ABSTRACT
The Streptococcus mutans lrgAB and cidAB operons have been previously described as a potential model system to dissect the complexity of biofilm development and virulence of S. mutans. Herein, we have attempted to further characterize the Cid/Lrg system by focusing on CidB, which has been shown to be critical for the ability of S. mutans to survive and persist in a nonpreferred oxygen-enriched condition. We have found that the expression level of cidB is critical to oxidative stress tolerance of S. mutans, most likely by impacting lrg expression. Intriguingly, the impaired aerobic growth phenotype of the cidB mutant could be restored by the additional loss of either CidA or LrgA. Growth-dependent expression of cid and lrg was demonstrated to be tightly under the control of both CcpA and the VicKR two-component system (TCS), regulators known to play an essential role in controlling major catabolic pathways and cell envelope homeostasis, respectively. RNA sequencing (RNA-Seq) analysis revealed that mutation of cidB resulted in global gene expression changes, comprising major domains of central metabolism and virulence processes, particularly in those involved with oxidative stress resistance. Loss of CidB also significantly changed the expression of genes related to genomic islands (GI) TnSmu1 and TnSmu2, the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas system, and toxin-antitoxin (T/A) modules. Taken together, these data show that CidB impinges on the stress response, as well as the fundamental cellular physiology of S. mutans, and further suggest a potential link between Cid/Lrg-mediated cellular processes, S. mutans pathogenicity, and possible programmed growth arrest and cell death mechanisms.
IMPORTANCE The ability of Streptococcus mutans to survive a variety of harmful or stressful conditions and to emerge as a numerically significant member of stable oral biofilm communities are essential elements for its persistence and cariogenicity. In this study, the homologous cidAB and lrgAB operons, previously identified as being highly balanced and coordinated during S. mutans aerobic growth, were further characterized through the functional and transcriptomic analysis of CidB. Precise control of CidB levels is shown to impact the expression of lrg, oxidative stress tolerance, major metabolic domains, and the molecular modules linked to cell death and lysis. This study advances our understanding of the Cid/Lrg system as a key player in the integration of complex environmental signals (such as oxidative stress) into the regulatory networks that modulate S. mutans virulence and cell homeostasis.
INTRODUCTION
Streptococcus mutans, a significant constituent of cariogenic oral biofilms, is capable of withstanding a variety of stressors encountered in the oral cavity. The ability of this bacterium to efficiently and rapidly adjust to the dynamic oral environment is essential for its pathogenic lifestyle. Oxidative stress is one of the most important environmental variables affecting the pathogenic potential of S. mutans, as high oxygen concentrations disfavor growth of S. mutans and other oral bacteria (1–4). Therefore, in order to fulfill its role as a major component of cariogenic oral biofilms and to sustain its virulence, S. mutans must overcome oxidative stress. Oxidation sensing and response constitute a complex yet highly regulated process, particularly because it is cross-regulated with other metabolic pathways, stress responses, and virulence physiology (4, 5). A more complete appreciation of how S. mutans survives and persists in a suboptimal aerobic environment is critical to understanding the pathogenicity of cariogenic biofilms and ultimately to uncovering novel therapeutic targets. In this respect, the two paralogously related dicistronic operons, lrgAB (SMU_575c-SMU_574c) and cidAB (SMU_1701c-SMU_1700c) have been previously identified to be highly regulated and finely tuned when S. mutans grows with aeration (6). The cid and lrg operons encode predicted membrane-associated proteins CidA, CidB, LrgA, and LrgB, and their single or combinatory mutations affect a comprehensive “hit list” of virulence traits, such as autolysis, biofilm development, and the oxidative stress response (6). Specifically, when cultured in an aerobic incubator on agar plates, S. mutans ΔlrgAB, ΔcidAB, and ΔcidB mutants were almost completely inhibited in terms of growth (6), while ΔcidA, ΔlrgA, and ΔlrgB mutants showed no inhibition compared with the wild-type strain. Additionally, expression levels of lrg and cid have been shown to be counterbalanced throughout the growth cycle and in response to the availability of oxygen and glucose, environmental stimuli that have a profound effect on plaque biofilm development (6). This opposing pattern of lrg and cid expression was also previously demonstrated in microarray data of S. mutans cells exposed to blood plasma, whereby lrg expression was upregulated 17-fold and cid expression was downregulated 3-fold (7). These observations imply that the lrg and cid operons may operate as a tightly regulated system, possibly in response to nonpreferred environmental stimuli.
Another important facet of the Cid/Lrg system is the potential ability of these proteins to mediate cell death or lysis in a programmed fashion (8, 9). This is analogous to apoptosis (programmed cell death [PCD]), which occurs during development of multicellular organisms. PCD would be beneficial to the cell population within the biofilm by (i) eliminating distinct subpopulations of cells as part of the “normal” course of biofilm development and/or (ii) as an altruistic response to eliminate bacterial cells damaged by environmental or antibiotic stresses (8). Further support comes from the predicted structural similarities between CidA/LrgA and the bacteriophage-encoded holin family of proteins (6, 10–12). Holins are small membrane proteins that induce formation of holes in the cytoplasmic membrane, which allow for cell leakage and access of phage endolysins to the cell wall, which is rapidly cleaved, leading to cell lysis. Antiholins function to inhibit and finely tune the timing of holin action. Although it has been proposed that the Cid/Lrg system mediates cell death in a holin-like manner (13–15), the molecular details of how Cid and Lrg function to control cell death and lysis, as well as many fundamental questions about the Cid/Lrg system, have not yet been completely elucidated. Our previous studies have shown that the S. mutans LrgA/B and CidA/B proteins do have a substantial influence on properties of this organism that would affect its ability to colonize and persist in dental plaque biofilm. In fact, our previous analysis of cid and lrg mutant phenotypes in S. mutans suggests a complex interplay between the gene products of these operons (6, 16). For example, lrgA and lrgB mutations had opposing effects on S. mutans autolysis and sucrose-dependent biofilm, whereas lrgAB, cidB, and cidAB mutants each displayed severe aerobic growth inhibition (6). These data have led us to hypothesize that Cid/Lrg may function as a potentially redundant system in S. mutans, which contributes to the integration of complex environmental signals such as oxidative stress into the regulatory networks that modulate virulence and homeostasis. The present study further characterizes the S. mutans Cid/Lrg system, particularly by providing additional evidence for a potential functional link between cid and lrg, as well as for the central role of CidB in the ability of S. mutans to respond to excessive oxygen. In order to identify Cid/Lrg-associated mechanisms and pathways, we also analyzed the global impact of CidB on the transcriptomes of S. mutans cells, grown anaerobically and aerobically, using RNA sequencing (RNA-Seq). Bioinformatics analysis of functional genes of interest provided further insight into how the Cid/Lrg system may contribute to the cellular physiology and virulence of S. mutans. Overall, the data obtained herein address the question of how the Cid/Lrg system is integrated into the strategy of S. mutans to survive and persist under nonpreferred oxidative stress conditions.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. All S. mutans strains were grown in brain heart infusion (BHI) broth (Difco) as static cultures for overnight cultures or on BHI agar plates at 37°C in a 5% (vol/vol) CO2 atmosphere. For selection of antibiotic-resistant colonies after genetic transformation, erythromycin (10 μg ml−1), kanamycin (1 mg ml−1), or spectinomycin (1 mg ml−1) was added to the medium, when needed. For aerobic growth assays, overnight cultures were diluted 1:100 in fresh medium and the optical density at 600 nm (OD600) was monitored using a Bioscreen C lab system (Helsinki, Finland) as detailed elsewhere (3). Plain BHI broth (without antibiotics) was used in growth experiments in order to avoid additional stress. To achieve an anaerobic condition, sterile mineral oil (50 μl per well) was additionally placed on top of the cultures. For RNA-Seq experiments, S. mutans cells were cultured in three biological replicates. For aerobic growth, an overnight culture was diluted 1:50 into a 250-ml conical flask containing 50 ml of BHI broth and cultures were grown on a rotary shaker (110 rpm) at 37°C. For anaerobic growth, cultures were incubated without agitation in a BBL GasPak Plus anaerobic system (BD, Franklin Lakes, NJ), as described previously (1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| UA159 | Wild type | |
| ΔcidA mutant | ΔcidA::npKm | 6 |
| ΔcidB mutant | ΔcidB::npKm | 6 |
| ΔcidAB mutant | ΔcidAB::npKm | This study |
| ΔcidA::ΩKm mutant | ΔcidA::ΩKm | 6 |
| ΔlrgA mutant | ΔlrgA::npSp | 6 |
| ΔlrgB mutant | ΔlrgB::npEm | 6 |
| ΔlrgAB mutant | ΔlrgAB::ΩKm | 6 |
| ΔlrgA ΔcidB mutant | ΔlrgA::npSp ΔcidB::npKm | This study |
| ΔlrgB ΔcidB mutant | ΔlrgB::npEm ΔcidB::npKm | This study |
| ccpA-deficient mutant | ΔccpA::ΩKm | 29 |
| vicK-deficient mutant | ΔvicK::npKm | 3 |
| UA159 ΩPldh-cidAB | Pldh-cidAB integrated into the chromosome of UA159 (strain constitutively expressing cidAB) | This study |
| UA159 ΩPldh-lrgAB | Pldh-cidAB integrated into the chromosome of UA159 (strain constitutively expressing lrgAB) | This study |
| 184-cidAB/UA159 | UA159 carrying pIB184::cidAB (cidAB-overexpressing strain) | This study |
| 184-cidA/UA159 | UA159 carrying pIB184::cidB (cidA-overexpressing strain) | This study |
| 184-lrgAB/UA159 | UA159 carrying pIB184::lrgAB (lrgAB-overexpressing strain) | This study |
| 184-lrgA/UA159 | UA159 carrying pIB184::lrgA (lrgA-overexpressing strain) | This study |
| Plasmid | ||
| pIB184 | Shuttle expression plasmid with constitutive P23 promoter, Em | 20 |
np, nonpolar; p, polar; Km, kanamycin resistant; Em, erythromycin resistant; Sp, spectinomycin resistant.
