Abstract
Individuals with type 2 diabetes display metabolic abnormalities, such as hyperglycemia, increased free fatty acids, insulin resistance, and altered ceramide levels, that contribute to vascular dysfunctions and compromised oxygen delivery. Caenorhabditis elegans fed a glucose-supplemented diet or with altered ceramide metabolism, due to a hyl-2 mutation, are sensitive to oxygen deprivation (anoxia). Our experiments showed that the combination of these factors further decreased the anoxia survival. RNA-sequencing analysis was performed to assess how a glucose-supplemented diet and/or a hyl-2 mutation altered the transcriptome. Comparison analysis of transcripts associated with anoxia-sensitive animals [hyl-2(tm2031) mutation or a glucose diet] revealed 199 common transcripts encoded by genes with known or predicted functions involving innate immunity, cuticle function (collagens), or xenobiotic and endobiotic phase I and II detoxification system. Use of RNA interference (RNAi) to target gene products of the xenobiotic and endobiotic phase I and II detoxification system (UDP-glycosyltransferase and Cytochrome p450 genes; ugt-15, ugt-18, ugt-19, ugt-41, ugt-63, cyp-13A12, cyp-25A1, and cyp-33C8) increased anoxia survival in wild-type animals fed a standard diet. Anoxia sensitivity of the hyl-2(tm2031) animals was suppressed by RNAi of cyp-25A1 or cyp-33C8 genes. A glucose diet fed to the P0 hermaphrodite decreased the anoxia survival of its F1 embryos; however, the RNAi of ugt-63 and cyp-33C8 suppressed anoxia sensitivity. These studies provide evidence that the detoxification system impacts oxygen deprivation responses and that C. elegans can be used to model the conserved detoxification system.
Keywords: lipid biosynthesis, glucose toxicity, xenobiotic and endobiotic detoxification, innate immunity, collagen
Reduced oxygen delivery is central to may human health issues including stroke, cardiac or pulmonary dysfunction, ischemia, and trauma resulting in blood loss or suffocation. Organisms have evolved mechanisms to respond to a lack of oxygen by altering physiological, metabolic, and molecular functions (Hochachka and Lutz 2001; Jonz et al. 2016; Nathaniel et al. 2015). Despite response and rescue efforts initiated during oxygen deprivation, a continual lack of oxygen can irreversibly damage tissues and cause death. Vascular dysfunction will also reduce blood flow and oxygen delivery, which in turn compromises tissue functions. Individuals with type 2 diabetes display metabolic abnormalities (hyperglycemia, increased free fatty acids, and insulin resistance) that contribute to both microvascular and macrovascular dysfunctions eventually leading to organ dysfunction, tissue damage, blindness, and amputations (Ogden et al. 2012; Paneni et al. 2013; Beckman et al. 2013; Creager et al. 2003). There is evidence that individuals with type 2 diabetes have a worse recovery when challenged with an ischemic event. For example, the human diabetic heart is more vulnerable to ischemic injury (Paulson 1997). Additionally, a human clinical study of patients (4537 patients, 937 diabetic patients) hospitalized for a first-time stroke, addressed if stroke subtype, severity, and prognosis differed between diabetic and nondiabetic patients 3 months after stroke (Megherbi et al. 2003). Overall, the diabetic patients, compared to nondiabetic patients, were significantly more likely to have motor deficits, limb weaknesses, dysarthria, a higher handicap and disability index, and a significant increase in ischemic stroke. Although studies indicate that type 2 diabetes may worsen the outcome of an ischemic event, it is not yet completely understood why this is the case.
Altered ceramide levels can mediate insulin resistance and mitochondrial dysfunction in diabetics (Xia et al. 2014; Novgorodov et al. 2016; Turpin et al. 2014; Galadari et al. 2013). Ceramides, produced by the sphingolipid/ceramide biosynthetic pathway in the endoplasmic reticulum (ER), function within the ER and mitochondria (Hannun and Obeid 2011). This complex set of lipids has roles in various cellular processes including apoptosis, stress, and the mitochondrial unfolded protein response (Deng et al. 2008; Liu et al. 2014; Hannun and Obeid 2011; Xia et al. 2014; Turpin et al. 2014; Larsen and Tennagels 2014; Novgorodov and Gudz 2011; Argraves et al. 2011; Zigdon et al. 2013; Li and Zhang 2013). The finding that specific ceramide species are associated with diseases involving mitochondrial dysfunction has fueled further investigation of these complex molecules (Lopez et al. 2013; Larsen and Tennagels 2014; Xia et al. 2014). Furthermore, the ability to conduct phenotype analysis of gene mutations impacting the sphingolipid/ceramide pathway is valuable to understanding the functional role of these lipids (Zhang et al. 2013; Voelzmann and Bauer 2010). For example, in Caenorhabditis elegans, genetic analyses of mutants impacting ceramide/sphingomyelin biosynthesis showed changes in cellular ceramide species (Menuz et al. 2009), oxygen deprivation (anoxia) responses (Menuz et al. 2009; Garcia et al. 2015), the mitochondrial surveillance system (Liu et al. 2014), apoptotic signals in the germline (Deng et al. 2008), and dietary restriction-induced autophagy and lifespan (Mosbech et al. 2013; Cutler et al. 2014). Genetic mutations in the ceramide biosynthetic genes alter ceramide species length such that efficient synthesis of C20 to C22 ceramide and sphingomyelin species requires hyl-2 and efficient synthesis of C24 and C26 ceramide and sphingomyelin requires hyl-1 (Menuz et al. 2009). A mutation in hyl-2 results in oxygen deprivation sensitivity and suppresses the anoxia resistance observed in the daf-2(e1370) insulin receptor mutant (Menuz et al. 2009; Garcia et al. 2015). Conversely, a mutation in hyl-1 moderately increases oxygen deprivation survival (Menuz et al. 2009). The mechanism linking ceramide species and anoxia survival remains to be determined.
C. elegans is a well-established model for studying oxygen deprivation at the molecular level since various stages of development are capable of surviving a broad range of oxygen levels (Van Voorhies and Ward 2000; Padilla et al. 2002). In fact, the worm is capable of surviving severe oxygen deprivation (anoxia) by entering into a hypometabolic state, referred to as suspended animation, in which observable biological processes (cell division, development, and movement) reversibly arrest (Padilla et al. 2002; Padilla and Ladage 2012). Several labs have identified genetic mutations affecting various biological processes (e.g., insulin signaling, ovulation, HIF-1 signaling, and metabolism), that either increase or decrease the oxygen deprivation survival rate (Scott et al. 2002; Mendenhall et al. 2009, 2006; Jiang et al. 2001; Anderson et al. 2009). The C. elegans nervous system can sense small changes in oxygen levels (Zimmer et al. 2009; Gray et al. 2004) and methodologies have been developed to use C. elegans as a model for hypoxic injury and ischemia/reperfusion (Queliconi et al. 2014; Fawcett et al. 2015; Mao et al. 2016). Interestingly, a sugar-supplemented diet reduces oxygen deprivation survival and feeding the wild-type worms the diabetic drug metformin enhances anoxia survival (LaRue and Padilla 2011; Garcia et al. 2015). A glucose-supplemented diet can also be considered an obesity mimetic as it increases lipid accumulation as seen by oil red O staining (Garcia et al. 2015). Furthermore, a glucose diet decreases membrane fluidity in worms with altered fatty acid metabolism (paqr-2 and iglr-2 mutants), alters gene expression, impacts lifespan, and increases cellular ROS levels and protein glycosylation (Svensk et al. 2016; Mondoux et al. 2011; Choi 2011; Schlotterer et al. 2009; Lee et al. 2009; Garcia et al. 2015; Liggett et al. 2015). Dietary glucose impacts various physiological and molecular processes in C. elegans, which makes it a good model for understanding hyperglycemia and obesity.
In this study, we begin to address why a glucose diet or a mutation in the hyl-2 ceramide synthase gene reduces anoxia survival. The hyl-2(tm2031) mutant, compared to wild-type controls, is more sensitive to anoxia when fed a glucose diet. RNA-sequencing (RNA-Seq) analysis indicates the hyl-2(tm2031) and N2 animals fed a glucose diet have overlapping but not identical gene expression responses and that the transcript levels of lipid metabolic genes are altered. Comparison analysis of the transcriptome profiles associated with anoxia sensitive animals (hyl-2(tm2031) mutation, wild-type fed a glucose diet) revealed altered expression levels of 199 common transcripts involved with various functions including innate immunity, cuticle function, and the xenobiotic and endobiotic phase I and II detoxification system (referred to here as the detoxification system). The detoxification system is understudied in C. elegans, but has been well characterized in humans, especially in the context of drug metabolism. These enzymes regulate the levels of potentially toxic lipophilic molecules through oxidation and sugar conjugation, which facilitates elimination by decreasing the hydrophobicity of the target molecule. RNAi knockdown of cyp (cytochrome P450, phase I) and ugt (UDP-glucuronlytransferases, phase II) detoxification genes increased anoxia survival and in some cases suppressed anoxia sensitivity induced by the hyl-2 mutation or a glucose diet. These are the first studies that provide evidence that changes in the detoxification system suppress oxygen deprivation sensitivity induced by a glucose diet or altered ceramide species.
