Abstract
The tegument protein ppUL82 (pp71) of human cytomegalovirus (HCMV) has previously been shown to activate the immediate-early transcription of HCMV and to enhance the infectivity of viral DNA. This is concordant with its localization adjacent to promyelocytic leukemia oncogenic domains (PODs) immediately after infection. In a yeast two-hybrid screen, we identified the tegument protein ppUL35 as an interacting partner of ppUL82. The interaction could be confirmed in transfected and infected cells. The domain responsible for interaction was narrowed down to amino acids 447 to 516 within ppUL35, thus allowing both forms of ppUL35 to interact with ppUL82. Immunofluorescence experiments showed a relocalization of ppUL35 from a diffuse nuclear pattern when expressed alone to PODs when expressed together with ppUL82. In accordance with this observation and the role of ppUL82 as a transactivator, we observed a cooperative activation of the HCMV major immediate-early enhancer but not of heterologous viral enhancer elements. These results suggest an important role for this interaction in the stimulation of viral immediate-early gene expression and viral infection.
Human cytomegalovirus (HCMV) is a herpesvirus belonging to the Betaherpesvirinae subfamily, which is characterized by a narrow host range. Postnatal primary infection is usually asymptomatic, and prevalence in normal populations is high, ranging from about 40% in developed countries to 100% in less developed countries. However, in individuals with an immature or compromised immune system, HCMV can cause considerable morbidity and mortality after primary infection or reactivation from a latent stage (32).
The virion of HCMV has a tripartite structure composed of the DNA-containing capsid, the envelope, and an interspersed proteinaceous tegument layer, which is characteristic for herpesviruses. The composition of the particle is complex, consisting of a minimum of 40 proteins (1, 28). At least 20 of these proteins can be found in the tegument.
Besides their role in the formation of the virion (27), tegument proteins have been proposed to act in several other processes, such as intracellular particle transport (2, 40), immune evasion (7, 16), and stimulation of immediate-early gene expression. The activation of the HCMV major immediate-early enhancer-promoter (MIEP) by virion components was first reported in 1985, when it was observed that HCMV virions could activate transiently or stably transfected reporter genes (36, 39). The first tegument protein for which such a transacting activity was reported was ppUL82 (23). Later, we found that another regulatory protein of HCMV, pUL69, was incorporated into the tegument and was able to activate the MIEP cooperatively with ppUL82 tegument proteins (45, 46). These observations have been reproduced and extended by other groups, thereby identifying at least three other proteins with the ability to enhance MIEP activity (24, 34, 38). Of these additional proteins, pIRS1/pTRS1 and ppUL35 have also been reported to act cooperatively with ppUL82. By use of deletion mutants, the genes UL82 and UL69 were shown to be essential for viral growth, at least when a low multiplicity of infection (MOI) was used (4, 17). However, while infection of fibroblasts with a virus carrying a deletion of the UL82 gene led to a strongly reduced transcription of immediate-early genes, in accordance with its role as a transactivator of the MIEP, the virus with a deletion of UL69 showed almost-wild-type levels of immediate-early transcription. Therefore, pUL69 appeared to be more important for the regulation of later phases of the infection or other, more indirect effects, such as cell cycle arrest (17).
The precise mechanism of action of the regulatory tegument proteins is largely unknown. So far, only ppUL82 has been investigated in some detail. Immediately after infection, the protein could be detected in promyelocytic leukemia (PML) oncogenic domains (PODs) (19). This same localization was also observed after expression from plasmids or by recombinant herpes simplex virus (18, 26). The physical interaction between ppUL82 and human Daxx (hDaxx), a component of PODs, appeared to be necessary for its recruitment to PODs (18). In addition, cotransfection of ppUL82 and hDaxx showed a cooperative activation of the MIEP. In accordance with this, infection of murine Daxx knockout cells did not result in localization of ppUL82 to PODs and led to reduced immediate-early gene expression (19). It has been shown that ppUL82 acts via 19-bp elements of the MIEP (23). Interactions of ppUL82 with other viral proteins have not been reported so far.
The UL35 open reading frame has been identified as an early-late gene (24). It is transcribed into two coterminal transcripts, guiding the synthesis of two phosphorylated protein products, ppUL35 and a shorter form, ppUL35A, which corresponds to the carboxy-terminal 193 amino acids of ppUL35. Whereas the synthesis of the small protein starts in the early phase, the full-length UL35 protein is a late product that is specifically incorporated into viral particles. A differential localization and function of both proteins has been proposed.
Here we report for the first time an interaction between two cooperatively transactivating tegument proteins, ppUL35 and ppUL82. Cotransfection of UL35 and UL82 leads to an accumulation in or close to PODs, similar to the situation during the initiation of an infection. In addition, we observed a cooperative activation of the MIEP by both proteins and we show that this is a highly specific process.
MATERIALS AND METHODS
Oligonucleotides.
The following oligonucleotides were obtained from MWG (Ebersberg, Germany) or Sigma/ARK (Darmstadt, Germany): UL35-5, GTGTCTCGAGGCCTCATGGCCCAAGGATCGCGA; UL35-3, CTCATCGATTCAGAGATGCCGTAGGTT; UL82-5, GTGGATCCATGTCTCAGGCATCGTCC; UL82-3, GATATCCTAGATGCGGGGTCGACT; PPClinkA, TCGAACGTCGACGAATTCCCGGGATATCA; PPClinkB, GATCTGATATCCCGGGAATTCGTCGACTG; UL35_se, CATGTCGACAGATATCCAATGGCCCAAGGATCGCGA; UL35-d1C, GGTGCGGCCGCTCACTGCTCCAGGTCCGCGGAGC; UL35-del2CA, GGAGCGGCCGCTCAGCTATAGATGCGCTGGAG; UL35-d3C, GGAGCGGCCGCTCAGCGCGCCGTGGGATCGGCTT; UL35-d4C, GGAGCGGCCGCTCAGAAGGCCGGACTGCTGTGTT; UL35-d5C, GGAGCGGCCGCTCAGGACAGGTTCTCGGCGCACA; UL35-d6C, GGAGCGGCCGCTCAGGTTTGCGCAGCGCGGCAGA; UL35-del1N, GGCATGTCGACAGATATCCAGCTGCGCAAACCGGCGTCA; UL35-del2N, GGCATGTCGACAGATATCCACTGTCCGACGTAACACAAC; UL35-del3N, GGCATGTCGACAGATATCCACAACACAGCAGTCCGGCC; UL35-del4N, GGCATGTCGACAGATATCCACTCGAAGCCGATCCCACG; ul35d5na5b, GGCATGTCGACAGATATCCAATGATGATCGAGGGCGCC; and UL35-del6N, GGCATGTCGACAGATATCCAAGCTCCGCGGACCTGGAGC
Plasmid constructions.
