ABSTRACT
Precise control of microbial gene expression resulting in a defined, fast, and homogeneous response is of utmost importance for synthetic bio(techno)logical applications. However, even broadly applied biotechnological workhorses, such as Corynebacterium glutamicum, for which induction of recombinant gene expression commonly relies on the addition of appropriate inducer molecules, perform moderately in this respect. Light offers an alternative to accurately control gene expression, as it allows for simple triggering in a noninvasive fashion with unprecedented spatiotemporal resolution. Thus, optogenetic switches are promising tools to improve the controllability of existing gene expression systems. In this regard, photocaged inducers, whose activities are initially inhibited by light-removable protection groups, represent one of the most valuable photoswitches for microbial gene expression. Here, we report on the evaluation of photocaged isopropyl-β-d-thiogalactopyranoside (IPTG) as a light-responsive control element for the frequently applied tac-based expression module in C. glutamicum. In contrast to conventional IPTG, the photocaged inducer mediates a tightly controlled, strong, and homogeneous expression response upon short exposure to UV-A light. To further demonstrate the unique potential of photocaged IPTG for the optimization of production processes in C. glutamicum, the optogenetic switch was finally used to improve biosynthesis of the growth-inhibiting sesquiterpene (+)-valencene, a flavoring agent and aroma compound precursor in food industry. The variation in light intensity as well as the time point of light induction proved crucial for efficient production of this toxic compound.
IMPORTANCE Optogenetic tools are light-responsive modules that allow for a simple triggering of cellular functions with unprecedented spatiotemporal resolution and in a noninvasive fashion. Specifically, light-controlled gene expression exhibits an enormous potential for various synthetic bio(techno)logical purposes. Before our study, poor inducibility, together with phenotypic heterogeneity, was reported for the IPTG-mediated induction of lac-based gene expression in Corynebacterium glutamicum. By applying photocaged IPTG as a synthetic inducer, however, these drawbacks could be almost completely abolished. Especially for increasing numbers of parallelized expression cultures, noninvasive and spatiotemporal light induction qualifies for a precise, homogeneous, and thus higher-order control to fully automatize or optimize future biotechnological applications.
INTRODUCTION
Corynebacterium glutamicum represents one of the most important biotechnological platform organisms and massively contributes to the industrial production of amino acids (1–5), but it has also been engineered, for instance, for the production of lower alcohols (6–9), organic acids (10–13), diamines (14, 15), and carotenoids (16–18). However, currently applied expression setups show only moderate performance regarding both precise control and expression homogeneity. Population heterogeneity affecting both growth and expression may strongly impact biotechnological processes (19). For instance, a distinct population heterogeneity has recently been described for C. glutamicum cultures producing l-valine (20, 21). While cell-to-cell variations within isogenic populations may constitute an overall fitness advantage over cohabiting competitors (22, 23) in natural environments, such heterogeneity is highly unfavorable in biotechnological production processes (19).
Therefore, an emerging need for novel synthetic expression tools exists that enable homogeneous and higher-order control over gene expression. Optogenetics, originally devised to control cells, typically neurons in living tissue, that have been genetically modified to express light-sensitive ion channels, now also include the light-mediated control and specific triggering of gene expression in a noninvasive and highly resolving spatiotemporal fashion (24). In contrast to other induction signals, light involves a seemingly homogeneous signal perception for both photosensory modules (25, 26) and chemically synthesized phototriggers, such as photocaged compounds (27, 28), given that cultures are uniformly exposed. Photocaged molecules are rendered biologically inactive through the addition of a photoremovable protection group, also designated a photocaging group or photocage. Specific functionality can be restored easily and noninvasively (i.e., without exerting manipulations that may alter the physiology of cells during ongoing cultivation) by light-mediated release (uncaging) of the bioactive molecule (29, 30).
By means of photocaged isopropyl-β-d-thiogalactopyranoside (IPTG), a conventional lac promoter-based gene expression system was recently redirected to enable accurately controlled and homogeneous gene expression by UV-A light in Escherichia coli (27). Traditional lac-based gene regulation in C. glutamicum, however, has limitations, e.g., due to the absence of a lactose uptake system and poor permeability of the C. glutamicum membrane for IPTG (31). Therefore, derepression of the lac or tac promoter using IPTG is often conducted during inoculation (32–34), which prohibits tight and temporally accurate regulation, so that the precise control of toxic gene products or metabolic fluxes is distinctly impeded. In a recent study, for instance, the IPTG-induced production of (+)-valencene, a natural constituent of the essential oils of citrus fruits, faced serious growth impairment in C. glutamicum (34). Here, only moderate production could be obtained of this bicyclic sesquiterpenoid, which is used as an additive by the food and beverage industry, due to its pleasant orange-like odor (35, 36).
Within this work, we demonstrate the limitations of IPTG induction in C. glutamicum and how to circumvent them during tac-based gene expression by using light-responsive photocaged inducer molecules. The established optogenetic expression setup was finally applied to improve production of the sesquiterpene (+)-valencene, despite its growth-impeding properties.
MATERIALS AND METHODS
DNA manipulation and construction of plasmids.
DNA techniques and molecular biology methods were performed basically as described previously (37).
Bacterial strains, plasmids, and media.
All bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. E. coli strains were cultured at 37°C under constant agitation in lysogeny broth (LB) (Luria/Miller; Carl Roth, Karlsruhe, Germany) (37) and supplemented with kanamycin (50 μg · ml−1) and spectinomycin (250 μg · ml−1), if required. C. glutamicum ATCC 13032 was used as the wild-type strain (38). Cultivations were performed using brain heart infusion (BHI) complex medium (Difco; BD, Heidelberg, Germany) or CGXII minimal medium (39, 40). If appropriate, media were supplemented with 25 μg · ml−1 kanamycin and 100 μg · ml−1 spectinomycin.
TABLE 1.