Materials and Methods
Strains and culture conditions
The wild-type Bristol (N2) and hyl-2(tm2031) strains were cultured using NGM plates seeded with Escherichia coli (OP50 or HT115) and raised at 20°. To obtain animals of the specified stage of development for each experiment as noted below, 1 d old adult hermaphrodites were allowed to lay eggs on a plate (1–2 hr), and anatomical markers such as gonad morphology and time post molting stage were used to identify the developmental stage of the subsequent offspring. Animals were fed a glucose-supplemented diet as described previously (Garcia et al. 2015). Glucose (Sigma) solution was spread evenly, fully covering the entire plate, and allowed to dry before being seeded with bacteria (OP50, HT115, or appropriate RNAi strain). To minimize differences between the bacterial lawns, freshly made plates were used within a 7 d period. To examine how a glucose diet impacts the F1 generation, hermaphrodites were raised on the glucose diet to adulthood, placed onto a fresh NGM glucose plate, allowed to lay eggs for approximately 2 hr and removed, so that the F1 generation of embryos could be phenotypically examined.
RNAi analysis
RNA interference (RNAi) was conducted as described (Kamath and Ahringer 2003; LaRue and Padilla 2011). The HT115 and RNAi E. coli strains were obtained from the MRC Geneservice or Source BioSciences (Cambridge, UK). Embryos were placed on to NGM-IPTG plates (200 mg/ml ampicillin, 12.5 mg/ml tetracycline, and 0.5 mg/ml IPTG) seeded with or without glucose and either HT115 control food (contains L4440 plasmid with no insert) or the appropriate E. coli strain for RNAi of a specified gene. Embryos were grown to 1 d old adults at 20° and then exposed to anoxia as described below.
Anoxia experiments
Animals were placed into anoxia as previously described (Padilla et al. 2002, 2011). Briefly, animals at the indicated developmental state were exposed to anoxia (20°) for the time indicated for each experiment (1 or 2 d of exposure) using the BD Biobag type A anaerobic environmental chamber (BD Biosciences, Rockville, MD). Animals were given 24 hr of recovery in air (20°) before survivors were examined for an unimpaired or impaired phenotype, as previously described (LaRue and Padilla 2011; Garcia et al. 2015). The survivors were scored as unimpaired if there were no detectable defects in morphology, behavior, or motility using a standard stereomicroscope, whereas impaired animals displayed a defect in morphology, behavior, or motility. A minimum of three independent experiments of approximately 50 animals each was conducted. Statistical analysis of total survival rate relative to control was conducted as indicated for each experiment. Tests conducted include one-way ANOVA, Bonferonni or Tukey multiple comparisons or two-way ANOVA, and Sidak’s multiple comparisons test if genotype and developmental stage or genotype and glucose concentration were analyzed (Prism 6.0).
Fecundity assays
The number of progeny, produced by hyl-2(tm2031) or N2 animals, was determined as previously described (Mendenhall et al. 2009). Briefly, four synchronized animals were collected at the L4-to-adult molt, placed as individuals onto a NGM plate, and allowed to lay eggs over a 24 hr period. The adult worms were moved every 24 hr and the progenies produced during each 24 hr interval were counted after hatching. Animals were examined until no progenies were produced. For statistical analysis, a two-way ANOVA, Sidak’s multiple comparisons test was conducted relative to genotype and adult stage. The total numbers of offspring, for N2 and hyl-2(tm2031) animals, was statistically analyzed using an unpaired t-test (two-tailed). For all statistical analyses Prism 6.0 was used.
RNA extraction and sequencing
Young nongravid adults were collected for mRNA isolation as previously described (Garcia et al. 2015). RNA sampling was done in three biological replicates for hyl-2(tm2031) animals fed either OP50 or glucose-supplemented OP50 diet (0.5% glucose). Briefly, gravid adults were allowed to lay eggs on several NGM plates for 2–4 hr, adults were then washed off with M9, and the embryos, which remained on the plate, were allowed to hatch. L1 animals were collected and placed on respective media plates (OP50 only or OP50 supplemented with 0.5% glucose). Approximately 10,000 animals (per trial) were grown to young adulthood, past the L4 molt but prior to egg production, before collection by gravity separation with M9 in 15 ml conical tubes. The animals were washed 3 times with M9 and allowed to settle by gravity separation to remove bacteria. Excess M9 was removed, and 4 vol of Trizol per vol of animals were added. The tubes were immediately frozen in liquid nitrogen, thawed and briefly vortexed; this step was repeated and then followed by a 5 min incubation at room temperature. To each tube, 0.2 ml of chloroform per 1 ml of Trizol was added, mixed gently for 15 sec, incubated at room temperature for 2–3 min, and then centrifuged at 12,000 × g for 15 min at 4°. The colorless upper phase was transferred to an RNase free tube. RNA was then purified using an Ambion PureLink RNA Mini Kit (Cat# 12183018A). Following RNA purification, the RNA was treated with DNase I (Life Technologies, Cat# 12185010), quantified using a NanoDrop Spectrophotometer and stored at −80°. RNA was isolated from three independent experiments. Next-generation RNA sequencing (RNA-Seq) experiments and analysis was done as previously described (Garcia et al. 2015). The RNA-Seq was done at the University of Texas Southwestern Genomics Core facility, which resulted in generation of approximately 30.86 M, 35.42 M, and 30.78 M single-end reads of length 50 bp for the three hyl-2(tm2031) OP50-fed replicates (mean 32.35 million reads), and 30.31 M, 29.79 M, and 31.56 M single-end reads of length 50 bp for the three hyl-2(tm2031) glucose-fed replicates (mean 30.56 million reads). These deep sequencing raw transcriptome data were then used for a comprehensive quantitative analysis of differential gene regulation in hyl-2(tm2031) and glucose-fed C. elegans. The publicly available Tuxedo suite of programs was used for RNA-Seq analysis, which includes the software Bowtie (Langmead et al. 2009), Tophat (Trapnell et al. 2009), and Cufflinks (Trapnell et al. 2010). The C. elegans genome build WS195 at NCBI was used as the reference genome for obtaining the read alignment using Bowtie and the splice junctions using Tophat. Cufflinks was used to identify the genes in glucose-fed C. elegans with significant alteration in their expressions compared to wild-type C. elegans. Cufflinks performs a statistical significance test for differential expression of each transcript based on a negative binomial model estimated from the data (Trapnell et al. 2010). The transcripts passing the significance test [False discovery rate (FDR) adjusted P-value to be < 0.05, Trapnell et al. 2010] were examined further for their abundance fold change between the conditions. Custom programs were developed for further parsing of this data and classifying the differentially expressed genes based on functional annotation at Wormbase database (http://www.wormbase.org/).
Gene ontology (GO) and enrichment of functional gene classes
Genes differentially regulated by a glucose diet and/or a hyl-2(tm2031) mutation were classified and evaluated using PANTHER (http://www.pantherdb.org) and Wormbase annotation (http://www.wormbase.org) (Mi et al. 2013b; Chen et al. 2005). The PANTHER classification system combines gene function, ontology, and pathways to analyze large data sets (Mi et al. 2013a). The transcripts involved with lipid metabolism or the 199 transcripts common to the anoxia sensitive animals (hyl-2(tm2031) and N2 glucose-fed) were analyzed using the web-based Babelomics 5 platform (Al-Shahrour et al. 2005; Alonso et al. 2015). Single enrichment analysis using Fatigo was performed with the entire C. elegans genome as the comparative background (Al-Shahrour et al. 2007). GO biological processes, molecular function, and cellular component databases were simultaneously queried. For gene set enrichment analysis, a logistic model method available within the Fatiscan tool was utilized (Al-Shahrour et al. 2007). Genes were ranked for analysis using their fragments per kb of transcript per million mapped reads (FPKM) expression values for both the hyl-2(tm2031) and N2 supplemented with glucose RNA-Seq experimental datasets.