For yeast two-hybrid screening, we developed a novel vector, pPC86EmL. First, we modified the polylinker of pPC86 (10) by inserting the phosphorylated and annealed linker oligonucleotides PPClinkA/PPClinkB into the SalI site, to generate the plasmid pPC86m. To adapt this plasmid to other yeast two-hybrid vectors, we exchanged the yeast selection marker. For this, we isolated the ApaI/SpeI fragment containing the GAL4 activation domain from pPC86m and inserted it into the ApaI- and SpeI-treated pPC97E to generate pPC86EmL. Plasmid pPC97E was generated from pPC97 (42) by digestion of pPC97 with ClaI and EcoRI followed by replacement with the ClaI/EcoRI fragment derived from pGAD424. This removed the EcoRV site from the LEU2 marker.
The pAS-based bait plasmid pHM677 for the yeast two-hybrid screen has been described and used previously (18). As this plasmid showed residual background activation, an alternative bait plasmid, pGBT9-UL82, was generated by cloning the BamHI/EcoRI UL82 fragment from pHM677 into BamHI- and EcoRI-cut pGBT9. The resulting plasmid, pGBT-UL82, did not show background activation in yeast.
The eukaryotic expression vector for pUL69 (pHM160) has been described previously (44). Eukaryotic expression vectors for ppUL32 (pp150) (pp150-CB6) and pIE1 (pHM494) contain the open reading frames in the context of pCB6 (5) and pcDNA3 (Invitrogen, Karlsruhe, Germany), respectively. The UL35 and UL82 open reading frames were PCR amplified by using the oligonucleotides UL35-5/UL35-3 and UL82-5/UL82-3 and the cosmids pCM1017 and pCM1193 as templates (14). PCR products were cloned into pUC18 by using the Sureclone kit (Amersham Biosciences, Freiburg, Germany). To generate a eukaryotic expression vector for UL35, the StuI/ClaI fragment from pUC18-UL35 was treated with Klenow polymerase and cloned into EcoRV-cut pcDNA3. The short form of UL35, UL35A, was generated by inserting the BglII/NotI fragment from pcDNA3-UL35 into BamHI- and NotI-digested pcDNA3. To generate a eukaryotic expression vector for ppUL82, the UL82 fragment was excised from pUC18-UL82 with BamHI and EcoRV and ligated with pcDNA3. A hemagglutinin (HA)-tagged version of ppUL82 was generated by inserting the UL82 fragment into BamHI- and EcoRV-cut pcDNA3-HA-N, a pcDNA3 vector modified to allow generation of amino-terminally HA-tagged genes. The carboxy-terminal deletions of UL35 were generated by PCR with primer UL35_se in combination with primer UL35-d1C, UL35-del2CA, UL35-d3C, UL35-d4C, UL35-d5C, or UL35-d6C and pCM1017 as the template. Similarly, for the amino-terminal deletions, primer UL35-3 was used together with primer UL35-del1N, UL35-del2N, UL35-del3N, UL35-del4N, ul35d5na5b, or UL35-del6N and pCM1017 as the template. These fragments were cloned into EcoRV- and NotI-digested vector pcDNA3-HA-N or pcDNA3-myc-N to obtain eukaryotic expression vectors for amino-terminally HA- or Myc-tagged proteins. For use in the yeast two-hybrid system, the respective fragments were cloned into SalI- and NotI-digested plasmid pPC86EmL. The sequences of all clones were determined to conform to the published HCMV AD169 sequence (8).
Luciferase reporter constructs were generated by replacing the chloramphenicol acetyltransferase reporter genes of plasmids pRR57, pRR55, pRR56/3, and pRR56/5 (13, 37) with the Photinus pyralis luciferase gene. For this, the chloramphenicol acetyltransferase gene was removed from the respective plasmids by digestion with BamHI, followed by insertion of the luciferase cassette of p19luc (41). This led to plasmids pHM287, pHM284, pHM289, and pHM286 containing the HCMV modulator-enhancer-promoter (position −1228 to 53), enhancer-promoter (−671 to +53 or −188 to +53), or promoter (−65 to +53) regions, respectively. The luciferase reporter constructs pHIVluc (human immunodeficiency virus [HIV] long terminal repeat [LTR] upstream of luciferase) and pSV40luc (simian virus 40 [SV40] enhancer-promoter upstream of luciferase) have been described previously (44).
Library construction.
For generation of a genomic library of HCMV, we used an overlapping set of cosmids (11, 17, 20, 26, 62, °17, °60, °63) derived from the HCMV strain TB40E (35). Cosmid DNA was sonicated to a medium size of 0.5 to 1 kb and blunt ended with mung bean nuclease. Thereafter, a single A nucleotide was added by treatment with Taq DNA polymerase in the presence of dATP. Fragments were size fractionated on an agarose gel. The fraction of fragments between 0.5 and 0.7 kb was ligated into the vector pPC86EmL that had been digested with SmaI and treated with Taq DNA polymerase in the presence of dTTP to generate single T overhangs (25). The ligation reaction mixture was electroporated into Escherichia coli DH10B. Colonies were harvested, and library DNA was isolated by CsCl gradient purification (22).
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed by using GAL4 fusion proteins as described previously (11). Saccharomyces cerevisiae strain Y153 was transformed by the lithium acetate method with the bait plasmid pHM677 (18). The plasmid pHM677 was stably maintained by selection for tryptophan prototrophy. The yeast strain containing pHM677 was subsequently transformed with a genomic library of the HCMV strain TB40E fused to the GAL4 activation domain in the pPC86EmL vector. Primary transformants (500,000) were selected for growth on His-Leu-Trp dropout plates containing 20 mM 3-aminotriazole. His+ colonies were thereafter analyzed for β-galactosidase activity by filter lift experiments. Interactor plasmids from clones positive in both assays were recovered in E. coli and sequenced. Subsequently, these plasmids were cotransformed into yeast strain Y153 together with pGBT9-UL82 or control plasmids in order to confirm the interaction in filter lift experiments.
Cell lines and culture.
Human foreskin fibroblasts (HFF) were maintained in minimal essential medium (Gibco/BRL, Eggenstein, Germany) and used before passage 20. U373-MG and 293T cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in Dulbecco's modified Eagle medium (Gibco/BRL). Murine hybridoma cells (p63-27, 28-77, 69-66, 12CA5, and 9E10 [see below]) were grown in RPMI 1640 medium (Gibco/BRL). All media were supplemented with 10% fetal calf serum, 2 mM l-glutamine (Biochrom AG, Berlin, Germany), 100 U of penicillin and 100 μg of streptomycin (Gibco/BRL) per ml, and 1× nonessential amino acids (Biochrom AG). The cells were grown at 37°C in a humidified 5% CO2 atmosphere.
Antibodies and coimmunoprecipitation.
