Abstract
The induction of alpha/beta interferon (IFN-α/β) is a powerful host defense mechanism against viral infection, and many viruses have evolved strategies to overcome the antiviral effects of IFN. In this study, we found that IFN-α had only some degree of antiviral activity against Japanese encephalitis virus (JEV) infection, in contrast to another flavivirus, dengue virus serotype 2, which was highly sensitive to IFN-α in the cultured cell system. JEV infection appeared to render cells resistant to IFN-α since the IFN-α-induced luciferase reporter activity driven by the IFN-stimulated response element (ISRE) was gradually reduced as the JEV infection progressed. Since the biological activities of IFNs are triggered by the Janus kinase (Jak) signal transducer and activation of transcription (Stat) signaling cascade, we then studied the activation of Jak-Stat pathway in the virus-infected cells. The IFN-α-stimulated tyrosine phosphorylation of Stat1, Stat2, and Stat3 was suppressed by JEV in a virus replication and de novo protein synthesis-dependent manner. Furthermore, JEV infection blocked the tyrosine phosphorylation of IFN receptor-associated Jak kinase, Tyk2, without affecting the expression of IFN-α/β receptor on the cell surface. Consequently, expression of several IFN-stimulated genes in response to IFN-α stimulation was also reduced in the JEV-infected cells. Overall, our findings suggest that JEV counteracts the effect of IFN-α/β by blocking Tyk2 activation, thereby resulting in inhibition of Jak-Stat signaling pathway.
The alpha/beta interferons (IFN-α/β) are directly produced by most types of cells in response to viral infection, and play an important role in the first line of host defense in mammals (50). To initiate viral replication and production of progeny virus, many viruses have evolved different strategies to circumvent the host IFN-α/β response, such as inhibition of IFN production and signaling and blocking of the functions of IFN-induced proteins (15, 28, 41, 50, 52). In fact, it is quite common for certain viruses to encode more than one mechanism in order to evade the IFN response at one or more levels. The speed and efficiency with which a given virus circumvents the IFN response may be critical determinants in the virus host range and pathogenicity.
Since the discovery of IFN nearly half a century ago, much has been learned about the molecular composition of IFN, as well as its mode of induction and action. The biological activities of IFNs are triggered by the binding of IFNs to their cognate receptors on the cell surface to initiate a signaling cascade, known as the Janus kinase (Jak)-signal transducer and activation of transcription (Stat) pathways (18, 35, 50, 55). The large family of IFN-α/β proteins all bind to a single type of receptor, which is composed of two chains: IFNAR1 and IFNAR2. The intracellular domain of IFNAR1 associates with a member of the Jak kinase family, Tyk2, whereas IFNAR2 associates with Jak1. The major substrates for tyrosine phosphorylation subsequent to IFN receptor binding are members of the Stat family of transcription factors. These proteins are normally latent and reside in the cytoplasm in unstimulated cells. Once phosphorylated, Stat1 and Stat2 dimerize and assemble with another protein, p48 (IRF-9), to form the multimeric transcription factor, ISGF3. ISGF3 binds to the IFN-stimulated response element (ISRE) of IFN-stimulated genes (ISGs) in the nucleus and activates their transcription. IFNs can induce the synthesis of more than 300 cellular proteins, including enzymes, signaling proteins, chemokines, antigen presentation proteins, transcription factors, heat shock proteins, and apoptotic proteins (11). Of these proteins, the best-characterized IFN-inducible components of the antiviral response are the double-stranded RNA-activated protein kinase (PKR), the 2′,5′-oligoadenylate synthetases (2′-5′-OAS), and the Mx protein(s).
A number of viruses have been found to impair the activity of the Jak-Stat signaling pathway by using various mechanisms. Several poxviruses encode a soluble IFN receptor homologue that acts as a decoy to inhibit the biological activity of IFN (9, 56, 60). The adenovirus E1A protein can inhibit both IFN-α/β and IFN-γ signaling by mechanisms such as blocking the ISGF3 transcriptional complex formation (27), decreasing Stat1 and p48 protein levels (34), and competing for the CREB-binding protein (CBP)/p300 with Stat1 (65) or Stat2 (3), as well as by suppressing Stat1 through a CBP/p300-independent mechanism (40). Several members of Paramyxoviridae are also capable of blocking IFN-α/β signaling (19), although through distinct mechanisms. For instance, simian virus 5 (14) and the mumps virus (31) may target Stat1 for degradation. Human parainfluenza virus type 2, in contrast, causes Stat2 degradation (46, 47). Sendai virus may interact with Stat1 and thereby inhibit the IFN-α/β-stimulated tyrosine phosphorylation of Stat (16, 20, 29). Besides the paramyxoviruses, other RNA viruses, i.e., Ebola virus (21), hepatitis C virus (HCV) (24), and dengue virus serotype 2 (DEN-2) (45) have also been reported to inhibit IFN signaling.
The Flavivirus genus comprises over 70 viruses, many of which are important human pathogens and may cause severe encephalitic, hemorrhagic, hepatic, and febrile illnesses (4, 39). Of particular importance for public health are the mosquito-borne flaviviruses such as DEN, yellow fever virus, West Nile virus (WNV) and Japanese encephalitis virus (JEV). Despite the major clinical impact of flaviviruses, no vaccine (besides those for yellow fever virus and JEV) or specific antiviral drug is available to treat infections with these viruses. A role for IFN in the modulation of DEN infection has been demonstrated in IFN receptor-deficient mice (26) and in cultured cell systems. IFN-α/β inhibits DEN by blocking the accumulation of negative-sense viral RNA (13) in a PKR-independent manner (12). JEV also appears to be sensitive to IFN (10, 22, 23, 62) and potent IFN inducers (17, 59). However, a recent randomized double-blind placebo-controlled trial showed that IFN-α2a did not improve the outcome of patients with Japanese encephalitis (JE) (53). Recently, expression of the DEN-2 nonstructural proteins NS4B and, to a lesser extent, NS2A and NS4A has been found to result in downregulation of IFN-β-stimulated gene expression and to enhance replication of an IFN-sensitive virus in human A549 cells (45). Moreover, cells expressing NS4B or infected with DEN-2 do not exhibit nuclear Stat1 translocation upon treatment with IFN-β or IFN-γ, indicating that NS4B might be involved in blocking IFN signaling during DEN infections (45).