Network analysis, clustering, and heatmap generation
To generate the functional-association dataset the 199 transcripts, common to the anoxia sensitive animals, were processed through the GeneMANIA web interface (Warde-Farley et al. 2010; Mostafavi et al. 2008). The 199 differentially expressed genes were input as query genes to GeneMANIA, which queried against the C. elegans organism database. Network weighting was based on biological processes as in GO. Edge weights are automatically determined by GENEMANIA based on the input gene list, and vary by association type. Association types include gene coexpression, protein–protein interaction, shared domains, shared pathways, and colocalization. The parameters were set such that only the 199 differentially expressed genes were analyzed for network interactions and thus no predicted genes outside of this data set were included. To identify gene clusters, the generated network of weighted edges and nodes was downloaded in text format directly from GeneMANIA and converted into the SIF file format using custom awk commands (Aho et al. 1987). A protein–protein interaction network for the 199 differentially expressed genes was generated using yeast two-hybrid data available at the Worm Interactome Database (Simonis et al. 2009). The interaction dataset thus obtained was converted to SIF file format using custom awk commands for use with the Cytoscape program (Version 3.3.), which allows the visualization and interpretation of interactions (Shannon et al. 2003; Simonis et al. 2009; Montojo et al. 2014). Here, the genes were represented as nodes and the positive yeast two-hybrid interactions as edges between interacting nodes. All networks were further analyzed for functional modules using the Markov Clustering Algorithm (Enright et al. 2002). Custom awk commands were used to convert the network data to “abc” format specified in the MCL algorithm manual. The inflation parameter was tested at values ranging from 1 to 6 in order to identify the optimal granularity for clustering (Mao et al. 2009). For the GeneMANIA network, an inflation parameter of value 2 was found to produce the clusters for further analysis. Heatmaps, for the 199 common transcripts list and for individual network clusters, were generated with unsupervised hierarchical clustering using the “aheatmap” function of the NMF R library (Gaujoux and Seoighe 2010). The heatmap settings were set such that the differentially expressed fold change value determined the transcript order. Hierarchical clustering was performed using Euclidian distance and complete linkage analysis (Gaujoux and Seoighe 2010).
Quantitative RT-PCR
Animals were collected as young adults for mRNA isolation and RNA extraction was performed as described above. Reverse transcription to generate cDNA was performed using SuperScript III first-strand synthesis system for RT-PCR (Invitrogen, Cat# 18080-051). Quantitative RT-PCR was carried out using a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) and SsoFast EvaGreen Supermix (Bio-Rad). Following primer validation, results were analyzed using the 2−ΔΔCT Method (Livak and Schmittgen 2001). The mRNA level of Y45F10D.4 was used for normalization (Hoogewijs et al. 2008). The average of at least three technical replicates was used for each independent experiment. Primer sequences are available upon request. Statistical analysis was conducted using one-way ANOVA, Bonferroni multiple comparisons.
Data availability
The data have been archived in NCBI GEO under the series record number GSE83887.
Results
Phenotypes associated with the hyl-2(tm2031) animal
To examine the role that ceramides have in anoxia survival, phenotypic analysis and transcriptomic analysis of the hyl-2(tm2031) mutant were conducted. Consistent with what others have observed, the hyl-2(tm2031) 1 d old adult hermaphrodite exposed to 2 d of anoxia had a decreased survival rate (Menuz et al. 2009) (Figure 1A). We also examined the anoxia survival rate of the hyl-2(tm2031) animal at other developmental stages and in the adult male, and found that hermaphrodites at the L4 larvae stage were also sensitive to anoxia (Figure 1A). This suggests that a physiological attribute in L4 and 1 d old adult hermaphrodites in combination with altered ceramide metabolism compromises anoxia survival. We noticed that the hyl-2(tm2031) mutant also displayed a decrease in average brood size relative to wild-type animals (287.8 ± 6.3 compared to 341.8 ± 15.8, P < 0.05). However, the reduced offspring observed in hyl-2(tm2031) hermaphrodites is due to a decrease in egg laying in 3 d old adults (Figure 1B). Thus, it is probably not egg production per se that is contributing to the anoxia sensitivity observed in 1 d old adult hermaphrodites. Previously, we showed that wild-type animals fed a glucose-supplemented diet have a reduced 1 d anoxia survival rate (Garcia et al. 2015). Here we found that the hyl-2(tm2031) adult hermaphrodite, fed a low concentration of glucose, has a reduced 1 d anoxia survival rate in comparison to the wild type fed the same glucose diet (Figure 1C). Note that the hyl-2(tm2031) mutant fed a standard diet will survive 1 d, but not 2 d, of anoxia exposure. These results indicate that both altered ceramide levels and a glucose diet have an additive and negative impact on anoxia survival and that a glucose diet, more so than the hyl-2 mutation, reduces the survival of 1 d of anoxia.
Figure 1.
Phenotype of the hyl-2(tm2031) mutant. (A) Wild-type and hyl-2(tm2031) animals, at larval stages, 1 d or 5 d old adult hermaphrodites, and 1 d old males, were exposed to 2 d of anoxia. The hyl-2(tm2031) L4 larvae and adult animals had a significant decrease in anoxia survival relative to N2 controls of the same developmental stage (* indicates P < 0.05, *** indicates P < 0.0001, two-way ANOVA, Sidak’s multiple comparisons test). (B) The average number of progeny produced in 1–5 d old hyl-2(tm2031) adults relative to N2 controls of the same developmental stage. There was a significant difference in the number of progeny produced in 3 d old hyl-2(tm2031) adults (** indicates P < 0.0001, two-way ANOVA, Sidak’s multiple comparisons test). There was a significant decrease in the total number of offspring produced per hyl-2(tm2031) hermaphrodite relative to N2 controls (N2 = 341.8 ± 15.8, hyl-2(tm2031) = 287.8 ± 6.3, P < 0.05, two-tailed unpaired t-test). (C) The hyl-2(tm2031) 1 d old adult hermaphrodite is more sensitive to one day of anoxia, after being fed a diet supplemented with glucose prior to anoxia exposure, relative to N2 control animals. Bar indicates a significant decrease in the number of hyl-2(tm2031) animals alive in comparison to N2 animals fed the same concentrations of glucose (0.0625%, 0.125%) prior to anoxia exposure (two-way ANOVA, Sidak’s multiple comparisons test). ANOVA, analysis of variance.
Transcriptional changes induced by an altered ceramide metabolism and a glucose diet
Previously, we reported the transcript profile of wild-type animals fed a glucose-supplemented diet relative to those fed an E. coli OP50-only diet and determined that the expression of 2370 genes were altered in response to the glucose-supplemented diet (Garcia et al. 2015). Given that ceramide changes (hyl-2 mutant) and a glucose-supplemented diet negatively impact anoxia survival, we hypothesized that there is an overlap in the transcriptional profile between these two anoxia sensitive animals (hyl-2(tm2031) and N2 glucose-fed animals). Furthermore, since the hyl-2(tm2031) mutant, relative to N2, is more sensitive to anoxia after being fed a glucose-supplemented diet, we hypothesized that the transcriptome profile of N2 animals fed a glucose diet and that of hyl-2(tm2031) animals fed a glucose diet are overlapping but not identical. To test these hypotheses, we used RNA-Seq to identify transcriptional changes in the hyl-2 mutant fed a standard and a glucose-supplemented diet (0.5%) and compared these datasets to our RNA-Seq results of N2 animals fed a standard diet and those fed a glucose-supplemented diet (Garcia et al. 2015). Note that the previously reported transcriptome profiles for the N2 animals and the hyl-2(tm2031) transcriptome profiles reported here were conducted at the same time (Garcia et al. 2015).
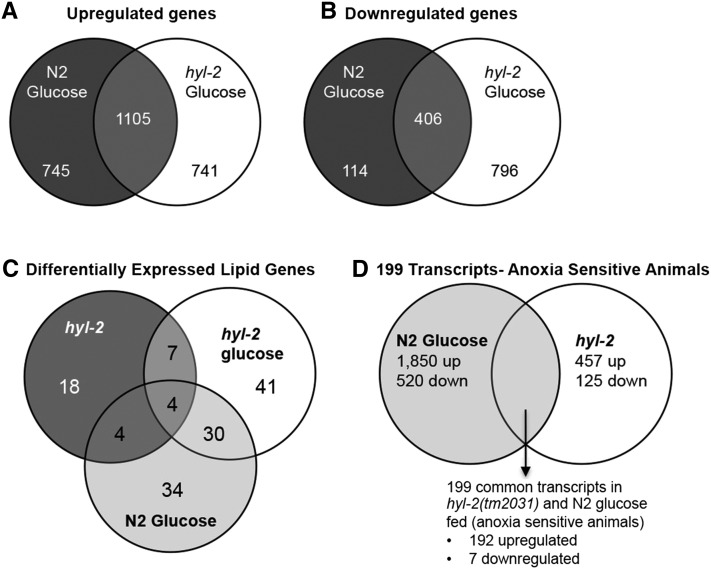
The hyl-2(tm2031) animals had 457 genes that were significantly upregulated and 125 genes that were significantly downregulated in comparison to the N2 animals (Supplemental Material, Figure S1A and Table S1). The hyl-2(tm2031) animals fed a glucose-supplemented diet had 1846 genes that were significantly upregulated and 1202 genes that were significantly downregulated in comparison to the hyl-2(tm2031) animals fed an OP50 only diet (Figure S1B and Table S2). Based on the PANTHER biological processes classification (http://www.pantherdb.org), genes involved with metabolic or cellular processes were the gene classes that displayed the highest number of gene expression changes in the hyl-2 mutant (Figure S2). We used the Gene List Venn Diagram program (http://genevenn.sourceforge.net) to assess the overlap between the RNA-Seq results of hyl-2(tm2031) animals fed a glucose diet to that of N2 wild-type animals fed a glucose diet. The genes that were differentially regulated in response to a glucose diet (upregulated, Figure 2A and Table S3; downregulated, Figure 2B and Table S4) were overlapping but not identical between N2 and hyl-2(tm2031) animals. Together, these data indicate that the hyl-2(tm2031) mutation impacts gene expression profiles and that the hyl-2(tm2031) and N2 animals fed a glucose-supplemented diet do not have identical gene expression responses.
