The transcription factor NFATC3 binds to IRF7 and to type 1 IFN promoters, regulating IRF7-mediated IFN expression in pDCs.
Abstract
Plasmacytoid dendritic cells (pDCs) rapidly produce large amounts of type 1 interferon (IFN) after Toll-like receptor 7 and 9 engagements. This specialized function of type 1 IFN production is directly linked to the constitutive expression of IRF7, the master transcription factor for type 1 IFN production. However, the IRF7 regulatory network in pDCs remains largely unknown. In this study, we identify that the transcription factor NFATC3 specifically binds to IRF7 and enhances IRF7-mediated IFN production. Furthermore, knockout of NFATC3 greatly reduced the CpG DNA–induced nuclear translocation of IRF7, which resulted in impaired type 1 IFN production in vitro and in vivo. In addition, we found that NFATC3 and IRF7 both bound to type 1 IFN promoters and that the NFAT binding site in IFN promoters was required for IRF7-mediated IFN expression. Collectively, our study shows that the transcription factor NFATC3 binds to IRF7 and functions synergistically to enhance IRF7-mediated IFN expression in pDCs.
INTRODUCTION
Plasmacytoid DCs (pDCs) are specialized cells that rapidly produce large amounts of type 1 IFN in response to microbial RNA or DNA through TLR7 and TLR9 (Liu, 2005). In pDCs, IRF7 is constitutively expressed and plays a key role in type 1 IFN production, whereas IRF3 is dispensable for type 1 IFN production (Honda et al., 2005b). Upon stimulation with TLR7/9 ligands, IRF7 is activated and translocates to the nuclei through a signaling cascade of MyD88-IRAK4-IRAK1-TRAF6, leading to the production of large amounts of type 1 IFN. Recent studies have shown that IRF7 is tightly regulated by many different mechanisms (Bao and Liu, 2013). At the mRNA level, Dcp2 promotes the degradation of IRF7 mRNA, whereas the translational repressors 4EBP1/2 inhibit IRF7 mRNA translation (Colina et al., 2008; Li et al., 2012). The biological activity of IRF7 protein can be repressed through TRIM28-mediated SUMOylation, and the RTA-associated E3 ubiquitin ligase directly catalyzes K48-linked polyubiquitination of IRF7 and promotes its degradation (Yu and Hayward, 2010; Liang et al., 2011). IKK-α is involved in the phosphorylation and activation of IRF7 (Hoshino et al., 2006). The IRF7 promoter is negatively regulated by the transcription factor FOXO3 that forms a complex with nuclear corepressor 2 and histone deacetylase 3 (Litvak et al., 2012). In addition, it has been shown that the transcription factor ELF4 binds to type 1 IFN promoters and synergizes with IRF3, IRF7, and NF-κB to induce IFN expression. However, CpG DNA–induced IFN production is not affected in Elf4−/− pDCs, indicating that ELF4 is not involved in type 1 IFN signaling in pDCs (You et al., 2013). Despite these extensive studies, the regulation of IRF7 in pDCs is still far from being fully elucidated.
The NFAT family, including NFAT cell (NFATC) 1, NFATC2, NFATC3, NFATC4, and NFAT5, plays important roles in the regulation of T cells, B cells, mast cells, and other immune cells. NFATC family members pair with other transcription factors to regulate the expression of cytokine genes and other inducible genes (Crabtree and Olson, 2002). In T cells, cooperation between NFAT and AP-1 is required for IL-2 and IL-4 gene transcription (Müller and Rao, 2010). It has also been shown that FOXP3 forms a cooperative complex with NFAT to repress NFAT–AP-1–dependent transcription in regulatory T cells (Wu et al., 2006). In B cells, NFATC1 is necessary for the activation and function of splenic B cells upon BCR stimulation (Bhattacharyya et al., 2011). Despite their roles in the regulation of T cells and B cells, recent studies have demonstrated that NFATC members are also involved in the regulation of innate immune responses in conventional DCs, mast cells, and macrophages (Zanoni and Granucci, 2012). However, the functions of the NFAT family in pDCs have not been studied.
In this study, by immunoprecipitation and mass spectrometry analyses, we find that NFATC3 selectively binds to IRF7 and positively regulates type 1 IFN production in pDCs. NFATC3 deficiency leads to reduced type 1 IFN production in pDCs both in vitro and in vivo. Furthermore, we find that NFATC3 and IRF7 bind to IFN promoters directly and synergistically induce IFN transcription. Thus, these results indicate that NFATC3 functions as a novel cotranscriptional factor for IRF7 in pDCs.
RESULTS
NFATC3 selectively binds to IRF7 and enhances IRF7-mediated type 1 IFN transcriptional activities
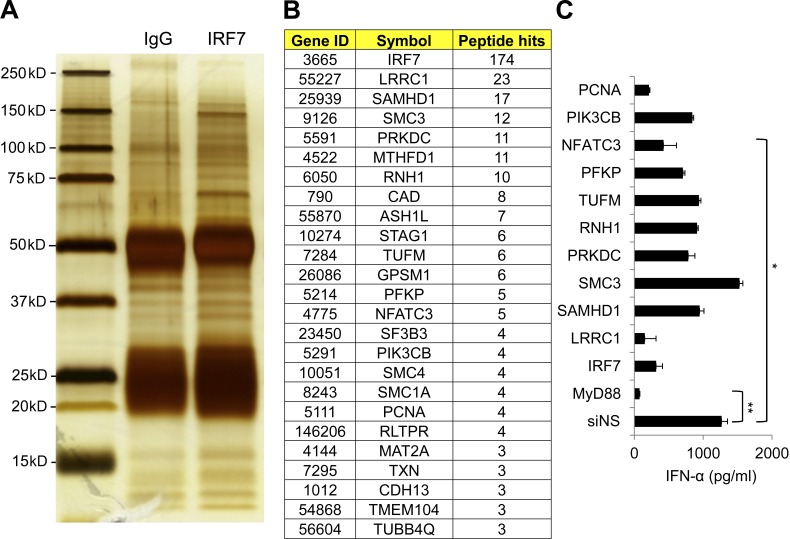
IRF7 is constitutively expressed by pDCs. However, the regulatory network of IRF7 in pDCs remains elusive. Because of the rareness of pDCs in peripheral blood (0.2–0.8%), it is difficult to decipher the regulatory network of IRF7 in pDCs. We took advantage of Gen2.2 cells, a human pDC cell line that shares all the key features of human primary pDCs (Chaperot et al., 2006). By immunoprecipitation with anti-IRF7 antibodies in Gen2.2 cell lysates followed by mass spectrometry analyses, we identified molecules that showed specific binding to IRF7 (Fig. 1, A and B). By using siRNA targeting the candidate genes, we knocked down the candidate molecules and checked their responses to CpG A (Fig. 1 C). After the screening, we identified NFATC3 as a novel IRF7 binding protein that was required for type 1 IFN production.
Figure 1.
Identifying IRF7 binding proteins and investigating their roles in IFN-α production. (A) Gen2.2 cells were left untreated or treated with 1 µM CpG A for 6 h, and then the cells were pooled together, lysed, and immunoprecipitated with anti-IRF7 antibody or isotype control. IRF7 binding proteins were analyzed by mass spectrometry. (B) Selected IRF7 binding proteins and the peptides hit in mass spectrometry are listed. (C) IRF7-associated genes were knocked down by using siRNA targeting specific molecules or nonspecific siRNA control (siNS), and the knockdown cells were stimulated with 1 µM CpG A for 20 h. IFN-α production in the supernatants was analyzed by ELISA. Data are representative of two independent experiments and are presented as the mean ± SEM of triplicate wells. *, P < 0.05; **, P < 0.01 by Student’s t test.
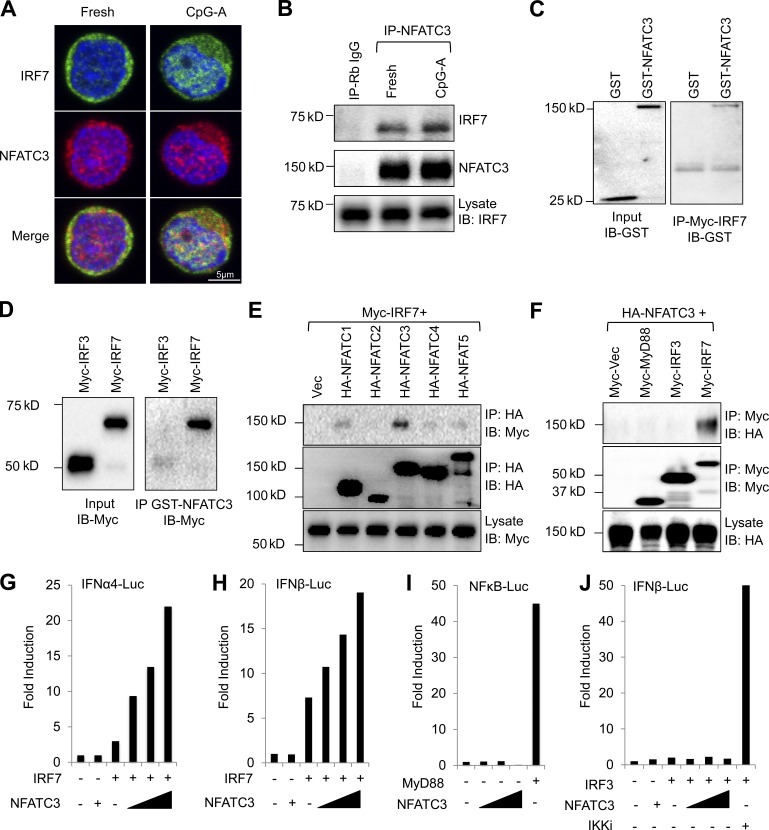
To confirm the binding between NFATC3 and IRF7 in human primary pDCs, we isolated peripheral blood pDCs and stimulated them with CpG A. The interaction between NFATC3 and IRF7 was investigated with confocal microscopy. We found that NFATC3 associated with IRF7 in fresh isolated pDCs and that the association was increased and colocalized in the cell nucleus after CpG A stimulation (Fig. 2 A). In addition, the association between NFATC3 and IRF7 was also confirmed by coimmunoprecipitation in human primary pDCs (Fig. 2 B). By using purified Myc-tagged IRF7 proteins, we found that IRF7 bound to glutathione S-transferase (GST)–tagged NFATC3 proteins but not to GST proteins (Fig. 2 C). We also used GST-tagged NFATC3 fusion proteins to pull down purified Myc-tagged IRF7 and Myc-tagged IRF3 proteins and found that NFATC3 directly bound to IRF7 but not IRF3 (Fig. 2 D).