Strains, plasmid, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristics or sequence (5′ →3′)a | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− thi-1 endA1 hsdR17(r− m−) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 relA1 | 41 |
| C. glutamicum ATCC 13032 | Biotin auxotroph, wild-type strain | 38, 42 |
| C. glutamicum EYFP | ATCC 13032(pEKEx2-EYFP) | 32 |
| C. glutamicum VLC3 | ATCC 13032 ΔcrtE ΔidsA(pVWEx1-CnVS)(pEKEx3-ispA) | 34 |
| C. glutamicum VLC4 | ATCC 13032 ΔcrtE ΔidsA(pVWEx1-oCnVS)(pEKEx3-ispA) | This study |
| C. glutamicum VLC5 | ATCC 13032 ΔcrtE ΔidsA(pVWEx1)(pEKEx3-ispA-oCnVS) | This study |
| C. glutamicum VLC6 | ATCC 13032 ΔcrtE ΔidsA(pVWEx1-dxs-idi)(pEKEx3-ispA-oCnVS) | This study |
| Plasmids | ||
| pEKEx2-EYFP | Kmr; pEKEx2 containing EYFP with an artificial RBS under the control of Ptac | 32 |
| pVWEx1 | Kmr; E. coli/C. glutamicum shuttle vector for regulated gene expression (Ptac, lacIq, pCG1 oriVCg) | 45 |
| pVWEx1-CnVS | pVWEx1 derivative for IPTG-inducible expression of (+)-valencene synthase gene CnVS of Callitropsis nootkatensis containing an artificial ribosome binding site | 34 |
| pVWEx1-oCnVS | pVWEx1 derivative for IPTG-inducible expression of codon optimized (+)-valencene synthase gene oCnVS from Callitropsis nootkatensis containing an artificial ribosome binding site | This work |
| pVWEx1-dxs-idi | pVWEx1 derivative for IPTG-inducible expression of 1-deoxy-d-xylulose 5-phosphate synthase (dxs) and the isopentenyl pyrophosphate isomerase (idi) genes of Corynebacterium glutamicum | This work |
| pEKEx3 | Specr; E. coli/C. glutamicum shuttle vector for regulated gene expression (Ptac, lacIq, pBL1 oriVCg) | 46 |
| pEKEx3-ispA | pEKEx3 derivative for IPTG-inducible expression of FPP synthase gene ispA from E. coli containing an artificial ribosome binding | 34 |
| pEKEx3-ispA-oCnVS | pEKEx3 derivative for IPTG-inducible expression of FPP synthase gene ispA from E. coli and the codon-optimized (+)-valencene synthase gene oCnVS from Callitropsis nootkatensis | This work |
| Oligonucleotides | ||
| (1) dxs_fwd | CCTGCAGGTCGACTCTAGAGAGGAGGCCCTTCAGATGGGAATTCTGAACAG | |
| (2) dxs_rev | CCCTAAGCTTAGACATCTGAAGGGCCTCCTTTATTCCCCGAACAGGG | |
| (3) idi_fwd | CCCTGTTCGGGGAATAAAGGAGGCCCTTCAGATGTCTAAGCTTAGGG | |
| (4) idi_rev | CGAGCTCGGTACCCGGGGATCTTACTCTGCGTCAAACGCTTCC | |
| (5) ispA_fwd | CCTGCAGGTCGACTCTAGAGAGGAGGCCCTTCAGATGGACTTTCCGCAGC | |
| (6) ispA_rev | CGTTGAACATTTCCGCCATATGAAGGGCCTCCTTTATTTATTACGCTGGATGATG | |
| (7) oCnVS_fwd | CATCATCCAGCGTAATAAATAAAGGAGGCCCTTCATATGGCGGAAATGTTCAACG | |
| (8) oCnVS_rev | CGAGCTCGGTACCCGGGGATCTTACGGGATGATCGGTTCCACG |
Artificial ribosome binding site sequences are in bold type within oligonucleotide sequences. Kmr, kanamycin resistance; Specr, spectinomycin resistance; RBS, ribosome binding site.
C. glutamicum EYFP expression cultures.
Enhanced yellow fluorescent protein (EYFP) expression cultures were cultivated (800 μl at 1,500 rpm, 85% relative humidity, and 30°C) in a BioLector microbioreactor system (m2p-labs, Germany) under constant monitoring of biomass accumulation and EYFP fluorescence development. Expression cultures were inoculated to cell densities corresponding to an optical density at 600 nm (OD600) of 1.0 for CGXII medium and 0.05 for BHI medium.
Chemical synthesis of NP-photocaged IPTG.
6-Nitropiperonyl (NP)-photocaged IPTG was synthesized in a one-step reaction from IPTG and 6-nitropiperonal, as previously described (27).
Conventional and light induction of gene expression.
Conventional IPTG induction was conducted after 0, 2, 4, 6, or 8 h of cultivation via small-volume pipetting. Light-induced expression cultures were directly supplemented with photocaged IPTG (from dimethyl sulfoxide [DMSO] stock solutions, just as for IPTG) prior to cultivation and noninvasively exposed to UV-A light (VL-315.BL hand lamp, 45 W; Vilber Lourmat, France; distance to FlowerPlate, 1.5 cm, approximately 0.9 mW · cm−2) at desired time points. Light exposure was varied by using different exposure times (0 to 30 min) or different light intensities experimentally created by dimming the reaction wells with various layers of diffusion foils (White Diffusion Lee 216; Lee Filters, USA). Here, an exposure time of 30 min resulted in full induction. Light intensities were quantified using a thermal power sensor (S302C; Thorlabs, Inc., USA).
(+)-Valencene production cultures.
Seed cultures for (+)-valencene production and growth experiments with C. glutamicum were performed in 10 ml of LB supplemented with 50 mM glucose at 30°C and 120 rpm in 100-ml nonbaffled flasks. For adaptation of the cells to production conditions, a second seed culture in CGXII minimal medium with 4% glucose monohydrate was inoculated from the LB seed culture and grown for 5 to 6 h (50 ml of CGXII, 500-ml baffled flasks, 30°C, 120 rpm). Both the second seed and the main cultures were inoculated to an OD600 of 1. Production and growth experiments were conducted in 48-well FlowerPlates (m2p-labs, Germany) in 800 μl of CGXII medium with 4% glucose monohydrate as the source of carbon at 30°C and 1,200 rpm offline for production or in a BioLector system (m2p-labs) for online monitoring of growth. Production was induced with IPTG 0, 2, 4, and 6 h after inoculation and for NP-photocaged IPTG (cIPTG)-based light induction (see above) 4 and 6 h after inoculation using either 0.1 or 0.25 mM each compound, respectively. After induction, 200 μl of n-dodecane was added aseptically to the cultures, which were grown for additional 24 h. After cultivation, the n-dodecane layer was harvested by centrifugation at 4°C for 1 to 2 h at 24,000 × g. Subsequently, the (+)-valencene content of the n-dodecane phase was determined by gas chromatography-mass spectrometry (GC-MS) measurements.
Flow cytometry analysis.
