Jiang et al. generated Dpy30 conditional knockout mice to determine what role Dpy30 and its associated H3K4 methylation may play in the fate determination of tissue-specific stem cells such as HSCs.
Abstract
As the major histone H3K4 methyltransferases in mammals, the Set1/Mll complexes play important roles in animal development and are associated with many diseases, including hematological malignancies. However, the role of the H3K4 methylation activity of these complexes in fate determination of hematopoietic stem and progenitor cells (HSCs and HPCs) remains elusive. Here, we address this question by generating a conditional knockout mouse for Dpy30, which is a common core subunit of all Set1/Mll complexes and facilitates genome-wide H3K4 methylation in cells. Dpy30 loss in the adult hematopoietic system results in severe pancytopenia but striking accumulation of HSCs and early HPCs that are defective in multilineage reconstitution, suggesting a differentiation block. In mixed bone marrow chimeras, Dpy30-deficient HSCs cannot differentiate or efficiently up-regulate lineage-regulatory genes, and eventually fail to sustain for long term with significant loss of HSC signature gene expression. Our molecular analyses reveal that Dpy30 directly and preferentially controls H3K4 methylation and expression of many hematopoietic development-associated genes including several key transcriptional and chromatin regulators involved in HSC function. Collectively, our results establish a critical and selective role of Dpy30 and the H3K4 methylation activity of the Set1/Mll complexes for maintaining the identity and function of adult HSCs.
INTRODUCTION
The stability and plasticity of cell identity is ultimately controlled at the level of gene expression, which is profoundly influenced by the global and local chromatin and epigenetic status of the cell. Hematological diseases, including leukemias, can be caused by perturbation of epigenetic pathways that leads to dysregulated maintenance, proliferation, and differentiation of hematopoietic stem and/or progenitor cells (HSCs and HPCs, or HSPCs; Chung et al., 2012; Shih et al., 2012; Issa, 2013). On the other hand, targeting epigenetic modulators has shown promising efficacy against certain hematopoietic diseases, especially cancer, even if no major genetic lesions are found in the genes encoding the modulators (Dawson and Kouzarides, 2012).
Histone H3K4 methylation is one of the most prominent of epigenetic modifications that are generally associated with gene activation (Martin and Zhang, 2005; Kouzarides, 2007). As the major histone H3K4 methylation enzyme in mammals, the Set1/Mll complexes comprise Set1a, Set1b, Mll1 (Mll, Kmt2a), Mll2 (Kmt2b), Mll3 (Kmt2c), or Mll4 (Kmt2d) as the catalytic subunit, and Wdr5, Rbbp5, Ash2l, and Dpy30 as integral core subunits necessary for the full methylation activity (Dou et al., 2006; Shilatifard, 2008, 2012; Ernst and Vakoc, 2012). The functional role of their H3K4 methylation activity, however, remains largely unclear in various physiological processes, including hematopoiesis and fate determination of somatic stem cells such as HSCs. Moreover, whereas genetic lesions and altered expression of several subunits in the Set1/Mll complexes have been increasingly associated with developmental disorders and cancers, including blood cancers (Lüscher-Firzlaff et al., 2008; Ng et al., 2010; Jones et al., 2012; Kim et al., 2014; Takata et al., 2014; Lee et al., 2015; Rao and Dou, 2015), the role of their H3K4 methylation activity in these diseases remains elusive, creating a barrier to a better understanding and potential pharmacological targeting of these modulators in diseases.
Our understanding of roles of Set1/Mll complexes in hematopoiesis is largely limited to genetic studies of Mll1, which is essential for hematopoiesis and self-renewal of adult HSCs, as indicated by the rapid loss of HSCs upon Mll1 deletion in the hematopoietic system (Jude et al., 2007; Gan et al., 2010) or after transplantation (Jude et al., 2007; McMahon et al., 2007; Gan et al., 2010). The H3K4 methylation activity of Mll1, however, was recently shown to be dispensable for hematopoiesis or HSC function, whereas Mll1’s other activities, such as its recruitment of H4K16 acetyltransferase, are critically required (Mishra et al., 2014). Acute Mll1 deletion has no impact on global or gene-specific H3K4 methylation (Mishra et al., 2014), possibly as a result of compensation by other Set1/Mll enzymes, and is thus not suitable for studying the role of H3K4 methylation for hematopoiesis. Similarly, although roles of a few other integral subunits of the Set1/Mll complexes (Chen et al., 2014; Chun et al., 2014; Santos et al., 2014; Zhang et al., 2015) in mammalian hematopoiesis have been reported, the role of the associated H3K4 methylation activities was not clearly established in hematopoiesis and HSC function.
We have previously established a direct role for the Dpy30 subunit of the Set1/Mll complexes in facilitating genome-wide H3K4 methylation (Jiang et al., 2011). Through direct binding to Ash2l, the Dpy30 core subunit is believed to facilitate the H3K4 methylation activities of all Set1/Mll complexes (Ernst and Vakoc, 2012). This allows an effective interrogation of the role of H3K4 methylation activity in stem cells through genetic manipulation of Dpy30. Interestingly, Dpy30-facilitated H3K4 methylation is not essential for self-renewal of mouse embryonic stem cells (ESCs) or the expression of the pluripotency genes in ESCs, but is crucial for the activation potential of the bivalently marked developmental genes and the efficient differentiation of ESCs (Jiang et al., 2011). It remains unknown what role, if any, Dpy30 may play in the maintenance and differentiation of somatic stem cells, and in particular, the HSCs. We recently showed that Dpy30 depletion reduced proliferation and affected the proper differentiation of mobilized human CD34+ HPCs in ex vivo culture (Yang et al., 2014). We then showed that Dpy30-depleted zebrafish had reduced global H3K4 methylation, disrupted hematopoiesis, and surprisingly, elevated levels of early hematopoietic markers, suggesting an accumulation of early HSPCs (Yang et al., 2014). These results prompted us to further study the role of Dpy30 in hematopoiesis and regulation of HSPC function in a mouse model.
RESULTS
Dpy30 deletion in the mouse hematopoietic system results in pancytopenia
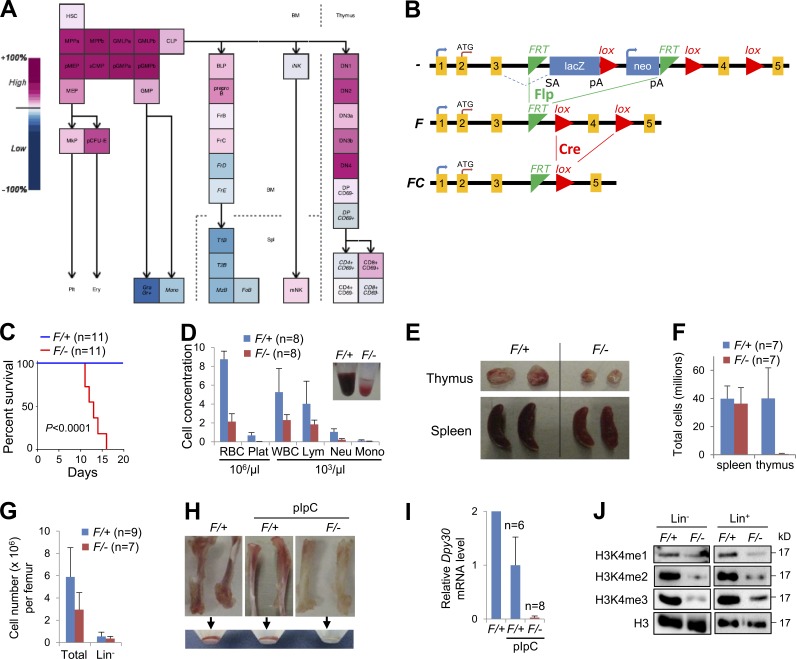
In the mouse hematopoietic system, Dpy30 expression is modest in HSCs and highest in multipotent progenitors and myeloid progenitor (MyePro) cells; it is also high in precursor erythroid, B, and T cells, but very low in certain mature cells, including granulocytes, monocytes, and splenic B cells (Fig. 1 A). Although HSCs are overall more active in transcription than their progeny (Chambers et al., 2007), Dpy30 expression is actually increased from HSCs to HPCs, suggesting that Dpy30 does not merely control the general transcription level, but may play a role in regulating genes involved in lineage determination.
Figure 1.
Dpy30 KO in the hematopoietic system results in depletion of H3K4 methylation and pancytopenia. (A) Dpy30 expression levels in the mouse hematopoietic system, shown as output from Gene Expression Commons. (B) The Dpy30 KO strategy, showing different status of the Dpy30 allele. Dpy30 protein has 99 aa in total, and the domain responsible for facilitating methylation is at the C-terminal half, which is encoded starting within the targeted exon 4, and is thus deleted by Cre-mediated recombination. (C–J) pIpC was injected into Mx1-Cre; Dpy30F/+ (F/+, control) and Mx1-Cre; Dpy30F/− (F/−, KO) mice for 4 times, except in H and I where no pIpC is labeled. All mice were examined 4 d after last pIpC injection. (C) Kaplan-Meier curves for survival of animals after pIpC injections (first pIpC injection on day 0). n = 11 mice for each genotype. P < 0.0001 by log-rank test. (D) Profiles of PB. Insert shows comparison of typical PB samples. n = 8 mice for each genotype. (E) Thymus and spleen in the control and KO mice. (F) Numbers of total splenocytes and thymocytes in the control and KO mice. n = 7 mice for each genotype. (G) Total and Lin− BM cell numbers. n = 9 mice for F/+; n = 7 mice for F/−. (H) Representative long bones and cells flushed from these bones. (I) Relative Dpy30 mRNA levels in BM by RT-qPCR and normalized to Actb. n = 6 mice for F/+; n = 8 mice for F/−; both with pIpC injection. (J) Western blotting for total H3 and different levels of H3K4 methylation in sorted Lin− and Lin+ BM. Data are shown as mean ± SD for D, F, G, and I.
