ABSTRACT
There are an estimated 8 million users of smokeless tobacco products (STPs) in the United States, and yet limited data on microbial populations within these products exist. To better understand the potential microbiological risks associated with STP use, a study was conducted to provide a baseline microbiological profile of STPs. A total of 90 samples, representing 15 common STPs, were purchased in metropolitan areas in Little Rock, AR, and Washington, DC, in November 2012, March 2013, and July 2013. Bacterial populations were evaluated using culture, pyrosequencing, and denaturing gradient gel electrophoresis (DGGE). Moist-snuff products exhibited higher levels of bacteria (average of 1.05 × 106 CFU/g STP) and diversity of bacterial populations than snus (average of 8.33 × 101 CFU/g STP) and some chewing tobacco products (average of 2.54 × 105 CFU/g STP). The most common species identified by culturing were Bacillus pumilus, B. licheniformis, B. safensis, and B. subtilis, followed by members of the genera Oceanobacillus, Staphylococcus, and Tetragenococcus. Pyrosequencing analyses of the 16S rRNA genes identified the genera Tetragenococcus, Carnobacterium, Lactobacillus, Geobacillus, Bacillus, and Staphylococcus as the predominant taxa. Several species identified are of possible concern due to their potential to cause opportunistic infections and reported abilities to reduce nitrates to nitrites, which may be an important step in the formation of carcinogenic tobacco-specific N′-nitrosamines. This report provides a microbiological baseline to help fill knowledge gaps associated with microbiological risks of STPs and to inform potential regulations regarding manufacture and testing of STPs.
IMPORTANCE It is estimated that there 8 million users of smokeless tobacco products (STPs) in the United States; however, there are limited data on microbial populations that exist within these products. The current study was undertaken to better understand the potential microbiological risks associated with STP use and provide a baseline microbiological profile of STPs. Several bacterial species were identified that are of possible concern due to their potential to cause opportunistic infections. In addition, some species have abilities to reduce nitrates to nitrites, which may be an important step in the formation of carcinogenic tobacco-specific N′-nitrosamines. Overall, this report provides a microbiological baseline to help fill knowledge gaps related to the microbiological risks of STPs and to inform potential regulations regarding the manufacture and testing of STPs.
INTRODUCTION
The use of tobacco products is a major health concern throughout the world. It is estimated that 3.5% of U.S. adults and 6.1% of high school students are users of smokeless tobacco products (STPs) (1, 2). As with many consumer products, there is the potential for the presence of microorganisms in tobacco products that may be of human health concern (3). Very limited data on the microbial populations present in STPs are available, and the data that are publicly available are mostly more than 20 years old (4). Tobacco processing and STP manufacturing methods can greatly affect the microbial content of STPs (5). Thus, there is a strong need to gain an understanding of the types and numbers of microorganisms that may be present in commercially available STPs for the protection of public health.
The use of smokeless tobacco products has been considered by some consumers to be a “less risky” alternative to smoking cigarettes (6), even though the use of STPs has been linked with an increased risk for the development of oral, pancreatic, and esophageal cancers. There are multiple types of STPs sold in the United States, including snuff, snus, chewing tobacco, and some forms of dissolvable tobacco (6, 7). Snuff is a finely ground tobacco product that is sold loose, moist, or dry or packaged in a pouch (Fig. 1). Moist snuff represents over 75% of the STP market share in the United States (8). Users of loose snuff typically place a pinch (dip) of the tobacco product between the gum and lip, where the tobacco compounds are absorbed through the oral mucosa. Loose leaf chewing tobacco, which has a much larger particle (cut) size, is also placed between the gum and lip, and the users of both types of products typically spit out saliva that is generated during tobacco usage. In contrast, snus, which is typically very fine, dry tobacco purchased in a pouch, does not induce the need to spit. Other STPs include newer dissolvable products derived from tobacco, such as orbs, strips, and sticks, which are ingested during use (6). As the name implies, STPs are consumed without smoking (heating), which could lead to greater potential for the consumer to be exposed to viable microorganisms or their toxic metabolites during use, if the product is contaminated or if the manufacturing process fails to reduce the levels of the microorganisms commonly found in tobacco.
FIG 1.
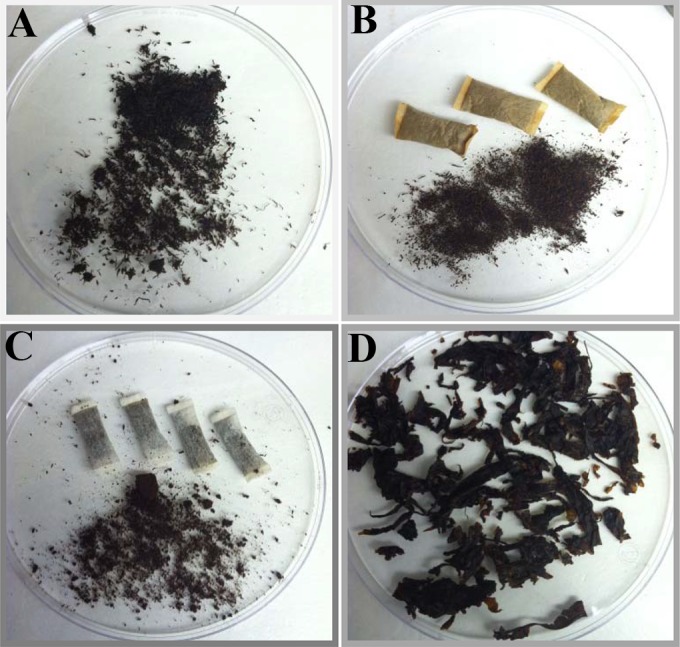
Types of smokeless tobacco products evaluated in the study. (A) Loose moist snuff. (B) Moist snuff in pouches. (C) Snus. (D) Chewing tobacco.
Few studies have been undertaken to address the types or quantities of microorganisms and their by-products (e.g., proteins, lipids, DNA, and cell walls) in tobacco products. Past studies were primarily focused on cigarette products and were mostly done before the 1990s (9). More-recent studies that have focused on cigarettes using modern molecular methods have reported the detection of a wide diversity of bacterial genera, including Gram-positive organisms (Bacillus, Clostridium, Enterococcus, and Staphylococcus) and Gram-negative organisms (Acinetobacter, Burkholderia, Campylobacter, Klebsiella, and Pseudomonas) (3). Many of these genera include species that have been associated with human diseases and that may be present in STPs. A case report and a follow-up study from the 1950s indicated that snuff was the likely source of Pseudomonas aeruginosa in a patient who developed chronic bronchitis (10). In the follow-up investigation into the source of the P. aeruginosa, the medical team sampled multiple previously unopened packages of snuff and found a wide range of potential bacterial pathogens, including Staphylococcus aureus, S. epidermidis, Bacillus subtilis, Proteus vulgaris, and P. aeruginosa (10). In 2002, Rubinstein and Pedersen isolated five different Bacillus species from chewing tobacco sold in the United States (11). Two of the species detected, B. licheniformis and B. pumilus, have been associated with pulmonary inflammation and opportunistic infections. Culture supernatants from these bacilli caused tissue edema and dysfunction when inoculated into the oral cavity of hamsters (11). B. pumilus and B. subtilis have also been identified as causative agents in spice-associated outbreaks and may produce a mild toxin that can result in illness after growing to a large population in a food (12, 13).