Figure 2.
Venn diagram depicts the number of (A) upregulated transcripts or (B) downregulated transcripts identified between N2 and hyl-2(tm2031) animals fed a glucose diet. (C) Venn diagram depicts the transcripts that are differentially expressed and predicted to produce lipid metabolism proteins. (D) Venn diagram depicts that 199 transcripts are common between the anoxia sensitive animals (N2 glucose-fed and hyl-2(tm2031) animals).
Previously, we determined that, relative to wild-type controls, wild-type animals fed a glucose-supplemented diet have an increase in lipid droplets and hyl-2(tm2031) animals fed a standard OP50 diet have a reduction in lipid droplets (as detected by Oil Red O staining) (Garcia et al. 2015). Thus, it is not simply that a general increase or decrease of lipid levels results in anoxia sensitivity, and perhaps the anoxia sensitivity is tied to alteration of specific lipid pathways. There are 471 known or predicted genes involved with lipid metabolism in the C. elegans genome (Zhang et al. 2013). We used the Gene List Venn Diagram program (http://genevenn.sourceforge.net) to identify and compare the lipid metabolism genes that were differentially regulated in hyl-2(tm2031) animals, glucose-fed hyl-2(tm2031) animals, and glucose-fed wild-type animals (Figure 2C, Table S1, and Table S2). The hyl-2 mutation and a glucose diet alter the expression profile of lipid genes in an overlapping but not identical manner. Four lipid metabolism genes (cyp-25A1, cyp-13A12, cyp-33C8, and acs-2) were differentially expressed, relative to N2 controls fed a standard diet, in the hyl-2(tm2031) animals fed a standard or glucose diet, and glucose-fed wild-type animals (Figure 2C and Table S5). We used the Babelomics 5 web interface (babelomics.bioinfo.cipf.es) to conduct an enrichment analysis to find GO terms that were overrepresented in the lipid metabolism gene datasets (Table S5). The GO terms, namely, membrane lipid metabolism, sphingolipid metabolic process, and cellular lipid metabolic process were overrepresented in the hyl-2(tm2031), hyl-2(tm2031) fed glucose, and N2 fed glucose animals (Table S5). Genes within these GO term classes include asm-2, cgt-1, gba-4, and fat-5 (Table S5).
The transcriptome of anoxia sensitive animals
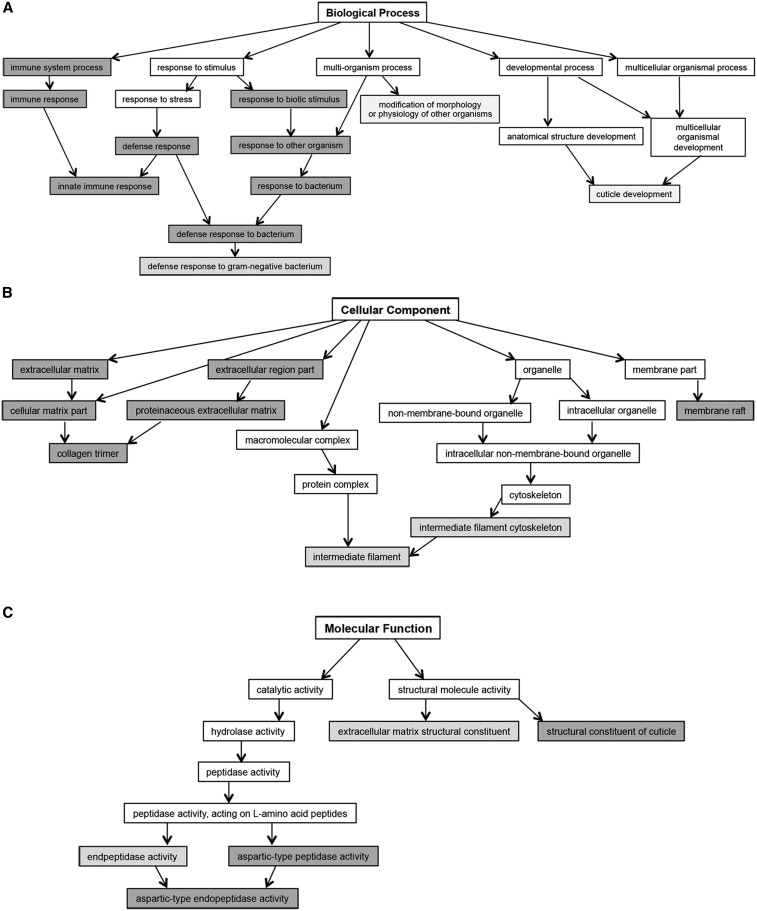
Sensitivity to anoxia is observed in wild-type animals fed a glucose diet and in hyl-2 mutants, indicating that both diet and genotype impact oxygen deprivation responses to varying degrees. To identify transcripts associated with anoxia sensitivity, we compared the RNA-Seq data for the anoxia sensitive animals (N2 glucose-fed, hyl-2(tm2031) OP50 diet) relative to wild-type N2 control animals fed a standard OP50 diet (Figure 2D). This revealed 199 common transcripts in the anoxia sensitive animals; 192 transcripts were upregulated and 7 transcripts were downregulated (Figure 2D and Table S6). To identify GO terms that were overrepresented in the 199 common transcripts the Babelomics 5 web interface was used (babelomics.bioinfo.cipf.es). The classification of transcripts by biological processes identified GO terms involved with bacterial defense, innate immunity, and cuticle development (Figure 3A and Table S7); cell component included GO terms associated with the extracellular matrix (ECM), intermediate filament, and membrane raft (Figure 3B and Table S7) and; molecular cell function identified GO terms associated with peptidase activity and the structural constituent of the cuticle (Figure 3C and Table S7).
Figure 3.
Babelomics 5 was used to identify the predominant significantly enriched GO terms associated with specific (A) biological processes, (B) cellular components, or (C) molecular functions, identified by FatiGO and FatiScan enrichment analysis for the transcriptional profiles of the 199 genes associated with anoxia sensitivity in the N2 animals fed a glucose diet and in the hyl-2(tm2031) mutant. Significantly enriched GO terms are represented by shaded nodes with the intensity of shading increasing as adjusted P-values decrease below 0.05 using the FDR procedure of Benjamini and Hochberg (1995). FDR, false discovery rate.
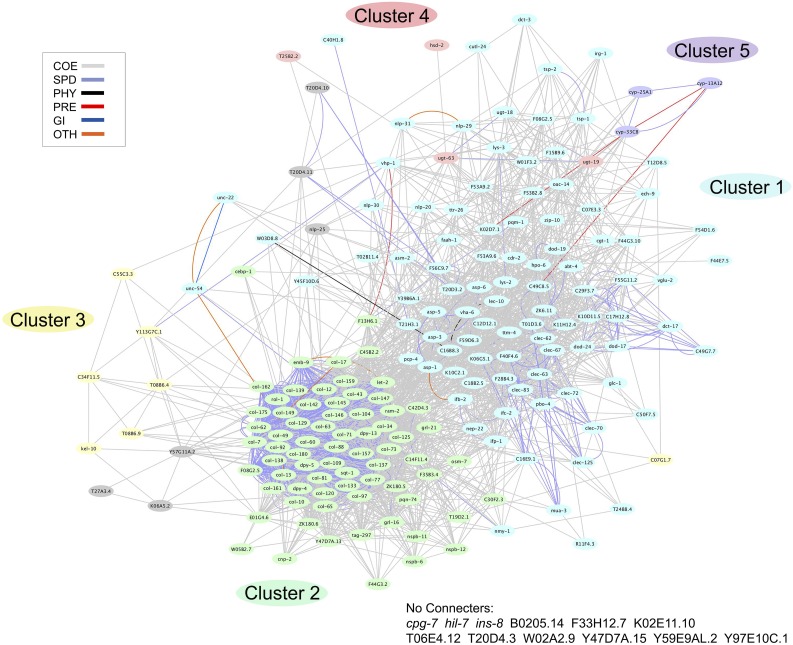
Visualization of the transcript data, using a heatmap, indicates that many of the 199 common transcripts were upregulated in a similar manner in the anoxia sensitive animals (Figure S3). However, there were some transcripts that were at higher levels in the glucose-fed wild-type animals (Figure S3). The transcripts that were downregulated were at similar levels in the anoxia sensitive animals (Figure S3). The GeneMANIA web interface (http://www.genemania.org) was used to generate a composite functional association network of the 199 genes, which was then analyzed for modularity using a Markov Clustering Algorithm (Enright et al. 2002). The composite network consists of various individual functional association networks derived from publicly available genomic and proteomic datasets by GeneMANIA. Weighted connective edges link the 199 genes, referred to as the nodes of the network, based on the Pearson correlation of each gene pair’s corresponding source data. All but 12 of the 199 genes identified were part of an interacting network, which included two large gene clusters (cluster 1 and 2) and three smaller gene clusters (cluster 3–5) (Figure 4 and Table S8). Cluster 1, with 86 genes, includes genes known to be involved in innate immunity, while Cluster 2, with 71 genes, includes genes that code for collagens or are involved in cuticle formation. Cluster 4 and Cluster 5 include genes encoding members of the xenobiotic and endobiotic phase I and II detoxification system (Table S8). Gene expression heatmaps were generated for genes in Cluster 1 (Figure S3B), Cluster 2 (Figure S3C), and combined for Cluster 3, 4, and 5 (Figure S3D). Cluster 1 contains a few genes with higher expression in the hyl-2(tm2031) animal relative to the glucose-fed wild-type animal (Figure S3B). Cluster 2 contains many genes displaying higher expression in the glucose-fed wild-type animal relative to the hyl-2(tm2031) animal (Figure S3C). Since the Clusters 3, 4, and 5 were smaller, we combined the transcript data into a single gene expression heatmap; these clusters contain genes that were upregulated or downregulated in a similar manner (Figure S3D). Taken together, the use of two independent methods for transcript analysis (GeneMANIA and Babelomics 5) indicates that immune responses and collagen functions could be altered in animals that are sensitive to anoxia exposure.
