Abstract
Transvection—pairing-dependent interallelic regulation resulting from enhancer action in trans—occurs throughout the Drosophila melanogaster genome, likely as a result of the extensive somatic homolog pairing seen in Dipteran species. Recent studies of transvection in Drosophila have demonstrated important qualitative differences between enhancer action in cis vs. in trans, as well as a modest synergistic effect of cis- and trans-acting enhancers on total tissue transcript levels at a given locus. In the present study, we identify a system in which cis- and trans-acting GAL4-UAS enhancer synergism has an unexpectedly large quantitative influence on gene expression, boosting total tissue transcript levels at least fourfold relative to those seen in the absence of transvection. We exploit this strong quantitative effect by using publicly available UAS-shRNA constructs from the TRiP library to assay candidate genes for transvection activity in vivo. The results of the present study, which demonstrate that in trans activation by simple UAS enhancers can have large quantitative effects on gene expression in Drosophila, have important new implications for experimental design utilizing the GAL4-UAS system.
Keywords: transvection, enhancer, somatic pairing, GAL4-UAS
The nuclear genome is often pictured to function as a linear arrangement of nucleotides grouped into genes and regulatory elements operating more or less locally in cis . However, physical interactions between distant genomic sites, including interactions between distinct chromosomes, are increasingly understood to play important roles in eukaryotic genome function (Cavalli and Misteli 2013). For example, in trans interactions between distinct chromosomes have been shown to play a critical role in mammalian genomes during the process of X-inactivation (Masui et al. 2011) and the monogenic expression of olfactory receptors (Lomvardas et al. 2006).
The paired somatic genomes of Drosophila melanogaster and other Dipteran species are a striking example of nuclear organization in which there is widespread physical interaction between maternal and paternal homologous chromosomes (McKee 2004; Metz 1916; Stevens 1908). That the physical pairing of homologous somatic chromosomes in Drosophila could have functional consequences was hypothesized over 100 years ago (Stevens 1908), but genetic interaction between physically paired alleles at the same genomic locus was first demonstrated in 1954 by Lewis (1954), who established the term transvection to describe such interactions.
Classically, transvection has been characterized in relatively circumscribed settings in which genetic complementation between two mutant alleles at the same locus can be eliminated by the introduction of chromosomal rearrangements (Lewis 1954). However, the relatively recent development of site-directed transgenesis technologies (Fish et al. 2007; Markstein et al. 2008) has opened up vast new possibilities for the study of transvection phenomena. Using such technologies, it was recently shown that transvection appears to be a pervasive feature of the Drosophila genome, with transcriptional activation in trans occurring at essentially all loci tested, independent of enhancer identity, hypothetical pairing sequences, and even strict homology between paired genetic elements (Bateman et al. 2012a; Blick et al. 2016; Mellert and Truman 2012). A recent study has also demonstrated transvection-like phenomena in budding yeast (Zhang and Bai 2016).
Despite these significant recent advances, transvection remains poorly understood at the mechanistic as well as functional level. One interesting observation made in prior studies of transvection in Drosophila has been the detection of quantitatively significant increases in transcription at loci for which transcriptional activation can be driven by regulatory elements that are both in cis and/or in trans (Bateman et al. 2012a; Gibson et al. 1999; Goldsborough and Kornberg 1996; Lum and Merritt 2011). Though the effect sizes observed have been modest (Bateman et al. 2012a; Gibson et al. 1999; Lum and Merritt 2011) or difficult to quantify (Goldsborough and Kornberg 1996), previous authors have speculated that quantitative changes in gene expression as a result of transvection could play important roles in normal and/or pathological genome function. Indeed, somatic chromosomal pairing appears to drive dysregulated gene expression in at least one type of human neoplasm (Koeman et al. 2008).
In addition to affecting gene expression quantitatively, transcriptional activation in trans also appears to be qualitatively different from transcriptional activation in cis (Bateman et al. 2012a; Blick et al. 2016; Mellert and Truman 2012). Recent studies have shown that in trans transcriptional activation occurs stochastically and with variable intensity among neighboring cells within a tissue, while activation in cis occurs predictably and at consistent levels within those same tissues (Bateman et al. 2012a; Blick et al. 2016; Mellert and Truman 2012). These qualitative differences suggest that in cis and in trans transcriptional activation may be mechanistically different, and raise the interesting question of whether they might utilize different sets of regulatory or effector molecules.
To date, very few molecular factors required for transvection have been identified. Transvection is thought to require the physical proximity of homologous chromosomes, which is maintained by specific cellular factors (Hartl et al. 2008), and high throughput screens recently undertaken in Drosophila cell culture have identified numerous genes that affect somatic chromosomal pairing (Bateman et al. 2012b; Joyce et al. 2012). However, such screens do not assess the functional role that identified pairing factors might play in transvection, and an efficient method for assaying chromosomal pairing factors for transvection activity in vivo has not yet been developed.
In the present study, using a Drosophila model of neurodegeneration, we have identified conditions under which transvection between allelic GAL4-UAS-driven transgenes produces a dramatic (approximately fourfold) increase in transcriptional efficacy that is readily detected by semiquantitative RT-PCR and Western blot. We find that quantitatively similar interallelic increases in transcription can be caused by publicly available UAS-tagged transgenes, including those from the TRiP shRNA library (Ni et al. 2011). Finally, by using interallelic enhancement of transcription as a sensitive functional measure of transvection, we utilize TRiP shRNA constructs to test a panel of candidate genes for transvection activity in vivo.
Materials and Methods
Fly stocks and husbandry
Flies carrying the UAS-mouse (Mo)PrP insert at attP2 have been previously described (Murali et al. 2014). The GAL4 driver lines w1118;P{ChAT-GAL4.7.4}/CyO,P{sevRas1.V12}FK1, w*;P{GAL4-elav.L}3, and y1 w*;P{tubP-GAL4}LL7/TM3, Sb1 Ser1, referred to as Cha-GAL4, Elav-GAL4, and α-tubulin-GAL4, respectively, were obtained from the Bloomington Drosophila Stock Center (BDSC) (Bloomington, IN). Cha-GAL4 drives UAS-tagged transgene expression in cholinergic neurons at all developmental stages (Salvaterra and Kitamoto 2001), while Elav-GAL4 drives transgene expression in all postmitotic neurons as well as in certain neuroblasts and embryonic glial cells (Berger et al. 2007; Robinow and White 1988). Flies were maintained at 25°, 70% relative humidity on standard Drosophila cornmeal, yeast, molasses, and agar medium with methylparaben as a mold inhibitor.
Construction of plasmids and generation of transgenic fly lines
The vector pUASTattB MoPrP was generated previously (Murali et al. 2014). To avoid the introduction of mutations into the pUASTattB (Bischof et al. 2007) targeting vector, the MoPrP insert was transferred as a BglII/XhoI restriction fragment into the cloning vector pCombo3 (Mike Scott, UCSF) and site-directed mutagenesis performed using the GeneTailor system (Invitrogen, Grand Island, NY) to create pCombo3 P101L, Q171R, and D177N MoPrP constructs. The P101L, Q171R, and D177N MoPrP inserts were then cloned into pUASTattB as BglII/XhoI fragments and the plasmid sequence confirmed by sequencing prior to phiC31-integrase-mediated insertion at attP2 and attP40 performed by Genetic Services, Inc. (Cambridge, MA). Isogenic lines carrying UAS-P101L, Q171R, and D177N MoPrP were isolated, balanced, and maintained using standard Drosophila genetic techniques.