Construction of S. mutans mutant strains.
Standard DNA manipulation techniques were used to engineer plasmids and strains (17, 18). S. mutans mutants were created using a PCR ligation mutagenesis approach (19) to replace nearly all of the target open reading frame (ORF) with a nonpolar resistance cassette of erythromycin (Em), kanamycin (Km), or spectinomycin (Sp). Transformants were selected on BHI agar containing appropriate antibiotics. Double-crossover recombination into each gene was confirmed by PCR and sequencing to ensure that no mutations were introduced into flanking genes. For construction of strains constitutively expressing either cid or lrg, we first generated a fragment (ΩKm-Pldh) containing a polar kanamycin resistance gene (ΩKm) and an ldh promoter region (Pldh), replacing the promoter region of either cid (Pcid) or lrg (Plrg). Briefly, two ∼0.5-kb fragments flanking the −35 and −10 sequences of the cidA or lrgA promoter were amplified by PCR, ligated into the ΩKm-Pldh cassette, and used to transform S. mutans. For construction of overexpression strains, the genes of interest were amplified from UA159 and cloned into a shuttle expression plasmid, pIB184 (20), in which the gene is driven by a constitutive P23 promoter. All S. mutans stock cultures were maintained at −80°C in 50% glycerol. For each experiment, UA159 and isogenic mutants were streaked on BHI agar plates, with supplementation of appropriate antibiotics. Agar plates were grown for 48 h at 37°C and 5% CO2 prior to subculturing of individual colonies in BHI broth.
RNA-Seq.
Total RNA was isolated and purified from wild-type strain UA159 and ΔcidB cell cultures, grown aerobically and anaerobically in BHI broth, and harvested in mid-exponential phase (OD600 = 0.5). To remove 16S and 23S rRNAs from each sample, 10 μg of high-quality total RNA (A260/280 > 2.0) was processed twice using the MICROBExpress bacterial mRNA enrichment kit (Ambion, Life Technologies, Grand Island, NY, USA), followed by ethanol precipitation and resuspension in 25 μl of nuclease-free water. cDNA libraries were created from the enriched mRNA samples by using the NEBNext Ultra directional RNA library prep kit for Illumina and NEBNext multiplex oligonucleotides for Illumina (New England BioLabs, Ipswich, MA), according to the instructions from the supplier. The concentration and final quality of cDNA libraries were analyzed on an Agilent TapeStation (Agilent Technologies, Santa Clara, CA, USA). Deep sequencing was performed using the Illumina NextSeq500 platform in single-read format by the NextGen DNA Sequencing Core Laboratory, ICBR, at the University of Florida (Gainesville, FL). Read mapping was performed on a Galaxy server hosted by the research computing center at the University of Florida, using Bowtie for Illumina (version 1.1.2).
Data analyses.
Mapped reads per gene were then counted from BAM files using htseq-count, as described previously (21). Count-based differential expression analysis of RNA-Seq data was performed with the R package edgeR on RStudio, as described elsewhere (22). Briefly, a table of read counts of all the open reading frames, excluding all rRNA and tRNA genes, was uploaded into RStudio, and then the statistical procedure of edgeR, which is available as packages of the Bioconductor software development project (23), was employed to call differentially expressed genes. To confirm a subset of the differential analysis results obtained using RNA-Seq, conventional quantitative real-time (qRT)-PCR was employed to measure changes in the mRNA level of each ORF as described above. For first-strand cDNA synthesis, 1 μg of total RNA without rRNA depletion treatment was used in the reverse transcription reaction mix for analysis and validation. For further functional analysis of differentially expressed genes, we first conducted gene annotation enrichment analysis using the DAVID bioinformatics resources 6.7 (http://david.abcc.ncifcrf.gov/), which currently searches for over 40 annotation categories, including gene ontology (GO) terms, protein functional domains, and the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway (24, 25). To remove and cluster redundant GO terms from the results, we used REVIGO (26), and the results were presented in a table (see Tables 4 and 5 and Table S2 in the supplemental material).
TABLE 4.
Gene ontology enrichment analysis of the DEGs from anaerobic RNA-Seq of UA159 versus ΔcidB mutant using DAVID and REVIGO toolsa
| Gene category | Term ID and GO | Functional description | EASE score |
|---|---|---|---|
| Upregulated | [GOTERM_CC] GO:0030312 | External encapsulating structure | 2.25E−02 |
| [GOTERM_MF] | |||
| GO:0019842 | Vitamin binding | 6.40E−03 | |
| GO:0043176 | Amine binding | 3.87E−04 | |
| GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process | 6.46E−09 | |
| GO:0048037 | Cofactor binding | 1.20E−04 | |
| GO:0031406 | Carboxylic acid binding | 2.86E−03 | |
| GO:0016597 | Amino acid binding | 3.87E−04 | |
| GO:0031177 | Phosphopantetheine binding | 5.77E−06 | |
| Downregulated | [GOTERM_BP] GO:0055114 | Oxidation reduction | 3.38E−02 |
EASE score, a modified Fisher exact P value; GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; ACP, acyl carrier protein. Bold indicates cluster-representative terms.
TABLE 5.
Gene ontology and KEGG pathway enrichment analysis of the DEGs from aerobic RNA-Seq of UA159 versus ΔcidB mutant using DAVID and REVIGO toolsa
| Gene category | Term ID and GO | Functional description | EASE score |
|---|---|---|---|
| Upregulated | [GOTERM_BP] | ||
| GO:0046352 | Disaccharide catabolic process | 2.17E−06 | |
| GO:0005990 | Lactose catabolic process | 2.31E−05 | |
| GO:0019512 | Lactose catabolic process via tagatose-6-phosphate | 2.31E−05 | |
| GO:0009313 | Oligosaccharide catabolic process | 2.17E−06 | |
| GO:0005988 | Lactose metabolic process | 7.55E−05 | |
| GO:0005984 | Disaccharide metabolic process | 2.26E−05 | |
| GO:0006084 | Acetyl-CoA metabolic process | 1.01E−02 | |
| GO:0009401 | Phosphoenolpyruvate-dependent sugar phosphotransferase system | 2.20E−05 | |
| GO:0006457 | Protein folding | 1.04E−02 | |
| GO:0045333 | Cellular respiration | 3.90E−02 | |
| GO:0006099 | Tricarboxylic acid cycle | 3.90E−02 | |
| GO:0009060 | Aerobic respiration | 3.90E−02 | |
| GO:0030001 | Metal ion transport | 4.82E−03 | |
| GO:0006812 | Cation transport | 4.26E−02 | |
| GO:0051187 | Cofactor catabolic process | 3.90E−02 | |
| GO:0009109 | Coenzyme catabolic process | 3.90E−02 | |
| GO:0046356 | Acetyl-CoA catabolic process | 3.90E−02 | |
| GO:0006811 | Ion transport | 4.24E−02 | |
| GO:0009311 | Oligosaccharide metabolic process | 2.26E−05 | |
| GO:0016052 | Carbohydrate catabolic process | 1.27E−02 | |
| GO:0008643 | Carbohydrate transport | 1.19E−04 | |
| GO:0006814 | Sodium ion transport | 2.06E−02 | |
| GO:0044275 | Cellular carbohydrate catabolic process | 3.62E−03 | |
| [GOTERM_CC] | |||
| GO:0031224 | Intrinsic component of membrane | 3.13E−02 | |
| GO:0016021 | Integral component of membrane | 4.76E−02 | |
| [GOTERM_MF] | |||
| GO:0009024 | Tagatose-6-phosphate kinase activity | 2.03E−02 | |
| GO:0015294 | Solute:cation symporter activity | 3.89E−06 | |
| GO:0005351 | Sugar:proton symporter activity | 7.08E−05 | |
| GO:0005402 | Cation:sugar symporter activity | 7.08E−05 | |
| GO:0015295 | Solute:proton symporter activity | 7.08E−05 | |
| GO:0015293 | Symporter activity | 3.89E−06 | |
| GO:0008982 | Protein-N(PI)-phosphohistidine-sugar phosphotransferase activity | 2.07E−05 | |
| GO:0051119 | Sugar transmembrane transporter activity | 7.08E−05 | |
| [KEGG_PATHWAY] | |||
| smu02060 | Phosphotransferase system (PTS) | 7.78E−06 | |
| smu00020 | Citrate cycle (TCA cycle) | 1.21E−04 | |
| smu00052 | Galactose metabolism | 2.94E−04 | |
| smu00620 | Pyruvate metabolism | 8.86E−04 | |
| smu00051 | Fructose and mannose metabolism | 1.90E−02 | |
| smu00010 | Glycolysis/gluconeogenesis | 3.14E−02 | |
| Downregulated | [GOTERM_BP] | ||
| GO:0008643 | Carbohydrate transport | 6.28E−04 | |
| GO:0009401 | Phosphoenolpyruvate-dependent sugar phosphotransferase system | 3.29E−03 | |
| [KEGG_PATHWAY] | |||
| smu00051 | Fructose and mannose metabolism | 4.95E−04 | |
| smu02060 | Phosphotransferase system (PTS) | 7.54E−04 | |
| smu00520 | Amino sugar and nucleotide sugar metabolism | 9.64E−03 |
EASE score, a modified Fisher exact P value; GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes. Bold indicates cluster-representative terms.
