Age-related lymphoid cell decline is due to a cell-autonomous progressive loss of LMPP/MPP4 cells associated with increased cycling and differentiation potential toward the myeloid lineage.
Abstract
Declining immune function with age is associated with reduced lymphoid output of hematopoietic stem cells (HSCs). Currently, there is poor understanding of changes with age in the heterogeneous multipotent progenitor (MPP) cell compartment, which is long lived and responsible for dynamically regulating output of mature hematopoietic cells. In this study, we observe an early and progressive loss of lymphoid-primed MPP cells (LMPP/MPP4) with aging, concomitant with expansion of HSCs. Transcriptome and in vitro functional analyses at the single-cell level reveal a concurrent increase in cycling of aging LMPP/MPP4 with loss of lymphoid priming and differentiation potential. Impaired lymphoid differentiation potential of aged LMPP/MPP4 is not rescued by transplantation into a young bone marrow microenvironment, demonstrating cell-autonomous changes in the MPP compartment with aging. These results pinpoint an age and cellular compartment to focus further interrogation of the drivers of lymphoid cell loss with aging.
Introduction
Age-induced alterations in hematopoiesis, including reduction in functional B and T lymphocytes and expansion of myeloid cells, are associated with numerous hematopoietic pathologies (Wahlestedt et al., 2015). These cellular changes are associated with and can be driven by age-dependent decline in hematopoietic stem cell (HSC) function (Morrison et al., 1996) and biased HSC fate toward myeloerythroid lineages at the expense of lymphoid (Rossi et al., 2005; Beerman et al., 2010; Dykstra et al., 2011). The hierarchical structure of hematopoiesis defines the production of multipotent progenitors (MPPs) from HSCs (Christensen and Weissman, 2001), which serve as effector cells to tailor output of myeloid and lymphoid lineages. Recently, a major role for the MPP compartment in long-term blood production during steady-state hematopoiesis has been uncovered by in vivo lineage-tracing studies (Sun et al., 2014; Busch et al., 2015), highlighting the importance of further study of this compartment and its contribution to hematopoietic aging and pathology.
Within the heterogeneous MPP compartment, the brightest ∼25% of Flk2-expressing cells represent lymphoid-primed MPPs (LMPPs; Adolfsson et al., 2005). Additionally, differential expression of CD150, CD48, and Flk2 defines myeloid-biased MPP2 and MPP3 and lymphoid-primed MPP4 (Wilson et al., 2008; Cabezas-Wallscheid et al., 2014; Pietras et al., 2015). It remains undetermined as to whether the process of aging dynamically alters the composition and functional output of the MPP compartment.
To identify age-dependent cellular and molecular changes in the MPP compartment, we systematically examined MPP composition with aging and combined single-cell transcriptome and functional studies of MPP4/LMPP. We found that aging induces increased cycling, loss of lymphoid priming, and differentiation potential of MPP4/LMPP cells. In vivo transplantation of aged LMPPs into a young BM microenvironment demonstrates cell-autonomous defects in lymphoid production and skewing toward myeloid cell production. Together, this suggests that early alterations in the MPP compartment may be the effectors of lymphoid cell loss in aging hematopoiesis.
Results and discussion
Aging-induced loss of LMPPs
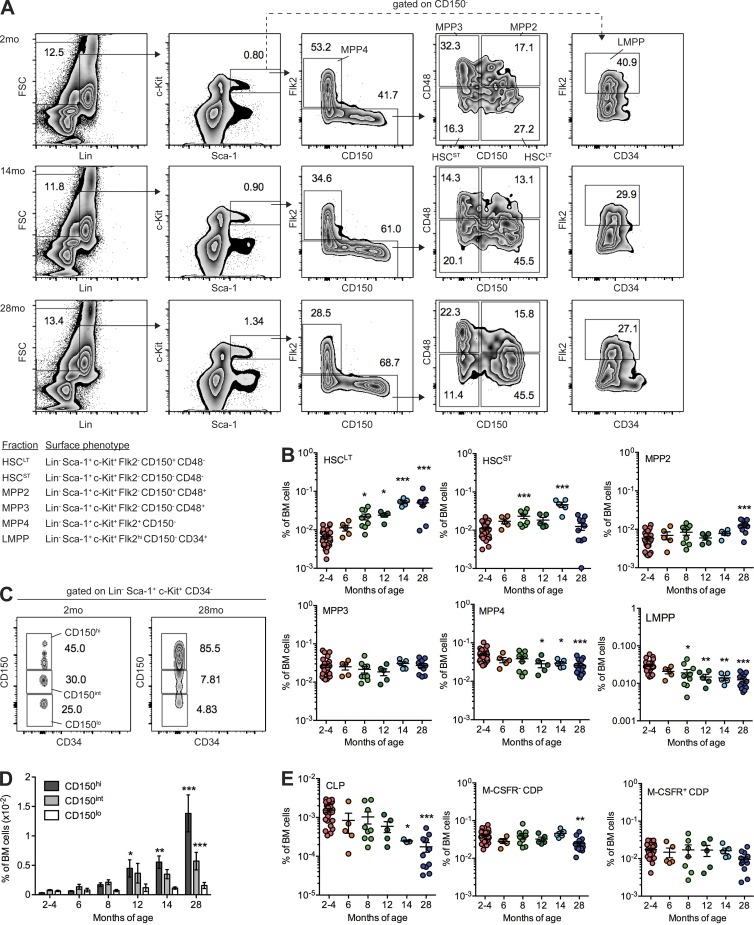
We began by examining alterations in BM frequency of long-term HSCs (LT-HSC), short-term HSCs (ST-HSCs), MPP2, MPP3, MPP4, and LMPPs with age using defined markers (Fig. 1 A; Adolfsson et al., 2005; Wilson et al., 2008; Pietras et al., 2015). Analysis of C57BL/6J female mice between 2 and 28 months old (mo) revealed a significant increase in BM frequency of LT-HSCs and ST-HSCs as early as 8 mo (Fig. 1 B), consistent with known phenotypic HSC expansion with aging (Rossi et al., 2005). Increased frequency of MPP2 was observed at 28 mo, consistent with reported molecular and functional megakaryocyte/erythroid bias of aged HSCs (Grover et al., 2016; Rundberg Nilsson et al., 2016). In contrast, a significant, progressive decline in BM frequencies of MPP4 and LMPPs was observed by 12 and 8 mo, respectively. To compare this phenotype with previous studies of an aging-induced shift in lineage-biased HSC composition (Beerman et al., 2010; Challen et al., 2010; Dykstra et al., 2011), we examined CD150hi (myeloid biased), CD150int (balanced), and CD150lo (lymphoid biased) HSCs (Fig. 1 C; Beerman et al., 2010; Morita et al., 2010). We observed significant increase in frequency of CD150hi HSCs by 12 mo and of CD150int HSCs by 28 mo (Fig. 1 D). Although this defines an overall myeloid skewing of the HSC compartment mediated by expansion of CD150hi HSCs, we find that lymphoid-biased HSCs (CD150lo) are not specifically depleted with aging. These data suggest that MPP4/LMPP loss with aging may be independent of alterations in the lymphoid-biased CD150lo HSC compartment.