Climbing assays
Locomotor function of flies was assessed using an assay similar to that described previously (Ganetzky and Flanagan 1978; Gavin et al. 2006; Le Bourg and Lints 1992) and performed at room temperature (22°). Briefly, groups of 15 male flies were collected and allowed to recover from CO2 anesthesia for at least 12 hr (at 25°) before being transferred, without anesthesia, into fresh assay vials containing fly media. Flies were allowed to acclimatize for at least 10 min in the assay vial before being tested. To measure locomotor function, flies were knocked to the bottom of the vial and the number climbing 4 cm above the surface of the media within 30 sec was counted. Each group of flies was tested in triplicate and the mean number of flies passing the assay was recorded for that group. At least six independent groups of 15 flies were tested in this way for each genotype, and the mean and standard deviation of the number of flies passing the assay was taken. These values were then normalized to give a percent passing rate for each genotype, which is reported below. Finally, it was noticed that flies with poor locomotor function often displayed improved climbing activity with successive trials, so for all groups of flies tested, six pretrials were performed without a break, followed by the three experimental trials, also without a break. All assays were performed on male flies aged 5 d at 25° after eclosion.
Preparation of fly head homogenates for protein quantification by Western blotting
Under light CO2 anesthesia and using a fresh razor blade, 20 male fly heads were collected and transferred into 50 μl ice-cold lysis buffer (0.5% v/v Triton X-100, 0.5% w/v deoxycholate, 10 mM NaCl, and 50 mM Tris, pH 7.5) in a 0.2 ml glass Kontes micro tissue grinder. A smooth suspension was generated by several strokes of the glass pestle and debris pelleted by a 1 min centrifugation at ∼9500 rcf (relative centrifugal force). 40 μl of the resulting supernatant was mixed with 40 μl of ice-cold 2× SDS-PAGE loading buffer and the sample boiled at 95° for 5 min, then frozen at −80° before electrophoresis and Western blotting. All samples were taken from male flies aged 5 d at 25° after eclosion.
SDS-PAGE was performed as described previously (Deleault et al. 2007). After electrophoresis, proteins were transferred to a methanol-charged PVDF membrane using a Hoefer wet transfer apparatus set at a constant current of 1 A for 2 hr. After electroblotting, membranes were blocked for 1 hr at 4° in 15% w/v powdered skim milk dissolved in TBST. The following primary antibodies were used at the indicated dilutions: anti-PrP, mAb 27/33 (Deleault et al. 2012) at 1:15,000 in TBST; anti-GFP, JL-8 (Clontech, Mountainview, CA) at 1:10,000 in TBST + milk; anti-kinesin heavy chain, and AKIN01 (Cytoskeleton, Denver, CO) at 1:10,000 in TBST + milk. Membranes were incubated in primary antibody overnight at 4° then washed four times with TBST and incubated for 1 hr at 4° in HRP-conjugated sheep-anti-mouse (GE Biosciences, Piscataway, NJ) or goat-anti-rabbit (Bio-Rad, Hercules, CA) secondary antibody prior to four TBST washes and detection by enhanced chemiluminescence. MoPrP expressed in Drosophila appears as a quadruplet of ∼20–25 kDa Mr (Gavin et al. 2006). For quantification of MoPrP expression, all four bands were measured in total.
RNA extraction and semiquantitative RT-PCR
Under light CO2 anesthesia and using a fresh razor blade, 30 male fly heads were collected and transferred into 250 μl ice-cold Trizol reagent (Life Technologies, Carlsbad, CA), then frozen at −80° for no more than 1 wk prior to RNA isolation. Total RNA was isolated using the Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA) modified with an in-solution DNase digestion. Briefly, after washing the MiniPrep column with the provided RNA Wash Buffer, 100 μl of DNase digestion solution [∼1 U/ μl RQ1 RNase-free DNase (Promega, Madison, WI) in provided buffer] was added to the column and centrifuged for 30 sec at 500 rcf into an RNase-free 1.5 ml tube. Both column and flow-through were incubated at 37° for 30 min, followed by collection of residual digestion solution in the column matrix by a brief maximal speed centrifugation. The collected ∼100 μl of digestion solution was mixed with 300 μl RNA Binding Buffer (Zymo Research) and 400 μl 100% ethanol, and the column was reloaded and carried through the remaining wash and elution steps. Purity and concentration of the isolated RNA was measured by spectrophotometry.
cDNA synthesis was performed using an oligo dT primer (Roche, Indianapolis, IN) and M-MLV reverse transcriptase (Life Technologies) in reactions containing Protector RNase Inhibitor (Roche). Amplification of cDNA for transcripts encoding PrP or RpL32 was performed simultaneously using 20 cycles of PCR and the following primer pairs: 5′-GGTGGTGGTGACCGTGTGCTGCTT-3′, 5′-CGAACCTTGGCTACTGGCTGCTG-3′ and 5′-AGGCCCAAGATCGTGAAGAA-3′, 5′-TTGTGCACCAGGAACTTCTTGAA-3′, respectively. Samples were stained directly with SybrGold (Life Technologies) at a 1:1000 final dilution and electrophoresed on 1% w/v TBE agarose gels prior to detection by UV transillumination. Under the conditions used, the intensity of the respective amplified bands correlated approximately linearly with the amount of cDNA template added over a 26-fold range spanning the highest and lowest band intensities observed.
Statistical analysis
Unless otherwise stated, comparisons were performed using one-way ANOVA with Tukey’s HSD post-test. All statistical analyses were performed with GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). Standard deviations are shown as indicated by error bars. P < 0.05 was considered statistically significant; * indicates P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, and ns denotes not significant.
Data and reagent availability
UAS-PrP fly strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
While utilizing the GAL4-UAS system in Drosophila to model prion protein (PrP)-induced neurodegeneration, we have observed that certain transgenic lines express a neurodegenerative phenotype when the UAS-PrP transgene is homozygous, but not when it is hemizygous, at a given locus (Gavin et al. 2006). In these transgenic lines, the neurodegenerative phenotype seen in homozygous UAS-PrP flies is accompanied, and presumably caused, by a dramatic increase in PrP protein levels disproportionate to a twofold increase in UAS-PrP gene dosage (Gavin et al. 2006).