Many of the previous microbial characterization studies looked at relatively small sample sets, primarily from cigarettes, and did not analyze the types or quantities of microorganisms in STPs. Thus, there are data gaps in understanding the microorganisms present in the STPs which are potentially important to public health for the prevention of infectious diseases (9). Therefore, the goal of this study was to conduct a microbiological survey of STPs, which should help to fill scientific knowledge gaps associated with microbiological risks of currently marketed STPs that are considered important in FDA's tobacco product reviews and help to create a microbiological baseline for science-based regulation of tobacco product manufacturing and testing.
MATERIALS AND METHODS
Sample collection.
Fifteen smokeless tobacco products (STPs) were purchased from retail locations in the metropolitan areas of Little Rock, AR, and Washington, DC, in November 2012, March 2013, and July 2013. The same brands and products were collected at each time and location. The loose moist-snuff products included in the study consisted of brand A straight (where “straight” signifies “unflavored”), brand A wintergreen, brand B straight, brand B wintergreen, brand C wintergreen, and brand D straight. The snuff pouch products included in the study consisted of brand B pouches, brand A pouches, and brand B mint pouches. The snus products included in the study consisted of brand E snus mellow, brand F snus mint, and brand B snus mint. The chewing tobacco products included in the study consisted of brand G chewing tobacco, brand H chewing tobacco, and brand G Golden Blend chewing tobacco. For each of the 90 total product samples collected, information on the manufacturer, lot, purchase location, date of purchase, and sell-by date (if available) were recorded. The STPs were purchased and placed unopened in zip top bags; the samples from Little Rock, AR, were hand carried, and those from Washington, DC, were shipped by a common carrier to the laboratory in Jefferson, AR, to conduct the experimental analyses. The unopened packages were stored at room temperature, as they were in the retail setting, until processing. Prior to sampling, the STP packages were placed under UV light in a biological safety cabinet for at least 30 min to decontaminate the exterior packaging and minimize potential contamination by exterior microorganisms. The date when the package was initially opened for sample analyses was considered day 1.
Sample characterization. (i) Moisture content.
The tobacco products were tested for pH and moisture content after appropriate equilibration at room temperature. The moisture content was determined using the Karl Fischer coulometric titration method with samples prepared as described in document ISO 6488 (14). Samples were prepared by adding a 0.5-g sample of the tobacco product to 25 ml of methanol in a desiccated Erlenmeyer flask; the flask was then sealed with Parafilm M and shaken for 30 min at room temperature. After the shaking step, the tobacco samples were left at room temperature for 24 h to allow moisture extraction from the samples. A 1.0-g aliquot was transferred to a Karl Fischer titration apparatus for sample analysis, and the water content (expressed in milligrams of water per gram of sample) was calculated. Each of the samples was analyzed a minimum of two times.
(ii) pH determination.
The pH was determined by weighing 1.0 g of tobacco and adding it to a 50-ml polypropylene container with 10 ml of deionized distilled water. The samples were stirred, and the pH of the solution was measured over a 15-min time interval to get a stable pH value.
(iii) Particle size determination.
The particle size was evaluated for each product during the first sampling period by measuring the length and width to the nearest millimeter. A total of 100 individual particles were measured and their dimensions recorded, and the mode particle size range was determined for each product. The results determined in the second and third sampling periods were compared to the results determined in the first period to verify that the products had the same particle sizes.
(iv) Microbial loads.
A 0.10-g tobacco sample was weighed aseptically and placed in a sterile tube, to which 10 ml of sterile water was added. The samples were subjected to thorough vortex mixing. Total aerobic plate counts were determined for each product during the second and third samplings by spreading 100 μl of serially diluted tobacco suspension onto sheep's blood agar (SBA) plates and incubating the plates overnight at 37°C. After incubation, the colonies that grew on the plates were counted. For each sample, the suspensions were plated in triplicate and the numbers of aerobic bacteria present in the samples were determined by multiplying the number of organisms counted by the dilution factor. To assess the differences in microbial loads associated with different products, data from the four samples analyzed for each product were combined and assessed using the Kruskal-Wallis one-way analysis of variance on ranks approach and the SigmaPlot program (version 13.0; Systat Software, Inc., Chicago, IL). Statistical significance in differences between the microbial loads of products was represented by P values of <0.05.
(v) Sample recovery experiments.
To determine whether there were likely inhibitory substances or product conditions in tobacco products that limited the ability of microorganisms to grow, three representative STP types (brand C wintergreen moist snuff, brand H chewing tobacco, and brand F snus mint) were evaluated by bacterial seeding and recovery experiments. With the exception of a subset of brand F snus mint samples, the STP samples were autoclaved to kill the resident microbial populations and inoculated with sterile phosphate-buffered saline (PBS) or approximately 103 CFU/ml of S. aureus, P. aeruginosa, or B. subtilis as described previously (15). After a 2-h incubation, 3 ml of PBS was added to the STPs and 100-μl aliquots were plated on tryptic soy agar plates and incubated overnight at 37°C. The following day, the plates were counted and results compared to those obtained with the other STP samples and the initial cell suspensions that were diluted and plated. For each experiment, the samples were plated in triplicate and the experiments repeated at least twice. The recovery efficiency was calculated by dividing the average number of organisms recovered from the samples by the average CFU count of the initial inoculum. To assess the differences in recovery, the Kruskal-Wallis one-way analysis of variance on ranks approach was employed using SigmaPlot. Significant difference was represented by P values of <0.05.
Detection of culturable bacteria. (i) Culturing.
The viable and culturable bacteria were identified by growth on three different media. Culturing of bacteria was done on the day of initial sampling (day 1) and subsequently on days 3, 5, 8, and 15. Between samplings, the STPs were stored at 25°C under ambient conditions. For culturing, 100-μl aliquots of the tobacco suspension (prepared as described above) were transferred to (a) SBA, (b) mannitol salt agar (MSA), and (c) MacConkey agar plates (MAC). For the initial sampling, the plates were incubated at 25, 37, and 42°C and observed for growth (11). Because there were no distinguishable differences in the colonies growing at the different temperatures, the 37°C incubation temperature was used for subsequent cultures. Visually unique bacterial colonies were selected and described; representative colonies were subcultured, identified (as described below), and archived in brain heart infusion (BHI) broth with 20% glycerol at −80°C for long-term storage.
(ii) Bacterial identification.
Bacterial colonies were picked from the subculture plates and added to sterile water in PCR tubes or 96-well plates and thoroughly mixed. The suspensions were heated to 99°C for 10 min to lyse the bacterial cells and to liberate the DNA template for amplification and sequencing of the 16S rRNA gene. For the 16S rRNA gene analyses, the template DNA was combined with 2× PCR Mastermix (Promega, Madison, WI) and PCR primers 91E (5′-GGAATTCAAAKGAATTGACGGGGGC-3′) and 13B (5′-CGGGATCCCAGGCCCGGGAACGTATTCAC-3′) to amplify a 440-bp fragment of the 16S rRNA gene using a PCR protocol with an initial denaturation at 95°C, followed by 40 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, with a final 5-min extension at 72°C (16). The PCR products were separated by agarose gel electrophoresis (2% E-gel; Life Technologies, Grand Island, NY) to verify the presence of a product, purified by membrane filtration, and prepared for DNA sequencing with a BigDye Terminator kit (Applied Biosystems, Foster City, CA). The amplified products were sent to the core sequencing facility at the University of Arkansas for Medical Sciences (UAMS) for sequencing using BigDye chemistry on an ABI 3130XL sequencer (Applied Biosystems). The resultant sequences were visually inspected and compared to sequences in the GenBank database to identify the bacterial genus and, in most cases, species (17).
Identification and specific bacterial numbers present in the STPs. (i) Sample preparation.