Flow cytometry analyses of C. glutamicum (43) were performed with a FACSAria II flow cytometer (Becton Dickinson, Heidelberg, Germany) using a blue solid-state laser (Sapphire 488-20) with an excitation wavelength of 488 nm. Cytometer setup and performance tracking were performed with Cytometer Setup 0026 and Tracking Beads (bright [3 mm], mid [3 mm], and dim [2 mm] beads) labeled with a mixture of fluorochromes (Becton Dickinson). Forward-scatter characteristics (FSC) and side-scatter characteristics were detected as small-angle and orthogonal scatters of the 488-nm laser, respectively. EYFP fluorescence was detected using a 502-nm long-pass and a 530/30-nm band-pass filter set. FACSDiva software 6.0 was used to record the measurements. During analyses, thresholding on FSC was applied to remove background noise. Data were analyzed with the FlowJo version 10.0.8 analysis software (Tree Star, Ashland, OR, USA). To stain dead cells, cells with an OD600 of approximately 0.05 were incubated with 20 μM propidium iodide (PI) (stock solution, 20 mM in DMSO) (Molecular Probes, Leiden, The Netherlands) for 15 min at room temperature (RT). For validation of the protocol, intact cells and cells with injured membranes (treated with 70% isopropyl alcohol for 30 min) were mixed in different ratios. Afterwards, cells were stained as described above. For the detection of the fluorescent dye PI, a 595-nm long-pass and a 610/20-nm band-pass filter set was used.
Strain construction.
The genes dxs and idi coding for 1-deoxy-d-xylulose-5-phosphate synthase and isopentenyl pyrophosphate (IPP) isomerase, respectively, were PCR amplified using oligonucleotides 1 to 4 (Table 1) from genomic DNA of C. glutamicum ATCC 13032, which was isolated as previously described (44). For improved expression, the (+)-valencene synthase gene CnVS of Callitropsis nootkatensis was codon optimized for C. glutamicum ATCC 13032 using a codon usage protocol provided by http://www.kazusa.or.jp/codon/ and synthesized by GeneArt/Life Technologies (Darmstadt, Germany), yielding the gene oCnVS (see the supplemental material). The farnesyl pyrophosphate synthase gene ispA was PCR amplified from genomic DNA of E. coli as described previously (34). All genes were cloned into the expression vector pVWEx1 (45) or pEKEx3 (46) by Gibson assembly (47) using the BamHI restriction site and the respective oligonucleotides shown in Table 1. Primers were constructed such that an artificial ribosomal binding site (AGGAGG) was added 8 bp upstream of the translational start codon of each gene. The integrity of all inserts was confirmed by sequencing (Sequencing Core Facility, Bielefeld University).
GC-MS measurements.
The harvested n-dodecane phases of the production cultures were analyzed using a Thermo Scientific Trace GC Ultra connected to a Thermo Scientific ISQ single quadrupole mass spectrometer using a TG-5MS column (length, 30 m; inside diameter [i.d.], 0.25 mm; film thickness, 0.25 μm) (Thermo Scientific, Waltham, MA, USA). After splitless injection of 1 μl, the initial temperature of 40°C was increased by 10°C/min to 160°C and then by 15°C/min to 300°C, with a 2-min ramp at 300°C at the end of the measurement and a constant helium gas flow rate of 1 ml/min. The MS operating parameters were ionization voltage, 70 eV (electron impact ionization); and ion source and interface temperature, 230°C. (+)-Valencene was identified by the comparison of retention time and mass spectrum to technical (+)-valencene (Sigma-Aldrich, Steinheim, Germany). For quantitative analysis of (+)-valencene, a calibration curve with technical (+)-valencene was used.
RESULTS
Establishing NP-photocaged IPTG-based light induction in C. glutamicum.
In a previous study, NP-photocaged IPTG (cIPTG) was employed in E. coli to noninvasively control gene expression by light in a gradual and homogeneous fashion (27). Here, we aimed to transfer the photoswitch to the biotechnological workhorse C. glutamicum to generate an easily light-addressable induction system applicable for various biotechnological purposes. The frequently applied IPTG-inducible tac promoter-based pEKEx expression vector (44, 48) was chosen as a target system for cIPTG-mediated light induction. To characterize the light responsiveness of gene expression in C. glutamicum, the pEKEx2-EYFP (32) expression vector was employed, allowing online monitoring of induction processes in batch cultures, as well as at the single-cell level by means of EYFP reporter fluorescence.
We first determined key system specifications, namely, (i) the inducibility of the tac promoter at different time points during cultivation, (ii) the maximum expression levels, and (iii) the respective dynamic range of induction. cIPTG, which efficiently releases IPTG in a two-step photocleavage reaction (49) upon short UV-A light exposure and subsequent enzymatic hydrolysis of the photoproduct esters (Fig. 1A), was added to EYFP expression cultures. To test the light induction of EYFP expression at different growth phases of C. glutamicum, cultures were illuminated at different time points of cell growth (0 to 8 h) during cultivation in BHI complex and CGXII minimal medium. Moreover, the irradiation dose was increased stepwise by extending the time of light exposure (0 to 30 min) in order to evaluate the gradual responsiveness of the chosen expression system. The EYFP production profiles clearly demonstrated that cIPTG can be used as a photoswitch enabling light induction of gene expression in C. glutamicum in a gradual manner and over a long period of cultivation (Fig. 1B). In BHI complex medium, full inducibility and maximal dynamic range (i.e., an expression range of 0 to 100%) were obtained by increasing the exposure time after 4 to 6 h of cultivation (corresponding to the mid-exponential-growth phase; see Fig. S1 in the supplemental material). Induction in the lag or early exponential phase (0 to 2 h; 0 to 80% expression output) as well as in the late-exponential phase (8 h; 0 to 50% expression output) still resulted in a gradual light response of EYFP expression but impaired the dynamic range. In CGXII minimal medium, light induction produced expression outputs corresponding properly to the exposure time at all monitored induction time points. UV-A light exposure times of 20 to 25 min at moderate intensities (0.9 mW · cm−2) were sufficient to fully induce gene expression in BHI and CGXII medium.
FIG 1.
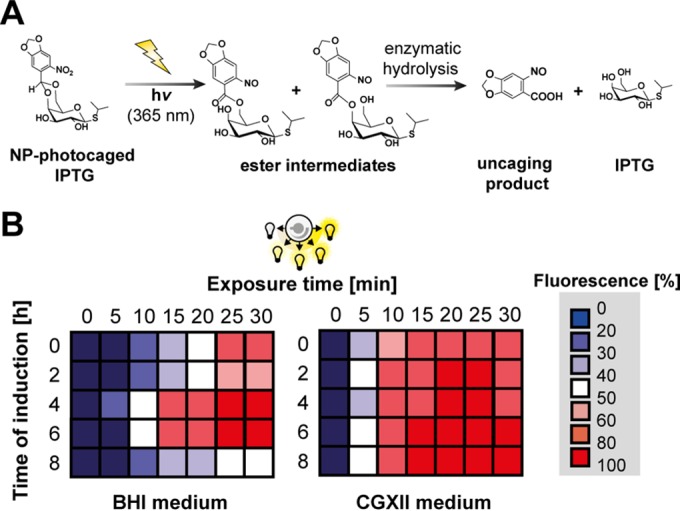
Light-controlled gene expression in C. glutamicum using cIPTG as a photoswitch. (A) Two-step release of IPTG from cIPTG by UV-A light-mediated photocleavage and enzymatic hydrolysis of photoproduct esters as described by Young and Deiters (49). (B) Gradual upregulation of EYFP expression in C. glutamicum ATCC 13032(pEKEx2-EYFP) depending on the time of UV-A light exposure (λmax = 365 nm, 0.9 mW · cm−2) using BHI complex (left) or CGXII-glucose minimal medium (right) supplemented with 100 μM cIPTG. Relative EYFP fluorescence values originate from biomass-normalized triplicates and depict low (blue) to high (red) EYFP fluorescence intervals after 20 h of overexpression. Color gradations represent differential expression outputs obtained by variation of induction time or UV-A light exposure. Maximum biomass-normalized fluorescence values obtained in both media are means of triplicates and were arbitrarily set to 100%. hv, light energy.
