Abstract
Drosophila melanogaster is a powerful model organism for dissecting the molecular mechanisms that regulate sleep, and numerous studies in the fly have identified genes that impact sleep–wake cycles. Conditional genetic analysis is essential to distinguish the mechanisms by which these genes impact sleep: some genes might exert their effects developmentally, for instance by directing the assembly of neuronal circuits that regulate sleep; other genes may regulate sleep in adulthood; and yet other genes might influence sleep by both developmental and adult mechanisms. Here we have assessed two ligand-inducible expression systems, Geneswitch and the Q-system, for conditional and neuronally restricted manipulations of sleep in Drosophila. While adult-specific induction of a neuronally expressed Geneswitch transgene (elav-GS) is compatible with studies of sleep as shown previously, developmental induction of elav-GS strongly and nonspecifically perturbs sleep in adults. The alterations of sleep in elav-GS animals occur at low doses of Geneswitch agonist and in the presence of transgenes unrelated to sleep, such as UAS-CD8-GFP. Furthermore, developmental elav-GS induction is toxic and reduces brood size, indicating multiple adverse effects of neuronal Geneswitch activation. In contrast, the transgenes and ligand of the Q-system do not significantly impact sleep–wake cycles when used for constitutive, developmental, or adult-specific neuronal induction. The nonspecific effects of developmental elav-GS activation on sleep indicate that such manipulations require cautious interpretation, and suggest that the Q-system or other strategies may be more suitable for conditional genetic analysis of sleep and other behaviors in Drosophila.
Keywords: Geneswitch, RU486, Q-system, insomniac, Cul3, FlyBook
The establishment of Drosophila as a model organism for studies of sleep (Hendricks et al. 2000; Shaw et al. 2000) has facilitated unbiased and candidate-based screens for genes that impact sleep–wake cycles (e.g., Cirelli et al. 2005; Kume et al. 2005; Koh et al. 2008). While these studies have revealed an increasing number of genes that influence sleep, the underlying mechanisms are in most cases poorly defined. One critical aspect of elucidating these mechanisms is defining the temporal window in which genes function in relation to sleep. Some genes might function principally in a developmental manner, for example by contributing to the assembly of neuronal circuits relevant to sleep, while other genes might function specifically in adulthood and regulate sleep in a sustained or dynamic manner. Given that most Drosophila genes are expressed at multiple stages of the life cycle (Graveley et al. 2011) and that many genes have pleiotropic functions, individual genes may impact sleep by both developmental and adult mechanisms. Temporally restricted genetic manipulations are therefore essential to distinguish among these mechanisms and to shape hypotheses for how various genes exert their effects on sleep.
Strategies for temporally restricted genetic manipulations in Drosophila use temperature, chemical ligands, and light as conditional triggers. Temperature-regulated systems include heat-inducible promoters fused directly to heterologous genes (Lis et al. 1983) or to the Gal4 activator (Brand et al. 1994), and the Gal80ts system in which a temperature-sensitive form of the Gal80 suppressor restricts Gal4 activity (McGuire et al. 2003). Ligand-inducible systems include tetracycline-dependent activators (Bello et al. 1998; Bieschke et al. 1998; Stebbins et al. 2001), steroid-activated forms of Gal4 (Osterwalder et al. 2001; Roman et al. 2001), and the Q-system derived from Neurospora (Potter et al. 2010; Riabinina et al. 2015). More recently, photosensitive transcriptional activators have been described (Chan et al. 2015). Of these strategies, the steroid-inducible Geneswitch system (Osterwalder et al. 2001; Roman et al. 2001) has been the most frequently employed to study sleep and has been used in nearly two dozen such studies to date (Yuan et al. 2006; Joiner et al. 2006; Seugnet et al. 2008, 2011; Bushey et al. 2009; Donlea et al. 2009; Wu et al. 2009; Kuo et al. 2010, 2012; Crocker et al. 2010; Ishimoto and Kitamoto 2010; Pfeiffenberger and Allada 2012; Erion et al. 2012; Ueno et al. 2012; Vanderheyden et al. 2013; Tulina et al. 2014; Liu et al. 2014; Kayser et al. 2014; Oh et al. 2014; Chen et al. 2015; Tabuchi et al. 2015; Dissel et al. 2015; Afonso et al. 2015).
Geneswitch is a tripartite fusion protein containing the Gal4 DNA-binding domain, the progesterone receptor ligand-binding domain, and the p65 transcriptional activation domain (Burcin et al. 1999). In the presence of RU486, a progesterone receptor agonist, Geneswitch induces the transcription of genes located downstream of the upstream activating sequence (UAS) element bound by Gal4 (Burcin et al. 1999). In Drosophila, spatial restriction of Geneswitch activity is conferred by tissue-specific regulatory elements, while temporal control is achieved by delivery of RU486 in a restricted manner during development or adulthood (Osterwalder et al. 2001; Roman et al. 2001).
Nearly all of the reported uses of Geneswitch to manipulate sleep in Drosophila have utilized adult-specific induction, achieved by feeding RU486-containing food to adult flies (Yuan et al. 2006; Joiner et al. 2006; Seugnet et al. 2008, 2011; Bushey et al. 2009; Donlea et al. 2009; Wu et al. 2009; Kuo et al. 2010, 2012; Crocker et al. 2010; Ishimoto and Kitamoto 2010; Pfeiffenberger and Allada 2012; Erion et al. 2012; Ueno et al. 2012; Vanderheyden et al. 2013; Tulina et al. 2014; Liu et al. 2014; Kayser et al. 2014; Oh et al. 2014; Chen et al. 2015; Tabuchi et al. 2015; Dissel et al. 2015; Afonso et al. 2015). RU486 is well tolerated in adults with no detectable toxic effects at high concentrations (500 µM) (Osterwalder et al. 2001; Roman et al. 2001), and studies of sleep have typically used concentrations at or below this threshold to activate Geneswitch transgenes expressed neuronally (elav-GS) (e.g., Joiner et al. 2006; Seugnet et al. 2008; Bushey et al. 2009), in mushroom bodies (MB-GS) (Yuan et al. 2006; Joiner et al. 2006; Wu et al. 2009; Ishimoto and Kitamoto 2010), in fat bodies (S1106-GS) (Kuo et al. 2010), and ubiquitously (da-GS) (Tabuchi et al. 2015; Dissel et al. 2015). In adult animals, Geneswitch and RU486 do not alter sleep in the absence of effector transgenes (e.g., Wu et al. 2009; Afonso et al. 2015), permitting use of the Geneswitch system for various adult-specific manipulations of sleep.