Quantitative real-time PCR (qRT-PCR) analysis.
To measure the expression of genes using qRT-PCR, S. mutans UA159 and its derivatives were grown in BHI broth at 37°C in a 5% (vol/vol) CO2 atmosphere. For validation of the RNA-Seq data, cells were harvested in mid-exponential phase (OD600 = 0.5). To measure the growth-dependent expression of cid and lrg, cells were harvested in early (OD600 = 0.2) and late (OD600 = 0.9) exponential or stationary phase. Extraction of RNA, qRT-PCR, and data analysis were performed as described elsewhere (27). Expression was normalized against an internal standard (16S rRNA).
Statistical analysis.
All assays were performed in triplicate with RNA isolated from three independent biological replicates. Student's t test was used to compare the data for two groups, and data were considered significantly different if the P value was at least ≤0.01.
RESULTS
The cid and lrg operons appear to be transcriptionally cross-regulated.
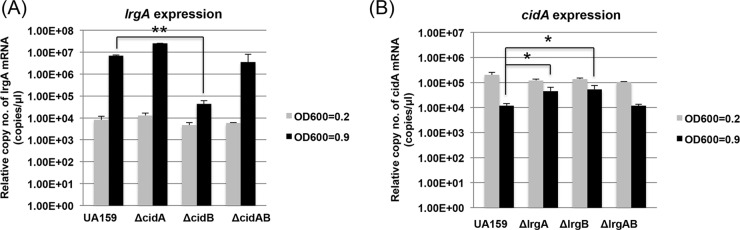
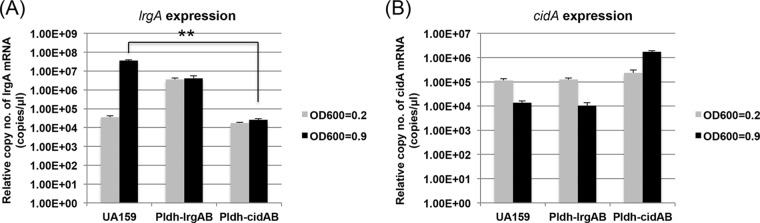
One of the most interesting findings from our previous studies was that the cid and lrg operons respond to various environmental stimuli, including oxidative stress, in opposing manners (6, 7), suggesting a potential functional relationship between cid and lrg. We therefore assumed that understanding the inverse correlation between cid and lrg expression may be important to characterizing the Cid/Lrg system. To further elucidate the interrelated nature of these two operons, we first measured the level of lrg and cid expression in various cid and lrg mutants, respectively, using qRT-PCR, in comparison to that of the wild-type strain. To further clarify the functional interrelatedness of cid and lrg, we also created an additional cidAB-deficient mutant (ΔcidAB::npKm) in which both cidA and cidB were deleted. Given the inverse correlation between lrg and cid expression throughout the growth cycle (6), expression of lrg and cid was measured at both early (OD600 = 0.2) and late (OD600 = 0.9) exponential growth phases of each strain. Interestingly, induction of lrg expression at late exponential phase (typically observed in the wild-type strain) was lost in the ΔcidB mutant but not in the ΔcidA and ΔcidAB mutants (Fig. 1A). As observed in previously published studies (6, 16), growth phase repression of cid expression occurred in the wild-type strain, but this late-exponential-phase repression was modestly lost in the lrgA and lrgB mutants (Fig. 1B). However, expression of cid in the lrgAB mutant resembled that in the wild-type strain (Fig. 1B). Transcriptional perturbation of cid and lrg in the wild-type strain by constitutively expressing either cidAB or lrgAB from the ldh promoter also resulted in altered expression of the native lrg and cid operons. As shown in Fig. 2A, when cidAB was constitutively expressed from the ldh promoter of S. mutans, late-exponential-phase induction of lrg expression was almost completely repressed, while the effect of the constitutive expression of lrgAB on cid expression was minimal (Fig. 2B). Taken together, these results suggest that growth-dependent expression of cid and lrg genes may be transcriptionally linked.
FIG 1.
Transcriptional cross-regulation between lrg and cid operons. The level of lrg or cid expression was measured using qRT-PCR in cid and lrg derivative strains, respectively, in comparison to that of the wild-type strain. (A) The expression of lrgA was measured at early (OD600 = 0.2) and late (OD600 = 0.9) exponential growth phases of UA159 (wild type [WT]) and various cid mutant strains (ΔcidA, ΔcidB, and ΔcidAB). (B) The expression of cidA was measured under the same conditions as those for panel A with UA159 (WT) and various lrg mutant strains (ΔlrgA, ΔlrgB, and ΔlrgAB). Data are averages of three independent biological replicates. Differences in relative gene expression between strains were evaluated for statistical significance by Student's t test. *, P < 0.05; **, P < 0.005.
FIG 2.
Effect of overexpression of lrg or cid on cid or lrg, respectively. The expression of lrgA (A) and cidA (B) was measured at early (OD600 = 0.2) and late (OD600 = 0.9) exponential growth phases of UA159 (WT), UA159 ΩPldh-lrg, and UA159 ΩPldh-cid strains by qRT-PCR. Expression of cid and lrg genes was constitutively driven by the constitutive ldh promoter of S. mutans in UA159 ΩPldh-lrg and UA159 ΩPldh-cid strains, respectively. Data are averages of three independent biological replicates. Differences in relative gene expression between strains were evaluated for statistical significance by Student's t test. **, P < 0.005.
Response of S. mutans to oxygen is tightly regulated by CidB.
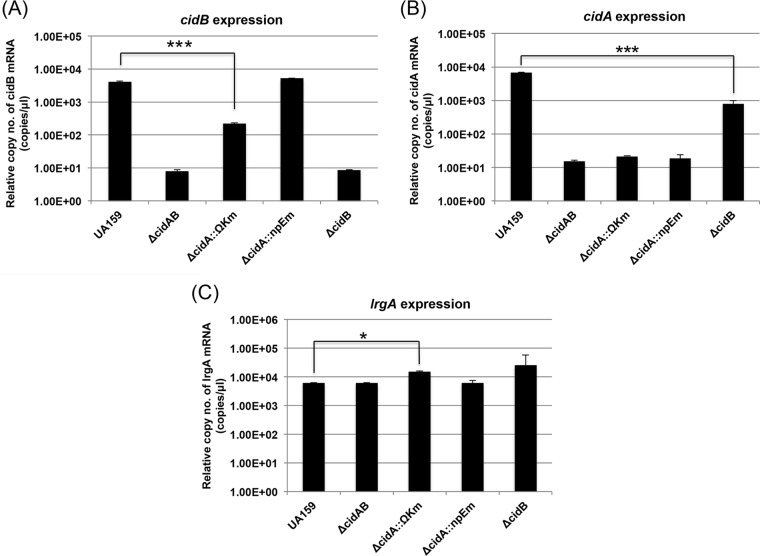
We have previously shown that deficiency of CidAB, or of CidB alone, critically impaired the capability of S. mutans to grow under aerobic or oxidative stress conditions, while cidA deficiency had no discernible effect on aerobic growth (6), highlighting the dominant role of CidB in the cid operon. Interestingly, however, the effect of ΔcidB and ΔcidAB mutations on lrg expression did not follow the same pattern (Fig. 1). Specifically, the cidAB deletion mutant had no impact the expression of lrg, unlike the cidB mutation (Fig. 1A). In fact, the cidAB-deficient mutant (ΔcidAB::npKm) used above was different from that (ΔcidA::ΩKm) used in the previous study (6). Thus, to resolve this issue, we next evaluated the read-through of the terminator in the ΩKm element by measuring the expression of the cidB gene downstream of the polar marker (ΩKm) that replaced cidA in the ΔcidA::ΩKm mutant (6). As shown Fig. 3A, qRT-PCR analysis revealed a 1.2-log reduction in cidB expression in the cidA polar mutant (ΔcidA::ΩKm), whereas a 2.7-log reduction in cidB expression was observed in both the ΔcidB and ΔcidAB mutants, relative to that of the wild-type strain. When cidA was replaced by a nonpolar erythromycin marker (ΔcidA::npEm), cidB expression was identical to that of the wild-type strain. These results suggest that although cidB expression was substantially reduced by the ΩKm terminator in the ΔcidA::ΩKm strain, the read-through might be sufficient to impact the dissection of phenotypes specific to cidA and cidB. Measurement of cidA expression in each of the cid derivative strains confirmed loss of cidA in the ΔcidAB, ΔcidA::ΩKm, and ΔcidA::npEm mutants (Fig. 3B). Intriguingly, cidA expression was also reduced by about 1 log in the ΔcidB mutant (Fig. 3B), suggesting possible autoregulation within the cid operon for a balance between cidA and cidB. More interestingly, either a decrease or a lack of cidB expression in the ΔcidA::ΩKm and ΔcidB mutants, respectively, could upregulate expression of lrg (Fig. 3C). Although the changes in lrg expression were subtle and not statistically significant in the ΔcidB mutant, this result further supports the potential transcriptional linkage between the cid and lrg operons. Next, to determine whether the differences between the two cidAB-deficient strains (ΔcidA::ΩKm and ΔcidAB::npKm) influence the ability of S. mutans in coping with oxidative stress, we reevaluated aerobic growth of these two different cidAB-deficient mutants and other lrg- or cid-deficient strains in BHI liquid medium, as described previously (6). As reported previously, both ΔcidB and ΔcidA::ΩKm mutants were defective in aerobic growth (Fig. 4A) (6). Interestingly, however, when both cidA and cidB were completely deleted (ΔcidAB), growth of this mutant was identical to that of the wild-type strain (Fig. 4A). Furthermore, mutation of either lrgA or lrgB alone had no effect on aerobic growth of S. mutans, but deficiency in both lrgA and lrgB critically impaired aerobic growth (Fig. 4B), as described previously (6). These results suggest that additional loss of CidA may reverse the effect of a cidB mutation or, alternatively, that CidA may be functionally connected with CidB.