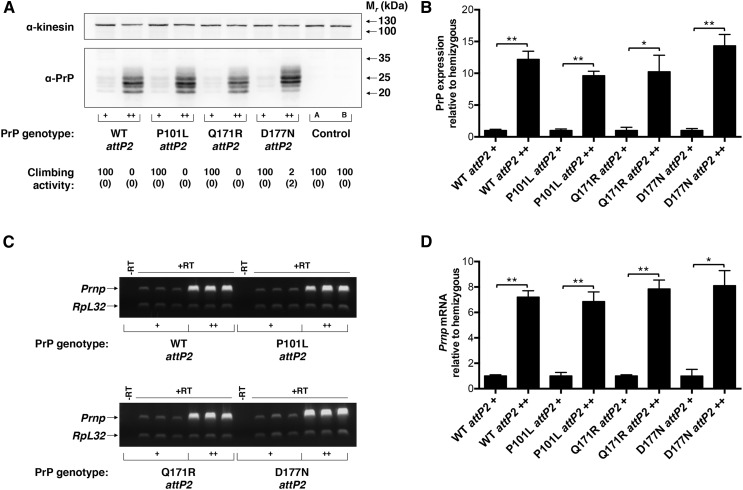
To better characterize this phenomenon, we used the site-directed phiC31 integrase system (Fish et al. 2007; Markstein et al. 2008) to generate transgenic Drosophila lines expressing wild-type, pathogenic (P101L and D177N) (Hsiao et al. 1990; Jackson et al. 2009), and protective (Q171R) (Geoghegan et al. 2009; Kaneko et al. 1997) PrP variants at the widely used attP2 locus (Markstein et al. 2008) (Figure 1). Unexpectedly, we found that, independent of the misfolding propensity of the UAS-PrP variant expressed, each transgenic line was exquisitely sensitive to UAS-PrP gene dosage at attP2; hemizygous UAS-PrP flies passed a simple climbing assay at a rate of 100% and were phenotypically normal, while homozygous UAS-PrP flies of the same line displayed dramatic locomotor defects, with essentially no flies passing the climbing assay (Figure 1A). Moreover, these locomotor defects were accompanied by a 10- to 14-fold increase in PrP protein expression in homozygous UAS-PrP flies relative to hemizygous flies of the same line (Figure 1, A and B). To understand the mechanism of PrP accumulation in these homozygous transgenic lines, we performed semiquantitative RT-PCR on fly head RNA extracts (Figure 1C). Interestingly, we found that PrP mRNA levels were increased approximately seven- to eightfold in homozygous flies relative to hemizygous flies of the same line (Figure 1, C and D), which accounts for the majority of PrP accumulation in these animals and suggests that homozygosity at attP2 is associated with increased transcriptional efficacy of the UAS-PrP transgene. For all subsequent experiments we focused on the Q171R PrP variant, which we refer to simply as PrP.
Figure 1.
Homozygosity of UAS-PrP at attP2 increases Prnp transcriptional efficacy. Four different UAS-PrP constructs, representing wild-type, pathogenic (P101L and D177N), and protective (Q171R) PrP sequences were inserted at attP2 and driven by Cha-GAL4. (A) Western blot of fly head homogenates from flies hemi- and homozygous for UAS-PrP constructs at attP2 (+, hemizygous; ++, homozygous). Experiment was performed in triplicate with a representative blot shown. Climbing activity of the various lines is shown below the corresponding lane of the Western blot and expressed as percent passing the climbing assay (see Materials and Methods) with standard deviation shown below in brackets. Control A is driver alone; control B is UAS-PrP alone. (B) Densitometry of Western blot signals in (A) and associated replicates. PrP signals were normalized to the kinesin loading control and then to the respective hemizygous UAS-PrP attP2 lines. (C) Semiquantitative RT-PCR performed on fly head mRNA of hemi- and homozygous attP2 UAS-PrP constructs driven by Cha-GAL4 (+, hemizygous; ++, homozygous). Prnp and RpL32 transcripts were coamplified from total cDNA, and each lane represents a biological replicate. Control samples, labeled –RT, represent PCR reactions of input RNA without addition of RT or oligo dT primer during the cDNA synthesis step. (D) Densitometry of RT-PCR signals in (C). For each sample, the ratio of Prnp to RpL32 mRNA levels was taken and then normalized to the average value for the respective hemizygous UAS-PrP attP2 line. Error bars in (B) and (D) represent standard deviations, with n = 3 for each group; ns denotes not significant, * P < 0.05, and ** P < 0.01, two-tailed Student’s t-test. cDNA, complementary DNA; mRNA, messenger RNA; PrP, prion protein; RT, reverse transcriptase; RT-PCR, reverse transcription polymerase chain reaction; WT, wild-type.
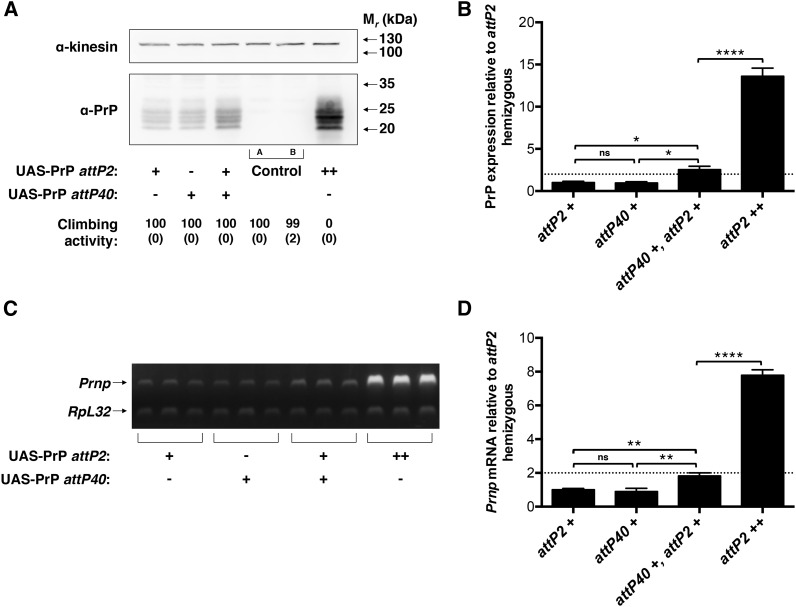
Previous investigations in Drosophila have demonstrated that the pairing of cis- and trans-acting enhancers can synergistically increase transcription at a given genomic locus (Bateman et al. 2012a; Gibson et al. 1999; Goldsborough and Kornberg 1996; Lum and Merritt 2011), but the effect size reported has been modest (Bateman et al. 2012a; Gibson et al. 1999; Lum and Merritt 2011) or difficult to quantify (Goldsborough and Kornberg 1996). In these prior studies, mutual transcriptional enhancement between paired alleles has been thought to represent one manifestation of transvection. To test whether the dramatic sensitivity to PrP gene dosage observed in our system (Figure 1) might be a pairing-dependent (i.e., transvection) effect, we created a transgenic Drosophila line expressing PrP on a different chromosome, at the attP40 genomic locus. We found that UAS-PrP transgene insertion at the original attP2 locus (third chromosome) and at the new attP40 locus (second chromosome) each yielded almost identical PrP expression levels (Figure 2A, left two lanes and Figure 2B, left two groups). The functional equivalence of these two loci for UAS-PrP expression allowed us to then examine the effect of doubling UAS-PrP gene dosage using copies of UAS-PrP at allelic vs. nonallelic loci (Figure 2). We found that, when UAS-PrP transgenes were expressed at nonallelic loci, PrP protein expression was additive and locomotor behavior was normal (Figure 2A, three left lanes). In contrast, when UAS-PrP transgenes were expressed at the same attP2 genomic site, protein expression was synergistic and flies demonstrated profound locomotor defects (Figure 2A, third lane vs. sixth lane). We found analogous results at the mRNA level (Figure 2B), suggesting that homozygosity of UAS-PrP alleles increases the transcriptional efficacy of each UAS-PrP transgene by approximately fourfold (Figure 2D). Synergistic expression of allelic UAS-PrP transgenes was also observed at the attP40 locus (Supplemental Material, Figure S1). Expression at attP40 was driven by Elav-GAL4 rather than Cha-GAL4 due to the technical requirement of a third chromosome driver.
Figure 2.