In contrast to the many studies of sleep that have used adult-specific Geneswitch induction, few have used constitutive or developmental-specific induction (Kuo et al. 2010; Pfeiffenberger and Allada 2012). One relevant concern is the developmental toxicity of RU486 at concentrations lower than those tolerated in adulthood (Osterwalder et al. 2001; van Swinderen 2007; Shen et al. 2009; Landis et al. 2015). In the absence of Geneswitch transgenes, high concentrations of RU486 are intrinsically toxic to early Drosophila development, as indicated by reduced numbers of larvae arising from parents fed 233 µM RU486 (Osterwalder et al. 2001). The toxicity of RU486 appears to be increased in the presence of elav-GS and effector transgenes, as suggested by developmental lethality from exposure to RU486 concentrations greater than 25 µM (van Swinderen 2007). While the threshold for developmental toxicity of Geneswitch is not well defined, low concentrations of RU486 (11.6 µM = 5 µg/ml) were found to be permissive for larval development in the context of ∼200 different Geneswitch drivers (Nicholson et al. 2008). In addition to its acute effects on development, early Geneswitch activation can have adverse consequences later in life, as indicated by reduced lifespan of elav-GS animals exposed to RU486 during development (Shen et al. 2009).
The impact of developmental Geneswitch activation on sleep and other adult behaviors has not been assessed comprehensively. Constitutive activation of the elav-GS and S1106-GS drivers was reported in a study of the immune response and sleep (Kuo et al. 2010), though sleep was assessed in a narrow window postinjury and effects on total daily sleep were not determined. A second study reported developmental-specific elav-GS induction, by setting crosses on RU486-containing food and moving adults to food lacking RU486 (Pfeiffenberger and Allada 2012). While developmental toxicity was not addressed, sleep was reported to be near normal in elav-GS animals exposed developmentally to 50 µM RU486 (Pfeiffenberger and Allada 2012). Additional studies are required to assess the general utility of developmental Geneswitch activation, and to compare this strategy to other conditional manipulations of sleep.
The Q-system is a more recently developed ligand-inducible system that utilizes components of the Neurospora crassa quinic acid gene cluster (Potter et al. 2010): the QF transcriptional activator, the QS suppressor that binds and inhibits QF in a quinic acid-dependent manner, and the QUAS regulatory element bound by QF. A refinement of the Q-system is the hybrid Gal4QF activator, in which the Gal4 DNA-binding domain replaces that of QF, enabling activation of UAS transgenes while preserving suppression by QS and derepression by quinic acid (Riabinina et al. 2015). Quinic acid has no obvious adult toxicity (Potter et al. 2010) and panneuronal QF expression does not significantly alter circadian rhythms or sleep (Riabinina et al. 2015). However, the effects of quinic acid exposure on sleep have not been determined in animals lacking or expressing Gal4QF and QS, components necessary for conditional activation of UAS transgenes. Thus, whether the Q-system is compatible with developmental and adult manipulations of sleep is not yet known.
Here we have evaluated Geneswitch and the Q-system for conditional, neuronally restricted manipulations of sleep. While adult-specific induction of the panneuronal elav-GS driver is compatible with assessing sleep as reported in earlier studies, developmental-specific or constitutive induction of elav-GS causes developmental defects and nonspecific reductions of sleep in adulthood. In contrast, the constituent transgenes and inducing ligand of the Q-system do not alter sleep in developmental-specific, adult-specific, and constitutive neuronal manipulations. Our findings indicate that nonspecific perturbations of sleep caused by developmental elav-GS induction preclude such manipulations for studies of sleep, and that the Q-system may have broader utility for systematically defining the temporal windows in which genes impact sleep and other behaviors in Drosophila.
Materials and Methods
Stocks and transgenes
elav-GS (Osterwalder et al. 2001), UAS-dcr2 (Bloomington #24651; Dietzl et al. 2007), UAS-CD8-GFP (Bloomington #5137; Lee and Luo 1999), tub-QS (Potter et al. 2010), nsyb-Gal4QF (Riabinina et al. 2015), and UAS-inc-RNAi (VDRC 18225; Dietzl et al. 2007) were described previously. elav-GS in the iso31 background (Ryder et al. 2004) was described previously (Crocker and Sehgal 2008). A third chromosome insertion of nsyb-Gal4QF and two different second chromosome tub-QS insertions were each backcrossed eight generations to the iso31 background. After backcrossing, both tub-QS insertions were separately combined with nsyb-Gal4QF to yield tub-QS; nsyb-Gal4QF stocks. The elav-GS, UAS-dcr2 stock was obtained by meiotic recombination; two independently derived recombinants were verified by PCR and behaved similarly. UAS-CD8-GFP was used in its existing genetic background. All experiments utilized male animals bearing one copy of indicated transgenes.
Drosophila culture and conditional induction of Geneswitch and the Q-system
For all experiments, crosses were performed with five virgin females and three males and supplemented with yeast in standard fly vials (28.5 mm outer diameter × 95 mm height) and cultured at 25° in alternating 12 hr cycles of light and darkness (LD). Flies were cultured on food containing the following ingredients: 1800 g cornmeal (Labscientific, FLY-8010-20), 1800 ml molasses (Labscientific, FLY-8008-16), 744 g yeast (Labscientific, FLY-8040-20F), 266 g agar (Mooragar, 41084), 56 g Tegosept (Sigma, H3647), 560 ml alcohol (Fisher, A962P4), 190 ml propionic acid (Fisher, A258500), and 47 l of water. To prepare food for conditional induction experiments, solid food was melted in a microwave oven and cooled before addition of RU486, quinic acid, or appropriate vehicle.
For developmental (ON→OFF) Geneswitch induction, crosses were set on food containing RU486 (Sigma, M8046) at 11.6, 50, or 500 μM. One- to four-day-old young adults eclosing from these cultures were moved to vehicle-containing food (ethanol at a maximum 0.43% concentration) in Drosophila Activity Monitoring (DAM) tubes (Trikinetics) for behavioral assay. For constitutive (ON) Geneswitch induction, crosses were set on 11.6 μM RU486, and young adult progeny were moved to DAM tubes containing 500 µM RU486. For adult-specific Geneswitch induction (OFF→ON), crosses were set on food containing vehicle, and young adult progeny were moved to DAM tubes containing 500 µM RU486. For the negative control condition (OFF), crosses were set, and young adults were assayed, on vehicle-containing food.
Q-system induction was performed similarly, using food containing quinic acid or vehicle as dictated by the four different induction regimens. Quinic acid (Sigma, 138622) was freshly prepared by dissolving 1 g of quinic acid in 3 ml of water, and 330 μl of this solution was added per 10 ml of fly food. An equal volume of water was used for vehicle controls.
For assessing developmental toxicity of Geneswitch, vials were photographed and total numbers of pupae above the food surface were counted for all genotypes 10 or 11 d after setting crosses, as noted in the figure legends.