We generated a Dpy30 conditional KO mouse using ESCs of C57BL/6 background (Pettitt et al., 2009) harboring a Dpy30 knockout first allele. The knockout first allele is initially nonexpressive (–) as a result of the trapping cassette, but can be converted to a conditional allele (F) via Flp recombination, and further to a null allele via Cre recombination by causing a frameshift mutation and likely triggering nonsense-mediated decay of the mutant transcript (Fig. 1 B). Consistent with a recent study showing that Dpy30−/− mouse embryos die between E7.5 and E9.5 (Bertero et al., 2015), we never recovered any Dpy30−/− mice from intercrossing of Dpy30+/− mice. We then bred Dpy30+/− with Rosa26-Flpe mice to generate Dpy30F/+ and additional Dpy30F/F mice. For corroboration, we used three different Cre lines: Mx1-Cre (Kühn et al., 1995), in which the Cre expression is mainly induced in the hematopoietic system by injection of polyinosinic-polycytidylic acid (pIpC), and CAG-CreER (Hayashi and McMahon, 2002) and Rosa26-CreER (Ventura et al., 2007), in both of which the Cre expression is induced in most tissues by tamoxifen injection. To avoid potential effects from Cre expression, we bred the Dpy30F/F with Cre; Dpy30+/− mice to produce and compare Cre; Dpy30F/+ and Cre; Dpy30F/− littermates.
All Mx1-Cre; Dpy30F/− (KO) mice died 11–18 d after the start of pIpC injections (Fig. 1 C). Moribund animals were pancytopenic with reduced peripheral blood (PB) counts of all cell types (Fig. 1 D). These animals had virtually no thymocytes (Fig. 1, E and F), although the splenocyte number was not significantly affected (Fig. 1 F). Similar reduction of the white blood cell number was also seen in the tamoxifen-injected CAG-CreER; Dpy30F/− and Rosa26-CreER; Dpy30F/− mice, although the red blood cell number was not affected (not depicted), presumably as a result of the less efficient Dpy30 deletion in these mice (Fig. 2 F and not depicted). Long bones of Dpy30 KO mice had 50% fewer nucleated cells (Fig. 1 G) and a very low number of red blood cells (Fig. 1 H). We confirmed Dpy30 deletion in BM by genomic PCR (not depicted), and showed that Dpy30 expression was nearly abolished at both RNA and protein levels (Fig. 1 I; see also Fig. 7 D) in the BM of the pIpC-injected Mx1-Cre; Dpy30F/− mice. Consistent with a role of Dpy30 in regulating all three levels of H3K4 methylation (Jiang et al., 2011), global H3K4me2 and H3K4me3 were drastically reduced, and H3K4me1 was also diminished in both Lin− and Lin+ fractions of the Dpy30 KO BM (Fig. 1 J). These results indicate that Dpy30 and its associated H3K4 methylation are critical for mouse hematopoiesis, likely at early stages.
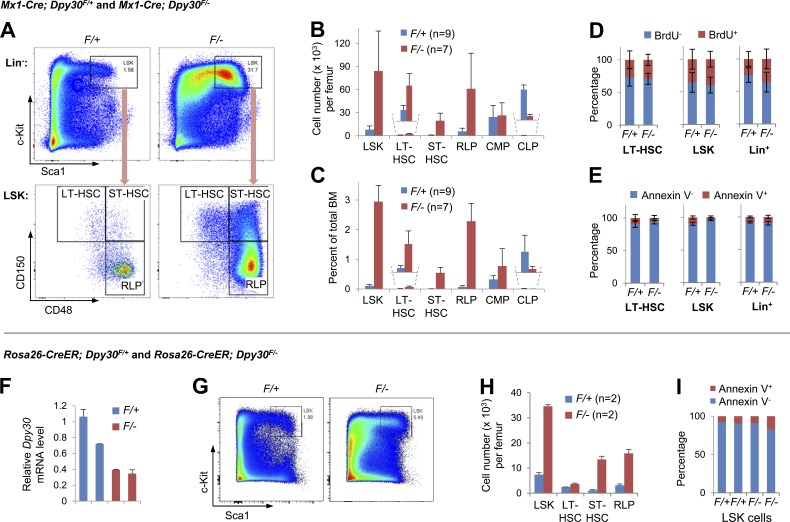
Figure 2.
Dpy30 KO results in accumulation of early HSPCs. (A–E) Analyses of BM from Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice 4 d after the last pIpC injection. (A) Representative FACS analysis of BM cells. (B and C) Absolute numbers of BM cell populations (B) and percentages in total BM cells (C). n = 9 mice for F/+; n = 7 mice for F/-, except for n = 4 for CMP and CLP of each genotype. P < 0.001 for LSK, LT-HSCs, ST-HSCs, and CLP; P < 0.01 for RLP, by Student’s t test. (D) BrdU incorporation assay for BM. n = 10 mice for each genotype. P > 0.05 for all cell types by Student’s t test. (E) Annexin V staining for BM. n = 7 mice for F/+; n = 9 mice for F/−. P > 0.05, except for LSK cells by Student’s t test. (F–I) Analyses of BM from Rosa26-CreER; Dpy30F/+ and Rosa26-CreER; Dpy30F/− mice 4 d after the last tamoxifen injection in a series of seven injections. (F) Relative Dpy30 mRNA levels in BM of individual mice (each by a bar) was determined by RT-qPCR of duplicate measurements and normalized to Actb. (G) Representative FACS analysis of Lin− BM. (H) Absolute numbers of indicated BM cell populations. n = 2 mice per group. P < 0.05 for all by Student’s t test. (I) Annexin V staining for BM LSK cells. Note that the F/− LSK cells still accumulate in H, despite the mild increase of Annexin V+ percentage in these cells. Data are shown as mean ± SD for B–F and H.
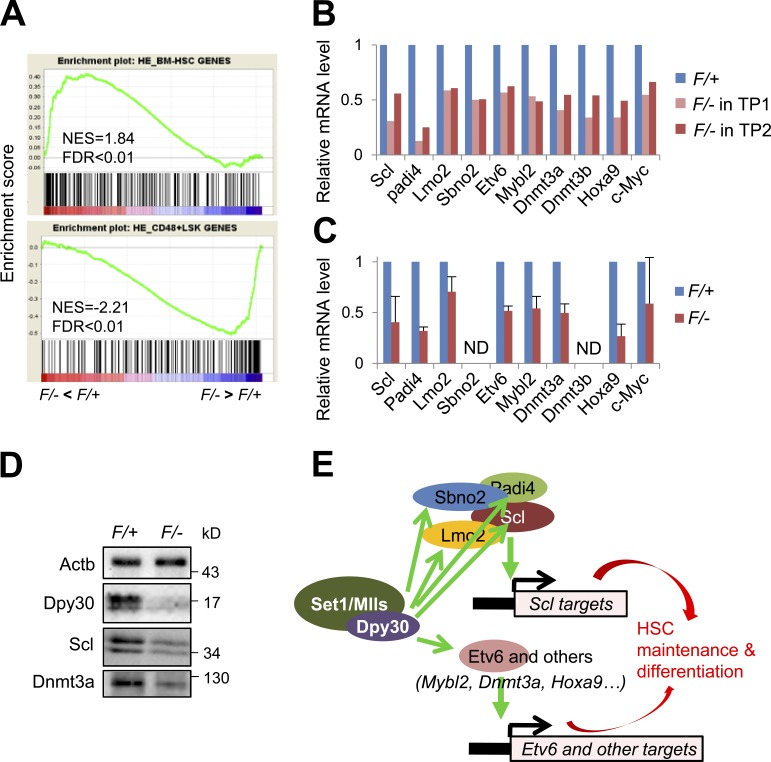
Figure 7.
Dpy30 KO results in loss of HSC identity and down-regulation of key genes for HSC maintenance and function. (A–C) Gene expression analyses of donor-derived LT-HSCs in BM chimeras 2 wk after pIpC injections following the schemes in Fig. 4 A and Fig. S1. (A) GSEA for genes affected in the Dpy30 KO (F/–) HSCs in the BM chimeras, after RNA-seq analyses. The top panel shows an enrichment of LT-HSC (LSK CD48−CD150+) gene set in genes down-regulated, whereas the bottom panel shows an enrichment of RLP and ST-HSC (LSK CD48+) gene set in genes up-regulated, in Dpy30 KO HSCs. Both gene sets are from (He et al., 2011). (B) Relative expression of key HSC-regulatory genes in the control (F/+) versus Dpy30 KO (F/–) HSCs in the BM chimeras, as analyzed by RNA-seq in two independent BM transplantations (TP1 and TP2). (C) Relative expression of the same genes as in B, as analyzed by RT-qPCR and normalized to Actb using HSCs from three independent BM transplantations and shown as mean ± SD. ND, not done due to insufficient RNAs. P < 0.05 for all genes by Student’s t test, except for c-Myc. (D) Western blotting for indicated proteins in whole BMs from pIpC-injected Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/– mice. (E) A model illustrating HSC regulation by Dpy30 via multiple genes and pathways.
Strong accumulation of phenotypic early HSPCs in the Dpy30 KO BM
Unexpectedly, and in contrast to the rapid HSC loss upon pIpC injection in the Mx1-Cre; Mll1F/F BM (Jude et al., 2007), we consistently detected a striking accumulation, in absolute number and frequency, of HSPCs including LT-HSCs, ST-HSCs, and restricted lineage progenitors (RLPs; Kiel et al., 2005) in the Dpy30 KO BM (Fig. 2, A–C; and Table S1 for cell surface markers). Although common MyePros (CMPs) were unaffected, common lymphoid progenitors (CLPs) were markedly reduced (Fig. 2, B and C). Similar accumulation of early HSPCs was also seen in the BM of tamoxifen-injected CAG-CreER; Dpy30F/− and Rosa26-CreER; Dpy30F/− mice (Fig. 2, F–H; and not depicted), and thus was not an artifact of pIpC injection. These phenotypes in mouse are consistent with those in Dpy30-depleted zebrafish, which has severe anemia yet possibly increased early HSPCs (Yang et al., 2014).
Dpy30 loss had no significant effect on the proliferation or apoptosis of HSC, LSK, and Lin+ BM cells (Fig. 2, D and E), except for a significant (but mild) reduction of apoptosis for the KO LSK cells (Fig. 2 E). LSK cell accumulation, however, was not likely to be dependent on reduction of LSK apoptosis, as LSK cells did not show reduction in apoptosis after Rosa26-CreER–mediated Dpy30 KO (Fig. 2 I), but still showed marked accumulation (Fig. 2, G and H). Therefore, although the mildly reduced LSK apoptosis upon Mx1-Cre–mediated Dpy30 KO may contribute to their accumulation, it is unlikely that the early HSPC accumulation is a result of abnormal cell proliferation or inhibition of cell death.