Allelic interaction between UAS-PrP transgenes is required for enhanced Prnp transcriptional efficacy. UAS-PrP inserted at attP2 and/or attP40 was expressed using the Cha-GAL4 driver. (A) Western blot of fly head homogenates (+, hemizygous; ++, homozygous at the indicated locus). Note that PrP is independently expressed at approximately equal levels from attP2 and attP40. Experiment was performed in triplicate with a representative blot shown. Climbing activity of each line is indicated below the corresponding lane of the Western blot and expressed as percent passing the climbing assay (see Materials and Methods) with standard deviation shown below in brackets. Control A is driver alone; control B is UAS-PrP alone. (B) Densitometry of Western blot signals in (A) and associated replicates. PrP signals were normalized to the kinesin loading control and then to the hemizygous attP2 UAS-PrP line. (C) Semiquantitative RT-PCR performed on fly head mRNA of attP2 and attP40 UAS-PrP constructs driven by Cha-GAL4 (+, hemizygous; ++, homozygous at the indicated locus). Prnp and RpL32 transcripts were coamplified in the same PCR reactions, and each lane represents a biological replicate. (D) Densitometry of RT-PCR signals in (C). For each sample, the ratio of Prnp to RpL32 mRNA levels was taken and then normalized to the average value for the hemizygous attP2 UAS-PrP line. Error bars in (B) and (D) represent standard deviations, with n = 3 for each group; ns denotes note significant, ** P < 0.01, and **** P < 0.0001. Dotted line emphasizes a twofold increase in expression relative to hemizygous. mRNA, messenger RNA; PrP, prion protein; RT-PCR, reverse transcription polymerase chain reaction.
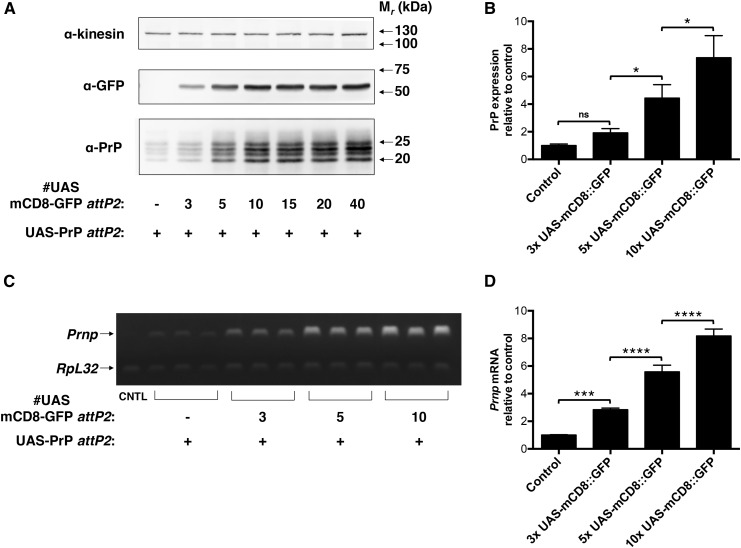
To further characterize the phenomenon of interallelic transcriptional enhancement, we next used the series of attP2 UAS-mCD8::GFP lines created by Pfeiffer et al. (2010), which carry a varying number of UAS elements upstream of the mCD8::GFP reporter construct, to test: a) Whether enhancement of PrP expression could result from the interaction of UAS-PrP and a non-PrP UAS-driven transgene at the same genomic locus, and b) whether the degree of in trans enhancement of PrP transcription depends upon the strength of the UAS enhancer on the paired transgene (Figure 3). We found that, indeed, UAS-mCD8::GFP substantially boosted UAS-PrP expression at the protein and mRNA levels when both transgenes were paired at the attP2 locus (Figure 3, A and C). In addition, this trans-acting transcriptional enhancement was approximately proportional to the number of UAS elements on the paired UAS-mCD8::GFP transgene (Figure 3, B and D), though expression of both mCD8::GFP and PrP appeared to saturate for those lines with mCD8::GFP enhancers containing more than 10 UAS elements (Figure 3A). We believe that the above experiments provide compelling evidence that observed interallelic transcriptional enhancement of UAS-PrP expression is a manifestation of transvection.
Figure 3.
Increasing UAS enhancer strength at the attP2 locus in trans increases attP2 UAS-PrP expression. UAS-mCD8::GFP constructs with variable UAS number were expressed at attP2 in trans to UAS-PrP, with expression driven by Cha-GAL4. (A) Western blot of fly head homogenates. Experiment was performed in triplicate with a representative blot shown. (B) Densitometry of Western blot signals in (A) and associated replicates. PrP signals were normalized to the kinesin loading control and then to the hemizygous attP2 UAS-PrP line lacking UAS-mCD8::GFP in trans. (C) Semiquantitative RT-PCR performed on fly head mRNA. Prnp and RpL32 transcripts were coamplified in the same PCR reactions, and each lane represents a biological replicate. CNTL is of mRNA prepared from UAS-PrP flies that do not express GAL4. (D) Densitometry of RT-PCR signals in (C). For each sample, the ratio of Prnp to RpL32 mRNA levels was taken and then normalized to the average value for the attP2 UAS-PrP line lacking UAS-mCD8::GFP in trans. Error bars in (B) and (D) represent standard deviations, with n = 3 for each group; ns denotes not significant, * P < 0.05, *** P < 0.001, and **** P < 0.0001. Control sample, CNTL; GFP, green fluorescent protein; mRNA, messenger RNA; PrP, prion protein; RT-PCR, reverse transcription polymerase chain reaction.
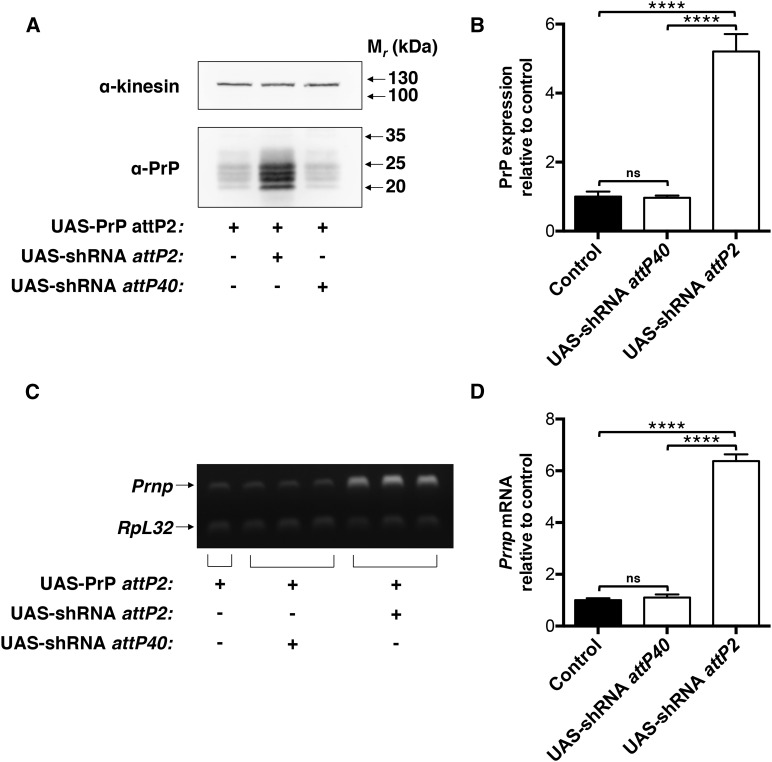
Having identified PrP expression as a sensitive measure of cross talk between allelic enhancers in our system, we next sought to determine whether such a measure could be used to identify or study factors required for chromosomal pairing and/or transvection in vivo. A powerful tool available for the in vivo dissection of biological processes in Drosophila is the TRiP library, a genome-scale collection of UAS-shRNA constructs inserted at either the attP2 or attP40 sites (Ni et al. 2011). To determine whether TRiP constructs could be used in combination with our UAS-PrP lines to study chromosomal pairing and transvection, we obtained TRiP lines expressing identical UAS-shRNA hairpins against an irrelevant gene (GFP) inserted at either attP2 or attP40, and crossed these lines to our attP2 UAS-PrP line (Figure 4). As predicted, we observed a robust increase in PrP protein and mRNA levels, approximately five- to sixfold, when UAS-PrP and UAS-shRNA were paired at attP2, but not when UAS-shRNA was expressed from the distant attP40 site (Figure 4, A–D).