In the present study, we examined the antiviral effects of IFN-α/β on JEV and DEN-2 infection. We found that pretreatment of cells with IFN-α greatly reduced virus titers and virus-induced cytopathic effects (CPE) in DEN-2 infection but only slightly in JEV infection. We further demonstrated that JEV interfered with Jak-Stat signaling through the blocking of Tyk2 tyrosine phosphorylation and Stat activation. Our findings indicate that JEV is capable of counteracting the signal transduction of IFN-α.
MATERIALS AND METHODS
Viruses and cell lines. A plaque-purified neurovirulent Taiwanese JEV strain, RP-9 (8), was used throughout the present study. A local Taiwanese strain of DEN-2, PL406 (38), isolated from a dengue fever patient was generously provided by the National Institute of Preventive Medicine, Taiwan, Republic of China (ROC). The virus was propagated by using C6/36 cells in RPMI 1640 medium containing 5% fetal bovine serum (FBS; Gibco-BRL). UV irradiation of JEV was done by using a Stratalinker 2400 (Stratagene, La Jolla, Calif.) with a short wavelength of 254 nm at a distance of ca. 5 to 10 cm for 15 min on ice. The virus inactivation was verified by plaque assay and immunofluorescence assay with anti-JEV antibodies. The baby hamster kidney fibroblast cell line, BHK-21, was grown in RPMI 1640 medium containing 5% FBS. The human lung carcinoma cell line, A549, was cultured in F-12 medium (Gibco) supplemented with 10% FBS.
Chemicals and antibodies.
Cycloheximide and human recombinant IFN-αA/D (BglII) were obtained from Sigma. The protease inhibitor cocktail was obtained from Roche. Rabbit polyclonal anti-Stat1, anti-phospho-Stat1 (Tyr701), anti-Stat3, anti-phospho-Stat3 (Tyr705), and anti-phospho-Tyk2 (Tyr1054/1055) antibodies were purchased from Cell Signaling Technology. Rabbit polyclonal Anti-Tyk2 and anti-PKR antibodies were obtained from BD Transduction Laboratories. Rabbit polyclonal anti-IRF-9 and anti-Stat2 antibodies were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-phospho-Stat2 (Tyr689) antibodies were obtained from Upstate Biotechnology. Mouse monoclonal antibodies specific for JEV NS1 were acquired as described previously (63).
Virus infection.
For virus infection, monolayers of the cells grown in 6- or 12-well plates were initially adsorbed with virus at the indicated multiplicity of infection (MOI) for 1 h at 37°C. After 1 h of adsorption, the unbound virus was removed from cells by gentle washing with RPMI 1640 medium. At indicated time points postinfection (p.i.), culture media were harvested for the plaque-forming assay and the lactate dehydrogenase release assay to determine the virus titer and cytotoxicity, as described previously (63). The virus infection rate was monitored by immunofluorescent assay by using the virus-specific monoclonal antibodies as previously described (7, 38) to show a 100% infection rate when an MOI of 5 was used.
Measurement of ISRE activity by the luciferase reporter assay.
A permanent cell line expressing an ISRE-dependent luciferase reporter was established to avoid the varied transfection efficiencies from different experiments. BHK-21 cells were stably transfected with a reporter plasmid, pISRE-Luc (Stratagene), carrying the luciferase gene downstream of five ISRE (5′-TAGTTTCACTTTCCC-3′) binding elements derived from IFI-56, and pcDNA3, a neomycin-resistant gene containing plasmid. The cell lysates were harvested at various experimental time points, and the luciferase activity was determined by using a luciferase assay system kit purchased from Promega. Luciferase activity was expressed as counts per second per milligram of protein.
Western immunoblot analysis.
Cells were lysed in sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 50 mM dithiothreitol, 0.1% bromophenol blue) containing a cocktail of protease inhibitors. Similar amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Hybond-C Super; Amersham). The nonspecific antibody-binding sites were blocked with 5% skim milk in TBS-T (25 mM Tris, 0.8% NaCl, and 2.68 mM KCl [pH 7.4], with 0.1% Tween 20) prior to the addition of the primary antibody. The blots were then treated with a horseradish peroxidase-conjugated secondary antibody (Amersham) and developed with an ECL system (Amersham). For reblotting, the membrane was washed with 1× ReBlot plus strong antibody stripping solution (Chemicon) for 15 min at room temperature. The membrane was then blocked twice with 5% skim milk in TBS-T for 5 min, followed by reprobing with the primary antibody. The bands intensities were quantified by using MetaMorph software from Universal Imaging Corp.
For detection of phosphorylated Jak1, immunoprecipitation was performed by using the Catch-and-Release Immunoprecipitation System from Upstate with an anti-phospho-tyrosine monoclonal antibody (P-Tyr-100 antibody from Cell Signaling Technology). The eluted proteins were then analyzed by Western blotting with anti-Jak1 antibody (BD Transduction Laboratories).
Flow cytometry analysis.
Cells were detached with 5 mM EDTA in phosphate-buffered saline (PBS), washed with PBS, and resuspended in ice-cold F-12 medium containing 10% FBS and 0.1% sodium azide. For each sample, 105 cells were incubated with 10 μg of goat anti-human IFNAR1 (R&D Systems) or rabbit anti-human IFNAR2 (PBL Biomedical Laboratories) polyclonal antibodies/ml in F-12 medium containing 10% FBS on ice for 1 h. After a wash with F-12 medium containing 10% FBS, cells were incubated with biotin-SP-conjugated rabbit anti-goat or goat anti-rabbit immunoglobulin G antibodies (H+L; Jackson ImmunoResearch Laboratories) in a 1:75 dilution on ice for 45 min. Cells were washed again and incubated on ice for 30 min in the dark with R-phycoerythrin-conjugated streptavidin (Jackson ImmunoResearch Laboratories) in a 1:200 dilution. Subsequently, the stained cells were washed, resuspended, and analyzed by FACScaliber (Becton Dickinson). Negative controls consisted of cells that were left unstained or stained with an unrelated primary antibody.
RNA extraction and RT-PCR.