Sleep analysis
For measurements of sleep, 1–4-d-old male animals eclosing from LD-entrained cultures raised at 25° were loaded into glass DAM tubes (5 mm diameter × 65 mm length) containing food, and RU486, quinic acid, or appropriate vehicle as dictated by induction conditions. Animals were assayed for 7 d at 25° in LD cycles using DAM2 monitors (Trikinetics) and sleep was measured beginning 36–48 hr after animals were transferred to tubes, to permit acclimation to tubes and to allow feeding for conditional induction experiments. Locomotor data were collected in 1 min bins, and inactivity greater than 5 min (Shaw et al. 2000; Huber et al. 2004) was used to define sleep. Sleep parameters were analyzed with custom MATLAB (Mathworks) software (Stavropoulos and Young 2011). Dead animals were excluded from analysis by a combination of automated filtering and visual inspection of locomotor traces.
Statistical analysis
For analysis of RU486 toxicity, one-way ANOVA and Tukey-Kramer post hoc tests were used to compare within each genotype. For statistical analysis of total sleep, daytime sleep, and nighttime sleep, one-way ANOVA and Tukey–Kramer post hoc tests were used to compare between induction conditions for each genotype. For comparisons of sleep bout length, nonparametric Kruskal–Wallis tests followed by Dunn’s post hoc tests were used.
Immunohistochemistry
For preparation of larval brains, crosses of tub-QS; nsyb-Gal4QF and UAS-CD8-GFP animals were set on food containing quinic acid. Brains of third instar larvae were dissected in PBS, fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, and washed 3 × 15 min at room temperature in PBS containing 0.2% Triton X-100 (PBST). For preparation of adult brains, crosses of tub-QS; nsyb-Gal4QF and UAS-CD8-GFP animals were set on standard fly food. One- to four-day-old young adult males eclosing from these crosses were moved to food containing quinic acid for 1 wk, fixed with 4% paraformaldehyde in PBST at 4° for 3 hr, and washed 3 × 15 min in PBST at room temperature before brain dissection. Dissected larval and adult brains were blocked with 5% normal donkey serum (NDS) (Lampire Biological, 7332100) in PBST at room temperature for 30 min, and incubated in primary antibody cocktail overnight at 4°, followed by 3 × 15 min PBST washes at room temperature. Brains were subsequently incubated in secondary antibody cocktail overnight at 4°, washed 3 × 15 min at room temperature in PBST, and mounted on microscope slides in Vectashield (Vector Labs, H-1000). Antibody cocktails were prepared in 5% NDS in PBST. Primary antibodies were rabbit anti-GFP (Life Technologies, A11122) used at 1:500, and mouse nc82 anti-Bruchpilot (DSHB) used at 1:50. Secondary antibodies were Alexa 488 donkey anti-rabbit and Alexa 647 donkey anti-mouse (Life Technologies, A21206 and A31573), both used at 1:1000. Brains were imaged on a Zeiss LSM800 confocal microscope at 512 × 512 or 1024 × 1024 pixel resolution with 0.95 μM z-steps. Confocal images were processed in ImageJ by collapsing z-stacks into single images using maximum intensity projection.
Data availability
Fly stocks and locomotor data from this study are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Constitutive, developmental-specific, and adult-specific neuronal induction using Geneswitch and the Q-system
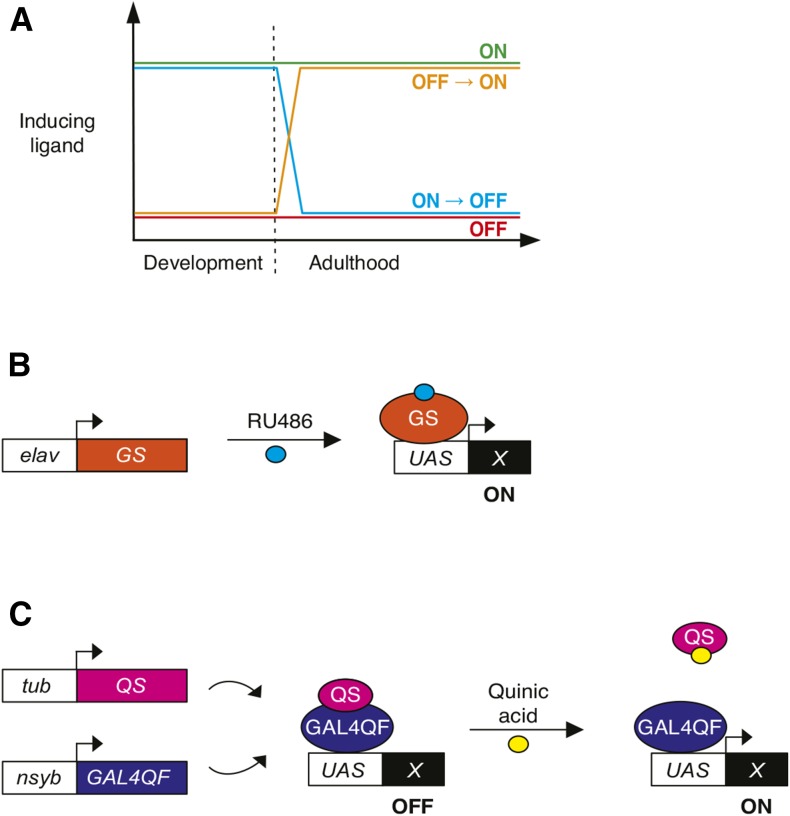
To determine whether Geneswitch and the Q-system can be used to assess developmental and adult contributions of genes relevant to sleep, we tested these systems in four different regimens of ligand exposure (Figure 1A). The nomenclature of these regimens indicates the presence or absence of inducing ligand during development and subsequently during adulthood and behavioral assay. In the noninduced control condition (OFF), animals developed on vehicle-containing food and were maintained in the absence of ligand throughout adulthood and behavioral assay. For adult-specific induction (OFF→ON), animals developed in the presence of vehicle but were moved as young adults to ligand-containing food for behavioral assay. For developmental-specific induction (ON→OFF), ligand was present during development and young adults were moved to vehicle-containing food for behavioral assay. Finally, for constitutive induction (ON), ligand was present throughout development, adulthood, and behavioral assay. Because many genes implicated in regulating sleep function within neurons, we used panneuronally expressed drivers for our conditional manipulations: elav-GS for the Geneswitch system (Figure 1B), and nsyb-Gal4QF for the Q-system (Figure 1C).
Figure 1.
Conditional neuronal manipulations using ligand-inducible systems. (A) Schematic depicting regimens for vehicle control (OFF), and constitutive (ON), developmental-specific (ON→OFF), and adult-specific (OFF→ON) induction by temporally restricted delivery of inducing ligands. See Materials and Methods for additional details. (B) The elav-GS transgene encodes a neuronally expressed, RU486-inducible form of the Gal4 activator, which activates a UAS-driven effector transgene (X) in the presence of RU486. (C) The Q-system is composed of the chimeric GAL4QF activator, the quinic acid-sensitive QS suppressor, and a UAS-driven effector transgene (X). Ubiquitous and constitutive tub-QS expression suppresses neuronally expressed GAL4QF. Quinic acid derepresses the UAS transgene.
