Abstract
The effects of human papillomavirus type 18 (HPV-18) E6 and E7 proteins on global patterns of host gene expression in primary human keratinocytes grown in organotypic raft culture system were assessed. Primary human keratinocytes were infected with retroviruses that express the wild-type HPV-18 E6 and E7 genes from the native differentiation-dependent HPV enhancer-promoter. Total RNA was isolated from raft cultures and used to generate probes for querying Affymetrix U95A microarrays, which contain >12,500 human gene sequences. Quadruplicate arrays of each E6/E7-transduced and empty vector-transduced samples were analyzed by 16 pairwise comparisons. Transcripts altered in ≥12 comparisons were selected for further analysis. With this approach, HPV-18 E6/E7 expression significantly altered the expression of 1,381 genes. A large increase in transcripts associated with DNA and RNA metabolism was observed, with major increases noted for transcription factors, splicing factors, and DNA replication elements, among others. Multiple genes associated with protein translation were downregulated. In addition, major alterations were found in transcripts associated with the cell cycle and cell differentiation. Our study provides a systematic description of transcript changes brought about by HPV-18 E6/E7 in a physiologically relevant model and should furnish a solid source of information to guide future studies.
Human papillomaviruses (HPVs) are small DNA viruses containing a circular double-stranded DNA genome. The genome contains eight open reading frames located on the same strand of the DNA (24). More than 100 human papillomavirus genotypes infecting both cutaneous and mucosal epithelia have been identified to date. The mucosotropic viruses are usually classified into high-risk and low-risk groups, based on their association with the potential development of anogenital carcinomas. Greater than 99% of cervical cancers contain viral sequences derived from high-risk viruses (50, 53).
The productive phase of the HPV life cycle is closely linked to differentiation of the infected keratinocytes within squamous epithelia and in cultured keratinocytes (22). The virus initially infects basal cells, where it is maintained and replicated as low-copy, extrachromosomal plasmids along with the host DNA. Viral genome amplification, late gene expression, and virion packaging occur in the differentiated keratinocytes in vivo (10, 24). The dependence of viral reproduction on keratinocyte differentiation is paradoxical because differentiated keratinocytes are normally unable to reenter the cell cycle. Papillomaviruses possess few genes and therefore require the participation of the host cell DNA replication machinery to execute viral DNA replication (30). Accordingly, the virus must reprogram the differentiated cells to restore S phase to facilitate viral DNA replication, and this is mediated by the viral E6 and E7 proteins.
The high-risk HPV E6 protein (150 amino acids) and E7 protein (98 amino acids) are under the control of the upstream regulatory region containing the enhancer and promoter (URR-E6 promoter). E6 intragenic splicing allows efficient translation reinitiation for the downstream E7 protein (1). Squamous epithelial differentiation of primary human keratinocytes or cell lines has been recapitulated in vitro in organotypic cultures at the air-medium interface on collagen rafts (11). Expression of the HPV-18 E7 gene is sufficient to induce S-phase reentry in a subset of postmitotic differentiating keratinocytes in raft cultures for one or more rounds of unscheduled cellular DNA synthesis. Another subset of the cells are prevented from doing so by accumulating high levels of cyclin E in kinase-inactive complexes with cyclin-dependent kinase 2 (cdk2) and the cdk 2 inhibitor p21cip1 or p27kip1 (7, 9, 26, 37). Possible molecular mechanisms underlying this E7 function include binding and destabilization of the retinoblastoma protein, which controls the G1-to-S transition (33, 42) and direct activation of cyclin/cdk2 (23). These target proteins are well-characterized regulators of S-phase entry and progression. Concordantly, mutations in E7 abolishing retinoblastoma protein interaction eliminate unscheduled cellular DNA synthesis and viral DNA amplification in differentiated keratinocytes (8, 20). However, strong evidence exists for additional E7 functions that may facilitate viral DNA amplification (3, 12, 25, 33).
Alone, E6 expression does not promote unscheduled DNA synthesis in differentiated keratinocytes (7, 26). However, E6 is necessary to support plasmid viral DNA maintenance in submerged cultures (47, 39). In addition, E6 also possesses additional activities in submerged cultures, including activation of hTERT, which is critical for immortalization of primary human keratinocytes, downregulation of keratinocyte differentiation-specific transcripts, which might slow down programmed cell death of the differentiated keratinocytes, and inhibition of CBP and p300, whose role in HPV infection is not yet understood (17, 29, 41, 49).
The current study examines global changes in the pattern of gene expression brought about by HPV-18 E6/E7 expression in organotypic “raft” cultures of human keratinocytes. This system is a physiologically relevant model because it supports the entire program of keratinocyte differentiation and HPV reproduction (13, 32). The complexity of raft cultures hampers meaningful microarray analysis with standard approaches because it produces high levels of biological “noise” due to random and variable gene activity in cells in various stages of differentiation. Rather, it demands the implementation of statistical methods to identify those genes whose expression is reproducibly altered by E6/E7. In this study, 16 pairwise comparisons were conducted. The results show that E6/E7 causes broad changes in the transcript profile, affecting multiple cellular regulation and metabolism profiles. In some cases, clustering of transcript alterations within pathways suggests pivotal points of cell regulation where the E6 and E7 proteins are likely to have a major impact. This investigation is the first to attempt a comprehensive description of pathways altered by HPV-18 E6 and E7 in keratinocyte raft cultures.
MATERIALS AND METHODS
Retroviruses and organotypic raft cultures of primary human keratinocytes.