Figure 2.
NFATC3 selectively binds to IRF7 and enhances its transcriptional activity. (A) Fresh isolated human primary pDCs or pDCs stimulated with CpG A for 4 h. Cells were fixed with paraformaldehyde, permeablized, stained with anti-IRF7 (green) and anti-NFATC3 (red), and mounted with Prolong gold antifade reagent with DAPI (blue). Cells were analyzed using confocal microscopy. (B) Untouched human pDCs were isolated as described in the Confocal microscopy section of Materials and methods. pDCs were left untreated or treated with 1 µM CpG A for 3 h, and pDCs were lysed and immunoprecipitated (IP) with NFATC3 antibody. IRF7 was detected by immunoblotting (IB). (C) Purified GST or GST-tagged NFATC3 were incubated with Myc-tagged IRF7. Myc-tagged IRF7 was immunoprecipitated with Myc–agarose beads. GST was detected by anti-GST HRP conjugate antibody. (D) Purified Myc-tagged IRF3 or IRF7 was incubated with GST-tagged NFATC3, and GST-tagged NFATC3 was immunoprecipitated with glutathione Sepharose beads. Then, Myc-tagged proteins were detected by anti-Myc antibody. (E) HEK-293T cells were transfected with HA-tagged NFAT plasmids and Myc-tagged IRF7; 36 h after transfection, cell lysates were immunoprecipitated with anti-HA beads, and then the immunoprecipitates and lysates were analyzed with anti-HA or anti-Myc antibodies. (F) HEK-293T cells were transfected with HA-tagged NFATC3 plasmids and Myc-tagged MyD88, IRF3, and IRF7 plasmids; 36 h after transfection, cell lysates were immunoprecipitated with anti-HA beads, and then the immunoprecipitates and lysates were analyzed with anti-HA or anti-Myc antibodies. (G and H) HEK-293T cells were transfected with 100 ng IFN-α4 luciferase reporter plasmid (IFNα4-Luc) or IFN-β luciferase reporter plasmid (IFNβ-Luc) and 0.5 ng IRF7 expression vector together with an increasing amount of NFATC3 expression vector (25, 50, and 100 ng). 0.1 ng renilla luciferase reporter plasmid was transfected simultaneously as an internal control. Results are presented as fold induction relative to the activity of renilla luciferase. (I) HEK-293T cells were transfected with 100 ng NF-κB luciferase reporter plasmid (NFκB-Luc) together with 5 ng MyD88 expression vector or an increasing amount of NFATC3 expression vector. (J) HEK-293T cells were transfected with 100 ng IFN-β luciferase reporter plasmid (IFNβ-Luc) together with 5 ng IKKi expression vector or an increasing amount of NFATC3 expression vector. (G–J) Data are representative of four independent experiments.
The NFAT family contains five members. To investigate whether other family members are also involved in regulating IRF7, we first investigated the binding capacities between NFAT family members and IRF7. The hemagglutinin (HA)-tagged NFAT and Myc-tagged IRF7 expression plasmids were transfected into HEK-293T cells. The binding between NFAT proteins and IRF7 was checked by coimmunoprecipitation. Among the five NFAT members, NFATC3 showed specific binding to IRF7 (Fig. 2 E). IRF3 and IRF7 are critical transcriptional factors for type 1 IFN expression. In most mammalian cell types, nucleic acid sensing will activate IRF3, which in turn activates the IFN-β gene. However, in pDCs, type 1 IFN production depends on IRF7, but not on IRF3. To investigate whether NFATC3 binds to IRF3 and regulates its activation, HA-tagged NFATC3 and Myc-tagged MyD88, IRF3, and IRF7 plasmids were transfected into HEK-293T cells, and the binding was checked by coimmunoprecipitation. NFATC3 was found to specifically bind to IRF7 (Fig. 2 F).
The finding that NFATC3 selectively binds to IRF7 prompts us to investigate whether NFATC3 regulates the transcriptional activities of IRF7. HEK-293T cells were transiently transfected with IRF7 plasmids and a luciferase reporter plasmid containing an IFNA4 or IFNB1 promoter together with an increasing amount of plasmid-encoding NFATC3. The luciferase activity was measured 36 h after transfection. Transfection of IRF7 alone resulted in activation of the IFNA4 promoter. Transfection of NFATC3 alone did not increase IFNA4 or IFNB1 promoter activity. However, the IRF7-induced IFNA4 promoter activation was further increased when transfected with an increased dose of NFATC3 (Fig. 2 G). Similarly, the IRF7-induced IFNB1 promoter activation was also increased in an NFATC3 dose-dependent manner (Fig. 2 H). In contrast, NFATC3 did not induce the activation of NF-κB (Fig. 2 I). Also, NFATC3 did not show the enhancement of the IRF3-induced IFNB1 promoter activation (Fig. 2 J), which was consistent with the results that NFATC3 did not bind to IRF3. These results indicate that NFATC3 selectively enhances IRF7 activities.
NFATC3 is required for type 1 IFN production induced by CpG A
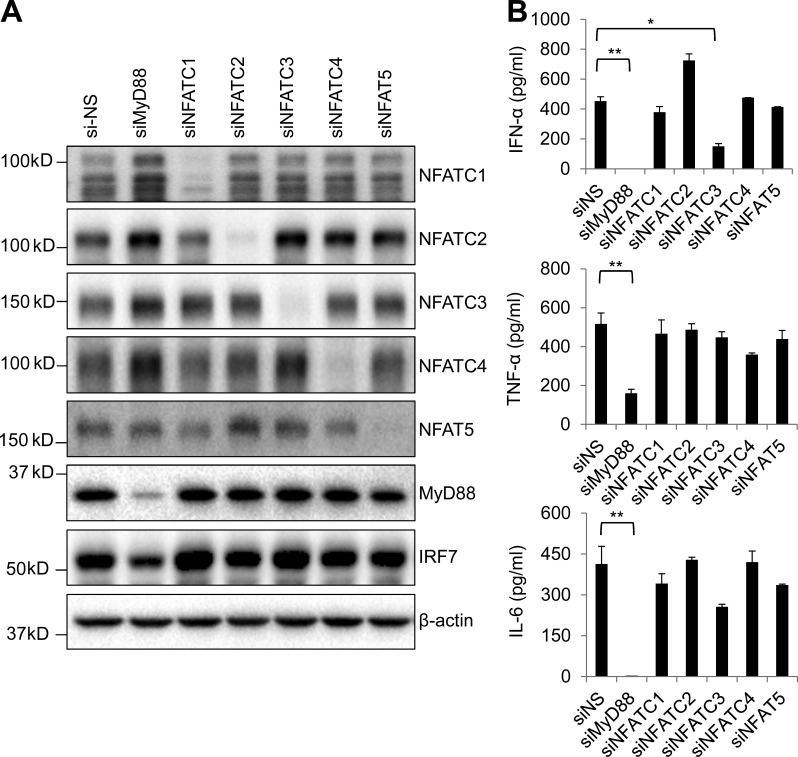
pDCs constitutively express IRF7 and produce large amounts of type 1 IFN in response to TLR9 ligands. To further determine the function of NFATC3 in pDCs, we knocked down NFATC3 as well as other NFAT family molecules in Gen2.2 cells by using siRNA. The knockdown effects were assessed by immunoblotting (Fig. 3 A). Then, the siRNA knockdown Gen2.2 cells were stimulated with CpG A, and cytokines including IFN-α, TNF-α, and IL-6 were measured. Knockdown of NFATC3, but not of other NFAT family members, led to a substantial reduction of IFN-α. However, TNF-α and IL-6 were not affected (Fig. 3 B). Therefore, NFATC3 is required for the activation of IRF7 to induce type 1 IFN production in response to CpG A.
Figure 3.
Knockdown of NFATC3 reduces CpG DNA–induced IFN-α in Gen2.2 cells. (A) Gen2.2 cells were knocked down by using nonspecific siRNA (si-NS) or siRNA targeting MyD88, NFATC1–4, and NFAT5. The knockdown efficiency was analyzed by immunoblotting. (B) siRNA knockdown Gen2.2 cells were stimulated with 1 µM CpG A for 24 h. IFN-α, TNF-α, and IL-6 in supernatants were analyzed by ELISA. Data are representative of four independent experiments and are presented as the mean ± SEM of triplicate wells. *, P < 0.05; **, P < 0.01 by Student’s t test.