FIG 3.
Gene expression determined by qRT-PCR in the cid derivative strains. Strains were grown in BHI broth at 37°C in a 5% (vol/vol) CO2 atmosphere, and cells were harvested in mid-exponential phase (OD600 = 0.5). Expression levels of cidB (A), cidA (B), and lrgA (C) were measured and normalized against an internal standard (16S rRNA). Further experimental details are described in Materials and Methods. Data are averages of three independent biological replicates. Differences in relative gene expression between strains were evaluated for statistical significance by Student's t test. *, P < 0.05; ***, P < 0.001.
FIG 4.
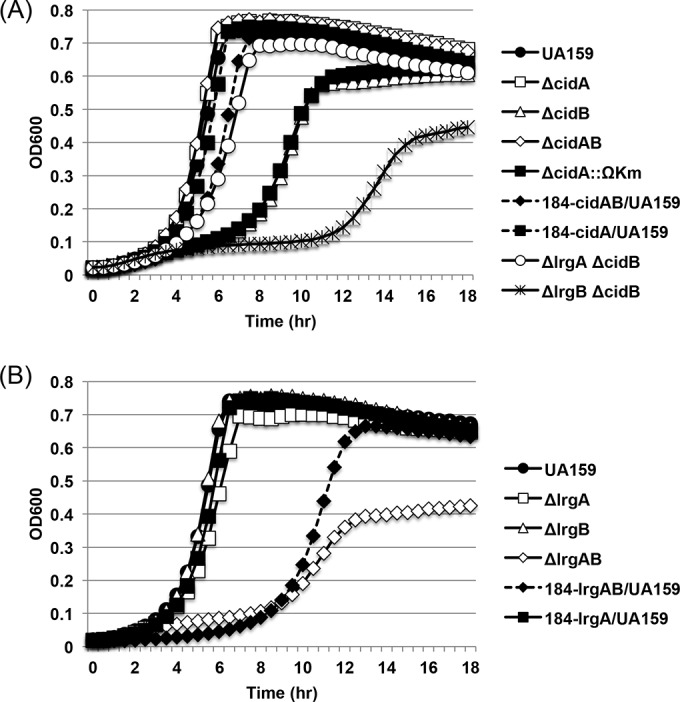
Aerobic growth curves of S. mutans UA159 (wild type) and cid derivatives (A) or lrg derivatives (B). Strains were grown in plain BHI medium. Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C lab system. To achieve relatively aerobic conditions, no sterile mineral oil was placed on top of the cultures. The results are representative of three independent experiments, each performed in triplicate. (A) UA159, wild type; ΔcidA, nonpolar cidA mutant; ΔcidB, nonpolar cidB mutant; ΔcidAB, nonpolar cidAB mutant; ΔcidA::ΩKm, polar cidA mutant; 184-cidAB/UA159, cidAB overexpression strain in the wild-type background; 184-cidA/UA159, cidA overexpression strain in the wild-type background; ΔlrgA ΔcidB, lrgA cidB double nonpolar mutant; and ΔlrgB ΔcidB, lrgB cidB double nonpolar mutant. (B) UA159, wild type; ΔlrgA, nonpolar lrgA mutant; ΔlrgB, nonpolar lrgB mutant; ΔlrgAB, nonpolar lrgAB mutant; 184-lrgAB/UA159, lrgAB overexpression strain in the wild-type background; and 184-lrgA/UA159, lrgA overexpression strain in the wild-type background.
A potential functional linkage of CidB and LrgA.
Given the potential functional relationship between the cid and lrg operons, we next addressed whether the cidB mutation could be influenced by mutation of lrg. For this, we additionally mutated either lrgA (ΔlrgA ΔcidB) or lrgB (ΔlrgB ΔcidB) in the cidB background by replacement with nonpolar Sp or Em resistance cassettes, respectively, and then monitored the ability of these mutants to grow under aerobic conditions. Intriguingly, inhibition of aerobic growth by the cidB mutation could be substantially rescued by additional mutation of lrgA but not as much as when cidA was mutated (Fig. 4A). In contrast, mutation of both cidB and lrgB inhibited aerobic growth of S. mutans even more than the cidB mutation alone. Therefore, these results suggest that the function of “B” components (CidB and LrgB) might be functionally checked by the “A” components (CidA and LrgA). Although LrgB was not required for aerobic growth of S. mutans (Fig. 4A), it was previously shown to affect its growth in the presence of paraquat, a superoxide anion-generating agent (6).
Another possible scenario is that unbalanced expression between the “A” and “B” components of each operon may be toxic under aerobic growth conditions. This interpretation is in line with the previously published observation that S. mutans displayed impaired aerobic growth when deficient in either CidAB or CidB alone (6): CidA activity may be “unchecked” in the absence of CidB and may be responsible for this decreased fitness under oxidative stress. To test this, the cidA and lrgA genes were each cloned under the control of a constitutive promoter (P23) into the shuttle vector pIB184 in the wild-type genetic background. However, the uncontrolled overexpression of cidA or lrgA had no discernible effect on the aerobic growth of S. mutans (Fig. 4A and B). We also investigated the effect of overexpression of cidAB (184-cidAB/UA159) and lrgAB (184-lrgAB/UA159) operons in the aerobic growth of the organism. Interestingly, our results showed that overexpression of the lrgAB operon caused severe aerobic growth impairment (Fig. 4B) and a moderate difference was found in the cidAB-overexpressed strain (Fig. 4A). Therefore, these results suggest that either lack or overexpression of lrgAB could be critical to the ability of S. mutans to grow in an aerobic environment. In the cid operon, only overexpression of cidAB seemed to negatively influence the ability of the organism to grow aerobically.
Impact of CidB on the fundamental cellular physiology of S. mutans.
Next, to provide further insights into the role and regulation of the Cid/Lrg system, we employed an RNA-Seq approach to compare the transcriptomes of S. mutans wild-type and cidB mutant strains under both anaerobic and aerobic growth conditions. To validate the RNA-Seq data, qRT-PCR was performed on a subset of the differentially expressed genes using the same total RNAs of UA159 and cidB cells as those used for the RNA-Seq experiment described below. As shown in Table 2, the expression ratio (ΔcidB mutant to UA159) for each gene obtained by qRT-PCR was similar to the RNA-Seq results.
TABLE 2.
qRT-PCR validation of RNA-Seq results
| Gene ID | Gene namea | Expression fold change (ΔcidB mutant/UA159) |
|||
|---|---|---|---|---|---|
| RNA-Seq |
qRT-PCR |
||||
| Anaerobic | Aerobic | Anaerobic | Aerobic | ||
| SMU_1363c | tpn | 24.97 | 22.2 | 45.22 | 18.87 |
| SMU_574c | lrgB | 1.2 | 3.78 | 1.79 | 1.99 |
| SMU_1396 | gbpC | 1.25 | 1.51 | ||
| SMU_1914c | cipB | 0.82 | 0.93 | ||
| SMU_423 | nlmD | 0.74 | 0.82 | ||
| SMU_490 | pflC | 0.46 | 0.55 | ||
| SMU_1752 | NA | 0.4 | 0.5 | ||
| SMU_191 | NA | 0.19 | 0.13 | ||
| SMU_40 | NA | 7.08 | 1.43 | ||
| SMU_1421 | pdhC | 3.55 | 1.92 | ||
| SMU_609 | NA | 3.33 | 1.68 | ||
| SMU_41 | NA | 3.2 | 3.02 | ||
| SMU_82 | dnaK | 2.54 | 1.39 | ||
| SMU_925 | cipI | 0.64 | 0.39 | ||
| SMU_508 | NA | 0.45 | 0.44 | ||
| SMU_940c | patB | 0.44 | 0.38 | ||
NA, not available.
UA159 versus ΔcidB mutant expression profiles during anaerobic growth.