Cell culture supernatant of human monoclonal antibody X2-16 directed against pUL48 was obtained from S. Foung (3). Murine monoclonal antibodies against IE1 (p63-27), pUL69 (69-66), ppUL82 (CMV355), ppUL83 (pp65) (28-77), pUL86 (MCP) (28-4), HA tag (12CA5), and Myc tag (9E10) were described previously and used as cell culture supernatants (6, 12, 30, 31, 33, 43). Rabbit antisera directed against ppUL35, pUL69, and IE1 and -2 were described previously (15, 24, 44).
For coimmunoprecipitation, 293T cells were seeded in 6-cm-diameter dishes at 70% confluency the day before transfection; 24 h later, the cells were transfected by calcium phosphate coprecipitation with 2 μg of each plasmid (9). After incubation overnight, the cells were washed two times with phosphate-buffered saline (PBS) to remove the precipitate and incubated with fresh medium. After 24 h, cells were washed again two times with PBS and harvested in 1 ml of PBS.
After centrifugation, the cell pellet was lysed in 500 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1× protease inhibitor mix [Roche, Mannheim, Germany]) and incubated for 20 min on ice with occasional vortexing. For salt titration, cell pellets were resuspended in lysis buffer containing different concentrations of NaCl (150, 300, and 500 mM). After clearing of the lysate by centrifugation (16,000 × g, 10 min), an 80-μl aliquot was removed as a lysate control. The cleared lysate was supplemented with 300 μl of lysate buffer and antibody and incubated for 1.5 h at 4°C. Then, 40 μl of protein A-Sepharose CL-4B (30 mg/ml; Amersham Biosciences) was added, followed by additional shaking for 2 h at 4°C. Precipitated protein complexes were collected (2,300 × g, 5 min, 4°C) and washed three times with 700 μl of lysis buffer. After a final centrifugation at 16,000 × g, excess liquid was removed and the pellet was resuspended in 2× sodium dodecyl sulfate (SDS) buffer (80 mM Tris-HCl [pH 8.8], 2 mM EDTA, 400 mM sucrose, 0.4 mM dithiothreitol, 0.4% SDS, 0.1% bromphenol blue). Precipitated proteins were separated on 10 or 12% polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes by using a Transblot SD (Bio-Rad, Munich, Germany). To avoid nonspecific antibody binding, filters were blocked for 1 h in 3% skim milk powder-PBS. For detection of specific proteins, filters were incubated with primary antibodies for 1 h at room temperature. Rabbit antisera (anti-UL35, anti-UL69, and anti-IE1/2) were diluted 1:5,000 in PBS-0.1% Tween 20 containing 1% skim milk powder, whereas hybridoma supernatant was used undiluted. After the filters were washed three times for 10 min each in PBS-0.1% Tween 20 containing 1% skim milk powder, they were incubated for 1 h with horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibodies (DAKO, Hamburg, Germany) diluted 1:5,000 in PBS-0.1% Tween 20 containing 1% skim milk powder. Finally, the filters were washed two times for 10 min each in PBS-0.1% Tween 20 containing 1% skim milk powder and 10 for min in PBS-0.1% Tween 20. Bound antibodies were detected by using the ECL Western blotting detection reagents (Amersham Biosciences) according to the instructions of the manufacturer.
For coimmunoprecipitations from human fibroblasts, cells were seeded at 80% confluency and infected with HCMV strain AD169 1 day later. For precipitation of complexes from late infected cells, cells were infected at an MOI of 1. At 60 h postinfection (hpi), cells were washed twice with PBS, trypsinized, and harvested in 1 ml of PBS. After centrifugation, the cell pellet was lysed in 300 μl of lysis buffer and processed as described above. For precipitation during the immediate-early phase, cells were infected at an MOI of 10. After 4 h, cells were trypsinized, pelleted, and washed twice with PBS-EDTA. The pellet was lysed in 300 μl of lysis buffer for 20 min on ice. Insoluble material was pelleted by centrifugation (5 min, 13,000 rpm, 4°C). The supernatant was then centrifuged at 23,000 × g for 2 h at 4°C to remove viral particles. The remaining supernatant was subjected to coimmunoprecipitation as described above.
Indirect immunofluorescence analysis.
The day before transfection, HFF were seeded onto glass coverslips in 24-well plates at 4 × 104 cells per well. Cells were transfected by using the Superfect transfection reagent (Qiagen, Hilden, Germany); 48 h later, transfected cells were washed three times with PBS and fixed for 10 min in 4% paraformaldehyde, followed by permeabilization with 0.2% Triton X-100 for 2 min. To block nonspecific binding, the coverslips were first incubated for 20 min at 37°C in 10% normal horse serum. Subsequently, the primary antibodies anti-UL35 rabbit serum (1:200 dilution), anti-UL69 rabbit serum (1:2,000 dilution), anti-PML murine monoclonal antibody PG-M3 (1:200 dilution; Santa Cruz Biotechnology, Santa Cruz, Calif.), and anti-pp71 murine monoclonal antibody CMV355 (undiluted cell culture supernatant) were added and incubated for 2 h at 4°C. After washing three times for 5 min with PBS, coverslips were incubated for 2 h at 4°C with secondary antibodies and DAPI (4′,6′-diamidino-2-phenylindole) (final concentration, 1 μg/ml). As secondary antibodies, goat anti-mouse-fluorescein isothiocyanate and swine anti-rabbit-tetramethyl rhodamine isocyanate sera (DAKO, Hamburg, Germany) were used at a dilution of 1:50. After two final washing steps in PBS, the coverslips were mounted on slides with Vectashield H-1000 (Vector, Burlingame, Calif.). As controls, mock-transfected cells were fixed and stained as described above. Stainings with all combinations of antibodies used gave no signal. Images were taken with a TCS laser scanning system and the Leica TCS NT software (Leitz, Bensheim, Germany).
Reporter gene assays.
For luciferase assays, U373-MG and HFF cells were plated onto six-well dishes at 3 × 105 cells per well the day before transfection. Plasmid transfection was performed by the DEAE-dextran method as described previously. Briefly, medium was replaced with 750 μl of fresh medium per well before transfection. Plasmid DNAs were mixed with 8 μl of DEAE-dextran (50 mg/ml) and 250 μl of medium and added to each well. After 3 h of incubation, the cells were washed two times with PBS and finally incubated in fresh medium. After 48 h, cells were lysed with 300 μl of lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM dithiothreitol, 2 mM CDTA [trans-1,2-diaminocyclohexane-N,N,N′,N′-tetra-acetic acid], 1% Triton-X100, 10% glycerol) with vigorous shaking for 30 min. The supernatant was collected without scraping, and nonsoluble cell debris was pelleted by centrifugation. One hundred microliters of the final supernatant was mixed with an equal amount of cold assay buffer (100 mM potassium phosphate [pH 7.8], 15 mM MgSO4, 5 mM ATP) and assayed for luciferase activity by injection of luciferin solution (1 mM d-Luciferin [Roche] in assay buffer), using a Lumat LB9507 instrument (Berthold, Wildbad, Germany).