Comparative analysis of EYFP expression in C. glutamicum using conventional IPTG and cIPTG-based light induction.
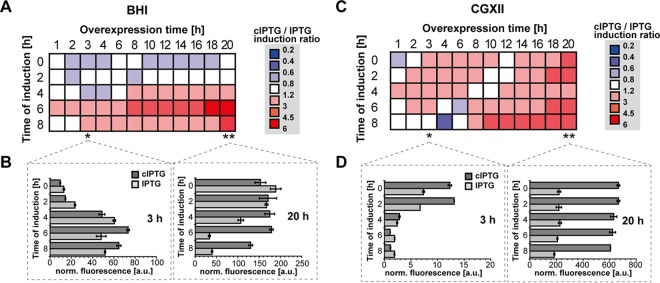
In a next step, cIPTG-based light induction of gene expression was compared to conventional IPTG induction using the same EYFP reporter system (pEKEx2-EYFP). EYFP output signals were measured depending on different induction time points (0 to 8 h) over the course of a following overexpression period (0 to 20 h) in both BHI and CGXII media. To directly compare conventional and light induction, the EYFP fluorescence ratios determined after cIPTG (30-min light exposure, 100 μM) and IPTG induction (100 μM) are shown as heat maps (Fig. 2). In BHI medium (Fig. 2A), IPTG induction was similar to or slightly outperformed light-dependent cIPTG induction in the lag and early exponential-growth phases (0 to 2 h). In contrast, upon induction in the mid- or late-exponential phase (4 to 8 h), cIPTG outperformed conventional IPTG, particularly after overexpression periods longer than 8 h. Significant effects were observed for induction after 6 h of cultivation, where light induction produced an expression output up to 6-fold higher than induction with conventional IPTG after 18 h of overexpression. Besides the evaluation of relative effects, a description of individual absolute fluorescence outputs provides insights into system-inherent specifications. In this sense, the initial inducibility of the tac promoter in C. glutamicum cultures during early to late-logarithmic-growth phases, which was determined 3 h after induction (Fig. 2B, left), showed that the velocity of induction increases with ongoing cultivation time for both IPTG and cIPTG. Here, only a slight improvement in light induction could be observed for mid- to late-logarithmic-growth phases (6 to 8 h). For longer expression times (in particular after 20 h of target gene expression), however, light-mediated induction of mid- to late-logarithmic C. glutamicum cultures (6 to 8 h) using cIPTG resulted in much higher EYFP expression than that with conventional induction (Fig. 2B, right).
FIG 2.
Comparative analysis of IPTG and cIPTG induction of tac promoter-mediated EYFP expression in C. glutamicum ATCC 13032(pEKEx2-EYFP). Fluorescence ratio intervals of cIPTG- (30 min UV-A, 100 μM) and IPTG-induced (100 μM) EYFP fluorescence are shown during cultivation in BHI complex (A) and CGXII-glucose minimal medium (C) depending on the time of induction and on overexpression times. Fluorescence ratios originate from biomass-normalized triplicates and depict low (blue, superior IPTG induction) to high (red, superior cIPTG induction) ratio intervals in color gradations. Bar plots indicate individual biomass-normalized (norm.) fluorescence values (in arbitrary units [a.u.]) as means of triplicates after 3 h (left) and 20 h (right) of IPTG- (gray) and cIPTG-induced (dark gray) EYFP expression in BHI complex (B) and CGXII-glucose minimal medium (D). Error bars indicate the respective standard deviations.
In CGXII medium (Fig. 2C), light induction can effectively be applied for a broad range of induction time points from early to late-exponential phases and outperformed conventional IPTG induction up to 4-fold for overexpression periods greater than 12 h. For late-exponential induction (6 to 8 h), IPTG initially performed well but was outpaced by cIPTG with increasing cultivation times. In contrast to BHI medium, the expression response in CGXII medium is significantly slower for both IPTG and cIPTG, as depicted by low fluorescence levels after 3 h of EYFP overexpression (Fig. 2D, left). After 20 h of overexpression (Fig. 2D, right), however, considerable EYFP fluorescence was observed especially for light induction, which outperformed conventional IPTG induction up to 3-fold. Strikingly, expression outputs for both cIPTG- and IPTG-mediated induction were, contrary to cultivations in BHI medium and irrespective of the time of induction, highest at the end of the experiment (Fig. 2D, right).
Basal background levels (see Fig. S2 in the supplemental material) using nonexposed cIPTG were found to be moderate in BHI medium (up to 1.5-fold increase in comparison to control strains) and CGXII medium (up to 2.2-fold). As reported in the literature (50), basal expression levels of the system used here were found to be elevated in CGXII minimal medium and increased with ongoing cultivation times. The dynamic range of induction, however, was high and comparable in BHI medium (63-fold for cIPTG and 61-fold for IPTG). In CGXII minimal medium, light induction was higher than conventional IPTG induction (239-fold compared to 46-fold, respectively; see Fig. S3 in the supplemental material).
In summary, the comparative analysis of cIPTG and IPTG induction in BHI and CGXII media showed that noninvasive light induction is broadly applicable in C. glutamicum. Particularly, light induction during mid- to late-exponential growth significantly outperformed conventional IPTG induction up to 6-fold. Moreover, for long overexpression periods of up to 20 h, cIPTG persistently proved to be as efficient as equimolar amounts of IPTG. Interestingly, despite a slightly delayed induction response caused by the essential enzymatic cleavage of the photoproducts (Fig. 1A), cIPTG-based light induction was comparable to IPTG induction in C. glutamicum and temporally even outperformed the conventional induction response for late induction in BHI and early induction in CGXII medium.
Single-cell analysis of EYFP expression after induction with IPTG and cIPTG.