An approximately 0.4-g sample of each tobacco product was collected, and the bacterial DNA was extracted using an UltraClean soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer's instructions. The DNA was quantified using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and used as a template for PCRs to amplify the V1/V2 and V6 variable regions of the 16S rRNA genes. To amplify the V1/V2 16S rRNA region, PCRs were prepared with the purified DNA, PCR master mix, and associated primers presented in Table 1.
TABLE 1.
PCR and 454 sequencing primers for pyrosequencinga
| Primer | Sequence (5′→3′) | Direction |
|---|---|---|
| V1/V2 region primers | ||
| 27F | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGTCAGAGTTTGATCCTGGCTCAG | Forward |
| 338R | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXCATGCTGCCTCCCGTAGGAGT | Reverse |
| V6 region primers | ||
| 967F-PP | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXCNACGCGAAGAACCTTANC | Forward |
| 967F-UC1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXCAACGCGAAAAACCTTACC | Forward |
| 967F-UC2 | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXCAACGCGCAGAACCTTACC | Forward |
| 967F-UC3 | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXATACGCGARGAACCTTACC | Forward |
| 967F-AQ | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXCTAACCGANGAACCTYACC | Forward |
| 1046R | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGACAGCCATGCANCACCT | Reverse |
| 1046R-PP | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGACAACCATGCANCACCT | Reverse |
| 1046R-AQ1 | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGACGGCCATGCANCACCT | Reverse |
| 1046R-AQ2 | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGACGACCATGCANCACCT | Reverse |
The primers contained 454-specific adaptors (26 bases at the beginning of each primer), linker nucleotides (underlined), an STP sample-specific 6-base barcode (Xs, in either the reverse or forward primers), and the specific 16S rRNA bacterial primers (bold).
To amplify the V1/V2 regions, one set of primers was used that contain 454-specific adaptors (Roche/454 Life Sciences; Branford, CT), linker nucleotides (underlined in Table 1), a unique STP-specific 6-base barcode (indicated by Xs in the reverse primer), and the specific V1/V2 bacterial primers (bolded). The PCRs were carried out as previously described (18). The PCRs were quantified using a PicoGreen assay (Invitrogen, Carlsbad, CA) and the reaction products pooled in equimolar amounts of PCR products for 454 Life Sciences Titanium sequencing at the David H. Murdock Research Institute (Kannapolis, NC).
To amplify the V6 regions, PCRs were carried out using the multiplex sets of primers described by Huber et al. (19). The sets include 5 forward and 4 reverse primers containing 454-specific adaptors, linker nucleotides (underlined in Table 1), a unique 6-base barcode (indicated by Xs in the forward primers), and the specific V6 bacterial primers (bolded). The PCRs were carried out as previously described (19). The PCRs were quantified, pooled, and sequenced as described above for the V1/V2 region.
Following the sequencing runs, the DNA sequence data were checked for quality and the pooled sequence runs were then sorted on the basis of their unique barcode and primer sequences using the Newbler program (Roche/454 Life Sciences). The sorted sequence flowgram files were submitted to the MG-RAST sequence analysis pipeline (Argonne National Laboratory, Argonne, IL) and converted to FASTQ format. The taxonomic identifications were done using MG-RAST's Best Hit Classification algorithm to compare the sequences to those in the GreenGenes databases to determine the number of sequences corresponding to specific operational taxonomic units (OTU) for each of the STPs (20, 21). The resultant OTU data (numbers of sequence reads per OTU) were exported to Microsoft Excel to determine the relative percentages of the different taxa present in each sample. Sample populations were evaluated in MG-RAST, and rarefaction curves were generated and subsequent alpha diversity (Shannon diversity) values calculated to evaluate the population composition of each sample (20, 21). Principal coordinate analysis (PCoA) was carried out in MG-RAST, and phylogenetic analysis using the Euclidean distance (BioNumerics; Applied Maths, Austin, TX) was used to evaluate population differences between the samples at the class level.
(ii) Denaturing gradient gel electrophoresis.
Denaturing gradient gel electrophoresis (DGGE) was used to evaluate potential changes in the bacterial population in the STPs over the sampling period. Total bacterial DNA was isolated from each tobacco sample, using an UltraClean soil DNA isolation kit, and the DNA concentration was quantified using an ND-1000 spectrophotometer as described above. Isolated DNA was stored at −80°C to facilitate normalization of DNA concentrations across all samples (days 1, 3, 5, 8, and 15) for a particular STP. For each of the DNA samples for a product, the DNA concentrations were normalized and subjected to PCR using a common master mix. PCR primers (GC-clamp-340F [5′-TCCTACGGGAGGCAGCAG-3′] and 518R [5′-ATTACCGCGGCTGCTGG-3′]) were used to amplify the V3 region of the 16S rRNA gene as previously described (22, 23). The DNA concentrations of the PCR products were normalized, and 500-ng volumes of each sample from a particular tobacco product were separated on a single gel using the denaturing conditions of Muyzer et al. (24) as modified by Martín et al. (25). This approach, which included the use of each of the samples from each tobacco product from each of the time points on a single gel, allowed detection of potential changes in the microbial populations over the sampling period (24, 25). The DGGE gels were stained using GelStar nucleic acid gel stain (Lonza, Basel, Switzerland) and digitally photographed. Banding patterns were compared to identify changes in band position and intensity, which indicate population changes.
RESULTS
Tobacco characterization.
A total of 90 individual tobacco samples representing 15 different smokeless tobacco products were tested. These sample groups have been named AR1, DC1, AR2, DC2, AR3, and DC3, where “AR” represents Arkansas, “DC” represents Washington, DC, and the numbers represent the sampling periods as follows: November 2012 (sampling period 1), March 2013 (sampling period 2), and July 2013 (sampling period 3) (see Materials and Methods).
Moisture.
The moisture levels were relatively consistent across the study for the same products, with the exception of the DC1 samples, for which moisture levels were significantly higher than those seen with the other sample sets and those previously reported in the literature (26, 27). Based on troubleshooting and additional testing, a problem with one of the reagents was identified and the reagent was replaced for subsequent testing; therefore, the DC1 samples were not included in the averaged results (Table 2). Overall, the moist-snuff products, including both the loose and pouch types, had the highest moisture content, followed by the snus and the chewing tobacco. The moist-snuff products on average had moisture content levels of between 522 and 583 mg water/g of tobacco. The snus products averaged between 269 and 315 mg water/g of product and the chewing tobacco products between 252 and 271 mg water/g of product.
TABLE 2.
