Abstract
To investigate the genetic diversity of the Plasmodium falciparum apical membrane antigen 1 (PfAMA1) gene in Southeast Asia, we determined PfAMA1 sequences from 135 field isolates collected from the China-Myanmar border area and compared them with 956 publically available PfAMA1 sequences from seven global P. falciparum populations. This analysis revealed high genetic diversity of PfAMA1 in global P. falciparum populations with a total of 229 haplotypes identified. The genetic diversity of PfAMA1 gene from the China-Myanmar border is not evenly distributed in the different domains of this gene. Sequence diversity in PfAMA1 from the China-Myanmar border is lower than that observed in Thai, African and Oceanian populations, but higher than that in the South American population. This appeared to correlate well with the levels of endemicity of different malaria-endemic regions, where hyperendemic regions favor genetic cross of the parasite isolates and generation of higher genetic diversity. Neutrality tests show significant departure from neutrality in the entire ectodomain and Domain I of PfAMA1 in the China-Myanmar border parasite population. We found evidence supporting a substantial continent-wise genetic structure among P. falciparum populations, with the highest genetic differentiation detected between the China-Myanmar border and the South American populations. Whereas no alleles were unique to a specific region, there were considerable geographical differences in major alleles and their frequencies, highlighting further necessity to include more PfAMA1 alleles in vaccine designs.
Keywords: Plasmodium falciparum, PfAMA1, genetic diversity, China-Myanmar border, malaria
1. Introduction
Plasmodium falciparum is the most deadly of five Plasmodium species that infect humans, with an estimated 584,000 deaths (90% from sub-Saharan Africa and 78% children < 5 years of age) in 2013 (WHO, 2014). Increasing drug resistance of the parasites and insecticide resistance of the vector mosquitoes have made malaria control difficult, and there is a strong demand for an effective vaccine to contain this deadly disease. However, antigen polymorphism of many asexual stage vaccine candidates such as merozoite surface protein 1 (MSP1), MSP2, and MSP3 (Jordan et al., 2009; Koukouikila-Koussounda et al., 2012; Sakihama et al., 2001) hamper the development of vaccines effective against all parasite populations. Thus, successful interventions targeting different developmental stages will require better understanding of genetic variations of target antigens within and between parasite populations.
Apical membrane antigen 1 (AMA1) is conserved in apicomplexans, and P. falciparum apical membrane antigen-1 (PfAMA1) is one of the leading blood stage vaccine candidates in human trials (Dutta et al., 2003; Kocken et al., 2002; Mitchell et al., 2004). PfAMA1 is an 83 kDa antigen synthesized by mature blood stages of the parasite and is initially localized in the micronemes of the merozoite, an apical organelle that plays a key role in erythrocyte invasion (Healer et al., 2002; Narum and Thomas, 1994; Peterson et al., 1989). Prior to merozoite invasion, PfAMA1 is processed into a 66 kDa product and released onto the merozoite surface, where they participate in the formation of tight junctions by interacting with the RON proteins and transducing the force generated by the parasite motor during internalization (Bargieri et al., 2012; Cao et al., 2009; Richard et al., 2010). The complete 1686 bp PfAMA1 coding region contains an ectodomain with three sub-domains (Domain I – III) (Silvie et al., 2004). A higher rate of mutations and level of diversifying selection have been shown in Domain I (Cortes et al., 2003; Figtree et al., 2000; Garg et al., 2007; Thakur et al., 2008). However, Domain II presents a high-degree of amino acid sequence conservation. A loop region within this domain has been shown to contain an epitope recognized by an invasion-inhibitory monoclonal antibody (Chesne-Seck et al., 2005).
The ectodomain of AMA1 are highly immunogenic. Antibodies raised against AMA1 inhibit merozoite invasion in vitro (Crewther et al., 1996; Healer et al., 2004; Hodder et al., 2001; Kennedy et al., 2002; Kocken et al., 2002; Kusi et al., 2009; Remarque et al., 2008). Immunization with recombinant AMA1 has been shown to elicit protective antibody response against homologous strain challenges in both rodent and primate models, but the response is less protective against a heterologous parasite line (Healer et al., 2004; Remarque et al., 2008). Antibodies to AMA1 are typically highly prevalent amongst malaria endemic populations (Thomas et al., 1994). A systematic meta-analysis of data that met rigorous quality criteria detected a tendency towards a protective association amongst studies of AMA1 (Fowkes et al., 2010). These results support the development of AMA1 as a malaria vaccine, but also highlight the need to better understand genetic diversity of PfAMA1.
The Greater Mekong Subregion (GMS) is one of the most threatening foci of malaria in Southeast Asia (Cui et al., 2012). The recent emergence of artemisinin resistant P. falciparum parasites has posed a serious problem for both regional and global malaria control (Dondorp et al., 2011). Within the GMS, more than half of the malaria cases and an estimated 75% of the malaria deaths occurred in Myanmar (Wang et al., 2015). Moreover, international border regions, such as the one shared by China and Myanmar have the highest malaria incidence in the GMS (Cui et al., 2012). Border malaria is extremely difficult to monitor and control, largely due to frequent human population movements across the porous international borders. In the present study, we investigated the genetic diversity in the ectodomain of PfAMA1 gene in the China-Myanmar border area. By comparing with global P. falciparum populations, we identified important differences in the PfAMA1 alleles and their frequencies, which bear important implications for the design of PfAMA1 based vaccines.
2. Materials and methods
2.1. Sample collection
A total of 171 field isolates were collected by finger-prick from patients attending malaria clinics in northwest Myanmar along the China-Myanmar border between April 2011 and October 2013. Blood samples were confirmed for P. falciparum infections based on microscopy of Giemsa-stained thick smears (Li et al., 2013). Use of the samples for this study was approved by the Biomedical Research Ethics Review Board of China Medical University.
