov SA, Lee PW, Pokorski JK. Biomacromolecules. 2016;17:641–648. doi: 10.1021/acs.biomac.5b01582. [DOI] [PubMed] [Google Scholar]
- 108.Chen L, Zhao X, Lin Y, Huang Y, Wang Q. Chem. Commun. (Camb) 2013;49:9678–9680. doi: 10.1039/c3cc45559a. [DOI] [PubMed] [Google Scholar]
- 109.Hu J, Wang P, Zhao X, Lv L, Yang S, Song B, Wang Q. Chem. Commun. (Camb) 2014;50:14125–14128. doi: 10.1039/c4cc05195e. [DOI] [PubMed] [Google Scholar]
- 110.Yildiz I, Lee KL, Chen K, Shukla S, Steinmetz NF. J. Control. Release. 2013;172:568–578. doi: 10.1016/j.jconrel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wen AM, Le N, Zhou X, Steinmetz NF, Popkin DL. ACS Biomater. Sci. Eng. 2015;1:1050–1054. doi: 10.1021/acsbiomaterials.5b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao J, Guenther RH, Sit TL, Lommel SA, Opperman CH, Willoughby JA. ACS Appl. Mater. Interfaces. 2015;7:9546–9553. doi: 10.1021/acsami.5b00940. [DOI] [PubMed] [Google Scholar]
- 113.Honarbakhsh S, Guenther RH, Willoughby JA, Lommel SA, Pourdeyhimi B. Adv. Healthc. Mater. 2013;2:1001–1007. doi: 10.1002/adhm.201200434. [DOI] [PubMed] [Google Scholar]
- 114.Loo L, Guenther RH, Lommel SA, Franzen S. Chem. Commun. (Camb) 2008:88–90. doi: 10.1039/b714748a. [DOI] [PubMed] [Google Scholar]
- 115.Mao C, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM. Science. 2004;303:213–217. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- 116.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 117.Douglas T, Strable E, Willits D, Aitouchen A, Libera M, Young M. Adv. Mater. 2002;14:415–418. [Google Scholar]
- 118.Aljabali AA, Sainsbury F, Lomonossoff GP, Evans DJ. Small. 2010;6:818–821. doi: 10.1002/smll.200902135. [DOI] [PubMed] [Google Scholar]
- 119.Knez M, Bittner AM, Boes F, Wege C, Jeske H, Maiβ E, Kern K. Nano Letters. 2003;3:1079–1082. [Google Scholar]
- 120.Altintoprak K, Seidenstucker A, Welle A, Eiben S, Atanasova P, Stitz N, Plettl A, Bill J, Gliemann H, Jeske H, Rothenstein D, Geiger F, Wege C. Beilstein J. Nanotechnol. 2015;6:1399–1412. doi: 10.3762/bjnano.6.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lim J-S, Kim S-M, Lee S-Y, Stach EA, Culver JN, Harris MT. J. Nanomater. 2010:2010. [Google Scholar]
- 122.Aljabali AA, Lomonossoff GP, Evans DJ. Biomacromolecules. 2011;12:2723–2728. doi: 10.1021/bm200499v. [DOI] [PubMed] [Google Scholar]
- 123.Daniel MC, Tsvetkova IB, Quinkert ZT, Murali A, De M, Rotello VM, Kao CC, Dragnea B. ACS Nano. 2010;4:3853–3860. doi: 10.1021/nn1005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, Kao CC, Dragnea B. Proc. Natl. Acad. Sci. USA. 2007;104:1354–1359. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luque D, Escosura A. d. l., Snijder J, Brasch M, Burnley RJ, Koay MST, Carrascosa JL, Wuite GJL, Roos WH, Heck AJR, Cornelissen JJLM, Torres T, Caston JR. Chem. Sci. 2014;5:575–581. [Google Scholar]
- 126.Huang X, Bronstein LM, Retrum J, Dufort C, Tsvetkova I, Aniagyei S, Stein B, Stucky G, McKenna B, Remmes N, Baxter D, Kao CC, Dragnea B. Nano Lett. 2007;7:2407–2416. doi: 10.1021/nl071083l. [DOI] [PubMed] [Google Scholar]
- 127.Loo L, Guenther RH, Lommel SA, Franzen S. J. Am. Chem. Soc. 2007;129:11111–11117. doi: 10.1021/ja071896b. [DOI] [PubMed] [Google Scholar]
- 128.van Eldijk MB, Wang JC, Minten IJ, Li C, Zlotnick A, Nolte RJ, Cornelissen JJ, van Hest JC. J. Am. Chem. Soc. 2012;134:18506–18509. doi: 10.1021/ja308132z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schoonen L, Pille J, Borrmann A, Nolte RJ, van Hest JC. Bioconjug. Chem. 2015 doi: 10.1021/acs.bioconjchem.5b00485. DOI: 10.1021/acs.bioconjchem.5b00485. [DOI] [PubMed] [Google Scholar]
- 130.Minten IJ, Nolte RJ, Cornelissen JJ. Macromol. Biosci. 2010;10:539–545. doi: 10.1002/mabi.201000030. [DOI] [PubMed] [Google Scholar]
- 131.Patterson DP, Schwarz B, Waters RS, Gedeon T, Douglas T. ACS Chem. Biol. 2014;9:359–365. doi: 10.1021/cb4006529. [DOI] [PubMed] [Google Scholar]
- 132.Sanchez-Sanchez L, Tapia-Moreno A, Juarez-Moreno K, Patterson DP, Cadena-Nava RD, Douglas T, Vazquez-Duhalt R. J. Nanobiotechnology. 2015;13:66. doi: 10.1186/s12951-015-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miest TS, Cattaneo R. Nat. Rev. Microbiol. 2014;12:23–34. doi: 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kay MA. Nat. Rev. Genet. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 135.Mullard A. Nat. Rev. Drug Discov. 2015;14:811. doi: 10.1038/nrd.2016.259. [DOI] [PubMed] [Google Scholar]
- 136.Wen AM, Rambhia PH, French RH, Steinmetz NF. J. Biol. Phys. 2013;39:301–325. doi: 10.1007/s10867-013-9314-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kaiser CR, Flenniken ML, Gillitzer E, Harmsen AL, Harmsen AG, Jutila MA, Douglas T, Young MJ. Int. J. Nanomedicine. 2007;2:715–733. [PMC free article] [PubMed] [Google Scholar]
- 138.Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Virology. 2014;449:163–173. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. J. Control. Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Prasuhn DE, Jr., Singh P, Strable E, Brown S, Manchester M, Finn MG. J. Am. Chem. Soc. 2008;130:1328–1334. doi: 10.1021/ja075937f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Molenaar TJ, Michon I, de Haas SA, van Berkel TJ, Kuiper J, Biessen EA. Virology. 2002;293:182–191. doi: 10.1006/viro.2001.1254. [DOI] [PubMed] [Google Scholar]
- 142.Vitiello CL, Merril CR, Adhya S. Virus Res. 2005;114:101–103. doi: 10.1016/j.virusres.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 143.Shukla S, Ablack AL, Wen AM, Lee KL, Lewis JD, Steinmetz NF. Mol. Pharm. 2013;10:33–42. doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Esfandiari N, Arzanani MK, Soleimani M, Kohi-Habibi M, Svendsen WE. Tumour Biol. 2015 doi: 10.1007/s13277-015-3867-3. DOI: 10.1007/s13277-015-3867-3. [DOI] [PubMed] [Google Scholar]
- 145.Wu Z, Chen K, Yildiz I, Dirksen A, Fischer R, Dawson PE, Steinmetz NF. Nanoscale. 2012;4:3567–3576. doi: 10.1039/c2nr30366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jain RK, Stylianopoulos T. Nat. Rev. Clin. Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee KL, Hubbard LC, Hern S, Yildiz I, Gratzl M, Steinmetz NF. Biomater. Sci. 2013;1:581–588. doi: 10.1039/C3BM00191A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bruckman MA, Jiang K, Simpson EJ, Randolph LN, Luyt LG, Yu X, Steinmetz NF. Nano Lett. 2014;14:1551–1558. doi: 10.1021/nl404816m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wen AM, Wang Y, Jiang K, Hsu GC, Gao H, Lee KL, Yang AC, Yu X, Simon DI, Steinmetz NF. J. Mater. Chem. B. 2015;3:6037–6045. doi: 10.1039/C5TB00879D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wen AM, Steinmetz NF. Adv. Healthc. Mater. 2014;3:1739–1744. doi: 10.1002/adhm.201400141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, Eiben S, Twyman RM, Wege C, Steinmetz NF. Adv. Healthc. Mater. 2015;4:874–882. doi: 10.1002/adhm.201400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kwon OJ, Kang E, Choi JW, Kim SW, Yun CO. J. Control. Release. 2013;169:257–265. doi: 10.1016/j.jconrel.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 153.Tesfay MZ, Kirk AC, Hadac EM, Griesmann GE, Federspiel MJ, Barber GN, Henry SM, Peng KW, Russell SJ. J. Virol. 2013;87:3752–3759. doi: 10.1128/JVI.02832-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shukla S, Wen AM, Ayat NR, Commandeur U, Gopalkrishnan R, Broome AM, Lozada KW, Keri RA, Steinmetz NF. Nanomedicine. 2014;9:221–235. doi: 10.2217/nnm.13.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fisher KD, Stallwood Y, Green NK, Ulbrich K, Mautner V, Seymour LW. Gene Ther. 2001;8:341–348. doi: 10.1038/sj.gt.3301389. [DOI] [PubMed] [Google Scholar]
- 156.Carlisle RC, Benjamin R, Briggs SS, Sumner-Jones S, McIntosh J, Gill D, Hyde S, Nathwani A, Subr V, Ulbrich K, Seymour LW, Fisher KD. J. Gene Med. 2008;10:400–411. doi: 10.1002/jgm.1161. [DOI] [PubMed] [Google Scholar]
- 157.Grunwald GK, Vetter A, Klutz K, Willhauck MJ, Schwenk N, Senekowitsch-Schmidtke R, Schwaiger M, Zach C, Wagner E, Goke B, Holm PS, Ogris M, Spitzweg C. Mol. Ther. Nucleic Acids. 2013;2:e131. doi: 10.1038/mtna.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shao W, Paul A, Abbasi S, Chahal PS, Mena JA, Montes J, Kamen A, Prakash S. Int. J. Nanomedicine. 2012;7:1575–1586. doi: 10.2147/IJN.S26891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pitek AS, Jameson SA, Veliz FA, Shukla S, Steinmetz NF. Biomaterials. 2016;89:89–97. doi: 10.1016/j.biomaterials.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rodriguez PL, Harada T, Christian DA, Pantano DA, Tsai RK, Discher DE. Science. 2013;339:971–975. doi: 10.1126/science.1229568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chariou PL, Lee KL, Wen AM, Gulati NM, Stewart PL, Steinmetz NF. Bioconjug. Chem. 2015;26:262–269. doi: 10.1021/bc500545z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pokorski JK, Hovlid ML, Finn MG. Chembiochem. 2011;12:2441–2447. doi: 10.1002/cbic.201100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zeng Q, Wen H, Wen Q, Chen X, Wang Y, Xuan W, Liang J, Wan S. Biomaterials. 2013;34:4632–4642. doi: 10.1016/j.biomaterials.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 164.Destito G, Yeh R, Rae CS, Finn MG, Manchester M. Chem. Biol. 2007;14:1152–1162. doi: 10.1016/j.chembiol.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Adv. Mater. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pitek AS, Wen AM, Shukla S, Steinmetz NF. Small. 2016;12:1758–1769. doi: 10.1002/smll.201502458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu Y, Zhao Y, Sun B, Chen C. Acc. Chem. Res. 2013;46:702–713. doi: 10.1021/ar300028m. [DOI] [PubMed] [Google Scholar]
- 168.Semmler-Behnke M, Kreyling WG, Lipka J, Fertsch S, Wenk A, Takenaka S, Schmid G, Brandau W. Small. 2008;4:2108–2111. doi: 10.1002/smll.200800922. [DOI] [PubMed] [Google Scholar]