The retroviral vector for 18URR-E6/E7 and the empty retroviral vector pLC were described previously (7, 8). Amphotropic recombinant Moloney murine leukemia viruses containing HPV-18 URR-E6/E7 and pLC were generated as described (7, 40). Primary human keratinocytes were isolated from several pooled neonatal foreskins as described (52), acutely infected with recombinant Moloney murine leukemia viruses, and selected for 2 days at 300 μg/ml of geneticin (Invitrogen). All keratinocytes surviving selection were infected (7, 8). Each batch of keratinocytes transduced with one of the retroviruses was used to seed 12 to 15 collagen plugs embedded with mouse 3T3 J2 fibroblasts. The cultures were raised to the air-medium interface for 9 days, and raft culture medium was changed every other day. One culture of each group was exposed to 50 μg of bromodeoxyuridine per ml for the final 12 h of culturing, subsequently fixed in 10% buffered formalin, paraffin embedded, and cut into 4-μm sections. The sections were used for immunohistochemical analysis of bromodeoxyuridine incorporation to examine S-phase entry. The remaining raft cultures were washed with 1x phosphate-buffered saline, removed from collagen, frozen in dry ice-ethanol, and stored at −80°C until RNA purification or protein extraction.
Immunohistochemical detection of bromodeoxyuridine incorporation.
Immunohistochemical detection of bromodeoxyuridine incorporation was conducted with sections of epithelial raft cultures, and images were captured as described (37) with mouse monoclonal antibromodeoxyuridine antibody (18-0103; Zymed Laboratories Inc., South San Francisco, Calif.; 1:50 dilution) in conjunction with the Histostain-SP broad-spectrum kit following the manufacturer's instructions (Zymed Laboratories).
Sample preparation and array hybridization.
Total RNA was extracted from raft cultures by homogenizing with a motorized tissue grinder in 1 ml of Trizol reagent (Life Technologies, Rockville, Md.). After removal of cellular debris by centrifugation and phase separation after the addition of chloroform, RNA was precipitated with isopropanol. In order to control for sample-to-sample variation that might occur due to RNA isolation or processing of cRNA, the following measures were employed. RNA was isolated from raft cultures at two separate times, and each RNA preparation was processed to cRNA twice, for a total of four cRNA samples from both pLC and E6/E7 rafts, as follows. After RNA clean-up with the RNeasy kit (Qiagen, Valencia, Calif.), double-stranded cDNA was synthesized with the Superscript Choice system (Life Technologies, Rockville, Md.) with a (dT)24-T7 promoter primer (GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG[dT]24). Approximately one-half of the cDNA reaction was transcribed into cRNA labeled with biotinylated CTP and UTP with the Bioarray RNA transcript labeling kit (Enzo, Farmingdale, N.Y.). The cRNA was fragmented by metal-induced hydrolysis, and human U95A arrays (Affymetrix, Santa Clara, Calif.) containing 12,626 probe sets (12,560 genes with 66 controls) were hybridized with 10 μg of fragmented cRNA per array overnight at 45°C with the procedures recommended by Affymetrix. The hybridization cocktail included control cRNAs of several Escherichia coli, Bacillus subtilis, and P1 bacteriophage genes (provided by the manufacturer). Probed arrays were then processed in an Affymetrix GeneChip Fluidics Station 400, and hybridization of labeled cRNA was detected by the amplification staining protocol provided by Affymetrix.
Briefly, a streptavidin-phycoerythrin conjugate (Molecular Probes, Eugene, Oreg.) and a biotinylated antistreptavidin antibody (Vector Labs, Burlingame, Calif.) were sequentially applied to the arrays, followed by an additional application of the streptavidin-phycoerythrin conjugate. The fluorescently amplified arrays were then scanned twice (at a wavelength of 570 nm and a pixel value of 3 μm) on an Affymetrix GeneArray scanner. The quality of the probe was determined by the intensity values for controls representing the 5′, middle, and 3′ regions of specific genes (i.e., glyceraldehhyde-3-phosphate dehydrogenase) with equivalent values indicating efficient and complete cDNA synthesis. The quality of the hybridization was determined by uniformity of visual appearance (absence of bubbles, spots, etc.), by the intensity levels of known controls (see above), and by the overall background and intensity levels of each array.
Microarray analysis.
For consistent comparison between all arrays, the average intensity of each array was scaled within the Affymetrix analysis software (Microarray Suite v4.0) to a set value of 700 prior to data analysis. This value was chosen because it resulted in the lowest scaling factor for all arrays. The average difference intensity (ADI) of each gene provided by the Affymetrix software analysis program was used in all subsequent data manipulations. The correlation of ADI values among arrays was examined with JMP statistical software (SAS Institute Inc., Cary, N.C.). To identify those genes with significant, reproducibly altered expression, the data were analyzed by a modification of previously published methods (2).
E6/E7 arrays were compared to the pLC (vector-only) arrays to determine cellular genes that were altered by the viral oncogenes. The baseline intensity level for detectable expression was set to an ADI of 100, based on the ADI levels found for negative control probe sets, and all values less than 100 were converted to the arbitrary value of 100. This was done to avoid zero ADI values, which would distort the mathematical analysis of any gene not expressed in either experimental or control samples. Next, the log ratio of the experimental ADI (E6/E7) over the control (pLC) ADI was determined for each gene. Two classes of altered transcripts were identified in the following manner. In the first, transcripts were dubbed as from “expressed genes” when they were detected in both E6/E7 and pLC samples but the expression was increased or decreased in the E6/E7 samples relative to the control. In the second, transcripts were dubbed as from “on/off” or “off/on” genes when their levels were undetectable in either the E6/E7 or the pLC samples.
To analyze changes in expressed genes consistently altered in replicate arrays, genes that were <100 in both arrays (experimental and control) were filtered out. The remaining genes were imported into the Spotfire DecisionSite software (Spotfire Inc., Somerville, Mass.) and binned by standard deviations of the log ratio for each experiment versus the control. Genes that were increased or decreased in E6/E7 over pLC by more than 1 standard deviation from the mean were selected as significantly changed. To analyze changes in on/off transcripts, genes that were expressed (>100 ADI) in the experimental but not expressed (<100 ADI) in the control were selected as on to off. Genes < 100 in the experimental but >100 in the control were selected as off to on. on/off or off/on genes with an average ADI of >200 were considered significantly changed. The analyses were conducted with chips in quadruplicate such that 16 pairwise comparisons were made between E6/E7 and control (pLC) samples and grouped according to reproducibility. Genes that were repeated in at least 12 of 16 comparisons were considered significant.