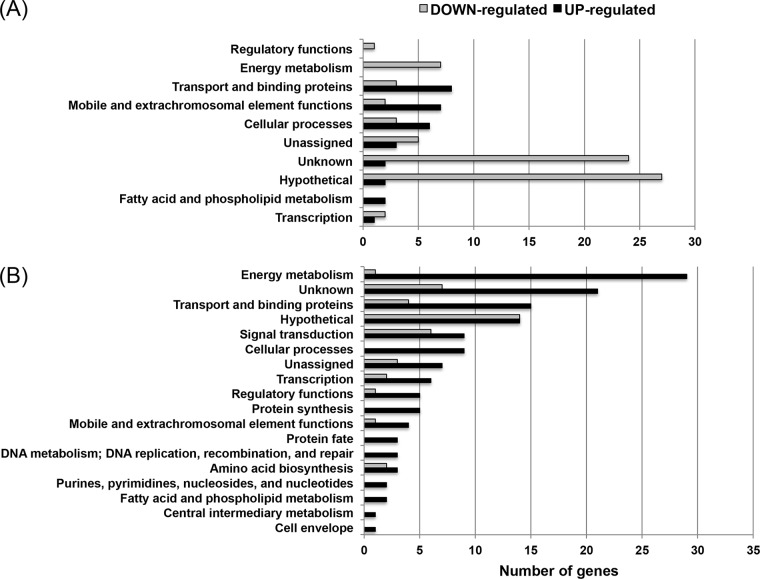
When we compared the transcriptomes of mid-exponential-phase UA159 and ΔcidB mutant anaerobic cultures, loss of CidB significantly affected the expression of 105 genes, and more genes were downregulated (n = 74) than upregulated (n = 31) (cutoff P value of ≤0.005) (Fig. 5A; see Table S1 in the supplemental material). The majority of differentially expressed genes (DEGs) encoded proteins of unknown function or hypothetical proteins (Fig. 5A). Notably, a large number of the DEGs belonged to very few functional groups, including the genomic islands (GIs) TnSmu1 and TnSmu2, the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas system, bacteriocin production, energy metabolism, and amino acid ABC transporters (Table 3). As shown in Table 3, the DEGs were also often structured as operon genes, which would be cotranscribed or coregulated as a single polycistronic mRNA. When the cutoff of ≥1.5-fold change was applied at the same P value, most DEGs belonged to the GIs TnSmu1 and TnSmu2 and CRISPR-Cas, with 11 genes upregulated and 35 genes downregulated (see Table S1). All 25 genes identified within TnSmu1 were 2-fold through 6-fold downregulated in the cidB mutant, compared to the wild-type UA159 strain under the anaerobic growth condition (Table 3). In contrast to TnSmu1 genes, all 21 genes identified within TnSmu2 were 1.2-fold or more upregulated, except for SMU_1367c, which was about 2-fold downregulated. Expression of six genes (SMU_1354c to SMU_1356c, SMU_1360c, and SMU_1361c, ≥4-fold upregulated; SMU_1363c, 25-fold upregulated), encoding putative transposes, hypothetical proteins, and a transcriptional regulator, were also highly upregulated in the cidB mutant during anaerobic growth (Table 3). These RNA-Seq results also revealed that the CRISPR2-associated cas genes, located between SMU_1752 and SMU_1764c, were about 2-fold downregulated in the ΔcidB mutant (Table 3; see Table S1), while the CRISPR1-Cas system, located between SMU_1398 and SMU_1405c, was not changed (data not shown). It is also notable that lrgB (SMU_574c) was 1.2-fold upregulated in the cidB mutant strain (Table 2; see Table S1), confirming the result observed in Fig. 3C and further supporting potential cross-regulation of cid and lrg expression. Furthermore, expression of SMU_1363c, the most highly upregulated gene in the cidB mutant, was measured by qRT-PCR in the other cid derivative strains to determine whether its observed differential expression can be corrected to the wild-type level in the ΔcidAB strain in the same manner as observed for lrg expression in Fig. 1A, 3C, 4A, and 6A. As expected, the SMU_1363c gene was also significantly upregulated in the oxygen-sensitive ΔcidA::ΩKm strain to the level observed in the ΔcidB strain (see Fig. S1 in the supplemental material). Expression of SMU_1363c was also restored to the wild-type levels in the ΔcidAB strain (see Fig. S1). These results suggest that TnSmu2 genes, including SMU_1363c, may participate in CidB-mediated physiological changes.
FIG 5.
Distribution of functions of genes affected by loss of CidB under anaerobic and aerobic conditions. The 105 (74 upregulated; 31 downregulated) and 180 (41 upregulated; 139 downregulated) genes differentially expressed at a P of ≤0.005 under anaerobic (A) or aerobic (B) conditions are grouped by functional classification according to the Los Alamos S. mutans genome database (http://www.oralgen.org/).
TABLE 3.
Interesting gene clusters differentially expressed in UA159 and ΔcidB cells when growing under anaerobic conditionsa
| Functional group and gene ID | Expression fold change (ΔcidB mutant/UA159) | Gene | Description |
|---|---|---|---|
| TnSmu1 | |||
| SMU_191c | 0.20 | NA | Phage-related integrase |
| SMU_193c | 0.15 | NA | Conserved hypothetical protein |
| SMU_194c | 0.21 | NA | Conserved hypothetical protein, phage related |
| SMU_195c | 0.13 | NA | Hypothetical protein |
| SMU_196c | 0.13 | NA | Immunogenic secreted protein (transfer protein) |
| SMU_197c | 0.14 | NA | Hypothetical protein |
| SMU_198c | 0.15 | tpn | Conjugative transposon protein |
| SMU_199c | 0.13 | NA | Hypothetical protein |
| SMU_200c | 0.14 | NA | Hypothetical protein |
| SMU_201c | 0.14 | NA | Conserved hypothetical protein |
| SMU_202c | 0.12 | NA | Conserved hypothetical protein; Streptococcus-specific protein |
| SMU_204c | 0.12 | NA | Hypothetical protein |
| SMU_205c | 0.10 | NA | Conserved hypothetical protein |
| SMU_206c | 0.14 | NA | Hypothetical protein |
| SMU_207c | 0.15 | NA | Transcriptional regulator, Cro/CI family |
| SMU_208c | 0.14 | NA | Conserved hypothetical protein, FtsK/SpoIIIE family |
| SMU_209c | 0.13 | NA | Hypothetical protein |
| SMU_210c | 0.15 | NA | Hypothetical protein |
| SMU_211c | 0.17 | NA | Hypothetical protein |
| SMU_212c | 0.19 | NA | Hypothetical protein |
| SMU_213c | 0.18 | NA | Hypothetical protein |
| SMU_214c | 0.21 | NA | Hypothetical protein |
| SMU_215c | 0.19 | NA | Hypothetical protein |
| SMU_216c | 0.22 | NA | Hypothetical protein |
| SMU_217c | 0.25 | NA | Conserved hypothetical protein; Streptococcus-specific protein |
| TnSmu2 | |||
| SMU_1339 | 1.26 | bacC | Bacitracin synthetase; surfactin synthetase; long-chain fatty acid–CoA ligase |
| SMU_1340 | 1.26 | bacA | Bacitracin synthetase 1/tyrocidin synthetase III |
| SMU_1341c | 1.26 | grs mycB | Gramicidin S synthase/mycosubtilin synthetase chain MycB; long-chain fatty acid–CoA ligase |
| SMU_1342 | 1.25 | bacA | Bacitracin synthetase; long-chain fatty acid–CoA ligase |
| SMU_1343c | 1.25 | pksC pksL | Putative polyketide synthase |
| SMU_1344c | 1.21 | fabD | Malonyl CoA-acyl carrier protein transacylase |
| SMU_1345c | 1.20 | ituA mycA | Peptide synthetase similar to MycA; long-chain fatty acid–CoA ligase |
| SMU_1347c | 1.91 | NA | Permease-ABC-type antimicrobial peptide transport system, permease component |
| SMU_1348c | 1.78 | NA | ABC transporter ATP-binding protein |
| SMU_1351 | 1.24 | NA | Transposase fragment |
| SMU_1353 | 1.33 | NA | Transposase; mobile element protein |
| SMU_1354c | 4.42 | NA | Putative transposase fragment |
| SMU_1355c | 5.18 | tnp | Transposase |
| SMU_1356c | 4.05 | tpn | Putative transposase fragment |
| SMU_1360c | 4.64 | NA | Hypothetical protein |
| SMU_1361c | 5.02 | yjjB | Transcriptional regulator, TetR family |
| SMU_1363c | 24.98 | tpn | Transposase fragment (IS605/IS200-like); mobile element protein |
| SMU_1365c | 2.01 | NA | Permease–FtsX-like permease |
| SMU_1366c | 1.79 | NA | ABC transporter ATP-binding protein |
| SMU_1367c | 0.54 | NA | Conserved hypothetical protein; methyltransferase |
| CRISPR/Cas associated | |||
| SMU_1750c | 0.58 | NA | Hypothetical protein |
| SMU_1752c | 0.40 | NA | Hypothetical protein |
| SMU_1753c | 0.41 | NA | CRISPR-associated protein Cas2 |
| SMU_1754c | 0.45 | NA | CRISPR-associated protein Cas1 |
| SMU_1755c | 0.45 | NA | CRISPR-associated protein Cas1 |
| SMU_1757c | 0.48 | NA | CRISPR-associated protein Cas1 |
| SMU_1758c | 0.47 | NA | CRISPR-associated RecB family exonuclease Cas4b |
| SMU_1760c | 0.49 | NA | CRISPR-associated protein, Csd2 family |
| SMU_1761c | 0.51 | NA | CRISPR-associated protein, Csd1 family |
| SMU_1762c | 0.54 | NA | CRISPR-associated protein, Csd1 family |
| SMU_1763c | 0.52 | NA | CRISPR-associated protein, CT1134 family |
| SMU_1764c | 0.50 | NA | CRISPR-associated helicase Cas3 |
| Bacteriocin associated | |||
| SMU_150 | 0.77 | nlmA | Nonlantibiotic mutacin IV A |
| SMU_151 | 0.74 | nlmB | Nonlantibiotic mutacin IV B |
| SMU_299c | 0.74 | NA | Bacteriocin peptide precursor |
| SMU_423 | 0.75 | nlmD | Possible bacteriocin |
| SMU_1914c | 0.82 | cipB | Possible bacteriocin |
| Amino acid ABC transporters | |||
| SMU_932 | 1.43 | NA | Conserved hypothetical protein |
| SMU_933 | 1.41 | atmA | Amino acid ABC transporter, amino acid substrate-binding protein |
| SMU_934 | 1.48 | NA | Amino acid ABC transporter, permease protein |
| SMU_935 | 1.52 | NA | Amino acid ABC transporter, permease protein |
| SMU_936 | 1.45 | NA | Amino acid ABC transporter, ATP-binding protein |
| Metabolic pathways | |||
| SMU_137 | 0.54 | mleS | Malolactic enzyme |
| SMU_138 | 0.58 | mleP | Malate permease/auxin efflux carrier |
| SMU_139 | 0.58 | oxdC | Oxalate decarboxylase |
| SMU_140 | 0.58 | gor gshR | Glutathione reductase |
| SMU_141 | 0.62 | NA | Conserved hypothetical protein; integral membrane protein |
| SMU_490 | 0.47 | act pflC | Pyruvate-formate lyase activating enzyme |
| SMU_491 | 0.65 | NA | Transcriptional regulator, DeoR family |
| SMU_493 | 0.52 | pfl pfl-2 | Formate acetyltransferase (pyruvate-formate lyase) |
| SMU_494 | 0.55 | mipB talC | Transaldolase family protein |
| SMU_495 | 0.54 | gldA | Glycerol dehydrogenase |
NA, not available; CoA, coenzyme A.