- 169.Jaganathan H, Godin B. Adv. Drug Deliv. Rev. 2012;64:1800–1819. doi: 10.1016/j.addr.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Koudelka KJ, Ippoliti S, Medina E, Shriver LP, Trauger SA, Catalano CE, Manchester M. Biomacromolecules. 2013;14:4169–4176. doi: 10.1021/bm401577f. [DOI] [PubMed] [Google Scholar]
- 171.Steinmetz NF, Bize A, Findlay KC, Lomonossoff GP, Manchester M, Evans DJ, Prangishvili D. Adv. Funct. Mater. 2008;18:3478–3486. [Google Scholar]
- 172.Wen AM, Infusino M, De Luca A, Kernan DL, Czapar AE, Strangi G, Steinmetz NF. Bioconjug. Chem. 2015;26:51–62. doi: 10.1021/bc500524f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gupta S, Chatni MR, Rao ALN, Vullev VI, Wang LV, Anvari B. Nanoscale. 2013;5:1772–1776. doi: 10.1039/c3nr34124k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Capehart SL, Coyle MP, Glasgow JE, Francis MB. Journal of the American Chemical Society. 2013;135:3011–3016. doi: 10.1021/ja3078472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rae CS, Khor IW, Wang Q, Destito G, Gonzalez MJ, Singh P, Thomas DM, Estrada MN, Powell E, Finn MG, Manchester M. Virology. 2005;343:224–235. doi: 10.1016/j.virol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 176.Koudelka KJ, Destito G, Plummer EM, Trauger SA, Siuzdak G, Manchester M. PLoS Pathog. 2009;5:e1000417. doi: 10.1371/journal.ppat.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Plummer EM, Thomas D, Destito G, Shriver LP, Manchester M. Nanomedicine. 2012;7:877–888. doi: 10.2217/nnm.11.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Steinmetz NF, Cho CF, Ablack A, Lewis JD, Manchester M. Nanomedicine. 2011;6:351–364. doi: 10.2217/nnm.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shriver LP, Koudelka KJ, Manchester M. J. Neuroimmunol. 2009;211:66–72. doi: 10.1016/j.jneuroim.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Nat. Med. 2006;12:354–360. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bardhan NM, Ghosh D, Belcher AM. J. Biophotonics. 2014;7:617–623. doi: 10.1002/jbio.201300010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Choi DS, Jin HE, Yoo SY, Lee SW. Bioconjug. Chem. 2014;25:216–223. doi: 10.1021/bc4003234. [DOI] [PubMed] [Google Scholar]
- 183.Hovlid ML, Steinmetz NF, Laufer B, Lau JL, Kuzelka J, Wang Q, Hyypia T, Nemerow GR, Kessler H, Manchester M, Finn MG. Nanoscale. 2012;4:3698–3705. doi: 10.1039/c2nr30571b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ren Y, Wong SM, Lim LY. Bioconjug. Chem. 2007;18:836–843. doi: 10.1021/bc060361p. [DOI] [PubMed] [Google Scholar]
- 185.Li K, Chen Y, Li S, Nguyen HG, Niu Z, You S, Mello CM, Lu X, Wang Q. Bioconjug. Chem. 2010;21:1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 186.Huang RK, Steinmetz NF, Fu CY, Manchester M, Johnson JE. Nanomedicine. 2011;6:55–68. doi: 10.2217/nnm.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yi H, Ghosh D, Ham M-H, Qi J, Barone PW, Strano MS, Belcher AM. Nano Letters. 2012;12:1176–1183. doi: 10.1021/nl2031663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.ElSohly AM, Netirojjanakul C, Aanei IL, Jager A, Bendall SC, Farkas ME, Nolan GP, Francis MB. Bioconjugate Chemistry. 2015;26:1590–1596. doi: 10.1021/acs.bioconjchem.5b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jin HE, Farr R, Lee SW. Biomaterials. 2014;35:9236–9245. doi: 10.1016/j.biomaterials.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 190.Ghosh D, Bagley AF, Na YJ, Birrer MJ, Bhatia SN, Belcher AM. Proc. Natl. Acad. Sci. USA. 2014;111:13948–13953. doi: 10.1073/pnas.1400821111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kelly KA, Waterman P, Weissleder R. Neoplasia. 2006;8:1011–1018. doi: 10.1593/neo.06610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Obermeyer AC, Capehart SL, Jarman JB, Francis MB. PLoS One. 2014;9:e100678. doi: 10.1371/journal.pone.0100678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Allen M, Bulte JW, Liepold L, Basu G, Zywicke HA, Frank JA, Young M, Douglas T. Magn Reson Med. 2005;54:807–812. doi: 10.1002/mrm.20614. [DOI] [PubMed] [Google Scholar]
- 194.Liepold L, Anderson S, Willits D, Oltrogge L, Frank JA, Douglas T, Young M. Magn Reson Med. 2007;58:871–879. doi: 10.1002/mrm.21307. [DOI] [PubMed] [Google Scholar]
- 195.Anderson EA, Isaacman S, Peabody DS, Wang EY, Canary JW, Kirshenbaum K. Nano Lett. 2006;6:1160–1164. doi: 10.1021/nl060378g. [DOI] [PubMed] [Google Scholar]
- 196.Hooker JM, Datta A, Botta M, Raymond KN, Francis MB. Nano Lett. 2007;7:2207–2210. doi: 10.1021/nl070512c. [DOI] [PubMed] [Google Scholar]
- 197.Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. J. Am. Chem. Soc. 2008;130:2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 198.Garimella PD, Datta A, Romanini DW, Raymond KN, Francis MB. Journal of the American Chemical Society. 2011;133:14704–14709. doi: 10.1021/ja204516p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Qazi S, Liepold LO, Abedin MJ, Johnson B, Prevelige P, Frank JA, Douglas T. Mol. Pharm. 2013;10:11–17. doi: 10.1021/mp300208g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Min J, Jung H, Shin HH, Cho G, Cho H, Kang S. Biomacromolecules. 2013;14:2332–2339. doi: 10.1021/bm400461j. [DOI] [PubMed] [Google Scholar]
- 201.Bruckman MA, VanMeter A, Steinmetz NF. ACS Biomater Sci Eng. 2015;1:13–18. doi: 10.1021/ab500059s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bruckman MA, Randolph LN, Gulati NM, Stewart PL, Steinmetz NF. J. Mater. Chem. B. 2015;3:7503–7510. doi: 10.1039/C5TB01014D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Qazi S, Uchida M, Usselman R, Shearer R, Edwards E, Douglas T. J. Biol. Inorg. Chem. 2014;19:237–246. doi: 10.1007/s00775-013-1075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huang X, Stein BD, Cheng H, Malyutin A, Tsvetkova IB, Baxter DV, Remmes NB, Verchot J, Kao C, Bronstein LM, Dragnea B. ACS Nano. 2011;5:4037–4045. doi: 10.1021/nn200629g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ghosh D, Lee Y, Thomas S, Kohli AG, Yun DS, Belcher AM, Kelly KA. Nat. Nanotechnol. 2012;7:677–682. doi: 10.1038/nnano.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Meldrum T, Seim KL, Bajaj VS, Palaniappan KK, Wu W, Francis MB, Wemmer DE, Pines A. J. Am. Chem. Soc. 2010;132:5936–5937. doi: 10.1021/ja100319f. [DOI] [PubMed] [Google Scholar]
- 207.Stevens TK, Palaniappan KK, Ramirez RM, Francis MB, Wemmer DE, Pines A. Magn. Reson. Med. 2013;69:1245–1252. doi: 10.1002/mrm.24371. [DOI] [PubMed] [Google Scholar]
- 208.Palaniappan KK, Ramirez RM, Bajaj VS, Wemmer DE, Pines A, Francis MB. Angew. Chem. Int. Ed. Engl. 2013;52:4849–4853. doi: 10.1002/anie.201300170. [DOI] [PubMed] [Google Scholar]
- 209.Hooker JM, O'Neil JP, Romanini DW, Taylor SE, Francis MB. Mol. Imaging Biol. 2008;10:182–191. doi: 10.1007/s11307-008-0136-5. [DOI] [PubMed] [Google Scholar]
- 210.Farkas ME, Aanei IL, Behrens CR, Tong GJ, Murphy ST, O'Neil JP, Francis MB. Mol. Pharm. 2013;10:69–76. doi: 10.1021/mp3003754. [DOI] [PubMed] [Google Scholar]
- 211.Flexman JA, Cross DJ, Lewellen BL, Miyoshi S, Kim Y, Minoshima S. IEEE Trans. Nanobioscience. 2008;7:223–232. doi: 10.1109/TNB.2008.2002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hajitou A, Lev DC, Hannay JA, Korchin B, Staquicini FI, Soghomonyan S, Alauddin MM, Benjamin RS, Pollock RE, Gelovani JG, Pasqualini R, Arap W. Proc. Natl. Acad. Sci. USA. 2008;105:4471–4476. doi: 10.1073/pnas.0712184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li Z, Jin Q, Huang C, Dasa S, Chen L, Yap LP, Liu S, Cai H, Park R, Conti PS. Theranostics. 2011;1:371–380. doi: 10.7150/thno/v01p0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Riedel S. Proc. (Bayl. Univ. Med. Cent.) 2005;18:21–25. doi: 10.1080/08998280.2005.11928028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Plotkin SA. Nat. Med. 2005;11:S5–11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Leleux J, Roy K. Adv. Healthc. Mater. 2013;2:72–94. doi: 10.1002/adhm.201200268. [DOI] [PubMed] [Google Scholar]
- 217.Aylward B, Tangermann R. Vaccine. 2011;29(Suppl 4):D80–85. doi: 10.1016/j.vaccine.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 218.Castillo-Solorzano CC, Matus CR, Flannery B, Marsigli C, Tambini G, Andrus JK. J. Infect. Dis. 2011;204(Suppl 1):S270–278. doi: 10.1093/infdis/jir166. [DOI] [PubMed] [Google Scholar]
- 219.Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Expert Rev. Vaccines. 2010;9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 220.Lee KL, Twyman RM, Fiering S, Steinmetz NF. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015 doi: 10.1002/wnan.1383. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Schiller J, Chackerian B. PLoS Pathog. 2014;10:e1004254. doi: 10.1371/journal.ppat.1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Benen TD, Tonks P, Kliche A, Kapzan R, Heeney JL, Wagner R. J. Biomed. Sci. 2014;21:79. doi: 10.1186/s12929-014-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Van Rompay KK, Hunter Z, Jayashankar K, Peabody J, Montefiori D, LaBranche CC, Keele BF, Jensen K, Abel K, Chackerian B. J. Virol. 2014;88:2011–2024. doi: 10.1128/JVI.02447-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, Investigators M-T. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 225.Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. J. Infect. Dis. 2007;196(Suppl 2):S430–437. doi: 10.1086/520583. [DOI] [PubMed] [Google Scholar]