Figure 1.
MPP composition is altered with aging. (A) FACS gating showing frequency of HSC and MPP subsets in representative 2-mo, 14-mo, and 28-mo mice. The inset table defines surface markers used for cell isolation. FSC, forward side scatter. (B) Frequency of HSC and MPP subsets in whole BM identified by FACS analysis. Bars denote the mean of 2–4 mo (n = 25), 6 mo (n = 5), 8 mo (n = 7), 12 mo (n = 5), 14 mo (n = 3), and 28 mo (n = 10) assessed in five independent experiments. (C) FACS gating showing frequency of CD150hi, CD150int, and CD150lo HSCs in representative 2-mo and 28-mo mice. (D) Frequency of CD150hi, CD150int, and CD150lo HSCs in whole BM identified by FACS analysis. Error bars denote mean ± SEM of 2–4 mo (n = 25), 6 mo (n = 5), 8 mo (n = 7), 12 mo (n = 5), 14 mo (n = 3), and 28 mo (n = 10) assessed in five independent experiments. (E) Frequency of CLP and CDP subsets in BM identified by FACS analysis. Bars denote the mean of 2–4 mo (n = 25), 6 mo (n = 5), 8 mo (n = 7), 12 mo (n = 5), 14 mo (n = 3), and 28 mo (n = 10) assessed in five independent experiments. (B, D, and E) P-values were generated by one-way ANOVA with Dunnett’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. HSCLT, LT-HSC; HSCST, ST-HSC.
Downstream lymphoid-committed progenitors including common lymphoid progenitors (CLPs) and common dendritic cell progenitors (CDPs) are depleted with aging (Miller and Allman, 2003; Min et al., 2006; Grover et al., 2016; Xiao et al., 2016). We observed significant decreases in BM frequency of CLPs and M-CSFR− CDPs at 14 and 28 mo, respectively (Fig. 1 E and Fig. S1), older ages than our observed loss of MPP4/LMPP cells, suggesting that loss of MPP4/LMPP may underlie reduction in output of committed progenitors and mature lymphoid cell types with aging.
Single-cell transcriptome analysis identifies altered cycling and lymphoid priming of aged LMPPs
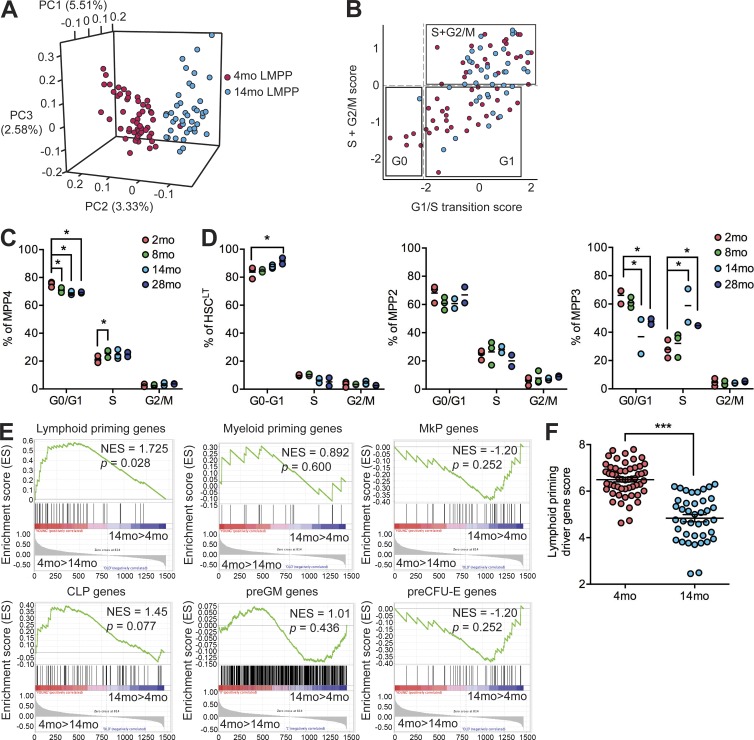
We performed single-cell RNA sequencing (scRNA-seq) using the C1 Autoprep system on LMPPs isolated by FACS from 4- and 14-mo mice. After stringent filtering, we retained 54 and 40 libraries from 4-mo and 14-mo LMPPs, respectively. Mean gene expression in single cells was well correlated with expression in bulk LMPP cells (Pearson; r = 0.701 ± 0.026; n = 4). Principal component analysis revealed separation between 4- and 14-mo LMPPs (Fig. 2 A). Gene set enrichment analysis (GSEA) focusing on hallmark gene sets (Subramanian et al., 2005) identified significant enrichment (P < 0.05; false discovery rate [FDR] < 0.25) of cell cycle–related gene signatures in 14-mo LMPPs. To further dissect cell cycle state, we scored cells via mean expression of G1/S transition, S, and G2/M phase–specific signatures to define three groups of cells: G0, G1, and S/G2/M (Fig. 2 B; Kowalczyk et al., 2015; Tsang et al., 2015). The frequency of cells in S/G2/M was significantly increased in 14- versus 4-mo LMPPs (67.5% vs. 42.5%; P = 0.0097; hypergeometric test), suggesting increased cycling of LMPPs with aging. Short-term (1 h) in vivo 5-ethynyl–2’-deoxyuridine (EdU) incorporation experiments directly confirmed increased proliferation of MPP4 cells by 8 mo (Fig. 2 C and Fig. S2). Consistent with previously reported reduction in BrdU incorporation by aged HSCs (Janzen et al., 2006), we observed increased quiescence of LT-HSCs at 28 mo (Fig. 2 D). In MPP3 cells, we observed increased cycling by 14 mo, demonstrating additional cell cycle changes within the MPP compartment.