2.2. DNA isolation, PCR amplification and sequencing of the PfAMA1 gene
Plasmodium DNA was extracted from filter papers or whole blood using QIAamp DNA Blood Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Parasites were genotyped at three polymorphic loci to exclude multi-clonal infections (Meng et al., 2010). Extracellular coding region contain Domain I–III (DI–III) of PfAMA1 were amplified using the following primer pairs: Pfama1_F (5′-GAAGTTCATGGTTCAGGTATAAG-3′), Pfama1_R (5′-GTATGGTTTTTCCATCAGAACTGG-3′), Pfama1_NF (5′-GATGCTGAAGTAGCTGGAACTC-3′), and Pfama1_NR (5′-GTGATGCTCTTTTTTCTTCCCCCC-3′). The PCR reaction contained 2 µl of 10×KOD-Plus-Neo buffer, 2 µl of 2 mM dNTPs, 0.8 µl of 25 mM MgSO4, 0.5 µl of 10 µM of each primer, 0.5 units of KOD-Plus-Neo DNA polymerase (Toyobo, Osaka, Japan), and 1.0 µl genomic DNA template in a final volume of 20 µl. The cycling parameters for the primary PCR were as follows: initial denaturation at 94°C for 2 min, 45 cycles of 94°C for 15 sec, 56°C for 15 sec, and 68°C for 90 sec, and a final extension at 68°C for 5 min. The amplification conditions for nested PCR were the same as those for primary PCR except that the number of cycles was 35 cycles. Nested PCR products were agarose gel purified using the QIAquick Gel Extraction Kit (QIAGEN, CA, USA), as per manufacturer protocol for preparation of templates for sequencing. Purified PCR products were sequenced with primers Pfama1_NF and Pfama1_NR in both directions using the ABI Prism® BigDye™ cycle sequencing kit (Applied Biosystems, Foster City, CA, USA).
2.3. Sequence alignment, polymorphism and statistical analyses
Of the 141 P. falciparum infections amplified by PCR, PfAMA1 was successfully sequenced from 135 samples (sequences submitted to GenBank under accession nos: KT897327-KT897461). To evaluate the polymorphism of PfAMA1, the AMA1 gene (PF3D7_1133400) from the P. falciparum 3D7 strain was used as the reference gene. A single contiguous 1347 bp of PfAMA1 (nucleotide region 445–1791 and codons 149–597) was derived for each of the 135 PfAMA1 sequences, which include DI (codons 149–302), DII (codons 320–418) and DIII (codons 443–509). These sequences were aligned using the CLUSTAL W program in MEGA6.0 (Tamura et al., 2013) and exported as a FASTA alignment for statistical analysis using the DnaSP5.10.01 software (Librado and Rozas, 2009). The number of segregating (polymorphic) sites (S), nucleotide diversity (π), the average number of nucleotide differences (k), the number of haplotypes (H) and haplotype diversity (Hd) were calculated by DnaSP (Librado and Rozas, 2009). The distribution of nucleotide diversity (π) across the DI–III of AMA1 gene was analyzed using the sliding window method with a window size of 90 bp and a step size of 3 bp.
2.4. Haplotype network construction, linkage disequilibrium (LD) and Fst analysis
The haplotype network was constructed using the program NETWORK Version 4.6.1.3 with the Median-Joining method (Bandelt et al., 1999). A total of 956 publically available PfAMA1 sequences representing seven P. falciparum populations were retrieved from GenBank: Thailand (Polley et al., 2003), Papua New Guinea (Arnott et al., 2014), Gambia (Tetteh et al., 2009), Nigeria (Polley and Conway, 2001), Kenya (Osier et al., 2010), Mali (Takala et al., 2009), and Venezuela (Ord et al., 2008). Analysis of the minimum number of recombination events (RM) (Hudson and Kaplan, 1985), recombination parameter C (C = 4Nr, where N is the effective population size and r is the probability of recombination between adjacent nucleotides per generation) (Hudson, 1987), as well as linkage disequilibrium (LD) using the indices D’ (Lewontin, 1964) and R2 (Hill and Robertson, 1968) were calculated by DnaSP (Librado and Rozas, 2009). To assess the proportion of genetic variance due to population subdivision, the inter-population variance in allele frequency at the PfAMA1 locus was compared among populations by calculating the Wright’s Fst using DnaSP (Librado and Rozas, 2009).
2.5. Molecular evolutionary analysis
To test deviation from neutral evolution, the ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS) were calculated using the Nei and Gojobori method (Nei and Gojobori, 1986) with the Jukes and Cantor correction and were compared with the Z-test of selection (P < 0.05) in MEGA6.0 (Tamura et al., 2013). A significant excess of dN relative to dS indicates positive natural selection, whereas negative values indicate negative or purifying selection (Nei and Gojobori, 1986). Departure from neutrality was estimated by Tajima’s D test, Fu and Li’s D* and F* tests and McDonald-Kreitman test. In Tajima’s D test, departure from neutrality in the nucleotide frequency distributions is determined by divergence in the values of π (observed average pairwise nucleotide diversity) and θ (expected nucleotide diversity under neutrality derived from the number of segregating sites (S) (Tajima, 1989). Fu and Li’s D* and F* tests reflect the same trends as Tajima’s D test and evaluate departure from neutrality by comparing the number of mutations in the external (considered to be "new" mutations) and internal (considered to be "older" mutations) branches of the genealogy. The number of mutations in external phylogeny branches was estimated from singleton sites (S) and the total number of mutations giving the D* index, or between S and η giving the F* index (Fu and Li, 1993). A positive value of Tajima’s D and Fu and Li’s D* and F* tests corresponds to positive natural selection, whereas a negative value indicating population size expansion and/or negative/purifying selection. Sliding window plots with a window size of 90 bp and a step size of 3 bp were also performed for Tajima’s D and Fu and Li’s D* and F* tests by DnaSP5.10.01 (Librado and Rozas, 2009). McDonald-Kreitman test (McDonald and Kreitman, 1991) was applied by taking a single P. reichenowi AMA1 sequence as the outgroup (accession no. AJ252087; (Kocken et al., 2000)) using DnaSP. The ratio of interspecies fixed synonymous substitutions to nonsynonymous substitutions is compared to that of intraspecific synonymous substitutions to nonsynonymous substitutions. An excess of the ratio between species over within species suggests purifying selection. Two-tailed Fisher’s exact test was computed to determine the statistical significance (P < 0.05).