- 226.Sun Y, Carrion R, Jr., Ye L, Wen Z, Ro YT, Brasky K, Ticer AE, Schwegler EE, Patterson JL, Compans RW, Yang C. Virology. 2009;383:12–21. doi: 10.1016/j.virol.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Matassov D, Marzi A, Latham T, Xu R, Ota-Setlik A, Feldmann F, Geisbert JB, Mire CE, Hamm S, Nowak B, Egan MA, Geisbert TW, Eldridge JH, Feldmann H, Clarke DK. J. Infect. Dis. 2015;212(Suppl 2):S443–451. doi: 10.1093/infdis/jiv316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Cao Y, Lu Z, Sun J, Bai X, Sun P, Bao H, Chen Y, Guo J, Li D, Liu X, Liu Z. Vet. Microbiol. 2009;137:10–17. doi: 10.1016/j.vetmic.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stewart M, Dubois E, Sailleau C, Breard E, Viarouge C, Desprat A, Thiery R, Zientara S, Roy P. Vaccine. 2013;31:553–558. doi: 10.1016/j.vaccine.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 230.Thuenemann EC, Meyers AE, Verwey J, Rybicki EP, Lomonossoff GP. Plant Biotechnol. J. 2013;11:839–846. doi: 10.1111/pbi.12076. [DOI] [PubMed] [Google Scholar]
- 231.Rybicki EP. Plant Biotechnol. J. 2010;8:620–637. doi: 10.1111/j.1467-7652.2010.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lindblom N, de Villiers SH, Kalayanov G, Gordon S, Johansson AM, Svensson TH. Respiration. 2002;69:254–260. doi: 10.1159/000063629. [DOI] [PubMed] [Google Scholar]
- 233.Maurer P, Jennings GT, Willers J, Rohner F, Lindman Y, Roubicek K, Renner WA, Muller P, Bachmann MF. Eur. J. Immunol. 2005;35:2031–2040. doi: 10.1002/eji.200526285. [DOI] [PubMed] [Google Scholar]
- 234.Maurer P, Bachmann MF. Curr. Opin. Mol. Ther. 2006;8:11–16. [PubMed] [Google Scholar]
- 235.Jin H, Wang W, Zhao S, Yang W, Qian Y, Jia N, Feng G. Vaccine. 2014;32:4450–4456. doi: 10.1016/j.vaccine.2014.06.051. [DOI] [PubMed] [Google Scholar]
- 236.Farlow MR, Andreasen N, Riviere ME, Vostiar I, Vitaliti A, Sovago J, Caputo A, Winblad B, Graf A. Alzheimers Res. Ther. 2015;7:23. doi: 10.1186/s13195-015-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Cuzick J. Expert Rev. Vaccines. 2015;14:1047–1049. doi: 10.1586/14760584.2015.1051470. [DOI] [PubMed] [Google Scholar]
- 238.Tumban E, Muttil P, Escobar CA, Peabody J, Wafula D, Peabody DS, Chackerian B. Vaccine. 2015;33:3346–3353. doi: 10.1016/j.vaccine.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Shukla S, Wen AM, Commandeur U, Steinmetz NF. J. Mater. Chem. B. 2014;2:6249–6258. doi: 10.1039/c4tb00749b. [DOI] [PubMed] [Google Scholar]
- 240.Tegerstedt K, Lindencrona JA, Curcio C, Andreasson K, Tullus C, Forni G, Dalianis T, Kiessling R, Ramqvist T. Cancer Res. 2005;65:5953–5957. doi: 10.1158/0008-5472.CAN-05-0335. [DOI] [PubMed] [Google Scholar]
- 241.Miermont A, Barnhill H, Strable E, Lu X, Wall KA, Wang Q, Finn MG, Huang X. Chemistry. 2008;14:4939–4947. doi: 10.1002/chem.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yin Z, Comellas-Aragones M, Chowdhury S, Bentley P, Kaczanowska K, Benmohamed L, Gildersleeve JC, Finn MG, Huang X. ACS Chem. Biol. 2013;8:1253–1262. doi: 10.1021/cb400060x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yin Z, Nguyen HG, Chowdhury S, Bentley P, Bruckman MA, Miermont A, Gildersleeve JC, Wang Q, Huang X. Bioconjug. Chem. 2012;23:1694–1703. doi: 10.1021/bc300244a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kaufman HL, Kohlhapp FJ, Zloza A. Nat. Rev. Drug Discov. 2015;14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lichty BD, Breitbach CJ, Stojdl DF, Bell JC. Nat. Rev. Cancer. 2014;14:559–567. doi: 10.1038/nrc3770. [DOI] [PubMed] [Google Scholar]
- 246.Lizotte PH, Wen AM, Sheen MR, Fields J, Rojanasopondist P, Steinmetz NF, Fiering S. Nat. Nanotechnol. 2015 doi: 10.1038/nnano.2015.292. DOI: 10.1038/nnano.2015.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Friedmann T, Roblin R. Science. 1972;175:949–955. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- 248.Coune PG, Schneider BL, Aebischer P. Cold Spring Harb. Perspect. Med. 2012;2:a009431. doi: 10.1101/cshperspect.a009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Griesenbach U, Alton EW. Hum. Mol. Genet. 2013;22:R52–58. doi: 10.1093/hmg/ddt372. [DOI] [PubMed] [Google Scholar]
- 250.Chuah MK, Nair N, VandenDriessche T. Hum. Gene Ther. 2012;23:557–565. doi: 10.1089/hum.2012.088. [DOI] [PubMed] [Google Scholar]
- 251.Walther W, Schlag PM. Curr. Opin. Oncol. 2013;25:659–664. doi: 10.1097/CCO.0000000000000004. [DOI] [PubMed] [Google Scholar]
- 252.Morrison C. Nat. Biotechnol. 2015;33:217–218. doi: 10.1038/nbt0315-217. [DOI] [PubMed] [Google Scholar]
- 253.Schimmer J, Breazzano S. Hum. Gene Ther. Clin. Dev. 2015;26:144–149. doi: 10.1089/humc.2015.29001.sch. [DOI] [PubMed] [Google Scholar]
- 254.Verma IM. Mol. Ther. 2000;2:415–416. doi: 10.1006/mthe.2000.0213. [DOI] [PubMed] [Google Scholar]
- 255.Couzin J, Kaiser J. Science. 2005;307:1028. doi: 10.1126/science.307.5712.1028b. [DOI] [PubMed] [Google Scholar]
- 256.Schaffer DV, Koerber JT, Lim KI. Annu Rev Biomed Eng. 2008;10:169–194. doi: 10.1146/annurev.bioeng.10.061807.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kotterman MA, Schaffer DV. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wang D, Gao G. Discov. Med. 2014;18:151–161. [PMC free article] [PubMed] [Google Scholar]
- 259.Singh R, Kostarelos K. Trends Biotechnol. 2009;27:220–229. doi: 10.1016/j.tibtech.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 260.Westphal M, Yla-Herttuala S, Martin J, Warnke P, Menei P, Eckland D, Kinley J, Kay R, Ram Z, Group AS. Lancet Oncol. 2013;14:823–833. doi: 10.1016/S1470-2045(13)70274-2. [DOI] [PubMed] [Google Scholar]
- 261.Mowa MB, Crowther C, Ely A, Arbuthnot P. Biomed. Res. Int. 2014;2014:718743. doi: 10.1155/2014/718743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ferreira V, Petry H, Salmon F. Front. Immunol. 2014;5:82. doi: 10.3389/fimmu.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Rafii MS, Baumann TL, Bakay RA, Ostrove JM, Siffert J, Fleisher AS, Herzog CD, Barba D, Pay M, Salmon DP, Chu Y, Kordower JH, Bishop K, Keator D, Potkin S, Bartus RT. Alzheimers Dement. 2014;10:571–581. doi: 10.1016/j.jalz.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 264.Gardner MR, Kattenhorn LM, Kondur HR, von Schaewen M, Dorfman T, Chiang JJ, Haworth KG, Decker JM, Alpert MD, Bailey CC, Neale ES, Jr., Fellinger CH, Joshi VR, Fuchs SP, Martinez-Navio JM, Quinlan BD, Yao AY, Mouquet H, Gorman J, Zhang B, Poignard P, Nussenzweig MC, Burton DR, Kwong PD, Piatak M, Jr., Lifson JD, Gao G, Desrosiers RC, Evans DT, Hahn BH, Ploss A, Cannon PM, Seaman MS, Farzan M. Nature. 2015;519:87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Reid T, Warren R, Kirn D. Cancer Gene Ther. 2002;9:979–986. doi: 10.1038/sj.cgt.7700539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sumida SM, Truitt DM, Lemckert AA, Vogels R, Custers JH, Addo MM, Lockman S, Peter T, Peyerl FW, Kishko MG, Jackson SS, Gorgone DA, Lifton MA, Essex M, Walker BD, Goudsmit J, Havenga MJ, Barouch DH. J. Immunol. 2005;174:7179–7185. doi: 10.4049/jimmunol.174.11.7179. [DOI] [PubMed] [Google Scholar]
- 267.Bachtarzi H, Stevenson M, Subr V, Ulbrich K, Seymour LW, Fisher KD. J. Control. Release. 2011;150:196–203. doi: 10.1016/j.jconrel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Li S, Chen J, Xu H, Long J, Xie X, Zhang Y. PLoS One. 2014;9:e100670. doi: 10.1371/journal.pone.0100670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Prill JM, Subr V, Pasquarelli N, Engler T, Hoffmeister A, Kochanek S, Ulbrich K, Kreppel F. PLoS One. 2014;9:e82716. doi: 10.1371/journal.pone.0082716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hofherr SE, Mok H, Gushiken FC, Lopez JA, Barry MA. Hum. Gene Ther. 2007;18:837–848. doi: 10.1089/hum.2007.0051. [DOI] [PubMed] [Google Scholar]
- 271.Lanciotti J, Song A, Doukas J, Sosnowski B, Pierce G, Gregory R, Wadsworth S, O'Riordan C. Mol. Ther. 2003;8:99–107. doi: 10.1016/s1525-0016(03)00139-4. [DOI] [PubMed] [Google Scholar]
- 272.Singarapu K, Pal I, Ramsey JD. J. Biomed. Mater. Res. A. 2013;101:1857–1864. doi: 10.1002/jbm.a.34483. [DOI] [PubMed] [Google Scholar]
- 273.Burg MA, Jensen-Pergakes K, Gonzalez AM, Ravey P, Baird A, Larocca D. Cancer Res. 2002;62:977–981. [PubMed] [Google Scholar]
- 274.Tang KH, Yusoff K, Tan WS. J. Virol. Methods. 2009;159:194–199. doi: 10.1016/j.jviromet.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 275.Yata T, Lee KY, Dharakul T, Songsivilai S, Bismarck A, Mintz PJ, Hajitou A. Mol. Ther. Nucleic Acids. 2014;3:e185. doi: 10.1038/mtna.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Azizgolshani O, Garmann RF, Cadena-Nava R, Knobler CM, Gelbart WM. Virology. 2013;441:12–17. doi: 10.1016/j.virol.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 277.Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL, Brinker CJ, Caldeira Jdo C, Chackerian B, Wharton W, Peabody DS. ACS Nano. 2011;5:5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pan Y, Zhang Y, Jia T, Zhang K, Li J, Wang L. FEBS J. 2012;279:1198–1208. doi: 10.1111/j.1742-4658.2012.08512.x. [DOI] [PubMed] [Google Scholar]
- 279.Wei B, Wei Y, Zhang K, Wang J, Xu R, Zhan S, Lin G, Wang W, Liu M, Wang L, Zhang R, Li J. Biomed. Pharmacother. 2009;63:313–318. doi: 10.1016/j.biopha.2008.07.086. [DOI] [PubMed] [Google Scholar]
- 280.Everts M, Saini V, Leddon JL, Kok RJ, Stoff-Khalili M, Preuss MA, Millican CL, Perkins G, Brown JM, Bagaria H, Nikles DE, Johnson DT, Zharov VP, Curiel DT. Nano Lett. 2006;6:587–591. doi: 10.1021/nl0500555. [DOI] [PubMed] [Google Scholar]
- 281.Fontana J, Dressick WJ, Phelps J, Johnson JE, Rendell RW, Sampson T, Ratna BR, Soto CM. Small. 2014;10:3058–3063. doi: 10.1002/smll.201400470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Bromley KM, Patil AJ, Perriman AW, Stubbs G, Mann S. J. Mater. Chem. 2008;18:4796–4801. [Google Scholar]
- 283.Suci PA, Varpness Z, Gillitzer E, Douglas T, Young M. Langmuir. 2007;23:12280–12286. doi: 10.1021/la7021424. [DOI] [PubMed] [Google Scholar]