FIG 6.
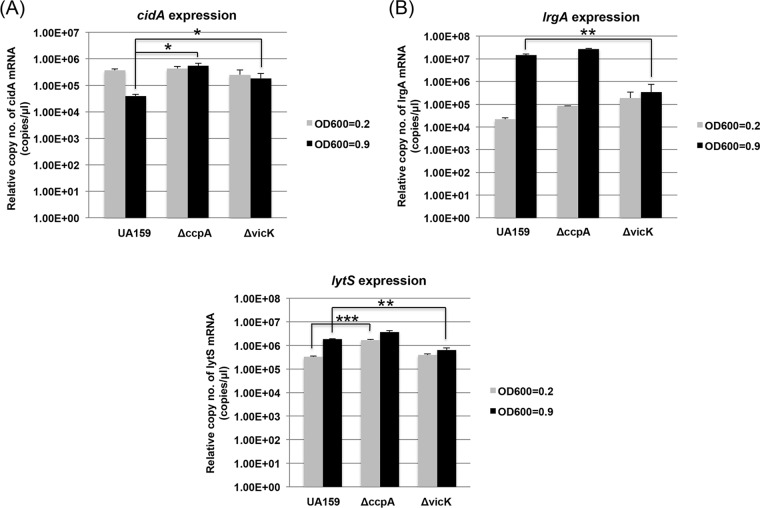
Effect of CcpA and VicK on expression of S. mutans cid, lrg, and lytS. The expression of cidA (A), lrgA (B), and lytS (C) was measured by qRT-PCR at early (OD600 = 0.2) and late (OD600 = 0.9) exponential growth phases of UA159 (WT), ccpA-deficient, and vicK-deficient strains. Data represent the averages of three independent biological replicates. Differences in relative gene expression levels between strains were evaluated for statistical significance by Student's t test. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
In order to gain further insights into the functional activities differentially represented in the absence or presence of CidB during anaerobic growth, we performed enrichment analysis on the DEGs using the DAVID bioinformatics tool (http://david.abcc.ncifcrf.gov/). During anaerobic growth, eight gene ontology (GO) terms, including acyl carrier activity, phosphopantetheine binding, cofactor binding, amine binding, amino acid binding, carboxylic acid binding, vitamin binding, and external encapsulating structure, were significantly (EASE score threshold, <0.05; a modified Fisher exact P value) enriched with 31 upregulated genes (see Table S2 in the supplemental material). Significantly changed protein domain terms are also listed in Table S2. In contrast, only one GO term, associated with oxidation reduction, and four protein domain terms were significantly enriched with 74 downregulated genes (see Table S2). No significant genes had a hit on KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway maps. To better understand and remove redundant GO terms from the results, we used REVIGO (26) and enriched GO terms were clustered with representative terms in Table 4.
UA159 versus ΔcidB mutant transcriptome changes during aerobic growth.
The transcriptomes of the UA159 and ΔcidB strains during aerobic growth were also compared, in an attempt to identify the molecular modules or pathways affected by CidB when cells cope with excessive oxygen. With a cutoff P value of ≤0.005, loss of CidB affected the expression of 159 total genes. In contrast to the DEGs under the anaerobic growth condition, more genes were upregulated (n = 140) than downregulated (n = 19) (Fig. 5B) during aerobic growth. As well, a more diverse group of genes was affected in the cidB mutant under aerobic growth than under the anaerobic growth condition. Notably, the cidB mutation affected a variety of major domains of metabolism, with pronounced changes associated with carbohydrate, amino acid, fatty acid/lipid, nucleotide metabolism, and transport (Fig. 5B; see Table S3 in the supplemental material). Of note, most of the TnSmu1 genes that were downregulated in the cidB mutant during anaerobic growth were upregulated under the aerobic condition, with the exception of SMU_191c and SMU_195c (Table 3; see Table S3). In addition, SMU_493 to SMU_495, encoding pyruvate-formate lyase, transaldolase, and glycerol dehydrogenase, respectively, and SMU_2024, encoding a putative transcriptional regulator, were also upregulated in the cidB mutant during aerobic growth, in contrast to the anaerobic condition (see Tables S1 and S3). This transcriptional “flipping” of gene expression between the anaerobic and aerobic growth conditions may be related to the elevated sensitivity of the ΔcidB mutant to oxidative stress. In contrast, TnSmu2 genes were upregulated in the cidB mutant regardless of the absence or presence of excessive oxygen (Table 3; see Table S3), suggesting an intimate functional linkage between CidB and TnSmu2. Under the aerobic condition, expression of five genes (SMU_101 to SMU_105) in another genomic island, GI-4 (28), was also about 2-fold downregulated in the cidB mutant strain (see Table S3). These genes largely encode the components of a phosphotransferase system (PTS). It is also notable that lrgA (SMU_575) and lrgB (SMU_574) expression levels were more than 3-fold upregulated in the cidB mutant, suggesting that CidB may have a greater impact on lrg expression during aerobic growth. Importantly, the aerobic RNA-Seq data also suggest that potential Cid/Lrg-mediated cell death mechanisms may be linked with the chromosomal toxin-antitoxin (T/A) modules in S. mutans, which are known to inhibit essential bacterial cell functions such as translation or DNA replication. As shown Table S3, two potential toxin-antitoxin (T/A) modules, SMU_173/SMU_172 and SMU_41/SMU_40, were highly upregulated during aerobic growth in the ΔcidB mutant in comparison to the wild type. As expected, mutation of cidB also led to altered expression of several stress-related loci. For example, SMU_80 to SMU_83, encoding HrcA, GrpE, DnaK, and DnaJ, were significantly upregulated (1.9- to 2.9-fold change). SMU_1954 and SMU_1955, encoding GroEL and GroES, were also upregulated more than 1.5-fold. As shown in Table 5, DAVID and REVIGO analysis of the aerobic growth DEGs revealed that 140 upregulated genes belonged to GO terms related to 23 biological processes (BP), 2 cellular components (CC), 8 molecular functions (MF), and 6 KEGG pathways, while two GO (BP) terms and three KEGG pathways were significantly enriched with 19 downregulated genes. Representative GO terms enriched with either upregulated or downregulated genes largely included carbohydrate transport and metabolism (listed in Table 5), suggesting that loss of CidB leads to a global adjustment in major domains of central metabolism when S. mutans copes with oxidative stress.
CcpA and VicKR are significantly involved in the expression of cid and lrg.
We have previously demonstrated that growth-dependent changes in lrgAB expression are regulated by the LytST two-component signal transduction system (TCS), which is located immediately upstream of lrgAB (6). As shown above (Fig. 1A, 2A, and 3C), alteration of cellular CidB levels can also affect expression of lrg. Nevertheless, the genetic regulatory circuits governing cid expression still remain poorly characterized. In an attempt to identify the regulatory mechanisms or pathways that control expression of cid, we decided to further explore the involvement of CcpA, known to serve as a major regulator of carbon metabolism in S. mutans and previously shown to be involved in the expression of cid and lrg by Northern blot analyses (6). We therefore quantified growth-dependent expression of cid and lrg in the wild type and ccpA (29) mutants using real-time qPCR, under the growth same conditions described above (Fig. 1 and 2). In contrast to the wild-type strain, increased expression of cidA at early exponential growth phase (OD600 = 0.2) was maintained through the late exponential growth phase (OD600 = 0.9) in the ccpA mutant (Fig. 6A). However, lrg expression was elevated at the late exponential growth phase (OD600 = 0.9), similar to that of the parent strain UA159. Nevertheless, the overall expression of lrg was moderately increased in the ccpA mutant (Fig. 6B), which might be due to elevated expression of lytST which has also been observed previously in the ccpA mutant (Fig. 6C). These results strongly suggest that expression of cid is regulated in a CcpA-dependent manner, which is in strong agreement with the aerobic RNA-Seq data showing a large number of DEGs involved in carbon/energy metabolic pathways (Table 5; see Table S3 in the supplemental material).
Analysis of the aerobic RNA-Seq data also revealed that expression of multiple genes involved in cell wall biosynthesis and metabolism were highly altered in the cidB mutant (see Table S3). Thus, we hypothesized that the VicKR TCS, well-known to control cell wall metabolism by effectively coordinating peptidoglycan plasticity with the cell division process in many Gram-positive bacteria (30–36), may be involved in regulation of the cid operon, the lrg operon, or both. To test if the cid and lrg genes are under the control of VicKR, we also compared the expression of lrgA and cidA in a vicK mutant (vicK deficient) (3) and wild-type strains in the early (OD600 = 0.2) and late (OD600 = 0.9) exponential phases using qRT-PCR. Interestingly, cid expression in the vicK mutant did not show the decrease in expression at late exponential phase that was noted in the wild-type strain (Fig. 6A). As well, there was no induction of the lrg genes in the vicK mutant as the cells entered late exponential phase (Fig. 6B), and lrg expression was significantly higher in early exponential phase in the vicK mutant than in UA159. The differences in expression patterns of the lrg operon in the vicK mutant may be explained, at least in part, by the observation that lytS operon expression was also altered in the VicK-deficient strain (Fig. 6C). Taken together, these data show that expression and function of cid and lrg are regulated in a more complex manner than previously thought, involving CcpA and at least two TCS (LytST and VicKR).