Total cellular RNA was isolated from 106 cells by using RNeasy Total RNA kit according to the manufacturer's recommendation (Qiagen). Reverse transcription-PCR (RT-PCR) was carried out with 1 μg of RNA by using the ThermoScript RT-PCR System (Invitrogen). The oligo(dT)-primed cDNA was serially diluted (1×, 10×, 100×, and 1,000×) and amplified by using the specific primers for human PKR (5′-CCAGGATACGGGAAGAAGAA-3′ and 5′-GATATGCAAGTTTAGCGGCC-3′), Stat1 (5′-CCCATGGAAATCAGACAGTACC-3 and 5′-ACTCTTTGCCACACCATTG-3′), IRF-9 (5′-CGGAGTGTGCTGGGATGAT-3′ and 5′-TCAGCAACATCCATGCGGC-3′), and β-actin (5′-TCCTGTGGCATCCACGAAACT-3′ and 5′-GAAGCATTTGCGGTGGACGAT-3′), respectively. Amplification reactions were run in a total volume of 25 μl for 30 cycles by using a PTC-200 Peliter Thermal Cycler (MJ Research), and then the amplified PCR products were analyzed by 2% agarose gel electrophoresis.
RESULTS
IFN-α readily inhibited replication of DEN-2 but only slightly inhibited JEV infection of BHK-21 cells.
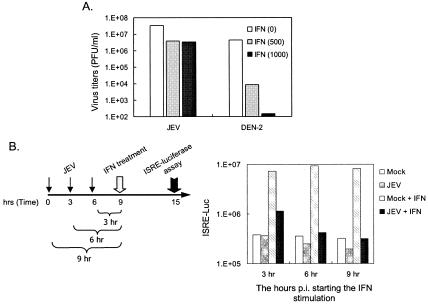
To examine the effects of IFN-α/β on JEV infection, we used a human recombinant IFN-αA/D (Sigma) that is known as a universal IFN-α/β and which crosses the species barrier. It has been previously reported that pretreatment of cells with IFN-α/β protected cells from DEN infection (13). Therefore, we included a DEN serotype 2 (DEN-2) here as a control. Baby hamster kidney BHK-21 cells were treated with or without IFN-α for 6 h prior to JEV or DEN-2 infection (MOI of 5). As shown in Fig. 1A, IFN-α strongly inhibited DEN-2 production (>3-log orders of magnitude) in a dose-dependent manner at 24 h p.i., whereas JEV production was only slightly affected (ca. 1 log order of magnitude) by the same IFN-α treatment. The effects of IFN-α on the CPE induced by JEV or DEN-2 were assessed by the release of a cytoplasmic enzyme, lactate dehydrogenase, into the culture medium. IFN-α decreased the JEV-induced CPE only slightly; in contrast, the DEN-2-induced CPE was effectively reduced by IFN-α (data not shown). These results indicate that flaviviruses DEN-2 and JEV have different sensitivities to IFN-α/β and that JEV appeared to be much more resistant than DEN-2 to the inhibitory effects of IFN.
FIG. 1.
Effects of IFN-α on JEV-infected cells. (A) The antiviral effects of IFN-α on JEV and DEN-2 infection of BHK-21 cells. BHK-21 cells were untreated or pretreated with recombinant human IFN-αA/D (500 and 1,000 U/ml) for 6 h before JEV or DEN-2 (MOI = 5) infection. At 24 h postinfection, virus titers shown as PFU per milliliter were determined by the plaque-forming assays. The representative results from three independent experiments are shown here. (B) Decrease in the IFN-α-induced ISRE activity in JEV-infected BHK-21 cells. The ISRE-Luc/BHK-21 cells were infected with JEV (MOI = 5) for 3, 6, and 9 h and then treated or not treated with IFN-αA/D (1,000 U/ml) for 6 h. The ISRE activation levels were determined by measuring the luciferase activity. The representative results of two independent experiments are shown here.
JEV blocked the IFN-α-induced Stat1 phosphorylation in BHK-21 cells.
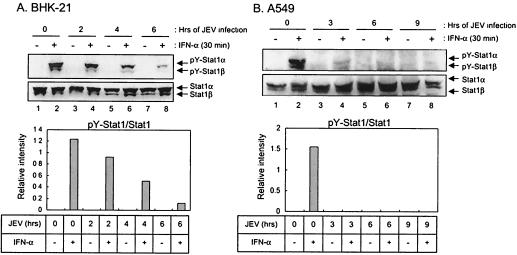
The inefficiency of IFN-α to block JEV production implies that JEV may be able to block IFN-α signaling. IFN-induced ISGF3, a transcription factor complex including activated Stat1, Stat2, and IRF-9 can specifically bind to the ISRE located in the promoter region of many, if not all, IFN-inducible genes. To monitor the activation of IFN-α signaling, a stable ISRE-Luc/BHK-21 cell line was established by transfection with a plasmid that carries an ISRE-containing promoter upstream of the luciferase gene (pISRE-Luc; Stratagene). As shown in Fig. 1B, this ISRE-Luc/BHK-21 cell line responded well to IFN-α treatment, as evidenced by the high luciferase activity in the mock-infected cells after IFN-α stimulation. This IFN-α responsiveness was gradually reduced to a basal level by JEV (MOI of 5) infection over 3, 6, and 9 h p.i. (Fig. 1B), and this suppression of ISRE-driving luciferase by JEV was infectious dose dependent (data not shown). Since JEV-induced CPE started to appear ca. 24 h p.i (37), the suppression of ISRE activity observed here was an early event of JEV infection and was not simply a consequence of virus-induced cytotoxicity. Because the Stat family of transcription factors is the major substrate for tyrosine phosphorylation subsequent to IFN receptor binding, we then analyzed the Stat1 phosphorylation by immunoblotting. JEV greatly reduced the levels of Stat1 (Tyr701) tyrosine phosphorylation (pY-Stat1) in an infectious time-dependent manner upon IFN-α stimulation of BHK-21 cells (Fig. 2A), although the total endogenous Stat1 protein levels were not significantly changed (Fig. 2A). As evident by a drop of pY-Stat1/Stat1 ratios (Fig. 2A, bottom panel), our results suggest that JEV possesses the ability to block IFN-α/β signaling in the infected cells.
FIG. 2.