- 284.Wen AM, Ryan MJ, Yang AC, Breitenkamp K, Pokorski JK, Steinmetz NF. Chem. Commun. (Camb) 2012;48:9044–9046. doi: 10.1039/c2cc34695h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Stephanopoulos N, Tong GJ, Hsiao SC, Francis MB. ACS Nano. 2010;4:6014–6020. doi: 10.1021/nn1014769. [DOI] [PubMed] [Google Scholar]
- 286.Rhee JK, Baksh M, Nycholat C, Paulson JC, Kitagishi H, Finn MG. Biomacromolecules. 2012;13:2333–2338. doi: 10.1021/bm300578p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Millan JG, Brasch M, Anaya-Plaza E, de la Escosura A, Velders AH, Reinhoudt DN, Torres T, Koay MS, Cornelissen JJ. J. Inorg. Biochem. 2014;136:140–146. doi: 10.1016/j.jinorgbio.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 288.Lammers T, Aime S, Hennink WE, Storm G, Kiessling F. Acc. Chem. Res. 2011;44:1029–1038. doi: 10.1021/ar200019c. [DOI] [PubMed] [Google Scholar]
- 289.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Adv. Drug Deliv. Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Gabizon AA. Cancer Invest. 2001;19:424–436. doi: 10.1081/cnv-100103136. [DOI] [PubMed] [Google Scholar]
- 291.Mizutani H, Tada-Oikawa S, Hiraku Y, Kojima M, Kawanishi S. Life Sci. 2005;76:1439–1453. doi: 10.1016/j.lfs.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 292.Bruckman MA, Czapar AE, VanMeter A, Rudolph LN, Steinmetz NF. J. Control. Release. 2016 doi: 10.1016/j.jconrel.2016.02.045. doi:10.1016/j.jconrel.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Ghosh D, Kohli AG, Moser F, Endy D, Belcher AM. ACS Synth. Biol. 2012;1:576–582. doi: 10.1021/sb300052u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lockney DM, Guenther RN, Loo L, Overton W, Antonelli R, Clark J, Hu M, Luft C, Lommel SA, Franzen S. Bioconjug. Chem. 2011;22:67–73. doi: 10.1021/bc100361z. [DOI] [PubMed] [Google Scholar]
- 295.Aljabali AA, Shukla S, Lomonossoff GP, Steinmetz NF, Evans DJ. Mol. Pharm. 2013;10:3–10. doi: 10.1021/mp3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Bar H, Yacoby I, Benhar I. BMC Biotechnol. 2008;8:37. doi: 10.1186/1472-6750-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Inoue T, Kawano MA, Takahashi RU, Tsukamoto H, Enomoto T, Imai T, Kataoka K, Handa H. J. Biotechnol. 2008;134:181–192. doi: 10.1016/j.jbiotec.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 298.DePorter SM, McNaughton BR. Bioconjug. Chem. 2014;25:1620–1625. doi: 10.1021/bc500339k. [DOI] [PubMed] [Google Scholar]
- 299.Wu W, Hsiao SC, Carrico ZM, Francis MB. Angew. Chem. Int. Ed. Engl. 2009;48:9493–9497. doi: 10.1002/anie.200902426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Min J, Moon H, Yang HJ, Shin H-H, Hong SY, Kang S. Macromolecular Bioscience. 2014;14:557–564. doi: 10.1002/mabi.201300401. [DOI] [PubMed] [Google Scholar]
- 301.Ganguly R, Wen AM, Myer AB, Czech T, Sahu S, Steinmetz NF, Raman P. Nanoscale. 2016;8:6542–6554. doi: 10.1039/c6nr00398b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Yacoby I, Bar H, Benhar I. Antimicrob. Agents Chemother. 2007;51:2156–2163. doi: 10.1128/AAC.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yacoby I, Shamis M, Bar H, Shabat D, Benhar I. Antimicrob. Agents Chemother. 2006;50:2087–2097. doi: 10.1128/AAC.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Czapar AE, Zheng YR, Riddell IA, Shukla S, Awuah SG, Lippard SJ, Steinmetz NF. ACS Nano. 2016 doi: 10.1021/acsnano.5b07360. DOI: 10.1021/acsnano.5b07360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Lin Y, Su Z, Niu Z, Li S, Kaur G, Lee LA, Wang Q. Acta Biomater. 2008;4:838–843. doi: 10.1016/j.actbio.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 306.Lee LA, Nguyen QL, Wu L, Horvath G, Nelson RS, Wang Q. Biomacromolecules. 2012;13:422–431. doi: 10.1021/bm2014558. [DOI] [PubMed] [Google Scholar]
- 307.Yoo SY, Oh JW, Lee SW. Langmuir. 2012;28:2166–2172. doi: 10.1021/la203840n. [DOI] [PubMed] [Google Scholar]
- 308.Wu L, Zang J, Lee LA, Niu Z, Horvatha GC, Braxtona V, Wibowo AC, Bruckman MA, Ghoshroy S, zur Loye H-C, Li X, Wang Q. J. Mater. Chem. 2011;21:8550–8557. [Google Scholar]
- 309.Shin YC, Lee JH, Jin L, Kim MJ, Oh JW, Kim TW, Han DW. Biomater. Res. 2014;18:14. doi: 10.1186/2055-7124-18-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Rong J, Lee LA, Li K, Harp B, Mello CM, Niu Z, Wang Q. Chem. Commun. (Camb) 2008 doi: 10.1039/b811039e. DOI: 10.1039/b811039e, 5185-5187. [DOI] [PubMed] [Google Scholar]
- 311.Wu L, Lee LA, Niu Z, Ghoshroy S, Wang Q. Langmuir. 2011;27:9490–9496. doi: 10.1021/la201580v. [DOI] [PubMed] [Google Scholar]
- 312.Lin Y, Balizan E, Lee LA, Niu Z, Wang Q. Angew. Chem. Int. Ed. Engl. 2010;49:868–872. doi: 10.1002/anie.200904993. [DOI] [PubMed] [Google Scholar]
- 313.Zan X, Feng S, Balizan E, Lin Y, Wang Q. ACS Nano. 2013;7:8385–8396. doi: 10.1021/nn403908k. [DOI] [PubMed] [Google Scholar]
- 314.Yoo SY, Chung W-J, Kim TH, Le M, Lee S-W. Soft Matter. 2011;7:363–368. [Google Scholar]
- 315.Chung W-J, Merzlyak A, Lee S-W. Soft Matter. 2010;6:4454–4459. [Google Scholar]
- 316.Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, Bronk LF, Ananta JS, Mandelin J, Georgescu MM, Bankson JA, Gelovani JG, Killian TC, Arap W, Pasqualini R. Nat. Nanotechnol. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Souza GR, Christianson DR, Staquicini FI, Ozawa MG, Snyder EY, Sidman RL, Miller JH, Arap W, Pasqualini R. Proc. Natl. Acad. Sci. USA. 2006;103:1215–1220. doi: 10.1073/pnas.0509739103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Souza GR, Yonel-Gumruk E, Fan D, Easley J, Rangel R, Guzman-Rojas L, Miller JH, Arap W, Pasqualini R. PLoS One. 2008;3:e2242. doi: 10.1371/journal.pone.0002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Chung WJ, Merzlyak A, Yoo SY, Lee SW. Langmuir. 2010;26:9885–9890. doi: 10.1021/la100226u. [DOI] [PubMed] [Google Scholar]
- 320.Feng S, Lu L, Zan X, Wu Y, Lin Y, Wang Q. Langmuir. 2015;31:9402–9409. doi: 10.1021/acs.langmuir.5b02226. [DOI] [PubMed] [Google Scholar]
- 321.Yoo SY, Merzlyak A, Lee S-W. Soft Matter. 2011;7:1660–1666. [Google Scholar]
- 322.Wu Y, Feng S, Zan X, Lin Y, Wang Q. Biomacromolecules. 2015;16:3466–3472. doi: 10.1021/acs.biomac.5b00884. [DOI] [PubMed] [Google Scholar]
- 323.Kaur G, Valarmathi MT, Potts JD, Wang Q. Biomaterials. 2008;29:4074–4081. doi: 10.1016/j.biomaterials.2008.06.029. [DOI] [PubMed] [Google Scholar]
- 324.Kaur G, Valarmathi MT, Potts JD, Jabbari E, Sabo-Attwood T, Wang Q. Biomaterials. 2010;31:1732–1741. doi: 10.1016/j.biomaterials.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Sitasuwan P, Lee LA, Bo P, Davis EN, Lin Y, Wang Q. Integr. Biol. (Camb) 2012;4:651–660. doi: 10.1039/c2ib20041d. [DOI] [PubMed] [Google Scholar]
- 326.Kaur G, Wang C, Sun J, Wang Q. Biomaterials. 2010;31:5813–5824. doi: 10.1016/j.biomaterials.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 327.He T, Abbineni G, Cao B, Mao C. Small. 2010;6:2230–2235. doi: 10.1002/smll.201001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lee LA, Muhammad SM, Nguyen QL, Sitasuwan P, Horvath G, Wang Q. Mol. Pharm. 2012;9:2121–2125. doi: 10.1021/mp300042t. [DOI] [PubMed] [Google Scholar]
- 329.Sitasuwan P, Lee LA, Li K, Nguyen HG, Wang Q. Front. Chem. 2014;2:31. doi: 10.3389/fchem.2014.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wang J, Wang L, Li X, Mao C. Sci. Rep. 2013;3:1242. doi: 10.1038/srep01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Yoo SY, Kobayashi M, Lee PP, Lee SW. Biomacromolecules. 2011;12:987–996. doi: 10.1021/bm1013475. [DOI] [PubMed] [Google Scholar]
- 332.Zhu H, Cao B, Zhen Z, Laxmi AA, Li D, Liu S, Mao C. Biomaterials. 2011;32:4744–4752. doi: 10.1016/j.biomaterials.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Chung WJ, Oh JW, Kwak K, Lee BY, Meyer J, Wang E, Hexemer A, Lee SW. Nature. 2011;478:364–368. doi: 10.1038/nature10513. [DOI] [PubMed] [Google Scholar]
- 334.Wang J, Yang M, Zhu Y, Wang L, Tomsia AP, Mao C. Adv. Mater. 2014;26:4961–4966. doi: 10.1002/adma.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Li H, Zhang FL, Shi WJ, Bai XJ, Jia SQ, Zhang CG, Ding W. PLoS One. 2015;10:e0129013. doi: 10.1371/journal.pone.0129013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Yoo SY, Jin HE, Choi DS, Kobayashi M, Farouz Y, Wang S, Lee SW. Adv. Healthc. Mater. 2015 doi: 10.1002/adhm.201500179. DOI: 10.1002/adhm.201500179. [DOI] [PubMed] [Google Scholar]
- 337.Luckanagul JA, Lee LA, You S, Yang X, Wang Q. J. Biomed. Mater. Res. A. 2015;103:887–895. doi: 10.1002/jbm.a.35227. [DOI] [PubMed] [Google Scholar]
- 338.Smith GP. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 339.Bazan J, Calkosinski I, Gamian A. Hum. Vaccin. Immunother. 2012;8:1817–1828. doi: 10.4161/hv.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Church WB, Dastmalchi S. Drug Discov. Today. 2013;18:1144–1157. doi: 10.1016/j.drudis.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 341.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. Curr. Issues Mol. Biol. 2011;13:51–76. [PubMed] [Google Scholar]
- 342.Arap, Marco A. Genet. Mol. Biol. 2005 DOI: 10.1590/s1415-47572005000100001. [Google Scholar]
- 343.Gray BP, Brown KC. Chem. Rev. 2014;114:1020–1081. doi: 10.1021/cr400166n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Loset GA, Roos N, Bogen B, Sandlie I. PLoS One. 2011;6:e17433. doi: 10.1371/journal.pone.0017433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Smith GP, Scott JK. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 346.Jespers LS, Messens JH, De Keyser A, Eeckhout D, Van den Brande I, Gansemans YG, Lauwereys MJ, Vlasuk GP, Stanssens PE. Bio/Technology. 1995;13:378–382. doi: 10.1038/nbt0495-378. [DOI] [PubMed] [Google Scholar]
- 347.Loset GA, Bogen B, Sandlie I. PLoS One. 2011;6:e14702. doi: 10.1371/journal.pone.0014702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Loset GA, Sandlie I. Methods. 2012;58:40–46. doi: 10.1016/j.ymeth.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 349.Fagerlund A, Myrset AH, Kulseth MA. Appl. Microbiol. Biotechnol. 2008;80:925–936. doi: 10.1007/s00253-008-1630-z. [DOI] [PubMed] [Google Scholar]