Average initial characterization results from the smokeless tobacco products used in the studya
| Product | Moisture content (mg/g) (avg) |
pH (steady state) |
Particle size (mm) (mode) |
|||
|---|---|---|---|---|---|---|
| AR | DC | AR | DC | AR | DC | |
| Brand A straight loose moist snuff | 561 | 542 | 7.7 | 7.8 | ≤1 × 2 | ≤1 × 2 |
| Brand A wintergreen loose moist snuff | 538 | 576 | 7.8 | 7.6 | 1 × 4 | 1 × 6 |
| Brand B straight loose moist snuff | 550 | 633 | 7.4 | 7.4 | 1 × 4 | 1 × 6 |
| Brand B wintergreen loose moist snuff | 537 | 564 | 7.4 | 7.4 | 1 × 6 | 1 × 4 |
| Brand C wintergreen loose moist snuff | 561 | 507 | 8.1 | 8.2 | 1 × 4 | 1 × 4/1 × >8 |
| Brand D straight loose moist snuff | 566 | 458 | 7.9 | 7.6 | 1 × 4 | ≤1 × 2 |
| Brand B pouches | 550 | 596 | 7.6 | 7.6 | ≤1 × ≤1 | ≤1 × ≤1 |
| Brand A pouches | 557 | 527 | 7.8 | 7.8 | ≤1 × 2 | ≤1 × 2 |
| Brand B mint pouches | 527 | 558 | 7.6 | 7.8 | ≤1 × ≤1 | ≤1 × ≤1 |
| Brand E snus mellow | 320 | 308 | 7.5 | 7.6 | ≤1 × 2 | ≤1 × 2 |
| Brand F snus mint | 261 | 282 | 6.0 | 5.8 | ≤1 × ≤1 | ≤1 × ≤1 |
| Brand B snus mint | 258 | 306 | 6.1 | 6.6 | ≤1 × ≤1 | ≤1 × ≤1 |
| Brand G chewing tobacco | 258 | 244 | 5.9 | 6.0 | >2 × >8 | >2 × >8 |
| Brand H chewing tobacco | 242 | 277 | 5.5 | 5.6 | >2 × >8 | >2 × >8 |
| Brand G Golden Blend chewing tobacco | 255 | 295 | 5.9 | 5.9 | >2 × >8 | >2 × >8 |
AR and DC indicate that the samples were purchased in the metropolitan areas of Little Rock, AR, and Washington, DC, respectively.
pH.
There were also some differences among the product types in the pH levels of the products (Table 1), with moist snuff having higher pH levels (pH 7.4 to 8.2) and the chewing tobacco samples having among the lowest (pH 5.5 to 6.0). The snus samples were more variable, with the brand E snus mellow samples having an average pH of 7.6, while the brand F snus mint (pH 5.9) and brand B snus mint (pH 6.4) samples had considerably lower average pH values.
Particle sizes.
To determine the product sizes of the individual fragments, 100 individual particles were measured and classified into size categories (ranging from ≤1 by ≤1 mm to >2 by >8 mm) based on the length and width of the particles due to challenges in measuring some of the smallest fragments accurately. The mode (most commonly detected) particle sizes were determined for each product (Table 2). Figure 1 shows a representative of each type of product sampled. The chewing tobacco had by far the largest particles (>2 by >8 mm), followed by the loose moist snuff (modes ranging from ≤1 by 2 mm to 1 by >8 mm), and the pouched moist snuff and snus had the smallest particle sizes (modes from ≤1 by ≤1 mm to ≤1 by 2 mm) (Table 2). The pouches provide a means to keep the very fine particles contained for the user.
Microbial loads.
The results from the initial sampling period indicated that there were differences in the diversity and number of microorganisms in the different smokeless tobacco samples. Therefore, total aerobic plate counts were carried out for each sample during the second and third sampling periods to determine the number of CFU of bacteria per gram of product. The moist-snuff products had the highest microbial loads (Table 3), followed by some of the chewing tobacco samples (the brand G products had significantly higher loads than the brand H samples; P < 0.05), and the lowest loads were in the snus samples. Two of the products (brand E snus mellow and brand F snus mint) did not have any growth during the aerobic plate count studies, and the third product (brand B snus mint) had only a single colony in one sampling. Table 3 shows the mean numbers of CFU per gram of product across the four sample sets tested. In general, the moist-snuff samples averaged around 106 CFU/g, which was considerably higher than the levels seen with the other products, with the exception of the brand G chewing tobacco, which averaged approximately 7 × 105 CFU/g. All of the moist-snuff samples and all but brand H among the chewing tobacco samples had significantly higher microbial loads (P < 0.05) than the brand B snus mint sample, which was the only snus sample with microbial growth.
TABLE 3.
Mean total aerobic plate counts for the smokeless tobacco products used in the studya
| Product | Bacterial count (CFU/g) (sheep's blood agar) |
|||
|---|---|---|---|---|
| AR2 | DC2 | AR3 | DC3 | |
| Brand A straight | 8.20 × 105 | 4.00 × 106 | 1.80 × 105 | 8.10 × 105 |
| Brand A wintergreen | 2.00 × 106 | 1.05 × 106 | 9.20 × 105 | 7.00 × 105 |
| Brand B straight | 1.47 × 106 | 1.14 × 106 | 1.23 × 106 | 7.30 × 105 |
| Brand B wintergreen | 8.00 × 105 | 9.90 × 105 | 1.32 × 106 | 7.00 × 105 |
| Brand C wintergreen | 2.70 × 106 | 1.40 × 106 | 1.35 × 106 | 3.00 × 106 |
| Brand D straight | 4.20 × 105 | 1.57 × 105 | 1.53 × 106 | 2.90 × 105 |
| Brand B pouches | 1.95 × 105 | 4.80 × 105 | 4.10 × 105 | 4.00 × 105 |
| Brand A pouches | 2.30 × 105 | 1.90 × 105 | 1.01 × 106 | 8.50 × 105 |
| Brand B mint pouches | 4.80 × 104 | 5.10 × 104 | 1.67 × 105 | 4.00 × 106 |
| Brand E snus mellow | ND | ND | ND | ND |
| Brand F snus mint | ND | ND | ND | ND |
| Brand B snus mint | ND | ND | 1.00 × 103 | ND |
| Brand G chewing tobacco | 7.20 × 105 | 3.90 × 104 | 1.37 × 106 | 6.60 × 105 |
| Brand H chewing tobacco | 1.00 × 103 | 1.00 × 103 | 1.00 × 103 | ND |
| Brand G Golden Blend chewing tobacco | 4.40 × 104 | 4.00 × 104 | 6.50 × 104 | 1.09 × 105 |
AR and DC indicate that the samples were purchased in the metropolitan areas of Little Rock, AR, and Washington, DC, respectively, during the second sampling period. ND, none detected.
Sample recovery experiments.
Initial experiments were conducted to determine whether there were likely inhibitory substances or product conditions that limited the ability of microorganisms to grow in some of the samples. Three sample types that had different bacterial loads in the earlier portions of the study, including moist snuff (brand C wintergreen) with high levels, chewing tobacco (brand H chewing tobacco) with intermediate levels, and snus (brand F snus mint) with low levels, were evaluated. The samples were autoclaved, with the exception of a subset of snus samples that had a low background of microorganisms, and were included to see if autoclaving affected growth. Overall, when the products were seeded with S. aureus, there was significantly higher recovery of organisms from the moist-snuff samples (62%) than from the other samples (<5%; P < 0.05). The lowest recovery was from the chewing tobacco samples, where the average recovery level was approximately 1.1%; however, that level was not significantly lower than the level of recovery from snus samples. There appeared to be a slightly higher level of recovery from the nonautoclaved sample of snus than from the autoclaved sample (4.9% versus 1.4%); however, this may have been due to the background microflora of the product (there were low levels of growth in some of the nonautoclaved samples). The levels of recovery of P. aeruginosa were much lower in general, with the highest average level of recovery from moist snuff (3.8%), and the average level of recovery from each of the samples was less than 1.0%. Likewise, the levels of recovery of B. subtilis were low, with the highest average recovery at 1.2% for the moist-snuff sample and the lowest at 0.4% for the chewing tobacco. The observed differences in the levels of recovery of P. aeruginosa and B. subtilis from the various products did not reach the level of significance.
Detection of culturable bacteria.