Early RU486 exposure causes developmental defects in flies bearing elav-GS
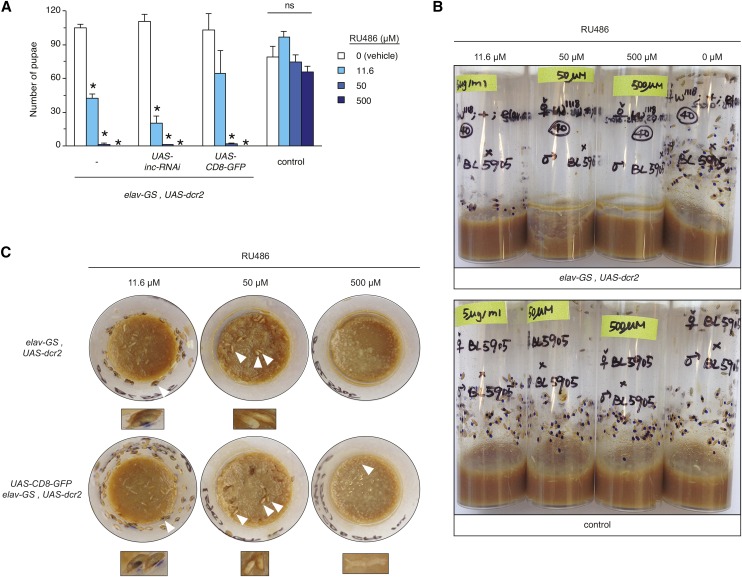
Conditional loss-of-function enables the temporal requirements of genes to be assessed with respect to sleep and can be achieved by driving RNAi with the Geneswitch system. We therefore first assessed animals which carry elav-GS and the UAS-dcr2 transgene expressing the Dcr2 ribonuclease that enhances RNAi (Dietzl et al. 2007), both of which are used routinely for studies of sleep (e.g., Yuan et al. 2006; Stavropoulos and Young 2011; Pfeiffenberger and Allada 2012; Ueno et al. 2012). To test whether continuous elav-GS induction is compatible with development and with behavioral assay of sleep, we set crosses on RU486 concentrations (11.6, 50, and 500 μM) that span a ∼50-fold range and encompass concentrations typically used in studies of sleep. While we observed similar numbers of pupae for elav-GS, UAS-dcr2/+ and isogenic w1118 control animals exposed developmentally to ethanol vehicle, the presence of RU486 had sharply contrasting effects on the two genotypes (Figure 2A). The number of control pupae was not significantly reduced in the presence of RU486 (Figure 2, A and B), indicating that these animals tolerate high levels of RU486 through pupal development. In contrast, the numbers of pupae bearing elav-GS were reduced in a dose-dependent manner when RU486 was present throughout development (Figure 2, A and B). Pupae bearing elav-GS failed to develop at 500 μM RU486, with abundant eggs or embryos and a lack of larvae suggesting lethality at early developmental stages (Figure 2, A–C). At 50 μM RU486, few elav-GS-bearing larvae and pupae were observed, and at 11.6 μM RU486, the number of pupae was less than half of that observed for vehicle controls (Figure 2, A–C). We observed similar developmental toxicity in elav-GS, UAS-dcr2/+ animals that also carried UAS-inc-RNAi or UAS-CD8-GFP transgenes, with >98% reductions in numbers of pupae for both genotypes at 500 and 50 μM RU486, and 82 and 37% reductions respectively at 11.6 μM RU486 (Figure 2, A and C). Similar developmental toxicity was observed when elav-GS was inherited maternally (Figure 2) or paternally (Supplemental Material, Figure S1), indicating that these effects are unlikely to reflect parent-specific contributions or reproductive deficits, and are instead likely to arise in progeny bearing the elav-GS driver.
Figure 2.
Developmental RU486 exposure is toxic to elav-GS animals. (A) Pupal number is shown for indicated genotypes exposed developmentally to vehicle or to indicated RU486 concentrations. Mean ± SEM is shown; * P < 0.01, and ns denotes P > 0.05, for comparisons to vehicle control within each genotype. Data are averaged from two independently derived elav-GS, UAS-dcr2 recombinant lines. (B and C) Side view (B) and top-down (C) photographs of vials containing progeny of indicated genotypes. Photographs were taken 11 d after crosses were initiated. Pupal cases are marked with blue dots to facilitate counting and to distinguish them from adults that have eclosed. In (C), white arrowheads indicate mature pigmented pupae on vial walls (11.6 µM RU486), immature unpigmented pupae located on the food surface (50 µM), and undeveloped eggs and embryos (500 µM). Magnifications are shown underneath top-down photographs.
In addition to reduced brood size, we observed additional developmental defects that were dependent on the presence of elav-GS and the dose of RU486. Experimental genotypes exposed to 50 µM RU486 exhibited developmental delays, as indicated by a lack of pupal pigmentation in comparison to animals exposed to 11.6 µM RU486, or to control animals scored at the same time point (Figure 2, B and C). Furthermore, elav-GS-bearing animals exposed to 50 µM RU486 pupated primarily on the surface of food rather than on vial walls (Figure 2, B and C). Control animals lacking elav-GS pupated normally when exposed developmentally to RU486 (Figure 2, A and B). Taken together, these findings strongly suggest that developmental elav-GS activation is toxic and that compromised neuronal function may underlie this toxicity.
Developmental induction of elav-GS causes strong and nonspecific alterations of sleep
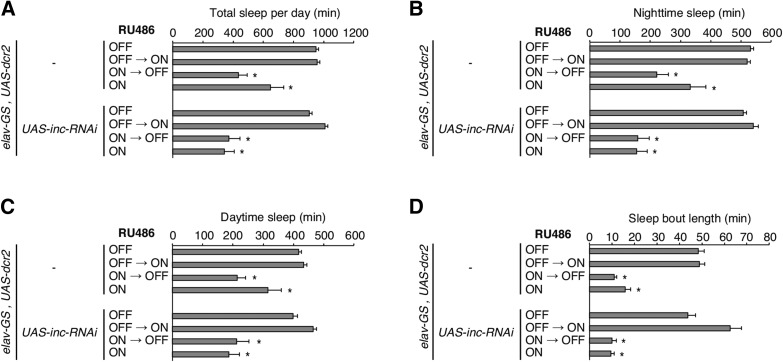
While developmental exposure to moderate and high concentrations of RU486 blocked development to adulthood, the lowest concentration we tested (11.6 μM) permitted eclosion of animals bearing elav-GS and additional UAS transgenes (Figure 2, A–C). We therefore used this concentration for developmental-specific induction (ON→OFF), and for the developmental portion of the constitutive induction (ON) regimen (Figure 1A); adult animals in the latter regimen were fed higher RU486 concentrations (500 µM) after eclosion as in prior studies. We assessed the four induction conditions in animals carrying elav-GS, UAS-dcr2, and an RNAi transgene directed against insomniac (inc), a gene we previously isolated in a chemical mutagenesis screen for short sleep mutants (Stavropoulos and Young 2011). inc null mutants exhibit severely curtailed sleep, and this phenotype is recapitulated in animals in which inc RNAi is driven by elav-Gal4, indicating that inc is required neuronally (Stavropoulos and Young 2011; Pfeiffenberger and Allada 2012).