Figure 2.
Increased cycling and loss of lymphoid priming in aged LMPP/MPP4. (A) Loading plot of principal components (PC) 1–3 from principal component analysis of 4-mo (n = 54) and 14-mo (n = 40) LMPP scRNA-seq data. Percent contribution of each principal component to total variation is shown. (B) Mean expression of G1/S transition genes (x axis) and S + G2/M genes (y axis) in each 4-mo (n = 54) and 14-mo (n = 40) LMPP scRNA-seq library. Boxed groups represent predicted G0, G1, and S + G2/M cell cycle phases. (C) Frequency of MPP4 in G0/G1, S, or G2/M defined by in vivo EdU incorporation and FACS analysis. (D) Frequency of LT-HSCs, MPP2, and MPP3 in G0/G1, S, or G2/M defined by in vivo EdU incorporation and FACS analysis. (C and D) Bars denote the mean of 2 mo (n = 3), 8 mo (n = 3), 14 mo (n = 2), and 28 mo (n = 2) in two independent experiments. (E) Enrichment of lymphoid-priming, myeloid-priming, MkP, CLP, preGM, and preCFU-E gene signatures in 4-mo (n = 54) versus 14-mo (n = 40) LMPP scRNA-seq data. NES, normalized enrichment score. (F) Mean expression of lymphoid-priming driver genes in each scRNA-seq library. Bars denote the mean of 4 mo (n = 54) and 14 mo (n = 40). (C, D, and F) P-values were generated by unpaired, two-tailed Student’s t tests. *, P < 0.05; ***, P < 0.001.
To interrogate lineage priming and specification in our LMPP scRNA-seq data, we performed bulk RNA-seq to define a lymphoid-priming signature (genes up-regulated in CLPs or LMPPs vs. granulocyte-macrophage progenitors [GMPs] at log fold change >2; FDR < 0.05) and a myeloid-priming signature (genes up-regulated in GMPs vs. CLPs or LMPPs). Testing enrichment of these signatures in our scRNA-seq data revealed significant enrichment of the lymphoid-priming signature in 4-mo LMPPs (Fig. 2 E). Published lymphoid (CLP), pre–granulocyte-macrophage (preGM), megakaryocyte progenitor (MkP), and pre-CFU erythroid (preCFU-E) gene signatures independently confirmed enrichment of a lymphoid (CLP) signature in 4-mo LMPPs. To compare relative lymphoid priming between single cells, we quantitated the mean expression of lymphoid driver genes identified by GSEA in each cell, including genes up-regulated during B lymphocyte differentiation from LMPPs (Fos, Jun, Gm2a, Tmem173, and Pde4b; Ng et al., 2009; Mercer et al., 2011) and the Ebf1-activated gene Dusp2 (Vilagos et al., 2012). We observed that the lymphoid-priming driver gene score was significantly decreased in 14-mo LMPPs (Fig. 2 F). On a per-cell basis, 14 out of 40 (35%) sampled 14-mo LMPPs had a lymphoid priming driver gene score below the range observed in 4-mo LMPPs. These results suggest a deficiency in expression of lymphoid-priming genes as a consequence of aging in the LMPP compartment.