RESULTS
ppUL82 and ppUL35 interact in yeast and mammalian cells.
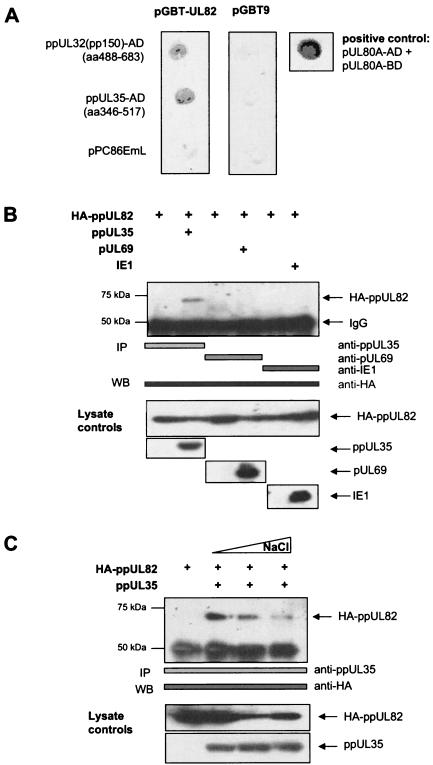
To identify viral interaction partners of ppUL82 (pp71), we performed yeast two-hybrid screening. For this, we generated a random genomic library of HCMV strain TB40E fused to the GAL4 activation domain, with an average insert size of 0.5 to 0.7 kb, encompassing about 200 amino acids. This size should ensure good coverage of protein domains, which for eukaryotic proteins comprise about 130 amino acids. Our library consisted of 80,000 independent clones, thus scanning the HCMV genome with an average spacing of about 13 nucleotides. Using this library, we performed yeast two-hybrid screening with a bait plasmid encoding a fusion of the GAL4 DNA binding domain (GAL4-BD) with the UL82 open reading frame. We obtained two independent interacting clones corresponding to amino acids 346 to 516 of ppUL35 and 488 to 683 of ppUL32 (pp150). After isolation, the interactor clones were retested with plasmids encoding GAL4-BD alone or GAL4-BD fused to UL82 (pGBT9-UL82). In a filter lift assay, both interactor clones showed a strong interaction with pGBT9-UL82 but not with the empty vector pGBT9 (Fig. 1A). As positive control, we tested the interaction of UL80a with itself, which has been reported previously (47).
FIG. 1.
ppUL82 and ppUL35 interact in yeast and mammalian cells. (A) Interactor clones containing fragments of ppUL32 (pp150) (amino acids [aa] 488 to 683) and ppUL35 (aa 346 to 517) fused to the GAL4 activation domain, as well as the empty vector pPC86EmL, were cotransformed either with a plasmid containing ppUL82 fused to the GAL4-BD (pGBT9-UL82) or with a control vector (pGBT9). Yeast colonies were analyzed for expression of the reporter gene β-galactosidase by filter lift assays. The known homodimerization of HCMV UL80a was used as a positive control for the filter lift assay. (B) 293T cells were transfected with eukaryotic expression vectors encoding HA-ppUL82, ppUL35, pUL69, and ppUL123 (IE1) as indicated. Immunoprecipitation (IP) was performed with antibodies as indicated by bars. Precipitates were separated by SDS-polyacrylamide gel electrophoresis, and coprecipitated HA-ppUL82 was detected in immunoblot analysis (WB) with the monoclonal anti-HA antibody. Lysates of all transfected cells used for immunoprecipitation were separated by SDS-polyacrylamide gel electrophoresis, and proteins were detected with the indicated antibodies. (C) 293T cells were cotransfected with plasmids encoding either HA-ppUL82 or ppUL35 as indicated. Coimmunoprecipitations were performed with different concentrations (150, 300, and 500 mM) of NaCl in the lysis buffer and the anti-ppUL35 antibody. Coprecipitated HA-ppUL82 proteins were detected by immunoblotting with the monoclonal anti-HA antibody. Lysates of all transfected cells used for immunoprecipitation were analyzed for comparable protein expression as detected by the indicated antibodies.
To confirm the interaction between ppUL82 and ppUL35 in human cells, coimmunoprecipitation assays were performed. We transfected 293T cells with expression plasmids for HA-tagged ppUL82 and ppUL35 or control vectors. Protein complexes were precipitated with a rabbit serum directed against ppUL35. After separation in a protein gel, coprecipitated HA-tagged ppUL82 was detected in an immunoblot with a monoclonal antibody against the HA tag. As shown in Fig. 1B, ppUL82 could be precipitated only when coexpressed with ppUL35. As controls, we performed similar experiments after cotransfection of HA-tagged UL82 with the HCMV regulatory proteins pUL69 and ppUL123 (pIE1). Despite strong expression of all proteins, ppUL82 did not coprecipitate with pUL69 or ppUL123 (pIE1) (Fig. 1B).
Finally, we investigated the salt stability of the ppUL82-ppUL35 complex by coimmunoprecipitation. Protein extracts were prepared in the presence of a standard salt concentration (150 mM NaCl) or 300 and 500 mM NaCl. As shown in Fig. 1C, the interaction could be detected up to the highest concentration of NaCl. However, the amount of ppUL82 decreased with increasing amounts of NaCl, showing some salt sensitivity of the complex. In summary, we could detect a novel and specific interaction between the two HCMV tegument proteins ppUL82 and ppUL35, which could be due to electrostatic forces.
ppUL82 interacts with both isoforms of ppUL35.