Ontology.
Expression data from the significantly changed genes were merged with the functional and pathway ontologies for these genes found in the Incyte database (Incyte Genomics, Inc., Palo Alto, Calif.). Incomplete entries were edited on an individual basis and ontologies were assigned whenever possible.
Real-time RT-PCR.
A subset of genes were selected for quantitative reverse transcription (RT)-PCR. The selected sequences (provided by Affymetrix) associated with the probe sets located on the array were tested against public databases to confirm the identity of the genes. Once confirmed, primer and probe sets were designed with Primer Expressä software (Applied Biosystems, Foster City, Calif.). Amplicons of approximately 70 bp in the 3′ translated region were chosen.
Total RNA isolated from selected raft cultures (empty vector and E6/E7) used for synthesis of microarray probes (above) was treated with DNase for 30 min at 37oC, followed by heat inactivation prior to RT-PCR, to remove any contaminating genomic DNA. RT-PCR was performed with Taqman RT-PCR technology on the ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, Calif.). Sense and antisense primers were synthesized and high-pressure liquid chromatography (HPLC) purified by Oligos, Etc. (Wilsonville, Oreg.). Fluorescent probes labeled with the 5′ reporter dye FAM (6-carboxyfluorescein) and the 3′ quencher dye TAMRA (6-carboxytetramethylrhodamine) were synthesized and purified by Applied Biosystems (Foster City, Calif.). Triplicate reactions, performed with the Taqman One-Step RT-PCR Master Mix kit (Applied Biosystems, Foster City, Calif.), contained final concentrations of 25 ng of total RNA, 1× Universal Master Mix, 1× MultiScribe and RNase Inhibitor mix, 300 nM each primer, and 200 nM probe in a reaction volume of 50 μl.
Both reverse transcription and PCR were performed sequentially in an ABI Prism 7700 sequence detector. Reverse transcription was carried out for 30 min at 48oC, followed by activation of the AmpliTaq Gold enzyme (in the Universal Master Mix) for 10 min at 95°C. Real-time PCR sequence detection occurred for the duration of 40 cycles of 15 s at 95°C, followed by 1 min at 60°C. Data were analyzed by averaging the triplicate Ct (cycle threshold = cycle at which amplification signal increases above background) values for each RNA sample. Fold change was determined with the formula 2(pLC − E6/E7). Each E6/E7 average Ct was subtracted from each empty vector control average Ct. The fold changes were then averaged, and standard deviations were determined. Glyceraldehhyde-3-phosphate dehydrogenase was used as an internal control as its RNA levels, as expected, were found to remain constant between control and E6/E7 raft RNAs. Control reactions lacking reverse transcriptase were run for all genes to rule out the possible amplification of contaminating genomic DNA. These reactions were always negative.
RESULTS
Global transcript changes identified by pairwise analysis of gene expression.
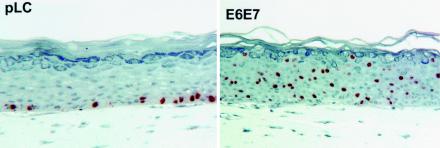
Keratinocytes infected with the appropriate retroviruses were selected with geneticin and used for construction of raft cultures. The quality of the raft cultures was confirmed by bromodeoxyuridine uptake and histochemical analysis (Fig. 1). As reported previously, E6/E7 dramatically induced unscheduled DNA synthesis in a fraction of cells within the differentiated compartment. For each experimental E6/E7 and control series of raft cultures, a total of four cRNA samples were used to probe separate microarrays. Comparison of all four E6/E7 microarrays to all four control (pLC) microarrays was then carried out so that the degree of reproducibility in the expression level of each gene could be ascertained.
FIG. 1.
Micrographs showing bromodeoxyuridine incorporation and morphology of pLC and HPV-18 E6/E7 raft cultures. Note stimulation of bromodeoxyuridine incorporation in the differentiating compartment in the E6/E7 culture.
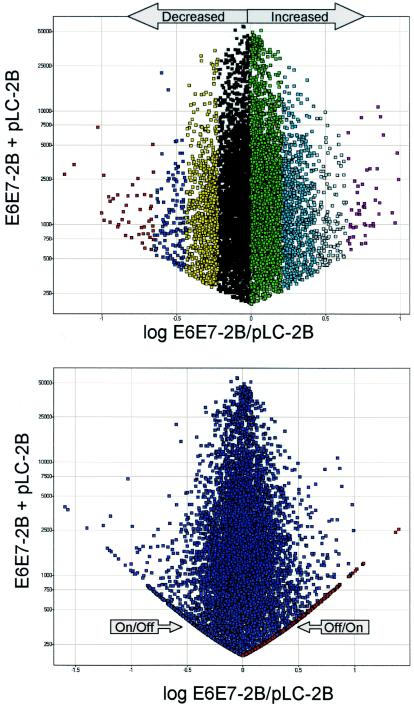
All transcripts significantly altered in the E6/E7 samples (± >1 standard deviation relative to pLC) were identified by comparing log experimental/control to the sum of experimental and control (Fig. 2). This analysis allowed identification of all transcripts from E6/E7 microarrays that were either increased or decreased or turned on or off relative to the same genes in the pLC controls. By comparing all altered genes (either significantly increased or decreased or turned on or off relative to pLC) in every possible comparison (n = 16), the degree of reproducibility of each altered gene was assessed. A summary of this strategy is provided as supplemental material.
FIG. 2.
Scatter plot of a representative graph of the log ratio of experimental (E6/E7-2B)/control(pLC-2B) plotted against the sum of experimental E6/E7-2B and pLC-2B. (Top) Only genes in the increased/decreased categories are shown. (Bottom) Off/on and on/off genes are also shown and appear as arms of the graph. In each graph, every point represents an individual transcript. Each color stripe to the left or right of 0 represents >1, 1, 2, and 3 standard deviations from the mean.