Figure 4.
Control UAS-shRNA constructs from the Drosophila TRiP library enhance UAS-PrP transcriptional efficacy in trans. In flies expressing UAS-PrP from attP2 driven by Cha-GAL4, an shRNA hairpin against GFP was expressed at the paired attP2 or at the unpaired attP40 locus to assess the suitability of the TRiP library as an in vivo screening tool for transvection activity. (A) Western blot of fly head homogenates. Experiment was performed in triplicate with a representative blot shown. (B) Densitometry of Western blot signals in (A) and associated replicates. PrP signals were normalized to the kinesin loading control and then to the hemizygous attP2 UAS-PrP line lacking UAS-shRNA. (C) Semiquantitative RT-PCR performed on fly head mRNA. Prnp and RpL32 transcripts were coamplified in the same PCR reactions, and each lane represents a biological replicate. (D) Densitometry of RT-PCR signals in (C). For each sample, the ratio of Prnp to RpL32 mRNA levels was taken and then normalized to the average value for the attP2 UAS-PrP line lacking UAS-shRNA. Error bars in (B) and (D) represent standard deviations (n = 2 for UAS-PrP alone mRNA quantification, n = 3 for all other groups); ns denotes not significant and **** P < 0.0001. GFP, green fluorescent protein; mRNA, messenger RNA; PrP, prion protein; RT-PCR, reverse transcription polymerase chain reaction; shRNA, short hairpin RNA; TRiP, Transgenic RNA Interference Project.
Taking our results together, we next designed an experiment to test candidate genes for transvection activity in vivo, which is depicted schematically in Figure 5. Briefly, TRiP UAS-shRNA constructs integrated at the attP2 site were expressed in trans to UAS-PrP. In this experiment, hairpins that do not target genes required for transvection (Figure 5C) would be expected, through the presence of their UAS enhancers, to boost UAS-PrP transcription at the allelic attP2 locus five- to sixfold, as seen with control anti-GFP hairpins (Figure 4). On the other hand, hairpins targeting essential transvection factors would be expected to reduce or eliminate interallelic transcriptional enhancement (Figure 5D). In this system, the degree to which target gene knockdown impacts transvection could be simply quantified by a Western blot for PrP protein levels.
Figure 5.
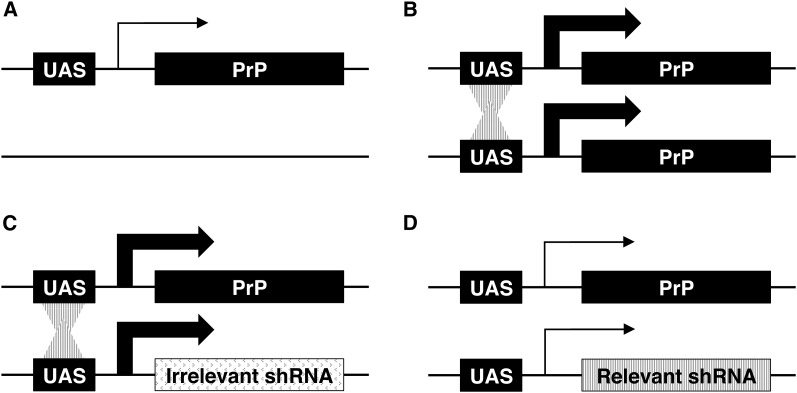
Schematic of an in vivo assay for transvection activity based upon interallelic transcriptional enhancement. All transgenes are inserted at the attP2 locus and the size of the arrow represents the rate of transcription at the associated locus. (A) Hemizygous UAS-PrP is expressed at a low level. (B) Homozygous UAS-PrP is expressed at a high level due to the in trans interaction between UAS enhancers at the same locus. (C) UAS-PrP in combination with UAS-shRNAs targeting genes lacking transvection activity (irrelevant shRNA) also leads to high level PrP expression due to the preserved interaction between UAS enhancers in trans. (D) UAS-PrP in combination with UAS-shRNAs targeting genes with transvection activity (relevant shRNA) interfere with the trans interaction between UAS enhancers, reducing PrP expression. The degree to which target gene knockdown impacts transvection can be simply quantified in this system by a Western blot of PrP levels. PrP, prion protein; shRNA, short hairpin RNA.
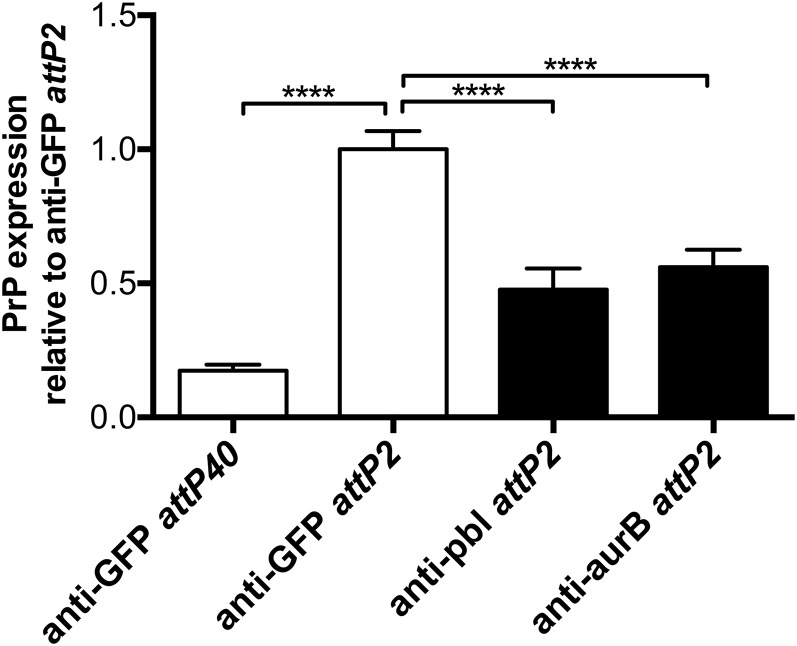
Therefore, we obtained TRiP UAS-shRNA constructs targeting 25 different genes identified previously in in vitro screens (Bateman et al. 2012b; Joyce et al. 2012) or limited in vivo studies (Fritsch et al. 2006; Nguyen et al. 2015) as playing a role in somatic homolog pairing (Table 1), which is believed to be a prerequisite for transvection. shRNAs targeting 16 of these pairing factors produced viable adult flies when expression was driven by Cha-GAL4 (Table 1), allowing us to test whether the targets of those shRNAs played a role in transvection-mediated interallelic transcriptional enhancement. Interestingly, we found that hairpins targeting just two of these 16 candidate genes, pbl and aurB, significantly reduced interallelic transcriptional enhancement of UAS-PrP expression in vivo (Figure 6). Given that in vivo knockdown validation is challenging when shRNA expression is limited to select cells within a tissue of interest, we attempted to get around this problem by expressing each of the shRNAs assayed for transvection-modifying activity ubiquitously with an α-tubulin-GAL4 driver. With the exception of the hairpins against Hsc70Cb and su(Hw), all of the shRNAs that produced viable adults when expressed with Cha-GAL4 were lethal when expressed with α-tubulin-GAL4 (Table 1), implying successful knockdown of an essential endogenous gene product. As a control to demonstrate that shRNA sequence specificity was required for this lethality, α-tubulin-GAL4-driven expression of an shRNA against the nonessential w gene produced viable adult flies with white eyes (Table 1 and G. P. Noble, unpublished data).