Inhibition of tyrosine phosphorylation of Stat1 in JEV-infected cells. BHK-21 (A) or A549 (B) cells were mock-infected or infected with JEV (MOI = 5) for various time periods (in hours) as shown on top of each panel prior to stimulation with IFN-αA/D (1,000 U/ml) for 30 min or left unstimulated. The cell lysates were examined by Western blot analyses with the antibodies to Stat1 (Tyr701) tyrosine phosphorylation (pY-Stat1) or Stat1 as indicated on the right side of each panel. The bands intensities were quantified by MetaMorph (Universal Imaging Corp.), and the ratios of pY-Stat1 to Stat1 are shown.
Blocking of IFN-α signaling by JEV was not limited to one single cell type.
To exclude the possibility that this inhibition of IFN-α signaling by JEV infection was only restricted to BHK-21 cells, the human lung carcinoma A549 cell line, which is capable of producing and is responsive to IFN-α/β, was used to study the effect of JEV infection on IFN signaling. Based on the virus titration and immunoassay, A549 cells were found to be susceptible to both JEV and DEN-2 infections (data not shown). The suppression of IFN signaling by JEV in A549 cells was revealed by Western blot analyses as shown in Fig. 2B, and JEV readily inhibited IFN-α-stimulated tyrosine phosphorylation of Stat1 as early as 3 h p.i. (Fig. 2B). Together, these results suggest that JEV infection interferes with the IFN-α/β-induced Jak-Stat signaling pathway regardless of host cell type.
Suppression of the IFN-α signal transduction required viral replication and newly synthesized protein.
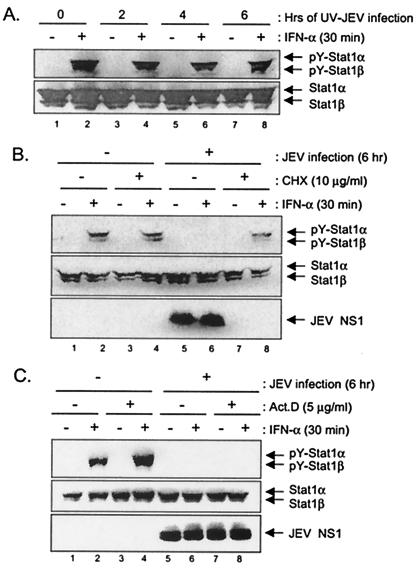
Viral proteins acting as IFN antagonists to inhibit the IFN signal transduction pathway have been identified for several viruses (50, 52). To further characterize whether JEV replication was essential for blocking IFN-α signal transduction, we used the UV-inactivated JEV to infect BHK-21 cells (at an MOI of 5). The levels of Stat1 (Tyr701) phosphorylation upon IFN-α stimulation were not affected by UV-inactivated JEV at 2, 4, or 6 h p.i. (Fig. 3A). An even higher MOI of UV-inactivated JEV (MOI of 50) was also found not to affect the Stat1 phosphorylation (data not shown). In the presence of the potent protein synthesis inhibitor, cyclocheximide (10 μg/ml), Stat1 phosphorylation was still detectable in the mock-infected BHK-21 cells upon IFN-α stimulation (Fig. 3B, top panel, lane 4), indicating that no newly synthesized host protein was required for Stat1 activation. However, in the presence of cycloheximide, which blocked JEV protein expression (Fig. 3B, bottom panel, compare lanes 5 to 6 to lanes 7 to 8), the blocking of Stat1 phosphorylation was no longer seen in the JEV-infected cells (Fig. 3B, top panel, compare lane 6 to lane 8), suggesting that newly synthesized protein was required for JEV to block IFN signaling. The addition of actinomycin D, which blocks the DNA-dependent RNA polymerase but not the viral transcription, did not hamper the JEV ability to block Stat1 phosphorylation (Fig. 3C), supporting the notion that host transcription was not involved in the JEV's suppression of IFN signaling. Together, these results suggest that inhibition of the IFN-α signaling by JEV likely requires productive viral replication to synthesize its viral proteins.
FIG. 3.
Blocking of Stat1 phosphorylation requires JEV replication and newly synthesized proteins. (A) BHK-21 cells infected with UV-inactivated JEV (MOI = 5) for 2, 4, and 6 h were stimulated with IFN-αA/D (1,000 U/ml) for 30 min or left unstimulated. The cell lysates were then harvested for Western blotting with anti-phospho-Stat1 (Tyr701) and anti-Stat1 antibodies. (B) BHK-21 cells pretreated with or without cyclocheximide (10 μg/ml) for 1 h were mock infected or infected with JEV (MOI = 5) in the presence or absence of cyclocheximide (10 μg/ml) for 6 h. The cells were then stimulated with IFN-αA/D (1,000 U/ml) for 30 min before the cell lysates were harvested for Western blot analyses with the antibodies as indicated on the right side of each panel. (C) Actinomycin D (5 μg/ml) treatment was started 1 h prior the infection and maintained throughout the experiment in the indicated samples. BHK-21 cells were mock infected or infected with JEV (MOI = 5) for 6 h in the presence or absence of actinomycin D. Before they were harvested for Western blotting, the cells were stimulated with IFN-αA/D (1,000 U/ml) for 30 min. The antibodies used for each panels are indicated on the right side of the figure.
DEN-2 infection suppressed the IFN-α-induced Stat1 tyrosine phosphorylation in a much slower fashion.
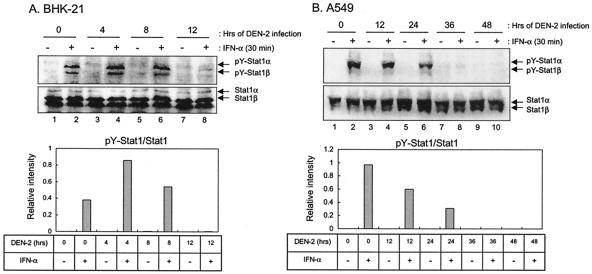
In agreement with a previous report by Diamond et al. (13), we also found that IFN-α efficiently blocked the replication of DEN-2 in our cell culture system (Fig. 1A). The discrepancy of JEV and DEN-2 in their sensitivities to IFN-α treatment suggests that the IFN-α/β signaling events in DEN-2-infected cells might not be blocked in the same efficiency as in the JEV-infected cells. This notion is confirmed in Fig. 4, which demonstrated that during the early hours of infection, DEN-2 did not appear to reduce the levels of IFN-α-stimulated Stat1 phosphorylation in either BHK-21 (up to 8 h p.i. in Fig. 4A) or A549 (up to 24 h p.i. in Fig. 4B) cells. However, at later time points of DEN-2 infection, significant reduction of Stat1 phosphorylation in both of the cell types was observed. Therefore, our results suggest that different flaviviruses, such as JEV and DEN-2 might possess varied abilities to block the IFN-induced Jak-Stat activation.