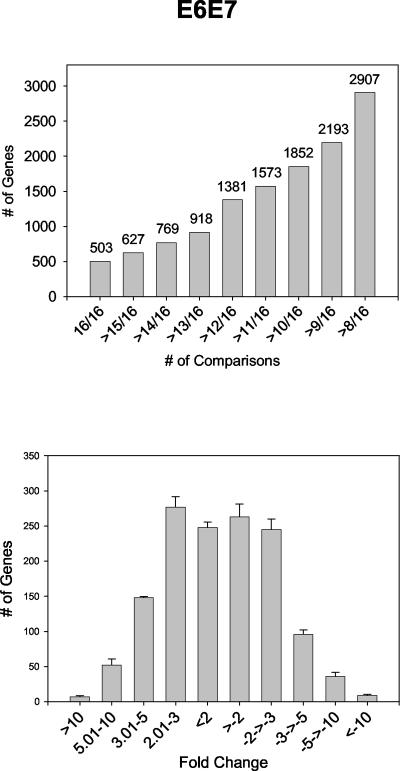
Given the complex nature of raft cultures, a high degree of sample-to-sample variability was expected to exist, even within the pLC (empty vector control) and E6/E7 experimental groups. This outlook was confirmed in initial analyses. However, scatter plots and correlation coefficients clearly demonstrated that greater variability existed between rafts from different groups than between rafts within a group (see the supplemental material). Those genes that were reproducibly and significantly altered between the E6/E7 samples and the empty vector control (pLC) were then identified by pairwise comparisons. The number of times a transcript was altered in E6/E7 cultures (with a standard deviation of >1) relative to control cultures was recorded for each gene. A total of 503 genes were altered by >1 SD in 16 of 16 comparisons in cultures expressing E6/E7 relative to control (pLC) samples, while 2,907 genes were significantly changed in at least half (8 of 16) of the comparisons (Fig. 3A). The relative degree of change of these genes produced a normal distribution, with the most significantly changed transcripts in the two- to fivefold range (Fig. 3B).
FIG. 3.
Histograms depicting numbers of reproducible transcript changes versus total number of comparisons (top). The 1,381 genes that were found altered in ≥12 of 16 comparisons were selected for further analysis. Histograms on the bottom show the distribution of 1,381 transcripts, altered in ≥12 of 16 comparisons, according to fold change.
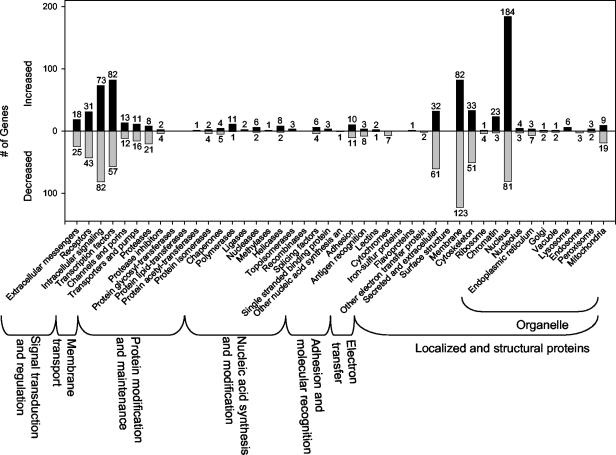
Genes whose expression was significantly changed in ≥12 out of 16 comparisons were assigned to specific Incyte functional and pathway hierarchies (Fig. 4 and 5). The results clearly show that a net gain of genes with increased expression was generally associated with nuclear events (Fig. 4). Conversely, a net sum of transcripts with reduced levels was associated with the cell membrane, mitochondria, and cytoskeleton (Fig. 4). Among the cell membrane transcripts with the greatest decrease were the voltage-dependent calcium channel β4 subunit, fatty acid desaturase, claudin 5, and flotillin 2. Strongly downregulated mitochondrial transcripts included BCL-xL, alanine/glyoxylate aminotransferase, glutamate dehydrogenase-2, and H+-ATP synthase subunit b. Highly decreased transcripts included primarily those encoding actin-associated (plectin 1, drebrin 1, and gelosin) and tubulin (tubulin α3 and tubulin β4) cytoskeletal elements.
FIG. 4.
Incyte functional hierarchies of transcripts reproduced in ≥12 of 16 comparisons. This and the following figure are meant to provide an overall view of transcript changes. Note that because increased/decreased genes may appear multiple times within these hierarchies, the number of genes provided in this and the following figure is relative, not absolute.
FIG. 5.
Incyte pathway hierarchies of transcripts reproduced in at least 12 of 16 comparisons. Note that because increased/decreased genes may appear multiple times within these hierarchies, the number of genes provided in this and the previous figure is relative, not absolute.
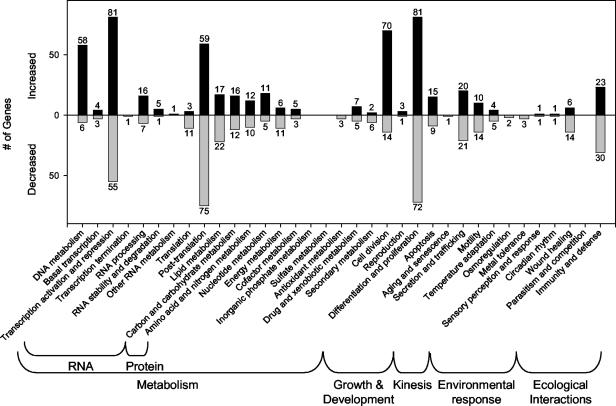
Mapping of transcripts into Incyte pathway hierarchies reveals major changes in a variety of cell processes (Fig. 5). In many such groupings, the number of genes increased in expression is similar to the number of genes with decreased expression. This is not the case, however, with transcripts associated with DNA metabolism (Fig. 5, far left). Fifty-eight DNA metabolism transcripts were increased, while only eight transcripts associated with this pathway were decreased. A similar large net increase in transcripts was noted for genes in the cell division hierarchy. A summary of E6/E7 effects on some notable pathways follows. The fully annotated gene lists are available as supplemental material.