DISCUSSION
The present study was geared toward understanding the molecular and cellular bases for the contribution of CidB to the ability of S. mutans to cope with oxidative stress, as this may improve our understanding of how the Cid/Lrg system contributes to this pathogen's persistence and survival in the environmentally dynamic oral cavity. The presented comparisons of wild-type and cidB mutant transcriptomes have yielded important clues that will help make potential connections between the cid and lrg operons, resistance to oxidative stress, and possibly regulation of cell death. Overall, these findings also highlight a potential functional interplay between cid and lrg, which was previously implicated by the counterbalanced expression of these genes in response to growth phase, glucose, and oxygen levels (6). Notably, loss of either lrgA (or lrgB) or cidB influenced the expression of cid or lrg, respectively, and constitutive expression of cid strongly repressed the stationary-phase induction of lrg expression. These results suggest that the Cid/Lrg system may be controlled by a potential cross-regulatory mechanism, resulting in a rapid and balanced modulation of Cid and Lrg activities at the transcriptional level once the organism encounters environmental stresses. If this idea is correct, any resulting unbalance between lrg and cid may result in a profound impact on S. mutans cellular functions, including the ability to grow and survive under stress conditions. A more complex picture emerged when we observed that the cidB mutation could be completely or partly restored by additional mutation of either cidA or lrgA, respectively, and mutation of both “B” components of each operon works additively to increase sensitivity to oxygen, further suggesting that the individual “A” and “B” components of CidAB and LrgAB may also be regulated within the system. Nevertheless, it is still unclear why the ΔcidAB and ΔcidA::ΩKm strains display opposite phenotypes. In fact, the phenotypes of the ΔcidA::ΩKm strains are much closer to those of the ΔcidB strains than the ΔcidAB strains in that both ΔcidA::ΩKm and ΔcidB strains have increased sensitivity to oxygen and could induce expression of lrg. Interestingly, we also found that the cidB mutation positively regulated expression of the cid operon, since expression of cidA was significantly decreased in the ΔcidB strain. Thus, either increased expression of cidA or unbalanced expression between cidA and cidB might be crucial to the phenotypic effects of Cid/Lrg. However, no discernible phenotype, including sensitivity to oxygen, was observed in the cidA-overexpressed strain. Although further investigation is needed to mechanistically understand the differences between the ΔcidAB and ΔcidA::ΩKm strains, the data obtained herein strongly suggest that CidB is tightly regulated in the S. mutans Cid/Lrg system. A possible scenario for regulation of cid may be that expression of this operon is regulated by positive-feedback autoregulation of the cid operon or, alternatively, the Cid/Lrg system and that the level of expression of cidB modulates the strength of this potential feedback.
Clearly, the transcriptional linkage between cid and lrg adds another layer of complexity to the regulation that occurs within the Cid/Lrg system. Given that both ΔcidA::ΩKm and ΔcidB strains were able to induce lrg expression, their phenotypes may be, at least in part, attributed to the elevated level of lrg expression. These phenotypes also imply that LrgAB is more active during aerobic growth, which is in line with our previous finding that the lrg operon is more highly expressed during aerobic growth (1, 6). Further considering that either lack or overexpression of lrg increases sensitivity of S. mutans to oxygen, lrg expression levels may be an actual determinant of the Lrg/Cid-mediated phenotypes. Therefore, these data support the hypothesis that an optimal balance between the levels of cid and lrg expression is required for ideal aerobic growth and that these operons are functionally interconnected in as yet unknown ways. Based on these and previously published results, it is likely that both Cid and LytST (6, 16) contribute to regulation of lrg expression. In fact, we have previously shown that expression of lrg is not completely under the control of LytST, suggesting the presence of an additional regulator(s) that controls lrg expression (16). The detailed mechanism for how expression of lrg is coordinated by Cid and LytST appears to be an important part of understanding the Cid/Lrg system and warrants further investigation.
From a mechanistic standpoint, it seems significant that lrg and, to an even greater extent, cid are under the tight control of CcpA, reinforcing the idea that the Cid/Lrg system is coordinated with the metabolic status of S. mutans. This concept correlates well with the RNA-Seq data presented in this study of wild-type versus ΔcidB aerobic cultures grown in BHI broth (containing 11 mM glucose). This analysis revealed that mutation of cidB largely altered the expression of genes associated with major metabolic pathways, including phosphotransferase systems (PTS), the citrate cycle (tricarboxylic acid [TCA] cycle), galactose metabolism, pyruvate metabolism, fructose and mannose metabolism, glycolysis/gluconeogenesis, and amino sugar and nucleotide sugar metabolism. More recently, expression of cidB was also shown to be 3-fold upregulated in fructose-grown UA159 cells compared to glucose-grown cells (21). Collectively, these findings suggest that the Cid/Lrg system functions primarily under unfavorable environmental conditions such as glucose limitation and aerobic growth. In the same context, it is also noteworthy that expression of cid and lrg is under the tight control of the VicKR TCS. VicKR and its homologous TCSs have been reported to be directly and intimately involved in regulating bacterial life and death, primarily by controlling cell wall metabolism (30, 37). Intriguingly, VicKR homologues also share the unusual feature that this TCS is essential for viability in most bacteria possessing this regulatory system (30–36). In line with these studies, a vicK-deficient strain was recently shown to enhance cell death and lysis in S. mutans (38). Therefore, it is not surprising that the Cid/Lrg pathway, previously postulated to control bacterial cell lysis and death (8, 9), is regulated by VicKR, an attractive target for determining if a controlled cell lysis/death system functions in S. mutans. Importantly, VicKR has also been reported to play an important role in oxidative stress tolerance (3, 31, 33, 39), antibiotic resistance (40, 41), and infection and virulence (34–36, 42, 43). In line with this logic, it is highly possible that both the VicKR and holin-like Cid/Lrg systems may contribute to a controlled cell death mechanism, potentially involved in orchestrating the cell wall response to environmental stress. This idea is further supported by a possible role for VicKR in sensing and responding to oxygen levels and/or redox potential, consistent with the presence of a PAS domain (44) in VicK. Combined with our previous and current observations (6, 16), lrg expression and cid expression appear to be controlled by at least two TCSs (LytST and VicKR) and CcpA. LytST regulates lrg, but not cid, while VicKR regulates both. In contrast, CcpA is largely involved in cid regulation. And as described above, Cid is also able to regulate the function of Lrg. Therefore, our future studies will focus on evaluating the relative contributions of LytT, VicR, and CcpA in governing expression of lrg and cid, providing a deeper insight for the balanced expression pattern observed between these two operons.
As implicated by the data presented here, it is likely that CidB is a central player in the Cid/Lrg system. Specifically, the RNA-Seq transcriptomic studies comparing the wild type and the ΔcidB mutant have provided further insights into the role and regulation of the Cid/Lrg system. These results have shown that the bulk of genes differentially expressed in the ΔcidB strain during anaerobic growth belong to several genomic islands (GIs), such as TnSmu1, TnSmu2, and CRISPR-Cas. Notably, the TnSmu2 genes were previously shown to be downregulated in the strain lacking lytS, located upstream of lrgAB and known to positively regulate lrgAB expression (16). GIs are ubiquitous in prokaryotes and are acquired by horizontal gene transfer (45, 46). They normally carry genes that encode a variety of functions responsible for adapting to a particular set of environmental conditions. TnSmu1 corresponds to a large region of 23 kb spanning from SMU_191c to SMU_226c in strain UA159 and carries the genes that encode predicted integrases, bacteriophage-associated proteins, and transposon proteins (28). Particularly, TnSmu1 genes encode many functionally unknown hypothetical proteins, suggesting that the role and regulation of the GI in S. mutans are still far from fully elucidated. TnSMu2 is the largest GI (57 kb) found in the S. mutans genome, is reported to contain about 47 genes (28) involved in bacitracin and gramicidin synthesis, and is responsible for nonribosomal peptide and polyketide (NRP/PK) biosynthesis of a pigment that enhances aerobic growth and tolerance to H2O2 challenge in UA159 (47). It is also noteworthy that the Cid/Lrg system is connected to the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) system, known to provide a sequence-based adaptive immunity against mobile genetic elements, such as phages, invasive conjugative plasmids, and transposable elements in bacteria (48–52). The S. mutans UA159 genome contains two distinct CRISPR-Cas systems: a type II-A CRISPR1-Cas system and a type I-C CRISPR2-Cas system (53, 54). The cas genes were recently reported by Serbanescu et al. to contribute to the ability of S. mutans UA159 to persist under various stressful conditions, including cell membrane and oxidative stress, heat shock, and DNA-damaging conditions (53). Given the potential that CidA and LrgA bear a resemblance to phage holins, this observation further suggests that perturbation of the Cid/Lrg system may mimic a phage-induced response. Further efforts are under way to investigate how the CRISPR-Cas is linked with CidB and the Cid/Lrg system in coping with unfavorable environmental conditions. Intriguingly, Serbanescu et al. also reported that the expression of cas genes within the CRISPR-Cas systems could be differentially regulated by the VicKR TCS (53), which was also shown to regulate cid and lrg gene expression in this study. Collectively, these data support a role for the Cid/Lrg system in coping with environmental stressors and mediating cell death and lysis, and the three GIs that were transcriptionally altered in the cidB mutant appear to be tightly linked with the Cid/Lrg system.