FIG. 4.
Effects of DEN-2 infection on IFN-α-stimulated tyrosine phosphorylation of Stat1. BHK-21 (A) and A549 (B) cells were mock infected or infected with DEN-2 (MOI = 5) for various hours as indicated and then stimulated with IFN-αA/D (1,000 U/ml) for 30 min. Cell lysates were analyzed by Western blotting with anti-phospho-Stat1 (Tyr701) and anti-Stat1 antibodies. The bands intensities were quantified by MetaMorph (Universal Imaging Corp.), and the ratios of pY-Stat1 to Stat1 are shown.
Tyk2 phosphorylation was blocked in JEV-infected cells.
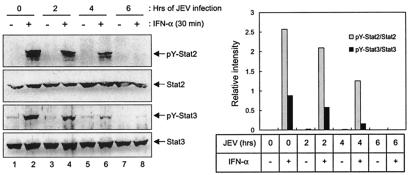
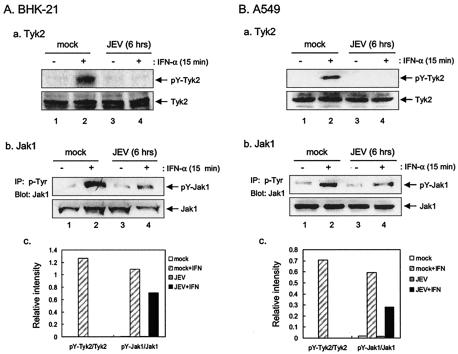
In addition to Stat1, we also examined the phosphorylation levels of other components in the Jak-Stat signaling pathway, such as Stat2, a component of ISGF3, and Stat3, which is one of the downstream molecules of the IFN-α signal transduction but not a component of ISGF3. Western blot analyses revealed an apparent inhibition of IFN-α-stimulated tyrosine phosphorylation of not only Stat1 but also Stat2 (Tyr689) and Stat3 (Tyr705) in the JEV-infected BHK-21 cells (Fig. 5). To further trace the upstream event, we examined whether the receptor-associated kinases, Jak1 and Tyk2, which are responsible for the activation of Stats, were also suppressed by JEV infection. At 6 h after JEV infection, the levels of tyrosine-phosphorylated Tyk2 (Tyr1054/1055) in response to IFN-α stimulation were completely blocked in the JEV-infected BHK-21 cells (Fig. 6Aa and c). Similarly, the IFN-α-induced Tyk2 phosphorylation was also inhibited in JEV-infected A549 cells (Fig. 6Ba and c). The levels of Jak1 tyrosine phosphorylation as revealed by immunoprecipitation-Western immunoblotting were slightly reduced in the JEV-infected BHK-21 (Fig. 6Ab and c) and A549 cells (Fig. 6Bb and c). These results indicate that JEV infection blocks the tyrosine phosphorylation of the IFN-α/β receptor-associated kinase, Tyk2.
FIG. 5.
Suppression of IFN-α-stimulated tyrosine phosphorylation of Stat2 and Stat3 in JEV-infected BHK-21 cells. BHK-21 cells were mock infected or infected with JEV (MOI = 5) for 2, 4, or 6 h prior to stimulation with IFN-αA/D (1000 U/ml) for 30 min or left unstimulated. The cell lysates were examined by Western blot analyses with the antibodies indicated on the right side of each panel. The bands intensities were quantified by MetaMorph (Universal Imaging Corp.), and the ratios of pY-Stat2 to Stat2 and of pY-Stat3 to Stat3 are shown.
FIG. 6.
Suppression of IFN-α-stimulated tyrosine phosphorylation of Tyk2 in JEV-infected cells. BHK-21 (A) or A549 (B) cells were mock or infected with JEV (MOI = 5) for 6 h and then stimulated with IFN-αA/D (1,000 U/ml) for 15 min or left untreated before harvest for Western blotting with anti-phospho-Tyk2 (Tyr1054/1055) and anti-Tyk2 antibodies. The tyrosine phosphorylation of Jak1 was analyzed by immunoprecipitation-Western as described in Materials and Methods. The protein levels of Jak1 were detected by Western immunoblotting. The bands intensities were quantified by MetaMorph (Universal Imaging Corp.), and the ratios of pY-Tyk2 to Tyk2 and of pY-Jak1 to Jak1 are shown.
JEV infection did not affect the expression levels of IFNAR1 and IFNAR2 on the cell surface.
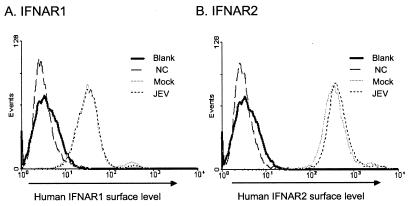
To examine whether the blocking of the receptor-associated kinase by JEV was due to downregulation of cell surface IFN-α/β receptor, we checked the surface expression levels of IFNAR1 and IFNAR2. At 6 h after JEV infection, when the Tyk2 phosphorylation was completely blocked, the surface expression levels of IFNAR1 and IFNAR2 on A549 cells were analyzed by flow cytometry. As shown in Fig. 7, the surface levels of both IFNAR1 and IFNAR2 in JEV-infected A549 cells were comparable to those in mock-infected A549 cells. Therefore, our results imply that the blocking of Tyk2 phosphorylation by JEV infection is not mediated by the suppression of IFNAR1 and IFNAR2 expression on the cell surface.
FIG. 7.
IFNAR1 and IFNAR2 cellular surface levels on JEV-infected cells. The A549 cell surface levels of IFNAR1 (A) and IFNAR2 (B) were determined by flow cytometry analysis. The mock- and JEV-infected (MOI = 5 for 6 h) A549 cells were stained with anti-human IFNAR1 or anti-IFNAR2 antibodies as described in the Materials and Methods. For controls, A549 cells were unstained (blank) or stained with an unrelated antibody (NC).
Suppression of ISG expression by JEV infection.