Table 1. Testing viability of chromosomal pairing factor knockdown in vivo.
| Adult Viability with shRNA Expression | |||
|---|---|---|---|
| Gene | BDSC shRNA Stock Number | Cha-GAL4 > shRNA | α-tubulin-GAL4 > shRNA |
| CG2469 | 33,736 | − | N/a |
| chb | 34,669 | − | N/a |
| Dhc64C | 36,698 | − | N/a |
| Klp61f | 33,685 | − | N/a |
| Nlp | 33,688 | − | N/a |
| shtd | 38,531 | − | N/a |
| SkpA | 32,991 | − | N/a |
| slmb | 33,986 | − | N/a |
| Tlk | 33,983 | − | N/a |
| Arp1 | 32,032 | + | − |
| aurB | 28,691 | + | − |
| cal1 | 41,716 | + | − |
| CG7236 | 27,505 | + | − |
| CKIα | 25,786 | + | − |
| Cul1 | 29,520 | + | − |
| Det | 51,837 | + | − |
| E(bx) | 33,658 | + | − |
| fzy | 40,933 | + | − |
| Hsc70Cb | 33,742 | + | + |
| ida | 34,552 | + | − |
| Lis-1 | 35,043 | + | − |
| Nc73EF | 33,686 | + | − |
| pbl | 28,343 | + | − |
| polo | 33,042 | + | − |
| su(Hw) | 34,006 | + | + |
| w | 33,623 | + | + |
shRNAs from the TRiP library against the listed genes, which have been identified in previous studies as playing a role in somatic homolog pairing, were obtained from the BDSC. shRNA expression was driven in cholinergic neurons with the Cha-GAL4 driver, or ubiquitously with the α-tubulin-GAL4 driver. shRNA against the w gene was included as a positive control. shRNA, short hairpin RNA; BDSC, Bloomington Drosophila Stock Center; −, no viable adult flies recovered; N/a, cross not performed; +, viable adult flies recovered; TRiP, Transgenic RNA Interference Project.
Figure 6.
Expression of shRNAs against two putative chromosomal pairing factors, pbl and aurB, reduces interallelic transcriptional enhancement in vivo. Flies expressing UAS-PrP in trans to UAS-shRNAs directed against 16 different genes thought to be involved in chromosomal pairing were generated. Transgene expression was driven in cholinergic neurons by Cha-GAL4 and interallelic enhancement of PrP expression was measured by Western blot of fly head homogenates. PrP expression levels were normalized to the level seen in the presence of an anti-GFP control hairpin in trans. Error bars represent standard deviations, with n = 3 for experimental samples and n = 4 for control samples; **** P < 0.0001. We found that shRNAs against pbl and aurB significantly decreased interallelic enhancement of PrP expression. GFP, green fluorescent protein; PrP, prion protein; shRNA, short hairpin RNA.
Discussion
Transvection—the in trans transcriptional activation or repression of an allele by regulatory elements present on a physically paired homologous allele—has recently been shown to be a pervasive feature of the Drosophila genome (Bateman et al. 2012a; Chen et al. 2002; Mellert and Truman 2012), likely due to the existence of somatic homolog pairing in Drosophila and other Dipteran species (Hartl et al. 2008; McKee 2004; Metz 1916; Stevens 1908). Although the existence of transvection throughout the Drosophila genome is now firmly established, relatively little is known about this genetic regulatory mechanism and its implications for genome function. With the development of site-directed transgenesis technologies (Fish et al. 2007; Markstein et al. 2008), it is now possible to readily and precisely study transvection phenomena in vivo.
In the present study, we have identified a strong interallelic interaction in which in trans transcriptional activation boosts overall transcription at the attP2 locus by at least fourfold relative to levels achieved in its absence (Figure 2). Although we have not performed chromosomal rearrangements in order to formally define this interallelic interaction as transvection, the fact that the effect is observed only when the transgenes tested are inserted at homologous loci (Figure 2 and Figure 4) suggests strongly that transvection is involved. This, to our knowledge, is the largest relative increase in transcription at a tissue level that has been attributed to transvection to date, and a striking example in which apparent synergism, as opposed to additivity, between cis- and trans-acting enhancers has been observed (Figure 2).
Quantitative increases in transcription at a given locus that can be attributed to the interallelic interaction of enhancers have been reported in prior studies (Bateman et al. 2012a; Gibson et al. 1999; Goldsborough and Kornberg 1996; Lum and Merritt 2011). To the best of our knowledge, the largest such increases have been observed at the Gpdh (Gibson et al. 1999) and Men (Lum and Merritt 2011) loci, at which up to approximately 100% of an enhancer’s activity in cis could be directed toward transcriptional activation in trans. However, in these two prior studies, trans-acting enhancer activity was measured using alleles with disrupted promoters in cis, which may have exaggerated the trans-acting effect (Geyer et al. 1990; Martinez-Laborda et al. 1992). In another study at the Ubx locus, quantitatively significant pairing-dependent increases in transcription were also observed, but the relative magnitude of the observed effect is challenging to assess due to widely different activities of the paired cis- and trans-acting enhancers that were studied (Goldsborough and Kornberg 1996).
To the best of our knowledge, the clearest prior quantitative comparisons to the present study, which examines the effect of pairing on transcription at the tissue level of intact, fully functional alleles, are provided by Bateman et al. (2012a) and Schoborg et al. (2013). In Bateman et al. (2012a), tissue transcript levels of a single allele at the 53F locus were increased modestly (∼15%) when combined with a second functional allele in trans. In Schoborg et al. (2013) homozygous alleles exhibited at most a ∼50% increase in transcriptional efficacy compared to hemizygous alleles. In contrast, in the present study, we observe that interaction between fully functional alleles in trans produces at least a ∼300% increase in the transcriptional output of each allele when measured at the tissue level (Figure 2).
The magnitude of the effect observed in the present study, many-fold higher than in previous reports, raises the question: why is the action of in trans enhancers (i.e., transvection) so strong in the present system? We have shown that the effect does not appear to be a unique feature of the Cha-GAL4 driver or of the attP2 locus, given that interallelic transcriptional enhancement was also observed using the attP40 locus and the Elav-GAL4 driver (Figure S1). We have further shown that the magnitude of the effect is highly dependent on the strength of the enhancer located in trans (Figure 3) and is independent of strict homology between the paired alleles (Figure 3 and Figure 4). These results provide confirmation of the findings of Mellert and Truman, who demonstrated that the presence of GAL4 bound to UAS in trans to a functional promoter is sufficient to drive transvection (Mellert and Truman 2012). These results also support the recent findings of Blick et al. (2016), who showed that the propensity of many endogenous Drosophila enhancers to act in trans is related to the strength of the enhancer’s activity in cis, and is markedly increased by enhancer multimerization (Blick et al. 2016). Of note, the exogenous UAS enhancer used to drive PrP expression in the present study contains five tandem repeats of the GAL4 transcription factor binding site (Bischof et al. 2007), which may be one reason why it is an unusually efficient in trans activator.