3. Results
3.1. Sequence diversity of the PfAMA1 gene among the China-Myanmar border isolates
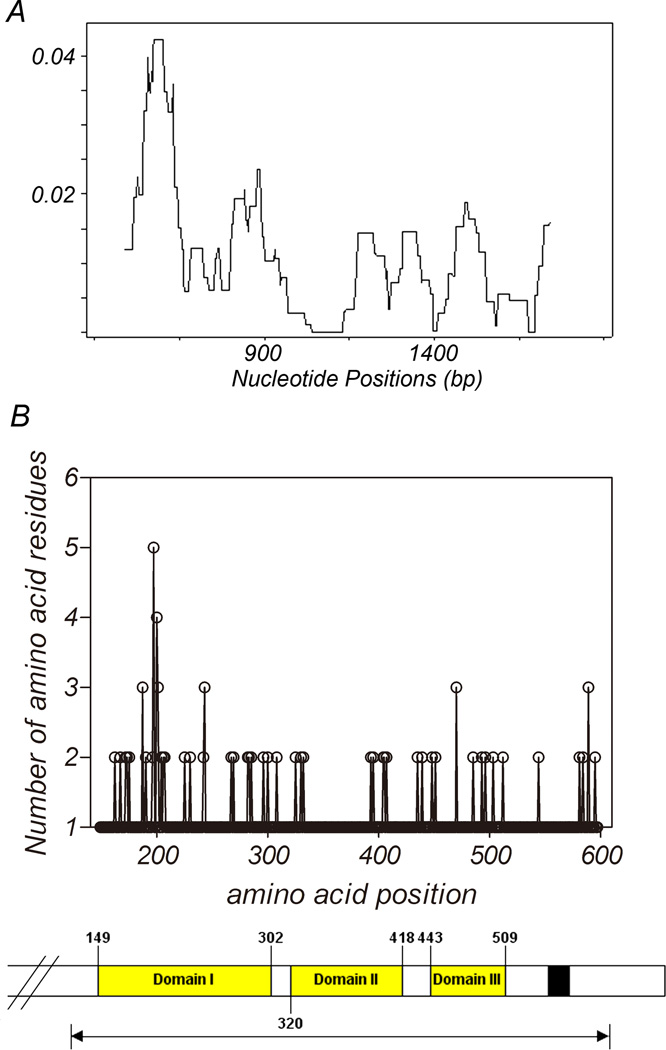
For the 135 PfAMA1 sequences obtained from the China-Myanmar border isolates, the average number of nucleotide differences (k) for the entire 1347 bp sequenced region, DI, DII, and DIII were 15.15, 8.16, 1.55, and 2.06, respectively (Table 1). A total of 34 distinct haplotypes (H) were identified. The haplotype diversity (Hd) was relatively high, with an average of 0.859 in the entire sequenced region of all isolates (Table 1). Within the entire sequenced region (codons 149–597), there were 57 polymorphic sites. These polymorphic sites are distributed unevenly within the sequenced region, with 31, 7 and 8 being located in DI, were DII, and DIII, respectively (Table 1). This is further illustrated in a sliding window plot of π across the sequenced region (Fig. 1A). Likewise, the pairwise nucleotide diversity (π) is highest in DI (π= 0.018), and lower in DII (π= 0.005) and DIII (π= 0.010). Also illustrated by the locations of the substitutions (Fig. 1B), DI (nt 445–906 and aa 149–302) is more polymorphic than DII (nt 958–1254 and aa 320–418) and DIII (nt 1327–1527 and aa 443–509).
Table 1.
Estimation of nucleotide diversity and summary statistics of PfAMA1 in 135 P. falciparum isolates from the China-Myanmar border area§.
| S | k | π±SD | H | Hd±SD | dN/dS | D | D* | F* | MK | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total | 57 | 15.157 | 0.011±0.001 | 34 | 0.859±0.025 | 16.459*** | 1.198 | 1.911** | 1.929* | 0.000*** |
| D I | 31 | 8.167 | 0.018±0.001 | 20 | 0.752±0.036 | 18.909** | 1.058 | 2.030** | 1.964* | 0.008** |
| D II | 7 | 1.550 | 0.005±0.000 | 9 | 0.705±0.037 | - | 0.477 | 1.171 | 1.107 | 0.417 |
| D III | 8 | 2.058 | 0.010±0.000 | 11 | 0.699±0.037 | - | 0.605 | −0.271 | 0.045 | 0.308 |
The total sequenced region includes codons 149 to 597, Domain I codons 149 to 302, Domain II codons 320 to 418, and Domain III codons 443 to 509 (Hodder et al., 1996). S, number of polymorphic (segregating) sites; k; the average number of nucleotide differences, π, pairwise nucleotide diversity; H, number of haplotypes; Hd, haplotype diversity; dN/dS, the ratio of non-synonymous to synonymous mutations; D, Tajima’s D test; D*, Fu and Li’s D* value; F*, Fu and Li’s F* value; MK, Mcdonald-Kreitman Test.
; P< 0.05,
; P< 0.02,
; P< 0.001.
SD = Standard deviation.
Fig. 1.
Sliding window plot of nucleotide diversity and amino acid polymorphism of PfAMA1 ectodomain in China-Myanmar border isolates. Nucleotide diversity is plotted with a window length of 90 bp and step size of 3 bp (A) and the number of the amino acid residues at each amino acid position is plotted, also to visualize the location sites showing high diversity, a scheme of the ectodomain of ama1 is shown below (B). Black arrow indicates analyzed regions. A total of 135 sequences from China-Myanmar border are used. Nucleotide and amino acid positions are after the 3D7 line sequences.
3.2. Recombination and LD
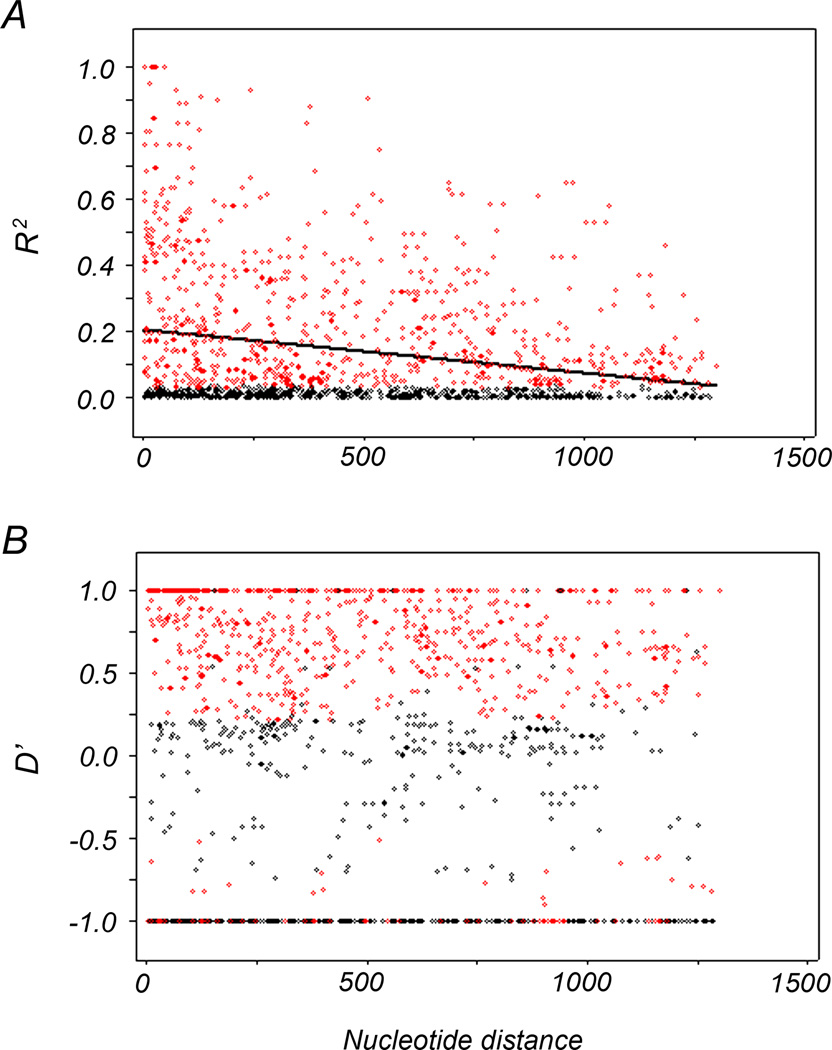
We compared the recombination and LD of PfAMA1 between populations from the China-Myanmar border area and other geographic regions. A minimum of 16 recombination events were detected in the China-Myanmar border and in Thai samples (Polley et al., 2003), compared to over 20 recombinant events detected in Papua New Guinea (Arnott et al., 2014) and African populations (Osier et al., 2010; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009) (Table 2). The recombination parameter (C) between adjacent nucleotide sites and for the whole sequence had a value of 0.003 and 3.8, respectively in the China-Myanmar border population. This value was much lower than that from other geographic areas analyzed in this study but higher than that of the Venezuelan population (Ord et al., 2008) (Table 2). The LD index R2 decreased rapidly with increasing nucleotide distance for the China-Myanmar border samples, indicating a high meiotic recombination rate (Fig. 2).