Figure 4.
The network for the 199 transcripts associated with anoxia sensitivity. This network of 199 transcripts was analyzed by GeneMANIA to identify genes that are: coexpressed (COE), have similar protein domains (SPD), physically interact (PHY), predicted to interact (PRE), genetically interact (GI), or interact in some other manner (OTH). Further bioinformatics analysis identified two large gene clusters (cluster 1 and 2) and three smaller gene clusters (cluster 3, cluster 4, and cluster 5) within the network.
Xenobiotic and endobiotic phase I and phase II detoxification system
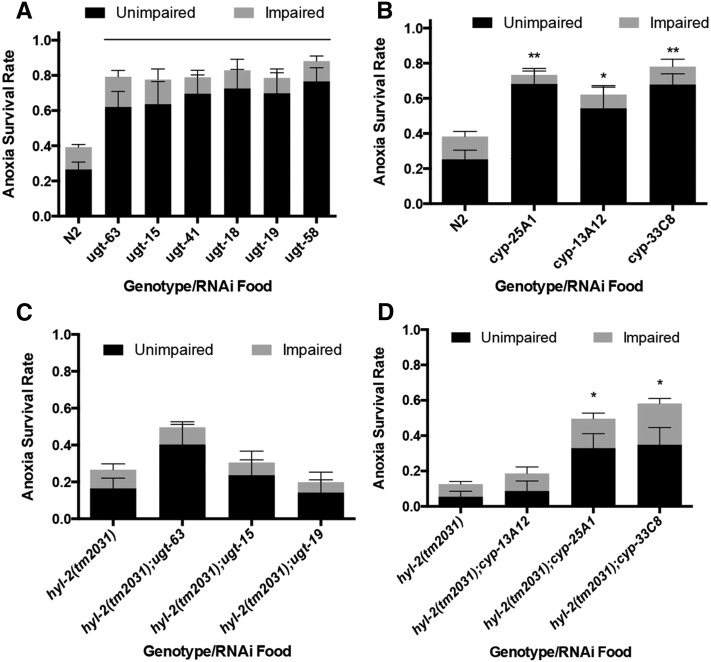
To determine if some of the transcriptional changes revealed by our RNA-Seq analysis alter anoxia responses, we used RNAi to target some genes differentially expressed in the anoxia sensitive animals. First, we examined the phenotype associated with RNAi knockdown of the seven genes downregulated in the anoxia sensitive animals (Table S6). RNAi knockdown of ugt-63 and cyp-25A1 resulted in a significant increase in anoxia survival relative to N2 animals, (Figure 5, A and B) but RNAi of the other five genes did not result in a significant difference in anoxia survival (Figure S4). The ins-8(RNAi) and F44E7.5(RNAi) animals exposed to anoxia had an inconsistent and variable anoxia survival rate; additional analysis would be needed to determine if changes in these genes impact the anoxia response (Figure S4).
Figure 5.
Knockdown of genes predicted to be involved with the xenobiotic and endobiotic phase I and phase II system increases anoxia survival. Relative to N2 wild-type controls, RNAi of ugt (A) and cyp (B) genes increases survival in 1 d old adult animals exposed to 2 d of anoxia (line and ** indicates P < 0.001, * indicates P < 0.05). Relative to hyl-2(tm2031) control, RNAi of ugt did not significantly increase anoxia survival (C), but RNAi of cyp-25A1 and cyp-33C8 (D) did increase anoxia survival (* indicates P < 0.05). For (A) and (B), animals were raised on the RNAi food or HT115 E. coli. with no added glucose. For all, statistical analysis was done using one-way ANOVA, Bonferonni multiple comparisons test; at least three independent experiments, with n ≥ 100. ANOVA, analysis of variance; RNAi, RNA interference.
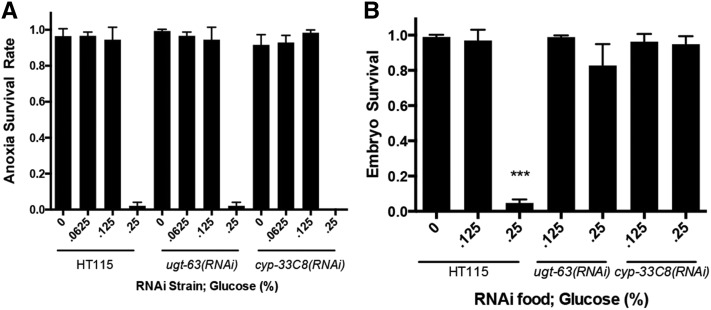
Quantitative RT-PCR verified that a glucose diet and hyl-2 mutation decreased the gene expression of cyp-25A1 and ugt-63 genes (Figure S5). Notably, it is not the decreased expression of cyp-25A1 and ugt-63 that contributes to anoxia sensitivity since the knockdown of these genes using RNAi actually enhanced anoxia survival. The ugt-63 and cyp-25A1 genes, part of gene Cluster 4 and 5 (Figure 4, B and E), code for proteins that show homology to detoxification system proteins (Ma et al. 2013; Keller et al. 2014; Gems and McElwee 2005). The RNA-Seq data indicate that several ugt and cyp gene family members have a differential gene expression profile in the anoxia sensitive animals relative to controls (Figure 4A and Table S6). The ugt and cyp genes (ugt-15, ugt-18, ugt-19, ugt-41, ugt-63, cyp-13A12, cyp-25A1, and cyp-33C8), which are differentially expressed in N2 animals fed glucose and in hyl-2(tm2031) animals, were knocked-down via RNAi and the anoxia phenotype was examined. In all cases, relative to controls fed HT115 E. coli, the knockdown of the ugt genes and cyp genes increased the survival of 2 d of anoxia exposure (Figure 5, A and B). However, only cyp-25A1(RNAi) and cyp-33C8(RNAi) suppressed the sensitivity to 2 d of anoxia in the hyl-2(tm2031) animals (Figure 5, C and D). These data indicate that knockdown of some ugt and cyp genes enhances anoxia survival in animals fed a standard E. coli diet (no glucose supplement). We next examined if RNAi knockdown of ugt-63 or cyp-33C8 suppressed anoxia sensitivity in animals fed a glucose diet. The ugt-63(RNAi) or cyp-33C8(RNAi) animals fed a glucose diet did not survive anoxia exposure significantly differently than the HT115 control animals (Figure 6A). Note that the E. coli strain used here was HT115 and not OP50; this may account for a different survival rate in the N2 animals fed a 0.125% glucose-supplemented diet (Figure 6A and Figure 1C). In our analysis of the wild-type adult animals fed glucose we noted that the embryos they produced did not survive anoxia. The embryos of N2 hermaphrodites fed a 0.25% glucose-supplemented diet were very sensitive to anoxia, indicating that diet can impact the next generation’s capacity to respond to stress (Figure 6B). However, knockdown of ugt-63 and cyp-33C8 suppressed the anoxia sensitivity in the embryos from hermaphrodites fed a glucose diet (Figure 6B). Together, these data indicate that a glucose diet in the P0 generation reduces anoxia survival in the F1 generation embryo and that the knockdown of genes predicted to be a part of the xenobiotic and endobiotic phase I and II detoxification system suppresses this anoxia sensitivity.
Figure 6.