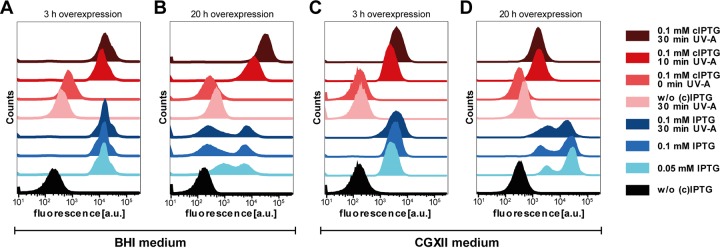
As cIPTG-dependent light induction was successfully demonstrated for standard bulk cultivations in both BHI complex and CGXII minimal media, light- and IPTG-induced gene expression was next analyzed at the single-cell level in order to analyze the homogeneity of expression behavior within C. glutamicum batch cultures. To this end, the fluorescence of single cells from conventional and light-induced cultures was monitored by flow cytometry. Single-cell EYFP fluorescence values for cultures induced with light after 5 h of cultivation (light- to deep-red-colored histograms) showed a homogeneous distribution after both 3 h (Fig. 3A and C) and 20 h (Fig. 3B and D) of overexpression in BHI (Fig. 3A and B) and CGXII media (Fig. 3C and D). For IPTG induction (light- to deep-blue-colored histograms), however, a heterogeneous fluorescence distribution was observed with increasing expression time in both media. Surprisingly, the two media differently influenced the expression phenotype of the IPTG-induced C. glutamicum expression strain. In BHI medium, a second population with lower fluorescence intensity occurred after 20 h of overexpression, whereas prolonged expression in CGXII medium obviously resulted in the formation of a second population with increased YFP accumulation. In principle, expression heterogeneity has recently been described for a similar expression setup in C. glutamicum (50). Notably, flow cytometric single-cell analysis using propidium iodide-based LIVE/DEAD staining (43) further suggested that light induction did not affect membrane integrity and thus cell viability (see Fig. S4 in the supplemental material).
FIG 3.
Flow cytometric single-cell analysis of gene expression induced by IPTG and cIPTG in C. glutamicum ATCC 13032(pEKEx2-EYFP). Distribution of EYFP fluorescence intensities was plotted against the number of cells (counts). Expression cultures were compared in BHI complex (A and B) and CGXII-glucose minimal medium (C and D), and fluorescence was measured after 3 h (A and C) and 20 h (B and D) of EYFP overexpression and induction with IPTG and light after 5 h of cultivation. For light induction, cells were exposed to UV-A light (λmax = 365 nm) for 10 and 30 min, respectively, with a light intensity of 0.9 mW · cm−2.
In summary, cIPTG-based light control of gene expression in C. glutamicum was shown to distinctly outperform conventional IPTG induction with respect to maximum expression levels, responsiveness, homogeneity, and inducibility, especially for longer expression periods.
Employing cIPTG for the production of the toxic sesquiterpene (+)-valencene.
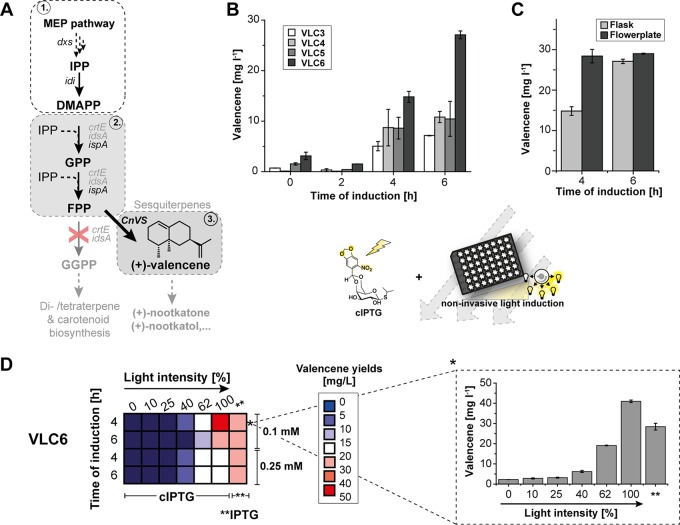
As a challenging task, we tried to apply the light-controlled expression setup to improve the biosynthesis of the bacterial growth-inhibiting terpenoid (+)-valencene. A C. glutamicum strain producing (+)-valencene was recently constructed by metabolic engineering (34). Here, we first analyzed whether (+)-valencene production could be further elevated via metabolic engineering (Fig. 4A). The initial strain C. glutamicum VLC3 carries deletions of the crtE and idsA genes to preclude formation of the undesired geranylgeranyl pyrophosphate (GGPP). Farnesyl pyrophosphate (FPP) was synthesized by the FPP synthase IspA from E. coli. FPP, in turn, is converted to (+)-valencene via the (+)-valencene synthase CnVS from Callitropsis nootkatensis (35). In the newly constructed (+)-valencene producer strain VLC4, the CnVS-encoding gene was expressed after its adaptation to the codon usage of C. glutamicum. In strain VLC6, the genes coding for IspA and CnVS were combined on a single vector, allowing the introduction of a second IPTG-inducible vector for overexpression of the genes encoding the 1-deoxy-d-xylulose 5-phosphate synthase (dxs) and isopentenyl pyrophosphate isomerase (idi) to enhance the supply of the precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) (18), and consequently, of FPP. Since CnVS gene expression perturbed growth, we presumed that the optimization and timing of CnVS expression would be one key aspect to elevate (+)-valencene production (Fig. 4A) and hence, an appealing target for cIPTG-based light control. Therefore, (+)-valencene accumulation was first monitored in flask cultivations using the primary VLC3 producer strain, as well as our newly constructed producers VLC4 to -6 (Table 1) upon conventional induction of gene expression after different cultivation times (0 to 6 h). Cultivation in shake flasks revealed that the time of induction was crucial for (+)-valencene productivity, as the (+)-valencene titers in all four tested strains were largely improved when induced after 4 and 6 h (Fig. 4B). Moreover, after 6 h of induction, VLC4 with the codon-optimized valencene synthase (oCnVS) showed slightly increased production of (+)-valencene up to 1.5-fold (10.8 ± 1.1 mg · liter−1) compared to that with VLC3 (7.2 ± 0.6 mg · liter−1). In contrast, the coexpression of ispA and oCnVS genes from a single plasmid in VLC5 showed only a negligible influence on titer (10.5 ± 3.5 mg · liter−1).
FIG 4.