Dpy30 KO BM is functionally defective in multilineage reconstitution
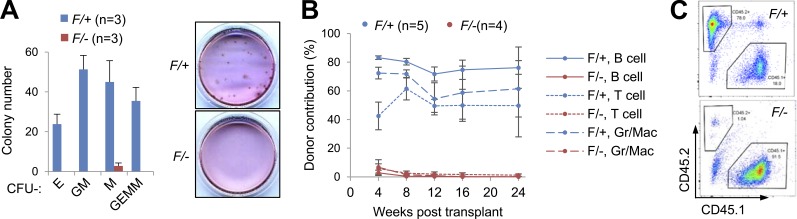
We showed that the ability of BM cells to form all different types of colonies was nearly abolished after Dpy30 loss by Mx1-Cre or Rosa26-CreER induction (Fig. 3 A and not depicted). We then evaluated HSC function in competitive BM transplantation assays, using donor BM with Dpy30 deleted by either CAG-CreER or Mx1-Cre. Although the unaffected donor contribution to recipient BM at 18 h after transplantation indicated an intact homing capacity (unpublished data), BM with Dpy30 deleted by either Cre was severely defective in reconstituting multiple PB lineages (Fig. 3, B and C; and not depicted). Together with early HSPC accumulation and largely unaffected cell proliferation/death, these functional defects are most consistent with a HSPC differentiation block after Dpy30 loss.
Figure 3.
Dpy30 KO HSPCs are functionally defective. (A) Colony formation assay using 105 BM cells from Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice 4 d after the last pIpC injection. n = 3 mice for each genotype. A representative image of the plates with formed colonies is shown on the right. (B) CAG-CreER; Dpy30F/+ and CAG-CreER; Dpy30F/− mice were injected with tamoxifen seven times and whole BMs were used in competitive transplantation (donor: competitor = 10:1), and donor contribution to indicated PB cells was determined at different times after transplantation. n = 5 mice for F/+; n = 4 mice for F/−. P < 0.001 for all by Student’s t test. (C) A representative FACS analysis of PB B cells 8 wk after transplantation shown in B. Data are shown as mean ± SD for A and B.
Dpy30 KO BM is defective in differentiation
To more rigorously study the cell-autonomous effects of Dpy30 loss, we established mixed BM chimeras and then deleted Dpy30 after stable engraftment of the donor BM (Fig. 4 A). This approach minimizes potential effects of HSC homing/lodging and provides a largely normal cellular microenvironment, with phenotypes being evaluated relative to the competitors in the same BM.
Figure 4.
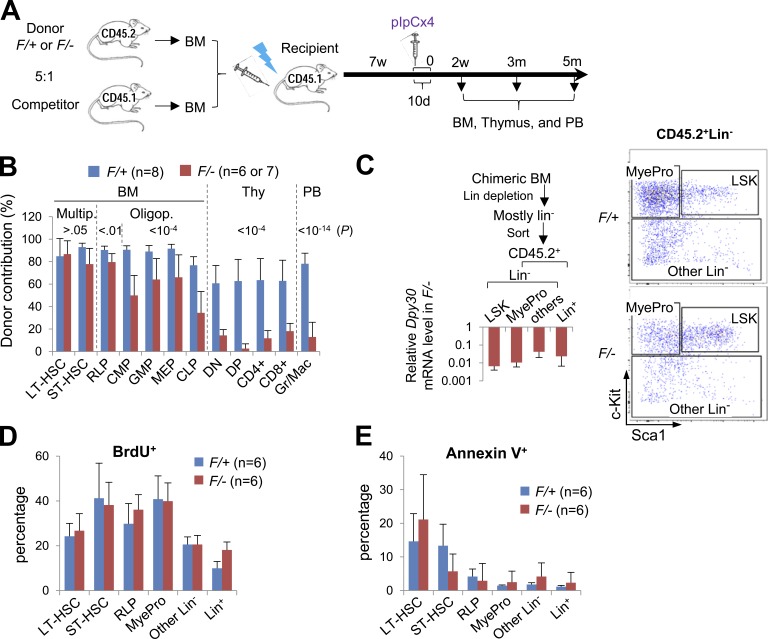
Short-term BM chimera system further reveals differentiation defects of Dpy30 KO HSPCs. (A) Scheme for the mixed BM chimera system using whole BM from Mx1-Cre; Dpy30F/+ or Mx1-Cre; Dpy30F/− mice as donors and from CD45.1+ mice as competitors (donor: competitor = 5:1). This scheme is for all panels in Figs. 4, 5, 6,and 7, except for Fig. 6 (C and D) and Fig. 7 D. (B–E) Analyses of donor-derived cells 2 wk after pIpC injections. (B) Donor contribution to different cell populations in BM chimeras. n = 8 (recipient) mice for F/+; n = 6 or 7 mice for F/-. P > 0.05 for multipotent cells; P < 0.01 for RLP; P < 10−4 for CMP, GMP, MEP, and thymocytes; P < 10−14 for PB, by Student’s t test. (C) Different populations of donor-derived cells were isolated following the scheme on the left. Dpy30 mRNA levels in sorted F/− relative to F/+ cells (set as 1) of the same cell types were determined by RT-qPCR and normalized to Actb. n = 5 mice for each genotype. Shown on the right is a representative FACS sorting of CD45.2+Lin− cells. (D) BrdU incorporation assay for different (donor-derived) cell populations in chimera BM. n = 6 mice for each genotype. P > 0.2 for all cell types except Lin+ (P = 0.002) by Student’s t test. (E) Annexin V staining for different (donor-derived) cell populations in chimera BM. n = 6 mice for each genotype. P > 0.1 for all cell types except ST-HSC (P = 0.05) by Student’s t test. Data are shown as mean ± SD for B–E.
As shown by our analyses 2 wk after pIpC injections, Dpy30 loss had minimal effects on donor contribution to HSCs, but significantly reduced donor contribution to oligopotent HPCs, peripheral Gr1+/Mac1+ cells, and developing T cells in thymus (Fig. 4 B), consistent with a multilineage differentiation defect. The observed effect is likely underestimated by the half-life of cells generated before Dpy30 deletion, as we noted no reduction (unpublished data) in long-lived mature B and T cells (Förster and Rajewsky, 1990; Kondo et al., 1997) at this time point. Importantly, the progressive loss of donor cell frequency was not due to an increased KO efficiency along specific differentiation pathways, as we sorted donor-derived cell populations into LSK, MyePro, Other Lin− (including lymphoid progenitors), and Lin+ cells (Fig. 4 C) and found that Dpy30 deletion was actually most efficient in the LSK cells (less than 1% mRNA remaining in the KO LSK cells; Fig. 4 C). Moreover, Dpy30 loss did not significantly affect the proliferation or death of most of the donor-derived HSPC populations (Fig. 4, D and E). The progressive loss of hematopoietic cells is thus most consistent with a defect in multi-stage differentiation. Our data show important divergence of the Dpy30 KO and Mll1 KO phenotypes at the same time point in the same chimera system, the latter showing reduced contribution to HSPCs, but not to mature cells including granulocytes (Jude et al., 2007).
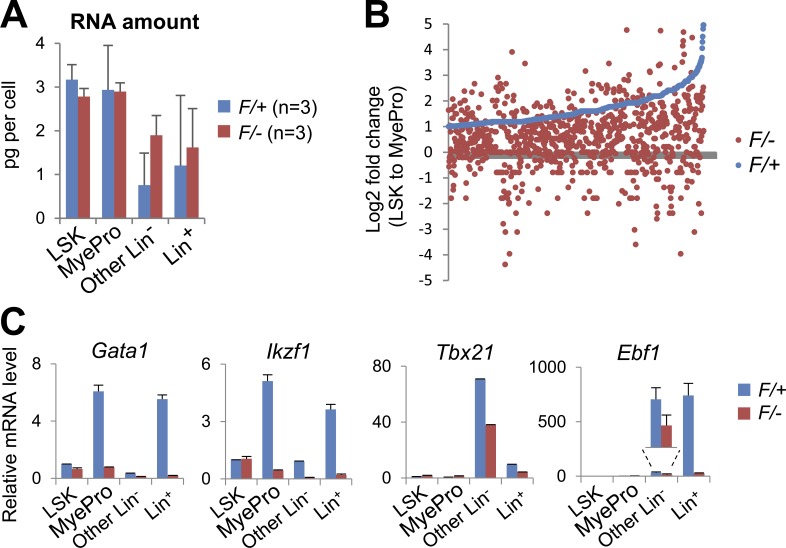
Because cell identity is ultimately defined by its gene expression program, we sought to determine the KO effect on gene expression by RNA sequencing (RNA-seq), using the donor-derived cell populations sorted based on their surface markers (Fig. 4 C). Importantly, Dpy30 loss did not significantly affect the overall RNA levels in any of these cell populations (Fig. 5 A). As we focused on the transition from LSK to MyePro cells in the control genetic background, we found that many genes in the genome were markedly up-regulated, consistent with the gradual commitment to the myeloid lineage. Such up-regulation, however, was greatly impaired in the same transition in the Dpy30 KO background (Fig. 5 B and Table S2). Further analyses by qPCR assays (Fig. 5 C) showed that, in the transition from LSK to either MyePro cells or lymphoid progenitor–containing population (the Other Lin– population), Dpy30 loss resulted in an impaired up-regulation of several genes encoding transcription factors known to regulate myeloid and/or lymphoid lineage development, such as Gata1 (Wolff and Humeniuk, 2013), Ikzf1/Ikaros (Ng et al., 2007), Tbx21/Tbet (Jayaraman, 2013; Lazarevic et al., 2013), and Ebf1 (Boller and Grosschedl, 2014). These results indicate that, in addition to the reduced number of immunophenotypic HPCs, Dpy30 loss resulted in an intrinsic block in transition of transcription programs, even in cells that (temporally) displayed correct surface markers.
Figure 5.
Impaired induction of lineage-regulatory genes after loss of Dpy30. (A–C) Gene expression analyses were performed for Mx1-Cre; Dpy30F/+ or Mx1-Cre; Dpy30F/– donor-derived cells 2 wk after pIpC injections after the schemes in Fig. 4 (A and C). (A) RNA amount per cell in sorted cell populations. n = 3 mice for each genotype. P > 0.1 for all by Student’s t test. Data are shown as mean ± SD. (B) Expression change (shown as Log2 fold change) from LSK to MyePro cells in F/− (red) and in F/+ (blue) backgrounds, as determined by RNA-seq of sorted cells. Genes (represented by each dot) were ranked according to their fold changes in F/+ cells, and only genes with fold change >2 (Log2 fold change >1) in F/+ cells are shown. Note that most of these genes show a smaller fold change in F/− than in F/+ cells. See Table S2 for the gene lists. (C) Expression levels of indicated genes in the sorted cell populations were determined by RT-qPCR and normalized to Actb, and shown as mean ± SD of duplicate measurements. The expression levels in F/+ LSK cells were set as 1. Shown are representative results from one out of three independent chimera transplantation experiments. Results from two more independent transplantation experiments (not depicted) are consistent with these results.