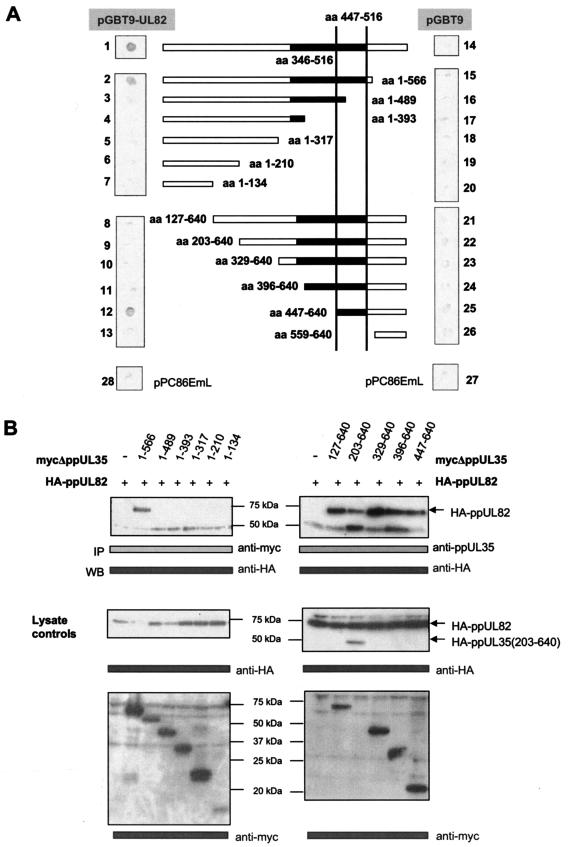
To further characterize the interaction, we generated amino- and carboxy-terminal deletions of ppUL35 and then performed analyses in the yeast two-hybrid system and by coimmunoprecipitation. As shown in Fig. 2, the deletion of the last 74 amino acids did not affect the interaction with ppUL82. However, further deletion down to amino acid 489 abolished this interaction in both the yeast two-hybrid system and coimmunoprecipitation. Concerning the amino-terminal deletions, only the polypeptide corresponding to ppUL35A (amino acids 447 to 640) showed interaction with ppUL82 in the yeast two-hybrid system (Fig. 2A). The failure to detect the interaction with the other amino-terminal deletions of ppUL35 is likely due to conformational distortion or instability in yeast cells, as all forms down to ppUL35A could be coprecipitated with ppUL82 after transient expression in 293T cells (Fig. 2B). As expression of the shortest form of UL35, comprising amino acids 559 to 640, could not be detected, we did not perform any immunoprecipitation experiment with this polypeptide. Thus, our deletion analysis narrowed down the interacting domain to amino acids 447 to 566, which is in agreement with the results of the yeast two-hybrid screening. Both approaches taken together show that the domain responsible for binding of ppUL82 involves amino acids 447 to 516 of ppUL35, indicating that both isoforms of ppUL35 can bind to ppUL82.
FIG.2.
Identification of the ppUL82 binding domain in ppUL35. (A) Amino- and carboxy-terminal deletion mutants of ppUL35 were generated by PCR and either cloned into yeast vector pPC86EmL to generate a fusion with the GAL4 activation domain or inserted into eukaryotic expression vectors to generate Myc-tagged versions. The deletion mutant containing amino acids (aa) 447 to 640 is equivalent to the short protein isoform ppUL35A. Yeast strain Y153 was cotransformed with the indicated constructs containing a UL35 deletion fused to the GAL4 activation domain together with either a plasmid containing ppUL82 fused to the GAL4-BD (pGBT9-UL82) (panels 2 to 13) or pGBT9 as control (panels 15 to 26). Colonies were analyzed for the expression of β-galactosidase by filter lift assay. The interaction of the full-length protein ppUL35 with ppUL82 was used as a positive control (panel 1). Empty vectors pPC86EmL and pGBT9 were used as negative controls (panels 14, 27, and 28). (B) 293T cells were cotransfected with a plasmid containing HA-ppUL82 and an expression vector encoding distinct C- or N-terminally truncated, Myc-tagged ppUL35 mutants. The N-terminal deletion mutant comprising amino acids 203 to 640 was expressed as HA-tagged protein. Immunoprecipitations (IP) were performed with the anti-Myc monoclonal antibody (C-terminal ppUL35 deletion mutants) (left panels) or the anti-ppUL35 antibody (N-terminal ppUL35 deletion mutants) (right panels), and coprecipitated HA-ppUL82 was detected with the monoclonal anti-HA antibody. Lysates of all transfected cells used for immunoprecipitation were analyzed for comparable protein expression and detected with the indicated antibodies. WB, immunoblotting.
ppUL82 and ppUL35/ppUL35A form a complex in infected cells.
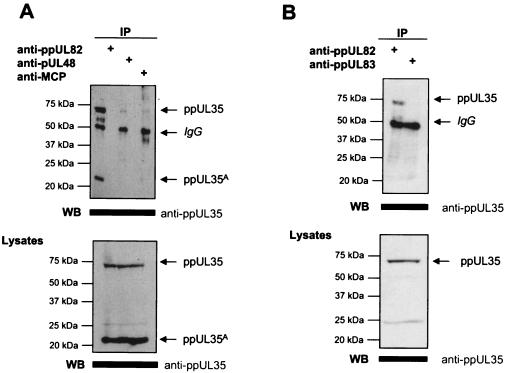
To observe the interaction between ppUL82 and ppUL35 under physiologic conditions, we infected human fibroblasts with HCMV strain AD169. Extracts from late infected cells (60 hpi) were precipitated with monoclonal antibodies recognizing ppUL82, pUL48, and the major capsid protein pUL86 (MCP). Precipitated proteins were analyzed by immunoblotting with a rabbit serum against ppUL35. As shown in Fig. 3A, both forms of ppUL35 could be clearly precipitated with the monoclonal antibody CMV355 against ppUL82. Some minor signals were seen in coprecipitations with monoclonal antibodies against pUL48 or pUL86 (MCP) of HCMV. This is could be due to the precipitation of tegumented capsids that start to accumulate during the late phase. This experiment shows that a strong interaction between ppUL35 and ppUL82 can also be detected in infected cells.
FIG. 3.
Specific interaction between ppUL82 and ppUL35 as determined in infected HFF cells. (A) HFF cells were infected with HCMV strain AD169 (MOI of 2), followed by preparation of cell extracts after 60 hpi. Immunoprecipitations (IP) were performed with the anti-ppUL82 (CMV355), anti-pUL48 (X2-16), or anti-pUL86 (MCP) monoclonal antibody as indicated. Coprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis, followed by detection of UL35 proteins with the polyclonal anti-ppUL35 serum. Lysates of infected cells used for immunoprecipitation were separated by SDS-polyacrylamide gel electrophoresis, and expression of both isoforms of ppUL35 was detected by immunoblotting (WB). (B) HFF cells were infected with HCMV strain AD169 (MOI of 10), followed by preparation of cell extracts after 4 hpi. Intracellular virus particles were removed by centrifugation. Immunoprecipitations were performed with the anti-ppUL82 (CMV355) and anti-ppUL83 (pp63) (28-77) monoclonal antibodies. Coprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis, and UL35 proteins were detected by immunoblot analysis with the polyclonal anti-ppUL35 serum. Lysates of infected cells were separated by SDS-polyacrylamide gel electrophoresis, followed by detection of ppUL35 by immunoblotting.
As both ppUL35 and ppUL82 are delivered into the cell by the incoming virion, we also attempted to demonstrate the interaction between these proteins during the immediate-early phase. For this, we infected human fibroblasts with HCMV strain AD169 at an MOI of 10 and harvested cellular extracts at 4 hpi. Signals from precipitated intracellular virions were avoided by removal of particles from the extract by high-speed centrifugation. Thereafter, protein complexes were precipitated with monoclonal antibodies directed against ppUL82 or ppUL83 (pp65). Coprecipitated ppUL35 was detected by immunoblotting with a specific rabbit serum. As shown in Fig. 3B, we could clearly observe an interaction between ppUL82 and ppUL35. No interaction was seen after precipitation of ppUL83 (pp65). In addition, we did not detect a signal at the size of ppUL35A, as this isoform is not incorporated into the virion.
ppUL82 and ppUL35 cooperatively transactivate the HCMV enhancer-promoter.