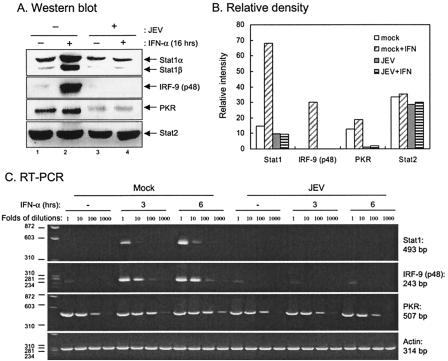
Since the Jak-Stat pathway is an essential signaling pathway for the transcription of many ISGs, whose protein products mediate a variety of specific IFN-dependent antiviral responses, we then examined whether ISGs such as Stats, PKR, and p48 (IRF-9) were consequently suppressed in JEV-infected cells. In A549 cells, of the three known ISGs tested, Stat1, IRF-9, and PKR (to a lesser extent) but not Stat2 were found to be induced by IFN-α in the mock-infected cells (Fig. 8A, lane 2, and B). In contrast, this induction was no longer detectable in the JEV-infected cells (Fig. 8A, lane 4, and B). Furthermore, the RNA levels of these ISGs were determined by RT-PCR with serially diluted samples and, as shown in Fig. 8C, the IFN-α-induced Stat1, IRF-9 (noticeable at every dilution), and PKR (noticeable only at 100- and 1,000-fold dilutions) RNA levels were lower in the JEV-infected cells, suggesting that the reduction of ISG protein level changes likely took place in the transcriptional level. These results demonstrated that JEV infection blocked the IFN-α/β receptor-associated kinase and the downstream events such as Stat phosphorylation, ISRE-dependent promoter activation, and ISG protein expression.
FIG. 8.
Expression of ISGs in JEV-infected cells. (A) A549 cells were mock infected or infected with JEV (MOI = 5) for 6 h and then cultured in the presence or absence of IFN-αA/D (1,000 U/ml) for 16 h. The cell lysates were analyzed by Western blotting with the antibodies indicated on the right side of each panels. (B) The bands intensities were quantified by MetaMorph (Universal Imaging Corp.), and the relative intensities for each protein bands are shown. (C) A549 cells were mock infected or infected with JEV (MOI = 5) for 6 h and then cultured in the presence or absence of IFN-αA/D (1,000 U/ml) for 3 or 6 h. The total cellular RNA was analyzed by RT-PCR as described in Materials and Methods. The expected sizes of PCR fragments for each gene are shown on the right sides of the panels.
DISCUSSION
In the present study, we found that IFN-α (500 U/ml) could only reduce JEV titer by 10-fold (90% reduction in viral production), much less effectively than DEN-2 infection (Fig. 1A). Most significantly, we also found that JEV is likely to have acquired the ability to target the Jak-Stat signaling cascade (Fig. 2, 5, and 6) as a countermeasure for evading the IFN response. IFN has been identified as the most promising antiviral candidate against JEV (10, 22, 23, 62), however, a recent randomized double-blind placebo-controlled clinical trial, which was the first randomized controlled antiviral trial for any flavivirus, showed that intramuscular IFN-α2a did not improve the outcome of children with JE (53). Moreover, it has been revealed that in patients with acute JE, locally synthesized IFN-α appears to have no discernible beneficial effect, since IFN-α was detectable in the cerebrospinal fluid from 9 of 10 patients who died, compared to only 4 of 10 who survived (5). Our findings in the present study that JEV might counteract the IFN actions could provide an explanation as to why JEV may not be sensitive to IFN in the clinical trial and in JE patients.
The interplay between IFN and the virus might affect viral pathogenicity, clearance, and immunity, and viruses encoding IFN antagonists often display increased virulence. The effects of IFN-α/β on flavivirus infection have not been extensively studied, resulting in a very limited understanding of IFN-induced pathways during flavivirus infection. Several flaviviruses, including JEV and DEN (25, 32) can induce IFN-α/β in vivo and in cell cultures (61). The noncytopathic biotype of bovine viral diarrhea virus (ncpBVDV), a pestivirus belonging to the family Flaviviridae, does not induce IFN-α/β in vitro (1) and blocks the induction of IFN by other activators (51), whereas cytopathic BVDV (cpBVDV) has been shown to induce IFN-α/β. However, in contrast to the in vitro findings, ncpBVDV was found to stimulate potent IFN-α/β responses in vivo (6). Recently, the Flv gene, which confers the differential susceptibility to flavivirus-induced morbidity and mortality in mice (61), has been mapped to a cluster of genes encoding the IFN-induced 2′-5′-OAS in chromosome 5 (48). In another mouse model, which has been established to study WNV, the phenotype of resistance or susceptibility is determined by a major locus, Wnv, which was also mapped to cluster of genes encoding 2′-5′-OAS in chromosome 5 (42). In both of these studies, the fourth exon of the L1 isoform of OAS (OAS1B) was found to contain a stop codon, leading to a protein lacking 30% of the C-terminal sequence, in susceptible strains of mice, whereas all of the resistant mice studied thus far have been found to carry an intact variant of the same gene (42, 48). These studies suggest that the IFN-induced 2′-5′-OAS might be critical in restricting WNV replication and play a role in its pathogenesis. In the present study, we further suggest that flavivirus such as JEV might interfere the IFN-induced Jak-Stat signaling and provide another possible mechanism for flavivirus to counteract the IFN host defense system.
HCV, a member of the Flaviviridae family, is a leading cause of liver disease worldwide, and IFN-α alone or in combination with ribavirin is now being used for the treatment of chronic hepatitis C. The HCV genotype is an important predictor of the response to IFN-based antiviral therapy; for example, the genotype 1 is resistant to the antiviral effects of IFN-α (33). At least two HCV proteins, i.e., the envelope glycoprotein E2 and the nonstructural protein NS5A, have been reported to be potential inhibitors of the IFN response (57, 58). Four or probably five genotypes of JEV have been recognized based on their nucleotide sequences (54). The JEV strain, RP-9, used throughout the present study is a neurovirulent strain that causes a high mortality rate in experimental mice (8) and belongs to genotype III of JEV (54). Further studies are required to determine whether the ability of JEV to block IFN signaling is associated with its pathogenicity and whether different strains or genotypes of JEV might exhibit varied sensitivity to IFN inhibition.