Dpy30 KO BM fails to sustain in long-term transplantation
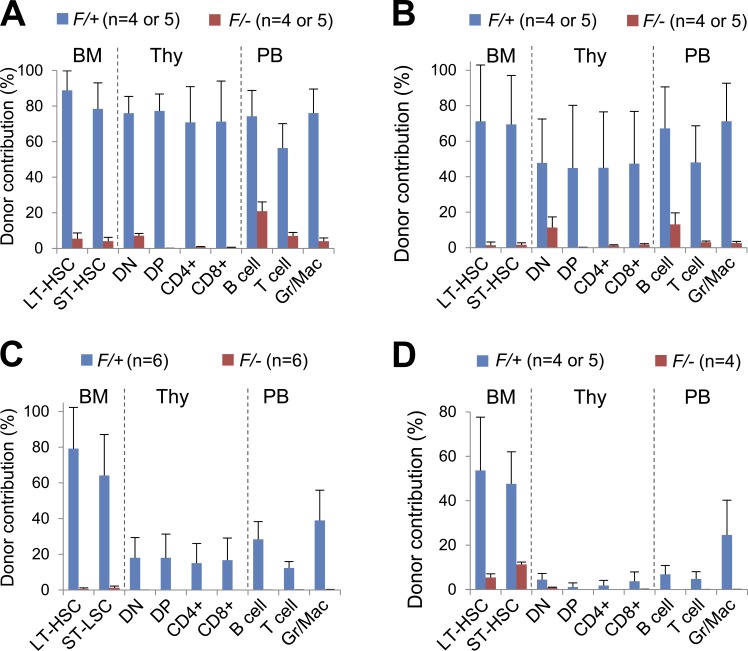
As we followed the BM chimeras for a longer period of time, we found that Dpy30 deletion greatly reduced donor frequency for HSCs, developing T cells in thymus, and mature multilineage hematopoietic cells in periphery at 3 and 5 mo after pIpC injections (Fig. 6, A and B). To complement the whole BM transplantation, we also purified LT-HSCs from Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice after pIpC injections and competitively transplanted these cells into recipients. Compared with the whole BM transplant, transplant using the purified Dpy30 KO HSCs showed an even more profound loss of HSCs and multilineage reconstitution (Fig. 6 C). To further demonstrate a defect in long-term maintenance of Dpy30 KO HSCs, we performed secondary transplantation using immunophenotypically sorted donor-derived LT-HSCs from primary recipients at 2 mo after pIpC injections, supplemented with whole BM competitor cells. Collectively, our analyses at 3 mo after secondary transplant showed a severe loss of HSCs and multilineage mature blood cells with a Dpy30 KO background (Fig. 6 D). These results indicate an essential role of Dpy30 in the long-term maintenance of HSCs, in addition to their differentiation as revealed at the early time point.
Figure 6.
Dpy30 KO results in loss of long-term maintenance of HSCs. (A and B) Donor contribution in BM chimeras 3 (A) and 5 (B) mo after pIpC injections following the scheme in Fig. 4 A. n = 10 mice of each genotype for PB; n = 4 or 5 mice of each genotype for BM and thymus. P < 0.001 for all cell populations by Student’s t test. (C) Donor contribution in recipients 3 mo after transplantation of LT-HSCs purified from pIpC-injected Mx1-Cre; Dpy30F/+ or Mx1-Cre; Dpy30F/– mice. n = 6 mice for each genotype. P < 0.01 for all cell populations by Student’s t test. (D) Donor contribution 3 mo after secondary transplantation, in which donor-derived HSCs sorted from pIpC-injected BM chimeras (primary recipients) were competitively transplanted into secondary recipient mice. n = 4 or 5 mice for each genotype. P < 0.05 for all cell populations by Student’s t test, except for CD4+ and CD8+ thymocytes, for which the levels in control genotype were already extremely low and highly variable, preventing a manifestation of a statistically significant reduction in the KO background. Data are shown as mean ± SD for all panels.
Dpy30 KO results in loss of HSC identity and down-regulation of key pathways regulating HSC maintenance and function
To understand the molecular mechanisms underlying HSC regulation by Dpy30 in a cell-autonomous manner, we performed RNA-seq using immunophenotypically purified donor-derived HSCs 2 wk (to ensure sufficient numbers of the KO HSCs) after pIpC injections into the mixed BM chimeras (Fig. S1). Gene Set Enrichment Analysis (GSEA) revealed that the HSC signature genes (Chambers et al., 2007; He et al., 2011) were significantly enriched among down-regulated genes, whereas RLP signature genes were significantly enriched among genes up-regulated in the Dpy30 KO HSCs (Fig. 7 A and not depicted). This suggests that, despite the (temporal) expression of the HSC surface markers in the analyzed cells, the HSC identity (as more intrinsically reflected by their signature gene expression programs) was significantly impaired in the absence of Dpy30. Further examination of the RNA-seq results (Fig. 7 B and Table S3), and subsequent validation by qPCR assays (Fig. 7 C), showed that the KO HSCs significantly down-regulated several genes known to be critically involved in HSC maintenance and/or differentiation. These genes encode for key transcription factors (Scl/Tal1, Etv6, Mybl2/B-Myb, Hoxa9, and c-Myc) and co-regulators (Padi4, Lmo2, and Sbno2), and epigenetic regulators (Dnmt3a and Dnmt3b, and Padi4; see Discussion). We also confirmed the down-regulation of Scl and Dnmt3a protein levels in the Dpy30 KO BM (Fig. 7 D).
Preferential control of H3K4me3 at hematopoietic genes by Dpy30 in BM cells
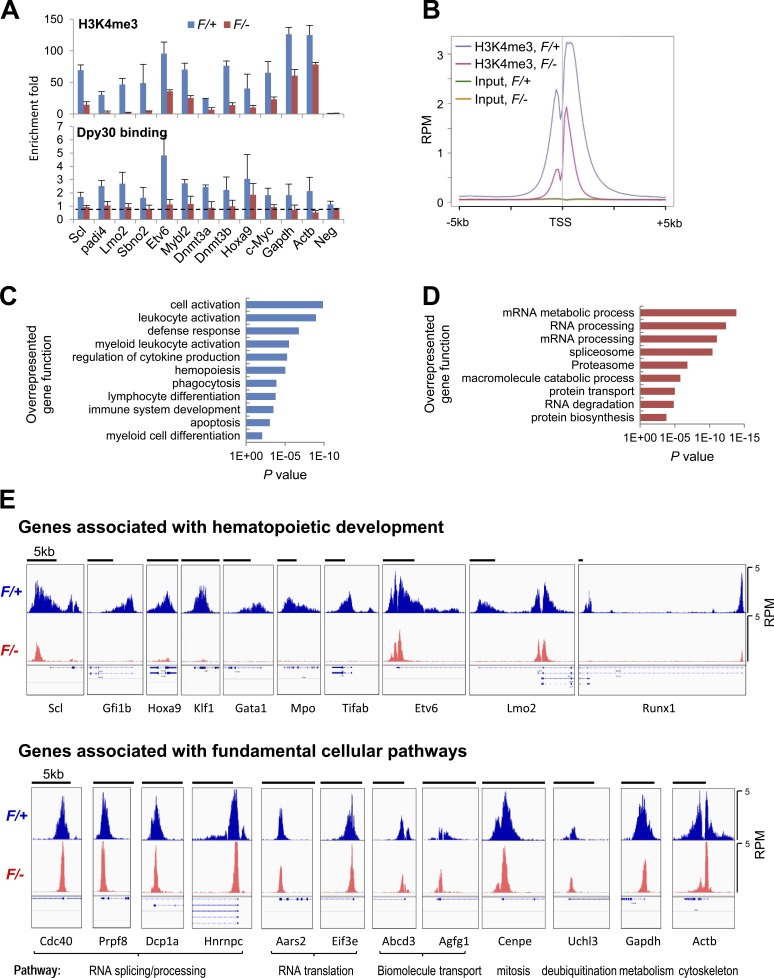
We next sought to determine the epigenetic impact of Dpy30 loss on the affected HSC-regulatory genes by chromatin immunoprecipitation (ChIP) assays on Lin− BM cells from Mx1-Cre; Dpy30F/+ (control) and Mx1-Cre; Dpy30F/− (KO) mice after pIpC injections. Our qPCR assays showed that abundant H3K4me3 (comparable to the house-keeping genes Gapdh and Actb) was enriched at the transcription start sites (TSSs) of these genes in control BM cells, and this modification was markedly reduced at all of the examined genes after Dpy30 loss (Fig. 8 A, top). We also determined that Dpy30 directly bound to the TSSs of all of these genes, and its binding was greatly reduced after Dpy30 loss (Fig. 8 A, bottom). Interestingly, although the H3K4me3 signal level was the highest for Gapdh and Actb, Dpy30 binding to these genes was not higher than most of the HSC regulatory genes. Furthermore, the impact of Dpy30 loss on promoter H3K4me3 appears to be stronger at those HSC regulatory genes than at Gapdh and Actb (Fig. 8 A).
Figure 8.
Differential effect of Dpy30 KO on genomic H3K4me3. (A) ChIP for H3K4me3 (top) and Dpy30 (bottom) using Lin− BM cells from Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice 4 d after the last pIpC injection, and shown as mean ± SD from three independent injection experiments. The dotted line in the bottom panel represents the approximate level of nonspecific ChIP signal that needs to be subtracted when assessing the Dpy30-specific binding level, given the thorough depletion of Dpy30 in the KO cells. (B) The composite profiles of indicated ChIP-seq results for all genes that has an H3K4me3 peak within 5 kb up- or downstream of its TSS. (C and D) Gene ontology analysis by DAVID for genes with >90% reduction (C) or <50% reduction (D) of H3K4me3 at TSS regions in the F/– (KO) compared with the F/+ (control) cells (see Table S4 for the gene lists). (E) H3K4me3 ChIP-seq profiles shown in Integrated Genomic Viewer for the representative genes associated with hematopoietic development (top) and genes associated with various fundamental cellular pathways (bottom). The black bars on top of each panel show 5-kb scale. All panels have the same signal scale of 0–5 RPM on the y axis. The expanded RefSeq genes are shown below each panel to show the transcript isoforms, including those with alternative TSSs (and thus multiple H3K4me3 peaks).