Novel culture conditions to concurrently support lymphoid and myeloid differentiation
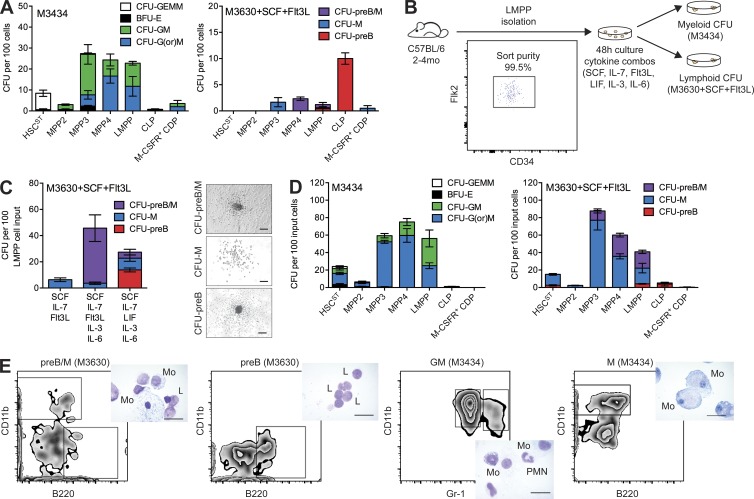
Although myeloid differentiation potential of MPP4/LMPPs can be interrogated using methylcellulose-based assays (M3434), lymphoid differentiation of de novo isolated MPP4/LMPPs is not robust in methylcellulose (M3630 with stem cell factor [SCF] + flt-3 ligand [Flt3L]; Fig. 3 A). To develop a unified in vitro system to interrogate both lymphoid and myeloid differentiation potential, we cultured LMPPs for 48 h with several defined growth factor combinations (Fig. 3 B). The results of this screen identified a combination of five growth factors (SCF, IL-7, leukemia inhibitory factor [LIF], IL-3, and IL-6) that stimulated B cell differentiation of LMPPs to a greater degree than the previously defined combination of SCF, IL-7, and Flt3L (Månsson et al., 2007) or when Flt3L was substituted for LIF (SCF, IL-7, Flt3L, IL-3, and IL-6; Fig. 3 C). This assay allowed assessment of lymphoid potential from ST-HSCs, MPP3, MPP4, and LMPPs, also simultaneously supporting myeloid differentiation (Fig. 3, D and E).
Figure 3.
Novel in vitro culture conditions supporting concurrent B cell and myeloid cell production. (A) CFUs per 100 de novo isolated HSC and progenitor cell subsets plated into myeloerythroid-promoting M3434 (left) or preB-promoting M3630 (right) methylcellulose media. Results are shown as mean ± SEM of ST-HSCs (n = 3), MPP2 (n = 3), MPP3 (n = 3), MPP4 (n = 3), LMPPs (n = 4), CLPs (n = 4), and CDPs (n = 2) in two independent experiments. (B) Schematic of in vitro assay design. (C) CFUs per 100 LMPPs after 48-h culture with denoted cytokine cocktails. Results are mean ± SEM of n = 3 in three independent experiments. (Right) Representative CFU images. Bars, 200 µm. (D) CFUs per 100 input HSC and progenitor cell subsets after 48-h culture followed by plating in M3434 (left) or M3630 (right). Results are shown as mean ± SEM of ST-HSCs (n = 5), MPP2 (n = 3), MPP3 (n = 3), MPP4 (n = 3), LMPPs (n = 6), CLPs (n = 6), and CDPs (n = 3) in three independent experiments. GEMM, granulocyte, erythroid, macrophage, and megakaryocyte. (E) Representative FACS analysis and Wright-Giemsa staining (inset) of cells isolated from single colonies. Cell surface markers against macrophages (CD11b), granulocytes (Gr-1), and B cells (B220) were used. Bars, 10 µm. L, lymphocyte; Mo, macrophage; PMN, granulocyte.
Single-cell functional analysis reveals lymphoid defect in aged LMPPs
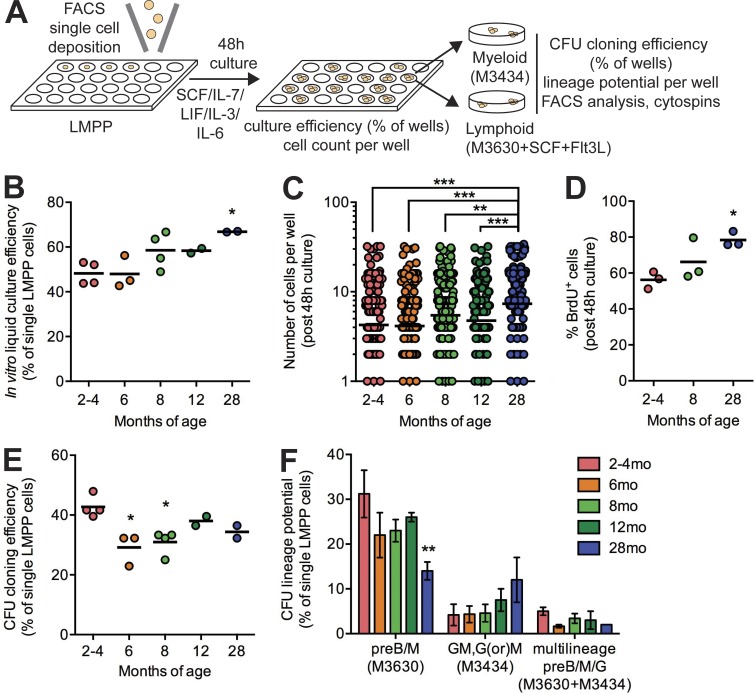
To assess functional alterations in LMPPs with aging, we adapted our 48-h culture system to a single-cell assay (Fig. 4 A). The proportion of single LMPP cells proliferating in culture increased with aging (Fig. 4 B), and we observed significantly higher median number of cells generated from single LMPPs from 28-mo mice (Fig. 4 C). Short-term BrdU incorporation (1 h) at the end of the 48-h culture period confirmed increased cycling of 28-mo LMPPs (Fig. 4 D). After 48-h culture, seeding cells into myeloid and lymphoid methylcellulose assays revealed a reduction in differentiation potential with aging (Fig. 4 E). Interrogating the lineage potential of single LMPPs in this assay revealed that cells with lymphoid potential (preB/M in M3630 assay) were significantly reduced in frequency with aging, and cells with myeloid-restricted potential (GM, G, or M in M3434 assay) trended toward increased frequency with aging (Fig. 4 F). Together, these data suggest that increased proliferation in the LMPP/MPP4 compartment is accompanied by functional lineage skewing, impairing lymphoid differentiation and also promoting myeloid differentiation.