- 350.Giebel LB, Cass RT, Milligan DL, Young DC, Arze R, Johnson CR. Biochemistry. 1995;34:15430–15435. doi: 10.1021/bi00047a006. [DOI] [PubMed] [Google Scholar]
- 351.Kay BK, Adey NB, He YS, Manfredi JP, Mataragnon AH, Fowlkes DM. Gene. 1993;128:59–65. doi: 10.1016/0378-1119(93)90153-t. [DOI] [PubMed] [Google Scholar]
- 352.Kalnina Z, Silina K, Meistere I, Zayakin P, Rivosh A, Abols A, Leja M, Minenkova O, Schadendorf D, Line A. J. Immunol. Methods. 2008;334:37–50. doi: 10.1016/j.jim.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 353.Krumpe LR, Atkinson AJ, Smythers GW, Kandel A, Schumacher KM, McMahon JB, Makowski L, Mori T. Proteomics. 2006;6:4210–4222. doi: 10.1002/pmic.200500606. [DOI] [PubMed] [Google Scholar]
- 354.Qi H, Lu H, Qiu HJ, Petrenko V, Liu A. J. Mol. Biol. 2012;417:129–143. doi: 10.1016/j.jmb.2012.01.038. [DOI] [PubMed] [Google Scholar]
- 355.Ruigrok VJ, Levisson M, Eppink MH, Smidt H, van der Oost J. Biochem. J. 2011;436:1–13. doi: 10.1042/BJ20101860. [DOI] [PubMed] [Google Scholar]
- 356.Gungormus M, Fong H, Kim IW, Evans JS, Tamerler C, Sarikaya M. Biomacromolecules. 2008;9:966–973. doi: 10.1021/bm701037x. [DOI] [PubMed] [Google Scholar]
- 357.Naik RR, Stringer SJ, Agarwal G, Jones SE, Stone MO. Nat. Mater. 2002;1:169–172. doi: 10.1038/nmat758. [DOI] [PubMed] [Google Scholar]
- 358.Cao B, Mao C. Biomacromolecules. 2009;10:555–564. doi: 10.1021/bm801224q. [DOI] [PubMed] [Google Scholar]
- 359.Kolodziej AF, Nair SA, Graham P, McMurry TJ, Ladner RC, Wescott C, Sexton DJ, Caravan P. Bioconjug. Chem. 2012;23:548–556. doi: 10.1021/bc200613e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Koivunen E, Wang B, Ruoslahti E. Bio/Technology. 1995;13:265–270. doi: 10.1038/nbt0395-265. [DOI] [PubMed] [Google Scholar]
- 361.Roberts BL, Markland W, Ley AC, Kent RB, White DW, Guterman SK, Ladner RC. Proc. Natl. Acad. Sci. USA. 1992;89:2429–2433. doi: 10.1073/pnas.89.6.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Lu G, Zheng M, Zhu Y, Sha M, Wu Y, Han X. Int. J. Biol. Sci. 2012;8:650–662. doi: 10.7150/ijbs.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Popkov M, Rader C, Barbas CF., 3rd J. Immunol. Methods. 2004;291:137–151. doi: 10.1016/j.jim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 364.Zehnder-Fjallman AH, Marty C, Halin C, Hohn A, Schibli R, Ballmer-Hofer K, Schwendener RA. Oncol. Rep. 2007;18:933–941. [PubMed] [Google Scholar]
- 365.Gao C, Mao S, Lo CH, Wirsching P, Lerner RA, Janda KD. Proc. Natl. Acad. Sci. USA. 1999;96:6025–6030. doi: 10.1073/pnas.96.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Forrer P, Jung S, Pluckthun A. Curr. Opin. Struct. Biol. 1999;9:514–520. doi: 10.1016/S0959-440X(99)80073-6. [DOI] [PubMed] [Google Scholar]
- 367.Demartis S, Huber A, Viti F, Lozzi L, Giovannoni L, Neri P, Winter G, Neri D. J. Mol. Biol. 1999;286:617–633. doi: 10.1006/jmbi.1998.2476. [DOI] [PubMed] [Google Scholar]
- 368.Barry MA, Dower WJ, Johnston SA. Nat. Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 369.Thapa N, Hong HY, Sangeetha P, Kim IS, Yoo J, Rhee K, Oh GT, Kwon IC, Lee BH. J. Control. Release. 2008;131:27–33. doi: 10.1016/j.jconrel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 370.Shadidi M, Sioud M. FASEB J. 2003;17:256–258. doi: 10.1096/fj.02-0280fje. [DOI] [PubMed] [Google Scholar]
- 371.Lo A, Lin CT, Wu HC. Mol. Cancer Ther. 2008;7:579–589. doi: 10.1158/1535-7163.MCT-07-2359. [DOI] [PubMed] [Google Scholar]
- 372.Kim Y, Lillo AM, Steiniger SC, Liu Y, Ballatore C, Anichini A, Mortarini R, Kaufmann GF, Zhou B, Felding-Habermann B, Janda KD. Biochemistry. 2006;45:9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 373.Zhou C, Kang J, Wang X, Wei W, Jiang W. BMC Cancer. 2015;15:889. doi: 10.1186/s12885-015-1891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Loi M, Di Paolo D, Soster M, Brignole C, Bartolini A, Emionite L, Sun J, Becherini P, Curnis F, Petretto A, Sani M, Gori A, Milanese M, Gambini C, Longhi R, Cilli M, Allen TM, Bussolino F, Arap W, Pasqualini R, Corti A, Ponzoni M, Marchio S, Pastorino F. J. Control. Release. 2013;170:233–241. doi: 10.1016/j.jconrel.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Samli KN, McGuire MJ, Newgard CB, Johnston SA, Brown KC. Diabetes. 2005;54:2103–2108. doi: 10.2337/diabetes.54.7.2103. [DOI] [PubMed] [Google Scholar]
- 376.Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Nat. Med. 2008;14:454–458. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Pasqualini R, Ruoslahti E. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 378.Newton-Northup JR, Dickerson MT, Kumar SR, Smith GP, Quinn TP, Deutscher SL. Mol. Imaging Biol. 2014;16:854–864. doi: 10.1007/s11307-014-0762-z. [DOI] [PubMed] [Google Scholar]
- 379.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Nat. Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 380.Mao C, Liu A, Cao B. Angew. Chem. Int. Ed. Engl. 2009;48:6790–6810. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Schmelcher M, Loessner MJ. Bacteriophage. 2014;4:e28137. doi: 10.4161/bact.28137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Voller A, Bartlett A, Bidwell DE. J. Clin. Pathol. 1978;31:507–520. doi: 10.1136/jcp.31.6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Brigati J, Williams DD, Sorokulova IB, Nanduri V, Chen IH, Turnbough CL, Jr., Petrenko VA. Clin. Chem. 2004;50:1899–1906. doi: 10.1373/clinchem.2004.038018. [DOI] [PubMed] [Google Scholar]
- 384.Tan WS, Tan GH, Yusoff K, Seow HF. J. Clin. Virol. 2005;34:35–41. doi: 10.1016/j.jcv.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 385.Tinazzi E, Merlin M, Bason C, Beri R, Zampieri R, Lico C, Bartoloni E, Puccetti A, Lunardi C, Pezzotti M, Avesani L. Front. Plant Sci. 2015:6. doi: 10.3389/fpls.2015.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Aljabali AA, Barclay JE, Steinmetz NF, Lomonossoff GP, Evans DJ. Nanoscale. 2012;4:5640–5645. doi: 10.1039/c2nr31485a. [DOI] [PubMed] [Google Scholar]
- 387.Koch C, Wabbel K, Eber FJ, Krolla-Sidenstein P, Azucena C, Gliemann H, Eiben S, Geiger F, Wege C. Front Plant Sci. 2015;6:1137. doi: 10.3389/fpls.2015.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Goldman ER, Pazirandeh MP, Mauro JM, King KD, Frey JC, Anderson GP. J. Mol. Recognit. 2000;13:382–387. doi: 10.1002/1099-1352(200011/12)13:6<382::AID-JMR511>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 389.Sapsford KE, Soto CM, Blum AS, Chatterji A, Lin T, Johnson JE, Ligler FS, Ratna BR. Biosens. Bioelectron. 2006;21:1668–1673. doi: 10.1016/j.bios.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 390.Soto CM, Blaney KM, Dar M, Khan M, Lin B, Malanoski AP, Tidd C, Rios MV, Lopez DM, Ratna BR. Biosens. Bioelectron. 2009;25:48–54. doi: 10.1016/j.bios.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 391.Jin H, Won N, Ahn B, Kwag J, Heo K, Oh JW, Sun Y, Cho SG, Lee SW, Kim S. Chem. Commun. (Camb) 2013;49:6045–6047. doi: 10.1039/c3cc42032a. [DOI] [PubMed] [Google Scholar]
- 392.Liu A, Abbineni G, Mao C. Adv. Mater. 2009;21:1001–1005. [Google Scholar]
- 393.Nanduri V, Bhunia AK, Tu SI, Paoli GC, Brewster JD. Biosens. Bioelectron. 2007;23:248–252. doi: 10.1016/j.bios.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 394.Lee JH, Xu PF, Domaille DW, Choi C, Jin S, Cha JN. Adv. Funct. Mater. 2014;24:2079–2084. [Google Scholar]
- 395.Oh JW, Chung WJ, Heo K, Jin HE, Lee BY, Wang E, Zueger C, Wong W, Meyer J, Kim C, Lee SY, Kim WG, Zemla M, Auer M, Hexemer A, Lee SW. Nat. Commun. 2014;5:3043. doi: 10.1038/ncomms4043. [DOI] [PubMed] [Google Scholar]
- 396.Yang LM, Tam PY, Murray BJ, McIntire TM, Overstreet CM, Weiss GA, Penner RM. Anal. Chem. 2006;78:3265–3270. doi: 10.1021/ac052287u. [DOI] [PubMed] [Google Scholar]
- 397.Bruckman MA, Liu J, Koley G, Li Y, Benicewicz B, Niu Z, Wang Q. J. Mater. Chem. 2010;20:5715–5719. [Google Scholar]
- 398.Zang F, Gerasopoulos K, Fan XZ, Brown AD, Culver JN, Ghodssi R. Chemical Communications. 2014;50:12977–12980. doi: 10.1039/c4cc06735e. [DOI] [PubMed] [Google Scholar]
- 399.Kaushik A, Tiwari S, Dev Jayant R, Marty A, Nair M. Biosens. Bioelectron. 2016;75:254–272. doi: 10.1016/j.bios.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Mahony JB. Expert Rev. Anti Infect. Ther. 2010;8:1273–1292. doi: 10.1586/eri.10.121. [DOI] [PubMed] [Google Scholar]
- 401.Maurin M. Expert Rev. Mol. Diagn. 2012;12:731–754. doi: 10.1586/erm.12.53. [DOI] [PubMed] [Google Scholar]
- 402.Schrader C, Schielke A, Ellerbroek L, Johne R. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 403.Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, DuBois DB. J. Clin. Microbiol. 1998;36:3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.WalkerPeach CR, Winkler M, DuBois DB, Pasloske BL. Clin. Chem. 1999;45:2079–2085. [PubMed] [Google Scholar]