Light-controlled (+)-valencene production in C. glutamicum using CGXII-glucose medium. (A) Biosynthetic route for (+)-valencene production (3) based on the MEP pathway (1) and appropriate isoprenoid pathway gene deletions for improved FPP precursor supply (2). To improve the metabolic flux toward FPP, the heterologous gene ispA and, at a later stage, the endogenous genes dxs and idi, were overexpressed. (B) Screening of engineered C. glutamicum strains VLC3 to VLC6 for (+)-valencene productivity upon IPTG induction (0.1 mM) after different induction time points. (C) (+)-Valencene production upon IPTG induction (0.1 mM) by strain VLC6 grown for 24 h in a flask (light gray) and FlowerPlate (dark gray). (D) (+)-Valencene productivity profiles (left) indicating valencene titer intervals from low (blue) to high (red) in milligrams per liter after 24 h of production with VLC6 using different times of induction as well as different (c)IPTG concentrations (0.1 and 0.25 mM). Light intensities were incremented in a stepwise manner (100% here correlates to 0.9 mW · cm−2). The results obtained for induction after 4 h with 0.1 mM cIPTG marked with an asterisk are shown in more detail on the right. Double asterisks indicate control experiments with IPTG induction. All averaged data originated from the results of at least three independent biological triplicates. MEP, methylerythritol phosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate. Error bars indicate the respective standard deviations.
Notably, additional overexpression of dxs and idi in VLC6 led to 3.8-fold-improved (+)-valencene production (27.1 ± 0.6 mg · liter−1). The production of (+)-valencene by VLC6 was further characterized at a microtiter plate scale using 48-well FlowerPlates (51, 52). With this approach, (+)-valencene production was further enhanced when induced after 4 h and slightly after 6 h (Fig. 4C). Here, improved valencene titers could be attributed to the fact that oxygen-unlimited FlowerPlate cultivations yielded up to 2-fold-higher biomass formation (see Table S1 in the supplemental material). Next, we analyzed whether light-mediated induction with cIPTG could further optimize the (+)-valencene production level by applying different light regimes, cIPTG concentrations, and induction time points (Fig. 4D, left).
Variable light control of (+)-valencene production led to 1.4-fold elevated (+)-valencene titers compared to those with conventional IPTG induction. Optimal productivity was found for full light induction (i.e., 100% light intensity = 0.9 mW · cm−2) after 4 h of cultivation using 0.1 mM cIPTG (Fig. 4D, left) and yielded final titers of 41.0 ± 0.1 mg · liter−1. During (+)-valencene production, IPTG-induced dxs-idi overexpression in VLC6 was found to significantly lower growth rates up to 30% (μmax = 0.29 ± 0.01 · h−1) compared to those with noninduced cultures (μmax = 0.42 ± 0.01 · h−1). Compared to expression cultures that have been induced by IPTG, growth impairment could partly be abolished using cIPTG (μmax = 0.34 ± 0.01 · h−1). However, cell growth was still hampered by cIPTG induction, since noninduced cultures still exhibited an approximately 20% higher growth rate (see Fig. S5 in the supplemental material). Much larger growth differences in light and IPTG-induced production cultures were observed for early induction (see Fig. S6 in the supplemental material).
By applying cIPTG-based light-controlled screening for optimized expression parameters, we were able to significantly elevate (+)-valencene titers in C. glutamicum approximately 6-fold, from an initial 7.2 mg · liter−1 to 41.0 mg · liter−1. The combination of metabolic engineering and light-controlled expression resulted in an about 17-fold improved (+)-valencene production compared to a titer of 2.4 mg · liter−1 reported in our previous study.
DISCUSSION
This study demonstrates optogenetic control of microbial gene expression as a valuable tool for synthetic bio(techno)logical applications. Photocaged carbohydrates (27, 28), for instance, can be readily transferred to different biotechnological production hosts and employed to precisely control gene expression in a straightforward and spatiotemporal fashion. Moreover, photocaged carbohydrate inducers seem to abrogate native expression heterogeneity because their increased membrane permeability may supersede uptake by specific transport systems or by poor diffusion. A homogeneous expression response was shown here for NP-photocaged IPTG in C. glutamicum, as well as in a recent study employing photocaged arabinose in E. coli (28). Furthermore, the fact that the strong light induction response was independent of cellular states enables appropriate biomass production, which is essential for several complex biosynthetic procedures and especially those leading to the production of toxic compounds. Noninvasive and gradual upregulation of gene expression by light, moreover, provides a fast and easy option to screen for optimized expression conditions, rendering time-consuming and invasive inducer supplementation obsolete. The established optogenetic expression modules can thus be applied in novel photomicrobioreactors (53) and single-cell cultivation platforms (19) to precisely control the expression of target genes and thereby fully automatize the optimization of microbial production processes in a high-throughput fashion. Notably, cIPTG-based light induction requires elevated expenditures of costs and labor and is further unsuited for large-scale fermentations where light exposure poses an additional challenge. However, especially for closed (e.g., anaerobic) systems and increasing numbers of parallelized expression cultures, noninvasive and spatiotemporal light induction will provide a higher-order control.
For the light-controlled (+)-valencene production in this study, light induction facilitated the optimal balance between growth and production (see Fig. S6 in the supplemental material) and provided stronger gene expression levels in CGXII (Fig. 2D) and thus efficient gene expression for late induction. Noteworthy, those improvements could not be reproduced by optimizing conventional IPTG induction with respect to inducer concentration (see Fig. S7 in the supplemental material). Finally, for the light-controlled expression system using the best (+)-valencene-producing strain, C. glutamicum VLC6, a titer of 41.0 mg · liter−1 and volumetric productivity of 1.46 mg · liter−1 · h−1 were obtained (see Table S1 in the supplemental material). These values are in the same range as those obtained with other bacteria and higher than those obtained with eukaryotic microorganisms. To the best of our knowledge, the highest titers and volumetric (+)-valencene productivities so far were described for Rhodobacter sphaeroides expressing the mevalonate operon from Paracoccus zeaxanthinifaciens and the codon-optimized (+)-valencene synthase CnVS, which reached a titer of 352 mg · liter−1 and a productivity of 4.88 mg · liter−1 · h−1 (35). The yeasts Saccharomyces cerevisiae (titer, 1.36 mg · liter−1; productivity, 18 μg · liter−1 · h−1), Schizophyllum commune (titer, 16.6 mg · liter−1; productivity, 115 μg · liter−1 · h−1), and Pichia pastoris (titer, 51 mg · liter−1; productivity, 1 mg · liter−1 · h−1), however, proved suitable for the in situ conversion of (+)-valencene to more valuable products, such as (+)-nootkatone (35, 54, 55).