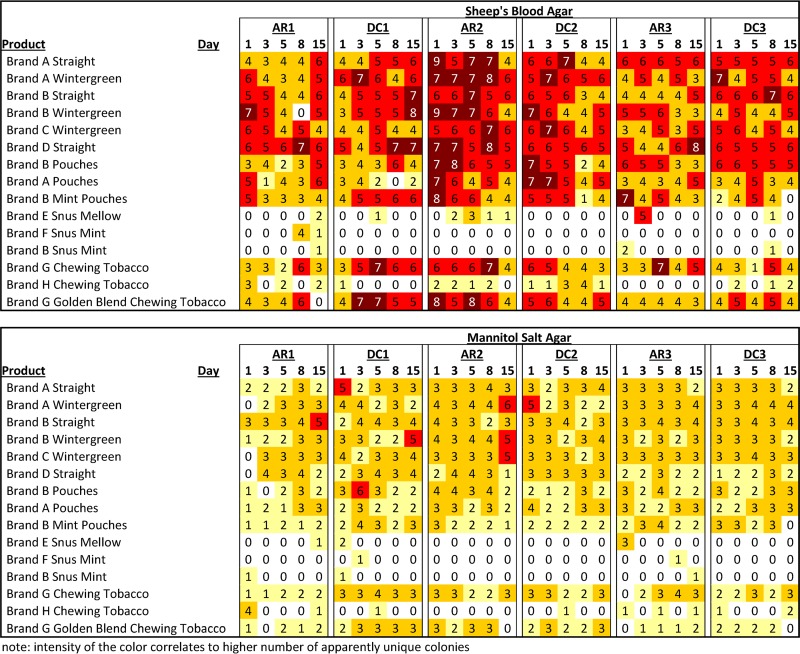
The containers of STPs were sampled over a 15-day period during each set of experiments. Over 2,500 different bacterial colonies that appeared to have unique morphologies on the individual sample plates (individual STP extract spread on each plate) were selected. At most sampling periods, the moist-snuff samples had higher numbers and higher apparent diversity of bacterial colonies than the snus samples and some chewing tobacco samples. Figure 2 shows which samples were positive for bacterial growth and the numbers of colonies selected for the different culture media, sampling days, and periods. There was no growth observed on any of the MacConkey agar plates, indicating the likely absence of Gram-negative enteric bacteria. On a majority of sampling days, no bacteria were grown from most snus samples. This intermittent positivity is likely due to a low level of bacterial content or unequal distribution of organisms, such that there were not always bacteria in the suspensions that were plated. Additionally, there did not appear to be an increase in the number of positive samples in the snus products over the sampling period, indicating that there was likely no active growth of the bacteria in the products. The chewing tobacco samples were more variable, with the brand G products having a higher percentage of positive samples than the brand H products. These findings mirror the microbial load studies described above.
FIG 2.
Number of apparently distinct bacterial colonies detected in the samples. Note that colors with higher intensity correlate to higher numbers of apparently unique colonies.
To identify the bacterial organisms cultured, 16S rRNA gene sequencing indicated that over 90% of bacteria isolated belonged to the genus Bacillus. The most common species identified (in order of frequency) were B. pumilus, B. licheniformis, B. safensis, and B. subtilis. Among the non-Bacillus species, Oceanobacillus, Staphylococcus (including S. epidermidis and S. hominis), and Tetragenococcus were the most common genera (see Table S1 in the supplemental material). The Oceanobacillus strains were most commonly isolated from the chewing tobacco samples, whereas most of the staphylococci were isolated from the snus samples.
Detection of total bacterial populations.
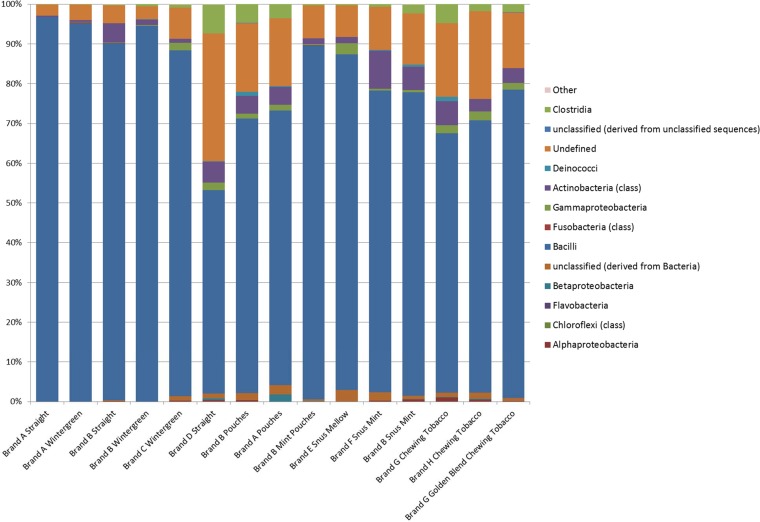
16S rRNA gene sequencing was done to determine the populations of bacteria, including uncultured bacteria, associated with the STPs. Sequences of approximately 330 bases were obtained from the V1/V2 region, which in most cases allowed classification of taxa down to the genus level; however, for comparisons among the samples, the populations were evaluated at the class level (Fig. 3 and Fig. S1 in the supplemental material). Across all three sampling periods, the average number of sequencing reads per sample per run was 20,966; however, sample DC1-5 (brand C, wintergreen) did not provide adequate sequence data for efficient analysis (26 reads). The predominant species in most of the samples were members of the class of Bacilli, while some of the others had a higher proportion of Gammaproteobacteria. The alpha (Shannon) diversity of the samples ranged from 1.80 (removing sample DC1-5, with very low sequence coverage) to 19.07 (see Table S2 in the supplemental material). There was little similarity of alpha diversity values calculated across the same product types. Likewise, when the results from the samples were compared to one another using PCoA and phylogenetic analyses, the samples from the same types of products generally did not cluster together; however, several of the samples from the third sample period appeared to form separate groupings (see Fig. S2 and S3 in the supplemental material). Examining further those samples with a high proportion of members of the Bacilli, the predominant bacterial DNA sequences identified were from the genera Tetragenococcus, Carnobacterium, Lactobacillus, Geobacillus, Bacillus, and Staphylococcus. Some of these genera were also cultured from the STP samples; however, others were not detected by culturing under aerobic conditions. Thus, during the third sampling period, a set of tobacco suspensions plated on brain heart infusion (BHI) agar were incubated anaerobically to determine whether there were differences in the bacterial species isolated. The organisms that grew were sequenced and determined to be members of the genus Bacillus and matched the aerobic culture results (data not shown). The data for the V1/V2 sequencing are publically available through the MG-RAST program (http://metagenomics.anl.gov) and were deposited as project 10176 with accession numbers 4573000.3 to 4573089.3 and 4574738.8 (see Table S2 in the supplemental material).
FIG 3.
Cumulative pyrosequencing results for the V1/V2 region of the 16S rRNA analysis of each of the different types of smokeless tobacco samples analyzed. The bar graph indicates the percentages of sequences detected that belonged to a particular bacterial class. The classes of organisms and color schemes are presented on the right side of the figure. The data corresponding to the DNA sequences identified as chloroplasts have been removed due to their likely representing plant origins rather than microbial populations.