Animals bearing elav-GS, UAS-dcr2, and UAS-inc-RNAi exhibited a severe reduction of sleep when they were exposed to RU486 specifically during development (ON→OFF) or constitutively (ON); no change in sleep was observed when RU486 was absent (OFF) or fed to adults (OFF→ON) (Figure 3A). These results are consistent with earlier findings, in which reduced sleep was reported for developmental-specific induction of inc RNAi using the same elav-GS driver (Pfeiffenberger and Allada 2012). Surprisingly, however, control animals lacking UAS-inc-RNAi showed similarly decreased sleep when these animals were exposed to RU486 developmentally (ON→OFF) or constitutively (ON), but not when RU486 was absent (OFF) or present only in adulthood (OFF→ON) (Figure 3A). In addition to exhibiting strong reductions in total sleep, both genotypes displayed qualitatively and quantitatively similar decreases in daytime sleep, nighttime sleep, and in sleep bout length after developmental (ON→OFF) or constitutive (ON) RU486 exposure (Figure 3, B–D). These results indicate that the reductions in sleep observed in animals carrying UAS-inc-RNAi, elav-GS, and UAS-dcr2 are unlikely to arise from the depletion of inc, but rather, from nonspecific alterations of sleep that are similarly observed in control animals lacking the UAS-inc-RNAi transgene.
Figure 3.
Developmental or continuous RU486 exposure reduces sleep in elav-GS adults. (A–D) Sleep parameters are shown for animals of indicated genotypes and regimens of RU486 exposure. Total sleep per day (A), nighttime sleep (B), daytime sleep (C), and sleep bout length (D) are plotted. Mean ± SEM is shown; n = 18–38; * P < 0.01 for comparisons to vehicle control condition within each genotype.
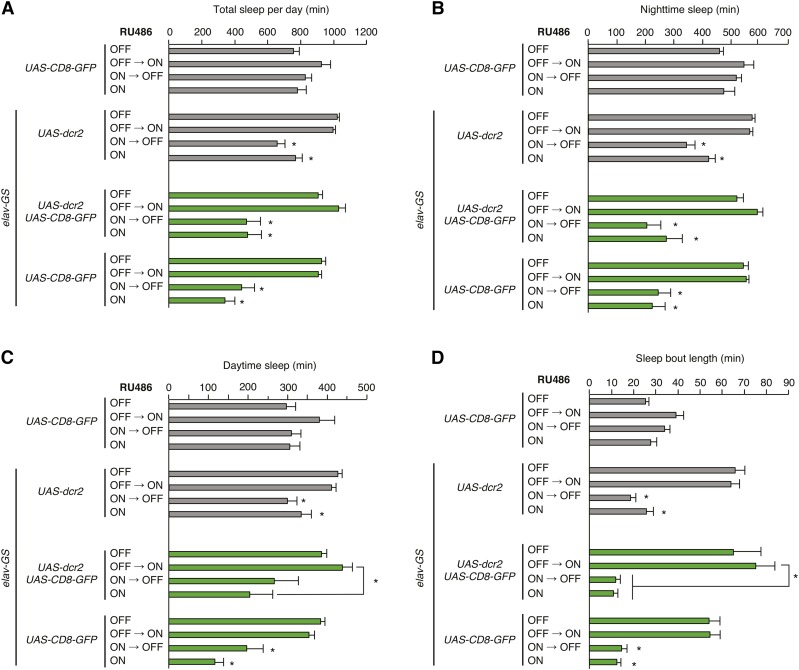
To further test whether elav-GS elicits nonspecific alterations in sleep when induced developmentally or constitutively, we assessed sleep in animals bearing elav-GS, UAS-dcr2, and UAS-CD8-GFP, a transgene unrelated to sleep. Surprisingly, we observed strong reductions in sleep when these animals were exposed to RU486 developmentally (ON→OFF) or constitutively (ON); animals exposed to vehicle (OFF) or to RU486 specifically in adulthood (OFF→ON) exhibited wild-type levels of sleep (Figure 4A). We performed additional experiments to assess whether reductions of sleep were dependent on the elav-GS, UAS-dcr2, and UAS-CD8-GFP transgenes. Perturbations of sleep were similarly severe in animals carrying only elav-GS and UAS-CD8-GFP, indicating that these effects do not depend on the presence of UAS-dcr2 (Figure 4A). Animals bearing only UAS-CD8-GFP showed no significant alterations of sleep in all four regimens of RU486 exposure, indicating that reductions in sleep require elav-GS (Figure 4A). All genotypes with decreased sleep exhibited strongly reduced sleep during the daytime and at night, as well as reduced sleep bout length (Figure 4, B–D). These findings indicate that developmental induction of elav-GS strongly and nonspecifically perturbs sleep. As was the case for developmental toxicity (Figure 2 and Figure S1), altered sleep in elav-GS animals exposed developmentally to RU486 was observed regardless of whether the elav-GS transgene was inherited maternally or paternally (Figure 4 and Figure S2). These data strongly suggest that sleep defects are caused by altered nervous system function as a consequence of developmental elav-GS activation.
Figure 4.
Perturbations of sleep elicited by developmental RU486 exposure are dependent on elav-GS and independent of effector transgenes. (A–D) Sleep parameters are shown for animals of indicated genotypes and regimens of RU486 exposure. Total sleep per day (A), nighttime sleep (B), daytime sleep (C), and sleep bout length (D) are plotted. Mean ± SEM is shown; n = 6–40; * P < 0.01 for comparisons to vehicle control condition within each genotype except where indicated.
In further experiments, we extended our analysis to additional genotypes carrying elav-GS and UAS-cDNA or UAS-RNAi transgenes. In total, we assayed ∼1800 animals representing 19 different genotypes in the four induction regimens described above. For all of these genotypes, we consistently observed strongly reduced sleep when animals bearing elav-GS were exposed to RU486 developmentally or constitutively (data not shown). These results strongly support the conclusion that these reductions of sleep are independent of particular effector transgenes and genetic background, and that they occur instead from developmental elav-GS activation. While our findings confirm that elav-GS and RU486 are well tolerated in adults and are compatible with adult-specific conditional manipulations of sleep, they indicate that developmental induction of elav-GS, even at low concentrations of RU486, alters sleep acutely and nonspecifically during adulthood, confounding the use of this driver for developmental manipulations of sleep.