Figure 4.
Impaired lymphoid differentiation of aged LMPPs in vitro. (A) Schematic of single-cell assay design. (B) Frequency of wells found to contain one or more cells after 48-h culture of single LMPP cells. Dots represent a single plate of n = 96 single cells. Bars represent the mean of n = 2 independent experiments. (C) Number of cells counted per well after 48-h culture of single LMPP cells. Dots represent individual wells. Bars represent the mean of n = 288 collected in three independent experiments. (D) Frequency of BrdU+ cells after 48-h culture of LMPP cells by FACS analysis. Dots represent independent experiments, and bars represent the mean of n = 3. (E) Frequency of wells found to form CFUs after 48-h culture of single LMPP cells and plating in methylcellulose assays. Dots represent a single plate of n = 96 single cells. Bars represent the mean of n = 2 independent experiments. (B–E) P-values were generated by one-way ANOVA with Holm-Sidak’s multiple comparisons test. (F) Frequency of wells found to form CFU-preB/M, CFU-GM, or CFU-preB/M/G after 48-h culture of single LMPP cells and plating in methylcellulose assays. Results are shown as mean ± SEM of n = 2–4 in two independent experiments. P-values were generated by two-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Cell-autonomous in vivo lineage skewing of aged LMPPs
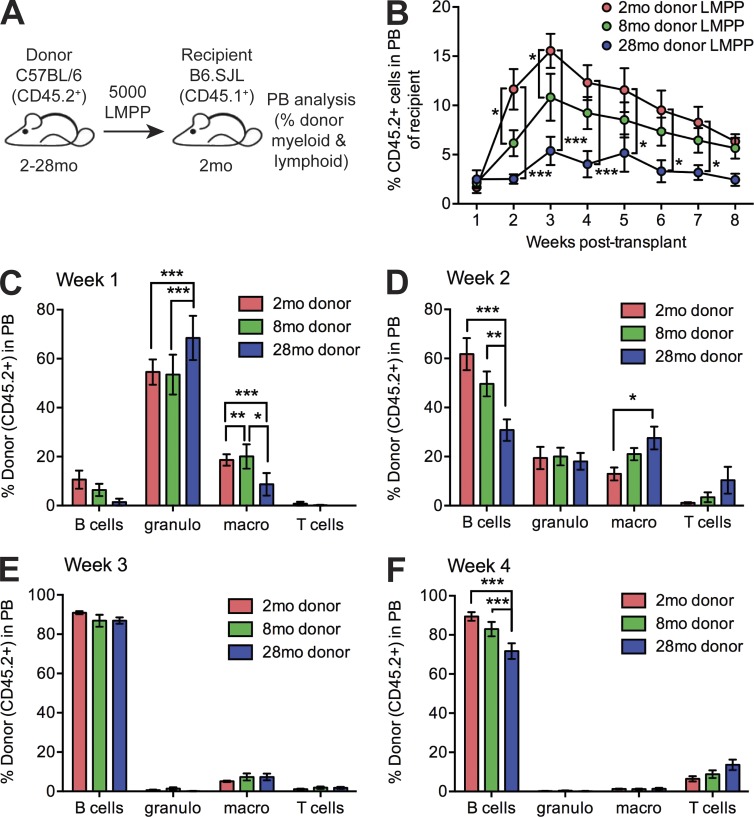
To interrogate functional changes in LMPPs in vivo, we transplanted 5,000 LMPPs isolated from 2-mo, 8-mo, or 28-mo CD45.2 mice into sublethally irradiated 2-mo CD45.1 recipient mice (Fig. 5 A). We observed significant impairment in peripheral blood reconstitution of 8-mo and 28-mo LMPPs versus 2-mo LMPPs (Fig. 5 B), confirming an age-dependent intrinsic decrease in the ability of LMPPs to sustain production of mature progeny. The reconstitution dynamics of LMPPs (Fig. S3) demonstrate predominant granulocyte and macrophage production at 1 wk after transplant (Fig. 5 C), a shift to B cell production at 2 wk (Fig. 5 D) that is dominant at 3 wk (Fig. 5 E), and a shift to T cell production initiated at 4 wk (Fig. 5 F). We observed that granulocyte production was significantly increased from 28-mo mice, whereas B cell production was significantly decreased, supporting a cell-intrinsic lineage skewing of aging LMPPs toward myeloid cell production at the expense of B cell production. Together, our data demonstrate an early and progressive loss of LMPP/MPP4 cells with aging that may be initiated independently of alterations in HSCs. Within aged LMPP/MPP4 cells, increased cycling is concomitant with loss of molecular lymphoid priming, and differentiation potential is skewed toward the myeloid lineage at the expense of lymphoid lineage production. Although defects in LMPP/MPP4 are cell autonomous and not fully rescued by transplantation into a young microenvironment, future investigation into the cell-intrinsic and cell-extrinsic alterations initiating LMPP/MPP4 loss and lineage skewing with aging will be a critical to define the drivers of hematopoietic aging.
Figure 5.