Mapping alterations induced by E6/E7 to specific pathways.
The pathway category showing the greatest total number of significantly altered transcripts was differentiation and proliferation (Fig. 5). This broad category, which contains a broad variety of genes, including those encoding transcription factors, kinases, and cytoskeletal elements, contained 153 transcripts that were significantly altered: 81 were significantly increased, and 72 were decreased. The transcript showing the greatest increase was PCNA (+10.84-fold), while the transcripts showing the greatest decrease were insulin-like growth factor binding protein 6 (−11.39) and JunB proto-oncogene (−8.15). Other notable, reproducibly elevated transcripts included those encoding the cyclin-dependent kinase inhibitors p27kip1 and p16ink4A. Six kinase transcripts of the canonical mitogen-activated protein/Janus-associated kinase signaling pathway were also increased. These transcripts represent each level of the pathways as follows: mitogen-activated protein kinase kinase kinase (MAP3K4 and MAP3K5) → mitogen-activated protein kinase kinase (MAP2K5) → mitogen-activated protein kinase (p38β2, JNK2). Also affected in this pathway were the mitogen-activated protein kinase/Janus-associated kinase substrates mitogen-activated protein kinase activated protein kinase 5 (increased) and JunB (decreased), as just described.
DNA replication.
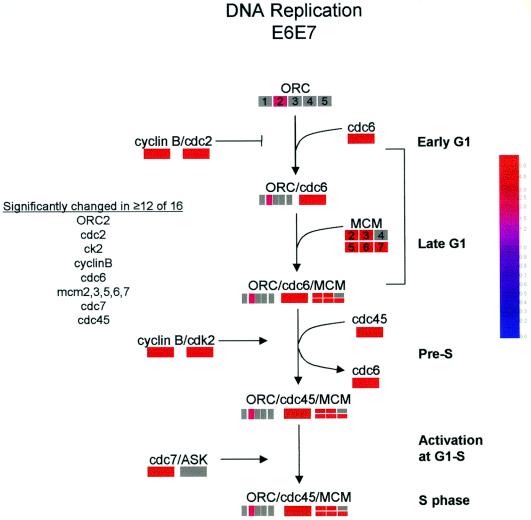
While not all genes in the DNA replication pathway are upregulated by E6/E7, virtually every aspect of the pathway is positively affected in one way or another (Fig. 6). Five of six MCM proteins were strongly upregulated by E6/E7. Other increased transcripts included ORC6, cdc6, and cdc45 (Fig. 6). Additional genes regulating the DNA replication complex were also upregulated, including the cell cycle kinases cdc2/cyclin B and cdk2/cyclin E that phosphorylate members of the replication complex. Thus, these extensive alterations in the DNA replication pathway reflect a general induction of DNA synthesis by E6/E7 to create a cellular milieu supporting viral replication. Additional genes in the DNA synthesis category narrowly missed inclusion in the list due to the stringent criteria employed in this study. The sixth MCM transcript (MCM4) was upregulated in half (8 of 16) of all comparisons. Likewise, ASK, the partner of cdc7 (Fig. 6), was also increased in 8 of 16 comparisons.
FIG. 6.
Changes in transcripts associated with the DNA synthesis pathway. A grey bar indicates that gene did not achieve reproducibility in ≥12 of 16 comparisons. The colored bars indicate the fold difference relative to controls (see inset) of transcripts that were reproducibly changed in ≥12 of 16 comparisons.
Spindle and G2/M checkpoints.
The spindle checkpoint is only partially understood in mammalian cells, but it is apparent from our studies that two of the proximal elements of this pathway were affected by E6/E7 (Table 1). Bub1 and TTK, the homolog of the Mps1 gene, were both strongly upregulated by E6/E7. Both cdc2 and cyclin B, partners in the kinase complex that directly regulates progression into M phase, were strongly and reproducibly upregulated by E6/E7 (Table 1). Immunohistochemical analysis indeed showed a significant increase in cells containing cytoplasmic cyclin B1 (data not shown). Also upregulated, however, was the cdc2/cyclin B inhibitor Wee1. The Plk1 and Myt1 transcripts also showed evidence of upregulation (but did not make the stringency cutoff), with significant transcripts significantly up in 10 of 16 and 4 of 16 comparisons, respectively (data not shown).
TABLE 1.
Transcripts in both the G2/M and spindle checkpoints are significantly increased in ≥12 of 16 comparisons of raft cultures by E6E7a
| Transcript | Pathway | Increase (fold) |
|---|---|---|
| cdc2 | G2/M checkpoint | 4.07 |
| Cyclin B1 | G2/M checkpoint | 4.80 |
| Wee1 | G2/M checkpoint | 2.91 |
| TTK | Spindle checkpoint | 4.42 |
| BUB 1 | Spindle checkpoint | 1.40 |
Both cdc2 and cyclin B1 are upregulated along with the mitotic inhibitor Wee1. Both TTK and BUB 1 transcripts from the spindle checkpoint are upregulated.
Translation.
A significant number of transcripts within the translation hierarchy were decreased in ≥12 of 16 comparisons by E6/E7 relative to the pLC controls (Fig. 5). Among these transcripts were small and large ribosomal protein transcripts and translation initiation factors. Transcripts decreased in E6/E7 rafts relative to the pLC controls included mitochondrial ribosomal protein S12 (BE407202), eukaryotic translation initiation factor 5A (U17969), ribosomal protein S30 (X65923), initiation factor 4E bp 1 (L36055), ribosomal protein L35 (X55954), and ribosomal protein S13 (AA013087).