Table 2.
Comparison of different estimates of recombination events in PfAMA1 among global isolates.
| Locality (no.) | Ra | Rb | Rm |
|---|---|---|---|
| Myanmar (135) | 0.003 | 3.8 | 16 |
| Thailand (50) | 0.051 | 66.5 | 16 |
| PNG (76) | 0.050 | 58.7 | 21 |
| Gambia (114) | 0.089 | 106 | 26 |
| Nigeria (51) | 0.160 | 207 | 25 |
| Kenya (129) | 0.123 | 146 | 26 |
| Mali (506) | 0.112 | 133 | 28 |
| Venezuela (30) | 0.000 | 0.001 | 1 |
Ra, recombinant parameter between adjacent sites; Rb, recombinant parameter for the whole gene; Rm, minimum number of recombination events.
Fig. 2.
Linkage disequilibrium (LD) across the PfAMA1 gene of China-Myanmar border isolates was calculated by using the (A) R2 and (B) D’ index. Those pairs of sites that show significant linkage disequilibrium as calculated by Fisher’s exact test is shown as red circles, while all others are shown by black circles. Trace line represents the regression line.
3.3. Fst analysis of PfAMA1 in the China-Myanmar border area and other countries
In order to understand the distribution of diversity across geographically different populations, Fst values of the China-Myanmar border population and seven worldwide populations with full-length ectodomain sequences were evaluated. Pairwise population comparisons showed a high level of genetic differentiation (Fst=0.47) between the South American population (Venezuela) with the China-Myanmar border population, and other worldwide populations (range from 0.23–0.35). A moderate range of Fst value (0.13–0.16) was detected when comparing the China-Myanmar border populations with the Thai and African populations, but a much lower genetic difference was revealed when the China-Myanmar border population was compared with the Oceania population (Fst=0.02) (Table 3).
Table 3.
Genetic differentiation (Fst) of the PfAMA1 gene among 10 geographic different populations
| Locality (no.) | Myanmar | Thailand | PNG | Gambia | Nigeria | Kenya | Mali |
|---|---|---|---|---|---|---|---|
| Myanmar (135) | |||||||
| Thailand (50) | 0.14 | ||||||
| PNG (76) | 0.02 | 0.05 | |||||
| Gambia (114) | 0.13 | 0.03 | 0.05 | ||||
| Nigeria (51) | 0.16 | 0.04 | 0.07 | 0.01 | |||
| Kenya (129) | 0.15 | 0.03 | 0.07 | 0.01 | 0.01 | ||
| Mali (506) | 0.15 | 0.02 | 0.06 | 0.00 | 0.01 | 0.01 | |
| Venezuela (30) | 0.47 | 0.27 | 0.35 | 0.25 | 0.23 | 0.26 | 0.25 |
Note: Myanmar, China-Myanmar border population; PNG, Papua New Guinea population.
3.4. Positive diversifying selection on PfAMA1 gene
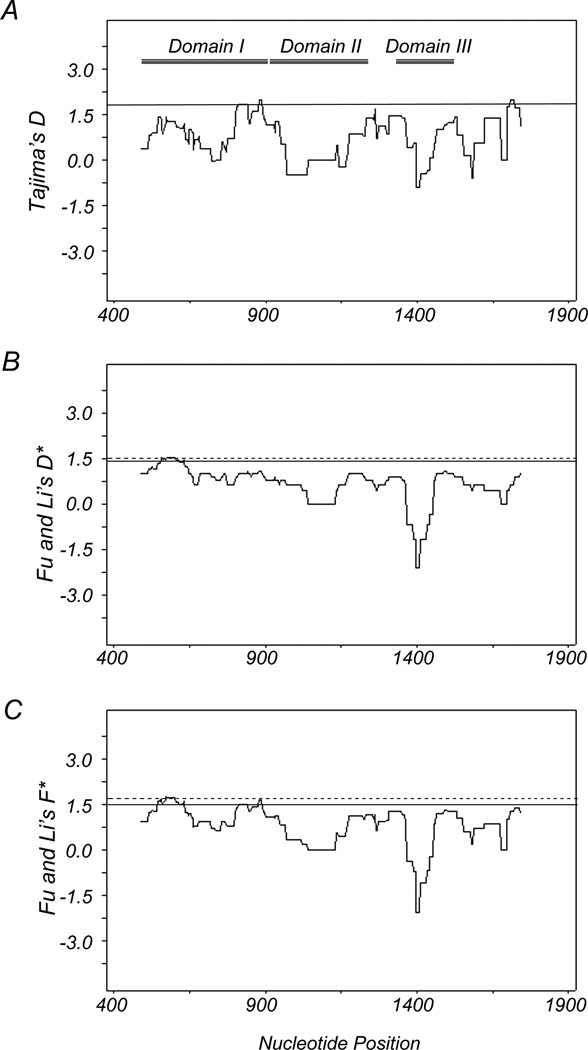
Because of the observed high genetic diversity of PfAMA1, we evaluated signatures of selection on PfAMA1 in the China-Myanmar border isolates (Table 1). A significant excess of nonsynonymous substitutions over synonymous substitutions was detected when the entire sequenced region was evaluated (P<0.001). The same analysis performed against DI, DII, and DIII detected a significant excess of nonsynonymous substitutions over synonymous substitutions in DI, but not DII or DIII (Table 1). The results suggest that positive selection is acting on DI of PfAMA1. Tajima’s D test shows positive values of D for the region as a whole and each domain separately (Table 1). Although not significant, sliding window plot depicted positive D values in DI and linker regions of each domain, suggesting natural selection in these regions (Fig. 3). Furthermore, Table 1 also shows significant values of Fu and Li's D* (1.911) and F* (1.929) for the entire sequenced region (Fu and Li's D*; P<0.02 and F*; P<0.05). When the three domains were separately assessed, significant deviations greater than zero were detected for DI (D*=2.030, P<0.02 and F*=1.964; P<0.05), but not DII or DII. Sliding window plot analysis of and Li's D* and F* also revealed a significant larger deviation than in DI (Fig. 3). Finally, the McDonald-Kreitman test showed a significant excess of intraspecific nonsynonymous substitutions over synonymous substitutions in the entire sequenced region and DI as compared with interspecies fixed differences of nonsynonymous and synonymous changes, suggesting positive selection (P<0.001 and P<0.02). Although not statistically significant, DII and III show the same trend as DI, suggesting that positive selection occurred across the ectodomain of PfAMA1. Collectively, all evolutionary tests for neutrality detected a signature of positive diversifying selection on the whole sequenced region and DI at the 98% confidence level.