Cell-autonomous in vivo lymphoid differentiation defect of aged LMPPs. (A) Schematic of experimental design for in vivo analysis of peripheral blood reconstitution by transplanted LMPP cells. (B) Frequency of donor-derived (CD45.2+) cells in peripheral blood of recipient mice at 1–8 wk after transplant by FACS analysis. Results are shown as mean ± SEM of 2-mo donor LMPPs (n = 7), 8-mo donor LMPPs (n = 9), and 28-mo donor LMPPs (n = 7) in three independent experiments. (C–F) Frequency of donor-derived B cells (CD45.2+ B220+), granulocytes (granulo; CD45.2+ CD11b+ Gr1hi), macrophages (macro; CD45.2+ CD11b+ Gr1lo), and T cells (CD45.2+ CD3+) in peripheral blood of recipient mice at 1–4 wk after transplant by FACS analysis. Results are shown as mean ± SEM of 2-mo donor LMPPs (n = 7), 8-mo donor LMPPs (n = 9), and 28-mo donor LMPPs (n = 7) in three independent experiments. P-values were generated by two-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Materials and methods
Mice
C57BL/6J and B6.SJL female mice were obtained from and aged within The Jackson Laboratory, with some contributed by D. Harrison, S. Ackerman, and G. Howell (The Jackson Laboratory, Bar Harbor, ME). For all experiments, mice were used at ages 2–4, 6, 8, 12, 14, or 28 mo. The Animal Care and Use Committee at The Jackson Laboratory approved all animal studies.
Isolation of hematopoietic stem and progenitor cells
BM mononuclear cells from the iliac crest, femur, and tibiae were isolated by Ficoll-Paque density centrifugation (GE Healthcare). Mononuclear cells were stained with fluorochrome-conjugated anti–c-Kit (clone 2B8), Sca-1 (clone E13-161.7), CD16/32 (clone 93), CD34 (clone RAM34), CD150 (clone mShad150), Flk2 (clone A2F10), CD48 (clone HM48-1), CD127 (clone A7R34), CD115 (clone AFS98), mature Lin markers (CD11b, Gr-1, B220, CD3, and Ter119), and viability stain (propidium iodide or DAPI). Cells were isolated with a FACSAria flow cytometer (BD) with doublet discrimination or data were collected on an LSRII flow cytometer (BD). The purity of sorted populations was ≥97%. FACS data were analyzed using FlowJo (Tree Star). For single-cell assays, single cells were sorted into a 96-well plate using an advanced cell deposition unit and visually confirmed by microscopy.
scRNA-seq library preparation and analysis
LMPPs isolated from pooled 4-mo or 14-mo mice were resuspended at a concentration of 200 cells/µl and loaded onto the C1 Single-Cell Auto Prep Integrated Fluidic Circuit for capturing 5–10 µm cells (Fluidigm), with loading buffer including LIVE/DEAD Viability/Cytotoxicity staining (Thermo Fisher Scientific). Images of captured cells were collected with a microscope (AxioObserver.Z1; ZEISS) in brightfield, GFP, and CY3 channels using ZenPro software. Single-cell RNA extraction and mRNA amplification were performed following manufacturer’s recommendations including exogenous spike-in controls. cDNA was quantified using the Quant-iT PicoGreen dsDNA Assay kit (Thermo Fisher Scientific) and a high-sensitivity DNA chip (Agilent Technologies) and was diluted to a final concentration of 0.15–0.3 ng/µl. Diluted cDNA reaction products were used to generate scRNA-seq libraries using the Nextera XT DNA Sample Preparation kit (Illumina). After PCR amplification, all samples were pooled, purified, and quantified using a high-sensitivity DNA chip. Libraries were paired-end sequenced using a HiSeq 2500 sequencing system (Illumina). We sequenced 96 libraries per rapid-run flow cell, generating ∼15 × 106 reads per library. An index for RNA-seq by expectation maximization (RSEM) was generated on the basis of the Ensembl mm10 transcriptome downloaded from the University of California, Santa Cruz Genome Browser (23,637 total genes). Read data were aligned directly to this index using RSEM/bowtie. Quantification of gene expression levels in tags per million (TPM) for all genes in all samples was performed using RSEM (v1.2.8). Before all subsequent analyses, we filtered the data as previously described (Kowalczyk et al., 2015). First, we filtered out cells with <2,500 genes with log2(TPM + 1) > 2. Second, we excluded genes that were log2(TPM + 1) < 4 in aggregated data. Third, we centered the data by subtracting from each gene its mean expression (log2[TPM + 1]) across all cells. After filtering, our dataset included 94 single cell transcriptomes, 54 representing 4-mo LMPPs and 40 representing 14-mo LMPPs, and 1,467 genes. These libraries had <20% of counts mapping to mitochondrial genes (4.64 ± 3.13%; mean ± SD). Principal component analysis was performed using the R stats prcomp() function with variables scaled for unit variance and a 0.05 tolerance. GSEA was performed with the javaGSEA application (version 2.0.14) with the default settings (Subramanian et al., 2005). Enrichment was considered significant if FDR < 25% and P < 0.05. Cell cycle genes were defined as those cycling (G1/S, S, and G2/M) in synchronized HeLa cells (Whitfield et al., 2002; Kowalczyk et al., 2015). We refined this list by only including genes present in our filtered dataset. We plotted the mean of the G1/S transition signature versus the mean of the S + G2/M signatures for each cell. Lineage-specific gene sets (CLP, PreGM, MkP, and preCFU-E) were previously published (Sanjuan-Pla et al., 2013). Comparison with bulk RNA-seq libraries was performed by calculating the geometric mean of all 4-mo or 14-mo LMPP scRNA-seq data (log2[TPM + 1]) and comparing with the bulk RNA-seq log2(TPM + 1). Pearson correlation coefficients were calculated based on linear fit.