To determine the significance of this particular cluster of decreased transcripts, we asked if similar results occurred in a separate experiment conducted in an identical manner with a different vector. In this experiment, an even greater number of transcripts in the translation hierarchy were decreased, including ribosomal protein SA (M14199), ribosomal protein S21 (AI526028), ribosomal protein L37a (L06499), ribosomal protein S15 (J02984), initiation factor 4E bp 1 (L36055), ribosomal protein S2 (X17206), ribosomal protein S17 (M13932), translation initiation factor (W28892), ribosomal protein L29 (Z49148), ribosomal protein L38 (Z26876), ribosomal protein L3 (AL022326), ribosomal protein S9 (U14971), ribosomal protein S3 (X55715), ribosomal protein, large, P1 (M17886), elongation factor 1 alpha 1 (L41498, J04617), and ribosomal protein, large, P0 (M17885). Together, the data suggest a significant effect by E6/E7 upon transcripts involved in protein synthesis.
Real-time PCR quantification of transcript levels.
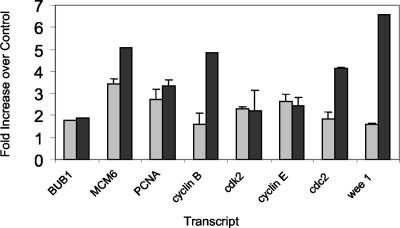
Transcripts were chosen for quantification via real-time (Taqman) PCR to determine if alternative means of measuring transcript levels would support alterations measured by microarray ADI values. In most cases the Taqman values were either very close to or less than those obtained via microarray ADI values (Fig. 7). However, in general, transcripts found to be increased in microarray analyses were also found to be increased in PCR studies (Fig. 7).
FIG. 7.
Bar graph comparing Taqman PCR data (grey bars) to Affymetrix array (black bars) ADIs.
DISCUSSION
The results reported here represent the first comprehensive microarray analysis to assess the consequences of HPV E6/E7 oncogene activities in the normal tissue environment that supports viral reproduction: noncycling, differentiated keratinocytes in organotypic epithelial raft cultures. To meet the challenge of obtaining biologically and statistically significant microarray data from the highly complex cultures, we used pairwise comparisons of quadruplet microarrays, thereby establishing the reproducibility of transcript alterations. As a result, we are able to report a comprehensive and reproducible data set of genes regulated by HPV-18 E6/E7. These data delineate major changes in host cell physiology which form a foundation for understanding how the HPV E6 and E7 proteins act to promote viral DNA replication and other aspects of the HPV reproductive program. These changes involve reprogramming of the differentiating keratinocyte in multiple ways that include dramatic alterations in nucleic acid metabolism, promotion of cell cycle and signaling events, and suppression of protein synthesis and ribosomal biogenesis.
It is worth commenting on the suppression of protein metabolism. The studies reported here show that E6/E7 can have a dramatic effect upon transcripts that encode elements of the DNA synthetic machinery. Together with the observation that genes of DNA metabolism, including DNA synthesis, are strongly upregulated by E6/E7, it appears that E6/E7 causes a shift away from protein synthesis toward DNA synthesis that is consistent with an important role in promoting viral replication within differentiated keratinocytes. Interestingly, a growing body of evidence points to an interaction between cell signaling networks that regulate cell proliferation via mitogens and cell growth regulated by nutrients (45).
Exit from the cell cycle accompanied by concomitant growth in cell size and differential expression of proteins is normally associated with the terminal differentiation, or cornification, of the keratinocyte. HPV genomic DNA and HPV E6/E7 are able to subvert this balance and allow DNA synthesis to occur in a subset of the most differentiated cells of raft cultures (7, 21, 20). While it has long been understood that cell growth is required for cell cycle progression to occur, recent progress in understanding this complex relationship indicates that ribosome biogenesis may have a suppressive effect upon cell cycle progression (46). The relationship between protein synthesis and cell proliferation is likely to be important for epithelial homeostasis and for the stability of the committed, differentiated phenotype which is characteristic of the suprabasal keratinocyte (18). For this reason, it will be interesting to determine whether a cause-and-effect relationship exists between the loss of transcripts of the protein synthesis apparatus caused by E6/E7 and entry into S phase in differentiated keratinocytes.
Previous studies with marker proteins such as PCNA or incorporation of bromodeoxyuridine have clearly shown that expression of HPV E7 alone or E6 and E7 together is able to induce DNA synthesis within the differentiating compartment of keratinocyte raft cultures (7, 26, 20). Through all stages of our analyses, we followed PCNA expression because this gene is strongly induced by E6/E7 and E7 (7, 9. 26, 27). As our analyses progressed, the reproducible upregulation of PCNA by E6/E7 provided assurance that we were measuring a physiologically relevant subset of genes. It is now clear that PCNA is but one member of a large subset of DNA synthesis genes whose expression is directly or indirectly upregulated by E6/E7. Other genes in this category with upregulated expression include multiple MCM transcripts, cdc6, and cdc7, among others. These findings are in agreement with the direct assessment of host DNA and viral DNA replication in raft cultures reported previously. In addition, cell cycle kinase proteins that are involved in regulation of the DNA synthetic machinery, including cdc2, cyclin B, cdk2 and cyclin E, are also upregulated. Together, these data provide the most thorough description of E6/E7 effects on DNA synthesis to date. Equally, these data verify that our methods are able to reveal cellular changes brought about by HPV oncogenes with high precision.