Knockdown of genes predicted to be involved with the xenobiotic and endobiotic phase I and phase II system increases anoxia survival in the offspring of hermaphrodites fed a glucose diet. (A) Relative to N2 wild-type controls, RNAi of ugt-63 or cyp-33C8 did not increase anoxia survival in 1 d old adult animals fed a glucose diet. (B) Wild-type adult animals fed a glucose diet (0.25%) produce embryos that are unable to survive anoxia exposure. RNAi of ugt-63 or cyp-33C8 suppressed the anoxia sensitivity observed in embryos from adults fed a glucose diet (*** indicates P < 0.0001 using one-way ANOVA, Tukey multiple comparisons test; at least three independent experiments, with n > 50 embryos per experiment). ANOVA, analysis of variance; RNAi, RNA interference.
Discussion
Studies show that hyperglycemia worsens the outcome of an ischemic event such as stroke (Masrur et al. 2015; Sprafka et al. 1994; Megherbi et al. 2003; Bruno et al. 1999; Weir et al. 1997). However, it is not clear if specific physiological or molecular factors associated with type 2 diabetes impede oxygen deprivation responses. Using C. elegans we observed that a glucose-supplemented diet or altered ceramide metabolism (hyl-2 mutant) decreases oxygen deprivation survival, and when these factors are combined there is a further decrease in anoxia survival. We used RNA-Seq to examine the transcriptomes of hyl-2(tm2031) animals fed a standard OP50 and OP50 glucose-supplemented diet and found that both the hyl-2 mutation and glucose diet induces gene expression changes. The impact a glucose-supplemented diet has on the transcriptome of the wild-type animal is overlapping but not identical to that in the hyl-2(tm2031) animal (Garcia et al. 2015). Our approach to identify molecular changes that impact the oxygen deprivation response was based on comparative transcriptomics of anoxia sensitive animals (N2 glucose-fed and hyl-2 mutant). This comparison identified 199 common transcripts that are differentially regulated in anoxia sensitive animals. Bioinformatics analysis of these transcripts shed a light on their functional significance revealing, in particular, the roles of their products in cuticle/collagen and ECM formation, innate immunity, and the xenobiotic and endobiotic phase I and II detoxification system.
A large proportion of the 199 transcripts that are differentially regulated in the anoxia sensitive animals represent the known or predicted collagen genes involved with the structural constituent of the cuticle and/or ECM (Figure 3 and Table S7). The C. elegans skin, composed of an epidermal epithelium and cuticle, provides a barrier for the animal and its environment (Page and Johnstone 2007; Chisholm and Hsiao 2012). Collagens make up a large portion of the protein component of the cuticle (Chisholm and Xu 2012) and the epidermis is also an organ that stores lipids (O’Rourke et al. 2009; Chisholm and Xu 2012). Collagen genes are typically expressed in particular larval stages or in a cyclic manner during cuticle synthesis for each molt (Johnstone 2000). It is not clear why many collagen genes are differentially regulated in animals fed a glucose diet or with altered ceramide biosynthesis. However, these findings are of interest in the context of complications associated with type 2 diabetes. Patients with diabetic nephropathy display changes to the ECM and an increase in collagen within the kidney (Kolset et al. 2012). Additionally, the skin problems associated with diabetics could result from several cellular changes including fragmentation of connective tissue and collagen, an increase in matrix metalloproteinases responsible for the fragmentation, and glycosylation of ECM proteins (Demirseren et al. 2014; Argyropoulos et al. 2016). Furthermore, in diabetic retinopathy, the ECM undergoes many structural changes including increases in collagens (Roy et al. 2015). C. elegans can serve as a model system to study how hyperglycemia or altered ceramide levels impact the structure and function of the ECM.
Interestingly, genes involved in innate immunity were also differentially regulated in the anoxia sensitive animals. Many of the genes that are involved with innate immunity in C. elegans have been identified by the study of the bacterial food and intestinal pathogens (e.g., Pseudomonas aeruginosa and Salmonella typhimurium) (Gravato-Nobre and Hodgkin 2005; Ewbank and Pujol 2016). The standard E. coli strains (OP50 and HT115) used in our studies are not pathogenic per se to C. elegans; however, it is possible that if C. elegans is fed these bacteria along with glucose the animal might initiate an antimicrobial response. Some of the innate immunity genes are also differentially expressed in the hyl-2(tm2031) animals fed the standard E. coli diet. Previous studies have reported that the disruption of mitochondrial activity induces the pathogen-response and endobiotic and xenobiotic detoxification pathways (Liu et al. 2014; Melo and Ruvkun 2012; Govindan et al. 2015). Furthermore, the animals with altered regulation of ceramide biosynthesis pathways were deficient in responding to mitochondrial dysfunction (Liu et al. 2014). Thus, it is possible that an altered ceramide metabolism or a glucose diet disrupts mitochondrial functions leading to the expression of innate immunity genes. Another explanation for the expression of innate immunity genes in the anoxia sensitive animals is related to changes in cuticle structure; genetic mutations leading to abnormal cuticle development can trigger innate immune responses (Taffoni and Pujol 2015). Thus, it is possible that changes in cuticle components may be triggering downstream effects on genes involved with innate immunity. Further experiments would be needed to examine if these two processes are independently induced by a glucose diet or altered ceramide species or if they are mechanistically linked.
A recent study demonstrated that a glucose diet causes decreased membrane fluidity in worms with altered fatty acid metabolism (paqr-2 and iglr-2 mutants) (Svensk et al. 2016). PAQR-2, a homolog to the human AdipoR1/AdipoR2 proteins that regulate fatty acid metabolism, appears to work with IGLR-2 to maintain membrane homeostasis. The iglr-2 transcript is upregulated in the hyl-2(tm2031) animals fed a glucose diet but not in wild-type animals fed the same diet. It is evident from the study by Svensk et al. (2016) that both paqr-2 and iglr-2 protect against glucose toxicity; perhaps the regulation of these genes in response to glucose is not at the transcriptional level. We found that a number of lipid metabolism genes are differentially expressed in the hyl-2(tm2031) mutant and the glucose-fed animals (Garcia et al. 2015). The GO terms—membrane lipid metabolism, sphingolipid metabolic process and cellular lipid metabolic process—were overrepresented in the hyl-2(tm2031), hyl-2(tm2031) fed glucose, and N2 fed glucose animals. Ceramides are known to localize to mitochondria in C. elegans (Liu et al. 2014). Also, since there is an increase in lipids in glucose-fed animals, a glucose diet likely impacts the lipidomic profile (Garcia et al. 2015). Thus, it is possible that membrane fluidity of cells and perhaps organelles such as the mitochondria or ER are altered in glucose-fed animals and hyl-2(tm2031) mutants. Additional experiments could determine if this is the case and if this results in a compromised ability to survive anoxia exposure.
We hypothesized that the gene expression changes observed in an animal with altered ceramide metabolism or fed a glucose diet could negatively impact anoxia survival. We used RNAi to screen a subset of these genes and determined that the knockdown of the genes encoding the xenobiotic and endobiotic phase I and II detoxification system increased anoxia survival. The detoxification system is involved in the conversion of exogenous and endogenous molecules into hydrophilic products for excretion. The phase I detoxification system involves cytochrome P450s, which are largely responsible for the addition of oxygen to substrate molecules. The phase II detoxification system encompasses a broad range of enzymes including UDP-glycosyltransferases (UGTs), sulfotransferases, glutathione S-transferases, and N-acetyltransferases. The UGT enzymes reside in the ER membrane and catalyze the addition of the sugar glucuronic acid to lipophilic endobiotics and xenobiotics. In C. elegans, these detoxification gene families are large but very few functional analysis studies have been conducted. Interestingly, a member of the cyp gene family (cyp-13A12) was implicated in the reoxygenation response in C. elegans (Ma et al. 2013). Furthermore, there is evidence that C. elegans have a surveillance system that monitors core cellular activities (e.g., translation, respiration, and secretory pathways) and engage in behavioral, immune, and detoxification responses when these core cellular activities are compromised (Melo and Ruvkun 2012). The cyp-35B1::GFP reporter, expressed in the intestine, is induced by the inactivation of genes involved in protein synthesis, mitochondrial function, metabolism, vascular trafficking, and cuticle specific functions, suggesting that there is some signaling between different organ types (Melo and Ruvkun 2012). In our study, most of the detoxification genes differentially regulated in the glucose-fed and hyl-2(tm2031) animals were upregulated.
It is not yet clear why knockdown of the detoxification genes via RNAi increases anoxia survival. It is possible that core cellular processes are disrupted by a glucose diet or altered ceramide metabolism and this, in turn, induces expressional changes in the detoxification genes, which ultimately leads to an increased flux of the detoxification pathway. Perhaps, an increased flux of the detoxification pathway compromises anoxia survival by utilizing molecules (such as stored oxygen) needed to survive anoxia. In humans, UGTs promote the excretion of toxic metabolites, including pharmaceuticals, in bile or urine and facilitate the homeostasis of endogenous molecules such as bilirubin (end-product of heme metabolism), some steroid hormones, and fatty acids (Guillemette 2003). Bilirubin, in the past thought of only as a toxic waste product, may also possess potent antioxidant capabilities at physiologically relevant oxygen concentrations. Though a sustained high concentration of bilirubin is neurotoxic, low to moderately increased levels have been shown to be cytoprotective (Stocker et al. 1987; Hegyi et al. 1994; Erdogan et al. 2012). This supports the idea that the detoxification system can modulate the stress response through secondary mechanisms, and that it is not necessarily paradoxical that knockdown of some detoxification genes may be beneficial in some circumstances. Thus, it is possible that knockdown of the detoxification genes in C. elegans alters the level of a target molecule which, in turn, alters oxygen deprivation responses. However, since the detoxification genes are understudied in C. elegans, their substrates and/or targets are currently unclear.