We were also interested in how the transcriptome of the cidB mutant changes under aerobic growth conditions, due to the requirement of CidB for S. mutans growth under harsh oxidative conditions. Notably, we found that mutation of cidB led to a global readjustment in central metabolism and virulence processes, particularly in coping with oxidative stress. The fact that major carbon metabolic pathways were especially active opens the possibility that CidB or the Cid/Lrg system may serve to prime the cellular response to oxidative stress and/or to ensure survival of a subpopulation in a metabolically adjusted manner. In this regard, it is also possible that CidB may be involved in persister formation, possibly through modulating cellular processes within a population. Persisters constitute a subpopulation of phenotypic variants that are genetically identical and metabolically slow-growing cells in a clonal bacterial population, largely in response to antibiotics (55–57). Recently, other stress responses and particularly oxidative stress were also reported to contribute to persister formation in Escherichia coli (58). Thus, this idea is consistent with the potential of the Cid/Lrg system to regulate cell death and lysis in a programmed fashion within a population. The idea is further supported by our RNA-Seq data showing that genes encoding two potential toxin/antitoxin (T/A) modules, SMU_173/SMU_172 and SMU_41/SMU_40, were significantly upregulated in the ΔcidB mutant compared to the wild type during aerobic growth. SMU_173/SMU_172 (mazE/mazF) has been previously well characterized as a functional type II toxin-antitoxin (T/A) addiction system, consisting of a pair of genes that encode a stable toxin and a labile antitoxin (59). However, the function of SMU_41/SMU_40 is currently unknown. SMU_41 lacks canonical T/A features by sequence homology analysis, although SMU_40 is predicted to encode a putative antitoxin by RASTA-Bacteria (Rapid Automated Scan for Toxins and Antitoxins in Bacteria), a Web-based tool for identifying toxin-antitoxin loci in prokaryotes (60). T/A modules are reported to play a central role in persister formation and/or the persister state (61–63). T/A modules are also known to function as “regulatory switches” under adverse conditions, allowing cells to enter a state that confers protection against a severe stressful condition or nutrient limitation, ultimately contributing to programmed cell death as a bacterial survival strategy (57, 64–66). Given that many genes related to PTS, the major route for internalization of carbohydrates in S. mutans, were differentially and highly regulated in the cidB mutant strain, it is also possible that the Cid/Lrg system fine-tunes cellular physiology during periods of stress to help preserve normal cellular homeostasis (e.g., during periods of “feast or famine” in oral plaque) through the potential T/A modules. Therefore, in order to improve our understanding of the connections between the Cid/Lrg system and metabolic pathways, a complete metabolomics analysis and comparisons of wild-type and ΔcidB cultures constitute an attractive approach and are under way in our research group.
In conclusion, our data suggest that the Cid/Lrg system is regulated in a more complex manner than previously expected, involving at least two TCSs (LytST and VicKR), CcpA, and potential cross talk/regulation between the cid and lrg operons themselves. However, due to the functional and genetic interplay between cid and lrg operons, it is difficult to establish direct correlations between the observed phenotypes of some of these mutants. Nevertheless, it seems that both cid and lrg work together when S. mutans is challenged by environmental stresses, although we still do not understand how the cid and lrg gene products are functionally related. In the present study, we were able to probe changes in the transcriptome of S. mutans in great depth and with great sensitivity as a function of exposure to oxygen, by using RNA-Seq. The transcriptomic analysis of wild-type versus cidB mutant strains revealed potential effects of CidB on various predicted cellular processes. Given the global effects of CidB on the metabolic pathways and stress response of the organism, future study of the Cid/Lrg system has the potential to disclose how complex environmental signals are integrated into the regulatory networks modulating S. mutans virulence and homeostasis at the cellular and/or community level.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Burne laboratory, including Lin Zeng, for technical assistance with RNA-Seq experiments.
This work was supported by NIH-NIDCR grants R03 DE023604 (S.-J.A.) and R01 DE025237 (S.-J.A.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01499-16.
REFERENCES
- 1.Ahn SJ, Wen ZT, Burne RA. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J Bacteriol 189:8519–8527. doi: 10.1128/JB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn SJ, Ahn SJ, Browngardt CM, Burne RA. 2009. Changes in biochemical and phenotypical properties of Streptococcus mutans growing with aeration. Appl Environ Microbiol 75:2517–2527. doi: 10.1128/AEM.02367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn SJ, Burne RA. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol 189:6293–6302. doi: 10.1128/JB.00546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marquis RE. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol 15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 5.Bowden GH, Hamilton IR. 1998. Survival of oral bacteria. Crit Rev Oral Biol Med 9:54–85. doi: 10.1177/10454411980090010401. [DOI] [PubMed] [Google Scholar]
- 6.Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147. doi: 10.1099/mic.0.039586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung CJ, Zheng QH, Shieh YH, Lin CS, Chia JS. 2009. Streptococcus mutans auto AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol Microbiol 74:888–902. doi: 10.1111/j.1365-2958.2009.06903.x. [DOI] [PubMed] [Google Scholar]
- 8.Bayles KW. 2007. The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 9.Bayles KW. 2014. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 11.Young R. 2002. Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol 4:21–36. [PubMed] [Google Scholar]
- 12.Young R, Blasi U. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev 17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 13.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol 182:1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice KC, Bayles KW. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol 50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- 15.Rice KC, Firek BA, Nelson JB, Yang SJ, Patton TG, Bayles KW. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol 185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. 2012. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol 12:187. doi: 10.1186/1471-2180-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 19.Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–205. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 20.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng L, Burne RA. 2015. Sucrose- and fructose-specific effects on the transcriptome of Streptococcus mutans, as determined by RNA sequencing. Appl Environ Microbiol 82:146–156. doi: 10.1128/AEM.02681-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Supek F, Bosnjak M, Skunca N, Smuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn SJ, Lemos JA, Burne RA. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol 187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen ZT, Burne RA. 2002. Analysis of cis- and trans-acting factors involved in regulation of the Streptococcus mutans fructanase gene (fruA). J Bacteriol 184:126–133. doi: 10.1128/JB.184.1.126-133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubrac S, Msadek T. 2008. Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv Exp Med Biol 631:214–228. doi: 10.1007/978-0-387-78885-2_15. [DOI] [PubMed] [Google Scholar]
- 31.Echenique JR, Trombe MC. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J Bacteriol 183:4599–4608. doi: 10.1128/JB.183.15.4599-4608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M, Hanks TS, Zhang J, McClure MJ, Siemsen DW, Elser JL, Quinn MT, Lei B. 2006. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 152:967–978. doi: 10.1099/mic.0.28706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin PK, Li T, Sun D, Biek DP, Schmid MB. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol 181:3666–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Throup JP, Koretke KK, Bryant AP, Ingraham KA, Chalker AF, Ge Y, Marra A, Wallis NG, Brown JR, Holmes DJ, Rosenberg M, Burnham MK. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol 35:566–576. [DOI] [PubMed] [Google Scholar]
- 36.Wagner C, de Saizieu A, Schonfeld HJ, Kamber M, Lange R, Thompson CJ, Page MG. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect Immun 70:6121–6128. doi: 10.1128/IAI.70.11.6121-6128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 38.Senadheera DB, Cordova M, Ayala EA, Chavez de Paz LE, Singh K, Downey JS, Svensater G, Goodman SD, Cvitkovitch DG. 2012. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng DM, Liu MJ, ten Cate JM, Crielaard W. 2007. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res 86:606–610. doi: 10.1177/154405910708600705. [DOI] [PubMed] [Google Scholar]
- 40.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jansen A, Turck M, Szekat C, Nagel M, Clever I, Bierbaum G. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol 297:205–215. doi: 10.1016/j.ijmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Kadioglu A, Echenique J, Manco S, Trombe MC, Andrew PW. 2003. The MicAB two-component signaling system is involved in virulence of Streptococcus pneumoniae. Infect Immun 71:6676–6679. doi: 10.1128/IAI.71.11.6676-6679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubrac S, Boneca IG, Poupel O, Msadek T. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novick RP, Ram G. 2016. The floating (pathogenicity) island: a genomic dessert. Trends Genet 32:114–126. doi: 10.1016/j.tig.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langille MG, Hsiao WW, Brinkman FS. 2010. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol 8:373–382. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 47.Wu C, Cichewicz R, Li Y, Liu J, Roe B, Ferretti J, Merritt J, Qi F. 2010. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl Environ Microbiol 76:5815–5826. doi: 10.1128/AEM.03079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 49.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 50.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 51.Louwen R, Staals RH, Endtz HP, van Baarlen P, van der Oost J. 2014. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev 78:74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 53.Serbanescu MA, Cordova M, Krastel K, Flick R, Beloglazova N, Latos A, Yakunin AF, Senadheera DB, Cvitkovitch DG. 2015. Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology. J Bacteriol 197:749–761. doi: 10.1128/JB.02333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Ploeg JR. 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- 55.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. [DOI] [PubMed] [Google Scholar]
- 56.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 57.Prax M, Bertram R. 2014. Metabolic aspects of bacterial persisters. Front Cell Infect Microbiol 4:148. doi: 10.1038/nprot.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Syed MA, Koyanagi S, Sharma E, Jobin MC, Yakunin AF, Levesque CM. 2011. The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J Bacteriol 193:1122–1130. doi: 10.1128/JB.01114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sevin EW, Barloy-Hubler F. 2007. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol 8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Page R, Peti W. 2016. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol 12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 62.Schuster CF, Bertram R. 2013. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett 340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol 77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood TK. 2016. Combatting bacterial persister cells. Biotechnol Bioeng 113:476–483. doi: 10.1002/bit.25721. [DOI] [PubMed] [Google Scholar]
- 65.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 66.Martinez LC, Vadyvaloo V. 2014. Mechanisms of post-transcriptional gene regulation in bacterial biofilms. Front Cell Infect Microbiol 4:38. doi: 10.3389/fcimb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.