In addition, multiple other pathways and functional categories of genes whose expression could not be predicted are affected by E6/E7. Previous studies used microarray analyses to understand cellular changes induced by full-length HPV-11 and HPV-31 genomes (6, 48, 42), by HPV oncogenes (35, 44, 17), or by cell cycle arrest brought about by HPV E2 in E6/E7-positive cells (51). These studies, generally conducted in cycling cells in monolayer cultures, have found a variety of changes connected to HPV. For instance, interferon-regulated genes and STAT-1 are decreased in cells that maintain episomal copies of the high-risk HPV-31 genome (6). High-risk HPV-16 E6/E7, especially E6 but also E7, was likewise found to decrease interferon-regulated gene expression, suggesting that evasion of interferon pathways by HPV might be attributed to E6/E7 (35). This result is in concordance with previously published results showing E7 abolition of alpha interferon signaling (4, 5). However, given the limitations of microarray analyses that attempt to monitor expression of thousands of genes in a differentiating system, it is not surprising that other studies have not found genes involved in immune response such as STAT-1 to be regulated by E6/E7 (17). While our own approach did not identify STAT-1 as altered by E6/E7, multiple immunity and defense genes, including interleukin-10 and interleukin-13 receptors, interleukin-5, and STAT-3, among others, were found to be downmodulated (see complete listing of genes). These results confirm and extend the previous observations in this category of function (6, 35).
Due to the number of pathways affected by E6/E7 expression, it is most difficult to discuss each in detail, but it is worthwhile to point out several interesting and novel insights that our results have provided. The HPV oncogenes E6 and E7 are sufficient to immortalize cells. However, it is well understood that an accumulation of genetic events following infection by high-risk HPVs is required for malignant progression. Recent publications indicate that abnormal centrosome duplication accounts for genomic instability in HPV lesions and is therefore an important contributor to malignant progression (14). The E6 and E7 oncogenes are sufficient to induce these abnormalities (15, 16).
A growing body of literature has begun to reveal the molecular machinery that underlies centrosome duplication. Many of the genes and their encoded proteins required for this process are members of the mitotic spindle checkpoint, which acts to ensure the faithful segregation of chromosome by ensuring that the mitotic spindle is fully and properly assembled before mitosis can proceed. We found that two members of this pathway, Bub1 and TTK (Mps1), are upregulated by E6/E7, suggesting that these genes may directly induce the centrosome abnormalities related to HPV. TTK (Mps1), which is strongly induced by E6/E7, is particularly interesting because its expression has been shown to be sufficient to promote centrosome duplication (19, 36). In postmitotic, differentiated cells, centrosome abnormality should have little effect on viral productive phase. However, if inappropriately expressed in the basal/parabasal cells as in dysplasias, upregulation of these genes could contribute to lesion progression if these cells escape apoptosis.
Another interesting observation is the strong upregulation of both cdc2 and cyclin B, two genes that together form the kinase required for progression into M phase. This observation is challenging because complete cell division is not consistent with the observation of polyploidy or multinucleated cells in papillomatous lesions or in E7 raft cultures (9). Thus, it is not surprising that a transcript encoding the well-characterized inhibitor of cdc2/cyclin B, the Wee1 kinase, is also upregulated. These data suggest that although cdc2/cyclin B may be upregulated, the kinase may be kept in an inactive state by Wee1.
In summary, our analyses have identified multiple pathways altered by the HPV oncoproteins. The challenge in the future will be to identify key functions and pathways that might be specifically targeted to block viral replication and host cell pathology.
Supplementary Material
Acknowledgments
David Elrod and Eileen Reardon are thanked for annotation of genes represented on the microarrays.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Baker, C., and C. Calef. 1997. Maps of papillomavirus mRNA transcripts. In G. Myers, F. Sverdrup, C. Baker, A. McBride, K. Münger, H.-U. Bernard, and J. Meissner (ed.), Human papillomaviruses, compendium, part III. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 2.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, L., C. Edmonds, and K. H. Vousden. 1990. Ability of the HPV16 E7 protein to bind RB and induce DNA synthesis is not sufficient for efficient transforming activity in NIH3T3 cells. Oncogene 5:1383-1389. [PubMed] [Google Scholar]
- 4.Barnard, P., and N. A. McMillan. 1999. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-α. Virology 259:305-313. [DOI] [PubMed] [Google Scholar]
- 5.Barnard, P., E. Payne, and N. A. McMillan. 2000. The human papillomavirus E7 protein is able to inhibit the antiviral and anti-growth functions of interferon-α. Virology 277:411-419. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 8.Chien, W.-M., J. N. Parker, D.-C. Schmidt-Grimminger, T. R. Broker, and L. T. Chow. 2000. Casein kinase II phosphorylation of the human papillomavirus-18 E7 protein is critical for promoting S-phase entry. Cell Growth Diff. 11:425-435. [PubMed] [Google Scholar]
- 9.Chien, W.-M., F. Noya, H. M. Benedict-Hamilton, T. R. Broker, and L. T. Chow. 2002. Alternative fates of keratinocytes transduced by human papillomavirus type 18 E7 during squamous differentiation. J. Virol. 76:2964-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, L. T., and T. R. Broker. 1997. Small DNA tumor viruses, p. 267-302. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 11.Chow, L. T., and T. R. Broker. 1997. In vitro experimental systems for HPV: Epithelial raft cultures for investigations of viral reproduction and pathogenesis and for genetic analyses of viral proteins and regulatory sequences. Clin. Dermatol. 15:217-227. [DOI] [PubMed] [Google Scholar]
- 12.Demers, G. W., E. Espling, J. B. Harry, B. G. Etscheid, and D. A. Galloway. 1996. Abrogation of growth arrest signals by human papillomavirus type 16 E7 is mediated by sequences required for transformation. J. Virol. 70:6862-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dollard, S. C., J. L. Wilson, L. M. Demeter, W. Bonnez, R. C. Reichman, T. R. Broker, and L. T. Chow. 1992. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes Dev. 6:1131-1142. [DOI] [PubMed] [Google Scholar]
- 14.Duensing, S., A. Duensing, E. R. Flores, A. Do, P. F. Lambert, and K. Mhnger. 2001. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J. Virol. 75:7712-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duensing, S., A. Duensing, C. P. Crum, and K. Mhnger. 2001. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61:2356-2360. [PubMed] [Google Scholar]
- 16.Duensing, A., J. Basile, S. Piboonniyom, S. Gonzalez, C. P. Crum, and K. Mhnger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffy, C. L., S. L. Phillips, and A. J. Klingelhutz. 2003. Microarray analysis identifies differentiation-associated genes regulated by human papillomavirus type 16 E6. Virology 314:196-205. [DOI] [PubMed] [Google Scholar]
- 18.Fisher, C. 1994. The cellular basis of development and differentiation in mammalian keratinizing epithelia, p. 131-150. In I. Leigh, B. Lane, and F. Watt (ed.), The keratinocyte handbook. Cambridge University Press, Cambridge, England.