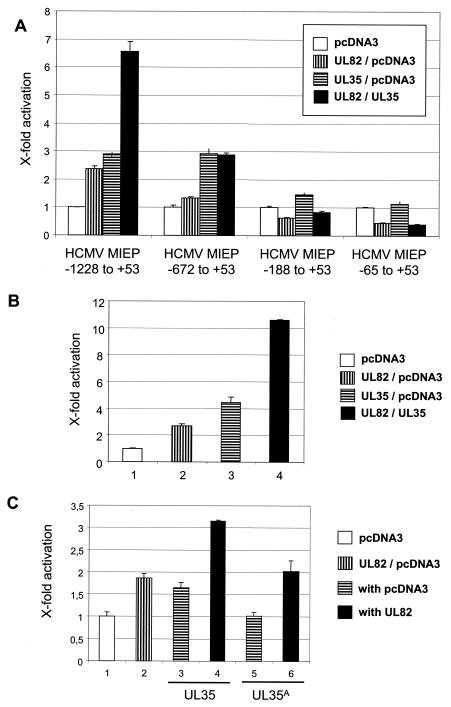
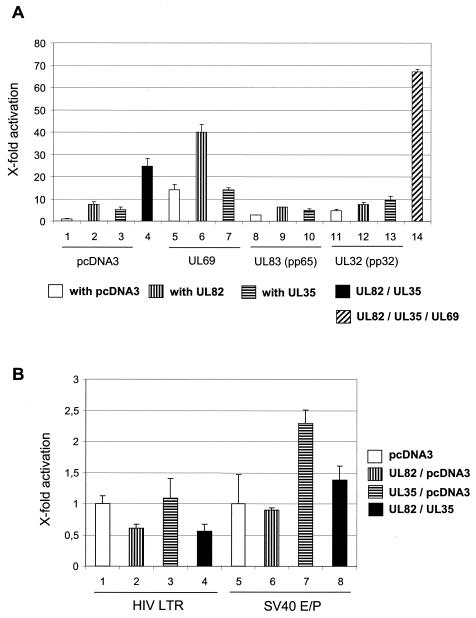
As ppUL82 has been previously shown to stimulate the MIEP of HCMV (23), we investigated whether the physical interaction between the two proteins would influence the activity of this regulatory element. For this, we transfected U373-MG cells with luciferase reporter constructs comprising different regions of the MIEP together with plasmids encoding ppUL82 and/or ppUL35 or pcDNA3 as a control (Fig. 4A). A reporter construct containing the modulator-enhancer (position −1228 to +53) was activated by ppUL82 or ppUL35 alone and showed a cooperative effect when both proteins were present. Essentially the same result was obtained when the transfection was repeated in fully permissive HFF cells (Fig. 4B). Therefore, the results obtained with both cell types are comparable, and for further experiments we relied on transfection of U373 cells. In contrast to the results obtained with the modulator-containing reporter plasmid, cotransfection with a construct containing the enhancer-promoter (position −672 to +53) showed activation only by ppUL35 and no cooperative effect. When constructs comprising only the promoter (−65 to +53) or part of the enhancer (−188 to +53) were used, we observed no activation after cotransfection with either UL82 or UL35 alone or in combination.
FIG. 4.
Cooperative activation by UL82 and UL35. Luciferase assays after transient transfections were performed in U373-MG and HFF cells. In each transfection, 0.5 μg of luciferase reporter construct was cotransfected with 2 μg of each effector plasmid; the total amount of transfected DNA was kept constant by adding pcDNA3. Each experiment was performed in triplicate and repeated at least three times. Error bars indicate standard deviations. (A) U373-MG cells were transfected with luciferase reporter constructs containing different deletions of the HCMV MIEP as indicated. Expression vector for UL35 or UL82 or empty vector pcDNA3 was cotransfected. (B) HFF cells were cotransfected with the luciferase reporter plasmid pHM287 carrying the luciferase reporter gene under control of the HCMV MIEP, including the modulator region, in combination with expression vector coding for ppUL82, ppUL35, or pUL69. The empty pcDNA3 vector was used as a control. (C) U373-MG cells were cotransfected with the luciferase reporter plasmid pHM287 and expression vectors encoding ppUL82 (bars 2, 4, and 6), ppUL35 (bars 3 and 4), and ppUL35A (bars 5 and 6). Empty vector pcDNA3 was used as a control.
As two isoforms of the UL35 protein, full-length ppUL35 and the shorter ppUL35A, are synthesized in infected cells and as both forms can interact with ppUL82, we also tested the UL35A gene in the luciferase assay. As can be seen in Fig. 4C, ppUL35A showed no activating effect on the modulator-containing HCMV MIEP construct, either alone or in combination with ppUL82.
Next, we wanted to test whether the cooperativity between ppUL35 and ppUL82 was specific by using two independent approaches. First, we employed expression vectors for additional tegument proteins, ppUL83 (pp65) and ppUL32 (pp150), as controls in cotransfections with the modulator-containing construct. As shown in Fig. 5A, both UL83 and UL32 showed only minor effects when transfected alone. Importantly, neither of these genes showed any cooperative effect with UL35 or UL82. In contrast, we again observed cooperativity between UL35 and UL82 and, as reported earlier, between UL69 and UL82 (Fig. 5A, bars 4 and 6) (45). When all three genes (UL35, UL69, and UL82) were transfected simultaneously, we found an even stronger activation (Fig. 5A, bar 14). However, we did not observe a cooperative effect after coexpression of ppUL35 and pUL69 (Fig. 5A, bar 7).
FIG. 5.
Specificity of cooperative activation by UL82 and UL35. (A) U373-MG cells were cotransfected with the luciferase reporter plasmid pHM287 and expression vectors encoding ppUL82 (bars 2, 4, 6, 9, 12, and 14), ppUL35 (bars 3, 4, 7, 10, 13, and 14), pUL69 (bars 5 to 7 and 14), ppUL83 (pp65) (bars 8 to 10), and ppUL32 (pp150) (bars 11 to 13). The empty pcDNA3 vector was used as a control. For bar 14, 1.7 μg DNA of each expression vector for ppUL82, ppUL35, and pUL69 was used to keep the total amount of effector plasmids at 4 μg. Error bars indicate standard deviations. (B) U373-MG cells were cotransfected with luciferase reporter plasmids carrying the luciferase reporter gene under control of the HIV LTR (bars 1 to 4) or the SV40 enhancer-promoter (bars 5 to 8) together with expression vectors encoding ppUL82 (bars 2, 4, 6, and 8) and ppUL35 (bars 3, 4, 7, and 8). The empty pcDNA3 vector was used as a control.