Whether transcriptional activation in cis and in trans are mechanistically similar or different is a question that remains unanswered. That transcription can be so dramatically increased at a particular locus as a result of transvection, as is seen in the present study, seems to suggest that the mechanisms of transcriptional activation in cis and in trans are fundamentally different. Indeed, recent studies have already shown that transcriptional activation in trans occurs stochastically (Bateman et al. 2012a; Blick et al. 2016; Mellert and Truman 2012) and at there can be widely varying levels among the cells of a particular tissue, with occasional “jackpot” cells exhibiting intense in trans activation (Bateman et al. 2012a; Blick et al. 2016). Thus, the strong apparent synergism seen between allelic enhancers in the present study (e.g., Figure 2) may represent the summation of infrequent jackpot expression events within the studied tissues, as opposed to a fourfold enhancement of gene expression across all PrP-expressing cells. Alternatively, in trans transcriptional activation could be mechanistically similar to in cis transcriptional activation, with the relative efficiency of each being determined largely by physical proximity of enhancer-promoter elements at a given locus in a given cell. This latter possibility is supported by the observation that homolog anti-pairing factors inhibit transvection (Hartl et al. 2008). However, it is not readily apparent how such a model would account for the significant interallelic enhancer synergism seen in the present study.
Taking the view that in cis and in trans transcriptional activation could be mechanistically different, it is not yet clear what might be occurring at a molecular level to make them so. One intriguing possibility is that these two transcriptional programs utilize different sets of effector molecules. To the best of our knowledge, no simple methods for identifying or screening for such transvection factors have yet been proposed. The system used in the present study, which a) provides a sensitive and readily quantified readout of transvection (Figure 2), and b) can be manipulated using a variety of publicly available Drosophila transgenes (Figure 3 and Figure 4), provides a unique opportunity to study the molecular basis of in trans transcriptional activation.
To that end, we utilized our UAS-PrP expression system in combination with shRNAs from the TRiP library (Ni et al. 2011) to assay candidate genes for transvection activity in vivo (Figure 5). We focused our investigation on genes previously implicated in homologous chromosome pairing, which is thought to be a prerequisite for transvection. Of 25 candidate genes known to be involved in chromosomal pairing (Bateman et al. 2012b; Fritsch et al. 2006; Joyce et al. 2012; Nguyen et al. 2015) for which shRNA lines were available, 16 produced viable adult flies when shRNAs against the gene in question were expressed within cholinergic neurons using the Cha-GAL4 driver (Table 1). Interestingly, of these 16 candidate genes that were subsequently assayed for transvection activity, shRNA-targeting of only two of the genes, pbl and aurB, led to a marked reduction in interallelic transcriptional enhancement (Figure 6).
It is important to acknowledge that both pbl and aurB play a role in the cell cycle (Bateman et al. 2012b; Joyce et al. 2012), and that knockdown of cell cycle genes may affect cell viability and/or cell fate. In the present assay, it is not possible to distinguish between decreased PrP expression due to inhibition of interallelic transcriptional enhancement vs. the loss of Cha-GAL4-expressing neurons secondary to pbl or aurB depletion.
Similarly, it is important to acknowledge that pairing of heterochromatin and euchromatin is known to occur with different frequencies in cell culture (Williams et al. 2007), and some authors have suggested that this observation may be a reflection of distinct hetero- and euchromatin pairing mechanisms. The majority of the genes tested in our screen (Table 1) were initially identified as playing a role specifically in heterochromatin pairing (Bateman et al. 2012b; Joyce et al. 2012). However, Joyce et al. (2012) have shown extensive overlap between pairing activities for heterochromatin and euchromatin. Indeed, several of the pairing factors targeted in the present screen (Table 1) have previously been shown to affect both hetero- and euchromatin pairing in cell culture (Joyce et al. 2012; Nguyen et al. 2015), suggesting that a lack of euchromatin pairing activity is unlikely to be responsible for the fact that only two of the 16 putative pairing factors targeted in the present study appear to affect transvection-mediated interallelic transcriptional enhancement (Figure 6).
Furthermore, to the best of our knowledge, it has not previously been determined whether Cha-GAL4 drives UAS-tagged transgene expression in mitotic cells of the neuronal lineage, though Cha-GAL4 is known to be expressed at all developmental stages (Salvaterra and Kitamoto 2001). Expression of shRNAs in predominately postmitotic cholinergic neurons may be one reason for the discrepancy between the in vivo screen results reported here and prior in vitro studies using cycling cultured cells.
Unfortunately, validation of shRNA knockdown by RT-PCR or Western blot in the present study is technically very challenging, due to the fact that shRNAs are only expressed in a subset of cells within the studied tissue. As one way around this problem, we attempted to express each of the shRNAs assayed for transvection-modifying activity ubiquitously with an α-tubulin-GAL4 driver (Table 1). Of note, with the exception of the hairpins against Hsc70Cb and su(Hw), all of the shRNAs that produced viable adults when expressed with Cha-GAL4 were lethal when expressed by the ubiquitous α-tubulin-GAL4 driver (Table 1), suggesting successful knockdown of an essential endogenous gene product. Given the limited number of appropriate, nonoverlapping TRiP constructs available targeting each candidate gene, we unfortunately cannot exclude the potential for off-target effects causing the observed lethality. However, as a control to demonstrate that some shRNA sequence specificity was required for the lethal effect, α-tubulin-GAL4-driven expression of an shRNA against the nonessential w gene produced viable adult flies with white eyes (Table 1 and data not shown).
Understanding the limitations of interpretation highlighted above, and under the assumption that pairing factor knockdown was likely successful in our proof of principle screen, the fact that transvection-mediated interallelic transcriptional enhancement was relatively robust to perturbation in the present study suggests the interesting possibility that transvection may be less dependent on the process of homolog pairing than previously thought. Alternatively, homolog pairing may itself be more robust in vivo than in Drosophila cell culture. Regardless, this preliminary study suggests that an analogous system—one that similarly takes advantage of robust interallelic transcriptional enhancement—might be useful in a reverse genetics approach to potentially identify as yet unknown molecular factors involved in transvection.
Due to the widespread existence of transvection within the Drosophila genome, prior investigators have cautioned against the use of multiple transgenes inserted at the same genomic locus when designing in vivo experiments (Mellert and Truman 2012). Those prior warnings were based largely on qualitative changes in gene expression that may result from transvection. The present study, which demonstrates that in trans activation by simple UAS enhancers can have large quantitative effects on gene expression in Drosophila, should be taken as a further cautionary tale for those designing experiments utilizing the GAL4-UAS system; not only should it be assumed that paired UAS enhancers boost overall transcription levels, one should also consider that unpaired UAS enhancers may confound experimental results by driving increased expression of endogenous genes, either in trans at the paired wild-type locus, as initially suggested by Mellert and Truman (2012) or locally in cis, as has been seen with the GMR enhancer (Hay et al. 1997).