Inhibition of NFAT reduces CpG DNA–induced type 1 IFN production in human primary pDCs
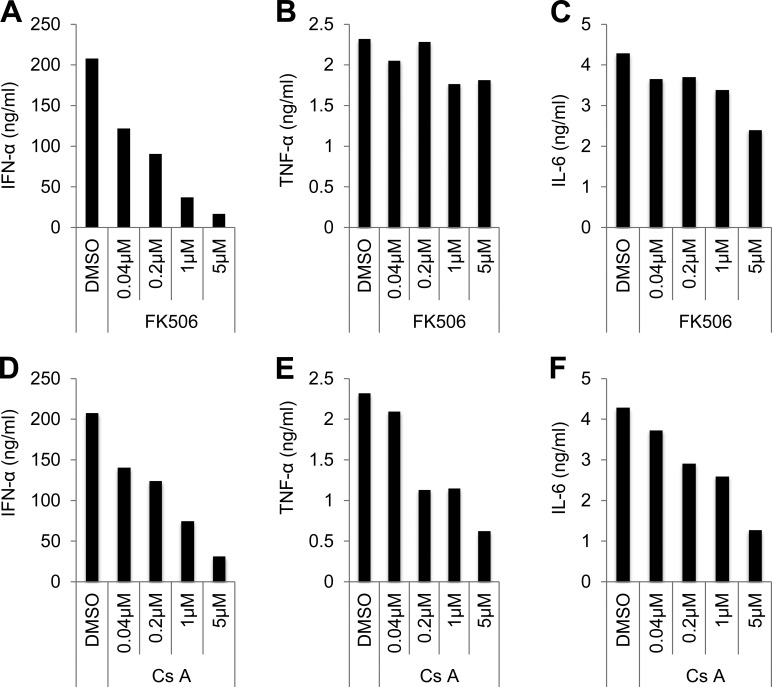
To investigate the role of NFAT in human primary pDCs, cyclosporin A (Cs A) and FK506 were used to inhibit the function of NFAT. Cs A and FK506 inhibit NFAT activation through inhibiting phosphatase calcineurin, which dephosphorylates and activates NFATC members (Ho et al., 1996). Fresh isolate human pDCs were treated with different concentrations of Cs A and FK506 for 1 h, and then pDCs were stimulated with CpG A or CpG B. Culture supernatants from CpG A–treated pDCs were analyzed for IFN-α, and supernatants from CpG B were analyzed for TNF-α and IL-6 secretion. FK506 and Cs A greatly reduced CpG A–induced IFN-α production (Fig. 4, A and D). FK506 did not show obvious inhibition on CpG B–induced TNF-α and IL-6 (Fig. 4, B and C). However, Cs A also showed inhibition on TNF-α and IL-6 (Fig. 4, E and F). FKBP12–FK506 and cyclophilin A–Cs A complexes both target calcineurin and potently inhibit its function; however, many studies suggest that these complexes display clinical and mechanistic differences (Schreiber and Crabtree, 1992; Hirano et al., 2008; Macartney et al., 2009; Kaxiras et al., 2014; Liu et al., 2015a). Our data also suggest that FK506 and Cs A function differently in regulating pDC function, which may contribute to the different clinical effects of FK506 and Cs A. These results indicate that NFAT family members play positive roles in regulating type 1 IFN production in human primary pDCs.
Figure 4.
Inhibition of NFAT reduces CpG DNA–induced IFN-α in human primary pDCs. (A) Human primary pDCs stimulated with 1 µM CpG A for 24 h together with FK506 or vehicle (DMSO). Production of IFN-α was measured by ELISA. (B and C) Human primary pDCs stimulated with 1 µM CpG B for 24 h together with FK506 or vehicle (DMSO). Production of TNF-α and IL-6 were measured by ELISA. (D) Human primary pDCs stimulated with 1 µM CpG A for 24 h together with Cs A or vehicle (DMSO). Production of IFN-α was measured by ELISA. (E and F) Human primary pDCs stimulated with 1 µM CpG A for 24 h together with Cs A or vehicle (DMSO). Production of TNF-α and IL-6 was measured by ELISA. Data are representative of four independent experiments.
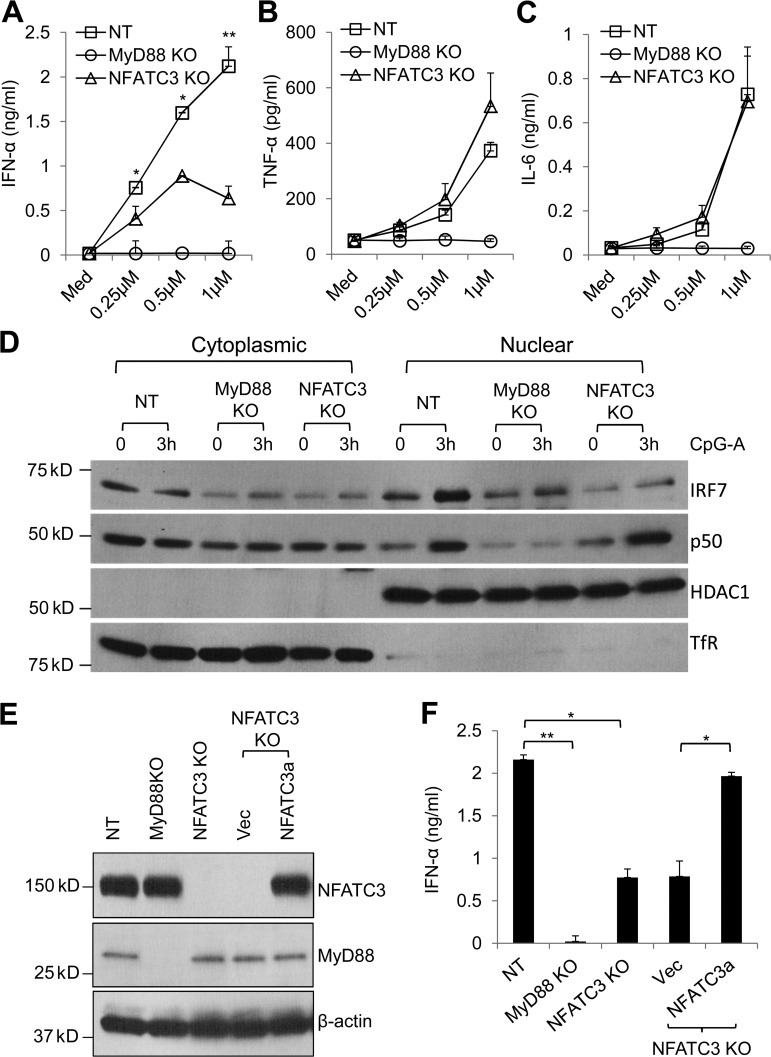
Knockout of NFATC3 in Gen2.2 cells by CRISPR impairs the CpG DNA–induced IRF7 signaling pathway
To further confirm the function of NFATC3 in pDCs, we used the genome-editing approach based on clustered regularly interspaced short palindromic repeats (CRISPR) and the endonuclease Cas9 to target the endogenous gene that encodes NFATC3. MyD88 knockout Gen2.2 cells and nontargeting (NT) guide RNA–transduced Gen2.2 cells were also generated as controls. Consistent with the previous results, knockout of NFATC3 showed selective reduction of IFN-α in response to CpG A (Fig. 5, A–C). Next, we investigated the CpG A–induced IRF7 and p50 (NF-κB1) nuclear translocation. Knockout of MyD88 completely abolished IRF7 and p50 nuclear translocation. However, knockout of NFATC3 inhibited IRF7 nuclear translocation while preserving p50 nuclear translocation (Fig. 5 D). These data are consistent with the results that NFATC3 selectively regulates IFN-α production but not that of TNF-α and IL-6.
Figure 5.
Knockout of NFATC3 reduces CpG DNA–induced IFN-α and inhibits IRF7 nuclear translocation, and NFATC3 restoration rescues the responses. (A–C) CRISPR knockout Gen2.2 cells were stimulated with different doses of CpG A. NT guide RNA–transduced cells were used as controls. Production of IFN-α, TNF-α, and IL-6 were measured by ELISA. P-values are NT compared with NFATC3 knockout Gen2.2 cells. (D) CRISPR knockout Gen2.2 cells were stimulated with CpG A for 3 h. IRF7 and p50 nuclear translocation were analyzed by immunoblotting. Anti-transferrin receptor (Tfr) antibody and anti-HDAC1 antibody were used to show the cytoplasmic fraction and nuclear fraction. (E) NFATC3 knockout Gen2.2 cells that were stably transduced with mutated NFATC3 expression lentiviral particles that contained the mutated guide RNA–targeting sequence (NFATC3a) to restore NFATC3 expression. The lentiviral particles that contained empty lentiviral vector were used as a control. NFATC3 expression level was measured by immunoblotting. (F) NFATC3 knockout and NFATC3-restored Gen2.2 cells were stimulated with CpG A for 24 h. Production of IFN-α was measured by ELISA. Data are presented as the mean ± SEM of triplicate wells and are representative of three independent experiments. *, P < 0.05; **, P < 0.01 by Student’s t test.