We found that P0 animals fed a glucose diet led to a reduced anoxia survival in the F1 generation indicating that the parental diet negatively impacted anoxia resistance in offspring. Knockdown of ugt-63 or cyp-33C8 in the P0 generation fed a glucose diet suppressed anoxia sensitivity observed in the F1 embryo. However, the mechanism involved with this transgenerational preconditioning effect is not understood. Others have analyzed the impact that a high glucose diet has on subsequent generations (Tauffenberger and Parker 2014). They found that a 2% glucose diet fed to the P0 generation reduced progeny production but increased resistance to the oxidative stress inducer juglone in the F1 generation. We have not observed glucose-induced stress resistance so perhaps differences in methodology (e.g., glucose concentration or specific type of stress) impacts how the animal responds to the stress. Future studies are needed to understand the functional role the detoxification genes have in oxygen deprivation survival, particularly relative to altered ceramide metabolism and elevated glucose level, in both the parental and subsequent generations.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis elegans Genetics Stock Center (CGC), which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a grant from the NIH [DK109524-01, NIH/National Institute of Diabetes and Digestive Kidney Diseases (NIDDK)] to P.A.P. and R.K.A.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.031583/-/DC1
Communicating editor: M. Walhout
Literature Cited
- Aho A. F., Kernighan B. W., Weinbeger P. J., 1987. The AWK Programming Language, Addison-Wesley Longman Publishing Co., Inc., Boston, MA. [Google Scholar]
- Al-Shahrour F., Minguez P., Vaquerizas J.M., Conde L., Dopazo J., 2005. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 33: W460–W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F., Minguez P., Tarraga J., Medina I., Alloza E., et al. , 2007. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 35: W91–W96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R., Salavert F., Garcia-Garcia F., Carbonell-Caballero J., Bleda M., et al. , 2015. Babelomics 5.0: functional interpretation for new generations of genomic data. Nucleic Acids Res. 43(W1): W117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. L., Mao X., Scott B. A., Crowder C. M., 2009. Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science 323(5914): 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argraves K. M., Sethi A. A., Gazzolo P. J., Wilkerson B. A., Remaley A. T., et al. , 2011. S1P, dihydro-S1P and C24:1-ceramide levels in the HDL-containing fraction of serum inversely correlate with occurrence of ischemic heart disease. Lipids Health Dis. 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyropoulos A. J., Robichaud P., Balimunkwe R. M., Fisher G. J., Hammerberg C., et al. , 2016. Alterations of Dermal Connective Tissue Collagen in Diabetes: Molecular Basis of Aged-Appearing Skin. PLoS One 11(4): e0153806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. A., Paneni F., Cosentino F., Creager M. A., 2013. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part II. Eur. Heart J. 34(31): 2444–2452. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 57(1): 289–300. [Google Scholar]
- Bruno A., Biller J., Adams H. P., Jr, Clarke W. R., Woolson R. F., et al. , 1999. Acute blood glucose level and outcome from ischemic stroke. Trial of ORG 10172 in Acute Stroke Treatment (TOAST) Investigators. Neurology 52(2): 280–284. [DOI] [PubMed] [Google Scholar]
- Chen N., Harris T. W., Antoshechkin I., Bastiani C., Bieri T., et al. , 2005. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 33(Database issue): D383–D389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. D., Hsiao T. I., 2012. The Caenorhabditis elegans epidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip. Rev. Dev. Biol. 1(6): 861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. D., Xu S., 2012. The Caenorhabditis elegans epidermis as a model skin. II: differentiation and physiological roles. Wiley Interdiscip. Rev. Dev. Biol. 1(6): 879–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. S., 2011. High glucose diets shorten lifespan of Caenorhabditis elegans via ectopic apoptosis induction. Nutr. Res. Pract. 5(3): 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creager M. A., Luscher T. F., Cosentino F., Beckman J. A., 2003. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: Part I. Circulation 108(12): 1527–1532. [DOI] [PubMed] [Google Scholar]
- Cutler R. G., Thompson K. W., Camandola S., Mack K. T., Mattson M. P., 2014. Sphingolipid metabolism regulates development and lifespan in Caenorhabditis elegans. Mech. Ageing Dev. 143–144: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirseren D. D., Emre S., Akoglu G., Arpaci D., Arman A., et al. , 2014. Relationship between skin diseases and extracutaneous complications of diabetes mellitus: clinical analysis of 750 patients. Am. J. Clin. Dermatol. 15(1): 65–70. [DOI] [PubMed] [Google Scholar]
- Deng X., Yin X., Allan R., Lu D. D., Maurer C. W., et al. , 2008. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science 322(5898): 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S., Ouzounis C. A., 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7): 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan T., Cicek Y., Kocaman S. A., Canga A., Cetin M., et al. , 2012. Increased serum bilirubin level is related to good collateral development in patients with chronic total coronary occlusion. Intern. Med. 51(3): 249–255. [DOI] [PubMed] [Google Scholar]
- Ewbank J. J., Pujol N., 2016. Local and long-range activation of innate immunity by infection and damage in C. elegans. Curr. Opin. Immunol. 38: 1–7. [DOI] [PubMed] [Google Scholar]
- Fawcett E. M., Hoyt J. M., Johnson J. K., Miller D. L., 2015. Hypoxia disrupts proteostasis in Caenorhabditis elegans. Aging Cell 14(1): 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galadari S., Rahman A., Pallichankandy S., Galadari A., Thayyullathil F., 2013. Role of ceramide in diabetes mellitus: evidence and mechanisms. Lipids Health Dis. 12: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A. M., Ladage M. L., Dumesnil D. R., Zaman K., Shulaev V., et al. , 2015. Glucose Induces Sensitivity to Oxygen Deprivation and Modulates Insulin/IGF-1 Signaling and Lipid Biosynthesis in Caenorhabditis elegans. Genetics 200(1): 167–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R., Seoighe C., 2010. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D., McElwee J. J., 2005. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 126(3): 381–387. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Jayamani E., Zhang X., Breen P., Larkins-Ford J., et al. , 2015. Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans. Nat. Cell Biol. 17(10): 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato-Nobre M. J., Hodgkin J., 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7(6): 741–751. [DOI] [PubMed] [Google Scholar]
- Gray J. M., Karow D. S., Lu H., Chang A. J., Chang J. S., et al. , 2004. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430(6997): 317–322. [DOI] [PubMed] [Google Scholar]
- Guillemette C., 2003. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 3(3): 136–158. [DOI] [PubMed] [Google Scholar]
- Hannun Y. A., Obeid L. M., 2011. Many ceramides. J. Biol. Chem. 286(32): 27855–27862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi T., Goldie E., Hiatt M., 1994. The protective role of bilirubin in oxygen-radical diseases of the preterm infant. J. Perinatol. 14(4): 296–300. [PubMed] [Google Scholar]
- Hochachka P. W., Lutz P. L., 2001. Mechanism, origin, and evolution of anoxia tolerance in animals. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 130(4): 435–459. [DOI] [PubMed] [Google Scholar]
- Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J. R., 2008. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Guo R., Powell-Coffman J. A., 2001. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. USA 98(14): 7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I. L., 2000. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 16(1): 21–27. [DOI] [PubMed] [Google Scholar]
- Jonz M. G., Buck L. T., Perry S. F., Schwerte T., Zaccone G., 2016. Sensing and surviving hypoxia in vertebrates. Ann. N. Y. Acad. Sci. 1365(1): 43–58. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30(4): 313–321. [DOI] [PubMed] [Google Scholar]
- Keller J., Ellieva A., Ma D. K., Ju J., Nehk E., et al. , 2014. CYP-13A12 of the nematode Caenorhabditis elegans is a PUFA-epoxygenase involved in behavioural response to reoxygenation. Biochem. J. 464(1): 61–71. [DOI] [PubMed] [Google Scholar]
- Kolset S. O., Reinholt F. P., Jenssen T., 2012. Diabetic nephropathy and extracellular matrix. J. Histochem. Cytochem. 60(12): 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3): R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P. J., Tennagels N., 2014. On ceramides, other sphingolipids and impaired glucose homeostasis. Mol. Metab. 3(3): 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue B. L., Padilla P. A., 2011. Environmental and genetic preconditioning for long-term anoxia responses requires AMPK in Caenorhabditis elegans. PLoS One 6(2): e16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Murphy C. T., Kenyon C., 2009. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 10(5): 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P.L., and Y. Zhang, 2013 Cross talk between ceramide and redox signaling: implications for endothelial dysfunction and renal disease. Handb. Exp. Pharmacol. 216: 171–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett M. R., Hoy M. J., Mastroianni M., Mondoux M. A., 2015. High-glucose diets have sex-specific effects on aging in C. elegans: toxic to hermaphrodites but beneficial to males. Aging (Albany, N.Y.) 7(6): 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Samuel B. S., Breen P. C., Ruvkun G., 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508(7496): 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4): 402–408. [DOI] [PubMed] [Google Scholar]