In a second experiment, we cotransfected UL35 and UL82 with reporter constructs containing the HIV LTR or the SV40 enhancer-promoter (Fig. 5B). In contrast to the cooperative activation of the HCMV MIEP, both heterologous enhancer elements were not cooperatively stimulated. Interestingly, UL82 showed a slight but consistent reduction of activity of the HIV LTR (Fig. 5B, bars 2 and 4). A similar reduction was also seen on the promoter region of the HCMV MIEP (Fig. 4A, HCMV MIEP −65 to +53). UL35 was only able to activate the SV40 enhancer-promoter and did not show any effect on the HIV LTR (Fig. 5B, bar 7). This activation was reduced by UL82 (Fig. 5B, bar 8).
ppUL82 concentrates ppUL35 to PODs.
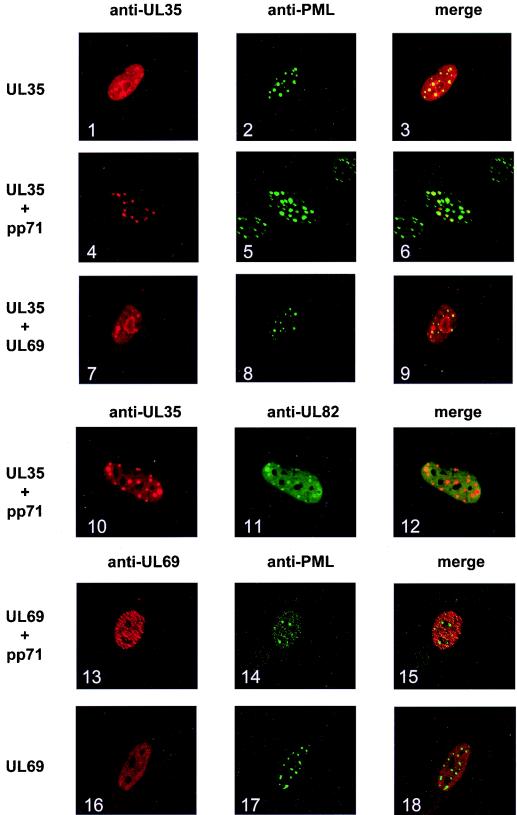
Having shown the interaction between ppUL82 and ppUL35, we wondered whether cotransfection of both genes would affect their subnuclear localization. When ppUL35 was transiently expressed in HFF cells, it showed a dispersed nuclear staining with some accumulation at PODs as visualized with an antibody against PML (Fig. 6, panels 1 to 3). Interestingly, for cotransfection with UL82, we detected a clear accumulation of ppUL35 in PODs (Fig. 6, panels 4 to 6), with almost no ppUL35 remaining in other parts of the nucleus. In addition, ppUL35 perfectly colocalized with ppUL82 (Fig. 6, panels 10 to 12). As a control, we used pUL69, which was able to cooperate with ppUL82 (Fig. 5A) but did not physically interact (Fig. 1B). When expressed alone, pUL69 showed disperse nuclear staining (Fig. 6, panels 16 to 18), which was not altered after coexpression of UL82 (pp71) (panels 13 to 15). Likewise, coexpression of pUL69 and ppUL35 did not lead to a change in the nuclear distribution of ppUL35 (Fig. 6, panels 7 to 9), demonstrating that the ppUL82-induced relocalization of ppUL35 is a specific effect.
FIG. 6.
Localization of ppUL35 in transfected HFF cells. HFF cells were cotransfected with expression vector encoding ppUL82, ppUL35, or pUL69, as indicated on the left of each row. The amount of transfected DNA was kept constant by adding the empty expression vector pcDNA3. Cells were stained with antibody directed against ppUL35, ppUL82, pUL69, or PML as indicated. Merged images are shown in the right column.
DISCUSSION
Several tegument proteins of HCMV have been shown to activate the MIEP either alone or cooperatively. For most of these proteins, the mechanism of activation and cooperation remains elusive. In this report, we provide for the first time a physical basis for the functional interaction between two of these tegument proteins, namely, ppUL35 and ppUL82.
The interaction between the two proteins was identified in a yeast two-hybrid screen with ppUL82 as bait and a genomic library of HCMV. Physical interaction was confirmed by coimmunoprecipitation of both proteins from transfected and infected cells. This interaction was specific, as other nuclear proteins of HCMV, i.e., pUL69 and pUL123 (IE1), could not coprecipitate ppUL82. In addition, the interaction between ppUL35 and ppUL82 could be detected in the presence of high salt concentrations, making it unlikely that the proteins form an unspecific complex. Nevertheless, higher salt concentrations reduced binding, indicating that the interaction could be due to electrostatic binding forces. This is in agreement with the observation that many heteromeric interactions rely mostly on electrostatic forces (21).
In addition to the physical interaction, we could demonstrate a colocalization of ppUL35 and ppUL82 by immunofluorescence analysis from transiently transfected HFF cells. Whereas ppUL35 showed a largely diffuse nuclear staining when expressed alone, almost all ppUL35 accumulated in nuclear dot structures in the presence of ppUL82. When we analyzed for colocalization with PML, which is responsible for the formation and maintenance of PODs, we observed that the dot-like structures formed by colocalization of ppUL82 and ppUL35 overlapped with PODs. The association of ppUL82 with PODs in both transfected and infected cells had previously been described to be dependent on interaction with hDaxx, a component of PODs (18, 19). Furthermore, upon initiation of infection, ppUL82 and viral DNA localized to or in close proximity to PODs, and this localization appeared to be necessary for efficient initiation of immediate-early transcription (19). These observations make it likely that ppUL82 is a component that brings together both viral and cellular protein components (ppUL35 and hDaxx) for efficient initiation of the viral replication cycle. However, final evidence for delivery of ppUL35 into PODs directly after infection is still lacking, as we were not able to detect the ppUL35 tegument component of incoming virus particles immediately after infection by immunofluorescence.
In our hands, the full-length form of ppUL35 localized to the nucleus. This is in contrast to a previous report, where it was proposed that full-length ppUL35 is present in the cytoplasm and only ppUL35A is localized to the nucleus (24). At present, we have no explanation for these discrepant results. However, the described cytoplasmic distribution of full-length ppUL35 is based on an enhanced green fluorescent protein fusion protein, where the enhanced green fluorescent protein domain could lead to a change in localization. Alternatively, HCMV strain differences could be responsible for altered properties of ppUL35. We note that our nuclear localization of ppUL35 also fits with the activation of the HCMV MIEP, which was not observed in the previous study (24). Furthermore, we would assume that ppUL35 relocalizes to the cytoplasm during virus maturation due to incorporation into the tegument.