Neuronal induction of the Q-system is broadly compatible with assessing sleep
We next sought to assess the suitability of the Q-system for conditional neuronal manipulations of sleep. For these experiments we used the panneuronally expressed Gal4QF activator (nsyb-Gal4QF), the ubiquitously expressed tubulin-QS suppressor (tub-QS), and quinic acid (Figure 1C). While quinic acid has no obvious toxicity in Drosophila (Potter et al. 2010), its effects on sleep have not been assessed. Similarly, animals bearing QF and QS transgenes have not been tested in the presence and absence of quinic acid to assess compatibility with behavioral assays of sleep.
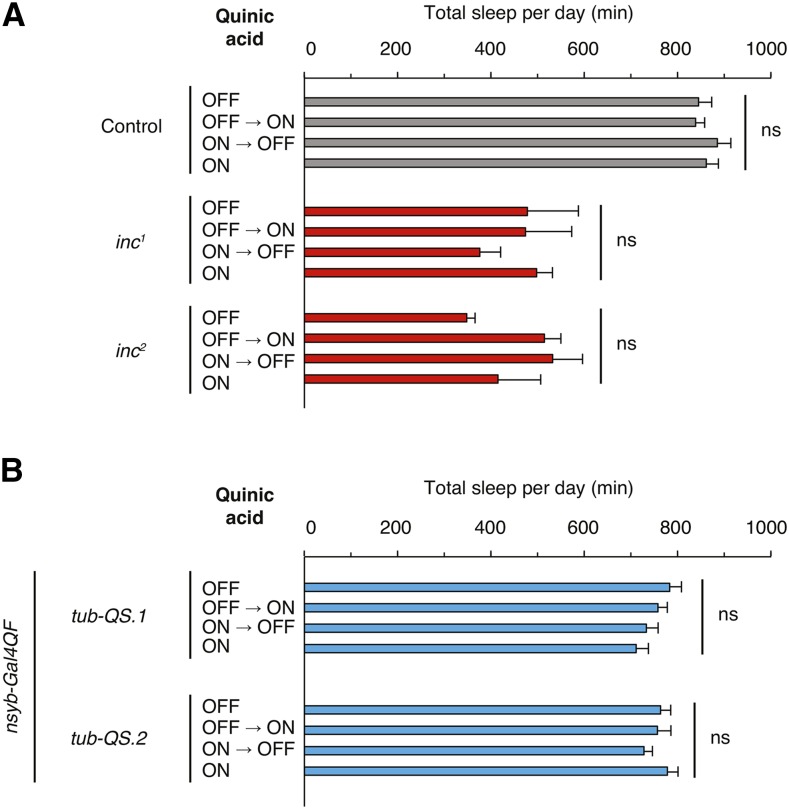
First, we assessed whether quinic acid exposure alters sleep in control animals lacking Q-system transgenes. In these animals, quinic acid exposure during development, adulthood, or both did not alter sleep with respect to vehicle control (Figure 5A). We next assessed whether quinic acid exposure alters the sleep of inc1 and inc2 animals, which bear null alleles of insomniac that strongly curtail sleep (Stavropoulos and Young 2011). Quinic acid supplied developmentally or during adulthood did not significantly change the short sleep phenotype of either mutant, indicating that quinic acid exposure does not perturb the levels of sleep in a short-sleeping genetic background (Figure 5A).
Figure 5.
The Q-system is compatible with assessing sleep. (A and B) Total sleep per day is plotted for animals of indicated genotypes and regimens of quinic acid exposure. Mean ± SEM is shown; n = 5–16 for (A); n = 22–24 for (B); ns denotes P > 0.05, for comparisons across induction conditions within each genotype.
Next, we tested animals carrying the nsyb-Gal4QF and tub-QS transgenes necessary for conditional, neuronally restricted Q-system manipulations. The presence of quinic acid during development, adulthood, or both, did not significantly alter sleep with respect to vehicle control (Figure 5B and Figure S3), and this was the case for two stocks bearing different insertions of the tub-QS transgene. These findings contrast with those obtained with neuronally expressed Geneswitch (Figure 3 and Figure 4), and indicate that persistent quinic acid exposure is well tolerated and does not alter sleep in the presence of Q-system components (Figure 5, A and B). Furthermore, quinic acid exposure does not cause any obvious developmental abnormalities (data not shown).
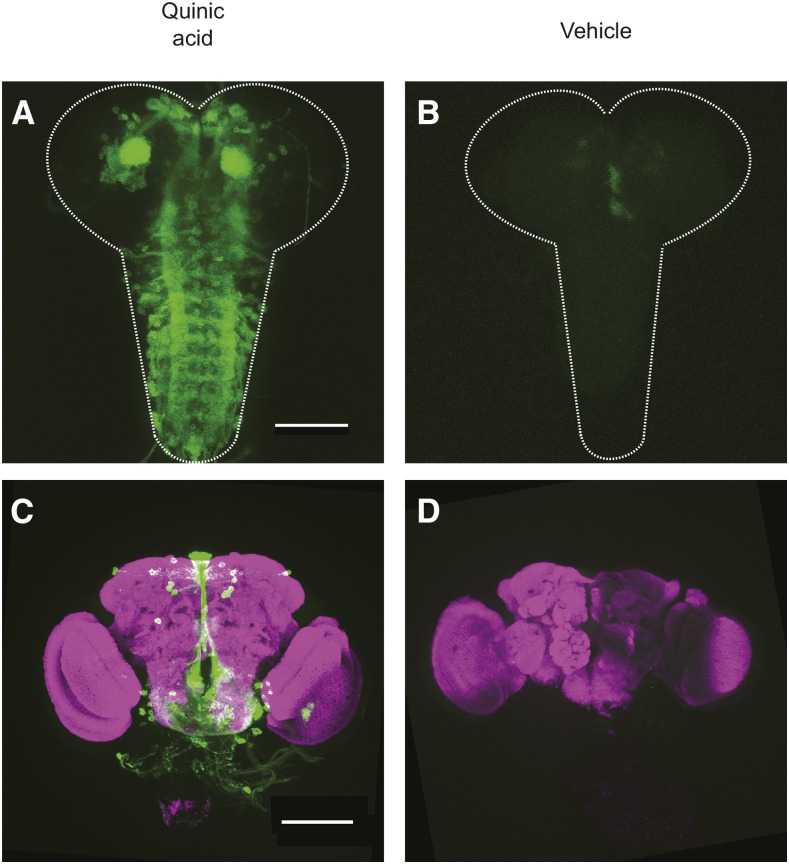
To test whether conditions compatible with assessing sleep enable conditional induction of transgenes with the Q-system, we assessed expression of a UAS-CD8-GFP reporter in animals bearing nsyb-Gal4QF and tub-QS and exposed to quinic acid developmentally or during adulthood. Developmental exposure to quinic acid elicited UAS-CD8-GFP expression, as assessed in the brains of third instar larvae (Figure 6A). Animals developing in the presence of vehicle exhibited little to no GFP signal (Figure 6B). Similarly, the brains of adult animals exposed to quinic acid in adulthood exhibited GFP signal (Figure 6C) similar to that reported previously (Riabinina et al. 2015), while vehicle controls exhibited no signal (Figure 6D). While the kinetics and efficacy of Q-system induction are likely to vary with each effector transgene, the conditional induction of GFP expression and the compatibility of inducing conditions with behavioral analysis of sleep (Figure 5) suggest that the Q-system has considerable potential for dissecting the temporal contributions of genes that impact sleep.