In the present study, we have identified an example of transvection in Drosophila highlighting the significant quantitative effect that interallelic enhancer interactions can have on gene expression at the tissue level. We have utilized this system to test previously identified chromosomal pairing factors for transvection activity in vivo. Our findings add new data to the growing body of work suggesting that transvection may profoundly affect genome function in Drosophila and other organisms.
Supplementary Material
Acknowledgments
We would like to thank Giovanni Bosco for helpful discussions. We thank Hassina Benchabane for assistance with semiquantitative reverse transcription polymerase chain reaction experiments and Katelyn Byrne for assistance with figure preparation. We thank the Transgenic RNA interference (RNAi) Project (TRiP) at Harvard Medical School [National Institutes of Health/National Institute of General Medical Science (NIH/NIGMS) R01-GM084947] for providing transgenic RNAi fly stocks used in this study. This work was funded by a research grant from the NIH (2R01 NS046478) to S.S.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.032300/-/DC1
Communicating editor: J. A. Birchler
Literature Cited
- Bateman J. R., Johnson J. E., Locke M. N., 2012a Comparing enhancer action in cis and in trans. Genetics 191: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Larschan E., D’Souza R., Marshall L. S., Dempsey K. E., et al. , 2012b A genome-wide screen identifies genes that affect somatic homolog pairing in Drosophila. G3 (Bethesda) 2: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Renner S., Luer K., Technau G. M., 2007. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev. Dyn. 236: 3562–3568. [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blick A. J., Mayer-Hirshfeld I., Malibiran B. R., Cooper M. A., Martino P. A., et al. , 2016. The Capacity To Act in trans Varies Among Drosophila Enhancers. Genetics 201: 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., Misteli T., 2013. Functional implications of genome topology. Nat. Struct. Mol. Biol. 20: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. L., Huisinga K. L., Viering M. M., Ou S. A., Wu C. T., et al. , 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 3723–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault N. R., Harris B. T., Rees J. R., Supattapone S., 2007. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. USA 104: 9741–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault N. R., Walsh D. J., Piro J. R., Wang F., Wang X., et al. , 2012. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. USA 109: E1938–E1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish M. P., Groth A. C., Calos M. P., Nusse R., 2007. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2: 2325–2331. [DOI] [PubMed] [Google Scholar]
- Fritsch C., Ploeger G., Arndt-Jovin D. J., 2006. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 14: 451–464. [DOI] [PubMed] [Google Scholar]
- Ganetzky B., Flanagan J. R., 1978. On the relationship between senescence and age-related changes in two wild-type strains of Drosophila melanogaster. Exp. Gerontol. 13: 189–196. [DOI] [PubMed] [Google Scholar]
- Gavin B. A., Dolph M. J., Deleault N. R., Geoghegan J. C., Khurana V., et al. , 2006. Accelerated accumulation of misfolded prion protein and spongiform degeneration in a Drosophila model of Gerstmann-Straussler-Scheinker syndrome. J. Neurosci. 26: 12408–12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. C., Miller M. B., Kwak A. H., Harris B. T., Supattapone S., 2009. Trans-dominant inhibition of prion propagation in vitro is not mediated by an accessory cofactor. PLoS Pathog. 5: e1000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G., 1990. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 9: 2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. B., Reed D. S., Bartoszewski S., Wilks A. V., 1999. Structural changes in the promoter region mediate transvection at the sn-glycerol-3-phosphate dehydrogenase gene of Drosophila melanogaster. Biochem. Genet. 37: 301–315. [DOI] [PubMed] [Google Scholar]
- Goldsborough A. S., Kornberg T. B., 1996. Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381: 807–810. [DOI] [PubMed] [Google Scholar]
- Hartl T. A., Smith H. F., Bosco G., 2008. Chromosome alignment and transvection are antagonized by condensin II. Science 322: 1384–1387. [DOI] [PubMed] [Google Scholar]
- Hay B. A., Maile R., Rubin G. M., 1997. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 94: 5195–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K. K., Scott M., Foster D., Groth D. F., DeArmond S. J., et al. , 1990. Spontaneous neurodegeneration in transgenic mice with mutant prion protein. Science 250: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Jackson W. S., Borkowski A. W., Faas H., Steele A. D., King O. D., et al. , 2009. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron 63: 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Williams B. R., Xie T., Wu C. T., 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8: e1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K., Zulianello L., Scott M., Cooper C. M., Wallace A. C., et al. , 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. USA 94: 10069–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeman J. M., Russell R. C., Tan M. H., Petillo D., Westphal M., et al. , 2008. Somatic pairing of chromosome 19 in renal oncocytoma is associated with deregulated EGLN2-mediated [corrected] oxygen-sensing response. PLoS Genet. 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourg E., Lints F. A., 1992. Hypergravity and aging in Drosophila melanogaster. 4. Climbing activity. Gerontology 38: 59–64. [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The Theory and Application of a New Method of Detecting Chromosomal Rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239. [Google Scholar]
- Lomvardas S., Barnea G., Pisapia D. J., Mendelsohn M., Kirkland J., et al. , 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Lum T. E., Merritt T. J., 2011. Nonclassical regulation of transcription: interchromosomal interactions at the malic enzyme locus of Drosophila melanogaster. Genetics 189: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Laborda A., Gonzalez-Reyes A., Morata G., 1992. Trans regulation in the Ultrabithorax gene of Drosophila: alterations in the promoter enhance transvection. EMBO J. 11: 3645–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui O., Bonnet I., Le Baccon P., Brito I., Pollex T., et al. , 2011. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell 145: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677: 165–180. [DOI] [PubMed] [Google Scholar]
- Mellert D. J., Truman J. W., 2012. Transvection is common throughout the Drosophila genome. Genetics 191: 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C. W., 1916. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance. J. Exp. Zool. 21: 213–279. [Google Scholar]
- Murali A., Maue R. A., Dolph P. J., 2014. Reversible symptoms and clearance of mutant prion protein in an inducible model of a genetic prion disease in Drosophila melanogaster. Neurobiol. Dis. 67: 71–78. [DOI] [PubMed] [Google Scholar]
- Nguyen H. Q., Nye J., Buster D. W., Klebba J. E., Rogers G. C., et al. , 2015. Drosophila casein kinase I alpha regulates homolog pairing and genome organization by modulating condensin II subunit Cap-H2 levels. PLoS Genet. 11: e1005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Ngo T. T., Hibbard K. L., Murphy C., Jenett A., et al. , 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics 186: 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S., White K., 1988. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev. Biol. 126: 294–303. [DOI] [PubMed] [Google Scholar]
- Salvaterra P. M., Kitamoto T., 2001. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns 1: 73–82. [DOI] [PubMed] [Google Scholar]
- Schoborg T., Kuruganti S., Rickels R., Labrador M., 2013. The Drosophila gypsy insulator supports transvection in the presence of the vestigial enhancer. PLoS One 8: e81331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N. M., 1908. A study of the germ cells of certain diptera, with reference to the heterochromosomes and the phenomena of synapsis. J. Exp. Zool. 5: 359–374. [Google Scholar]
- Williams B. R., Bateman J. R., Novikov N. D., Wu C. T., 2007. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics 177: 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Bai L., 2016. Interallelic interaction and gene regulation in budding yeast. Proc. Natl. Acad. Sci. USA 113: 4428–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.