- 405.Meng S, Li J. Virol. J. 2010;7:117. doi: 10.1186/1743-422X-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Dreier J, Stormer M, Kleesiek K. J. Clin. Microbiol. 2005;43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Yu XF, Pan JC, Ye R, Xiang HQ, Kou Y, Huang ZC. J. Clin. Microbiol. 2008;46:837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Bressler AM, Nolte FS. J. Clin. Microbiol. 2004;42:987–991. doi: 10.1128/JCM.42.3.987-991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Stevenson J, Hymas W, Hillyard D. J. Virol. Methods. 2008;150:73–76. doi: 10.1016/j.jviromet.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Song L, Sun S, Li B, Pan Y, Li W, Zhang K, Li J. J. Clin. Microbiol. 2011;49:3591–3595. doi: 10.1128/JCM.00686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Beld M, Minnaar R, Weel J, Sol C, Damen M, van der Avoort H, Wertheim-van Dillen P, van Breda A, Boom R. J. Clin. Microbiol. 2004;42:3059–3064. doi: 10.1128/JCM.42.7.3059-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Eisler DL, McNabb A, Jorgensen DR, Isaac-Renton JL. J. Clin. Microbiol. 2004;42:841–843. doi: 10.1128/JCM.42.2.841-843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Hietala SK, Crossley BM. J. Clin. Microbiol. 2006;44:67–70. doi: 10.1128/JCM.44.1.67-70.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Zhan S, Li J, Xu R, Wang L, Zhang K, Zhang R. J. Clin. Microbiol. 2009;47:2571–2576. doi: 10.1128/JCM.00232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Wei B, Wei Y, Zhang K, Yang C, Wang J, Xu R, Zhan S, Lin G, Wang W, Liu M, Wang L, Zhang R, Li J. Intervirology. 2008;51:144–150. doi: 10.1159/000141707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Huang J, Yang CM, Wang LN, Meng S, Deng W, Li JM. Intervirology. 2008;51:42–49. doi: 10.1159/000119119. [DOI] [PubMed] [Google Scholar]
- 417.Huang Q, Cheng Y, Guo Q, Li Q. Clin. Chem. 2006;52:1446–1448. doi: 10.1373/clinchem.2006.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Stocher M, Berg J. Clin. Chem. 2004;50:2163–2166. doi: 10.1373/clinchem.2004.035519. [DOI] [PubMed] [Google Scholar]
- 419.Meng S, Zhan S, Li J. Virol. J. 2009;6:226. doi: 10.1186/1743-422X-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Gotsch A, Schubert A, Bombis A, Wiedmann M, Zauke M, Schorling S. J. Clin. Microbiol. 2007;45:2570–2574. doi: 10.1128/JCM.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Zhang L, Sun Y, Chang L, Jia T, Wang G, Zhang R, Zhang K, Li J. Appl. Microbiol. Biotechnol. 2015;99:7047–7057. doi: 10.1007/s00253-015-6664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.King DP, Montague N, Ebert K, Reid SM, Dukes JP, Schadlich L, Belsham GJ, Lomonossoff GP. J. Virol. Methods. 2007;146:218–225. doi: 10.1016/j.jviromet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 423.Lam P, Gulati NM, Stewart PL, Keri RA, Steinmetz NF. Sci. Rep. 2016 doi: 10.1038/srep23803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Carette N, Engelkamp H, Akpa E, Pierre SJ, Cameron NR, Christianen PC, Maan JC, Thies JC, Weberskirch R, Rowan AE, Nolte RJ, Michon T, Van Hest JC. Nat. Nanotechnol. 2007;2:226–229. doi: 10.1038/nnano.2007.76. [DOI] [PubMed] [Google Scholar]
- 425.Comellas-Aragones M, Engelkamp H, Claessen VI, Sommerdijk NA, Rowan AE, Christianen PC, Maan JC, Verduin BJ, Cornelissen JJ, Nolte RJ. Nat. Nanotechnol. 2007;2:635–639. doi: 10.1038/nnano.2007.299. [DOI] [PubMed] [Google Scholar]
- 426.Minten IJ, Claessen VI, Blank K, Rowan AE, Nolte RJM, Cornelissen JJLM. Chem. Sci. 2011;2:358–362. [Google Scholar]
- 427.Fiedler JD, Brown SD, Lau JL, Finn MG. Angew. Chem. Int. Ed. Engl. 2010;49:9648–9651. doi: 10.1002/anie.201005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Patterson DP, Prevelige PE, Douglas T. ACS Nano. 2012;6:5000–5009. doi: 10.1021/nn300545z. [DOI] [PubMed] [Google Scholar]
- 429.Patterson DP, Schwarz B, El-Boubbou K, van der Oost J, Prevelige PE, Douglas T. Soft Matter. 2012;8:10158–10166. [Google Scholar]
- 430.Glasgow JE, Asensio MA, Jakobson CM, Francis MB, Tullman-Ercek D. ACS Synth. Biol. 2015;4:1011–1019. doi: 10.1021/acssynbio.5b00037. [DOI] [PubMed] [Google Scholar]
- 431.O'Neil A, Prevelige PE, Douglas T. Biomater. Sci. 2013;1:881–886. doi: 10.1039/c3bm60063g. [DOI] [PubMed] [Google Scholar]
- 432.Patterson DP, LaFrance B, Douglas T. Chem. Commun. (Camb) 2013;49:10412–10414. doi: 10.1039/c3cc46517a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Patterson DP, McCoy K, Fijen C, Douglas T. J. Mater. Chem. B. 2014;2:5948–5951. doi: 10.1039/c4tb00983e. [DOI] [PubMed] [Google Scholar]
- 434.Jordan PC, Patterson DP, Saboda KN, Edwards EJ, Miettinen HM, Basu G, Thielges MC, Douglas T. Nat. Chem. 2016;8:179–185. doi: 10.1038/nchem.2416. [DOI] [PubMed] [Google Scholar]
- 435.Atkinson HJ, Lilley CJ, Urwin PE. Curr. Opin. Biotechnol. 2012;23:251–256. doi: 10.1016/j.copbio.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 436.Gleba YY, Tuse D, Giritch A. Curr. Top. Microbiol. Immunol. 2014;375:155–192. doi: 10.1007/82_2013_352. [DOI] [PubMed] [Google Scholar]
- 437.Lico C, Chen Q, Santi L. J. Cell. Physiol. 2008;216:366–377. doi: 10.1002/jcp.21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Kumagai MH, Turpen TH, Weinzettl N, della-Cioppa G, Turpen AM, Donson J, Hilf ME, Grantham GL, Dawson WO, Chow TP, et al. Proc. Natl. Acad. Sci. USA. 1993;90:427–430. doi: 10.1073/pnas.90.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Lindbo JA. BMC Biotechnol. 2007;7:52. doi: 10.1186/1472-6750-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y. Proc. Natl. Acad. Sci. USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Klimyuk V, Pogue G, Herz S, Butler J, Haydon H. Curr. Top. Microbiol. Immunol. 2014;375:127–154. doi: 10.1007/82_2012_212. [DOI] [PubMed] [Google Scholar]
- 442.Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y. Nat. Biotechnol. 2005;23:718–723. doi: 10.1038/nbt1094. [DOI] [PubMed] [Google Scholar]
- 443.Santi L, Batchelor L, Huang Z, Hjelm B, Kilbourne J, Arntzen CJ, Chen Q, Mason HS. Vaccine. 2008;26:1846–1854. doi: 10.1016/j.vaccine.2008.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Huang Z, Santi L, LePore K, Kilbourne J, Arntzen CJ, Mason HS. Vaccine. 2006;24:2506–2513. doi: 10.1016/j.vaccine.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 445.Bendandi M, Marillonnet S, Kandzia R, Thieme F, Nickstadt A, Herz S, Frode R, Inoges S, Lopez-Diaz de Cerio A, Soria E, Villanueva H, Vancanneyt G, McCormick A, Tuse D, Lenz J, Butler-Ransohoff JE, Klimyuk V, Gleba Y. Ann. Oncol. 2010;21:2420–2427. doi: 10.1093/annonc/mdq256. [DOI] [PubMed] [Google Scholar]
- 446.Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y. Proc. Natl. Acad. Sci. USA. 2006;103:14701–14706. doi: 10.1073/pnas.0606631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Santi L, Giritch A, Roy CJ, Marillonnet S, Klimyuk V, Gleba Y, Webb R, Arntzen CJ, Mason HS. Proc. Natl. Acad. Sci. USA. 2006;103:861–866. doi: 10.1073/pnas.0510014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Nausch H, Mikschofsky H, Koslowski R, Meyer U, Broer I, Huckauf J. PLoS One. 2012;7:e48938. doi: 10.1371/journal.pone.0048938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Gils M, Kandzia R, Marillonnet S, Klimyuk V, Gleba Y. Plant Biotechnol. J. 2005;3:613–620. doi: 10.1111/j.1467-7652.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- 450.Lindbo JA. Plant Physiol. 2007;145:1232–1240. doi: 10.1104/pp.107.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Li C, Jiang Y, Guo W, Liu Z. Hum. Immunol. 2013;74:531–537. doi: 10.1016/j.humimm.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 452.Toth RL, Chapman S, Carr F, Santa Cruz S. FEBS Lett. 2001;489:215–219. doi: 10.1016/s0014-5793(01)02091-9. [DOI] [PubMed] [Google Scholar]
- 453.Uhde-Holzem K, Schlosser V, Viazov S, Fischer R, Commandeur U. J. Virol. Methods. 2010;166:12–20. doi: 10.1016/j.jviromet.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 454.Avesani L, Marconi G, Morandini F, Albertini E, Bruschetta M, Bortesi L, Pezzotti M, Porceddu A. Transgenic Res. 2007;16:587–597. doi: 10.1007/s11248-006-9051-1. [DOI] [PubMed] [Google Scholar]
- 455.Dickmeis C, Fischer R, Commandeur U. Biotechnol. J. 2014;9:1369–1379. doi: 10.1002/biot.201400347. [DOI] [PubMed] [Google Scholar]
- 456.Komarova TV, Skulachev MV, Zvereva AS, Schwartz AM, Dorokhov YL, Atabekov JG. Biochemistry (Mosc) 2006;71:846–850. doi: 10.1134/s0006297906080049. [DOI] [PubMed] [Google Scholar]
- 457.Larsen JS, Curtis WR. BMC Biotechnol. 2012;12:21. doi: 10.1186/1472-6750-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Azhakanandam K, Weissinger SM, Nicholson JS, Qu R, Weissinger AK. Plant Mol. Biol. 2007;63:393–404. doi: 10.1007/s11103-006-9096-9. [DOI] [PubMed] [Google Scholar]
- 459.Minato N, Komatsu K, Okano Y, Maejima K, Ozeki J, Senshu H, Takahashi S, Yamaji Y, Namba S. Arch. Virol. 2014;159:885–896. doi: 10.1007/s00705-013-1860-y. [DOI] [PubMed] [Google Scholar]
- 460.Liu Z, Kearney CM. BMC Biotechnol. 2010;10:88. doi: 10.1186/1472-6750-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Sainsbury F, Sack M, Stadlmann J, Quendler H, Fischer R, Lomonossoff GP. PLoS One. 2010;5:e13976. doi: 10.1371/journal.pone.0013976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Vardakou M, Sainsbury F, Rigby N, Mulholland F, Lomonossoff GP. Protein Expr. Purif. 2012;81:69–74. doi: 10.1016/j.pep.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 463.Canizares MC, Liu L, Perrin Y, Tsakiris E, Lomonossoff GP. Plant Biotechnol. J. 2006;4:183–193. doi: 10.1111/j.1467-7652.2005.00170.x. [DOI] [PubMed] [Google Scholar]
- 464.Sainsbury F, Lavoie PO, D'Aoust MA, Vezina LP, Lomonossoff GP. Plant Biotechnol. J. 2008;6:82–92. doi: 10.1111/j.1467-7652.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 465.Sainsbury F, Lomonossoff GP. Plant Physiol. 2008;148:1212–1218. doi: 10.1104/pp.108.126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Sainsbury F, Thuenemann EC, Lomonossoff GP. Plant Biotechnol. J. 2009;7:682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- 467.D'Aoust MA, Couture MM, Charland N, Trepanier S, Landry N, Ors F, Vezina LP. Plant Biotechnol. J. 2010;8:607–619. doi: 10.1111/j.1467-7652.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 468.Sainsbury F, Saxena P, Aljabali AA, Saunders K, Evans DJ, Lomonossoff GP. Methods Mol. Biol. 2014;1108:139–153. doi: 10.1007/978-1-62703-751-8_11. [DOI] [PubMed] [Google Scholar]
- 469.Dugdale B, Mortimer CL, Kato M, James TA, Harding RM, Dale JL. Nat. Protoc. 2014;9:1010–1027. doi: 10.1038/nprot.2014.068. [DOI] [PubMed] [Google Scholar]