Figure 6.
Conditional neuronal induction of a GFP reporter with the Q-system. (A and B) Anti-GFP signal (green) in third instar larval brains prepared from tub-QS/+; nsyb-Gal4QF/UAS-CD8-GFP animals exposed developmentally to (A) quinic acid or (B) vehicle. (C and D) anti-GFP (green) and anti-Bruchpilot (magenta) signal in adult brains prepared from tub-QS/+; nsyb-Gal4QF/UAS-CD8-GFP animals exposed during adulthood to (C) quinic acid or (D) vehicle. Scale bars represent 100 µm.
Discussion
Conditional genetic manipulations provide important information for defining the temporal window in which genes exert their phenotypic consequences. For genes implicated in regulating sleep, temporal requirements vis-à-vis sleep have been assessed chiefly in an adult-specific manner using the Geneswitch system. While our results confirm that the panneuronally expressed elav-GS transgene is compatible with adult-specific manipulations as reported previously (Yuan et al. 2006; Joiner et al. 2006; Seugnet et al. 2008, 2011; Bushey et al. 2009; Donlea et al. 2009; Wu et al. 2009; Kuo et al. 2010, 2012; Crocker et al. 2010; Ishimoto and Kitamoto 2010; Pfeiffenberger and Allada 2012; Erion et al. 2012; Ueno et al. 2012; Vanderheyden et al. 2013; Tulina et al. 2014; Liu et al. 2014; Kayser et al. 2014; Oh et al. 2014; Chen et al. 2015; Tabuchi et al. 2015; Dissel et al. 2015; Afonso et al. 2015), they indicate that developmental elav-GS induction elicits strong, nonspecific alterations of sleep later in adulthood. Notably, these sleep deficits occur in animals exposed to low concentrations (11.6 µM) of RU486, nearly 50-fold below those typically used for adult manipulations (500 µM). Transcriptome profiling studies have identified genes whose expression is altered by RU486 exposure (Etter et al. 2005; Landis et al. 2015), but whether these changes underlie elav-GS-dependent alterations of sleep or mediate developmental toxicity awaits further investigation.
While many studies of sleep have employed adult-specific elav-GS induction, few have reported developmental manipulations, despite the obvious value of such manipulations for assessing the contributions that genes may make outside of adulthood. We suspect that this relative dearth of studies reflects complications of developmental induction that have been observed but not widely reported. This notion is supported by both published and unpublished findings that suggest a threshold for developmental toxicity of RU486 in elav-GS animals similar to what we have observed. In one study, developmental lethality was reported above 25 µM RU486 for animals bearing elav-GS and effector transgenes (van Swinderen 2007). In unpublished findings that parallel our results, concentrations above 50 µM RU486 were found to be developmentally lethal to elav-GS animals; at 50 µM RU486, some animals were observed to eclose, but viability was reduced (W. Joiner, personal communication). In other unpublished studies, developmental lethality and alterations of sleep in some genotypes bearing elav-GS were observed with developmental exposure to 25 µM RU486 (J. Williams, personal communication). Though our results suggest that developmental toxicity in elav-GS animals is significantly reduced at 11.6 µM RU486, a concentration permissive for the development of animals carrying ∼200 different larvally expressed Geneswitch transgenes (Nicholson et al. 2008), nonspecific alterations of sleep persist at this concentration across a wide range of genotypes.
Our findings suggest that developmental manipulations using elav-GS need to be interpreted cautiously in the context of assessing sleep, and that some earlier results may need to be revisited. While our findings do not preclude developmental functions of inc and Cul3 with respect to sleep, the interpretational difficulties associated with developmental elav-GS induction suggest that conditional systems other than Geneswitch may be required to resolve the temporal requirements of inc and Cul3 conclusively. Our results emphasize the necessity of validating the kinetics and efficacy of conditional manipulations in parallel to behavioral assays, in order to interpret phenotypes of these manipulations. This point is underscored by the finding that Geneswitch activation depends upon the age and sex of animals, and that some Geneswitch drivers are expressed in unanticipated locations or in the absence of RU486 inducer (Poirier et al. 2008). Finally, we note that sleep is a behavior sensitive to a number of factors, and that genetic background, food composition, or environmental conditions may play a role in the developmental toxicity and nonspecific alterations of sleep that result from elav-GS activation during development.
More broadly, our findings are relevant for conditional neuronal manipulations of other behaviors using ligand-inducible expression systems. Developmental activation of elav-GS may cause persistent changes in the structure or activity of the nervous system and perturb other behaviors nonspecifically, potentially limiting the use of this driver to adulthood. While the Q-system requires validation in additional behavioral contexts, our findings suggest that it may have broader potential for conditional neuronal manipulations throughout the Drosophila life cycle and for systematically defining the temporal contributions of genes that underlie various behaviors.
Supplementary Material
Acknowledgments
We thank S. Harbison for communicating similar unpublished findings with Geneswitch; W. Joiner, K. Koh, B. White, and J. Williams for sharing unpublished observations regarding elav-GS animals; J. Tower for discussions of Geneswitch-dependent alterations in longevity; C. Potter and O. Riabinina for sharing information regarding the Q-system prior to publication; C. Potter for stocks; and N. Ringstad, M. Shirasu-Hiza, G. Suh, and members of the Stavropoulos lab for comments on the manuscript. This research was supported by an International Student Research Fellowship from the Howard Hughes Medical Institute (Q.L.) and by grants from the Whitehall, Alfred P. Sloan, Leon Levy, and Mathers Foundations, a National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Award from the Brain and Behavior Research Foundation, a New York University Whitehead Fellowship, and the J. Christian Gillin, M.D. Research Award from the Sleep Research Society Foundation (N.S.).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.034132/-/DC1.