Fig. 3.
Sliding window plot of Tajima’s D test (A), Fu and Li’s D* (B) and F* (C) tests for PfAMA1 isolates from China-Myanmar border area. Nucleotide numbers are those of 3D7. Window length is 90 bp, and step size is 3 bp. The three domains of AMA1 are represented within the first plot as lines labeled Domain I, Domain II, and Domain III, respectively. Regions outside of the dash lines indicate the region with a significant departure from neutrality (P< 0.05, one-tailed). Regions outside the solid lines indicate regions with positive values (P< 0.1, one-tailed).
3.5. Haplotype network reconstruction
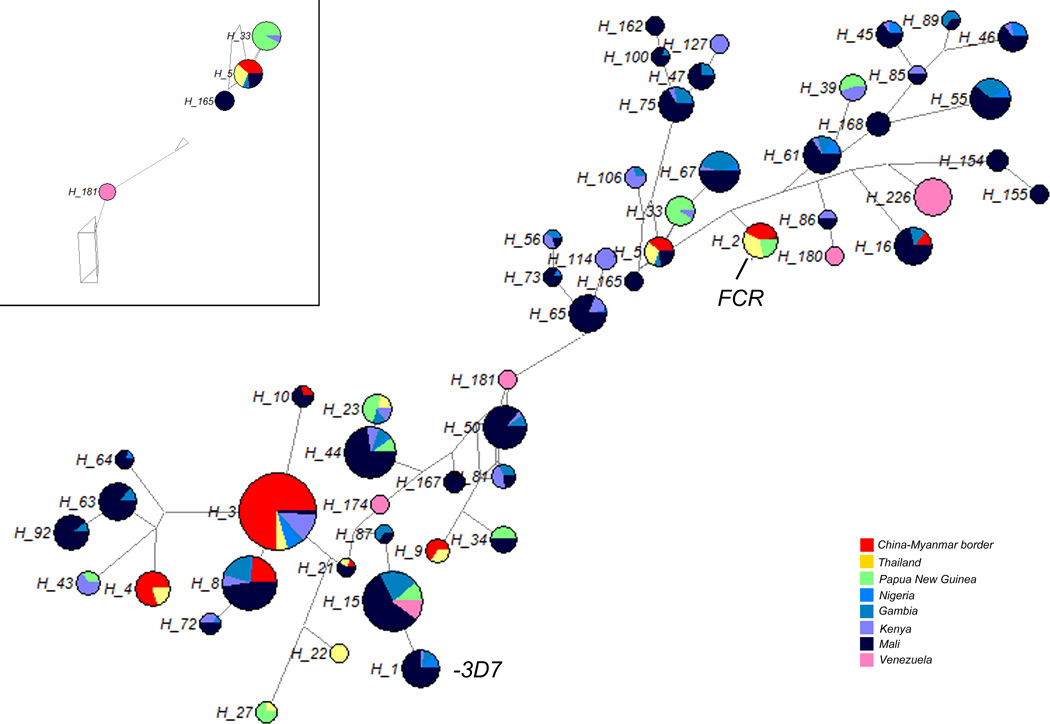
A haplotype network was constructed to establish the relationships among the PfAMA1 haplotypes from global P. falciparum populations. A total of 229 haplotypes were identified in 1091 sequences, of which 45.4% was singleton. Haplotype prevalence ranged from 0.4 to 38.9%. Haplotypes with sequences matching the P. falciparum vaccine strains 3D7 and FVO (GenBank accession no AJ277646.1) were 9.6% and 7.8% respectively. Haplotypes 3, 5, 8, 15 16 and 21 are shared between the China-Myanmar border population and African populations. Among which, haplotype 3 has an observed frequency of 38.9%, and it is the dominant haplotype of the China-Myanmar border population with a frequency of 60.4%. Haplotype 15 is the only haplotype shared among populations from the four continents (Asia, Africa, Oceania, and South America), with a frequency of 23.1% in total sequences analyzed. This haplotype occurred at 0.4% in the China-Myanmar border, 18.3% in African, 2.2% in Oceanian, and 2.2% in South American populations, respectively. To obtain a haplotype network of major haplotypes, we excluded the 104 singletons and frequency ≤2% haplotypes from analysis. When only the network torso is displayed, a clustered distribution of haplotypes correlated with the continents of origin and clearly separated the South American population from the rest of worldwide populations (Fig. 4).
Fig. 4.
The proportion of PfAMA1 haplotypes variation observed in different geographic populations. The size of the pies reflects the frequency of a particular haplotype. The lengths of the lines connecting the pies, measured from their centers, are in proportion to the number of base pair substitutions separating the haplotypes. Color of each pie represents different country. The torso of haplotype network is shown up-left.