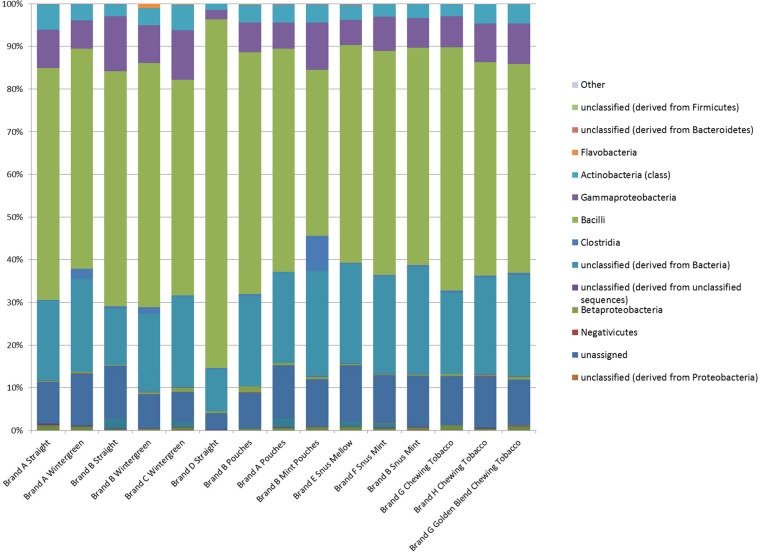
The analyzed V6 region fragment is smaller and is typically around 80 bases in size. There were an average of 19,552 sequence reads per sample sequenced; however, two samples (DC2-8 [brand A pouches] and DC3-1 [brand A straight]) did not provide adequate sequence data for efficient analysis (20 and 45 reads, respectively). As with the V1/V2 sampling, the predominant classes identified were members of the Bacilli and Gammaproteobacteria; however, a relatively high proportion of the sequences could not be fully discriminated at the class level and were listed as unclassified bacteria or unclassified members of Firmicutes (the phylum that includes the class Bacilli) (Fig. 4; see also Fig. S4 in the supplemental material). The most commonly identified genera from the V6 analyses and the comparison to the Greengenes database were Bacillus, Aeribacillus, Geobacillus, Tetragenococcus, Corynebacterium, Halomonas, Staphylococcus, and Anoxybacillus. The alpha (Shannon) diversity of the samples ranged from 1.37 to 21.27 (see Table S2 in the supplemental material). Compared to the results from the V1/V2 analyses, there was little consistency in the alpha diversity detected, such that samples with comparatively low diversity with respect to the V1/V2 region had higher levels detected from the V6 region and vice versa. As with the V1/V2 results, PCoA and phylogenetic analysis showed little clustering based on product type or sampling period (see Fig. S5 and S6 in the supplemental material). The data for the V6 sequencing are publicly available through the MG-RAST program (http://metagenomics.anl.gov) and were deposited as project 10910 with accession numbers 4582827.3 to 4582916.3 (see Table S2 in the supplemental material).
FIG 4.
Cumulative pyrosequencing results for the V6 region of the 16S rRNA analysis of each of the different types of smokeless tobacco samples analyzed. The bar graph indicates the percentages of sequences detected that belonged to a particular bacterial class. The classes of organisms and color schemes are presented on the right side of the figure. The DNA sequences identified as chloroplasts have been removed due to their their likely representing plant origins rather than microbial populations.
Evaluation of bacterial population changes in open product.
The comparison of the DGGE profiles of the total bacteria isolated indicated that the bacterial populations in some of the open product samples varied during the sampling periods (see Fig. S7 in the supplemental material). Overall, at least 21 (23.3%) of the 90 individual samples analyzed displayed changes in the banding profiles of the samples, indicative of population changes. In all but five instances, the products with the apparent bacterial population changes had variability in two or more sampling batches. The only samples that did not display any population changes in any of the samples during the sampling periods were the three snus products (see Table S3 in the supplemental material).
DISCUSSION
This project was undertaken to gain a better understanding of the microbial populations that may be present in STPs. Microbial populations in STPs are an important consideration for the protection of public health and for the FDA as it develops new regulations under section 906 of the Federal Food, Drug, and Cosmetic Act. A potential risk for STP users is that the products could carry pathogenic or opportunistic microorganisms that might result in the development of an infectious disease. This is a concern in part because STPs are typically held for extended periods of time in close contact with the oral mucosa. Another potential risk associated with microbial contamination is the development of microbial metabolic by-products that may be harmful to consumers, such as microbial toxins and carcinogens. Thus, the goals of the present study were to determine the types and numbers of microorganisms present in the STPs to create a microbiological baseline for STPs and to lay the foundation for further studies to evaluate the potential microbial risks of smokeless tobacco use.
The major objectives of the study were to identify the bacterial microorganisms that were present in a convenience sample of STPs available to consumers in two geographically distinct locations and to determine the impact of temporal changes on the microbial communities present in STPs. Fifteen different products representing four major types of STPs were analyzed over the study period. A potential limitation of the study was that for each type of product, a small cross-section of the available products was tested; however, efforts were made to include a variety of products and manufacturers to minimize this potential concern. For example, the moist-snuff samples, which have the highest market share of users, had the highest number of products sampled. The most common bacteria identified among the STPs in the culture-based experiments were members of the genus Bacillus (see Table S1 in the supplemental material). These organisms are able to form endospores, which resist drying, high temperature, and other factors that inhibit the growth and reproduction of vegetative microorganisms. The species of Bacillus that we detected are also fairly common in the environment and are not often associated with acute illness in humans; however, B. licheniformis and B. pumilus are potential causes of pulmonary inflammation and opportunistic infections (11). In addition, B. pumilus and B. subtilis have been identified as causative agents in spice-associated outbreaks and can produce a mild toxin that may result in illness after growing to a large population (12, 13). Another potential concern regarding the bacilli is that some of the Bacillus species, including B. pumilus, B. licheniformis, and B. subtilis, are able to reduce nitrates to nitrites (28). Nitrites are important precursors for the nitrosation of nicotine to form the carcinogenic tobacco-specific N′-nitrosamines (TSNAs) (29, 30). The presence of these microorganisms in the STPs suggests the possibility of TSNA formation after packaging. This phenomenon would be consistent with earlier studies that found that the levels of TSNAs in certain STPs stored under ambient conditions increased over time (31, 32).
Strains of S. epidermidis and S. hominis are also able to reduce nitrate and present potential health concerns as opportunistic pathogens, especially for immunocompromised users. Both staphylococcal species have been reported to cause bacterial endocarditis (heart valve infection), which arises due to transmission to the heart via the bloodstream (33). STP users often have problems with gingivitis and other oral health issues which may allow bacterial entry into the bloodstream (34). However, to our knowledge no studies have yet shown a causal link between STP use and bacterial endocarditis.
Tetragenococcus and Oceanobacillus are members of the families Enterococcaceae and Bacillaceae, respectively, and some species are associated with fermented foods. Thus, it is possible that these organisms may be present in the production of the different tobacco products. Oceanobacillus was most often detected in the chewing tobacco samples. Interestingly, some of the better characterized Oceanobacillus isolates in the literature have been identified as being more alkaliphilic (35) and yet the chewing tobacco samples had the lowest pH levels.
A potential concern with the experimental results was the lack of positive cultures from the snus samples. The other STP types had relatively high microbial loads; thus, experiments were conducted to determine whether there were inhibitory substances or product conditions that limited the ability of microorganisms to grow in the snus samples. In the seeding and recovery experiment, both the snus and chewing tobacco appeared to inhibit bacterial growth. These findings suggest that this may have resulted from the lower pH or moisture content in the snus and chewing tobacco samples or from the presence of antimicrobial compounds in these products. For example, nicotine has been shown to impact the growth of certain bacteria associated with dental caries (36, 37).