Bulk RNA-seq
Four independent biological replicates of bulk LMPPs (20,000–32,000 cells; Lin−Sca-1+c-Kit+Flk2+CD150−CD34+), GMPs (30,000–61,000 cells; Lin−Sca-1−c-Kit+CD150−CD34+CD16/32+), or CLPs (8,500–28,142 cells; Lin−Sca-1loc-KitloFlk2+CD127+) isolated from 4-mo C57BL/6J female mice were sorted directly into 350 µl RLT buffer (QIAGEN) and flash frozen. Total RNA was isolated (QIAGEN), including DNase treatment. RNA was processed using an Ovation RNA-Seq kit (V2; NuGen). After shearing, a TruSeq DNA sample prep kit (v2; Illumina) was used to prepare libraries. Libraries were sequenced on the HiSeq 2000 platform (Illumina) at a sequencing depth of >35 million reads per sample. Transcript abundances were estimated for each RNA-seq sample using RSEM. Read counts estimated for each gene by RSEM were given as input to the R package edgeR for differential expression analysis (Robinson et al., 2010). Genes were considered differentially expressed among LMPPs, GMPs, and CLPs based on log fold change >2 and FDR <0.05 criteria.
In vivo cell cycle analysis
Mice were injected with 50 mg/kg EdU for 1 h before BM harvest. Sorted LT-HSCs and MPP2, MPP3, and MPP4 cells were processed using a Click-iT Edu Flow Cytometry Assay kit (Thermo Fisher Scientific).
Liquid culture
Isolated populations were cultured in a 96-well plate in a total volume of 50 µl (single cells) or 100 µl (bulk culture) of IMDM supplemented with 10% FCS and combinations of the cytokines rmIL-3 (10 ng/ml), rmIL-6 (10 ng/ml), rmSCF (100 ng/ml), rmIL-7 (20 ng/ml), rmFlt3L (25 ng/ml; PeproTech), and rmLIF (20 ng/ml; Gibco) for 48 h at 37°C and 5% CO2. After 48 h, the cellular contents of each well were harvested, washed, resuspended in IMDM with 2% FCS, and split in equal volume into the CFU assay media described in the next paragraph. For single-cell culture, before seeding in the CFU assay, each well was examined under an inverted light microscope to count the number of cells in each well. For BrdU experiments, cells were cultured for 1 h with 10 µM BrdU and analyzed using a BrdU Flow kit (BD).
CFU assay
De novo isolated or post–48-h liquid culture cells were plated in MethoCult GF M3434, supporting myeloid and erythroid differentiation, or MethoCult GF M3630, supporting preB cell differentiation (STEMCELL Technologies), supplemented with 0–20 ng/ml rmFlt-3L and 0–100 ng/ml rmSCF (PeproTech), and cultured at 37°C and 5% CO2. Colonies were scored between days 7 and 10 after plating, and images were captured using an inverted microscope (Diaphot 200; Nikon) at 4× with SPOT imaging software (v.5.0.15). Single representative colonies were isolated and used to confirm cell lineage by FACS analysis for the surface markers CD11b (M1/70), Gr-1 (RB6-8C5), and B220 (RA3-6B2) and Wright-Giemsa staining of fixed cytospins. Cytospin images were captured using an inverted microscope (Eclipse E200-LED; Nikon) at 100× with SPOT imaging software.
In vivo transplantation analysis
B6.SJL (CD45.1) recipient mice were sublethally irradiated (600 rads) and were reconstituted by sorted donor LMPPs (CD45.2) using retroorbital injection. Multilineage peripheral blood reconstitution was monitored weekly based on flow cytometric analysis of CD45.2, CD45.1, CD11b, Gr-1, B220, and CD3 staining.
Statistical analysis
Unpaired, two-tailed Student’s t tests, one-way ANOVA (Kruskal-Wallis test) followed by Dunnett’s multiple comparisons test or Holm-Sidak’s multiple comparisons test, or two-way ANOVA followed by Tukey’s multiple comparisons test were used for determining significance of the results as appropriate for the data type. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Data access
Bulk and scRNA-seq data are deposited in Gene Expression Omnibus (accession nos. GSE77736 and GSE77740).
Online supplemental material
Fig. S1 shows the flow cytometry gating strategy for committed lymphoid progenitor cells. Fig. S2 shows the flow cytometry gating strategy for EdU incorporation and cell cycle analysis. Fig. S3 shows the flow cytometry gating strategy for lineage composition of peripheral blood in transplanted mice.
Supplementary Material
Acknowledgments
We thank Kevin Mills, Kyuson Yun, Robert Braun, Gavin Schnitzler, and members of the Trowbridge laboratory for advice and input on this work; David Harrison, Susan Ackerman, and Gareth Howell for contributing aged mice; Tara Murphy for technical support; and Will Schott for cell sorting.