- Lopez X., Goldfine A. B., Holland W. L., Gordillo R., Scherer P. E., 2013. Plasma ceramides are elevated in female children and adolescents with type 2 diabetes. J. Pediatr. Endocrinol. Metab. 26(9–10): 995–998. [DOI] [PubMed] [Google Scholar]
- Ma D. K., Rothe M., Zheng S., Bhatla N., Pender C. L., et al. , 2013. Cytochrome P450 drives a HIF-regulated behavioral response to reoxygenation by C. elegans. Science 341(6145): 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Van Hemert J. L., Dash S., Dickerson J. A., 2009. Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X. R., Kaufman D. M., Crowder C. M., 2016. Nicotinamide mononucleotide adenylyltransferase promotes hypoxic survival by activating the mitochondrial unfolded protein response. Cell Death Dis. 7: e2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrur S., Cox M., Bhatt D. L., Smith E. E., Ellrodt G., et al. , 2015. Association of Acute and Chronic Hyperglycemia With Acute Ischemic Stroke Outcomes Post-Thrombolysis: Findings From Get With The Guidelines-Stroke. J. Am. Heart Assoc. 4(10): e002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megherbi S. E., Milan C., Minier D., Couvreur G., Osseby G. V., et al. , 2003. Association between diabetes and stroke subtype on survival and functional outcome 3 months after stroke: data from the European BIOMED Stroke Project. Stroke 34(3): 688–694. [DOI] [PubMed] [Google Scholar]
- Melo J. A., Ruvkun G., 2012. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149(2): 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall A. R., LaRue B., Padilla P. A., 2006. Glyceraldehyde-3-phosphate dehydrogenase mediates anoxia response and survival in Caenorhabditis elegans. Genetics 174(3): 1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall A. R., LeBlanc M. G., Mohan D. P., Padilla P. A., 2009. Reduction in ovulation or male sex phenotype increases long-term anoxia survival in a daf-16-independent manner in Caenorhabditis elegans. Physiol. Genomics 36(3): 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I., et al. , 2009. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324(5925): 381–384. [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., 2013a Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8(8): 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P. D., 2013b PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41(Database issue): D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux M. A., Love D. C., Ghosh S. K., Fukushige T., Bond M., et al. , 2011. O-linked-N-acetylglucosamine cycling and insulin signaling are required for the glucose stress response in Caenorhabditis elegans. Genetics 188(2): 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Rodriguez H., Bader G. D., Morris Q., 2014. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000 Res. 3: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech M. B., Kruse R., Harvald E. B., Olsen A. S., Gallego S. F., et al. , 2013. Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PLoS One 8(7): e70087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q., 2008. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9(Suppl 1): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathaniel T. I., Williams-Hernandez A., Hunter A. L., Liddy C., Peffley D. M., et al. , 2015. Tissue hypoxia during ischemic stroke: adaptive clues from hypoxia-tolerant animal models. Brain Res. Bull. 114: 1–12. [DOI] [PubMed] [Google Scholar]
- Novgorodov S. A., Gudz T. I., 2011. Ceramide and mitochondria in ischemic brain injury. Int. J. Biochem. Mol. Biol. 2(4): 347–361. [PMC free article] [PubMed] [Google Scholar]
- Novgorodov S. A., Riley C. L., Yu J., Keffler J. A., Clarke C. J., et al. , 2016. Lactosylceramide contributes to mitochondrial dysfunction in diabetes. J. Lipid Res. 57(4): 546–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden C. L., Carroll M. D., Kit B. K., Flegal K. M., 2012. Prevalence of Obesity in the United States, 2009–2010. NCHS Data Brief; 82: 1–8. [PubMed] [Google Scholar]
- O’Rourke E. J., Soukas A. A., Carr C. E., Ruvkun G., 2009. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10(5): 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla P. A., Ladage M. L., 2012. Suspended animation, diapause and quiescence: Arresting the cell cycle in C. elegans. Cell Cycle 11(9): 1672–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla P. A., Nystul T. G., Zager R. A., Johnson A. C., Roth M. B., 2002. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol. Biol. Cell 13(5): 1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla P. A., Goy J. M., Hajeri V. A., 2011. Anoxia-Induced Suspended Animation in Caenorhabditis elegans, pp. 25–58 in Anoxia, edited by P. A. Padilla . InTech, Rijeka. [Google Scholar]
- Page A. P., Johnstone I. L., 2007. The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.138.1, http://www.wormbook.org. [Google Scholar]
- Paneni F., Beckman J. A., Creager M. A., Cosentino F., 2013. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur. Heart J. 34(31): 2436–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson D. J., 1997. The diabetic heart is more sensitive to ischemic injury. Cardiovasc. Res. 34(1): 104–112. [DOI] [PubMed] [Google Scholar]
- Queliconi B. B., Kowaltowski A. J., Nehrke K., 2014. An anoxia-starvation model for ischemia/reperfusion in C. elegans. J. Vis. Exp. 85. Available at: http://www.jove.com/video/51231/an-anoxia-starvation-model-for-ischemiareperfusion-in-c-elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Bae E., Amin S., Kim D., 2015. Extracellular matrix, gap junctions, and retinal vascular homeostasis in diabetic retinopathy. Exp. Eye Res. 133: 58–68. [DOI] [PubMed] [Google Scholar]
- Schlotterer A., Kukudov G., Bozorgmehr F., Hutter H., Du X., et al. , 2009. C. elegans as model for the study of high glucose- mediated life span reduction. Diabetes 58(11): 2450–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B. A., Avidan M. S., Crowder C. M., 2002. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296(5577): 2388–2391. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11): 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N., Rual J. F., Carvunis A. R., Tasan M., Lemmens I., et al. , 2009. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods 6(1): 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprafka J. M., Virnig B. A., Shahar E., McGovern P. G., 1994. Trends in diabetes prevalence among stroke patients and the effect of diabetes on stroke survival: the Minnesota Heart Survey. Diabet. Med. 11(7): 678–684. [DOI] [PubMed] [Google Scholar]
- Stocker R., Yamamoto Y., McDonagh A. F., Glazer A. N., Ames B. N., 1987. Bilirubin is an antioxidant of possible physiological importance. Science 235(4792): 1043–1046. [DOI] [PubMed] [Google Scholar]
- Svensk E., Devkota R., Stahlman M., Ranji P., Rauthan M., et al. , 2016. Caenorhabditis elegans PAQR-2 and IGLR-2 Protect against Glucose Toxicity by Modulating Membrane Lipid Composition. PLoS Genet. 12(4): e1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffoni C., Pujol N., 2015. Mechanisms of innate immunity in C. elegans epidermis. Tissue Barriers 3(4): e1078432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauffenberger A., Parker J. A., 2014. Heritable transmission of stress resistance by high dietary glucose in Caenorhabditis elegans. PLoS Genet. 10(5): e1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9): 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28(5): 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin S. M., Nicholls H. T., Willmes D. M., Mourier A., Brodesser S., et al. , 2014. Obesity-Induced CerS6-Dependent C16:0 Ceramide Production Promotes Weight Gain and Glucose Intolerance. Cell Metab. 20(4): 678–686. [DOI] [PubMed] [Google Scholar]
- Van Voorhies W. A., Ward S., 2000. Broad oxygen tolerance in the nematode Caenorhabditis elegans. J. Exp. Biol. 203(Pt 16): 2467–2478. [DOI] [PubMed] [Google Scholar]
- Voelzmann A., Bauer R., 2010. Ceramide synthases in mammalians, worms, and insects: emerging schemes. Biomol. Concepts 1(5–6): 411–422. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., et al. , 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38: W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir C. J., Murray G. D., Dyker A. G., Lees K. R., 1997. Is hyperglycaemia an independent predictor of poor outcome after acute stroke? Results of a long-term follow up study. BMJ 314(7090): 1303–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J. Y., Morley T. S., Scherer P. E., 2014. The adipokine/ceramide axis: key aspects of insulin sensitization. Biochimie 96: 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zou X., Ding Y., Wang H., Wu X., et al. , 2013. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC Genomics 14: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigdon H., Kogot-Levin A., Park J. W., Goldschmidt R., Kelly S., et al. , 2013. Ablation of ceramide synthase 2 causes chronic oxidative stress due to disruption of the mitochondrial respiratory chain. J. Biol. Chem. 288(7): 4947–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M., Gray J. M., Pokala N., Chang A. J., Karow D. S., et al. , 2009. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61(6): 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been archived in NCBI GEO under the series record number GSE83887.