- 19.Fisk, H. A., and M. Winey. 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell 106:95-104. [DOI] [PubMed] [Google Scholar]
- 20.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garner-Hamrick, P., and C. Fisher. 2002. HPV episomal copy number closely correlates with cell size in keratinocyte monolayers. Virology 301:334-341. [DOI] [PubMed] [Google Scholar]
- 23.He, W., and C. Fisher. 2003. Direct activation of cyclin-dependent kinase 2 by human papillomavirus E7. J. Virol. 77:10566-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Jewers, R. J., P. Hildebrandt, J. W. Ludlow, B. Kell, and D. J. McCance. 1992. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J. Virol. 66:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jian, Y. C., D.-C. Schmidt-Grimminger, W.-M. Chien, X. Wu, T. R. Broker, and L. T. Chow. 1998. Post-transcriptional induction of p21cip1 protein by human papillomavirus E7 inhibits unscheduled DNA synthesis reactivated in differentiated keratinocytes. Oncogene 17:2027-2038. [DOI] [PubMed] [Google Scholar]
- 27.Jian, Y., B. A. Van Tine, W.-M. Chien, G. M. Shaw, T. R. Broker, and L. T. Chow. 1999. Concordant induction of cyclin E and p21cip1 in differentiated keratinocytes by the HPV E7 protein inhibits cellular and viral DNA synthesis. Cell Growth Differ. 10:101-111. [PubMed] [Google Scholar]
- 28.Jones, D. L., R. M. Alani, and K. Münger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, S.-R., J.-S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 31.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers, C., M. G. Frattini, J. B. Hudson, and L. A. Laimins. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973. [DOI] [PubMed] [Google Scholar]
- 33.McIntyre, M. C., M. G. Frattini, S. R. Grossman, and L. A. Laimins. 1993. Human papillomavirus type 18 E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. J. Virol. 67:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Münger, K., and A. L. Halpern. 1997. HPV 16 E7: Primary structure and biological properties, p. 17-36. In G. Myers, F. Sverdrup, C. Baker, A. McBride, K. Münger, H.-U. Bernard, and J. Meissner, (ed.), Human papillomaviruses compendium, part III. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 35.Nees, M., J. M. Geoghegan, T. Hyman, S. Frank, L. Miller, and C. D. Woodworth. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J. Virol. 75:4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norman, T. C., D. L. Smith, P. K. Sorger, B. L. Drees, S. M. O'Rourke, T. R. Hughes, C. J. Roberts, S. H. Friend, S. Fields, and A. W. Murray. 1999. Genetic selection of peptide inhibitors of biological pathways. Science 285:591-595. [DOI] [PubMed] [Google Scholar]
- 37.Noya, F., W.-M. Chien, T. R. Broker, and L. T. Chow. 2001. p21cip1 degradation in differentiated keratinocytes is abrogated by costabilization with cyclin E induced by human papillomavirus E7. J. Virol. 75:6121-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noya, F., W.-M., Chien, X. Wu, N. S. Banerjee, J. C. Kappes, T. R. Broker, and L. T. Chow. 2002. The promoter of the human proliferating cell nuclear antigen gene is not sufficient for cell cycle-dependent regulation in organotypic cultures of keratinocytes. J. Biol. Chem. 277:17271-17280. [DOI] [PubMed] [Google Scholar]
- 39.Park, R. B., and E. J. Androphy. 2002. Genetic analysis of high-risk E6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J. Virol. 76:11359-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker, J. N., W. Zhao, K. J. Askins, T. R. Broker, and L. T. Chow. 1997. Mutational analyses of differentiation-dependent human papillomavirus type-18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ. 11:751-762. [PubMed] [Google Scholar]
- 41.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruutu, M., P. Peitsaro, B. Johansson, and S. Syrjänen. 2002. Transcriptional profiling of a human papillomavirus 33-positive squamous epithelia cell line which acquired a selective growth advantage after viral integration. Int. J. Cancer 100:318-326. [DOI] [PubMed] [Google Scholar]
- 43.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 44.Schwarze, S. R., S. E. DePrimo, L. M. Grabert, V. X. Fu, J. D. Brooks, and D. Jarrod. 2002. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J. Biol. Chem. 277:14877-14883. [DOI] [PubMed] [Google Scholar]
- 45.Shamji, A. F., P. Nghiem, and S. L. Schreiber. 2003. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell 12:271-280. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2:E71-E72. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Hum. papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, J. T., S. T. Oh, S. S. Terhune, and L. A. Laimins. 2001. Cellular changes induced by low-risk human papillomavirus type 11 in keratinocytes that stably maintain viral episomes. J. Virol. 75:7564-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Muoz. 1999. Hum. papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 51.Wells, S. I., B. J. Arinow, T. M. Wise, S. S. Williams, J. A. Couget, and P. M. Howley. 2003. Transcriptome signature of irreversible senescence in human-papillomavirus-positive cervical cancer cells. Proc. Natl. Acad. Sci. USA 100:7093-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, J. L., Dollard, S. C., Chow, L. T., and T. R. Broker. 1992. Epithelial-specific gene expression during differentiation of stratified primary human keratinocyte cultures. Cell Growth Differ. 3:471-483. [PubMed] [Google Scholar]
- 53.zur Hausen, H. 1999. Viruses in human cancers. Eur. J. Cancer 35:1878-1885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.