The interaction between the proteins ppUL82 and ppUL35 was extended to the functional level by reporter gene assays. Using transient luciferase experiments, we could show a strong cooperative activation of the HCMV MIEP by UL82 and UL35, whereas each gene alone showed only a weak stimulation. This effect was seen both in fully permissive primary human fibroblasts and in the glioblastoma cell line U373-MG and is in accordance with a previous report (24). As several other tegument proteins have been shown to activate the HCMV MIEP alone and in various combinations, we used two sets of experiments to demonstrate the specificity of cooperation between UL82 and UL35. First, two abundant tegument proteins, ppUL83 (pp65) and ppUL32 (pp150), had no effect on the activity of the MIEP, independent of whether ppUL82 or ppUL35 was present or not. Second, the synergism of UL35 and UL82 was highly specific for the HCMV MIEP and could not be detected by using heterologous enhancer-promoter elements, such as the SV40 enhancer-promoter and the HIV LTR. This is in agreement with published data that ppUL82 was not able to transactivate the SV40 enhancer-promoter or the HIV LTR (23). To get closer to the situation during infection, we also performed infectivity assays, in which viral DNA was cotransfected with expression vectors for certain viral genes and the number of plaques originating from this transfection was scored. We found a cooperative enhancement of infectivity when UL82 and UL35 were cotransfected with viral DNA (K. Schierling and M. Winkler, unpublished observation). Furthermore, we have observed that the deletion of UL35 from the viral genome, like the deletion of UL82, resulted in a growth defect which was more pronounced at low MOI (K. Schierling and M. Winkler, unpublished data). MOI-dependent growth defects have been described for a variety of HCMV mutants associated with immediate-early gene expression (20, 29).
In summary, we could show an interaction between the HCMV tegument proteins ppUL35 and ppUL82, leading to the accumulation of ppUL35 in PODs, which are essential for initiation of viral IE transcription. As both proteins cooperatively activated the HCMV MIEP, this interaction could contribute to an effective onset of lytic replication and virus production.
Acknowledgments
We thank H. Ertle and N. Uschkurat for technical assistance, L. von Müller for help with the confocal microscope, and H. Hofmann for critically reading the manuscript. We also thank M. Vidal for yeast strains and vectors, M. Mach for TB40E cosmids and MAb 28-4, B. Biegalke for UL35 rabbit serum, S. Foung for human MAb X2-16, and U. Grawunder for purified 12CA5 MAb.
This work was supported by grants from the BMBF (BEO 0311685/1406) and the Deutsche Forschungsgemeinschaft (DFG and SFB473) to T. Stamminger and by grants from the DFG (Wi 1725/2-1), IZKF (01KS9605/2), and Universität Ulm (P.680) to M. Winkler.
REFERENCES
- 1.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearer, E. L., X. O. Breakefield, D. Schuback, T. S. Reese, and J. H. LaVail. 2000. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc. Natl. Acad. Sci. USA 97:8146-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradshaw, P. A., M. R. Duran-Guarino, S. Perkins, J. I. Rowe, J. Fernandez, K. E. Fry, G. R. Reyes, L. Young, and S. K. Foung. 1994. Localization of antigenic sites on human cytomegalovirus virion structural proteins encoded by UL48 and UL56. Virology 205:321-328. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer, C. B. 1994. Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol. 43:233-245. [DOI] [PubMed] [Google Scholar]
- 6.Britt, W. J., and L. Vugler. 1987. Structural and immunological characterization of the intracellular forms of an abundant 68,000 Mr human cytomegalovirus protein. J. Gen. Virol. 68:1897-1907. [DOI] [PubMed] [Google Scholar]
- 7.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:126-169. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevray, P. M., and D. Nathans. 1992. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl. Acad. Sci. USA 89:5789-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 12.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fickenscher, H., T. Stamminger, R. Ruger, and B. Fleckenstein. 1989. The role of a repetitive palindromic sequence element in the human cytomegalovirus major immediate early enhancer. J. Gen. Virol. 70:107-123. [DOI] [PubMed] [Google Scholar]
- 14.Fleckenstein, B., J. Müller, and J. Collins. 1982. Cloning of the complete cytomegalovirus genome in cosmids. Gene 18:39-46. [DOI] [PubMed] [Google Scholar]
- 15.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert, M. J., S. R. Riddell, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720-722. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, H., H. Sindre, and T. Stamminger. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76:5769-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isomura, H., and M. F. Stinski. 2003. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J. Virol. 77:3602-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingston, R. E., C. A. Chen, H. Okayama, and J. K. Rose. 1996. Transfection of DNA into eukaryotic cells, p. 1.1-1.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Strahl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Boston, Mass.
- 23.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Y., and B. J. Biegalke. 2002. The human cytomegalovirus UL35 gene encodes two proteins with different functions. J. Virol. 76:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchuk, D., M. Drumm, A. Saulino, and F. S. Collins. 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall, K. R., K. V. Rowley, A. Rinaldi, I. P. Nicholson, A. M. Ishov, G. G. Maul, and C. M. Preston. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 83:1601-1612. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 29.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niman, H. L., R. A. Houghten, L. E. Walker, R. A. Reisfeld, I. A. Wilson, J. M. Hogle, and R. A. Lerner. 1983. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 80:4949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325-338. [DOI] [PubMed] [Google Scholar]
- 32.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 33.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642-652. [DOI] [PubMed] [Google Scholar]
- 34.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinzger, C., K. Schmidt, J. Knapp, M. Kahl, R. Beck, J. Waldman, H. Hebart, H. Einsele, and G. Jahn. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol. 80:2867-2877. [DOI] [PubMed] [Google Scholar]
- 36.Spaete, R. R., and E. S. Mocarski. 1985. Regulation of cytomegalovirus gene expression: alpha and beta promoters are trans activated by viral functions in permissive human fibroblasts. J. Virol. 56:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamminger, T., H. Fickenscher, and B. Fleckenstein. 1990. Cell type-specific induction of the major immediate early enhancer of human cytomegalovirus by cyclic AMP. J. Gen. Virol. 71:105-113. [DOI] [PubMed] [Google Scholar]
- 38.Stamminger, T., M. Gstaiger, K. Weinzierl, K. Lorz, M. Winkler, and W. Schaffner. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J. Virol. 76:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stinski, M. F., and T. J. Roehr. 1985. Activation of the major immediate-early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J. Virol. 55:431-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Leeuwen, H., G. Elliott, and P. O'Hare. 2002. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J. Virol. 76:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Zonneveld, A. J., S. A. Curriden, and D. J. Loskutoff. 1988. Type 1 plasminogen activator inhibitor gene: functional analysis and glucocorticoid regulation of its promoter. Proc. Natl. Acad. Sci. USA 85:5525-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal, M., R. K. Brachmann, A. Fattaey, E. Harlow, and J. D. Boeke. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93:10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkler, M., T. Aus dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler, M., S. Schmolke, B. Plachter, and T. Stamminger. 1995. The pUL69 protein of human cytomegalovirus (HCMV), a homologue of the herpes simplex virus ICP27, is contained within the tegument of virions and activates the major immediate-early enhancer of HCMV in synergy with the tegument protein pp71 (ppUL82). Scand. J. Infect. Dis. Suppl. 99:8-9. [Google Scholar]
- 46.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]