4. Discussion
The current study analyzed the extent of genetic polymorphism in and diversifying selection on PfAMA1 in the China-Myanmar border P. falciparum population. In this first attempt to determine PfAMA1 diversity from this region, we detected high levels of genetic diversity in the PfAMA1 ectodomain from 135 parasite isolates. This observation is comparable with results published on global isolates (Arnott et al., 2014; Ord et al., 2008; Osier et al., 2010; Polley et al., 2003; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009). Yet, the level of genetic diversity of PfAMA1 from the China-Myanmar border population is lower than that from the African populations (π = 0.026~0.028) (Osier et al., 2010; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009)), the Southeast Asian population (Thai, π=0.025 (Polley et al., 2003)), and the Oceania population (Papua New Guinea, π=0.026 (Arnott et al., 2014)), but higher than that from the South American population (π=0.012 (Ord et al., 2008)). The lower genetic diversity in P. falciparum population at the China-Myanmar border isolates is consistent with the epidemiologic characteristics of the study area, where malaria transmission is seasonal and unstable (Li et al., 2013) as compared to the holoendemic African regions. Though the number of analyzed sequences varied in parasite populations used in the comparison, the majority of mutation sites are shared among these populations, suggesting that similar selective forces (host immunity) act in various geographical regions.
Recombination and positive selection are possibly two major factors responsible for the evolution and genetic variation in PfAMA1 (Eisen et al., 1999). In several populations, recombination is frequently observed, as shown by a high estimate of the recombination parameter, C, and very rapid decline in linkage disequilibrium with increasing distance between nucleotide sites (Osier et al., 2010; Polley et al., 2003; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009). Though higher than that in the South American population (Ord et al., 2008), the estimated recombinant parameters of our study population are much lower than those of the African populations (Osier et al., 2010; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009). Within Southeast Asia, the recombination rate of the China-Myanmar border population is lower than that in the Thai-Myanmar border population (Polley et al., 2003). Considering the average recombination rate in the P. falciparum genome as 1cM per 17 kb of sequence (Su et al., 1999), which equates to r=6×10−7, the estimated genetically effective population size N in the current study would be 1.3×103, which is much lower than the estimate for African populations at ~6.6×104 (Polley et al., 2003). This again may be due to the lower malaria endemicity in the China-Myanmar border area, which restricts the opportunity of multiclonal infections, subsequent cross-fertilization, and recombination in mosquitoes (Walliker, 2000). Our recent epidemiological studies at the international borders of the GMS showed much lower P. falciparum endemicity at the China-Myanmar border than at the Thai-Myanmar border (Baum et al., 2015; Zhou et al., 2014). We also obtained evidence indicating limited genetic differentiation of PfAMA1 (overall Fst value 0.09) among parasite populations. However, the Fst value between the China-Myanmar border and the Venezuelan population (Fst=0.47) showed evidence of strong population division (Ord et al., 2008). In addition, Fst value for the China-Myanmar border population in comparison with other global populations showed moderate levels of genetic differentiation, which is largely due to geographic area-specific SNPs (Fst = 0.13 – 0.16 (Osier et al., 2010; Polley et al., 2003; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009). However, it is intriguing that the China-Myanmar border population showed little genetic differentiation from the Papua New Guinea population (Arnott et al., 2014). While this may resulted from similar host immune selection forces imposed on PfAMA1 in these geographically separated parasite populations, it nonetheless raised hope that the final PfAMA1-based vaccine design targeting a limited number of PfAMA1-haplotypes may be effective for different endemic regions (Drew et al., 2012).
The significant ratio of nonsynonymous to synonymous substitutions (dN/dS) observed in our study and the other 7 P. falciparum populations suggest that amino acid replacements are generally favored in PfAMA1 and this adaptive evolution is presumably due to host immune pressure. Additional neutrality tests provided corroborating findings indicative of positive diversifying selection operating at PfAMA1 DI and possibly the entire ectodomain. Though the value of Tajima’s D test was not significant in either total region or DI–III, the positive values of this test also indicated departure from neutral evolution and the tendency of positive diversifying selection, together with other studies, reflecting the importance of PfAMA1 as a target of host protective immunity (Cortes et al., 2003; Healer et al., 2004; Remarque et al., 2008; Terheggen et al., 2014).
The number of haplotypes in DI (445–906 bp) identified in this study (20 haplotypes in 135 isolates) was larger than the numbers of Venezuelan (5 haplotypes in 30 isolates) and Thai (18 haplotypes in 50 isolates), but fewer than that from Papua New Guinea (27 haplotypes in 50 isolates), Gambian (51 haplotypes in 114 isolates), Nigerian (35 haplotypes in 51 isolates), Kenya (64 haplotypes in 129 isolates), and the Mali population (123 haplotypes in 506 isolates) (Arnott et al., 2014; Ord et al., 2008; Osier et al., 2010; Polley et al., 2003; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009). Moreover, besides the Venezuelan population, haplotype diversity in DI (445–906 bp) in the China-Myanmar border population was lower than other analyzed populations (Arnott et al., 2014; Ord et al., 2008; Osier et al., 2010; Polley et al., 2003; Polley and Conway, 2001; Takala et al., 2009; Tetteh et al., 2009). Nonetheless, we identified 8 haplotypes that were unique from China-Myanmar border isolates. As one of the few vaccine candidates reaching clinical trials, all current AMA1 vaccines are based on sequences from one or two P. falciparum strains, 3D7 and FVO (Ellis et al., 2012; Laurens et al., 2013; Thera et al., 2010; Thera et al., 2011). In our study, only 5% of the China-Myanmar border PfAMA1 haplotypes was identical to the FVO strain, and none of them was identical to the 3D7 strain. Thus general vaccine design based on these two variants may offer much lower protection. The inclusion of other PfAMA1 alleles needs to be considered given that the protective efficacy of AMA1 is allele specific.
In conclusion, our study presented baseline data of the PfAMA1 polymorphism in 135 P. falciparum isolates from the China-Myanmar border area. Understanding polymorphism and host immune response is an important aspect for malaria vaccine development in light of extensive antigenic diversity and allele-specific protective immunity.