To eliminate the potential off-target effects of CRISPR, we restored the NFATC3 expression in NFATC3 knockout Gen2.2 cells. First, we generated a guide RNA–resistant NFATC3 (NFATC3a) lentiviral vector that expressed wild-type NFATC3; then, NFATC3a and the empty lentiviral vector were used to generate lentiviruses. The NFATC3 knockout Gen2.2 cells were infected with NFATC3a lentiviruses or control lentiviruses. NFATC3a-transduced cells successfully restored the NFATC3 expression in the NFATC3 knockout cells (Fig. 5 E). More importantly, the IFN-α production in NFATC3 knockout Gen2.2 cells was also rescued (Fig. 5 F).
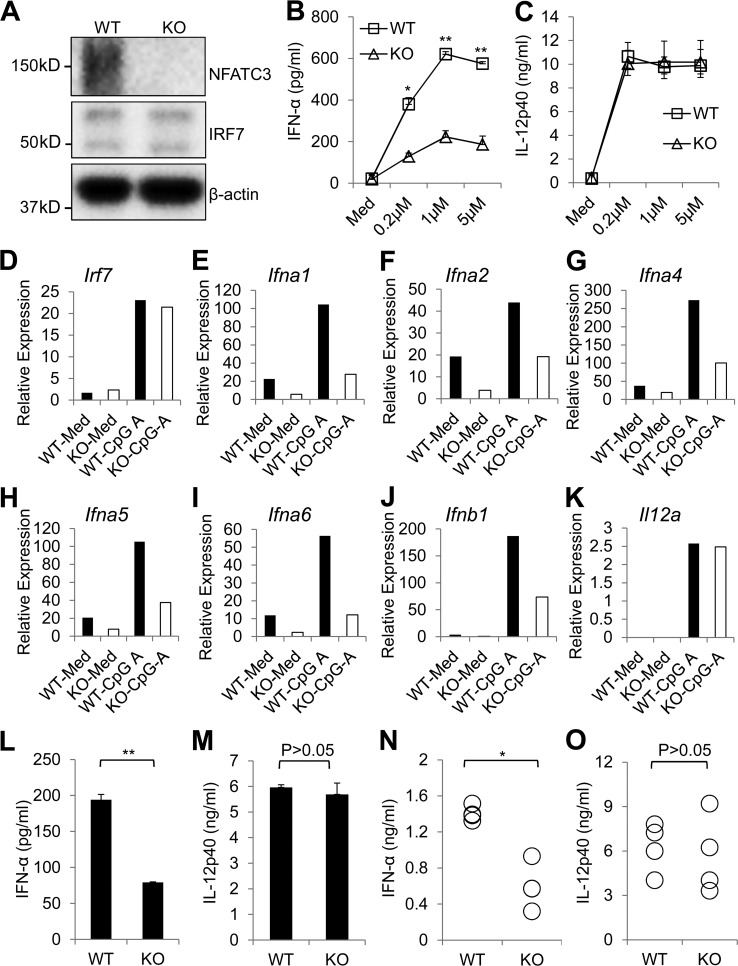
Type 1 IFN production is impaired in Nfatc3-deficent mice
Human and mouse NFATC3 display a high degree of similarity. We found that mouse NFATC3 could also enhance IRF7-mediated IFNA4 luciferase activity (Fig. S1 A). In addition, NFAT inhibitors FK506 and Cs A greatly reduced CpG A–induced IFN-α production in mouse pDCs. However, CpG A–induced IL-12p40 production was not changed (Fig. S1, B and C). To further confirm the function of NFATC3, pDCs from wild-type and Nfatc3-deficient mice were isolated, and the response to CpG A was investigated. Nfatc3-deficient mice did not show any obvious defects in pDC development (Fig. S2). pDCs from the Nfatc3-deficient mice showed normal IRF7 expression. However, the IFN-α induced by CpG A was greatly reduced (Fig. 6, A and B). In contrast, CpG A–induced IL-12p40 was not influenced (Fig. 6 C). Bone marrow–derived pDCs were stimulated with CpG A, and the expression levels of different subtypes of type 1 IFN were analyzed by RT-PCR (Table S1). As shown in Fig. 6, the type 1 IFN mRNA expressions were greatly reduced, whereas Il12a mRNA expression was not changed. Interestingly, the Irf7 mRNA expression level was not changed even after CpG A stimulation, which indicates that Nfatc3 is not involved in regulating Irf7 transcription. Bone marrow–derived pDCs were stimulated with R848 to test whether TLR7-mediated IFN production was also affected by NFATC3. Similar to TLR9 ligand stimulation, R848-induced IFN-α was greatly reduced in pDCs from Nfatc3−/− mice (Fig. 6 L), whereas IL-12p40 was not affected (Fig. 6 M).
Figure 6.
pDCs from Nfatc3−/− mice show impaired IFN-α production in response to CpG A. (A) Mouse primary pDCs were isolated from wild-type or Nfatc3−/− mice. Expression of NFATC3 and IRF7 were analyzed by immunoblotting. (B and C) pDCs from wild-type or Nfatc3−/− mice stimulated with different concentrations of CpG A (D19) for 24 h. IFN-α and IL-12p40 in supernatants were measured by ELISA. (D–K) Bone marrow–derived pDCs from wild-type or Nfatc3−/− mice were left unstimulated or stimulated with 1 µM CpG A for 20 h, and Irf7, Ifna1, Ifna2, Ifna4, Ifna5, Ifna6, Ifnb1, and IL12a gene expression were detected by RT-PCR. (L and M) Bone marrow–derived pDCs from wild-type or Nfatc3−/− mice were left unstimulated or stimulated with 10 µg/ml R848 for 20 h, and IFN-α and IL-12p40 were measured by ELISA. (N and O) Wild-type or Nfatc3−/− mice were injected with CpG A for 6 h. IFN-α and IL-12p40 in serum were measured by ELISA. Data are representative of three independent experiments and are presented as the mean ± SEM of triplicate wells. *, P < 0.05; **, P < 0.01 by Student’s t test.
Next, we examined the in vivo relevance of Nfatc3 deficiency in IRF7 signaling. Mice were injected intravenously with CpG A, and serum concentrations of IFN were detected. As shown in Fig. 6, serum concentrations of IFN-α were greatly reduced in Nfatc3−/− mice. However, the production of IL-12p40 was not changed in either group (Fig. 6, N and O). These data further confirm that NFATC3 is required for the activation of IFN but not for proinflammatory cytokine genes.
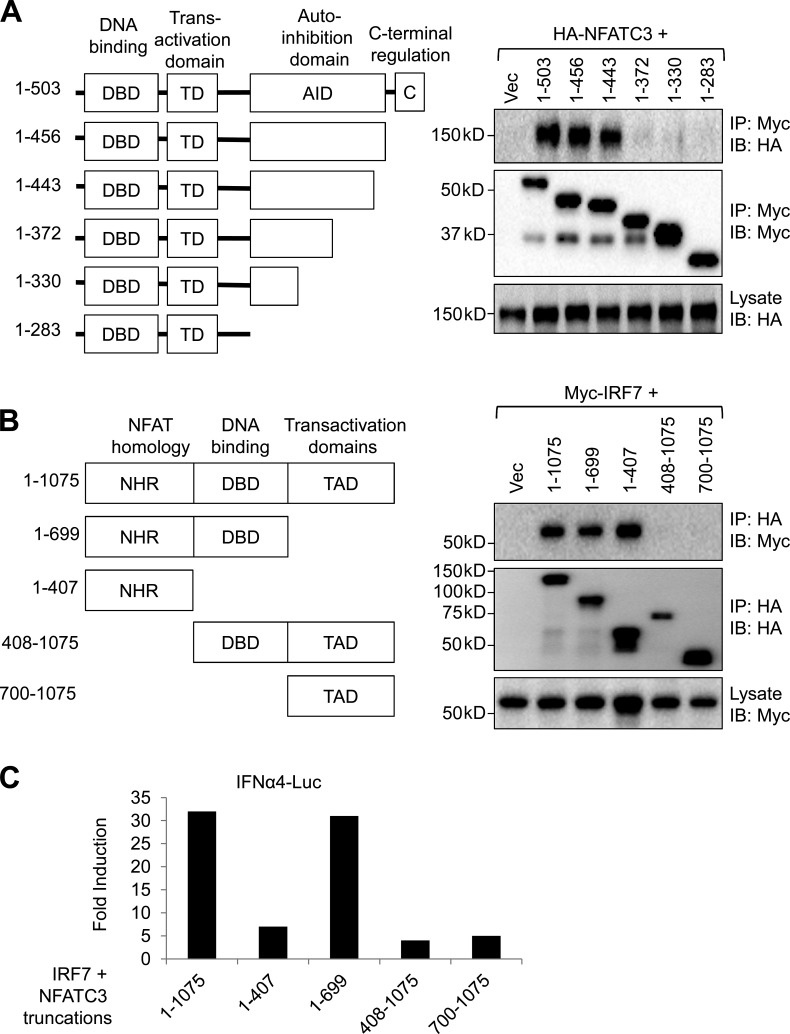
Interaction of NFATC3 with IRF7
IRF7 contains the N-terminal DNA-binding domain, a trans-activation domain, an autoinhibitory domain, and a C-terminal regulatory domain (Fig. 7 A). To investigate the mechanism that NFATC3 enhances IRF7 activation, we first mapped the domains of IRF7 that mediate the binding to NFATC3. HA-tagged NFATC3 and truncations of Myc-tagged IRF7 were expressed in HEK-293T cells. Then, truncations of IRF7 were immunoprecipitated and NFATC3 was detected. NFATC3 binds to the autoinhibitory domain (aa 373–443) of IRF7 (Fig. 7 A). Next, we investigated the domains of NFATC3 required for IRF7 binding. NFATC3 contains an N-terminal NFAT homology region (NHR) that is highly phosphorylated by NFAT kinases and is important for NFAT nuclear translocation, a DNA-binding domain, and a C-terminal trans-activation domain (Fig. 7 B). HA-tagged NFATC3 truncations and Myc-tagged IRF7 were cotransfected into HEK-293T cells. HA-tagged NFATC3 truncations were immunoprecipitated, and the IRF7 was detected. The results showed that the NHR domain of NFATC3 was required to bind to IRF7 (Fig. 7 B).