Future improvements in (+)-valencene production by C. glutamicum might include further engineering of FPP biosynthesis and process optimization by, e.g., fed-batch cultivation with glucose or alternative carbon sources feeding directly into the methylerythritol phosphate (MEP) pathway. Moreover, the temporal decoupling and thus independent regulation of FPP and (+)-valencene biosynthetic pathways, e.g., by means of multichromatic optogenetic control (26, 56, 57), might offer potential for debottlenecking, further optimizing the metabolic flux toward (+)-valencene biosynthesis. To realize future metabolic flux engineering and uncoupled just-in-time gene expression, novel molecular tools have to be developed (58, 59). Here, alternative C. glutamicum expression systems based on anhydrotetracycline, arabinose, or propionate might be interesting targets for future light-controlled expression setups (60–62).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Federal Ministry of Education and Research (OptoSys, FKZ 031A167) and the Ministry of Innovation, Science and Research of North-Rhine Westphalia and German Research Foundation (INST 208/654-1 FUGG).
We thank Nadine Katzke and Katrin Troost for advice on the initial GC measurements and terpenoid biosynthesis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01457-16.
REFERENCES
- 1.Park SH, Kim HU, Kim TY, Park JS, Kim S-S, Lee SY. 2014. Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun 5:4618. doi: 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- 2.Mahr R, Gätgens C, Gätgens J, Polen T, Kalinowski J, Frunzke J. 2015. Biosensor-driven adaptive laboratory evolution of l-valine production in Corynebacterium glutamicum. Metab Eng 32:184–194. doi: 10.1016/j.ymben.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Wendisch VF. 2014. Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol 30:51–58. doi: 10.1016/j.copbio.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Eggeling L, Bott M. 2015. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol 99:3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- 5.Jensen JVK, Wendisch VF. 2013. Ornithine cyclodeaminase-based proline production by Corynebacterium glutamicum. Microb Cell Fact 12:63. doi: 10.1186/1475-2859-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niimi S, Suzuki N, Inui M, Yukawa H. 2011. Metabolic engineering of 1,2-propanediol pathways in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:1721–1729. doi: 10.1007/s00253-011-3190-x. [DOI] [PubMed] [Google Scholar]
- 8.Siebert D, Wendisch VF. 2015. Metabolic pathway engineering for production of 1,2-propanediol and 1-propanol by Corynebacterium glutamicum. Biotechnol Biofuels 8:91. doi: 10.1186/s13068-015-0269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jojima T, Noburyu R, Sasaki M, Tajima T, Suda M, Yukawa H, Inui M. 2015. Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl Microbiol Biotechnol 99:1165–1172. doi: 10.1007/s00253-014-6223-4. [DOI] [PubMed] [Google Scholar]
- 10.Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. 2013. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol 6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahoor A, Otten A, Wendisch VF. 2014. Metabolic engineering of Corynebacterium glutamicum for glycolate production. J Biotechnol 192:366–375. [DOI] [PubMed] [Google Scholar]
- 13.Otten A, Brocker M, Bott M. 2015. Metabolic engineering of Corynebacterium glutamicum for the production of itaconate. Metab Eng 30:156–165. doi: 10.1016/j.ymben.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Kind S, Jeong WK, Schröder H, Wittmann C. 2010. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab Eng 12:341–351. doi: 10.1016/j.ymben.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Schneider J, Wendisch VF. 2010. Putrescine production by engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol 88:859–868. doi: 10.1007/s00253-010-2778-x. [DOI] [PubMed] [Google Scholar]
- 16.Heider SAE, Peters-Wendisch P, Wendisch VF. 2012. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol 12:198. doi: 10.1186/1471-2180-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heider SAE, Peters-Wendisch P, Netzer R, Stafnes M, Brautaset T, Wendisch VF. 2013. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:1223–1235. [DOI] [PubMed] [Google Scholar]
- 18.Heider SAE, Wolf N, Hofemeier A, Peters-Wendisch P, Wendisch VF. 2014. Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front Bioeng Biotechnol 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grünberger A, Wiechert W, Kohlheyer D. 2014. Single-cell microfluidics: opportunity for bioprocess development. Curr Opin Biotechnol 29:15–23. doi: 10.1016/j.copbio.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Mustafi N, Grünberger A, Kohlheyer D, Bott M, Frunzke J. 2012. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng 14:449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Mustafi N, Grünberger A, Mahr R, Helfrich S, Nöh K, Blombach B, Kohlheyer D, Frunzke J. 2014. Application of a genetically encoded biosensor for live cell imaging of l-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS One 9:e85731. doi: 10.1371/journal.pone.0085731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 23.Eldar A, Elowitz MB. 2010. Functional roles for noise in genetic circuits. Nature 467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drepper T, Krauss U, Meyer zu Berstenhorst S, Pietruszka J, Jaeger K-E. 2011. Lights on and action! Controlling microbial gene expression by light. Appl Microbiol Biotechnol 90:23–40. doi: 10.1007/s00253-011-3141-6. [DOI] [PubMed] [Google Scholar]
- 25.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A. 2012. From dusk till dawn: one-plasmid systems for light-regulated gene expression. J Mol Biol 416:534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Tabor JJ, Levskaya A, Voigt CA. 2011. Multichromatic control of gene expression in Escherichia coli. J Mol Biol 405:315–324. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binder D, Grünberger A, Loeschcke A, Probst C, Bier C, Pietruszka J, Wiechert W, Kohlheyer D, Jaeger K-E, Drepper T. 2014. Light-responsive control of bacterial gene expression: precise triggering of the lac promoter activity using photocaged IPTG. Integr Biol (Camb) 6:755–765. doi: 10.1039/C4IB00027G. [DOI] [PubMed] [Google Scholar]
- 28.Binder D, Bier C, Grünberger A, Drobietz D, Hage-Hülsmann J, Wandrey G, Büchs J, Kohlheyer D, Loeschcke A, Wiechert W, Jaeger K-E, Pietruszka J, Drepper T. 2016. Photocaged arabinose–a novel optogenetic switch for rapid and gradual control of microbial gene expression. Chembiochem 17:296–299. doi: 10.1002/cbic.201500609. [DOI] [PubMed] [Google Scholar]