Acknowledgments
This work was supported by a grant from NIAID, National Institutes of Health (U19AI089672) and a National Natural Science Foundation of China (Grant no. 81301455)
References
- Arnott A, Wapling J, Mueller I, Ramsland PA, Siba PM, Reeder JC, Barry AE. Distinct patterns of diversity, population structure and evolution in the AMA1 genes of sympatric Plasmodium falciparum and Plasmodium vivax populations of Papua New Guinea from an area of similarly high transmission. Malaria journal. 2014;13:233. doi: 10.1186/1475-2875-13-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular biology and evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bargieri D, Lagal V, Tardieux I, Menard R. Host cell invasion by apicomplexans: what do we know? Trends Parasitol. 2012;28:131–135. doi: 10.1016/j.pt.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Baum E, Sattabongkot J, Sirichaisinthop J, Kiattibutr K, Davies DH, Jain A, Lo E, Lee MC, Randall AZ, Molina DM, Liang X, Cui L, Felgner PL, Yan G. Submicroscopic and asymptomatic Plasmodium falciparum and Plasmodium vivax infections are common in western Thailand - molecular and serological evidence. Malaria journal. 2015;14:95. doi: 10.1186/s12936-015-0611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Kaneko O, Thongkukiatkul A, Tachibana M, Otsuki H, Gao Q, Tsuboi T, Torii M. Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitol Int. 2009;58:29–35. doi: 10.1016/j.parint.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Chesne-Seck ML, Pizarro JC, Vulliez-Le Normand B, Collins CR, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. Structural comparison of apical membrane antigen 1 orthologues and paralogues in apicomplexan parasites. Mol Biochem Parasitol. 2005;144:55–67. doi: 10.1016/j.molbiopara.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infection and immunity. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crewther PE, Matthew ML, Flegg RH, Anders RF. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect Immun. 1996;64:3310–3317. doi: 10.1128/iai.64.8.3310-3317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Yan G, Sattabongkot J, Cao Y, Chen B, Chen X, Fan Q, Fang Q, Jongwutiwes S, Parker D, Sirichaisinthop J, Kyaw MP, Su XZ, Yang H, Yang Z, Wang B, Xu J, Zheng B, Zhong D, Zhou G. Malaria in the Greater Mekong Subregion: heterogeneity and complexity. Acta Trop. 2012;121:227–239. doi: 10.1016/j.actatropica.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Fairhurst RM, Slutsker L, Macarthur JR, Breman JG, Guerin PJ, Wellems TE, Ringwald P, Newman RD, Plowe CV. The threat of artemisinin-resistant malaria. The New England journal of medicine. 2011;365:1073–1075. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew DR, Hodder AN, Wilson DW, Foley M, Mueller I, Siba PM, Dent AE, Cowman AF, Beeson JG. Defining the antigenic diversity of Plasmodium falciparum apical membrane antigen 1 and the requirements for a multi-allele vaccine against malaria. PloS one. 2012;7:e51023. doi: 10.1371/journal.pone.0051023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Haynes JD, Moch JK, Barbosa A, Lanar DE. Invasion-inhibitory antibodies inhibit proteolytic processing of apical membrane antigen 1 of Plasmodium falciparum merozoites. Proc Natl Acad Sci U S A. 2003;100:12295–12300. doi: 10.1073/pnas.2032858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen DP, Marshall VM, Billman-Jacobe H, Coppel RL. A Plasmodium falciparum apical membrane antigen-1 (AMA-1) gene apparently generated by intragenic recombination. Molecular and biochemical parasitology. 1999;100:243–246. doi: 10.1016/s0166-6851(99)00054-7. [DOI] [PubMed] [Google Scholar]
- Ellis RD, Wu Y, Martin LB, Shaffer D, Miura K, Aebig J, Orcutt A, Rausch K, Zhu D, Mogensen A, Fay MP, Narum DL, Long C, Miller L, Durbin AP. Phase 1 study in malaria naive adults of BSAM2/Alhydrogel(R)+CPG 7909, a blood stage vaccine against P. falciparum malaria. PLoS One. 2012;7:e46094. doi: 10.1371/journal.pone.0046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A. Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA 1 and MSP 1 genes. Mol Biochem Parasitol. 2000;108:53–66. doi: 10.1016/s0166-6851(00)00204-8. [DOI] [PubMed] [Google Scholar]
- Fowkes FJ, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: A systematic review and meta-analysis. PLoS Med. 2010;7:e1000218. doi: 10.1371/journal.pmed.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Alam MT, Das MK, Dev V, Kumar A, Dash AP, Sharma YD. Sequence diversity and natural selection at domain I of the apical membrane antigen 1 among Indian Plasmodium falciparum populations. Malaria journal. 2007;6:154. doi: 10.1186/1475-2875-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healer J, Crawford S, Ralph S, McFadden G, Cowman AF. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect Immun. 2002;70:5751–5758. doi: 10.1128/IAI.70.10.5751-5758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman AF, Batchelor A. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Molecular microbiology. 2004;52:159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect Immun. 2001;69:3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. The disulfide bond structure of Plasmodium apical membrane antigen-1. The Journal of biological chemistry. 1996;271:29446–29452. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Estimating the recombination parameter of a finite population model without selection. Genetical research. 1987;50:245–250. doi: 10.1017/s0016672300023776. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SJ, Branch OH, Castro JC, Oster RA, Rayner JC. Genetic diversity of the malaria vaccine candidate Plasmodium falciparum merozoite surface protein-3 in a hypoendemic transmission environment. Am J Trop Med Hyg. 2009;80:479–486. [PMC free article] [PubMed] [Google Scholar]
- Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, Long CA, Miller LH, Stowers AW. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70:6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocken CH, Narum DL, Massougbodji A, Ayivi B, Dubbeld MA, van der Wel A, Conway DJ, Sanni A, Thomas AW. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Molecular and biochemical parasitology. 2000;109:147–156. doi: 10.1016/s0166-6851(00)00250-4. [DOI] [PubMed] [Google Scholar]