Figure 7.
The NFAT homology domain and DNA-binding domain of NFATC3 are required to enhance IRF7 activity. (A) HEK-293T cells were transfected with Myc-tagged IRF7 or truncated IRF7 and HA-tagged NFATC3 plasmids; 36 h after transfection, cell lysates were immunoprecipitated (IP) with anti-Myc beads, and then the immunoprecipitates and lysates were analyzed with anti-HA or anti-Myc antibodies. Scheme represents the IRF7 domain structure. (B) HEK-293T cells were transfected with HA-tagged NFATC3 or truncated NFATC3 and Myc-tagged IRF7 plasmids; cell lysates were immunoprecipitated with anti-Myc beads, and then the immunoprecipitates and lysates were analyzed with anti-HA or anti-Myc antibodies. Scheme represents the NFATC3 domain structure. (C) HEK-293T cells were transfected with 100 ng IFN-α4 luciferase reporter plasmid, 0.5 ng IRF7 expression vector, and 100 ng of full-length NFATC3 or truncated NFATC3 expression vectors. 0.1 ng renilla luciferase reporter plasmid was transfected simultaneously as an internal control. Results are presented as fold induction relative to the activity of renilla luciferase. Data are representative of four independent experiments.
Our data suggest that the highly phosphorylated N-terminal NHR is required to bind to the autoinhibitory domain of IRF7. Next, we investigated whether the binding of the NHR domain to IRF7 was enough to enhance IRF7 activity by using the luciferase reporter plasmid containing the IFNA4 promoter. Interestingly, the NHR domain alone was not enough to activate IRF7. We found that both the NHR domain and the DNA-binding domain were required to enhance IRF7 activity (Fig. 7 C).
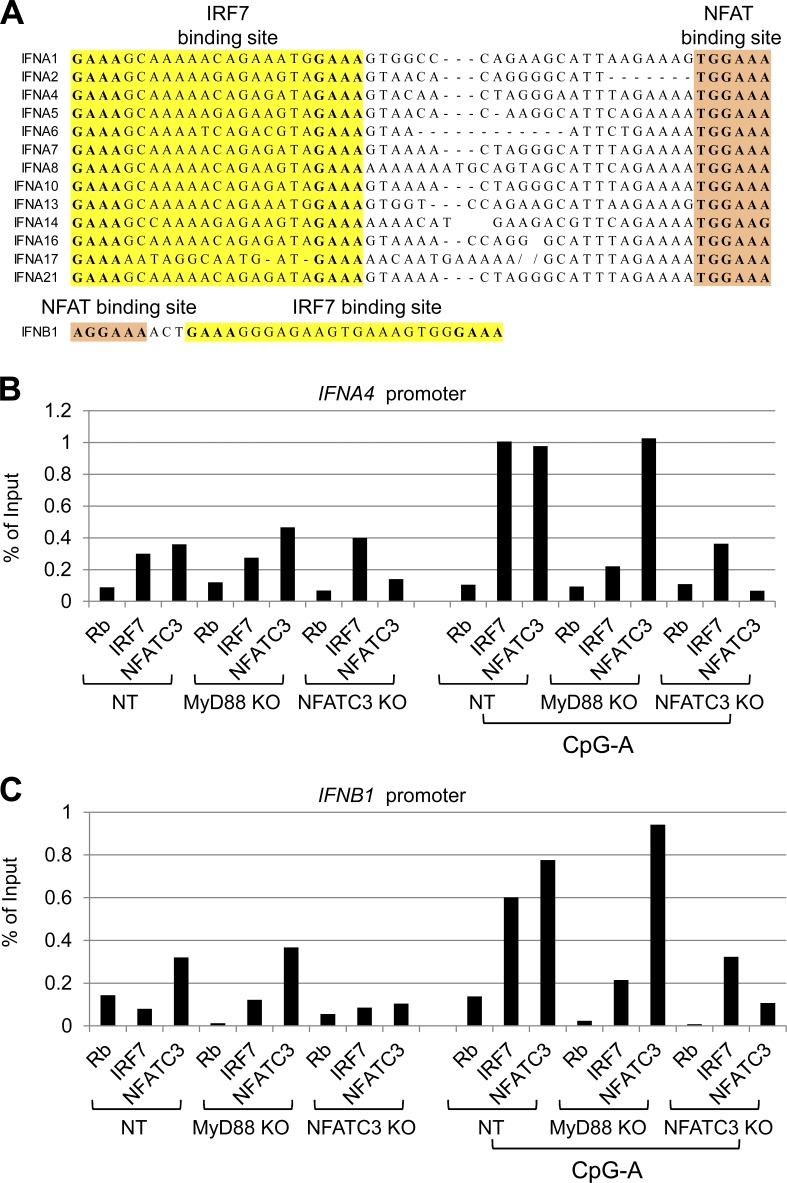
NFATC3 and IRF7 bind to IFNA4 and IFNB1 promoters, and NFATC3 is required for IRF7 transcriptional activities
Based on our data, the NHR domain and the DNA-binding domain of NFATC3 are indispensable to enhance IRF7 activity. This indicates that both IRF7 and NFATC3 need to bind to an IFN promoter to induce optimized IFN production. NFATC family molecules are transcription factors that bind to the consensus sequence (T/AGGAAA) and regulate gene transcription together with other transcription factors (Rao et al., 1997). First, we investigated the NFAT binding sequence in IFN promoter regions. As shown in Fig. 8 A, we could identify the NFAT binding sites in all type 1 IFN promoters.
Figure 8.
NFATC3 and IRF7 bind to type 1 IFN promoters. (A) Sequence alignment of IRF7 and NFATC3 binding sites in type 1 IFN promoters and sequence illustration of IRF7 and NFATC3 binding sites in the IFNB1 promoter. (B and C) Gen2.2 cells were fixed, lysed, and sonicated. Precleared lysates were incubated with rabbit IgG, anti-IRF7, and anti-NFATC3. Immunocomplexes were collected and purified. IFNA4 and IFNB1 promoter DNA were detected by real-time PCR. Data are representative of three independent experiments.
Next, we performed chromatin immunoprecipitation experiments to examine whether NFATC3 and IRF7 are associated with IFN promoters endogenously. Because of the rareness of primary pDCs, we took advantage of the CRISPR knockout Gen2.2 cells. CRISPR knockout Gen2.2 cells were resting in medium or stimulated with CpG A for 1 h. The cells were fixed and sonicated. NFATC3 antibody and IRF7 antibody and isotype control were used to precipitate the protein/DNA complexes. The immunoprecipitated DNA was purified and subjected to real-time PCR analysis with primers for the endogenous IFNA4 or IFNB1 promoter proximal region. We found that both NFATC3 antibody and IRF7 antibody precipitated the IFNA4 promoter and IFNB1 promoter in Gen2.2 cells after CpG A stimulation (Fig. 8, B and C). MyD88 deficiency led to greatly reduced IRF7 binding to IFN promoters but did not affect NFATC3 binding. However, NFATC3 deficiency led to reduced IRF7 binding to IFNA4 or IFNB1 promoters. These results demonstrate that NFATC3 and IRF7 associate with the endogenous IFN promoters.