Signaling derived from the IFN receptor through the Jak-Stat pathway mediates rapid and robust transcriptional induction of genes encoding antiviral proteins. This process is a highly evolved system that protects animals from viral infection. Using various strategies, both DNA and RNA viruses can inhibit IFN-induced host defense mechanisms. Some viruses reduce the basal levels of particular molecules of the Jak-Stat pathway, for example, there was a significant decrease of Jak1 protein in the human cytomegalovirus-infected cells (43, 44). Inactivation of components of the Jak-Stat pathway has also been demonstrated by several types of viruses. For example, herpes simplex virus type 1 viral gene expression blocks the IFN-stimulated phosphorylation levels of Jaks and Stats (64). Our results indicate that JEV apparently inhibits the IFN-α-stimulated tyrosine phosphorylation but not the total protein expression levels of Stats and Tyk2 (Fig. 2, 5 and 6). Negative regulation of the Jak-Stat pathway can occur by a common mechanism such as receptor internalization or, alternatively, by more specific intracellular inhibition signals such as SH2-containing phosphatases (e.g., SHP-1), the protein inhibitors of activated Stats (PIAS), and suppressor of cytokine signaling (SOCS) proteins (2, 30). Since the surface expression of both chains (IFNAR1 and IFNAR2) of the IFN-α/β receptor was not affected by JEV infection (Fig. 7), it is more likely that an intracellular event blocking the Tyk2 phosphorylation was induced by JEV infection. Human papillomavirus type 18 E6 proteins have been found to physically interact with Tyk2 through a domain, which is important for its binding to the cytoplasmic portion of IFNAR1 and therefore impairs the Tyk2 activation upon IFN-α stimulation (36). A partial inhibition of Tyk2 but not Jak1 tyrosine phosphorylation has been suggested to contribute to the failure of Stat activation in Sendai virus-infected cells (29). The possible molecular mechanisms adapted by JEV to block Tyk2 phosphorylation, for instance, the potential involvement of a tyrosine phosphatase specific for Tyk2 but not Jak1 (49) or a direct protein-protein interaction of certain viral protein(s) with Tyk2, warrant further study.
Similar to the observations on herpes simplex virus type 1 (64), UV-inactivated JEV also failed to block Jak-Stat signaling, indicating that virus replication was a process required for JEV to manifest its anti-IFN effects (Fig. 3A). The addition of a protein synthesis inhibitor, cyclocheximide, suppressed the ability of JEV to block Jak-Stat signaling, indicating that newly synthesized protein(s) was required for this event (Fig. 3B). These results imply that certain unidentified viral protein(s) might possess the IFN antagonist activity. The identity of these JEV proteins, which are able to block IFN signaling as suggested for DEN-2 NS4B (45), will be further studied. The search for this viral antagonist, which renders cells unresponsive to IFN, might provide a better understanding as to how JEV counteracts the antiviral effects of IFN. This information will also be of benefit for the rational design of vaccines and the development of antiviral medicines.
In the present study, we found that JEV infection blocked the IFN-α-stimulated Jak-Stat signaling pathway and resulted in reduced levels of ISRE activity and ISG production. The mechanism adapted by JEV to interfere with IFN-α/β signal transduction was to block the tyrosine phosphorylation of receptor-associated kinase Tyk2 and Stats but not to downregulate the IFN-α/β receptor or the protein expression levels of components in the signaling cascade. These results have added JEV to a growing list of viruses that evolve strategies to counteract the actions of IFN-α/β. This unprecedented study not only shows suppression of IFN-α/β-induced Jak-Stat signaling in JEV-infected cells but also gives further consideration to the potential clinical use of IFN on JE patients.
Acknowledgments
Y.-L.L. was supported by grants from the National Health Research Institute (NHRI-EX92-9028SP) and the National Science Council (NSC91-2320-B-001-060) and by Academia Sinica, Taiwan, Republic of China.
REFERENCES
- 1.Adler, B., H. Adler, H. Pfister, T. W. Jungi, and E. Peterhans. 1997. Macrophages infected with cytopathic bovine viral diarrhea virus release a factor(s) capable of priming uninfected macrophages for activation-induced apoptosis. J. Virol. 71:3255-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, W. S. 2002. Suppressors of cytokine signalling (SOCS) in the immune system. Nat. Rev. Immunol. 2:410-416. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 4.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 5.Burke, D. S., and J. C. Morill. 1987. Levels of interferon in the plasma and cerebrospinal fluid of patients with acute Japanese encephalitis. J. Infect. Dis. 155:797-799. [DOI] [PubMed] [Google Scholar]
- 6.Charleston, B., L. S. Brackenbury, B. V. Carr, M. D. Fray, J. C. Hope, C. J. Howard, and W. I. Morrison. 2002. Alpha/beta and gamma interferons are induced by infection with noncytopathic bovine viral diarrhea virus in vivo. J. Virol. 76:923-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L. K., C. L. Liao, C. G. Lin, S. C. Lai, C. I. Liu, S. H. Ma, Y. Y. Huang, and Y. L. Lin. 1996. Persistence of Japanese encephalitis virus is associated with abnormal expression of the nonstructural protein NS1 in host cells. Virology 217:220-229. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L. K., Y. L. Lin, C. L. Liao, C. G. Lin, Y. L. Huang, C. T. Yeh, S. C. Lai, J. T. Jan, and C. Chin. 1996. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223:79-88. [DOI] [PubMed] [Google Scholar]
- 9.Colamonici, O. R., P. Domanski, S. M. Sweitzer, A. Larner, and R. M. Buller. 1995. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J. Biol. Chem. 270:15974-15978. [DOI] [PubMed] [Google Scholar]
- 10.Crance, J. M., N. Scaramozzino, A. Jouan, and D. Garin. 2003. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antivir. Res. 58:73-79. [DOI] [PubMed] [Google Scholar]
- 11.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 13.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 16.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, S. N., M. K. Goverdhan, P. S. Sathe, S. C. Chelliah, S. V. Naik, P. V. Godbole, and K. Banerjee. 1990. Protective effect of 6-MFA, a fungal interferon inducer against Japanese encephalitis virus in bonnet macaques. Indian J. Med. Res. 91:408-413. [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 20.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 21.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1999. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1β, in endothelial cells. J. Virol. 73:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harinasuta, C., C. Wasi, and S. Vithanomsat. 1984. The effect of interferon on Japanese encephalitis virus in vitro. Southeast Asian J. Trop. Med. Public Health. 15:564-568. [PubMed] [Google Scholar]