- 470.Dugdale B, Mortimer CL, Kato M, James TA, Harding RM, Dale JL. Plant Cell. 2013;25:2429–2443. doi: 10.1105/tpc.113.113944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Huang Z, Phoolcharoen W, Lai H, Piensook K, Cardineau G, Zeitlin L, Whaley KJ, Arntzen CJ, Mason HS, Chen Q. Biotechnol. Bioeng. 2010;106:9–17. doi: 10.1002/bit.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Regnard GL, Halley-Stott RP, Tanzer FL, Hitzeroth II, Rybicki EP. Plant Biotechnol. J. 2010;8:38–46. doi: 10.1111/j.1467-7652.2009.00462.x. [DOI] [PubMed] [Google Scholar]
- 473.Maclean J, Koekemoer M, Olivier AJ, Stewart D, Hitzeroth II, Rademacher T, Fischer R, Williamson AL, Rybicki EP. J. Gen. Virol. 2007;88:1460–1469. doi: 10.1099/vir.0.82718-0. [DOI] [PubMed] [Google Scholar]
- 474.Fang J, Soto CM, Lin T, Johnson JE, Ratna BR. Langmuir. 2002;18:308–310. [Google Scholar]
- 475.He J, Niu Z, Tangirala R, Wang JY, Wei X, Kaur G, Wang Q, Jutz G, Boker A, Lee B, Pingali SV, Thiyagarajan P, Emrick T, Russell TP. Langmuir. 2009;25:4979–4987. doi: 10.1021/la803533n. [DOI] [PubMed] [Google Scholar]
- 476.Kuncicky DM, Naik RR, Velev OD. Small. 2006;2:1462–1466. doi: 10.1002/smll.200600399. [DOI] [PubMed] [Google Scholar]
- 477.Lee SW, Woods BW, Belcher AM. Langmuir. 2003;19:1592–1598. [Google Scholar]
- 478.Niu Z, Bruckman MA, Li S, Lee LA, Lee B, Pingali SV, Thiyagarajan P, Wang Q. Langmuir. 2007;23:6719–6724. doi: 10.1021/la070096b. [DOI] [PubMed] [Google Scholar]
- 479.Steinmetz NF, Findlay KC, Noel TR, Parker R, Lomonossoff GP, Evans DJ. Chembiochem. 2008;9:1662–1670. doi: 10.1002/cbic.200800070. [DOI] [PubMed] [Google Scholar]
- 480.BESAC 2012 http://science.energy.gov/bes/news-and-resources/reports/basic-research-needs/
- 481.Pimpin A, Srituravanich W. Eng. J. 2012;16 10.4186/ej.2012.4116.4181.4137. [Google Scholar]
- 482.Qin D, Xia Y, Whitesides GM. Nature protocols. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 483.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 484.Lohmuller T, Aydin D, Schwieder M, Morhard C, Louban I, Pacholski C, Spatz JP. Biointerphases. 2011;6:MR1–12. doi: 10.1116/1.3536839. [DOI] [PubMed] [Google Scholar]
- 485.Bang J, Jeong U, Ryu du Y, Russell TP, Hawker CJ. Adv. Mater. 2009;21:4769–4792. doi: 10.1002/adma.200803302. [DOI] [PubMed] [Google Scholar]
- 486.Malloy M, Litt LC. J. Micro/Nanolith. 2011;10:032001. [Google Scholar]
- 487.Salaita K, Wang Y, Mirkin CA. Nature nanotechnology. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 488.Ginger DS, Zhang H, Mirkin CA. Angewandte Chemie. 2004;43:30–45. doi: 10.1002/anie.200300608. [DOI] [PubMed] [Google Scholar]
- 489.Yun JM, Kim KN, Kim JY, Shin DO, Lee WJ, Lee SH, Lieberman M, Kim SO. Angewandte Chemie. 2012;51:912–915. doi: 10.1002/anie.201106198. [DOI] [PubMed] [Google Scholar]
- 490.Liu W, Zhong H, Wang R, Seeman NC. Angewandte Chemie. 2011;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Sacca B, Niemeyer CM. Angewandte Chemie. 2012;51:58–66. doi: 10.1002/anie.201105846. [DOI] [PubMed] [Google Scholar]
- 492.Rajendran A, Endo M, Sugiyama H. Curr. Protoc. Nucleic Acid Chem. 2012 doi: 10.1002/0471142700.nc1209s48. Chapter 12, Unit 12 19 11-18. [DOI] [PubMed] [Google Scholar]
- 493.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 494.Endo M, Sugiyama H. Curr. Protoc. Nucleic Acid Chem. 2011 doi: 10.1002/0471142700.nc1208s45. Chapter 12, Unit12 18. [DOI] [PubMed] [Google Scholar]
- 495.Doerr A. Nat. Methods. 2011;8:453. doi: 10.1038/nmeth0611-454. [DOI] [PubMed] [Google Scholar]
- 496.Shih WM, Lin C. Current opinion in structural biology. 2010;20:276–282. doi: 10.1016/j.sbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Majumder U, Rangnekar A, Gothelf KV, Reif JH, LaBean TH. Journal of the American Chemical Society. 2011;133:3843–3845. doi: 10.1021/ja1108886. [DOI] [PubMed] [Google Scholar]
- 498.Lin C, Liu Y, Rinker S, Yan H. Chemphyschem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 499.Park SH, Pistol C, Ahn SJ, Reif JH, Lebeck AR, Dwyer C, LaBean TH. Angewandte Chemie. 2006;45:735–739. doi: 10.1002/anie.200503797. [DOI] [PubMed] [Google Scholar]
- 500.Serin JM, Brousmiche DW, Frechet JM. Chemical communications. 2002:2605–2607. doi: 10.1039/b207905d. [DOI] [PubMed] [Google Scholar]
- 501.Furuta P, Brooks J, Thompson ME, Frechet JM. Journal of the American Chemical Society. 2003;125:13165–13172. doi: 10.1021/ja0371348. [DOI] [PubMed] [Google Scholar]
- 502.Mynar JL, Lowery TJ, Wemmer DE, Pines A, Frechet JM. Journal of the American Chemical Society. 2006;128:6334–6335. doi: 10.1021/ja061735s. [DOI] [PubMed] [Google Scholar]
- 503.Nantalaksakul A, Dasari RR, Ahn TS, Al-Kaysi R, Bardeen CJ, Thayumanavan S. Org. Lett. 2006;8:2981–2984. doi: 10.1021/ol0608956. [DOI] [PubMed] [Google Scholar]
- 504.Bergamini G, Ceroni P, Fabbrizi P, Cicchi S. Chemical communications. 2011;47:12780–12782. doi: 10.1039/c1cc16000a. [DOI] [PubMed] [Google Scholar]
- 505.Miura Y, Momotake A, Takeuchi K, Arai T. Photochem. Photobiol. Sci. 2011;10:116–122. doi: 10.1039/c0pp00272k. [DOI] [PubMed] [Google Scholar]
- 506.Zhao Y, Thorkelsson K, Mastroianni AJ, Schilling T, Luther JM, Rancatore BJ, Matsunaga K, Jinnai H, Wu Y, Poulsen D, Frechet JM, Alivisatos AP, Xu T. Nature materials. 2009;8:979–985. doi: 10.1038/nmat2565. [DOI] [PubMed] [Google Scholar]
- 507.Stevens MM, Flynn NT, Wang C, Tirrel DA, Langer R. Adv. Mater. 2004;16:915–918. [Google Scholar]
- 508.Wagner J, Stephan T, Kalla R, Bruckmann H, Strupp M, Brandt T, Jahn K. Exp. Brain Res. 2008;191:247–255. doi: 10.1007/s00221-008-1520-8. [DOI] [PubMed] [Google Scholar]
- 509.Chen P-Y, Dang X, Klug MT, Courchesne N-MD, Qi J, Hyder MN, Belcher AM, Hammond PT. Chem. Mater. 2015;27:1531–1540. [Google Scholar]
- 510.Nam KT, Kim DW, Yoo PJ, Chiang CY, Meethong N, Hammond PT, Chiang YM, Belcher AM. Science. 2006;312:885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- 511.Nam YS, Park H, Magyar AP, Yun DS, Pollom TS, Belcher AM. Nanoscale. 2012;4:3405–3409. doi: 10.1039/c2nr30115f. [DOI] [PubMed] [Google Scholar]
- 512.French RH, Parsegian VA, Podgornik R, Rajter RF, Jagota A, Luo J, Asthagiri D, Chaudhury MK, Chiang Y.-m., Granick S, Kalinin S, Kardar M, Kjellander R, Langreth DC, Lewis J, Lustig S, Wesolowski D, Wettlaufer JS, Ching W-Y, Finnis M, Houlihan F, von Lilienfeld OA, van Oss CJ, Zemb T. Rev. Mod. Phys. 2010;82:1887–1944. [Google Scholar]
- 513.Hamley IW. Introduction to soft matter: synthetic and biological self-assembling materials. revised edition Wiley; Hoboken, N.J.: 2007. [Google Scholar]
- 514.Shenton W, Douglas T, Young M, Stubbs G, Mann S. Adv. Mater. 1999;11:253–256. [Google Scholar]
- 515.Hess GT, Guimaraes CP, Spooner E, Ploegh HL, Belcher AM. ACS Synth. Biol. 2013;2:490–496. doi: 10.1021/sb400019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Dogic Z, Fraden S. Curr. Opin. Colloid Interface Sci. 2006;11:47–55. [Google Scholar]
- 517.Onsager L. Science. 1969;166:1359–1364. doi: 10.1126/science.166.3911.1359. [DOI] [PubMed] [Google Scholar]
- 518.Onsager C. Ann. N.Y. Acad. Sci. 1994;51:627. [Google Scholar]
- 519.Lapointe J, Marvin DA. Mol. Cryst. Liq. Cryst. 1973;19:269–278. [Google Scholar]
- 520.Ermolina I, Milner J, Morgan H. Electrophoresis. 2006;27:3939–3948. doi: 10.1002/elps.200500928. [DOI] [PubMed] [Google Scholar]
- 521.Hirai M, Koizumi M, Han R, Hayakawa T, Sano Y. J. Appl. Crystallogr. 2003;36:520–524. [Google Scholar]
- 522.Yang L, Wang S, Fukuto M, Checco A, Niu Z, Wang Q. Soft Matter. 2009;5:4951–4961. [Google Scholar]
- 523.Wang S, Fukuto M, Checco A, Niu Z, Wang Q, Yang L. J. Colloid Interface Sci. 2011;358:497–505. doi: 10.1016/j.jcis.2011.03.048. [DOI] [PubMed] [Google Scholar]
- 524.Adams M, Fraden S. Biophys. J. 1998;74:669–677. doi: 10.1016/S0006-3495(98)77826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Dogic Z, Frenkel D, Fraden S. Phys. Rev. E. 2000;62:3925–3933. doi: 10.1103/physreve.62.3925. [DOI] [PubMed] [Google Scholar]
- 526.Nedoluzhko A, Douglas T. J. Inorg. Biochem. 2001;84:233–240. doi: 10.1016/s0162-0134(01)00174-x. [DOI] [PubMed] [Google Scholar]
- 527.Lee SY, Culver JN, Harris MT. J. Colloid Interface Sci. 2006;297:554–560. doi: 10.1016/j.jcis.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 528.Li T, Winans RE, Lee B. Langmuir. 2011;27:10929–10937. doi: 10.1021/la202121s. [DOI] [PubMed] [Google Scholar]
- 529.Lee SW, Belcher AM. Nano Lett. 2004;4:387–390. [Google Scholar]
- 530.Cheung CL, Chung SW, Chatterji A, Lin T, Johnson JE, Hok S, Perkins J, De Yoreo JJ. J. Am. Chem. Soc. 2006;128:10801–10807. doi: 10.1021/ja0616884. [DOI] [PubMed] [Google Scholar]
- 531.Cheung CL, Camarero JA, Woods BW, Lin T, Johnson JE, De Yoreo JJ. J. Am. Chem. Soc. 2003;125:6848–6849. doi: 10.1021/ja034479h. [DOI] [PubMed] [Google Scholar]
- 532.Suci PA, Klem MT, Arce FT, Douglas T, Young M. Langmuir. 2006;22:8891–8896. doi: 10.1021/la0612062. [DOI] [PubMed] [Google Scholar]
- 533.Steinmetz NF, Calder G, Lomonossoff GP, Evans DJ. Langmuir. 2006;22:10032–10037. doi: 10.1021/la0621362. [DOI] [PubMed] [Google Scholar]
- 534.Steinmetz NF, Bock E, Richter RP, Spatz JP, Lomonossoff GP, Evans DJ. Biomacromolecules. 2008 doi: 10.1021/bm700797b. DOI: 10.1021/bm700797b. [DOI] [PubMed] [Google Scholar]
- 535.Suci PA, Klem MT, Douglas T, Young M. Langmuir. 2005;21:8686–8693. doi: 10.1021/la050217c. [DOI] [PubMed] [Google Scholar]
- 536.Lvov Y, Haas H, Decher G, Moehwald H, Mikhailov A, Mtchedlishvily B, Morgunova E, Vainshtein B. Langmuir. 1994;10:4232–4236. [Google Scholar]
- 537.Gerasopoulos K, McCarthy M, Banerjee P, Fan X, Culver JN, Ghodssi R. Nanotechnology. 2010;21:055304. doi: 10.1088/0957-4484/21/5/055304. [DOI] [PubMed] [Google Scholar]
- 538.Tiu BDB, Tiu SB, Wen AM, Steinmetz NF, Advincula RC. in review.