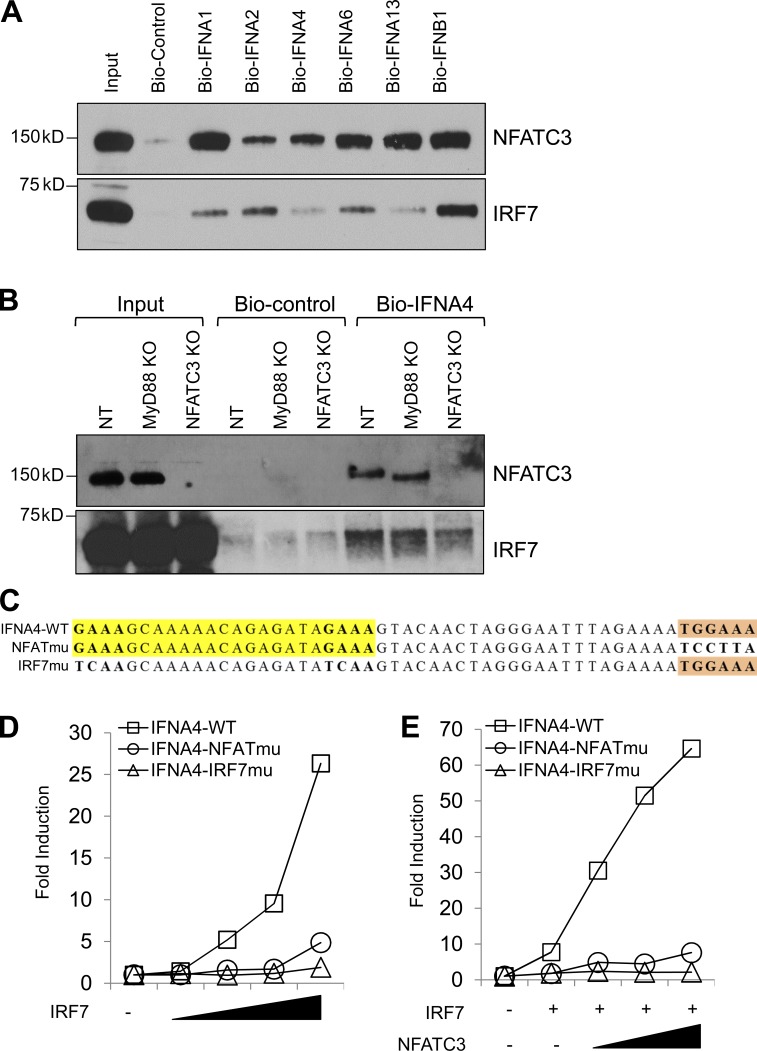
To further confirm our observation that IRF7 and NFATC3 bind to IFN promoters, we used biotin-labeled IFN promoters to pull down IRF7 and NFATC3 (Table S2). The HA-tagged NFAT and Myc-tagged IRF7 expression plasmids were transfected into HEK-293T cells. The cell lysate was incubated with biotin-labeled IFN promoters, and the binding of IRF7 and NFATC3 was detected by immunoblotting. All of the tested IFN promoter oligonucleotides except control oligonucleotides showed binding to NFATC3 and IRF7 (Fig. 9 A). Interestingly, IFNA1 and IFNA13 promoters, which contain another NFATC3 binding site, within the IRF7 binding site showed higher NFATC3 binding affinity, whereas IFNA2 promoters showed higher IRF7 binding affinity, indicating that IFNA promoters may be differentially regulated by NFATC3 and IRF7. Furthermore, we used the CRISPR knockout Gen2.2 cells to test whether knockout of NFATC3 would affect IRF7 binding to IFNA4 promoters. The CRISPR knockout Gen2.2 cells were stimulated with CpG A for 1 h, cell lysates were incubated with biotin-labeled IFNA4 promoters or control oligonucleotides, and the binding of IRF7 and NFATC3 was detected. Knockout of MyD88 abolished CpG A signaling that led to reduced IRF7 binding to the IFNA4 promoter without affecting NFATC3 binding. Knockout NFATC3 also greatly reduced IRF7 binding to IFNA4 promoters (Fig. 9 B). Collectively, these data suggest that NFATC3 binds to IFN promoters and NFATC3 binding is important for IRF7 activity.
Figure 9.
NFATC3 binds to type 1 IFN promoters, and NFATC3 is required for their transcription. (A) HEK-293T cells were transfected with HA-tagged NFATC3 and Myc-tagged IRF7. Cells were lysed, cell lysates were incubated with biotin-labeled oligonucleotides, and NFATC3 and IRF7 were detected by immunoblotting. (B) CRISPR knockout Gen2.2 cells were stimulated with CpG A for 1 h, and cell lysates were precleared, incubated with biotin-labeled oligonucleotides, and precipitated with NeutrAvidin agarose beads. NFATC3 and IRF7 were detected by immunoblotting. (C) Illustration of mutations in IRF7 and NFATC3 binding sites in IFNA4–Luc reporter constructs. (D) HEK-293T cells were transfected with plasmids encoding wild-type or the indicated mutant IFNA4–Luc and an increasing amount of IRF7 expression vector. 0.1 ng renilla luciferase reporter plasmid was transfected simultaneously as an internal control. Results are presented as fold induction relative to the activity of renilla luciferase. (E) HEK-293T cells transfected with wild type or the indicated mutant IFNA4–Luc plasmids and IRF7 expression vector together with an increasing amount of NFATC3 expression vector (25, 50, and 100 ng). 0.1 ng renilla luciferase reporter plasmid was transfected simultaneously as an internal control. Results are presented as fold induction relative to the activity of renilla luciferase. Data are representative of three independent experiments.
To determine whether these NFAT binding sites are necessary for IFN production, we generated IFNA4 promoter luciferase constructs containing mutated NFAT binding sites or mutated IRF7 binding sites (Fig. 9 C). Then, the IRF7-induced IFNA4 luciferase activity was investigated. Compared with wild-type IFNA4 promoters, mutation of IRF7 binding sites completely abolished IRF7-induced IFNA4 luciferase activity, and mutation of NFAT binding sites also showed greatly reduced IRF7-induced IFNA4 luciferase activity (Fig. 9 D). In addition, mutation of NFAT binding sites abolished the NFATC3-induced enhancement of IRF7 activity (Fig. 9 E). Collectively, our data suggest that NFATC3 represents a novel transcription factor that complexes with IRF7 and binds to IFN promoters to synergistically induce optimal IFN production.
DISCUSSION
IRF7 is the key transcriptional factor for type 1 IFN expression. Unlike all the other cell types in the body, pDCs constitutively express IRF7, which endows pDCs to rapidly produce large amounts of type 1 IFN upon TLR7/9 engagement. Previous studies have shown that activated IRF7 forms a transcriptional complex with IRF3, NF-κB, and ATF2/c-Jun on the promoter region of IFNB1 genes to induce IFN-β production (Panne et al., 2007). However, pDCs from IRF3 knockout mice produce normal type 1 IFN in response to TLR7/9 ligands, which indicates that IRF7 is uniquely regulated in pDCs (Honda et al., 2005b). By using a direct biochemical approach in a pDC cell line (Gen2.2 cells), we have identified that transcriptional factor NFATC3 specifically binds to and positively regulates IRF7 activities in pDCs. NFATC3 deficiency led to impaired IRF7 activation and reduced type 1 IFN production but had no effects on proinflammatory cytokine production. We further demonstrated that the NFATC3/IRF7 complex directly bound to the promoter regions of type 1 IFN genes and that the NFAT binding sites were necessary for type 1 IFN production. We therefore have revealed a previously unknown aspect of NFATC3 that functions as a novel IRF7 cotranscriptional factor in pDCs.
IRF3 is the closest family member to IRF7 and plays a key role in regulating IFN-β production in most of the cell types except pDCs. IRF3 is activated through RIG-I–MAVS, cGAS–STING, and TLR3/4–TRIF signaling pathways (Liu et al., 2015b). The CREB binding protein/p300 acetyl transferase has been identified as a cotranscriptional activator that associates with activated IRF3 to induce IFN-β production (Yoneyama et al., 1998). IFN-β can further induce IRF7 expression via a type 1 IFNR signaling-positive feedback loop to induce more robust IFN-α. Our data demonstrated that NFATC3 specifically bound to IRF7 but not to IRF3. Also, NFATC3 enhanced IRF7 activity but was dispensable for the IRF3-mediated IFN-β production. A previous study shows that IRF4 synergizes with NFATC2 to augment IL4 promoter activity (Rengarajan et al., 2002a). Therefore, this indicates that each NFAT family molecule may associate with a different IRF member to show different regulatory effects.
NFATC family members are activated by Ca2+ mobilization, which leads to the activation of phosphatase calcineurin that dephosphorylates NFATC proteins, promoting their nuclear translocation and activation (Crabtree and Olson, 2002). Previous studies indicate that different NFAT proteins have redundant and specific roles in regulating immune responses (Crabtree and Olson, 2002). NFATC2 and NFATC3 play redundant roles in maintaining thresholds for T cell activation (Rengarajan et al., 2002b; Bopp et al., 2005). NFATC1-deficient T cells display reduced proliferation and IL-4 production, whereas mice deficient in NFATC2s mount enhanced Th2 responses (Ranger et al., 1998; Yoshida et al., 1998). In master cells, NFATC1 and NFATC2 regulate the expression of TNF-α and IL-13, whereas NFATC3 is dispensable (Klein et al., 2006). In myeloid DCs, NFATC2 is activated by LPS, leading to the apoptotic death of terminally differentiated DCs (Zanoni et al., 2009). These studies indicate that the functions of NFAT proteins are isoform specific and cell type specific. Nevertheless, the functions of NFATC proteins in pDCs are unknown. It has been postulated that NFATCs are not involved in IFN production based on evolutionary analysis (Zanoni and Granucci, 2012). In contrast, our study demonstrates that NFATC3 is a unique member of the NFATC family that plays a critical role in regulating type 1 IFN but not proinflammatory production in pDCs. NFATC3 selectively bound to IRF7, which endows NFATC3 the unique ability to enhance IRF7-mediated IFN production. A previous study also shows that NFATC3 exhibits distinct DNA binding specificity compared with NFATC1 and NFATC2 (Ho et al., 1995), which indicates that NFATC3 may play a unique role in regulating gene expressions. Collectively, our study suggests that NFATC3, but not other NFAT molecules, can be activated by CpG A and functions as a cotranscriptional factor for IRF7 to promote type 1 IFN production in pDCs.
pDCs sense CpG A in the early endosomes through TLR9 that is coupled with IRF7 activation and leads to type 1 IFN production. pDCs sense CpG B in the late endosomes through TLR9 that is coupled to NF-κB activation and leads to proinflammatory production and pDC maturation (Honda et al., 2005a). Our data indicate that CpG DNA induced type 1 IFN through synergistic signaling pathways in pDCs. CpG DNA activates IRF7 in the early endosome; meanwhile, CpG DNA also induces NFATC3 activation. Then, the activated IRF7 and NFATC3 translocate to the cell nucleus, bind to IFN promoters, and function synergistically to induce maximal type 1 IFN production. In all, our data revealed NFATC3 as a novel cotranscription factor of IRF7, and this finding will pave new avenues for pDC studies.