The pyrosequencing results from the V1/V2 region of the 16S rRNA genes in the total DNA isolated from STPs were compared to the culturing results, and there were many instances of incongruence. While members of the genera Tetragenococcus, Geobacillus, Bacillus, and Staphylococcus were among the top taxa identified by both sequencing of the V1/V2 region and culturing (Fig. 3; see also Fig. S1 in the supplemental material), members of other genera, including Carnobacterium, Lactobacillus, and Corynebacterium, were identified by sequencing but not detected by culture. There are multiple possible explanations for these differences between the pyrosequencing and culture results, including nonoptimal culturing conditions or amplification of DNA from nonviable organisms. For example, many lactobacilli are facultative anaerobes or microaerophiles and may not grow efficiently under the aerobic culture conditions used for isolation and culture. When culture plates from STP samples were incubated under anaerobic conditions, the bacteria that grew were the same Bacillus species that were detected with the standard aerobic culturing methods. The probable explanation for the discrepant results is that the STPs had fairly high loads of lactobacilli which may have grown during the fermentation process in the production of the finished STP but were killed during further processing. In such a case, their DNA could remain in the product and be amplified in the initial steps of the sequencing experiments. In future experiments, it may be preferential to initially isolate bacterial RNA, which is much less stable than DNA and therefore may provide a more positive correlation with viability status, and to use it as the starting material for sequencing (38). This approach would minimize the carryover detection of dead organisms and maintain the population ratios of organisms in the sample. An alternative approach would be to perform a short enrichment step with the samples before bacterial isolation and DNA extraction. This approach would minimize the impact of the dead organisms and potentially activate Bacillus spores, making DNA extraction more efficient, but unfortunately would likely distort the ratios of bacterial populations.
The V6 pyrosequencing results in general were similar to those seen in the V1/V2 analyses; however, a greater proportion of sequences were “unclassified” at each of the taxonomic levels using both Ribosomal Database Project (RDP) and GreenGenes comparisons. These ambiguous results were likely due to the short length of the sequence that was amplified, sequenced, and searched. The V6 region was only about 80 bp in size. This target worked well for intestinal bacteria (19), where the data sets are likely much richer; however, some of the microorganisms in STPs may not be well represented in the sequence database, thus preventing specific matches and leading to the higher degree of “unclassified” samples. Representative sequence reads from the unclassified bacteria category were submitted for BLAST searching, and the highest-scoring matches returned generally identified the lactobacilli, which is consistent with the V1/V2 findings. As with the V1/V2 results, Bacillus, Geobacillus, Tetragenococcus, Corynebacterium, Halomonas, and Staphylococcus were among the most commonly identified genera (Fig. 4; see also Fig. S4 in the supplemental material).
Interestingly, the V6 sequencing results provided added resolution for quantification the Proteobacteria, especially the Alpha-, Beta-, and Deltaproteobacteria, in the samples. This outcome was likely the result of the choice of PCR primer sets used to amplify the products. The approach for amplification of the V6 region used five forward and four reverse primers that included several degenerate bases that likely allowed a more efficient amplification of a wider array of the DNA templates present in the tobacco samples. The approach for amplification of V1/V2 used a single pair of primers, which likely impacted the efficiency of amplification of some taxa. These observations are consistent with the generally higher alpha diversity calculations for the V6 sequences than for the V1/V2 samples (average Shannon diversity index of 9.85 versus 7.00, respectively).
Amplification of enough DNA templates for sequencing was difficult for several samples, even though there was ample DNA template based on the DNA concentrations and the results of the DGGE experiments. Because the DGGE primers were outside the region of the V1/V2 primers, for the third sampling, the DNA template was initially amplified with the 27F forward primer and the 518R DGGE primer followed by amplification with the V1/V2 primers (27F and 338R), and the resultant sequence results were more robust. These results included increased amplification of the Proteobacteria species that were poorly amplified using the V1/V2 primers but were detected using the V6 sequences. Related to these findings, the values representing the alpha diversity results were higher for several of the samples from the third sampling period. Sixteen of the 30 samples with the highest Shannon diversity values among the V1/V2 samples were from the third sampling period (see Table S2 in the supplemental material). Additionally, the result of this amplification change can likely be observed in results of both the PCoA and phylogenetic analyses, whereby several samples from the third sampling period (the AR3 and DC3 samples) were separated from many of the samples from sampling periods 1 and 2 (see Fig. S2 and S3 in the supplemental material). This divergence is likely attributable to the amplification of increased proportions of the Proteobacteria, Actinobacteria, and Clostridia, which can be observed in the bottom cluster shown in Fig. S3.
Several genera identified by sequence analyses from the various STPs have been shown to be part of the oral microbiome. Among the suspected periodontal pathogens, some STPs were positive for sequences representing the genera Eubacterium, Prevotella, and Porphyromonas (39), although they were present in relatively low numbers and in a limited number of samples. Other genera identified by pyrosequencing, including Lactobacillus and Actinomyces, have been suggested to play roles in the pathogenesis of dental caries (40). Sequences of lactobacilli were among those most commonly identified among the V1/V2 sequences but were at a much lower proportion among the V6 sequences. This finding may have been due to the increased ability of the V1/V2 sequencing to map to specific sequences in the data sets, which was likely due to their larger fragment size. Overall, many of the taxa detected in the STP samples, including many of the members of Firmicutes, Proteobacteria, and Actinobacteria, have been reported to be part of the normal microbiota of the oral cavity (41). The STP samples had much lower proportions of Bacteroides and Fusobacterium than have been reported as part of the normal oral microbiota (40, 42).
There did not appear to be significant changes in the bacterial populations over the sampling periods. Similar numbers of distinct colonies (Fig. 2; see also Table S1 in the supplemental material) were detected across the sampling period, and the morphologies detected were consistent. Sequencing of the isolates resulted in the identification of similar species across the sampling days. With DGGE, about 21 of 90 samples appeared to have different patterns across the sampling periods; bands present on earlier days (e.g., 1 and 3) often gradually diminished over the sampling period. These results could reflect the degradation of DNA from dead microorganisms present in the samples, such as those detected with the pyrosequencing studies. More work will need to be done to verify this hypothesis. Based on the culture findings and the DGGE results, the bacterial populations appeared to be fairly consistent during the sampling periods.
In summary, the moisture and pH levels that we detected in the STP samples are consistent with data reported elsewhere for similar products (26, 27). The moist-snuff samples had the highest levels of bacterial contamination (both in diversity and number of organisms), which may coincide with their more neutral pH and higher moisture levels. The snus samples and some of the chewing tobacco samples harbored significantly fewer bacteria than the moist-snuff samples, potentially indicating that the relatively high levels of bacteria in moist snuff may warrant greater attention in future studies. The study findings provide baseline data on the microbial content of STPs, which is important to assess the potential risks to the users, aid in the assessment of manufacturing processes, and assist the FDA in establishing regulations. The fact that many of the bacterial species identified have the ability to reduce nitrate to nitrite and thus potentially contribute to the production of highly carcinogenic TSNAs also warrants further attention in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the FDA Center for Tobacco Products for funding this project. Jing Han and Yasser Sanad were supported through the Oak Ridge Institute for Science and Education.
We also thank Carl E. Cerniglia, Brad Schnackenberg, Michael Koenig, Rajesh Nayak, and Huizhong Chen for their assistance with this project, helpful reviews, and critiques of the manuscript.
The findings and conclusions in this report are ours and do not necessarily represent the official views or positions of the FDA or the Department of Health and Human Services.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01612-16.