To examine the impact of Dpy30 loss on genome-wide epigenetic modifications, we sequenced the H3K4me3 and H3K27me3 ChIP DNAs from the control and Dpy30 KO Lin− BM cells. Consistent with the global reduction of H3K4me3 detected by Western blotting (Fig. 1 J), both the intensity and the breadth of the genome-wide H3K4me3 domains were markedly reduced in the KO cells (Fig. 8 B), whereas the H3K27me3 ChIP signal was minimally affected (unpublished data). We note that because the total H3K4me3-enriched DNA amount per cell was greatly reduced in the KO sample, the KO effect on local H3K4me3 was most likely underestimated throughout the genome, as our ChIP-seq counts of reads in any specific regions were normalized to total counts of reads in the sample. Nevertheless, the relative effects for different gene loci are preserved and were further analyzed. We ranked genes by the fold change of H3K4me3 at their TSS regions in the Dpy30 KO versus control cells and focused on the group of genes with severe (>90%) reduction of H3K4me3 in the KO cells, as well as the group with modest (<50%) reduction of H3K4me3 (Table S4). As revealed by gene ontology analyses or KEGG pathway analysis, the group with severe reduction of H3K4me3 was significantly enriched with genes associated with hematopoietic development (Fig. 8 C and not depicted), whereas the group with modest reduction was significantly enriched with genes involved in fundamental cellular pathways, including RNA/protein processing and degradation (Fig. 8 D and not depicted). Comparison of the H3K4me3 profiles for representatives of hematopoietic development–associated genes and fundamental pathway–associated genes on the Integrated Genomic Viewer also clearly demonstrates a generally more profound effect of Dpy30 loss on the former group of genes than the latter, although the difference was not absolute (Fig. 8 E). Consistent with a recent study showing that H3K4me3 breadth is linked to cell identity (Benayoun et al., 2014), hematopoietic development–associated genes often appeared to have broader H3K4me3 domains than the fundamental pathway-associated genes (Fig. 8 E). Moreover, Dpy30 loss greatly reduced both the intensity and the breadth of the H3K4me3 domains at the hematopoietic genes, yet it often affected the breadth, but not much of the intensity, of the H3K4me3 domains at the fundamental pathway-associated genes (Fig. 8 E). These results suggest that Dpy30 may preferentially regulate genes critically involved in developmental processes, consistent with the profound effects on HSC function and yet marginal effect on general cell survival after its loss.
DISCUSSION
Multiple lines of evidence collectively support an important role of Dpy30 in hematopoietic differentiation: (1) a severe reduction in multilineage mature hematopoietic cells is accompanied by a strong accumulation of early HSPCs in the Dpy30 KO BM, which cannot be sufficiently explained by effects on cell proliferation and apoptosis; (2) a progressive loss of more downstream donor-derived HPCs in the mixed BM chimeras with little effects on cell proliferation and apoptosis shortly after Dpy30 loss; (3) the impaired induction of key lineage-master regulators in the absence of Dpy30 not only reflects lineage abnormality, but may also functionally disrupt lineage differentiation. In addition to differentiation, it is clear that Dpy30 plays a crucial role in maintaining HSC identity, as demonstrated by the down-regulation of HSC signature gene profile and the eventual loss of the Dpy30 KO HSCs in the mixed BM chimeras. We note that HSC defects in self-renewal and differentiation are not mutually exclusive, as exemplified by Eed KO (Xie et al., 2014). We also do not exclude a possible regulatory role of Dpy30 in more downstream lineages, for example, the developing T cells in thymus (with a high Dpy30 expression level in the double negative T cells; Fig. 1 A), and this is consistent with the dramatic effects of Dpy30 loss on thymocytes (Figs. 1 F, 4 B, and 6, A–C). However, we note that the defective HSC function is a critical reason for the thymocytes loss upon Dpy30 deletion, as purified Dpy30 KO HSCs failed to reconstitute thymocytes in the transplant recipients (Fig. 6 C).
Our findings here are also consistent with our previous work (Yang et al., 2014) on the requirement of Dpy30 for zebrafish hematopoiesis and for proper differentiation of human CD34+ HPCs. Some different outcomes in lineage differentiation (e.g., erythroid differentiation) could be explained by the different developmental stages in which Dpy30 depletion is initiated—the Mx1-cre–mediated Dpy30 excision occurs in all stages of hematopoiesis, including the most immature HSCs, and leads to functionally diminished early HPCs, and thus will obscure possible consequences of Dpy30 loss in a later stage as seen in the Dpy30-depleted human CD34+ HPCs. It was somewhat surprising that Dpy30 loss had no effect on the in vivo proliferation of the mouse hematopoietic cells, but greatly reduced the ex vivo proliferation of the human CD34+ HPCs. In addition to the species difference, the very different cellular environments for the ex vivo versus in vivo assays may account for the different effects on proliferation seen here, as well as in some precedents, including L3mbtl1, where depletion of the gene inhibits human CD34+ cell proliferation (Perna et al., 2010), but its genetic loss in mice has no effect on hematopoiesis (Qin et al., 2010).
Our results here show that Dpy30 directly and epigenetically controls multiple key chromatin and transcription regulators that may contribute to the control of HSC maintenance and differentiation (Fig. 7 E). Scl/Tal1 (Lacombe et al., 2010; Rojas-Sutterlin et al., 2014), Etv6 (Hock et al., 2004), Mybl2/B-Myb (Baker et al., 2014), and Hoxa9 (Lawrence et al., 1997; Magnusson et al., 2007) are all transcription factors with established roles in regulating HSC maintenance and/or function. Notably, Dpy30 appears to exert comprehensive regulation of the Scl-transcriptional pathway, as it directly promotes the H3K4 methylation and expression of not only Scl itself, but also genes encoding for cofactors for Scl in transcription regulation, such as Lmo2 (Wadman et al., 1997), Sbno2 (Maruyama et al., 2013), and Padi4 (Kolodziej et al., 2014). Moreover, Padi4 also positively regulates stem cell gene expression in pluripotent cells through citrullinating histone H1 and decondensing chromatin (Christophorou et al., 2014), and thus may also contribute to chromatin and gene regulation in HSCs. Dnmt3a (Challen et al., 2011), Dnmt3b (Challen et al., 2014), and c-Myc (Wilson et al., 2004) are all important regulators for HSC differentiation, and thus may contribute to Dpy30’s role in regulating HSC differentiation. Extensive cross-regulation may also exist among these Dpy30-regulated pathways. For example, Padi4 has been shown to stabilize Dnmt3a protein (Deplus et al., 2014) and also regulate c-Myc expression (Nakashima et al., 2013). Moreover, it is almost certain that many other Dpy30-regulated genes (Table S3) may also functionally contribute to HSC regulation by Dpy30. In light of the new findings (Benayoun et al., 2014) linking the breadth of H3K4me3 domains with the consistency or stability of transcription, we note that Dpy30 also regulates the breadth of the genomic H3K4me3 domains (Fig. 8, B and E), thereby suggesting an important role for Dpy30 in controlling transcriptional consistency. As transcriptional consistency is considered to play an important role in cellular decision making and metazoan development (Raj et al., 2010; Balázsi et al., 2011), it is possible that Dpy30 may regulate HSPC function partly through maintaining the transcriptional consistency of many genes, in addition to regulating the expression levels of genes involved in HSPC function as shown by our results here.
Despite its profound role in HSPC function, including differentiation and long-term maintenance, its loss has little effect on hematopoietic cell proliferation or survival in vivo. Consistently, Dpy30-mediated efficient global H3K4 methylation is not essential for general gene expression, and its severe reduction does not lead to crash of global RNA expression (Fig. 5 A), but significantly affects expression of genes associated with HSPC function. Moreover, Dpy30 loss results in an overall more profound depletion of H3K4 methylation at developmental genes than at genes involved in fundamental cellular pathways. The molecular mechanisms underlying the differential regulation is unknown. One reason could be that the genes in fundamental pathways have generally higher H3K4me3 levels than the hematopoietic genes, and thus may retain more methylation after the Dpy30 loss. There are, however, abundant examples of fundamental pathway-associated genes (e.g., Aars2, Uchl3) at which the H3K4me3 level is comparable to that at the hematopoietic genes and is still only modestly reduced upon Dpy30 loss (Fig. 8 E and Table S4). Although it is possible that our observations may have resulted from selective elimination of a subpopulation of Dpy30 KO cells in which H3K4me3 was greatly reduced at fundamental pathway-associated genes, this possibility is low because we observed accumulation of Lin− BM cells and no significant increase of BM cell death after Dpy30 loss. Regardless of the underlying mechanisms, our studies provide direct evidence that, although considered a global epigenetic modulator, the functional impact of Dpy30 on local epigenetic modification is not uniform throughout the genome, but rather differential on selective pathways depending on the specific cellular context. We think that such differential epigenetic regulation has important implications for developing potential inhibitors of this modulator to preferentially target disease-associated rather than fundamental cellular pathways.
Another interesting finding is that while Dpy30 is crucial for differentiation of both ESCs and HSCs, it is essential for the maintenance of HSCs, but not ESCs (Jiang et al., 2011). The reason for this differential effect is unclear. It is plausible that as the stem cells get more committed from ESCs to HSCs, the more restricted chromatin environment renders the stem cell maintenance genes more dependent on active epigenetic modifications including H3K4 methylation.
The phenotypes of the Dpy30 KO mice are markedly different (even opposite in some aspect) from the Mll1 KO mice (Jude et al., 2007), and we also did not detect a significant enrichment of the Mll1 KO-affected gene set (Artinger et al., 2013) in the genes down-regulated by Dpy30 KO in HSCs (unpublished data). The differential effects of Mll1 and Dpy30 on HSC function may be explained by the many different catalytic subunits of the Set1/Mll complexes that Dpy30 regulates, as well as the nonenzymatic activities of Mll1 (Mishra et al., 2014). It is worth noting that although Mll1 regulates hematopoietic genes through mechanisms other than its H3K4 methylation activity, it does not rule out a possible role and requirement of H3K4 methylation by other enzymes in regulating their expression. It is likely that the correct expression of some hematopoietic genes requires the nonmethylation activities of Mll1, such as recruiting the H4K16 acetylation activity (Mishra et al., 2014), as well as efficient H3K4 methylation catalyzed by other Dpy30-dependent methyltransferases, considering a genome-wide correlation (Bernstein et al., 2005) and functional coupling of histone acetylation and H3K4 methylation in regulating gene expression (Dou et al., 2005; Jiang et al., 2013; Tang et al., 2013).