This work was funded by the National Institutes of Health (grant no. P30 AG038070 to J.J. Trowbridge and T32 HD007065 to K. Young), the Ellison Medical Foundation (grant no. AG-NS-0993-13 J.J. Trowbridge), and the Pyewacket Fund (K. Young).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CDP
- common dendritic cell progenitor
- CLP
- common lymphoid progenitor
- EdU
- 5-ethynyl–2’-deoxyuridine
- FDR
- false discovery rate
- GMP
- granulocyte-macrophage progenitor
- GSEA
- gene set enrichment analysis
- HSC
- hematopoietic stem cell
- LIF
- leukemia inhibitory factor
- LMPP
- lymphoid-primed MPP
- LT-HSC
- long-term HSC
- MkP
- megakaryocyte progenitor
- mo
- months old
- MPP
- multipotent progenitor
- preCFU-E
- pre-CFU erythroid
- preGM
- pre–granulocyte-macrophage
- RSEM
- RNA-seq by expectation maximization
- SCF
- stem cell factor
- scRNA-seq
- single-cell RNA sequencing
- ST-HSC
- short-term HSC
- TPM
- tags per million
References
- Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.J., Thoren L.A., et al. 2005. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: A revised road map for adult blood lineage commitment. Cell. 121:295–306. 10.1016/j.cell.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I.L., Bryder D., and Rossi D.J.. 2010. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. USA. 107:5465–5470. 10.1073/pnas.1000834107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., and Rodewald H.R.. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 518:542–546. 10.1038/nature14242 [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., et al. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 15:507–522. 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Challen G.A., Boles N.C., Chambers S.M., and Goodell M.A.. 2010. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-β1. Cell Stem Cell. 6:265–278. 10.1016/j.stem.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J.L., and Weissman I.L.. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546. 10.1073/pnas.261562798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B., Olthof S., Schreuder J., Ritsema M., and de Haan G.. 2011. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208:2691–2703. 10.1084/jem.20111490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A., Sanjuan-Pla A., Thongjuea S., Carrelha J., Giustacchini A., Gambardella A., Macaulay I., Mancini E., Luis T.C., Mead A., et al. 2016. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 7:11075 10.1038/ncomms11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen V., Forkert R., Fleming H.E., Saito Y., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., and Scadden D.T.. 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 443:421–426. 10.1038/nature05159 [DOI] [PubMed] [Google Scholar]
- Kowalczyk M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., Schneider R.K., Wagers A.J., Ebert B.L., and Regev A.. 2015. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25:1860–1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Månsson R., Hultquist A., Luc S., Yang L., Anderson K., Kharazi S., Al-Hashmi S., Liuba K., Thorén L., Adolfsson J., et al. 2007. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 26:407–419. 10.1016/j.immuni.2007.02.013 [DOI] [PubMed] [Google Scholar]
- Mercer E.M., Lin Y.C., Benner C., Jhunjhunwala S., Dutkowski J., Flores M., Sigvardsson M., Ideker T., Glass C.K., and Murre C.. 2011. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 35:413–425. 10.1016/j.immuni.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.P., and Allman D.. 2003. The decline in B lymphopoiesis in aged mice reflects loss of very early B-lineage precursors. J. Immunol. 171:2326–2330. 10.4049/jimmunol.171.5.2326 [DOI] [PubMed] [Google Scholar]
- Min H., Montecino-Rodriguez E., and Dorshkind K.. 2006. Effects of aging on the common lymphoid progenitor to pro-B cell transition. J. Immunol. 176:1007–1012. 10.4049/jimmunol.176.2.1007 [DOI] [PubMed] [Google Scholar]
- Morita Y., Ema H., and Nakauchi H.. 2010. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 207:1173–1182. 10.1084/jem.20091318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Wandycz A.M., Akashi K., Globerson A., and Weissman I.L.. 1996. The aging of hematopoietic stem cells. Nat. Med. 2:1011–1016. 10.1038/nm0996-1011 [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Yoshida T., Zhang J., and Georgopoulos K.. 2009. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 30:493–507. 10.1016/j.immuni.2009.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Reynaud D., Kang Y.A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., and Passegué E.. 2015. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., and Weissman I.L.. 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 102:9194–9199. 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundberg Nilsson A., Soneji S., Adolfsson S., Bryder D., and Pronk C.J.. 2016. Human and murine hematopoietic stem cell aging is associated with functional impairments and intrinsic megakaryocytic/erythroid bias. PLoS One. 11:e0158369 10.1371/journal.pone.0158369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan-Pla A., Macaulay I.C., Jensen C.T., Woll P.S., Luis T.C., Mead A., Moore S., Carella C., Matsuoka S., Bouriez Jones T., et al. 2013. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 502:232–236. 10.1038/nature12495 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., and Camargo F.D.. 2014. Clonal dynamics of native haematopoiesis. Nature. 514:322–327. 10.1038/nature13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J.C., Yu Y., Burke S., Buettner F., Wang C., Kolodziejczyk A.A., Teichmann S.A., Lu L., and Liu P.. 2015. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol. 16:178 10.1186/s13059-015-0739-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilagos B., Hoffmann M., Souabni A., Sun Q., Werner B., Medvedovic J., Bilic I., Minnich M., Axelsson E., Jaritz M., and Busslinger M.. 2012. Essential role of EBF1 in the generation and function of distinct mature B cell types. J. Exp. Med. 209:775–792. 10.1084/jem.20112422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt M., Pronk C.J., and Bryder D.. 2015. Concise review: hematopoietic stem cell aging and the prospects for rejuvenation. Stem Cells Transl. Med. 4:186–194. 10.5966/sctm.2014-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., and Botstein D.. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000. 10.1091/mbc.02-02-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129. 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Xiao J., Zhou H., Wu N., and Wu L.. 2016. The non-canonical Wnt pathway negatively regulates dendritic cell differentiation by inhibiting the expansion of Flt3+ lymphocyte-primed multipotent precursors. Cell. Mol. Immunol. 13:593–604. 10.1038/cmi.2015.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.