- 23.Hasegawa, H., Y. Satake, and Y. Kobayashi. 1990. Effect of cytokines on Japanese encephalitis virus production by human monocytes. Microbiol. Immunol. 34:459-466. [DOI] [PubMed] [Google Scholar]
- 24.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho, L. J., J. J. Wang, M. F. Shaio, C. L. Kao, D. M. Chang, S. W. Han, and J. H. Lai. 2001. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J. Immunol. 166:1499-1506. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, A. J., and J. T. Roehrig. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalvakolanu, D. V., S. K. Bandyopadhyay, M. L. Harter, and G. C. Sen. 1991. Inhibition of interferon-inducible gene expression by adenovirus E1A proteins: block in transcriptional complex formation. Proc. Natl. Acad. Sci. USA 88:7459-7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs, D. L., and D. J. Hilton. 2001. SOCS proteins: negative regulators of cytokine signaling. Stem Cells 19:378-387. [DOI] [PubMed] [Google Scholar]
- 31.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C-terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 32.Kurane, I., and F. A. Ennis. 1987. Induction of interferon alpha from human lymphocytes by autologous, dengue virus-infected monocytes. J. Exp. Med. 166:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 34.Leonard, G. T., and G. C. Sen. 1996. Effects of adenovirus E1A protein on interferon-signaling. Virology 224:25-33. [DOI] [PubMed] [Google Scholar]
- 35.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 36.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 37.Liao, C. L., Y. L. Lin, J. J. Wang, Y. L. Huang, C. T. Yeh, S. H. Ma, and L. K. Chen. 1997. Effect of enforced expression of human bcl-2 on Japanese encephalitis virus-induced apoptosis in cultured cells. J. Virol. 71:5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, Y. L., C. L. Liao, L. K. Chen, C. T. Yeh, C. I. Liu, S. H. Ma, Y. Y. Huang, Y. L. Huang, C. L. Kao, and C. C. King. 1998. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 72:9729-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 40.Look, D. C., W. T. Roswit, A. G. Frick, Y. Gris-Alevy, D. M. Dickhaus, M. J. Walter, and M. J. Holtzman. 1998. Direct suppression of Stat1 function during adenoviral infection. Immunity 9:871-880. [DOI] [PubMed] [Google Scholar]
- 41.Mahalingam, S., J. Meanger, P. S. Foster, and B. A. Lidbury. 2002. The viral manipulation of the host cellular and immune environments to enhance propagation and survival: a focus on RNA viruses. J. Leukoc. Biol. 72:429-439. [PubMed] [Google Scholar]
- 42.Mashimo, T., M. Lucas, D. Simon-Chazottes, M. P. Frenkiel, X. Montagutelli, P. E. Ceccaldi, V. Deubel, J. L. Guenet, and P. Despres. 2002. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl. Acad. Sci. USA 99:11311-11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, D. M., B. M. Rahill, J. M. Boss, M. D. Lairmore, J. E. Durbin, J. W. Waldman, and D. D. Sedmak. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, D. M., Y. Zhang, B. M. Rahill, W. J. Waldman, and D. D. Sedmak. 1999. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J. Immunol. 162:6107-6113. [PubMed] [Google Scholar]
- 45.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 48.Perelygin, A. A., S. V. Scherbik, I. B. Zhulin, B. M. Stockman, Y. Li, and M. A. Brinton. 2002. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. USA 99:9322-9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petricoin, E., III, M. David, K. Igarashi, C. Benjamin, L. Ling, S. Goelz, D. S. Finbloom, and A. C. Larner. 1996. Inhibition of alpha interferon but not gamma interferon signal transduction by phorbol esters is mediated by a tyrosine phosphatase. Mol. Cell. Biol. 16:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer, M., and E. Peterhans. 2001. Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J. Virol. 75:4692-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 53.Solomon, T., N. M. Dung, B. Wills, R. Kneen, M. Gainsborough, T. V. Diet, T. T. Thuy, H. T. Loan, V. C. Khanh, D. W. Vaughn, N. J. White, and J. J. Farrar. 2003. Interferon alfa-2a in Japanese encephalitis: a randomised double-blind placebo-controlled trial. Lancet 361:821-826. [DOI] [PubMed] [Google Scholar]
- 54.Solomon, T., H. Ni, D. W. Beasley, M. Ekkelenkamp, M. J. Cardosa, and A. D. Barrett. 2003. Origin and evolution of Japanese encephalitis virus in southeast Asia. J. Virol. 77:3091-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 56.Symons, J. A., A. Alcami, and G. L. Smith. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81:551-560. [DOI] [PubMed] [Google Scholar]
- 57.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 58.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 59.Taylor, J. L., C. Schoenherr, and S. E. Grossberg. 1980. Protection against Japanese encephalitis virus in mice and hamsters by treatment with carboxymethylacridanone, a potent interferon inducer. J. Infect. Dis. 142:394-399. [DOI] [PubMed] [Google Scholar]
- 60.Upton, C., K. Mossman, and G. McFadden. 1992. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science 258:1369-1372. [DOI] [PubMed] [Google Scholar]
- 61.Urosevic, N. 2003. Is flavivirus resistance interferon type I-independent? Immunol. Cell Biol. 81:224-229. [DOI] [PubMed] [Google Scholar]
- 62.Vithanomsat, S., C. Wasi, C. Harinasuta, and P. Thongcharoen. 1984. The effect of interferon on flaviviruses in vitro: a preliminary study. Southeast Asian J. Trop. Med. Public Health 15:27-31. [PubMed] [Google Scholar]
- 63.Wu, S. F., C. J. Lee, C. L. Liao, R. A. Dwek, N. Zitzmann, and Y. L. Lin. 2002. Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 76:3596-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokota, S., N. Yokosawa, T. Kubota, T. Suzutani, I. Yoshida, S. Miura, K. Jimbow, and N. Fujii. 2001. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and janus kinases during an early infection stage. Virology 286:119-124. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, J. J., U. Vinkemeier, W. Gu, D. Chakravarti, C. M. Horvath, and J. E. Darnell, Jr. 1996. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl. Acad. Sci. USA 93:15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]