MATERIALS AND METHODS
Reagents
CpG DNA were purchased from Sigma-Aldrich. The ON-TARGETplus siRNA were from GE Healthcare. The following antibodies were used for immunoblot analysis and/or immunoprecipitation: anti-IRF7 (sc-74472), anti-IRF7 (sc-9083), anti-NFATC1 (sc-13033), anti-NFATC2 (sc-7296), anti-NFATC3 (sc-8321), anti-NFATC4 (sc-13036), and anti-NFAT5 (sc-13035) were from Santa Cruz Biotechnology, Inc.; anti-IRF7 (51–3300) was from Thermo Fisher Scientific; anti-MyD88 (4283), anti–NF-κB1 p105/p50, and anti-HDAC1 (5356) were from Cell Signaling Technology; and anti-NFATC3 (A303-572A) was from Bethyl Laboratories, Inc. HRP-conjugated anti-HA (ab1265), anti-Myc (ab62928), and anti-transferrin receptor antibody (ab84036) were from Abcam; anti–β-actin, anti-HA antibodies, anti-GST HRP conjugate antibody, HA–agarose beads, and Myc–agarose beads were purchased from Sigma-Aldrich. Glutathione Sepharose 4B beads were from GE Healthcare. Human IFN-α, TNF-α, and IL-6 ELISA kits were from Mabtech. The mouse IFN-α ELISA kit was from eBioscience. The mouse IL-12p40 ELISA kit was from R&D Systems.
Cells and stimulation
Mouse pDCs were isolated from mouse bone marrow from Nfatc3−/− or wild-type mice. pDCs were sorted as Lin-PDCA1+ B220+ CD11c+ MHCII+ cells and stimulated as described previously (Esashi et al., 2012). Bone marrow–derived pDCs were induced by culture total bone marrow cells with mouse Flt3 ligands for 7 d. Gen2.2 cells were cultured, transfected, and stimulated as described previously (Bao et al., 2012). The concentrations of IFN-α, IL-6, and TNF-α in culture supernatants were measured by ELISA.
Confocal microscopy
Human pDCs were isolated using the Plasmacytoid Dendritic Cell Isolation Kit (Miltenyi Biotec) and sorted as CD3-, CD14-, CD16-, CD56-, CD19-, CD20-, and CD11c-. The purity of pDCs was checked by BDCA2 staining (purity >95%). Fresh pDCs were placed on the coverslip, fixed, and permeabilized for staining. The cells were incubated with anti-IRF7 antibody and anti-NFATC3 antibody. Then, the cells were stained with Alexa Fluor–conjugated secondary antibodies (Invitrogen). The stained cells were analyzed using a confocal microscope (Leica Biosystems).
Mice
Nfatc3−/− mice were purchased from The Jackson Laboratory. We bred Nfatc3+/− heterozygous mice to generate age-matched Nfatc3−/− and wild-type littermates. Animals were housed in specific pathogen-free barrier facilities. All experiments were conducted according to institutional guidelines of Baylor Health Care Systems. Age- and sex-matched mice were used for all experiments.
Luciferase assay
HEK-293T cells were inoculated in 24-well plates and transfected with the indicated plasmids and with pRL-TK renilla vector (Promega). Empty control vector was added so that an equal amount of DNA was transfected into each well. At 36 h after transfection, cells were lysed and luciferase activity was detected with a dual-luciferase reporter assay kit (Promega). Human IFN-α4 luciferase vectors containing mutated NFAT binding sites or IRF7 binding sites were generated by PCR site-directed mutagenesis.
Immunoprecipitation and immunoblot analysis
HEK-293T cells were transfected with HA-tagged or Myc-tagged plasmids as indicated. Immunoprecipitation and immunoblotting experiments in transfected HEK-293T cells or fresh isolated pDCs were performed as described previously (Bao et al., 2012). For the purified protein–binding assay, GST-tagged NFATC3 proteins or GST proteins were incubated with purified Myc-tagged IRF7 proteins. Myc–agarose beads were used to pull down Myc-tagged IRF7, and anti-GST HRP-conjugated antibody was used to detect the binding of GST and GST-NFATC3. GST-tagged NFATC3 protein was also incubated with Myc-tagged IRF7 and Myc-tagged IRF3. Glutathione Sepharose 4B beads were used to pull down GST-tagged NFATC3. Myc antibody was used to detect the binding of IRF7 and IRF3.
Real-time PCR
Total RNA was extracted, and RT of total RNA to cDNA was performed with an iScript RT supermix kit (Bio-Rad Laboratories). Real-time PCR was performed using the SYBR green fluorescence system. Primers were synthesized by Integrated DNA Technologies (Table S1).
In vivo CpG challenge
Mice were injected intravenously with 5 µg CpG A (D19) complexed with DOTAP (Roche; 30 µl DOTAP/150 µl of total volume). 6 h later, blood was collected and serum was separated. IFN-α and IL-12p40 levels in serum were measured by ELISA.
Knockout with lentivirus-delivered CRISPR and restoration
Gen2.2 cells were transduced with Cas9 lentiviral particles, and Cas9-expressing stable Gen2.2 cells were selected. Cas9-expressing Gen2.2 cells were further transduced with lentiviral particles containing guide RNA targeting MyD88 and NFATC3. Then, cells were selected and clonal expanded. The knockout efficiency was determined by immunoblotting. The Cas9 and guide RNA lentiviral vectors were bought from System Biosciences, Inc. The sequences of the guide RNA target sequences were as follows: non-targeting (sense), 5′-ACGGAGGCTAAGCGTCGCAA-3′; MyD88 (sense), 5′-CTAGTGAGCTCATCGAAAAG-3′; and NFATC3 (sense), 5′-GATCAAGCTGCCATACTACC-3′. To restore NFATC3 expression in NFATC3 knockout cells, an NFATC3 expression lentiviral vector (NFATC3a) that contains mutated NFATC3 guide RNA targeting sequence was used to generate NFATC3a stable–expressing NFATC3 knockout Gen2.2 cells.
Chromatin immunoprecipitation
Gen2.2 cells were fixed, lysed, and sonicated. Precleared lysates were incubated overnight at 4°C with anti-IRF7 (sc-74472; Santa Cruz Biotechnology, Inc.), anti-NFATC3 (A303-572A; Bethyl Laboratories, Inc.), or control rabbit IgG (Cell Signaling Technology). Immunocomplexes were collected, purified, and analyzed by real-time PCR. Primers for IFNA4 promoter detection were forward, 5′-CCATAAAAGCCTTTGAGTGCAGG-3′, and backward, 5′-GAAGACTTTGCTCTGTGCATAGG-3′. Primers for IFNB1 promoter detection were forward, 5′-GACATAGGAAAACTGAAAGGGAG-3′, and backward, 5′-TGAGATGGTCCTCTCTCTATTCA-3′.
Pull-down assay
Biotin-labeled oligonucleotides were synthesized by Integrated DNA Technologies (Table S2). HEK-293T cells were transfected with HA-tagged NFATC3 and Myc-tagged IRF7. Cells were lysed and precleared with NeutrAvidin agarose beads (Thermo Fisher Scientific). Cell lysate was incubated with biotin-labeled oligonucleotides at 4°C overnight with rotation followed by 2-h incubation with NeutrAvidin agarose beads. The beads were washed extensively, and NFATC3 and IRF7 were detected by immunoblotting. CRISPR knockout Gen2.2 cells were stimulated with CpG A, and cell lysates were precleared, incubated with biotin-labeled oligonucleotides, and precipitated with NeutrAvidin agarose beads. NFATC3 and IRF7 were detected by immunoblotting.
Statistical analysis
Data were analyzed statistically and are shown as mean ± SD. The p-values were calculated using an unpaired two-tailed Student’s t test; a p-value of <0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows that mouse Nfatc3 enhances mouse Ifn-α4 transcriptional activity and inhibition of Nfat activity reduces CpG DNA–induced IFN-α in mouse primary pDCs. Fig. S2 shows that Nfatc3 deficiency does not affect pDC differentiation. Table S1 shows a list of primers used for real-time PCR. Table S2 shows a list of biotin-labeled oligonucleotides used for pull-down assay.
Supplementary Material
Acknowledgments
M. Bao, H. Lu, L. Weng, and S. Hanabuchi are full-time employees of MedImmune, LLC. Y. Wang, W. Sha, and Y.-J. Liu were employees of MedImmune, LLC. This work was done at both Baylor Institute for Immunology Research and MedImmune, LLC.
The authors declare no further competing financial interests.
Author contributions: M. Bao performed experiments and analyzed the data. M. Bao and Y.-J. Liu wrote the manuscript. Y. Wang, Y.-J. Liu, L. Weng, Z. Zhang, and H. Lu performed experiments. P. Shi, W. Sha, and S. Hanabuchi provided technical support. J. Qin performed mass spectrometry analysis. J. Plumas and L. Chaperot provided the Gen2.2 cell line.