- 29.Deiters A. 2009. Light activation as a method of regulating and studying gene expression. Curr Opin Chem Biol 13:678–686. doi: 10.1016/j.cbpa.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. 2012. Light-controlled tools. Angew Chem Int Ed Engl 51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- 31.Pátek M, Nesvera J, Guyonvarch A, Reyes O, Leblon G. 2003. Promoters of Corynebacterium glutamicum. J Biotechnol 104:311–323. doi: 10.1016/S0168-1656(03)00155-X. [DOI] [PubMed] [Google Scholar]
- 32.Hentschel E, Will C, Mustafi N, Burkovski A, Rehm N, Frunzke J. 2013. Destabilized eYFP variants for dynamic gene expression studies in Corynebacterium glutamicum. Microb Biotechnol 6:196–201. doi: 10.1111/j.1751-7915.2012.00360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binder S, Schendzielorz G, Stäbler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. 2012. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol 13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frohwitter J, Heider SAE, Peters-Wendisch P, Beekwilder J, Wendisch VF. 2014. Production of the sesquiterpene (+)-valencene by metabolically engineered Corynebacterium glutamicum. J Biotechnol 191:205–213. doi: 10.1016/j.jbiotec.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Beekwilder J, van Houwelingen A, Cankar K, van Dijk ADJ, de Jong RM, Stoopen G, Bouwmeester H, Achkar J, Sonke T, Bosch D. 2014. Valencene synthase from the heartwood of Nootka cypress (Callitropsis nootkatensis) for biotechnological production of valencene. Plant Biotechnol J 12:174–182. doi: 10.1111/pbi.12124. [DOI] [PubMed] [Google Scholar]
- 36.Fraatz MA, Berger RG, Zorn H. 2009. Nootkatone–a biotechnological challenge. Appl Microbiol Biotechnol 83:35–41. doi: 10.1007/s00253-009-1968-x. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 39.Keilhauer C, Eggeling L, Sahm H. 1993. Isoleucine Synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175:5595–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unthan S, Grünberger A, van Ooyen J, Gätgens J, Heinrich J, Paczia N, Wiechert W, Kohlheyer D, Noack S. 2013. Beyond growth rate 0.6: what drives Corynebacterium glutamicum to higher growth rates in defined medium. Biotechnol Bioeng 111:359–371. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 42.Abe S, Takayama K-I, Kinoshita S. 1967. Taxonomical studies on glutamicum acid-producing bacteria. J Gen Appl Microbiol 13:279–301. doi: 10.2323/jgam.13.279. [DOI] [Google Scholar]
- 43.Neumeyer A, Hübschmann T, Müller S, Frunzke J. 2013. Monitoring of population dynamics of Corynebacterium glutamicum by multiparameter flow cytometry. Microb Biotechnol 6:157–167. doi: 10.1111/1751-7915.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eikmanns BJ, Thum-Schmitz N, Eggeling L, Lüdtke KU, Sahm H. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817–1828. doi: 10.1099/13500872-140-8-1817. [DOI] [PubMed] [Google Scholar]
- 45.Peters-Wendisch PG, Schiel B, Wendisch VF, Katsoulidis E, Möckel B, Sahm H, Eikmanns BJ. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J Mol Microbiol Biotechnol 3:295–300. [PubMed] [Google Scholar]
- 46.Stansen C, Uy D, Delaunay S, Eggeling L, Goergen J-L, Wendisch VF. 2005. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol 71:5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93–98. doi: 10.1016/0378-1119(91)90545-M. [DOI] [PubMed] [Google Scholar]
- 49.Young DD, Deiters A. 2007. Photochemical activation of protein expression in bacterial cells. Angew Chem Int Ed Engl 46:4290–4292. doi: 10.1002/anie.200700057. [DOI] [PubMed] [Google Scholar]
- 50.Kortmann M, Kuhl V, Klaffl S, Bott M. 2014. A chromosomally encoded T7 RNA polymerase-dependent gene expression system for Corynebacterium glutamicum: construction and comparative evaluation at the single-cell level. Microb Biotechnol 8:253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Funke M, Diederichs S, Kensy F, Müller C, Büchs J. 2009. The baffled microtiter plate: increased oxygen transfer and improved online monitoring in small scale fermentations. Biotechnol Bioeng 103:1118–1128. doi: 10.1002/bit.22341. [DOI] [PubMed] [Google Scholar]
- 52.Schlepütz T, Büchs J. 2014. Scale-down of vinegar production into microtiter plates using a custom-made lid. J Biosci Bioeng 117:485–496. doi: 10.1016/j.jbiosc.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 53.Wandrey G, Bier C, Binder D, Hoffmann K, Jaeger K-E, Pietruszka J, Drepper T, Büchs J. 2016. Light-induced gene expression with photocaged IPTG for induction profiling in a high-throughput screening system. Microb Cell Fact 15:63. doi: 10.1186/s12934-016-0461-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scholtmeijer K, Cankar K, Beekwilder J, Wösten HA, Lugones BLG, Bosch D. 2014. Production of (+)-valencene in the mushroom-forming fungus S. commune. Appl Microbiol Biotechnol 98:5059–5068. doi: 10.1007/s00253-014-5581-2. [DOI] [PubMed] [Google Scholar]
- 55.Wriessnegger T, Augustin P, Engleder M, Leitner E, Müller M, Kaluzna I, Schürmann M, Mink D, Zellnig G, Schwab H, Pichler H. 2014. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab Eng 24:18–29. doi: 10.1016/j.ymben.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Velema WA, van der Berg JP, Szymanski W, Driessen AJM, Feringa BL. 2014. Orthogonal control of antibacterial activity with light. ACS Chem Biol 9:1969–1974. doi: 10.1021/cb500313f. [DOI] [PubMed] [Google Scholar]
- 57.Hansen MJ, Velema WA, Lerch MM, Szymanski W, Feringa BL. 2015. Wavelength-selective cleavage of photoprotecting groups: strategies and applications in dynamic systems. Chem Soc Rev 44:3358–3377. doi: 10.1039/C5CS00118H. [DOI] [PubMed] [Google Scholar]
- 58.Medema MH, Breitling R, Bovenberg R, Takano E. 2011. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol 9:131–137. doi: 10.1038/nrmicro2478. [DOI] [PubMed] [Google Scholar]
- 59.Wendisch VF, Jorge JMP, Pérez-García F, Sgobba E. 2016. Updates on industrial production of amino acids using Corynebacterium glutamicum. World J Microbiol Biotechnol 32:105. doi: 10.1007/s11274-016-2060-1. [DOI] [PubMed] [Google Scholar]
- 60.Lausberg F, Chattopadhyay AR, Heyer A, Eggeling L, Freudl R. 2012. A tetracycline inducible expression vector for Corynebacterium glutamicum allowing tightly regulable gene expression. Plasmid 68:142–147. doi: 10.1016/j.plasmid.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Shang X, Lai S, Zhang G, Liang Y, Wen T. 2012. Development and application of an arabinose-inducible expression system by facilitating inducer uptake in Corynebacterium glutamicum. Appl Environ Microbiol 78:5831–5838. doi: 10.1128/AEM.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plassmeier JK, Busche T, Molck S, Persicke M, Pühler A, Rückert C, Kalinowski J. 2013. A propionate-inducible expression system based on the Corynebacterium glutamicum prpD2 promoter and PrpR activator and its application for the redirection of amino acid biosynthesis pathways. J Biotechnol 163:225–232. doi: 10.1016/j.jbiotec.2012.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.