It remains unknown which catalytic subunits of the Set1/Mll complexes are primarily responsible for the Dpy30-regulated H3K4 methylation that is involved in hematopoiesis and HSC function. Most of these subunits appear to be possible candidates based on their largely ubiquitous expression patterns in the mouse hematopoietic system (Gene Expression Commons; unpublished data). Several lines of evidence as follows, however, suggest that the H3K4 methylation activities of Set1a/b as well as Mll3 and Mll4, but not Mll1 or Mll2, might be the major contributor to the control of key gene expression involved in HSC function: (1) the phenotypes (pancytopenia and accumulation of early HSPCs) of the Dpy30 KO mice resemble those of the mice lacking Cfp1 (Chun et al., 2014), an important subunit of Set1a/b complexes, and knockdown of Wdr82, a subunit restricted to Set1a/b complexes, modestly reduced Hoxa9 expression (at least in mouse embryonic fibroblasts; Mishra et al., 2014); (2) no hematopoietic abnormalities were found in functional assays for mice with specific deletion of the SET catalytic domain in Mll1 (Mishra et al., 2014); (3) postnatal ablation of Mll2 produced mice with no overt somatic phenotypes (Glaser et al., 2009), although Mll2 loss impairs macrophage function, but not differentiation (Austenaa et al., 2012); and (4) depletion of Mll3 (Chen et al., 2014) or ablation of or Mll4 (Santos et al., 2014) in the hematopoietic system resulted in increased HSPCs with altered differentiation capacity, although no hematopoietic abnormalities were reported in mice with specific deletion of the SET domain in Mll3 (Lee et al., 2006). In addition, a role of Mll4 for cell differentiation in nonhematopoietic systems has also been reported (Lee et al., 2013). Meanwhile, we cannot exclude the possibility of a synthetic lethality situation in which a modest contribution from the H3K4 methylation activity of each Mll protein, albeit in itself dispensable, may collectively lead to the dramatic phenotypes if all were impaired, as in the case of loss of the Dpy30 core subunit. More definitive understanding of selective roles of the specific catalytic subunits will require genetic studies targeting the SET domains in individual enzymatic subunits. We also cannot formally exclude the possible contribution from potential nonmethylation related activities of Dpy30.
In summary, our results here reveal a profound and previously unrecognized role of Dpy30 and Set1/Mll complexes-associated H3K4 methylation in HSPC fate determination. Further molecular and functional studies using this Dpy30 KO mouse model will help provide a better understanding of how such a prominent epigenetic activity regulates normal hematopoiesis, as well as certain HSPC-based hematopoietic diseases.
MATERIALS AND METHODS
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham (UAB). All mice were maintained under specific pathogen-free conditions and housed in individually ventilated cages. ESCs on C57BL/6 background that harbor a Dpy30tm1a(KOMP)Wtsi (knockout first) allele were purchased from the Knock Out Mouse Project Repository (KOMP), and were injected into blastocysts to generate chimeras at the University of Alabama (UAB) Transgenic Mouse Facility (Birmingham, Alabama). Dpy30+/− mice with germline transmitted Dpy30-targeted allele (−) allele were crossed to the Rosa26-Flpe mice (The Jackson Laboratory; provided by the UAB Transgenic Mouse Facility) to generate Dpy30F/+ mice, which were further crossed to produce Dpy30F/F mice. Dpy30+/− mice were also crossed to Mx1-Cre (The Jackson Laboratory), CAG-CreER (The Jackson Laboratory), and Rosa26-CreER (National Cancer Institute Mouse Repository, Bethesda, MD) mice to produce Cre; Dpy30+/− mice. These mice were further crossed to Dpy30F/F mice to produce Cre; Dpy30F/+ and Cre; Dpy30F/− littermates for experimental use.
Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− littermates at 6–8 wk of age were injected with pIpC (7.5 µg/g; InvivoGen) once every other day for a total of four times. 6–8-wk-old CAG-CreER; Dpy30F/+ and CAG-CreER; Dpy30F/− littermates and Rosa26-CreER; Dpy30F/+ and Rosa26-CreER; Dpy30F/− littermates were injected with 1 mg tamoxifen (T5648; Sigma-Aldrich) once every other day for a total of seven times. PB profiles were measured using Hemavet 950 (DREW Scientific Inc.).
Transplantation assays
All transplant recipient mice in this work were C57BL/6J and CD45.1+ and purchased from Charles River. They were all female and 10–11 wk old at the time of transplantation. They were lethally irradiated with a split dose of 1,100 rad 3 h apart before transplantation.
For competitive transplantation assays, the donor BMs were obtained from mice 4 d after last pIpC or tamoxifen injections by flushing marrow out of the bone in PBS supplemented with 3% heat-inactivated FBS, followed by removal of red blood cells using ACK lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 nM EDTA, pH 7.2). 106 donor cells (CD45.2+) and 106 whole BM cells (C57BL/6J, CD45.1+) for a ratio of 1:1 or 2 × 106 donor cells (CD45.2+) and 2 × 105 whole BM cells (C57BL/6J, CD45.1+) for a ratio 10:1 of were transplanted via tail vein injection into irradiated recipient mice (CD45.1+).
For mixed BM chimera assays, the donor whole BMs were prepared from uninjected mice in the same way as described above, and 1.25 × 106 donors cells (CD45.2+) and 2.5 × 105 competitor cells (C57BL/6J and CD45.1+) were transplanted into irradiated recipient mice (CD45.1+). Recipients were then injected with pIpC (7.5 µg/g; InvivoGen) four times over 10 d starting at 7 wk after transplantation. The first two injections were 2 d apart and last injections were 4 d apart. Reconstitution levels were assessed at different time points after pIpC injection.
For HSC transplantation, 500 HSCs (LSK CD48−CD150+) FACS-sorted from pIpC-injected Mx1-Cre; Dpy30F/+ or Mx1-Cre; Dpy30F/− donor mice and 2.5 × 105 competitor cells (whole BM cells from C57BL/6J, CD45.1+ mice) were transplanted into lethally irradiated recipient mice (CD45.1+).
For secondary transplantation, donor-derived HSCs (LSK CD48−CD150+CD45.2+) were FACS-sorted from mixed BM chimeras (primary recipients) at 2 mo after last pIpC injection into the chimeras. 500 donor-derived HSCs and 2.5 × 105 competitor cells (whole BM cells from C57BL/6J, CD45.1+ mice) were transplanted into lethally irradiated recipient mice (CD45.1+).
Donor contribution was determined by flow cytometry at different time points and calculated by CD45.2+/(CD45.1+ + CD45.2+) × 100% within the indicated cell type.
Flow cytometry for analysis and sorting
Single-cell suspensions were prepared from BM, thymus, spleen, or PB. Red blood cells were removed using ACK lysis buffer. BM was stained with a lineage antibody cocktail (Mouse Hematopoietic Stem and progenitor Cell Isolation kit; BD) containing APC-conjugated antibodies against CD3e (145-2C11), CD45R (RA3-6B2), CD11b (M1/70), Gr1 (RB6-8C5), and Ter119 (TER-119), and the following antibodies to discriminate the cell types of interest: Sca1 (D7)-PE-Cy7; CD117 (cKit 2B8)-PE; CD135 (Flt3 A2F10)-biotin; CD127 (A7R34)-FITC, and PerCP-Cy5.5; CD48 (HM48.1)-FITC and Pacific blue; CD150 (TC15-12F12.2)-biotin; CD16/32 (FcγRIII/II; 93)-biotin; CD45.1 (A20)-BV510 and Pacific blue; CD45.2 (104)-PerCP-Cy5.5. For splenocyte and thymocyte analysis, cells were stained with antibodies for CD4 (GK1.5)-FITC; CD8 (53–6.7)-PE; CD45R (RA3-6B2)-APC; CD45.1 (A20)–Pacific blue, and CD45.2 (104)-PerCP-Cy5.5. For PB analysis, cells were stained with antibodies for CD3e (145-2C11)-PerCP-Cy5.5; CD45R (RA3-6B2)-APC; CD11b (M1/70)-biotin; Gr1 (RB6-8C5)-biotin; CD45.1 (A20)-PE, and CD45.2 (104)-FITC. FACS analysis was performed on LSRFortessa (Becton Dickinson), and data were analyzed using FlowJo software (Tree Star).
FACS sorting was performed on LSRAria II (BD). Using pooled BM from two to three chimera mice at 2 wk after the last pIpC injection into the chimeras, lin− BMs were enriched with magnetic Lineage depletion beads (Miltenyi Biotec) following manufacturer's instructions. Note that our procedure used less amount of lineage antibody cocktail than recommended to leave some Lin+ cells un-depleted for later assays. These Lin− BMs were double sorted by FACS into the four donor-derived cell populations including LSK, MyePro, Other Lin− (including lymphoid progenitors), and Lin+ cells. In separate BM chimeras, BMs from seven to nine mice were pooled to enrich for lin− BMs, and then to FACS sort 6,000-20,000 donor-derived LT-HSCs using procedures shown in Fig. S1.
Colony formation assays
Indicated numbers of bone cells were plated in duplicate into 1.1 ml Mouse Methylcellulose Complete Media (R&D System, HSC007), which includes recombinant human insulin, human transferrin, recombinant mouse SCF, IL-3, and IL-6, and recombinant human Epo. Colonies were scored at 14 d after plating.
Assays for HSPC proliferation and apoptosis
HSPC proliferation and apoptosis were determined by BrdU incorporation and Annexin V staining assays, respectively, as previously described (Jude et al., 2007; Yang et al., 2014). In brief, mice were injected with pIpC or tamoxifen, as described in the Animals section. For BrdU analysis, 1.5 mg BrdU was intraperitoneally injected into each mouse at 12 and 6 h before sacrifice. Cells were stained with cell surface antibodies as described above and then processed with BrdU staining using the FITC-BrdU Flow kit (BD) following the manufacturer’s instructions.