Footnotes
Abbreviations used:
- CRISPR
- clustered regularly interspaced short palindromic repeats
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- NFATC
- NFAT cell
- NHR
- NFAT homology region
- NT
- nontargeting
- pDC
- plasmacytoid DC
References
- Bao M., and Liu Y.J.. 2013. Regulation of TLR7/9 signaling in plasmacytoid dendritic cells. Protein Cell. 4:40–52. 10.1007/s13238-012-2104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M., Hanabuchi S., Facchinetti V., Du Q., Bover L., Plumas J., Chaperot L., Cao W., Qin J., Sun S.C., and Liu Y.J.. 2012. CD2AP/SHIP1 complex positively regulates plasmacytoid dendritic cell receptor signaling by inhibiting the E3 ubiquitin ligase Cbl. J. Immunol. 189:786–792. 10.4049/jimmunol.1200887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Deb J., Patra A.K., Thuy Pham D.A., Chen W., Vaeth M., Berberich-Siebelt F., Klein-Hessling S., Lamperti E.D., Reifenberg K., et al. 2011. NFATc1 affects mouse splenic B cell function by controlling the calcineurin–NFAT signaling network. J. Exp. Med. 208:823–839. 10.1084/jem.20100945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp T., Palmetshofer A., Serfling E., Heib V., Schmitt S., Richter C., Klein M., Schild H., Schmitt E., and Stassen M.. 2005. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J. Exp. Med. 201:181–187. 10.1084/jem.20041538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaperot L., Blum A., Manches O., Lui G., Angel J., Molens J.P., and Plumas J.. 2006. Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J. Immunol. 176:248–255. 10.4049/jimmunol.176.1.248 [DOI] [PubMed] [Google Scholar]
- Colina R., Costa-Mattioli M., Dowling R.J., Jaramillo M., Tai L.H., Breitbach C.J., Martineau Y., Larsson O., Rong L., Svitkin Y.V., et al. 2008. Translational control of the innate immune response through IRF-7. Nature. 452:323–328. 10.1038/nature06730 [DOI] [PubMed] [Google Scholar]
- Crabtree G.R., and Olson E.N.. 2002. NFAT signaling: choreographing the social lives of cells. Cell. 109(2, Suppl):S67–S79. 10.1016/S0092-8674(02)00699-2 [DOI] [PubMed] [Google Scholar]
- Esashi E., Bao M., Wang Y.H., Cao W., and Liu Y.J.. 2012. PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells. Eur. J. Immunol. 42:573–579. 10.1002/eji.201142045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Ichikawa T., Nakao K., Matsumoto A., Miyaaki H., Shibata H., Eguchi S., Takatsuki M., Ikeda M., Yamasaki H., et al. 2008. Differential effects of calcineurin inhibitors, tacrolimus and cyclosporin a, on interferon-induced antiviral protein in human hepatocyte cells. Liver Transpl. 14:292–298. 10.1002/lt.21358 [DOI] [PubMed] [Google Scholar]
- Ho S.N., Thomas D.J., Timmerman L.A., Li X., Francke U., and Crabtree G.R.. 1995. NFATc3, a lymphoid-specific NFATc family member that is calcium-regulated and exhibits distinct DNA binding specificity. J. Biol. Chem. 270:19898–19907. 10.1074/jbc.270.34.19898 [DOI] [PubMed] [Google Scholar]
- Ho S., Clipstone N., Timmermann L., Northrop J., Graef I., Fiorentino D., Nourse J., and Crabtree G.R.. 1996. The mechanism of action of cyclosporin A and FK506. Clin. Immunol. Immunopathol. 80:S40–S45. 10.1006/clin.1996.0140 [DOI] [PubMed] [Google Scholar]
- Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., and Taniguchi T.. 2005a Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 434:1035–1040. 10.1038/nature03547 [DOI] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T.. 2005b IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Hoshino K., Sugiyama T., Matsumoto M., Tanaka T., Saito M., Hemmi H., Ohara O., Akira S., and Kaisho T.. 2006. IκB kinase-α is critical for interferon-α production induced by Toll-like receptors 7 and 9. Nature. 440:949–953. 10.1038/nature04641 [DOI] [PubMed] [Google Scholar]
- Kaxiras A., Yamamoto S., Soderdahl G., Wernerson A., Axelsson R., and Ericzon B.G.. 2014. Cyclosporin A, but not tacrolimus, negatively affects the hepatic extraction fraction of hepatobiliary scintigraphy in liver transplant recipients. EJNMMI Res. 4 10.1186/s13550-014-0073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M., Klein-Hessling S., Palmetshofer A., Serfling E., Tertilt C., Bopp T., Heib V., Becker M., Taube C., Schild H., et al. 2006. Specific and redundant roles for NFAT transcription factors in the expression of mast cell–derived cytokines. J. Immunol. 177:6667–6674. 10.4049/jimmunol.177.10.6667 [DOI] [PubMed] [Google Scholar]
- Li Y., Dai J., Song M., Fitzgerald-Bocarsly P., and Kiledjian M.. 2012. Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7. Mol. Cell. Biol. 32:1164–1172. 10.1128/MCB.06328-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Deng H., Li X., Wu X., Tang Q., Chang T.H., Peng H., Rauscher F.J. III, Ozato K., and Zhu F.. 2011. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 187:4754–4763. 10.4049/jimmunol.1101704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V., Ratushny A.V., Lampano A.E., Schmitz F., Huang A.C., Raman A., Rust A.G., Bergthaler A., Aitchison J.D., and Aderem A.. 2012. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 490:421–425. 10.1038/nature11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275–306. 10.1146/annurev.immunol.23.021704.115633 [DOI] [PubMed] [Google Scholar]
- Liu L.S., Li J., Chen X.T., Zhang H.X., Fu Q., Wang H.Y., Xiong Y.Y., Liu S., Liu X.M., Li J.L., et al. 2015a Comparison of tacrolimus and cyclosporin A in CYP3A5 expressing Chinese de novo kidney transplant recipients: a 2-year prospective study. Int. J. Clin. Pract. Suppl. 183:43–52. 10.1111/ijcp.12666 [DOI] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., and Chen Z.J.. 2015b Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Macartney C., Freilich M., Odame I., Charpentier K., and Dror Y.. 2009. Complete response to tacrolimus in a child with severe aplastic anemia resistant to cyclosporin A. Pediatr. Blood Cancer. 52:525–527. 10.1002/pbc.21751 [DOI] [PubMed] [Google Scholar]
- Müller M.R., and Rao A.. 2010. NFAT, immunity and cancer: a transcription factor comes of age. Nat. Rev. Immunol. 10:645–656. 10.1038/nri2818 [DOI] [PubMed] [Google Scholar]
- Panne D., Maniatis T., and Harrison S.C.. 2007. An atomic model of the interferon-beta enhanceosome. Cell. 129:1111–1123. 10.1016/j.cell.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger A.M., Hodge M.R., Gravallese E.M., Oukka M., Davidson L., Alt F.W., de la Brousse F.C., Hoey T., Grusby M., and Glimcher L.H.. 1998. Delayed lymphoid repopulation with defects in IL-4–driven responses produced by inactivation of NF-ATc. Immunity. 8:125–134. 10.1016/S1074-7613(00)80465-3 [DOI] [PubMed] [Google Scholar]
- Rao A., Luo C., and Hogan P.G.. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707–747. 10.1146/annurev.immunol.15.1.707 [DOI] [PubMed] [Google Scholar]
- Rengarajan J., Mowen K.A., McBride K.D., Smith E.D., Singh H., and Glimcher L.H.. 2002a Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195:1003–1012. 10.1084/jem.20011128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J., Tang B., and Glimcher L.H.. 2002b NFATc2 and NFATc3 regulate TH2 differentiation and modulate TCR-responsiveness of naïve TH cells. Nat. Immunol. 3:48–54. 10.1038/ni744 [DOI] [PubMed] [Google Scholar]
- Schreiber S.L., and Crabtree G.R.. 1992. The mechanism of action of cyclosporin A and FK506. Immunol. Today. 13:136–142. 10.1016/0167-5699(92)90111-J [DOI] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387. 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., and Fujita T.. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087–1095. 10.1093/emboj/17.4.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Nishina H., Takimoto H., Marengère L.E., Wakeham A.C., Bouchard D., Kong Y.Y., Ohteki T., Shahinian A., Bachmann M., et al. 1998. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity. 8:115–124. 10.1016/S1074-7613(00)80464-1 [DOI] [PubMed] [Google Scholar]
- You F., Wang P., Yang L., Yang G., Zhao Y.O., Qian F., Walker W., Sutton R., Montgomery R., Lin R., et al. 2013. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol. 14:1237–1246. 10.1038/ni.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., and Hayward G.S.. 2010. The ubiquitin E3 ligase RAUL negatively regulates type 1 interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity. 33:863–877. 10.1016/j.immuni.2010.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I., and Granucci F.. 2012. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs). Eur. J. Immunol. 42:1924–1931. 10.1002/eji.201242580 [DOI] [PubMed] [Google Scholar]
- Zanoni I., Ostuni R., Capuano G., Collini M., Caccia M., Ronchi A.E., Rocchetti M., Mingozzi F., Foti M., Chirico G., et al. 2009. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature. 460:264–268. 10.1038/nature08118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.