REFERENCES
- 1.Substance Abuse and Mental Health Services Administration. 2010. Results from the 2009 National Survey on Drug Use and Health: detailed tables. Substance Abuse and Mental Health Services Administration, Office of Applied Studies, Rockville, MD. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2010. Tobacco use among middle and high school students—United States, 2000–2009. MMWR Morb Mortal Wkly Rep 59:1063–1068. [PubMed] [Google Scholar]
- 3.Sapkota AR, Berger S, Vogel TM. 2010. Human pathogens abundant in the bacterial metagenome of cigarettes. Environ Health Perspect 118:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cockrell W, Roberts J, Kane B, Fulghum R. 1989. Microbiology of oral smokeless tobacco products. Tob Sci 33:55–57. [Google Scholar]
- 5.National Cancer Institute and Centers for Disease Control and Prevention. 2014. Smokeless tobacco and public health: a global perspective. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Institutes of Health, Bethesda, MD. [Google Scholar]
- 6.Krisberg K. 2010. New types of smokeless tobacco present growing risks for youth: survey: products mistaken for candy. The Nation's Health 40:1–14. [Google Scholar]
- 7.Critchley JA, Unal B. 2003. Health effects associated with smokeless tobacco: a systematic review. Thorax 58:435–443. doi: 10.1136/thorax.58.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 4 August 2011. Smokeless tobacco facts. Centers for Disease Control and Prevention. http://www.cdc.gov/tobacco/data_statistics/fact_sheets/smokeless/products_marketing/index.htm Accessed 2 May 2016.
- 9.Pauly JL, Paszkiewicz G. 2011. Cigarette smoke, bacteria, mold, microbial toxins, and chronic lung inflammation. J Oncol 2011:819129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dygert HP. 1957. Snuff—a source of pathogenic bacteria in chronic bronchitis. N Engl J Med 257:311–313. doi: 10.1056/NEJM195708152570704. [DOI] [PubMed] [Google Scholar]
- 11.Rubinstein I, Pedersen GW. 2002. Bacillus species are present in chewing tobacco sold in the United States and evoke plasma exudation from the oral mucosa. Clin Diagn Lab Immunol 9:1057–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Food and Drug Administration. 2013. FDA draft risk profile: pathogens and filth in spices. Center for Food Safety and Applied Nutrition, U.S. Department of Health and Human Services, College Park, MD. [Google Scholar]
- 13.Logan NA. 2012. Bacillus and relatives in foodborne illness. J Appl Microbiol 112:417–429. doi: 10.1111/j.1365-2672.2011.05204.x. [DOI] [PubMed] [Google Scholar]
- 14.International Organization for Standardization. 2004. ISO 6488: tobacco and tobacco products—determination of water content—Karl Fischer method, 2nd ed International Organization for Standardization, Technical Committee, Geneva, Switzerland. [Google Scholar]
- 15.U.S. Pharmacopeial Convention. 2012. Microbiological examination of nonsterile products: microbial enumeration tests, p 56–60. In United States Pharmacopeia and The National Formulary 35. United States Pharmacopeial Convention, Rockville, MD. [Google Scholar]
- 16.Mignard S, Flandrois JP. 2006. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods 67:574–581. doi: 10.1016/j.mimet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 20.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ben Omar N, Ampe F. 2000. Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl Environ Microbiol 66:3664–3673. doi: 10.1128/AEM.66.9.3664-3673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, VanderGheynst J, Mills DA. 2003. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:6801–6807. doi: 10.1128/AEM.69.11.6801-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín R, Heilig GH, Zoetendal EG, Smidt H, Rodríguez JM. 2007. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol 103:2638–2644. doi: 10.1111/j.1365-2672.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- 26.Lawler TS, Stanfill SB, Zhang L, Ashley DL, Watson CH. 2013. Chemical characterization of domestic oral tobacco products: total nicotine, pH, unprotonated nicotine and tobacco-specific N-nitrosamines. Food Chem Toxicol 57:380–386. doi: 10.1016/j.fct.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter P, Hodge K, Stanfill S, Zhang L, Watson C. 2008. Surveillance of moist snuff: total nicotine, moisture, pH, un-ionized nicotine, and tobacco-specific nitrosamines. Nicotine Tob Res 10:1645–1652. doi: 10.1080/14622200802412937. [DOI] [PubMed] [Google Scholar]
- 28.Gregory LG, Bond PL, Richardson DJ, Spiro S. 2003. Characterization of a nitrate-respiring bacterial community using the nitrate reductase gene (narG) as a functional marker. Microbiol 149:229–237. doi: 10.1099/mic.0.25849-0. [DOI] [PubMed] [Google Scholar]
- 29.Sleiman M, Gundel LA, Pankow JF, Jacob P III, Singer BC, Destaillats H. 2010. Formation of carcinogens indoors by surface-mediated reactions of nicotine with nitrous acid, leading to potential thirdhand smoke hazards. Proc Natl Acad Sci U S A 107:6576–6581. doi: 10.1073/pnas.0912820107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klus H, Kunze M, Koenig S, Poeschl E. 2009. Smokeless tobacco—an overview. Contributions Tob Res 23:248–276. [Google Scholar]
- 31.Djordjevic MV, Fan J, Bush LP, Brunnemann KD, Hoffmann D. 1993. Effects of storage conditions on levels of tobacco-specific N-nitrosamines and N-nitrosamino acids in U.S. moist snuff. J Agric Food Chem 41:1790–1794. doi: 10.1021/jf00034a051. [DOI] [Google Scholar]
- 32.Fisher MT, Bennett CB, Hayes A, Kargalioglu Y, Knox BL, Xu D, Muhammad-Kah R, Gaworski CL. 2012. Sources of and technical approaches for the abatement of tobacco specific nitrosamine formation in moist smokeless tobacco products. Food Chem Toxicol 50:942–948. doi: 10.1016/j.fct.2011.11.035. [DOI] [PubMed] [Google Scholar]
- 33.Demitrovicova A, Hricak V, Karvay M, Krcmery V. 2007. Endocarditis due to coagulase-negative staphylococci: data from a 22-years national survey. Scand J Infect Dis 39:655–656. doi: 10.1080/00365540701299632. [DOI] [PubMed] [Google Scholar]
- 34.Greer RO., Jr 2011. Oral manifestations of smokeless tobacco use. Otolaryngol Clin North Am 44:31–56, v. doi: 10.1016/j.otc.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Yumoto I, Hirota K, Nodasaka Y, Nakajima K. 2005. Oceanobacillus oncorhynchi sp. nov., a halotolerant obligate alkaliphile isolated from the skin of a rainbow trout (Oncorhynchus mykiss), and emended description of the genus Oceanobacillus. Int J Syst Evol Microbiol 55:1521–1524. doi: 10.1099/ijs.0.63483-0. [DOI] [PubMed] [Google Scholar]
- 36.Li M, Huang R, Zhou X, Qiu W, Xu X, Gregory RL. 2016. Effect of nicotine on cariogenic virulence of Streptococcus mutans. Folia Microbiol (Praha) doi: 10.1007/s12223-016-0465-8. [DOI] [PubMed] [Google Scholar]
- 37.Huang R, Li M, Ye M, Yang K, Xu X, Gregory RL. 2014. Effects of nicotine on Streptococcus gordonii growth, biofilm formation, and cell aggregation. Appl Environ Microbiol 80:7212–7218. doi: 10.1128/AEM.02395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keer JT, Birch L. 2003. Molecular methods for the assessment of bacterial viability. J Microbiol Methods 53:175–183. doi: 10.1016/S0167-7012(03)00025-3. [DOI] [PubMed] [Google Scholar]
- 39.Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A 111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simon-Soro A, Pignatelli M, Mira A. 2012. The oral metagenome in health and disease. ISME J 6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wade WG. 2013. The oral microbiome in health and disease. Pharmacol Res 69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.