- 539.Yi H, Nisar S, Lee SY, Powers MA, Bentley WE, Payne GF, Ghodssi R, Rubloff GW, Harris MT, Culver JN. Nano Lett. 2005;5:1931–1936. doi: 10.1021/nl051254r. [DOI] [PubMed] [Google Scholar]
- 540.Yi H, Rubloff GW, Culver JN. Langmuir. 2007;23:2663–2667. doi: 10.1021/la062493c. [DOI] [PubMed] [Google Scholar]
- 541.Eber FJ, Eiben S, Jeske H, Wege C. Angew. Chem. Int. Ed. Engl. 2013;52:7203–7207. doi: 10.1002/anie.201300834. [DOI] [PubMed] [Google Scholar]
- 542.Bittner AM, Alonso JM, Gorzny ML, Wege C. Subcell. Biochem. 2013;68:667–702. doi: 10.1007/978-94-007-6552-8_22. [DOI] [PubMed] [Google Scholar]
- 543.Cheung CL, Rubinstein AI, Peterson EJ, Chatterji A, Sabirianov RF, Mei WN, Lin T, Johnson JE, DeYoreo JJ. Langmuir. 2010;26:3498–3505. doi: 10.1021/la903114s. [DOI] [PubMed] [Google Scholar]
- 544.Kewalramani S, Wang S, Lin Y, Nguyen HG, Wang Q, Fukuto M, Yang L. Soft Matter. 2011;7:939–945. [Google Scholar]
- 545.Fukuto M, Nguyen QL, Vasilyev O, Mank N, Washington-Hughes CL, Kuzmenko I, Checco A, Mao Y, Wang Q, Yang L. Soft Matter. 2013;9:9633–9642. doi: 10.1039/c3sm51853a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Casselyn M, J., Perez A, Tardieu P, Vachette J, Witz, Delacroix H. Acta Crystallogr. 2001;D57:799–1812. doi: 10.1107/s0907444901014949. [DOI] [PubMed] [Google Scholar]
- 547.Cigler P, Lytton-Jean AK, Anderson DG, Finn MG, Park SY. Nat. Mater. 2010;9:918–922. doi: 10.1038/nmat2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Rosilo H, McKee JR, Kontturi E, Koho T, Hytonen VP, Ikkala O, Kostiainen MA. Nanoscale. 2014;6:11871–11881. doi: 10.1039/c4nr03584d. [DOI] [PubMed] [Google Scholar]
- 549.Doni G, Kostiainen MA, Danani A, Pavan GM. Nano Lett. 2011;11:723–728. doi: 10.1021/nl103857e. [DOI] [PubMed] [Google Scholar]
- 550.Kostiainen MA, Kasyutich O, Cornelissen JJ, Nolte RJ. Nat. Chem. 2010;2:394–399. doi: 10.1038/nchem.592. [DOI] [PubMed] [Google Scholar]
- 551.Mikkilä J, Rosilo H, Nummelin S, Seitsonen J, Ruokolainen J, Kostiainen MA. ACS Macro Lett. 2013;2:720–724. doi: 10.1021/mz400307h. [DOI] [PubMed] [Google Scholar]
- 552.Kostiainen MA, Hiekkataipale P, Laiho A, Lemieux V, Seitsonen J, Ruokolainen J, Ceci P. Nat. Nanotechnol. 2013;8:52–56. doi: 10.1038/nnano.2012.220. [DOI] [PubMed] [Google Scholar]
- 553.Liljeström V, Mikkilä J, Kostiainen MA. Nat. Commun. 2014:5. doi: 10.1038/ncomms5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Mikkila J, Anaya-Plaza E, Liljestrom V, Caston JR, Torres T, Escosura Ade L, Kostiainen MA. ACS Nano. 2016;10:1565–1571. doi: 10.1021/acsnano.5b07167. [DOI] [PubMed] [Google Scholar]
- 555.Chiang C-Y, Epstein J, Brown A, Munday JN, Culver JN, Ehrman S. Nano Letters. 2012;12:6005–6011. doi: 10.1021/nl303579z. [DOI] [PubMed] [Google Scholar]
- 556.Liu Y, Xu Y, Zhu Y, Culver JN, Lundgren CA, Xu K, Wang C. ACS Nano. 2013;7:3627–3634. doi: 10.1021/nn400601y. [DOI] [PubMed] [Google Scholar]
- 557.Liu Y, Zhang W, Zhu Y, Luo Y, Xu Y, Brown A, Culver JN, Lundgren CA, Xu K, Wang Y, Wang C. Nano Letters. 2012;13:293–300. doi: 10.1021/nl304104q. [DOI] [PubMed] [Google Scholar]
- 558.Nuraje N, Dang X, Qi J, Allen MA, Lei Y, Belcher AM. Adv. Mater. 2012;24:2885–2889. doi: 10.1002/adma.201200114. [DOI] [PubMed] [Google Scholar]
- 559.Oh D, Dang X, Yi H, Allen MA, Xu K, Lee YJ, Belcher AM. Small. 2012;8:1006–1011. doi: 10.1002/smll.201102036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Dang X, Yi H, Ham M-H, Qi J, Yun DS, Ladewski R, Strano MS, Hammond PT, Belcher AM. Nat. Nanotechnol. 2011;6:377–384. doi: 10.1038/nnano.2011.50. [DOI] [PubMed] [Google Scholar]
- 561.Tseng RJ, Tsai C, Ma L, Ouyang J, Ozkan CS, Yang Y. Nature nanotechnology. 2006;1:72–77. doi: 10.1038/nnano.2006.55. [DOI] [PubMed] [Google Scholar]
- 562.Portney NG, Martinez-Morales AA, Ozkan M. ACS Nano. 2008;2:191–196. doi: 10.1021/nn700240z. [DOI] [PubMed] [Google Scholar]
- 563.Steinmetz NF, Lomonossoff GP, Evans DJ. Small. 2006;2:530–533. doi: 10.1002/smll.200500453. [DOI] [PubMed] [Google Scholar]
- 564.Steinmetz NF, Lomonossoff GP, Evans DJ. Langmuir. 2006;22:3488–3490. doi: 10.1021/la060078e. [DOI] [PubMed] [Google Scholar]
- 565.Nam KT, Wartena R, Yoo PJ, Liau FW, Lee YJ, Chiang Y-M, Hammond PT, Belcher AM. Proc. Natl. Acad. Sci. USA. 2008 doi: 10.1073/pnas.0711620105. DOI: 10.1073/pnas.0711620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Oh D, Qi J, Lu YC, Zhang Y, Shao-Horn Y, Belcher AM. Nat. Commun. 2013;4:2756. doi: 10.1038/ncomms3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Lee YJ, Lee Y, Oh D, Chen T, Ceder G, Belcher AM. Nano letters. 2010;10:2433–2440. doi: 10.1021/nl1005993. [DOI] [PubMed] [Google Scholar]
- 568.Oh D, Qi J, Han B, Zhang G, Carney TJ, Ohmura J, Zhang Y, Shao-Horn Y, Belcher AM. Nano Letters. 2014;14:4837–4845. doi: 10.1021/nl502078m. [DOI] [PubMed] [Google Scholar]
- 569.Lee Y, Kim J, Yun DS, Nam YS, Shao-Horn Y, Belcher AM. Energy Environ. Sci. 2012;5:8328–8334. doi: 10.1039/C2EE21156D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Lee BY, Zhang J, Zueger C, Chung W-J, Yoo SY, Wang E, Meyer J, Ramesh R, Lee S-W. Nat. Nanotechnol. 2012;7:351–356. doi: 10.1038/nnano.2012.69. [DOI] [PubMed] [Google Scholar]
- 571.Nelson N, Ben-Shem A. Nat. Rev. Mol. Cell. Biol. 2004;5:971–982. doi: 10.1038/nrm1525. [DOI] [PubMed] [Google Scholar]
- 572.Endo M, Fujitsuka M, Majima T. Chemistry. 2007;13:8660–8666. doi: 10.1002/chem.200700895. [DOI] [PubMed] [Google Scholar]
- 573.Ma YZ, Miller RA, Fleming GR, Francis MB. J. Phys. Chem. B. 2008;112:6887–6892. doi: 10.1021/jp8006393. [DOI] [PubMed] [Google Scholar]
- 574.Nam YS, Shin T, Park H, Magyar AP, Choi K, Fantner G, Nelson KA, Belcher AM. Journal of the American Chemical Society. 2010;132:1462–1463. doi: 10.1021/ja908812b. [DOI] [PubMed] [Google Scholar]
- 575.Park H, Heldman N, Rebentrost P, Abbondanza L, Iagatti A, Alessi A, Patrizi B, Salvalaggio M, Bussotti L, Mohseni M, Caruso F, Johnsen HC, Fusco R, Foggi P, Scudo PF, Lloyd S, Belcher AM. Nat. Mater. 2016;15:211–216. doi: 10.1038/nmat4448. [DOI] [PubMed] [Google Scholar]
- 576.Patrick R, Masoud M, Ivan K, Seth L, Alán A-G. New J. Phys. 2009;11:033003. [Google Scholar]
- 577.Lee YM, Kim YH, Lee JH, Park JH, Park N-G, Choe W-S, Ko MJ, Yoo PJ. Adv. Funct. Mater. 2011;21:1160–1167. [Google Scholar]
- 578.Chen P-Y, Ladewski R, Miller R, Dang X, Qi J, Liau F, Belcher AM, Hammond PT. J. Mater. Chem. A. 2013;1:2217–2224. [Google Scholar]
- 579.Chen PY, Dang X, Klug MT, Qi J, Dorval Courchesne NM, Burpo FJ, Fang N, Hammond PT, Belcher AM. ACS Nano. 2013;7:6563–6574. doi: 10.1021/nn4014164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Nuraje N, Lei Y, Belcher A. Catal. Commun. 2014;44:68–72. [Google Scholar]
- 581.Alu A, Engheta N. Phys. Rev. Lett. 2009;103:043902. doi: 10.1103/PhysRevLett.103.043902. [DOI] [PubMed] [Google Scholar]
- 582.Cai W, Shalaev Springer WM. 2010 [Google Scholar]
- 583.Engheta N, Ziolkowski RW. Wiley-IEEE Press. 2006 [Google Scholar]
- 584.Xiao S, Drachev VP, Kildishev AV, Ni X, Chettiar UK, Yuan HK, Shalaev VM. Nature. 2010;466:735–738. doi: 10.1038/nature09278. [DOI] [PubMed] [Google Scholar]
- 585.Khlebtsov B, Khanadeev V, Khlebtsov N. Phys. Chem. Chem. Phys. 2010;12:3210–3218. doi: 10.1039/b925102b. [DOI] [PubMed] [Google Scholar]
- 586.Daniel MC, Astruc D. Chem. Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 587.Blaber MG, Henry A-I, Bingham JM, Schatz GC, Van Duyne RP. J. Phys. Chem. C. 2012;116:393–403. [Google Scholar]
- 588.Agrawal A, Huang S, Wei Haw Lin A, Lee MH, Barton JK, Drezek RA, Pfefer TJ. J. Biomed. Opt. 2006;11:041121. doi: 10.1117/1.2339071. [DOI] [PubMed] [Google Scholar]
- 589.Kah JC, Chow TH, Ng BK, Razul SG, Olivo M, Sheppard CJ. Appl. Opt. 2009;48:D96–D108. doi: 10.1364/ao.48.000d96. [DOI] [PubMed] [Google Scholar]
- 590.Chen YS, Frey W, Kim S, Kruizinga P, Homan K, Emelianov S. Nano Lett. 2011;11:348–354. doi: 10.1021/nl1042006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Lee HE, Lee HK, Chang H, Ahn HY, Erdene N, Lee HY, Lee YS, Jeong DH, Chung J, Nam KT. Small. 2014;10:3007–3011. doi: 10.1002/smll.201400527. [DOI] [PubMed] [Google Scholar]
- 592.Stern JM, Stanfield J, Kabbani W, Hsieh JT, Cadeddu JA. J. Urol. 2008;179:748–753. doi: 10.1016/j.juro.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 593.Zhang G, Kirk B, Jauregui LA, Yang H, Xu X, Chen YP, Wu Y. Nano Lett. 2012;12:56–60. doi: 10.1021/nl202935k. [DOI] [PubMed] [Google Scholar]
- 594.Bardhan R, Chen W, Bartels M, Perez-Torres C, Botero MF, McAninch RW, Contreras A, Schiff R, Pautler RG, Halas NJ, Joshi A. Nano Lett. 2010;10:4920–4928. doi: 10.1021/nl102889y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Fan JA, Wu C, Bao K, Bao J, Bardhan R, Halas NJ, Manoharan VN, Nordlander P, Shvets G, Capasso F. Science. 2010;328:1135–1138. doi: 10.1126/science.1187949. [DOI] [PubMed] [Google Scholar]
- 596.Bardhan R, Lal S, Joshi A, Halas NJ. Acc. Chem. Res. 2011;44:936–946. doi: 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Gobin AM, Lee MH, Halas NJ, James WD, Drezek RA, West JL. Nano Lett. 2007;7:1929–1934. doi: 10.1021/nl070610y. [DOI] [PubMed] [Google Scholar]
- 598.Stockman MI. Opt. Express. 2011;19:22029–22106. doi: 10.1364/OE.19.022029. [DOI] [PubMed] [Google Scholar]
- 599.Ray K, Badugu R, Lakowicz JR. Chem. Mater. 2007;19:5902–5909. doi: 10.1021/cm071510w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Sreekanth KV, Biaglow T, Strangi G. J. Appl. Phys. 2013;114:134306. [Google Scholar]
- 601.Sreekanth KV, De Luca A, Strangi G. Sci. Rep. 2013;3:3291. doi: 10.1038/srep03291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Wang D, Capehart SL, Pal S, Liu M, Zhang L, Schuck PJ, Liu Y, Yan H, Francis MB, De Yoreo JJ. ACS Nano. 2014;8:7896–7904. doi: 10.1021/nn5015819. [DOI] [PubMed] [Google Scholar]