For apoptosis assays, cells were harvested and stained with antibodies as described above. After washing twice with cold PBS containing 3% heat-inactivated FBS, the cells were then incubated with FITC-Annexin V (BD) and 7-amino-actinomycin D (7-AAD) for 15 min in binding buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2) at room temperature in dark. The stained cells were analyzed immediately by flow cytometry.
RNA extraction, chromatin immunoprecipitation (ChIP), and quantitative PCR (qPCR)
Total RNAs were isolated using RNeasy Plus Mini (for non-HSCs) or Micro (for HSCs) kit (QIAGEN). Some RNAs were reverse transcribed with SuperScript III (Invitrogen). ChIP assays were performed as previously described (Jiang et al., 2011). qPCR was performed with SYBR Advantage qPCR Premix (Takara Bio Inc.) on a ViiA7 Real-Time PCR System (Applied Biosystems). Primers used are listed in Table S5. Relative expression levels were normalized to Actb. For ChIP results, percent input was first calculated from ChIP qPCR results as previously described (Jiang et al., 2011), and ChIP enrichment fold was calculated as the ratio of the percent input value for each locus over that for the negative control site in the control cells (Mx1-Cre; Dpy30F/+).
RNA-seq, ChIP-seq, and data analyses
Total RNAs and ChIP (and their corresponding input) DNAs were sent to the Genomic Services Lab at the HudsonAlpha Institute for Biotechnology (Huntsville, AL) for sequencing, including library construction. In brief, the quality of the total RNA and DNA was assessed using the Agilent 2100 Bioanalyzer. Two rounds of polyA+ selection was performed for RNA samples, followed by conversion to cDNAs. Due to the very low amounts, RNAs from the donor-derived LT-HSCs were amplified using Ovation RNA-seq System V2 (Nugen) before library preparation. No amplification was performed for RNAs from non-HSCs. We used the mRNA library generation kits (Agilent) and TruSeq ChIP Sample Prep kit (Illumina) per the manufacturer’s instructions. The indexed DNA libraries were quantitated using qPCR in a Roche LightCycler 480 with the Kapa Biosystems kit for library quantitation (Kapa Biosystems) before cluster generation. Clusters were generated to yield ∼725–825 K clusters/mm2. Cluster density and quality were determined during the run after the first base addition parameters were assessed. Sequencing was performed on Illumina HiSeq2500 with sequencing reagents and flow cells providing up to 300 Gb of sequence information per flow cell.
For RNA-seq, we obtained 35–55 million (for non-HSCs) or >60 million (for HSCs) 51-bp paired-end reads for each RNA sample. All the reads were mapped to the mouse reference genome (mm10) using TopHat (v2.0.13). Low quality mapped reads (MQ < 30) were removed from the analysis. Read count tables were generated using HTSeq (v0.6.0; Anders et al., 2015) and differential expression (DE) analyses were performed using DESeq (v3.0; Anders and Huber, 2010). All of the downstream statistical analyses and generating plots were performed in R (v3.1.1). GSEA was performed using Gene Set Enrichment Analyses.
For ChIP-seq, we obtained 25–35 million of 50-bp single-end reads for each DNA sample. All of the reads were mapped to the mouse reference genome (mm10/GRCm38.74) using the Bowtie2 (v2.2.4) gapped alignment algorithm (Langmead and Salzberg, 2012), and duplicate reads were removed using Picard tools (v.1.126). Peak calling was performed using MACS (v1.4.2; Zhang et al., 2008) and peak count tables were created using BEDTools (Quinlan and Hall, 2010). The ChIPseeker (v1.8.0; Yu et al., 2015) R package was used for peak annotations (ngs.plot v2.47; Shen et al., 2014) and ChIPseeker was used for TSS-binding site visualizations and quality controls. BigWig files were generated using MACS and normalized by reads per million (RPM). Gene ontology analysis was performed with DAVID (http://david.abcc.ncifcrf.gov/), and KEGG pathway analysis was performed using clusterProfiler R package (v3.0.0; Yu et al., 2012).
Our RNA-seq and ChIP-seq datasets have been deposited in the Gene Expression Omnibus database under GSE81390.
Western blotting
Lineage fractionation of BM was performed using magnetic Lineage depletion beads (Miltenyi Biotec) following the manufacturer's instructions, and both Lin− and Lin+ fractions were retained for Western blotting. Whole or lineage-fractionated BM cells were lysed by lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-Cl, pH 7.5) with fresh added Protease Inhibitor Cocktail (Roche) and boiled for 5 min. Proteins were resolved on SDS-PAGE gel followed by Western blotting using previously described antibodies (Jiang et al., 2011) for Dpy30, H3, H3K4me1, H3K4me2, and H3K4me3, or from Santa Cruz Biotechnology for Actb (sc-47778), Dnmt3a (sc-20703), and Scl/Tal1 (sc-12984).
Statistics
The unpaired two-tailed Student’s t test was used to calculate P-values and evaluate the statistical significance of the difference between control and KO samples.
Online supplemental material
Fig. S1 shows flow cytometry gating strategies. Tables S1–S5 are available as Excel files. Table S1 lists cell surface phenotypes used in this work. Table S2 lists gene expression in LSK and MyePro cell populations in control and Dpy30 KO background. Table S3 lists genes that were changed in expression in Mx1-Cre; Dpy30F/− versus Mx1-cre; Dpy30F/+ donor-derived LT-HSCs in BM chimeras in two independent transplantations. Table S4 lists H3K4me3 at TSS regions of genes in control and Dpy30 KO background. Table S5 lists primers used in this study.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UAB transgenic core facility for help in generating our knockout mice and for providing the Rosa26-Flpe mouse. The Dpy30 KO mouse was created from ES cell clone EPD0714_4_A06 generated by the Wellcome Trust Sanger Institute and obtained from KOMP Repository. We also thank Yudong Liu for generous help with initial intravenous injections, and Christopher Klug for valuable scientific advice.
The X-RAD 320 irradiator was purchased by the UAB animal facility using National Institutes of Health grant G20RR022807, and the Comprehensive Flow Cytometry Core at UAB are supported by National Institutes of Health core grants P30 AR048311 and P30 AI027767. This work was supported by a Start-up fund from the State of Alabama and National Institutes of Health grant R01DK105531. H. Jiang is a recipient of the American Society of Hematology Scholar Award and American Cancer Society Research Scholar Award.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- ESC
- embryonic stem cell
- GSEA
- gene set enrichment analysis
- HPC
- hematopoietic progenitor cell
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem and progenitor cell
- LT-HSC
- long-term HSC
- MyePro
- myeloid progenitor
- PB
- peripheral blood
- pIpC
- polyinosinic-polycytidylic acid
- qPCR
- quantitative PCR
- RLP
- restricted lineage progenitor
- RNA-seq
- RNA sequencing
- ST-HSC
- short-term HSC
- TSS
- transcription start site
References
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinger E.L., Mishra B.P., Zaffuto K.M., Li B.E., Chung E.K., Moore A.W., Chen Y., Cheng C., and Ernst P.. 2013. An MLL-dependent network sustains hematopoiesis. Proc. Natl. Acad. Sci. USA. 110:12000–12005. 10.1073/pnas.1301278110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa L., Barozzi I., Chronowska A., Termanini A., Ostuni R., Prosperini E., Stewart A.F., Testa G., and Natoli G.. 2012. The histone methyltransferase Wbp7 controls macrophage function through GPI glycolipid anchor synthesis. Immunity. 36:572–585. 10.1016/j.immuni.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Baker S.J., Ma’ayan A., Lieu Y.K., John P., Reddy M.V., Chen E.Y., Duan Q., Snoeck H.W., and Reddy E.P.. 2014. B-myb is an essential regulator of hematopoietic stem cell and myeloid progenitor cell development. Proc. Natl. Acad. Sci. USA. 111:3122–3127. 10.1073/pnas.1315464111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi G., van Oudenaarden A., and Collins J.J.. 2011. Cellular decision making and biological noise: from microbes to mammals. Cell. 144:910–925. 10.1016/j.cell.2011.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun B.A., Pollina E.A., Ucar D., Mahmoudi S., Karra K., Wong E.D., Devarajan K., Daugherty A.C., Kundaje A.B., Mancini E., et al. . 2014. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 158:673–688. 10.1016/j.cell.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J. III, Gingeras T.R., et al. . 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 120:169–181. 10.1016/j.cell.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Bertero A., Madrigal P., Galli A., Hubner N.C., Moreno I., Burks D., Brown S., Pedersen R.A., Gaffney D., Mendjan S., et al. . 2015. Activin/nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes Dev. 29:702–717. 10.1101/gad.255984.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller S., and Grosschedl R.. 2014. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol. Rev. 261:102–115. 10.1111/imr.12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., et al. . 2011. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44:23–31. 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen G.A., Sun D., Mayle A., Jeong M., Luo M., Rodriguez B., Mallaney C., Celik H., Yang L., Xia Z., et al. . 2014. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 15:350–364. 10.1016/j.stem.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Boles N.C., Lin K.Y., Tierney M.P., Bowman T.V., Bradfute S.B., Chen A.J., Merchant A.A., Sirin O., Weksberg D.C., et al. . 2007. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 1:578–591. 10.1016/j.stem.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Y., Rappaport A.R., Kitzing T., Schultz N., Zhao Z., Shroff A.S., Dickins R.A., Vakoc C.R., Bradner J.E., et al. . 2014. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 25:652–665. 10.1016/j.ccr.2014.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou M.A., Castelo-Branco G., Halley-Stott R.P., Oliveira C.S., Loos R., Radzisheuskaya A., Mowen K.A., Bertone P., Silva J.C., Zernicka-Goetz M., et al. . 2014. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 507:104–108. 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun K.T., Li B., Dobrota E., Tate C., Lee J.H., Khan S., Haneline L., HogenEsch H., and Skalnik D.G.. 2014. The epigenetic regulator CXXC finger protein 1 is essential for murine hematopoiesis. PLoS One. 9:e113745 10.1371/journal.pone.0113745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y.R., Schatoff E., and Abdel-Wahab O.. 2012. Epigenetic alterations in hematopoietic malignancies. Int. J. Hematol. 96:413–427. 10.1007/s12185-012-1181-z [DOI] [PubMed] [Google Scholar]
- Dawson M.A., and Kouzarides T.. 2012. Cancer epigenetics: from mechanism to therapy. Cell. 150:12–27. 10.1016/j.cell.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Deplus R., Denis H., Putmans P., Calonne E., Fourrez M., Yamamoto K., Suzuki A., and Fuks F.. 2014. Citrullination of DNMT3A by PADI4 regulates its stability and controls DNA methylation. Nucleic Acids Res. 42:8285–8296. 10.1093/nar/gku522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y., Milne T.A., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., and Roeder R.G.. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 121:873–885. 10.1016/j.cell.2005.04.031 [DOI] [PubMed] [Google Scholar]
- Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., and Roeder R.G.. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13:713–719. 10.1038/nsmb1128 [DOI] [PubMed] [Google Scholar]
- Ernst P., and Vakoc C.R.. 2012. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief. Funct. Genomics. 11:217–226. 10.1093/bfgp/els017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster I., and Rajewsky K.. 1990. The bulk of the peripheral B-cell pool in mice is stable and not rapidly renewed from the bone marrow. Proc. Natl. Acad. Sci. USA. 87:4781–4784. 10.1073/pnas.87.12.4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan T., Jude C.D., Zaffuto K., and Ernst P.. 2010. Developmentally induced Mll1 loss reveals defects in postnatal haematopoiesis. Leukemia. 24:1732–1741. 10.1038/leu.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S., Lubitz S., Loveland K.L., Ohbo K., Robb L., Schwenk F., Seibler J., Roellig D., Kranz A., Anastassiadis K., and Stewart A.F.. 2009. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2:5 10.1186/1756-8935-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., and McMahon A.P.. 2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244:305–318. 10.1006/dbio.2002.0597 [DOI] [PubMed] [Google Scholar]
- He S., Kim I., Lim M.S., and Morrison S.J.. 2011. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 25:1613–1627. 10.1101/gad.2052911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H., Meade E., Medeiros S., Schindler J.W., Valk P.J., Fujiwara Y., and Orkin S.H.. 2004. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18:2336–2341. 10.1101/gad.1239604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa J.P. 2013. The myelodysplastic syndrome as a prototypical epigenetic disease. Blood. 121:3811–3817. 10.1182/blood-2013-02-451757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman S. 2013. T-bet in the spot light: roles in distinct T-cell fate determination. Cell. Mol. Immunol. 10:289–291. 10.1038/cmi.2013.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Shukla A., Wang X., Chen W.Y., Bernstein B.E., and Roeder R.G.. 2011. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 144:513–525. 10.1016/j.cell.2011.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Lu X., Shimada M., Dou Y., Tang Z., and Roeder R.G.. 2013. Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95. Nat. Struct. Mol. Biol. 20:1156–1163. 10.1038/nsmb.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W.D., Dafou D., McEntagart M., Woollard W.J., Elmslie F.V., Holder-Espinasse M., Irving M., Saggar A.K., Smithson S., Trembath R.C., et al. . 2012. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91:358–364. 10.1016/j.ajhg.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude C.D., Climer L., Xu D., Artinger E., Fisher J.K., and Ernst P.. 2007. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 1:324–337. 10.1016/j.stem.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., and Morrison S.J.. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121. 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Banerjee T., Vinckevicius A., Luo Q., Parker J.B., Baker M.R., Radhakrishnan I., Wei J.J., Barish G.D., and Chakravarti D.. 2014. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol. Cell. 54:613–625. 10.1016/j.molcel.2014.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej S., Kuvardina O.N., Oellerich T., Herglotz J., Backert I., Kohrs N., Buscató E., Wittmann S.K., Salinas-Riester G., Bonig H., et al. . 2014. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat. Commun. 5:3995 10.1038/ncomms4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., and Akashi K.. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128:693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., and Rajewsky K.. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Lacombe J., Herblot S., Rojas-Sutterlin S., Haman A., Barakat S., Iscove N.N., Sauvageau G., and Hoang T.. 2010. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. 115:792–803. 10.1182/blood-2009-01-201384 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence H.J., Helgason C.D., Sauvageau G., Fong S., Izon D.J., Humphries R.K., and Largman C.. 1997. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 89:1922–1930. [PubMed] [Google Scholar]
- Lazarevic V., Glimcher L.H., and Lord G.M.. 2013. T-bet: a bridge between innate and adaptive immunity. Nat. Rev. Immunol. 13:777–789. 10.1038/nri3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Wang C., Xu S., Cho Y.W., Wang L., Feng X., Baldridge A., Sartorelli V., Zhuang L., Peng W., and Ge K.. 2013. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2:e01503 10.7554/eLife.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee D.K., Dou Y., Lee J., Lee B., Kwak E., Kong Y.Y., Lee S.K., Roeder R.G., and Lee J.W.. 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl. Acad. Sci. USA. 103:15392–15397. 10.1073/pnas.0607313103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Han M.E., Baek S.J., Kim S.Y., and Oh S.O.. 2015. Roles of DPY30 in the Proliferation and Motility of Gastric Cancer Cells. PLoS One. 10:e0131863 10.1371/journal.pone.0131863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher-Firzlaff J., Gawlista I., Vervoorts J., Kapelle K., Braunschweig T., Walsemann G., Rodgarkia-Schamberger C., Schuchlautz H., Dreschers S., Kremmer E., et al. . 2008. The human trithorax protein hASH2 functions as an oncoprotein. Cancer Res. 68:749–758. 10.1158/0008-5472.CAN-07-3158 [DOI] [PubMed] [Google Scholar]
- Magnusson M., Brun A.C., Lawrence H.J., and Karlsson S.. 2007. Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Exp. Hematol. 35:1421–1428. 10.1016/j.exphem.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Martin C., and Zhang Y.. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838–849. 10.1038/nrm1761 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Uematsu S., Kondo T., Takeuchi O., Martino M.M., Kawasaki T., and Akira S.. 2013. Strawberry notch homologue 2 regulates osteoclast fusion by enhancing the expression of DC-STAMP. J. Exp. Med. 210:1947–1960. 10.1084/jem.20130512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon K.A., Hiew S.Y., Hadjur S., Veiga-Fernandes H., Menzel U., Price A.J., Kioussis D., Williams O., and Brady H.J.. 2007. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 1:338–345. 10.1016/j.stem.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Mishra B.P., Zaffuto K.M., Artinger E.L., Org T., Mikkola H.K., Cheng C., Djabali M., and Ernst P.. 2014. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Reports. 7:1239–1247. 10.1016/j.celrep.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Arai S., Suzuki A., Nariai Y., Urano T., Nakayama M., Ohara O., Yamamura K., Yamamoto K., and Miyazaki T.. 2013. PAD4 regulates proliferation of multipotent haematopoietic cells by controlling c-myc expression. Nat. Commun. 4:1836 10.1038/ncomms2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.Y., Yoshida T., and Georgopoulos K.. 2007. Ikaros and chromatin regulation in early hematopoiesis. Curr. Opin. Immunol. 19:116–122. 10.1016/j.coi.2007.02.014 [DOI] [PubMed] [Google Scholar]
- Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. . 2010. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42:790–793. 10.1038/ng.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F., Gurvich N., Hoya-Arias R., Abdel-Wahab O., Levine R.L., Asai T., Voza F., Menendez S., Wang L., Liu F., et al. . 2010. Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells: possible role in 20q- polycythemia vera. Blood. 116:2812–2821. 10.1182/blood-2010-02-270611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt S.J., Liang Q., Rairdan X.Y., Moran J.L., Prosser H.M., Beier D.R., Lloyd K.C., Bradley A., and Skarnes W.C.. 2009. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods. 6:493–495. 10.1038/nmeth.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Van Buren D., Huang H.S., Zhong L., Mostoslavsky R., Akbarian S., and Hock H.. 2010. Chromatin protein L3MBTL1 is dispensable for development and tumor suppression in mice. J. Biol. Chem. 285:27767–27775. 10.1074/jbc.M110.115410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Rifkin S.A., Andersen E., and van Oudenaarden A.. 2010. Variability in gene expression underlies incomplete penetrance. Nature. 463:913–918. 10.1038/nature08781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.C., and Dou Y.. 2015. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 15:334–346. 10.1038/nrc3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Sutterlin S., Lecuyer E., and Hoang T.. 2014. Kit and Scl regulation of hematopoietic stem cells. Curr. Opin. Hematol. 21:256–264. 10.1097/MOH.0000000000000052 [DOI] [PubMed] [Google Scholar]
- Santos M.A., Faryabi R.B., Ergen A.V., Day A.M., Malhowski A., Canela A., Onozawa M., Lee J.E., Callen E., Gutierrez-Martinez P., et al. . 2014. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 514:107–111. 10.1038/nature13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Shao N., Liu X., and Nestler E.. 2014. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 15:284 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.H., Abdel-Wahab O., Patel J.P., and Levine R.L.. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 12:599–612. 10.1038/nrc3343 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20:341–348. 10.1016/j.ceb.2008.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81:65–95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata A., Xu B., Ionita-Laza I., Roos J.L., Gogos J.A., and Karayiorgou M.. 2014. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 82:773–780. 10.1016/j.neuron.2014.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Chen W.Y., Shimada M., Nguyen U.T., Kim J., Sun X.J., Sengoku T., McGinty R.K., Fernandez J.P., Muir T.W., and Roeder R.G.. 2013. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 154:297–310. 10.1016/j.cell.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., and Jacks T.. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature. 445:661–665. 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grütz G.G., Agulnick A.D., Westphal H., Forster A., and Rabbitts T.H.. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157. 10.1093/emboj/16.11.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., and Trumpp A.. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763. 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff L., and Humeniuk R.. 2013. Concise review: erythroid versus myeloid lineage commitment: regulating the master regulators. Stem Cells. 31:1237–1244. 10.1002/stem.1379 [DOI] [PubMed] [Google Scholar]
- Xie H., Xu J., Hsu J.H., Nguyen M., Fujiwara Y., Peng C., and Orkin S.H.. 2014. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 14:68–80. 10.1016/j.stem.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Augustin J., Chang C., Hu J., Shah K., Chang C.W., Townes T., and Jiang H.. 2014. The DPY30 subunit in SET1/MLL complexes regulates the proliferation and differentiation of hematopoietic progenitor cells. Blood. 124:2025–2033. 10.1182/blood-2014-01-549220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., and He Q.Y.. 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 31:2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zhang J., Dominguez-Sola D., Hussein S., Lee J.E., Holmes A.B., Bansal M., Vlasevska S., Mo T., Tang H., Basso K., et al. . 2015. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 21:1190–1198. 10.1038/nm.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.