Communicating editor: J. C. Dunlap
Literature Cited
- Afonso D. J. S., Liu D., Machado D. R., Pan H., Jepson J. E. C., et al. , 2015. TARANIS functions with Cyclin A and Cdk1 in a novel arousal center to control sleep in Drosophila. Curr. Biol. 25: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B., Resendez-Perez D., Gehring W. J., 1998. Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system. Development 125: 2193–2202. [DOI] [PubMed] [Google Scholar]
- Bieschke E. T., Wheeler J. C., Tower J., 1998. Doxycycline-induced transgene expression during Drosophila development and aging. Mol. Gen. Genet. 258: 571–579. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Manoukian A. S., Perrimon N., 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44: 635–654. [DOI] [PubMed] [Google Scholar]
- Burcin M. M., Schiedner G., Kochanek S., Tsai S. Y., O’Malley B. W., 1999. Adenovirus-mediated regulable target gene expression in vivo. Proc. Natl. Acad. Sci. USA 96: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C., 2009. The Drosophila fragile X mental retardation gene regulates sleep need. J. Neurosci. 29: 1948–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-B., Alekseyenko O. V., Kravitz E. A., 2015. Optogenetic control of gene expression in Drosophila. PLoS One 10: e0138181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-F., Maguire S., Sowcik M., Luo W., Koh K., et al. , 2015. A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol. Psychiatry 20: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Bushey D., Hill S., Huber R., Kreber R., et al. , 2005. Reduced sleep in Drosophila Shaker mutants. Nature 434: 1087–1092. [DOI] [PubMed] [Google Scholar]
- Crocker A., Sehgal A., 2008. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J. Neurosci. 28: 9377–9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A., Shahidullah M., Levitan I. B., Sehgal A., 2010. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65: 670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Dissel S., Angadi V., Kirszenblat L., Suzuki Y., Donlea J., et al. , 2015. Sleep restores behavioral plasticity to Drosophila mutants. Curr. Biol. 25: 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Ramanan N., Shaw P. J., 2009. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion R., DiAngelo J. R., Crocker A., Sehgal A., 2012. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. J. Biol. Chem. 287: 32406–32414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter P. D., Narayanan R., Navratilova Z., Patel C., Bohmann D., et al. , 2005. Synaptic and genomic responses to JNK and AP-1 signaling in Drosophila neurons. BMC Neurosci. 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks J. C., Finn S. M., Panckeri K. A., Chavkin J., Williams J. A., et al. , 2000. Rest in Drosophila is a sleep-like state. Neuron 25: 129–138. [DOI] [PubMed] [Google Scholar]
- Huber R., Hill S. L., Holladay C., Biesiadecki M., Tononi G., et al. , 2004. Sleep homeostasis in Drosophila melanogaster. Sleep 27: 628–639. [DOI] [PubMed] [Google Scholar]
- Ishimoto H., Kitamoto T., 2010. The steroid molting hormone Ecdysone regulates sleep in adult Drosophila melanogaster. Genetics 185: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J., Crocker A., White B. H., Sehgal A., 2006. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441: 757–760. [DOI] [PubMed] [Google Scholar]
- Kayser M. S., Yue Z., Sehgal A., 2014. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344: 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Joiner W. J., Wu M. N., Yue Z., Smith C. J., et al. , 2008. Identification of SLEEPLESS, a sleep-promoting factor. Science 321: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K., Kume S., Park S. K., Hirsh J., Jackson F. R., 2005. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S.-Y., Tu C.-H., Hsu Y.-T., Wang H.-D., Wen R.-K., et al. , 2012. A hormone receptor-based transactivator bridges different binary systems to precisely control spatial-temporal gene expression in Drosophila. PLoS One 7: e50855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.-H., Pike D. H., Beizaeipour Z., Williams J. A., 2010. Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NFkappaB Relish. BMC Neurosci. 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G. N., Salomon M. P., Keroles D., Brookes N., Sekimura T., et al. , 2015. The progesterone antagonist mifepristone/RU486 blocks the negative effect on life span caused by mating in female Drosophila. Aging (Albany, N.Y.) 7: 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Lis J. T., Simon J. A., Sutton C. A., 1983. New heat shock puffs and beta-galactosidase activity resulting from transformation of Drosophila with an hsp70-lacZ hybrid gene. Cell 35: 403–410. [DOI] [PubMed] [Google Scholar]
- Liu S., Lamaze A., Liu Q., Tabuchi M., Yang Y., et al. , 2014. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768. [DOI] [PubMed] [Google Scholar]
- Nicholson L., Singh G. K., Osterwalder T., Roman G. W., Davis R. L., et al. , 2008. Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics 178: 215–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Yoon S.-E., Zhang Q., Chae H.-S., Daubnerová I., et al. , 2014. A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS Biol. 12: e1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T., Yoon K. S., White B. H., Keshishian H., 2001. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98: 12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C., Allada R., 2012. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet. 8: e1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier L., Shane A., Zheng J., Seroude L., 2008. Characterization of the Drosophila Gene-Switch system in aging studies: a cautionary tale. Aging Cell 7: 758–770. [DOI] [PubMed] [Google Scholar]
- Potter C. J., Tasic B., Russler E. V., Liang L., Luo L., 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141: 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M. N., et al. , 2015. Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12: 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G., Endo K., Zong L., Davis R. L., 2001. P{Switch}, a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98: 12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Vine L., Gottschalk L., Shaw P. J., 2008. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr. Biol. 18: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Merlin G., Gottschalk L., Duntley S. P., et al. , 2011. Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr. Biol. 21: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J., Tononi G., 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287: 1834–1837. [DOI] [PubMed] [Google Scholar]
- Shen J., Curtis C., Tavaré S., Tower J., 2009. A screen of apoptosis and senescence regulatory genes for life span effects when over-expressed in Drosophila. Aging (Albany, N.Y.) 1: 191–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N., Young M. W., 2011. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72: 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins M. J., Urlinger S., Byrne G., Bello B., Hillen W., et al. , 2001. Tetracycline-inducible systems for Drosophila. Proc. Natl. Acad. Sci. USA 98: 10775–10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M., Lone S. R., Liu S., Liu Q., Zhang J., et al. , 2015. Sleep interacts with Aβ to modulate intrinsic neuronal excitability. Curr. Biol. 25: 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N. M., Chen W.-F., Chen J. H., Sowcik M., Sehgal A., 2014. Day-night cycles and the sleep-promoting factor, Sleepless, affect stem cell activity in the Drosophila testis. Proc. Natl. Acad. Sci. USA 111: 3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Tomita J., Tanimoto H., Endo K., Ito K., et al. , 2012. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci. 15: 1516–1523. [DOI] [PubMed] [Google Scholar]
- Vanderheyden W. M., Gerstner J. R., Tanenhaus A., Yin J. C., Shaw P. J., 2013. ERK phosphorylation regulates sleep and plasticity in Drosophila. PLoS One 8: e81554.24244744 [Google Scholar]
- van Swinderen B., 2007. Attention-like processes in Drosophila require short-term memory genes. Science 315: 1590–1593. [DOI] [PubMed] [Google Scholar]
- Wu M. N., Ho K., Crocker A., Yue Z., Koh K., et al. , 2009. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J. Neurosci. 29: 11029–11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Joiner W. J., Sehgal A., 2006. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16: 1051–1062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fly stocks and locomotor data from this study are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.