- Kocken CH, Withers-Martinez C, Dubbeld MA, van der Wel A, Hackett F, Valderrama A, Blackman MJ, Thomas AW. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infection and immunity. 2002;70:4471–4476. doi: 10.1128/IAI.70.8.4471-4476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukouikila-Koussounda F, Malonga V, Mayengue PI, Ndounga M, Vouvoungui CJ, Ntoumi F. Genetic polymorphism of merozoite surface protein 2 and prevalence of K76T pfcrt mutation in Plasmodium falciparum field isolates from Congolese children with asymptomatic infections. Malar J. 2012;11:105. doi: 10.1186/1475-2875-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusi KA, Faber BW, Thomas AW, Remarque EJ. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One. 2009;4:e8110. doi: 10.1371/journal.pone.0008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurens MB, Thera MA, Coulibaly D, Ouattara A, Kone AK, Guindo AB, Traore K, Traore I, Kouriba B, Diallo DA, Diarra I, Daou M, Dolo A, Tolo Y, Sissoko MS, Niangaly A, Sissoko M, Takala-Harrison S, Lyke KE, Wu Y, Blackwelder WC, Godeaux O, Vekemans J, Dubois MC, Ballou WR, Cohen J, Dube T, Soisson L, Diggs CL, House B, Bennett JW, Lanar DE, Dutta S, Heppner DG, Plowe CV, Doumbo OK. Extended safety, immunogenicity and efficacy of a blood-stage malaria vaccine in malian children: 24-month follow-up of a randomized, double-blinded phase 2 trial. PLoS One. 2013;8:e79323. doi: 10.1371/journal.pone.0079323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Parker DM, Yang Z, Fan Q, Zhou G, Ai G, Duan J, Lee MC, Yan G, Matthews SA, Cui L, Wang Y. Risk factors associated with slide positivity among febrile patients in a conflict zone of north-eastern Myanmar along the China-Myanmar border. Malar J. 2013;12:361. doi: 10.1186/1475-2875-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Meng H, Zhang R, Yang H, Fan Q, Su X, Miao J, Cui L, Yang Z. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrobial agents and chemotherapy. 2010;54:4306–4313. doi: 10.1128/AAC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect Immun. 2004;72:154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum DL, Thomas AW. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1994;67:59–68. doi: 10.1016/0166-6851(94)90096-5. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular biology and evolution. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ord RL, Tami A, Sutherland CJ. ama1 genes of sympatric Plasmodium vivax and P. falciparum from Venezuela differ significantly in genetic diversity and recombination frequency. PLoS One. 2008;3:e3366. doi: 10.1371/journal.pone.0003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier FH, Weedall GD, Verra F, Murungi L, Tetteh KK, Bull P, Faber BW, Remarque E, Thomas A, Marsh K, Conway DJ. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect Immun. 2010;78:4625–4633. doi: 10.1128/IAI.00576-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol Cell Biol. 1989;9:3151–3154. doi: 10.1128/mcb.9.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends in parasitology. 2008;24:74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Richard D, MacRaild CA, Riglar DT, Chan JA, Foley M, Baum J, Ralph SA, Norton RS, Cowman AF. Interaction between Plasmodium falciparum apical membrane antigen 1 and the rhoptry neck protein complex defines a key step in the erythrocyte invasion process of malaria parasites. J Biol Chem. 2010;285:14815–14822. doi: 10.1074/jbc.M109.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakihama N, Kaneko A, Hattori T, Tanabe K. Limited recombination events in merozoite surface protein-1 alleles of Plasmodium falciparum on islands. Gene. 2001;279:41–48. doi: 10.1016/s0378-1119(01)00748-x. [DOI] [PubMed] [Google Scholar]
- Silvie O, Franetich JF, Charrin S, Mueller MS, Siau A, Bodescot M, Rubinstein E, Hannoun L, Charoenvit Y, Kocken CH, Thomas AW, Van Gemert GJ, Sauerwein RW, Blackman MJ, Anders RF, Pluschke G, Mazier D. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem. 2004;279:9490–9496. doi: 10.1074/jbc.M311331200. [DOI] [PubMed] [Google Scholar]
- Su X, Ferdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, Escalante AA, Ouattara A, Traore K, Niangaly A, Djimde AA, Doumbo OK, Plowe CV. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Science translational medicine. 2009;1:2ra5. doi: 10.1126/scitranslmed.3000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terheggen U, Drew DR, Hodder AN, Cross NJ, Mugyenyi CK, Barry AE, Anders RF, Dutta S, Osier FH, Elliott SR, Senn N, Stanisic DI, Marsh K, Siba PM, Mueller I, Richards JS, Beeson JG. Limited antigenic diversity of Plasmodium falciparum apical membrane antigen 1 supports the development of effective multi-allele vaccines. BMC Med. 2014;12:183. doi: 10.1186/s12916-014-0183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh KK, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, Marsh K, Weedall GD, Conway DJ. Prospective identification of malaria parasite genes under balancing selection. PLoS One. 2009;4:e5568. doi: 10.1371/journal.pone.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A, Alam MT, Bora H, Kaur P, Sharma YD. Plasmodium vivax: sequence polymorphism and effect of natural selection at apical membrane antigen 1 (PvAMA1) among Indian population. Gene. 2008;419:35–42. doi: 10.1016/j.gene.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Kone AK, Guindo AB, Traore K, Sissoko M, Diallo DA, Diarra I, Kouriba B, Daou M, Dolo A, Baby M, Sissoko MS, Sagara I, Niangaly A, Traore I, Olotu A, Godeaux O, Leach A, Dubois MC, Ballou WR, Cohen J, Thompson D, Dube T, Soisson L, Diggs CL, Takala SL, Lyke KE, House B, Lanar DE, Dutta S, Heppner DG, Plowe CV. Safety and immunogenicity of an AMA1 malaria vaccine in Malian children: results of a phase 1 randomized controlled trial. PloS one. 2010;5:e9041. doi: 10.1371/journal.pone.0009041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, Guindo AB, Traore K, Traore I, Kouriba B, Diallo DA, Diarra I, Daou M, Dolo A, Tolo Y, Sissoko MS, Niangaly A, Sissoko M, Takala-Harrison S, Lyke KE, Wu Y, Blackwelder WC, Godeaux O, Vekemans J, Dubois MC, Ballou WR, Cohen J, Thompson D, Dube T, Soisson L, Diggs CL, House B, Lanar DE, Dutta S, Heppner DG, Jr, Plowe CV. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–1013. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AW, Trape JF, Rogier C, Goncalves A, Rosario VE, Narum DL. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am J Trop Med Hyg. 1994;51:730–740. doi: 10.4269/ajtmh.1994.51.730. [DOI] [PubMed] [Google Scholar]
- Walliker D. Malaria. 2000 [Google Scholar]
- Wang Y, Zhong D, Cui L, Lee MC, Yang Z, Yan G, Zhou G. Population dynamics and community structure of Anopheles mosquitoes along the China-Myanmar border. Parasites & vectors. 2015;8:445. doi: 10.1186/s13071-015-1057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Health Organization; 2014. World Malaria Report 2014; p. 242. [Google Scholar]
- Zhou G, Sun L, Xia R, Duan Y, Xu J, Yang H, Wang Y, Lee MC, Xiang Z, Yan G, Cui L, Yang Z. Clinical malaria along the China-Myanmar border, Yunnan Province, China, January 2011–August 2012. Emerging infectious diseases. 2014;20:675–678. doi: 10.3201/eid2004.130647. [DOI] [PMC free article] [PubMed] [Google Scholar]